
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Genet. , 24 February 2022
Sec. Genetics of Common and Rare Diseases
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.821226
This article is part of the Research Topic Genetic and Environmental Factors in the Occurrence of Paediatric Disorders – Volume II View all 10 articles
 Hairui Sun1†
Hairui Sun1† Xiaowei Liu1†
Xiaowei Liu1† Xiaoyan Hao1†
Xiaoyan Hao1† Xiaoxue Zhou1
Xiaoxue Zhou1 Jingyi Wang1
Jingyi Wang1 Jiancheng Han1
Jiancheng Han1 Mengmeng Liang2
Mengmeng Liang2 Hongjia Zhang3*†
Hongjia Zhang3*† Yihua He1*
Yihua He1*Background: Left ventricular noncompaction (LVNC) is a rare cardiomyopathy, long QT syndrome (LQTS) is a rare ion channel disease, and simultaneous occurrence of both is even rarer. Further clinical reports and studies are needed to identify the association between LVNC and LQTS and the underlying mechanism.
Methods and Results: A 26-year-old primigravida was referred at 25 weeks gestation for prenatal echocardiography due to fetal bradycardia detected during the routine ultrasound examination. The echocardiographic findings were consistent with biventricular noncompaction cardiomyopathy (BVNC) with pulmonary stenosis and suspected LQTS. After detailed counseling, the couple decided to terminate the pregnancy, and subsequent postmortem examination confirmed BVNC and pulmonary stenosis. Then, A trio (fetus and the parents) whole-exome sequencing (WES) and copy number variation sequencing (CNV-seq) were performed. CNV-seq identified no aneuploidy or pathogenic CNV. A de novo missense variant in KCNH2 (NM_000238.3:c.1847A > G,p.Tyr616Cys) was identified by WES. This KCNH2 missense mutation was classified as pathogenic according to the American College of Medical Genetics and Genomics and the Association for Molecular Pathology variant interpretation guidelines.
Conclusion: We report the first prenatal case of KCNH2 mutation presenting with LVNC combined with bradycardia and second-degree 2:1 atrioventricular block. Importantly, this case reminds clinicians to systematically search ion channel gene mutations in patients with LVNC and arrhythmia.
Left ventricular noncompaction (LVNC) is rare genetic cardiomyopathy (Towbin et al., 2015). Genes associated with LVNC usually include those encoding sarcomere, ion channels, nuclear envelope, and chaperone proteins. Many ion-channel genes, such as SCN5A, RYR2, KCNQ1, and HCN4, have been associated with LVNC, but the underlying molecular mechanisms are unknown (Milano et al., 2014; Nakashima et al., 2013; Schweizer et al., 2014; Towbin, 2014). KCNH2, and an ion-channel gene, encodes the pore-forming subunit of a rapidly activating-delayed rectifier potassium channel that plays a critical role in the final repolarization of the ventricular action potential (Gianulis and Trudeau, 2011). Mutations in the KCNH2 gene cause long QT syndrome type 2 (LQTS2, MIM:613688) (Amin et al., 2008). The combination of LVNC with LQTS is scarce, and clinical reports of KCNH2 variants in such cases are even rarer. Due to the scarcity of clinical reports, LVNC has not been recognized as a feature of LQTS2. Here we report the first fetal case, to our knowledge, with KCNH2 mutation presenting with LVNC, LQTS, and sinus bradycardia. We also reviewed the literature to identify additional cases of KCNH2 mutation with LVNC-LQTS combined phenotype.
This study was approved by the Ethics Committee of Beijing Anzhen Hospital, Capital Medical University and adhered to the tenets of the Declaration of Helsinki. Informed written consent was obtained from the parents of the fetus.
A complete fetal echocardiographic examination, including twodimensional (2D), M-mode, color, and pulse Doppler echocardiography, was performed using the General Electric Voluson E8 ultrasound system with transabdominal 2- to 4- MHz curvilinear transducers (GE Healthcare Ultrasound, Milwaukee, WI, United States) according to the American Society of Echocardiography guidelines and standards for performance of the fetal echocardiogram (Rychik et al., 2004).
