- National Key Laboratory of Crop Biology, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai’an, China
Potassium channels are important ion channels that are responsible for the absorption of potassium in the plant nutrient uptake system. In this study, we used homologous molecular cloning to obtain 8 K+ channel genes from the superior apple rootstock line 12-2 (self-named): MsAKT1-1, MsKAT3-2, MsKAT1-3, MsK2P3-4, MsK2P3-5, MsK2P5-6, MsK2P3-7, and MsK2P3-8. Their lengths varied from 942 bp (MsK2P5-6) to 2625 bp (MsAKT1-1), and the number of encoded amino acids varied from 314 (MsK2P5-6) to 874 (MsAKT1-1). Subcellular localization predictions showed that MsAKT1-1, MsKAT3-2, and MsKAT1-3 were localized on the plasma membrane, and MsK2P3-4, MsK2P3-5, MsK2P5-6, MsK2P3-7, and MsK2P3-8 were localized on the vacuole and plasma membrane. The 8 K+ channel proteins contained α helices, extended strands, β turns, and random coils. MsKAT1-3 had four transmembrane structures, MsKAT3-2 had six, and the other six K+ channel genes had five. Protein structure domain analysis showed that MsAKT1-1 contained nine protein domains, followed by MsKAT3-2 with four, MsKAT1-3 with three, and the other five two-pore domain K+ channel proteins with two. Semi-quantitative RT-PCR detection of the K+ channel genes showed that their expression levels were high in roots. qRT-PCR analysis showed that the relative expression levels of the 8 genes changed after exposure to ARD stress. The above results provide a theoretical basis for further research on the functions of potassium channel genes in 12-2 and a scientific basis for the breeding of ARD-resistant rootstock.
Introduction
Apple replant disease (ARD) refers to the general phenomenon in which the same or closely related crops are continuously planted on the same piece of land, leading to yield reductions, quality deterioration, and impaired growth under normal management (Chen et al., 2020). Apple is grown throughout the world; many countries list apple as a major consumer product because of its strong ecological adaptability, high nutritional value, good storage characteristics, and long supply cycle (Mao et al., 2021c). Limited by land resources, the replanting of old orchards is becoming more and more common, and major apple-producing areas in the world are facing ARD (Mazzola and Manici, 2012). About 50% of the apple orchards in the United Kingdom, New Zealand, and Poland have ARD (Manici et al., 2013). ARD can lead to poor growth of new roots, slow growth, short plants, reduced resistance, disease, and death of the entire plant, leading to severe economic repercussions (Narwal 2010). Strategies for effectively ameliorating the effects of ARD on apple are urgently needed.
The ARD pathogen complex consists of oomycetes, including Pythium and Phytophthora, and fungi such as Ilyonectria and Rhizoctonia, at times acting in concert with the lesion nematode Pratylenchus penetrans (Zhu et al., 2016). However, the specific pathogen complex may vary across geographic regions, or even between orchards in the same region (Zhou et al., 2021). Previous research has shown that Fusarium is the main pathogen that causes ARD in the Bohai Bay area (Wang et al., 2018). The specialized Fusarium proliferatum strain MR5 (MW600437.1) associated with ARD has recently been screened and identified (in review); it is highly pathogenic to apple roots.
The development of resistant rootstocks has always been the focus of research for prevention and control of ARD. Leinfelder and Merwin (2006) documented the growth of four rootstocks (M26, M7, CG6210, and G30) after planting in replant soil for 4 years. The growth of the G30 and CG6210 plants increased significantly, and the average life span of the CG6210 root system was five times that of M7. Rumberger et al. (2004) grafted the Royal Empire variety onto three CG rootstocks (CG16, CG30, and CG210) and the M7 and M26 conventional rootstocks. Over three consecutive years, the CG210 and CG30 rootstocks were more resistant to ARD. Mazzola and Zhao (2010) reported that the rate of Rhizopus rot infection was significantly lower in the Geneva rootstock series than in M26, MM111, and MM166 in a Washington State replant orchard. Nevertheless, these rootstocks have not been promoted in China.
