- 1Dr. Panjwani Center for Molecular Medicine and Drug Research (PCMD), International Center for Chemical and Biological Sciences (ICCBS), University of Karachi, Karachi, Pakistan
- 2Department of Human Genetics and Molecular Biology, University of Health Sciences, Lahore, Pakistan
- 3Atomic Energy Medical Centre (AEMC), Jinnah Postgraduate Medical Centre (JPMC), Karachi, Pakistan
Demographics for breast cancers vary widely among nations. The frequency of germline mutations in breast cancers, which reflects the hereditary cases, has not been investigated adequately and accurately in highly-consanguineous Pakistani population. In the present discovery case series, germ-line mutations in twenty-seven breast cancer candidate genes were investigated in eighty-four sporadic breast cancer patients along with the clinical correlations. The germ-line variants were also assessed in two healthy gender-matched controls. The clinico-pathological features were evaluated by descriptive analysis and Pearson χ2 test (with significant p-value <0.05). The most frequent parameters associated with hereditary cancer cases are age and ethnicity. Therefore, the analyses were stratified on the basis of age (≤40 years vs. >40 years) and ethnicity. The breast cancer gene panel assay was carried out by BROCA, which is a genomic capture, massively parallel next generation sequencing assay on Illumina Hiseq2000 with 100bp read lengths. Copy number variations were determined by partially-mapped read algorithm. Once the mutation was identified, it was validated by Sanger sequencing. The ethnic analysis stratified on the basis of age showed that the frequency of breast cancer at young age (≤40 years) was higher in Sindhis (n = 12/19; 64%) in contrast to patients in other ethnic groups. Majority of the patients had stage III (38.1%), grade III (50%), tumor size 2–5 cm (54.8%), and invasive ductal carcinoma (81%). Overall, the analysis revealed germ-line mutations in 11.9% of the patients, which was not significantly associated with younger age or any particular ethnicity. The mutational spectrum was restricted to three genes: BRCA1, BRCA2, and TP53. The identified mutations consist of seven novel germ-line mutations, while three mutations have been reported previously. All the mutations are predicted to result in protein truncation. No mutations were identified in the remaining twenty-four candidate breast cancer genes. The present study provides the framework for the development of hereditary-based preventive and treatment strategies against breast cancers in Pakistani population.
Introduction
Epidemiological studies have shown ethnic and geographic differences in breast cancer etiology. The increased susceptibility to breast cancers has been attributed to socio-economic, environmental, and genetic factors (Zeeshan et al., 2019). Hereditary breast cancers comprise a significant number. In the US population, these constitute 10–15% of the cases. There is paucity of data from low- to middle- income countries (LMIC) (Akarolo-Anthony et al., 2010; Jazayeri et al., 2015).
It has been estimated that half of all breast cancer cases occur in the 12% women who are at the maximum genetic risk (El Saghir et al., 2007). BRCA1 and BRCA2 are high penetrant breast cancer genes. These have been especially associated with hereditary breast and/or ovarian cancers. The mutations in these genes are considered to increase the life-time risks of breast cancer by 82% (Antoniou and Easton, 2006; Shiovitz and Korde, 2015). Other highly penetrant but rare genes include PTEN, TP53, CDH1, and STK11. Moderate penetrance genes, which increase the risk for breast cancer by twofold, include genes involved in DNA repair such as ATM, BRIP1 (BACH1), CHEK2, and PALB2. Still other genes are considered to confer a low but significant risk for breast cancers (Walsh et al., 2017).
In Pakistani population, some studies have analyzed BRCA1 and BRCA2 mutations (Liede et al., 2002; Rashid et al., 2006; Moatter et al., 2011; Ahmad et al., 2012; Aziz et al., 2016; Rashid et al., 2016; Rashid et al., 2017; Torres et al., 2017), mainly through conventional methodologies. In case of other putative breast cancer susceptibility genes, scarce or no data is available from this region (Khaliq et al., 2000; Baig et al., 2011; Rashid et al., 2014; Baloch et al., 2016; Rashid et al., 2019). The introduction of next generation sequencing (NGS) including multi-gene testing necessitates re-assessment of the available information and generation of missing data.
Pakistani population comprises distinct ethnic groups. These include Sindhis, Balochis, Brahui, Makrani, and Parsis from Southern Pakistan. The other ethnicities Punjabis, Pathans, Hindko, Hazara, Kalash, Kashmiri and Burusho are from Northern Pakistan (Qamar et al., 1999; Khaliq et al., 2000). The data for these genetically distinct ethnic groups has not been incorporated in the few relevant regional-based publications on the breast cancers. The location of present study is a metropolitan city, situated in Southern Pakistan. The population comprises multiple ethnic communities from all over Pakistan. In addition, a self-defined Urdu-speaking ethnicity, comprising immigrants from India, is also a major group residing in the city.
The present study investigates molecular epidemiology of breast cancers from Southern Pakistan. It is the first such report from this region. The study investigates the inherited contribution to breast cancers by next-generation sequencing. A panel of twenty-seven breast cancer-associated candidate genes, has been analysed in breast cancer patients belonging to genetically distinct Pakistani ethnic groups.
Materials and Methods
Patients
Participants in the present study included 82 females and 02 males from Southern Pakistan. In total, 84 diagnosed cases of breast cancer were included in the study. The participants visited a tertiary care hospital: the Atomic Energy Medical Centre (AEMC), Jinnah Post-Graduate Medical Centre (JPMC), Karachi, Pakistan, from July 2016–July 2017. The patients were treated for primary invasive breast cancer post-mastectomy.
As the present study was carried out for the discovery purposes at the preliminary stage, all the clinically diagnosed primary breast cancer cases were included regardless of the age and/or family history. All the participants signed an informed consent form. Independent ethical review boards of the participating institutions approved the protocol.
