- 1CAS Key Laboratory of Mountain Ecological Restoration and Bioresource Utilization and Ecological Restoration and Biodiversity Conservation Key Laboratory of Sichuan Province, Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences, Yezin Nay Pyi Taw, Myanmar
Animal olfactory systems evolved with changes in habitat to detect odor cues from the environment. The aquatic environment, as a unique habitat, poses a formidable challenge for olfactory perception in animals, since the higher density and viscosity of water. The olfactory system in snakes is highly specialized, thus providing the opportunity to explore the adaptive evolution of such systems to unique habitats. To date, however, few studies have explored the changes in gene expression features in the olfactory systems of aquatic snakes. In this study, we carried out RNA sequencing of 26 olfactory tissue samples (vomeronasal organ and olfactory bulb) from two aquatic and two non-aquatic snake species to explore gene expression changes under the aquatic environment. Weighted gene co-expression network analysis showed significant differences in gene expression profiles between aquatic and non-aquatic habitats. The main olfactory systems of the aquatic and non-aquatic snakes were regulated by different genes. Among these genes, RELN may contribute to exploring gene expression changes under the aquatic environment by regulating the formation of inhibitory neurons in the granular cell layer and increasing the separation of neuronal patterns to correctly identify complex chemical information. The high expression of TRPC2 and V2R family genes in the accessory olfactory systems of aquatic snakes should enhance their ability to bind water-soluble odor molecules, and thus obtain more information in hydrophytic habitats. This work provides an important foundation for exploring the olfactory adaptation of snakes in special habitats.
Introduction
Vertebrates have evolved complex phenotypes to adapt to different ecotopes driven by various selection regimes. The olfactory sensory system has played a crucial role in this process due to its involvement in food acquisition, mate and offspring recognition, predator avoidance, and habitat detection (Nei et al., 2008; Gomez-Diaz et al., 2018). Olfactory systems perform critical chemical communication functions, which are highly dependent on environmental media. Notably, propagation media and molecules differ markedly between terrestrial and aquatic environments (Freitag et al., 1998). Higher density and viscosity of water, coupled with a much lower diffusion speed in comparison to air make olfactory perception difficult underwater environment (Weiss et al., 2021). Therefore, the olfactory systems of animals have evolved to adapt to aquatic habitats. For instance, unlike for terrestrial mammals, in aquatic mammals the vomeronasal system is inactivated and the olfactory nervous system may be lost (Thewissen and Nummela, 2008; Kishida, 2021).
Snakes are a special group of animals that have evolved a suite of complex traits that enable them to survive in a variety of habitats worldwide (Peng et al., 2020; Ren et al., 2022). Their sensory system is unique among vertebrates, with severely degraded visual and acoustic faculties (Christensen et al., 2012; Lillywhite, 2014; Cronin, 2020) but highly developed and specialized olfactory systems. Snakes possess two olfactory systems: i.e., the main olfactory system (MOS), with the olfactory bulb as the core, and the accessory olfactory system (AOS), consisting of the vomeronasal organ and tongue. These two olfactory systems are separated completely and function independently. The MOS is sensitive to airborne molecules and detects volatile chemicals, while the AOS is specifically stimulated by non-volatile and water-soluble macromolecular chemicals, such as pheromones (Wang and Halpern, 1980a; Wang and Halpern, 1980b; Lohman and Smeets, 1993). The highly developed olfaction of snakes, accompanied by the degeneration of other senses, suggests that olfactory system evolution plays an important role in environmental adaptation.
The two olfactory systems of snakes are regulated by different receptors. Odorant receptors (ORs) are localized in the MOS and vomeronasal receptors (V1Rs and V2Rs) play a role in the AOS (Buck and Axel, 1991; Herrada and Dulac, 1997; Mombaerts, 2004). V1Rs and class II ORs are thought to bind to chemicals in the air, while water-soluble chemical molecules are recognized by V2Rs and class I ORs (Shi and Zhang, 2007; Nei et al., 2008; Saito et al., 2009). Snakes contain many OR and V2R genes (Halpern, 2003; Hogan et al., 2021). Compared with non-aquatic snakes, the MOS of sea snakes is degenerated and OR genes are rarely expressed, but gigantic mapping of V2Rs genes exists (Kishida et al., 2019; Li et al., 2021). While previous studies have focused on receptor copy number variation, the comprehensive expression patterns of the olfactory in aquatic snakes remain to be parsed.
To explore the molecular basis of aquatic adaptation of olfaction in snakes, we performed transcriptome sequencing and weighted gene co-expression network analysis (WGCNA) of 26 samples (main olfactory bulb and vomeronasal organ) from two aquatic and two non-aquatic snake species: i.e., Hypsiscopus plumbea and Opisthotropis zhaoermii are typical groups of aquatic snakes (Murphy and Voris, 2014; Ren et al., 2017), and Ahaetulla prasine and Pareas menglaensis perched on bushwood (Amber et al., 2017; Wang et al., 2021), thus the four species are practicable models for exploring olfactory characteristics associated with habitat types. We observed considerable environment-dependent differences in overall expression profiles. In the MOS of aquatic snakes, the unique co-expression network regulated by RELN may facilitate the accuracy of chemical information identification to adapt to complex underwater environments. In the AOS of aquatic snakes, the expression of TRPC2 gene and V2R family is associated with the ability to bind water-soluble molecules. These findings contribute to our understanding of the physiological mechanisms underpinning olfactory adaptation to the environment.
Materials and Methods
Sample Collection
This study involved four species: Ahaetulla prasina, Hypsiscopus plumbea, Opisthotropis zhaoermii and Pareas menglaensis. We defined two categories of habitat types based on propagation medium: 1) aquatic; 2) non-aquatic. More than 50% of their lifetime is in the water (Sheehy et al., 2015), and they are assumed as aquatic snakes. If not, they are considered as non-aquatic snakes. H. plumbea and O. zhaoermii were seen commonly in ponds or streams, and A. prasina and P. menglaensis were discovered in wooded areas frequently (Zhao, 2006). Therefore, H. plumbea and O. zhaoermii were assumed as aquatic snakes, A. prasina and P. menglaensis were considered as non-aquatic snakes (Figure 1).

FIGURE 1. Four species of snakes in the study. (A) Hypsiscopus plumbea (aquatic). (B) Opisthotropis zhaoermii (aquatic). (C) Ahaetulla prasina (non-aquatic). (D) Pareas menglaensis (non-aquatic). Photos by Jin-Long Ren and Jun-Jie Huang.
Based on phylogenetic relationships, four distantly related snake species were selected from two different habitats for transcriptome sequencing (Supplementary Figure S1). A total of 13 healthy and adult snakes were collected, including three biological duplications of A. prasine, H. plumbea, and O. zhaoermii, respectively, and four individuals of P. menglaensis (Supplementary Table S1). The vomeronasal organ that is an important component of the AOS and the main olfactory bulb that is integral to the MOS of specimens were harvested. The vomeronasal organ and main olfactory bulb tissues were collected using stereo microscope according to Kardong (2012) (Supplementary Figure S2). After sampling, all the tissues were immediately frozen in liquid nitrogen for ten minutes and stored at –80°C prior until RNA isolation.
RNA Isolation, cDNA Library Construction and mRNA Sequencing
Total RNA was extracted from olfactory tissues using QIAGEN® RNA Mini Kit following the manufacturer’s protocol. After degradation and contamination monitored on 1% agarose gel, RNA purity was checked using the NanoPhotometer® spectrophotometer (IMPLEN, Calabasas, CA, United States). We used the Qubit®RNA Assay Kit in Qubit®2.0 Flurometer (Life Technologies, Carlsbad, CA, United States) to measure RNA concentration, and used the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2,100 system (Agilent Technologies, Santa Clara, CA, United States) to evaluate the RNA integrity number (RIN). Each sample has extracted 1.5 μg RNA. NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, United States) was used to construct sequencing libraries following the manufacturer’s instruction. Add index code to the attribute sequence of each sample. TruSeq PE Cluster Kit v3-cBot-HS (Illumina) was used to cluster index-coded samples on a cBot Cluster Generation System. RNA sequencing was performed on an Illumina NovaSeq 6,000 platform and 150 bp paired-end reads were generated.
