
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 18 May 2022
Sec. Evolutionary and Population Genetics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.819941
This article is part of the Research TopicEpigenomic Polymorphisms: the Drivers of Diversity and HeterogeneityView all 11 articles
 Priyanka Kumari1
Priyanka Kumari1 Sajid Khan1
Sajid Khan1 Ishfaq Ahmad Wani1
Ishfaq Ahmad Wani1 Renu Gupta2
Renu Gupta2 Susheel Verma3*
Susheel Verma3* Pravej Alam4
Pravej Alam4 Abdullah Alaklabi5
Abdullah Alaklabi5Epigenetics are the heritable changes in gene expression patterns which occur without altering DNA sequence. These changes are reversible and do not change the sequence of the DNA but can alter the way in which the DNA sequences are read. Epigenetic modifications are induced by DNA methylation, histone modification, and RNA-mediated mechanisms which alter the gene expression, primarily at the transcriptional level. Such alterations do control genome activity through transcriptional silencing of transposable elements thereby contributing toward genome stability. Plants being sessile in nature are highly susceptible to the extremes of changing environmental conditions. This increases the likelihood of epigenetic modifications within the composite network of genes that affect the developmental changes of a plant species. Genetic and epigenetic reprogramming enhances the growth and development, imparts phenotypic plasticity, and also ensures flowering under stress conditions without changing the genotype for several generations. Epigenetic modifications hold an immense significance during the development of male and female gametophytes, fertilization, embryogenesis, fruit formation, and seed germination. In this review, we focus on the mechanism of epigenetic modifications and their dynamic role in maintaining the genomic integrity during plant development and reproduction.
Epigenetics is the study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in the DNA sequence (Waddington 1957; Waddington, 2012; Iwasaki and Paszkowski, 2014). Epigenetic changes are induced through behavioral or environmental factors that may affect the way the genes work (Niederhuth and Schmitz, 2017; Parker et al., 2021). These modifications are reversible and do not change the sequences of DNA but alters the way in which DNA sequences are read. Epigenetic modifications include DNA methylation (Bouyer et al., 2017), histone modifications (Zhou, 2009; Liu et al., 2010), ubiquitination of histone N-tails, and posttranscriptional silencing through small noncoding RNAs and RNA-mediated mechanisms (Slotkin and Martienssen, 2007; Matzke and Mosher, 2014). Gene and transposon activity get affected by epigenetic changes in the DNA present within a chromatin (Lippman et al., 2004; Rodrigues and Zilberman, 2015). Epigenetics regulate flowering time in plants through transposon silencing, paramutation, and genomic imprinting (Yaish et al., 2011; Ay et al., 2014; Blüumel et al., 2015; Parker et al., 2021). Epigenetic changes are inherited through alleles or epialleles. Epialleles are the sites that get transmitted to the next generations after being retained stably in the chromatin state. Epialleles act as supplementary sources of variation to regulate phenotypic diversity. In plant species, epialleles affect floral morphology, time of flowering, resistance against diseases, and leaf senescence (Brukhin and Albertini, 2021). The present review is a summary of the information available on different epigenetic modifications that appear to be important in growth, development, and reproduction of plants.
Plants being sessile in nature are invariably affected by changing environmental conditions. However, they have the ability to adapt their biological processes according to the changing environments. They interact with their environment through consistent adjustments at the molecular level by modifying the patterns of gene expression (Yaish et al., 2011). Epigenetic regulations assist plants in increased tolerance against different environmental stresses by reprogramming their developmental stages, such as flowering time (Barozai and Aziz, 2018). In transgenic plants, epigenetics helps to understand the problems related to suitable expression of newly introduced transgenic segments (Madhusudhan, 2015). Epigenetic changes are conserved in plants and influence the structure of the chromatin which in turn regulates the gene expression. Epigenetic mechanisms are important to regulate various biological processes and disruption of any one of the epigenetic mechanisms leads to developmental abnormalities in plants. Therefore, epigenetic changes play dynamic roles in the growth and development of plants (Zhang et al., 2018).
Different biological pathways such as phytohormone signaling, photoperiodism, and vernalization in combination with the environmental signals (epigenetic changes) regulate flowering time by integrating internal state of development of plants (An et al., 2004; He, 2009; Amasino, 2010; Andrés and Coupland, 2012; Sun et al., 2014; Burgarella et al., 2016). This network of flowering regulation involves FLOWERING LOCUS D (FLD) and FLOWERING LOCUS C (FLC) gene transcription which are controlled through epigenetic mechanisms such as ubiquitination, acetylation/deacetylation, and methylation/demethylation concealed by hormone signaling. HISTONE DEACETYLASE 6 (HDA6) protein increases the rate of expression of FLC gene but ethylene sets off HDA6 expression. FLC is suppressed by FLOWERING LOCUS D through demethylation of H3K4me2 which facilitates H4 deacetylation in the same locus. H3K27me3 is added by PRC2 marking into FLD chromatin. This also engages PICKLE to link with DELLA which in turn facilitates the repression of FLC (Bastow et al., 2004; De Lucia et al., 2008; Jiang et al., 2008; Choi et al., 2009).
Phenotypic plasticity within the plant species plays an important role in adaptation to different environmental conditions allowing different cultivars to adjust and grow. Plasticity maintains the homeostasis within changing environmental conditions that allows better gene expression to adapt to different biotic or abiotic constrains (disease, herbivory, plant–plant competition, altitude, soil type, seasonal, day length, rain, and ambient temperature) (Gratani, 2014). Genetic plasticity within the plants is inferred by different epigenetic modifications that are regulated by DNA methylation, histone modification, transposon modification, noncoding RNAs, and chromatin modulation (Zhang et al., 2018; Zhang et al., 2008; Zhang et al., 2013).
DNA methylation is a chromatin modification in plants and is conceivably inherited mitotically or meiotically over generations. DNA methylation is catalyzed by cytosine methyltransferases. It involves the addition of a CH3 group (methyl group) at the fifth carbon position on cytosine residue generating 5-methyl cytosine in a sequence-specific manner. The methyl group acts as a platform for various protein complexes to attach and modify the chromatin scaffolds causing altered gene expression (Niederhuth and Schmitz, 2017). On the basis of the target sequence, methylation is of two types: asymmetrical and symmetrical methylation. Symmetrical methylation is CG and CHG methylation and asymmetrical methylation is CHH methylation (where H denotes any nucleotide other than guanine). Both symmetric, i.e., CG and CHG, and asymmetric methylation, i.e., CHH, exist in plants (Jacobsen and Meyerowitz, 1997). Only some genes are methylated in plants within a gene body, and methylation is restricted only to CG sites (Niederhuth and Schmitz, 2017). DNA methylation is found to be higher at repetitive sequences than genic regions in case of plant species. To maintain genome stability, silencing of TEs is important which can be mediated through RNA-directed DNA methylation (RdDM) (Slotkin and Martienssen, 2007; Matzke and Mosher, 2014). Despite having a pivotal role in different biological processes, DNA methylation applications in crop improvement are not fully investigated.
DNA methylation is induced biochemically as an epigenetic heritable change initiated through enzymes. It entails a shift of a methyl group to the fifth position on the cytosine residue and is catalyzed by DNA methyltransferases utilizing S-adenosylmethionine (Thapa and Shrestha, 2020). DNA methylation occurs at cytosine regions, viz., CG, CHG, and CHH (H stands in for A, T, or C) (Figure 1). METHYLTRANSFERASE 1 (MET1) enzyme catalyzes the methylation of CG. After the completion of DNA replication process, MET1 identifies hemi-methylated CG dinucleotides and methylates the unchanged cytosine in the daughter strand (Kankel et al., 2003; He et al., 2011). DNA methyltransferases CHROMOMETHYLASE 3 (CMT3) and CHROMOMETHYLASE 2 (CMT2) are said to catalyze CHG methylation (Lindroth et al., 2001; Stroud et al., 2014). Depending on the chromosomal region, CHH methylation is catalyzed through DOMAINS REARRANGED methyltransferase 2 (DRM2) or CMT2 methyltransferases. Methylation through CHROMOMETHYLASE 2 is catalyzed at histone H1–containing heterochromatin sites, while DRM2 catalyzes methylation at RdDM target areas (Zemach et al., 2013; Zhang et al., 2018) (Figure 1). There are two types of DNA methylation mechanisms: active and passive DNA methylation. A particular protein participates in the active process and demethylates the DNA sequence. The base excision repair pathway is involved. During DNA replication, methylation of cytosine is replaced with unmodified cytosine in a passive process. The reduction of activity of DNA methylases such as METHYLTRANSFERASE 1 (MET1) and CHROMOMETHYLASE 3 (CMT3) causes the addition of non-modified cytosine during DNA replication (Ibarra et al., 2012). In the case of plants, cytosine methylation has been extensively researched (Ruffini Castiglione et al., 2002). Cytosine alterations are not constant and vary greatly depending on the stages of development in plants and environmental factors (Burn et al., 1993).
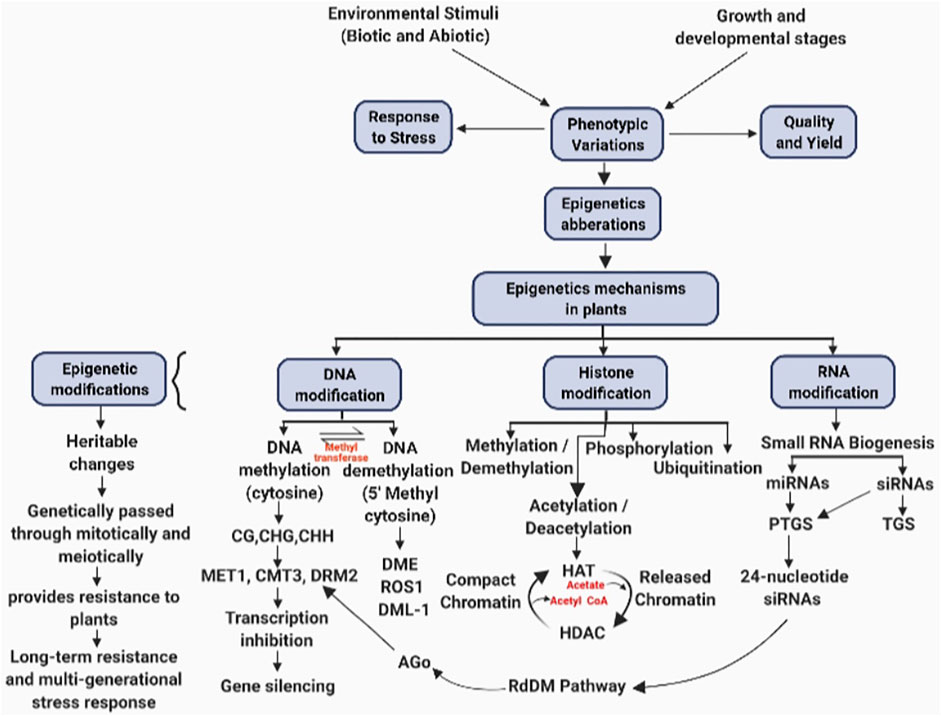
FIGURE 1. Epigenetic changes in response to stress management during growth and development of plant.
DNA methylation is recruited de novo via DRM2 at all sequence contexts, and its role in CHH methylation is more prominent because CHH methylation depends only on de novo methylation. The RdDM pathway regulates the DRM2 activity. The RdDM pathway contains two plant-specific DNA-dependent RNA polymerases. The large subunit of these polymerases consists of proteins, i.e., NRPD1 and NRPE1, and functions specifically in transcriptional gene silencing and de novo DNA methylation. Single-stranded RNAs are produced through DNA-dependent RNA polymerase IV (Pol IV) in DNA repeat sites and in transposon regions. Pol IV transcription is facilitated by chromatin remodeling protein CLASSYs (CLSYs; CLSY1-4) (Yang et al., 2018; Zhou et al., 2018). Single-stranded RNA is converted to double-stranded RNAs via RNA-dependent RNA polymerase (RDR2). Double-stranded RNA is then cleaved by Dicer-like protein (DCL3) into 24-nt siRNA. This 24-nt siRNA binds to ARGONAUTE proteins (AGO4, AGO6, and AGO9). Loading of siRNA to ARGONAUTE proteins require KOW CONTAINING TRANSCRIPTION FACTOR 1 (KTF1)/SPT5-like protein. KTF1 (RNA-binding protein) binds to noncoding RNA transcripts produced from Pol V forming RNA-directed DNA methylation effector complex. Pol V generates single-stranded RNA transcripts in intergenic noncoding (IGN) regions. RNA transcripts generation via Pol V requires DRD1, DMS3, RDM1 and RDM4. RDM1 binds to single-stranded methylated DNA and leads to the recruitment of Pol V to chromatin regions. To catalyze new DNA methylation, de novo DNA methyltransferases DRM2 are directed to specific chromatin regions via effector complex (He et al., 2011; Xie et al., 2012). It has been reported that six homologous proteins act in a redundant way in the RdDM pathway. These proteins are found in Arabidopsis and named as FACTOR of DNA METHYLATION 1–5 (FDM1-5) and INVOLVED IN DE NOVO (IDN2/RDM12). They belong to the SGS3-like plant-specific protein family, and their rice homolog is X1. They have an important role in transcriptional gene silencing like the SGS3 protein family (Xie et al., 2012).
Histone modifications comprise an interesting part in epigenetics (Pfluger and Wagner, 2007). Histone proteins act as winder around which the segment of DNA gets wrapped and leads to the formation of a structural unit called as nucleosome. Nucleosomes comprise histone octamers consisting of two copies of each of the H2A, H2B, H3, and H4 histone proteins. The N-terminal tail of these histone proteins undergoes different modifications such as acetylation, methylation, sumoylation, ubiquitination, and phosphorylation. These histone modifications are correlated with either gene activation or repression. Several histone variants and enzymes are present in plants that modify histones posttranslationally and regulate gene expression (Zhou, 2009; Liu et al., 2010). Gene expression is brought about by the process of acetylation and phosphorylation, whereas gene expression is reduced through sumoylation and biotinylation. In the case of plants, deacetylation and methylation of H3K27 and H3K9 repress genes, while H3K4 and H3K36 acetylation and methylation activate gene expression. Epigenetic modifications participate in several biological processes like transcription, replication, and DNA repair by recruiting specific proteins needed in such processes (Jiang et al., 2009; Iwasaki and Paszkowski, 2014). Epigenetic modifications not only consist histone marks/modifications but also consist replacement with histone variants having different properties to influence gene expression (Liu et al., 2010). Chromatin immunoprecipitation applications following deep sequencing provide an insight for the genome-wide association studies regarding variants of histones and their posttranscriptional modifications (Butterbrodt et al., 2006). Responding to various biotic and abiotic stresses, histone modifications regulate the DNA transcription by interfering with the packaging structure either by activating the DNA to transcribe or making condensed structures thereby deactivating transcription machinery.
N-terminal tails of histones are the sites where most of the histone modifications take place. These modifications specify the function of chromatin and transcriptional activities (Jenuwein and Allis, 2001; Zhao et al., 2019). Histone modifications include methylation, acetylation, ubiquitination, and phosphorylation and occur at lysine and arginine residues. Histone methyltransferases (writers) are a group of SET domain which catalyzes histone lysine methylation, and downstream events are mediated by proteins (readers) which recognize specific lysine methylation. Two histone demethylases, i.e., Jumonji C (Jmj C) and lysine-specific demethylase 1 (LSD 1) catalyze the removal of histone lysine methylation (Zhao et al., 2019). In eukaryotes, the involvement of histone acetylation and methylation in gene expression regulation was first identified by Allfrey et al. (1964). It has been demonstrated that increase in histone acetylation leads to poor separation of sister chromatids in human fibroblasts and causes chromosomal defects during cell cycle in tobacco. Trichostatin A (TSA) is identified to have negative pleiotropic effects and has been found to decrease global histone deacetylation, resulting in an increase in acetylated histones (Cimini et al., 2003; Li et al., 2005).
The overexpression of antisense of the histone deacetylase gene (AtHD1) induces histone acetylation activity in Arabidopsis thaliana. The AtHD1 gene gives rise to pleiotropic phenotypes having a variety of developmental defects such as the suppression of apical dominance, ectopic expression of silenced genes, floral structure abnormalities, male and female sterility, heterochronic shift toward juvenility (Tian and Chen, 2001). In plant genome, the repressive state of heterochromatic regions is marked by H3K9me1 and H3K9me2. Heterochromatic regions are enriched with transposable elements and repetitive sequences. Heterochromatic regions in Arabidopsis are enriched with H3K27me1, and the association of H3K27me1 is catalyzed by plant-specific histone methyltransferases ARABIDOPSIS TRITHORAX-RELATED PROTEINS, i.e., ATXR 5 and ATXR 6. Mutation in H3K27me1 results in de-condensation of heterochromatin and the release of transposable silencing (Zhao et al., 2019). H3K27me3 deposition on chromatin is catalyzed by polycomb repressive complex 2 (PRC2) via histone methyltransferases. The subunits of PRC2 were first identified in Drosophila melanogaster (Shen et al., 2021). In plants, PRC2 deposits the H3K27me3 methylation and plays an important role in growth and developmental phases of plants. This modification is found in protein-coding genes and is regulated dynamically during the growth stages of plants. PRC2 consists of four components, viz., histone methyltransferases enhancer of zeste [E(z)], extra sex combs (Esc), suppressor of zeste 12 [Su(z)12], and the histone-binding nucleosome-remodeling factor 55 kDa (Nurf55, also called p55). One component of PRC2, i.e., E(z), specifically belongs to the SET [Su(var)3-9; E(z); trithorax] domain family and is responsible for histone H3 tri-methylation at Lys27 (Czermin et al., 2002; Butenko and Ohad, 2011; Simon and Kingston, 2013; Shen et al., 2021). In case of Arabidopsis, PRC2 components have multiple duplications, and there exists three homologs of E(z), viz., CURLY LEAF (CLF), SWINGER (SWN), and MEDEA (MEA); three homologs of Su(z), viz., EMBRYONIC FLOWER 2 (EMF2), VERNALIZATION 2 (VRN2), and FERTILIZATION-INDEPENDENT SEED 2 (FIS2); and one homolog of Esc, viz., COPY SUPPRESSOR OF IRA 1–5 (MSI1-5). FIS2 regulates mega-gametogenesis and endosperm development in plants during postfertilization events and EMF, and VRN polycomb repressive complexes regulate the development of sporophyte and phase transition, i.e., vegetative to reproductive in plants (Shen et al., 2021). Histone demethylases found in Arabidopsis, i.e., JUMONJI 13 (JMJ13), JUMONJI 30 (JMJ30), JUMONJI 32 (JMJ32), EARLY FLOWERING 6 (ELF6), and RELATIVE OF EARLY FLOWERING 6 (REF6), demethylate H3K27 and depress genes temporally or spatially for processes like flowering, signaling of hormones, and circadian clock control (Sanchez et al., 2020).
