- 1College of Food Science and Biology, Hebei University of Science and Technology, Shijiazhuang, China
- 2College of Life Sciences, Yantai University, Yantai, China
- 3School of Computer and Control Engineering, Yantai University, Yantai, China
- 4Dezhou Agricultural Technology Extension and Seed Industry Center, Dezhou, China
Powdery mildew induced by Blumeria graminis f. sp. Tritici (Bgt) has a devastating impact on global wheat yield and quality. Host resistance is the most effective and economical means to control this disease. In this study, Heng 4568, an elite wheat cultivar, shows high resistance to 12 Bgt isolates from different regions in China at the seedling stage. Genetic analysis demonstrates that the powdery mildew resistance in Heng 4568 is conferred by a single dominant locus, temporarily designated PmH4568. Furthermore, PmH4568 is mapped to the reported Pm2 interval on chromosome 5DS with five Pm2 linked markers and flanked by the markers Bwm20 and Bwm21 with a genetic distance of 0.3 and 0.6 cM, respectively. To further investigate the relationship between PmH4568 and Pm2, the diagnostic marker Pm2b-map-3 of Pm2 is used to genotype the F2:3 population derived from the cross Heng 4568 × Daimai 2173. Notably, there is no recombination found, indicating that PmH4568 is also probably a Pm2 allele. In addition, five closely linked markers as well as one diagnostic marker are successfully developed and tested in 16 wheat cultivars from different agro-ecological areas in China, which have potential applications in molecular breeding by marker-assisted selection.
Introduction
Wheat powdery mildew incited by the biotrophic fungus Blumeria graminis f. sp. tritici (Bgt) is a foliar disease worldwide (Wu et al., 2019). The rapid spread of powdery mildew will cause severe wheat yield losses in a short time, especially in the winter wheat-growing regions with high inputs of irrigation and fertilizers (Luo et al., 2009; Hao et al., 2014). In China alone, the area of winter wheat affected annually by powdery mildew has exceeded 6 mha during recent decades, causing 300,000 tons of crop loss each year (http://cb.natesc.gov.cn/sites/cb/). With the climate getting warmer, the epidemics of wheat powdery mildew in China are growing more severe, which will always be a serious threat to national food security.
Given the significant yield-limiting effects of powdery mildew, the research and exploration of effective prevention and control technology has become urgent in wheat production. Currently, chemical control, biological control, and cultivation of disease-resistant varieties are common means. Chemical control is mainly by spraying fungicides to kill the Bgt isolates; however, it always pollutes the environment and accelerates variation of the Bgt isolates (Ma et al., 2018, 2019). Biological control mainly relies on some natural beneficial microorganisms and/or some existing substances in nature, to act as a natural antagonist of pathogen to resist other plant pathogen (Curtis et al., 2012). In comparison, host resistance is relatively the most effective, economical, and environmental way to control wheat powdery mildew, including broadening wheat resistance sources, polymerizing disease-resistant genes, and spreading disease-resistant cultivars (Felsenstein et al., 2010; Ma et al., 2018, Ma et al., 2019).
To date, more than 100 powdery mildew resistance (Pm) genes/alleles have been identified at 63 loci in wheat and its relatives (Li et al., 2019; McIntoshet al., 2019; He et al., 2021a; He et al., 2021b). Although several Pm genes have been widely used in production and provided high protection at both seedling and adult plant stages, more and more Pm genes are no longer effective against powdery mildew due to virulent mutants of the Bgt isolates (Xiao et al., 2013). Therefore, it is necessary to increase the genetic diversity of the resistant genes and characterize more effective alleles in wheat germplasms.
When a Pm gene was identified, its utilization efficiency in wheat production was mainly decided by its effectiveness and the agronomic performance of its donor (Zhao et al., 2013; Ma et al., 2018). There are some reports that several genes cannot be easily used for genetic improvement of powdery mildew resistance because of the linkage drag, competition drag, and adverse pleiotropism (Jia et al., 2020). One example is the gene Pm16, which is able to provide high resistance to different Bgt isolates, but will cause severe reduction of 15% in yield (Summers and Brown, 2013). Usually, commercial wheat cultivars have excellent agronomic performance without significantly bad traits and could be used as donors of valuable genes. In fact, several Pm genes have been identified from the cultivars with broad-spectrum resistance, such as PmJM23 from Jimai 23 (Jia et al., 2020), Pm52 from Liangxing 99 (Zhao et al., 2013), PmW14 from Wennong 14 (Song et al., 2014), and PmTm4 from Tangmai 4 (Xie et al., 2017). Therefore, characterization of powdery mildew resistance in the elite cultivars is important for isolating the underlying genes, which could be rationally used in breeding.
Marker-assisted selection (MAS) has enormous potential to improve the efficiency and precision in wheat breeding. Compared to the conventional breeding, MAS can combine several functional alleles from several individuals into one single genotype more precisely, with less unintentional losses and in fewer selection cycles (Xu and Crouch. 2008; Jiang et al., 2017). To perform MAS, closely linked molecular markers play a key role in tracing the targeted genes in the breeding population. Up to now, many molecular markers closely linked to Pm genes have been developed for MAS and efficiently used in different genetic backgrounds, thereby generating a large number of wheat breeding lines or resistant cultivars (Ma et al., 2018; Shah et al., 2018; Yu et al., 2018; Ye et al., 2019; Jia et al., 2020; Zhang et al., 2021). For instance, using tightly linked markers to Pm2 and Pm21 or co-segregate with Pm4a, three two-gene combinations, namely, Pm2 + Pm4a, Pm2 + Pm21, and Pm4a + Pm21, were successfully transferred into the commercial wheat cultivar “Yangmai 158” and double homozygotes were selected from the F2 population (Liu et al., 2000). In addition, MAS was also applied for other disease resistant in wheat, such as Fusarium head blight and stripe rust (Steiner et al., 2017; Nsabiyera et al., 2018; Su et al., 2018; Randhawa et al., 2019; Maré et al., 2020).
