- 1Department of Agriculture and Life Industry, Kangwon National University, Chuncheon, South Korea
- 2Department of Agricultural Biotechnology/National Academy of Agricultural Science, Rural Development Administration, Jeonju, South Korea
In cultivated plants, shoot morphology is an important factor that influences crop economic value. However, the effects of gene expression patterns on shoot morphology are not clearly understood. In this study, the molecular mechanism behind shoot morphology (including leaf, stem, and node) was analyzed using RNA sequencing to compare weedy (creeper) and cultivar (stand) growth types obtained in F7 derived from a cross of wild and cultivated soybeans. A total of 12,513 (in leaves), 14,255 (in stems), and 11,850 (in nodes) differentially expressed genes were identified among weedy and cultivar soybeans. Comparative transcriptome and expression analyses revealed 22 phytohormone-responsive genes. We found that GIBBERELLIN 2-OXIDASE 8 (GA2ox), SPINDLY (SPY), FERONIA (FER), AUXIN RESPONSE FACTOR 8 (ARF8), CYTOKININ DEHYDROGENASE-1 (CKX1), and ARABIDOPSIS HISTIDINE KINASE-3 (AHK3), which are crucial phytohormone response genes, were mainly regulated in the shoot of weedy and cultivar types. These results indicate that interactions between phytohormone signaling genes regulate shoot morphology in weedy and cultivar growth type plants. Our study provides insights that are useful for breeding and improving crops to generate high-yield soybean varieties.
Introduction
Soybean [Glycine max (L.) Merr.] is a major species of legume crop grown for human and animal consumption. Although Glycine soja (wild soybean) is genetically close to cultivated G. max, it is a divergent phenotype (Singh and Hymowitz, 1988; Stupar, 2010; Joshi et al., 2013; Kofsky et al., 2018). The cultivated soybean has an erect growth type, a strong primary stem, and large variable seeds, whereas the wild soybean is a creeper and has a weedy growth type, a long and thin vine stem, and small seeds (Liu et al., 2007). Soybean plants morphology varies according to its domestication characteristics which are wild, semi wild, and cultivated soybeans (Qiu et al., 2014). Agronomical traits are influenced by stem growth habit. According to stem termination, soybean growth types are classified into determinate as the cultivar type and indeterminate, the weedy type (Bernard, 1972). In weedy types, the vegetative activity continues with the terminal bud resembling long and thin stem (Liu et al., 2010). In major crops, shoot morphology is the prime factor for grain yield (Kurepin et al., 2013; Chen et al., 2020; Zhang et al., 2021); desirable vertical shoot structure with beneficial traits, plant height, leaf size and stem structure lead to improved economic return (Khush, 2001; Mencuccini, 2015). (Kilgore-Norquest and Sneller, 2000) reported that cultivar soybeans had greater yield than the wild soybeans at the late planting dates. Therefore, unraveling the genetic mechanism behind plant architecture aids the development of high-yield cultivars (Wang and Li, 2008).
Development and defense systems in plants are mainly controlled by plant hormones (Bari and Jones, 2009; Huot et al., 2014; Davière and Achard, 2016). Phytohormones also regulate plant morphology (Hepworth and Lenhard, 2014; Yue et al., 2016). The major plant hormones viz, gibberellin, auxin, and cytokinin influence cell differentiation, proliferation, and elongation in plants and are responsible for divergent shoot architectures (Mok, 1988; Gaspar et al., 1996; Azizi et al., 2015; Haruta and Sussman, 2017). Gibberellin regulates stem elongation in plants (Jones and Kaufman, 1983; Harberd et al., 1998; Kende et al., 1998; Olszewski et al., 2002). Gibberellic acid (GA) also commonly called gibberellins, a tetracyclic di-terpenoid compound stimulating plant hormone (Gupta and Chakrabarty, 2013). Deregulation of GA-responsive genes, gibberellin-oxidases (GA20oxs, GA3oxs, and GA2oxs)results in altered phenotypes in plants (Thomas and Sun, 2004; Dai et al., 2007; Rieu et al., 2008; Wuddineh et al., 2015; Wang et al., 2017). Moreover, GA20ox and GA3ox promotes bioactive GA through the biosynthetic signaling pathway (Busov et al., 2008; Yang et al., 2012) whereas GA2ox involves in inactivation of GA (Thomas et al., 1999; Olszewski et al., 2002). Functional deficit of GA20x and GA3ox results in semi dwarf plants in Arabidopsis (Yamaguchi et al., 1998) and rice (Sakamoto et al., 2001). However, mutants of GA2ox responses opposite of GA20ox and GA3ox, resulting long and slender stem in pea (Martin et al., 1999) and switchgrass (Wuddineh et al., 2015). In soybean, Glyma18g06870 (GAox), 2-oxoglutarate-dependent dioxygenase is identified as a candidate gene responsible for vine growth habit (VGH) (Wang et al., 2019).
Shoot organogenesis and overall plant architecture is also determined by auxin (Gallavotti, 2013; Taylor-Teeples et al., 2016). Auxin response factors (ARFs) and Indole Acetic acid (IAA) plays an important role in regulation of auxin response genes (Ma and Li, 2019). ARF transcription factor interacts with PHYTOCHROME INTERACTING FACTOR (PIF) to increase auxin biosynthesis and promote cell elongation in the stem of Arabidopsis (Oh et al., 2014). All physiological mechanisms in A. thaliana, such as cell division, shoot apical dominance, tropism, and root initiation, are governed by auxin (Hagen and Guilfoyle, 2002). Upstream of auxin responsive genes promote cell elongation of the hypocotyls and plant organs (Chae et al., 2012). Shoot apical balance is controlled by cytokinin via the regulation of cell division and cell proliferation (Mok and Mok, 2001). The level of active cytokinin is suppressed by cytokinin dehydrogenase (CKX) (Motyka et al., 1996; Werner et al., 2001). For example, high regulation of CKX gene in Arabidopsis inhibit cytokinin level, indicating a decrease in overall shoot morphology (Schmülling et al., 2003). Three cytokinin receptors (ARABIDOPSIS HISTIDINE KINASEs; AHK2, 3, 4) and eleven response regulators (ARABIDOSPIS RESPONSE REGULATORs, ARRs) have been identified in Arabidopsis determining the specificity of cytokinin in plant growth (Kim et al., 2006). The receptor gene AHK involve in activating cell division and meristem maintenance (Jeon et al., 2010).
Interaction among plant hormones controls overall plant growth and development (Ohri et al., 2015). Auxin promotes bioactive gibberellins like gibberellic acid-1 (GA1), and is most important for cell division and internode elongation (Ross et al., 2001). Both cytokinin and gibberellin act mutually in the early and late developmental stages of shoot meristem elongation (Richards et al., 2001; Schmülling, 2002). Auxin interacts with cytokinin antagonistically. For example, apical dominance is regulated by auxin and cytokine in an antagonistic manner (Shimizu-Sato et al., 2009). Reduction of auxin signaling gene expression induces cytokinin signaling in most tissues (Kurepa et al., 2019).
Growth and development are continuous processes influenced by phytohormone interactions that produce diverse phenotypes. To date, few studies have been conducted to explore the genes that determine the weedy and cultivar growth types in soybean. Comparative transcriptome analysis by RNA sequencing is an efficient method for identifying important differences in gene expression between two plant relatives (Koenig et al., 2013). Here, we used the transcriptomes of leaves, stems, and nodes to identify differentially expressed genes (DEGs) associated with the weedy and cultivar growth types in soybean. We performed high-throughput Illumina sequencing to comprehensively characterize the transcriptomes. To investigate how the expression of these genes affects soybean phenotypes, we compared the expression profiles of the leaves, stems, and nodes and the crosstalk of DEGs. The results of this research provide insights into the relevant genes responsible for the weedy and cultivar growth types in soybean and facilitate a better understanding of the molecular mechanism behind the differences in growth and development of the two soybean relatives.
Materials and Methods
Plant Materials
KB000001 (Glycine max), a cultivated soybean and KB000002 (a hybrid of G. max and Glycine soja), a wild type of soybean seeds was collected from Rural Development Administration (RDA), South Korea. The parents, KB000001 with an erect phenotype and KB000002 with the weedy phenotype, were crossed to generate recombinant inbred lines (RILs) by single seed descent (SSD) method. A single was obtained from the first filial generation (F1) grown in the field in Chuncheon-Si (Gangwon-do, South Korea). To generate an inbred population, the F2 seeds were harvested and grown by selfing further until the F5 generation. Both the weedy and cultivar growth types were observed in every filial generation. From F1 to F6, two lines were targeted, with each displaying one of the distinct phenotypes of the parental lines, i.e., the weedy (creeper) and the cultivar (stand) growth types. The selected plant’s seeds from F6 were harvested and grown both in the field and the greenhouse. The shoot morphology in F7 was the same as F6, so they were selected for the rest of the study. To confirm the phenotype, the plants were grown in the growth room with optimal temperature (25°C) and 60–70% humidity. Finally, these two lines were selected to study the molecular mechanism underlying the weedy and cultivar growth types. The leaves, stems, and nodes from 12-week-old plants were sampled and harvested immediately in frozen liquid nitrogen (N) for RNA isolation.
RNA Extraction and RNA-Seq Library Construction
Total RNA was isolated from the leaves, stems, and nodes from the respective samples using GeneAll® Ribospin™Plant (Geneall Biotechnology Co., Ltd., Seoul, Korea) following the manufacturer’s instructions. Genomic DNA digestion was performed with DNase I (Sigma, St. Louis, MO, United States of America). The RNA quality was observed on 1% agarose gel electrophoresis. The total RNA integrity was checked using an Agilent 2100 BioAnalyzer with RNA integrity number (RIN) value >6. For the total mRNA library, sequencing was performed using the Illumina Nova Seq high-throughput sequencing kit and platform (Illumina Ltd, San Diego, CA, United States) according to the manufacturer’s protocol and performed by Theragen Etex Bio Institute, a professional DNA sequencing service provider (Theragen Etex Inc. Suwon, South Korea).
