
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 28 March 2022
Sec. Human and Medical Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.822832
This article is part of the Research Topic Next Generation Sequencing (NGS) for Rare Diseases Diagnosis - Volume II View all 17 articles
 Jingying Zhang1,2,3
Jingying Zhang1,2,3 Xiao-Jun Xu1,2,3
Xiao-Jun Xu1,2,3 Lixia Liu4
Lixia Liu4 Hua Song1,2,3
Hua Song1,2,3 Heping Shen1,2,3
Heping Shen1,2,3 Weiqun Xu1,2,3
Weiqun Xu1,2,3 Fenying Zhao1,2,3
Fenying Zhao1,2,3 Juan Liang1,2,3
Juan Liang1,2,3 Chan Liao1,2,3
Chan Liao1,2,3 Yan Wang1,2,3
Yan Wang1,2,3 Tian Xia1,2,3
Tian Xia1,2,3 Shanbo Cao4
Shanbo Cao4 Yongmin Tang1,2,3*
Yongmin Tang1,2,3* Jiayue Qin4*
Jiayue Qin4* Diying Shen1,2,3*
Diying Shen1,2,3*Acute lymphoblastic leukemia (ALL) is a malignancy associated with altered lymphoid precursor hyperplasia and accompanied with different genetic mutations. Few studies have been reported on the association between gene mutations and clinical features of IKZF1 mutation in children with B-cell ALL (B-ALL). We investigated clinical and genetic characteristics in 200 newly diagnosed pediatric B-ALL through multiplex ligation-dependent probe amplification (MLPA) and targeted next-generation sequencing (NGS) method. We found that IKZF1 mutations, including large segment deletions, small insertions or deletions (InDels) and single nucleotide variations (SNVs), were detected in 22 patients with a positive mutation rate of 11.0%. IKZF1 mutation was significantly associated with higher WBC count (19.38 × 109/L vs. 5.80 × 109/L, p = 0.002). Compared with IKZF1 wild-type cases, a higher frequency of IL7R gene mutation was discovered in IKZF1 mutant cases (9.1% vs. 0.0%, p = 0.012). Patients with IKZF1 mutation were less sensitive to glucocorticoid induction than patients without IKZF1 mutation (63.6% vs. 9.0%, p < 0.001). On the 15th day of induction, minimal residual disease (MRD) > 10−3 level were higher in IKZF1 mutant patients than wild-type patients (45.5% vs. 22.3%, p = 0.018). In conclusion, our study reveals the association between genetic mutations and clinical features in Chinese children with B-ALL, which might contribute to molecular classification, risk stratification and prognosis evaluation, and provide new ideas for targeted therapy in ALL.
Acute lymphoblastic leukemia (ALL) is a malignancy associated with altered lymphoid precursor hyperplasia, and about 75% of children with ALL develop chromosomal changes, such as aneuploidy, translocation, copy number changes, or gene rearrangements (Holmfeldt et al., 2013). With the wide development of genome-wide analysis, some ALL children have IKZF1 mutation, including large segment deletion, small insertions or deletions (InDels) and single nucleotide variations (SNVs), which is considered to be a marker of poor prognosis in pediatric ALL.
IKZF1 gene is located on chromosome 7p12.2 band and consists of 8 exons, encoding transcription factor IKAROS, which plays a key regulatory role in lymphocyte production (Rebollo & Schmitt, 2003). IKAROS contains six zinc finger structures, four of which are located in DNA binding domains encoded by exons 4 to 6 and are essential for maintaining IKAROS tumor suppressor function. The remaining 2 zinc fingers are encoded by exon 8 and mediate IKAROS as homologous dimerization or heterodimerization with other transcription factors of the family, such as AIOLOS and spirochetes (Stanulla et al., 2020). The presence of IKZF1 deletion was associated with older age at diagnosis, higher white blood cell count, and higher minimal residual disease (MRD) levels after induction and consolidation (Mullighan et al., 2009; Waanders et al., 2011; Asai et al., 2013; Dorge et al., 2013; Palmi et al., 2013; Volejnikova et al., 2013; Yamashita et al., 2013; Vrooman et al., 2018; Yeoh et al., 2018). However, the distribution of IKZF1 mutation in Chinese children with B-cell ALL (B-ALL) has been relatively poorly studied.
Here, we systematically analyzed the clinical and genetic characteristics of Chinese B-ALL children with IKZF1 mutation in our single center. These data may provide evidence for risk stratification and individualized treatment for B-ALL.
A retrospective analysis was performed on 200 newly diagnosed patients with B-ALL aged 0–16 years who were admitted to the Children Hospital of Zhejiang University School of Medicine from 1 October 2017 to 31 August 2020. The diagnosis of B-ALL was based on the 2016 World Health Organization (WHO) classification criteria for hematopoietic and lymphoid tissue tumors (Arber et al., 2016). All patients were confirmed by comprehensive diagnosis of cytomorphology, immunology, cytogenetics and molecular biology, and complete medical history could be traced. Exclusion criteria: untraceable biological samples; unable to obtain necessary biological information; acute promyelocytic leukemia; other hematologic or non-hematologic tumors. The study was approved by the institutional review board of the Children’s Hospital of Zhejiang University Medical College and informed consents were obtained from patients and/or their legal guardians in accordance with the Declaration of Helsinki.