Heng 4568, an elite wheat cultivar, shows high resistance to powdery mildew. A previous study indicated that Heng 4568 most likely carries the known Pm52 inherited from Liangxing 99 (Zou et al., 2017). Notably, Heng 4568 showed a broader resistant spectrum to different Bgt isolates than Liangxing 99 in our evaluation of disease resistance. Therefore, the objectives of this study include (1) analyzing its powdery mildew resistance using different Bgt isolates, (2) clarifying the presence of other Pm genes in Heng 4568 besides Pm52, and (3) developing molecular markers of the new identified Pm gene for MAS.
Materials and Methods
Plant Materials
The winter wheat cultivar Heng 4568 crossed by Hengyou 18 and Liangxing 99 was provided by the Institute of Dry Farming Agriculture, Hebei Academy of Agriculture and Forestry Sciences, and used as the donor of resistant gene(s) against powdery mildew. Wheat cultivar Daimai 2173 served as a susceptible parent that was crossed with Heng 4568 to produce F1 hybrids, F2 population, and F2:3 families for genetic analysis and molecular mapping of the Pm gene(s) in Heng 4568. Five wheat cultivars/lines with known Pm genes, namely, Liangxing99 (Pm52), Wennong14 (PmW14), Zhongmai155 (PmZ155), Jimai22 (Pm52 + PmJM23), and Ulka/8*Cc (Pm2a), were used to compare their reaction patterns to different Bgt isolates with that of Heng 4568. Susceptible wheat cultivar Huixianhong was used as the susceptible control for phenotypic assessment. Sixteen susceptible wheat cultivars from different ecological regions in China (Hebei, Shandong, Henan, Shaanxi, Beijing, Anhui, and Jiangsu provinces in China) were used to evaluate the availability of closely linked markers for MAS.
Assessment of Disease Resistance at the Seedling Stage
From 2019 to 2021, the assessment of disease resistance at the seedling stage was carried out in the greenhouse at Hebei University of Science and Technology (Shijiazhuang, China). Twelve Bgt isolates were collected from different wheat production regions in China. They were used to determine the reaction patterns of Heng 4568 and wheat genotypes with known Pm genes. For each Bgt isolate, at least 20 seeds for each genotype were sown in 128-cell rectangular trays in a growth chamber. The susceptible control Huixianhong was randomly planted in the tray. When the first leaves were unfolded, the seedlings were inoculated by Bgt conidiospores that were previously increased on Huixianhong seedlings. Then, the inoculated seedlings were incubated in an airtight dark environment for 24 h and then allowed disease symptom development in a greenhouse with a daily cycle of 14 h of light at 22°C and 10 h of darkness at 18°C (Qu et al., 2020). To perform genetic analysis, Bgt isolate KD07 was selected to inoculate seeding of Heng 4568, the susceptible cultivar Daimai 2173, and their F1, F2, and F2:3 progenies. For F1 hybrids, 10 plants were sown; for F2 population, 177 plants were sown; for F2:3 families, 172 families and 20–30 plants per family were sown. Two weeks after inoculation when the spores were fully developed on the susceptible controls, infection types (ITs) on the primary leaves of plants were rated with a scale of 0, 0;, 1, 2, 3, and 4. The leaves that displayed ITs 0–2 and 3–4 were regarded as resistant and susceptible, respectively (Liu et al., 1999). Three repeated experiments were carried out using the same procedure.
Molecular Marker Analysis
Genomic DNA was extracted from the young leaf tissues following the cetyltriethylammonium bromide method (Saghai-Maroof et al., 1984). Resistant and susceptible DNA bulks were created by separately mixing equal amount of DNA from 10 homozygously resistant and 10 homozygously susceptible plants, respectively. Forty-eight molecular markers closely linked to 37 known Pm genes were firstly screened for their polymorphisms between Heng 4568, Daimai 2173, and their derived resistant and susceptible bulks (Supplementary Table S1). When the Pm gene in Heng 4568 was preliminarily to Pm2 locus, other Pm2 linked markers Cfd81, Bwm20, Bwm21, Bwm25, and Swgi067 (Lu et al., 2015; Ma et al., 2018) and the diagnostic marker Pm2b-map-3 (Jin et al., 2021) were also used to add the marker density for conducting the linkage map (Supplementary Table S2). PCR was performed in a 10-μl reaction volume containing 1 µl of 40–50 ng/μl template genomic DNA, 4.5 µl of 2×Taq Master Mix (Vazyme, China), and 0.5 µl of 10 μM/μl primer mix. The PCR program used was 95°C for 5 min; 36 cycles of 95°C for 30 s, 50–60°C (depending on specific primers) for 40 s, final extension at 72°C for 5 min; and storage at 4°C. PCR products were separated in 8.0% nondenaturing polyacrylamide gels with 29:1 ratio of acrylamide and bis-acrylamide with 1 × TBE buffer and then silver-stained and visualized as previously described (Santos et al., 1993).
Statistical Analysis and Linkage Map Construction
After confirming genotypes of F2:3 families of Heng 4568 and Daimai 2173, the deviations of the observed phenotypic data from theoretically expected segregation ratios for goodness of fit were assessed using χ2 test. MAPMAKER 3.0 and the Kosambi function were performed to construct the linkage map of the powdery mildew resistance gene in Heng 4568.
Results
Evaluation of Powdery Mildew Resistance in Heng 4568
Heng 4568 was highly resistant to 12 Bgt isolates with the ITs 0–2, whereas Daimai 2173 and susceptible control Huixianhong were all highly susceptible to all the tested Bgt isolates (Table 1). Compared with the Pm52 donor Liangxing 99, Heng 4568 was resistant to the Bgt isolates KD03, KD07, KD08, and KD11, while Pm52 was susceptible to these four Bgt isolates, indicating that Heng 4568 contains other Pm gene(s).
