
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Genet., 04 February 2022
Sec. Cancer Genetics and Oncogenomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.829283
This article is part of the Research TopicAdvances in AI‐Based Tools for Personalized Cancer Diagnosis, Prognosis and TreatmentView all 11 articles
Background: The interleukin10 (IL-10) gene polymorphisms have been indicated to be associated with breast cancer (BC) risk, but the findings are still controversial. To derive a more precise evaluation, we performed a comprehensive meta-analysis.
Methods: A systematic literature search was conducted using PubMed, Embase, CNKI, China biomedical (CBM), and Google Scholar to 29 March 2020. Revman5.3 and Stata 12.0 software analyzed the data, and the strength of the association was identified using the odds ratio (OR) and the corresponding 95% confidence interval (CI).
Results: A total of 23 studies (7,250 cancer cases and 7,675 case-free controls) were included in this meta-analysis. The results show that IL-10 gene polymorphisms were significantly correlated with BC risk based on subgroup analysis by ethnicity. The IL-10 rs1800896 polymorphism was significantly associated with the risk of BC in Asians (G vs. A: OR = 0.78, 95% CI = 0.65–0.95, p = 0.01; GG vs. AA: OR = 0.51, 95% CI = 0.31–0.84, p = 0.007; GA vs. AA: OR = 0.6, 95% CI = 0.44–0.81, p = 0.0009; GG + GA vs. AA: OR = 0.6, 95% CI = 0.45–0.81, p = 0.0007); Moreover, an increased BC risk in Asians were also associated with the IL-10 rs1800872 polymorphism (AA vs CC: OR = 0.74, 95% CI = 0.55–0.99, p = 0.04; A vs C: OR = 0.85, 95% CI = 0.74–0.98, p = 0.03). In addition, The IL-10 rs1800871 (CT vs. TT: OR = 1.8, 95% CI = 1.03–3.13, p = 0.04) and rs1800872 polymorphism (A vs C: OR = 0.65, 95% CI 0.43–0.98, p = 0.04) were associated with BC risk in Caucasians.
Conclusion: Collectively, this meta-analysis demonstrated that IL-10 rs1800896 and rs1800872 (AA vs. CC; A vs. C) polymorphisms significantly increased the risk of BC in Asians, while the rs1800871 and rs1800872 (A vs. C) were associated with the risk of BC in Caucasians. Therefore, this may provide new ideas for predicting and diagnosing BC susceptibility through the detection of IL-10 gene polymorphism.
Systematic Review Registration: [https://www.crd.york.ac.uk/ PROSPERO], identifier [CRD42021266635].
Breast cancer (BC) is the leading cause of female cancer-related death worldwide and is one of the most common cancer forms (Anastasiadi et al., 2017). BC incidence varies widely, ranging from 27/100,0002 (Central-East Asia and Africa) to 85–94/100,0002 (Australia, North America, and Western Europe). And the incidence of BC in France is the highest in Europe (Sancho-Garnier and Colonna, 2019). In Asian countries, the incidence rate of BC has also been increasing rapidly (Mubarik et al., 2020; Oblak et al., 2020). The pathogenesis of BC is multifactorial. Hereditary BC accounts for only 5–10% of all BC cases and germline mutations, with the two significant BC susceptibility genes, BRCA1 and BRCA2 is responsible for approximately 2–3% of all cases (Kwong et al., 2016). Besides gene tests for identifying high-risk BRCA1 or BRCA2 mutations carriers (Ha et al., 2017), the ability to predict BC development is not well established yet. Although genetic, environmental, and lifestyle factors are associated with BC occurrence, the biological mechanism that causes BC remains unclear.
Inflammation plays a significant role in BC development and is an important part of the BC microenvironment (Mohamed et al., 2018). Interleukin-10 (IL-10) is an important anti-inflammatory and immunomodulatory cytokine in the human immune response. IL-10 is located on chromosome 1 (1q31-1q32), composed of five exons and four introns (Roh et al., 2002). Single nucleotide polymorphism (SNP) is the most common genetic variation. In the SNP database (http://www.ncbi.nlm.nih.gov/snp), three promoter SNPs of IL-10, rs1800896 (-1082A/G), rs1800871 (-819T/C), and rs1800872 (-592A/C) were extensively investigated in many diseases. Because they might affect IL-10 gene transcription and translation, resulting in abnormal cell proliferation and cancer development (Howell and Rose-Zerilli, 2007). The possible mechanism is that IL-10 is activated by the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathways through its receptor IL-10 R1 which binds to STAT3. Then STAT3 is translocated into the nucleus, where it binds to STAT-binding elements in the promoters of proliferation-related genes. It has been reported that IL-10 gene polymorphism plays an important role in the occurrence and development of cancers such as BC, gastric cancer, lung cancer (Bhattacharjee et al., 2016; Chen et al., 2019; Zhao et al., 2019). And some studies reported the high IL-10 expression levels in the BC paraffin section and its expression is correlated with worse outcomes in patients with malignant tumors (Li et al., 2014; Zhao et al., 2015).
In recent years, several studies have reported the relationship between IL-10 polymorphisms and BC susceptibility. A study found that: rs1800896 (-1082A/G) polymorphism was correlated with cancer staging and associated with the progression of BC at AA genotype (Abedinzadeh et al., 2018). In the research based on Caucasians, it was found that there was a significant association between the IL10-1082 G/G genotype and the increased risk of BC (Zhu et al., 2020). Another study found that the rs1800871 (-819T/C) polymorphism increased the risk of BC in Han Chinese women (Li et al., 2020). And a study shows the rare allele of rs1800872 (-592A/C) polymorphism may be a potential prognostic indicator of disease-free survival in BC patients (Gerger et al., 2010). These suggest that IL-10 gene polymorphism may affect the risk of human BC (Setrerrahmane and Xu, 2017). However, these results are inconsistent. Moreover, IL-10 polymorphism and BC susceptibility studies are constantly updated, and adjustments vary between included studies based on race, age, lifestyle, and other covariates (Patricia Gallegos-Arreola et al., 2019). Considering the critical role of IL-10 in the development of BC, we conducted this systematic review. And compared with previous meta-analyses, we comprehensively included the latest relevant studies to evaluate the association of IL-10 rs1800896, rs1800871, and rs1800872 polymorphisms with the risk of BC in different ethnic groups. It will provide theoretical evidence for the genetic mechanism of BC.