Chromosomes were tested by Adicon Clinical Laboratory (Hangzhou, China). Leukemia fusion genes were sequenced by Kindstar Globalgene Technology (Wuhan, China). Flow cytometry (FCM) MRD were detected by the Children’s Hospital Leukemia Laboratory affiliated to the Children Hospital of Zhejiang University School of Medicine (Hangzhou, China). Hazard groups refer to CCLG-ALL-2008 scheme criteria (Brown et al., 2019; Brown et al., 2020). MRD detection for children examined by FCM: residual status of bone marrow tumor cells after induction (D15) and before consolidation (D33) treatment.
Targeted copy number screening of the IKZF1 gene was performed by multiplex ligation-dependent probe amplification (MLPA). The children’s mono-nuclear cells were retained at the initial diagnosis. DNA was extracted and analyzed using the SALSA MLPA KIT P335-B1 ALL-IKZF1 probemix according to the manufacturer’s instructions. This SALSA contained a probe for each IKZF1 exon. All MLPA reactions, including DNA denaturation, hybridization, ligation, and PCR, were carried out in a 96-well PCR thermocycler. The amplification products were quantified and identified by capillary electrophoresis. Normalization of the data was carried out by dividing the peak area of each probe by the average peak area of the control probes. This normalized peak pattern was divided by the average peak pattern of all the samples in the same experiment. The resulting values were 0–1 for every wild-type peak, 0.5 for heterozygous deletions and 1.5 for heterozygous duplications.
DNA was extracted from whole bone marrow collected at diagnosis. Based on next-generation sequencing (NGS) of targeted capture, the mutation hotspots or entire coding region of 185 genes known to mutate frequently in hematological malignancies were sequenced (Supplementary Table S1). The following criteria were used to filter raw variant results: average effective sequencing depth on target per sample ≥1,000×; mapping quality ≥30; and base quality ≥30; variant allele frequency (VAF) ≥1% for SNVs and small InDels. Burrows-Wheeler alignment (BWA, version 0.7.12) was performed to align the trimmed reads. MarkDuplicates tool from Picard was used to mark the PCR duplicates. IndelRealigner and BaseRecalibrator from Genome Analysis Toolkit (GATK, version 3.8) were applied for realignment and recalibration of the BWA data, respectively. Variant calling, including SNVs and small InDels, was performed in Mutect2. ANNOVAR software was used to annotate all the variants including 1000G projects, COSMIC, SIFT, and PolyPhen.
Statistical analyses were carried out using R (version 3.5.2) or SPSS software (version 22.0). Mann-Whiney U test was used to compare the continuous variables. Chi-square test or Fisher’s exact test was used to compare the categorical variables. p < 0.05 was considered to indicate a statistically significant difference.
A total of 200 B-ALL patients were enrolled in our study, including 106 males and 94 females, with a median age of 3.71 years (range, 0.05–16.25), as shown in Table 1. The median white blood cell (WBC) count, hemoglobin (Hb) concentration, and platelet (PLT) count was 6.49 × 109/L, 81.00 g/L and 60.00 × 109/L, respectively. Sixty-eight patients with a hyperdiploid chromosome karyotype (34.0%) were discovered in our cohort. On the 15th day of induction, 49 cases were with MRD >10−3 (24.9%). On the 33rd day of induction, 12 cases were with MRD >10−4 (6.0%). According to hazard groups from CCLG-ALL-2008 scheme criteria, 80 patents were assigned to the low-risk group (40%), 70 cases were in the intermediate risk group (35.0%) and 50 cases in the high risk group (25.0%). In 200 B-ALL patients, the overall rate of mutation prevalence was 82.0% (164/200) (Figure 1). A total of 88 mutated genes were detected, and the most common mutated gene was NRAS (25.0%), followed by KRAS (19.0%) and FLT3 (13.5%) (Figure 2A). In total, 553 mutation sites were detected, and nonsynonymous SNV (65.0%) was the most common mutation type (Figure 1). Significant associations were discovered between mutated SETD2 and mutations in TP53, PCLO, PIK3R1 and FAT1, and between mutated ASXL1 and CHD2 and NF1 mutations (Figure 2B).