Both CNV-seq and WES were done in the setting of a purely research-based protocol, and performed using methods as previously described on genomic DNA from the deceased fetus and the parents (Sun et al., 2019). Briefly, genomic DNA was extracted, hybridized and enriched for whole-exome sequencing. The captured libraries were sequenced using Illumina NovaSeq 6,000 (Illumina, Inc., San Diego, CA, United States). Then, the sequencing data were aligned to the human reference genome (hg38/GRCh38) using BWA (http://bio-bwa.sourceforge.net/) and PCR duplicates were removed by using Picard v1.57 (http://picard.sourceforge.net/). GATK (https://software.broadinstitute.org/gatk/) was applied for variant calling. ANNOVAR (http://wannovar.wglab.org/) was used for variant annotation and interpretation. We determined the frequency of each variant in the dbSNP150 (https://www.ncbi.nlm.nih.gov/snp/), 1,000 Genomes Project (http://www.internationalgenome.org/) and gnomAD (https://gnomad.broadinstitute.org/) to remove common SNPs (minor allele frequency >0.1%). Then, non-synonymous, splicing, frameshift and non-frameshift variants, as well as variants located in splice sites within 20 base pairs of an exon, were prioritized for evaluation. SIFT (http://sift.jcvi.org), PolyPhen-2 (http://genetics.bwh. harvard. edu/pph2), MutationTaster (http://www.mutation taster.org) and CADD (http://cadd.gs.washington.edu) were used to predict the pathogenicity of missense variants, while HSF (http://www.umd.be/HSF3/), and MatEntScan (Yeo and Burge, 2004) were used to evaluate the effects on splicing. Missense variants not presenting damaging results in any protein function prediction from SIFT, Polyphen2, MutationTaster, and CADD were excluded. Intronic variants not presenting damaging results in any prediction from HSF and MatEntScan were excluded. Pathogenicity of variants was determined according to current ACMG guidelines that recommend classifying variants into five categories: pathogenic, likely pathogenic, uncertain significance, likely benign or benign (Richards et al., 2015). Sanger sequencing was used to validate the presence of positive genetic results.
A 26-year-old primigravida was referred at 25 weeks’ gestation for prenatal echocardiography due to fetal bradycardia detected during the routine ultrasound examination. The woman was healthy with no significant family history and did not take any medication. Her anti-Ro/SSA and anti-La/SSB antibody status were both negative. She and her partner were non-consanguineous.
The fetal echocardiography identified noncompacted layers in both ventricles. An extensive trabeculated layer, with multiple deep intratrabecular recesses filled with blood directly from the ventricular cavity, was seen (Figure 1A). Moreover, a thickened, stenotic pulmonary valve was noted with a peak velocity of 158 cm/s shown by spectral Doppler (Figure 1B). There was fetal bradycardia; second-degree 2:1 atrioventricular block was seen by spectral Doppler and M-mode Echocardiography (Figures 1C,D). The fetal echocardiographic findings were consistent with biventricular noncompaction cardiomyopathy (BVNC) with pulmonary stenosis, second-degree 2:1 atrioventricular block, and sinus bradycardia.