Screening of Differentially Expressed Genes by RNA-Seq
The sequenced data from Illumina were preprocessed using Trimmomatic in which raw data were trimmed and converted to total read bases. The adapters were used for trimming contaminant sequences and for library construction in Trimmomatic v0.36 (http://www.usadellab.org/cms/index.php?page=trimmomatic). Low-quality reads were removed by applying Trimmomatic’s sliding window (4:20) with average quality (30) and minimum read size (36 bp). The sequence was read in units of 4 bp with a PHRED score less than 20. Reads with average length of less than 20 bases were removed. Each sample was aligned to the reference sequence, Glycine max Wm82.a2. v1, using the HISAT2 v2.1.0 program (https://ccb.jhu.edu/software/hisat2/index.shtml). The processed sequences consisting of paired-end reads were mapped to the reference sequence. The read count (expression level) was calculated using StringTie v1.3.4d software (https://ccb.jhu.edu/software/stringtie/index.shtml). Based on the read count of the StringTie calculated at the transcript level, a comparative analysis between the samples was performed using DESeq V1.36.0. In DESeq, read count was normalized through size factor and dispersion, DEG analysis was performed using a log2 fold change value, and false discovery rate (FDR) was calculated after normalization. Calculation of unigene expression was performed using the fragments per kilobase of transcript per million mapped reads (FPKM) method. FPKM values were calculated for each of the genes and only unigenes with FPKM >1.0 were used for the further analysis. Visualization analyses, i.e., heat maps, Venn diagrams, and Minus versus Add (MA) plots, of the DEGs were performed by an in-house R script.
Gene Annotation and Functional Analysis
The genomic reference information of Glycine max was obtained from Soybase, which includes Gene Ontology (GO) annotations for each gene. The Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations were obtained from the KEGG database. The BLASTp (e-value 1e-3) analysis was performed using NCBI’s Refseq (http://www.ncbi.nlm.nih.gov) plant protein sequence, TAIR, and Uniprot as databases (DB). Gene Ontology analysis with biological process, cellular component, and molecular function was done using InterProscan and BLAST. The KEGG pathway was performed by BLAST2GO (https://www.blast2go.com/). A p-value of ≤.05 was used as a significance threshold for the GO and KEGG pathway enrichment analysis.
Reverse-Transcription Quantitative PCR Analysis
RT-qPCR analysis was conducted using the SyBr Green PCR kit on a PIKOREAL 96 Real-Time PCR system to validate RNA-Seq data. Candidate reference RT-qPCR gene primers were designed using IDT (Integrated DNA technologies). The GmAct6 (Actin-6) soybean gene was selected as the endogenous reference. All the primers were synthesized by Bionicsro.co.kr. One-step RT-qPCR analysis was performed on the extracted total RNA in three biological replicates and three technical replicates of the leaves, stems, and nodes. A total of 10 μl RNA was used for cDNA synthesis using the HyperScript™ Rt master mix (with oligo dT) protocol for RT-qPCR expression analysis. In this experiment, each reaction contained 2 µl of diluted cDNA, 1 µl of each primer, 5 µl of 2X SYBR Green mix, and 2 µl of RNase free water. All the RT-qPCRs were performed with the following conditions: denaturation at 95°C for 1 min, followed by 40 cycles of 95°C for 5 s, 57°C for 10 s, 72°C for 10 s, and final extension at 72°C for 5 min. To verify product specificity, melting curve analysis was performed after each amplification. The relative expression level of each unigene was calculated using the 2−ΔΔCT approach (Livak and Schmittgen, 2001).
Results
Phenotype Selection During Generation of the Recombinant Inbred Lines
To understand phenotypic variation among two plant relatives at the transcriptome level, we crossed representative parent soybeans, cultivated type and a wild type, to generate recombinant inbred lines (RILs) (Figure 1A). We obtained independent RILs over seven generations by selfing. The F2–F5 generation plants, generated using single seed descent from F1, could be segregated by plant height, maturing time, and plant structure (weedy and cultivar types). We generated F6 plants with the weedy phenotype from KB000003-47 RIL (F5). We obtained F7 seeds from a weedy type of plant (KB000003_47) and a cultivar type plant (KB000003_47.4) in F6. In the F7 generation, the phenotype of plants did not differ between normal field and greenhouse conditions (Figures 1A,B). The weedy type (KB000003_47.5) typically showed a thin stem, leaf senescence, and reduced node number compared to the cultivar type (KB000003_47.4). In all the generations (F6-F7), there were clear differences in stem morphology between the weedy type and cultivar type (Figure 1C). These results indicate that the stem can be used to characterize developmental differences among weedy type and cultivar type plants.
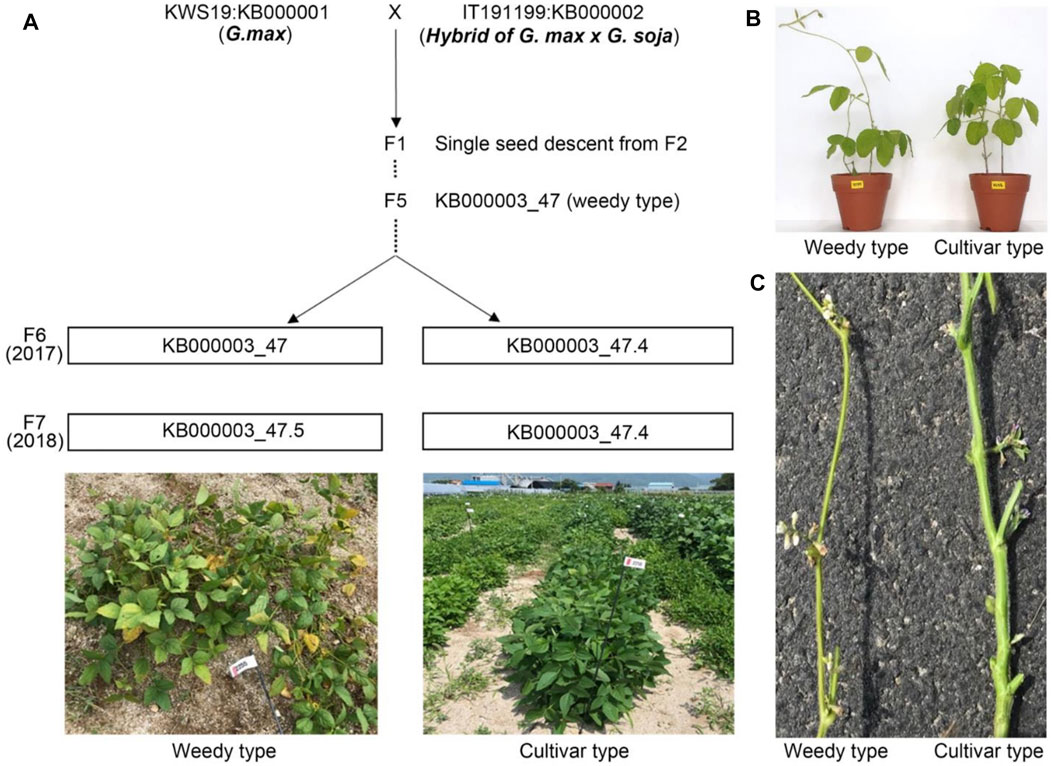
FIGURE 1. Population phenotypes and crossing scheme used to generate the recombinant inbred lines (RILs). (A) Standard parents G. max (KWS: KB000001) and a hybrid of G. max and G soja (KWS: KB000001) were used for crossing, and their phenotype distribution descended from F2 to F7. Individual lines from F6 displaying parental phenotype, i.e., weedy (KB000003_47.5) and cultivar type (KB000003_47.4), continues in F7. (B) Morphology of weedy and cultivar growth type plants (F7 generation). Eight-week-old plants were grown in a greenhouse. (C) The stems of weedy type plants and cultivar type plants have different morphology. The F7 generation weedy and cultivar type plants were grown in the field under normal conditions.
Assembly of RNA Sequencing Data
To identify differences in development-related gene expression in RILs, we analyzed the transcriptomes of the leaves, stems, and nodes of weedy and cultivar growth type plants obtained in F7 using RNA sequencing (Figure 2; Table 1). There were 224,804,226 sequencing reads in the raw data and 213,047,794 reads in the clean data (94.77% of the total) (Table 1). The trimmed sequence reads were mapped to the reference genome (Glycine max Wm82. a2. v1), and the average percentage of mapped reads amongst all the samples was 84.69%. The average ratios of mapped reads in stems, nodes, and leaves of weedy and cultivar type plants were 85.5%, 89.17%, and 79.4%, respectively (Figure 2A). A total of 73,722 unigenes were generated for DEG analysis. The results indicate that the quality of sequencing was satisfactory for further analysis.
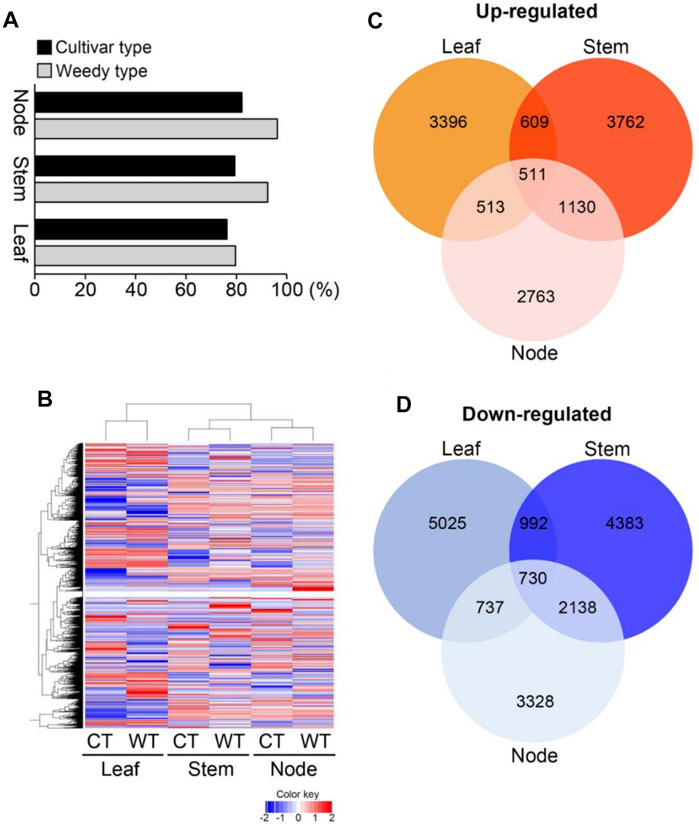
FIGURE 2. Transcriptome analysis of leaves, stems, and nodes of weedy type (WT) and cultivar type (CT) soybean. Total RNA was prepared from leaves, stems, and nodes of 12-week-old plants. (A) Mapping of sequence reads to the reference Glycine max Wm82.a2. v1. (B) Hierarchical cluster analysis of differentially expressed genes (DEGs). Color key indicates gene expression FPKM values from −2 to 2: blue = lowest and red = highest among the DEGs between leaves, stems, and nodes of WT and CT. (C) and (D) Comparisons of the number and overlapping relationship of the DEGs between leaves, stems, and nodes of WT and CT. Red circles indicate up-regulated genes (C) and blue circles represent down-regulated genes (D) in cultivar type with respect to weedy type (>log2 fold change).