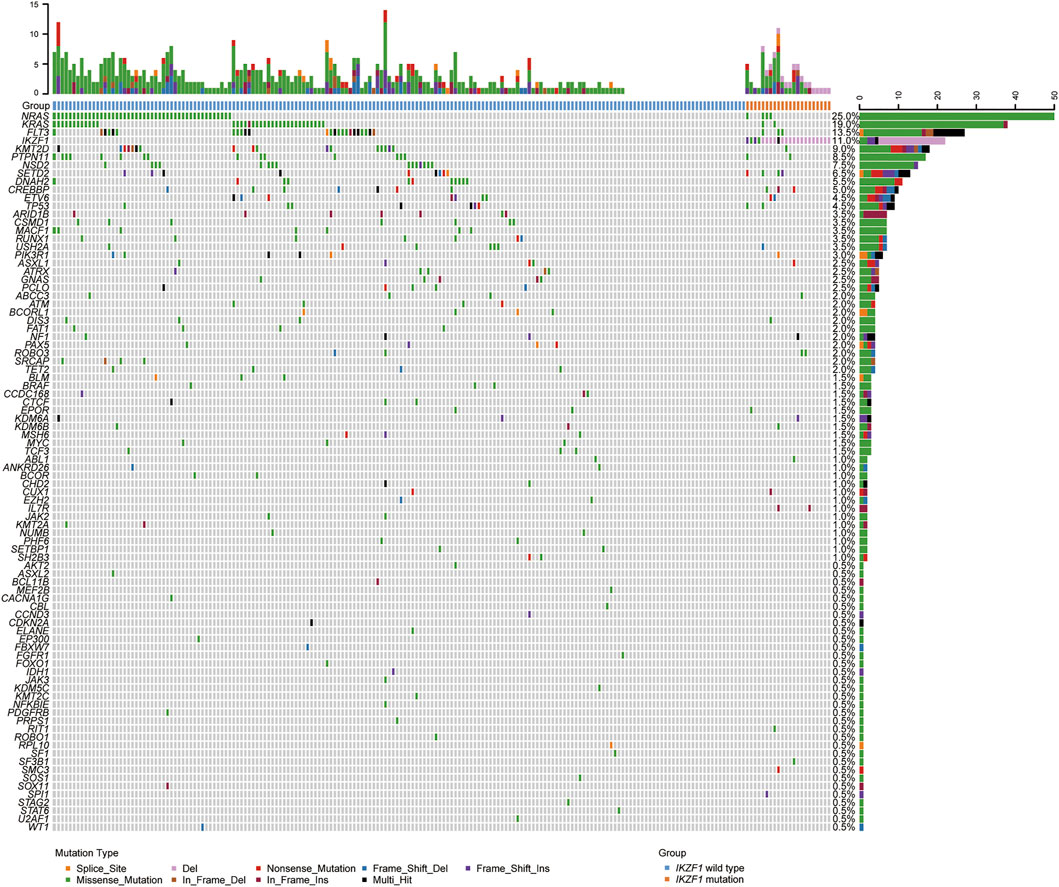
FIGURE 1. Overview of the gene mutations identified by targeted next-generation sequencing and multiplex ligation-dependent probe amplification in 200 B-ALL patients. Heatmap shows the specific mutations in each patient based on different gene mutation types, including large segment deletions, small insertions or deletions, and single nucleotide variations.
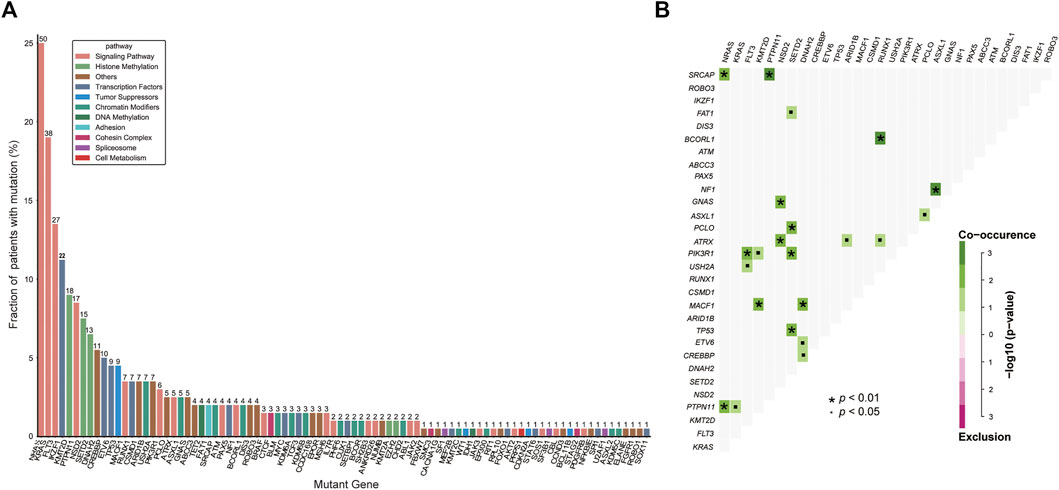
FIGURE 2. Genetic analyses in the whole cohort. (A) Histogram shows the frequency of gene mutations detected in the whole cohort according to the different functional groups assigned to each gene. (B) Diagram shows pairwise gene mutation correlations on the basis of the mutated genes detected in ≥2% patients. The odds ratio of the correlation is coded by different colors, and the significance level is marked by the symbol in each field.
IKZF1 mutations, including large segment deletions, small InDels and SNVs, were detected in 22 of 200 B-ALL children, with a positive mutation rate of 11.0%. The median WBC count in IKZF1 mutant children was 19.38 × 109/L, and was about 4 times higher than that in IKZF1 wild-type children (p = 0.002). Both the median hemoglobin levels and platelet counts were not significantly different between IKZF1 mutant and wild-type patients. More than half of cases with IKZF1 mutation were not sensitive to glucocorticoid induction, and the proportion was more than 5 times higher than that of wild-type cases (63.6% vs. 9.0%, p < 0.001). On the 15th day of induction, 10 IKZF1 mutant cases were MRD >10−3 by FCM, while 39 IKZF1 wild-type cases were MRD >10−3 (45.5% vs. 22.3%, p = 0.018) (Table 2).
