- 1College of Horticulture and Plant Protection, Yangzhou University, Yangzhou, China
- 2Joint International Research Laboratory of Agriculture and Agri-Product Safety of Ministry of Education of China, Yangzhou University, Yangzhou, China
Water dropwort (Oenanthe javanica) is a popular vegetable with high nutritional value and distinctive flavor. The flavor is mainly correlate with the biosynthesis of terpenoids. Shading cultivation was used to improve the flavor in the production of water dropwort. However, the changes of terpenoids and the genes involved in terpenoids biosynthesis under shading treatment remains unclear. In this study, the long- and short-reads transcriptomes of water dropwort were constructed. In total, 57,743 non-redundant high-quality transcripts were obtained from the transcriptome. 28,514 SSRs were identified from non-redundant transcripts and the mono-nucleotide repeats were the most abundant SSRs. The lncRNAs of water dropwort were recognized and their target genes were predicted. The volatile compound contents in petioles and leaf blades of water dropwort were decreased after the shading treatment. The DEGs analysis was performed to identify the terpenoids biosynthesis genes. The results indicated that 5,288 DEGs were differentially expressed in petiole, of which 22 DEGs were enriched in the terpenoids backbone biosynthesis pathway. A total of 12 DEGs in terpenoids biosynthesis pathway were selected and further verified by qRT-PCR assay, demonstrating that the terpenoids biosynthesis genes were down-regulated under shading treatment. Here, the full-length transcriptome was constructed and the regulatory genes related to terpenoids biosynthesis in water dropwort were also investigated. These results will provide useful information for future researches on functional genomics and terpenoids biosynthesis mechanism in water dropwort.
Introduction
Water dropwort (Oenanthe javanica) is a perennial aquatic herb belongs to Apiaceae family, which is mainly cultivated in tropical and temperate regions (Lu and Li, 2019). It is a popular vegetable rich in vitamins, proteins, dietary fibers, and other nutrients (Park et al., 2015; Feng et al., 2018b). Water dropwort contains many bioactive substances and it is well known to have various medicinal effects, such as antithrombotic (Ku et al., 2013), hepatoprotective (Yang et al., 2014), neuroprotective (Ma et al., 2010), anti-inflammatory (Ahn and Lee, 2017), antioxidant (Her et al., 2019), antiviral (Han et al., 2008), and anti-senescence (Moon et al., 2009).
Water dropwort is a vegetable crop with distinctive flavor. It is known that the accumulation of high concentrations of terpenoids in water dropwort produce undesirable flavor (Seo and Baek, 2005). The flavor of water dropwort can be improved by shading cultivation (Ye et al., 2009). In the production of water dropwort, one of the shading cultivation patterns is using the soil to form a soil wall to protect the water dropwort from sunlight. The distinctive flavor and tastes of water dropwort were attributed to the biosynthesis of volatile components. A previous study has reported that the volatile components that affect the taste of water dropwort are mainly terpenoids (Deng et al., 2003). Solid phase microextraction-gas chromatography-olfactometry (SPME-GC-O) assay indicated that the terpenoids, including α-terpinolene, p-cymene, β-caryophyllene, and α-terpinene, were the characteristic aroma components in water dropwort (Seo and Baek, 2005). The biosynthesis of terpenoids in water dropwort was controlled by multiple genetic factors, whereas the related regulatory genes remain unknown.
Terpenoids are an important secondary metabolite and one of the most diverse natural products in chemistry and structure (Christianson, 2017). Terpenoids are widely distributed in plants and participate in a variety of physiological processes, such as photosynthesis, ion transport, growth regulation, and stress response (Velikova et al., 2015). Terpenoids also showed high economic value. The β-caryophyllene, linalool, myrcene, and other terpenoids can be used in spices production (Behr and Johnen, 2009; Tholl, 2015). In addition, terpenoids were investigated to have many medicinal functions for humans, including anticancer, anti-inflammatory, and hypoglycemic effects (Amato et al., 2002; Ukiya et al., 2002; Juergens, 2014; Singh and Sharma, 2015). All the terpenoids are synthesized from the common C5 isoprene precursors, isopentenyl diphosphate (IPP) or its allylic isomer dimethylallyl diphosphate (DMAPP). The biosynthesis of IPP and DMAPP in plants mainly depends on two pathways, the mevalonate (MVA) pathway located in the cytoplasm and the 2-C-methylerythritol 4-phosphate (MEP) pathway located in the plastid (Vranova et al., 2013). The trans/cis-prenyltransferases catalyzes the conversion of IPP and DMAPP to polyprenyl diphosphates, such as geranyl/neryl diphosphate (GPP/NPP, C10), farnesyl diphosphate (FPP, C15), and geranylgeranyl diphosphate (GGPP, C20). Then, the diverse terpene backbones, including isoprene, monoterpenes, sesquiterpenes, diterpenes, and sesterterpenes, were synthesized under the catalysis of terpene synthase (TPS) (Nagegowda and Gupta, 2020). These terpene backbones are modified by a series of glycosylation, methylation, and hydroxylation to form different terpenoids (Pateraki et al., 2015).
The distinctive aroma and taste of water dropwort was related to the terpenoids biosynthesis and accumulation (Seo and Baek, 2005). The information on molecular mechanism and biosynthesis pathway of terpenoid in water dropwort was still limited. The transcriptome sequencing is considered to be an efficient technology to investigate the gene regulatory network and molecular mechanism in plants (Yang et al., 2012). The next generation sequencing (NGS) has been conducted to identify the genes and miRNA in water dropwort under abiotic stress (Jiang et al., 2015). With the development of sequencing technology, the genome of O. javanica was also released recently (Liu et al., 2021). In view of the full-length sequences and transcript structures, the PacBio single molecule real-time (SMRT) technology gradually takes the place of NGS sequencing and become the mainstream of transcriptome sequencing (Li et al., 2017). In this study, the PacBio SMRT and Illumina RNA sequencing was employed to investigate the regulatory genes of terpenoid biosynthesis in water dropwort. The results in this study will provide a novel insight into the terpenoid biosynthesis in water dropwort and give a reference for improving the flavor by molecular breeding.
Materials and Methods
Plant Materials and Treatments
“Liyang baiqin,” a water dropwort variety in China, was used as plant material in this study. The plants of water dropwort were grown in the field of Liyang, Jiangsu province (31°42′N, 119°48′E) under natural growth. The shading cultivation was performed. We used soil to form a barrier for preventing sunlight from water dropwort. After shading cultivation for 30 days, the green petioles (GP), green leaf blades (GL), white petioles (WP), and white leaf blade (WL), were collected for further experiments. Three biological replicates of water dropwort plants were prepared.
Extraction and Measurement of Volatile Compounds
The volatile compounds of water dropwort were extracted and measured using auto-head space-solid phase microextraction-gas chromatography-mass spectrometry (HS-SPME-GC-MS) method (Seo and Baek, 2005), with some modifications. The collected water dropwort was washed and cut into pieces. A total of 5 g of water dropwort samples were placed in flask applied and 3-heptanol (Sigma-Aldrich) was added. The volatile compound was extracted and collected by placing the SPME fiber (65 μm PDMS/DVB) in the headspace at 50°C for 30 min. The collected volatile compound was then injected to GC-MS (DSQ-II Thermo) equipped with DB-5 mass spectrometry column (30 m × 0.25 mm × 0.25 µm) for further analysis. Helium was used as carrier gas and its constant flow rate was 0.8 ml/min. The oven temperature was set at 50°C for 2 min, raised to 75°C at 5°C/min and keep 3 min, then raised to 155°C at 10°C/min and keep 4 min, and finally raised to 210°C at 5 °C/min and keep 2 min. The mass spectra were performed with 70 eV of ionization energy and 33–350 amu of scan range. The kinds of volatile compounds in water dropwort were identified by comparing the mass spectra with the NIST 05 library. 3-heptanol was used as internal standard, and the relative contents of volatile compounds in water dropwort were calculated based on the peak areas (Seo and Baek, 2005).
