
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 30 March 2022
Sec. Evolutionary and Population Genetics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.816123
 Chao Luo1,2,3,4
Chao Luo1,2,3,4 Wulue Huang1,2,3
Wulue Huang1,2,3 Huseyin Yer4,5
Huseyin Yer4,5 Troy Kamuda4
Troy Kamuda4 Xinyi Li1,2,3
Xinyi Li1,2,3 Yang Li1,2,3
Yang Li1,2,3 Yuhong Rong1,2,3
Yuhong Rong1,2,3 Bo Yan1,2,3
Bo Yan1,2,3 Yonghui Wen1,2,3
Yonghui Wen1,2,3 Qiong Wang1,2,3
Qiong Wang1,2,3 Meijuan Huang1,2,3*
Meijuan Huang1,2,3* Haiquan Huang1,2,3*
Haiquan Huang1,2,3*Impatiens L., the largest genus in the family Balsaminaceae with approximately 1,000 species, is a controversial genus. Due to the conflict of morphological features and insufficient genomic resources, the studies of systematic evolution and understanding of taxonomic identification are considered to be very limited. Hence, we have sequenced the complete chloroplast genomes of three ornamental species (Impatiens balsamina, I. hawkeri, and I. walleriana), and compared them with previously published wild species data. We performed a detailed comparison of a highly similar basic structure, size, GC content, gene number, order, and functional array among them. Similarly, most divergent genes were detected from previous work in the literature. The mutational regions containing highly variable nucleotide hotspots were identified and may be used as potential markers for species identification and taxonomy. Furthermore, using whole chloroplast genome data to analysis the phylogenetic relationship of the Balsaminaceae species, we found that they were all part of a single clade. The three phenotypically different ornamental species were clustered together, suggesting that they were very likely to be closely related. We achieved and characterized the plastid genome structure, identified the divergence hotspots, and determined the phylogenetic and taxonomic positions of the three cultivated species in the Impatiens genus. The results may show that the chloroplast genome can be used to solve phylogenetic problems in or between the Impatiens genus and also provide genomic resources for the study of the Balsaminaceae family’s systematics and evolution.
The Balsaminaceae family consists of only two genera; the species-rich Impatiens L. and the monospecific Hydrocera triflora with substantial similarity in morphology and molecular biology datasets (Chen, 2001; Janssens et al., 2012). The controversial and complex flowering genus Impatiens, consists of approximately 1,000 species, which are distributed from the tropics to subtropics and extend to temperate regions of tropical Africa, Southwest Asia, Southern China, Europe, Russia, and North America (Grey-Wilson, 1989; Yu, 2012). Tropical Africa, Madagascar, Sri Lanka, Himalayas, and Southeast Asian are the five biodiversity hotspots for the endemic Impatiens (Grey-Wilson, 1980; Chen, 2001). Due to the diverse flowering and morphological variables, many cultivars (Impatiens balsamina, I. hawkeri, and I. walleriana) are widely used as urban ornamental and gardening plants (Jiang et al., 2017; Torrecilha et al., 2013; Kim et al., 2017). I. balsamina was also called “zhijiahua” in ancient China, the plant can be mashed and directly applied on the nails (Chen et al., 2007). I. hawkeri and I. walleriana are annual flowering plants with a high value, they become extremely popular bedding plants (Cafa et al., 2020), and are also used as annual herbs for the treatment of rheumatism, beriberi, bruises, pain, snakebites, fingernail inflammation and onychomycosis in traditional Asian regions (Thakur et al., 2009; Bhaskar, 2012; Szewczyk, 2018). The derivatives of 1,4-naphthoquinones (impatienol and balsaquinone) were proven to be significant in nonsteroidal, anti-inflammatory drug development (Fan et al., 2013; Li et al., 2015). Additionally, previous research has demonstrated that the Impatiens species have the potential to accumulate high levels of metals such as copper, zinc, chromium, and nickel (Torrecilha et al., 2013; Lai and Cai, 2016; Campos et al., 2017).
Previous publications have primarily focused on specific geographical regions and divided species into groups by purely descriptive traditional morphology, palynology, and anatomy characters, such as flower, stem, and spur (Yuan et al., 2004; Chen et al., 2007). To date, molecular classification for Impatiens was based on morphological characters, several chloroplast plastids (such as coding gene rbcL, matK, trnK and intergenic regions atpB-rbcL and trnL-trnF) (Yuan et al., 2004; Janssens et al., 2006a; Ruchisansakun et al., 2015; Shajitha P. P. et al., 2016). Both of the inter-simple sequence repeat (ISSR) and the nuclear ribosomal ITS markers were utilized in identifying the genetic diversity of populations and the phylogenetic and evolutionary relationships between the Impatiens species (Yuan et al., 2004; Shajitha P. P. et al., 2016). The present published data is based on a few samples which only provide regional characteristics with conflicting results, adequate phylogenetic information for an examination of phylogenetic relationships amongst the Balsaminaceae species is currently missing (Yu et al., 2016; Li Y. et al., 2018). Sequencing whole chloroplast genomes may remarkably increase the resolution and clarify poorly defined phylogenetic relationships.
The nuclear, chloroplast, and mitochondrial genomes are the three major genetic systems (Yuan et al., 2004; Li ZZ. et al., 2018). Unlike the other genomes, the whole chloroplast genome has a self-replication mechanism, relatively independent evolution, slow evolving nature, and unique maternal inheritance (Park and Lee, 2016; Li et al., 2019). It is feasible for the reconstruction of plant phylogeny and the construction of taxonomy between families and genera from the perspective of population genetics to investigate deep comparisons of angiosperm, gymnosperm, and fern families (Huang et al., 2019). Furthermore, the chloroplast genomes of most land plants are highly conserved in terms of conserved structural regions, size, gene content, and gene types. The conservative and differential gene characteristics can provide vital information for the identification, classification, and phylogenetic reconstruction of relationships among species and families. Chloroplast genomes are also useful in genetic engineering, molecular markers, barcoding identification, and plant evolution (Gu et al., 2018).
Based on medicinal and ornamental values, it is essential to analyze and explore the genetic characteristics of the Balsaminaceae species. In the study, we analyzed the chloroplast genome of six phenotypically different species, including three previously published plastid genomes (I. piufanensis, I. glandulifera, and H. triflora) and three newly sequenced ornamental Impatiens species (I. balsamina, I. hawkeri, and I. walleriana) by using Illumina sequencing technology. The study aimed to: 1) characterize the plastid genome structure of three Impatiens species; 2) identify divergence spots among the genomes; 3) reconstruct a plastid genome-based phylogenetic relationships among the available sequences. The present investigation is a novel attempt to reveal and identify the phylogenetic relationship and taxonomic position of the six species based on chloroplast genes. This study will not only contribute to further research on the phylogeny of Impatiens species but also provide partly insights into the chloroplast genome evolutionary history of the order Balsaminaceae.
No specific permits were required for the collection of specimens for this study. This research was carried out in compliance with the relevant laws of China.
All leaf samples were collected and identified by Prof. Haiquan Huang, the samples were deposited in the plant Laboratory of the College of Landscape Architecture and Horticulture Science, Southwest Forestry University, Kunming, Yunnan, China (Table 1). The I. hawkeri was only sequenced in the previously work, we didn’t analysis it and lack of a well comparion with other species (Luo et al., 2021). Fresh leaves were collected and stored in liquid nitrogen. Total DNA was extracted using the Tiangen DNA Reagent Extraction Kit, and an approximate 5–10 µg of genomic DNA quality was checked (Doyle et al., 1987).
The purified genomic DNA was sequenced by using an Illumina MiSeq sequencer (Biozeron, Shanghai, China) (Bankevich et al., 2012; Langmead and Salzberg, 2012). The clean data were assembled and manually corrected using GetOrganelle version 1.6.2 software (Jin et al., 2018). Each assembled genome was annotated with the GenSeq software (Tillich et al., 2017) and the online Dual Organellar Genome Annotator (DOGMA) (Wyman et al., 2004), the start and stop codon positions were searched by gene identification. The position of tRNAs was confirmed with tRNAscan v1.23 software (Schattner et al., 2005). The notes were manually corrected and verified using Geneious R8.0.2 by realigning with references (Kearse et al., 2012). The reference plastid used is from a closely related species I. piufanensis (GenBank MG162586.1). Additionaly, the sequences of the Balsaminaceae plants used in this study were downloaded from GenBank as follows: I. glandulifera (GenBank MK358447.1), I. piufanensis, and H. triflora (GenBank MG162585). The online program OGDrawV1.2 generated the circular chloroplast genome maps.