The incidence of IKZF1 mutation was shown in Figure 3A. 36.0% of IKZF1 wild-type cases carried a hyperdiploid chromosome karyotype, while only 18.2% of IKZF1 mutant cases were with hyperdiploid (p = 0.097). 97.9% of TEL-AML1 positive B-ALL children had the wild-type IKZF1, and only one case had abnormal IKZF1 (2.1%). For 11 MLL rearrangement positive B-ALL cases, IKZF1 mutations were detected in 2 cases, and wild-type IKZF1 was in 9 cases (18.2% vs. 81.8%). IKZF1 mutation was not detected in the E2A-HLF subgroup (0%). Of the 9 E2A-PBX1 positive cases, only one case had abnormal IKZF1, while of the 10 Ph+/Ph-like cases, 4 (40.0%) had IKZF1 mutations. Compared with IKZF1 wild-type cases, the interleukin (IL)-7 receptor (IL7R) gene mutation only occurred in IKZF1 mutant cases, and the difference was statistically significant (9.1% vs. 0.0%, p = 0.012) (Figure 3B). SETD2 and ROBO3 mutations were found in 18.2% and 9.1% of IKZF1 mutant cases, respectively, which seemed higher than that in wild-type cases (18.2% vs. 5.1%; 9.1% vs. 1.1%, respectively), but no statistical difference was discovered. Furthermore, based on the analysis of the number of mutated genes, there was no significant difference between IKZF1 mutant and wild-type patients (p = 0.753) (Figure 3C).
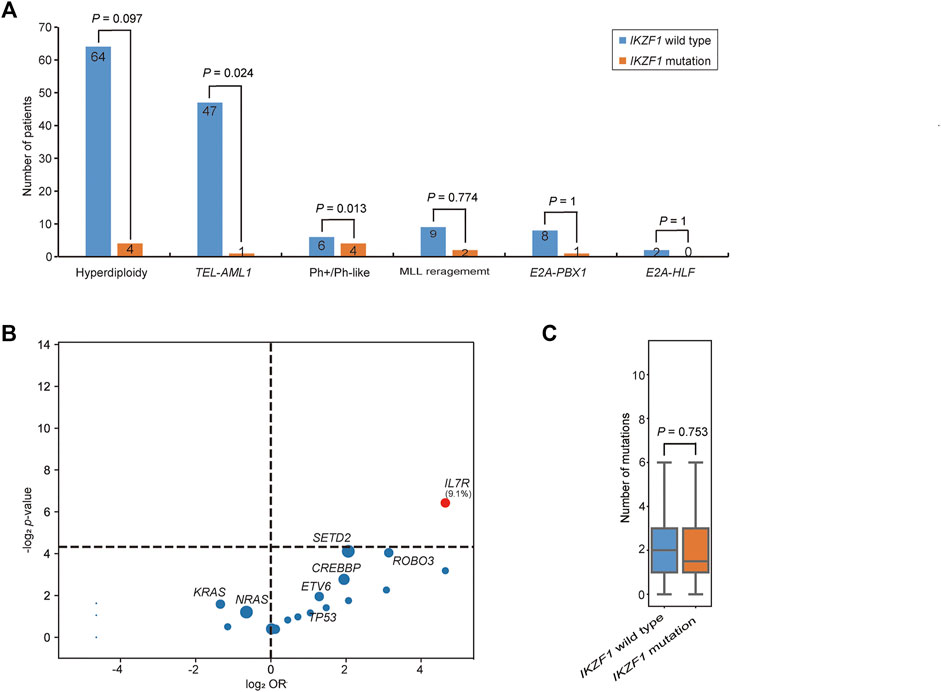
FIGURE 3. Comparison of genetic characteristics between IKZF1 mutant and wild-type patients. (A) Bar chart shows the associations between IKZF1 mutations and different cytogenetics or genetic aberrations. (B) Volcano plot shows the distribution of genetic characteristics according to IKZF1 mutant and wild-type patients. The x axis indicates the magnitude of association (log2 odds ratio), and the y axis represents the −log2 p value. Each circle shows a mutated gene and the size of each circle represents the frequency of the mutated gene. (C) Box plot shows the comparison of the number of mutations between IKZF1 mutant and wild-type patients.
Among 22 IKZF1 mutant cases, 17 cases carried only IKZF1 large segment deletion, 4 cases had SNV or small InDel mutations in the IKZF1 gene, and 1 case had both IKZF1 large segment deletion and SNV mutation. For large segment deletion, 4 (18.2%) cases involved exon 1–8 deletion of the entire gene, while 14 (63.6%) cases involved focal gene deletion, including exon 4–7 deletion in 8 cases (36.4%), exon 2–7 deletion in 3 cases (13.6%), exon 4–8 deletion in 2 cases (9.1%), and exon 2–8 deletion in one case (4.5%) (Figure 4A). For IKZF1 SNV and small InDel mutations, the main types were frameshift and missense mutations, two of which were located in the zinc finger structure of exon 4–7, including IKZF1 G158S and L161fs (Figure 4B). Based on the analysis of two different IKZF1 mutation types, including large segment deletion, and SNV or small InDel, the correlations between paired genes were revealed (Figure 4C).