Library Preparation and Transcriptome Sequencing
The total RNA was extracted from the three biological replicates of WP, GP, WL, and GL samples of water dropwort, respectively. The purity and quality of RNA were estimated by Nanodrop, Agilent 2,100, and electrophoresis. A total of 12 Illumina RNA sequencing libraries was constructed using the NEBNext® Ultra™ RNA Library Prep Kit (NEB, Massachusetts, United States) according to the manufacturer’s protocols. The mixed RNA samples, F01 (WP and WL) and F02 (GP and GL), were used to construct the PacBio Iso-Seq library. The full-length cDNA was synthesized by SMARTer™ PCR cDNA Synthesis Kit (TaKaRa, Dalian, China) following the operating instructions. Bluepippin (Sage Science Beverly, MA, USA) was used to screen full-length cDNA fragments and construct cDNA libraries with different sizes (1–2, 2–3, and >3 kb). The constructed libraries were evaluated by Qubit 2.0 Fluorometer and Agilent 2,100. The SMRT sequencing and NGS was performed with Pacific Bioscience RS II (Pacific Biosciences, California, United States) platform and Illumina HiSeq4000 platform (San Diego, CA, United States) at Biomarker Technology Co. (Biomarker, Beijing, China), respectively.
Data Filtering and de Novo Assembly
The raw polymerase reads were filtered and the reads of insert (ROI) were extracted with set parameters (full passes of ≥0 and quality of >0.75). The ROI sequences were classified into FLNC and non-full-length reads (NFL) based on the filtering of cDNA primers and polyA tail signal. The FLNC sequences from the same isoform were clustered into one consensus sequence using SMRT Analysis (v2.3.0) software with iterative clustering for error correction (ICE) algorithm. The Quiver program was used to polish the consensus sequences and generate the high-quality isoforms with an accuracy rate of >99%. The quality and accuracy of Illumina RNA-seq data were detected and the raw reads with adapter and low-quality reads were filtered. Subsequently, the low-quality isoforms were further corrected by the Illumina RNA-seq data using proovread (Hackl et al., 2014). The CD-HIT program (identity >0.99) was used to de-redundancy the high-quality isoforms and corrected low-quality isoforms (Li and Godzik, 2006). Finally, the non-redundant transcripts with high-quality were constructed.
Analysis of AS Events, SSR, and lncRNA
The non-redundant transcripts with high-quality were obtained for further alternative splicing (AS) events, simple sequence repeat (SSR), and long non-coding RNAs (lncRNAs) analysis. The IsoSeq AS de novo script was used to identity the AS events (Liu et al., 2017). The FLNC transcripts of water dropwort were clustered by Cogent software (v1.0). The General feature format (GFF) file was constructed by GMAP. The AS events were detected in SUPPA with the default settings (Wu and Watanabe, 2005). MIcroSAtellite identification tool (MISA) was used to conduct the SSR analysis (http://pgrc.ipk-gatersleben.de/misa/). Seven types of SSRs were identified from the sequences of transcriptome, including mono-nucleotide repeats, di-nucleotide repeats, tri-nucleotide repeats, tetra-nucleotide repeats, penta-nucleotide repeats, hexa-nucleotide repeats, and compound SSRs. CREMA and RNAplonc were used to predict the lncRNA in plants (Simopoulos et al., 2018; Negri et al., 2019). In this study, four common computational approaches, including coding potential calculator (CPC) (Kong et al., 2007), coding-non-coding index (CNCI) (Sun et al., 2013), coding potential assessment tool (CPAT) (Wang et al., 2013), and Pfam (Finn et al., 2014), were combined to distinguish the lncRNAs in water dropwort. Based on these four approaches, lncRNAs have been successfully identified from long-reads transcriptomes in many species (Cui et al., 2020; Hou et al., 2021). LncRNAs works through mRNA binding, and the target genes of lncRNAs were predicted by LncTar (Li et al., 2015). As an efficient tool for predicting the target genes of lncRNAs, LncTar has been widely used to identify the targets in many plants, such as rice (Leng et al., 2020), Gnetum luofuense (Hou et al., 2021), and Carex breviculmis (Teng et al., 2019).
Gene Function Annotation
The coding sequences (CDS) was identified by TransDecoder (v3.0.0) online software (https://github.com/TransDecoder/TransDecoder/releases) based on the length of open reading frame (ORF), log-likelihood score, and alignments of sequences with Pfam database. The function annotation of non-redundant transcripts was conducted using BLAST software (v2.2.26) against seven databases, including NCBI non-redundant protein database (NR), Swiss-prot, gene ontology (GO), clusters of orthologous groups (COG), eukaryotic orthology groups (KOG), protein family (Pfam), kyoto encyclopedia of genes and genomes (KEGG).
Phylogenetic Analysis
Using PF03936 and PF01397 as queries, the TPS family genes in water dropwort were identified by HMMER 3.0 software with an expected threshold value <10−4 (Eddy, 2011). The TPS family sequences of Arabidopsis thaliana was obtained from the previous study (Aubourg et al., 2002). The phylogenetic tree was constructed by MEGA 7.0 using neighbor-joining method with 1,000 bootstraps (Kumar et al., 2016; Chen et al., 2017). The TPS genes of water dropwort were divided into different subfamilies according to the phylogenetic relationships with the known A. thaliana TPS proteins.
DEGs Analysis
The high-quality Illumina sequencing reads were aligned with non-redundant transcripts by Bowtie2 (Langmead and Salzberg, 2012). The expression abundance was quantified by RSEM based on the transcripts per million (TPM) (Li and Dewey, 2011). The identification of differentially expressed genes (DEGs) in different samples were conducted by DESeq R package based on the read counts (Anders and Huber, 2010). The transcripts that meet the set parameters (Fold Change ≥2 and FDR <0.01) are identified as DEGs. The GO enrichment analysis of DEGs was conducted using GOseq-R package (Tian et al., 2016). Kolmogorov-Smirnov test was employed to correct the p-value, and the corrected p-value (≤0.05) was considered as significantly enriched. The enrichment of DEGs in KEGG pathway were analyzed by KOBAS (Xie et al., 2011). The heatmap were performed based on the TPM values to investigate the expression abundance of DEGs in different samples.
qRT-PCR Assay
To validate the availability of transcriptome, qRT-PCR assay was conducted to further investigate the expression level of DEGs. The qRT-PCR was carried out based on our previous study (Feng et al., 2018a). The primers of selected DEGs were designed by Primer 6.0 software (Supplementary Table S1). OjPP2A was selected as internal reference gene. The relative expression levels of DEGs were calculated according to the 2−ΔΔCT method (Schmittgen and Livak, 2008; Jiang et al., 2014). The expression of F02.PB35028 in WP sample was used as the calibrator for qRT-PCR analysis.
Statistical Analysis
Statistical analysis was performed by SPSS 17.0 software. Correlation analysis between gene expression and volatile contents in water dropwort were conducted using the Pearson’s correlation analysis.
Results
Volatile Compound Contents of Water Dropwort
The volatile compound contents of water dropwort were detected by SPME-GC-MS equipment (Figure 1A). The results indicated that the volatile compounds in water dropwort were mainly composed of terpenoids. The total volatile compound contents of water dropwort were significantly decreased after the shading treatment (Figure 1B).
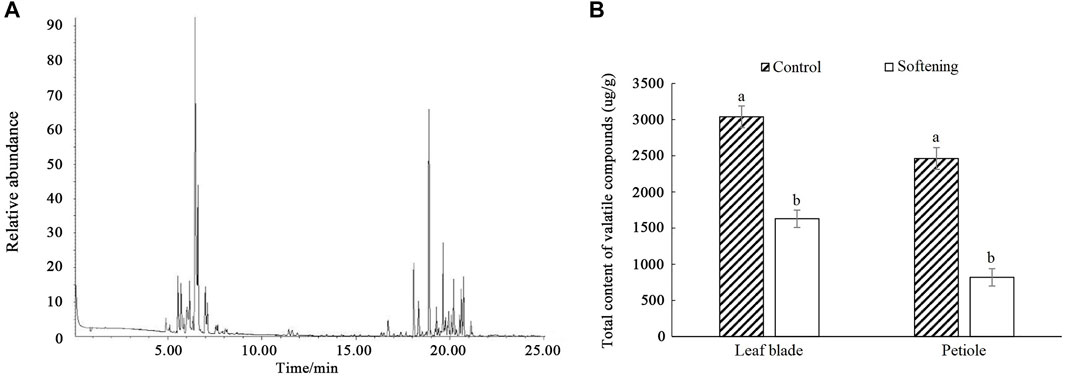
FIGURE 1. The measurement of volatile compound contents in water dropwort. (A) The gas chromatography of volatile compound in water dropwort. (B) The total volatile compound contents of water dropwort. The bar represents the mean values of three independent experiments ±SD. Different lowercase letters indicate significant differences at p < 0.05.