This meta-analysis was conducted according to the PRISMA reporting criteria (Moher et al., 2009).
Research articles on the relationship between IL-10 gene polymorphisms and BC risk were searched in different databases, including PubMed, Web of Knowledge, Embase, CNKI, CBM, and Google Scholar up to 29 March 2020. And we retrieved with the keywords: (“breast cancer” or “breast tumor” or “breast neoplasm” or “malignant breast tumor” or “breast carcinoma”) and (“Interleukin-10” or “IL-10”) and (“polymorphism” or “SNP” or “single nucleotide polymorphism” or “variation” or “mutation”).
Inclusion criteria: (1) Clinical BC patients were selected as the case group and healthy people as the control group; (2) Case-control or cohort studies about associations between IL-10 gene polymorphism and BC in humans; (3) Full manuscript in English or Chinese is retrievable; (4) Reporting the number of cases and controls for each genotype and detailed genotyping data, or knowing the odds ratio (OR) helped to calculate the 95% confidence interval (CI).
Exclusion criteria: (1) Abstracts, reviews; (2) Studies on the relationship between IL-10 gene polymorphism and prognosis of BC; (3) studies on the apparent imbalance of baseline between the case group and control group; (4) The cases and control sources were not provided; (5) Repeatedly published literature. If multiple studies from the same case series were available, the one including the most individuals were used in the analysis.
Two researchers selected the literature according to the inclusion and exclusion criteria, extracted the data, and cross-checked them independently into a standard data collection form. If there were any disputes, we would reach an agreement by discussion or by a third party and strive to reach a consensus on each project. Data were collected from each article included: the first author, year of publication, study location, type of study, ethnicity (classified as Asian, Caucasian, or mixed descent), total number of cases and controls, genotype frequency, genotype detection method, and the source of authority.
Sensitivity analysis was performed to assess the stability of the results. The Funnel plot, Begg’s test, and Egger’s test were used to evaluate publication bias. RevMan5.3 and Stata 12.0 software was used for the above statistical analysis.
The correlation between IL-10 gene polymorphisms and BC risk was evaluated by OR and 95% CI as the effect size. 95% CI without one and P(OR) < 0.05 was considered statistically significant. The Z-test determines the significance of the OR value. The effects of heterogeneity were quantified by I2 and P(H) values. In addition, the I2 value is used to quantify the degree of heterogeneity (I2 < 25%: low/no heterogeneity; 25 < I2 < 75%: moderate heterogeneity; I2 > 75%: extreme high heterogeneity). The fixed-effect model is adopted when the I2 < 25%; otherwise, the random effect model is adopted. We further carried out subgroup analyses by ethnicity to get ethnic-specific results.
We had a total of 78 articles after removing three duplicated pieces. After the layer-by-layer screening, a total of 23 articles finally met the criteria for inclusion in this meta-analysis. Eligible papers were published between 2004 and 2019. This meta-analysis updated three 2019 case-control studies compared to previous meta-analyses (Dai et al., 2014; Abedinzadeh et al., 2018; Moghimi et al., 2018). A flow diagram schematizing the inclusion and exclusion process of identified articles with the inclusion criteria is presented in Figure 1.
The 23 eligible articles had a total sample size of 14,925 participants, including 7,250 BC patients and 7,675 healthy controls (Smith et al., 2004; Abdolrahim-Zadeh et al., 2005; Guzowski et al., 2005; Langsenlehner et al., 2005; Balasubramanian et al., 2006; Onay et al., 2006; Scola et al., 2006; Gonullu et al., 2007; Pharoah et al., 2007; Kong et al., 2010; He et al., 2012; Pooja et al., 2012; Meijiang, 2014; Wang et al., 2014; Vinod et al., 2015; AlSuhaibani et al., 2016; Atoum, 2016; Maruthi et al., 2017; Sabet et al., 2017; Tian et al., 2017; Azher et al., 2019; Fanyu et al., 2019; Patricia Gallegos-Arreola et al., 2019). The samples were involved in three IL-10 polymorphism sites: rs1800896, rs1800871, and rs1800872. There were 17 studies on rs1800896 (3,308 cases and 3,425 controls), twelve studies on rs1800871 (2,530 cases and 2,698 controls), and 13 studies on rs1800872 (4,702 cases and 4,818 controls). Ten studies were based on Caucasians, six were based on Asians, and the remaining seven studies were mixed-race in the 23 criteria studies. Of these studies, eighteen were hospital-based, and five were population-based. The Newcastle-Ottawa Scale (NOS) was used to assess the quality of the included articles (Stang, 2010). And NOS scores ranged from zero to nine. We considered the study’s methodological quality good if the score was ≥ seven. Two authors independently completed our data extraction and quality evaluation. Table 1 and Table 2 show the basic characteristics of the included literature, the distribution of polymorphisms at the studied gene sites, allele frequency, and the quality assessment of the included studies.
The association between IL-10 gene polymorphisms (rs1800896, rs1800871, and rs1800872) and BC is shown in Table 3 and Figures 2–5. Squares and horizontal lines correspond to study-specific OR and 95% CI. The area of a square reflects the weight (inversely proportional to the variance). The diamond represents the sum of OR and 95% CI.