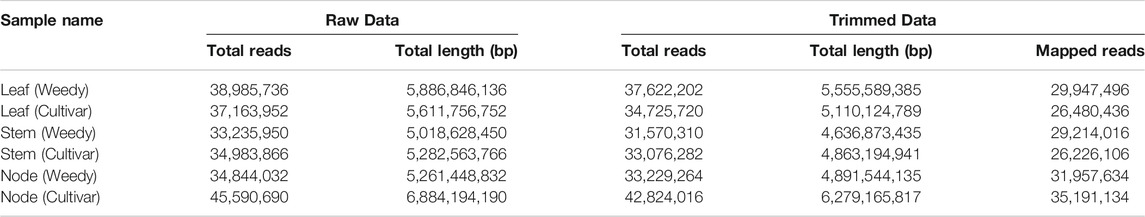
TABLE 1. The summary of the sequencing data by a high-throughput DNA sequencing platform in the weedy type and cultivar type soybeans.
DEG Analysis Between Weedy and Cultivar Type Plants
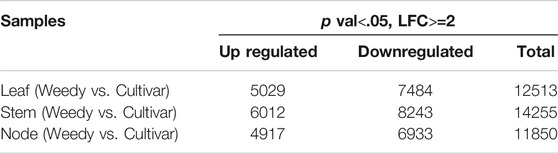
To investigate the gene expression levels of each sample, the transcripts were assembled and normalized by fragments per kilobase of transcript per million mapped reads (FPKM), and DEGs in each weedy type vs. cultivar type pair (leaves, stems, and nodes) were selected with p-value < 0.05 and | log2 (fold change) | ≥ 1. A total of 38,618 DEGs were visualized using a heat map (Figure 2B). The heat map showed that the color of the plots for stems and leaves was more visually different compared to the nodes of weedy type and cultivar type plants. A total of 12,513, 14,255, and 11,850 DEGs in leaves, stems, and nodes, respectively, were screened. Specifically, 5,029, 6,012, and 4,917 upregulated genes and 7,484, 8,243, and 6,933 downregulated genes were identified in leaves, stems, and nodes, respectively (Table 2). Furthermore, the Venn diagram of DEGs showed that 609, 513, and 1,130 upregulated genes (Figure 2C) and 992, 737, and 2,138 downregulated genes (Figure 2D) overlap in leaves and stems, leaves and nodes, and stems and nodes, respectively. Also, 511 upregulated genes and 730 downregulated genes overlapped in all samples (leaves, stems, and nodes) (Figures 2C,D). We constructed a Minus-versus-Add (MA) plot and a Volcano plot of the DEGs, which showed that the stem of the weedy and cultivar types had the greatest number of DEGs (Supplementary Figure S2), suggesting that the stem has more contribution for phenotype variation in plants. The analyses indicate that gene expression changes are involved in the characteristic developmental differences of weedy type and cultivar type plants.
Pathway Enrichment Analysis of DEGs for GO Annotation and KEGG. To determine the associations of DEGs in the development of leaves, stems, and nodes in weedy and cultivar type soybeans, we performed GO annotation and KEGG pathway enrichment analysis using DEGs. A total of 73,722 DEGs were annotated in three major categories: biological processes, cellular components, and molecular function (Figure 3A). The total of 21,618, 8,107, and 33,821 were assigned to biological process, cellular component, and molecular function, respectively. The molecular function category showed a greater number of DEGs compared to the other categories. A total of 12,513 DEGs in leaves, 14,255 DEGs in stems, and 11,850 DEGs in nodes were classified into GO terms. Of the total, 1,574 DEGs in leaves, 4,762 DEGs in stems, and 3,866 DEGs in nodes were classified as being involved in biological processes; 554 DEGs in leaves, 1,619 DEGs in stems, and 1,407 DEGs in nodes were classified as being involved in cellular components; and 2,401 DEGs in leaves, 7,119 DEGs in stems, and 5,865 DEGs in nodes were classified as being involved in molecular function. Among them, the number of DEGs in stems was significantly enriched in GO terms. The enriched DEGs in stems are associated with protein phosphorylation, oxidation-reduction, transcription factor regulation, carbohydrate metabolism, and transmembrane transport in biological process; with nucleus, membrane, and integral components of membranes in cellular component; and with catalytic activity, DNA binding, protein binding, and protein and nucleic acid binding in molecular function. These GO terms might be related to the phenotype of weedy type and cultivar type plants.
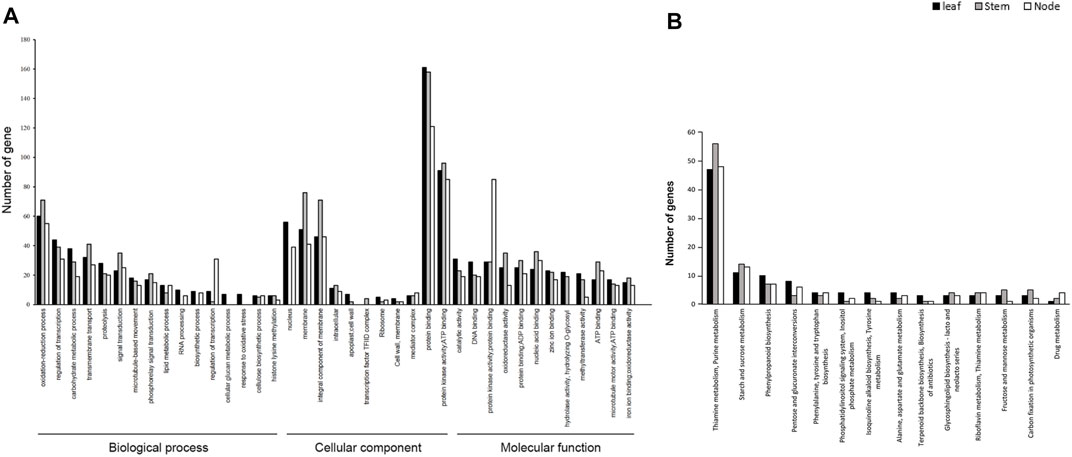
FIGURE 3. Gene Ontology (GO) functional classification and Kyoto Encyclopedia of Genes and Genomes (KEGG) classifications of DEGs of leaves, stems, and nodes. (A) GO analysis of differentially expressed genes (DEGs). GO describes number of most enriched DEGs with three independent categories: biological process, cellular component, and molecular function. The x-axis represents the GO terms and the y-axis the number of unigenes. (B) KEGG pathway of DEG terms grouped into 15 categories. The x-axis represents KEGG terms and the y-axis number of unigenes.
To better understand the molecular difference between weedy and cultivar types, the upregulated and downregulated DEGs were grouped by KEGG. The pathways of the DEGs were analyzed using the KEGG database (http://www.genome.jp/kegg/pathway.html) (Figure 3B). A total of 814 unigenes were annotated to various pathways, of which 159, 172, and 149 DEGs of leaves, stems, and nodes, respectively, were assigned to a KEGG pathway. The KEGG classification showed that the DEGs were mainly involved in thiamine metabolism in leaves, stems, and nodes. For DEGs in the leaf, 47 were assigned to thiamine and purine metabolism, 11 to starch and sucrose metabolism, and 10 to phenylpropanoid biosynthesis. In DEGs in the stem, 56 were assigned to thiamine and purine metabolism, 14 to starch and sucrose metabolism, and 7 to phenylpropanoid biosynthesis. For DEGs in the node, 48 were assigned to thiamine and purine metabolism, 13 to starch and sucrose metabolism, and 7 to phenylpropanoid biosynthesis. Unlike in the leaves and nodes, 14 DEGs of the stems were highly enriched in starch and sucrose metabolism and porphyrin and chlorophyll metabolism. These pathways were related to differences in weedy type and cultivar type plant height, indicating that several genes regulate the growth of soybean, through controlling biological processes.
Identification of Genes Related to Traits Between Weedy and Cultivar Type Soybeans
The above results suggest that several genes play important roles in the antagonistic regulation of plant development to produce weedy type and cultivar type plants. The development of leaves, stems, and nodes is regulated by the plant hormones gibberellin, auxin, and cytokinin. We analyzed and compared the DEGs that respond to gibberellin (GA), auxin, and cytokinin in the leaves, stems, and nodes (Table 3). The GA-related genes were upregulated in the leaves, stems, and nodes of the cultivar type plant compared to the weedy type of plant. SPINDLY (SPY) and Gibberellin 2-oxidase (GA2Ox8) genes were upregulated in the cultivar type plant. Homeobox protein (HAZ1) and Kaurene Oxidase (KO) genes were down regulated in the leaves, stems, and nodes of the cultivar type compared to the weedy type. These genes are related to the gibberellin signaling pathway that regulates the level of GA. The overexpression of SPY and GA2Ox8 in A. thaliana showed reduced plant height and thick stems when compared to control (Thornton et al., 1999). These results indicate that upregulation of GA2ox in plants leads to short and thick stem. Most of the auxin-related genes are downregulated in cultivar type soybean. Auxin response factor 5 (ARF5) and FERONIA (FER) showed remarkable upregulation, whereas POPCORN (PCN), PIN like proteins (PINS), Auxin response element (AUX22E), Indole Acetic acid (IAA30), and Auxin response factor (ARF8) were downregulated in leaves, stems, and nodes of the cultivar type compared to the weedy type. These genes are known as auxin signaling genes. Reduction in FER expression in A. thaliana results in reduced cell elongation in various tissues (Guo et al., 2009). These genes play a prominent role in producing a thick stem in the cultivar type and a thin stem in the weedy type in soybean. Cytokinin dehydrogenase-1 (CKX1), Arabidopsis Histidine kinase (AHK3), Arabidopsis histidine phosphotransfer (AHP1 and AHP5), Arabidopsis response regulator (ARR2), and KORRIGAN (KOR) are up regulated in the cultivar type compared to the weedy type. Overexpression of cytokinin response gene (CKX) in A. thaliana results in shorter internodes with reduced growth (Werner et al., 2001), and changes in the expression of cytokinin response genes result in variation of meristem size (Schmülling et al., 2003). The expression of the regulator and receptor genes in the stem of weedy and cultivar type, response to the major phytohormones is shown in Supplementary Table S1. The catalyzing enzyme of Gibberellin, cytochrome P450, called ent-kaurenoic acid oxidase (KAO) were up regulated in the stem of weedy type. Similarly, the DELLA proteins response to GA signaling were up regulated in the stem of cultivar types. DELLA proteins are the negative regulator of gibberellin signaling (Yoshida et al., 2014). Most of the auxin and cytokinin response regulatory genes are up regulated in the stem of cultivar types (Supplementary Table S1). These findings show that the phytohormone response genes play an important role in the stem of the shoot compared to the leaf and node.