The online tool REPuter detected the size and location of repeat types (Kurtz et al., 2017). The Geneious R8.0.2 software was utilized to calculate GC content (Kearse et al., 2012). The online MISA software was used to detect SSRs (Beier et al., 2017). The software CodonW investigated the distribution of codon usage, the distribution of codon usage was investigated with the RSCU ratio (Sharp and Li, 1987).
The multiple alignment of conserved genomic sequence with rearrangements was aligned with the previously published monospecific H. triflora chloroplast genome, using the MAUVE software (Darling et al., 2004). MAFFT version was used to detect divergence hotspots (Katoh et al., 2019). The software mVISTA was used to align the whole genome and other species (Brudno et al., 2003; Frazer et al., 2004). The DnaSP v5.10 software was used to calculate the nucleotide divergence values by using the sliding window length of 800 bp and a 200 bp step size (Rozas et al., 2017; R Development Core Team, 2017).
The MAFFT version 7.222 software was used to align the complete chloroplast genomes with the default parameters (Katoh and Toh, 2010). The Maximum likelihood (ML) and Bayesian Inference (BI) were conducted for the topologies. The ML analysis was implemented in RAxML v.8.2.9. The best-fitting model was a GTR + F + I + G4 substitution with 1,000 bootstrap replicates based on the Akaike information criterion (AIC) (Posada, 2008). The Bayesian inference (BI) tree was implemented in MrBayes version 3.2 (Ronquist et al., 2012). Based on the Markov chain Monte Carlo (MCMC) algorithm, the best-fitting model was a TVM + F + I + G4 substitution within one million generations with four independent heated chains sampled after every 1,000 generations. The FigTree ver1.4.2 software visualized the output trees (Ranbaut. 2014).
The total DNA of I. balsamina, I. hawkeri, and I. walleriana were sequenced using next-generation sequencing technology. As a result, the genomic libraries had a total of 28.6 GB. Contigs mapped to the I. piufanensis reference were then used to reconstruct the chloroplast DNA of Impatiens where the sizes of I. balsamina, I. hawkeri, and I. walleriana were 152,271 bp, 151,691 bp, and 151,953 bp, respectively (Table 2 and Supplementary Table S1). The length ranged from 151,691 bp (I. hawkeri) to 154,189 bp (H. triflora), which consists of a large single copy (LSC, 82,906–83,497 bp), a small single copy (SSC, 17,493–18,276 bp) and a pair of inverted repeats (IRs, 25,249–25,710 bp) (Table 2 and Figure 1). The lengths of I. hawkeri and I. walleriana were close with I. balsamina showing the longest length. The whole guanine-cytosine (GC) contents in the Balsaminaceae species ranged from 36.7 to 36.9%, with I. balsamina having the lowest and I. glandulifera and H. triflora having the highest GC content (Table 2). The GC contents in the LSC, IR, and SSC regions were average with 34.4, 43.2, 29.5%, respectively (Supplementary Table S1 and Figure 1).

FIGURE 1. Chloroplast genome structure of three Impatiens species (I. balsamina, I. hawkeri, and I. walleriana).
The genetic physical maps of the I. balsamina, I. hawkeri, and I. walleriana closely resembled the previously published I. piufanensis, but the trnG-UCC gene was annotated as a pseudogene in H. triflora resulting in a total number of 114 genes compared to the other five Impatiens species (Figure 1 and Supplementary Figure S1). Another exception is that the genes ycf15 and trnfM-CAU are interchanged due to the incorrect annotation in I. glandulifera.
Like other typical angiosperms, the chloroplast genomes of the Balsaminaceae species encoded 114 total distinct genes except for I. glandulifera and H. triflora including 81 protein-coding, 29 transfer RNA genes (tRNA), and 4 ribosomal RNA genes (rRNA) (Table 2 and Supplementary Table S2). Most genes of this genus appear in the form of a single copy in the LSC or SSC region with 20 gene duplications in the IR regions, including rpl2, ycf1, ndhB, rps7, rps12, rps19, ycf2, rpl23, ycf15, trnA-UGC, trnV-GAC, trnI-GAU, trnL-CAA, trnI-CAU, trnR-ACG, trnN-GUU, rrn23 rrn4.5, rrn16, and rrn5 (Table 3).
Introns are missing in the annotations of I. piufanensis and H. triflora, namely the trnG-GCC tRNA gene. 16 unique genes were annotated to include introns, whereas, with 14 genes containing one intron (rps12, trnI-GAU, trnA-UGC, rpoC1, ndhB, trnK-UUU, trnG-GCC, ndhA, rpl2, petB, atpF, rps16, trnv-UAC, and trnI-UAA); and the ycf3 and clpP genes each containing two introns (Table 3 and Supplementary Table S3). The rpoC1 gene had the longest exon and the rps12 gene had the longest intron.
To analyze the genetic information and the relationship between evolution and phylogeny of Impatiens, we examined the codons in its coding region. The total number of codons was 304,804. The significant number of codons identified in the different species was as follows: 50,757 (I. balsamina), 50,503 (I. hawkeri), 50,651 (I. walleriana), 50,745 (I. piufanensis), 50,753 (I. glandulifera), and 51,395(H. triflora) (Supplementary Table S4). Among the 20 AAs, the most abundant AA was leucine (29,142, 9.56%), followed by isoleucine (25,482, 8.36%). Tryptophan had the lowest frequency AA in the Balsaminaceae species and was encoded by only 3,960 codons (1.2%). Among species, codon usage based on the relative synonymous codon usage value (RSCU) had not changed, except for some reductions found in five AAs of I. piufanensis, I. glandulifera, I. balsamina, I. hawkeri, and I. walleriana. H. triflora had 36 codons which were more frequently used than the expected usage at equilibrium (RSCU>1). I. glandulifera had 30 codons which were less frequently used than the expected usage at equilibrium (RSCU<1).
A total of 141 unique forward, complement, reverse, and palindromic repeats were examined among the six Balsaminaceae species using REPuter software. I. balsamina contained a total of 28 repeats including 18 palindromic repeats, 9 forward repeats, and 1 reverse repeat (Figure 2). In I. hawkeri, I. walleriana, I. piufanensis, I. glandulifera, and H. triflora, 24, 22, 18, 20, and 20 total repeat pairs were detected, respectively (Supplementary Table S5). Among all six species, the most common repeat types were palindromic and forward repeats, compliment repeats were not identified, and reverse repeats were only found in the I. balsamina and I. hawkeri species, respectively. Most of the repeat lengths were less than 40 bp, however, the I. balsamina and I. hawkeri chloroplasts had 2 forward or palindromic repeats with a length of between 41 and 50 bp.