Demographic and Clinico-Pathological Information
Patients were interviewed about their family history of cancers (breast cancer and/or any other cancer), ethnicity, age at menarche and menopause (if applicable), and gynecological and obstetrics history. The medical records were reviewed for breast cancer diagnosis, staging, grading, and tumour size. According to the clinical reports, staging was carried out based on American Joint Committee on Cancer (AJCC), while grading was performed with Nottingham Histologic Score System based on Scarff-Bloom-Richardson Grading System.
Genomics
The participants contributed 5–8 ml of blood samples for DNA extraction. Germ-line DNA was extracted from the patient’s WBCs by standard phenol-chloroform method (Sambrook and Russell, 2001). DNA was quantified spectrophotometrically (Beckman Coulter™, DU® 530). Sufficient DNA was available for 84 subjects. BROCA, a targeted capture and multiplexed massively parallel sequencing gene panel assay was performed (Walsh et al., 2017). Briefly, it is a next-generation sequencing assay on Illumina Hiseq2000 with 100bp read lengths. The copy number variations (CNVs) were determined by partially-mapped read algorithm (Nord et al., 2011). It enables detection of all types of mutations for candidate and established breast cancer genes. Twenty-seven genes, which are highly associated with breast cancers, were investigated in the project: BRCA1, BRCA2, TP53, ATR, BARD1, BRIP1, FAM175A, FANCM, GEN1, MRE11A, NBN, RAD51B, RAD51C, RAD51D, RECQL, RINT1, SLX4, BAP1, PALB2, PTEN, STK11, XRCC2, ATM, CHEK1, CHEK2, CDH1, and CTNNA1.
Validation of Mutations
After the NGS investigations, the identified mutations were validated by Sanger sequencing. Previously published protocols were used for the amplification of exons (Friedman et al., 1994; Friedman et al., 1997; Olivier et al., 2002) followed by standard method for Sanger sequencing.
Statistical and Bio-Informatic Analysis
Data were entered, encoded and analysed using SPSS, version 17.0 (IBM™, United States). Breast cancer cases in the present study were grouped into three categories: age at sampling (≤40years vs >40 years); receptor (estrogen, progesterone, and HER2/Neu) status; and ethnicities. Descriptive analysis was carried out for the evaluation of demographics and clinico-pathological features. Groups were compared by Pearson χ2 test of independence for the clinico-pathological features: tumour size, grade, and stage. The p-values <0.05 were considered to be statistically significant.
The mutations were compared against BIC (BIC, 2019) and ExAC (ExAC, 2019) databases for novelty.
Results
Demographic Analysis
Total study included 84 breast cancer patients, with a diagnosis of primary breast cancer. Patients’ demographics are shown in Table 1. In the present cohort, majority of the breast cancer cases belonged to Urdu-speaking (25%) and Sindhi (24%) ethnicities. The frequency of breast-cancer cases among young patients (≤40 years) was higher in Hindko (n = 4; 75%) and Sindhi (n = 19; 64%) ethnicities, in contrast to other ethnic groups. Among other ethnic groups, the numbers of breast cancer cases in older patients (i.e., > 40 years) exceeded those who were ≤40 years.
Clinicopathological Evaluation
Pathology records were sought for the patients. Data were available for 70.9, 95, and 82.5% patients in case of tumour stage, grade and hormonal status, respectively. Among patients with available pathology data, the distribution of tumor stage was 5% Stage I, 33% Stage II, 56% Stage III, and 6% Stage IV. The distribution of tumour grade was 1.2% Grade I, 48.7% Grade II, and 50% Grade III. Overall, the distribution among stages and grades (low vs high) varied significantly (p <0.01). In case of tumours with available hormone profiles, 26% were triple negative (TNBC). Supplementary Table S1 lists the available clinicopathological information.
Novel Germ-Line Mutations
A total of 84 samples were analyzed based upon sufficient DNA quantity. Genomic analysis of known breast cancer genes revealed that 11.9% (10/84) patients carried an unambiguously pathogenic germline mutation in three genes: BRCA1, BRCA2, and TP53 (Table 2). Novel germ-line mutations were identified in seven patients (3 in BRCA1, 3 in BRCA2, and 1 in TP53) (Table 2 and Supplementary Figure S1). The predicted outcome of all the identified mutations is protein truncation.
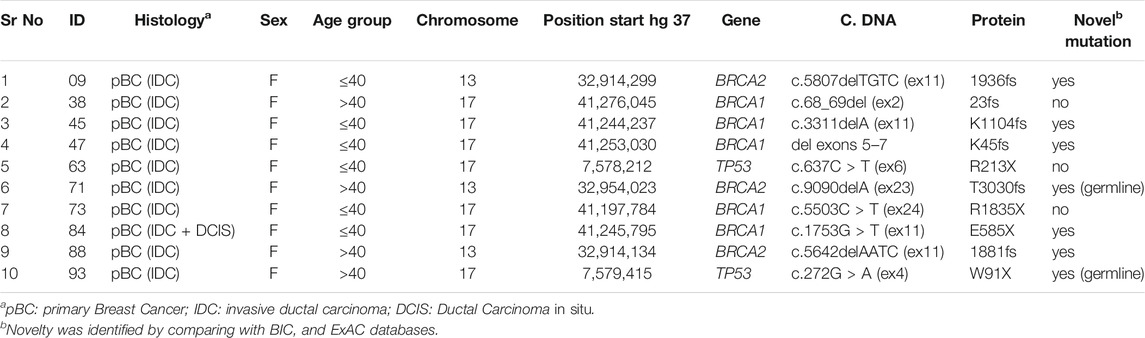
TABLE 2. Summary of germline mutations identified in BRCA1, BRCA2, and TP53 genes in Pakistani breast cancer patients.