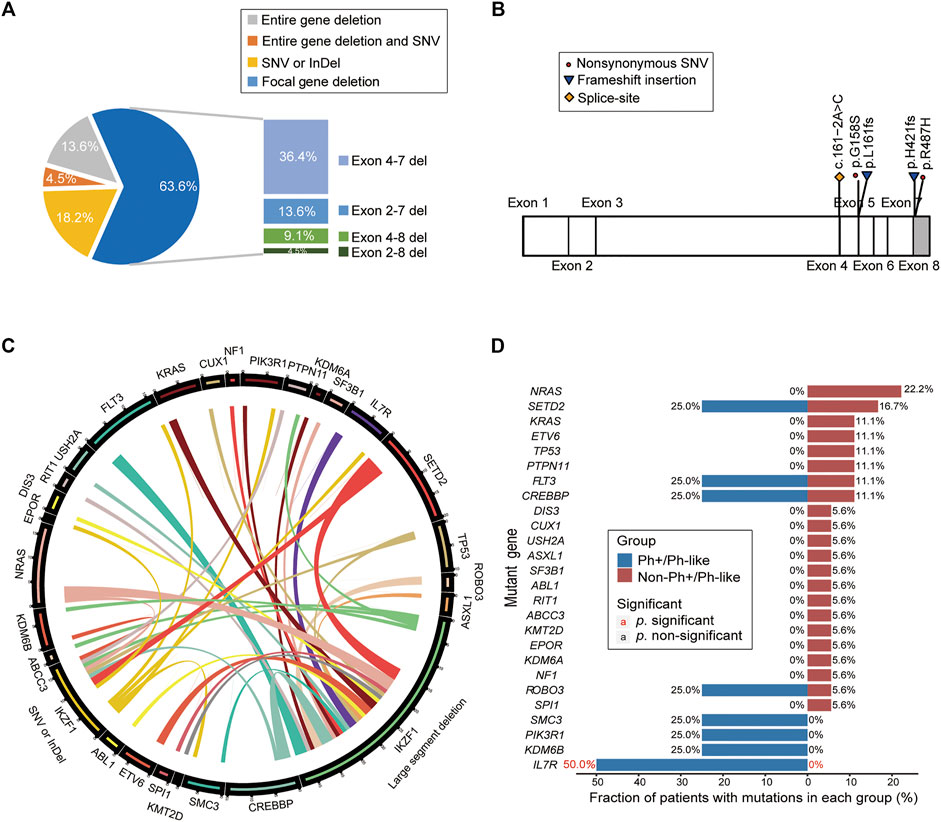
FIGURE 4. Analysis of gene mutation characteristics in patients with IKZF1 mutation. (A) Pie chart shows specific types of IKZF1 mutations, including the entire gene deletions, focal gene deletions, single nucleotide variations, and small insertions or deletions. (B) Schematic representation of the mutations detected in the IKZF1 gene, only for single nucleotide variations, and small insertions or deletions. (C) Circos plot shows all the genetic mutations in the IKZF1 mutation cohort, corresponding to the relative frequency and pairwise co-occurrence of gene mutations. The length of the arc indicates the frequency of mutations in the first gene, and the width of the ribbon represents the percentage of patients carrying the second gene mutation. (D) Comparison of the mutational genotypes of Ph+/Ph-like positive and Ph+/Ph-like negative B-ALL with IKZF1 mutation. Percentage frequencies in each group are depicted.
According to the distribution of mutations in 22 IKZF1 mutant patients, 26 mutant genes were discovered, and NRAS, SETD2, FLT3, CREBBP were common detected genes (Supplementary Figure S1A). The cluster analysis based on gene function pathways showed that the mutant genes were mainly related to signaling pathway (40.5%) and transcription factor (16.7%) (Supplementary Figure S1B). The IL7R mutation accounted for 50.0% of B-ALL cases with Ph+/Ph-like combined with IKZF1 abnormalities (50.0% vs. 0%, p = 0.03) (Figure 4D).
In this study, we systematically identified clinical and genetic characteristics of Chinese B-ALL children with IKZF1 mutation. IKZF1 mutation has been a hot topic in the field of leukemia since Mullighan et al. firstly reported in ALL patients in 2008 that single allele focal deletion affects its coding region (Mullighan et al., 2008). IKZF1 mutation will lead to the obstruction of lymphocyte differentiation and development, resulting in leukemia. Based on MLPA to determine the partial or complete IKZF1 large segment deletions, our study found IKZF1 large segment deletion frequency was 9.0%, which was consistent with the result that Asai et al. reported 19 of 202 (9.4%) patients were carrying IKZF1 large segment deletions (Asai et al., 2013). However, other studies observed differences in IKZF1 large segment deletion frequency: 12% in German patients, 20.6% in Mexico patients, 16% in Swedish patients, and 28.6% in American patients. IKZF1 SNV and small InDel mutation in our study was 2.5%, almost close to the proportion reported in the literature (<1%) (Mullighan et al., 2008; Dorge et al., 2013; Ofverholm et al., 2013; Ayon-Perez et al., 2019; Stanulla et al., 2020).