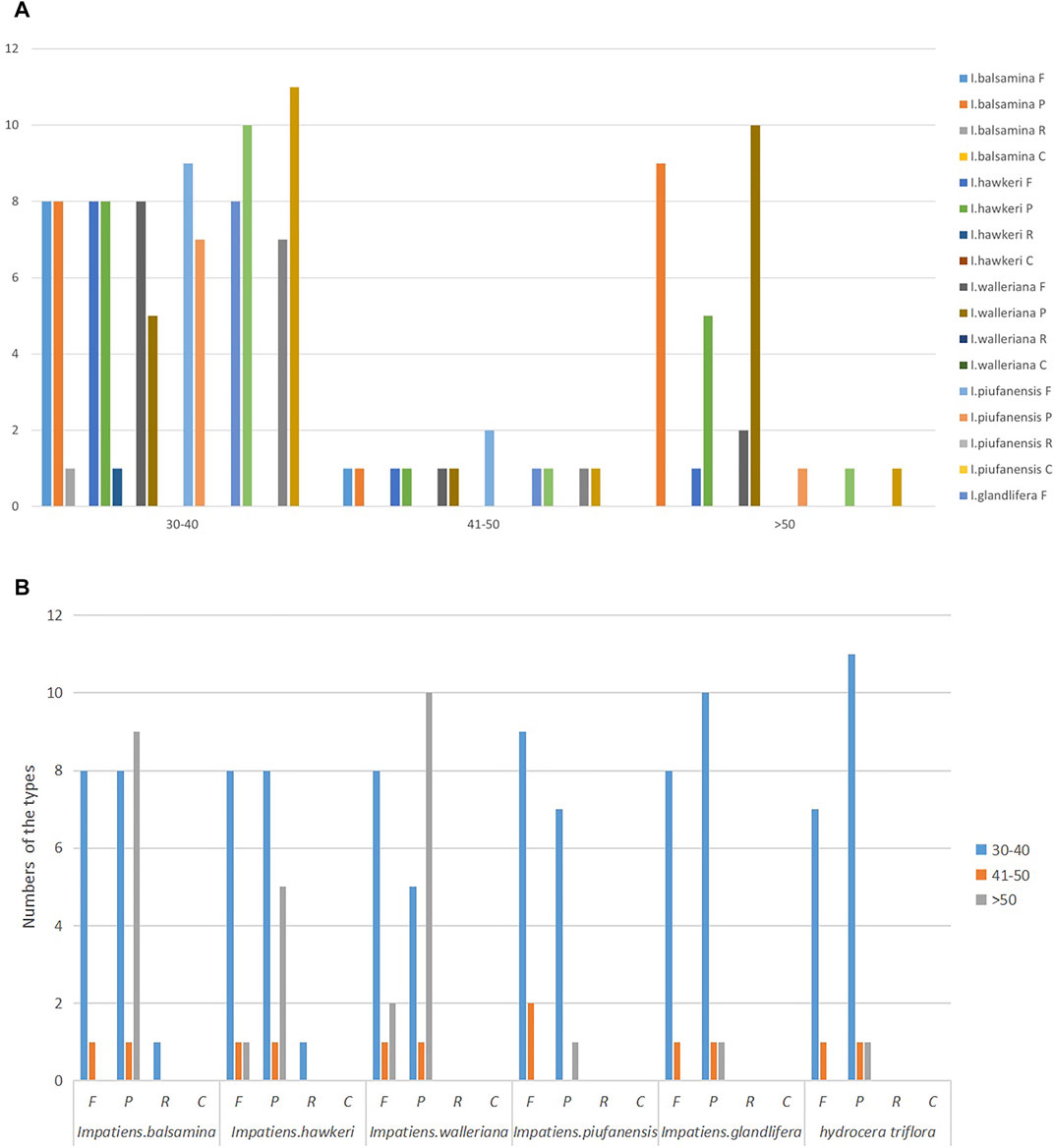
FIGURE 2. Analysis of repeated sequences in the I. balsamina, I. hawkeri, I. walleriana, I. piufanensis, I. glandulifera, and H. triflora chloroplast genomes. (A) A total of six species of four repeat types by length; (B) Total six species of four repeat types.
Among the six Balsaminaceae species, there were 97, 90, 91, 95, 96, and 51 SSRs in the I. balsamina, I. hawkeri, I. walleriana, I. piufanensis, I. glandulifera, and H. triflora chloroplast genomes, respectively (Figure 3 and Supplementary Table S6). Mononucleotide repeats were more abundant with A/T repeats being the most highly represented repeats with a size of 33–79, which accounted for about 64.7–81.44% of the total SSRs, while poly C/G repeats were rather rare (0–3.15%). Among the dinucleotide repeat motifs, AT/AT were the most abundant, while AG/CT only found in I. glandulifera. Three trinucleotide motifs (AAC/GTT, AAG/GTT, AAT/ATT), six tetranucleotide (AAAT/ATTT, AAGT/ACTT, AATG/ATTC, AATT/AATT, AAAG/CTTT), three pentanucleotide (AATAC/ATTGT, AAAAG/CTTTT, AATAG/ATTCT) were identified (Figure 4). However, only one hexanucleotide (AATCCC/ATTGGG) repeat was found in the H. triflora.
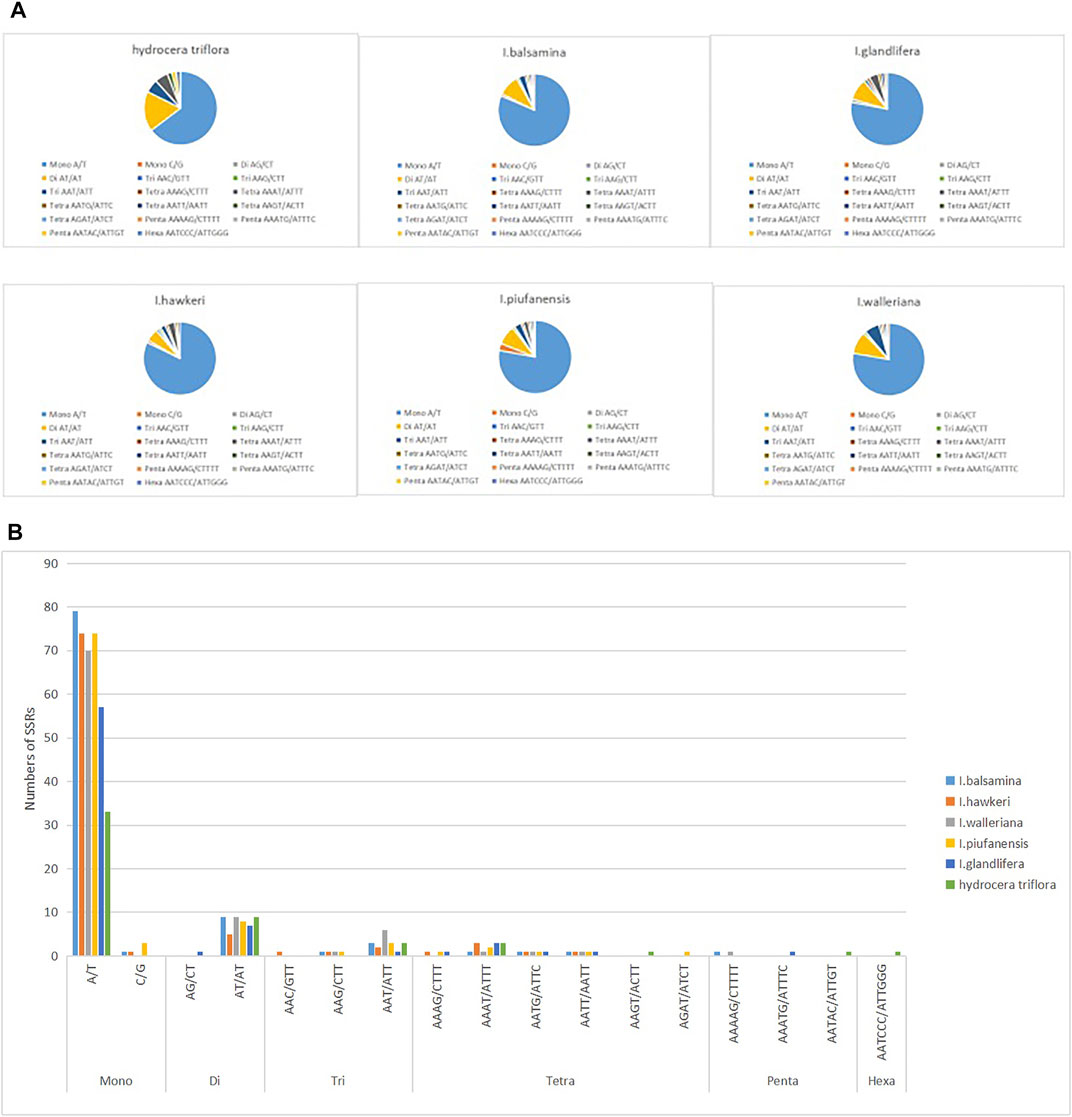
FIGURE 3. Analysis of simple sequence repeats (SSRs) in the chloroplast genomes of I. balsamina, I. hawkeri, I. walleriana, I. piufanensis, I. glandulifera, and H. triflora. (A) The number of different SSR types detected in each species; (B) type and frequency of each identified SSR.
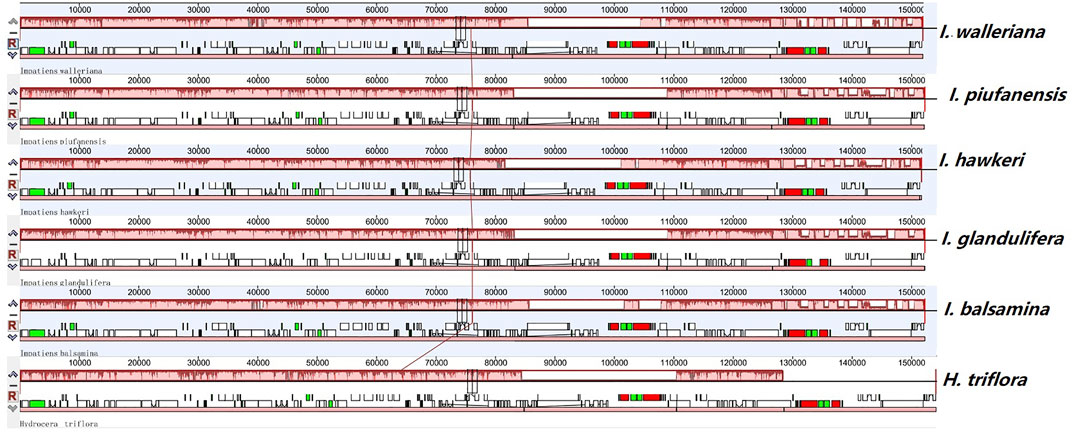
FIGURE 4. Comparison of sequence arrangement in the chloroplast genomes of six Balsaminaceae species.
