
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 30 August 2022
Sec. Evolutionary and Population Genetics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.827655
This article is part of the Research Topic Advancements in Genetic Control and Surveillance of Disease Vectors View all 6 articles
 Heng-Duan Zhang1†
Heng-Duan Zhang1† Jian Gao1†
Jian Gao1† Dan Xing1
Dan Xing1 Xiao-Xia Guo1
Xiao-Xia Guo1 Chun-Xiao Li1
Chun-Xiao Li1 Yan-De Dong1
Yan-De Dong1 Zhong Zheng1
Zhong Zheng1 Zu Ma1
Zu Ma1 Zhi-Ming Wu1
Zhi-Ming Wu1 Xiao-Juan Zhu1
Xiao-Juan Zhu1 Ming-Hui Zhao1
Ming-Hui Zhao1 Qin-Mei Liu1
Qin-Mei Liu1 Ting Yan1
Ting Yan1 Hong-Liang Chu2*
Hong-Liang Chu2* Tong-Yan Zhao1*
Tong-Yan Zhao1*Background: Aedes albopictus is an indigenous primary vector of dengue and Zika viruses in China. Wolbachia is a gram-negative and common intracellular bacteria, which is maternally inherited endosymbionts and could expand their propagation in host populations by means of various manipulations. Compared with research on the dispersion of Ae. albopictus at the macrospatial level (mainly at the country or continent level), little is known about its variation and Wolbachia infection at the microspatial level, which is essential for its management. Meanwhile, no local cases of dengue fever have been recorded in the history of Nanjing, which implies that few adulticides have been applied in the city. Thus, the present study examines how the Ae. albopictus population varies and the Wolbachia infection status of each population among microspatial regions of Nanjing City.
Methods: The genetic structure of 17 Aedes albopictus populations collected from urban, urban fringe, and rural regions of Nanjing City was investigated based on 9 microsatellite loci and the mitochondrial coxI gene. The Wolbachia infection status of each population was also assessed with Wolbachia A- and Wolbachia B-specific primers.
Results: Nine out of 58 tested pairs of microsatellite markers were highly polymorphic, with a mean PIC value of 0.560, and these markers were therefore chosen for microsatellite genotyping analysis. The Na value of each Ae. albopictus population was very high, and the urban area populations (7.353 ± 4.975) showed a lower mean value than the urban fringe region populations (7.866 ± 5.010). A total of 19 coxI haplotypes were observed among 329 Ae. albopictus individuals via haplotype genotyping, with the highest diversity observed among the urban fringe Ae. albopictus populations (Hd = 0.456) and the lowest among the urban populations (Hd = 0.277). Each Ae. albopictus population showed significant departure from HWE, and significant population expansion was observed in only three populations from the urban (ZSL), urban fringe (HAJY), and rural areas (HSZY) (p < 0.05). Combined with DAPC analysis, all the Ae. albopictus populations were adequately allocated to two clades with significant genetic differences according to population structure analysis, and the best K value was equal to two. AMOVA results showed that most (96.18%) of the genetic variation detected in Ae. albopictus occurred within individuals (FIT = 0.22238, p < 0.0001), while no significant positive correlation was observed via isolation by distance (IBD) analysis (R2 = 0.03262, p = 0.584). The TCS network of all haplotypes showed that haplotype 1 (H1) and haplotype 4 (H4) were the most frequent haplotypes among all populations, and the haplotype frequency significantly increased from urban regions (36.84%) to rural regions (68.42%). Frequent migration was observed among Ae. albopictus populations from rural to urban regions via the urban fringe region, with four direct migration routes between rural and urban regions. Furthermore, Wolbachia genotyping results showed that most of the individuals of each population were coinfected with Wolbachia A and Wolbachia B. The independent infection rate of Wolbachia A was slightly higher than that of Wolbachia B, and no significant differences were observed among different regions.
Conclusion: In the microspatial environment of Nanjing City, the urban fringe region is an important region for the dispersion of Ae. albopictus populations between rural and urban areas, and Wolbachia A and Wolbachia B coinfection is the most common Wolbachia infection status in all Ae. albopictus populations among different regions.
Aedes albopictus, which is distributed in southern Chaoyang, Liaoning Province and to eastern Tianshui in Gansu Province, China, is an important vector of several arboviruses, including dengue virus (DENV), Zika virus (ZIKV), and chikungunya (CHIKV), in mainland China and many other tropical and subtropical countries (Baolin, 1990; Li et al., 2018; Kraemer et al., 2019). Considering the frequent outbreaks of dengue fever among southern regions of China during the past few years, Ae. albopictus is regarded as the major target of the current integrated mosquito control and vector management program because it is widespread among urban and rural regions of southern China. The proposal of new reliable mosquito prevention and control strategies, especially for monitoring the dynamic changes and dispersion of mosquito populations among urban and rural regions also seems extremely important.
Source reduction and space spraying policies are the current strategies for the control of Aedes albopictus in China (Baolin, 1990). As the main strategy for Ae. albopictus control, the goal of source reduction is to reduce the population density of Ae. albopictus and eliminate its breeding sites without adulticide application; this approach is implemented when there is no outbreak of dengue fever. The insecticide space spraying policy is implemented as a supplementary control method when dengue fever emerges (Baolin, 1990; Hongliang, 2013; Yan-de et al., 2016). Source reduction may cause habitat fragmentation because the flight distance of Ae. albopictus is limited to 500 m (Taissa PDS et al., 2018) and could influence gene flow. Space spraying may decrease the densities of adult mosquitoes and the numbers of individuals sensitive to the insecticide (KO et al., 2017). The control of Ae. albopictus by source reduction is a component of the National Hygienic City program, which focuses on the built-up area of cities. The rural-urban continuum is a buffer zone where space spraying could prevent adult mosquitoes from invading built-up areas (Hongliang, 2013; Yan-de et al., 2016). It is crucial to study the population genetic structure of Ae. albopictus in the built-up areas, the rural-urban continuum, and rural areas of a city to achieve precise integrated mosquito management at the fine scale.
To understand the population genetic structure and dynamics of the gene flow patterns in habitats with a source reduction status based on mosquito population genetics studies, microsatellites are more frequently employed as markers in population studies of recent evolutionary events among subpopulations within an individual species or among closely related species relative to other biological markers, such as isoenzymes, random amplified polymorphic DNA (RAPD), restriction fragment length polymorphism (RFLP), single-strand conformation polymorphisms (SSCPs), and single nucleotide polymorphisms (SNPs), because of their per-locus advantages and high rate of polymorphism (Feldman et al., 1997; Sciarretta et al., 2001; Tsitrone et al., 2001; PORRETTA et al., 2006; Chambers et al., 2007; Tabachnick, 2010; Karl et al., 2012; Beebe et al., 2013; Delatte et al., 2013). These markers have been successfully used in studying many important vector mosquitos, such as Ae. albopictus, Aedes aegypti, Anopheles sinensis, Culex pipiens quinquefasciatus, Cx. pipiens pallens and Anopheles gambiae (Fonseca et al., 1998; Ma et al., 2011; Gélin et al., 2016; Miles et al., 2017; Wilke et al., 2018; Carvajal et al., 2020a; Gao et al., 2021). Such efforts revealed the fine-scale population genetic structure of the dengue mosquito vector Ae. aegypti in Metropolitan Manila, in the Philippines, based on microsatellite analysis, in low genetic differentiation was observed across mosquito populations, and high gene flow and/or weak genetic drift was found among mosquito populations, implying the passive or long-distance dispersal capability of Ae. aegypti, possibly through human-mediated transportation (Carvajal et al., 2020b). The structure of the studied populations of Ae. albopictus revealed using 12 microsatellite loci was divided into two groups, with restricted gene flow between these groups and no evidence of isolation by distance in the city of São Paulo (Multini et al., 2019).
Wolbachia is a rickettsia microorganism that is widely found in arthropods and can cause cytoplasmic incompatibility (CI), resulting in the decreasing densities of the host (Zhou et al., 1998a). The CI of mosquitos caused by Wolbachia was first discovered in Cx. pipiens and was later found in mosquitos such as Ae. albopictus and Armigeres subalbatus (Hertig and Wolbach, 1924; Hertig, 1936; Johnson, 2015; Nugapola et al., 2017). Numerous studies have shown that most widespread Ae. albopictus populations are infected with Wolbachia strain A or B alone, or coinfection by these strains. CI caused by different types of Wolbachia strains can affect the offspring of widespread Ae. albopictus populations, which in turn hinders the genetic communication of Ae. albopictus populations and affect the natural population genetic structure of the species (Sinkins et al., 1995a; Sinkins et al., 1995b; Zhou et al., 1998b; Das et al., 2014). Thus, the detection of Wolbachia infections in natural Ae. albopictus populations is another way to monitor the population dynamics of genetic structure.