Previous studies showed that the different type of IKZF1 mutations, including large segment deletion, small InDel and SNV, produced different molecular results. Deficiencies from these sites, such as the entire gene (including exons 1–8), or focal gene (including exons 2 and/or 8), or untranscriptional regulatory regions (including exon 1), can lead to IKZF1 hypofunction (Iacobucci & Mullighan, 2017). The deletion of exons 4–7, lacking the ability to bind DNA, was a negative domain and thus led to leukemia (Mullighan et al., 2008; Iacobucci et al., 2009; Chiaretti et al., 2016), which was the most common deletion pattern of IKZF1 in our cohort (36%), consistent with results in Germany, Japan, Sweden and the US and different from results in Mexico, where the deletion of exon 1 (85%) occurred most frequently (Ayon-Perez et al., 2019). Although SNV or small InDel mutations of IKZF1 were infrequent and present, their molecular consequences could be either haploid insufficiency or dominant negative effects, as with deletions. Given this, the molecular effects of these types of mutations can be further judged by gene expression. Recent research discovered that IKZF1 missense mutation (p.N159Y) affect the DNA binding domain and validated by gene expression profile (Li et al., 2018; Gu et al., 2019). Five IKZF1 mutations, including SNV or small InDel mutations, were also detected in our study, whose final molecular effects need to be further clarified by transcriptome sequencing.
However, IKZF1 is controversial as an independent risk factor for patient prognostic stratification. Some studies suggested IKZF1 large segment deletion was closely related to the high recurrence and low survival of pediatric B-ALL (Kuiper et al., 2010; Yang et al., 2011; Buitenkamp et al., 2012). Boer et al. showed that any kind of IKZF1 large segment deletion increased risk compared to patients with wild-type IKZF1, based on their high WBC count >50,000/µl (Boer et al., 2016). Indeed, we observed this phenomenon for 22 patients with IKZF1 mutations in this study, who had higher levels of leukocytes at the time of initial diagnosis, insensitivity to glucocorticoid, and higher levels of MRD on day 15th of induction remission.
We also found that the partner mutant genes associated with the IKZF1 mutations are closely related to the signaling pathway and transcription factor function (NRAS, SETD2, FLT3 and CREBBP). In particular, the mutation in IL7R was only found in IKZF1 mutation cases, suggesting the IL7R mutation may be synergistic with the IKZF1 mutation and participate in the occurrence of the B-ALL. In our study, these two patients with IL7R mutation were insensitive to glucocorticoid therapy. Several research suggested that IL7R functional acquired mutations made IL7R highly expressed, and IKZF1 deletion deprived the IKAROS of its inhibitory effect on the promoter region. IKZF1 and IL7R synergistically activated downstream JAK/STAT5 and PI3K/Akt/mTOR signaling pathways to promote leukemia (Ge et al., 2016). Thomas et al. (2021) showed that IL7R mutation led to B-ALL alone in a mouse model and IKZF1 mutation contributed to the process of leukemogenesis.
Our study has several limitations. First of all, due to the lack of follow-up data, the long-term prognostic value of IKZF1 mutation remains to be explored. Secondly, this study is a single-center result, which may not fully reflect the distribution of clinical and genetic characteristics in the Chinese population. Therefore, large-scale multi-center studies and long-term follow-up should be included in the future.
Inconclusion, our research showed clinical and genetic characteristics of IKZF1 mutation in Chinese Children with B-ALL. This study reveals the association between genetic mutations and clinical features. These investigations might contribute to molecular classification, risk stratification and prognosis evaluation, and provide new ideas for targeted therapy.
The data that support the findings of this study have been deposited into CNGB Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb) with accession number CNP0002707 https://db.cngb.org/search/project/CNP0002707.
The studies involving human participants were reviewed and approved by the Children’s Hospital of Zhejiang University Medical College. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
DS, JQ and YT designed the study and approved the final manuscript. JZ, XX, HS, HS, WX, FZ, JL, CL, YW and TX collected the clinical sample and data. LL and SC performed the NGS platform and statistical analysis. JZ, YT, JQ, and DS wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by National Natural Science Foundation (No. 81770202), Zhejiang Provincial Science and Technology Department Fund (No. 2019C03032) and Pediatric Leukemia Diagnostic and Therapeutic Technology Research Center of Zhejiang Province (No. JBZX-201904).