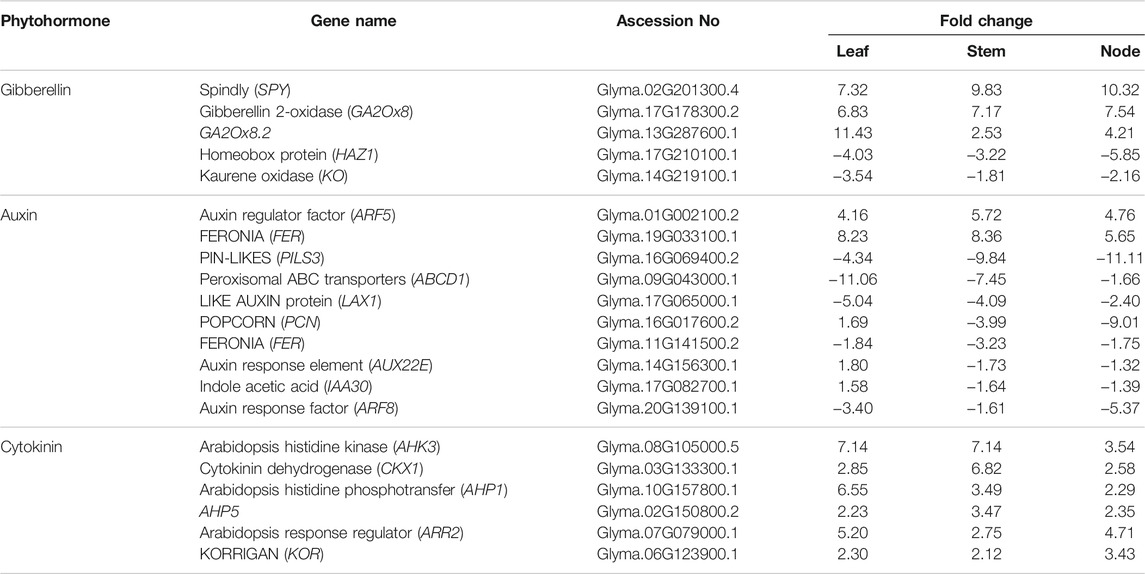
TABLE 3. Significantly differentially expressed gene response to phytohormones in cultivar types with respect to weedy types.
Validation by Reverse-Transcription Quantitative PCR
To test the reliability of FPKM expression patterns (RNA-Seq) of the selected DEGs, we checked the expression level by RT-qPCR. A total of 22 DEGs that respond to plant phytohormones were selected from the weedy and cultivar types. The genes were classified under the three major plant hormones: gibberellin, auxin, and cytokinin (Figure 4; Table 3). Most of the gibberellin response genes were found to be up regulated in the cultivar type with respect to the weedy type. The expression of GA response genes, SPY and GA2ox, was found to be significantly higher in the stem of the cultivar type compared to the weedy type (Figure 4A). The expression of genes encoding Homeobox protein (HAZ1) and Kaurene oxidase (KO) was down regulated in the cultivar growth type in soybean (Supplementary Figure S1). Based on the RT-qPCR, auxin response genes were downregulated in the leaves, stems, and nodes of the cultivar type relative to the weedy type. Here, the auxin response gene FER1 was upregulated and ARF5 was downregulated in the cultivar type compared to the weedy type (Figure 4B). Cytokinin-responsive genes were found to be upregulated in cultivar types. AHK3 and CKX1 showed higher expression in the leaves, stems, and nodes of the cultivar type with respect to the weedy type (Figure 4C). According to the RT-qPCR expression and transcriptome data, we found that auxin and cytokinin are antagonistically expressed in leaves, stems, and nodes among cultivar and weedy type soybeans. The expression profile of the remaining genes is described in Supplementary Figures S1A–C. Here, we focused on the stem phenotype, and the genes studied were most differentially expressed in the stem compared to the leaf and the node of weedy and cultivar type soybean. The genes and their expression patterns were mostly consistent with transcriptome data; hence, the results indicate the transcriptome data we presented were reliable and accurate.
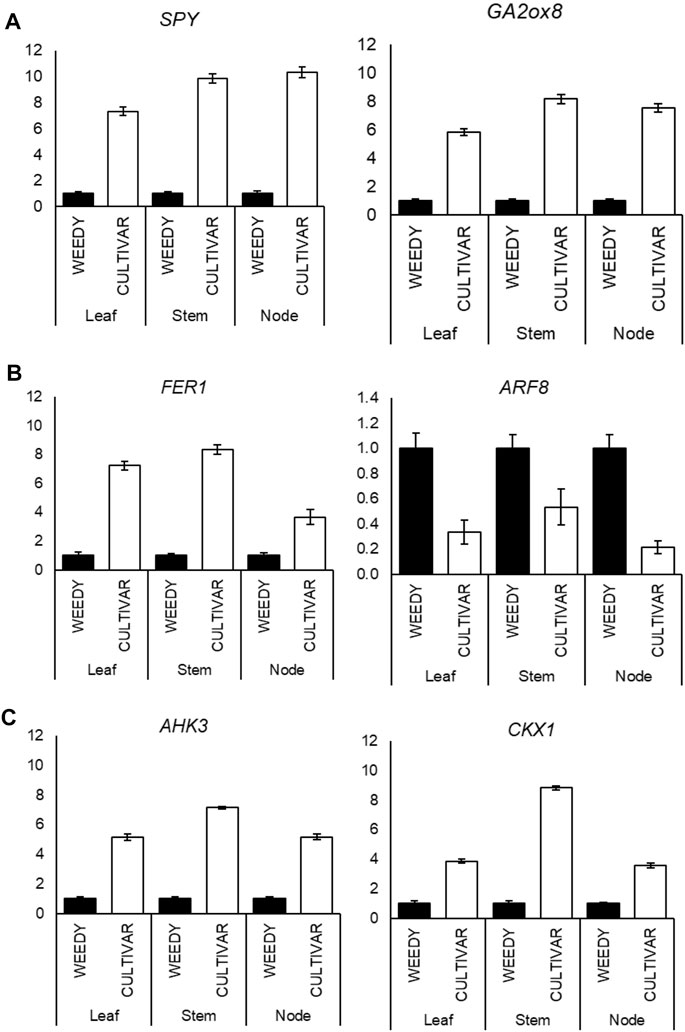
FIGURE 4. (A–C) Reverse-transcription quantitative PCR (RT-qPCR) analysis shows changes in gibberellin response genes: gibberellin 2-oxidase 8 (GA2ox8) and SPINDLY (SPY) (A); auxin response genes: FERONIA (FER) and auxin response factor (ARF8) (B); and cytokinin response genes: Arabidopsis histidine kinase-3 (AHK3) and cytokinin dehydrogenase-1 (CKX1). (C) Expression levels in the leaves, stems, and nodes of weedy type (WT) and cultivar type (CT). Total RNA was prepared from the 12-week-old plants for the indicated samples. Control plants were weedy type (WT). Error bars indicate SD of triplicate measurements. GmAct6 was used as internal control, and relative expression levels are shown as fold values.
Discussion
Plant morphology and architecture are an important economic characteristic that impact soybean yield. Although several factors that affect soybean growth and development are known, many aspects of this process remain to be elucidated. Therefore, studying gene expression and function with respect to phenotype provides an efficient way to reveal the hidden mechanism. In addition, comparative RNA sequencing technology allows us to investigate the molecular mechanism between two plant relatives. In this study, we performed a transcriptome analysis between weedy and cultivar growth types in soybean obtained from the F7 RIL population derived from a cross of wild and cultivated soybean. Our findings show that the weedy type has a longer stem and creeps on the ground, whereas the cultivar type exhibits the upright standing growth type. From the result, we hypothesize the characteristic of the stem determines if the plant is the weedy or cultivar type in soybean. Furthermore, the stem transcriptome was investigated to understand the relationship between gene regulation and phenotype.
Phytohormones participate in many physiological processes in plants (Bari and Jones, 2009; Luo et al., 2016). Auxin, gibberellin, cytokinin, and ethylene play abundant roles in shaping the morphological structure of plants (Fahad et al., 2015). Gibberellin 2-oxidase (GA2ox) is involved in degradation of bioactive GA and controls GA levels (Martin et al., 1999; Thomas et al., 1999). GA2ox modulate under the GA biosynthesis and inactivation pathway which converts the active form of GAs to inactive form (Yamaguchi, 2008). Upstream of GA2ox inactivates bioactive GA resulted in short internode plants whereas its downstream induce stem elongation (Supplementary Figure S3). Overexpression of two switchgrass GA2ox genes showed abnormal shoot architecture with extremely reduced internodes (Wuddineh et al., 2015). Similarly, overexpression of A. thaliana GA2ox8 causes a decrease in GA levels, which showed a reduction in stem length (Schomburg et al., 2003). Additionally, GA2ox1 transformed Solanum species exhibited low GA with phenotype alteration, reduced plant height, and internode distance (Dijkstra et al., 2008). Downstream of GA level in plants results in semi dwarf, while increased GA concentration promote taller plants (Busov et al., 2008). Elongated stem and vine growth habit are characteristic of the wild phenotype in soybean (Tian et al., 2010). In our study, the cultivar type has short internodes and thick stems, whereas the weedy type has thin and long stems (Figure 1C). RNA-Seq data revealed that GA2ox8 is differentially expressed between weedy and cultivar types of soybeans. The expression of GA2ox8 was found to be up regulated in the stem of the cultivar type, which might reduce the GA level and result in a short and thick stem. Conversely, the expression of GA2ox was down regulated in weedy type, which resulted in a thin stem with long internodes. The RT-qPCR analysis showed that the transcription level of GA2ox in the cultivar type was higher in comparison to the weedy type (Figure 4A). SPINDLY (SPY) is the GA-signaling protein that also functions as a negative regulator of GA during seed germination (Qin et al., 2011) and is characterized by slender phenotype (Martin et al., 1999) Altered SPY expression affects overall plant growth (Swain et al., 2001). Thus, the mutant plant lacking SPY showed suppressed phenotype plants (Thornton et al., 1999). Here, SPY is upregulated in the cultivar type plant and downregulated in the weedy type of plant (Figure 4A). This indicates that deregulation of SPY results in an altered phenotype. Additionally, SPY genes positively interact with cytokinin, which plays a major role in plant growth and development (Greenboim-Wainberg et al., 2005). The activity of cytosolic SPY stimulates cytokinin and inhibits GA signaling (Greenboim-Wainberg et al., 2005; Maymon et al., 2009). Two other GA response genes, homeobox protein (HAZ1) and kaurene oxidase (KO), are down regulated in cultivar type plants (Supplementary Figure S1A). KO also called cytochrome P450, catalyzes successive oxidation of enzymes in gibberellin biosynthesis pathway (Morrone et al., 2010), (Supplementary Figure S3). Therefore, GA-responsive genes might contribute to phenotype variation to produce either the weedy or cultivar type.