Nanjing is an important tourism and economic centre in China and serves as an essential hub connecting the southern and northern regions of China via air transport, the Yangtze River, and railway transportation. Ae. albopictus is also the dominant mosquito in this city, and human-aided transmission is the main reason for its diffusion in natural conditions (Zhou et al., 1998b). Additionally, Nanjing is referred to as a National Hygienic City, and source reduction is carried out each year. The study of Ae. albopictus population genetics in Nanjing City to reveal the characteristics of genetic population structure provides a model of urban mosquito management-based source reduction and can contribute to an accurate approach for mosquito control. In the present study, a total of 17 Ae. albopictus habitats, distributed in either urban or rural areas, were investigated within the Nanjing region from July to September. Nine microsatellite loci in combination with the wsp gene were employed to analyse the genetic variability and Wolbachia infection status of each Ae. albopictus population. The results can provide a reference for subsequent mosquito management.
Aedes albopictus individuals were collected between July and September 2013 from 17 independent habitats within the Nanjing region, including urban, urban fringe and rural areas (Figure 1; Table 1). Larvae were surveyed using the container method (Zhang and Zeng, 2014); small indoor and outdoor water receptacles were examined for larvae at each sampling site. If larvae were found at a sampling site, a 1 km zone around that site was surveyed to confirm the essential interval between the different Ae. albopictus breeding habitats. Larvae were collected in the field with pipette and raised in a laboratory, and adults were identified as Ae. albopictus after emergence under a microscope (OLYMPUS, Japan) before DNA isolation (Lu, 2003).
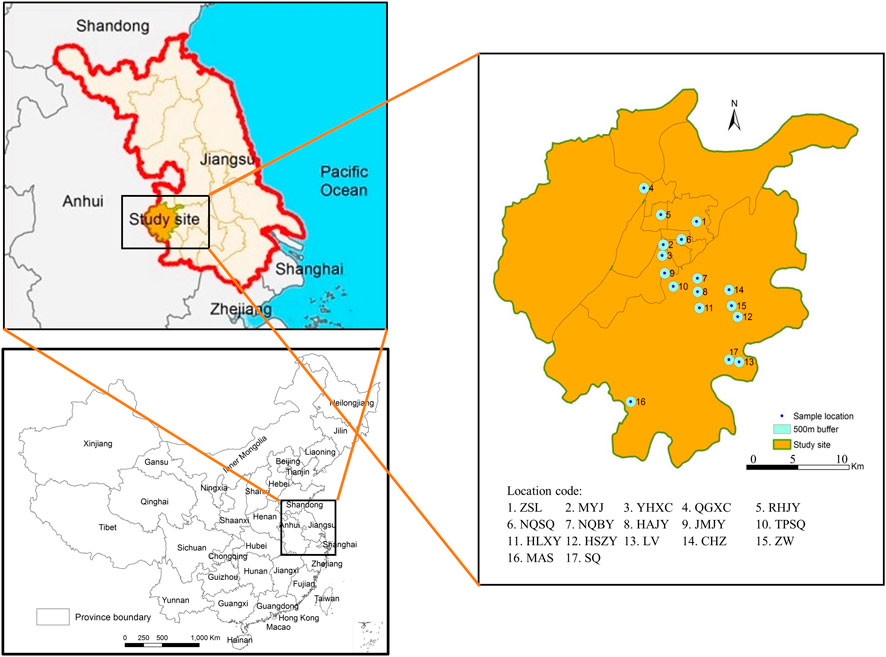
FIGURE 1. A province and county-level digital map was obtained from China Resource and Environment Science and Data Center (https://www.resdc.cn/), the detailed location of sample sites were mapped and showed using ArcGIS 10.7 (Environmental Systems Research Institute Inc., Redlands, CA, USA).
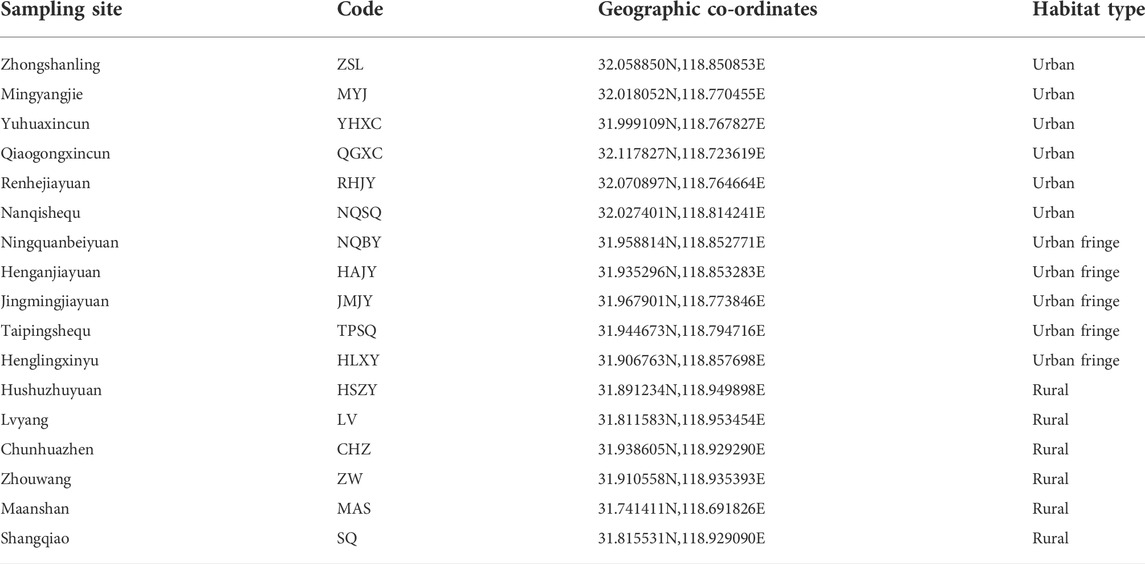
TABLE 1. Location names, geographic coordinates and habitat type of all 17 Ae. albopictus sampling sites.
We used GPS Pathfinder Office 3.10 to differentially correct the coordinates of Aedes albopictus breeding sites recorded with hand-held GPS, and the converted data were exported to an ESRI Shapefile. We used ArcGIS to delineate a 500 m radius buffer zone around each sampling site and to calculate the distance between each pair of sampling sites.
We genotyped a total of 510 mosquitoes, including 30 specimens (half male and female) from each of the 17 sample sites. The DNA of individual mosquitoes was extracted following the steps indicated in a TAKARA® DNA kit and stored in a -40 °C freezer until required. Individuals were genotyped based on the variation at nine microsatellite loci (PORRETTA et al., 2006; Chambers et al., 2007; Beebe et al., 2013; Delatte et al., 2013) (Table 2). Approximately 1–2 μL of each PCR product was diluted with 10 μL of autoclaved distilled water for DNA genotyping. Two microlitres of the diluted product was then added to 7.75 μL of Hi-Di™ Formamide and 0.25 μL of the Gene Scan 500 LIZ™ size standard. The mixture was heated at 94 °C for 5 min and then immediately chilled on ice for 2 min. Genotyping was carried out in a Genetic Analyzer 3130 xl (Applied Biosystems, United States). According to Bonacum J et al. (Bonacum et al., 2001), coxI sequence polymorphisms were investigated among 329 individuals. Briefly, the DNA amplification of a 550-bp fragment of coxI was performed on a T100 Thermal Cyclerin a 50 µL reaction mix containing 10 µL PCR buffer, 4 µL dNTPs, 1 µL primers and 0.5 µL PrimeSTAR® HS DNA polymerase (TAKARA, Japan). The PCR amplification program was set as follows: predenaturation at 94°C for 3 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 45 s and elongation at 72°C for 1 min, with a final extension at 72°C for 10 min. All the PCR products were detected and separated by 2% agarose gel electrophoresis. The coxI target fragments were then cut from the gel under UV light and purified with the GenElute™ PCR Clean-Up Kit (TAKARA, Japan). Each purified PCR product was subsequently cloned into the pCR™2.1 vector with a TA Cloning™ Kit (TAKARA, Japan) and selected by bacterial liquid PCR with T7 promoter primers. Finally, at least 20 positive clones for each PCR product were subjected to the sequencing of both strands using an ABI 3730XL automatic sequencer (Applied Biosystems, United States).