De novo Transcriptome Assembly and Sequence Annotation
The low-quality reads of raw reads data were filtered by using seqtk v1.3-r106, and the remaining were considered as clean reads data. Following preprocessing, the high-quality reads data of 26 samples were employed for de novo assembly using Trinity v2.4.0 (Grabherr et al., 2013) with parameters “-group_pairs_distance 230 -min_contig_length 600 -min_glue 4” (Tang et al., 2021), and the resulting sequences were identified as unigenes. TransDecoder v3.0.1 was used to extract the open reading frame (ORF) and predict coding DNA sequence (CDS) (Lee et al., 2015), and BUSCO v3.0.2 (Benchmarking Universal Single-Copy Orthologs) was used to assess the completeness of transcriptome (Simão et al., 2015). We used the BLASTP algorithm for searching longest coding protein genes against the non-redundant SWISS-Port database1 (-line 2 -word_size 4 -evalue 1e-5 -top 3) to match the best-retrieved result. Additionally, the same protocol was used against the NCBI non-redundant protein (NR) database and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Gene ontology (GO) functional annotation of unigenes was obtained by using an internal Perl script, which is based on SWISS-Prot annotated results. GO annotation results were counted by WEGO 2.02 (Ye et al., 2018).
Orthologous Genes Identification and Quantification of Gene Expression
BLAST algorithm was used to identify orthologous genes based on amino acid sequences of four species. Sequence alignments were conducted for transcripts between four species (E-value = 1e-5). We obtained reciprocal best hits in each four and retained orthologous genes shared by four species. The TPM (Transcripts Per Million) value was calculated to determine the expression level of each transcript. Gene and isoform abundances were calculated with RSEM (Li and Dewey, 2011).
Principal Component Analysis and Weighted Gene Co-expression Network Analysis
Principal component analysis (PCA) was applied with the R package FactoMineR (Lê et al., 2008) and factoextra (Kassambara and Mundt, 2017) to gain more insight into the cluster of samples based on expression data of orthologous genes. The co-expression networks of the orthologous genes were constructed using the WGCNA v1.69 package (Langfelder and Horvath, 2008) in R based on the TPM. We calculated expression correlation coefficients of the orthologous genes to select a suitable soft threshold using a scale-free topology model (Wan et al., 2018). The module was defined by setting the minimum number of modules to 150 and the hierarchal clustering dendrogram was cut using the dynamic tree-cutting algorithm (mergeCutHeight = 0.2). Genes were clustered with similar expression patterns into a co-expression module and the module eigengenes (ME) were calculated for each module. The olfactory systems of snakes in two types of habitats were defined as different phenotypes, and we focused on the modules that are most relevant to the traits. Subsequently, we constructed a co-expression network, and visualized using Cytoscape v3.7.2 (Shannon et al., 2003) based on the genes in the corresponding module and identified hub genes of the co-expression network according to the degree method using cytoHubba (Chin et al., 2014).
Gene Ontology Enrichment Analysis
To understand the potential functions of the module eigengenes, GO enrichment analysis was performed using the R package clusterProfiler v3.14.3 (Yu et al., 2012). The GO annotations with all annotated unigenes were seen as background set to identify significantly enriched GO terms. Enriched terms with p-values < 0.05 were considered significant (Liu et al., 2020). And GO enrichment results were completed redundancy removal by REVIGO3 (Supek et al., 2011).
Results
De novo Transcriptome Assembly and Functional Annotation
To compare the olfactory gene expression profiles between aquatic and non-aquatic snakes, we collected 26 samples (main olfactory bulb and vomeronasal organ) from four species of snakes inhabiting two types of habitats. A total of 797.88 million 150-bp paired-end raw reads were generated, and 762.97 million clean reads (95.7%) were retained after quality control. Among the samples, the Q30 scores were >90% and GC content ranged from 42.95% to 46.95% (Supplementary Table S1). The clean reads were assembled using Trinity v2.4.0, generating a total of 207,427 transcripts with an N50 of 3,447 bp for A. prasina, 243,737 transcripts with an N50 of 3,512 bp for H. plumbea, 217,064 transcripts with an N50 of 3,506 bp for O. zhaoermii, and 270,112 transcripts with an N50 of 3,598 bp for P. menglaensis (Supplementary Table S2). Based on nucleotide mode and lineage data from vertebrates, BUSCO v3.0.2 (Simão et al., 2015) was used to assess transcriptome assembly, which showed that more than 77% of protein-coding genes were complete and the proportion of fragmented genes was under 10% (Supplementary Table S3).
Among the transcripts, 71,526 for A. prasina, 74,197 for H. plumbea, 77,549 for O. zhaoermii, and 94,479 for P. menglaensis contained open reading frames (ORFs) that predicted peptides >100 amino acids (aa) in length, including 36,058, 37,947, 38,713 and 44,937 full-length transcripts for predicting protein-coding sequences. After removing redundancy and clustering, 25,839, 27,413, 27,793, and 31,844 of the longest protein-coding transcripts were retained for orthologous gene analysis. The longest protein-coding transcripts were searched against the NR, Gene Ontology (GO), SWISSProt, and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Results showed that 17,963 (69.5%) in A. prasina, 18,032 (65.8%) in H. plumbea, 19,082 (68.7%) in O. zhaoermii, and 18,424 (57.9%) in P. menglaensis were significantly matched to known genes in the databases (Figures 2A–D). Functional annotation analysis divided these genes into cellular components, molecular functions, and biological process categories, with the annotated GO terms shown in Figure 2E.
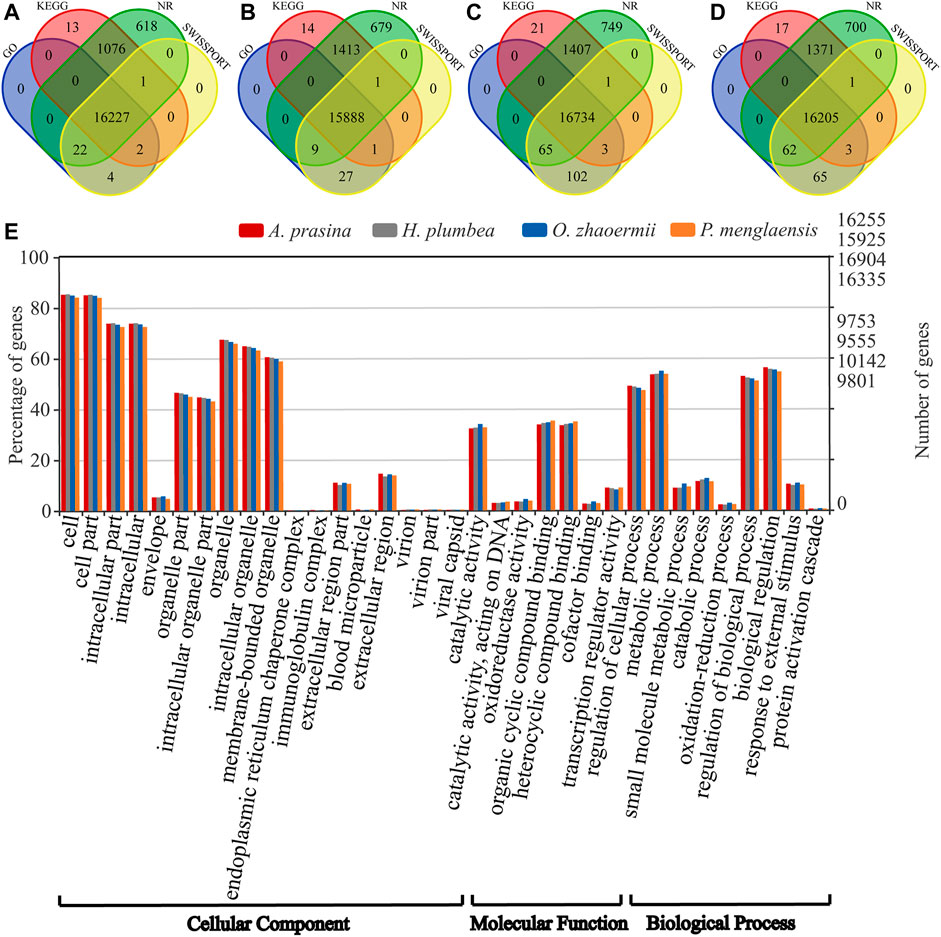
FIGURE 2. Overview of annotative summary of de novo transcriptome assembly. (A–D) Venn diagram showing the number of specific and overlapping annotated unigenes between annotated results from four public databases. (A) Functional annotation of transcripts in of A. prasina. (B) Functional annotation of transcripts of H. plumbea. (C) Functional annotation of transcripts of O. zhaoermii. (D) Functional annotation of transcripts of P. menglaensis. (E) Comparison of Gene Ontology (GO) classifications based on de novo transcriptome assembly of four species of snakes.