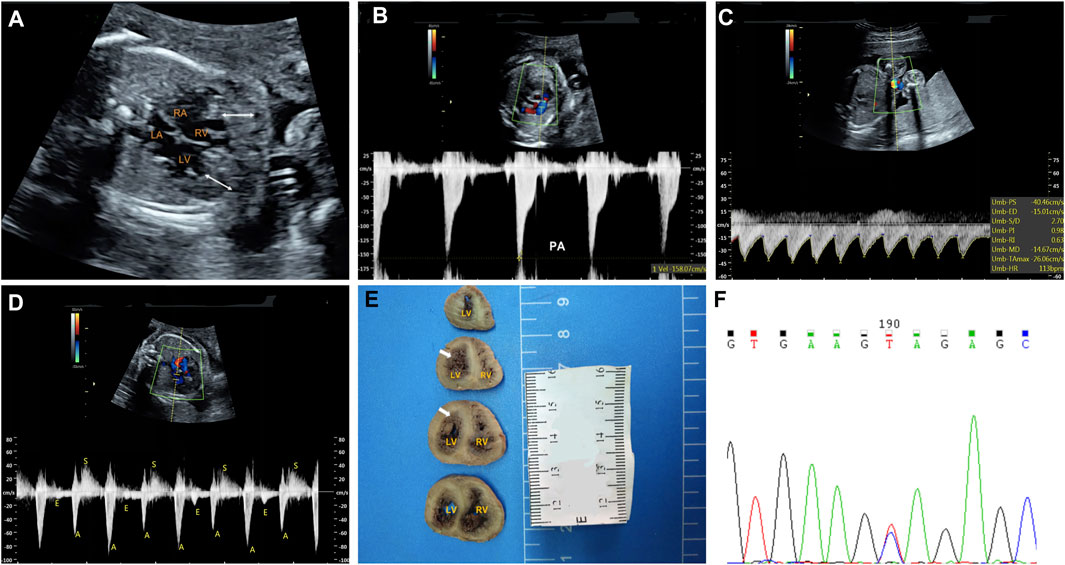
FIGURE 1. Clinical phenotypes and molecular findings of the fetus. (A–D): Echocardiography of the fetus at 25 weeks’ gestation. The phenotype includes biventricular noncompaction cardiomyopathy (A), pulmonary stenosis (B), sinus bradycardia (C) and second-degree 2:1 atrioventricular block (D). A. The area between the white arrows indicate numerous ventricular trabeculae (C). Sinus bradycardia was seen with ventricular rate of 113 bpm by umbilical artery blood flow spectrum (D). Mitral inflow and left ventricular outflow spectrum shows a 2:1 second-degree atrioventricular block (E): Pathological anatomy shows that the noncompaction myocardium below the level of the left ventricular papillary muscle is obvious. The white arrows indicate the noncompaction myocardium (F): Sanger sequencing shows that the mutation is heterozygous in the fetus. LA: left atrium; LV: left ventricle; RA: right atrium; RV: right ventricle.
At the time of the fetal diagnosis, the family was counseled about the potential overall poor prognosis for this fetus related to the BVNC with the second-degree atrioventricular block and pulmonary stenosis. Finally, the family decided to terminate the pregnancy and undergo genetic testing. The pregnancy was terminated at 26 weeks’ gestation. Subsequent postmortem examination confirmed BVNC and pulmonary stenosis (Figure 1E).
A trio (fetus and the parents) CNV-seq and WES were performed to determine the underlying genetic cause of the fetal cardiac phenotype. CNV-seq analysis identified no chromosomal abnormalities. The WES analysis initially identified 83,186 initial variants. The filtering cascades for WES data are listed in Supplementary Table S1. After four filters of the variants data for WES data, 91 variants were kept (Supplementary Table S2). Finally, we identified a de novo missense variant in KCNH2 (NM_000238.3:c.1847A > G,p.Tyr616Cys) in the fetus (Figure 1F), while no pathogenic variants in other known genes associated with cardiomyopathy or arrhythmias were identified. This KCHN2 variant was not found in the biggest general population database (gnomAD, https://gnomad.broadinstitute.org) and in-house control database and showed a deleterious effect by multiple in silico algorithms. The variant has been reported previously in several individuals with LQTS (Kapplinger et al., 2009; Ware et al., 2012), and ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar) contains an entry for this variant (Variation ID: 67295). In addition, the Tyr616 is located in the intramembrane pore-forming H5 domain of KCNH2, and mutations at surrounding codons (Leu615Phe, Leu615Val, Ala614Val, and Thr613Met) have also been reported in association with LQTS, supporting the functional importance of this region of the protein. Furthermore, functional studies carried out by Anderson et al. (2014) demonstrate Tyr616Cys generated minimal current, suggesting altered channel permeability as a mechanism that leads to Prolonged QT interval. In conclusion, the variant is classified as pathogenic according to the 2015 American College of Medical Genetics and Genomics guidelines (Richards et al., 2015).