Transcriptome Sequencing and Assembly
In this study, two mixed RNA samples (F01 and F02) were used to obtain the full-length transcripts with different libraries sizes (1–2, 2–3, and 3–6 kb). The paired-end reads of different libraries were listed in Supplementary Table S2. A total of 450,876 polymerase read sequences were obtained from F01, and 601,168 polymerase read sequences were obtained from F02. The polymerase reads (length >50 bp and accuracy >0.75) were filtered, yielding 3,539,289 subreads for F01 and 4,203,996 subreads for F02 samples, respectively. The reads of insert (ROI), ROI bases, mean read length of insert, and mean read quality of insert were analyzed in F01 and F02 samples (Supplementary Table S3). The ROI read length distributions of each size bins were shown in Figure 2
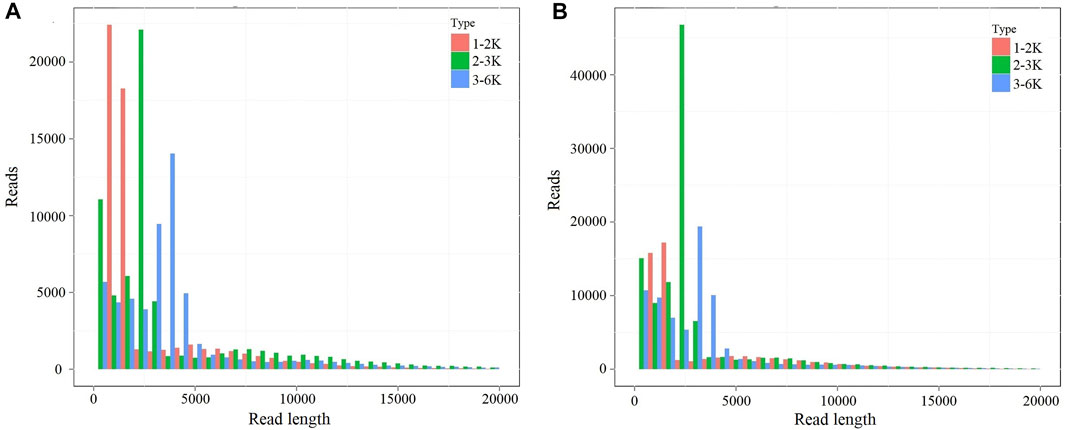
FIGURE 2. The ROI read length distribution of each size bins. (A) The ROI read length distribution of each size bins in F01 sample. (B) The ROI read length distribution of each size bins in F02 sample. The columns with different colors indicate different read length.
For F01 sample, a total of 77,470 FLNCs were extracted from the ROIs. The extracted 47,559 consensus FLNC reads includes 38,186 high-quality isoforms and 9,373 low-quality isoforms. Meanwhile, 103,548 FLNC reads were obtained from F02 sample. The extracted 59,819 consensus FLNC reads were composed of 48,096 high-quality isoforms and 11,723 low-quality isoforms (Supplementary Table S4). Finally, the high-quality and corrected low-quality transcripts of F01 and F02 generated 57,743 non-redundant transcripts.
AS Events, SSR, and lncRNA Analysis
In total, 664 and 870 AS events were detected from the full-length isoforms of F01 and F02 samples, respectively (Supplementary Table S5). 28,514 SSRs were identified from 57,700 non-redundant transcripts (>500 bp) (Supplementary Table S6). The identified SSRs can be divided into mono-nucleotide repeats (p1), di-nucleotide repeats (p2), tri-nucleotide repeats (p3), tetra-nucleotide repeats (p4), penta-nucleotide repeats (p5), hexa-nucleotide repeats (p6), and compound SSR(c). The density distribution of different SSR types indicated that the most abundant SSRs was mono-nucleotide repeats, followed by di-nucleotide repeats and tri-nucleotide repeats (Figure 3A). CPC, CNCI, CPAT, and Pfam databases were used to predict lncRNA. We found 612 lncRNAs were both present in all four prediction methods (Figure 3B). The target genes predication of lncRNAs demonstrated that 96 lncRNAs were predicted to target at least one gene, among which F02.PB15548 had the maximum target genes with 19 target genes (Supplementary Table S7).
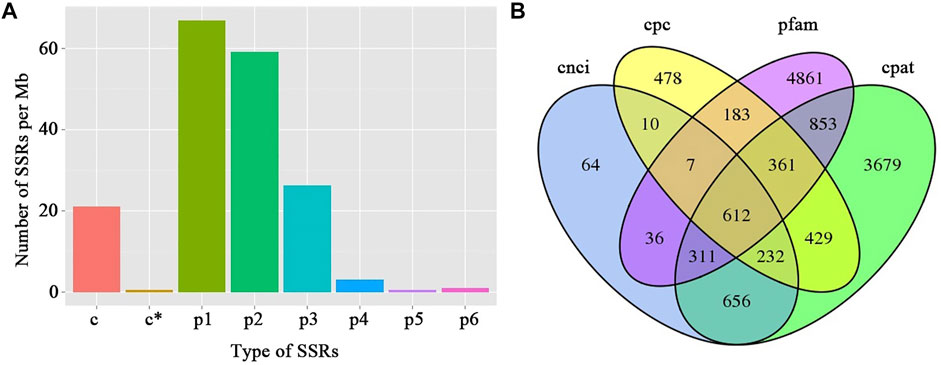
FIGURE 3. Statistic of SSR density and the prediction of lncRNAs in water dropwort. (A) Density analysis of the different SSR types. p1: mono-nucleotide repeats; p2: di-nucleotide repeats; p3: tri-nucleotide repeats; p4: tetra-nucleotide repeats; p5: penta-nucleotide repeats; p6: hexa-nucleotide repeats; c: compound SSR. (B) The venn diagram showing the number of lncRNAs using different prediction methods. CPC: coding potential calculator; CNCI: coding-non-coding index; CPAT: coding potential assessment tool; Pfam: protein family.
Coding Sequence Prediction and Functional Annotation
TransDecoder (v3.0.0) was used to predict the coding sequence (CDS) from the non-redundant transcripts. A total of 56,307 open reading frame (ORF) was obtained in water dropwort. The predicted length distribution of the ORF coding protein sequence indicated that the ORFs encoding 200–300 amino acid (aa) was the most, followed by that encoding 100–200 aa and 300–400 aa (Supplementary Figure S1). The transcripts were blast with the NR, Swiss-prot, GO, COG, KOG, Pfam, and KEGG databases to obtain the functional annotation. A total of 57,118 non-redundant transcripts were annotated, among which 27,100 were annotated in GO, 25,644 in KEGG, 37,227 in KOG, 48,433 in Pfam, 43,401 in Swiss-prot, 25,353 in COG, 55,815 in eggNOG, and 56,684 in NR (Supplementary Table S8).
Phylogenetic Analysis of TPS Family
TPS is the key enzyme in the terpenoid biosynthesis pathway and contributes to the formation of various terpenoid backbones (Bohlmann and Keeling, 2008). In this study, 25 TPS family genes were identified from the transcriptome of water dropwort. The phylogenetic tree was constructed using the TPS proteins from water dropwort and Arabidopsis. These TPS family genes were divided into five subfamilies, including TPS-a, TPS-b, TPS-c, TPS-e/f, and TPS-g (Figure 4). TPS-b subfamily has 13 TPS members in water dropwort, which accounts the highest proportion among all subfamilies.