K+ is the most abundant key cation in almost all organisms, and it plays a key role in the basic physiological processes of plants (Adams and Shin, 2014). Two major types of plant potassium ion transporter genes have been cloned, potassium ion transporter genes and potassium ion channel genes (Flügge and Schroeder, 2007). Potassium channel genes mainly include the Shaker family and the KCO family. One of the important features of the Shaker potassium channels is that they can form heterotetrameric structures that enable plants to regulate potassium ion transport activity in different cells (Gambale and Uozumi, 2006). AKT1 (Sentenac et al., 1992) and KAT1 (Anderson et al., 1992) were the first cloned plant potassium channels, and both are members of the Shaker family. In 1997, Dreyer et al. (1997) cloned the potassium channel genes AKT1, KAT1, and AtKAT3 in Arabidopsis. The potassium channel SKOR is expressed mainly in the central sheath and xylem parenchyma cells of Arabidopsis roots. It is responsible for the release of potassium ions from the column cells into the xylem and participates in potassium ion transport from the root to the crown (Gaymard et al., 1998). Czempinski was the first to find and clone the TPK family member TPK1/KCO1 from Arabidopsis using a homologous alignment method (Czempinski et al., 1997), and six additional members (TPK1–TPK5 and KCO3) were subsequently found in Arabidopsis (Czempinski et al., 1999). Two-pore domain potassium (K2P) channels are membrane proteins found in mammals, other animals (e.g., Drosophila), and multiple plant species (Goldstein et al., 2005; Enyedi and Czirják, 2010). Five genes encoding K2P channels have been found in the Arabidopsis genome (González et al., 2015).
K+ is also closely related to abiotic stress tolerance (Hasanuzzaman et al., 2018). K+ channels are ion channels responsible for the absorption of potassium in the plant nutrient uptake system. Xu et al. (2014) showed that the K+ channel gene GhAKT1 could regulate the uptake of K+ and increase the stress resistance of cotton. Kobayashi et al. (2016) used genome-wide association analysis to show that KATl was closely related to salt tolerance in Arabidopsis. Li et al. (2020) demonstrated that the expression of the KAT3 gene was induced by salt stress. A K+ uptake study of different apple rootstocks under ARD showed that the lower the ARD resistance, the more seriously the K+ absorption of the rootstocks was affected (Guo et al., 2015).
Previously, our research group selected and bred the new apple rootstock line 12-2 (self-named) through patented in situ breeding technology (Shen et al., 2015). A variety of physiological indexes were measured under ARD, and preliminary results showed that 12-2 had excellent resistance (Mao et al., 2021a). The use of non-damaging and non-destructive testing technology (Xu et al., 2020) to measure changes in K+ flow showed that ARD had little effect on K+ absorption in 12-2 (Mao et al., 2021b). Although numerous studies have shown that maintaining cell K+ balance is very important for plant resistance to various abiotic stresses (Assaha et al., 2017), there are few reports on how K+ channels affect plant resistance to ARD. In the present experiment, we used the K+ channel gene from Arabidopsis as a template and adopted a homologous cloning approach to identify K+ channel genes in 12-2. We then analyzed the relative expression of the K+ channel genes cloned in 12-2 under ARD-associated Fusarium proliferatum MR5 stress. Basic biochemical characterization of the multiple K+ channel genes and the results of quantitative reverse transcription-PCR (qRT-PCR) provide preliminary information on the molecular mechanisms by which K+ channel genes regulate root resistance to ARD in 12-2. They serve as an important theoretical basis for the breeding of ARD-resistant rootstocks.