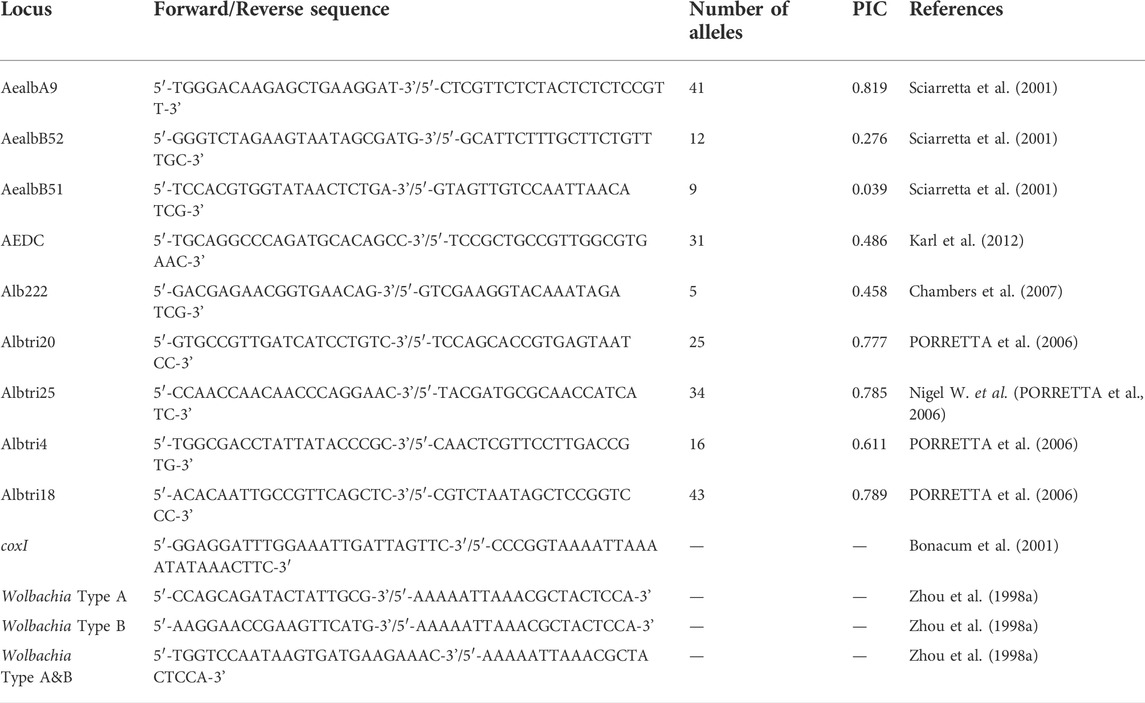
TABLE 2. Microsatellite primers and Wolbachia detection primers used to genotype Ae. albopictus specimens sampled from different regions of Nanjing City.
Genetic diversity (expected (He), observed (Ho) heterozygosity), the mean number of alleles (Na), and F-statistic parameters (FIS, FST, and FIT) were estimated from allele frequencies with FSTAT 2.9.3.2 (Goudet, 1995). For each locus-population combination in the global dataset and population groupings, we used Fisher’s exact test with Bonferroni correction to test for possible deviations from Hardy–Weinberg equilibrium (HWE) using POPGENE 1.32 (Rousset, 2008). Exact p-values were estimated using the Markov chain algorithm with 10,000 dememorizations, 500 batches, and 5000 iterations per batch. Pairwise differences between populations (Slatkin, 1995) and AMOVA analysis were displayed using Arlequin software 3.5.1.3 (Excoffier and Lischer, 2010) based on allelic frequency. In addition, the FST of each population was calculated from the mean value of the pairwise differences between populations. Bayesian clustering algorithms were implemented in STRUCTURE 2.3.3 (Pritchard et al., 2000; Falush et al., 2003) to infer population structure and explore the assignment of individuals and populations to specific genetic clusters. A UGPMA tree was built via NTsys software 2.10e. Excess heterozygosity at a significant number of loci is an indicator of a genetic bottleneck, whereas the converse situation (i.e., He, Heq) (Piry et al., 2017) may indicate population expansion. Estimates of Heq were computed using two mutation models for microsatellite evolution: a two-phase mutation model (T.P.M.) (Kimura and Ohta, 1978) and the stepwise mutation model (S.M.M.) (Nei, 1978). A Mantel test of the significance of genetic and geographic distances between each population was performed via the R package “adegenet 1.3”. Referring to the CoxI standard sequence MG198604.2, all the CoxI sequences were aligned with MAFFT 7.037 and credited via BioEdit 7.0.9.0. The haplotypes of all the populations were screened by DnaSP 6.0, and the TCS network was built via popART software 1.7, and the migration analysis was performed via R package “divMigrate” under R environment.
Following the Wolbachia wsp gene classification method established by Zhou et al. (Zhou et al., 1998a), we used three pairs of diagnostic primers to detect and identify Wolbachia.81F is a universal primer that can amplify a fragment of approximately 590–632 bp in all known Wolbachia strains. 328F amplifies an approximately 380 bp fragment specific to Wolbachia Type A, whereas 183F amplifies an approximately 501 bp fragment specific to Wolbachia Type B. The volume of amplification reaction was 50 μL, which included 25 µL Taq DNA polymerase, 16 µL ddH2O, 2 µL 10 μmol/L, the upstream and downstream primers, and a 5 µL DNA template. The amplification conditions were as follows: 95°C predenaturation for 3 min, followed by 35 cycles of amplification at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, and a final extension at 72°C for 7 min. Five microlitres of the above PCR product was used in 1.2% agarose gel electrophoresis. The results were examined under UV light, and the remaining products were sent to Tianyi Biotechnology Company Ltd.
In the present study, a total of 17 Aedes albopictus populations were collected from urban, urban fringe, and rural areas of Nanjing City (Table 1 and Figure 1). Each of the 17 breeding site buffer zones was isolated from each other, indicating the independence of each population. Additionally, 58 pairs of microsatellite markers were obtained from previous research, 9 of which were highly polymorphic and were therefore chosen for microsatellite genotyping analysis (Table 2). The PIC values of each locus ranged from 0.039 to 0.819, with a mean of 0.560. According to the definition of PIC values provided by Allah et al., nearly all the selected markers were highly informative (PIC value >0.5); the exception was AaelbB51, which had a value <0.25 and was considered to be a marker with low informativeness (Table 2). The linkage disequilibrium (LD) test showed that 286 pairs of loci among a total of 2412 (16.15%) across all locations were in significant LD after Bonferroni correction, and no consistency was found among them.
As show in Table 3, the observed number of alleles (Na) in each Aedes albopictus population was very high. The mean Na value of the urban area populations (7.353 ± 4.975) was lower than that of the rural area populations (7.387 ± 4.908), while the Na values of these two areas were all lower than those of the urban fringe area, which was 7.866 ± 5.010. The observed heterozygosity (Ho) values of all areas ranged from 0.543 ± 0.030 to 0.570 ± 0.030, which were lower than the expected heterozygosity (He) values (ranging from 0.566 ± 0.094 to 0.606 ± 0.096). The FIS value of each population ranged from −0.026 to 0.218, and each Ae. albopictus population showed significant departure from HWE (Table 3, Additional file 2: Supplementary Table S1). Pairwise FST analysis among different regions showed that the variations in the 17 populations were all significant and ranged from 0.003 to 0.092, and the FST value between urban and urban fringe regions (0.011) was greater than the FST value between the urban and rural regions (0.009) and the FST value between urban fringe and rural regions (0.007) (Additional file 3: Supplementary Table S2). Heterozygosity tests of all 17 Ae. albopictus populations based on the stepwise mutation model (S.M.M.) revealed that only three populations from the urban (ZSL), urban fringe (HAJY), and rural areas (HSZY) displayed significant bottleneck effects (p-values < 0.05), while four more populations from the urban (YHXC and NQSQ), urban fringe (TPSQ), and rural areas (CHZ) were also observed to have significant bottleneck effects under the two-phase model (T.P.M.) (Additional file 4 Supplementary Table S3).