Auxin regulates divergent developmental processes and helps in anisotropic cell expansion in plants (Kutschera et al., 1987; Zhao, 2010; Zazimalová et al., 2014). ARF8 helps in cell expansion and proliferation during plant development (Varaud et al., 2011). ARF8 is a positive regulator of auxin, and its overexpression resulted in a long hypocotyl in A. thaliana (Tian et al., 2004). Likewise, high expression of ARF8 in tobacco enhanced plant growth and development (Ge et al., 2016). Sometimes, deregulation of ARF8 causes developmental abnormalities in A. thaliana (Jay et al., 2011). ARF8 showed a conserved role in controlling the vegetative growth and shoot development of tomato (Liu et al., 2014). Functional deficient mutant of ARF8 in Arabidopsis showed auxin deficient phenotypes such as dwarfism than the wild types (Nagpal et al., 2005). Here, ARF8 was upregulated in the weedy type of soybean and downregulated in the cultivar type soybean. This might enhance stem elongation in weedy type and reduce stem length in cultivar type. Furthermore, ARF8 also promotes early flowering in A. thaliana (Nagpal et al., 2005; Goetz et al., 2007; Finet et al., 2010). In agreement with previous studies, the weedy type plants were observed to mature earlier compared to the cultivar type plants (Figure 1A). FERONIA (FER) is one of the auxin response genes known as a cell growth regulator (Shih et al., 2014; Oh et al., 2020). A study of an A. thaliana FER mutant showed that FER is essential for maximum cell elongation and proliferation (Guo et al., 2009). In addition, FER interacts with components of the cell wall to assist in monitoring of the cell walls in plants (Shih et al., 2014). In this study, FER was up regulated in the cultivar type compared to weedy type, indicating that more expansion of cells might result in thick stems in cultivar plants. RT-qPCR results showed that FER is highly expressed in cultivar plants (Figure 4B). There was differential expression in other auxin response genes as well; auxin response factor 5 (ARF5) was upregulated in the cultivar type, whereas PIN LIKES3 (PILS3) was downregulated in the cultivar type (Supplementary Figure S1B). PILS are the putative auxin carriers, its overexpression affects overall plant patterning (Feraru et al., 2012), while mutant leads to reduced plant growth in Arabidopsis (Barbez et al., 2012). A study has shown that ARF5 interacts with PIN1 in the formation of leaf primordia (Schuetz et al., 2019).
The Cytokinin oxidase/dehydrogenase (CKX) gene family regulates cytokinin in plants (Wang et al., 2014), playing a major role in metabolic cytokinin inactivation (Motyka et al., 1996; Mok and Mok, 2001). Overexpression of A. thaliana CKX1 resulted in short internodes with reduced growth compared to control (Werner et al., 2001). Also, similar results were obtained in transgenic tobacco, where a higher expression of CKX1 decreased the cytokinin level and resulted in reduced plant height (Yang et al., 2003). Here, CKX1 was found to be up regulated in cultivar type plants compared to weedy type plants, indicating that CKX1 plays a major role in making cultivar type plants exhibit thick stems and shorter plant height compared to the weedy type. Cytokinin signaling pathway consists of three proteins, histidine kinase receptor, histidine phosphotransfer and response regulator (Keshishian and Rashotte, 2015). Arabidopsis histidine kinase (AHK3) is an important cytokinin receptor responsible for secondary growth of vascular tissues (Hejátko et al., 2009). Loss of AHK3 results in reduction of overall shoot growth (Higuchi et al., 2004). This suggests that AHK3 is essential for overall shoot formation. Our transcriptome and expression analysis showed that CKX1 and AHK3 are highly expressed in the shoots of cultivar type plants (Figure 4C). Overexpression of Arabidopsis response regulator (ARR2) was shown in a previous study to result in dwarf plants and induce a cytokinin hypersensitive response (Shull et al., 2016). (Hwang and Sheen, 2001) reported ARR type-A proteins act as negative regulator of cytokinin signaling. Here, ARR2 was up regulated in the cultivar type (Supplementary Figure S1C). This result suggests that cytokinin signaling genes might be responsible for varying overall shoot morphology in plants.
Interaction of major plant hormones influence growth and development, which can elucidate the genetic basis and molecular mechanisms behind changes in plant physiology (Ross and O’Neill, 2001). Hormone interactions can increase or decrease the expression of responsive genes (Vanstraelen and Benkov, 2012). Auxin and gibberellins are essential plant hormones that perform different functions but exist at an equilibrium for normal stem elongation (Yang et al., 1996; Haga and Iino, 1998). In Chinese cabbage, an increase in biosynthesis of GA and indole acetic acid (IAA), which is an auxin, promotes stalk elongation (Kou et al., 2021). GA coordinates with auxin for cell division and growth of cambial derivatives in Poplar stems (Björklund et al., 2007). In gibberellin biosynthesis pathway, the biosynthesis of GA activates with the regulatory genes and interacts coordinately with the other phytohormone response genes which results in morphology variation (Supplementary Figure S3). We predict that the interaction of auxin and GA response genes in weedy type plants results in longer and thin stems compared to the cultivar type. In various developmental processes, gibberellin and cytokinin have opposite effects but are coordinately expressed (Greenboim-Wainberg et al., 2005). The SPY gene inhibits bioactive GA but promotes the cytokinin signaling pathway (Filardo and Swain, 2003; Eckardt, 2005). Cytokinin regulates auxin signaling metabolism; thus, higher expression of cytokinin response genes promotes the expression of auxin response genes (El-Showk et al., 2013). The expression of auxin is indirectly inhibited by the cytokinin signaling mechanism when cytokinin biosynthesis increases (Jasinski et al., 2005; Yanai et al., 2005). Arabidopsis response regulators (ARRs) work together to influence cytokinin and auxin signaling, which are the primary regulatory mechanisms at the shoot apical meristem (Zhao et al., 2010). DELLA proteins participate in complex crosstalk among plant hormone through interaction with many transcription factors (Hou et al., 2010; Yoshida et al., 2014; Li et al., 2015). The outcome of auxin and cytokinin interaction depends upon different cell-type specifications via different signaling pathways (Chandler and Werr, 2015). By means of synergistic or antagonistic action, phytohormones interact with developmental cues along with delivering environmental inputs as signaling crosstalk (Seif El-Yazal et al., 2015).
Conclusion
In summary, our study postulates candidate genes responsible for the weedy and cultivar growth types in soybean. Using the Illumina platform, transcriptome sequencing was performed and DEG analysis revealed that major phytohormone-responsive genes influence plant growth types. By comparing the gene expression of the shoots (leaves, stems, and nodes) of weedy and cultivar types, we found that the stem is the key modulator of phenotype variation in the F7 RIL population. GA2ox, SPY, ARF, CKX, and AHK are the major plant hormone signaling genes responsible for determining the weedy and cultivar growth types in soybean. These results show how the expression, regulation, and interaction of major plant hormone-responsive genes influences plant phenotype. Our study provides insights for future genetic breeding and facilitates target trait crop improvement for higher yield in soybean.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE188519.
Author Contributions
I-YC conceived and designed the project and edited the manuscript. PB and TU was a major contributor to the sample preparing, the data analysis and drafted the manuscript. NR and K-CP analyzed the data and performed the wet experiment.
Funding
This work was supported by Rural Development Administration (RDA) grant of the Korean government (No. PJ015326022021).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.805347/full#supplementary-material
Supplementary Figure 1 | Reverse-transcription quantitative PCR (RT-qPCR) analysis shows changes in gibberellin response genes: gibberellin 2-oxidase 8 (GA2ox8.2), Homeobox protein (HAZ1), and Kaurene oxidase (KO) (A); auxin response genes: Auxin response factor 5 (ARF5), PIN LIKES3 (PILS3), LIKE AUXIN protein (LAX1), Indole acetic acid 30 (IAA30), and Auxin response element (AUX22E) (B); and cytokinin response genes: Arabidopsis histidine phosphotransfer 1 (AHP1), Arabidopsis histidine phosphotransfer 5 (AHP5), Arabidopsis response regulator (ARR2), KORRIGAN (KOR), and Arabidopsis Histidine Kinase (AHK3) (C). Expression levels in leaves, stems, and nodes of weedy and cultivar types. Total RNA was prepared from the 12-week-old plants for the indicated samples. Control plants were weedy type (WT). Error bars indicate SD of triplicate measurements. GmAct6 was used as internal control, and relative expression levels are shown as fold values.
Supplementary Figure 2 | Expression profiles of the DEGs using Volcano and Minus-versus-Add (MA) plots. (A) Volcano plot of the upregulated and downregulated DEGs of the stems between the weedy and cultivar growth types. The down-regulated DEGs are on the left side, and up-regulated DEGs are on the right. Red indicates DEGs with p-value < .01 and LFC ≥ 1; green indicates p-value between .01 and .05, and LFC ≥ 1; yellow indicates p-value < .01 and LFC ≥ 1; and black indicates p-value < .05 and LFC ≤ 1. For each plot, the x-axis represents the log2 fold change (LFC), and y-axis represents −log 10 (p-values). (B) MA plot of the DEGs of the stem between the weedy and cultivar growth types. The x-axis represents the mean of the normalized counts of the genes, and the y-axis indicates the log2 fold change. Red points represent up-regulated DEGs, blue points represent down-regulated DEGs, and black points represent non-DEGs.
Supplementary Figure 3 | Relevant part of GA biosynthesis pathway and crosstalk. Solid boxes indicate the bioactive form of the GA, and the names with arrow headed are the catalyzing enzymes. The dashed box and line represent the inactive form of the GA. The upward headed arrow represents up regulation of the phytohormone response genes. After the subsequent process and interaction with other phytohormone response genes results into bioactive and inactivate GAs. (GA2ox- Gibberellin 2 oxidase, GA3ox – Gibberellin 3 oxidase, ARF – Auxin response factor, CKX – Cytokinin dehydrogenase, PILS – PIN Like proteins, KAO – ent-kaurenoic acid oxidase). Interaction of these genes in GA pathway results in morphology variation in soybean plants. Adapted and modified from (Ross et al., 2001; Yamaguchi, 2008; Nomura et al., 2013).