Burden of Mutations in Breast Cancer Genes
Genomic analysis of known breast cancer genes showed that 15.3% (6/39) of patients with age ≤40 years, whereas 8.5% (4/47) of patients with age >40 years carried a definitive pathogenic germline mutation in three identified genes (Table 3).
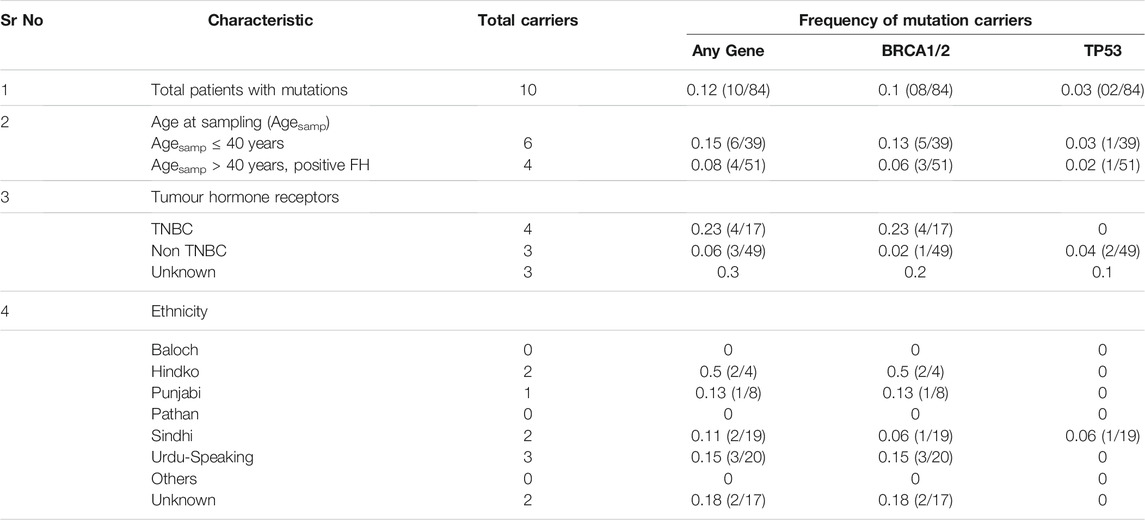
TABLE 3. Distribution of germ-line mutations in breast cancer patients. Data has been stratified on the basis of age, hormone receptor status, and ethnicities.
Among younger patients, 13% (5/39) carried a damaging mutation in BRCA1 and BRCA2 while 3% (1/39) carried a germ-line mutation in TP53. In the older patients, 5.8% (3/51) harboured a germ-line mutation in BRCA1 and BRCA 2, whereas the frequency of germ-line mutation in TP53 was 2% (1/51). Stratified analysis showed that the highest frequency of germ-line mutations in the investigated genes was in Hindko group (50%), followed by Urdu-speaking (15%), Punjabi (13%), and Sindhi (11%) ethnicities.
Among patients with known tumour hormone receptor status, 23% (4/17) TNBC patients carried a pathogenic germline mutation in BRCA1/2 genes (2 in BRCA1 and 2 in BRCA2).
Each identified mutation was present in only one family and no recurrent mutation was found in the present cohort. In the samples from two male patients, which were included in the study, no mutation in the selected genes was observed.
Further, no pathogenic germ-line variants were identified in the DNA from control samples.
Discussion
Breast cancer is the most frequently diagnosed malignancy among women and the leading cause of cancer-related mortality in developing countries (Ferlay J et al., 2018). It is estimated that globally 1 in 6 women is diagnosed with breast cancer and 1 in 8 women has invasive form of the cancer. However, substantial differences have been observed in breast cancer indices across different populations (Bray et al., 2015). The average age in Caucasians is 63 years. In the present study we report an average of 44.4 years, whereas from the same region it is reported in the range of 50–53 years from India, and 46–49 years from Iran, respectively (Ahmadi et al., 2015; Desmond et al., 2015; Mannan et al., 2016; Howlader et al., 2017).
Majority of familial aggregation in breast cancers is unexplained. Environmental factors are unlikely to explain the residual familial clustering (El Saghir et al., 2007). Among Caucasians, with the increasing application of next generation sequencing in the clinical setting, an upward trend (26%) in reports of hereditary breast and/or ovarian cancers is observed (Antoniou and Easton, 2006). A number of large scale studies report germ-line mutational frequencies ranging from 9 to 26% in critical breast cancer genes (Stratton and Rahman, 2008; Castéra et al., 2014; Buys et al., 2017; Eliade et al., 2017).
In case of non-Caucasian females, there is paucity of data regarding molecular basis of breast cancers. To the best of our knowledge the present study is the first report of a twenty-seven breast cancer gene panel analysis in a South-Asian population. The molecular investigation includes high and moderate penetrance genes (Walsh et al., 2017). Here we report germ-line mutations in three high penetrance genes: BRCA1, BRCA2, and TP53 in breast cancer patients from this population. The identified mutations consist of seven novel germ-line mutations, while three mutations have been reported previously. The location of inherited germ-line mutations the genes is shown in Figure 1.
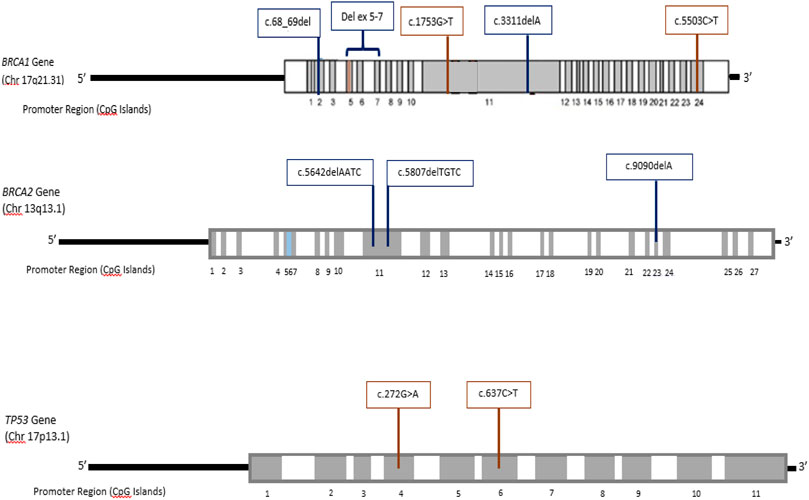
FIGURE 1. Exon-specific distribution of the identified germline mutations in BRCA1, BRCA2, and TP53 genes.