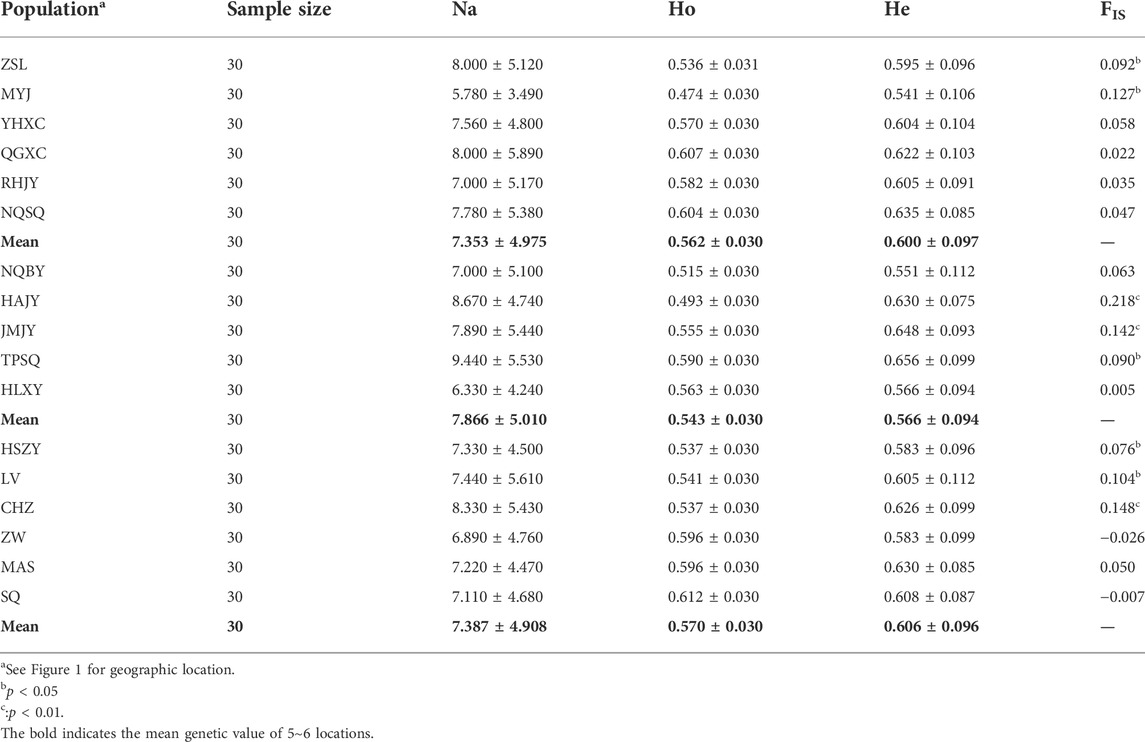
TABLE 3. Polymorphism and Inbreeding coefficient analysis at nine microsatellite loci in 17 Ae. albopictus specimens collected from 17 populations in Nanjing, China.
According to the population structure analysis results, all Aedes albopictus populations were adequately allocated to two clades with significant genetic differences, and the best K value, as determined via the ΔK method of Evanno et al., was also equal to two (Figures 2A,B). Additionally, a total of 88.7% of the variation was explained by 100 PCs in the discriminant analysis of principal components (DAPC) analysis; the structure analysis results revealed two genetically isolated groups (Figure 2C). The AMOVA results are presented in Table 4. From the total genetic variation partitioned, 5% could be attributed to differences among individuals within populations and 95% to differences within individuals (FIS = 0.02171, FIT = 0.22238, and all p < 0.0001). In contrast to the high individual variation observed, no significant positive correlation was observed among all individuals via isolation by distance (IBD) analysis (R2 = 0.03262, p = 0.584) (Figure 3).
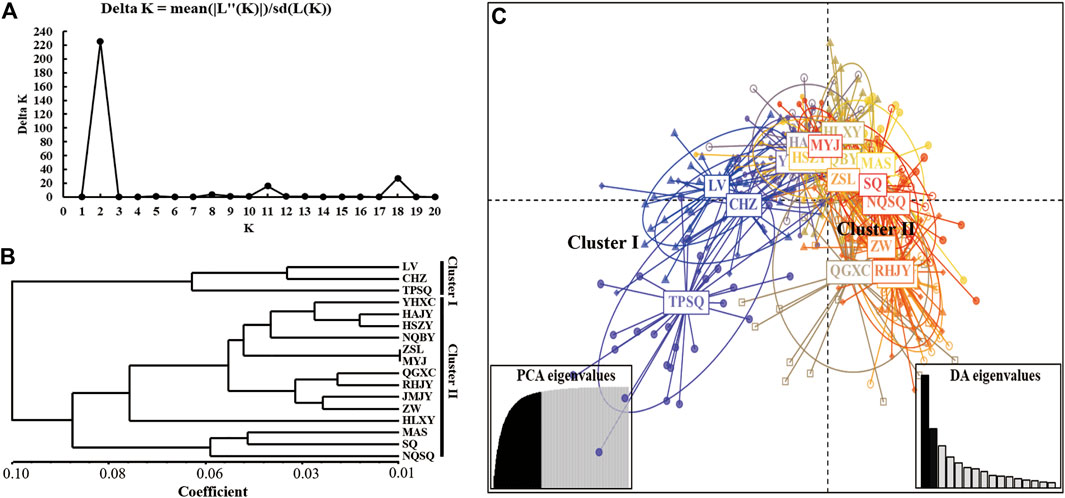
FIGURE 2. The population structure analysis of all 17 Ae. albopictus populations based on 9 microsatellite loci. (A) K values assessed via Evanno et al.‘s ΔK methods; (B) Bayesian clustering analysis of all Ae. albopictus populations; (C) DAPC analysis of all Ae. albopictus populations, and 86.4% of variation was explained by 50 PCs.
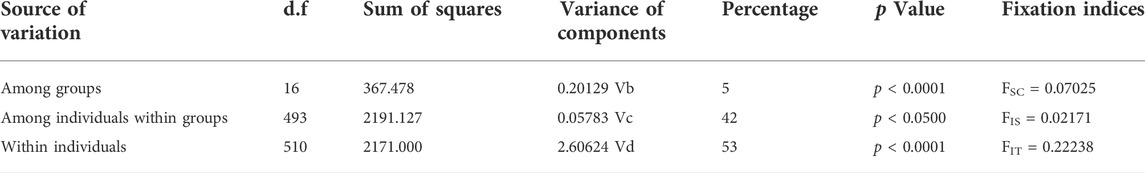
TABLE 4. AMOVA test of all 17 Ae. albopictus populations collected from different regions of Nanjing, China.
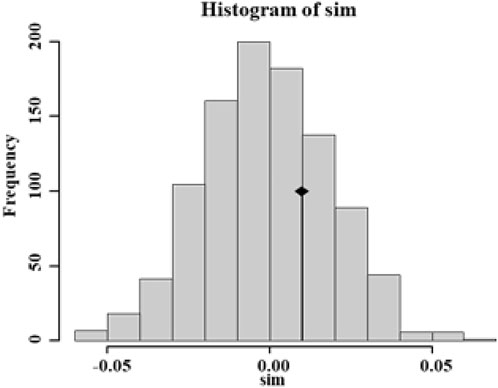
FIGURE 3. Isolation by distance (IBD) analysis of all 17 Ae. Albopictus populations from urban, urban fringe, and rural areas of Nanjing City.
A total of 19 coxI haplotypes were observed among 329 Aedes albopictus individuals based on an analysis of coxI sequences, and the haplotype indices changed dramatically across Ae. albopictus populations from different area (Table 5). The haplotype diversity (Hd) ranged from 0.0909 (RHJY, urban region) to 0.6727 (JMJY, urban fringe region), with nucleotide diversity (π) ranging from 0.0002 (RHJY, urban region) to 0.0024 (JMJY, urban fringe region). The average number of nucleotide differences (k) ranged from 0.0909 (RHJY, urban region) to 1.1636 (JMJY, urban fringe region), and the number of polymorphic sites ranged from 1 to 9 across all the populations. Overall, the urban fringe Ae. albopictus populations showed the highest diversity, with mean Hd, π, and k values of 0.4562, 0.0018 and 0.8836, respectively, whereas the urban populations showed the lowest diversity.