Histone methylation is influenced by environmental factors (Boyko and Kovalchuk, 2008; Kim et al., 2010). Global gene expression analysis and chromatin immunoprecipitation (ChIP) tests have revealed that histone H3 Lys4 methylation (H3K4) patterns in Arabidopsis respond dynamically to dehydration stress (van Dijk et al., 2010). The floral initiator SHK1 kinase BINDING PROTEIN 1 (SKB1) mutant line skb1 provides an example of the interaction between environmental stress and blooming. SKB1 attaches to chromatin and raises the quantity of histone 4 Arg3 (H4R3) symmetric dimethylation (H4R3sme2) and causes FLC expression and a number of stress-responsive genes to be downregulated. As a result, its mutant characteristics include salt hypersensitivity, late flowering, and stunted development (Zhang et al., 2011; Chen et al., 2013; Cheng et al., 2019). The standard ABC model determines flower architecture at the molecular level (Bowman et al., 1991; Bowman et al., 2012). The geographical bounds of each floral whorl are determined by precise union of gene expression and protein interactions in this model (sepals, petals, stamens, and carpel). The A class gene APETALA2 (AP2) regulates target gene expression as part of a complex that it forms with TOPLESS (TPL) and HISTONE DEACETYLASE 19 in Arabidopsis (HDA19). AGAMOUS (AG) and SEPALATA3 (SEP3), the C class and E class genes, respectively, are negatively regulated by the transcription repressor complex. Deacetylation of H4K16 in regulatory areas of AG and SEP3 mediates gene suppression (Krogan et al., 2012). Expression studies revealed and identified additional HDACs expressed in reproductive tissues in Arabidopsis, i.e., HDA5, HDA6, HDA7, HDA9, HDA15, and HDA18. Their function in fruit or flower development is unknown. Only the function of HDA6 has been reported, and it plays a role in the regulation of blooming time. Histone H3K4 demethylase, i.e., FLOWERING LOCUS D (FLD), interacts directly with HDA6. The complex represses the expression of three flowering repressors: FLD, MADS AFFECTING FLOWERING 4 (MAF4), and MAF5 by removing the acetyl and methyl groups from histone 3 at their loci (Yu et al., 2011). HDA6 is one of the HDACs engaged in RdDM. RdDM is a plant-specific epigenetic process and small interfering RNA (siRNA)–mediated epigenetic mechanism which regulates the chromatin silencing of developmental genes, transposable elements, and repetitive elements. The RdDM mechanism involves a large number of participants whose actions may be broken down into a few simple phases (Matzke and Mosher, 2014). The RdDM machinery involves two kinds of transcripts, viz., Pol IV and Pol V transcripts. Pol IV transcribes long noncoding RNAs (lncRNAs), and the lncRNAs gets transformed to double-stranded RNAs (dsRNAs) through RDR2 (Haag et al., 2012). dsRNAs then gets converted into siRNAs by DICER-like 3 (DCL3). The siRNAs are loaded into AGO4 and reimported into the nucleus after being exported to the cytoplasm. siRNA direct AGO4 to nascent scaffold transcripts of Pol V through precise base pairing. siRNA, AGO4, and lncRNA scaffold derived from Pol V recruit histone deacetylases (HDACs) and DNA methyltransferases, which in turn silence the genomic loci transcribed by Pol V by the process of histone deacetylation and DNA methylation. Histone deacetylation characterizes RdDM-silenced promoters, which is mediated by RPD3-type histone deacetylase AtHDA6 in Arabidopsis, which is homologous to SIHDA3 of tomato. Deacetylation is essential for subsequent methylation by histone methyltransferases (HMTs), and to control siRNA-dependent heterochromatin, there is a requirement of functional AtHDA6 (Li et al., 2005; Aufsatz et al., 2007). The mutants of AtHDA6 display the revival of RdDM-silenced promoters in spite of the presence of an RNA-silencing signal. Reduced cytosine methylation indicates that AtHDA6 plays an important role in methylation maintenance. The physical connection of AtHDA6 with DMTs, MET1, and CMT3 may facilitate this function. Acetylases and deacetylases of histones have an important role in flowering and fruit development of tomato plant (Aufsatz et al., 2002). Cigliano et al. (2013) identified potential histone modifiers of AU4 tomato genome using RNA sequencing data of tomato genome (Tomato Genome C 2012) generated by worldwide collaboration sequencing. Also, they analyzed the expression profiles of each histone modification in the sample tissues used by using RNA sequencing data from the same source. Two histone acetylases S1HAG18 and S1HAG6 showed peak expression in the floral samples used, which indicates their function in reproductive development of tomato plant. Recently, 15 histone deacetylases have been discovered in tomatoes. SlHDA3 was found to be the tomato homolog of AtHDA6 which is expressed in all tissues having the highest blossom stage expression (Zhao et al., 2014). Another tomato homolog of AtHDA19, i.e., S1HDA1, was found to be significantly expressed in the flowering stage, and its expression was repressed at the fruiting stage. In yeast two-hybrid tests, it has been found that histone deacetylases, including S1HDA1, S1HDA3, and S1HDA4, interact with MADS-box transcription factors, i.e., TOMATO AGAMOUS1 (TAG1) and TOMATO MADS-BOX (TM29) (Zhao et al., 2014). Transcription factor TAG1 is required for the expression of both ethylene-dependent and ethylene-independent ripening genes (Klee and Giovannoni, 2011). TM29 is a homolog of SEPALLATA, which when silenced, leads to the formation of aberrant flowers and parthenocarpic fruits (Ampomah-Dwamena et al., 2002).
Most species have transposon elements (TEs) in their chromosomes, and multicellular eukaryotes have TEs as a key component of their genome. The majority of transposable elements are silenced epigenetically, although certain transposable elements have active transcription in epigenetic regulation mutants. Furthermore, environmental stress can trigger TE transcription, a mechanism that occurs across the evolutionary spectrum from bacteria to mammals (Capy et al., 2000). McClintock (1984) was the first to report that stress might cause TEs to shift, a result that has been widely corroborated in subsequent research (Grandbastien, 1998). Tnt1 and Tto1 are LTR-type Class I retroelements in tobacco, whose transposition is triggered by injury or through pathogen attack (Takeda et al., 2001; Perez-Hormaeche et al., 2008). Also, Bs1 LTR-type Class I retroelement in maize was found to transpose after viral infection (Johns et al., 1985). In Arabidopsis, heat stress induces transcription of ONSEN (LTR-type Class I retroelement), and it transposes into siRNA-defective mutants (Ito et al., 2011). LTR of ONSEN has a heat-responsive region that is activated by transcriptional heat stress responses (Cavrak et al., 2014). As a result, genes near or containing newly inserted ONSEN copies become heat-responsive (Ito et al., 2011). All the above examples of transposons are of Class I DNA transposons, and these transpose via the “copy and paste” mechanism in response to stress. Class II DNA transposons transpose via a “cut and paste” process in response to stress. In Antirrhinum majus, low temperature increases the excision frequency of the Ac/Ds type transposon Tam3 (Harrison and Fincham, 1964; Carpenter et al., 1987). Transposable elements are a response of the genome toward environmental challenges and play a critical role in gene regulation and evolution of the genome (McClintock, 1984; Slotkin and Martienssen, 2007; Fedoroff, 2012). It has been proposed that TEs activation in response to environmental stress could provide epigenetic variability which could contribute toward the greater adaptive capacity of plants under stress conditions (Mirouze and Paszkowski, 2011; Bucher et al., 2012; Ashapkin et al., 2020). The active DNA transposon mPing has been found to preferentially insert into 50 flanking regions of genes rather than exons in rice. Cold and salt stress encourages transcription of a subset of genes by inserting mPing in the promoter region (Naito et al., 2009; Wang et al., 2016).
It has been reported that epigenetic reprogramming has an important role in transposon silencing and reprogramming in germ cells of plants (Feng et al., 2010). In Arabidopsis thaliana, one egg cell and one central cell containing two nuclei are produced during the process of female gametogenesis, and several accessory cells are also produced. The egg cell fuses with one sperm cell during double fertilization process forming an embryo, and the second sperm cell fuses with the central cell leading to the formation of an endosperm (triploid). A helix hairpin DNA glycosylase, DEMETER (DME) causes hypomethylation in the endosperm by removing methylated cytosine residues. Demethylation via DME activates expression of transposons through the RNAi pathway that introduces transcripts of transposons and produces additional siRNAs guiding DNA methylation. The siRNAs expression in the endosperm development indicates genome imprinting, and siRNAs production guides DNA methylation in egg cell reinforcing transposons silencing in the germ cells. Transposons silencing occurs in the germ cells, but mild activity of transposons in endosperm have no major effects as the endosperm is not inherited to next generation (Hsieh et al., 2009b; Gehring et al., 2009; Ibarra et al., 2012; Ito 2013). Transposons activity in the embryo has been suppressed via the RNAi pathway having a major role in reprogramming of paternal genome in Arabidopsis (Han et al., 2019). It has also been reported that sRNAs lead to hypomethylation of vegetative cells when they are transported to the sperm cell via the cytoplasm of the pollen grains. This accumulates siRNAs and activates de novo remethylation through the RdDM pathway leading to transposons silencing in the gametes. Genes responsible for biogenesis of siRNA and transposons silencing are expressed at a very low concentration in pollen. But the DDM1 gene is an exception, which is specifically expressed in the sperm cells of mature pollen. In the vegetative nucleus of wild-type, DNA demethylation and activation of transposons occur by the downregulation of the DDM1 gene. This activation of transposons in vegetative cells also has no effect on the fitness of the species, as vegetative cells are not inherited to the next generation and have no contribution of genetic material, i.e. DNA, to the fertilized embryo. This demonstrates that epigenetic inheritance and transposon silencing are contributed through genome reprogramming guided via RNAs (Ito, 2013; Dziegielewski and Ziolkowski, 2021).
miRNAs comprise 20–24 noncoding nucleotides that regulate gene expression after transcription and are also involved in the age pathway by regulating the time of flowering in plants by using RNA-directed DNA methylation (RdDM) (Matzke et al., 2001; Matzke et al., 2007; Pikaard, 2006; Teotia and Tang, 2015; Dziegielewski and Ziolkowski, 2021) (Figure 1). The miRNA molecules with the help of the RdDM pathway can bring about DNA methylation on a specific location (Teotia and Tang, 2015). Global gene expression analysis in Arabidopsis thaliana having genetic disorder in photoperiodic signaling pathway and system integrate genes suggesting the role of miRNAs in mediating the effects of floral induction (Schmid et al., 2003; Kinoshita and Richter, 2020). miR156 and miR172 are the two key miRNAs acting as the main elements in controlling the age pathways in plants by downregulating target genes and also effecting flowering time in many plant species (Figure 2). The level of miR156 increases during the vegetative stage of plants and decreases as plants proceed toward the reproductive stage, and at this stage, miR172 increases (Tanaka et al., 2011; Luo et al., 2013; Teotia and Tang, 2015). The expression of miRNAs is determined by environmental factors. miRNAs affect the expression of certain genes of plants when exposed to abiotic and biotic stress by frequently reprogramming genes involved in the developmental pathways (Covarrubias and Reyes, 2010; Hirayama and Shinozaki, 2010; Urano et al., 2010; Tiwari and Rajam, 2022) (Figure 1). Stress-inducible miRNAs and their expected targets have been discovered to be preserved in Arabidopsis (Sunkar and Zhu, 2004). It has been reported through global gene expression in rice plants that when exposed to stress conditions such as cold, drought, excessive salt, and ABA treatment, miRNAs modulate gene expression in the rice plants (Shen et al., 2010). Correlation between miRNA biogenesis mechanism proteins, response to stress, and flowering has been found in many mutant lines of Arabidopsis. For example, ABH1 and CBP20 encode cap-binding factors which are required for maturation of RNA (Papp et al., 2004). The abh1 mutant exhibits ABA hypersensitivity, and the cbp20 line exhibits both drought tolerance and ABA hypersensitivity (Hugouvieux et al., 2001; Kwak et al., 2005). In addition to the role of miRNA in stress responses, they are also important in controlling the flowering in Arabidopsis (Aukerman and Sakai, 2003; Chen and Li, 2004; Sunkar and Zhu, 2004). Long intronic noncoding RNA (COLDAIR) mediates interaction of H3K27me3 at FLC. This interaction of COLDAIR with FLOWERING LOCUS C (FLC) gene targets PRC2 to interact with FLC, resulting in FLC suppression during cold treatment, i.e., vernalization (Baulcombe and Dean, 2014). miRNA partially regulates the FL, and mutations in the miRNA biogenesis genes DCL1 and DCL3 cause delayed flowering in these mutant backgrounds due to overly high FLC expression (Schmitz et al., 2007) (Figure 1). Another mutant line HYPONASTIC LEAVES 1 (HYL1) was also found to show late flowering characteristics (Lu and Fedoroff, 2000). HYPONASTIC LEAVES 1 (HYL1) gene produces a protein that binds to double-stranded RNA (dsRNA) and mediates gene control via miRNA (Han et al., 2004). Scientists have reported that in addition to hyl1 mutants, many Arabidopsis mutants in miRNA biogenesis machinery genes have phenotype related to ABA and salt hypersensitivity, i.e., SERRATE (SE) gene, DCL1 gene, HUA-ENHANCER 1 (HEN1) gene, and HASTY gene (Lu and Fedoroff, 2000; Han et al., 2004; Rasia et al., 2010; Zhang et al., 2018).
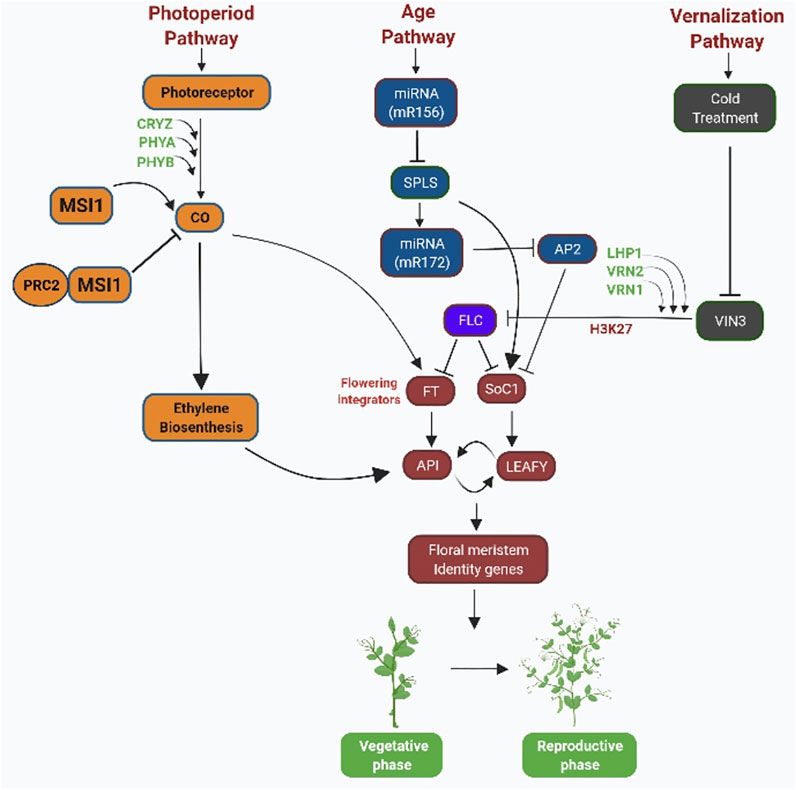
FIGURE 2. Role of epigenetic modifications in controlling flowering time in co-relation with biological pathways: flowering is induced by a number of molecular pathways that respond to external and internal signals. Flower integrator genes such as FT and SOC1 are regulated by flowering activators and repressors. In the photoperiodic pathway, chromatin modifications involve the well-conserved histone binding protein MULTICOPY SUPRESSORS OF IRA1 (MSI1)–like protein family. MSI1-like protein forms a complex with ubiquitous protein, i.e., POLYCOMB REPRESSIVE COMPLEX 2 in Arabidopsis and controls the switch to flowering. MSI1 acts in the normal expression of CO in long day (LD) plants. Reduced expression of CO in msi1 mutants leads to FT and SOC1 repression. In age pathway, miR156 and miR172 acts as positive regulators of SOC1 gene. Vernalization leads to the expression of VIN3. VIN3 represses FLC transcription by binding with PcG protein (VRN1, VRN2, LHP1). PcG proteins epigenetically modify chromatin of FLC by trimethylation of H3K27.
Stem cells are present in plant meristems and lead to the formation of all tissues and organs. The RdDM factor transcript levels are higher in tissues of meristem in A. thaliana than in cell expansion tissues, e. g., tissues of hypocotyl and differentiated leaves (Zhang et al., 2018). The DNA methylation level was found to be more in columella cells of root meristem because these cells are least condensed with pericentromeric chromatin. This allows more accessibility to RdDM factors. There have been no obvious reports of apparent meristem abnormalities in RdDM mutants in A. thaliana, but rice and maize mutants show severe developmental defects, indicating that these components play critical roles in meristem function (Zhu et al., 2012; Kawakatsu et al., 2017; Zhang et al., 2018). After the emergence of leaves from shoot apical meristem, many developmental genes get suppressed by the deposition of SET DOMAIN GROUP PROTEIN 711 (SDG711)–dependent H3K27me3 in rice. SDG711-dependent H3K27me3 synchronizes with DRM2-catalyzed non-CG DNA methylation in the gene body of Oryza sativa. SDG711 interacts physically with DRM2, and mutation in DRM2 decreases chromatin binding of SDG711 and deposition of H3K27me3 at repressed gene sites (Zhou et al., 2016; Zhang et al., 2018). During the growth of leaves in maize, DNA methyltransferases were found to be regulated variably resulting in distinct patterns of CG and CHG methylation in the division zone, transition, elongation, and mature zone reflecting the leaf’s spatial gradient of cells (Zhou et al., 2016). DNA methylation is required for pattern development in some leaf epidermal cells of Arabidopsis thaliana. DNA demethylation in Arabidopsis thaliana is dependent on genes of the ROS1 subfamily encoding 5-methylcytosine DNA glycosylases/lyases. DNA demethylation initiated via ROS1 leads to the expression of EPF2 gene. EPIDERMAL PATTERNING FACTOR 2 (EPF2) is a peptide ligand that inhibits stomatal development, resulting in an excess of stomatal lineage cells. Malfunction of ROS1 results in promoter hypermethylation and suppression of the gene encoding EPF2, resulting in stomatal cell development (Yamamuro et al., 2014; Zhou et al., 2016; Zhang et al., 2018). Similarly, the loss of function of H3K9 demethylase IBM1 (increase in BONSAI methylation 1) causes elevated H3K9me2 and CHG DNA methylation, as well as the suppression of three LRR RECEPTOR-LIKE SERINE/THREONINE-PROTEIN KINASE ERECTA family genes that encode EPF2 receptors, resulting in stomatal pattern formation problems as seen in ROS1 mutant plants. In ros1 plants, the mutation in RdDM factors, H3K9 methyltransferase SUVH4, and CMT3 in ibm1 plants can rescue the stomatal pattern formation resulting through abnormal epigenetic regulation because DNA methylation of EPF2 promoter and silencing of EPF2 depends on RNA-directed DNA methylation. Also, ROS1 acts against RdDM action leading to the expression of EPF2. This indicates that two DNA methylation-mediated mechanisms are responsible for regulating leaf epidermal cell patterning in A. thaliana (Table 1) (Candaele et al., 2014; Yamamuro et al., 2014; Wang et al., 2016; Zhou et al., 2016; Zhang et al., 2018). FLC, a MADS box transcriptional repressor that keeps Arabidopsis apices in the vegetative stage, is downregulated by vernalization. As a result, epigenetic alterations at the FLC locus hasten flowering (Kim et al., 2009).