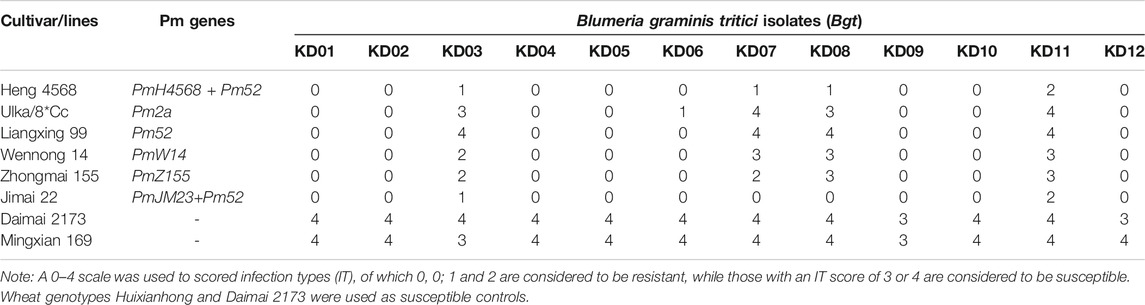
TABLE 1. Infection types of Heng 4568 and other genotypes with Pm2 alleles to 12 Blumeria graminis f. sp. tritici (Bgt) isolates at the seedling stage.
Genetic Analysis of Pm Genes in Heng 4568
To explore other Pm gene(s) besides Pm52 in Heng 4568, the isolate KD07 virulent to Liangxing 99 (with Pm52) and avirulent to Heng 4568 was selected to inoculate Heng 4568, Daimai 2173, and their derived F1 seeds, F2 population, and F2:3 families, respectively. All the tested F1 seedlings were resistant to KD07 similar to their parent Heng 4568. The F2 population fitted the segregation ratio of a single dominant gene (Table 2). The harvested F2:3 families from the F2 population confirmed the expected ratio of 1:2:1 (Table 2). Therefore, it was concluded that another dominant Pm gene is also involved in Heng 4568, which was temporarily designated PmH4568.

TABLE 2. Segregation ratios of F2 and F2:3 generations of Heng 4568 × Daimai 2173 following inoculation with Blumeria graminis f. s. tritici (Bgt) isolate KD07 at the seedling stage.
Molecular Mapping of PmH4568
To determine the genetic location of PmH4568, 48 molecular markers closely linked to the known Pm genes were firstly used to test their polymorphisms between the parents and the two contrasting bulks. The Pm2-linked marker Cfd81 showed consistent polymorphism between Heng 4568, Daimai 2173, and their derived contrasting bulks. Then, Cfd81 was used to genotype the F2:3 families of Heng 4568 and Daimai 2173 and confirmed its linkage relationship with PmH4568 (Figures 1, 2; Supplementary Table S2). This suggested that PmH4568 was most likely located in the Pm2 interval. To confirm this interval, four additional Pm2-linked markers, Bwm20, Bwm21, Bwm25, and Swgi067, were also proved to be closely linked to PmH4568 (Figure 1; Supplementary Table S2). A genetic linkage map was then conducted to locate PmH4568 to the Pm2 interval (Figure 2). To further confirm the relationship between PmH4568 and Pm2, Pm2b-map-3, the diagnostic marker of Pm2, was used to genotype the F2:3 families of Heng4568 and Daimai 2173 (Figure 1; Supplementary Table S2). No recombinants were found, suggesting that PmH4568 was located in the Pm2 locus and most likely a Pm2 allele.
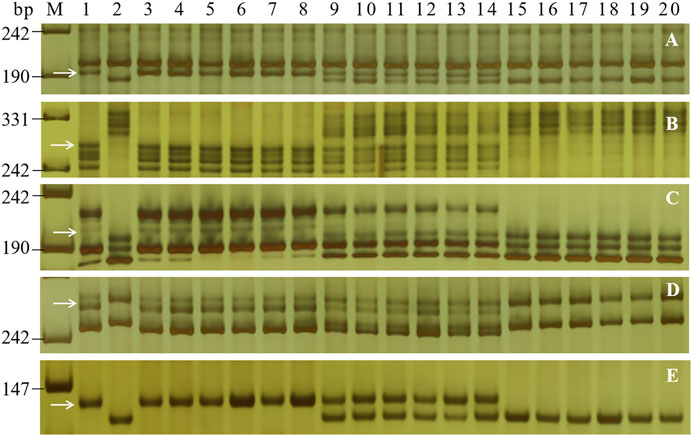
FIGURE 1. PCR amplification patterns of the selected markers Bwm20 (A), Bwm21(B), Bwm25(C), Cfd81 (D), and the diagnostic Pm2b-map-3 (E) in genotyping Heng 4568, Daimai 2173, and random selected F2:3 families of Heng 4568 × Daimai 2173. Lane M: pUC19 Msp I; lanes 1–2: Heng4568 and Daimai 2173; lanes 3–8: homozygous-resistant F2:3 families; lanes 9–14: heterozygous F2:3 families; lanes 15–20, homozygous susceptible F2:3 families. The white arrows indicate the polymorphic bands in Heng4568.
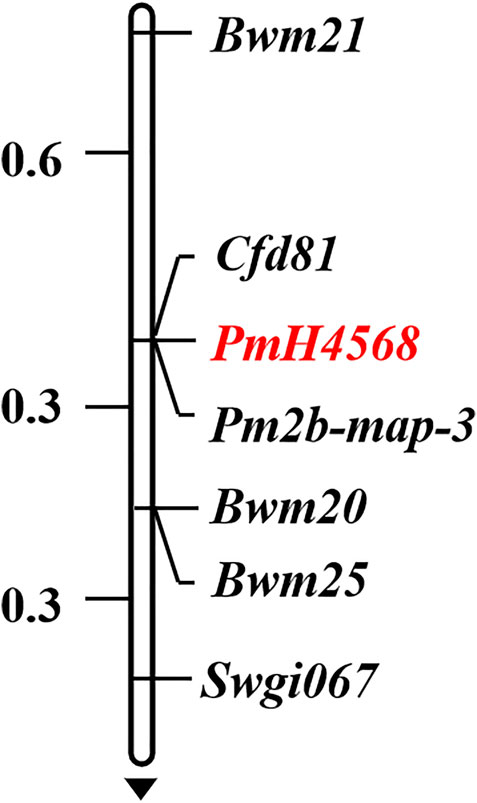
FIGURE 2. Linkage map of PmH4568 using the F2:3 families of Heng 4568 × Daimai 2173. Genetic distances in cM are showed to the left. The black arrow points to the centromere.