Principal Component Analysis and Weighted Gene Co-expression Network Analysis
PCA was performed based on transcripts per million (TPM) and revealed the presence of two distinct transcriptomic clusters, i.e., MOS and AOS, showing different expression patterns (Supplementary Figure S3A). Regarding to the MOS tissue, aquatic snakes clustered in a group and could be clearly distinguished from P. menglaensis (Figure 3A), as well as A. prasine (Supplementary Figure S3B). In the AOS data, the aquatic and non-aquatic samples clustered separately and the greatest source of variation was derived from habitat type by PC1 (Figure 3B), indicating considerable differences in olfaction gene expression patterns between aquatic and non-aquatic snakes.
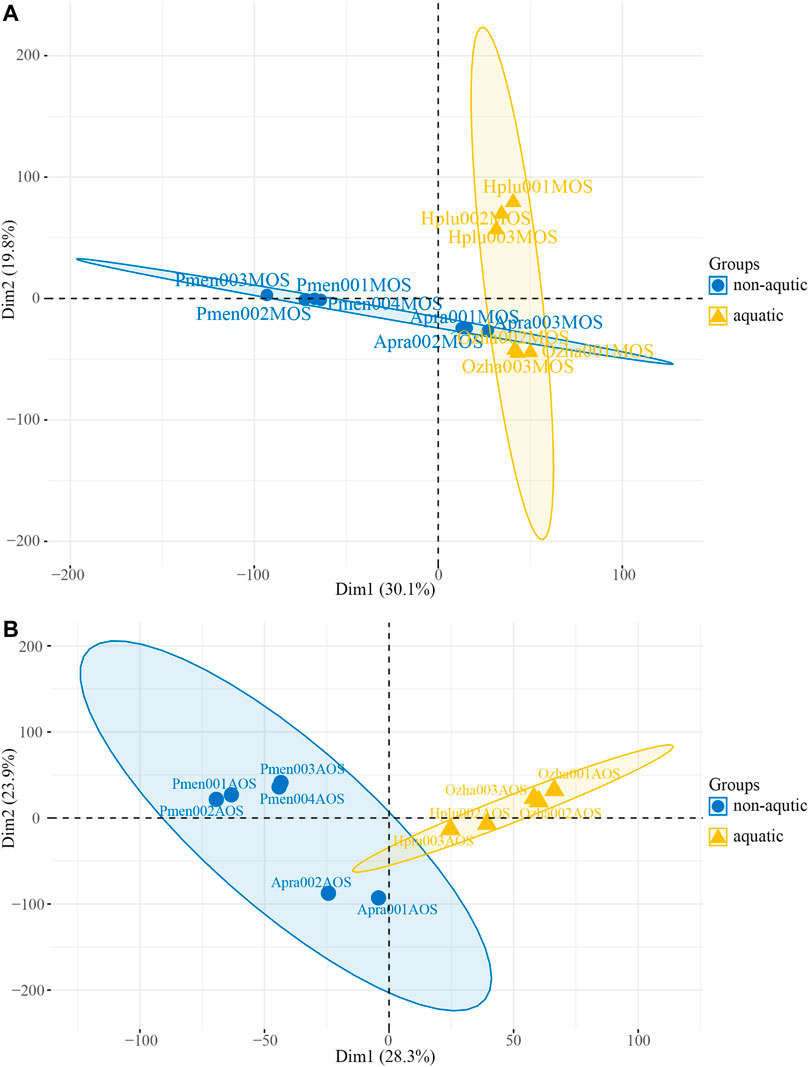
FIGURE 3. Plot of the principal component analysis (PCA) of four species of snakes. (A) PCA plot of main olfactory system (MOS) samples. (B) PCA plot of accessory olfactory system (AOS) samples.
A total of 8,618 orthologous genes were identified among the four species, which were used to construct weighted gene co-expression networks via the WGCNA v1.69 package using the R programming language. A beta power of 12 (Wan et al., 2018) was selected as the soft-thresholding power to ensure a scale-free network (Figure 4A). Strongly co-expressed genes in clusters were screened and assigned to a module, as shown in Figure 4B. The size of the modules depended on the number of genes contained therein, with the turquoise module containing the largest number of genes (861) and the light-yellow module containing the least (212). Genes assigned to the gray and gray60 modules were not co-expressed. The heatmap of the topological overlap matrix of all genes indicated that gene expression was relatively independent between modules (Figure 4C).
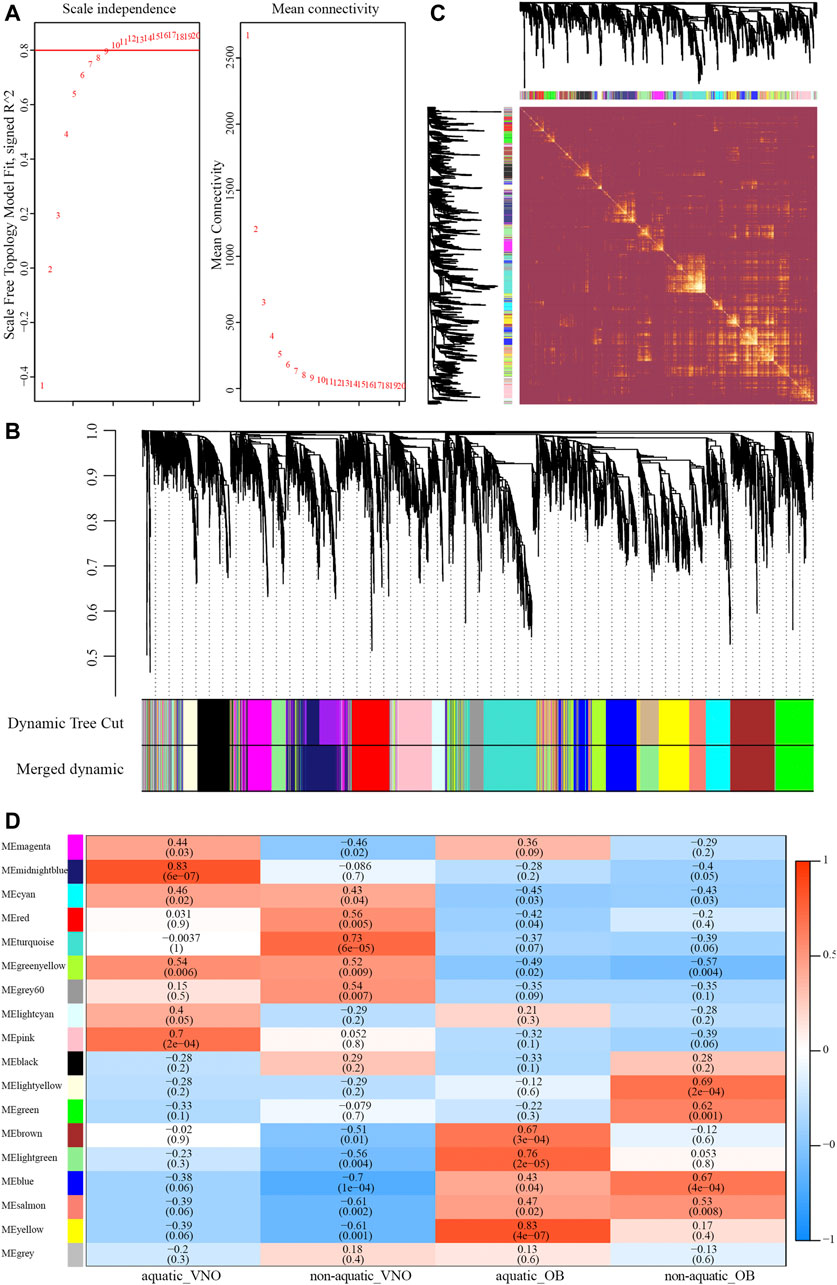
FIGURE 4. Construction of the weighted gene co-expression network (WGCNA). (A) Analysis of soft-thresholding powers based on scale independence (left) and mean connectivity (right). (B) The cluster dendrogram of orthologous genes. Each row corresponds to a module eigengene. (C) Network heatmap plot of orthologous genes. (D) Module-traits relationships identified by WGCNA. The color and the number (above) of each cell indicate the correlation and the numbers in parentheses represent p-value.