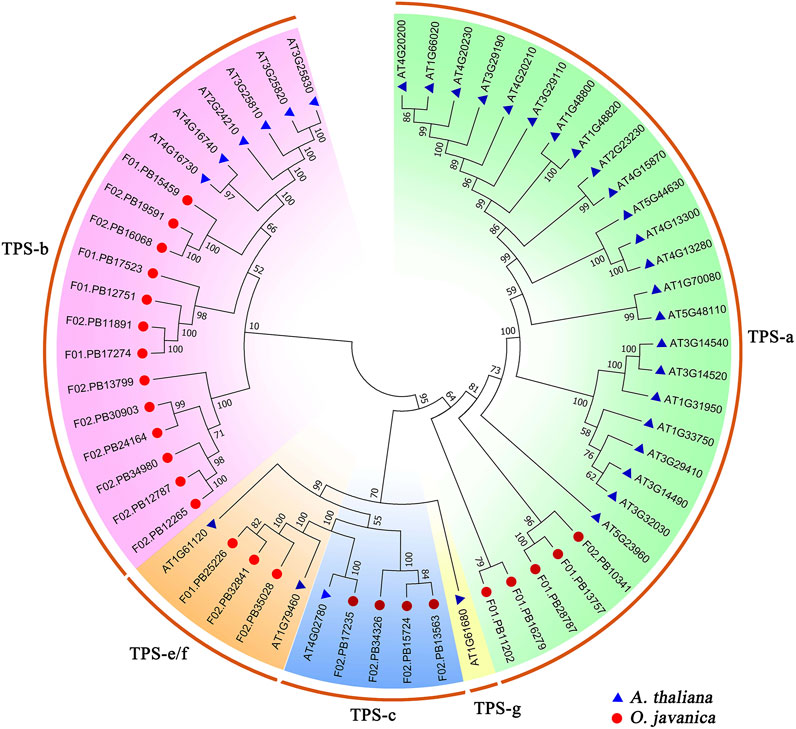
FIGURE 4. The phylogenetic analysis of TPS family proteins from water dropwort and Arabidopsis. The TPS family genes in water dropwort were identified by HMMER 3.0 software using PF03936 and PF01397 as queries. The TPS genes of water dropwort and Arabidopsis are represented by different colored shapes, respectively. The phylogenetic tree was constructed by MEGA 7.0 using neighbor-joining method with 1,000 bootstraps.
Analysis of Differentially Expressed Genes
The TPM values were used for visualization and correlation analysis. The distribution of TPM values in all samples were shown in Figure 5A. The correlation analysis of expression levels in different samples was conducted (Figure 5B). As the petioles of water dropwort were the main edible organs, the DEGs analysis was conducted in GP and WP. In this study, 5,288 differentially expressed genes (DEGs) were identified between GP and WP, including 1,253 up-regulated and 4,035 down-regulated DEGs (Supplementary Figure S2).
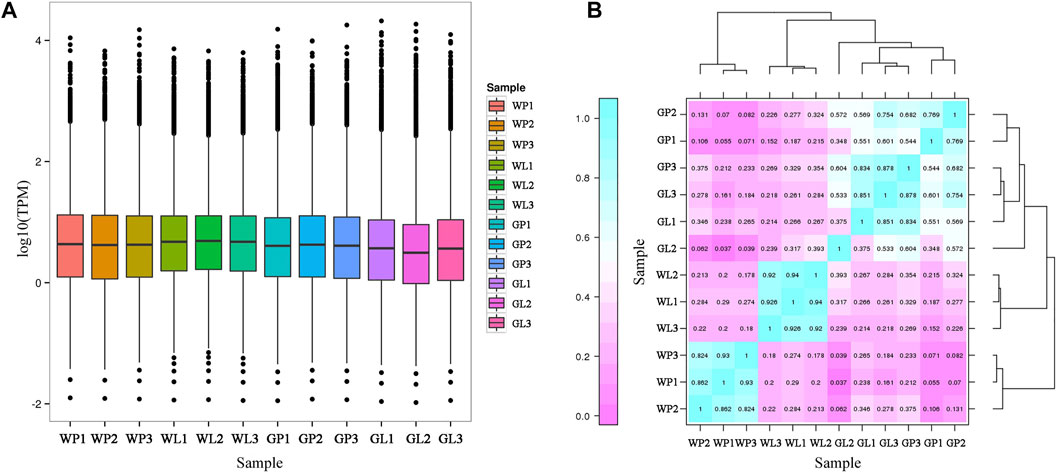
FIGURE 5. The statistic of TPM values among all water dropwort samples. (A) The distribution of TPM values in different water dropwort samples. (B)The heat map for correlation analysis of expression levels in different water dropwort samples. GP: green petioles; GL: green leaf blades; WP: white petioles; WL: white leaf blade.
The GO annotation and enrichment analysis suggested that the majority of DEGs were classified into three categories: “biological process,” “cellular component” and “molecular function” (Figure 6). As for ‘biological process’ category, the DEGs were the most enriched in ‘metabolic process’. The results indicated that the DEGs involved in metabolites biosynthesis play important roles in shading treatment of water dropwort. In order to investigate the DEGs of various metabolites pathways in water dropwort, the KEGG analysis was conducted. Carbon metabolism is the metabolic pathway with the most DEGs in photosynthetic organisms, followed by carbon fixation (Supplementary Figure S3). After the shading treatment, the volatile compound contents of water dropwort were decreased, and its flavor improved significantly. A total of 22 DEGs were enriched in terpenoid backbone biosynthesis. We speculated that these DEGs were involved in the regulation of terpenoid biosynthesis in water dropwort.
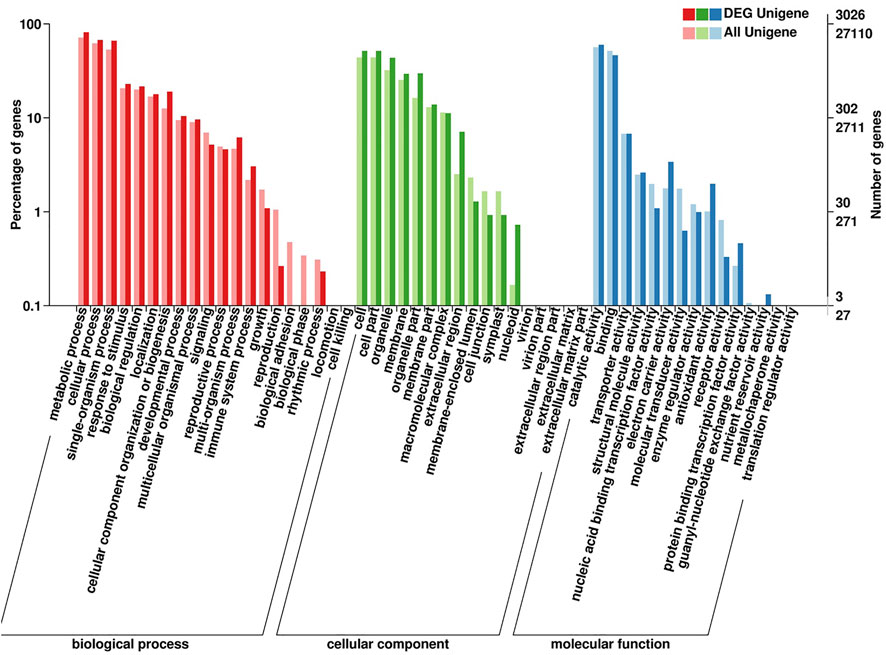
FIGURE 6. GO enrichment analysis of DEGs. Each gene was classified into at least one GO term, and all these genes were grouped into three categories, namely, molecular function, cellular component, and biological process.
Transcription Profiles of Genes Involved in Terpenoids Biosynthesis
The volatile terpenoids of water dropwort were decreased after the shading treatment. We analyzed the transcription profiles of DEGs in the terpenoids backbone biosynthesis pathway. The results indicated that the transcription levels of the DEGs were down-regulated after the shading treatment (Figure 7). To further verify the reliability of transcriptome data, 12 DEGs including DXS (F01.PB13304), HDS (F01.PB19227), HDR (F01.PB4573), GGPS (F01.PB9526, F02.PB11697, and F02.PB8158), SPS (F02.PB14607, F02.PB6913), and TPS (F02.PB12265, F02.PB17907, F02.PB13799, and F02.PB35028) in terpenoid biosynthesis pathway were selected to perform qRT-PCR assay (Figure 8). The expression levels of terpenoids biosynthesis genes in white petioles were down-regulated compared to green petioles, which were consistent with the results of RNA-Seq. Pearson’s correlation analysis indicated that the expression levels of terpenoids biosynthesis genes were positively correlated with the volatile contents in water dropwort (Supplementary Table S9).