Evaluation of Closely Linked Markers for Marker-Assisted Selection
To transfer PmH4568 to susceptible cultivars using MAS, five PmH4568-linked markers, Bwm20, Bwm21, Bwm25, Swgi067, and Cfd81, and the diagnostic marker Pm2b-map-3, were used to test Heng 4568 and 16 susceptible cultivars. The results showed that all the tested markers could amplify polymorphic bands between Heng 4568 and these susceptible cultivars, indicating that once PmH4568 is transferred into the susceptible cultivars through conventional hybridization, these markers can be used to detect PmH4568, especially the diagnostic marker Pm2b-map-3 (Figure 3; Table 3).
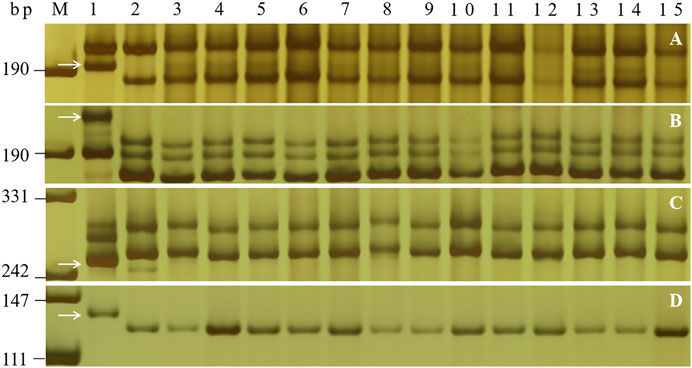
FIGURE 3. PCR amplification patterns of the markers Bwm20 (A), Bwm25 (B), Cfd81(C), and Pm2b-map-3 (D) in genotyping Heng 4568, Daimai 2173, and 13 selected wheat cultivars. Lanes M: pUC19 Msp I; lanes 1–2: Heng 4568 and Daimai 2173; lanes 3–15: Shannong 1538, Hanmai 13, Zhoumai 27, Xinong 979, Jimai 229, Jimai 21, Jimai 20, Zhongyu 9398, Womai 8, Shimai 15, Xinluo 4, Zhengmai 0856, and Wunong 6. The white arrows indicate the polymorphic bands in Heng 4568.
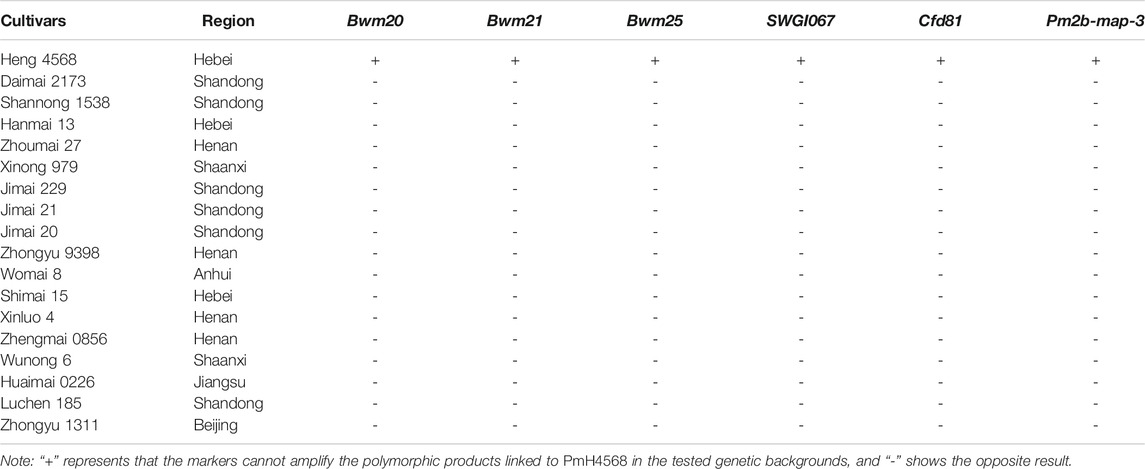
TABLE 3. Validation of PmH4568-linked and diagnostic markers on 16 Chinese wheat cultivars in marker-assisted selection (MAS) breeding.
Discussion
Heng 4568 is an elite winter wheat cultivar in Northern China. Due to its superior agronomic performance and powdery mildew resistance, Heng 4568 is considered as an attractive cultivar and serves as a favorable breeding parent for resistance improvement. A previous study indicated that the known Pm52 located on the chromosome 2BL was involved in Heng 4568, which may confer the powdery mildew resistance (Zou et al., 2017). Pm52 is a widely used Pm gene in Chinese cultivars, such as Hanong 2312, Zhongxinmai 99, Shimai 26, and DH51302. Heng 4568 was derived from the cross of Liangxing 99 with Hengyou 18, indicating that Pm52 in Heng 4568 may be derived from its parent Liangxing 99. However, our study demonstrated that Heng 4568 showed significantly broader resistant spectrum than the Pm52 donor Liangxing 99, suggesting that other Pm genes may also be involved in Heng 4568. To clarify the composition of the Pm genes in Heng 4568, a Bgt isolate virulent KD07 was used to identify other Pm gene(s) in Heng 4568. The result showed that another dominant Pm gene PmH4568 also contributed to the powdery mildew resistance in Heng 4568. This result, together with a previous study, provided an explicit genetic constitution for the powdery mildew resistance in Heng 4568, which contributes to scientific parental selection collocation.