Authors LL, SC, and JQ were employed by the company Acornmed Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank all the patients who participated in this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.822832/full#supplementary-material
Arber, D. A., Orazi, A., Hasserjian, R., Thiele, J., Borowitz, M. J., Le Beau, M. M., et al. (2016). The 2016 Revision to the World Health Organization Classification of Myeloid Neoplasms and Acute Leukemia. Blood 127 (20), 2391–2405. doi:10.1182/blood-2016-03-643544
Asai, D., Imamura, T., Suenobu, S. i., Saito, A., Hasegawa, D., Deguchi, T., et al. (2013). IKZF1 Deletion Is Associated with a Poor Outcome in Pediatric B-Cell Precursor Acute Lymphoblastic Leukemia in Japan. Cancer Med. 2 (3), 412–419. doi:10.1002/cam4.87
Ayón-Pérez, M. F., Pimentel-Gutiérrez, H. J., Durán-Avelar, M. d. J., Vibanco-Pérez, N., Pérez-Peraza, V. M., Pérez-González, Ó. A., et al. (2019). IKZF1 Gene Deletion in Pediatric Patients Diagnosed with Acute Lymphoblastic Leukemia in Mexico. Cytogenet. Genome Res. 158 (1), 10–16. doi:10.1159/000499641
Boer, J. M., van der Veer, A., Rizopoulos, D., Fiocco, M., Sonneveld, E., de Groot-Kruseman, H. A., et al. (2016). Prognostic Value of Rare IKZF1 Deletion in Childhood B-Cell Precursor Acute Lymphoblastic Leukemia: an International Collaborative Study. Leukemia 30 (1), 32–38. doi:10.1038/leu.2015.199
Brown, P. A., Wieduwilt, M., Logan, A., DeAngelo, D. J., Wang, E. S., Fathi, A., et al. (2019). Guidelines Insights: Acute Lymphoblastic Leukemia, Version 1.2019. J. Natl. Compr. Canc Netw. 17 (5), 414–423. doi:10.6004/jnccn.2019.0024
Brown, P., Inaba, H., Annesley, C., Beck, J., Colace, S., Dallas, M., et al. (2020). Pediatric Acute Lymphoblastic Leukemia, Version 2.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc Netw. 18 (1), 81–112. doi:10.6004/jnccn.2020.0001
Buitenkamp, T. D., Pieters, R., Gallimore, N. E., van der Veer, A., Meijerink, J. P. P., Beverloo, H. B., et al. (2012). Outcome in Children with Down's Syndrome and Acute Lymphoblastic Leukemia: Role of IKZF1 Deletions and CRLF2 Aberrations. Leukemia 26 (10), 2204–2211. doi:10.1038/leu.2012.84
Chiaretti, S., Gianfelici, V., O’Brien, S. M., and Mullighan, C. G. (2016). Advances in the Genetics and Therapy of Acute Lymphoblastic Leukemia. Am. Soc. Clin. Oncol. Educ. Book 35, e314–e322. doi:10.1200/edbk_156628
Dorge, P., Meissner, B., Zimmermann, M., Moricke, A., Schrauder, A., Bouquin, J.-P., et al. (2013). IKZF1 Deletion Is an Independent Predictor of Outcome in Pediatric Acute Lymphoblastic Leukemia Treated According to the ALL-BFM 2000 Protocol. Haematologica 98 (3), 428–432. doi:10.3324/haematol.2011.056135
Ge, Z., Gu, Y., Xiao, L., Han, Q., Li, J., Chen, B., et al. (2016). Co-existence of IL7R High and SH2B3 Low Expression Distinguishes a Novel High-Risk Acute Lymphoblastic Leukemia with Ikaros Dysfunction. Oncotarget 7 (29), 46014–46027. doi:10.18632/oncotarget.10014
Gu, Z., Churchman, M. L., Roberts, K. G., Moore, I., Zhou, X., Nakitandwe, J., et al. (2019). PAX5-driven Subtypes of B-Progenitor Acute Lymphoblastic Leukemia. Nat. Genet. 51 (2), 296–307. doi:10.1038/s41588-018-0315-5
Holmfeldt, L., Wei, L., Diaz-Flores, E., Walsh, M., Zhang, J., Ding, L., et al. (2013). The Genomic Landscape of Hypodiploid Acute Lymphoblastic Leukemia. Nat. Genet. 45 (3), 242–252. doi:10.1038/ng.2532
Iacobucci, I., and Mullighan, C. G. (2017). Genetic Basis of Acute Lymphoblastic Leukemia. J. Clin. Oncol. 35 (9), 975–983. doi:10.1200/JCO.2016.70.7836
Iacobucci, I., Storlazzi, C. T., Cilloni, D., Lonetti, A., Ottaviani, E., Soverini, S., et al. (2009). Identification and Molecular Characterization of Recurrent Genomic Deletions on 7p12 in the IKZF1 Gene in a Large Cohort of BCR-ABL1-Positive Acute Lymphoblastic Leukemia Patients: on Behalf of Gruppo Italiano Malattie Ematologiche dell'Adulto Acute Leukemia Working Party (GIMEMA AL WP). Blood 114 (10), 2159–2167. doi:10.1182/blood-2008-08-173963
Kuiper, R. P., Waanders, E., van der Velden, V. H. J., van Reijmersdal, S. V., Venkatachalam, R., Scheijen, B., et al. (2010). IKZF1 Deletions Predict Relapse in Uniformly Treated Pediatric Precursor B-ALL. Leukemia 24 (7), 1258–1264. doi:10.1038/leu.2010.87
Li, J.-F., Dai, Y.-T., Lilljebjörn, H., Shen, S.-H., Cui, B.-W., Bai, L., et al. (2018). Transcriptional Landscape of B Cell Precursor Acute Lymphoblastic Leukemia Based on an International Study of 1,223 Cases. Proc. Natl. Acad. Sci. USA 115 (50), E11711–E11720. doi:10.1073/pnas.1814397115