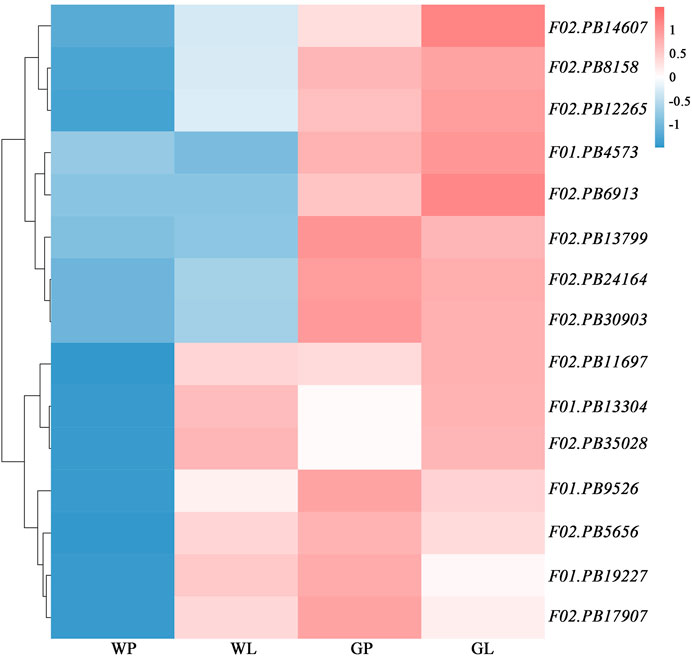
FIGURE 7. Heatmap of DEGs transcript abundances in terpenoids backbone biosynthesis pathway in water dropwort. GP: green petioles; GL: green leaf blades; WP: white petioles; WL: white leaf blade. Red and blue colors represent high and low transcript abundances, respectively.
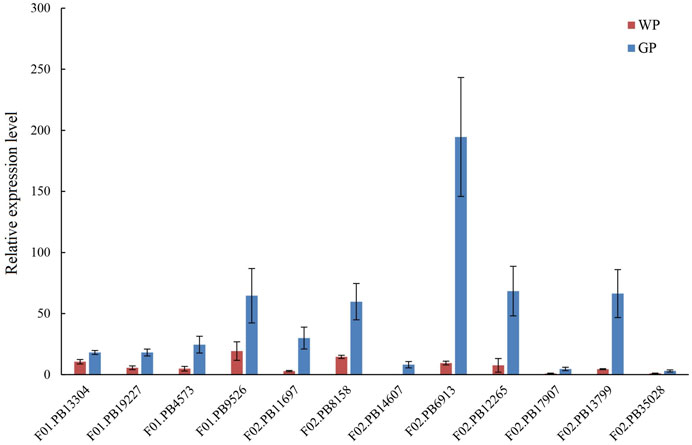
FIGURE 8. qRT-PCR assay of selected DEGs in terpenoids backbone biosynthesis pathway in water dropwort. The bar represents the mean values of three independent experiments ±SD. WP: white petioles; GP: green petioles.
Discussion
In view of the rising importance of aquatic vegetables, more and more studies on water dropwort have been reported in recent years (Park et al., 2015; Gao et al., 2020). However, the research on molecular mechanisms in water dropwort is still lagging far behind other crop species. In this work, the full-length transcriptome of water dropwort was constructed by the integrative analysis of the PacBio SMRT and Illumina RNA sequencing. A total of 57,743 non-redundant transcripts were obtained from the full-length transcriptome. The SSR, lncRNAs, and AS events were analyzed based on the full-length transcriptome. SSR is also known as microsatellites, which is a type of short tandem repetitive DNA sequences with 1–6 bases pairs (Toth et al., 2000). SSRs have been widely applied in population genetics, genetic linkage mapping, and molecular breeding (Kumar et al., 2015; Nachimuthu et al., 2015). A total of 28,514 SSRs were identified from the non-redundant transcripts, and the mono-nucleotide repeats were the most abundant SSRs. LncRNAs are a kind of non-coding RNA, which play important roles in biological processes by acting on target genes (Wang and Chang, 2011). Based on CPC, CNCI, CPAT, and Pfam databases, 612 lncRNAs were recognized and their target genes were predicted. The SSRs and lncRNAs obtained from the full-length transcriptome will provide useful information for future research on water dropwort.
Water dropwort was a popular vegetable in East Asia on account of its nutritional values and distinctive flavor (Ji et al., 2021). By using SPME-GC-O technology, terpenoids were investigated to be the characteristic aroma components in water dropwort. Terpenoids are widely existed in plants tissues and contribute to the formation of distinct smells (Dudareva et al., 2004). Water dropwort with a high concentration of terpenoids will emit undesirable flavor (Seo and Baek, 2005). Shading treatment was an effective approach to improve the flavor in the production of water dropwort. Whereas now, the changes of terpenoids and related molecular mechanism during shading treatment in water dropwort remains unknown. Current results demonstrated that the volatile compound contents of water dropwort were significantly decreased after shading treatment. Altered volatile substance content was related to the flavor improvement of water dropwort. Similarly, the high levels of flavonols resulted in the bitter flavor of tea, and the flavonols contents of tea were increased under UV-B and decreased after shading treatments (Zhao et al., 2021).
The key regulatory genes and related molecular mechanisms play an important role in plant growth and terpenoids biosynthesis (Feng et al., 2020; Nagegowda and Gupta, 2020). The DEGs analysis was conducted to investigate the molecular mechanism of water dropwort during shading treatment. In “biological process” category, the DEGs were the most enriched in “metabolic process,” suggesting the regulation of metabolites biosynthesis is of great importance in water dropwort during shading treatment (Young et al., 2010). KEGG analysis showed that DEGs in photosynthesis-related pathways resulted in the albino of water dropwort, which were induced by the lack of light after shading treatment. Previous study also reported that the photosynthetic pathway and chlorophyll content of Tetrastigma hemsleyanum Diels et Gilg were affected by shading treatment (Dai et al., 2009). A total of 22 DEGs were enriched in terpenoid backbone biosynthesis pathway based on the transcriptome analysis, suggesting these DEGs were involved in the terpenoids biosynthesis in water dropwort. Integrative analysis of transcriptome and metabolome demonstrated that the biosynthesis of sesquiterpene in Sindora glabra was regulated by the DEGs in terpenoid backbone biosynthesis pathway (Yu et al., 2021).
Terpenoids were synthesized using C5 isoprene as substrate under the catalysis of various enzymes in plants (Nagegowda, 2010). At present, the genes related to terpenoids biosynthesis in water dropwort are still unclear. In this work, qRT-PCR assay verified that the DEGs identified from transcriptome were down-regulated after shading treatment, such as DXS (F01.PB13304), HDS (F01.PB19227), HDR (F01.PB4573), and TPS (F02.PB12265, F02.PB17907, F02.PB13799, and F02.PB35028). DXS is the first rate-limiting enzyme in MEP pathway, which catalyze the condensation of glyceraldehyde 3-phosphate (G3P) and pyruvate to form 1-deoxy-d-xylulose-5-phosphate (DXP) (Rohdich et al., 2003). The DXS has been reported in pepper, rubber tree, potato, and masson pine (Bouvier et al., 1998; Seetang-Nun et al., 2008; Henriquez et al., 2016; Li et al., 2021). HDS is 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase, which is responsible for the biosynthesis of 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate (HMBPP) (Gutierrez-Nava et al., 2004). In the last step of MEP pathway, HDR catalyzed the conversion of HMBPP into IPP and DMAPP (Laule et al., 2003). The polyprenyl diphosphates generated from IPP and DMAPP was further converted into various terpenoid backbones by the enzymes of known TPS family (Bohlmann et al., 1998; Huang et al., 2017). In the future works, the above DEGs associated terpenoids biosynthesis will be selected for further functional verification in water dropwort.