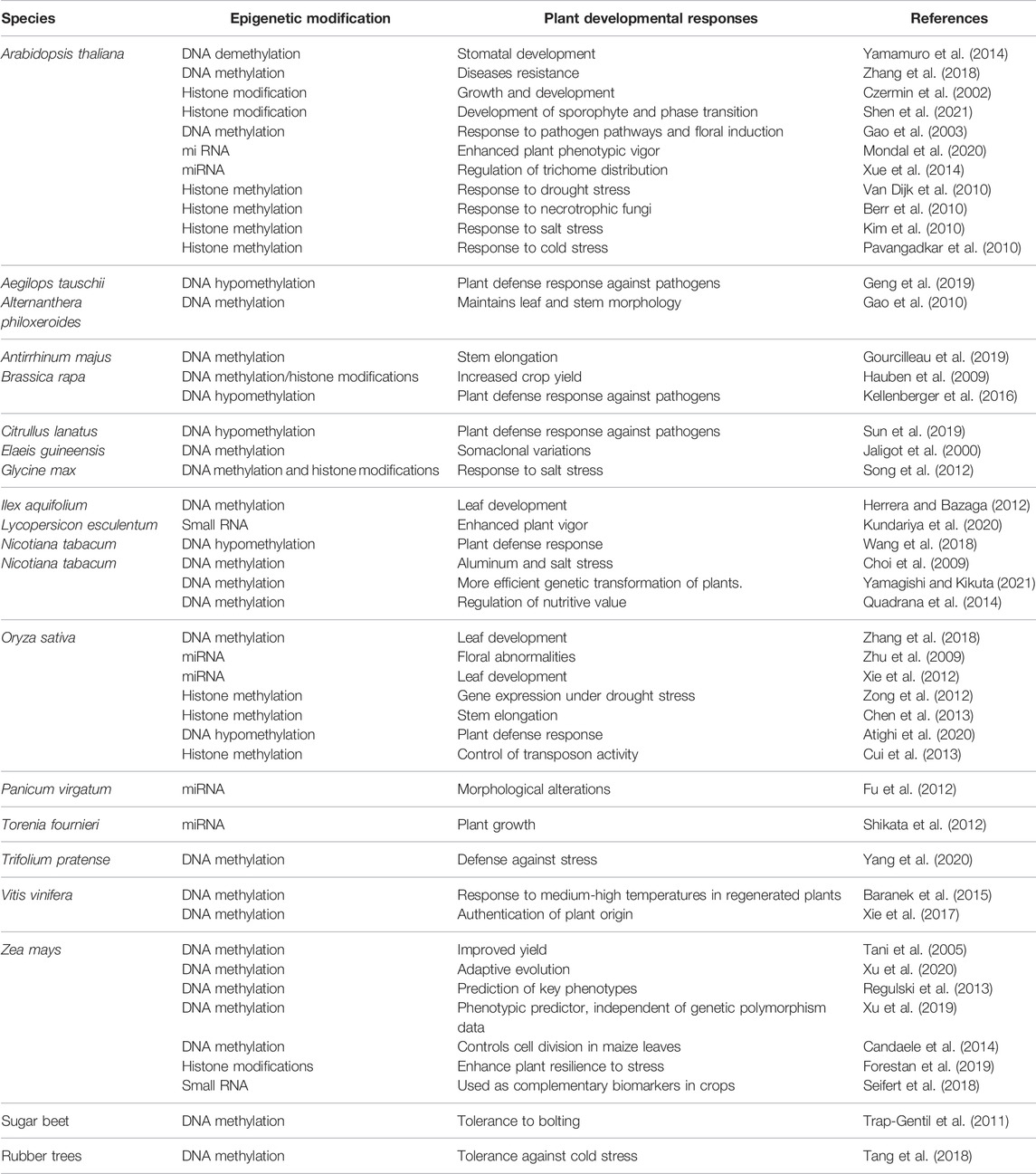
TABLE1. Role of epigenetically induced modifications in trait control, development, and morphogenesis of different plant species.
DNA methylation suppresses gene expression under stressful conditions, allowing the plant to conserve energy and strength for survival (Thapa and Shrestha, 2020). During water scarcity, CAM plants show the transition from C3 photosynthetic cycle to CAM pathways, which increases their resilience. This is accompanied by an increase in genomic methylation and hypermethylation of satellite DNA. Hypermethylation response is used to synthesize chromatin structure, which controls the expression of several genes and helps the plants to withstand stressful conditions. Hypermethylation was also discovered when the root tip of pea plants was exposed to water scarcity conditions (Thapa and Shrestha, 2020). The vernalization process, which involves prolonged exposure to cold conditions, initiates flowering in some plant species and is a well-studied example of how cold causes epigenetic changes that affect flowering. Epigenetic regulator, NRPD1, a DNA-binding bromodomain-containing protein, AtGCN5-related GNAT family 5 (acetyltransferase 5) and histone deacetylase were upregulated in Arabidopsis (Lee et al., 2005). Low temperature has been linked to DNA demethylation in Arabidopsis and other plant species like Zea mays (Steward et al., 2002), Antirrhinum majus, and Triticum aestivum (Sherman and Talbert, 2002; Hashida et al., 2003; Hashida et al., 2003) (Table 1). The Arabidopsis VERNALIZATION INSENSITIVE 3 gene (VIN3), a chromatin-remodeling plant homeodomain (PHD) finger protein that increases acetylation levels, is induced by cold exposure. This protein is essential for FLC repression and flowering enhancement. Because FLC expression is not lowered by cold treatment, the mutant lines for VIN3 do not respond to vernalization and so remain in a vegetative state for longer durations (Sung and Amasino, 2004; Soppe et al., 2021) (Table 1). During vernalization, this complex attaches to VIN3 locus chromatin (Schonrock et al., 2006). In Arabidopsis, on the other hand, a decrease in H3K27me3 modifications within the histones of the cold-responsive gene COR15A and the GALACTINOL SYNTHASE gene ATGOLS3 results in enhanced gene expression (Taji et al., 2002; Kwon et al., 2009). Similarly, during dehydration stress, the plant trithorax factor (ATX1) (Alvarez-Venegas et al., 2003) tri-methylates Lys4 residues of histone H3 (H3K4me3), regulating floral organ development and altering expression of transcription factor WRKY70 (Alvarez-Venegas et al., 2007). ATX1 mutations result in severe flaws in floral architecture (Alvarez-Venegas et al., 2003).
Various studies have reported that in A. thaliana and other plant species, including apples, Pharbitis nil, plant hormones such as auxin, cytokinin, Gibberellic acid, and abscisic acid interact to control flowering (Domagalska et al., 2010; Matsoukas, 2014). Salicylic acid (SA) is implicated in the control of CONSTANS, FLOWERING LOCUS C, FLOWERING LOCUS T, and MADS-box protein SOC1 transcription (Martinez et al., 2004). Interestingly, late-blooming phenotype of SA-deficient plants coincides with a 2- to 3-fold expression of FLC, lowering the FT levels in LD or SD circumstances as compared to wild-type plants. Furthermore, chromatin alterations are involved in the dynamic shift in the gene expression (Sun et al., 2014). For example, FLC and FT expression in A. thaliana is controlled epigenetically (Swiezewski et al., 2009; Ietswaart et al., 2012) (Figures 2, 3). It has been reported that under cold stress, Polycom Repressive Complex 2 (PRC2) is involved in silencing the FLC locus (floral repressor) through H3K27me3 (Yuan et al., 2016) (Figure 2). Also, some studies reported that the silencing of FLC successfully brought about through reducing H3K4me2 levels in FLC gene (Liu et al., 2010). PRC2 and Flowering Locus D (FLD) work in coordination to silence FLC (Shafiq et al., 2016; Campos-Rivero et al., 2017). Sumoylation/desumoylation action of the FLD gene can regulate acetylation/deacetylation of histones through the unspecified procedure. Histone demethylase is encoded by the FLD gene, thereby mediating H3K4me2 demethylation and facilitating H4 histone deacetylation in FLC chromatin (Jin et al., 2008). Exposure to various amounts of synthetic auxins results in epigenetic alterations that affect flower growth (Jaligot et al., 2000). The mantled phenotype of Elaeis guineensis Jacq (African oil palm) is characterized by anomalies in the development of flowers, leading to alteration in the auxin/cytokinin ratio (Table 2) (Eeuwens et al., 2002, Jaligot et al., 2011; Campos-Rivero et al., 2017). Mantled blooms were enhanced by applying a high amount of cytokinin (kinetin) and a low amount of auxin [1-naphthaleneacetic acid (NAA)], and there were less number of mantled flowers when a high amount of NAA and low amount of kinetin were applied (Eeuwens et al., 2002). Mantled phenotype resulted from DNA hypomethylation caused by kinetin, and the opposite phenotype of the plant resulted from DNA hypermethylation caused by NAA (Jaligot et al., 2000; Eeuwens et al., 2002, Jaligot et al., 2011). Pin-shaped inflorescence in Arabidopsis thaliana resulted from failed floral primordial initiation caused by Pin-formed mutant pin-1. This mutant diminishes the polar auxin transport thereby producing inflorescence devoid of flowers in A. thaliana (Okada et al., 1991). When IAA is given exogenously, it causes the production of flowers, which can be reversed (Reinhardt et al., 2000). Histone alterations have also been shown to have a role in transcriptional control of auxin target genes (Wu et al., 2015). mRNA accumulation increases in LFY and FILAMENTOUS FLOWER (FIL) when auxin is applied, resulting in initiation of the floral primordium. Auxin treatment leads to elevation of H3K9ac levels in LFY and FIL gene loci boosting flower primordial (Wu et al., 2015). In the absence of auxin, TOPLESS and HDA19 were reported to repress LFY and FIL loci through binding at their MP-sites, resulting in transcription inhibition of genes (Wu et al., 2015). Cytokinins play a major role in the division and differentiation of cells in the floral meristem (Schaller et al., 2015). Accumulation of cytokinins in A. thaliana meristem regulates the size of shoot apical meristem and the activity of cells in the shoot meristem. Cytokinin degradation is catalyzed by two cytokinin dehydrogenase enzymes CKX3 and CKX5 performing a regulatory function in the floral meristem of A. thaliana (Table 2) (Bartrina et al., 2011). In the central WUSCHEL (WUS) domain, CKX3 is expressed, and CKX5 expresses in the broad region of the apical meristem. Double mutants, i.e., ckx3 and ckx5, lead to the formation of large inflorescence and flower meristem. Phenotype developed by these double mutants indicates that cytokinin signaling precisely identifies the niche of stem cells and retards the development of cells (Bartrina et al., 2011). Meijon et al. (2011) reported that cytokinins work in coordination with epigenetic modifications and regulate flowering processes in plants. Cytokinin dihydrozeatin riboside and isopentenyladenine end the dormancy period and influence flowering through the DNA methylation process. The levels of DNA methylation decrease before the initiation of flowers, but after the formation of floral organs, the levels of DNA methylation increase (Table 2) (Meijon et al., 2011). Scientists state that cytokinin is an epigenetic component whose function is to regulate gene expression during the transition from the vegetative to reproductive form by instigating demethylation. Abscisic acid (ABA) acts as a floral repressor in A. thaliana where externally applied ABA affects the blooming time (Wang et al., 2013). ABSCISIC ACID-INSENSITIVE MUTANT 5 (AB15) overexpression retards floral initiation through upregulation of expression of FLC (Wang et al., 2013). ABA INSENSITIVE 3 and ABA INSENSITIVE 5 genes also regulate flowering by encoding two transcription factors, i.e., basic leucine zipper (bZIP)–type and B3-type (Hauser et al., 2011). ABA HYPERSENSITIVE 1 (HAB1) protein is also involved in controlling flowering in Arabidopsis by interconnecting with SWI/SNF chromatin-remodeling complex during transcription induced by ABA (Figure 3). In response to cold stress, chromatin-remodeling affects the histone core proteins by increasing the concentration of ABA, which leads to an increased level of histone H3 acetylation (Saez et al., 2004). Abscisic acid (ABA) and ABA-responsive factors play an important role in the maintenance of bud dormancy in perennial plants such as the Populus species. The PtAB13 gene is a homolog of AB13 of Arabidopsis, and its overexpression and downregulation regulate seed dormancy via ABA signaling. It causes alterations in bud developmental processes and misregulates the expression of genes during the process of bud dormancy (Rohde et al., 2002; Ruttink et al., 2007; Rios et al., 2014) (Table 2). There is some evidence that epigenetic regulation and ET synthesis are linked (Zhou et al., 2005). It is well known that during cold stress, ET levels rise, which may be associated with vernalization processes (Figure 2) (Chu and Lee, 1989; Zhao et al., 2014), which are often regulated by DNA methylation or demethylation (Burn et al., 1993; Sherman and Talbert, 2002). Cold temperatures cause a rise in ET in Arabidopsis, which delay bloom; nevertheless, if the temperature is again appropriate, blooming is stimulated. Ethylene, on the other hand, has been shown to promote the expression of HDA6 and HDA19 (Arabidopsis HDACs) (Zhou et al., 2005). HDA6 upregulates FLC expression in A. thaliana (Wu et al., 2008) (Table 2). HAD19 links the hormone response to pathogen pathways and floral induction in Brassica napus through a similar epigenetic mechanism in response to variations in ethylene sensitivity. HAD19 has been shown to interact with bnKCP1 (a putative factor with a kinase-inducible domain), a cold-inducible factor that is highly expressed in flowers (Gao et al., 2003). The function of ethylene in the control of floral development and blossoming time has been demonstrated in roses (Rosa hybrida Samantha). ET is involved in the regulation of petal cell expansion during the opening of a rose flower (Table 2) (Ma et al., 2008). In roses, changes in miRNA expression in response to ethylene have been documented, with five miRNAs (miR156, miR164, miR166, miR139, and rhy-miRC1) demonstrating a strong link between ethylene and petal growth control (Table 2) (Pei et al., 2013). Plants use Jasmonic Acid (JA) and jasmonate molecules as signaling molecules in response to stress, such as mechanical or biotic injuries produced by ozone exposure, dehydration, or pathogen infection (Overmyer et al., 2000; Berger, 2002; Farmer et al., 2003; Loyola-Vargas et al., 2012). These, on the other hand, are involved in a variety of developmental processes, including nitrogen storage, fruit ripening, senescence, and blooming (Creelman and Mullet, 1995; Creelman and Mullet, 1997). The study of the epigenetic involvement in the control of JA during blooming has been less thorough than the study of the genetic role. However, significant breakthroughs have been made. In Arabidopsis, for example, HDA6, a HISTONE DEACETYLASE, is essential for JA response and flowering (Wu et al., 2008). This deacetylase, in collaboration with MET1, governs locus-directed heterochromatin silencing, potentially by recruiting MET1 to certain loci and therefore sets the groundwork for later non-CG methylation (To et al., 2011). HDA6 also physically binds with FLD (Yu et al., 2011; Liu et al., 2012), which contributes in the deacetylation of FLC chromatin and hence represses gene expression (He et al., 2003; Wu et al., 2008). This shows that HDA6 is involved in Arabidopsis' JA response and blooming (Figure 3).
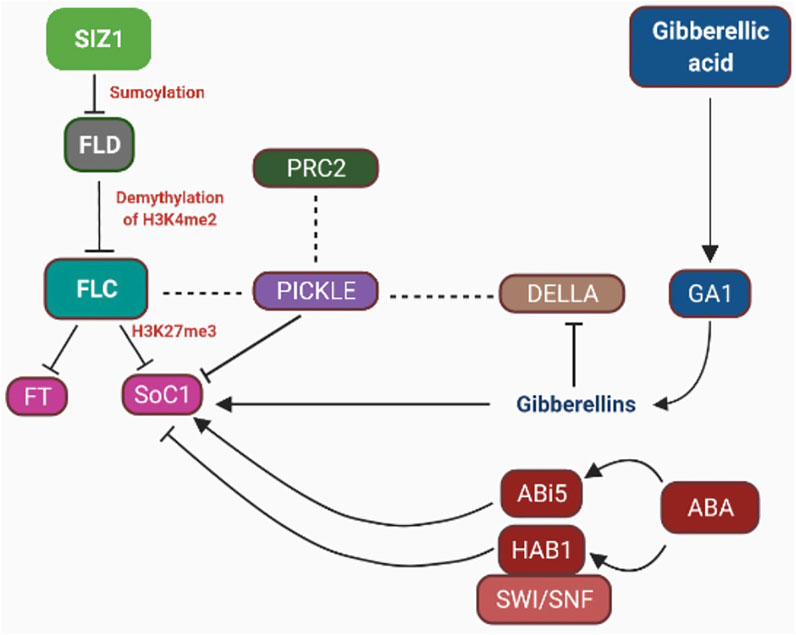
FIGURE 3. Epigenetic control on phytohormones: DELLA proteins are negative regulators of SOC1. DELLA proteins in combination with FLC interact with PICKLE (chromatin-remodeling protein) and PRC2 and repress the expression of SOC1. PRC2 regulates GA signaling by increasing H3K27me3-repressing histone protein. ABA INSENSITIVE MUTANT 5 overexpression delays flowering initiation by upregulating the expression of FLC. ABA HYPERSENSITIVE 1 (HAB1) is a negative regulator of flowering when combine with the chromatin-remodeling complex SWI/SNF. FLC is negatively regulated by lysine-specific demethylase 1–type histone demethylase (FLD). FLD causes demethylation of H3 histone (H3K4me3) in FLC. FLC downregulates SOC1 expression by trimethylation of lysine 27 on histone H3 protein via the formation of heterochromatic regions. SUMO E3 ligase (SIZ1) promotes expression of FLC by facilitating repression of FLD through sumoylation.
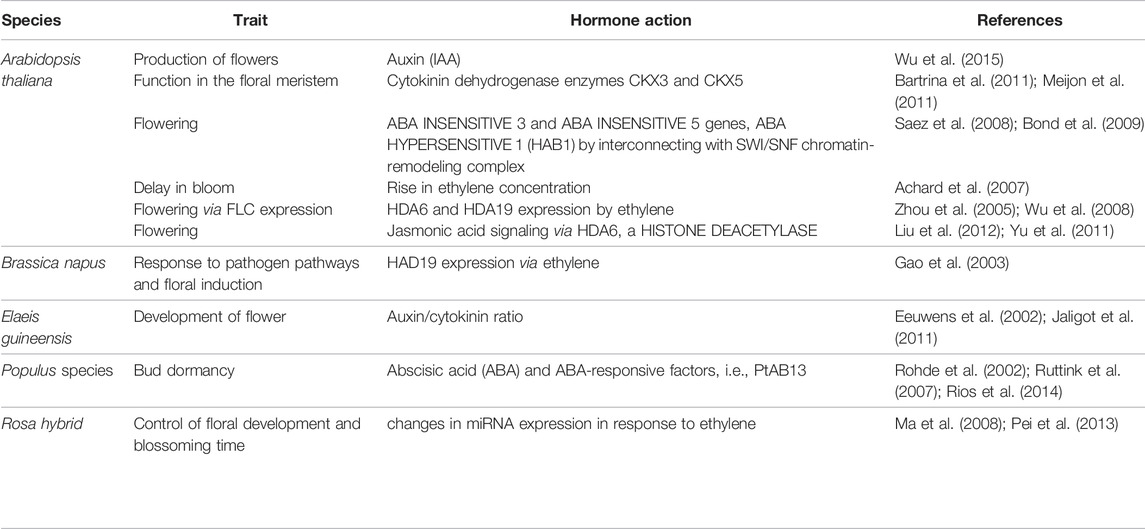
TABLE 2. Summary of the role of epigenetic regulation along with phytohormone action in plant species.