The identified mutations were heterozygous. Bi-allelic BRCA1 mutations are likely to be lethal at the embryonic stage, while such mutations in BRCA2 lead to Fanconi anemia type D1, with increased risk of childhood cancer (FitzGerald et al., 1998; Moatter et al., 2011). Germline mutations in TP53 are associated with Li-Fraumeni syndrome. In the present cohort, no syndromic cases were identified.
The founder mutations are expected in consanguineous populations like the present one. Although we did not find any recurrent mutation in the present cohort, comparison with previously published reports indicated that 185delAG may be a founder mutation (Liede et al., 2002; Rashid et al., 2006; Moatter et al., 2011; Ahmad et al., 2012; Aziz et al., 2016; Rashid et al., 2016; Rashid et al., 2017; Torres et al., 2017).
Germline mutations were not identified in the rest of reported high penetrant genes including PTEN, CDH1, and STK11 (Boardman, 1998; Pharoah et al., 2001; Meijers-Heijboer et al., 2002; Lim et al., 2004; Tan et al., 2012).
Moderate penetrance genes include additional DNA repair genes. These are CHEK2 (, BRIP1 (BACH1), ATM, PALB2 (Renwick et al., 2006; Seal et al., 2006; Rahman et al., 2007; Tanaka et al., 2012). These interact with BRCA1 and/or BRCA2. The mutations in these genes result in two-fold increase in breast cancer risk. In the present study, no germ-line mutation was identified in these genes.
Similarly, no germline mutations were identified in other candidate genes ATR, BARD1, FAM175A, FANCM, GEN1, MRE11A, NBN, RAD51B, RAD51C, RAD51D, RECQL, RINT1, SLX4, BAP1, XRCC2, CHEK1, and CTNNA1 (Varon et al., 1998; Stewart et al., 1999; Vahteristo et al., 2001; Bartkova et al., 2008; Turnbull et al., 2010; Loveday et al., 2011; Testa et al., 2011; Walsh et al., 2011; Wiesner et al., 2011; Loveday et al., 2012; Madanikia et al., 2012; Osher et al., 2012; Park et al., 2012; Ratajska et al., 2012; Solyom et al., 2012; Golmard et al., 2013; Majewski et al., 2013; Shah et al., 2013; Kiiski et al., 2014; Cybulski et al., 2015; Singh et al., 2018).
Interestingly, all the identified mutations are nonsense mutations and predicted to result in protein truncation. This corroborates the data for BRCA1 and BRCA2, but not for TP53 from India. Among South-Asian populations, the germ-line mutation rate (11.9%) in the present study is three folds less than reported for India (Mannan et al., 2016). In contrast to the reported observations that younger patients are likely to be the carriers of germ-line mutation, we report a lower frequency of such mutations as compared to the frequency of such mutations from India (36%).
Most studies on BRCA1 and BRCA2 mutations from Asia report a higher frequency for BRCA2 mutations than BRCA1, the exceptions being Pakistan and India (Kim and Choi, 2013). The pattern is also observed in the present study.
The present study is also the first report of higher frequency of younger breast cancer patients belonging to Hindko and Sindhi ethnicities as compared to the other ethnicities in the region. As the investigated genes do not account for all such cases, it is possible that as yet unidentified gene(s) may be involved in these ethnic groups. It is pertinent to mention that the present day Pakistan consists of more than 12 distinct ethnic and linguistic groups (Mehdi et al., 1999; Qamar et al., 2002).