In this report we present the first fetal case with KCNH2 mutation presenting with BVNC, pulmonary stenosis, second-degree 2:1 atrioventricular block, and sinus bradycardia. LVNC is an increasingly recognized type of cardiomyopathy characterized by excessive trabeculation of the ventricles with deep intertrabecular recesses. While LVNC was classified as distinct cardiomyopathy by the American Heart Association (Maron et al., 2006), the European Society of Cardiology categorizes it as unclassified cardiomyopathy (Elliott et al., 2007). In the early fetal period, about 12 weeks, the myocardium is widely formed by trabeculae. These trabeculations undergo a compaction process, mainly finished before the 16–18 weeks of pregnancy (Faber et al., 2021; Finsterer et al., 2017). According to the non-compaction theory, LVNC results from the arrest of endomyocardial morphogenesis, leading to trabecular compaction failure (Hussein et al., 2015).
In the absence of a known family history, the diagnosis of fetal LQTS is based on the correct recognize of the signature rhythms, such as second-degree AVB, and sinus bradycardia. Second-degree AVB is the signature rhythm of LQTS in the perinatal period, and have been reported in about 25% of fetal LQTS cases (Horigome et al., 2010; Mitchell et al., 2012). Sinus bradycardia is also a common manifestation of fetal LQTS, and has been reported in as many as 44–66% of fetuses diagnosed with LQTS (Cuneo et al., 2013; Greene et al., 2013; Horigome et al., 2010; Mitchell et al., 2012). In this fetus, a prenatal LQTS was highly suspected based on the following: second-degree AVB(Cuneo et al., 2013; Greene et al., 2013), sinus bradycardia (Cuneo et al., 2013; Greene et al., 2013; Mitchell et al., 2012), and the report of the same KCNH2 in several individuals with LQTS (Kapplinger et al., 2009; Ware et al., 2012). However, we were unable to make a definitive diagnosis because a QT prolongation was not proven.
Through systematic literature review, we identified several additional cases of KCNH2 mutation with LVNC-LQTS combined phenotype (AlSenaidi et al., 2014; Ogawa et al., 2009; Rammes et al., 2017). The first association between LVNC and KCNH2 mutations was described by Ogawa et al. (2009) reporting 2 unrelated individuals with isolated LVNC and LQTS carrying missense mutations in KCNH2. Subsequent AlSenaidi et al. (2014) reported a 5-year-old girl of consanguineous Oman parents carrying a KCNH2 homozygous frameshift mutation in association with phenotypes including LVNC, dilated ascending aorta and LQTS. Interestingly, both the parents of the girl carried this KCNH2 heterozygous mutation but neither presented with LVNC, indicating that LVNC is incomplete in LQTS patients with KCNH2 mutations. Recently, Rammes et al. (2017) also reported a familial case, in which both the proband and her son presented with LVNC, and LQTS carrying a published pathogenic variant in KCNH2. These increasing reports suggest that the coexistence of LVNC and arrhythmia may not be rare, significantly, as the detection rate of LVNC is gradually increasing with the advances of echocardiography and MRI.