Using Pm2-linked markers, PmH4568 was mapped to the known Pm2 interval on chromosome arm 5DS. According to previous studies, a series of Pm2 alleles have been reported in the Pm2 interval, such as Pm2a (Qiu et al., 2006), Pm2b (Ma et al., 2015a), Pm2c (Xu et al., 2015), PmLX66 (Huang et al., 2012), PmX3986-2 (Ma et al., 2014), PmWFJ (Ma et al., 2015b), PmYB (Ma et al., 2015c), PmZ155 (Sun H. et al., 2015), PmW14 (Sun Y. et al., 2015), PmWFJ (Ma et al., 2015a), PmSub (Jin et al., 2018), Pm10V-2 (Ma et al., 2018), PmJM23 (Jia et al., 2020), and PmFG (Ma et al., 2016). Using MutChromSeq (Mutant chromosome sequencing) (Sáchez-Martín et al., 2016) and analysis of the fine mapping interval (Chen et al., 2019), Pm2 was cloned and confirmed to encode a CC-NBS-LRR protein. Haplotype analysis of 48 hexaploid common wheat carrying Pm2 alleles showed that all these Pm2 donors have the perfectly consistent as the cloned sequence above. However, different Pm2 alleles derived from hexaploid common wheat have significantly different resistant spectra (Ma et al., 2018; Jia et al., 2020). This may be due to their different genetic backgrounds, and also complex genetic constitution or resistance mechanism may be involved in this interval. Anyway, Pm2 is an elite gene locus that is very valuable for resistance breeding, even though the complex Pm2 locus has not been fully characterized.
In wheat resistance breeding using MAS, the breeding potential of a certain gene depends not only on its resistance but also on the comprehensively agronomic traits, such as yield, quality, and high combining ability. Thus, although many Pm genes/alleles have been identified, only several have been widely used in breeding programs (Li et al., 2011). The main obstacle that limits the application of these genes is linkage drag in most resistance donors. After transferring these Pm genes into susceptible commercial cultivars, unfavorable traits linked to them will lead to poor agricultural yield or quality performances (Ma et al., 2015b). Therefore, the resistance donors with excellent agricultural performance are very popular for breeders. Fortunately, Heng 4568 is a cultivar with desirable comprehensively agronomic traits. For powdery mildew resistance, Heng 4568 also carries Pm52 besides the Pm2 allele PmH4568. In China, Pm2 and Pm52 are two major Pm genes in many resistance cultivars, such as Liangxing 66, Wennong 14, YingBo 700, Zhongmai 155, Jimai 23 (Jia et al., 2020), and Nongda 399, which all have the Pm2 allele, and Liangxing 99, Hannong 2312, Zhongxinmai 99, DH51302, and Zhimai 26, which all have Pm52 (Qu et al., 2020). Compared with these resistance cultivars, Heng 4568 has two resistance genes, which may show more durable resistance than a single resistance gene; such a situation also involved Jimai 22, a famous cultivar with the largest promotion area in the last 10 years (Jia et al., 2020). For the yield and quality, there was no significant defect in the recent years in our field. Particularly worth mentioning is its high combining ability in breeding; two famous wheat cultivars, Hengmai 28 and Jiamai 361, have been released in production using Heng 4568 as parent (https://www.Chinaseed114.com/seed/16/seed_77094.html; https://www.chinaseed114.com/seed/14/seed_69187.html), and in our lab, Heng 4568 is also a popular breeding parent for both resistance and yield improvement. Therefore, Heng 4568 can be not only directly popularized in region with high incidence of powdery mildew, but also used as a valuable breeding parent to improve powdery mildew resistance.
To transfer the Pm genes in Heng 4568, MAS is a rapid and effective way (William et al., 2007; Ashra and Foolad, 2013; Collard and Mackill, 2018). Since fine mapping of Pm52 has been carried out, many closely linked markers of Pm52 have been developed for MAS (Wu et al., 2019). In this study, the applicability of five closely linked markers and one diagnostic marker has been investigated in MAS with 16 susceptible wheat cultivars. In particular, the diagnostic marker Pm2b-map-3 is a functional marker designed by SNPs within the Pm2 sequence, suggesting that there is no recombinant in MAS. Therefore, resistance breeding using Heng 4568 as a parent is promising, and more trans-breeding studies using Heng 4568 are under way in our lab.
Conclusion
In this study, PmH4568, an effective Pm gene in the elite cultivar Heng 4568, has been identified and proved to be a Pm2 allele. We further clarify the genetic components of the powdery mildew resistance in Heng 4568. The applicability of closely linked markers, including the diagnostic marker, was validated in MAS. Overall, this work will accelerate the utilization of the powdery mildew resistance in Heng 4568.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
PM conceived the study. HG, XX, FL, PA, and PG performed the statistical analysis of the experiment. HG wrote the manuscript. All authors critically read, commented, and approved the final manuscript.