To identify modules significantly associated with habitat type, module-trait relationships were analyzed using correlations between module eigengenes and four traits (i.e., aquatic AOS, non-aquatic AOS, aquatic MOS, and non-aquatic MOS) (Figure 4D). The module with the highest correlation coefficient was defined as the habitat-specific module, however, results of enrichment analysis were not significant in the non-aquatic MOS module (light-yellow). Thus, another module that significantly related to the MOS of non-aquatic snakes and olfactory-related items could be significantly enriched was selected. A total of five habitat-specific modules were selected, including the midnight-blue (aquatic AOS, 779 genes), turquoise (non-aquatic AOS, 861 genes), yellow (aquatic MOS, 572 genes), and light-yellow and blue modules (non-aquatic MOS, 212 and 682 genes).
Differential Expression Patterns in Main Olfactory System Between Aquatic and Non-aquatic Snakes
For each gene expression profile, gene significance (GS) was calculated to measure the correlation between habitat and expression profiles, and module membership (MM) was calculated to measure the correlation between ME and expression profiles. Scatterplots of GS and MM in the aquatic and non-aquatic MOS-related modules are shown in Figures 5A,B. In the two modules, GS and MM were correlated, indicating that significant habitat-associated genes were important elements of the modules.
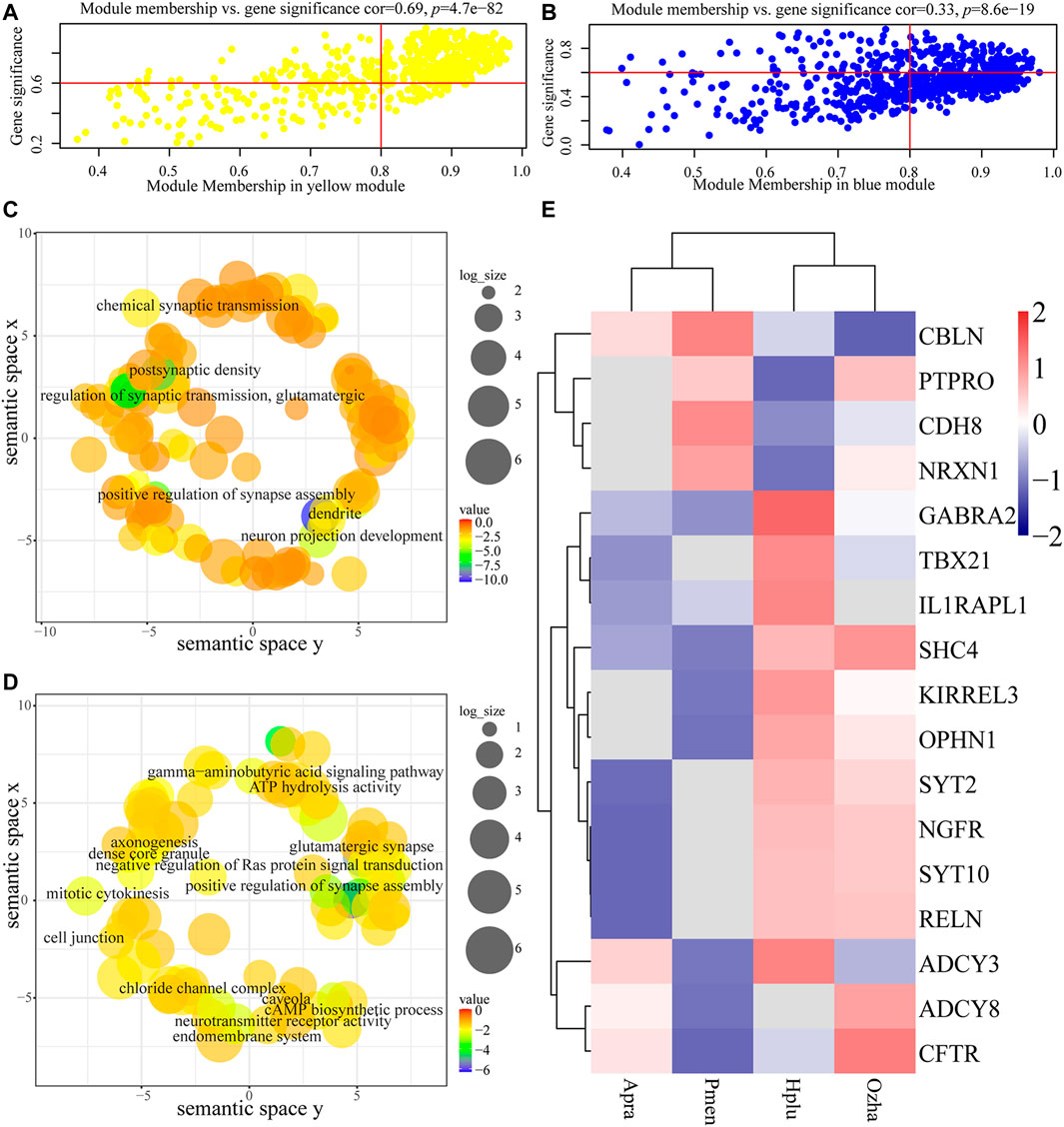
FIGURE 5. Analysis of the aquatic MOS (yellow) module and the non-aquatic MOS (blue) module. (A, B) The x-axis is module membership and the y-axis stand for gene significance. Gene significance (GS) was calculated to measure the correlation between habitat and expression profiles and module membership (MM) was calculated to measure the correlation between ME and expression profiles. (A) Scatterplots of gene significance (GS) for habitat traits versus module membership (MM) in the aquatic MOS module. (B) Scatterplots of gene significance (GS) for habitat traits versus module membership (MM) in the non-aquatic MOS module. (C, D) Each term was assigned x and y coordinates and more semantically similar GO terms were closer in the plot. The size of the circles indicates the number of child GO terms. (C) GO enrichment analysis of genes in the aquatic MOS module was visualized using REVIGO. (D) GO enrichment analysis of genes in the non-aquatic MOS module was visualized using REVIGO. (E) The expression of olfactory-related genes in the two modules.
Functional enrichment analysis revealed the biological significance of modules. Results showed that genes in the aquatic MOS-related modules (yellow) were mainly involved in the process of neuronal connection (e.g., KIRREL3, OPHN1, and SYT2), including dendrite (GO:0030425), postsynaptic density (GO:0014069), neuron projection development (GO:0031175), and chemical synaptic transmission (GO:0007268) (Figure 5C, Supplementary Table S5). In the non-aquatic MOS-related module (light-yellow), genes were mainly involved in the process of biosynthesis, including Cul3-RING ubiquitin ligase complex (GO:0031463) and cholesterol biosynthetic process (GO:0006695) (Supplementary Table S4). In the non-aquatic MOS-related module (blue), synapse assembly (GO:0051965) and glutamatergic synapse (GO:0098978) were the most abundantly represented GO terms, suggesting that the genes were highly associated with neuronal connection (e.g., CDH8, ADCY3, PTPRO, NRXN1, CBLN2, ADCY8, and CFTR) (Figure 5D, Supplementary Table S6). Intramodular connectivity was ranked according to the degree method using the cytoHubba plug-in, and the top 10 genes in each module of interest were used to visualize the specific modules (Supplementary Figure S4B). Among the hub genes, RELN, which is involved in the differentiation process of the granular cell layer in main olfactory bulb (Martin-Lopez et al., 2012), was highly expressed in aquatic snakes (Figure 5E).