Materials and Methods
Plant Materials and Experimental Treatments
The test material was the 12-2 rootstock that is tolerant to ARD and was selected through our patented in situ breeding technology. It was planted in the National Key Seedling Breeding Base of Shandong Agricultural University in Tai’an City, Shandong Province. At the beginning of May 2018, we selected sturdy twigs of 12-2 and cut them into 3-cm stem sections with new buds. After each section was rinsed with clean water for an hour, it was immersed in 75% alcohol for 30 s on an ultra-clean workbench that had been UV-sterilized for 30 min, then disinfected in 0.05% sodium hypochlorite solution for 10 min. It was washed three times with sterile water for 30 s each time. An oblique incision was made about 0.5 cm from the bottom of each stem segment with a sterilized scalpel, and the stems were inoculated on the induction medium. The medium was based on 1 L MS medium and contained 30 g L−1 sucrose (Sigma-Aldrich Co., Ltd., Shanghai, China), 7.5 g L−1 agar (Sigma-Aldrich), 0.6 mg L−1 6-BA (Sigma-Aldrich), and 0.2 mg L−1 IBA (Sigma-Aldrich) at pH 5.8. Five buds were inoculated in each vial of induction medium and placed in a tissue culture chamber at 25 ± 2°C with a 10-h light period and an illumination intensity of 1000 lx. Substitution took place every 40 days on average. After multiple generations, at the beginning of May 2020, the adventitious bud clusters that had grown to about 4 cm were cut into about 2-cm stems and then inoculated into the rooting medium. The medium was based on 1 L 1/2 MS medium and contained 20 g L−1 sucrose, 7.5 g L−1 agar, 0.2 mg L−1 6-BA, and 1.0 mg L−1 IBA at pH 5.8. After rooting, the collected roots were immediately frozen in liquid nitrogen and stored in a freezer at −80°C for RNA extraction. The extracted RNA was used for bioinformatics and semi-quantitative RT-PCR.
In mid-November 2021, one hundred 12-2 seedlings of similar size with 4–5 leaves were transplanted into black plastic pots (7.0 cm × 5.0 cm × 8.5 cm) filled with sterile substrate after seedling acclimatization. The specialized Fusarium proliferatum strain MR5 (MW600437.1) associated with ARD was recently screened and identified (in review); it is highly pathogenic to apple roots (Mao et al., 2021b) and was discovered by the research group of Professor Mao Zhiquan of Shandong Agricultural University. On November 24, a layer of pathogenic fungi was inoculated in liquid potato dextrose medium (PDB, Haibo, Qingdao, Shandong, China), cultured for 7 days, and then filtered through 8 layers of sterile gauze to obtain a spore suspension. The concentration was measured under a microscope (Nikon Ni-U, Tokyo, Japan) using a hemocytometer (Thermo Fisher Scientific, Waltham, MA, United States), and the final concentration was adjusted to 106 spores mL−1 with sterile water. In early December, fifty pots of 12-2 were irrigated with 20 ml ARD-associated Fusarium proliferatum MR5 (MR5) spore suspension, and the other fifty pots were irrigated with an equal volume of PDB medium to serve as controls. The seedlings were grown in a tissue culture room at 24 ± 2°C with a 16-h light photoperiod and a light intensity of 1000 lx. The pots were bottom-irrigated as needed to maintain 60% soil water content. In early December, before irrigation with MR5 spore solution, the roots of the controls that had not been irrigated with PDB medium were sampled and recorded as MR5 spore solution treatment for 0 days. Samples were subsequently collected from both treatments at 1, 3, 5, 7, and 9 days (Mao et al., 2021b; Xiang et al., 2021), quick frozen in liquid nitrogen, and stored in a freezer at −80°C for use in RNA extraction. The RNA extracted at this time was used for qRT-PCR analysis. Three biological replicates of each treatment were obtained at each sampling time point.
RNA Extraction and cDNA Synthesis
RNA was extracted from collected samples of 12-2 using the Invitrogen TRIzol Reagent kit (Thermo Fisher Scientific, Shanghai, China). The concentration and integrity of total RNA were determined by 0.8% agarose gel electrophoresis and a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Worcester, MA, United States). cDNA was synthesized by reverse transcription using the One-Step gDNA removal and cDNA synthesis kit (Transgen Biotech, Beijing, China).