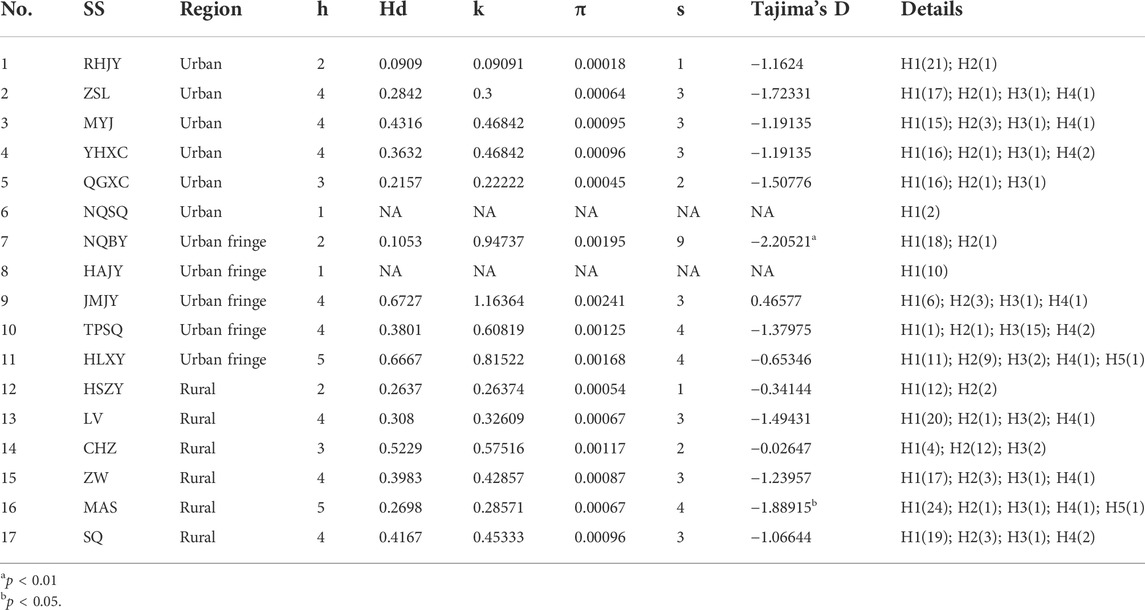
TABLE 5. The genetic diversity indices of all 17 Aedes albopictus populations via Mitochondrial coxI gene.
As shown in Figure 4, a TCS network was reconstructed with all 329 coxI sequences, and the results showed that haplotype 1 (H1) and haplotype 4 (H4) were the most frequent haplotypes and were distributed equally among all three sampling regions. Nearly all the other haplotypes were derived from H1 with one or more mutations, and the haplotype frequency increased significantly from the urban region (36.84%) to the rural area (68.42%). Relative to the urban regions (only H12 and H13), more exclusive haplotypes were detected among urban fringe regions (H6, H10, H17, and H19) and rural regions (H3, H15, H16, and H18). No additional shared haplotypes were detected between rural and urban fringe regions except H1 and H4.
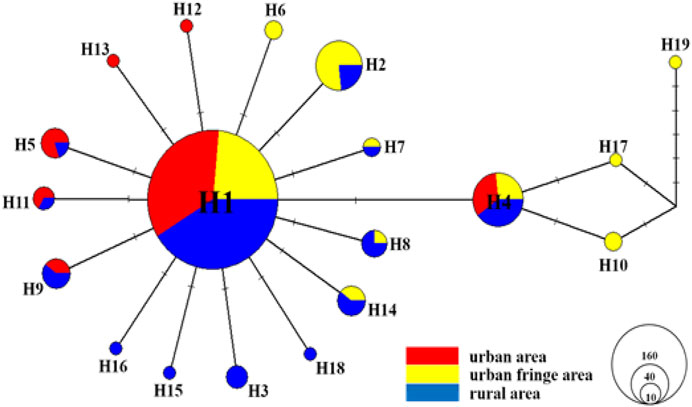
FIGURE 4. TCS haplotype network for the coxI gene of all Ae. albopictus individuals (n = 329) from urban, urban fringe, and rural areas of Nanjing City. The sizes of circles are proportional to haplotype frequency, and each line segment represents a single mutation.
Migration patterns were assessed using divMigrate networks representing all 17 Aedes albopictus populations. As expected, Ae. albopictus was observed to migrate frequently from rural to urban regions via the urban fringe region, and four direct migration routes between rural and urban regions were observed (between SQ and ZSL, ZSL and QGXC, SQ and NQSQ, and RHJY and QGXC, with Nm values as high as 0.68, 0.78, 0.55, and 0.83, respectively) (Figure 5). The obtained Tajima’s D values were negative, no statistically significant p-values were obtained except in population JMJY (urban fringe region), and the unimodal mismatch distributions were observed mainly among the urban fringe Ae. albopictus populations (Additional file 1: Supplementary Figure S1).
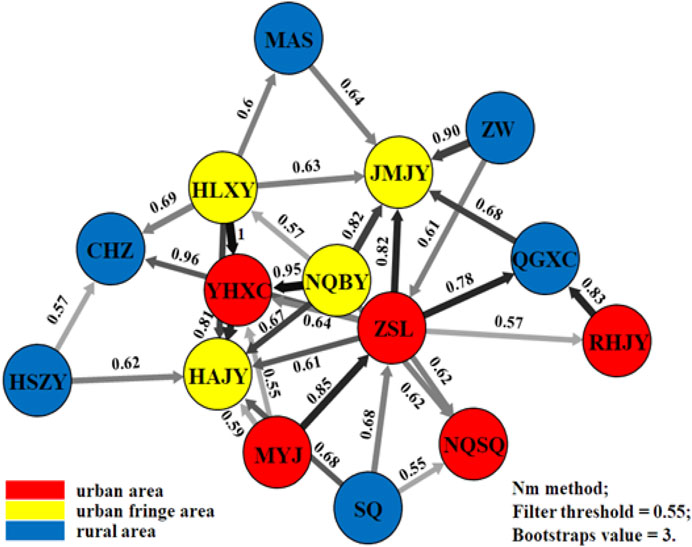
FIGURE 5. Migration patterns of Ae. albopictus among urban, urban fringe, and rural areas of Nanjing City.
Fragments of 500 bp and 380 bp were amplified from each of the 30 samples from all Aedes albopictus populations with the Wolbachia A- and Wolbachia B-specific primers, respectively, confirming Wolbachia infection. The sequencing results showed that most of the individuals of each population were coinfected with Wolbachia A and Wolbachia B, with infection rates (IR) ranging from 23.33% (ZSL, urban region) to 90.00% (HLXY, urban fringe region), and the mean infection rate of the rural region (70.56%) was significantly higher than that of the urban and urban fringe regions, which were 51.67 and 48.00%, respectively. Compared with the coinfection indices of Wolbachia A and Wolbachia B, the values of Wolbachia A and Wolbachia B for all individuals were low; the independent infection rate of Wolbachia A was slightly higher than that of Wolbachia B, and no significant differences were observed among different regions (Figure 6).
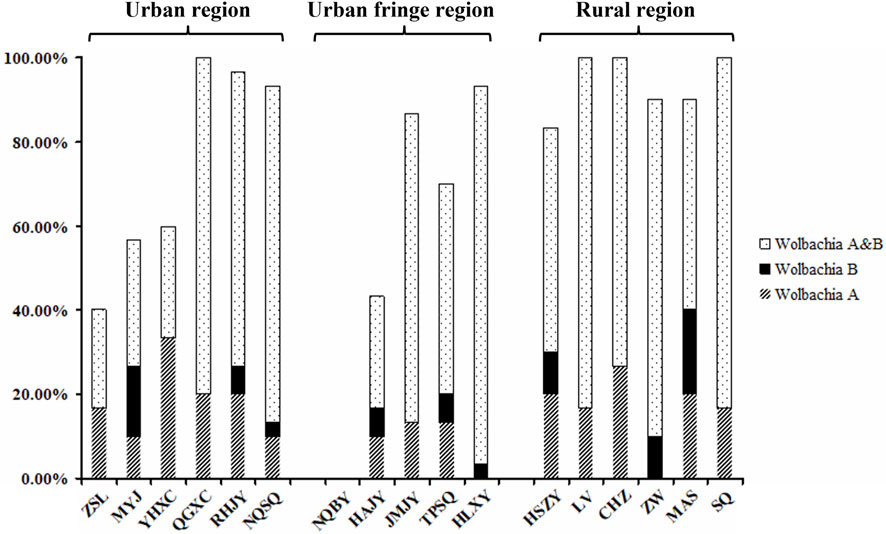
FIGURE 6. Independent infection rate of Wolbachia A and Wolbachia B among different Ae. albopictus populations from urban, urban fringe, and rural areas of Nanjing City.