Differential Expression Patterns in Accessory Olfactory System Between Aquatic and Non-aquatic Snakes
The GS and MM values of AOS-related modules were calculated to measure the correlation between module eigengenes and habitat type, which showed relatively high correlation (Figures 6A,B). Functional enrichment analysis indicated that the genes in the aquatic AOS-related module (midnight-blue) were involved in the process of olfactory signal transduction (e.g., TRPC2, V2R, and GPR180), including response to pheromone (GO:0019236), G protein-coupled olfactory receptor activity (GO:0038022), and neuron differentiation (GO:0030182) (Figure 6C, Supplementary Table S7). In the non-aquatic AOS-related module (turquoise), GO enrichment analysis indicated that the genes mainly participated in the regulation of cilium (e.g., BBS1, BBS4, and ARL13B), including cilium assembly (GO:0060271), cilium (GO:0005929), and motile cilium (GO:0031514) (Figure 6D, Supplementary Table S8).
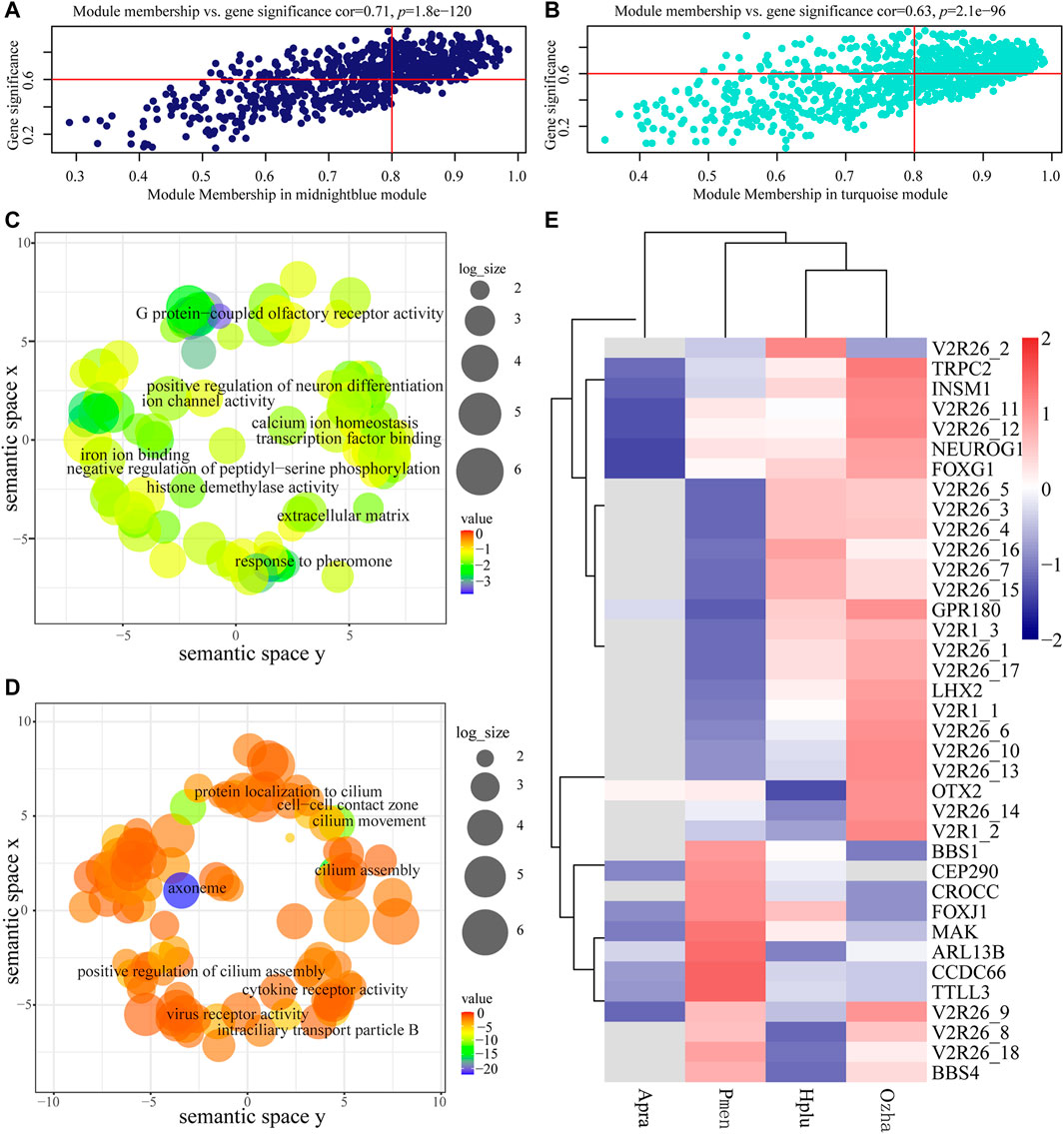
FIGURE 6. Analysis of the aquatic AOS (midnight-blue) module and the non-aquatic AOS (turquoise) module. (A) Scatterplots of gene significance (GS) for habitat traits versus module membership (MM) in the aquatic AOS module. (B) Scatterplots of gene significance (GS) for habitat traits versus module membership (MM) in the non-aquatic AOS module. (C) GO enrichment analysis of genes in the aquatic AOS module was visualized using REVIGO. (D) GO enrichment analysis of genes in the non-aquatic AOS module was visualized using REVIGO. (E) The expression of olfactory-related genes in the two modules.
The top 10 genes with the greatest connectivity were identified as hub genes, which may play roles in olfactory signal transduction (Supplementary Figure S4A). Among these genes, TRPC2 and V2R26_10 were identified as olfactory-related genes with involvement in response to pheromone (GO:0019236). A co-expression subnetwork containing TRPC2 and V2R26_10 was constructed, which showed that TRPC2 was co-expressed with 15 receptor genes (14 vomeronasal receptors and GPR180) and five neuronal development-related genes (OTX2, LHX2, NEUROG1, INSM1, and FOXG1). Of which, 11 vomeronasal receptor transcripts, GRP180, LHX2, NEUROG1, INSM1, and FOXG1, were highly expressed in the vomeronasal organ of aquatic snakes (Figure 6E), suggesting the potential vital role of the subnetwork in adaptations to the water environment.
Discussion
In animals, olfaction plays an important physiological function by sensing chemical cues in the environment, which provide key information on food, mates, and predators, thereby allowing organisms to make critical behavioral decisions at greater distances (Korsching, 2016). Different environments have different communication media, and it is hypothesized that transition to a new environment can result in olfactory system adaptations (Shichida et al., 2013). With the degeneration of their other senses (e.g., vision and hearing), snakes have developed a keen sense of smell. Together with their diverse habitats, snakes thus provide an excellent opportunity to explore the molecular basis and expression features of olfactory systems in vertebrates from different habitats (Lillywhite, 2014). In this study, we generated RNA-seq data from MOS and AOS tissues of aquatic and non-aquatic snakes and constructed a co-expression network related to habitat type. Results revealed unique gene expression profiles associated with environmental adaptation, especially key expression networks of aquatic adaptation. Given the lack of relevant published data, our findings provide an important resource for exploring the adaptation of vertebrate olfactory systems to special habitats.