Cloning and Screening of K+ Channel Genes From 12-2
There is no reference genome for 12-2, and in order to identify its K+ channel genes, we used 19 K+ channel genes (8 AKT, 3 KAT, 6 TPK, and 2 SKOR) from the Arabidopsis (Arabidopsis thaliana) genome (https://www.arabidopsis.org/) as queries. We used BLASTP (Gallin and Boutet, 2011) to retrieve 28 K+ channel protein sequences from apple (Malus domestica) at NCBI (9 AKT, 4 KAT, 4 SKOR, and 11 two-pore domain potassium channel genes). Taking these 28 apple K+ protein channel sequences as templates, we used Primer Premier 6.0 (Wang et al., 2020) to design specific primers and ultimately cloned 8 K+ channel genes from 12-2 (Supplementary Table S1). With cDNA from 12-2 as a template, we used the high-fidelity enzyme 2 × Phanta Max Master Mix (Vazyme Biotech, Nanjing, Jiangsu, China) for PCR amplification. The 50-μL total reaction system contained 20 μL ddH2O, 1 μL cDNA template (500 ng μL−1), 2 μL upstream and downstream primers, and 25 μL high-fidelity enzyme. The amplification procedure was 98°C for 3 min; 35 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 180 s; and 72°C for 10 min). A Gel Extraction Kit (CWBIO Biotech, Beijing, China) was used to recover bands of the appropriate lengths. These sequences were ligated into the p1300-GFP vector (Sangon Biotech, Shanghai, China). The resulting constructs were transformed into E. coli DH5α competent cells (Vazyme Biotech, Nanjing, Jiangsu, China), and the bacteria were cultured on LB solid medium containing kanamycin. The positive clones were screened by PCR, and the screened positive clones were sent to Shanghai Sangon Biotech Co., Ltd. for sequencing.
K+ Channel Gene Searches of the Pear, Poplar, Rice, Tobacco, and Tomato Genomes
To explore the relationships between K+ channel genes in 12-2 and those from other species and to compare their physical and chemical properties, we used 19 K+ channel genes (8 AKT, 3 KAT, 6 TPK, and 2 SKOR) from Arabidopsis as BLASTP queries at NCBI. We retrieved 9 K+ channel protein sequences from pear (Pyrus spp.) (2 AKT, 4 KAT, and 3 SKOR), 13 from poplar (Populus trichocarpa) (6 AKT, 3 KAT, and 4 SKOR), 14 from rice (Oryza sativa) (7 AKT, 5 KAT, and 2 SKOR), 25 from tobacco (Nicotiana tabacum L) (8 AKT, 10 KAT, and 7 SKOR), and 14 from tomato (Solanum lycopersicum) (5 AKT, 6 KAT, and 3 SKOR).
Multiple Sequence Alignment and Naming of K+ Channel Proteins in 12-2
DNAMAN software was used to construct a multiple sequence alignment of K+ channel proteins from 12-2 and apple, and the alignment results were combined with physical and chemical properties to predict the domain sequences and feature sites of each protein.
A phylogenetic tree was constructed from protein sequences of all species using MEGA7 software. The K+ channel proteins from 12-2 and the other species were grouped according to their cluster positions, and the K+ channel proteins from 12-2 were named according to the naming conventions of similar K+ channel proteins from apple.
Sequence Analysis of K+ Channel Genes and Proteins in 12-2
The molecular weight, base composition, and base distribution of amino acid sequences were analyzed using DNAMAN software. The ORF Finder program (https://www.ncbi.nlm.nih.gov/orffinder/) was used to predict the open reading frames of the K+ channel genes in 12-2. The motif distributions within the K+ channel proteins were analyzed by MEME (Multiple Em for Motif Elicitation, http://meme-suite.org/tools/meme) (Bailey et al., 2015). The protein sequence file for 12-2 was submitted to MEME, with the number of motifs set to 10 and the length of motifs set to 6–50.
Bioinformatics Analysis of K+ Channel Proteins in 12-2
ProtParam software (https://web.expasy.org/protparam/) (Garg, et al., 2016) was used to predict the isoelectric points, molecular masses, instability factors, fatty acid indexes, and GRAVY values of the 12-2 K+ channel proteins. Their subcellular localizations were predicted using ProtComp 9.0 (http://linux1.softberry.com/berry.phtml?topic=protcomppl&group=programs&subgroup=proloc) (Chou and Shen, 2010). DeepSig Server (https://deepsig.biocomp.unibo.it) (Savojardo et al., 2018) was used to predict the signal peptide sites of the proteins; their secondary structures, such as α helices, extended strands, β turns, and random coils, were predicted using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/). The TMHMM Server v2.0 (http://www.cbs.dtu.dk/services/TMHMM/) (Krogh et al., 2001) was used to predict the transmembrane structure of the K+ channel proteins. The PFAM website (El-Gebali et al., 2019) was used to obtain positional information on K+ channel protein domains. The domains were imported into TBtools (Chen et al., 2018), and the Visualize Pfam Domain Pattern tool was used to visualize the protein domains.