Funding
This research was financially supported by the Doctoral Research Start-up Fund Project of Hebei University of Science and Technology (1181438), the National Natural Science Foundation of China (32072053), the Key Research and Development Program of Shandong Province (2020CXGC010805), and Open Project Funding of the State Key Laboratory of Crop Stress Adaptation and Improvement (CX1130A0920014).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We appreciate the editor and reviewers for handling our manuscript and providing critical suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.819844/full#supplementary-material
References
Ashraf, M., and Foolad, M. R. (2013). Crop Breeding for Salt Tolerance in the Era of Molecular Markers and Marker-Assisted Selection. Plant Breed 132, 10–20. doi:10.1111/pbr.12000
Chen, F., Jia, H., Zhang, X., Qiao, L., Li, X., Zheng, J., et al. (2019). Positional Cloning of PmCH1357 Reveals the Origin and Allelic Variation of the Pm2 Gene for Powdery Mildew Resistance in Wheat. Crop J. 7, 771–783. doi:10.1016/j.cj.2019.08.004
Collard, B. C. Y., and Mackill, D. J. (2008). Marker-assisted Selection: an Approach for Precision Plant Breeding in the Twenty-First century. Phil. Trans. R. Soc. B 363, 557–572. doi:10.1098/rstb.2007.2170
De Curtis, F., De Cicco, V., and Lima, G. (2012). Efficacy of Biocontrol Yeasts Combined with Calcium Silicate or sulphur for Controlling Durum Wheat Powdery Mildew and Increasing Grain Yield Components. Field Crops Res. 134, 36–46. doi:10.1016/j.fcr.2012.04.014
Felsenstein, F., Semar, M., and Stammler, G. (2010). Sensitivity of Wheat Powdery Mildew (Blumeria Graminis f.Sp. Tritici) towards Metrafenone. Gesunde Pflanzen 62, 29–33. doi:10.1007/s10343-010-0214-x
Hao, Y., Parks, R., Cowger, C., Chen, Z., Wang, Y., Bland, D., et al. (2014). Molecular Characterization of a New Powdery Mildew Resistance Gene Pm54 in Soft Red winter Wheat. Theor. Appl. Genet. 128, 465–476. doi:10.1007/s00122-014-2445-1
He, H., Du, H., Liu, R., Liu, T., Yang, L., Gong, S., et al. (2021a). Characterization of a New Gene for Resistance to Wheat Powdery Mildew on Chromosome 1RL of Wild rye Secale Sylvestre. Theor. Appl. Genet. 134, 887–896. doi:10.1007/s00122-020-03739-1
He, H., Liu, R., Ma, P., Du, H., Zhang, H., Wu, Q., et al. (2021b). Characterization of Pm68, a New Powdery Mildew Resistance Gene on Chromosome 2BS of Greek Durum Wheat TRI 1796. Theor. Appl. Genet. 134, 53–62. doi:10.1007/s00122-020-03681-2
Huang, J., Zhao, Z., Song, F., Wang, X., Xu, H., Huang, Y., et al. (2012). Molecular Detection of a Gene Effective against Powdery Mildew in the Wheat Cultivar Liangxing 66. Mol. Breed. 30, 1737–1745. doi:10.1007/s11032-012-9757-0
Jia, M., Xu, H., Liu, C., Mao, R., Li, H., Liu, J., et al. (2020). Characterization of the Powdery Mildew Resistance Gene in the Elite Wheat Cultivar Jimai 23 and its Application in Marker-Assisted Selection. Front. Genet. 11, 241. doi:10.3389/fgene.2020.00241
Jiang, Y., Schulthess, A. W., Rodemann, B., Ling, J., Plieske, J., Kollers, S., et al. (2017). Validating the Prediction Accuracies of Marker-Assisted and Genomic Selection of Fusarium Head Blight Resistance in Wheat Using an Independent Sample. Theor. Appl. Genet. 130, 471–482. doi:10.1007/s00122-016-2827-7
Jin, Y., Shi, F., Liu, W., Fu, X., Gu, T., Han, G., et al. (2021). Identification of Resistant Germplasm and Detection of Genes for Resistance to Powdery Mildew and Leaf Rust from 2,978 Wheat Accessions. Plant Dis. doi:10.1094/PDIS-03-21-0532-RE
Jin, Y., Xu, H., Ma, P., Fu, X., Song, L., Xu, Y., et al. (2018). Characterization of a New Pm2 Allele Associated with Broad-Spectrum Powdery Mildew Resistance in Wheat Line Subtil. Sci. Rep. 8, 475. doi:10.1038/s41598-017-18827-4
Li, G., Cowger, C., Wang, X., Carver, B. F., and Xu, X. (2019). Characterization of Pm65, a New Powdery Mildew Resistance Gene on Chromosome 2AL of a Facultative Wheat Cultivar. Theor. Appl. Genet. 132, 2625–2632. doi:10.1007/s00122-019-03377-2
Li, H.-J., Wang, X.-M., Song, F.-J., Wu, C.-P., Wu, X.-F., Zhang, N., et al. (2011). Response to Powdery Mildew and Detection of Resistance Genes in Wheat Cultivars from China. A S 37, 943–954. doi:10.3724/sp.j.1006.2011.00943
Liu, J., Liu, D., Tao, W., Li, W., Wang, S., Chen, P., et al. (2000). Molecular Marker-Facilitated Pyramiding of Different Genes for Powdery Mildew Resistance in Wheat. Plant Breed. 119, 21–24. doi:10.1046/j.1439-0523.2000.00431.x
Liu, Z., Sun, Q., Ni, Z., Yang, T., and Mcintosh, R. A. (1999). Development of SCAR Markers Linked to the Pm21 Gene Conferring Resistance to Powdery Mildew in Common Wheat. Plant Breed. 118, 215–219. doi:10.1046/j.1439-0523.1999.118003215.x