In response to pathogen infection and symbiotic microbe colonization, plants show genome-wide DNA methylation alterations (Xiao et al., 2021). The first evidence of epigenetic regulations in response to biotic factors was reported to control virulence via posttranscriptional gene silencing (PTGS). Transcriptional gene silencing (TGS) is another permanent defense mechanism against DNA viruses via RdDM (Figure 1). It has been reported that virus infection in Arabidopsis is controlled via m6A-specific methylation of the RNA genome in the alfalfa mosaic virus (AMV). ALKBH9B, an Arabidopsis protein having demethylase activity, removes m6A from ssRNA molecules and accumulates in the cytoplasm of the siRNA bodies. This process suggests the role of m6A demethylase in mRNA silencing (Ramirez-Prado et al., 2018; Martinez-Perez et al., 2017; Ashapkin et al., 2020) (Table 1). The demethylase DME is required for nodulation in Medicago truncatula. Several hundred genomic sites, including a small proportion of nodule-specific symbiosis genes, are variably methylated during nodule growth. In cyst nematode–infected soybean and A. thaliana roots, widespread DNA hypomethylation was detected. Differential DNA methylation in A. thaliana has been produced by Pseudomonas syringae pv. tomato str. DC3000 (Pst DC3000) (Dowen et al., 2012; Sahu et al., 2013). Differentially methylated cytosines have been found in CG and CHH contexts in gene-rich areas, specifically at the 5′ and 3′ ends of protein-coding genes. Also, Pst DC3000–responsive DNA methylation correlates negatively with neighboring gene expression levels across the genome, indicating that at the gene borders, DNA methylation is regulated dynamically and may contribute to differential gene expression in response to pathogens. In cucumber leaves and pollen grains, DNA methylation is caused by plant pathogenic ncRNAs promoter areas and transcriptional activation of some ribosomal RNA (rRNA) genes, leading in an abundance of short RNAs produced from rRNA. In A. thaliana, external application with pathogenic resistance phytohormone, i.e., salicylic acid, resulted in megabase-scale DNA hypomethylation in pericentromeric areas. DNA methylation has been proved to be a protective mechanism against unwanted transposition and a defense system against endonuclease digestion (Yaish, 2013). After infection with the virus, genomic methylation increases, and gene methylation related to resistance decreases. An increase in methylation promotes stability of the genome when plants are attacked by a virus, but genetic recombination is caused by a decrease in gene methylation levels and ultimately new genes produced help in resistance against pathogens (Engler et al., 1993).
Plant susceptibility to certain infectious pathogens can be altered by mutations in DNA methylation or demethylation regulators (Yu et al., 2013; Zhang et al., 2018). Plant tolerance to the biotrophic fungus Hyaloperonospora arabidopsidis is similarly increased in DNA hypomethylation mutants like nrpe1 and diminished in DNA hypermethylation mutants like ros1 (Sanchez et al., 2016; Zhang et al., 2018). POL V mutations lower susceptibility to the fungal necrotrophic pathogens Botrytis cinerea and Plectosphaerella cucumerina, in addition to enhancing resistance to biotrophic pathogens. An nrpd1 (POL IV) mutant does not have altered resistance to Pst DC3000 or fungal infections, unlike POL V mutants, indicating that POL V can regulate plant immune responses independently of canonical RdDM (Lopez et al., 2011; Zhang et al., 2018). Plants with the AGO4 mutant alleles ago4-1 and ago4-2, on the other hand, showed higher vulnerability to Pst DC3000, suggesting that AGO4 plays a distinct role in plant disease resistance compared to the other RdDM components. The DNA demethylase triple mutant ros1–dml2–dml3 and wild-type A. thaliana were compared, and it was discovered that DNA hypermethylation occurred more frequently in the mutant at regions flanking the gene body, such as upstream promoter regions and three untranslated regions. In the ros1–dml2–dml3 plants, over 200 genes are repressed, a large number of which have known or potential involvement in biotic stress response and are enriched with tiny transposons in their promoters. The ros1–dml2–dml3 mutant is more susceptible to the fungal disease Fusarium oxysporum, which supports this theory (Zhang et al., 2018) (Table 1).
Epigenetic modifications display a significant contribution in the position and rate of crossovers; however, the mechanism of the molecular crossovers remains to be fully studied (Wibowo et al., 2016). The highly coordinated gene expression within the germ cells requires epigenetic reprogramming. Epigenetically induced molecular pathways play a vital role in essential chemical and physiological processes during plant meiosis. The transmission of epi-alleles produced in response to environmental pressures raises issues about how agronomic treatments and environmental circumstances may influence the expression of critical crop characteristics that are evaluated in particular genetic improvement techniques. Indeed, from the standpoint of agricultural genetic improvement, understanding the epigenetic regulation of plant reproduction and meiosis is of great importance.
Meiotic siRNAs play an important role in epigenetic control of meiotic chromosome condensation, with essential implications for crop genetic improvement. In maize, OUTER CELL LAYER 4 (OCL4) encodes a transcription factor HD-ZIP IV which is required for the biogenesis of small RNAs and the other 21-nt phasiRNA (Zhai et al., 2015). This transcription factor also induces the synthesis of other proteins belonging to pentatricopeptide repeat (PPR) proteins, NB-LRR, and MYB families in other species (Howell et al., 2007; Zhu et al., 2012). It has been established that the presence of the histone variation H2A.Z is favorably linked to the occurrence of crossovers (Shilo et al., 2015). Plant crossover hotspots are suppressed by DNA methylation and H3K9me2 (Yelina et al., 2015). In Arabidopsis, the loss of DNA methylation has also been demonstrated to change crossover distribution in a chromatin type–dependent way (Mirouze et al., 2012).
SPO11-1-oligonucleotides and SPO11 topoisomerase-like transesterases produce DNA double-strand breaks to generate a high-resolution method to profile meiotic double-strand break patterns genome-wide. SPO11-1-oligonucleotides have been mapped in the Arabidopsis genome, and their role in regulating chromatin, DNA, and crossover frequency has been studied (Choi et al., 2018). The identification and mapping of these short DNA sequences in crop genomes will be important to discover epigenetic markers associated with key epigenetic modulators. Another important gene involved in the epigenetic modulation of plant reproduction is DECREASE IN DNA METHYLATION 1 (DDM1). DDM1 has ATPase activity that controls DNA methylation linked to crossover occurrence (Castiglione et al., 2002; Higo et al., 2012).
In plants, the primary germ cells do not directly enter spermatogenesis and oogenesis. Instead, the pollen mother cells (PMCs) in the anthers and the megaspore mother cells (MMCs) in the ovaries are generated in the floral meristem as a consequence of two meiotic divisions followed by a series of mitotic divisions, resulting in haploid male and female gametophytes, respectively (Manning et al., 2006). The male and female gametophytes are good models for investigating cell polarity, morphogenesis, and epigenetic regulation of cell development and specialization, and signaling pathways in angiosperms, despite their modest size and limited number of cells. Microsporogenesis and microgametogenesis are two phases of pollen formation that take place in the anthers. The sporogenous layer of the anther produces diploid microsporocytes, or PMCs. PMCs produce a tetrad of four haploid cells after two meiotic divisions. The tetrads then split up into individual microspores. Following that, two mitotic divisions occur: the first produces a big vegetative cell and a smaller generative cell, followed by the generative cell division, which produces two sperm cells, while the vegetative cell does not divide (Brownfield et al., 2009).
Epigenetic alterations are critical in the development of both male and female gametophytes, as well as in fertilization (Ingouff et al., 2017). It was discovered in Arabidopsis that the methylation level of PMCs in a normal environment (CG and CHG) was greater than in an adverse situation (CHH) (Kumar and Mohapatra, 2021). Symmetric methylation is generally seen in transposable elements, whereas asymmetric hypermethylation is mostly found in protein-coding genes. Increased methylation in a symmetric situation is believed to facilitate TE activity suppression, which guarantees genome stability before and throughout meiosis. Inactivation of methylation in an asymmetric setting, on the other hand, increases the activation of genes required for sperm cell development and conception. A substantial remodeling of chromatin occurs during PMC maturation, in addition to DNA methylation, promoting the start of meiosis.
The transition from the mitotic to meiotic phase is accompanied by a decrease in restrictive chromatin (H3K27me1 and H3K27me3) and an increase in permissive chromatin (H3K4me3) (Borg et al., 2020). The vegetative cell becomes roundish during meiosis and asymmetric during mitotic division of the haploid microspore. It has more methylation in the CHH areas, but it loses centromere-specific histone H3 (CENH3) due to decondensation of pericentromeric heterochromatin, local hypomethylation due to DME/ROS1 demethylases, and transposable element activation (Calarco et al., 2012).
During mitotic and meiotic cell division, the centromeric histone H3 (CENH3) variation is critical for the assembly and function of kinetochores. The inclusion of CENH3 into centromeric nucleosomes is the first step in kinetochore formation. The amount of CENH3 deposited on the centromeres changes depending on the stage of the cell cycle. CENH3 is also required for vegetative cell division and the removal of additional DNA (Lermontova et al., 2015). Hypomethylation of TEs leads in the production of 21–22 nt siRNAs, which are transferred to sperm cells and used by RdDM methylation to repress their TEs (Slotkin et al., 2009). During the whole time of pollen generation and development, whole-genome cell-specific methylation profiling indicated a high degree of CG and CHG methylation in the DNA of microspores, sperm, and vegetative cells.
During the development of a megasporocyte, DNA methylation level remains unchanged in context to CG, while it decreases temporarily in the context of CHH (Ingouff et al., 2017). The MMC, as well as the functional megaspore, are specified and differentiated through intercellular interactions mediated by mobile trans activating siRNAs (tasiRNAs), which are produced in the nucellus' surrounding cells and transported to the MMC, where they implement transcriptional and translational silencing (Baulcombe and Dean, 2014). The AGO9, RDR6, and SDS3 (a suppressor of genetic silencing 3) enzymes have been demonstrated to control the generation of such siRNAs in Arabidopsis (Olmedo-Movolif et al., 2010). The onset of mega gametogenesis in Arabidopsis nucellus is hampered when AGO5 expression is disrupted. Throughout mega gametogenesis, methylation in the CG and CHH contexts stays constant. CG methylation inside genes and transposons of the central cell of the embryo sac was lower than that of sperm cells, as shown in Arabidopsis (Park et al., 2016). This suggests that even before fertilization, the potential transcription of male genes is repressed. Mobile noncoding tasiRNAs regulate epigenetic control during gametogenesis in the embryo sac, thus siRNAs from the central cell penetrate the egg cell and decrease transposable element activity.
In both the embryo and endosperm, fertilization eliminates CHH hypomethylation of the paternal genome (Ibarra et al., 2012). Remethylation of the paternal DNA is most likely mediated by maternal siRNAs. One rationale for the epigenetic suppression of the male genome during early embryogenesis might include maternal regulation of embryo and endosperm size, as well as detection of self-pollen, which is important in interspecific crosses (Creasey et al., 2014). For appropriate embryo development, proper and consistent methylation of the dividing egg cell DNA is critical. In comparison to mature embryos, young embryos and endosperm tissues are hypomethylated, which represents the high transcriptional activity of genes in the developing embryo and the preparation for the embryo’s dormancy (Bouyer et al., 2017; Kawakatsu et al., 2017).
The levels of DNA methylation are strictly controlled in distinct tissues and cell types throughout the life cycle of a plant. In comparison to the embryos, endosperms of Oryza sativa and A. thaliana show worldwide DNA hypomethylation. It is caused by DME-dependent active demethylation in the central cell before fertilization in A. thaliana (Gehring et al., 2009; Hseih et al., 2009a; Ibarra et al., 2012). MET1 transcriptional repression also occurs during female gametogenesis, but it is unable to play a role in demethylation. Because genome-wide CG hypomethylation was not found in wild-type endosperm, DNA methylation is recovered in the dme mutant endosperm (Ibarra et al., 2012; Park et al., 2016). The vegetative cells experience DME-dependent DNA demethylation, which is accompanied by significant DDM1 downregulation (Table 3) (Slotkin et al., 2009; Ibarra et al., 2012). As a result, demethylated and de-silenced transposons create siRNAs. The siRNAs reach the vegetative cell after passing through the sperm cells, fortifying the RdDM pathway. POL V and DRM2 but not POL IV of A. thaliana was found in egg cells. These are the requirements for generation of siRNA through the conventional RdDM pathway. Therefore, transposon siRNAs accumulation in the sperm cells may enhance transposon silencing following fertilization of egg cells. During seed development, there is a rise in global levels of CHH methylation, and during seed germination, the levels fall due to passive demethylation.
During seed germination, however, the metabolic and, as a result, transcriptional-genetic activity of the embryonic tissues increases again, accompanied by a reduction in methylation in the CHH context, which is linked to the activation of protein-coding gene expression (Papareddy et al., 2020). The epigenetic “memory” associated with the histone marks is removed and is not transferred to the following generations of cells because the histones inherited from the egg and sperm are not reproduced in the cells of the embryo but are synthesized afresh (Papareddy et al., 2020). Thus, embryogenesis comes after the meiosis checkpoint or clearing box, which eliminates the maternal sporophyte’s epigenetic markers from the DNA. Endosperm methylation is substantially lower than that of the embryo, reflecting its strong transcriptional and metabolic activity.
At the same time, paternal genomes (i.e., genomes transported by sperm cells into the egg cell and the central cell of the embryo sac) are more methylated than maternal genomes (Heish et al., 2009a). Endosperm demethylation appears to be necessary to diminish transposable element activity via the production of siRNAs, which are transported into neighboring embryo cells and methylate the terminal sections of transposons by RNA-directed DNA methylation, thus inactivating them (Ingouff et al., 2017).
About 1% of the DNA methylation at cytosine–phosphate–guanine sites in the fruit pericarp of tomato gets changed during fruit development (Lang et al., 2017; Zhang et al., 2018). Many fruit-ripening genes have active DNA demethylation because their promoter regions contain binding sites for RIPENING-INHIBITOR (RIN), a prominent ripening transcription factor. Most known ripening genes whose expressions were adversely linked with promoter DNA methylation levels had confirmed RIN binding to target promoters. Premature ripening of tomato fruits was induced by treatment with a chemical inhibitor of DNA methylation, which caused promoter hypomethylation and expression of the gene encoding COLORLESS NON-RIPENING (CNR), which is a critical RIN-targeted gene for fruit ripening (Gao et al., 2010; Liu et al., 2015; Lang et al., 2017). The gradual DNA demethylation that occur during fruit ripening in Solanum lycopersicum is mediated by DML2. The expression of DNA demethylase DME-LIKE 2 (DML2) increases rapidly in ripening fruits. In S. lycopersicum, DML2 targets both ripening-induced and ripening-repressed genes, implying that active DNA demethylation is necessary for both ripening-induced gene activation and ripening-repressed gene suppression (Telias et al., 2011; El-Sharkaw et al., 2015; Daccord et al., 2017; Zhang et al., 2018) (Table 3).
In perennial plants, epigenetic modifications regulate seasonal dormancy cycles. Meristem and bud growth are controlled by photoperiod, temperature, etc. Transcriptomic studies have revealed that bud dormancy events in plants such as Populus spp., Rubus idaeus, Euphorbia esula, Prunus mume, Vitis spp., Prunus persica, and many other perennial plant species have been triggered through changes in their gene expression. These changes affect regulation of cell cycle, perception of light, signaling of hormones, and response to stress (Rios et al., 2014). Characterization and identification of nondormant mutants of perennial plants, such as evergrowing (evg) mutant of Prunus persica, contributed toward increasing molecular work at the level of genes, thereby renewing the field of dormancy. Deletion in tandemly repeated sequences of MADS-box genes, i.e., DORMANCY-ASSOCIATED MADS-box gene (DAM1-6) leads to nondormant phenotype of evg mutant. DAM genes are expressed in buds and get affected by photoperiod and chilling temperatures, thereby affecting the developmental stages of plants (Lloret et al., 2021). The concentration of DAM genes, i.e., DAM5 and DAM6, was found to be high in dormant buds, and after chilling treatment, the level fell and released the dormancy of buds (Yamane et al., 2011). In many other perennial species, such as Rubus idaeus, Prunus mume, Pyrus pyrifolia, and Actinidia deliciosa, DAM-like genes have been found to have dormancy-dependent expressions. In transgenic plants, the heterologous expression of these genes has regulatory roles in flowering and dormancy. In Arabidopsis, the expression of DAM1 and of SVP1 and SVP3 in Actinidia deliciosa led to delay in the time of flowering (Horvath et al., 2010; Wu et al., 2012) (Table 3).
Plants being sessile in nature are invariably affected by changing environmental conditions. However, they have the ability to adapt their biological processes according to the changing environments. They interact with their environment through consistent adjustments at the molecular levels. Epigenetic mechanisms contribute to these adjustments. These changes within the plant species modify the gene expression and help different plant species to withstand the extremes of different biotic or abiotic constrains. Such changes are induced due to DNA methylation, histone modification, e.g. acetylation/deacetylation, methylation/demethylation, ubiquitination, phosphorylation, and sRNA/modifications, which work in tandem with respect to environmental signals, transposon silencing, and hormone signaling to control the expression of genes in plants. These alterations help to maintain the survival of plants and maximize the chances of sexual reproduction under stress conditions. The role of DNA methylation in plant improvement is the preferred mechanism to investigate gene function and manipulate plants for creating novel varieties having capability to survive under stress conditions. Therefore, improved knowledge of epigenetic mechanisms via thorough study at the molecular level will be helpful. FLC controls flowering time in Arabidopsis that involves many genes in FLC expression via chromatin modifications. Further studies have to be undertaken to know how these diverse epigenetic modifications interact with one another to regulate the expression of FLC. CRISPR-based systems can be useful in altering the expression of genes for research applications as well as crop improvement efforts. The generation of genetically viable agricultural varieties that can endure a warmer world relies heavily on information generated from the plants' epigenomic profiles subjected to environmental challenges. Understanding the dynamic management of histone methylation and how histone methylation controls plant growth would be expanded through biochemical and genetic studies. Identification of intrinsic histone demethylases in plants, particularly lysine and/or arginine demethylases, and elucidation of their functions in regulating plant development and genome stability would require biochemical and genetic evidence. The roles of regulation of H3K27me3 in plants are limited and more in-depth knowledge will enable new researchers to enhance productivity of crops under limiting environmental conditions.