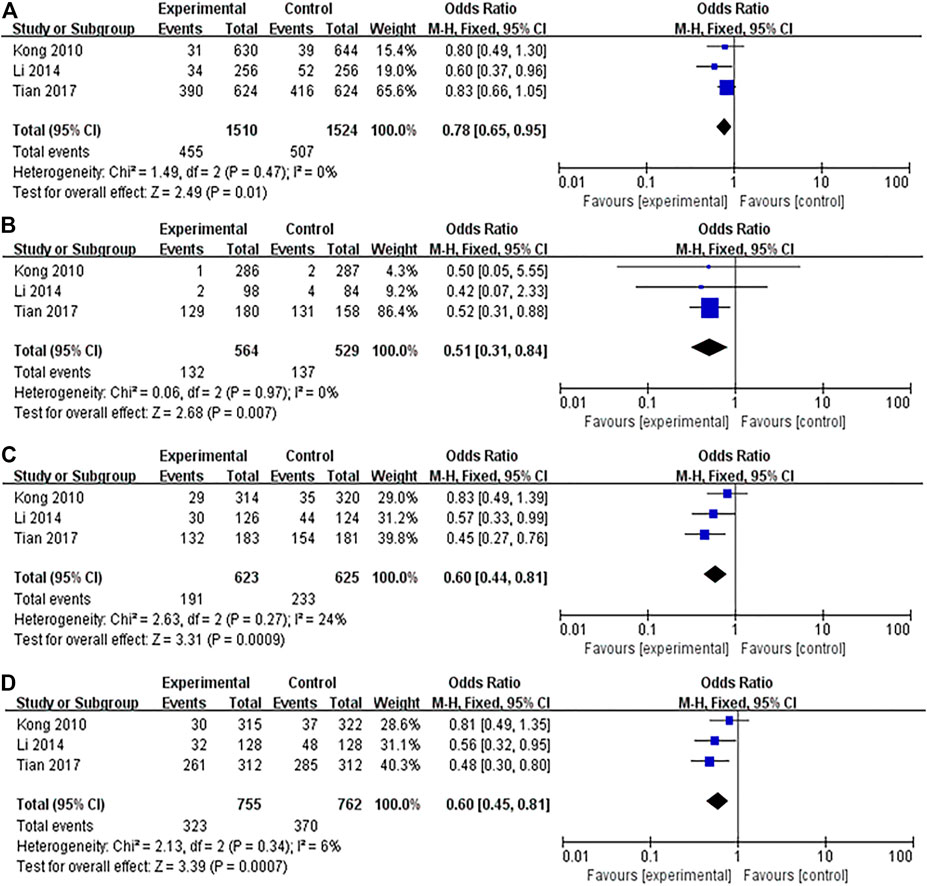
FIGURE 2. Forest plots showed a significant association of IL-10 rs1800896 polymorphism and breast cancer risk in Asians. (A) (allele model: G vs. A); (B) (homozygous model: GG vs. AA); (C) (heterozygous model: GA vs. AA); (D) (dominant model: GG + GA vs. AA). The squares and horizontal lines correspond to the study-specific odds ratio (OR) and 95% confidence interval (CI). The area of the squares reflects the weight (inverse of the variance). The diamond represents the summary OR and 95% CI.
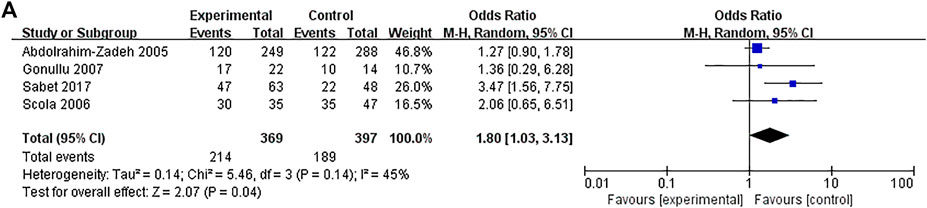
FIGURE 3. Forest plots showed a significant association of IL-10 rs1800871 polymorphism and breast cancer risk in the Caucasians (heterozygous model: CT vs. TT). The squares and horizontal lines correspond to the study-specific odds ratio (OR) and 95% confidence interval (CI). The area of the squares reflects the weight (inverse of the variance). The diamond represents the summary OR and 95% CI.
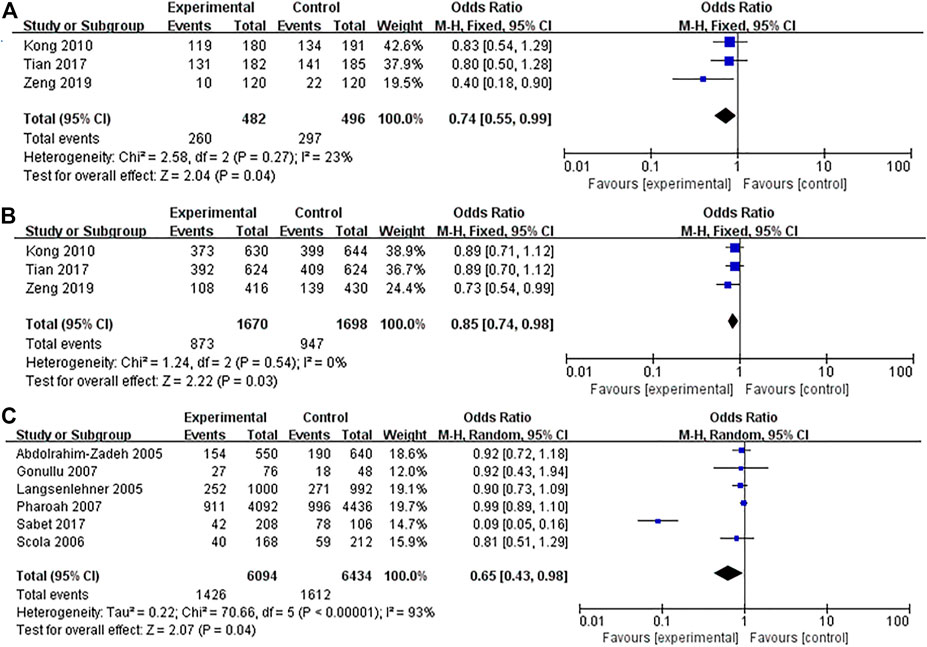
FIGURE 4. Forest plots showed a significant association of IL-10 rs1800872 polymorphism and breast cancer risk in Asians and Caucasians. (A) IL-10 rs1800872 polymorphism in Asians (homozygous model: AA vs. CC); (B) IL-10 rs1800872 polymorphism in Asians (allele model: A vs. C); (C) IL-10 rs1800872 polymorphism in Caucasians (allele model: A vs. C). The squares and horizontal lines correspond to the study-specific odds ratio (OR) and 95% confidence interval (CI). The area of the squares reflects the weight (inverse of the variance). The diamond represents the summary OR and 95% CI.