Lu, Y., Wu, X., Yao, M., Zhang, J., Liu, W., Yang, X., et al. (2015). Genetic Mapping of a Putative Agropyron Cristatum-Derived Powdery Mildew Resistance Gene by a Combination of Bulked Segregant Analysis and Single Nucleotide Polymorphism Array. Mol. Breed. 35, 96. doi:10.1007/s11032-015-0292-7
Luo, P. G., Luo, H. Y., Chang, Z. J., Zhang, H. Y., Zhang, M., and Ren, Z. L. (2009). Characterization and Chromosomal Location of Pm40 in Common Wheat: a New Gene for Resistance to Powdery Mildew Derived from Elytrigia Intermedium. Theor. Appl. Genet. 118, 1059–1064. doi:10.1007/s00122-009-0962-0
Ma, P., Han, G., Zheng, Q., Liu, S., Han, F., Wang, J., et al. (2020). Development of Novel Wheat-rye Chromosome 4R Translocations and Assignment of Their Powdery Mildew Resistance. Plant Dis. 104, 260–268. doi:10.1094/PDIS-01-19-0160-RE
Ma, P., Xu, H., Li, L., Zhang, H., Han, G., Xu, Y., et al. (2016). Characterization of a New Pm2 Allele Conferring Powdery Mildew Resistance in the Wheat Germplasm Line FG-1. Front. Plant Sci. 7, 88. doi:10.3389/fpls.2016.00546
Ma, P., Xu, H., Luo, Q., Qie, Y., Zhou, Y., Xu, Y., et al. (2014). Inheritance and Genetic Mapping of a Gene for Seedling Resistance to Powdery Mildew in Wheat Line X3986-2. Euphytica 200, 149–157. doi:10.1007/s10681-014-1178-1
Ma, P., Xu, H., Xu, Y., Li, L., Qie, Y., Luo, Q., et al. (2015a). Molecular Mapping of a New Powdery Mildew Resistance Gene Pm2b in Chinese Breeding Line KM2939. Theor. Appl. Genet. 128, 613–622. doi:10.1007/s00122-015-2457-5
Ma, P., Xu, H., Xu, Y., Song, L., Liang, S., Sheng, Y., et al. (2018). Characterization of a Powdery Mildew Resistance Gene in Wheat Breeding Line 10V-2 and its Application in Marker-Assisted Selection. Plant Dis. 102, 925–931. doi:10.1094/PDIS-02-17-0199-RE
Ma, P., Xu, H., Zhang, H., Li, L., Xu, Y., Zhang, X., et al. (2015b). The Gene PmWFJ Is a New Member of the Complex Pm2 Locus Conferring Unique Powdery Mildew Resistance in Wheat Breeding Line Wanfengjian 34. Mol. Breed. 35, 210. doi:10.1007/s11032-015-0403-5
Ma, P., Zhang, H., Xu, H., Xu, Y., Cao, Y., Zhang, X., et al. (2015c). The Gene PmYB Confers Broad-Spectrum Powdery Mildew Resistance in the Multi-Allelic Pm2 Chromosome Region of the Chinese Wheat Cultivar YingBo 700. Mol. Breed. 35, 124. doi:10.1007/s11032-015-0320-7
Maré, A., Boshoff, W. H. P., and Herselman, L. (2020). Molecular Breeding of Wheat Lines for Multiple Rust and Fusarium Head Blight Resistance. Euphytica 216, 1–12. doi:10.1007/s10681-020-02697-5
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Xia, X. C., and Raupp, W. J. (2019). “Catalogue of Gene Symbols for Wheat: 2019 Supplement,” in Annual Wheat Newsletter. Editor W. J. Raupp (Manhattan, USA, p98–113.
Nsabiyera, V., Bariana, H. S., Qureshi, N., Wong, D., Hayden, M. J., and Bansal, U. K. (2018). Characterisation and Mapping of Adult Plant Stripe Rust Resistance in Wheat Accession Aus27284. Theor. Appl. Genet. 131, 1459–1467. doi:10.1007/s00122-018-3090-x
Qiu, Y., Sun, X., Zhou, R., Kong, X., Zhang, S., and Jia, J. (2006). Identification of Microsatellite Markers Linked to Powdery Mildew Resistance genePm2in Wheat. Cereal Res. Commun. 34, 1267–1273. doi:10.1556/crc.34.2006.4.268
Qu, Y.-f., Wu, P.-p., Hu, J.-h., Chen, Y.-x., Shi, Z.-l., Qiu, D., et al. (2020). Molecular Detection of the Powdery Mildew Resistance Genes in winter Wheats DH51302 and Shimai 26. J. Integr. Agric. 19, 931–940. doi:10.1016/S2095-3119(19)62644-4
Randhawa, M. S., Bains, N. S., Sohu, V. S., Chhuneja, P., Trethowan, R. M., Bariana, H. S., et al. (2019). Marker Assisted Transfer of Stripe Rust and Stem Rust Resistance Genes into Four Wheat Cultivars. Agronomy 9, 4972073–4974395. doi:10.3390/agronomy9090497
Saghai-Maroof, M. A., Soliman, K. M., Jorgensen, R. A., and Allard, R. W. (1984). Ribosomal DNA Spacer-Length Polymorphisms in Barley: Mendelian Inheritance, Chromosomal Location, and Population Dynamics. Proc. Natl. Acad. Sci. 81, 8014–8018. doi:10.1073/pnas.81.24.8014
Sánchez-Martín, J., Steuernagel, B., Ghosh, S., Herren, G., Hurni, S., Adamski, N., et al. (2016). Rapid Gene Isolation in Barley and Wheat by Mutant Chromosome Sequencing. Genome Biol. 17, 221. doi:10.1186/s13059-016-1082-1
Santos, F. c., Pena, S. J., and Epplen, J. r. (1993). Genetic and Population Study of a Y-Linked Tetranucleotide Repeat DNA Polymorphism with a Simple Non-isotopic Technique. Hum. Genet. 90, 655–656. doi:10.1007/BF00202486
Shah, L., Rehman, S., Ali, A., Yahya, M., Riaz, M. W., Si, H., et al. (2018). Genes Responsible for Powdery Mildew Resistance and Improvement in Wheat Using Molecular Marker-Assisted Selection. J. Plant Dis. Prot. 125, 145–158. doi:10.1007/s41348-017-0132-6