PK conceptualized the review and wrote it in consultation with SV and SK. PK, IAW, and SK contributed to writing and editing various sections on epigenetic modifications in angiosperms. PK, SK, and IW contributed to the figures and table. AA and PA contributed to revision of the manuscript. SV contributed to supervision and review of the manuscript. All authors have read and approved the submitted version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Achard, P., Baghour, M., Chapple, A., Hedden, P., Van Der Straeten, D., Genschik, P., et al. (2007). The Plant Stress Hormone Ethylene Controls floral Transition via DELLA-dependent Regulation of floral Meristem-Identity Genes. Proc. Natl. Acad. Sci. 104 (15), 6484–6489. doi:10.1073/pnas.06107171010.1073/pnas.0610717104
Aiese Cigliano, R., Sanseverino, W., Cremona, G., Ercolano, M. R., Conicella, C., and Consiglio, F. M. (2013). Genome-wide Analysis of Histone Modifiers in Tomato: Gaining an Insight into Their Developmental Roles. BMC Genomics 14, 57. doi:10.1186/1471-2164-14-57
Allfrey, V. G., Faulkner, R., and Mirsky, A. E. (1964). Acetylation and Methylation of Histones and Their Possible Role in the Regulation of RNA Synthesis. Proc. Natl. Acad. Sci. 51 (5), 786–794. doi:10.1073/pnas.51.5.786
Alvarez-Venegas, R., Abdallat, A. A., Guo, M., Alfano, J. R., and Avramova, Z. (2007). Epigenetic Control of a Transcription Factor at the Cross Section of Two Antagonistic Pathways. Epigenetics 2 (2), 106–113. doi:10.4161/epi.2.2.4404
Alvarez-Venegas, R., Pien, S., Sadder, M., Witmer, X., Grossniklaus, U., and Avramova, Z. (2003). ATX-1, an Arabidopsis Homolog of Trithorax, Activates Flower Homeotic Genes. Curr. Biol. 13 (8), 627–637. doi:10.1016/s0960-9822(03)00243-4
Amasino, R. (2010). Seasonal and Developmental Timing of Flowering. Plant J. Cell Mol. Biol. 61 (6), 1001–1013. doi:10.1111/j.1365-313x.2010.04148.x
Ampomah-Dwamena, C., Morris, B. A., Sutherland, P., Veit, B., and Yao, J.-L. (2002). Down-Regulation of TM29, a TomatoSEPALLATA Homolog, Causes Parthenocarpic Fruit Development and Floral Reversion. Plant Physiol. 130, 605–617. doi:10.1104/pp.005223
An, H., Roussot, C., Suárez-López, P., Corbesier, L., Vincent, C., Piñeiro, M., et al. (2004). CONSTANS Acts in the Phloem to Regulate a Systemic Signal that Induces Photoperiodic Flowering ofArabidopsis. Development 131 (15), 3615–3626. doi:10.1242/dev.01231
Andrés, F., and Coupland, G. (2012). The Genetic Basis of Flowering Responses to Seasonal Cues. Nat. Rev. Genet. 13 (9), 627–639. doi:10.1038/nrg3291
Ashapkin, V. V., Kutueva, L. I., Aleksandrushkina, N. I., and Vanyushin, B. F. (2020). Epigenetic Mechanisms of Plant Adaptation to Biotic and Abiotic Stresses. Ijms 21 (20), 7457. doi:10.3390/ijms21207457
Atighi, M. R., Verstraeten, B., De Meyer, T., and Kyndt, T. (2020). Genome‐wide DNA Hypomethylation Shapes Nematode Pattern‐triggered Immunity in Plants. New Phytol. 227, 545–558. doi:10.1111/nph.16532
Aufsatz, W., Mette, M. F., van der Winden, J., et al. (2002). HDA6, a Putative Histone Deacetylase Needed to Enhance DNA Methylation Induced by Double-Stranded RNA. EMBO J. 21, 6832–6841. doi:10.1371/journal.pbio.000006710.1093/emboj/cdf663
Aufsatz, W., Stoiber, T., Rakic, B., et al. (2007). Arabidopsis Histone Deacetylase 6: a green Link to RNA 839 Silencing. Oncogene 26, 5477–5488. doi:10.1038/sj.onc.1210615
Aukerman, M. J., and Sakai, H. (2003). Regulation of Flowering Time and floral Organ Identity by a MicroRNA and its APETALA2-like Target Genes. Plant Cell 15 (11), 2730–2741. doi:10.1105/tpc.016238
Ay, N., Janack, B., and Humbeck, K. (2014). Epigenetic Control of Plant Senescence and Linked Processes. J. Exp. Bot. 65 (14), 3875–3887. doi:10.1093/jxb/eru132
Baránek, M., Čechová, J., Raddová, J., Holleinová, V., Ondrušíková, E., and Pidra, M. (2015). Dynamics and Reversibility of the DNA Methylation Landscape of Grapevine Plants (Vitis vinifera) Stressed by In Vitro Cultivation and Thermotherapy. PLoS ONE 10, e0126638. doi:10.1371/journal.pone.0126638
Barozai, M. Y. K., and Aziz, A. N. (2018). Recent Plant Growth and Stress Management Related Significant Advancements in Epigenetics. Ann. Agrarian Sci. 16 (4), 416–421. doi:10.1016/j.aasci.2018.07.002
Bartrina, I., Otto, E., Strnad, M., Werner, T., and Schmülling, T. (2011). Cytokinin Regulates the Activity of Reproductive Meristems, Flower Organ Size, Ovule Formation, and Thus Seed Yield inArabidopsis Thaliana. The Plant cell 23 (1), 69–80. doi:10.1105/tpc.110.079079
Bastow, R., Mylne, J. S., Lister, C., Lippman, Z., Martienssen, R. A., and Dean, C. (2004). Vernalization Requires Epigenetic Silencing of FLC by Histone Methylation. Nature 427 (6970), 164–167. doi:10.1038/nature02269
Baulcombe, D. C., and Dean, C. (2014). Epigenetic Regulation in Plant Responses to the Environment. Cold Spring Harbor Perspect. Biol. 6, a019471. doi:10.1101/cshperspectAcknowledgments.a019471
Berger, S. (2002). Jasmonate-related Mutants of Arabidopsis as Tools for Studying Stress Signaling. Planta 214 (4), 497–504. doi:10.1007/s00425-001-0688-y
Berr, A., McCallum, E. J., Alioua, A., Heintz, D., Heitz, T., and Shen, W.-H. (2010). Arabidopsis Histone Methyltransferase SET DOMAIN GROUP8 Mediates Induction of the Jasmonate/Ethylene Pathway Genes in Plant Defense Response to Necrotrophic Fungi. Plant Physiol. 154, 1403–1414. doi:10.1104/pp.110.161497
Blüumel, M., Dally, N., and Jung, C. (2015). Flowering Time Regulation in Crops—What Did We Learn from Arabidopsis? Curr. Opin. Biotechnol. 32, 121–129. doi:10.1016/j.copbio.2014.11.023
Bond, D. M., Dennis, E. S., Pogson, B. J., and Finnegan, E. J. (2009). Histone Acetylation, VERNALIZATION INSENSITIVE 3 , FLOWERING LOCUS C , and the Vernalization Response. Mol. Plant 2 (4), 724–737. doi:10.1093/mp/ssp021
Borrdorf, O., Arcuri, D., Richards, C. L., and Pigliucci, M. (2010). Experimental Alteration of DNA Methylation Affects the Phenotypic Plasticity of Ecologically Relevant Traits in Arabidopsis thaliana. Evol. Ecol 24, 541–553.
Borg, M., Jacob, Y., Susaki, D., LeBlanc, C., Buendía, D., Axelsson, E., et al. (2020). Targeted Reprogramming of H3K27me3 Resets Epigenetic Memory in Plant Paternal Chromatin. Nature Cell Biology 22, 1–9. doi:10.1038/s41556-020-0515-y
Bouyer, D., Kramdi, A., Kassam, M., Heese, M., Schnittger, A., Roudier, F., et al. (2017). DNA Methylation Dynamics during Early Plant Life. Genome Biol. 18, 179. doi:10.1186/s13059-017-1313-0
Bowman, J. L., Smyth, D. R., and Meyerowitz, E. M. (1991). Genetic Interactions Among floral Homeotic Genes of Arabidopsis. Development 112, 1–20. doi:10.1242/dev.08397210.1242/dev.112.1.1
Bowman, J. L., Smyth, D. R., and Meyerowitz, E. M. (2012). The ABC Model of Flower Development: Then and Now. Development 139, 4095–4098. doi:10.1242/dev.083972
Boyko, A., and Kovalchuk, I. (2008). Epigenetic Control of Plant Stress Response. Environ. Mol. Mutagen. 49 (1), 61–72. doi:10.1002/em.20347
Brownfield, L., Hafidh, S., Durbarry, A., Khatab, H., Sidorova, A., Doerner, P., et al. (2009). ArabidopsisDUO POLLEN3 Is a Key Regulator of Male Germline Development and Embryogenesis. The Plant cell 21 (7), 1940–1956. doi:10.1105/tpc.109.066373
Brukhin, V., and Albertini, M. (2021). Epigenetic Modification in Plant Development and Reproduction. Epigenomes 5, 25. doi:10.3390/epigenomes5040025
Bucher, E., Reinders, J., and Mirouze, M. (2012). Epigenetic Control of Transposon Transcription and Mobility in Arabidopsis. Curr. Opin. Plant Biol. 15, 503–510. doi:10.1016/j.pbi.2012.08.006
Burgarella, C., Chantret, N., Gay, L., Prosperi, J. M., Bonhomme, M., Tiffin, P., et al. (2016). Adaptation to Climate through Flowering Phenology: a Case Study in Medicago Truncatula. Mol. Ecol. 25 (14), 3397–3415. doi:10.1111/mec.13683
Burn, J. E., Bagnall, D. J., Metzger, J. D., Dennis, E. S., and Peacock, W. J. (1993). DNA Methylation, Vernalization, and the Initiation of Flowering. Proc. Natl. Acad. Sci. 90 (1), 287–291. doi:10.1073/pnas.90.1.287
Butenko, Y., and Ohad, N. (2011). Polycomb-group Mediated Epigenetic Mechanisms through Plant Evolution. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1809, 395–406. doi:10.1016/j.bbagrm.2011.05.013
Butterbrodt, T., Thurow, C., and Gatz, C. (2006). Chromatin Immunoprecipitation Analysis of the Tobacco PR-1a- and the Truncated CaMV 35S Promoter Reveals Differences in Salicylic Acid-dependent TGA Factor Binding and Histone Acetylation. Plant Mol. Biol. 61 (4-5), 665–674. doi:10.1007/s11103-006-0039-2
Calarco, J. P., Borges, F., Donoghue, M. T. A., Van Ex, F., Jullien, P. E., Lopes, T., et al. (2012). Reprogramming of DNA Methylation in Pollen Guides Epigenetic Inheritance via Small RNA. Cell 151, 194–205. doi:10.1016/j.cell.2012.09.001
Campos-Rivero, G., Osorio-Montalvo, P., Sánchez-Borges, R., Us-Camas, R., Duarte-Aké, F., and De-la-Peña, C. (2017). Plant Hormone Signaling in Flowering: An Epigenetic point of View. J. Plant Physiol. 214, 16–27. doi:10.1016/j.jplph.2017.03.018
Candaele, J., Demuynck, K., Mosoti, D., Beemster, G. T. S., Inzé, D., Nelissen, H., et al. (2014). Differential Methylation during maize Leaf Growth Targets Developmentally Regulated Genes. Plant Physiol. 164, 1350–1364. doi:10.1104/pp.113.233312
Capy, P., Gasperi, G., Biémont, C., and Bazin, C. (2000). Stress and Transposable Elements: Co-evolution or Useful Parasites? Heredity 85, 101–106. doi:10.1046/j.1365-2540.2000.00751.x
Carpenter, R., Martin, C., and Coen, E. S. (1987). Comparison of Genetic Behaviour of the Transposable Element Tam3 at Two Unlinked Pigment Loci in Antirrhinum majus. Mol. Gen. Genet. 207, 82–89. doi:10.1007/bf00331494
Castiglione, M. R., Cremonini, R., and Frediani, M. (2002). DNA Methylation Patterns on Plant Chromosomes. Caryologia 55 (4), 275–282. doi:10.1080/00087114.2002.10797876
Castiglione, M. R., Cremonini, R., and Frediani, M. (2002). DNA Methylation Patterns on Plant Chromosomes. Caryologia 55, 275–282. doi:10.1080/00087114.2002.10797876
Cavrak, V. V., Lettner, N., Jamge, S., Kosarewicz, A., Bayer, L. M., Mittelsten Scheid, O., et al. (2014). How a Retrotransposon Exploits the Plant's Heat Stress Response for its Activation. Plos Genet. 10 (1), e1004115. doi:10.1371/journal.pgen.1004115
Chen, Q., Chen, X., Wang, Q., Zhang, F., Lou, Z., Zhang, Q., et al. (2013). Structural Basis of a Histone H3 Lysine 4 Demethylase Required for Stem Elongation in rice. Plos Genet. 9, e1003239. doi:10.1371/journal.pgen.1003239
Chen, T., and Li, E. (2004). Structure and Function of Eukaryotic DNA Methyltransferases. Curr. Top. Dev Biol 60, 55–89. doi:10.1016/S0070-2153(04)60003-2
Cheng, Y.-H., Peng, X.-Y., Yu, Y.-C., Sun, Z.-Y., and Han, L. (2019). The Effects of DNA Methylation Inhibition on Flower Development in the Dioecious Plant Salix Viminalis. Forests 10, 173. doi:10.3390/f10020173
Choi, J., Hyun, Y., Kang, M.-J., In Yun, H., Yun, J.-Y., Lister, C., et al. (2009). Resetting and Regulation of Flowering Locus Cexpression during Arabidopsis Reproductive Development. The Plant J. Cell Mol. Biol. 57 (5), 918–931. doi:10.1111/j.1365-313x.2008.03776.x
Choi, K., Zhao, X., Tock, A. J., Lambing, C., Underwood, C. J., Hardcastle, T. J., et al. (2018). Nucleosomes and DNA Methylation Shape Meiotic DSB Frequency in Arabidopsis thaliana Transposons and Gene Regulatory Regions. Genome Res. 28, 532–546. doi:10.1101/gr.225599.117
Chu, C., and Lee, T. M. (1989). The Relationship between Ethylene Biosynthesis and Chilling Tolerance in Seedlings of rice (Oryza Sativa). Bot. Bull. Acad. Sin 30, 263–273.