Conclusion
In conclusion, we constructed the full-length transcriptome and obtained 57,743 non-redundant transcripts from water dropwort. To the best of our knowledge, this study is the first to report a full-length transcriptome in water dropwort. The AS events, SSR, and lncRNAs were predicted based on the transcriptome. The volatile compound contents of water dropwort were decreased after the shading treatment. 22 DEGs in terpenoid backbone biosynthesis pathway were differentially expressed. The current study identified the terpenoids biosynthesis genes and will be helpful for investigating the mechanism of terpenoids biosynthesis in water dropwort. Also, the full-length transcriptome in our study will establish a basis for functional genomics and genetic engineering breeding in water dropwort in the future.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: PRJNA780322.' 211112 SL: Assessed for PM, proceeding with review
Author Contributions
KF and L-JL initiated and designed the research. KF, X-YK, RL, and Y-JY, S-PZ, and PW performed the experiments. KF and RL analyzed the data. KF and LJL contributed reagents/materials/analysis tools. KF wrote the manuscript. L-JL revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was financially supported by the National Natural Science Foundation of China (32102368), Jiangsu Agricultural Science and Technology Innovation Fund (CX(21)3026), Natural Science Foundation of the Jiangsu Higher Education Institutions of China (21KJB210008), Jiangsu seed industry revitalization “Jie Bang Gua Shuai” project (JBGS(2021)017), and China Agriculture Research System (CARS-24).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank the reviewers for their helpful comments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.813216/full#supplementary-material
References
Ahn, H., and Lee, G.-S. (2017). Isorhamnetin and Hyperoside Derived from Water Dropwort Inhibits Inflammasome Activation. Phytomedicine 24, 77–86. doi:10.1016/j.phymed.2016.11.019
Amato, R. J., Perez, C., and Pagliaro, L. (2002). Irofulven, a Novel Inhibitor of DNA Synthesis, in Metastatic Renal Cell Cancer. Invest. New Drugs 20, 413–417. doi:10.1023/a:1020649827173
Anders, S., and Huber, W. (2010). Differential Expression Analysis for Sequence Count Data. Genome Biol. 11, R106. doi:10.1186/gb-2010-11-10-r106
Aubourg, S., Lecharny, A., and Bohlmann, J. (2002). Genomic Analysis of the Terpenoid Synthase (AtTPS) Gene Family of Arabidopsis thaliana. Mol. Gen. Genomics 267, 730–745. doi:10.1007/s00438-002-0709-y
Behr, A., and Johnen, L. (2009). Myrcene as a Natural Base Chemical in Sustainable Chemistry: A Critical Review. Chemsuschem 2, 1072–1095. doi:10.1002/cssc.200900186
Bohlmann, J., and Keeling, C. I. (2008). Terpenoid Biomaterials. Plant J. 54, 656–669. doi:10.1111/j.1365-313x.2008.03449.x
Bohlmann, J., Meyer-Gauen, G., and Croteau, R. (1998). Plant Terpenoid Synthases: Molecular Biology and Phylogenetic Analysis. Proc. Natl. Acad. Sci. U.S.A. 95, 4126–4133. doi:10.1073/pnas.95.8.4126
Bouvier, F., D'harlingue, A., Suire, C., Backhaus, R. A., and Camara, B. (1998). Dedicated Roles of Plastid Transketolases during the Early Onset of Isoprenoid Biogenesis in Pepper Fruits1. Plant Physiol. 117, 1423–1431. doi:10.1104/pp.117.4.1423
Chen, X., Yang, W., Zhang, L., Wu, X., Cheng, T., and Li, G. (2017). Genome-wide Identification, Functional and Evolutionary Analysis of Terpene Synthases in Pineapple. Comput. Biol. Chem. 70, 40–48. doi:10.1016/j.compbiolchem.2017.05.010
Christianson, D. W. (2017). Structural and Chemical Biology of Terpenoid Cyclases. Chem. Rev. 117, 11570–11648. doi:10.1021/acs.chemrev.7b00287
Cui, J., Shen, N., Lu, Z., Xu, G., Wang, Y., and Jin, B. (2020). Analysis and Comprehensive Comparison of PacBio and Nanopore-Based RNA Sequencing of the Arabidopsis Transcriptome. Plant Methods 16, 85. doi:10.1186/s13007-020-00629-x
Dai, Y., Shen, Z., Liu, Y., Wang, L., Hannaway, D., and Lu, H. (2009). Effects of shade treatments on the photosynthetic capacity, chlorophyll fluorescence, and chlorophyll content of Tetrastigma hemsleyanum Diels et Gilg. Environ. Exp. Bot. 65, 177–182. doi:10.1016/j.envexpbot.2008.12.008
Deng, C., Song, G., Zheng, X., Hu, Y., and Zhang, X. (2003). Analysis of the Volatile Constituents ofApium Graveolens L. andOenanthe L. By Gas Chromatography-Mass Spectrometry, Using Headspace Solid-phase Microextraction. Chromatographia 57, 805–809. doi:10.1007/bf02491769
Dudareva, N., Pichersky, E., and Gershenzon, J. (2004). Biochemistry of Plant Volatiles. Plant Physiol. 135, 1893–1902. doi:10.1104/pp.104.049981
Eddy, S. R. (2011). Accelerated Profile HMM Searches. Plos Comput. Biol. 7, e1002195. doi:10.1371/journal.pcbi.1002195
Feng, K., Hou, X.-L., Xing, G.-M., Liu, J.-X., Duan, A.-Q., Xu, Z.-S., et al. (2020). Advances in AP2/ERF Super-family Transcription Factors in Plant. Crit. Rev. Biotechnol. 40, 750–776. doi:10.1080/07388551.2020.1768509
Feng, K., Xu, Z.-S., Liu, J.-X., Li, J.-W., Wang, F., and Xiong, A.-S. (2018a). Isolation, Purification, and Characterization of AgUCGalT1, a Galactosyltransferase Involved in Anthocyanin Galactosylation in Purple Celery (Apium graveolens L.). Planta 247, 1363–1375. doi:10.1007/s00425-018-2870-5
Feng, K., Xu, Z.-S., Que, F., Liu, J.-X., Wang, F., and Xiong, A.-S. (2018b). An R2R3-MYB Transcription Factor, OjMYB1, Functions in Anthocyanin Biosynthesis in Oenanthe Javanica. Planta 247, 301–315. doi:10.1007/s00425-017-2783-8
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the Protein Families Database. Nucl. Acids Res. 42, D222–D230. doi:10.1093/nar/gkt1223
Gao, J., Yang, L., Zhong, R., Chen, Y., Zhang, J., Gao, J., et al. (2020). Comparison of Nitrogen and Phosphorus Removal Efficiency between Two Types of Baffled Vertical Flow Constructed Wetlands Planted with Oenanthe Javanica. Water Sci. Technol. 81, 2023–2032. doi:10.2166/wst.2020.265
Gutiérrez-Nava, M. D. L., Gillmor, C. S., Jiménez, L. F., Guevara-García, A., and León, P. (2004). CHLOROPLAST BIOGENESIS Genes Act Cell and Noncell Autonomously in Early Chloroplast Development. Plant Physiol. 135, 471–482. doi:10.1104/pp.103.036996
Hackl, T., Hedrich, R., Schultz, J., and Förster, F. (2014). Proovread : Large-Scale High-Accuracy PacBio Correction through Iterative Short Read Consensus. Bioinformatics 30, 3004–3011. doi:10.1093/bioinformatics/btu392