Semi-Quantitative RT-PCR Detection of K+ Channel Genes in Roots of 12-2
Using cDNA from 12-2 as a template, the 8 cloned potassium channel genes (Supplementary Table S2) were amplified using Taq Pro Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd., Nanjing, Jiangsu, China). The 20-μL reaction system contained 7 μL ddH2O, 1 μL cDNA template (500 ng μL−1), 1 μL upstream and downstream primers, and 10 μL Taq Pro Universal SYBR qPCR Master Mix. The amplification procedure was 95°C for 30 s; 40 cycles of 95°C for 10 s and 58°C for 30 s; 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. The MdActin (MD12G1140800) gene served as an internal control (Wang N. et al., 2015; Zhang et al., 2018). The amplification products were inspected and imaged using 1.0–1.5% agarose gel electrophoresis. ImageJ software (National Institutes of Health, United States) was used to analyze the gray values of the electrophoresis results. Three biological replicates of each sample were analyzed.
Relative Expression of K+ Channel Genes in Roots of 12-2 After Exposure to ARD-Associated Fusarium proliferatum MR5
The qRT-PCR amplification reactions were performed using Taq Pro Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd., Nanjing, Jiangsu, China). The reaction system and temperatures were the same as those described in the preceding Semi-Quantitative RT-PCR Detection of K+ Channel Genes in Roots of 12-2. Relative gene expression was normalized to that of the control treatment, and MdActin (MD12G1140800) served as the internal control (Wang N. et al., 2015; Zhang et al., 2018). The relative quantification of specific genes was performed using the cycle threshold (Ct) 2−ΔΔCt method (SoftwareIQ5 version 2.0). Three biological replicates of each sample were analyzed. The qRT-PCR primers are listed in Supplementary Table S2.
Results
Cloning and Identification of K+ Channel Genes in 12-2
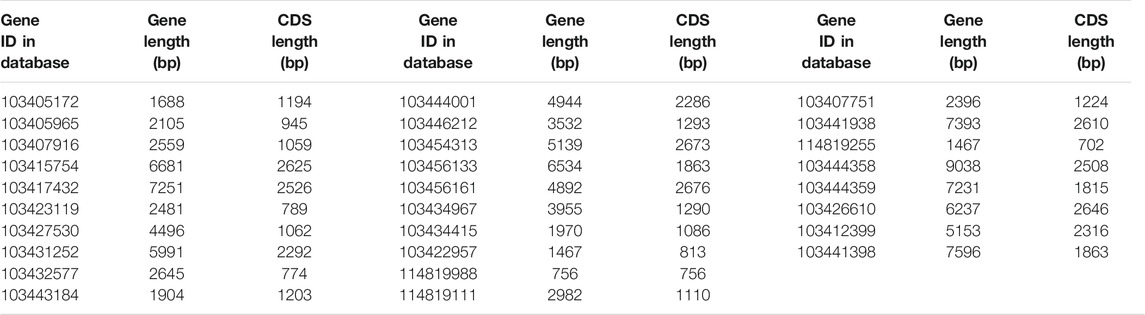
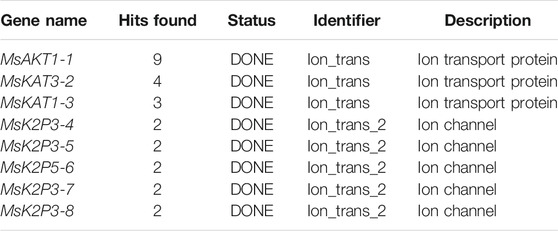
Using p1300-GFP as a vector for homologous cloning, 8 K+ channel genes were cloned based on 28 K+ channel gene sequences in apple (Table 1). Multiple sequence alignment (Supplementary Figures S1–S8) showed that the identity between the 8 K+ channel genes of 12-2 and their apple homologs ranged from 95.64 to 99.92%. This showed that the 8 K+ channel genes had been obtained from 12-2 by homologous cloning.