In conclusion, the present study while providing a framework for the investigation of genetic basis of breast cancers for cost-effective screening and management, raises many questions. The foremost is: as the germline mutations account for only 12% of the breast cancer cases, which other factors (genetic and/or environmental) are involved in the observed high incidence of breast cancers? It is expected that building on the present findings, a scientifically-focused approach may be developed for breast cancer research in a resource-limited setting.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the independent ethics review committee (ERC), International Center for Chemical and Biological Sciences (ICCBS), University of Karachi, Karachi, Pakistan (ICCBS/IEC-016-BS/HT-2016/Protocol/1.0), and the Atomic Energy Medical Centre (AEMC), Jinnah Postgraduate Medical Centre (JPMC), Karachi, Pakistan [Admin-3 (257)/2016]. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Conceptualization: SA Participant enrollment/Data collection: SA, S-ZZ, SMA, AS, and MAM. Benchwork: SA, S-ZZ, and SMA. Analysis: SA. Original draft: SA. Reviewing and editing: SA. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors wish to thank Prof. MC King and Prof. Tom Walsh for BROCA analysis, ICCBS for core facilities, and AEMC, JPMC staff for their co-operation. The authors are especially grateful to the participants in the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.820610/full#supplementary-material
References
Ahmad, J., Le Calvez-Kelm, F., Daud, S., Voegele, C., Vallée, M., Ahmad, A., et al. (2012). Detection ofBRCA1/2mutations in Breast Cancer Patients from Thailand and Pakistan. Clin. Genet. 82 (6), 594–598. doi:10.1111/j.1399-0004.2012.01869.x
Ahmadi, A. S., Mahdipour, L., Payandeh, M., and Sadeghi, M. (2015). Epidemiology, Pathology and Histochemistry Features in Women with Breast Cancer. Am. J. Cancer Prev. 3, 54–57. doi:10.12691/ajcp-3-3-2
Akarolo-Anthony, S. N., Ogundiran, T. O., and Adebamowo, C. A. (2010). Emerging Breast Cancer Epidemic: Evidence from Africa. Breast Cancer Res. 12 (12 Suppl. 4), S8. doi:10.1186/bcr2737
Antoniou, A. C., and Easton, D. F. (2006). Models of Genetic Susceptibility to Breast Cancer. Oncogene 25 (43), 5898–5905. doi:10.1038/sj.onc.1209879
Aziz, F., Fatima, W., Mahmood, S., and Khokher, S. (2016). Screening for Del 185 AG and 4627C>A BRCA1 Mutations in Breast Cancer Patients from Lahore, Pakistan. Asian Pac. J. Cancer Prev. 17 (4), 1725–1727. doi:10.7314/apjcp.2016.17.4.1725
Baig, R. M., Mahjabeen, I., Sabir, M., Masood, N., Hafeez, S., Malik, F. A., et al. (2011). Genetic Changes in the PTEN Gene and Their Association with Breast Cancer in Pakistan. Asian Pac. J. Cancer Prev. 12 (10), 2773–2778. http://journal.waocp.org/?sid=Entrez:PubMed&id=pmid:22320991&key=2011.12.10.2773
Baloch, A. H., Khosa, A. N., Bangulzai, N., Shuja, J., Naseeb, H. K., Jan, M., et al. (2016). Novel Nonsense Variants c.58C>T (p.Q20X) and c.256G>T (p.E85X) in the CHEK2 Gene Identified in Breast Cancer Patients from Balochistan. Asian Pac. J. Cancer Prev. 17 (7), 3623–3626. doi:10.7314/apjcp.2016.17.3.1089
Bartkova, J., Tommiska, J., Oplustilova, L., Aaltonen, K., Tamminen, A., Heikkinen, T., et al. (2008). Aberrations of the MRE11-RAD50-NBS1 DNA Damage Sensor Complex in Human Breast Cancer: MRE11 as a Candidate Familial Cancer-Predisposing Gene. Mol. Oncol. 2 (4), 296–316. doi:10.1016/j.molonc.2008.09.007
BIC (2019). BIC. Available at: https://research.nhgri.nih.gov/bic/ (Accessed June 8, 2019).
Boardman, L. A. (1998). Increased Risk for Cancer in Patients with the Peutz-Jeghers Syndrome. Ann. Intern. Med. 128 (11), 896–899. doi:10.7326/0003-4819-128-11-199806010-00004
Bray, F., Ferlay, J., Laversanne, M., Brewster, D. H., Gombe Mbalawa, C., Kohler, B., et al. (2015). Cancer Incidence in Five Continents: Inclusion Criteria, Highlights from Volume X and the Global Status of Cancer Registration. Int. J. Cancer 137 (9), 2060–2071. doi:10.1002/ijc.29670
Buys, S. S., Sandbach, J. F., Gammon, A., Patel, G., Kidd, J., Brown, K. L., et al. (2017). A Study of over 35,000 Women with Breast Cancer Tested with a 25‐gene Panel of Hereditary Cancer Genes. Cancer 123 (10), 1721–1730. doi:10.1002/cncr.30498
Castéra, L., Krieger, S., Rousselin, A., Legros, A., Baumann, J.-J., Bruet, O., et al. (2014). Next-generation Sequencing for the Diagnosis of Hereditary Breast and Ovarian Cancer Using Genomic Capture Targeting Multiple Candidate Genes. Eur. J. Hum. Genet. 22 (11), 1305–1313. doi:10.1038/ejhg.2014.16
Cybulski, C., Carrot-Zhang, J., Kluźniak, W., Rivera, B., Kashyap, A., Wokołorczyk, D., et al. (2015). Germline RECQL Mutations Are Associated with Breast Cancer Susceptibility. Nat. Genet. 47 (6), 643–646. doi:10.1038/ng.3284
Desmond, A., Kurian, A. W., Gabree, M., Mills, M. A., Anderson, M. J., Kobayashi, Y., et al. (2015). Clinical Actionability of Multigene Panel Testing for Hereditary Breast and Ovarian Cancer Risk Assessment. JAMA Oncol. 1 (7), 943–951. doi:10.1001/jamaoncol.2015.2690
El Saghir, N. S., Khalil, M. K., Eid, T., El Kinge, A. R., Charafeddine, M., Geara, F., et al. (2007). Trends in Epidemiology and Management of Breast Cancer in Developing Arab Countries: a Literature and Registry Analysis. Int. J. Surg. 5 (4), 225–233. doi:10.1016/j.ijsu.2006.06.015
Eliade, M., Skrzypski, J., Baurand, A., Jacquot, C., Bertolone, G., Loustalot, C., et al. (2017). The Transfer of Multigene Panel Testing for Hereditary Breast and Ovarian Cancer to Healthcare: What Are the Implications for the Management of Patients and Families? Oncotarget 8 (2), 1957–1971. doi:10.18632/oncotarget.12699
ExAC (2019). ExAC. Available at: http://exac.broadinstitute.org/ (Accessed June 8, 2019).
Ferlay, J., Ervik, M., Lam, F., Colombet, M., Mery, L., Piñeros, M., et al. (2018). Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. Available at: https://gco.iarc.fr/today (accessed December 24, 2018).