Our results showed that both aquatic and non-aquatic MOS-related modules were involved in synapse and neuron-related terms, indicating similar functions and the key role of synapses in the MOS. In the non-aquatic MOS module, seven genes participated in the regulation of synapse formation and olfactory neuron maturation and homeostasis. PTPRO is implicated in synapse growth and guidance during embryonic development (Kotani et al., 2010). ADCY3 plays an indispensable role in postnatal maturation of olfactory sensory neurons and the absence of ADCY3 leads to cumulative defects in olfactory sensory neuronsmaturation (Zhang et al., 2017). CFTR is localized in microvillar cells of the olfactory epithelium and regulates microvillar cells to maintain neuronal tissue homeostasis (Pfister et al., 2015). In the aquatic MOS module, synapse-related terms were enriched in different genes, including GABRA2, OPHN1, RELN, TBX21, IL1RAPL1, SHC4, KIRREL3, SYT2, and SYT10. GABRA2, OPHN1, and RELN are involved in the regulation of inhibitory neuronal formation, while the six other genes are involved in the regulation of neuronal maturation. RELN was also identified as a highly expressed hub gene in the aquatic MOS co-expression network, indicating its vital role in the MOS of aquatic snakes. In mice, RELN knockout results in disruption of the granular cell layer (Martin-Lopez et al., 2012). Granular cells inhibit mitral cells (MCs) through their dendrodendritic reciprocal connections, and inhibition of MCs can weaken odor representation and enhance the signal-to-noise ratio (Isaacson and Strowbridge, 1998; Koulakov and Rinberg, 2011). Therefore, RELN likely plays a vital role in aquatic snake MOS and may facilitate adaptation to aquatic environments by regulating the granular cell layer. Additionally, inhibitory neuron activation can reduce the excitability of MCs and increase MCs pattern separation, which enables the olfactory bulb to disambiguate sensory stimuli with overlapping features (Gschwend et al., 2015). Regulation of the maturation and formation of inhibitory neurons is regulated by OPHN1 and GABRA2 (Pallotto et al., 2012; Redolfi et al., 2016), which showed higher expression in the aquatic snakes. The nasal cavity in snakes bears a rich sensory epithelium that is sensitive to air-borne volatile chemicals (Lillywhite, 2014). In aquatic snakes, however, degradation of the MOS and large-scale reduction in olfactory receptors suggest degeneration of the aquatic MOS, which could be interpreted as an adaptation to environmental media (Kishida and Hikida, 2010; Kishida et al., 2019; Peng et al., 2020; Li et al., 2021). Our results suggest that aquatic snakes may increase neuronal pattern separation via the inhibitory neurons suppressing MCs to eliminate overlapping information from olfactory stimuli, thereby enabling the acquisition of valuable information within complex water environments. Note, in addition to the habitat types, diet may also affect the expression pattern of MOS, which may explain the clustering pattern of transcriptome data, i.e., in the MOS data aquatic snakes clustered in a group and could be clearly distinguished from P. menglaensis (Danaisawadi et al., 2016). P. menglaensis specializes in eating snails and the diet is unique to other snakes (Auer et al., 2020). Therefore, more species are required for exploring the effect of multifactors on olfactory system gene expression patterns in subsequent studies.
Changes in receptor molecule copy number may be related to animal evolution, with such changes possibly caused by habitat alteration and adaptation (Shichida et al., 2013). Previous comparative genomics analysis of seven vertebrates from terrestrial and aquatic environments found that the ratio of V1R to V2R genes increased during vertebrate evolution from water to land (Shi and Zhang, 2007). Here, in the aquatic AOS modules, TRPC2 and 22 receptor genes (21 V2Rs and GPR180) were enriched in the response to pheromone GO term. Among these genes, TRPC2 and V2R26_10 were identified as hub genes and were highly expressed with 11 other V2R transcripts in the aquatic snake AOS. V2R genes are putative pheromone receptors and are related to Ca2+-sensing receptors and metabotropic glutamate receptors (Halpern, 2003). In aquatic species, V2R genes are often found with a large number of copies. For example, previous research on two true sea snakes suggested that considerable expansion of the underwater olfaction-related V2R gene family likely occurred in ancestral species of the Hydrophiini clade, with >1,000 copies found in Hydrophis cyanocinctus and Hydrophis curtus, but only 389 found in the common viper (Li et al., 2021). Moreover, the aquatic species Xenopus tropicalis contains 330 V2R genes, while the semiaquatic red-legged salamander only contains 34 V2R genes (Ji et al., 2009; Kiemnec-Tyburczy et al., 2011; Herrel et al., 2014). These results suggest that aquatic snakes may have enhanced ability to bind water-soluble odor molecules in aqueous media by increased copy number and expression of V2Rs.
TRPC2 may also play a fundamental role in aquatic adaptation. TRPC2 drives the process of Na+ and Ca2+ influx-induced depolarization (Kaupp, 2010). In addition, TRPC2 is pseudogenized in the aquatic fin whale and semiaquatic harbor seal and river otter, which lack AOS, but is functionally intact in California sea lions, which possess AOS (Yu et al., 2010). In the current study, a subnetwork containing TRPC2 and V2R26_10 was constructed, which showed the co-expression of genes involved in neuronal differentiation (e.g., OTX2, LHX2, NEUROG1, INSM1, and FOXG1). Odor perception is caused by combining odor molecules with different receptors in sensory neurons (Ubeda-Banon et al., 2011). LHX2 is expressed in the olfactory epithelium and guides the differentiation of progenitor cells into a thousand neuronal subpopulations, each expressing a unique receptor gene (Kolterud et al., 2004). Insm1 is localized in the olfactory epithelium and promotes the transition of progenitors from apical and proliferative to basal, terminal, and neurogenic (Rosenbaum et al., 2011). Overall, aquatic snake adaptations to the water environment depended on changes in receptors and ion channels. Furthermore, the co-expression network with the TRPC2 hub gene played an essential role in adaptation, with TRPC2 potentially exerting its actions by regulating the five genes involved in neural differentiation.
In the current study, we comprehensively analyzed the olfactory system transcriptome of four snakes, considering different habitat conditions. Our findings indicated that the gene expression patterns of the olfactory systems differed between aquatic and non-aquatic snakes. In the MOS of aquatic snakes, RELN may improve the identification of valuable chemical information via the inhibitory neurons suppressing MCs to increase neuronal pattern separation, which may be in favour of aquatic adaptation. In the AOS of aquatic snakes, high expression of the TRPC2, together with vomeronasal receptor genes, may intensify their ability to bind water-soluble odor molecules to promote response to the aquatic environment. Our results provide a basis for future studies on olfactory systems and environmental adaptation in snakes.
Data Availability Statement
The data presented in the study are deposited in the China National GeneBank (CNGB) repository, accession number CNP0002425.
Ethics Statement
The animal study was reviewed and approved by the Animal Experiment Ethics Committee of the Chengdu Institution of Biology, Chinese Academy of Sciences.
Author Contributions