Mullighan, C. G., Miller, C. B., Radtke, I., Phillips, L. A., Dalton, J., Ma, J., et al. (2008). BCR-ABL1 Lymphoblastic Leukaemia Is Characterized by the Deletion of Ikaros. Nature 453 (7191), 110–114. doi:10.1038/nature06866
Mullighan, C. G., Su, X., Zhang, J., Radtke, I., Phillips, L. A. A., Miller, C. B., et al. (2009). Deletion ofIKZF1and Prognosis in Acute Lymphoblastic Leukemia. N. Engl. J. Med. 360 (5), 470–480. doi:10.1056/NEJMoa0808253
Öfverholm, I., Tran, A. N., Heyman, M., Zachariadis, V., Nordenskjöld, M., Nordgren, A., et al. (2013). Impact of IKZF1 Deletions and PAX5 Amplifications in Pediatric B-Cell Precursor ALL Treated According to NOPHO Protocols. Leukemia 27 (9), 1936–1939. doi:10.1038/leu.2013.92
Palmi, C., Valsecchi, M. G., Longinotti, G., Silvestri, D., Carrino, V., Conter, V., et al. (2013). What Is the Relevance of Ikaros Gene Deletions as a Prognostic Marker in Pediatric Philadelphia-negative B-Cell Precursor Acute Lymphoblastic Leukemia? Haematologica 98 (8), 1226–1231. doi:10.3324/haematol.2012.075432
Rebollo, A., and Schmitt, C. (2003). Ikaros, Aiolos and Helios: Transcription Regulators and Lymphoid Malignancies. Immunol. Cel Biol 81 (3), 171–175. doi:10.1046/j.1440-1711.2003.01159.x
Stanulla, M., Cavé, H., and Moorman, A. V. (2020). IKZF1 Deletions in Pediatric Acute Lymphoblastic Leukemia: Still a Poor Prognostic Marker. Blood 135 (4), 252–260. doi:10.1182/blood.2019000813
Thomas, K. R., Allenspach, E. J., Camp, N. D., Wray-Dutra, M. N., Khim, S., Zielinska-Kwiatkowska, A., et al. (2021). Activated Interleukin-7 Receptor Signaling Drives B-Cell Acute Lymphoblastic Leukemia in Mice. Leukemia 36, 42–57. doi:10.1038/s41375-021-01326-x
Volejnikova, J., Mejstrikova, E., Dörge, P., Meissner, B., Zimmermannova, O., Svojgr, K., et al. (2013). Ikaros (IKZF1 ) Alterations and Minimal Residual Disease at Day 15 Assessed by Flow Cytometry Predict Prognosis of Childhood BCR/ABL -negative Acute Lymphoblastic Leukemia. Pediatr. Blood Cancer 60 (3), 420–427. doi:10.1002/pbc.24299
Vrooman, L. M., Blonquist, T. M., Harris, M. H., Stevenson, K. E., Place, A. E., Hunt, S. K., et al. (2018). Refining Risk Classification in Childhood B Acute Lymphoblastic Leukemia: Results of DFCI ALL Consortium Protocol 05-001. Blood Adv. 2 (12), 1449–1458. doi:10.1182/bloodadvances.2018016584
Waanders, E., van der Velden, V. H., van der Schoot, C. E., van Leeuwen, F. N., van Reijmersdal, S. V., de Haas, V., et al. (2011). Integrated Use of Minimal Residual Disease Classification and IKZF1 Alteration Status Accurately Predicts 79% of Relapses in Pediatric Acute Lymphoblastic Leukemia. Leukemia 25 (2), 254–258. doi:10.1038/leu.2010.275
Yamashita, Y., Shimada, A., Yamada, T., Yamaji, K., Hori, T., Tsurusawa, M., et al. (2013). IKZF 1 and CRLF 2 Gene Alterations Correlate with Poor Prognosis in Japanese BCR - ABL 1 -negative High-Risk B-Cell Precursor Acute Lymphoblastic Leukemia. Pediatr. Blood Cancer 60 (10), 1587–1592. doi:10.1002/pbc.24571
Yang, Y.-L., Hung, C.-C., Chen, J.-S., Lin, K.-H., Jou, S.-T., Hsiao, C.-C., et al. (2011). IKZF1 Deletions Predict a Poor Prognosis in Children with B-Cell Progenitor Acute Lymphoblastic Leukemia: a Multicenter Analysis in Taiwan. Cancer Sci. 102 (10), 1874–1881. doi:10.1111/j.1349-7006.2011.02031.x
Yeoh, A. E. J., Lu, Y., Chin, W. H. N., Chiew, E. K. H., Lim, E. H., Li, Z., et al. (2018). Intensifying Treatment of Childhood B-Lymphoblastic Leukemia with IKZF1 Deletion Reduces Relapse and Improves Overall Survival: Results of Malaysia-Singapore ALL 2010 Study. J. Clin. Oncol. 36 (26), 2726–2735. doi:10.1200/JCO.2018.78.3050
Keywords: IKZF1 mutation, B-cell acute lymphoblastic leukemia, genetic characteristics, clinical features, targeted next-generation sequencing
Citation: Zhang J, Xu X-, Liu L, Song H, Shen H, Xu W, Zhao F, Liang J, Liao C, Wang Y, Xia T, Cao S, Tang Y, Qin J and Shen D (2022) Clinical and Genetic Characteristics of IKZF1 Mutation in Chinese Children With B-Cell Acute Lymphoblastic Leukemia. Front. Genet. 13:822832. doi: 10.3389/fgene.2022.822832
Received: 26 November 2021; Accepted: 22 February 2022;
Published: 28 March 2022.
Edited by:
Sadeq Vallian, University of Isfahan, IranReviewed by:
Xujie Zhao, St. Jude Children’s Research Hospital, United StatesCopyright © 2022 Zhang, Xu, Liu, Song, Shen, Xu, Zhao, Liang, Liao, Wang, Xia, Cao, Tang, Qin and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diying Shen, c2hlbmRpeWluZ0B6anUuZWR1LmNu; Jiayue Qin, anlxaW5AbGl2ZS5jbg==; Yongmin Tang, WV9NX1RhbmdAemp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.