Most chloroplast genomes in angiosperm plants are relatively stable. However, based on different evolutionary histories and genetic backgrounds, the chloroplast genome structure, size, and numbers can vary. Collinear blocks were used to analyze and compare the collinearity of chloroplast genomes. The mauve alignment for the six Balsaminaceae species revealed that the optimal collinearity within subgenus Impatiens is relatively conserved and lacks gene rearrangement (Figure 4). Compared with H. triflora, the linear relationships within genome structure and gene sequences indicated that there was high chloroplast genome homology.
Four junctions in regions of detailed structure were compared among the Balsaminaceae and subsequently presented (Figure 5). The IRb-LSC junction (JLB) was located in the rps19 coding region which was inserted between the IRB and LSC region in all six species. The length of the rps19 in the IRB region among the four species (I. walleriana, I. piufanensis, I. glandlifera, and H. triflora) had varied from 101 to 199 bp. Notably, the length of the rps19 in the IRB region of both I. balsamina and I. hawkeri was 0 bp. The SSC-IRB junction (JSB) was adjacent to gene rps19 and ndhF; JSB of six species except for I. walleriana were all located and adjoined the end of ycf1 from 933 bp to 1,189 bp. The overlap between ndhF and ycf1 was detected in I. hawkeri, with ndhF expanding into the IRB region for 1,161 bp. In the other five species, the distances between ndhF and JSB were 347, 41, 30, 62, and 7 bp, respectively. The IRA-SSC junction (JSA) was located in the ycf1 coding region which covered the IRA and SSC region. The length of ycf1 in the SSC region varied from 4,300 bp to 4,545 bp. However, six species overlap ycf1 in the IRA region were found 810, 1,179, 1,115, 1,101, 1,083, and 1,099 bp, respectively. The LSC-IRA junctions (JLA) were located between rpl12 and rps19 in I. balsamina and I. hawkeri, while in other four species, the distances between traH and rpl12 were 0 bp, 0 bp, 7 bp, 43 bp, respectively. In the JLA junction, the rps19 gene was 34 bp and 104 bp into the LSC region in I. balsamina and I. hawkeri, while the distances between rpl2 and JLA were 25, 46, 1, 1, 220, and 5 bp, respectively.
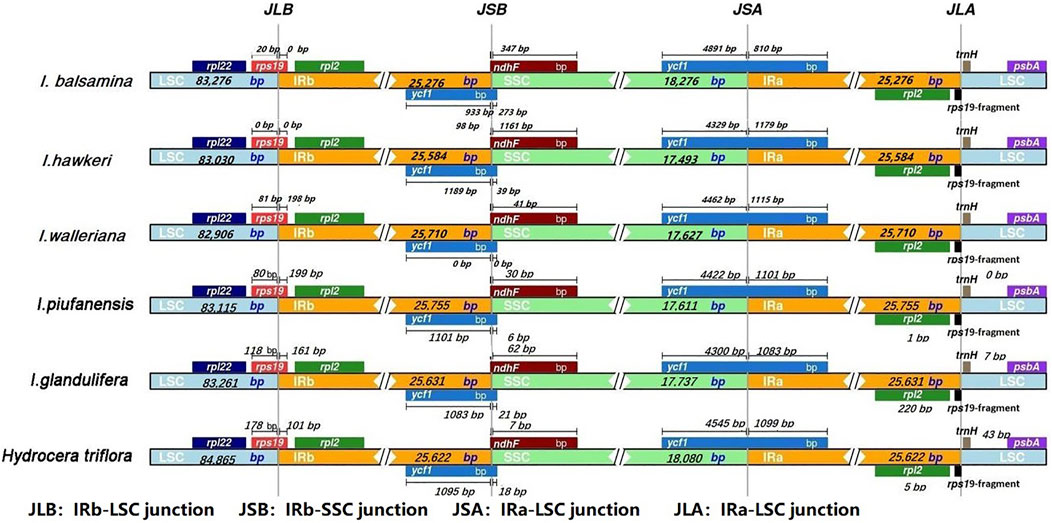
FIGURE 5. Comparison of the borders of four different regions (LSC, SSC, and IRs) among I. balsamina, I. hawkeri, I. walleriana, I. piufanensis, I. glandulifera, and H. triflora chloroplast genomes.
The mVISTA program was used to detect hyper-variable regions based on whole regions of chloroplast genomes. H. triflora and other Impatiens species showed sequence divergence in many regions such as rps3-rps19, matK, psbK, atpH-atpI, trnC-trnT, petN, psbM, atpE, rbcL, accD, psaL, ycf1, ndhG-ndhA,rpl16, rpoB, ndhB, ndhF, and ndhH (Figure 6). The three genes; ndhF, ycf1, and ndhH were detected in the SSC region. The psbK-psbI, atpI, and rps4-trnF genes showed some divergence in the LSC region of I. piufanensis, I. glandlifera, and H. triflora.
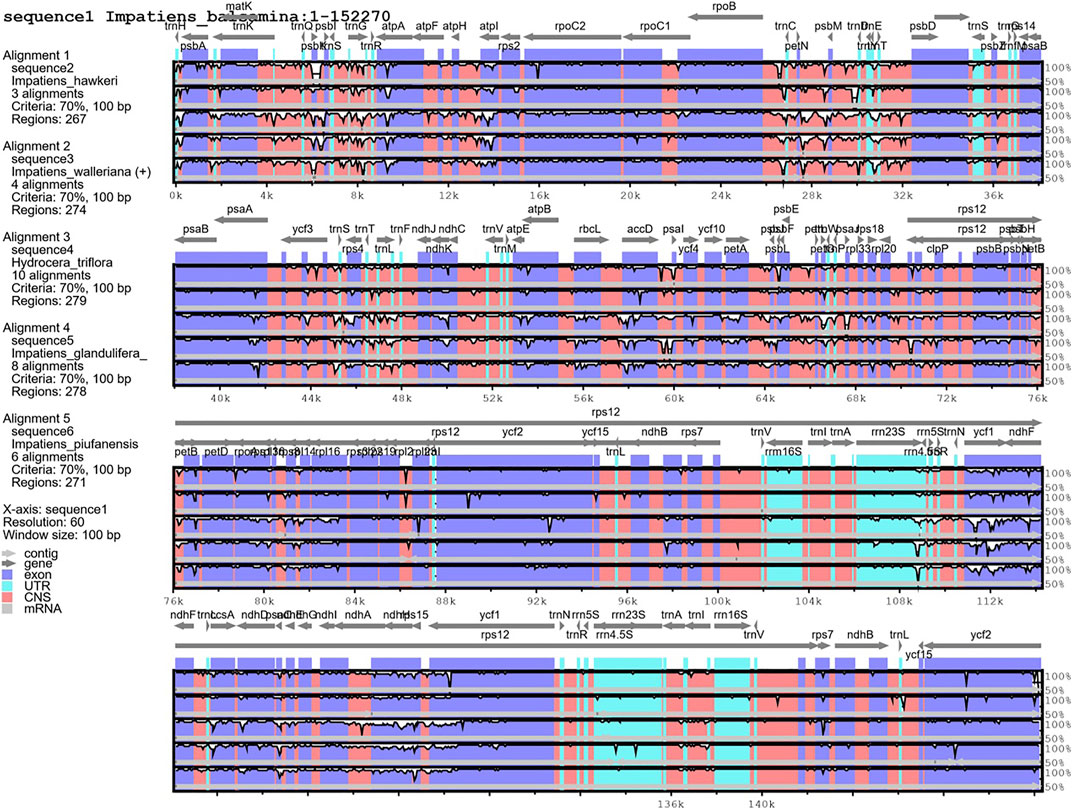
FIGURE 6. Alignment of the six chloroplast genomes. Sequence identity plot comparing the five chloroplast genomes with I. balsamina as a reference by using mVISTA.