Han, Y.-Q., Huang, Z.-M., Yang, X.-B., Liu, H.-Z., and Wu, G.-X. (2008). In Vivo and In Vitro Anti-hepatitis B Virus Activity of Total Phenolics from Oenanthe Javanica. J. Ethnopharmacology 118, 148–153. doi:10.1016/j.jep.2008.03.024
Henriquez, M. A., Soliman, A., Li, G., Hannoufa, A., Ayele, B. T., and Daayf, F. (2016). Molecular Cloning, Functional Characterization and Expression of Potato ( Solanum tuberosum ) 1-deoxy- D -xylulose 5-phosphate Synthase 1 ( StDXS1 ) in Response to Phytophthora Infestans. Plant Sci. 243, 71–83. doi:10.1016/j.plantsci.2015.12.001
Her, Y., Shin, B.-N., Lee, Y., Park, J., Kim, D., Kim, K., et al. (2019). Oenanthe Javanica Extract Protects Mouse Skin from UVB Radiation via Attenuating Collagen Disruption and Inflammation. Ijms 20, 1435. doi:10.3390/ijms20061435
Hou, C., Lian, H., Cai, Y., Wang, Y., Liang, D., and He, B. (2021). Comparative Analyses of Full-Length Transcriptomes Reveal Gnetum Luofuense Stem Developmental Dynamics. Front. Genet. 12, 615284. doi:10.3389/fgene.2021.615284
Huang, A. C., Kautsar, S. A., Hong, Y. J., Medema, M. H., Bond, A. D., Tantillo, D. J., et al. (2017). Unearthing a Sesterterpene Biosynthetic Repertoire in the Brassicaceae through Genome Mining Reveals Convergent Evolution. Proc. Natl. Acad. Sci. U S A. 114, E6005–E6014. doi:10.1073/pnas.1705567114
Ji, Q., Zhu, H., Huang, X., Zhou, K., Liu, Z., Sun, Y., et al. (2021). Uncovering Phylogenetic Relationships and Genetic Diversity of Water Dropwort Using Phenotypic Traits and SNP Markers. PLoS One 16, e0249825. doi:10.1371/journal.pone.0249825
Jiang, Q., Wang, F., Li, M.-Y., Ma, J., Tan, G.-F., and Xiong, A.-S. (2014). Selection of Suitable Reference Genes for qPCR Normalization under Abiotic Stresses in Oenanthe Javanica (BI.) DC. PLoS One 9, e92262. doi:10.1371/journal.pone.0092262
Jiang, Q., Wang, F., Tan, H.-W., Li, M.-Y., Xu, Z.-S., Tan, G.-F., et al. (2015). De Novo transcriptome Assembly, Gene Annotation, Marker Development, and miRNA Potential Target Genes Validation under Abiotic Stresses in Oenanthe Javanica. Mol. Genet. Genomics 290, 671–683. doi:10.1007/s00438-014-0953-y
Juergens, U. (2014). Anti-inflammatory Properties of the Monoterpene 1.8-cineole: Current Evidence for Co-medication in Inflammatory Airway Diseases. Drug Res. (Stuttg) 64, 638–646. doi:10.1055/s-0034-1372609
Kong, L., Zhang, Y., Ye, Z.-Q., Liu, X.-Q., Zhao, S.-Q., Wei, L., et al. (2007). CPC: Assess the Protein-Coding Potential of Transcripts Using Sequence Features and Support Vector Machine. Nucleic Acids Res. 35, W345–W349. doi:10.1093/nar/gkm391
Ku, S.-K., Kim, T. H., Lee, S., Kim, S. M., and Bae, J.-S. (2013). Antithrombotic and Profibrinolytic Activities of Isorhamnetin-3-O-Galactoside and Hyperoside. Food Chem. Toxicol. 53, 197–204. doi:10.1016/j.fct.2012.11.040
Kumar, M., Choi, J.-Y., Kumari, N., Pareek, A., and Kim, S.-R. (2015). Molecular Breeding in Brassica for Salt Tolerance: Importance of Microsatellite (SSR) Markers for Molecular Breeding in Brassica. Front. Plant Sci. 6, 688. doi:10.3389/fpls.2015.00688
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. doi:10.1093/molbev/msw054
Langmead, B., and Salzberg, S. L. (2012). Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 9, 357–359. doi:10.1038/nmeth.1923
Laule, O., Fürholz, A., Chang, H.-S., Zhu, T., Wang, X., Heifetz, P. B., et al. (2003). Crosstalk between Cytosolic and Plastidial Pathways of Isoprenoid Biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 100, 6866–6871. doi:10.1073/pnas.1031755100
Leng, Y., Sun, J., Wang, J., Liu, H., Zheng, H., Zhang, M., et al. (2020). Genome-wide lncRNAs Identification and Association Analysis for Cold-Responsive Genes at the Booting Stage in rice (Oryza Sativa L.). Plant Genome 13, e20020. doi:10.1002/tpg2.20020
Li, B., and Dewey, C. N. (2011). RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinformatics 12, 323. doi:10.1186/1471-2105-12-323
Li, J., Ma, W., Zeng, P., Wang, J., Geng, B., Yang, J., et al. (2015). LncTar: a Tool for Predicting the RNA Targets of Long Noncoding RNAs. Brief Bioinform 16, 806–812. doi:10.1093/bib/bbu048
Li, R., Chen, P., Zhu, L., Wu, F., Chen, Y., Zhu, P., et al. (2021). Characterization and Function of the 1-Deoxy-D-Xylose-5-Phosphate Synthase (DXS) Gene Related to Terpenoid Synthesis in Pinus Massoniana. Ijms 22, 848. doi:10.3390/ijms22020848
Li, W., and Godzik, A. (2006). Cd-hit: a Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 22, 1658–1659. doi:10.1093/bioinformatics/btl158
Li, Y., Dai, C., Hu, C., Liu, Z., and Kang, C. (2017). Global Identification of Alternative Splicing via Comparative Analysis of SMRT- and Illumina-Based RNA-Seq in Strawberry. Plant J. 90, 164–176. doi:10.1111/tpj.13462
Liu, J.-X., Jiang, Q., Tao, J.-P., Feng, K., Li, T., Duan, A.-Q., et al. (2021). Integrative Genome, Transcriptome, microRNA, and Degradome Analysis of Water Dropwort (Oenanthe Javanica) in Response to Water Stress. Hortic. Res. 8, 262. doi:10.1038/s41438-021-00707-8
Liu, X., Mei, W., Soltis, P. S., Soltis, D. E., and Barbazuk, W. B. (2017). Detecting Alternatively Spliced Transcript Isoforms from Single‐molecule Long‐read Sequences without a Reference Genome. Mol. Ecol. Resour. 17, 1243–1256. doi:10.1111/1755-0998.12670
Lu, C. L., and Li, X. F. (2019). A Review of Oenanthe Javanica (Blume) DC. As Traditional Medicinal Plant and its Therapeutic Potential. Evid. Based Complement. Alternat Med. 2019, 6495819. doi:10.1155/2019/6495819
Ma, C. J., Lee, K. Y., Jeong, E. J., Kim, S. H., Park, J., Choi, Y. H., et al. (2010). Persicarin from Water Dropwort (Oenanthe Javanica) Protects Primary Cultured Rat Cortical Cells from Glutamate-Induced Neurotoxicity. Phytother. Res. 24, 913–918. doi:10.1002/ptr.3065
Moon, S. C., Park, S. C., Yeo, E. J., and Kwak, C. S. (2009). Water Dropwort (Ostericum Sieboldii) and Sedum (Sedum Sarmentosum) Delay H2O2-Induced Senescence in Human Diploid Fibroblasts. J. Med. Food 12, 485–492. doi:10.1089/jmf.2008.1111
Nachimuthu, V. V., Muthurajan, R., Duraialaguraja, S., Sivakami, R., Pandian, B. A., Ponniah, G., et al. (2015). Analysis of Population Structure and Genetic Diversity in rice Germplasm Using SSR Markers: an Initiative towards Association Mapping of Agronomic Traits in Oryza Sativa. Rice 8, 30. doi:10.1186/s12284-015-0062-5
Nagegowda, D. A., and Gupta, P. (2020). Advances in Biosynthesis, Regulation, and Metabolic Engineering of Plant Specialized Terpenoids. Plant Sci. 294, 110457. doi:10.1016/j.plantsci.2020.110457
Nagegowda, D. A. (2010). Plant Volatile Terpenoid Metabolism: Biosynthetic Genes, Transcriptional Regulation and Subcellular Compartmentation. FEBS Lett. 584, 2965–2973. doi:10.1016/j.febslet.2010.05.045