Song, W., Sun, H.-G., Sun, Y.-L., Zhao, Z.-H., Wang, X.-M., Wu, X.-F., et al. (2014). Chromosomal Localization of the Gene for Resistance to Powdery Mildew in the Wheat Cultivar Wennong 14. Acta Agronomica Sinica 40, 798–804. doi:10.3724/SP.J.1006.2014.00798
Steiner, B., Buerstmayr, M., Michel, S., Schweiger, W., Lemmens, M., and Buerstmayr, H. (2017). Breeding Strategies and Advances in Line Selection for Fusarium Head Blight Resistance in Wheat. Trop. Plant Pathol. 42, 165–174. doi:10.1007/s40858-017-0127-7
Su, Z., Jin, S., Zhang, D., and Bai, G. (2018). Development and Validation of Diagnostic Markers for Fhb1 Region, a Major QTL for Fusarium Head Blight Resistance in Wheat. Theor. Appl. Genet. 131, 2371–2380. doi:10.1007/s00122-018-3159-6
Summers, R. W., and Brown, J. K. M. (2013). Constraints on Breeding for Disease Resistance in Commercially Competitive Wheat Cultivars. Plant Pathol. 62, 115–121. doi:10.1111/ppa.12165
Sun, H., Song, W., Sun, Y., Chen, X., Liu, J., Zou, J., et al. (2015a). Resistance of 'Zhongmai 155' Wheat to Powdery Mildew: Effectiveness and Detection of the Resistance Gene. Crop Sci. 55, 1017–1025. doi:10.2135/cropsci2014.05.0355
Sun, Y., Zou, J., Sun, H., Song, W., Wang, X., and Li, H. (2015b). PmLX66 and PmW14: New Alleles of Pm2 for Resistance to Powdery Mildew in the Chinese Winter Wheat Cultivars Liangxing 66 and Wennong 14. Plant Dis. 99, 1118–1124. doi:10.1094/PDIS-10-14-1079-RE
William, H. M., Trethowan, R., and Crosby-Galvan, E. M. (2007). Wheat Breeding Assisted by Markers: CIMMYT's Experience. Euphytica 157, 307–319. doi:10.1007/s10681-007-9405-7
Wu, P., Hu, J., Zou, J., Qiu, D., Qu, Y., Li, Y., et al. (2019). Fine Mapping of the Wheat Powdery Mildew Resistance Gene Pm52 Using Comparative Genomics Analysis and the Chinese spring Reference Genomic Sequence. Theor. Appl. Genet. 132, 1451–1461. doi:10.1007/s00122-019-03291-7
Xiao, M., Song, F., Jiao, J., Wang, X., Xu, H., and Li, H. (2013). Identification of the Gene Pm47 on Chromosome 7BS Conferring Resistance to Powdery Mildew in the Chinese Wheat Landrace Hongyanglazi. Theor. Appl. Genet. 126, 1397–1403. doi:10.1007/s00122-013-2060-6
Xie, J.-z., Wang, L.-l., Wang, Y., Zhang, H.-z., Zhou, S.-h., Wu, Q.-h., et al. (2017). Fine Mapping of Powdery Mildew Resistance Gene PmTm4 in Wheat Using Comparative Genomics. J. Integr. Agric. 16, 540–550. doi:10.1016/S2095-3119(16)61377-1
Xu, H., Yi, Y., Ma, P., Qie, Y., Fu, X., Xu, Y., et al. (2015). Molecular Tagging of a New Broad-Spectrum Powdery Mildew Resistance Allele Pm2c in Chinese Wheat Landrace Niaomai. Theor. Appl. Genet. 128, 2077–2084. doi:10.1007/s00122-015-2568-z
Xu, Y., and Crouch, J. H. (2008). Marker‐Assisted Selection in Plant Breeding: From Publications to Practice. Crop Sci. 48, 391–407. doi:10.2135/cropsci2007.04.0191
Ye, X., Zhang, S., Li, S., Wang, J., Chen, H., Wang, K., et al. (2019). Improvement of Three Commercial spring Wheat Varieties for Powdery Mildew Resistance by Marker-Assisted Selection. Crop Prot. 125, 104889. doi:10.1016/j.cropro.2019.104889
Yu, X., Ren, S., Zhao, L., Guo, J., Bao, Y., Ma, Y., et al. (2018). Molecular Mapping of a Novel Wheat Powdery Mildew Resistance Gene Ml92145E8-9 and its Application in Wheat Breeding by Marker-Assisted Selection. Crop J. 6, 621–627. doi:10.1016/j.cj.2018.04.004
Zhang, X., Wang, W., Liu, C., Zhu, S., Gao, H., Xu, H., et al. (2021). Diagnostic Kompetitive Allele-specific PCR Markers of Wheat Broad-Spectrum Powdery Mildew Resistance Genes Pm21, PmV, and Pm12 Developed for High-Throughput Marker-Assisted Selection. Plant Dis. 105, 2844–2850. doi:10.1094/PDIS-02-21-0308-RE
Zhao, Z., Sun, H., Song, W., Lu, M., Huang, J., Wu, L., et al. (2013). Genetic Analysis and Detection of the Gene Mllx99 on Chromosome 2bl Conferring Resistance to Powdery Mildew in the Wheat Cultivar Liangxing 99. Theor. Appl. Genet. 126, 3081–3089. doi:10.1007/s00122-013-2194-6
Keywords: wheat powdery mildew, Heng 4568, molecular markers, MAS, Pm2
Citation: Gao H, Xu X, Ai P, Luo F, Guo P and Ma P (2022) Identification of the Powdery Mildew Resistance in Chinese Wheat Cultivar Heng 4568 and its Evaluation in Marker-Assisted Selection. Front. Genet. 13:819844. doi: 10.3389/fgene.2022.819844
Received: 22 November 2021; Accepted: 17 January 2022;
Published: 17 February 2022.
Edited by:
Ahmed Sallam, Assiut University, EgyptReviewed by:
Amira M. I. Mourad, Assiut University, EgyptZixian Zeng, Sichuan Normal University, China
Copyright © 2022 Gao, Xu, Ai, Luo, Guo and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pengtao Ma, cHRtYUB5dHUuZWR1LmNu
 Huiming Gao
Huiming Gao Xiaozhe Xu2,3
Xiaozhe Xu2,3 Pengfei Ai
Pengfei Ai Fuyi Luo
Fuyi Luo Pengtao Ma
Pengtao Ma