The phylogenetic tree (Figure 1A) showed that the 130 plant K+ channel proteins could be divided into 5 categories based on sequence homology. There were 46 AKT proteins (red in Figure 1A), 37 KAT proteins (blue), 25 SKOR proteins (purple), 6 TPK proteins (yellow), and 16 two-pore domain potassium (K2P) channel proteins (green). The K+ channel genes of 12-2 and other species all originated from one ancestor. The 8 genes from 12-2 were highly similar to those from apple, Arabidopsis, pear, and rice. The K+ channel genes from 12-2 were named according to their closest apple homologs using the format ‘Ms’ + K+ channel name + sequence name. Finally, the 8 genes (MsAKT1-1, MsKAT3-2, MsKAT1-3, MsK2P3-4, MsK2P3-5, MsK2P5-6, MsK2P3-7, and MsK2P3-8) were cloned from 12-2. The two-pore domain potassium (K2P) channel genes of 12-2 were closely related to some AKT genes from apple, Arabidopsis, and rice.
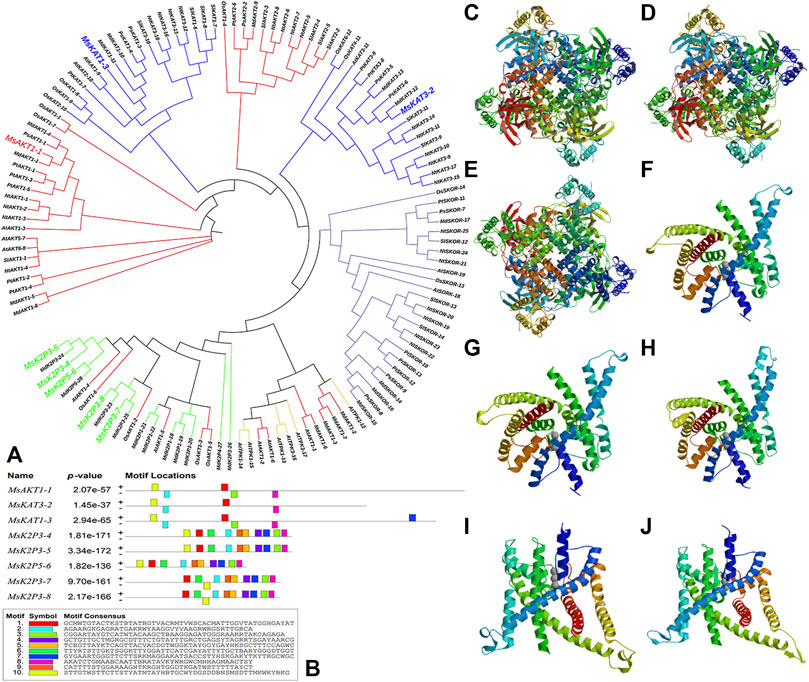
FIGURE 1. Phylogenetic tree of potassium channel genes in 12-2 and other species, conserved motifs in K+ channel genes, and secondary structure models of K+ channel proteins. (A): phylogenetic tree of K+ channel genes in 12-2 and other species. Red, AKT. Blue, KAT. Purple, SKOR. Yellow, TPK. Green, two-pore domain K+ channel protein (K2P). The 8 12-2 genes are enlarged and color-coded according to their respective family classifications. (B): conserved motifs in K+ channel genes of 12-2. (C–J): secondary structure models of K+ channel proteins in 12-2. (C): MsAKT1-1. (D): MsKAT3-2. (E): MsKAT1-3. (F): MsK2P3-4. (G): MsK2P3-5. (H): MsK2P5-6. (I): MsK2P3-7. (J): MsK2P3-8.