Similarly, we determined the average pairwise sequence divergence among three ornamental species of Impatiens chloroplast genomes. The nucleotide variability (Pi) of these 140 regions ranged from 0.1% (ycf2) to 5.6% (trnG-GCC) among three chloroplast genomes (Supplementary Table S7). Additionaly, ten different genes; psbA, trnS-trnG, trnG-GCC, atpH-atpL, trnE-trnT, psbD, cemA, ndhF, rpl32, ndhA, and ycf1 were sequenced within these genomes. The trnG-GCC gene demonstrated the highest average sequence divergence (0.056), followed by cemA (0.048), and ycf1 (0.046) (Figure 7). Sliding window analysis indicated that mutational hotspots included psbA, trnS-trnG, trnG-GCC, atpH-atpL, trnE-trnT, psbD, and cemA, which exhibited higher Pi values (>0.035) in the LSC and SSC regions. Single mutational hotspots in the IR regions with remarkably high PI values (>0.015) were not present.
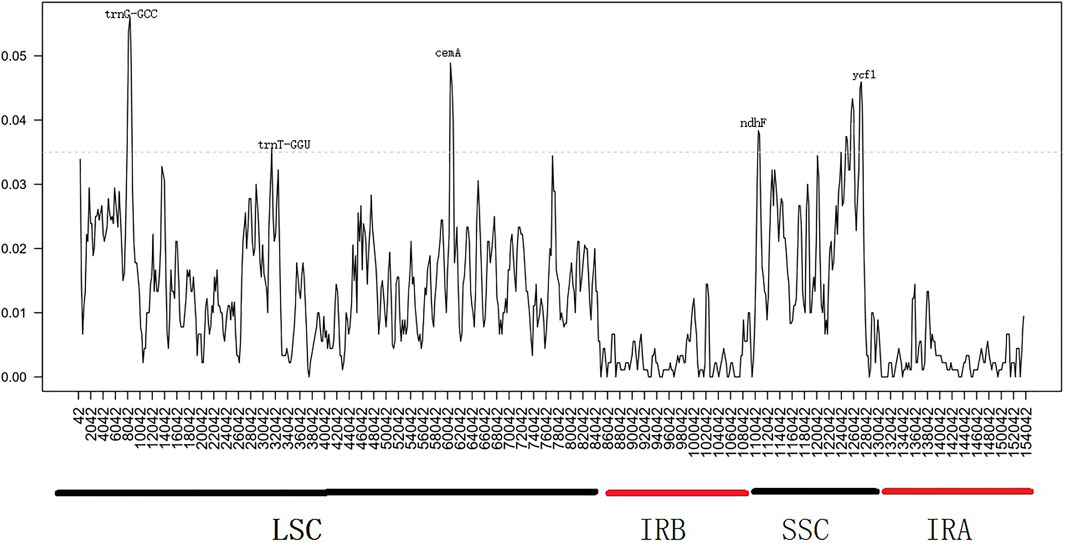
FIGURE 7. Sliding window analysis based on the chloroplast genomes of three Balsaminaceae species. Window length: 2000 bp; step size: 200 bp. X-axis: the position of the midpoint of a window. Y-axis: nucleotide diversity of each window.
An exploration of the phylogenetic positions and evolutionary relationships of Impatiens species based on the complete chloroplast genomes (Supplementary Table S8). The chloroplast genomes from seven families within six Balsaminaceae species, six Primulaceae species, five Ebenaceae species, four Theaceae species, two Saxifragaceae species, four Actinidiaceae species, and one Styracaceae species as outgroup. The topologies of the two datasets (ML and BI) yielded a similar structure. The seven families can be classified into five monophyletic clades (Figure 8). Actinidiaceae was the basal group in all phylogenetic trees. The Primulaceae and Ebenaceae were gathered into one clade and also the Balsaminaceae was a sister to Saxifragaceae. Most of the species from the same genus were clustered together. All Balsaminaceae species formed a monophyletic subclade in both trees. H. triflora was located at the bottom of the Balsaminaceae phylogenetic tree and clustered into a single clade. All Impatiens species were clustered into one clade, The cultivated species; I. balsamina, I. hawkeri, and I. walleriana were more closely related than the wild species I. piufanensis and I. glandulifera.
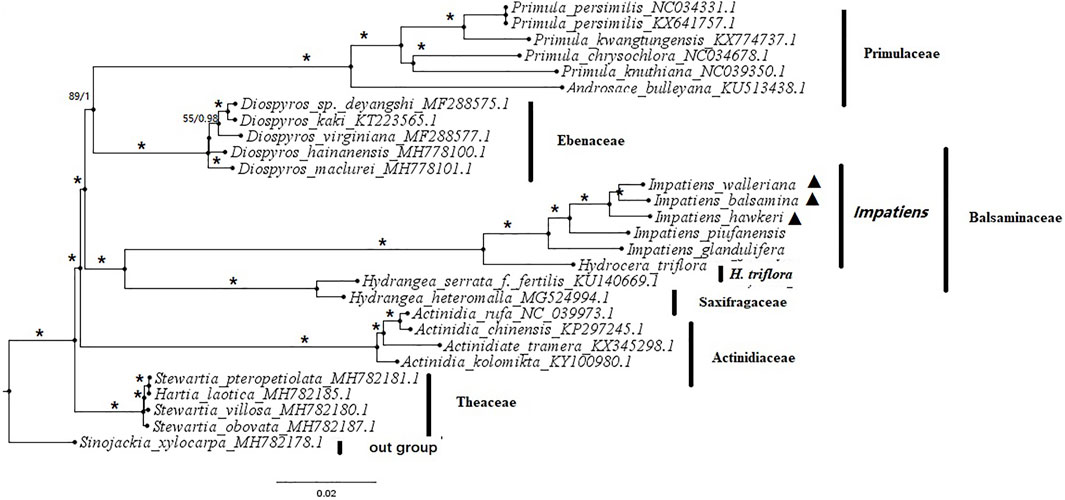
FIGURE 8. Phylogenetic tree based on whole chloroplast genome sequences from 6 Balsaminaceae species and 23 other species using maximum likelihood (ML) bootstraps and Bayesian posterior probabilities (PP). ML topology is shown with ML bootstrap support values/Bayesian PP given at each node. Asterisks indicate both of PP = 1 and LBS = 100%. Black triangles indicate the cp genomes of the three Impatiens species examined in this study.
Compared with the reported genome structures among Balsaminaceae, the family was slightly smaller in size with 151,691 bp (I. hawkeri) of the former to 154,189 bp (H. triflora) of the latter (Table 2 and Supplementary Table S1). There was a 2,498 bp difference in length between the Balsaminaceae species. Nevertheless, the basic structure and content of the genome were roughly similar (Yu et al., 2016; Li Y. et al., 2018). Chloroplast genomes were found to be highly conserved. The potential of ycf15, trnfM-CAU, and psbN genes had been annotated in all genomes of Impatiens species, while in H. triflora they were not excluded in this study. Likewise, the reading frames named the trnG-UCC gene which had been only annotated in I. glandulifera. Based on observations, their ability to encode proteins in angiosperms has not yet been confirmed. The results indicate homology in genome structure, therefore, that may be decisively resolves the systemic evolutionary relationship for species identification and taxonomy. The genes were divided into three categories based on function (Tanner et al., 2014). The first was related to photosynthesis and translation genes, such as Rubisco, ATP synthase, Cytochrome b/f complex, assembly, and stability of Photosystem I, II (Tamboli et al., 2018). The second category corresponds to Ribosomal and Transfer RNA (Beerling and Perrins, 1993); and the third category contained biosynthetic genes, such as Carbon metabolism gene cemA, Proteolysis gene clpP, fatty acid synthesis gene accD, and some unknown function genes (orf188, ycf1, ycf2, and ycf15) (Hulme and Bremner, 2006).
By detecting detailed boundary changes of the regions, we observed that the IR-SC boundary regions showed minimal differences (Figure 5). Some extensions or contractions were detected, with the IR regions ranging from 25,276 bp to 25,755 bp (in I. balsamina to I. piufanensis, respectively). Variations of rps19, ycf1, ndhF, and rpl2 genes were observed and partially duplicated genes were found at the beginnings and ends of the IR regions including 178 bp of rps19 in H. triflora, and the rps19 gene of I. hawkeri not extending into the IR region. The SSC and LSC regions showed higher sequence divergence than the IR regions. Moreover, the pairwise alignment of the I. balsamina showed high synteny with other species. Similarly, most divergent genes were detected, especially in psbA, trnS-trnG, trnG-GCC, atpH-atpL, trnE-trnT, psbD, cemA, ndhF, rpl32, ndhA, and ycf1 (Figure 7). The coding regions in all Balsaminaceae chloroplast genomes showed less divergence than the non-coding regions. As previously reported, trnG-GCC, cemA, and ycf1 genes possessed high variability as possible molecular markers. Therefore, these coding regions and non-coding genes may provide strong molecular evidence for resolving low-level phylogeny and phylogeography (Fujihashi et al., 2002; Li et al., 2015).