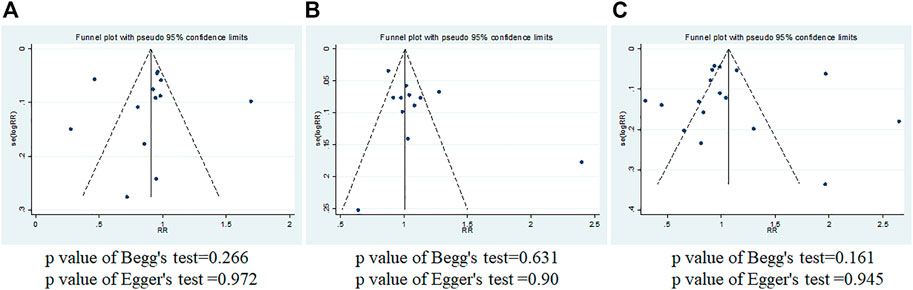
FIGURE 5. Begg’s and Egger’s funnel plots of IL-10 gene polymorphism and breast cancer risk for publication bias test. (A) rs1800896; (B) rs1800871; (C) rs1800872.
A total of 17 studies were conducted on the association between IL-10 rs1800896 polymorphism and BC risk, with a total sample size of 6,733 cases, including 3,308 patients and 3,425 healthy controls. Overall population heterogeneity test I2 was 82%. The random-effect model results showed that the comparison results of the five gene models showed no statistical significance between rs1800896 polymorphism and BC (Table 3). Subgroups by ethnicity showed that under the four genetic models (allele G vs. A: OR = 0.78, 95% CI = 0.65–0.95, p = 0.01; homozygous GG vs. AA: OR = 0.51, 95% CI = 0.31–0.84, p = 0.007; heterozygous GA vs. AA: OR = 0.6, 95% CI = 0.44–0.81, p = 0.0009; dominant GG + GA vs. AA: OR = 0.6, 95% CI = 0.45–0.81, p = 0.0007) (Figures 2A–D), rs1800896 polymorphism was significantly associated with BC risk in Asians. This result suggests that ethnicity is likely to be the source of heterogeneity, and the rs1800896 polymorphism is significantly associated with the BC risk in Asians.
A total of 12 studies with 2,530 patients and 2,698 controls evaluated the strength of the association between the IL-10 rs1800871 polymorphism and BC. There was no association between BC risk and rs1800871 polymorphism in any genetic model in the overall population. However, when stratified by ethnicity, the rs1800871 polymorphism was associated with BC risk in the heterozygous model in Caucasians (CT vs. TT: OR = 1.8, 95% CI = 1.03–3.13, p = 0.04) (Figure 3). This result indicates that Caucasians with the rs1800871 heterozygous model are more likely to develop BC than individuals with other genotypes.
Thirteen studies (4,702 cases and 4,818 controls) assessed the strength of the association between IL-10 rs1800872 polymorphism and BC susceptibility. As shown in Table 3, the five gene model comparison results showed that the association between IL-10 rs1800872 polymorphism and BC in the overall population was not statistically significant. However, after stratification by ethnicity, the homozygous model of rs1800872 polymorphism was associated with BC risk in Asians (AA vs. CC: OR = 0.74, 95% CI = 0.55–0.99, p = 0.04) (Figure 4A). Allele model of rs1800872 polymorphism was associated with the risk of BC in Asians (A vs. C: OR = 0.85, 95% CI = 0.74–0.98, p = 0.03) (Figure 4B) and Caucasians (A vs. C: OR = 0.65, 95% CI = 0.43–0.98, p = 0.04) (Figure 4C).
Funnel plot, Begg’s test, and Egger’s test were used to evaluate the publication bias (Stata12.0). As shown in Figure 5, the funnel plot was essentially symmetrical, and the p values of Begg’s test and Egger’s test are all greater than 0.05. It was indicated that there was almost no obvious publication bias at the three loci.
IL-10, known initially as cytokine synthesis inhibitory factor (CSIF), is a potent anti-inflammatory cytokine. IL-10 can stimulate the expression of carboxypeptidase B2 (CPB2) in inflammatory BC cells. Thus it increases the cancer cells’ aggressiveness (Mohamed et al., 2018). Moreover, IL-10 is involved in the abnormal proliferation of breast ducts and lobules and stimulates mitotic activity, leading to increased cancer risk (Kong et al., 2010; Moghimi et al., 2018). IL-10 can also induce tumor progression by inhibiting many cytokines such as IL-1a, IL-1b, IL-6, IL-8, IL-12, and IL-18. And IL-10 gene silencing down-regulates the expression of phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) and B cell lymphoma 2 (Bcl2) and increases the expression levels of BCL2 binding component 3(BBC3), Bax, and caspase3 (Alotaibi et al., 2018). Studies on the mechanism of IL-10 promoting BC have shown that the production of IL-10 may represent a new escape mechanism for BC cells to escape the destruction of the immune system. It might be closely related to the fact that polymorphic variations in the promoter sequences of the IL-10 gene might influence the gene expression and consequently play a specific role in susceptibility and the clinical course of BC. The IL-10 promoter region polymorphisms affected IL-10 gene transcription and translation, resulting in abnormal cell proliferation and cancer development (Moghimi et al., 2018; Sheikhpour et al., 2018).