Interestingly, KCNH2 mutations have also been reported in patients with isolated LVNC. Recent works of LVNC by the groups of Miszalski-Jamka and Wang have reported several individuals carrying KCNH2 mutations in association with LVNC(Miszalski-Jamka et al., 2017; Wang et al., 2017). Notably, no arrhythmia has been reported in these subjects. In addition, in the study by Wang et al. (2017), the number of rare variants in KCNH2 was significantly enriched in LVNC patients compared with the control group, further supporting the association between LVNC and KCNH2 mutation. In several recent independent LVNC cohorts, the variation burden of ion channel genes were as high as 8.8–14.7 (Hirono et al., 2020; Cambon-Viala et al., 2021; Miszalski-Jamka et al., 2017; Wang et al., 2017), significantly higher than in the general population. This suggests that ion-channel dysfunction may play a role in the pathogenesis of LVNC(Milano et al., 2014; Nakashima et al., 2013; Shan et al., 2008; Towbin, 2014). Although the exact mechanism is unclear, several hypotheses have been proposed. They suggest that direct protein-protein interaction between the ion channel gene products and the sarcomere may induce ventricular noncompaction or that ventricular noncompaction is an acquired adaptive remodeling feature in response to impaired conduction (Ergul et al., 2011; Brescia et al., 2013; Caliskan et al., 2012; Steffel and Duru, 2011; Zhang et al., 2013).
In summary, we present the first fetus with LVNC-LQTS combined phenotype and KCNH2 mutation. This case reminds clinicians that ion channel gene variants should be searched systematically in LVNC patients, especially in the arrhythmia phenotype.
The datasets for this article are not publicly available due to concerns regarding participant/patient anonymity. Requests to access the datasets should be directed to the corresponding author.
The studies involving human participants were reviewed and approved by the Ethics Committee of Beijing Anzhen Hospital, Capital Medical University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was obtained from the individual(s), and minor(s)' legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
HZ and YH designed the study. XH, XZ, and JH collected the clinical data and samples from the family. HS and ML completed the experiments. HS and XL analyzed and interpreted the data. HS and XL wrote the manuscript. HS and XL contributed equally to this work. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (No. 82170301 to YH), Beijing Advanced Innovation Center for Big Data-based Precision Medicine, Capital Medical University, Beijing, 100069, China (No. PXM 2021_014226_000026 to HZ) and Beijing Lab for Cardiovascular Precision Medicine, Beijing, China (No. PXM 2020_014226_000017_00377132_FCG to HZ).
Author ML was employed by Cipher Gene LLC.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.821226/full#supplementary-material
AlSenaidi, K. S., Wang, G., Zhang, L., Beer, D. A., AlFarqani, A. M., AlMaskaryi, S. N., et al. (2014). Long QT Syndrome, Cardiovascular Anomaly and Findings in ECG-Guided Genetic Testing. IJC Heart & Vessels 4, 122–128. doi:10.1016/j.ijchv.2014.06.001
Amin, A. S., Herfst, L. J., Delisle, B. P., Klemens, C. A., Rook, M. B., Bezzina, C. R., et al. (2008). Fever-induced QTc Prolongation and Ventricular Arrhythmias in Individuals with Type 2 Congenital Long QT Syndrome. J. Clin. Invest. 118 (7), 2552–2561. doi:10.1172/JCI35337