Negri, T. d. C., Alves, W. A. L., Bugatti, P. H., Saito, P. T. M., Domingues, D. S., and Paschoal, A. R. (2019). Pattern Recognition Analysis on Long Noncoding RNAs: a Tool for Prediction in Plants. Brief Bioinform 20, 682–689. doi:10.1093/bib/bby034
Park, J. H., Cho, J. H., Kim, I. H., Ahn, J. H., Lee, J.-C., Chen, B. H., et al. (2015). Oenanthe Javanica Extract Protects against Experimentally Induced Ischemic Neuronal Damage via its Antioxidant Effects. Chin. Med. J. (Engl). 128, 2932–2937. doi:10.4103/0366-6999.168063
Pateraki, I., Heskes, A. M., and Hamberger, B. (2015). Cytochromes P450 for Terpene Functionalisation and Metabolic Engineering. Adv. Biochem. Eng. Biotechnol. 148, 107–139. doi:10.1007/10_2014_301
Rohdich, F., Zepeck, F., Adam, P., Hecht, S., Kaiser, J., Laupitz, R., et al. (2003). The Deoxyxylulose Phosphate Pathway of Isoprenoid Biosynthesis: Studies on the Mechanisms of the Reactions Catalyzed by IspG and IspH Protein. Proc. Natl. Acad. Sci. U.S.A. 100, 1586–1591. doi:10.1073/pnas.0337742100
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 3, 1101–1108. doi:10.1038/nprot.2008.73
Seetang-Nun, Y., Sharkey, T. D., and Suvachittanont, W. (2008). Molecular Cloning and Characterization of Two cDNAs Encoding 1-Deoxy-D-Xylulose 5-phosphate Reductoisomerase from Hevea Brasiliensis. J. Plant Physiol. 165, 991–1002. doi:10.1016/j.jplph.2007.06.014
Seo, W. H., and Baek, H. H. (2005). Identification of Characteristic Aroma-Active Compounds from Water Dropwort (Oenanthe Javanica DC.). J. Agric. Food Chem. 53, 6766–6770. doi:10.1021/jf050150z
Simopoulos, C. M. A., Weretilnyk, E. A., and Golding, G. B. (2018). Prediction of Plant lncRNA by Ensemble Machine Learning Classifiers. BMC Genomics 19, 316. doi:10.1186/s12864-018-4665-2
Singh, B., and Sharma, R. A. (2015). Plant Terpenes: Defense Responses, Phylogenetic Analysis, Regulation and Clinical Applications. 3 Biotech. 5, 129–151. doi:10.1007/s13205-014-0220-2
Sun, L., Luo, H., Bu, D., Zhao, G., Yu, K., Zhang, C., et al. (2013). Utilizing Sequence Intrinsic Composition to Classify Protein-Coding and Long Non-coding Transcripts. Nucleic Acids Res. 41, e166. doi:10.1093/nar/gkt646
Teng, K., Teng, W., Wen, H., Yue, Y., Guo, W., Wu, J., et al. (2019). PacBio Single-Molecule Long-Read Sequencing Shed New Light on the Complexity of the Carex breviculmis Transcriptome. BMC Genomics 20, 789. doi:10.1186/s12864-019-6163-6
Tholl, D. (2015). Biosynthesis and Biological Functions of Terpenoids in Plants. Biotechnol. Isoprenoids 148, 63–106. doi:10.1007/10_2014_295
Tian, Z., Wang, C., Guo, M., Liu, X., and Teng, Z. (2016). An Improved Method for Functional Similarity Analysis of Genes Based on Gene Ontology. BMC Syst. Biol. 10, 119. doi:10.1186/s12918-016-0359-z
Tóth, G., Gáspári, Z., and Jurka, J. (2000). Microsatellites in Different Eukaryotic Genomes: Survey and Analysis. Genome Res. 10, 967–981. doi:10.1101/gr.10.7.967
Ukiya, M., Akihisa, T., Yasukawa, K., Tokuda, H., Toriumi, M., Koike, K., et al. (2002). Anti-inflammatory and Anti-tumor-promoting Effects of Cucurbitane Glycosides from the Roots of Bryonia Dioica. J. Nat. Prod. 65, 179–183. doi:10.1021/np010423u
Velikova, V., Müller, C., Ghirardo, A., Rock, T. M., Aichler, M., Walch, A., et al. (2015). Knocking Down of Isoprene Emission Modifies the Lipid Matrix of Thylakoid Membranes and Influences the Chloroplast Ultrastructure in Poplar. Plant Physiol. 168, 859–870. doi:10.1104/pp.15.00612
Vranová, E., Coman, D., and Gruissem, W. (2013). Network Analysis of the MVA and MEP Pathways for Isoprenoid Synthesis. Annu. Rev. Plant Biol. 64, 665–700. doi:10.1146/annurev-arplant-050312-120116
Wang, K. C., and Chang, H. Y. (2011). Molecular Mechanisms of Long Noncoding RNAs. Mol. Cel 43, 904–914. doi:10.1016/j.molcel.2011.08.018
Wang, L., Park, H. J., Dasari, S., Wang, S., Kocher, J. P., and Li, W. (2013). CPAT: Coding-Potential Assessment Tool Using an Alignment-free Logistic Regression Model. Nucleic Acids Res. 41, e74. doi:10.1093/nar/gkt006
Wu, T. D., and Watanabe, C. K. (2005). GMAP: a Genomic Mapping and Alignment Program for mRNA and EST Sequences. Bioinformatics 21, 1859–1875. doi:10.1093/bioinformatics/bti310
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a Web Server for Annotation and Identification of Enriched Pathways and Diseases. Nucleic Acids Res. 39, W316–W322. doi:10.1093/nar/gkr483
Yang, H., Tao, Y., Zheng, Z., Li, C., Sweetingham, M. W., and Howieson, J. G. (2012). Application of Next-Generation Sequencing for Rapid Marker Development in Molecular Plant Breeding: a Case Study on Anthracnose Disease Resistance in Lupinus Angustifolius L. BMC Genomics 13, 318. doi:10.1186/1471-2164-13-318
Yang, S.-A., Jung, Y.-S., Lee, S.-J., Park, S.-C., Kim, M.-J., Lee, E.-J., et al. (2014). Hepatoprotective Effects of Fermented Field Water-Dropwort (Oenanthe Javanica) Extract and its Major Constituents. Food Chem. Toxicol. 67, 154–160. doi:10.1016/j.fct.2014.02.010
Ye, Y., Ke, W., Sun, Y., Li, M., Zhu, H., and Huang, L. (2009). Cultivar Comparative Trial of Water Dropwort (Oenanthe Decumbens) under Different Intenerate Methods. J. Chang Veg 10, 45–46. doi:10.3865/j.issn.1001-3547.2009.16.016
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 11, R14. doi:10.1186/gb-2010-11-2-r14
Yu, N., Chen, Z., Yang, J., Li, R., and Zou, W. (2021). Integrated Transcriptomic and Metabolomic Analyses Reveal Regulation of Terpene Biosynthesis in the Stems of Sindora Glabra. Tree Physiol. 41, 1087–1102. doi:10.1093/treephys/tpaa168
Keywords: Oenanthe javanica, transcriptome, terpenoids, DEGs, regulation
Citation: Feng K, Kan X-Y, Li R, Yan Y-J, Zhao S-P, Wu P and Li L-J (2022) Integrative Analysis of Long- and Short-Read Transcriptomes Identify the Regulation of Terpenoids Biosynthesis Under Shading Cultivation in Oenanthe javanica. Front. Genet. 13:813216. doi: 10.3389/fgene.2022.813216
Received: 12 November 2021; Accepted: 21 March 2022;
Published: 07 April 2022.
Edited by:
Sunil Kumar Sahu, Beijing Genomics Institute (BGI), ChinaReviewed by:
Milan Kumar Lal, Central Potato Research Institute (ICAR), IndiaMuhammad Waseem, Hainan University, China
Copyright © 2022 Feng, Kan, Li, Yan, Zhao, Wu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang-Jun Li, bGpsaUB5enUuZWR1LmNu
†These authors have contributed equally to this work
 Kai Feng1†
Kai Feng1† Peng Wu
Peng Wu Liang-Jun Li
Liang-Jun Li