References
Azizi, P., Rafii, M. Y., Maziah, M., Abdullah, S. N. A., Hanafi, M. M., Latif, M. A., et al. (2015). Understanding the Shoot Apical Meristem Regulation: A Study of the Phytohormones, Auxin and Cytokinin, in rice. Mech. Dev. 135, 1–15. doi:10.1016/j.mod.2014.11.001
Barbez, E., Kubeš, M., Rolčík, J., Béziat, C., Pěnčík, A., Wang, B., et al. (2012). A Novel Putative Auxin Carrier Family Regulates Intracellular Auxin Homeostasis in Plants. Nature 485, 119–122. doi:10.1038/nature11001
Bari, R., and Jones, J. D. G. (2009). Role of Plant Hormones in Plant Defence Responses. Plant Mol. Biol. 69, 473–488. doi:10.1007/s11103-008-9435-0
Bernard, R. L. (1972). Two Genes Affecting Stem Termination in Soybeans 1. Crop Sci. 12, 235–239. doi:10.2135/cropsci1972.0011183x001200020028x
Björklund, S., Antti, H., Uddestrand, I., Moritz, T., and Sundberg, B. (2007). Cross-talk between Gibberellin and Auxin in Development of Populus wood: Gibberellin Stimulates Polar Auxin Transport and Has a Common Transcriptome with Auxin. Plant J. 52, 499–511. doi:10.1111/j.1365-313X.2007.03250.x
Busov, V. B., Brunner, A. M., and Strauss, S. H. (2008). Genes for Control of Plant Stature and Form. New Phytol. 177, 589–607. doi:10.1111/j.1469-8137.2007.02324.x
Chae, K., Isaacs, C. G., Reeves, P. H., Maloney, G. S., Muday, G. K., Nagpal, P., et al. (2012). Arabidopsis SMALL AUXIN UP RNA63 Promotes Hypocotyl and Stamen Filament Elongation. Plant J. 71, 684–697. doi:10.1111/j.1365-313X.2012.05024.x
Chandler, J. W., and Werr, W. (2015). Cytokinin-Auxin Crosstalk in Cell Type Specification. Trends Plant Sci. 20, 291–300. doi:10.1016/j.tplants.2015.02.003
Chen, Y., Dan, Z., Gao, F., Chen, P., Fan, F., and Li, S. (2020). Rice GROWTH-REGULATING FACTOR7 Modulates Plant Architecture through Regulating GA and Indole-3-Acetic Acid Metabolism. Plant Physiol. 184, 393–406. doi:10.1104/PP.20.00302
Dai, M., Zhao, Y., Ma, Q., Hu, Y., Hedden, P., Zhang, Q., et al. (2007). The RiceYABBY1Gene Is Involved in the Feedback Regulation of Gibberellin Metabolism. Plant Physiol. 144, 121–133. doi:10.1104/pp.107.096586
Davière, J.-M., and Achard, P. (2016). A Pivotal Role of DELLAs in Regulating Multiple Hormone Signals. Mol. Plant 9, 10–20. doi:10.1016/j.molp.2015.09.011
Dijkstra, C., Adams, E., Bhattacharya, A., Page, A. F., Anthony, P., Kourmpetli, S., et al. (2008). Over-expression of a Gibberellin 2-oxidase Gene from Phaseolus Coccineus L. Enhances Gibberellin Inactivation and Induces Dwarfism in Solanum Species. Plant Cel. Rep. 27, 463–470. doi:10.1007/s00299-007-0471-z
Eckardt, N. A. (2005). Cross Talk between Gibberellin and Cytokinin Signaling Converges on SPINDLY. Plant Cell 17, 1–3. doi:10.1105/tpc.104.170110
El-Showk, S., Ruonala, R., and Helariutta, Y. (2013). Crossing Paths: Cytokinin Signalling and Crosstalk. Dev 140, 1373–1383. doi:10.1242/dev.086371
Fahad, S., Hussain, S., Bano, A., Saud, S., Hassan, S., Shan, D., et al. (2015). Potential Role of Phytohormones and Plant Growth-Promoting Rhizobacteria in Abiotic Stresses: Consequences for Changing Environment. Environ. Sci. Pollut. Res. 22, 4907–4921. doi:10.1007/s11356-014-3754-2
Feraru, E., Vosolsobě, S., Feraru, M. I., Petrášek, J., and Kleine-Vehn, J. (2012). Evolution and Structural Diversification of PILS Putative Auxin Carriers in Plants. fpls 3, 1–13. doi:10.3389/fpls.2012.00227
Filardo, F. F., and Swain, S. M. (2003). SPYing on GA Signaling and Plant Development. J. Plant Growth Regul. 22, 163–175. doi:10.1007/s00344-003-0034-7
Finet, C., Fourquin, C., Vinauger, M., Berne-Dedieu, A., Chambrier, P., Paindavoine, S., et al. (2010). Parallel Structural Evolution of Auxin Response Factors in the Angiosperms. Plant J. 63, 952–959. doi:10.1111/j.1365-313X.2010.04292.x
Gallavotti, A. (2013). The Role of Auxin in Shaping Shoot Architecture. J. Exp. Bot. 64, 2593–2608. doi:10.1093/jxb/ert141
Gaspar, T., Kevers, C., Penel, C., Greppin, H., Reid, D. M., and Thorpe, T. A. (1996). Plant Hormones and Plant Growth Regulators in Plant Tissue Culture. In Vitro Cell.Dev.Biol.-Plant 32, 272–289. doi:10.1007/BF02822700
Ge, J., Li, B., Shen, D., Xie, J., Long, J., and Dong, H. (2016). Tobacco TTG2 Regulates Vegetative Growth and Seed Production via the Predominant Role of ARF8 in Cooperation with ARF17 and ARF19. BMC Plant Biol. 16, 1–17. doi:10.1186/s12870-016-0815-3
Goetz, M., Hooper, L. C., Johnson, S. D., Rodrigues, J. C. M., Vivian-Smith, A., and Koltunow, A. M. (2007). Expression of Aberrant Forms ofAUXIN RESPONSE FACTOR8Stimulates Parthenocarpy in Arabidopsis and Tomato. Plant Physiol. 145, 351–366. doi:10.1104/pp.107.104174
Greenboim-Wainberg, Y., Maymon, I., Borochov, R., Alvarez, J., Olszewski, N., Ori, N., et al. (2005). Cross Talk between Gibberellin and Cytokinin: The Arabidopsis GA Response Inhibitor SPINDLY Plays a Positive Role in Cytokinin Signaling. Plant Cell 17, 92–102. doi:10.1105/tpc.104.028472
Guo, H., Li, L., Ye, H., Yu, X., Algreen, A., and Yin, Y. (2009). Three Related Receptor-like Kinases Are Required for Optimal Cell Elongation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 106, 7648–7653. doi:10.1073/pnas.0812346106
Gupta, R., and Chakrabarty, S. K. (2013). Gibberellic Acid in Plant. Plant Signal. Behav. 8, e25504. doi:10.4161/psb.25504
Haga, K., and Iino, M. (1998). Auxin-growth Relationships in maize Coleoptiles and Pea Internodes and Control by Auxin of the Tissue Sensitivity to Auxin. Plant Physiol. 117, 1473–1486. doi:10.1104/pp.117.4.1473
Hagen, G., and Guilfoyle, T. (2002). Auxin-responsive Gene Expression: Genes, Promoters and Regulatory Factors. Plant Mol. Biol. 49, 373–385. doi:10.1007/978-94-010-0377-3_9
Harberd, N. P., King, K. E., Carol, P., Cowling, R. J., Peng, J., and Richards, D. E. (1998). Gibberellin: Inhibitor of an Inhibitor of? BioEssays 20, 1001–1008. doi:10.1002/(SICI)1521-1878(199812)20:12<1001:AID-BIES6>3.0.CO;2-O
Haruta, M., and Sussman, M. R. (2017). Ligand Receptor-Mediated Regulation of Growth in Plants. Curr. Top. Dev. Biol. 123, 331–363. 1st ed. Elsevier Inc. doi:10.1016/bs.ctdb.2016.11.007
Hejátko, J., Ryu, H., Kim, G.-T., Dobešová, R., Choi, S., Choi, S. M., et al. (2009). The Histidine Kinases CYTOKININ-INDEPENDENT1 and ARABIDOPSIS HISTIDINE KINASE2 and 3 Regulate Vascular Tissue Development inArabidopsisShoots. Plant Cell 21, 2008–2021. doi:10.1105/tpc.109.066696
Hepworth, J., and Lenhard, M. (2014). Regulation of Plant Lateral-Organ Growth by Modulating Cell Number and Size. Curr. Opin. Plant Biol. 17, 36–42. doi:10.1016/j.pbi.2013.11.005
Higuchi, M., Pischke, M. S., Mähönen, A. P., Miyawaki, K., Hashimoto, Y., Seki, M., et al. (2004). In Planta Functions of the Arabidopsis Cytokinin Receptor Family. Proc. Natl. Acad. Sci. 101, 8821–8826. doi:10.1073/pnas.0402887101
Hou, X., Lee, L. Y. C., Xia, K., Yan, Y., and Yu, H. (2010). DELLAs Modulate Jasmonate Signaling via Competitive Binding to JAZs. Dev. Cel. 19, 884–894. doi:10.1016/j.devcel.2010.10.024
Huot, B., Yao, J., Montgomery, B. L., and He, S. Y. (2014). Growth-defense Tradeoffs in Plants: A Balancing Act to Optimize Fitness. Mol. Plant 7, 1267–1287. doi:10.1093/mp/ssu049
Hwang, I., and Sheen, J. (2001). Two-component Circuitry in Arabidopsis Cytokinin Signal Transduction. Nature 413, 383–389. doi:10.1038/35096500
Jasinski, S., Piazza, P., Craft, J., Hay, A., Woolley, L., Rieu, I., et al. (2005). KNOX Action in Arabidopsis Is Mediated by Coordinate Regulation of Cytokinin and Gibberellin Activities. Curr. Biol. 15, 1560–1565. doi:10.1016/j.cub.2005.07.023
Jay, F., Wang, Y., Yu, A., Taconnat, L., Pelletier, S., Colot, V., et al. (2011). Misregulation of AUXIN RESPONSE FACTOR 8 Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis. Plos Pathog. 7, e1002035. doi:10.1371/journal.ppat.1002035
Jeon, J., Kim, N. Y., Kim, S., Kang, N. Y., Novák, O., Ku, S.-J., et al. (2010). A Subset of Cytokinin Two-Component Signaling System Plays a Role in Cold Temperature Stress Response in Arabidopsis. J. Biol. Chem. 285, 23371–23386. doi:10.1074/jbc.M109.096644
Jones, R. L., and Kaufman, P. B. (1983). The Role of Gibberellins in Plant Cell Elongation. Crit. Rev. Plant Sci. 1, 23–47. doi:10.1080/07352688309382170
Joshi, T., Valliyodan, B., Wu, J.-H., Lee, S.-H., Xu, D., and Nguyen, H. T. (2013). Genomic Differences between Cultivated Soybean, G. max and its Wild Relative G. Soja. BMC Genomics 14, 1–11. doi:10.1186/1471-2164-14-S1-S5
Kende, H., van der Knaap, E., and Cho, H.-T. (1998). Deepwater rice: A Model Plant to Study Stem Elongation. Plant Physiol. 118, 1105–1110. doi:10.1104/pp.118.4.1105
Keshishian, E. A., and Rashotte, A. M. (2015). Plant Cytokinin Signalling. Essays Biochem. 58, 13–27. doi:10.1042/BSE0580013
Khush, G. S. (2001). Green Revolution: The Way Forward. Nat. Rev. Genet. 2, 815–822. doi:10.1038/35093585
Kilgore-Norquest, L., and Sneller, C. H. (2000). Effect of Stem Termination on Soybean Traits in Southern U.S. Production Systems. Crop Sci. 40, 83–90. doi:10.2135/cropsci2000.40183x
Kim, H. J., Ryu, H., Hong, S. H., Woo, H. R., Lim, P. O., Lee, I. C., et al. (2006). Cytokinin-mediated Control of Leaf Longevity by AHK3 through Phosphorylation of ARR2 in Arabidopsis. Proc. Natl. Acad. Sci. 103, 814–819. doi:10.1073/pnas.0505150103
Koenig, D., Jiménez-Gómez, J. M., Kimura, S., Fulop, D., Chitwood, D. H., Headland, L. R., et al. (2013). Comparative Transcriptomics Reveals Patterns of Selection in Domesticated and Wild Tomato. Proc. Natl. Acad. Sci. 110, E2655–E2662. doi:10.1073/pnas.1309606110
Kofsky, J., Zhang, H., and Song, B.-H. (2018). The Untapped Genetic Reservoir: The Past, Current, and Future Applications of the Wild Soybean (Glycine Soja). Front. Plant Sci. 9, 949. doi:10.3389/fpls.2018.00949
Kou, E., Huang, X., Zhu, Y., Su, W., Liu, H., Sun, G., et al. (2021). Crosstalk between Auxin and Gibberellin during Stalk Elongation in Flowering Chinese Cabbage. Sci. Rep. 11, 1–9. doi:10.1038/s41598-021-83519-z
Kurepa, J., Shull, T. E., and Smalle, J. A. (2019). Antagonistic Activity of Auxin and Cytokinin in Shoot and Root Organs. Plant Direct 3, e00121. doi:10.1002/pld3.121
Kurepin, L. V., Ozga, J. A., Zaman, M., and Pharis, R. P. (2013). The Physiology of Plant Hormones in Cereal , Oilseed and Pulse Crops. Prairie Soils Crop Ejournal 6, 7–23. Available at: www.prairiesoilsandcrops.ca.