Cimini, D., Mattiuzzo, M., Torosantucci, L., and Degrassi, F. (2003). Histone Hyperacetylation in Mitosis Prevents Sister Chromatid Separation and Produces Chromosome Segregation Defects. MBoC 14, 3821–3833. doi:10.1091/mbc.e03-01-0860
Covarrubias, A. A., and Reyes, J. L. (2010). Post-transcriptional Gene Regulation of Salinity and Drought Responses by Plant microRNAs. Plant, cell envi 33 (4), 481–489. doi:10.1111/j.1365-3040.2009.02048.x
Creasey, K. M., Zhai, J., Borges, F., van Ex, F., Regulski, M., Meyers, B. C., et al. (2014). miRNAs Trigger Widespread Epigenetically Activated siRNAs from Transposons in Arabidopsis. Nature 508, 411–415. doi:10.1038/nature13069
Creelman, R. A., and Mullet, J. E. (1997). Biosynthesis and Action of Jasmonates in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 355–381. doi:10.1146/annurev.arplant.48.1.355
Creelman, R. A., and Mullet, J. E. (1995). Jasmonic Acid Distribution and Action in Plants: Regulation during Development and Response to Biotic and Abiotic Stress. Proc. Natl. Acad. Sci. 92 (10), 4114–4119. doi:10.1073/pnas.92.10.4114
Cubas, P., Vincent, C., and Coen, E. (1999). An Epigenetic Mutation Responsible for Natural Variation in floral Symmetry. Nature 401 (6749), 157–161. doi:10.1038/43657
Cui, X., Jin, P., Cui, X., Gu, L., Lu, Z., Xue, Y., et al. (2013). Control of Transposon Activity by a Histone H3K4 Demethylase in rice. Proc. Natl. Acad. Sci. 110, 1953–1958. doi:10.1073/pnas.1217020110
Czermin, B., Melfi, R., McCabe, D., Seitz, V., Imhof, A., and Pirrotta, V. (2002). Drosophila Enhancer of Zeste/ESC Complexes Have a Histone H3 Methyltransferase Activity that marks Chromosomal Polycomb Sites. Cell 111, 185–196. doi:10.1016/s0092-8674(02)00975-3
Daccord, N., Celton, J. M., Linsmith, G., Becker, C., Choisne, N., Schijlen, E., et al. (2017). High- Quality De Novo Assembly of the Apple Genome and Methylome Dynamics of Early Fruit Development. Nat. Genet. 49, 1099–1106. doi:10.1038/ng.3886
De Lucia, F., Crevillen, P., Jones, A. M. E., Greb, T., and Dean, C. (2008). A PHD-Polycomb Repressive Complex 2 Triggers the Epigenetic Silencing of FLC during Vernalization. Proc. Natl. Acad. Sci. 105 (44), 16831–16836. doi:10.1073/pnas.0808687105
Deal, R. B., Topp, C. N., McKinney, E. C., and Meagher, R. B. (2007). Repression of Flowering inArabidopsisRequires Activation ofFLOWERING LOCUS CExpression by the Histone Variant H2A.Z. Z. Plant Cell. 19, 74–83. doi:10.1105/tpc.106.048447
Domagalska, M. A., Sarnowska, E., Nagy, F., and Davis, S. J. (2010). Genetic Analyses of Interactions Among Gibberellin, Abscisic Acid, and Brassinosteroids in the Control of Flowering Time in Arabidopsis thaliana. PloS one 5, e14012. doi:10.1371/journal.pone.0014012
Dowen, R. H., Pelizzola, M., Schmitz, R. J., Lister, R., Dowen, J. M., Nery, J. R., et al. (2012). Widespread Dynamic DNA Methylation in Response to Biotic Stress. Proc. Natl. Acad. Sci. 109 (32), E2183–E2191. doi:10.1073/pnas.1209329109
Dziegielewski, W., and Ziolkowski, P. A. (2021). License to Regulate: Noncoding RNA Special Agents in Plant Meiosis and Reproduction. Front. Plant Sci. 12, 662185. doi:10.3389/fpls.2021.662185
Eeuwens, C. J., Lord, S., Donough, C. R., Rao, V., Vallejo, G., and Nelson, S. (2002). Effects of Tissue Culture Conditions during Embryoid Multiplication on the Incidence of ``mantled'' Flowering in Clonally Propagated Oil palm. Plant Cell, Tissue Organ. Cult. 70, 311–323. doi:10.1023/a:1016543921508
El-Sharkawy, I., Liang, D., and Xu, K. (2015). Transcriptome Analysis of an Apple (Malus×domestica) Yellow Fruit Somatic Mutation Identifies a Gene Network Module Highly Associated with Anthocyanin and Epigenetic Regulation. Exbotj 66, 7359–7376. doi:10.1093/jxb/erv433
Engler, P., Weng, A., and Storb, U. (1993). Influence of CpG Methylation and Target Spacing on V(D)J Recombination in a Transgenic Substrate. Mol. Cell Biol 13 (1), 571–577. doi:10.1128/mcb.13.1.571-577.1993
Farmer, E. E., Alméras, E., and Krishnamurthy, V. (2003). Jasmonates and Related Oxylipins in Plant Responses to Pathogenesis and Herbivory. Curr. Opin. Plant Biol. 6 (4), 372–378. doi:10.1016/s1369-5266(03)00045-1
Fedoroff, N. V. (2012). Transposable Elements, Epigenetics, and Genome Evolution. Science 338, 758–767. doi:10.1126/science.338.6108.758
Feng, S., Jacobsen, S. E., and Reik, W. (2010). Epigenetic Reprogramming in Plant and Animal Development. Science 330 (6004), 622–627. doi:10.1126/science.1190614
Forestan, C., Farinati, S., Zambelli, F., Pavesi, G., Rossi, V., and Varotto, S. (2019). Epigenetic Signatures of Stress Adaptation and Flowering Regulation in Response to Extended Drought and Recovery in Zea mays. Plant Cell Environ 43, 55–75. doi:10.1111/pce.13660
Fu, C., Sunkar, R., Zhou, C., Shen, H., Zhang, J. Y., Matts, J., et al. (2012). Overexpression of miR156 in Switchgrass ( Panicum Virgatum L.) Results in Various Morphological Alterations and Leads to Improved Biomass Production. Plant Biotechnol. J. 10, 443–452. doi:10.1111/j.1467-7652.2011.00677.x
Gao, L., Geng, Y., Li, B., Chen, J., and Yang, J. (2010). Genome-wide DNA Methylation Alterations of Alternanthera Philoxeroides in Natural and Manipulated Habitats: Implications for Epigenetic Regulation of Rapid Responses to Environmental Fluctuation and Phenotypic Variation. Plant Cell Environ. 33, 1820–1827. doi:10.1111/j.1365-3040.2010.02186.x
Gao, M.-J., Schäfer, U. A., Parkin, I. A. P., Hegedus, D. D., Lydiate, D. J., and Hannoufa, A. (2003). A Novel Protein fromBrassica Napushas a Putative KID Domain and Responds to Low Temperature. Plant J. 33 (6), 1073–1086. doi:10.1046/j.1365-313x.2003.01694.x
Gehring, M., Bubb, K. L., and Henikoff, S. (2009). Extensive Demethylation of Repetitive Elements during Seed Development Underlies Gene Imprinting. Science 324, 1447–1451. doi:10.1126/science.1171609
Geng, S., Kong, X., Song, G., Jia, M., Guan, J., Wang, F., et al. (2019). DNA Methylation Dynamics during the Interaction of Wheat Progenitor Aegilops Tauschii with the Obligate Biotrophic Fungus Blumeria Graminis F. Sp. Tritici. New Phytol. 221, 1023–1035. doi:10.1111/nph.15432
Ghoshal, B., and Gardiner, J. (2021). “CRISPR-dCas9-Based Targeted Manipulation of DNA Methylation in Plants,” in CRISPR-cas Methods. Editors M. T. Islam,, and K. A. Molla (Humana, New York: Springer Protocols Handbooks). doi:10.1007/978-1-0716-1657-4_5
Gourcilleau, D., Mousset, M., Latutrie, M., Marin, S., Delaunay, A., Maury, S., et al. (2019). Assessing Global DNA Methylation Changes Associated with Plasticity in Seven Highly Inbred Lines of Snapdragon Plants (Antirrhinum majus). Genes 10, 256. doi:10.3390/genes10040256
Grandbastien, M. (1998). Activation of Plant Retrotransposons under Stress Conditions. Trends Plant Sci. 3, 181–187. doi:10.1016/s1360-1385(98)01232-1
Gratani, L. (2014). Plant Phenotypic Plasticity in Response to Environmental Factors. Adv. Bot. 2014, 1–17. doi:10.1155/2014/208747
Haag, J. R., Ream, T. S., Marasco, M., Nicora, C. D., Norbeck, A. D., Pasa-Tolic, L., et al. (2012). In Vitro Transcription Activities of Pol IV, Pol V, and RDR2 Reveal Coupling of Pol IV and RDR2 for dsRNA Synthesis in Plant RNA Silencing. Mol. Cell 48, 811–818. doi:10.1016/j.molcel.2012.09.027
Han, M.-H., Goud, S., Song, L., and Fedoroff, N. (2004). The Arabidopsis Double-Stranded RNA-Binding Protein HYL1 Plays a Role in microRNA-Mediated Gene Regulation. Proc. Natl. Acad. Sci. 101 (4), 1093–1098. doi:10.1073/pnas.0307969100
Han, M. H., Goud, S., Song, L., and Fedoroff, N. (2019). The Arabidopsis Double-Stranded RNA-Binding Protein HYL1 Plays a Role in microRNA-Mediated Gene Regulation. Proc. Natl. Acad. Sci. U S A. 101, 1093–1098. doi:10.1073/pnas.0307969100
Harrison, B. J., and Fincham, J. R. S. (1964). Instability at the Pal Locus in Antirrhinum majus. Heredity 19, 237–258. doi:10.1038/hdy.1964.28
Hashida, S.-n., Kitamura, K., Mikami, T., and Kishima, Y. (2003). Temperature Shift Coordinately Changes the Activity and the Methylation State of Transposon Tam3 in Antirrhinum majus. Plant Physiol. 132 (3), 1207–1216. doi:10.1104/pp.102.017533
Hauben, M., Haesendonckx, B., Standaert, E., Van Der Kelen, K., Azmi, A., Akpo, H., et al. (200920109–20114). Energy Use Efficiency Is Characterized by an Epigenetic Component that Can Be Directed through Artificial Selection to Increase Yield. Proc. Natl. Acad. Sci. U S A. 106, 20109–20114. doi:10.1073/pnas.0908755106
Hauser, F., Waadt, R., and Schroeder, J. I. (2011). Evolution of Abscisic Acid Synthesis and Signaling Mechanisms. Curr. Biol. 21 (9), R346–R355. doi:10.1016/j.cub.2011.03.015
He, X.-J., Chen, T., and Zhu, J.-K. (2011). Regulation and Function of DNA Methylation in Plants and Animals. Cell Res 21, 442–465. doi:10.1038/cr.2011.23
He, Y. (2009). Control of the Transition to Flowering by Chromatin Modifications. Mol. Plant 2, 554–564. doi:10.1093/mp/ssp005
He, Y., Michaels, S. D., and Amasino, R. M. (2003). Regulation of Flowering Time by Histone Acetylation in Arabidopsis. Science 302 (5651), 1751–1754. doi:10.1126/science.1091109
Herrera, C. M., and Bazaga, P. (2012). Epigenetic Correlates of Plant Phenotypic Plasticity: DNA Methylation Differs between Prickly and Nonprickly Leaves in heterophyllousIlex aquifolium(Aquifoliaceae) Trees. Bot. J. Linn. Soc. 171, 441–452. doi:10.1111/boj.12007
Higo, H., Tahir, M., Takashima, K., Miura, A., Watanabe, K., Tagiri, A., et al. (2012). DDM1 (Decrease in DNA Methylation) Genes in rice (Oryza Sativa). Mol. Genet. Genomics 287, 785–792. doi:10.1007/s00438-012-0717-5
Hirayama, T., and Shinozaki, K. (2010). Research on Plant Abiotic Stress Responses in the post-genome Era: Past, Present and Future. Plant J. Cell Mol. Biol. 61 (6), 1041–1052. doi:10.1111/j.1365-313X.2010.04124.x
Horvath, D. P., Sung, S., Kim, D., Chao, W., and Anderson, J. (2010). Characterization, Expression and Function of DORMANCY ASSOCIATED MADS-BOX Genes from Leafy Spurge. Plant Mol. Biol. 73, 169–179. doi:10.1007/s11103-009-9596-5
Howell, M. D., Fahlgren, N., Chapman, E. J., Cumbie, J. S., Sullivan, C. M., Givan, S. A., et al. (2007). Genome-Wide Analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 Pathway inArabidopsisReveals Dependency on miRNA- and tasiRNA-Directed Targeting. Plant Cell. 19, 926–942. doi:10.1105/tpc.107.050062
Hsieh, D., Xia, Y., Wray, L., Qian, D., Pal, A., Dil, J. H., et al. (2009). Observation of Unconventional Quantum Spin Textures in Topological Insulators. Science 323, 919–922. doi:10.1126/science.1167733
Hsieh, T.-F., Ibarra, C. A., Silva, P., Zemach, A., Eshed-Williams, L., Fischer, R. L., et al. (2009). Genome-Wide Demethylation of Arabidopsis Endosperm. Science 324, 1451–1454. doi:10.1126/science.1172417
Hugouvieux, V., Kwak, J. M., and Schroeder, J. I. (2001). An mRNA Cap Binding Protein, ABH1, Modulates Early Abscisic Acid Signal Transduction in Arabidopsis. Cell 106 (4), 477–487. doi:10.1016/s0092-8674(01)00460-3
Ibarra, C. A., Feng, X., Schoft, V. K., Hsieh, T.-F., Uzawa, R., Rodrigues, J. A., et al. (2012). Active DNA Demethylation in Plant Companion Cells Reinforces Transposon Methylation in Gametes. Science 337 (6100), 1360–1364. doi:10.1126/science.1224839
Ietswaart, R., Wu, Z., and Dean, C. (2012). Flowering Time Control: Another Window to the Connection between Antisense RNA and Chromatin. Trends Genet. 28 (9), 445–453. doi:10.1016/j.tig.2012.06.002
Ingouff, M., Selles, B., Michaud, C., Vu, T. M., Berger, F., Schorn, A. J., et al. (2017). Live-cell Analysis of DNA Methylation during Sexual Reproduction in Arabidopsis Reveals Context and Sex-specific Dynamics Controlled by Noncanonical RdDM. Genes Dev. 31, 72–83. doi:10.1101/gad.289397.116
Ito, H., Gaubert, H., Bucher, E., Mirouze, M., Vaillant, I., and Paszkowski, J. (2011). An siRNA Pathway Prevents Transgenerational Retrotransposition in Plants Subjected to Stress. Nature 472 (7341), 115–119. doi:10.1038/nature09861
Ito, H. (2013). Small RNAs and Regulation of Transposons in Plants. Genes Genet. Syst. 88 (1), 3–7. doi:10.1266/ggs.88.3
Iwasaki, M., and Paszkowski, J. (2014). Identification of Genes Preventing Transgenerational Transmission of Stress-Induced Epigenetic States. Proc. Natl. Acad. Sci. 111 (23), 8547–8552. doi:10.1073/pnas.1402275111
Jacobsen, S. E., and Meyerowitz, E. M. (1997). Hypermethylated SUPERMAN Epigenetic Alleles in Arabidopsis. Science 277 (5329), 1100–1103. doi:10.1126/science.277.5329.1100
Jaligot, E., Adler, S., Debladis, É., Beulé, T., Richaud, F., Ilbert, P., et al. (2011). Epigenetic Imbalance and the floral Developmental Abnormality of the In Vitro-regenerated Oil palm Elaeis Guineensis. Ann. Bot. 108 (8), 1453–1462. doi:10.1093/aob/mcq266
Jaligot, E., Rival, A., Beulé, T., Dussert, S., and Verdeil, J.-L. (2000). Somaclonal Variation in Oil palm ( Elaeis Guineensis Jacq.): the DNA Methylation Hypothesis. Plant Cell Rep. 19, 684–690. doi:10.1007/s002999900177
Jenuwein, T., and Allis, C. D. (2001). Translating the Histone Code. Science 293, 1074–1080. doi:10.1126/science.1063127
Jiang, D., Gu, X., and He, Y. (2009). Establishment of the Winter-Annual Growth Habit via FRIGIDA-Mediated Histone Methylation at FLOWERING LOCUS C in Arabidopsis. The Plant cell 21 (6), 1733–1746. doi:10.1105/tpc.109.067967
Jiang, D., Wang, Y., Wang, Y., and He, Y. (2008). Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb Repressive Complex 2 Components. PLoS one 3 (10), e3404. doi:10.1371/journal.pone.0003404
Jin, J. B., Jin, Y. H., Lee, J., Miura, K., Yoo, C. Y., Kim, W. Y., et al. (2008). The SUMO E3 Ligase, AtSIZ1, Regulates Flowering by Controlling a Salicylic Acid-Mediated floral Promotion Pathway and through Affects on FLC Chromatin Structure. Plant J. 53 (3), 530–540. doi:10.1111/j.1365-313X.2007.03359.x
Johns, M. A., Mottinger, J., and Freeling, M. (1985). A Low Copy Number, Copia-like Transposon in maize. EMBO J. 4 (5), 1093–1101. doi:10.1002/j.1460-2075.1985.tb03745.x
Kankel, M. W., Ramsey, D. E., Stokes, T. L., Flowers, S. K., Haag, J. R., Jeddeloh, J. A., et al. (2003). Arabidopsis MET1 Cytosine Methyltransferase Mutants. Genetics 163 (3), 1109–1122. doi:10.1093/genetics/163.3.1109
Kawakatsu, T., Nery, J. R., Castanon, R., and Ecker, J. R. (2017). Dynamic DNA Methylation Reconfiguration during Seed Development and Germination. Genome Biol. 18, 171. doi:10.1186/s13059-017-1251-x
Kellenberger, R. T., Schlüter, P. M., and Schiestl, F. P. (2016). Herbivore-induced DNA Demethylation Changes floral Signalling and Attractiveness to Pollinators in Brassica Rapa. PLoS ONE 11, e0166646. doi:10.1371/journal.pone.0166646
Kim, J.-M., To, T. K., Nishioka, T., and Seki, M. (2010). Chromatin Regulation Functions in Plant Abiotic Stress Responses. Plant Cell Environ. 33, 604–611. doi:10.1111/j.1365-3040.2009.02076.x
Kim, M.-S., Kondo, T., Takada, I., Youn, M.-Y., Yamamoto, Y., Takahashi, S., et al. (2009). DNA Demethylation in Hormone-Induced Transcriptional Derepression. Nature 461, 1007–1012. doi:10.1038/nature08456
Kinoshita, A., and Richter, R. (2020). Genetic and Molecular Basis of floral Induction in Arabidopsis thaliana. J. Exp. Bot. 71, 2490–2504.
Klee, H. J., and Giovannoni, J. J. (2011). Genetics and Control of Tomato Fruit Ripening and Quality Attributes. 942. Annu. Rev. Genet. 45, 41–59. doi:10.1093/jxb/eraa057
Krogan, N. T., Hogan, K., and Long, J. A. (2012). APETALA2 Negatively Regulates Multiple floral Organ Identity Genes inArabidopsisby Recruiting the Co-repressor TOPLESS and the Histone Deacetylase HDA19. Development 139 (22), 4180–4190. doi:10.1242/dev.085407
Kumar, S., and Mohapatra, T. (2021). Dynamics of DNA Methylation and its Functions in Plant Growth and Development. Front. Plant Sci. 12. doi:10.3389/fpls.2021.596236
Kundariya, H., Yang, X., Morton, K., Sanchez, R., Axtell, M. J., Hutton, S. F., et al. (2020). MSH1-induced Heritable Enhanced Growth Vigor through Grafting Is Associated with the RdDM Pathway in Plants. Nat. Commun. 11, 5343–5414. doi:10.1038/s41467-020-19140-x
Kwak, K. J., Kim, Y. O., and Kang, H. (2005). Characterization of Transgenic Arabidopsis Plants Overexpressing GR-RBP4 under High Salinity, Dehydration, or Cold Stress. J. Exp. Bot. 56, 3007–3016. doi:10.1093/jxb/eri298
Kwon, C. S., Lee, D., Choi, G., and Chung, W.-I. (2009). Histone Occupancy-dependent and -independent Removal of H3K27 Trimethylation at Cold-Responsive Genes in Arabidopsis. Plant J. : Cel. Mol. Biol. 60 (1), 112–121. doi:10.1111/j.1365-313x.2009.03938.x
Lang, Z., Wang, Y., Tang, K., Tang, D., Datsenka, T., Cheng, J., et al. (2017). Critical Roles of DNA Demethylation in the Activation of Ripening-Induced Genes and Inhibition of Ripening-Repressed Genes in Tomato Fruit. Proc. Natl. Acad. Sci. USA 114, E4511–E4519. doi:10.1073/pnas.1705233114
Lee, B.-h., Henderson, D. A., and Zhu, J.-K. (2005). TheArabidopsisCold-Responsive Transcriptome and its Regulation by ICE1. The plant cell online 17 (11), 3155–3175. doi:10.1105/tpc.105.035568
Lermontova, I., Sandmann, M., Mascher, M., Schmit, A.-C., and Chabouté, M.-E. (2015). Centromeric Chromatin and its Dynamics in Plants. Plant J. 83, 4–17. doi:10.1111/tpj.12875
Li, X., Bian, H., Song, D., Ma, S., Han, N., Wang, J., et al. (2013). Flowering Time Control in Ornamental Gloxinia (Sinningia Speciosa) by Manipulation of miR159 Expression. Ann. Bot. 111, 791–799. doi:10.1093/aob/mct034
Li, Y., Butenko, Y., and Grafi, G. (2005). Histone Deacetylation Is Required for Progression through Mitosis in Tobacco Cells. Plant J. 41, 346–352. doi:10.1111/j.1365-313X.2004.02301.x
Lindroth, A. M., Cao, X., Jackson, J. P., Zilberman, D., McCallum, C. M., Henikoff, S., et al. (2001). Requirement of CHROMOMETHYLASE3 for Maintenance of CpXpG Methylation. Science 292 (5524), 2077–2080. doi:10.1126/science.1059745
Lippman, Z., Gendrel, A.-V., Black, M., Vaughn, M. W., Dedhia, N., Richard McCombie, W., et al. (2004). Role of Transposable Elements in Heterochromatin and Epigenetic Control. Nature 430 (6998), 471–476. doi:10.1038/nature02651
Liu, C., Lu, F., Cui, X., and Cao, X. (2010). Histone Methylation in Higher Plants. Annu. Rev. Plant Biol. 61, 395–420. doi:10.1146/annurev.arplant.043008.091939
Liu, G., Li, W., Zheng, P., Xu, T., Chen, L., Liu, D., et al. (2012). Transcriptomic Analysis of 'Suli' Pear (Pyrus Pyrifolia white Pear Group) Buds during the Dormancy by RNA-Seq. BMC Genomics 13, 700. doi:10.1186/1471-2164-13-700
Liu, R., How-Kit, , A., Stammitti, L., Teyssier, E., Rolin, D., Mortain-Bertrand, A., et al. (2015). A DEMETER-like DNA Demethylase Governs Tomato Fruit Ripening. Proc. Natl. Acad. Sci. USA 112, 10804–10809. doi:10.1073/pnas.1503362112
López, A., Ramírez, V., García-Andrade, J., Flors, V., and Vera, P. (2011). The RNA Silencing Enzyme RNA Polymerase V Is Required for Plant Immunity. Plos Genet. 7, e1002434. doi:10.1371/journal.pgen.1002434
López Sánchez, A., Stassen, J. H., Furci, L., Smith, L. M., and Ton, J. (2020). The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J. 88, 361–374. doi:10.1111/tpj.13252
López Sánchez, A., Stassen, J. H. M., Furci, L., Smith, L. M., and Ton, J. (2016). The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J. 88, 361–374. doi:10.1111/tpj.13252
Loyola-Vargas, V., Ruíz-May, E., Galaz-Ávalos, R., and De-la-Peña, C. (2012). The Role of Jasmonic Acid in Root Mitochondria Disruption. Plant Signaling Behav. 7 (6), 611–614. doi:10.4161/psb.20070
Lloret, A., Quesada-Traver, C., and Conejero, A. (2021). Regulatory Circuits Involving Bud Dormancy Factor PpeDAM6. Hortic. Res. 8 261. doi:10.1038/s41438-021-00706-9
Lu, C., and Fedoroff, N. (2000). A Mutation in the Arabidopsis HYL1 Gene Encoding a dsRNA Binding Protein Affects Responses to Abscisic Acid, Auxin, and Cytokinin. The Plant Cell 12, 2351–2366. doi:10.2307/3871234
Luo, Y., Guo, Z., and Li, L. (2013). Evolutionary Conservation of microRNA Regulatory Programs in Plant Flower Development. Developmental Biol. 380, 133–144. doi:10.1016/j.ydbio.2013.05.009
Ma, N., Xue, J., Li, Y., Liu, X., Dai, F., Jia, W., et al. (2008). Rh-PIP2;1, a Rose Aquaporin Gene, Is Involved in Ethylene-Regulated Petal Expansion. Plant Physiol. 148 (2), 894–907. doi:10.1104/pp.108.120154
Manning, K., Tör, M., Poole, M., Hong, Y., Thompson, A. J., King, G. J., et al. (2006). A Naturally Occurring Epigenetic Mutation in a Gene Encoding an SBP-Box Transcription Factor Inhibits Tomato Fruit Ripening. Nat. Genet. 38, 948–952. doi:10.1038/ng1841
Marfil, C. F., Camadro, E. L., and Masuelli, R. W. (2009). Phenotypic Instability and Epigenetic Variability in a Diploid Potato of Hybrid Origin, Solanum Ruiz-Lealii. BMC Plant Biol. 9, 21. doi:10.1186/1471-2229-9-21
Martínez, C., Pons, E., Prats, G., and León, J. (2004). Salicylic Acid Regulates Flowering Time and Links Defence Responses and Reproductive Development. Plant J. 37, 209–217. doi:10.1046/j.1365-313x.2003.01954.x
Martinez-Pérez, M., Aparicio, F., López-Gresa, M. P., Bellés, J. M., Sánchez-Navarro, J. A., and Pallás, V. (2017). Arabidopsis m6A Demethylase Activity Modulates Viral Infection of a Plant Virus and the m6A Abundance in its Genomic RNAs. Proc. Natl. Acad Sci. 114, 10755–10760.