Based on the analysis of various chloroplast genomes, repetitive sequences were essential for inducing indels and substitutions (Zuo et al., 2017; Yan et al., 2019). The sequences not only play a vital role in the rearrangement and stabilization of the chloroplast genome sequence but also affect the copy number differences between similar and different species (Xie et al., 2018; Wang et al., 2020). The Impatiens chloroplast genome had four different repetitive sequences. The forward repeats can be used as markers in phylogenetic studies due to the changes in genomic structure. Among all species, the most common type of repetition was a palindrome repeat. All species contained forward and palindromic repeats but compliment repeats were not identified in all species while reverse repeats were only found in I. balsamina and I. hawkeri (Figure 2 and Supplementary Table S5).
Simple sequence repeats (SSRs) have been recognized as a marker for having a high polymorphism rate and abundant variation at the species level (Wang et al., 2020). Moreover, SSRs can be used to detect genetic diversity, population, and polymorphisms at intraspecific, distant phylogenetic relationships and cultivar levels. Our analysis identified the distribution of 51–97 SSRs in the Balsaminaceae species ranging from 10 to 20 bp in size (Figure 3 and Supplementary Table S6). Furthermore, not all the SSR types were identified in all the species, hexanucleotide and pentanucleotide repeats were not detected in I. hawkeri and I. pinfanensis, while the hexanucleotide repeats were found only in H. triflora.
Analysis of the whole chloroplast genome can effectively solve the various problems in molecular evolution and the phylogeny of the same genus or family, hence it can enhance our understanding of molecular evolution (Janssens et al., 2009; Shajitha P. P. et al., 2016). The first molecular phylogeny of the genus was published by Fujihashi. However, due to limited taxon sampling and the use of a distant outgroup Tropaeolum (Tropaeolaceae), findings were limited information on the systemic evolutionary relationships (Fujihashi et al., 2002). Nuclear ribosomal internal transcribed spacer (ITS) and atpB-rbcL sequences for studying on 111 Balsaminaceae species, provided new phylogenetic insights, namely that the Impatiens had colonized from Southwest China to the African continent in three separate proliferation events (Janssens et al., 2006b; Shajitha P. P. et al., 2016). Subsequently, plastids, plastids and nuclear, or combined plastids and pollen data collected from the Impatiens were further analyzed (Yuan et al., 2004). A new classification of Impatiens based on morphological and molecular datasets divided them into two subgenera: Clavicarpa and Impatiens with Impatiens being further subdivided into seven sections based on morphological characteristics or combinations of the ITS results, atpB-rbcL, and trnL-F intergenic fragments, along with pollen data (Yu et al., 2016). Although the new schematic provided a robust basis for further research, all the published data contained only a few samples from obvious regional samples and the results were conflicted.
In the present study, based on the maximum likelihood (ML) and Bayesian Inference (BI) trees (Figure 8). Two phylogenetic trees showed the same results. The three selected families (Actinidiaceae, Theaceae, and Styracaceae) were clustered into a monophyletic branch, respectively. The Genus Primula and Androsace of the family Primulaceae were clustered into a clade, the family Theaceae also consisted of the Stewartia and the Hartia Dunn. The Balsaminaceae and Saxifragaceae were clustered into a clade. All Balsaminaceae species formed a subclade in both ML and BI trees. And H. triflora and Impatiens formed two different subclades (Figure 8). The I. balsamina, I. hawkeri, and I. walleriana species with the most similar morphological characteristics were clustered together, suggesting highly consistent phylogenetic relationships in morphology and genomics, and also were very likely to be derived from one species, and had the same ancestor (Yuan et al., 2004; Rahelivololona et al., 2018). The species I. piufanensis and I. glandulifera were closer to H. triflora in the Balsaminaceae, which may have experienced the same habitat and evolutionary process.
Similarly, the results of the similarities and differences identified the phylogenetic relationships between the Impatiens species by sequencing whole chloroplast genomes, traditional morphology and molecular classification indicated that the phylogenetic trees from the three cultivars of I. hawkeri, I. walleriana, and I. balsamina were in a relatively unique evolutionary position. Compared with the wild species, the cultivated species had a very high bootstrap value and an obvious evolutionary trend. Based on previous phylogenetic analyses using the ITS and matK fragments, the phylogenetic trees were divided into different clades (Yuan et al., 2014; Tamboli et al., 2018). In terms of morphology, except for I. balsamina, which is an annual herb, the other two were perennials (Chen, 2001); the stem was fleshy and the leaves of I. hawkeri were whorled and the other two were alternate, stalked (Yu, 2012; Yu et al., 2016); I. walleriana had ovate leaves, with the other two species having lanceolate leaves with sharp teeth on the edge (Chen et al., 2007; Yu, 2012); The three cultivars had the same morphology: solitary flowers without pedicels; two pieces lateral sepals; obliquely ovoid, round flag petals with keel-like protrusions, wing petals with short stalks, lip petals; boat-shaped; anthers spherical; fusiform ovary, and capsule fusiform (Cai et al., 2013). However, using the BI and ML morphology and molecular phylogenetic trees can be well integrated.
The resulting phylogenomic tree highly supported the clade of the Balsaminaceae species forming a monophyletic subclade, with the clusters of cultivated and wild species, confirming the validity of the assembled and annotated chloroplast genome of Balsaminaceae species, which is consistent with the results of plastid genes and supports the classification of Ericicales in the updated APG IV system (Janssens et al., 2009; Li ZZ. et al., 2018). The use of chloroplast genome data clearly reflects the evolutionary relationship between wild impatiens and cultivated species, and decisively resolves the systemic evolutionary relationship between wild species and cultivated Impatiens. The research shows that we have clearly identified the phylogenetic and taxonomic position of the three cultivated species in the Impatiens genus, and provides molecular evidence that the chloroplast genome can be applied to clarify phylogenetic questions within or between the Impatiens genus. The comparative analyses using whole chloroplast genomes provided an important new perspective into genome structure and resolved multiple inconsistencies in molecular evolution and genus phylogenetic relationships.
Three different ornamental species (I. balsamina, I. hawkeri, and I. walleriana) and three novel wild species of the genus Impatiens were analyzed in this study. They proved to be valuable genomic resources in the present examination of the Balsaminaceae family. The results showed a highly similar basic structure, size, GC content, gene number, order, and functional array. Similarly, most divergent genes were detected, mutational regions contained highly variable nucleotide hotspots that may be used as potential markers for species identification and taxonomy. Additionally, based on the ML and BI phylogenomic trees, the trees highly supported three different ornamental species forming a monophyletic subclade. The comparative analyses using whole chloroplast genomes provided an important new perspective into genome structure and resolved multiple inconsistencies in molecular evolution and genus phylogenetic relationships. However, the Impatiens consists of approximately 1,000 species, which makes it complicated to identify species by determining the whole genome of chloroplast. Future research on Balsaminaceae relationships needs a larger sampling of taxa, morphological characteristics combined with simple molecular markers, and genome-wide analyses to enhance our understanding of evolution.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
CL designed the experiment and wrote the manuscript. CL, WH, XL, and YL contributed to the sampling. CL, YR, BY, YW, QW, TK, and HY. analyzed the data. MH and HH proofed the manuscript. All authors have read and agreed to the published version of the manuscript.
This work was carried out with the support of the National Natural Science Foundation of China [32060364;32060366;31860230]; Major scientific and technological projects in Yunnan Province (202102AE090052); Key Research and Development Plan Program of Yunnan Province [2018BB013]; Young and Middle-aged Academic and Technical Leadership Training Project of Yunnan [2018HB024], Program for Innovative Research Team (in Science and Technology) in University of Yunnan Province; and Program for Doctoral Supervisors Team in Genetic Improvement and High-efficient Propagation of Landscape Plants in Yunnan Province.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Dan Zong helped to teach the software used for the experiments. Our sincere thanks are also to the reviewers for their comments and suggestions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.816123/full#supplementary-material
BI, Bayesian Inference; bp, base pairs; Gb, Gigabases; IGR, Intergenic region; IR, Inverted repeat; ITS, Internal transcribed spacer; LSC, Long single copy; LSR, Long sequence repeat; MCMC, Markov chain Monte Carlo; ML, Maximum likelihood; NCBI, National Center for Biotechnology Information; NGS, Next-generation sequencing; PCR, Polymerase chain reaction; PI, Parsimony informative; rRNA, ribosomal RNA; SSC, Short single copy; SSR, Simple sequence repeat; tRNA, transfer RNA.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a New Genome Assembly Algorithm and its Applications to Single-Cell Sequencing. J. Comput. Biol. 19, 455–477. doi:10.1089/cmb.2012.0021
Beerling, D. J., and Perrins, J. M. (1993). Impatiens Glandulifera Royle (Impatiens Roylei Walp.). J. Ecol. 81 (2), 367–382. doi:10.2307/2261507
Beier, S., Thiel, T., Münch, T., Scholz, U., and Mascher, M. (2017). MISA-web: A Web Server for Microsatellite Prediction. Bioinformatics 33, 2583–2585. doi:10.1093/bioinformatics/btx198
Bhaskar, V. (2012). Taxonomic Monograph on Impatiens L. (Balsaminaceae) of Western Ghats, South India: The Key Genus for Endemism. Karnataka, India: Centre for Plant Taxonomic Studies.