Studies have shown that the three most common single nucleotide polymorphisms (SNPs) play an important role in regulating IL-10 activity. They are located at the transcriptional starting point of rs1800896 (-1082A/G), rs1800871 (-819T/C), and rs1800872 (-592A/C). And they encode high (GCC), medium (ACC), and low (ATA) expression of IL-10, respectively (Westendorp et al., 1997; Zupin et al., 2014; Hofmann et al., 2018). Several other polymorphic loci of IL-10 (rs1800890, rs6703630, and rs6693899) are also controversial, but few relevant studies are present. Many studies have reported the relationship between race and IL-10 gene polymorphism and BC risk in recent years. For example, the IL-10 rs1800872 polymorphism was associated with BC susceptibility in the Mexican population (Patricia Gallegos-Arreola et al., 2019). Also, the mutant allele and genotypes of IL-10 rs1800896 were significantly associated with Indian postmenopausal BC (Pooja et al., 2012). Since the IL-10 gene polymorphisms were associated with the risk of BC, we hypothesized that race is the key to the association between IL-10 gene polymorphisms and BC. This meta-analysis conducted the most comprehensive analysis of the relationship between three IL-10 polymorphisms (rs1800896, rs1800871, and rs1800872) and the BC risk of different races. In a subgroup analysis by ethnicity (Asian and Caucasian/mixed race), the three IL-10 polymorphisms (rs1800896, rs1800871, and rs1800872) were significantly associated with BC. It showed that rs1800896 (allele G vs. A: OR = 0.78, 95% CI = 0.65–0.95, p = 0.01; homozygous GG vs. AA: OR = 0.51, 95% CI = 0.31–0.84, p = 0.007; heterozygous GA vs. AA: OR = 0.6, 95% CI = 0.44–0.81, p = 0.0009; dominant GG + GA vs. AA: OR = 0.6, 95% CI = 0.45–0.81, p = 0.0007) were significantly correlated with BC risk in Asians. The rs1800871 heterozygote model (CT vs. TT: OR = 1.8, 95% CI = 1.03–3.13, p = 0.04) was associated with BC risk in Caucasians. The rs1800872 homozygous model (AA vs CC: OR = 0.74, 95% CI = 0.55–0.99, p = 0.04) was associated with BC risk in Asians, and the allelic model (A vs. C: OR = 0.85, 95% CI = 0.74–0.98, p = 0.03) was associated with BC risk in Asians and Caucasians (A vs C: OR = 0.65, 95% = CI 0.43–0.98, p = 0.04). The above results indicate that the ethnic subgroup of IL-10 gene polymorphisms is the key factor affecting the susceptibility to BC. It is consistent with the results of previous studies: the relationship between IL-10 gene polymorphism and BC risk is strongly associated with ethnicity (Patricia Gallegos-Arreola et al., 2019).
Previously, three researchers (Dai et al., 2014; Abedinzadeh et al., 2018; Moghimi et al., 2018) have analyzed the correlation between IL-10 gene polymorphisms and BC risk, but their analysis is not comprehensive enough. Because there are few studies included and the ethnic division is not accurate enough in their articles. In addition, Xu and Wang did a meta-analysis on the relationship between various interleukins and BC. Still, their correlations between IL-10 gene polymorphisms and BC risk were inconsistent with ours (Xu and Wang, 2020). It may be related to the different criteria for inclusion and exclusion and quality assessment of the article. Because the quality, quantity, and new studies included in the meta-analysis will directly affect the credibility and stability of the results, we used a broad search strategy to capture all relevant information. This meta-analysis conducted a more comprehensive analysis of the relationship between three IL-10 polymorphisms (rs1800896, rs1800871, and rs1800872) and BC risk by including 23 studies (published between 2004 and 2019) and ruling out the researches with low quality. Moreover, this meta-analysis showed no significant publication bias, and the heterogeneity of the subgroups was small. Sensitivity analysis results were also stable. Therefore, the conclusion of the association between the three IL-10 gene polymorphisms (rs1800896, rs1800871, and rs1800872) and BC in this meta-analysis was reliable and had certain clinical guidance values.
However, this meta-analysis has several limitations that should be acknowledged. Firstly, due to the limited research on the interaction between these three polymorphic sites and their interaction with the environment, it is impossible to estimate the impact of gene-gene and gene-environment interaction on the study results. Secondly, we found that heterogeneity existed in the meta-analysis as indicated by the I2 values. Despite using a random-effects model in some studies, the heterogeneity remained. It is predictable because other factors that affect BC should be considered, such as staging and grading of tumors, age, genetic background, environment, and lifestyle. However, due to the lack of some qualified original data, we cannot calculate the impact of these factors on BC. Moreover, in the future we need to consider more factors influencing BC, such as age, menopausal state, environment, and lifestyle factors, to further validate gene-gene and gene-environment interactions on IL-10 polymorphisms and BC risk.
In summary, this meta-analysis provides a new idea for clinical, genetic, and epidemiological studies of BC. Our results show that alleles, homozygotes, and dominant genotypes of IL-10 rs1800896 are significantly associated with the risk of BC in Asians. The homozygous and allele patterns of rs1800872 increase the risk of BC in Asians, while the heterozygous pattern of rs1800871 and the allele pattern of rs1800872 increase the risk of BC in Caucasians. IL-10 gene polymorphisms may be a key regulator of BC susceptibility. Different ethnic groups can predict BC susceptibility by detecting other IL-10 polymorphisms locus. However, the etiology of BC is complex, so we strongly recommend further genetic association studies to explore the effects of gene-gene interactions on disease susceptibility. Large-scale multicenter studies can be conducted in the future to verify further the results of gene-gene and gene-environment interactions on IL-10 gene polymorphisms and BC risk in different environments.
LL designed and managed the study. WX interpreted the data and drafted the manuscript. LL and JC performed data analysis and wrote the manuscript. DL conceived the idea and supervised the study. All authors read and approved the final manuscript.