FitzGerald, M. G., Marsh, D. J., Wahrer, D., Bell, D., Caron, S., Shannon, K. E., et al. (1998). Germline Mutations in PTEN Are an Infrequent Cause of Genetic Predisposition to Breast Cancer. Oncogene 17 (6), 727–731. doi:10.1038/sj.onc.1201984
Friedman, L. S., Gayther, S. A., Kurosaki, T., Gordon, D., Noble, B., Casey, G., et al. (1997). Mutation Analysis of BRCA1 and BRCA2 in a Male Breast Cancer Population. Am. J. Hum. Genet. 60 (2), 313–319.
Friedman, L. S., Ostermeyer, E. A., Szabo, C. I., Dowd, P., Lynch, E. D., Rowell, S. E., et al. (1994). Confirmation of BRCA1 by Analysis of Germline Mutations Linked to Breast and Ovarian Cancer in Ten Families. Nat. Genet. 8 (4), 399–404. doi:10.1038/ng1294-399
Golmard, L., Caux-Moncoutier, V., Davy, G., Al Ageeli, E., Poirot, B., Tirapo, C., et al. (2013). Germline Mutation in the RAD51B Gene Confers Predisposition to Breast Cancer. BMC Cancer 13, 484. doi:10.1186/1471-2407-13-484
Howlader, N., Noone, A. M., Krapcho, M., Miller, D., Bishop, K., Kosary, C. L., et al. (2017). SEER Cancer Statistics Review, 1975–2014. Bethesda, MD: National Cancer Institute.
Jazayeri, S. B., Saadat, S., Ramezani, R., and Kaviani, A. (2015). Incidence of Primary Breast Cancer in Iran: Ten-Year National Cancer Registry Data Report. Cancer Epidemiol. 39 (4), 519–527. doi:10.1016/j.canep.2015.04.016
Khaliq, S., Hameed, A., Khaliq, T., Ayub, Q., Qamar, R., Mohyuddin, A., et al. (2000). P53 Mutations, Polymorphisms, and Haplotypes in Pakistani Ethnic Groups and Breast Cancer Patients. Genet. Test. 4 (1), 23–29. doi:10.1089/109065700316435
Kiiski, J. I., Pelttari, L. M., Khan, S., Freysteinsdottir, E. S., Reynisdottir, I., Hart, S. N., et al. (2014). Exome Sequencing Identifies FANCM as a Susceptibility Gene for Triple-Negative Breast Cancer. Proc. Natl. Acad. Sci. USA 111 (42), 15172–15177. doi:10.1073/pnas.1407909111
Kim, H., and Choi, D. H. (2013). Distribution ofBRCA1andBRCA2Mutations in Asian Patients with Breast Cancer. J. Breast Cancer 16 (4), 357–365. doi:10.4048/jbc.2013.16.4.357
Liede, A., Malik, I. A., Aziz, Z., Rios, P. d. l., Kwan, E., and Narod, S. A. (2002). Contribution of BRCA1 and BRCA2 Mutations to Breast and Ovarian Cancer in Pakistan. Am. J. Hum. Genet. 71 (3), 595–606. doi:10.1086/342506
Lim, W., Olschwang, S., Keller, J. J., Westerman, A. M., Menko, F. H., Boardman, L. A., et al. (2004). Relative Frequency and Morphology of Cancers in STK11 Mutation Carriers1 ☆. Gastroenterology 126 (7), 1788–1794. doi:10.1053/j.gastro.2004.03.014
Loveday, C., Turnbull, C., Ramsay, E., Hughes, D., Ruark, E., Frankum, J. R., et al. (2011). Germline Mutations in RAD51D Confer Susceptibility to Ovarian Cancer. Nat. Genet. 43 (9), 879–882. doi:10.1038/ng.893
Loveday, C., Turnbull, C., Ruark, E., Xicola, R. M. M., Ramsay, E., Hughes, D., et al. (2012). Germline RAD51C Mutations Confer Susceptibility to Ovarian Cancer. Nat. Genet. 44 (5), 475–476. doi:10.1038/ng.2224
Madanikia, S. A., Bergner, A., Ye, X., and Blakeley, J. O. N. (2012). Increased Risk of Breast Cancer in Women with NF1. Am. J. Med. Genet. 158a (12), 3056–3060. doi:10.1002/ajmg.a.35550
Majewski, I. J., Kluijt, I., Cats, A., Scerri, T. S., de Jong, D., Kluin, R. J., et al. (2013). An α-E-catenin (CTNNA1) Mutation in Hereditary Diffuse Gastric Cancer. J. Pathol. 229 (4), 621–629. doi:10.1002/path.4152
Mannan, A. U., Singh, J., Lakshmikeshava, R., Thota, N., Singh, S., Sowmya, T. S., et al. (2016). Detection of High Frequency of Mutations in a Breast And/or Ovarian Cancer Cohort: Implications of Embracing a Multi-Gene Panel in Molecular Diagnosis in India. J. Hum. Genet. 61 (6), 515–522. doi:10.1038/jhg.2016.4
Mehdi, S. Q., Qamar, R., Ayub, Q., Khaliq, S., Mansoor, A., Ismail, M., et al. (1999). “The Origins of Pakistani Populations,” in Genomic Diversity (Berlin: Springer), 83–90. doi:10.1007/978-1-4615-4263-6_7
Meijers-Heijboer, H., van den Ouweland, A., Klijn, J., Wasielewski, M., de Snoo, A., Oldenburg, R., et al. (2002). Low-penetrance Susceptibility to Breast Cancer Due to CHEK2(*)1100delC in Noncarriers of BRCA1 or BRCA2 Mutations. Nat. Genet. 31 (1), 55–59. doi:10.1038/ng879
Moatter, T., Aban, M., Khan, S., Azam, I., and Pervez, S. (2011). BRCA1 Status in Pakistani Breast Cancer Patients with Moderate Family History. J. Coll. Physicians Surg. Pak 21 (11), 680–684.