Brudno, M., Malde, S., Poliakov, A., Do, C. B., Couronne, O., Dubchak, I., et al. (2003). Glocal Alignment: Finding Rearrangements during Alignment. Bioinformatics 19, i54–i62. doi:10.1093/bioinformatics/btg1005
Cafa, G., Baroncelli, R., Ellison, C. A., and Kurose, D. (2020). Impatiens Glandulifera (Himalayan Balsam) Chloroplast Genome Sequence as a Promising Target for Populations Studies. PeerJ 8, e8739. doi:10.7717/peerj.8739
Cai, X. Z., Yi, R. Y., Zhuang, Y. H., Cong, Y. Y., Kuang, R. P., and Liu, K. M. (2013). Seed Coat Micromorphology Characteristics of Impatiens L. And its Systematic Significance. Acta Hort. Sin. 40, 1337–1348. doi:10.1111/j.1095-8339.2005.00436.x
Campos, V., Lessa, S. S., Ramos, R. L., Shinzato, M. C., and Medeiros, T. A. M. (2017). Disturbance Response Indicators of Impatiens Walleriana Exposed to Benzene and Chromium. Int. J. Phytoremediation 19 (8), 709–717. doi:10.1080/15226514.2017.1284745
Chen, Y. L. (2001). Balsaminaceae. Flora Reipublicae Popularis Sinica 47, 1–243. doi:10.3897/phytokeys.176.58825
Darling, A. C. E., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: Multiple Alignment of Conserved Genomic Sequence with Rearrangements. Genome Res. 14, 1394–1403. doi:10.1101/gr.2289704
Doyle, J., Doyle, J. L., Doyle, M., Doyle, J., Doyle, J. L., and Doyle, J. (1987). A Rapid DNA Isolation Procedure for Small Quantities of Leaf Tissue. Phytochem. Bull. 19, 11–15.
Fan, X., Reichling, J., and Wink, M. (2013). Antibacterial Activity of the Recombinant Antimicrobial Peptide Ib-AMP4 from Impatiens Balsamina and its Synergy with Other Antimicrobial Agents against Drug Resistant Bacteria. Pharmazie 68, 628–630. doi:10.1691/ph.2013.6512
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004). VISTA: Computational Tools for Comparative Genomics. Nucleic Acids Res. 32, W273–W279. doi:10.1093/nar/gkh458
Fujihashi, H., Akiyama, S., and Ohba, H. (2002). Origin and Relationships of the Sino-Himalayan Impatiens (Balsaminaceae) Based on Molecular Phylogenetic Analysis, Chromosome Numbers and Gross Morphology. J. Jap. Bot. 77, 284–295.
Grey-Wilson, C. (1989). A Revision of Sumatran Impatiens: Studies in Balsaminaceae: VIII. Kew Bull. 44, 67–105. doi:10.2307/4114646
Grey-Wilson, C. (1980). Impatiens in Papuasia: Studies in Balsaminaceae: I. Kew Bull. 34, 661–688. doi:10.2307/4119062
Gu, C., Tembrock, L., Zheng, S., and Wu, Z. (2018). The Complete Chloroplast Genome of Catha Edulis: A Comparative Analysis of Genome Features with Related Species. Ijms 19, 525. doi:10.3390/ijms19020525
Huang, Y., Yang, Z., Huang, S., An, W., Li, J., and Zheng, X. (2019). Comprehensive Analysis of Rhodomyrtus Tomentosa Chloroplast Genome. Plants 8, 89. doi:10.3390/plants8040089
Hulme, P. E., and Bremner, E. T. (2006). Assessing the Impact of Impatiens Glandulifera on Riparian Habitats: Partitioning Diversity Components Following Species Removal. J. Appl. Ecol. 43 (1), 43–50. doi:10.1111/j.1365-2664.2005.01102.x
Janssens, S., Geuten, K., Viaene, T., Yuan, Y. M., Song, Y., and Smets, E. (2006a). Phylogenetic Utility of the AP3/DEF K-Domain and its Molecular Evolution in Impatiens (Balsaminaceae). Mol. Phylogenet. Evol. 43, 225–239. doi:10.1016/j.ympev.11.01610.1016/j.ympev.2006.11.016
Janssens, S. B., Knox, E. B., Huysmans, S., Smets, E. F., and Merckx, V. S. F. T. (2009). Rapid Radiation of Impatiens (Balsaminaceae) during Pliocene and Pleistocene: Result of a Global Climate Change. Mol. Phylogenet. Evol. 52 (3), 806–824. doi:10.1016/j.ympev.2009.04.013
Janssens, S. B., Wilson, Y. S., Yuan, Y.-M., Nagels, A., Smets, E. F., and Huysmans, S. (2012). A Total Evidence Approach Using Palynological Characters to Infer the Complex Evolutionary History of the Asian Impatiens (Balsaminaceae). Taxon 61, 355–367. doi:10.1002/tax.612007
Janssens, S., Geuten, K., Yuan, Y.-M., Song, Y., Küpfer, P., and Smets, E. (2006b). Phylogenetics of Impatiens and Hydrocera (Balsaminaceae) Using Chloroplast atpB-rbcL Spacer Sequences. issn: 0363-6445 31, 171–180. doi:10.1600/036364406775971796
Jiang, H.-F., Zhuang, Z.-H., Hou, B.-W., Shi, B.-J., Shu, C.-J., Chen, L., et al. (2017). Adverse Effects of Hydroalcoholic Extracts and the Major Components in the Stems of Impatiens Balsamina L. On Caenorhabditis elegans. Evidence-Based Complement. Altern. Med. 2017, 1–10. doi:10.1155/2017/4245830
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., Yi, T. S., and Li, D. Z. (2018). GetOrganelle: a Simple and Fast Pipeline for De Novo Assembly of a Complete Circular Chloroplast Genome Using Genome Skimming Data. bioRxiv. 256479.