The present study was supported by the University-level Foundation of the University of South China (nk2020205).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The also authors would like to thank reviewers for their careful reading and valuable suggestions.
Abdolrahim-Zadeh, H., Hakkakian, N., Asadollahi, R., Gharesifard, B., Sarvari, J., Kamali, E., et al. (2005). Interleukin-10 Promoter Polymorphisms and Breast Cancer Risk in Iranian Women. Iranian J. Immunol. 2. 158–165.
Abedinzadeh, M., Neamatzadeh, H., Jafari, M., Forat-Yazdi, M., Nasiri, R., Farahnak, S., et al. (2018). Association of Interleukin-10 -1082A>G (Rs1800896) Polymorphism with Predisposition to Breast Cancer: a Meta-Analysis Based on 17 Case-Control Studies. Rev. Assoc. Med. Bras. 64, 756–764. doi:10.1590/1806-9282.64.08.756
Alotaibi, M. R., Hassan, Z. K., Al-Rejaie, S. S., Alshammari, M. A., Almutairi, M. M., Alhoshani, A. R., et al. (2018). Characterization of Apoptosis in a Breast Cancer Cell Line after IL-10 Silencing. Asian Pac. J. Cancer Prev. 19, 777–783. doi:10.22034/APJCP.2018.19.3.777
Alsuhaibani, E. S., Kizilbash, N. A., Malik, S., Dasti, J. I., Al Beladi, F., and El-Morshedi, N. (2016). Polymorphisms in Promoter Regions of IL-6 and IL-10 Genes in Breast Cancer: a Case-Control Study. Genet. Mol. Res. 15, gmr.150173. doi:10.4238/gmr.15017360
Anastasiadi, Z., Lianos, G. D., Ignatiadou, E., Harissis, H. V., and Mitsis, M. (2017). Breast Cancer in Young Women: an Overview. Updates Surg. 69, 313–317. doi:10.1007/s13304-017-0424-1
Atoum, M. (2016). ACC Interleukin-10 Gene Promoter Haplotype as a Breast Cancer Risk Factor Predictor Among Jordanian Females. Onco Targets Ther. 9, 3353–3357. doi:10.2147/OTT.S101628
Azher, A., Al-Ankoshy, M., Hamid, A., and Dawood Alatbee, A. (2019). The Impact of IL-10 Gene Polymorphism on Progressive Breast Cancer. J. Pharm. Sci. Res. 11 (1), 93–97.
Balasubramanian, S., Azmy, I., Higham, S., Wilson, A., Cross, S., Cox, A., et al. (2006). Interleukin Gene Polymorphisms and Breast Cancer: a Case Control Study and Systematic Literature Review. BMC Cancer 6, 188. doi:10.1186/1471-2407-6-188
Bhattacharjee, H. K., Bansal, V. K., Nepal, B., Srivastava, S., Dinda, A. K., and Misra, M. C. (2016). Is Interleukin 10 (IL10) Expression in Breast Cancer a Marker of Poor Prognosis? Indian J. Surg. Oncol. 7, 320–325. doi:10.1007/s13193-016-0512-6
Chen, L., Shi, Y., Zhu, X., Guo, W., Zhang, M., Che, Y., et al. (2019). IL‑10 Secreted by Cancer‑associated Macrophages Regulates Proliferation and Invasion in Gastric Cancer Cells via c‑Met/STAT3 Signaling. Oncol. Rep. 42, 595–604. doi:10.3892/or.2019.7206
Dai, Z.-J., Wang, X.-J., Zhao, Y., Ma, X.-B., Kang, H.-F., Min, W.-L., et al. (2014). Effects of Interleukin-10 Polymorphisms (Rs1800896, Rs1800871, and Rs1800872) on Breast Cancer Risk: Evidence from an Updated Meta-Analysis. Genet. Test. Mol. Biomarkers 18, 439–445. doi:10.1089/gtmb.2014.0012
Fanyu, Z., Wenhui, L., Shan, Z., Wei, T., Xiaofen, Z., and Qiujin, Z. (2019). Association of Interleukin-10 Gene Polymorphism with Susceptibility of Breast Cancer in Chinese Women of Guangxi Province. Guo ji mian yi xue za zhi 042, 464–467. doi:10.1016/j.jclinepi.2009.06.005
Gerger, A., Renner, W., Langsenlehner, T., Hofmann, G., Knechtel, G., Szkandera, J., et al. (2010). Association of Interleukin-10 Gene Variation with Breast Cancer Prognosis. Breast Cancer Res. Treat. 119, 701–705. doi:10.1007/s10549-009-0417-y
Gonullu, G., Basturk, B., Evrensel, T., Oral, B., Gozkaman, A., and Manavoglu, O. (2007). Association of Breast Cancer and Cytokine Gene Polymorphism in Turkish Women. Saudi Med. J. 28, 1728–1733.
Guzowski, D., Chandrasekaran, A., Gawel, C., Palma, J., Koenig, J., Wang, X. P., et al. (2005). Analysis of Single Nucleotide Polymorphisms in the Promoter Region of Interleukin-10 by Denaturing High-Performance Liquid Chromatography. J. Biomol. Tech. 16, 154–166.