Anderson, C. L., Kuzmicki, C. E., Childs, R. R., Hintz, C. J., Delisle, B. P., and January, C. T. (2014). Large-scale Mutational Analysis of Kv11.1 Reveals Molecular Insights into Type 2 Long QT Syndrome. Nat. Commun. 5 (1), 5535. [Journal Article; Research Support, N.I.H., Extramural; Research Support, Non-U.S. Gov't]. doi:10.1038/ncomms6535
Brescia, S. T., Rossano, J. W., Pignatelli, R., Jefferies, J. L., Price, J. F., Decker, J. A., et al. (2013). Mortality and Sudden Death in Pediatric Left Ventricular Noncompaction in a Tertiary Referral center. Circulation 127 (22), 2202–2208. doi:10.1161/CIRCULATIONAHA.113.002511
Caliskan, K., Ujvari, B., Bauernfeind, T., Theuns, A.M.J. D., Akca, F., Akca, F, et al. (2012). The Prevalence of Early Repolarization in Patients with Noncompaction Cardiomyopathy Presenting with Malignant Ventricular Arrhythmias. J. Cardiovasc. Electr. 23 (9), 938–944. doi:10.1111/j.1540-8167.2012.02325.x
Cambon-Viala, M., Gerard, H., Nguyen, K., Richard, P., Ader, F., Pruny, J.-F., et al. (2021). Phenotype/Genotype Relationship in Left Ventricular Noncompaction: Ion Channel Gene Mutations Are Associated with Preserved Left Ventricular Systolic Function and Biventricular Noncompaction. J. Card. Fail. 27 (6), 677–681. doi:10.1016/j.cardfail.2021.01.007
Cuneo, B. F., Etheridge, S. P., Horigome, H., Sallee, D., Moon-Grady, A., Weng, H.-Y., et al. (2013). Arrhythmia Phenotype during Fetal Life Suggests Long-QT Syndrome Genotype. Circ. Arrhythm Electrophysiol. 6 (5), 946–951. doi:10.1161/CIRCEP.113.000618
Elliott, P., Andersson, B., Arbustini, E., Bilinska, Z., Cecchi, F., Charron, P., et al. (2007). Classification of the Cardiomyopathies: a Position Statement from the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur. Heart J. 29 (2), 270–276. doi:10.1093/eurheartj/ehm342
Ergul, Y., Nisli, K., Varkal, M. A., Oner, N., Dursun, M., Dindar, A., et al. (2011). Electrocardiographic Findings at Initial Diagnosis in Children with Isolated Left Ventricular Noncompaction. Ann. Noninvas. Electro. 16 (2), 184–191. doi:10.1111/j.1542-474X.2011.00428.x
Faber, J. W., D'Silva, A., Christoffels, V. M., and Jensen, B. (2021). Lack of Morphometric Evidence for Ventricular Compaction in Humans. J. Cardiol. 78 (5), 397–405. doi:10.1016/j.jjcc.2021.03.006
Finsterer, J., Stöllberger, C., and Towbin, J. A. (2017). Left Ventricular Noncompaction Cardiomyopathy: Cardiac, Neuromuscular, and Genetic Factors. Nat. Rev. Cardiol. 14 (4), 224–237. doi:10.1038/nrcardio.2016.207
Gianulis, E. C., and Trudeau, M. C. (2011). Rescue of Aberrant Gating by a Genetically Encoded PAS (Per-Arnt-Sim) Domain in Several Long QT Syndrome Mutant Human Ether-Á-Go-Go-Related Gene Potassium Channels. J. Biol. Chem. 286 (25), 22160–22169. [Journal Article; Research Support, N.I.H., Extramural; Research Support, Non-U.S. Gov't]. doi:10.1074/jbc.M110.205948
Greene, E. A., Berul, C. I., and Donofrio, M. T. (2013). Prenatal Diagnosis of Long QT Syndrome: Implications for Delivery Room and Neonatal Management. Cardiol. Young 23 (1), 141–145. doi:10.1017/S1047951112000583
Hirono, K., Hata, Y., Miyao, N., Okabe, M., Takarada, S., Nakaoka, H., et al. (2020). Increased burden of Ion Channel Gene Variants Is Related to Distinct Phenotypes in Pediatric Patients with Left Ventricular Noncompaction. Circ. Genom Precis Med. 13 (4), e002940. doi:10.1161/CIRCGEN.119.002940