Nord, A. S., Lee, M., King, M.-C., and Walsh, T. (2011). Accurate and Exact CNV Identification from Targeted High-Throughput Sequence Data. BMC Genomics 12, 184–194. doi:10.1186/1471-2164-12-184
Olivier, M., Eeles, R., Hollstein, M., Khan, M. A., Harris, C. C., and Hainaut, P. (2002). The IARC TP53 Database: New Online Mutation Analysis and Recommendations to Users. Hum. Mutat. 19 (6), 607–614. doi:10.1002/humu.10081
Osher, D. J., De Leeneer, K., Michils, G., Hamel, N., Tomiak, E., Poppe, B., et al. (2012). Mutation Analysis of RAD51D in Non-brca1/2 Ovarian and Breast Cancer Families. Br. J. Cancer 106 (8), 1460–1463. doi:10.1038/bjc.2012.87
Park, D. J., Lesueur, F., Nguyen-Dumont, T., Pertesi, M., Odefrey, F., Hammet, F., et al. (2012). Rare Mutations in XRCC2 Increase the Risk of Breast Cancer. Am. J. Hum. Genet. 90 (4), 734–739. doi:10.1016/j.ajhg.2012.02.027
Pharoah, P. D. P., Guilford, P., and Caldas, C.International Gastric Cancer Linkage C (2001). Incidence of Gastric Cancer and Breast Cancer in CDH1 (E-Cadherin) Mutation Carriers from Hereditary Diffuse Gastric Cancer Families. Gastroenterology 121 (6), 1348–1353. doi:10.1053/gast.2001.29611
Qamar, R., Ayub, Q., Khaliq, S., Mansoor, A., Karafet, T., Mehdi, S. Q., et al. (1999). African and Levantine Origins of Pakistani YAP+ Y Chromosomes. Hum. Biol. 71 (5), 745–755.
Qamar, R., Ayub, Q., Mohyuddin, A., Helgason, A., Mazhar, K., Mansoor, A., et al. (2002). Y-chromosomal DNA Variation in Pakistan. Am. J. Hum. Genet. 70 (5), 1107–1124. doi:10.1086/339929
Rahman, N., Seal, S., Thompson, D., Kelly, P., Renwick, A., Elliott, A., et al. (2007). PALB2, Which Encodes a BRCA2-Interacting Protein, Is a Breast Cancer Susceptibility Gene. Nat. Genet. 39 (2), 165–167. doi:10.1038/ng1959
Rashid, M. U., Khan, F. A., Muhammad, N., Loya, A., and Hamann, U. (2019). Prevalence of PALB2 Germline Mutations in Early-Onset and Familial Breast/Ovarian Cancer Patients from Pakistan. Cancer Res. Treat. 51, 992–1000. doi:10.4143/crt.2018.356
Rashid, M. U., Muhammad, N., Amin, A., Loya, A., and Hamann, U. (2017). Contribution of BRCA1 Large Genomic Rearrangements to Early-Onset and Familial Breast/ovarian Cancer in Pakistan. Breast Cancer Res. Treat. 161 (2), 191–201. doi:10.1007/s10549-016-4044-0
Rashid, M. U., Muhammad, N., Bajwa, S., Faisal, S., Tahseen, M., Bermejo, J. L., et al. (2016). High Prevalence and Predominance of BRCA1 Germline Mutations in Pakistani Triple-Negative Breast Cancer Patients. BMC Cancer 16 (1), 673. doi:10.1186/s12885-016-2698-y
Rashid, M. U., Muhammad, N., Faisal, S., Amin, A., and Hamann, U. (2014). Deleterious RAD51C Germline Mutations Rarely Predispose to Breast and Ovarian Cancer in Pakistan. Breast Cancer Res. Treat. 145 (3), 775–784. doi:10.1007/s10549-014-2972-0
Rashid, M. U., Zaidi, A., Torres, D., Sultan, F., Benner, A., Naqvi, B., et al. (2006). Prevalence ofBRCA1 andBRCA2 Mutations in Pakistani Breast and Ovarian Cancer Patients. Int. J. Cancer 119 (12), 2832–2839. doi:10.1002/ijc.22269
Ratajska, M., Antoszewska, E., Piskorz, A., Brozek, I., Borg, Å., Kusmierek, H., et al. (2012). Cancer Predisposing BARD1 Mutations in Breast-Ovarian Cancer Families. Breast Cancer Res. Treat. 131 (1), 89–97. doi:10.1007/s10549-011-1403-8
Renwick, A., Thompson, D., Seal, S., Kelly, P., Chagtai, T., Ahmed, M., et al. (2006). ATM Mutations that Cause Ataxia-Telangiectasia Are Breast Cancer Susceptibility Alleles. Nat. Genet. 38 (8), 873–875. doi:10.1038/ng1837
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual, 2001. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
Seal, S., Thompson, D., Renwick, A., Elliott, A., Kelly, P., Barfoot, R., et al. (2006). Truncating Mutations in the Fanconi Anemia J Gene BRIP1 Are Low-Penetrance Breast Cancer Susceptibility Alleles. Nat. Genet. 38 (11), 1239–1241. doi:10.1038/ng1902
Shah, S., Kim, Y., Ostrovnaya, I., Murali, R., Schrader, K. A., Lach, F. P., et al. (2013). Assessment of SLX4 Mutations in Hereditary Breast Cancers. PLoS One 8 (6), e66961. doi:10.1371/journal.pone.0066961
Shiovitz, S., and Korde, L. A. (2015). Genetics of Breast Cancer: a Topic in Evolution. Ann. Oncol. 26 (7), 1291–1299. doi:10.1093/annonc/mdv022