Ha, S. M., Chae, E. Y., Cha, J. H., Kim, H. H., Shin, H. J., and Choi, W. J. (2017). Association of BRCA Mutation Types, Imaging Features, and Pathologic Findings in Patients with Breast Cancer with BRCA1 and BRCA2 Mutations. Am. J. Roentgenol. 209, 920–928. doi:10.2214/AJR.16.16957
He, J.-R., Chen, L.-J., Su, Y., Cen, Y.-L., Tang, L.-Y., Yu, D.-D., et al. (2012). Joint Effects of Epstein-Barr Virus and Polymorphisms in Interleukin-10 and Interferon-γ on Breast Cancer Risk. J. Infect. Dis. 205, 64–71. doi:10.1093/infdis/jir710
Hofmann, S. R., Laass, M. W., Fehrs, A., Schuppan, D., Zevallos, V. F., Salminger, D., et al. (2018). IL10 Promoter Haplotypes May Contribute to Altered Cytokine Expression and Systemic Inflammation in Celiac Disease. Clin. Immunol. 190, 15–21. doi:10.1016/j.clim.2018.02.010
Howell, W. M., and Rose-Zerilli, M. J. (2007). Cytokine Gene Polymorphisms, Cancer Susceptibility, and Prognosis. J. Nutr. 137, 194S–199S. doi:10.1093/jn/137.1.194S
Kong, F., Liu, J., Liu, Y., Song, B., Wang, H., and Liu, W. (2010). Association of Interleukin-10 Gene Polymorphisms with Breast Cancer in a Chinese Population. J. Exp. Clin. Cancer Res. 29, 72. doi:10.1186/1756-9966-29-72
Kwong, A., Shin, V. Y., Ho, J. C. W., Kang, E., Nakamura, S., Teo, S.-H., et al. (2016). Comprehensive Spectrum ofBRCA1andBRCA2deleterious Mutations in Breast Cancer in Asian Countries. J. Med. Genet. 53, 15–23. doi:10.1136/jmedgenet-2015-103132
Langsenlehner, U., Krippl, P., Renner, W., Yazdani-Biuki, B., Eder, T., Köppel, H., et al. (2005). Interleukin-10 Promoter Polymorphism Is Associated with Decreased Breast Cancer Risk. Breast Cancer Res. Treat. 90, 113–115. doi:10.1007/s10549-004-3607-7
Li, M., Yue, C., Zuo, X., Jin, G., Wang, G., Guo, H., et al. (2020). The Effect of Interleukin 10 Polymorphisms on Breast Cancer Susceptibility in Han Women in Shaanxi Province. PLoS One 15, e0232174. doi:10.1371/journal.pone.0232174
Li, Y., Yu, H., Jiao, S., and Yang, J. (2014). Prognostic Value of IL-10 Expression in Tumor Tissues of Breast Cancer Patients. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 30, 517–520.
Maruthi, G., Ramachander, V., Komaravalli, P. L., Sureka, T., and Jahan, P. (2017). Association of Il10 Gene Polymorphisms (Rs 1800896, Rs1800872) in Breast Cancer Patients. Int. J. Med. Sci. Clin. Invention 4, 3050–3061. doi:10.18535/ijmsci/v4i6.19
Meijiang, L. (2014). The Relationship between Interleukin Gene Polymorphism and Females Genetic Susceptibility to Breast Cancer in Western Guangxi. J. Youjiang Med. Univ. Nationalities 36, 339–343. doi:10.3969/j.issn.1001-5817.2014.03.007
Moghimi, M., Ahrar, H., Karimi-Zarchi, M., Aghili, K., Salari, M., Zare-Shehneh, M., et al. (2018). Association of IL-10 Rs1800871 and Rs1800872 Polymorphisms with Breast Cancer Risk: A Systematic Review and Meta-Analysis. Asian Pac. J. Cancer Prev. 19, 3353–3359. doi:10.31557/apjcp.2018.19.12.3353
Mohamed, H. T., El-Husseiny, N., El-Ghonaimy, E. A., Ibrahim, S. A., Bazzi, Z. A., Cavallo-Medved, D., et al. (2018). IL-10 Correlates with the Expression of Carboxypeptidase B2 and Lymphovascular Invasion in Inflammatory Breast Cancer: The Potential Role of Tumor Infiltrated Macrophages. Curr. Probl. Cancer 42, 215–230. doi:10.1016/j.currproblcancer.2018.01.009
Moher, D., Liberati, A., Tetzlaff, J., Altman, D. G., and Group, P. (2009). Preferred Reporting Items for Systematic Reviews and Meta-Analyses: the PRISMA Statement. J. Clin. Epidemiol. 62, 1006–1012. doi:10.1016/j.jclinepi.2009.06.005
Mubarik, S., Wang, F., Fawad, M., Wang, Y., Ahmad, I., and Yu, C. (2020). Trends and Projections in Breast Cancer Mortality Among Four Asian Countries (1990-2017): Evidence from Five Stochastic Mortality Models. Sci. Rep. 10, 5480. doi:10.1038/s41598-020-62393-1
Oblak, T., Zadnik, V., Krajc, M., Lokar, K., and Zgajnar, J. (2020). Breast Cancer Risk Based on Adapted IBIS Prediction Model in Slovenian Women Aged 40-49 Years - Could it Be Better? Radiol. Oncol. 54, 335–340. doi:10.2478/raon-2020-0040
Onay, V. Ü., Briollais, L., Knight, J. A., Shi, E., Wang, Y., Wells, S., et al. (2006). SNP-SNP Interactions in Breast Cancer Susceptibility. BMC Cancer 6, 114. doi:10.1186/1471-2407-6-114
Patricia Gallegos-Arreola, M., Zúñiga González, G. M., Figuera, L. E., Puebla Pérez, A. M., and Delgado Saucedo, J. I. (2019). Association of the IL-10 Gene Rs1800872 (-592 C>A) Polymorphism with Breast Cancer in a Mexican Population. J. BUON 24, 2369–2376.