The number of alleles, heterozygosity, and polymorphism information content (PIC) are common indices for evaluating the genetic diversity of populations (Nei et al., 1975; Botstein et al., 1980; Maudetr et al., 2002). The number of alleles is generally positively related to the level of genetic diversity among individual mosquitoes from the same population. Heterozygosity is also a measure of the genetic diversity of a population; the higher population heterozygosity is, the greater genetic diversity, polymorphism levels and the potential variation available for natural selection to act on will be. PIC is a relatively good index of gene fragment polymorphism. PIC values >0.5 indicate highly polymorphic loci. In earlier research, microsatellite loci and coxI have been widely used to evaluate the genetic variation and population structure of mosquitoes at a fine scale. The average number of alleles in this study was 7.387, the average expected heterozygosity was 0.606, and the average polymorphic information content was 0.560, which also indicated a similar status of Ae. aegypti in several areas. (Chambers et al., 2007; Beebe et al., 2013; Delatte et al., 2013; Monteiro et al., 2014; Multini et al., 2019; Carvajal et al., 2020a; Regilme et al., 2021). The variation of these nine microsatellites indicated that the 17 Ae. albopictus populations presented a relatively rich PIC and relatively even gene frequency distribution in Nanjing. This approach was shown to be a highly effective and reliable way of assessing genetic diversity. The results also indicated that our sample size was more than adequate. The analysis of the coxI polymorphism data of Ae. albopictus showed different haplotypes in each population; 19 haplotypes were found among the 329 samples, and the mean haplotype diversity was 0.359, which suggested relatively high genetic diversity.
A bottleneck effect will lead to excess heterozygosity in a population (Nei et al., 1975). It was shown herein that the Nanjing populations of Ae. albopictus, including ZSL, YHXC, NQSQ, HAJY, TPSQ, HSZY, and CHZ, may have experienced a bottleneck effect based on the results of the S.M.M. and T.P.M. model. FIS is an index of inbreeding within populations, where significant positive values indicate serious inbreeding within a population resulting in a loss of heterozygosity and significant negative values indicate outbreeding and an associated surplus of heterozygotes. The FIS values were all positive for the ZW and SQ populations, indicating that the degree of inbreeding in the populations was severe. The Ho values were lower than the He values except for the ZW and SQ populations, which indicated that the Ae. albopictus populations showed high gene consistency and suggested that the population might be experiencing inbreeding. The neutral test results for Tajima’s D showed that all populations except for NQSQ, HAJY, and JMJY presented negative values, suggesting that Ae. albopictus populations in Nanjing were selectively eliminated or were expanding (Tajima, 1989). The bottleneck effect and heterozygosity results showed that the Ae. albopictus populations in Nanjing were not in a state of expansion but were being selectively eliminated.
Implemented mosquito management (IMM) is aimed at reducing the density of mosquito species, which could change mosquito population genetics. An IMM program was effective in lowering the density of Ae. aegypti during an outbreak of dengue fever in Madeira in 2012, and a recent bottleneck effect of Ae. aegypti was simultaneously identified via heterozygosity tests (Seixas et al., 2019). The IMM of dengue vectors in China is focused on source reduction under conditions with no dengue cases. When the reported cases of dengue indicate an outbreak of the virus, the strategy of Ae. albopictus control involves both the chemical control of adult mosquitoes and source reduction (Hongliang, 2013). Nanjing is a national health city in China. Source reduction is a regular program applied every year to meet the standard of mosquito control in national health cities (General Administration of Quality Supervision IaQotPsRoC and Standardization Administration of China, 2011). Breeding sites of Ae. albopictus are regularly destroyed, the populations of Ae. albopictus are selectively eliminated, and the habitat of Ae. albopictus has become fragmented, which hinders gene flow and causes population inbreeding, leading to a bottleneck effect in some populations in Nanjing.
Aedes albopictus generally inhabits a home range with a distance of less than 50 m, and the flight distances of this species do not exceed 500 m (Hawley, 1988; Marini et al., 2010). Studies have shown that artificial landscapes such as urban roads are potential barriers to mosquito communication and hinder gene flow (Mondal et al., 2005; Gubler et al., 2010; Stoddard et al., 2019; Carvajal et al., 2020b; Regilme et al., 2021). However, urbanization substantially increases mosquito density and larval development. The Various populations in urban, suburban, and rural areas show differences in genetic differentiation, and the population in rural areas is more differentiated than other populations based on the results of FST analysis, which is different from the findings of Trinidad and Cairns et al. (Hemme et al., 2010; Schmidt et al., 2018; Regilme et al., 2021). Consistent with studies of Ae. albopictus conducted in Guangzhou (Li et al., 2014), our findings showed that although man-made landscapes such as urban roads are significantly more common in urban areas than in urban fringe and rural areas, urban areas are characterized by a higher human population density and more frequent human activities than urban fringe and rural areas, meaning that Ae. albopictus in urban areas still has more opportunities to feed on blood, mate, and reproduce. The genetic divergence index (FST) is an essential measure of population genetic divergence. Values within the range of 0.05–0.15 indicate moderate deviation, and values <0.05 indicate slight deviation (Balloux and Lugon-Moulin, 2002). The variation of the 17 populations was in the range of 0.003–0.092, indicating only slight genetic divergence among the 17 populations studied. Additionally, pairwise FST analysis among different regions showed that the FST value between urban and urban fringe regions (0.011) was greater than the FST value between urban and rural areas (0.009) and the FST value between urban fringe and rural areas (0.007), which indicates that the mosquitoes in the city come mainly from the urban fringes. Proliferation within cities is greater than direct movement from rural areas to cities, and the exchange of mosquito populations between cities and urban-rural junctions is greater than that between rural areas and urban-rural junctions. This may be because compared to rural areas, the urban-rural fringe is the main area where urban living resources are distributed, where the flow of people and logistics is intensive, with continuous transport to the city. At the same time, the corresponding resources in the city are transferred to other areas through the urban-rural fringe. Dispersal within the city has also been verified by migration analysis. The migration analysis showed that compared with the exchange of mosquitoes between the other two regions, there were 4 main mosquito communication paths between the city and the urban-rural junction, and the gene flow value along each path was greater than 0.8. A program of IMM can affect the gene flow of populations through area effects and isolation effects and ultimately lead to a decrease in the genetic diversity of populations. However, the results showed that 17 Ae. albopictus populations experienced genetic exchange, and there was no apparent loss of genetic diversity. This may have been due to the influence of the hosts, various vehicles and potential mates that different populations of Ae. albopictus may encounter.
Most populations of Aedes albopictus worldwide have been found to be infected with either Wolbachia Type A or B bacterial symbionts (Zhou et al., 1998b). CI can cause different populations of Ae. albopictus to be infected with different types of Wolbachia, causing them to produce few or no offspring, resulting in abnormal free mating between different populations and inbreeding within each population (Caragata et al., 2016; Schmidt et al., 2018; Xia et al., 2019; Zheng et al., 2019). Except for the NQBY population, all other populations were positive for Wolbachia. Wolbachia A and B sequencing results showed differences in Wolbachia infection status within each population. The genetic differentiation between populations is not obvious because Ae. albopictus individuals that are either infected with the same type of Wolbachia or not infected with Wolbachia can mate freely.
The present mosquito control strategy helps reduce the abundance and diversity of Aedes albopictus population, however, the active human-aided mosquito dispersion helps recover the Ae. albopictus population in a short period of time. Therefore, the continuous dynamic monitoring of Ae. albopictus population is important and will provide a solid foundation for the scientific prevention and control of Ae. albopictus.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
H-DZ, JG, H-LC, and T-YZ jointly designed and coordinated the study, with contributions from C-XL, X-XG, DX, and Y-DD. H-DZ drafted the article with contributions from JG. H-DZ, JG, ZZ, X-JZ, M-HZ, Q-ML, TY, and Z-MW collected samples from different climatic regions of China. H-DZ, JG, and ZM carried out the laboratory work and performed the statistical analysis. All authors read and approved the final manuscript.
This work was funded by grants from the Infective Diseases Prevention and Cure Project of China (No.2017ZX10303404).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.827655/full#supplementary-material
AMOVA test, Analysis of molecular variation test; coxI, Mitochondrial cytochrome c oxidase subunit 1; CI, Cytoplasmic Incompatibility; DAPC, Discriminant analysis of principal components; FIS, Inbreeding coefficient; FST, Genetic differences among populations; FIT: Genetic differences within individuals; Hd, Haplotype diversity; He, Excepted heterozygosity; Heq, Expected average heterozygosity; Ho, Observed heterozygosity; HWE, Hardy Weinberg equilibrium; IBD, Isolation by distance; IR, Infection Rates; IMM, Implemented mosquito management; LD, Linkage disequilibrium; Na, Observed number of alleles; Nm, Number of migrants; PIC, Polymorphism Information Content; S.M.M., Stepwise Mutation Model; T.P.M , Two phase model.
Balloux, F., and Lugon-Moulin, N. (2002). The estimation of population differentiation with microsatellite markers. Mol. Ecol. 11 (2), 155–165. doi:10.1046/j.0962-1083.2001.01436.x
Baolin, L. (1990). Dengue vector and dengue control in China: Guizhou People. Guizhou: s Publishing House.