J-TL conceived the study; Z-LP and J-LR collected experimental samples; Z-LP and C-YT performed data analyses; J-TL and Z-LP prepared the initial manuscript draft; J-TL, WW and DJ worked on the approval of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (CAS)(XDB31000000); the National Natural Science Foundation of China (32070410); the Second Tibetan Plateau Scientific Expedition and Research Program (STEP) (2019QZKK0501); Key Research Program of Frontier Sciences, CAS (QYZDB-SSW-SMC058); the International Partnership Program of Chinese Academy of Sciences (151751KYSB20190024); the Sichuan Science and Technology Program (2020YFH0005); the CAS “Light of West China” Program (2018XBZG_JCTD_001) and Talent Program from Organization Department of Sichuan Provincial Committee.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Jun-Jie Huang and Dan Wang for snake samples; and Changjun Peng, Zeng Wang and Meng-Huan Song for insightful suggestions; and Christine Watts for the linguistic modification.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.825974/full#supplementary-material
Footnotes
1^https://ftp.uniprot.org/pub/databases/uniprot/
References
Amber, E. D., Strine, C. T., Suwanwaree, P., and Waengsothorn, S. (2017). Intra-Population Color Dimorphism of Ahaetulla Prasina (Serpentes: Colubridae) in Northeastern Thailand. Curr. Herpetology 36 (2), 98–104. doi:10.5358/hsj.36.98
Auer, T. O., Khallaf, M. A., Silbering, A. F., Zappia, G., Ellis, K., Álvarez-Ocaña, R., et al. (2020). Olfactory Receptor and Circuit Evolution Promote Host Specialization. Nature 579 (7799), 402–408. doi:10.1038/s41586-020-2073-7
Buck, L., and Axel, R. (1991). A Novel Multigene Family May Encode Odorant Receptors: A Molecular Basis for Odor Recognition. Cell 65, 175–187. doi:10.1016/0092-8674(91)90418-X
Chin, C.-H., Chen, S.-H., Wu, H.-H., Ho, C.-W., Ko, M.-T., and Lin, C.-Y. (2014). cytoHubba: Identifying Hub Objects and Sub-networks from Complex Interactome. BMC Syst. Biol. 8 (S4), S11. doi:10.1186/1752-0509-8-S4-S11
Christensen, C. B., Christensen-Dalsgaard, J., Brandt, C., and Madsen, P. T. (2012). Hearing with an Atympanic Ear: Good Vibration and Poor Sound-Pressure Detection in the Royal python,Python Regius. J. Exp. Biol. 215 (Pt 2), 331–342. doi:10.1242/jeb.062539
Cronin, T. W. (2020). Sensory Ecology: In Sea Snake Vision, One Plus One Makes Three. Curr. Biol. 30 (13), R763–R766. doi:10.1016/j.cub.2020.05.012
Danaisawadi, P., Asami, T., Ota, H., Sutcharit, C., and Panha, S. (2016). A Snail-Eating Snake Recognizes Prey Handedness. Sci. Rep. 6, 23832. doi:10.1038/srep23832
Freitag, J., Ludwig, G., Andreini, I., Rössler, P., and Breer, H. (1998). Olfactory Receptors in Aquatic and Terrestrial Vertebrates. J. Comp. Physiol. A: Sensory, Neural Behav. Physiol. 183 (5), 635–650. doi:10.1007/s003590050287
Gomez-Diaz, C., Martin, F., Garcia-Fernandez, J. M., and Alcorta, E. (2018). The Two Main Olfactory Receptor Families in Drosophila, ORs and IRs: A Comparative Approach. Front. Cell. Neurosci. 12, 253. doi:10.3389/fncel.2018.00253
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Trinity: Reconstructing a Full-Length Transcriptome without a Genome from RNA-Seq Data. Nat. Biotechnol. 29, 644–652. doi:10.1038/nbt.1883
Gschwend, O., Abraham, N. M., Lagier, S., Begnaud, F., Rodriguez, I., and Carleton, A. (2015). Neuronal Pattern Separation in the Olfactory Bulb Improves Odor Discrimination Learning. Nat. Neurosci. 18 (10), 1474–1482. doi:10.1038/nn.4089
Halpern, M. (2003). Structure and Function of the Vomeronasal System: an Update. Prog. Neurobiol. 70 (3), 245–318. doi:10.1016/s0301-0082(03)00103-5
Herrada, G., and Dulac, C. (1997). A Novel Family of Putative Pheromone Receptors in Mammals with a Topographically Organized and Sexually Dimorphic Distribution. Cell 90 (4), 763–773. doi:10.1016/S0092-8674(00)80536-X
Herrel, A., Vasilopoulou-Kampitsi, M., and Bonneaud, C. (2014). Jumping Performance in the Highly Aquatic Frog, Xenopus Tropicalis: Sex-specific Relationships between Morphology and Performance. PeerJ 2, e661. doi:10.7717/peerj.661
Hogan, M. P., Whittington, A. C., Broe, M. B., Ward, M. J., Gibbs, H. L., and Rokyta, D. R. (2021). The Chemosensory Repertoire of the Eastern Diamondback Rattlesnake (Crotalus adamanteus) Reveals Complementary Genetics of Olfactory and Vomeronasal-type Receptors. J. Mol. Evol. 89 (4-5), 313–328. doi:10.1007/s00239-021-10007-3
Isaacson, J. S., and Strowbridge, B. W. (1998). Olfactory Reciprocal Synapses: Dendritic Signaling in the CNS. Neuron 20 (4), 749–761. doi:10.1016/S0896-6273(00)81013-2
Ji, Y., Zhang, Z., and Hu, Y. (2009). The Repertoire of G-Protein-Coupled Receptors in Xenopus Tropicalis. BMC Genomics 10, 263. doi:10.1186/1471-2164-10-263
Kardong, K. V. (2012). Vertebrates: Comparative Anatomy, Function, Evolution. New York: McGraw-Hill.
Kassambara, A., and Mundt, F. (2017). factoextra: Extract and Visualize the Results of Multivariate Data Analyses. Available at: https://cran.rproject.org/web/packages/factoextra/index.html.
Kaupp, U. B. (2010). Olfactory Signalling in Vertebrates and Insects: Differences and Commonalities. Nat. Rev. Neurosci. 11 (3), 188–200. doi:10.1038/nrn2789
Kiemnec-Tyburczy, K. M., Woodley, S. K., Watts, R. A., Arnold, S. J., and Houck, L. D. (2011). Expression of Vomeronasal Receptors and Related Signaling Molecules in the Nasal Cavity of a Caudate Amphibian (Plethodon shermani). Chem. Senses 37 (4), 335–346. doi:10.1093/chemse/bjr105
Kishida, T., Go, Y., Tatsumoto, S., Tatsumi, K., Kuraku, S., and Toda, M. (2019). Loss of Olfaction in Sea Snakes Provides New Perspectives on the Aquatic Adaptation of Amniotes. Proc. R. Soc. B. 286 (1910), 20191828. doi:10.1098/rspb.2019.1828
Kishida, T., and Hikida, T. (2010). Degeneration Patterns of the Olfactory Receptor Genes in Sea Snakes. J. Evol. Biol. 23 (2), 302–310. doi:10.1111/j.1420-9101.2009.01899.x
Kishida, T. (2021). Olfaction of Aquatic Amniotes. Cell Tissue Res 383 (1), 353–365. doi:10.1007/s00441-020-03382-8
Kolterud, A., Alenius, M., Carlsson, L., and Bohm, S. (2004). The Lim Homeobox Gene Lhx2 Is Required for Olfactory Sensory Neuron Identity. Development 131 (21), 5319–5326. doi:10.1242/dev.01416
Korsching, S. (2016). “Aquatic Olfaction,” in Chemosensory Transduction (Cambridge, USA: Academic Press), 81–100. doi:10.1016/b978-0-12-801694-7.00005-6
Kotani, T., Murata, Y., Ohnishi, H., Mori, M., Kusakari, S., Saito, Y., et al. (2010). Expression of PTPRO in the Interneurons of Adult Mouse Olfactory Bulb. J. Comp. Neurol. 518 (2), 119–136. doi:10.1002/cne.22239
Koulakov, A. A., and Rinberg, D. (2011). Sparse Incomplete Representations: A Potential Role of Olfactory Granule Cells. Neuron 72 (1), 124–136. doi:10.1016/j.neuron.2011.07.031
Langfelder, P., and Horvath, S. (2008). WGCNA: an R Package for Weighted Correlation Network Analysis. BMC Bioinformatics 9 (1), 559. doi:10.1186/1471-2105-9-559
Lê, S., Josse, J., and Husson, F. (2008). FactoMineR: AnRPackage for Multivariate Analysis. J. Stat. Soft. 25, 1–18. doi:10.18637/jss.v025.i01
Lee, B.-Y., Kim, H.-S., Choi, B.-S., Hwang, D.-S., Choi, A. Y., Han, J., et al. (2015). RNA-seq Based Whole Transcriptome Analysis of the Cyclopoid Copepod Paracyclopina Nana Focusing on Xenobiotics Metabolism. Comp. Biochem. Physiol. D: Genomics Proteomics 15, 12–19. doi:10.1016/j.cbd.2015.04.002
Li, A., Wang, J., Sun, K., Wang, S., Zhao, X., Wang, T., et al. (2021). Two Reference-Quality Sea Snake Genomes Reveal Their Divergent Evolution of Adaptive Traits and Venom Systems. Mol. Biol. Evol. 38 (11), 4867–4883. doi:10.1093/molbev/msab212
Li, B., and Dewey, C. N. (2011). RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinformatics 12 (1), 323. doi:10.1186/1471-2105-12-323