Kutschera, U., Bergfeld, R., and Schopfer, P. (1987). Cooperation of Epidermis and Inner Tissues in Auxin-Mediated Growth of maize Coleoptiles. Planta 170, 168–180. doi:10.1007/BF00397885
Li, K., Gao, Z., He, H., Terzaghi, W., Fan, L.-M., Deng, X. W., et al. (2015). Arabidopsis DET1 Represses Photomorphogenesis in Part by Negatively Regulating DELLA Protein Abundance in Darkness. Mol. Plant 8, 622–630. Elsevier Ltd. doi:10.1016/j.molp.2014.12.017
Liu, B., Fujita, T., Yan, Z.-H., Sakamoto, S., Xu, D., and Abe, J. (2007). QTL Mapping of Domestication-Related Traits in Soybean (Glycine max). Ann. Bot. 100, 1027–1038. doi:10.1093/aob/mcm149
Liu, B., Watanabe, S., Uchiyama, T., Kong, F., Kanazawa, A., Xia, Z., et al. (2010). The Soybean Stem Growth Habit Gene Dt1 Is an Ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol. 153, 198–210. doi:10.1104/pp.109.150607
Liu, N., Wu, S., Van Houten, J., Wang, Y., Ding, B., Fei, Z., et al. (2014). Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 Leads to floral Development Defects and Female Sterility in Tomato. J. Exp. Bot. 65, 2507–2520. doi:10.1093/jxb/eru141
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Luo, X., Zheng, J., Huang, R., Huang, Y., Wang, H., Jiang, L., et al. (2016). Phytohormones Signaling and Crosstalk Regulating Leaf Angle in rice. Plant Cel Rep 35, 2423–2433. doi:10.1007/s00299-016-2052-5
Ma, L., and Li, G. (2019). Auxin-dependent Cell Elongation during the Shade Avoidance Response. Front. Plant Sci. 10, 1–8. doi:10.3389/fpls.2019.00914
Martin, D. N., Proebsting, W. M., and Hedden, P. (1999). The SLENDER Gene of Pea Encodes a Gibberellin 2-Oxidase. Plant Physiol. 121, 775–781. doi:10.1104/pp.121.3.775
Maymon, I., Greenboim-Wainberg, Y., Sagiv, S., Kieber, J. J., Moshelion, M., Olszewski, N., et al. (2009). Cytosolic Activity of SPINDLY Implies the Existence of a DELLA-Independent Gibberellin-Response Pathway. Plant J. 58, 979–988. doi:10.1111/j.1365-313X.2009.03840.x
Mencuccini, M. (2015). Dwarf Trees, Super-sized Shrubs and Scaling: Why Is Plant Stature So Important? Plant Cel. Environ. 38, 1–3. doi:10.1111/pce.12442
Mok, D. W., and Mok, M. C. (2001). Cytokinin Metabolism and Action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 89–118. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11337393. doi:10.1146/annurev.arplant.52.1.89
Mok, M. C. (1988). Plant Hormones and Their Role in Plant Growth and Development. Peter J. Davies. Q. Rev. Biol. 63, 225. doi:10.1086/415875
Morrone, D., Chen, X., Coates, R. M., and Peters, R. J. (2010). Characterization of the Kaurene Oxidase CYP701A3, a Multifunctional Cytochrome P450 from Gibberellin Biosynthesis. Biochem. J. 431, 337–347. doi:10.1042/BJ20100597
Motyka, V., Faiss, M., Strand, M., Kamínek, M., and Schmülling, T. (1996). Changes in Cytokinin Content and Cytokinin Oxidase Activity in Response to Derepression of Ipt Gene Transcription in Transgenic Tobacco Calli and Plants. Plant Physiol. 112, 1035–1043. doi:10.1104/pp.112.3.1035
Nagpal, P., Ellis, C. M., Weber, H., Ploense, S. E., Barkawi, L. S., Guilfoyle, T. J., et al. (2005). Auxin Response Factors ARF6 and ARF8 Promote Jasmonic Acid Production and Flower Maturation. Development 132, 4107–4118. doi:10.1242/dev.01955
Nomura, T., Magome, H., Hanada, A., Takeda-Kamiya, N., Mander, L. N., Kamiya, Y., et al. (2013). Functional Analysis of Arabidopsis CYP714A1 and CYP714A2 Reveals that They Are Distinct Gibberellin Modification Enzymes. Plant Cel. Physiol. 54, 1837–1851. doi:10.1093/pcp/pct125
Oh, E., Zhu, J.-Y., Bai, M.-Y., Arenhart, R. A., Sun, Y., and Wang, Z.-Y. (2014). Cell Elongation Is Regulated through a central Circuit of Interacting Transcription Factors in the Arabidopsis Hypocotyl. Elife 3, 1–19. doi:10.7554/eLife.03031
Oh, M.-H., Honey, S. H., and Tax, F. E. (2020). The Control of Cell Expansion, Cell Division, and Vascular Development by Brassinosteroids: A Historical Perspective. Ijms 21, 1743–1815. doi:10.3390/ijms21051743
Ohri, P., Bhardwaj, R., Bali, S., Kaur, R., Jasrotia, S., Khajuria, A., et al. (2015). The Common Molecular Players in Plant Hormone Crosstalk and Signaling. Cpps 16, 369–388. doi:10.2174/1389203716666150330141922
Olszewski, N., Sun, T.-p., and Gubler, F. (2002). Gibberellin Signaling: Biosynthesis, Catabolism, and Response Pathways. Plant Cell 14, S61–S80. doi:10.1105/tpc.010476
Qin, F., Kodaira, K.-S., Maruyama, K., Mizoi, J., Tran, L.-S. P., Fujita, Y., et al. (2011). SPINDLY, a Negative Regulator of Gibberellic Acid Signaling, Is Involved in the Plant Abiotic Stress Response. Plant Physiol. 157, 1900–1913. doi:10.1104/pp.111.187302
Qiu, J., Wang, Y., Wu, S., Wang, Y.-Y., Ye, C.-Y., Bai, X., et al. (2014). Genome Re-sequencing of Semi-wild Soybean Reveals a Complex Soja Population Structure and Deep Introgression. PLoS One 9, e108479. doi:10.1371/journal.pone.0108479
Richards, D. E., King, K. E., Ait-ali, T., and Harberd, N. P. (2001). How Gibberellin Regulates Plant Growth and Development: A Molecular Genetic Analysis of Gibberellin Signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 67–88. doi:10.1146/annurev.arplant.52.1.67
Rieu, I., Eriksson, S., Powers, S. J., Gong, F., Griffiths, J., Woolley, L., et al. (2008). Genetic Analysis Reveals that C19-GA 2-Oxidation Is a Major Gibberellin Inactivation Pathway inArabidopsis. Plant Cell 20, 2420–2436. doi:10.1105/tpc.108.058818
Ross, J., and O'Neill, D. (2001). New Interactions between Classical Plant Hormones. Trends Plant Sci. 6, 2–4. doi:10.1016/S1360-1385(00)01795-7
Ross, J. J., O'Neill, D. P., Wolbang, C. M., Symons, G. M., and Reid, J. B. (2001). Auxin-gibberellin Interactions and Their Role in Plant Growth. J. Plant Growth Regul. 20, 346–353. doi:10.1007/s003440010034
Sakamoto, T., Kobayashi, M., Itoh, H., Tagiri, A., Kayano, T., Tanaka, H., et al. (2001). Expression of a Gibberellin 2-oxidase Gene Around the Shoot apex Is Related to Phase Transition in rice. Plant Physiol. 125, 1508–1516. doi:10.1104/pp.125.3.1508
Schmülling, T., Werner, T., Riefler, M., Krupková, E., and Bartrina Y Manns, I. (2003). Structure and Function of Cytokinin Oxidase/dehydrogenase Genes of maize, rice, Arabidopsis and Other Species. J. Plant Res. 116, 241–252. doi:10.1007/s10265-003-0096-4
Schmülling, T. (2002). New Insights into the Functions of Cytokinins in Plant Development. J. Plant Growth Regul. 21, 40–49. doi:10.1007/s003440010046
Schomburg, F. M., Bizzell, C. M., Lee, D. J., Zeevaart, J. A. D., and Amasino, R. M. (2003). Overexpression of a Novel Class of Gibberellin 2-oxidases Decreases Gibberellin Levels and Creates dwarf Plants. Plant Cell 15, 151–163. doi:10.1105/tpc.005975
Schuetz, M., Fidanza, M., and Mattsson, J. (2019). Identification of Auxin Response Factor-Encoding Genes Expressed in Distinct Phases of Leaf Vein Development and with Overlapping Functions in Leaf Formation. Plants 8, 242. doi:10.3390/plants8070242
Seif El-Yazal, S., Seif El-Yazal, M., Dwidar, E., and Rady, M. (2015). Phytohormone Crosstalk Research: Cytokinin and its Crosstalk with Other Phytohormones. Cpps 16, 395–405. doi:10.2174/1389203716666150330141159
Shih, H.-W., Miller, N. D., Dai, C., Spalding, E. P., and Monshausen, G. B. (2014). The Receptor-like Kinase FERONIA Is Required for Mechanical Signal Transduction in Arabidopsis Seedlings. Curr. Biol. 24, 1887–1892. doi:10.1016/j.cub.2014.06.064