Beebe, N. W., Ambrose, L., Hill, L. A., Davis, J. B., Hapgood, G., Cooper, R. D., et al. (2013). Tracing the tiger: Population genetics provides valuable insights into the Aedes (stegomyia) albopictus invasion of the australasian region. PLoS Negl. Trop. Dis. 7 (8), e2361. doi:10.1371/journal.pntd.0002361
Bonacum, J., DeSalle, R., O’Grady, P., Olivera, D., Wintermute, J., and Zilversmit, M. J. D. I. S. (2001). New nuclear and mitochondrial primers for systematics and comparative genomics in Drosophilidae. Dros. Inf. Serv. 84, 201–204.
Botstein, D., White, R. L., Skolnick, M., and Davis, R. W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32 (3), 314–331.
Caragata, E. P., Dutra, H. L. C., and Moreira, L. A. (2016). Exploiting intimate relationships: Controlling mosquito-transmitted disease with Wolbachia. Trends Parasitol. 32 (3), 207–218. doi:10.1016/j.pt.2015.10.011
Carvajal, T. M., Ogishi, K., Yaegashi, S., Hernandez, L. F. T., and Watanabe, K. J. P. N. T. D. (2020). Fine-scale population genetic structure of dengue mosquito. Aedes aegypti Metropolitan Manila, Philipp. 14 (5), e0008279.
Carvajal, T. M., Ogishi, K., Yaegeshi, S., Hernandez, L. F. T., Viacrusis, K. M., Ho, H. T., et al. (2020). Fine-scale population genetic structure of dengue mosquito vector, Aedes aegypti, in Metropolitan Manila, Philippines. PLoS Negl. Trop. Dis. 14 (5), e0008279. doi:10.1371/journal.pntd.0008279
Chambers, E. W., Meece, J. K., McGowan, J. A., Severson, D. W., Hemme, R. R., Chadee, D. D., et al. (2007). Microsatellite isolation and linkage group identification in the yellow fever mosquito Aedes aegypti. J. Hered. 98 (3), 202–210. doi:10.1093/jhered/esm015
Das, B., Satapathy, T., Kar, S. K., and Hazra, R. K. (2014). Genetic structure and Wolbachia genotyping in naturally occurring populations of Aedes albopictus across contiguous landscapes of Orissa, India. PLoS One 9 (4), e94094. doi:10.1371/journal.pone.0094094
Delatte, H., Toty, C., Boyer, S., Bouetard, A., Bastien, F., and Fontenille, D. (2013). Evidence of habitat structuring Aedes albopictus populations in Réunion Island. PLoS Negl. Trop. Dis. 7 (3), e2111. doi:10.1371/journal.pntd.0002111
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under linux and windows. Mol. Ecol. Resour. 10 (3), 564–567. doi:10.1111/j.1755-0998.2010.02847.x
Falush, D., Stephens, M., and Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164 (4), 1567–1587. doi:10.1093/genetics/164.4.1567
Feldman, M. W., Bergman, A., Pollock, D. D., and Goldstein, D. B. (1997). Microsatellite genetic distances with range constraints: Analytic description and problems of estimation. Genetics 145 (1), 207–216. doi:10.1093/genetics/145.1.207
Fonseca, D. M., Atkinson, C. T., and Fleischer, R. C. (1998). Microsatellite primers for Culex pipiens quinquefasciatus, the vector of avian malaria in Hawaii. Mol. Ecol. 7 (11), 1617–1619.
Gao, J., Zhang, H. D., Guo, X. X., Xing, D., Zhao, T. Y., Lan, C. J., et al. (2021). Dispersal patterns and population genetic structure of Aedes albopictus (Diptera: Culicidae) in three different climatic regions of China. Parasit. Vectors 14 (1), 12. doi:10.1186/s13071-020-04521-4
Gélin, P., Magalon, H., Drakeley, C., Maxwell, C., Magesa, S., Takken, W., et al. (2016). The fine-scale genetic structure of the malaria vectors Anopheles funestus and Anopheles gambiae (Diptera: Culicidae) in the north-eastern part of Tanzania. Int. J. Trop. Insect Sci. 36 (4), 161–170. doi:10.1017/s1742758416000175
General Administration of Quality Supervision IaQotPsRoC, Standardization Administration of China: Criteria for vector density control—mosquito. Beijing: China Standard Press; 2011: 8.
Goudet, J. (1995). FSTAT, a program to estimate and test gene diversities and fixation indices. LAUSANNE, Switzerland: Updated from Goudet.
Gubler, R. H. C. T. D. C. D. S. D., Thomas, C. L., Chadee, D. D., and Severson, D. W. (2010). Influence of urban landscapes on population dynamics in a short-distance migrant mosquito: Evidence for the dengue vector Aedes aegypti. PLoS Negl. Trop. Dis. 4 (3), e634. doi:10.1371/journal.pntd.0000634
Hemme, R. R., Thomas, C. L., Chadee, D. D., and Severson, D. W. (2010). Influence of urban landscapes on population dynamics in a short-distance migrant mosquito: Evidence for the dengue vector Aedes aegypti. PLoS Negl. Trop. Dis. 4 (3), e634. doi:10.1371/journal.pntd.0000634
Hertig, M. (1936). The Rickettsia, Wolbachia pipientis (gen. et sp.n.) and Associated Inclusions of the Mosquito, Culex pipiens. Parasitology 28 (4), 453–486. doi:10.1017/s0031182000022666
Hertig, M., and Wolbach, S. B. (1924). Studies on rickettsia-like micro-organisms in insects. J. Med. Res. 44 (3), 329327–329374.7.
Hongliang, Q. (2013). The pest control operation of health cities vector-biological control. Chin. J. Hyg. Insectides Equipments 19 (4), 4.
Johnson, K. N. (2015). The impact of Wolbachia on virus infection in mosquitoes. Viruses 7 (11), 5705–5717. doi:10.3390/v7112903
Karl, S. A., Toonen, R. J., Grant, W. S., and Bowen, B. W. (2012). Common misconceptions in molecular ecology: Echoes of the modern synthesis. Mol. Ecol. 21 (17), 4171–4189. doi:10.1111/j.1365-294X.2012.05576.x
Kimura, M., and Ohta, T. (1978). Stepwise mutation model and distribution of allelic frequencies in a finite population. Proc. Natl. Acad. Sci. U. S. A. 75 (6), 2868–2872. doi:10.1073/pnas.75.6.2868
Ko, C. Gunning, Astete, H., Vasquez, G. M., Morrison, A. C., Erhardt, E., Del Aguila, C., et al. (2017). Efficacy of Aedes aegypti control by indoor ultra low volume (ULV) insecticide spraying in iquitos, Peru. PLoS Negl. Trop. Dis. 12 (4), e0006378. doi:10.1371/journal.pntd.0006378
Kraemer, M., Messina, J., Faria, N., Pybus, O., Wint, W., Pigott, D. M., et al. (2019). Past and future spread of the arbovirus vectors Aedes aegypti and Aedes albopictus. Nat. Microbiol. 4, 854–863. doi:10.1038/s41564-019-0376-y
Li, C., Lu, Y., Liu, J., and Wu, X. (2018). Climate change and dengue fever transmission in China: Evidences and challenges. Sci. Total Environ. 622-623, 493–501. doi:10.1016/j.scitotenv.2017.11.326
Li, Y., Kamara, F., Zhou, G., Puthiyakunnon, S., Li, C., Liu, Y., et al. (2014). Urbanization increases Aedes albopictus larval habitats and accelerates mosquito development and survivorship. PLoS Negl. Trop. Dis. 8 (11), e3301. doi:10.1371/journal.pntd.0003301
Lu, B. W. H. (2003). Classification and identification of important medical insects of China: Henan: Henan science and technology. Henan: Pubulishing House.