Singh, J., Thota, N., Singh, S., Padhi, S., Mohan, P., Deshwal, S., et al. (2018). Screening of over 1000 Indian Patients with Breast And/or Ovarian Cancer with a Multi-Gene Panel: Prevalence of BRCA1/2 and Non-BRCA Mutations. Breast Cancer Res. Treat. 170 (1), 189–196. doi:10.1007/s10549-018-4726-x
Solyom, S., Aressy, B., Pylkäs, K., Patterson-Fortin, J., Hartikainen, J. M., Kallioniemi, A., et al. (2012). Breast Cancer-Associated Abraxas Mutation Disrupts Nuclear Localization and DNA Damage Response Functions. Sci. Transl Med. 4 (122), 122ra23. doi:10.1126/scitranslmed.3003223
Stewart, G. S., Maser, R. S., Stankovic, T., Bressan, D. A., Kaplan, M. I., Jaspers, N. G. J., et al. (1999). The DNA Double-Strand Break Repair Gene hMRE11 Is Mutated in Individuals with an Ataxia-telangiectasia-like Disorder. Cell 99 (6), 577–587. doi:10.1016/s0092-8674(00)81547-0
Stratton, M. R., and Rahman, N. (2008). The Emerging Landscape of Breast Cancer Susceptibility. Nat. Genet. 40 (1), 17–22. doi:10.1038/ng.2007.53
Tan, M.-H., Mester, J. L., Ngeow, J., Rybicki, L. A., Orloff, M. S., and Eng, C. (2012). Lifetime Cancer Risks in Individuals with Germline PTEN Mutations. Clin. Cancer Res. 18 (2), 400–407. doi:10.1158/1078-0432.ccr-11-2283
Tanaka, A., Weinel, S., Nagy, N., O'Driscoll, M., Lai-Cheong, J. E., Kulp-Shorten, C. L., et al. (2012). Germline Mutation in ATR in Autosomal- Dominant Oropharyngeal Cancer Syndrome. Am. J. Hum. Genet. 90 (3), 511–517. doi:10.1016/j.ajhg.2012.01.007
Testa, J. R., Cheung, M., Pei, J., Below, J. E., Tan, Y., Sementino, E., et al. (2011). Germline BAP1 Mutations Predispose to Malignant Mesothelioma. Nat. Genet. 43 (10), 1022–1025. doi:10.1038/ng.912
Torres, D., Bermejo, J. L., Rashid, M. U., Briceño, I., Gil, F., Beltran, A., et al. (2017). Prevalence and Penetrance of BRCA1 and BRCA2 Germline Mutations in Colombian Breast Cancer Patients. Sci. Rep. 7 (1), 4713. doi:10.1038/s41598-017-05056-y
Turnbull, C., Hines, S., Renwick, A., Hughes, D., Pernet, D., Elliott, A., et al. (2010). Mutation and Association Analysis of GEN1 in Breast Cancer Susceptibility. Breast Cancer Res. Treat. 124 (1), 283–288. doi:10.1007/s10549-010-0949-1
Vahteristo, P., Tamminen, A., Karvinen, P., Eerola, H., Eklund, C., Aaltonen, L. A., et al. (2001). p53, CHK2, and CHK1 Genes in Finnish Families with Li-Fraumeni Syndrome: Further Evidence of CHK2 in Inherited Cancer Predisposition. Cancer Res. 61 (15), 5718–5722.
Varon, R., Vissinga, C., Platzer, M., Cerosaletti, K. M., Chrzanowska, K. H., Saar, K., et al. (1998). Nibrin, a Novel DNA Double-Strand Break Repair Protein, Is Mutated in Nijmegen Breakage Syndrome. Cell 93 (3), 467–476. doi:10.1016/s0092-8674(00)81174-5
Walsh, T., Casadei, S., Lee, M. K., Pennil, C. C., Nord, A. S., Thornton, A. M., et al. (2011). Mutations in 12 Genes for Inherited Ovarian, Fallopian Tube, and Peritoneal Carcinoma Identified by Massively Parallel Sequencing. Proc. Natl. Acad. Sci. 108 (44), 18032–18037. doi:10.1073/pnas.1115052108
Walsh, T., Mandell, J. B., Norquist, B. M., Casadei, S., Gulsuner, S., Lee, M. K., et al. (2017). Genetic Predisposition to Breast Cancer Due to Mutations Other Than BRCA1 and BRCA2 Founder Alleles Among Ashkenazi Jewish Women. JAMA Oncol. 3 (12), 1647–1653. doi:10.1001/jamaoncol.2017.1996
Wiesner, T., Obenauf, A. C., Murali, R., Fried, I., Griewank, K. G., Ulz, P., et al. (2011). Germline Mutations in BAP1 Predispose to Melanocytic Tumors. Nat. Genet. 43 (10), 1018–1021. doi:10.1038/ng.910
Keywords: breast cancer, susceptibility, genomics, next-generating sequencing, candidate genes, Pakistani population
Citation: Ajaz S, Zaidi S-e-Z, Ali S, Siddiqa A and Memon MA (2022) Germline Mutation Analysis in Sporadic Breast Cancer Cases With Clinical Correlations. Front. Genet. 13:820610. doi: 10.3389/fgene.2022.820610
Received: 23 November 2021; Accepted: 08 February 2022;
Published: 09 March 2022.
Edited by:
Ata Abbas, Case Western Reserve University, United StatesReviewed by:
Kausar Jabbar, Beaumont Health, United StatesBabu Roshan Padmanabhan, University Hospitals Cleveland Medical Center, United States
Copyright © 2022 Ajaz, Zaidi, Ali, Siddiqa and Memon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sadia Ajaz, c2FkaWEuYWphekB1aHMuZWR1LnBr, c2FkaWFhamF6MUBnbWFpbC5jb20=
 Sadia Ajaz
Sadia Ajaz Sani-e-Zehra Zaidi
Sani-e-Zehra Zaidi Saleema Ali
Saleema Ali Aisha Siddiqa
Aisha Siddiqa Muhammad Ali Memon
Muhammad Ali Memon