Shimizu-Sato, S., Tanaka, M., and Mori, H. (2009). Auxin-cytokinin Interactions in the Control of Shoot Branching. Plant Mol. Biol. 69, 429–435. doi:10.1007/s11103-008-9416-3
Shull, T. E., Kurepa, J., and Smalle, J. A. (2016). Cytokinin Signaling Promotes Differential Stability of Type-B ARRs. Plant Signal. Behav. 11, e1169354–3. doi:10.1080/15592324.2016.1169354
Singh, R. J., and Hymowitz, T. (1988). The Genomic Relationship between Glycine max (L.) Merr. And G. Soja Sieb. and Zucc. as Revealed by Pachytene Chromosome Analysis. Theoret. Appl. Genet. 76, 705–711. doi:10.1007/BF00303516
Stupar, R. M. (2010). Into the Wild: The Soybean Genome Meets its Undomesticated Relative. Proc. Natl. Acad. Sci. 107, 21947–21948. doi:10.1073/pnas.1016809108
Swain, S. M., Tseng, T.-s., and Olszewski, N. E. (2001). Altered Expression of SPINDLY Affects Gibberellin Response and Plant Development. Plant Physiol. 126, 1174–1185. doi:10.1104/pp.126.3.1174
Taylor-Teeples, M., Lanctot, A., and Nemhauser, J. L. (2016). As above, So below: Auxin's Role in Lateral Organ Development. Dev. Biol. 419, 156–164. doi:10.1016/j.ydbio.2016.03.020
Thomas, S. G., and Sun, T.-p. (2004). Update on Gibberellin Signaling. A Tale of the Tall and the Short. Plant Physiol. 135, 668–676. doi:10.1104/pp.104.040279
Thomas, S. G., Phillips, A. L., and Hedden, P. (1999). Molecular Cloning and Functional Expression of Gibberellin 2- Oxidases, Multifunctional Enzymes Involved in Gibberellin Deactivation. Proc. Natl. Acad. Sci. 96, 4698–4703. doi:10.1073/pnas.96.8.4698
Thornton, T., Swain, S. M., and Olszewski, N. E. (1999). Gibberellin Signal Transduction Presents …the SPY Who O-GlcNAc'd Me. Trends Plant Sci. 4, 424–428. doi:10.1016/S1360-1385(99)01485-5
Tian, C. E., Muto, H., Higuchi, K., Matamura, T., Tatematsu, K., Koshiba, T., et al. (2004). Disruption and Overexpression Ofauxin Response Factor 8gene ofArabidopsisaffect Hypocotyl Elongation and Root Growth Habit, Indicating its Possible Involvement in Auxin Homeostasis in Light Condition. Plant J. 40, 333–343. doi:10.1111/j.1365-313X.2004.02220.x
Tian, Z., Wang, X., Lee, R., Li, Y., Specht, J. E., Nelson, R. L., et al. (2010). Artificial Selection for Determinate Growth Habit in Soybean. Proc. Natl. Acad. Sci. 107, 8563–8568. doi:10.1073/pnas.1000088107
Vanstraelen, M., and Benková, E. (2012). Hormonal Interactions in the Regulation of Plant Development. Annu. Rev. Cel Dev. Biol. 28, 463–487. doi:10.1146/annurev-cellbio-101011-155741
Varaud, E., Brioudes, F., Szécsi, J., Leroux, J., Brown, S., Perrot-Rechenmann, C., et al. (2011). AUXIN RESPONSE FACTOR8 RegulatesArabidopsisPetal Growth by Interacting with the bHLH Transcription Factor BIGPETALp. Plant Cell 23, 973–983. doi:10.1105/tpc.110.081653
Wang, Y., and Li, J. (2008). Molecular Basis of Plant Architecture. Annu. Rev. Plant Biol. 59, 253–279. doi:10.1146/annurev.arplant.59.032607.092902
Wang, Y., Liu, H., and Xin, Q. (2014). Genome-wide Analysis and Identification of Cytokinin Oxidase/dehydrogenase (CKX) Gene Family in Foxtail Millet (Setaria Italica). Crop J. 2, 244–254. doi:10.1016/j.cj.2014.05.001
Wang, Y., Zhao, J., Lu, W., and Deng, D. (2017). Gibberellin in Plant Height Control: Old Player, New story. Plant Cel. Rep. 36, 391–398. doi:10.1007/s00299-017-2104-5
Wang, R., Liu, L., Kong, J., Xu, Z., Akhter Bhat, J., and Zhao, T. (2019). QTL Architecture of Vine Growth Habit and Gibberellin Oxidase Gene Diversity in Wild Soybean (Glycine Soja). Sci. Rep. 9, 1–10. doi:10.1038/s41598-019-43887-z
Werner, T., Motyka, V., Strnad, M., and Schmülling, T. (2001). Regulation of Plant Growth by Cytokinin. Proc. Natl. Acad. Sci. 98, 10487–10492. doi:10.1073/pnas.171304098
Wuddineh, W. A., Mazarei, M., Zhang, J., Poovaiah, C. R., Mann, D. G. J., Ziebell, A., et al. (2015). Identification and Overexpression of Gibberellin 2‐oxidase ( GA 2ox ) in Switchgrass ( P Anicum Virgatum L.) for Improved Plant Architecture and Reduced Biomass Recalcitrance. Plant Biotechnol. J. 13, 636–647. doi:10.1111/pbi.12287
Yamaguchi, S., Sun, T.-p., Kawaide, H., and Kamiya, Y. (1998). The GA2 Locus of Arabidopsis thalianaEncodes Ent-Kaurene Synthase of Gibberellin Biosynthesis. Plant Physiol. 116, 1271–1278. doi:10.1104/pp.116.4.1271
Yamaguchi, S. (2008). Gibberellin Metabolism and its Regulation. Annu. Rev. Plant Biol. 59, 225–251. doi:10.1146/annurev.arplant.59.032607.092804
Yanai, O., Shani, E., Dolezal, K., Tarkowski, P., Sablowski, R., Sandberg, G., et al. (2005). Arabidopsis KNOXI Proteins Activate Cytokinin Biosynthesis. Curr. Biol. 15, 1566–1571. doi:10.1016/j.cub.2005.07.060
Yang, T., Davies, P. J., and Reid, J. B. (1996). Genetic Dissection of the Relative Roles of Auxin and Gibberellin in the Regulation of Stem Elongation in Intact Light-Grown Peas. Plant Physiol. 110, 1029–1034. doi:10.1104/pp.110.3.1029
Yang, S., Yu, H., Xu, Y., and Goh, C. J. (2003). Investigation of Cytokinin-Deficient Phenotypes in Arabidopsis by Ectopic Expression of Orchid DSCKX1. FEBS Lett. 555, 291–296. doi:10.1016/S0014-5793(03)01259-6
Yang, D.-L., Yao, J., Mei, C.-S., Tong, X.-H., Zeng, L.-J., Li, Q., et al. (2012). Plant Hormone Jasmonate Prioritizes Defense over Growth by Interfering with Gibberellin Signaling cascade. Proc. Natl. Acad. Sci. 109, E1192–E1200. doi:10.1073/pnas.1201616109
Yoshida, H., Hirano, K., Sato, T., Mitsuda, N., Nomoto, M., Maeo, K., et al. (2014). DELLA Protein Functions as a Transcriptional Activator through the DNA Binding of the INDETERMINATE DOMAIN Family Proteins. Proc. Natl. Acad. Sci. 111, 7861–7866. doi:10.1073/pnas.1321669111
Yue, J.-h., Zhang, D., Ren, L., and Shen, X.-h. (2016). Gibberellin and Auxin Signals Control Scape Cell Elongation and Proliferation in Agapanthus Praecox Ssp. Orientalis. J. Plant Biol. 59, 358–368. doi:10.1007/s12374-016-0056-x
Zazimalová, E., Petrasek, J., and Benková, E. (2014). Auxin and its Role in Plant Development. doi:10.1007/978-3-7091-1526-8
Zhang, X., Ding, W., Xue, D., Li, X., Zhou, Y., Shen, J., et al. (2021). Genome-wide Association Studies of Plant Architecture-Related Traits and 100-seed Weight in Soybean Landraces. BMC Genom Data 22, 1–14. doi:10.1186/s12863-021-00964-5
Zhao, Z., Andersen, S. U., Ljung, K., Dolezal, K., Miotk, A., Schultheiss, S. J., et al. (2010). Hormonal Control of the Shoot Stem-Cell Niche. Nature 465, 1089–1092. doi:10.1038/nature09126
Keywords: weedy type, cultivar type, transcriptome, differentially expressed genes, phytohormone
Citation: Basnet P, Um T, Roy NS, Cho WS, Park SC, Park K-C and Choi I-Y (2022) Identification and Characterization of Key Genes Responsible for Weedy and Cultivar Growth Types in Soybean. Front. Genet. 13:805347. doi: 10.3389/fgene.2022.805347
Received: 05 November 2021; Accepted: 17 January 2022;
Published: 24 February 2022.
Edited by:
Karthikeyan Adhimoolam, Jeju National University, South KoreaReviewed by:
Aditya Pratap, Indian Institute of Pulses Research (ICAR), IndiaErtugrul Filiz, Duzce University, Turkey
Copyright © 2022 Basnet, Um, Roy, Cho, Park, Park and Choi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ik-Young Choi, Y2hvaWlAa2FuZ3dvbi5hYy5rcg==
†These authors have contributed equally to this work
 Prakash Basnet1†
Prakash Basnet1† Ik-Young Choi
Ik-Young Choi