Ma, Y., Yang, M., Fan, Y., Wu, J., Ma, Y., and Xu, J. (2011). Population structure of the malaria vector Anopheles sinensis (Diptera: Culicidae) in China: Two gene pools inferred by microsatellites. PLoS One 6 (7), e22219. doi:10.1371/journal.pone.0022219
Marini, F., Caputo, B., Pombi, M., Tarsitani, G., Torre, A. D. J. M., Entomology, V., et al. (2010). Study of Aedes albopictus dispersal in Rome, Italy, using sticky traps in mark–release. Med. Vet. Entomol. 24 (4), 361–368. doi:10.1111/j.1365-2915.2010.00898.x
Maudetr, C., Miller, C., Bassano, B., Breitenmoser-Würsten, C., Gauthier, D., Obexer-Ruff, G., et al. (2002). Microsatellite DNA and recent statistical methods in wildlife conservation management: Applications in alpine ibex [Capra ibex(ibex)]. Mol. Ecol. 11 (3), 421–436. doi:10.1046/j.0962-1083.2001.01451.x
Miles, A., Harding, N. J., Bottà, G., Clarkson, C. S., Antão, T., Kozak, K., et al. (2017). Genetic diversity of the African malaria vector Anopheles gambiae. Nature 552 (7683), 96–100. doi:10.1038/nature24995
Mondal, R., Webb, C. E., Williams, C. R., and Ritchie, S. A. (2005). Mark–release–recapture study to measure dispersal of the mosquito Aedes aegypti in Cairns, Queensland, Australia. Med. Vet. Entomol. 19 (4), 451–457. doi:10.1111/j.1365-2915.2005.00589.x
Monteiro, F. A., Schama, R., Martins, A. J., Gloria-Soria, A., Brown, J. E., Powell, J. R., et al. (2014). Genetic diversity of Brazilian Aedes aegypti: Patterns following an eradication program. PLoS Negl. Trop. Dis. 8 (9), e3167. doi:10.1371/journal.pntd.0003167
Multini, L. C., de Souza, A., Marrelli, M. T., and Wilke, A. B. B. (2019). Population structuring of the invasive mosquito Aedes albopictus (Diptera: Culicidae) on a microgeographic scale. PLoS One 14 (8), e0220773. doi:10.1371/journal.pone.0220773
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89 (3), 583–590. doi:10.1093/genetics/89.3.583
Nei, M., Maruyama, T., and Chakraborty, R. (1975). The bottleneck effect and genetic variability in populations. Evolution 29 (1), 1–10. doi:10.1111/j.1558-5646.1975.tb00807.x
Nugapola, N., De Silva, W., and Karunaratne, S. (2017). Distribution and phylogeny of Wolbachia strains in wild mosquito populations in Sri Lanka. Parasit. Vectors 10 (1), 230. doi:10.1186/s13071-017-2174-9
Piry, S., Luikart, G., and Cornuet, J-M. (2017). Bottleneck : A computer program for detecting recent reductions in the effective population size using allele frequency data.
Porretta, D., Gargani, M., Bellini, R., Calvitti, M., and Urbanelli, S. (2006). Isolation of microsatellite markers in the tiger mosquito Aedes albopictus (Skuse). Mol. Ecol. Notes 6 (3), 880–881. doi:10.1111/j.1471-8286.2006.01384.x
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155 (2), 945–959. doi:10.1093/genetics/155.2.945
Regilme, M. A. F., Carvajal, T. M., Honnen, A. C., Amalin, D. M., and Watanabe, K. (2021). The influence of roads on the fine-scale population genetic structure of the dengue vector Aedes aegypti (Linnaeus). PLoS Negl. Trop. Dis. 15 (2), e0009139. doi:10.1371/journal.pntd.0009139
Rousset, F. (2008). genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 8 (1), 103–106. doi:10.1111/j.1471-8286.2007.01931.x
Schmidt, T. L., Filipović, I., Hoffmann, A. A., and Rašić, G. (2018). Fine-scale landscape genomics helps explain the slow spatial spread of Wolbachia through the Aedes aegypti population in Cairns, Australia. Hered. (Edinb) 120 (5), 386–395. doi:10.1038/s41437-017-0039-9
Sciarretta, A., Trematerra, P., and Baumgärtner, J. (2001). Geostatistical analysis of cydia funebrana (Lepidoptera: Tortricidae) pheromone trap catches at two spatial scales. Am. Entomologist 47 (3), 174–185. doi:10.1093/ae/47.3.174
Seixas, G., Salgueiro, P., Bronzato-Badial, A., Gonçalves, Y., Reyes-Lugo, M., Gordicho, V., et al. (2019). Origin and expansion of the mosquito Aedes aegypti in Madeira island (Portugal). Sci. Rep. 9 (1), 2241–2245. doi:10.1038/s41598-018-38373-x
Sinkins, S. P., Braig, H. R., and O'Neill, S. L. (1995). Wolbachia pipientis: Bacterial density and unidirectional cytoplasmic incompatibility between infected populations of Aedes albopictus. Exp. Parasitol. 81 (3), 284–291. doi:10.1006/expr.1995.1119
Sinkins, S. P., Braig, H. R., and O'Neill, S. L. (1995). Wolbachia superinfections and the expression of cytoplasmic incompatibility. Proc. Biol. Sci. 261 (1362), 325–330. doi:10.1098/rspb.1995.0154
Slatkin, M. (1995). A measure of population subdivision based on microsatellite allele frequencies. Genetics 139 (1), 457–462. doi:10.1093/genetics/139.1.457
Stoddard, S. G. Y. L. A. P. J. W. S., Lee, Y., Pierce, A. A., Wong, J., Chu, Y. Y., Morrison, A. C., et al. (2019). The genetic structure of Aedes aegypti populations is driven by boat traffic in the Peruvian Amazon. PLoS Negl. Trop. Dis. 13 (9), e0007552. doi:10.1371/journal.pntd.0007552
Tabachnick, W. J. (2010). Challenges in predicting climate and environmental effects on vector-borne disease episystems in a changing world. J. Exp. Biol. 213 (6), 946–954. doi:10.1242/jeb.037564
Taissa Pds, R. D., Vieira, S. D. A. F., Santos de Abreu, F. V., Luz, S. L. B., Santalucia, M., Jiolle, D., et al. (2018). Potential of Aedes albopictus as a bridge vector for enzootic pathogens at the urban-forest interface in Brazil. Emerg. Microbes Infect. 7, 191–198. doi:10.1038/s41426-018-0194-y
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 (3), 585–595. doi:10.1093/genetics/123.3.585
Tsitrone, A., Rousset, F., and David, P. (2001). Heterosis, marker mutational processes and population inbreeding history. Genetics 159 (4), 1845–1859. doi:10.1093/genetics/159.4.1845
Wilke, A. B. B., de Carvalho, G. C., and Marrelli, M. T. (2018). Retention of ancestral polymorphism in Culex nigripalpus (Diptera: Culicidae) from São Paulo, Brazil. Infect. Genet. Evol. 65, 333–339. doi:10.1016/j.meegid.2018.08.017
Xia, S., Baskett, M. L., and Powell, J. R. (2019). Quantifying the efficacy of genetic shifting in control of mosquito-borne diseases. Evol. Appl. 12 (8), 1552–1568. doi:10.1111/eva.12802
Yan-de, D., Wei-wei, L., Hong-liang, Q., Jing, L., Ying, T., Yong, Z., et al. (2016). Effect of creating national healthy counties on prevention and control of mosquito-borne disease. Chin. J. Hyg. Insectides Equipments 22 (04), 6.
Zhang, Y. T. L., and Zeng, X. P. (2014). Analysis of surveillance results and trends of mosquito density in Beijing from 2010 to 2012. Chin. J. Vector Biol. Control 9 (1), 5–12.
Zheng, X., Zhang, D., Li, Y., Yang, C., Wu, Y., Liang, X., et al. (2019). Incompatible and sterile insect techniques combined eliminate mosquitoes. Nature 572 (7767), 56–61. doi:10.1038/s41586-019-1407-9
Zhou, W., Rousset, F., and O'Neil, S. (1998). Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. Biol. Sci. 265 (1395), 509–515. doi:10.1098/rspb.1998.0324
Keywords: Aedes albopictus, genetic structure, haplotype, microspatial, urban region, microsatellite loci, wolbachia
Citation: Zhang H-D, Gao J, Xing D, Guo X-X, Li C-X, Dong Y-D, Zheng Z, Ma Z, Wu Z-M, Zhu X-J, Zhao M-H, Liu Q-M, Yan T, Chu H-L and Zhao T-Y (2022) Fine-scale genetic structure and wolbachia infection of aedes albopictus (Diptera: Culicidae) in Nanjing city, China. Front. Genet. 13:827655. doi: 10.3389/fgene.2022.827655
Received: 02 December 2021; Accepted: 27 July 2022;
Published: 30 August 2022.
Edited by:
Roya Haghighat-Khah, Imperial College London, United KingdomReviewed by:
Perran Ross, University of Melbourne, AustraliaCopyright © 2022 Zhang, Gao, Xing, Guo, Li, Dong, Zheng, Ma, Wu, Zhu, Zhao, Liu, Yan, Chu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-Liang Chu, bWVkY2h1QHZpcC4xNjMuY29t; Tong-Yan Zhao, dG9uZ3lhbnpoYW9AMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.