Matsoukas, I. G. (2014). Interplay between Sugar and Hormone Signaling Pathways Modulate floral Signal Transduction. Front. Genet. 5, 218. doi:10.3389/fgene.2014.00218
Matzke, M. A., Matzke, A. J. M., Pruss, G. J., and Vance, V. B. (2001). RNA-based Silencing Strategies in Plants. Curr. Opin. Genet. Development 11 (2), 221–227. doi:10.1016/s0959-437x(00)00183-0
Matzke, M. A., and Mosher, R. A. (2014). RNA-directed DNA Methylation: an Epigenetic Pathway of Increasing Complexity. Nat. Rev. Genet. 15 (6), 394–408. doi:10.1038/nrg3683
Matzke, M., Kanno, T., Huettel, B., Daxinger, L., and Matzke, A. J. (2007). Targets of RNA-Directed DNA Methylation. Current Opinion in Plant Biology 10 (5), 512–519. doi:10.1016/j.pbi.2007.06.007
Mazzitelli, L., Hancock, R. D., Haupt, S., Walker, P. G., Pont, S. D. A., McNicol, J., et al. (2007). Co-ordinated Gene Expression during Phases of Dormancy Release in Raspberry (Rubus Idaeus L.) Buds. J. Exp. Bot. 58, 1035–1045. doi:10.1093/jxb/erl266
McClintock, B. (1984). The Significance of Responses of the Genome to challenge. Science 226 (4676), 792–801. doi:10.1126/science.15739260
Meijón, M., Feito, I., Valledor, L., Rodríguez, R., and Cañal, M. J. (2011). Promotion of Flowering in Azaleas by Manipulating Photoperiod and Temperature Induces Epigenetic Alterations during floral Transition. Physiologia plantarum 143, 82–92. doi:10.1111/j.1399-3054.2011.01485.x
Mirouze, M., Lieberman-Lazarovich, M., Aversano, R., Bucher, E., Nicolet, J., Reinders, J., et al. (2012). Loss of DNA Methylation Affects the Recombination Landscape in Arabidopsis. Proc. Natl. Acad. Sci. 109, 5880–5885. doi:10.1073/pnas.1120841109
Mirouze, M., and Paszkowski, J. (2011). Epigenetic Contribution to Stress Adaptation in Plants. Curr. Opin. Plant Biol. 14, 267–274. doi:10.1016/j.pbi.2011.03.004
Mondal, P., Natesh, J., Penta, D., and Meeran, S. M. (2020). Progress and Promises of Epigenetic Drugs and Epigenetic Diets in Cancer Prevention and Therapy: A Clinical Update. Semin. Cancer Biol., S1044. doi:10.1016/j.semcancer.2020.12.006
Naito, K., Zhang, F., Tsukiyama, T., Saito, H., Hancock, C. N., Richardson, A. O., et al. (2009). Unexpected Consequences of a Sudden and Massive Transposon Amplification on rice Gene Expression. Nature 461, 1130–1134. doi:10.1038/nature08479
Niederhuth, C. E., and Schmitz, R. J. (2017). Putting DNA Methylation in Context: from Genomes to Gene Expression in Plants. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1860 (1), 149–156. doi:10.1016/j.bbagrm.2016.08.009
Okada, K., Ueda, J., Komaki, M. K., Bell, C. J., and Shimura, Y. (1991). Requirement of the Auxin Polar Transport System in Early Stages of Arabidopsis Floral Bud Formation. Plant Cell 3 (7), 677–684. doi:10.1105/tpc.3.7.677
Olmedo-Monfil, V., Durán-Figueroa, N., Arteaga-Vázquez, M., Demesa-Arévalo, E., Autran, D., Grimanelli, D., et al. (2010). Control of Female Gamete Formation by a Small RNA Pathway in Arabidopsis. Nature 464, 628–632. doi:10.1038/nature08828
Overmyer, K., Tuominen, H., Kettunen, R., Betz, C., Langebartels, C., Sandermann, H., et al. (2000). Ozone-sensitive Arabidopsis Rcd1 Mutant Reveals Opposite Roles for Ethylene and Jasmonate Signaling Pathways in Regulating Superoxide-dependent Cell Death. The Plant cell 12 (10), 1849–1862. doi:10.2307/3871197
Papareddy, R. K., Páldi, K., Paulraj, S., Kao, P., Lutzmayer, S., and Nodine, M. D. (2020). Chromatin Regulates Expression of Small RNAs to Help Maintain Transposon Methylome Homeostasis in Arabidopsis. Genome Biol. 21, 251. doi:10.1186/s13059-020-02163-4
Papp, I. n., Mur, L., Dalmadi, g., Dulai, S. n., and Koncz, C. (2004). A Mutation in the Cap Binding Protein 20 Gene Confers Drought. Plant Mol. Biol. 55, 679–686. doi:10.1007/s11103-004-1680-2
Park, K., Kim, M. Y., Vickers, M., Park, J.-S., Hyun, Y., Okamoto, T., et al. (2016). DNA Demethylation Is Initiated in the central Cells of Arabidopsis and rice. Proc. Natl. Acad. Sci. USA 113, 15138–15143. doi:10.1073/pnas.1619047114
Parker, A. H., Wilkinson, S. W., and Ton, J. (2021). Epigenetics: a Catalyst of Plant Immunity against Pathogens. New Phytol., 2338–2429. doi:10.1111/nph.17699
Pavangadkar, K., Thomashow, M. F., and Triezenberg, S. J. (2010). Histone Dynamics and Roles of Histone Acetyltransferases during Cold-Induced Gene Regulation in Arabidopsis. Plant Mol. Biol. 74, 183–200. doi:10.1007/s11103-010-9665-9
Pei, H., Ma, N., Chen, J., Zheng, Y., Tian, J., Li, J., et al. (2013). Integrative Analysis of miRNA and mRNA Profiles in Response to Ethylene in Rose Petals during Flower Opening. PLoS ONE 8 (5), e64290. doi:10.1371/journal.pone.0064290
Pérez-Hormaeche, J., Potet, F., Beauclair, L., Le Masson, I., Courtial, B., Bouché, N., et al. (2008). Invasion of the Arabidopsis Genome by the Tobacco Retrotransposon Tnt1 Is Controlled by Reversible Transcriptional Gene Silencing. Plant Physiol. 147 (3), 1264–1278. doi:10.1104/pp.108.117846
Pfluger, J., and Wagner, D. (2007). Histone Modifications and Dynamic Regulation of Genome Accessibility in Plants. Curr. Opin. Plant Biol. 10 (6), 645–652. doi:10.1016/j.pbi.2007.07.013
Pikaard, C. S. (2006). Cell Biology of the Arabidopsis Nuclear siRNA Pathway for RNA-Directed Chromatin Modification. Cold Spring Harbor symposia quantitative Biol. 71, 473–480. doi:10.1101/sqb.2006.71.046
Quadrana, L., Almeida, J., Asís, R., Duffy, T., Dominguez, P. G., Bermúdez, L., et al. (2014). Natural Occurring Epialleles Determine Vitamin E Accumulation in Tomato Fruits. Nat. Commun. 5, 4027. doi:10.1038/ncomms5027
Ramirez-Prado, J. S., Piquerez, S. J. M., Bendahmane, A., Hirt, H., Raynaud, C., and Benhamed, M. (2018). Modify the Histone to Win the Battle: Chromatin Dynamics in Plant–Pathogen Interactions. Front. Plant Sci. 9–355. doi:10.3389/fpls.2018.00355
Rasia, R. M., Mateos, J., Bologna, N. G., Burdisso, P., Imbert, L., Palatnik, J. F., et al. (2010). Structure and RNA Interactions of the Plant MicroRNA Processing-Associated Protein HYL1. Biochemistry 49, 8237–8239. doi:10.1021/bi100672x
Reinhardt, D., Mandel, T., and Kuhlemeier, C. (2000). Auxin Regulates the Initiation and Radial Position of Plant Lateral Organs. Plant Cell 12 (4), 507–518. doi:10.1105/tpc.12.4.507
Regulski, M., Lu, Z., Kendall, J., Donoghue, M. T., Reinders, J., Llaca, V., et al. (2013). The Maize Methylome Influences mRNA Splice Sites and Reveals Widespread Paramutation-Like Switches Guided by Small RNA. Genome Res. doi:10.1101/gr.153510.112
Ríos, G., Leida, C., Conejero, A., and Badenes, M. L. (2014). Epigenetic Regulation of Bud Dormancy Events in Perennial Plants. Front. Plant Sci. 5, 247. doi:10.3389/fpls.2014.00247
Rodrigues, J. A., and Zilberman, D. (2015). Evolution and Function of Genomic Imprinting in Plants. Genes Dev. 29 (24), 2517–2531. doi:10.1101/gad.269902.115
Rohde, A., Prinsen, E., De Rycke, R., Engler, G., Van Montagu, M., and Boerjan, W. (2002). PtABI3 Impinges on the Growth and Differentiation of Embryonic Leaves during Bud Set in poplar. Plant Cell 14, 1885–1901. doi:10.1105/tpc.003186
Ruttink, T., Arend, M., Morreel, K., Storme, V., Rombauts, S., Fromm, J., et al. (2007). A Molecular Timetable for Apical Bud Formation and Dormancy Induction in poplar. Plant Cell 19, 2370–2390. doi:10.1105/tpc.107.052811
Saez, A., Apostolova, N., Gonzalez‐Guzman, M., Gonzalez‐Garcia, M. P., Nicolas, C., Lorenzo, O., et al. (2004). Gain‐of‐function and Loss‐of‐function Phenotypes of the Protein Phosphatase 2C HAB1 Reveal its Role as a Negative Regulator of Abscisic Acid Signalling. Plant J. 37 (3), 354–369. doi:10.1046/j.1365-313x.2003.01966.x
Saez, A., Rodrigues, A., Santiago, J., Rubio, S., and Rodriguez, P. L. (2008). HAB1-SWI3B Interaction Reveals a Link between Abscisic Acid Signaling and Putative SWI/SNF Chromatin-Remodeling Complexes in Arabidopsis. The Plant cell 20 (11), 2972–2988. doi:10.1105/tpc.107.056705
Sahu, P. P., Pandey, G., Sharma, N., Puranik, S., Muthamilarasan, M., and Prasad, M. (2013). Epigenetic Mechanisms of Plant Stress Responses and Adaptation. Plant Cell Rep 32 (8), 1151–1159. doi:10.1007/s00299-013-1462-x
Schaller, G. E., Bishopp, A., and Kieber, J. J. (2015). The Yin-Yang of Hormones: Cytokinin and Auxin Interactions in Plant Development. Plant Cell 27 (1), 44–63. doi:10.1105/tpc.114.133595
Schmid, M., Uhlenhaut, N. H., Godard, F., Demar, M., Bressan, R., Weigel, D., et al. (2003). Dissection of floral Induction Pathways Using Global Expression Analysis. Development 130, 6001–6012. doi:10.1242/dev.00842
Schmitz, R. J., Hong, L., Fitzpatrick, K. E., and Amasino, R. M. (2007). DICER-LIKE 1 and DICER-LIKE 3 Redundantly Act to Promote Flowering via Repression of FLOWERING LOCUS C in Arabidopsis thaliana. Genetics 176, 1359–1362. doi:10.1534/genetics.107.070649
Schönrock, N., Bouveret, R., Leroy, O., Borghi, L., Köhler, C., Gruissem, W., et al. (2006). Polycomb-group Proteins Repress the floral Activator AGL19 in the FLC-independent Vernalization Pathway. Genes Dev. 20 (12), 1667–1678. doi:10.1101/gad.377206
Seifert, F., Thiemann, A., Schrag, T. A., Rybka, D., Melchinger, A. E., Frisch, M., et al. (2018). Small RNA-Based Prediction of Hybrid Performance in maize. BMC Genomics 19. doi:10.1186/s12864-018-4708-8
Shafiq, S., Li, J., and Sun, Q. (2016). Functions of Plants Long Non-coding RNAs. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1859 (1), 155–162. doi:10.1016/j.bbagrm.2015.06.009
Sheldon, C. C., Hills, M. J., Lister, C., Dean, C., Dennis, E. S., and Peacock, W. J. (2008). Resetting of FLOWERING LOCUS C Expression After Epigenetic Repression by Vernalization. Proc. Natl. Acad. Sci. U S A. 105, 2214–2219.
Shen, J., Xie, K., and Xiong, L. (2010). Global Expression Profiling of rice microRNAs by One-Tube Stem-Loop Reverse Transcription Quantitative PCR Revealed Important Roles of microRNAs in Abiotic Stress Responses. Mol. Genet. Genomics 284 (6), 477–488. doi:10.1007/s00438-010-0581-0
Shen, Q., Lin, Y., Li, Y., and Wang, G. (2021). Dynamics of H3K27me3 Modification on Plant Adaptation to Environmental Cues. Plants 10, 1165. doi:10.3390/plants10061165
Sherman, J. D., and Talbert, L. E. (2002). Vernalization-induced Changes of the DNA Methylation Pattern in winter Wheat. Genome 45 (2), 253–260. doi:10.1139/g01-147
Shi, J., Dong, A., and Shen, W. H. (2015). Epigenetic Regulation of rice Flowering and Reproduction. Front. Plant Sci. 5, 803. doi:10.3389/fpls.2014.00803
Shikata, M., Yamaguchi, H., Sasaki, K., and Ohtsubo, N. (2012). Overexpression of Arabidopsis miR157b Induces Bushy Architecture and Delayed Phase Transition in Torenia Fournieri. Planta 236, 1027–1035. doi:10.1007/s00425-012-1649-3
Shilo, S., Melamed-Bessudo, C., Dorone, Y., Barkai, N., and Levy, A. A. (2015). DNA Crossover Motifs Associated with Epigenetic Modifications Delineate Open Chromatin Regions in Arabidopsis. Plant Cell 27, 2427–2436. doi:10.1105/tpc.15.00391
Simon, J. A., and Kingston, R. E. (2013). Occupying Chromatin: Polycomb Mechanisms for Getting to Genomic Targets, Stopping Transcriptional Traffic, and Staying Put. Mol. Cell 49, 808–824. doi:10.1016/j.molcel.2013.02.013
Slotkin, R. K., and Martienssen, R. (2007). Transposable Elements and the Epigenetic Regulation of the Genome. Nat. Rev. Genet. 8 (4), 272–285. doi:10.1038/nrg2072
Slotkin, R. K., Vaughn, M., Borges, F., Tanurdžić, M., Becker, J. D., Feijó, J. A., et al. (2009). Epigenetic Reprogramming and Small RNA Silencing of Transposable Elements in Pollen. Cell 136, 461–472. doi:10.1016/j.cell.2008.12.038
Song, Q., Zhang, T., Stelly, D. M., and Chen, Z. J. (2017). Epigenomic and Functional Analyses Reveal Roles of Epialleles in the Loss of Photoperiod Sensitivity during Domestication of Allotetraploid Cottons.