Horigome, H., Nagashima, M., Sumitomo, N., Yoshinaga, M., Ushinohama, H., Iwamoto, M., et al. (2010). Clinical Characteristics and Genetic Background of Congenital Long-QT Syndrome Diagnosed in Fetal, Neonatal, and Infantile Life. Circ. Arrhythm Electrophysiol. 3 (1), 10–17. doi:10.1161/CIRCEP.109.882159
Hussein, A., Karimianpour, A., Collier, P., and Krasuski, R. A. (2015). Isolated Noncompaction of the Left Ventricle in Adults. J. Am. Coll. Cardiol. 66 (5), 578–585. doi:10.1016/j.jacc.2015.06.017
Kapplinger, J. D., Tester, D. J., Salisbury, B. A., Carr, J. L., Harris-Kerr, C., Pollevick, G. D., et al. (2009). Spectrum and Prevalence of Mutations from the First 2,500 Consecutive Unrelated Patients Referred for the FAMILION Long QT Syndrome Genetic testResearch Support, Non-U.S. Gov't]. [Journal ArticleHeart Rhythm 6 (9), 1297–1303. doi:10.1016/j.hrthm.2009.05.021
Maron, B. J., Towbin, J. A., Thiene, G., Antzelevitch, C., Corrado, D., Arnett, D., et al. (2006). Contemporary Definitions and Classification of the Cardiomyopathies. Circulation 113 (14), 1807–1816. doi:10.1161/CIRCULATIONAHA.106.174287
Milano, A., Vermeer, A. M. C., Lodder, E. M., Barc, J., Verkerk, A. O., Postma, A. V., et al. (2014). HCN4 Mutations in Multiple Families with Bradycardia and Left Ventricular Noncompaction cardiomyopathyResearch Support, Non-U.S. Gov't]. J. Am. Coll. CardiologyJ. Am. Coll. Cardiol. 64 (8), 745–756. doi:10.1016/j.jacc.2014.05.045
Miszalski-Jamka, K., Jefferies, J. L., Mazur, W., Głowacki, J., Hu, J., Lazar, M., et al. (2017). Novel Genetic Triggers and Genotype-Phenotype Correlations in Patients with Left Ventricular Noncompaction. Circ. Cardiovasc. Genetcirculation: Cardiovasc. Genet. 10 (4). doi:10.1161/CIRCGENETICS.117.001763
Mitchell, J. L., Cuneo, B. F., Etheridge, S. P., Horigome, H., Weng, H.-Y., and Benson, D. W. (2012). Fetal Heart Rate Predictors of Long QT Syndrome. Circulation 126 (23), 2688–2695. doi:10.1161/CIRCULATIONAHA.112.114132
Nakashima, K., Kusakawa, I., Yamamoto, T., Hirabayashi, S., Hosoya, R., Shimizu, W., et al. (2013). A Left Ventricular Noncompaction in a Patient with Long QT Syndrome Caused by a KCNQ1 Mutation: A Case Report. [Journal Article]Heart Vessels 28 (1), 126–129. doi:10.1007/s00380-012-0235-8
Ogawa, K., Nakamura, Y., Terano, K., Ando, T., Hishitani, T., and Hoshino, K. (2009). Isolated Non-compaction of the Ventricular Myocardium Associated with Long QT Syndrome A Report of 2 Cases. Circ. J. 73 (11), 2169–2172. doi:10.1253/circj.cj-08-0339
Rammes, S., Farr, M., Laser, T., and Dubowy, K. O. (2017). Left Ventricular Noncompaction Cardiomyopathy and Long QT Syndrome, a Common Cause? A Case Report. Thorac. Cardiovasc. Surg. 65 (S 02), S111–S142. doi:10.1055/s-0037-1598992
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J., et al. (2015). Standards and Guidelines for the Interpretation of Sequence Variants: a Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17 (5), 405–424. doi:10.1038/gim.2015.30
Rychik, J., Ayres, N., Cuneo, B., Gotteiner, N., Hornberger, L., Spevak, P. J., et al. (2004). American Society of Echocardiography Guidelines and Standards for Performance of the Fetal Echocardiogram. J. Am. Soc. Echocardiography 17 (7), 803–810. doi:10.1016/j.echo.2004.04.011