Pharoah, P. D. P., Tyrer, J., Dunning, A. M., Easton, D. F., Ponder, B. A. J., and Investigators, S. (2007). Association between Common Variation in 120 Candidate Genes and Breast Cancer Risk. Plos Genet. 3, e42. doi:10.1371/journal.pgen.0030042
Pooja, S., Chaudhary, P., Nayak, L. V., Rajender, S., Saini, K. S., Deol, D., et al. (2012). Polymorphic Variations in IL-1β, IL-6 and IL-10 Genes, Their Circulating Serum Levels and Breast Cancer Risk in Indian Women. Cytokine 60, 122–128. doi:10.1016/j.cyto.2012.06.241
Roh, J. W., Kim, M. H., Seo, S. S., Kim, S. H., Kim, J. W., Park, N. H., et al. (2002). Interleukin-10 Promoter Polymorphisms and Cervical Cancer Risk in Korean Women. Cancer Lett. 184, 57–63. doi:10.1016/s0304-3835(02)00193-3
Sabet, S., El-Sayed, S. K., Mohamed, H. T., El-Shinawi, M., and Mohamed, M. M. (2017). Inflammatory Breast Cancer: High Incidence of GCC Haplotypes (−1082A/G, −819T/C, and −592A/C) in the Interleukin-10 Gene Promoter Correlates with Over-expression of Interleukin-10 in Patients' Carcinoma Tissues. Tumour Biol. 39, 101042831771339. doi:10.1177/1010428317713393
Sancho-Garnier, H., and Colonna, M. (2019). Épidémiologie des cancers du sein. La Presse Médicale 48, 1076–1084. doi:10.1016/j.lpm.2019.09.022
Scola, L., Vaglica, M., Crivello, A., Palmeri, L., Forte, G. I., Macaluso, M. C., et al. (2006). Cytokine Gene Polymorphisms and Breast Cancer Susceptibility. Ann. N.Y Acad. Sci. 1089, 104–109. doi:10.1196/annals.1386.017
Setrerrahmane, S., and Xu, H. (2017). Tumor-related Interleukins: Old Validated Targets for New Anti-cancer Drug Development. Mol. Cancer 16, 153. doi:10.1186/s12943-017-0721-9
Sheikhpour, E., Noorbakhsh, P., Foroughi, E., Farahnak, S., Nasiri, R., and Neamatzadeh, H. (2018). A Survey on the Role of Interleukin-10 in Breast Cancer: a Narrative. Rep. Biochem. Mol. Biol. 7, 30–37.
Smith, K. C., Bateman, A. C., Fussell, H. M., and Howell, W. M. (2004). Cytokine Gene Polymorphisms and Breast Cancer Susceptibility and Prognosis. Eur. J. Immunogenet. 31, 167–173. doi:10.1111/j.1365-2370.2004.00462.x
Stang, A. (2010). Critical Evaluation of the Newcastle-Ottawa Scale for the Assessment of the Quality of Nonrandomized Studies in Meta-Analyses. Eur. J. Epidemiol. 25, 603–605. doi:10.1007/s10654-010-9491-z
Tian, K., Zhang, R., and Wang, X. (2017). Association of Interleukin-10 Polymorphisms and Haplotypes with the Risk of Breast Cancer in Northern China. Int. J. Clin. Exp. Pathol. 10, 6989–6996.
Vinod, C., Jyothy, A., Vijay Kumar, M., Raghu Raman, R., Nallari, P., and Venkateshwari, A. (2015). A Common SNP of IL-10 (-1082A/G) Is Associated with Increased Risk of Premenopausal Breast Cancer in South Indian Women. Iran J. Cancer Preven 8, e3434. doi:10.17795/ijcp-3434
Wang, Z., Liu, Q.-L., Sun, W., Yang, C.-J., Tang, L., Zhang, X., et al. (2014). Genetic Polymorphisms in Inflammatory Response Genes and Their Associations with Breast Cancer Risk. Croat. Med. J. 55, 638–646. doi:10.3325/cmj.2014.55.638
Westendorp, R. G., Langermans, J. A., Huizinga, T. W., Elouali, A. H., Verweij, C. L., Boomsma, D. I., et al. (1997). Genetic Influence on Cytokine Production and Fatal Meningococcal Disease. The Lancet 349, 170–173. doi:10.1016/s0140-6736(96)06413-6
Xu, G., and Wang, F. (2020). Associations of Polymorphisms in Interleukins with Susceptibility to Breast Cancer: Evidence from a Meta-Analysis. Cytokine 130, 154988. doi:10.1016/j.cyto.2020.154988
Zeng, F., Liu, W., Zhang, S., Tang, W., Zhao, X., and Zhang, Q. (2019). Correlation Analysis of IL-10 Gene Polymorphism and Breast Cancer Susceptibility in Women from Guangxi, China. Guo Ji Mian Yi Xue Za Zhi 42, 464–467. doi:10.3760/cma.j.issn.1673-4394.2019.05.004
Zhao, S., Wu, D., Wu, P., Wang, Z., and Huang, J. (2015). Serum IL-10 Predicts Worse Outcome in Cancer Patients: A Meta-Analysis. PLoS One 10, e0139598. doi:10.1371/journal.pone.0139598
Zhao, Y., Chen, S., Shen, F., Long, D., Yu, T., Wu, M., et al. (2019). In Vitro neutralization of Autocrine IL-10 Affects Op18/stathmin Signaling in Non-Small Cell Lung Cancer Cells. Oncol. Rep. 41, 501–511. doi:10.3892/or.2018.6795
Zhu, Z., Liu, J.-B., Liu, X., and Qian, L. (2020). Association of Interleukin 10 Rs1800896 Polymorphism with Susceptibility to Breast Cancer: a Meta-Analysis. J. Int. Med. Res. 48, 030006052090486. doi:10.1177/0300060520904863
Keywords: interleukin-10, gene polymorphism, breast cancer, meta-analysis, species variation
Citation: Li L, Xiong W, Li D and Cao J (2022) Association of Interleukin-10 Polymorphism (rs1800896, rs1800871, and rs1800872) With Breast Cancer Risk: An Updated Meta-Analysis Based on Different Ethnic Groups. Front. Genet. 13:829283. doi: 10.3389/fgene.2022.829283
Received: 05 December 2021; Accepted: 21 January 2022;
Published: 04 February 2022.
Edited by:
Rodrigo Drummond, A.C.Camargo Cancer Center, BrazilReviewed by:
Seyed Reza Mohebbi, Shahid Beheshti University of Medical Sciences, IranCopyright © 2022 Li, Xiong, Li and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiangang Cao, ODE0NzA5MjcwQHFxLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.