Song, Y., Ji, D., Li, S., Wang, P., Li, Q., and Xiang, F. (2012). The Dynamic Changes of DNA Methylation and Histone Modifications of Salt Responsive Transcription Factor Genes in Soybean. PLoS ONE 7, e41274. doi:10.1371/journal.pone.0041274
Soppe, W. J. J., Viñegra de la Torre, N., and Albani, M. C. (2021). The Diverse Roles of FLOWERING LOCUS C in Annual and Perennial Brassicaceae Species. Front. Plant Sci. 12, 627258. doi:10.3389/fpls.2021.627258
Steward, N., Ito, M., Yamaguchi, Y., Koizumi, N., and Sano, H. (2002). Periodic DNA Methylation in maize Nucleosomes and Demethylation by Environmental Stress. J. Biol. Chem. 277 (40), 37741–37746. doi:10.1074/jbc.M204050200
Stroud, H., Do, T., Du, J., Zhong, X., Feng, S., Johnson, L., et al. (2014). Non-CG Methylation Patterns Shape the Epigenetic Landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21 (1), 64–72. doi:10.1038/nsmb.2735
Sun, C., Chen, D., Fang, J., Wang, P., Deng, X., and Chu, C. (2014). Understanding the Genetic and Epigenetic Architecture in Complex Network of rice Flowering Pathways. Protein Cell 5 (12), 889–898. doi:10.1007/s13238-014-0068-6
Sun, Y., Fan, M., and He, Y. (2019). DNA Methylation Analysis of the Citrullus lanatus Response to Cucumber GreenMottle Mosaic Virus Infection by Whole-Genome Bisulfite Sequencing. Genes 10 (5), 344. doi:10.3390/genes10050344
Sung, S., and Amasino, R. M. (2004). Vernalization in Arabidopsis thaliana Is Mediated by the PHD finger Protein VIN3. Nature 427 (6970), 159–164. doi:10.1038/nature02195
Sunkar, R., and Zhu, J.-K. (2004). Novel and Stress-Regulated MicroRNAs and Other Small RNAs from Arabidopsis[W]. The Plant cell 16 (8), 2001–2019. doi:10.1105/tpc.104.022830
Swiezewski, S., Liu, F., Magusin, A., and Dean, C. (2009). Cold-induced Silencing by Long Antisense Transcripts of an Arabidopsis Polycomb Target. Nature 462 (7274), 799–802. doi:10.1038/nature08618
Taji, T., Ohsumi, C., Iuchi, S., Seki, M., Kasuga, M., Kobayashi, M., et al. (2002). Important Roles of Drought- and Cold-Inducible Genes for Galactinol Synthase in Stress Tolerance in Arabidopsis thaliana. Plant J. 29 (4), 417–426. doi:10.1046/j.0960-7412.2001.01227.x
Takeda, S., Sugimoto, K., Kakutani, T., and Hirochika, H. (2001). Linear DNA Intermediates of the Tto1 Retrotransposon in Gag Particles Accumulated in Stressed Tobacco and Arabidopsis thaliana. Plant J. : Cel. Mol. Biol. 28 (3), 307–317. doi:10.1046/j.1365-313x.2001.01151.x
Tamada, Y., Yun, J. Y., Woo, S. C., and Amasino, R. M. (2009). ARABIDOPSIS TRITHORAX-RELATED7 is Required for Methylation of Lysine 4 of Histone H3 and for Transcriptional Activation of FLOWERING LOCUS C. The Plant cell. 21 (10), 3257–3269. doi:10.1105/tpc.109.070060
Tanaka, N., Itoh, H., Sentoku, N., Kojima, M., Sakakibara, H., Izawa, T., et al. (2011). TheCOP1OrthologPPSRegulates the Juvenile-Adult and Vegetative-Reproductive Phase Changes in Rice. Plant Cell 23, 2143–2154. doi:10.1105/tpc.111.083436
Tang, X., Wang, Q., Yuan, H., and Huang, X. (2018). Chilling-induced DNA Demethylation Is Associated with the Cold Tolerance of Hevea Brasiliensis. BMC Plant Biol. 18, 70. doi:10.1186/s12870-018-1276-7
Tani, E., Polidoros, A. N., Nianiou-Obeidat, I., and Tsaftaris, A. S. (2005). DNA Methylation Patterns are Differentially Affected by Planting Density in Maize Inbreds and Their Hybrids. Maydica 50, 19–23.
Telias, A., Lin-Wang, K., Stevenson, D. E., Cooney, J. M., Hellens, R. P., Allan, A. C., et al. (2011). Apple Skin Patterning Is Associated with Differential Expression of MYB10. BMC Plant Biol. 11, 93. doi:10.1186/1471-2229-11-93
Teotia, S., and Tang, G. (2015). To Bloom or Not to Bloom: Role of MicroRNAs in Plant Flowering. Mol. Plant 8, 359–377. doi:10.1016/j.molp.2014.12.018
Thapa, B., and Shrestha, A. (2020). Epigenetic Mechanisms and its Role in Plant Growth and Development. J. Plant Biochem. Physiol. 8, 255.
Tian, L., and Chen, Z. J. (2001). Blocking Histone Deacetylation in Arabidopsis Induces Pleiotropic Effects on Plant Gene Regulation and Development. Proc. Natl. Acad. Sci. 98, 200–205. doi:10.1073/pnas.98.1.200
Tiwari, R., and Rajam, M. V. (2022). RNA- and miRNA-Interference to Enhance Abiotic Stress Tolerance in Plants. J. Plant Biochem. Biotechnol. doi:10.1007/s13562-022-00770-9
To, T. K., Kim, J.-M., Matsui, A., Kurihara, Y., Morosawa, T., Ishida, J., et al. (2011). Arabidopsis HDA6 Regulates Locus-Directed Heterochromatin Silencing in Cooperation with MET1. Plos Genet. 7 (4), e1002055. doi:10.1371/journal.pgen.1002055
Trap-Gentil, M.-V., Hébrard, C., Lafon-Placette, C., Delaunay, A., Hagège, D., Joseph, C., et al. (2011). Time Course and Amplitude of DNA Methylation in the Shoot Apical Meristem Are Critical Points for Bolting Induction in Sugar Beet and Bolting Tolerance between Genotypes. J. Exp. Bot. 62, 2585–2597. doi:10.1093/jxb/erq433
Urano, K., Kurihara, Y., Seki, M., and Shinozaki, K. (2010). 'Omics' Analyses of Regulatory Networks in Plant Abiotic Stress Responses. Curr. Opin. Plant Biol. 13 (2), 132–138. doi:10.1016/j.pbi.2009.12.006
van Dijk, K., Ding, Y., Malkaram, S., Riethoven, J.-J. M., Liu, R., Yang, J., et al. (2010). Dynamic Changes in Genome-wide Histone H3 Lysine 4 Methylation Patterns in Response to Dehydration Stress in Arabidopsis thaliana. BMC Plant Biol. 10, 238. doi:10.1186/1471-2229-10-238
Waddington, C. H. (1957). The Strategy of the Genes. A Discussion of Some Aspects of Theoretical Biology. London: George Allen & Unwin.
Wang, C., Wang, C., Xu, W., Zou, J., Qiu, Y., Kong, J., et al. (2018). Epigenetic Changes in the Regulation of Nicotiana Tabacum Response to Cucumber Mosaic Virus Infection and Symptom Recovery through Single-Base Resolution Methylomes. Viruses 10, 402. doi:10.3390/v10080402
Wang, Y., Xue, X., Zhu, J. K., and Dong, J. (2016). Demethylation of ERECTA Receptor Genes by IBM1 Histone Demethylase Affects Stomatal Development. Development 143, 4452–4461. doi:10.1242/dev.129932
Wang, Y., Li, L., Ye, T., Lu, Y., Chen, X., and Wu, Y. (2013). The Inhibitory Effect of ABA on floral Transition Is Mediated by ABI5 in Arabidopsis. J. Exp. Bot. 64 (2), 675–684. doi:10.1093/jxb/ers361
Wibowo, A., Becker, C., Marconi, G., Durr, J., Price, J., Hagmann, J., et al. (2016). Hyperosmotic Stress Memory in Arabidopsis Is Mediated by Distinct Epigenetically Labile Sites in the Genome and Is Restricted in the Male Germline by DNA Glycosylase Activity. eLife 5, e13546. doi:10.7554/eLife.13546
Wu, K., Zhang, L., Zhou, C., Yu, C.-W., and Chaikam, V. (2008). HDA6 Is Required for Jasmonate Response, Senescence and Flowering in Arabidopsis. J. Exp. Bot. 59 (2), 225–234. doi:10.1093/jxb/erm300
Wu, L., Liu, D., Wu, J., Zhang, R., Qin, Z., Liu, D., et al. (2013). Regulation of FLOWERING LOCUS T by a microRNA in Brachypodium Distachyon. The Plant Cell 25, 4363–4377. doi:10.1105/tpc.113.118620
Wu, M. F., Yamaguchi, N., Xiao, J., Bargmann, B., Estelle, M., Sang, Y., et al. (2015). Auxin-regulated Chromatin Switch Directs Acquisition of Flower Primordium Founder Fate. eLife 4, e09269. doi:10.7554/eLife.09269
Wu, R.-M., Walton, E. F., Richardson, A. C., Wood, M., Hellens, R. P., and Varkonyi-Gasic, E. (2012). Conservation and Divergence of Four Kiwifruit SVP-like MADS-Box Genes Suggest Distinct Roles in Kiwifruit Bud Dormancy and Flowering. J. Exp. Bot. 63, 797–807. doi:10.1093/jxb/err304
Xiao, D., Zhou, K., Yang, X., Yang, Y., Ma, Y., and Wang, Y. (2021). Crosstalk of DNA Methylation Triggered by Pathogen in Poplars with Different Resistances. Front. Microbiol. 12, 750089. doi:10.3389/fmicb.2021.750089
Xie, H., Konate, M., Sai, N., Tesfamicael, K. G., Cavagnaro, T., Gilliham, M., et al. (2017). Global DNA Methylation Patterns Can Play a Role in Defining Terroir in Grapevine (Vitis vinifera Cv. Shiraz). Front. Plant Sci. 8, 1860. doi:10.3389/fpls.2017.01860
Xie, K., Shen, J., Hou, X., Yao, J., Li, X., Xiao, J., et al. (2012). Gradual Increase of miR156 Regulates Temporal Expression Changes of Numerous Genes during Leaf Development in Rice. Plant Physiol. 158, 1382–1394. doi:10.1104/pp.111.190488
Xu, G., Lyu, J., Li, Q., Liu, H., Wang, D., Zhang, M., et al. (2020). Evolutionary and Functional Genomics of DNA Methylation in maize Domestication and Improvement. Nat. Commun. 11, 5539. doi:10.1038/s41467-020-19333-4
Xu, J., Chen, G., Hermanson, P. J., Xu, Q., Sun, C., Chen, W., et al. (2019). Population-level Analysis Reveals the Widespread Occurrence and Phenotypic Consequence of DNA Methylation Variation Not Tagged by Genetic Variation in maize. Genome Biol. 20, 243. doi:10.1186/s13059-019-1859-0
Xue, X.-Y., Zhao, B., Chao, L.-M., Chen, D.-Y., Cui, W.-R., Mao, Y.-B., et al. (2014). Interaction between Two Timing microRNAs Controls Trichome Distribution in Arabidopsis. Plos Genet. 10, e1004266. doi:10.1371/journal.pgen.1004266
Yaish, M. W., Colasanti, J., and Rothstein, S. J. (2011). The Role of Epigenetic Processes in Controlling Flowering Time in Plants Exposed to Stress. J. Exp. Bot. 62, 3727–3735. doi:10.1093/jxb/err177
Yaish, M. W. (2013). “DNA Methylation-Associated Epigenetic Changes in Stress Tolerance of Plants,” in Molecular Stress Physiology of Plants. Editors G. Rout, and A. Das (New Delhi: Springer), 427–440. doi:10.1007/978-81-322-0807-5_17
Yamagishi, K., and Kikuta, Y. (2021). Nucleoside Derivatives of 5-methylcytosine Suppress 5-Azacytidine-Induced Reactivation of a Silent Transgene in Suspension-Cultured Tobacco Cells. Plant Biotechnol. 38, 173–178. doi:10.5511/plantbiotechnology.20.1126b
Yamamuro, C., Miki, D., Zheng, Z., Ma, J., Wang, J., Yang, Z., et al. (2014). Overproduction of Stomatal Lineage Cells in Arabidopsis Mutants Defective in Active DNA Demethylation. Nat. Commun. 5, 4062. doi:10.1038/ncomms5062
Yamane, H., Ooka, T., Jotatsu, H., Hosaka, Y., Sasaki, R., and Tao, R. (2011). Expressional Regulation of PpDAM5 and PpDAM6, Peach (Prunus Persica) Dormancy-Associated MADS-Box Genes, by Low Temperature and Dormancy-Breaking Reagent Treatment. J. Exp. Bot. 62, 3481–3488. doi:10.1093/jxb/err028
Yang, X., Sanchez, R., Kundariya, H., Maher, T., Dopp, I., Schwegel, R., et al. (2020). Segregation of an MSH1 RNAi Transgene Produces Heritable Non-genetic Memory in Association with Methylome Reprogramming. Nat. Commun. 11, 2214–2217. doi:10.1038/s41467-020-16036-8
Yelina, N. E., Lambing, C., Hardcastle, T. J., Zhao, X., Santos, B., and Henderson, I. R. (2015). DNA Methylation Epigenetically Silences Crossover Hot Spots and Controls Chromosomal Domains of Meiotic Recombination in Arabidopsis. Genes Dev. 29, 2183–2202. doi:10.1101/gad.270876.115
Yu, A., Lepère, G., Jay, F., Wang, J., Bapaume, L., Yu Wang, Y., et al. (2013). Dynamics and Biological Relevance of DNA Demethylation in Arabidopsis Antibacterial Defense. Proc. Natl. Acad. Sci. 110, 2389–2394. doi:10.1073/pnas.1211757110
Yu, C.-W., Liu, X., Luo, M., Chen, C., Lin, X., Tian, G., et al. (2011). HISTONE DEACETYLASE6 Interacts with FLOWERING LOCUS D and Regulates Flowering in Arabidopsis. Plant Physiol. 156 (1), 173–184. doi:10.1104/pp.111.174417
Yuan, W., Luo, X., Li, Z., Yang, W., Wang, Y., Liu, R., et al. (2016). A Cis Cold Memory Element and a Trans Epigenome Reader Mediate Polycomb Silencing of FLC by Vernalization in Arabidopsis. Nat. Genet. 48, 1527–1534. doi:10.1038/ng.3712
Zemach, A., Kim, M. Y., Hsieh, P.-H., Coleman-Derr, D., Eshed-Williams, L., Thao, K., et al. (2013). The Arabidopsis Nucleosome Remodeler DDM1 Allows DNA Methyltransferases to Access H1-Containing Heterochromatin. Cell 153 (1), 193–205. doi:10.1016/j.cell.2013.02.033
Zhai, J., Zhang, H., Arikit, S., Huang, K., Nan, G.-L., Walbot, V., et al. (2015). Spatiotemporally Dynamic, Cell-type-dependent Premeiotic and Meiotic phasiRNAs in maize Anthers. Proc. Natl. Acad. Sci. USA 112, 3146–3151. doi:10.1073/pnas.1418918112
Zhang, H., Lang, Z., and Zhu, J. K. (2018). Dynamics and Function of DNA Methylation in Plants. Nat. Rev. Mol. Cell Biol 19, 489–506. doi:10.1038/s41580-018-0016-z
Zhang, J.-F., Yuan, L.-J., Shao, Y., Du, W., Yan, D.-W., and Lu, Y.-T. (2008). The Disturbance of Small RNA Pathways Enhanced Abscisic Acid Response and Multiple Stress Responses in Arabidopsis. Plant Cell Environ 31, 562–574. doi:10.1111/j.1365-3040.2008.01786.x
Zhang, M., Zhao, H., Xie, S., Chen, J., Xu, Y., Wang, K., et al. (2011). Extensive, Clustered Parental Imprinting of Protein-Coding and Noncoding RNAs in Developing maize Endosperm. Proc. Natl. Acad. Sci. 108, 20042–20047. doi:10.1073/pnas.1112186108
Zhang, Y. Y., Fischer, M., Colot, V., and Bossdorf, O. (2013). Epigenetic Variation Creates Potential for Evolution of Plant Phenotypic Plasticity. New Phytol. 197, 314–322. doi:10.1111/nph.12010
Zhao, M., Liu, W., Xia, X., Wang, T., and Zhang, W.-H. (2014). Cold Acclimation-Induced Freezing Tolerance ofMedicago Truncatulaseedlings Is Negatively Regulated by Ethylene. Physiol. Plantarum 152 (1), 115–129. doi:10.1111/ppl.12161
Zhao, T., Zhan, Z., and Jiang, D. (2019). Histone Modifications and Their Regulatory Roles in Plant Development and Environmental Memory. J. Genet. Genomics 46, 467–476. doi:10.1016/j.jgg.2019.09.005
Zheng, X. Y., and Gehring, M. (2019). Low-Input Chromatin Profiling in Arabidopsis Endosperm Using CUT&RUN. Plant Reprod 32, 63–75. doi:10.1007/s00497-018-00358-1
Zhong, W., Gao, Z., Zhuang, W., Shi, T., Zhang, Z., and Ni, Z. (2013a). Genome-wide Expression Profiles of Seasonal Bud Dormancy at Four Critical Stages in Japanese Apricot. Plant Mol. Biol. 83, 247–264. doi:10.1007/s11103-013-0086-4
Zhong, W., Gao, Z., Zhuang, W., Shi, T., Zhang, Z., and Ni, Z. (2013). Genome-wide Expression Profiles of Seasonal Bud Dormancy at Four Critical Stages in Japanese Apricot. Plant Mol. Biol. 83, 247–264. doi:10.1007/s11103-013-0086-4
Zhou, C., Zhang, L., Duan, J., Miki, B., and Wu, K. (2005). HISTONE DEACETYLASE19 Is Involved in Jasmonic Acid and Ethylene Signaling of Pathogen Response in Arabidopsis. Plant Cell 17 (4), 1196–1204. doi:10.1105/tpc.104.028514
Zhou, D.-X. (2009). Regulatory Mechanism of Histone Epigenetic Modifications in Plants. Epigenetics 4 (1), 15–18. doi:10.4161/epi.4.1.7371
Zhou, S., Liu, X., Zhou, C., Zhou, Q., Zhao, Y., Li, G., et al. (2016). Cooperation between the H3K27me3 Chromatin Mark and Non-CG Methylation in Epigenetic Regulation. Plant Physiol. 172, 1131–1141. doi:10.1104/pp.16.01238
Zhu, H., Xia, R., Zhao, B., An, Y.-Q., Dardick, C. D., Callahan, A. M., et al. (2012). Unique Expression, Processing Regulation, and Regulatory Network of Peach (Prunus Persica) miRNAs. BMC Plant Biol. 12, 149. doi:10.1186/1471-2229-12-149
Zhu, Q.-H., Upadhyaya, N. M., Gubler, F., and Helliwell, C. A. (2009). Over-expression of miR172 Causes Loss of Spikelet Determinacy and floral Organ Abnormalities in rice (Oryza Sativa). BMC Plant Biol. 9, 149. doi:10.1186/1471-2229-9-149
Keywords: epigenetic modification, DNA methylation, histone proteins, stress response, plant hormones, transposon silencing, miRNA
Citation: Kumari P, Khan S, Wani IA, Gupta R, Verma S, Alam P and Alaklabi A (2022) Unravelling the Role of Epigenetic Modifications in Development and Reproduction of Angiosperms: A Critical Appraisal. Front. Genet. 13:819941. doi: 10.3389/fgene.2022.819941
Received: 22 November 2021; Accepted: 14 February 2022;
Published: 18 May 2022.
Edited by:
Sajad Ahmad Dar, Jazan University, Saudi ArabiaCopyright © 2022 Kumari, Khan, Wani, Gupta, Verma, Alam and Alaklabi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Susheel Verma, ZXJlbXVydXNAcmVkaWZmbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.