Schweizer, P. A., Schröter, J., Greiner, S., Haas, J., Yampolsky, P., Mereles, D., et al. (2014). The Symptom Complex of Familial Sinus Node Dysfunction and Myocardial Noncompaction Is Associated with Mutations in the HCN4 channelResearch Support, Non-U.S. Gov't]. J. Am. Coll. CardiologyJ. Am. Coll. Cardiol. 64 (8), 757–767. doi:10.1016/j.jacc.2014.06.1155
Shan, L., Makita, N., Xing, Y., Watanabe, S., Futatani, T., Ye, F., et al. (2008). SCN5A Variants in Japanese Patients with Left Ventricular Noncompaction and Arrhythmia. Mol. Genet. Metab. 93 (4), 468–474. doi:10.1016/j.ymgme.2007.10.009
Steffel, J., and Duru, F. (2011). Rhythm Disorders in Isolated Left Ventricular Noncompaction. Ann. Med. 44 (2), 101–108. doi:10.3109/07853890.2011.554427
Sun, H., Yi, T., Hao, X., Yan, H., Wang, J., Li, Q., et al. (2020). Contribution of Single‐gene Defects to Congenital Cardiac Left‐sided Lesions in the Prenatal Setting. Ultrasound Obstet. Gynecol. 56, 225–232. doi:10.1002/uog.21883
Towbin, J. A. (2014). Ion Channel Dysfunction Associated with Arrhythmia, Ventricular Noncompaction, and Mitral Valve Prolapse. J. Am. Coll. Cardiol. 64 (8), 768–771. doi:10.1016/j.jacc.2014.06.1154
Towbin, J. A., Lorts, A., and Jefferies, J. L. (2015). Left Ventricular Non-compaction Cardiomyopathy. The Lancet 386 (9995), 813–825. doi:10.1016/S0140-6736(14)61282-4
Wang, C., Hata, Y., Hirono, K., Takasaki, A., Ozawa, S. W., Nakaoka, H., et al. (2017). A Wide and Specific Spectrum of Genetic Variants and Genotype-Phenotype Correlations Revealed by Next‐Generation Sequencing in Patients with Left Ventricular Noncompaction. Jaha 6 (9). doi:10.1161/JAHA.117.006210
Ware, J. S., Walsh, R., Cunningham, F., Birney, E., and Cook, S. A. (2012). Paralogous Annotation of Disease-Causing Variants in Long QT Syndrome Genes. Hum. Mutat. 33 (8), 1188–1191. doi:10.1002/humu.22114
Yeo, G., and Burge, C. B. (2004). Maximum Entropy Modeling of Short Sequence Motifs with Applications to RNA Splicing Signals. J. Comput. Biol. 11 (2-3), 377–394. [Journal Article; Research Support, Non-U.S. Gov't; Research Support, U.S. Gov't, Non-P.H.S.; Research Support, U.S. Gov't, P.H.S.]. doi:10.1089/1066527041410418
Zhang, W., Chen, H., Qu, X., Chang, C.-P., and Shou, W. (2013). Molecular Mechanism of Ventricular Trabeculation/compaction and the Pathogenesis of the Left Ventricular Noncompaction Cardiomyopathy (LVNC). Am. J. Med. Genet. 163 (3), 144–156. Research Support, N.I.H., Extramural; Research Support, Non-U.S. Gov't; Review]. doi:10.1002/ajmg.c.31369
Keywords: left ventricular noncompaction, long QT syndrome, congenital heart disease, prenatal diagnosis, KCNH2 gene mutation
Citation: Sun H, Liu X, Hao X, Zhou X, Wang J, Han J, Liang M, Zhang H and He Y (2022) Case Report: Biventricular Noncompaction Cardiomyopathy With Pulmonary Stenosis and Bradycardia in a Fetus With KCNH2 Mutation. Front. Genet. 13:821226. doi: 10.3389/fgene.2022.821226
Received: 24 November 2021; Accepted: 10 February 2022;
Published: 24 February 2022.
Edited by:
Rita Selvatici, University of Ferrara, ItalyReviewed by:
Seiko Ohno, National Cerebral and Cardiovascular Center, JapanCopyright © 2022 Sun, Liu, Hao, Zhou, Wang, Han, Liang, Zhang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongjia Zhang, emhhbmdob25namlhNzIyQGhvdG1haWwuY29t; Yihua He, aGV5aWh1YWVjaG9AaG90bWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.