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 20, 1160–1166. doi:10.1093/bib/bbx108
Kim, C. S., Bae, M., Oh, J., Subedi, L., Suh, W. S., and Choi, S. Z. (2017). Anti-Neurodegenerative Biflavonoid Glycosides From Impatiens Balsamina. J. Nat. Prod. 80, 471–478. doi:10.1021/acs.jnatprod.6b00981
Katoh, K., and Toh, H. (2010). Parallelization of the MAFFT Multiple Sequence Alignment Program. Bioinformatics 26, 1899–1900. doi:10.1093/bioinformatics/btq224
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 28, 1647–1649. doi:10.1093/bioinformatics/bts199
Kurtz, S., Bae, M., Oh, J., Subedi, L., Suh, W. S., Choi, S. Z., et al. (2017). Anti-neurodegenerative Biflavonoid Glycosides fromREPuter: the Manifold Applications of Repeat Analysis on a Genomic Scale. Impatiens Balsaminaj. Nat. Prod.Nucleic Acids Res. 8029, 4714633–4784642. doi:10.1093/nar/29.22.4633
Lai, H.-Y., and Cai, M.-C. (2016). Effects of Extended Growth Periods on Subcellular Distribution, Chemical Forms, and the Translocation of Cadmium inImpatiens Walleriana. Int. J. Phytoremediation 18 (3), 228–234. doi:10.1080/15226514.2015.1073677
Langmead, B., and Salzberg, S. L. (2012). Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 9, 357–359. doi:10.1038/nmeth.1923
Li, Q., Zhang, X., Cao, J., Guo, Z., Lou, Y., Ding, M., et al. (2015). Depside Derivatives with Anti-hepatic Fibrosis and Anti-diabetic Activities from Impatiens Balsamina L. Flowers. Fitoterapia 105, 234–239. doi:10.1016/j.fitote.2015.07.007
Li, W., Zhang, C., Guo, X., Liu, Q., and Wang, K. (2019). Complete Chloroplast Genome of Camellia Japonica Genome Structures, Comparative and Phylogenetic Analysis. PLoS ONE 14 (5), e0216645. doi:10.1371/journal.pone.0216645
Li, Y., Zhang, J., Li, L., Gao, L., Xu, J., and Yang, M. (2018a). Structural and Comparative Analysis of the Complete Chloroplast Genome of Pyrus hopeiensis-"Wild Plants with a Tiny Population"-And Three Other Pyrus Species. Ijms 19, 3262. doi:10.3390/ijms19103262
Li, Z. Z., Saina, J. K., Gichira, A. W., Kyalo, C. M., Wang, Q. F., and Chen, J. M. (2018b). Comparative Genomics of the Balsaminaceae Sister Genera Hydrocera Triflora and Impatiens Pinfanensis. Int. J. Mol. Sci. 19, 319. doi:10.3390/ijms19010319
Luo, C., Huang, W., Sun, H., Yer, H., Li, X., Li, Y., et al. (2021). Comparative Chloroplast Genome Analysis of Impatiens Species (Balsaminaceae) in the Karst Area of China: Insights into Genome Evolution and Phylogenomic Implications. BMC Genomics 22, 571. doi:10.1186/s12864-021-07807-8
Park, J. H., and Lee, J. (2016). The Complete Plastid Genome of Scopolia Parviflora (Dunn.) Nakai (Solanaceae). Korean J. Plant Taxonomy 46 (1), 60–64. doi:10.11110/kjpt.2016.46.1.60
Posada, D. (2008). jModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 25, 1253–1256. doi:10.1093/molbev/msn083
R Development Core Team. (2017). R: A Language and Environment for Statistical Computing. Available at: http://www.r-project.org (accessed December 2, 2018).
Rahelivololona, E. M., Fischer, E., Janssens, S. B., and Sylvain G Razafimandimbison, S. G. (2018). Phylogeny, Infrageneric Classification and Species Delimitation in the Malagasy Impatiens (Balsaminaceae). PhytoKeys 110, 51–67. doi:10.3897/phytokeys.110.28216
Ranbaut, A. (2014). FigTree Ver. 1.4.2. AvaliableAt: http://tree.bio.ed.ac.uk/soft ware/figtree (Accessed February 13, 2015).
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 61, 539–542. doi:10.1093/sysbio/sys029
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 34, 3299–3302. doi:10.1093/molbev/msx248
Ruchisansakun, S., Niet, T., Van Der, T., Janssens, S. B., Triboun, P., Jenjittikul, T., et al. (2015). Phylogenetic Analyses of Molecular Data and Reconstruction of Morphological Character Evolution in Asian Impatiens Section Semeiocardium (Balsaminaceae). Syst. Bot. 40, 1063–1074. doi:10.1600/036364415X690102
Schattner, P., Brooks, A. N., and Lowe, T. M. (2005). The tRNAscan-SE, Snoscan and snoGPS Web Servers for the Detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, W686–W689. doi:10.1093/nar/gki366
Shajitha, P. P., Dhanesh, N. R., Ebin, P. J., Joseph, L., Devassy, A., John, R., et al. (2016b). Molecular Phylogeny of Balsams (Genus Impatiens) Based on ITS Regions of Nuclear Ribosomal DNA Implies Two Colonization Events in South India. J. Appl. Biol. Biot. 4, 1–9. doi:10.7324/jabb.2016.40601
Shajitha, P. P., Dhanesh, N. R., Ebin, P. J., Laly, J., Aneesha, D., Reshma, J., et al. (2016a). A Combined Chloroplast atpB-rbcL and trnL-F Phylogeny Unveils the Ancestry of Balsams (Impatiens spp.) in the Western Ghats of India. 3 Biotech. 6, 258. doi:10.1007/s13205-016-0574-8
Sharp, P. M., and Li, W.-H. (1987). The Codon Adaptation index-a Measure of Directional Synonymous Codon Usage Bias, and its Potential Applications. Nucl. Acids Res. 15, 1281–1295. doi:10.1093/nar/15.3.1281
Szewczyk, K. (2018). Phytochemistry of the Genus Impatiens (Balsaminaceae): A Review. Biochem. Syst. Ecol. 80, 94–121. doi:10.1016/j.bse.2018.07.001
Tamboli, A. S., Dalavi, J. V., Patil, S. M., Yadav, S. R., and Govindwar, S. P. (2018). Implication of ITS Phylogeny for Biogeographic Analysis, and Comparative Study of Morphological and Molecular Interspecies Diversity in Indian Impatiens. Meta Gene 16, 108–116. doi:10.1016/j.mgene.2018.02.005
Tanner, R. A., Jin, L., Shaw, R., Murphy, S. T., and Gange, A. C. (2014). An Ecological Comparison of Impatiens Glandulifera Royle in the Native and Introduced Range. Plant Ecol. 215 (8), 833–843. doi:10.1007/s11258-014-0335-x
Thakur, G., Bag, M., Sanodiya, B., Bhadauriya, P., Debnath, M., Prasad, G., et al. (2009). Momordica Balsamina: a Medicinal and Neutraceutical Plant for Health Care Management. Cpb 10 (7), 667–682. doi:10.2174/138920109789542066
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq - Versatile and Accurate Annotation of Organelle Genomes. Nucleic Acids Res. 45, W6–W11. doi:10.1093/nar/gkx391
Torrecilha, J. K., Mariano, G. P., and Silva, P. S. C. (2013). Study of the “Impatiens Walleriana” for Phytoremediation of Chromium, Thorium, Uranium and Zinc Soil Contamination. Int. Nucl. Atla Conf. 46 (2), 24–29.
Wang, W., Yang, T., Wang, H.-L., Li, Z.-J., Ni, J.-W., Su, S., et al. (2020). Comparative and Phylogenetic Analyses of the Complete Chloroplast Genomes of Six Almond Species (Prunus Spp. L.). Sci. Rep. 10, 10137. doi:10.1038/s41598-020-67264-3
Wyman, S. K., Jansen, R. K., and Boore, J. L. (2004). Automatic Annotation of Organellar Genomes with DOGMA. Bioinformatics 20, 3252–3255. doi:10.1093/bioinformatics/bth352
Xie, D.-F., Yu, Y., Deng, Y.-Q., Li, J., Liu, H.-Y., Zhou, S.-D., et al. (2018). Comparative Analysis of the Chloroplast Genomes of the Chinese Endemic Genus Urophysa and Their Contribution to Chloroplast Phylogeny and Adaptive Evolution. Ijms 19, 1847. doi:10.3390/ijms19071847
Yan, M., Zhao, X., Zhou, J., Huo, Y., Ding, Y., and Yuan, Z. (2019). The Complete Chloroplast Genomes of Punica Granatum and a Comparison with Other Species in Lythraceae. Ijms 20, 2886. doi:10.3390/ijms20122886
Yu, S.-X., Janssens, S. B., Zhu, X.-Y., Lidén, M., Gao, T.-G., and Wang, W. (2016). Phylogeny ofImpatiens(Balsaminaceae): Integrating Molecular and Morphological Evidence into a New Classification. Cladistics 32 (2), 179–197. doi:10.1111/cla.12119
Yuan, Y.-M., Song, Y., Geuten, K., Rahelivololona, E., Wohlhauser, S., Fischer, E., et al. (2004). Phylogeny and Biogeography of Balsaminaceae Inferred from ITS Sequences. Taxon 53 (2), 391–404. doi:10.2307/4135617
Keywords: Impatiens, Balsaminaceae, chloroplast genome, comparative analysis, phylogenetic relationship
Citation: Luo C, Huang W, Yer H, Kamuda T, Li X, Li Y, Rong Y, Yan B, Wen Y, Wang Q, Huang M and Huang H (2022) Complete Chloroplast Genomes and Comparative Analyses of Three Ornamental Impatiens Species. Front. Genet. 13:816123. doi: 10.3389/fgene.2022.816123
Received: 16 November 2021; Accepted: 11 March 2022;
Published: 30 March 2022.
Edited by:
Sankar Subramanian, University of the Sunshine Coast, AustraliaReviewed by:
Saman Zulfiqar, University of Education Lahore, PakistanCopyright © 2022 Luo, Huang, Yer, Kamuda, Li, Li, Rong, Yan, Wen, Wang, Huang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meijuan Huang, eG1oaHEyMDAxQDE2My5jb20=; Haiquan Huang, aGFpcXVhbmxAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.