Liu, H., Zhang, J., Yuan, J., Jiang, X., Jiang, L., Li, Z., et al. (2020). Gene Coexpression Network Analysis Reveals a Novel Metabolic Mechanism of Clostridium Acetobutylicum Responding to Phenolic Inhibitors from Lignocellulosic Hydrolysates. Biotechnol. Biofuels 13, 163. doi:10.1186/s13068-020-01802-z
Lohman, A. H. M., and Smeets, W. J. A. J. (1993). Overview of the Main and Accessory Olfactory Bulb Projections in Reptiles. Brain Behav. Evol. 41 (3-5), 147–155. doi:10.1159/000113832
Martín-López, E., Corona, R., and López-Mascaraque, L. (2012). Postnatal Characterization of Cells in the Accessory Olfactory Bulb of Wild Type and Reeler Mice. Front. Neuroanat. 6, 15. doi:10.3389/fnana.2012.00015
Mombaerts, P. (2004). Genes and Ligands for Odorant, Vomeronasal and Taste Receptors. Nat. Rev. Neurosci. 5 (4), 263–278. doi:10.1038/nrn1365
Murphy, J. C., and Voris, H. K. (2014). A Checklist and Key to the Homalopsid Snakes (Reptilia, Squamata, Serpentes), with the Description of New Genera. Fieldiana Life Earth Sci. 8 (8), 1–43. doi:10.3158/2158-5520-14.8.1
Nei, M., Niimura, Y., and Nozawa, M. (2008). The Evolution of Animal Chemosensory Receptor Gene Repertoires: Roles of Chance and Necessity. Nat. Rev. Genet. 9 (12), 951–963. doi:10.1038/nrg2480
Pallotto, M., Nissant, A., Fritschy, J.-M., Rudolph, U., Sassoe-Pognetto, M., Panzanelli, P., et al. (2012). Early Formation of GABAergic Synapses Governs the Development of Adult-Born Neurons in the Olfactory Bulb. J. Neurosci. 32 (26), 9103–9115. doi:10.1523/JNEUROSCI.0214-12.2012
Peng, C., Ren, J.-L., Deng, C., Jiang, D., Wang, J., Qu, J., et al. (2020). The Genome of Shaw's Sea Snake (Hydrophis Curtus) Reveals Secondary Adaptation to its Marine Environment. Mol. Biol. Evol. 37 (6), 1744–1760. doi:10.1093/molbev/msaa043
Pfister, S., Weber, T., Härtig, W., Schwerdel, C., Elsaesser, R., Knuesel, I., et al. (2015). Novel Role of Cystic Fibrosis Transmembrane Conductance Regulator in Maintaining Adult Mouse Olfactory Neuronal Homeostasis. J. Comp. Neurol. 523 (3), 406–430. doi:10.1002/cne.23686
Redolfi, N., Galla, L., Maset, A., Murru, L., Savoia, E., Zamparo, I., et al. (2016). Oligophrenin-1 Regulates Number, Morphology and Synaptic Properties of Adult-Born Inhibitory Interneurons in the Olfactory Bulb. Hum. Mol. Genet. 23, ddw340. doi:10.1093/hmg/ddw340
Ren, J.-L., Wang, K., Jiang, K., Guo, P., and Li, J.-T. (2017). A New Species of the Southeast Asian Genus Opisthotropis (Serpentes: Colubridae: Natricinae) from Western Hunan, China. Zool. Res. 38 (005), 251–263. doi:10.24272/j.issn.2095-8137.2017.068
Ren, J.-L., Yan, C., Peng, Z. L., Yan, C., Peng, Z.-L., and Li, J.-T. (2022). Sichuan Hot-spring Snakes Imperiled: Reason, Situation, and protection. Zool Res. 43 (1), 95–97. doi:10.24272/j.issn.2095-8137.2021.321
Rosenbaum, J. N., Duggan, A., and García-Añoveros, J. (2011). Insm1promotes the Transition of Olfactory Progenitors from Apical and Proliferative to Basal, Terminally Dividing and Neuronogenic. Neural Dev. 6 (1), 6. doi:10.1186/1749-8104-6-6
Saito, H., Chi, Q., Zhuang, H., Matsunami, H., and Mainland, J. D. (2009). Odor Coding by a Mammalian Receptor Repertoire. Sci. Signal. 2 (60), ra9. doi:10.1126/scisignal.2000016
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 13 (11), 2498–2504. doi:10.1101/gr.1239303
Sheehy, C. M., Albert, J. S., Lillywhite, H. B., and Van Damme, R. (2015). The Evolution of Tail Length in Snakes Associated with Different Gravitational Environments. Funct. Ecol. 30 (2), 244–254. doi:10.1111/1365-2435.12472
Shi, P., and Zhang, J. (2007). Comparative Genomic Analysis Identifies an Evolutionary Shift of Vomeronasal Receptor Gene Repertoires in the Vertebrate Transition from Water to Land. Genome Res. 17 (2), 166–174. doi:10.1101/gr.6040007
Shichida, Y., Yamashita, T., Imai, H., and Kishida, T. (2013). Evolution and Senses. Tokyo: Springer.
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 31, 3210–3212. doi:10.1093/bioinformatics/btv351
Supek, F., Bošnjak, M., Škunca, N., and Šmuc, T. (2011). REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS One 6 (7), e21800. doi:10.1371/journal.pone.0021800
Tang, C.-Y., Song, M.-H., Peng, Z.-L., Wu, W., Peng, C., Yang, K., et al. (2021). Transcriptome Analyses Reveal Circadian-Related Expression Features in the Visual Systems of Two Snakes. Diversity 13 (12), 621. doi:10.3390/d13120621
Thewissen, J., and Nummela, S. (2008). “Sensory Evolution on the ThresholdAdaptations in Secondarily Aquatic Vertebrates,” in Comparative Anatomy and Physiology of Chemical Senses in Amphibians (Berkeley: University of California Press), 42–63. doi:10.1525/california/9780520252783.003.0004
Ubeda-Bañon, I., Pro-Sistiaga, P., Mohedano-Moriano, A., Saiz-Sanchez, D., de la Rosa-Prieto, C., Gutierrez-Castellanos, N., et al. (2011). Cladistic Analysis of Olfactory and Vomeronasal Systems. Front. Neuroanat. 5, 3. doi:10.3389/fnana.2011.00003
Wan, Q., Tang, J., Han, Y., and Wang, D. (2018). Co-expression Modules Construction by WGCNA and Identify Potential Prognostic Markers of Uveal Melanoma. Exp. Eye Res. 166, 13–20. doi:10.1016/j.exer.2017.10.007
Wang, P., Che, J., Liu, Q., Li, K., Jin, J. Q., Jiang, K., et al. (2021). A Revised Taxonomy of Asian Snail-Eating Snakes Pareas (Squamata, Pareidae): Evidence from Morphological Comparison and Molecular Phylogeny. Zk 939, 45–64. doi:10.3897/zookeys.939.49309
Wang, R. T., and Halpern, M. (1980a). Light and Electron Microscopic Observations on the normal Structure of the Vomeronasal Organ of Garter Snakes. J. Morphol. 164 (1), 47–67. doi:10.1002/9781119991038.ch910.1002/jmor.1051640105
Wang, R. T., and Halpern, M. (1980b). Scanning Electron Microscopic Studies of the Surface Morphology of the Vomeronasal Epithelium and Olfactory Epithelium of Garter Snakes. Am. J. Anat. 157 (4), 399–428. doi:10.1002/aja.1001570408
Weiss, L., Manzini, I., and Hassenklöver, T. (2021). Olfaction across the Water-Air Interface in Anuran Amphibians. Cell Tissue Res 383 (1), 301–325. doi:10.1007/s00441-020-03377-5
Ye, J., Zhang, Y., Cui, H., Liu, J., Wu, Y., Cheng, Y., et al. (2018). WEGO 2.0: a Web Tool for Analyzing and Plotting GO Annotations, 2018 Update. Nucleic Acids Res. 46 (W1), W71–W75. doi:10.1093/nar/gky400
Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012). clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS: A J. Integr. Biol. 16 (5), 284–287. doi:10.1089/omi.2011.0118
Yu, L., Jin, W., Wang, J.-x., Zhang, X., Chen, M.-m., Zhu, Z.-h., et al. (2010). Characterization of TRPC2, an Essential Genetic Component of VNS Chemoreception, Provides Insights into the Evolution of Pheromonal Olfaction in Secondary-Adapted marine Mammals. Mol. Biol. Evol. 27 (7), 1467–1477. doi:10.1093/molbev/msq027
Zhang, Z., Yang, D., Zhang, M., Zhu, N., Zhou, Y., Storm, D. R., et al. (2017). Deletion of Type 3 Adenylyl Cyclase Perturbs the Postnatal Maturation of Olfactory Sensory Neurons and Olfactory Cilium Ultrastructure in Mice. Front. Cell. Neurosci. 11, 1. doi:10.3389/fncel.2017.00001
Keywords: transcriptome, gene expression pattern, snakes, olfactory adaptation, aquatic adaptation
Citation: Peng Z-L, Wu W, Tang C-Y, Ren J-L, Jiang D and Li J-T (2022) Transcriptome Analysis Reveals Olfactory System Expression Characteristics of Aquatic Snakes. Front. Genet. 13:825974. doi: 10.3389/fgene.2022.825974
Received: 30 November 2021; Accepted: 05 January 2022;
Published: 25 January 2022.
Edited by:
Tao Ma, Sichuan University, ChinaCopyright © 2022 Peng, Wu, Tang, Ren, Jiang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia-Tang Li, bGlqdEBjaWIuYWMuY24=
 Zhong-Liang Peng1,2
Zhong-Liang Peng1,2 Jia-Tang Li
Jia-Tang Li