Nucleotide Sequence Analysis of K+ Channel Genes in 12-2
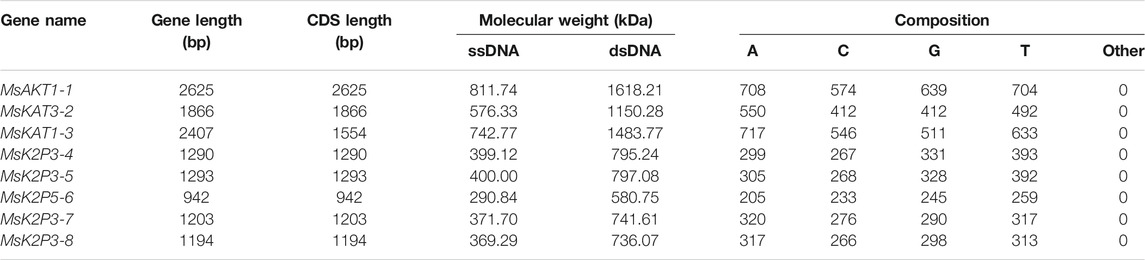
Table 2 shows that the gene length and CDS length of the 8 K+ channels varied from 942 bp (MsK2P5-6) to 2625 bp (MsAKT1-1), the molecular weight of single-stranded DNA (ssDNA) varied from 290.84 kDa (MsK2P5-6) to 811.74 kDa (MsAKT1-1), the molecular weight of double-stranded DNA (dsDNA) varied from 580.75 kDa (MsK2P5-6) to 1618.21 kDa (MsAKT1-1), and the base compositions of the sequences also differed. Numbers of bases ranged from 205 (A in MsK2P5-6) to 717 (A in MsKAT1-3).
Transcription factor binding site predictions (Figure 1B) showed that there were five transcription factor binding sites in MsAKT1-1, four in MsKAT3-2, six in MsKAT1-3, and 10 in each of the five two-pore domain potassium channel proteins.
Physicochemical Properties of K+ Channel Proteins
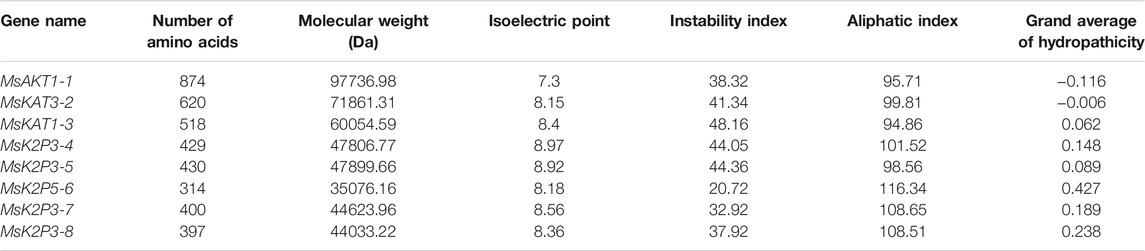
Table 3 shows that the number of amino acids in the 8 K+ channel proteins of 12-2 ranged from 314 (MsK2P5-6) to 874 (MsAKT1-1), with a minimum molecular weight of 35076.16 Da in MsK2P5-6, a maximum molecular weight of 97736.98 Da in MsAKT1-1, and an isoelectric point ranging from 7.3 (MsAKT1-1) to 8.97 (MsK2P3-4). The instability coefficients ranged from 20.72 (MsK2P5-6) to 48.16 (MsKAT1-3). MsAKT1-1, MsK2P5-6, MsK2P3-7, and MsK2P3-8 were stable proteins (instability coefficients less than 40), whereas the other four were unstable proteins with fatty acid coefficients ranging from 94.86 (MsKAT1-3) to 116.34 (MsK2P5-6). Except for MsKT1-1 and MsKAT3-2, the other six proteins were hydrophobic.
Subcellular Localization of K+ Channel Proteins
Subcellular localization predictions (Table 4) indicated that three of the 8 K+ channels of 12-2 were localized to the plasma membrane (MsAKT1-1, MsKAT3-2, and MsKAT1-3), whereas the other five two-pore domain potassium channels were localized mainly to the vacuole and secondarily to the plasma membrane.
Secondary Structures and Transmembrane Structures of K+ Channel Proteins
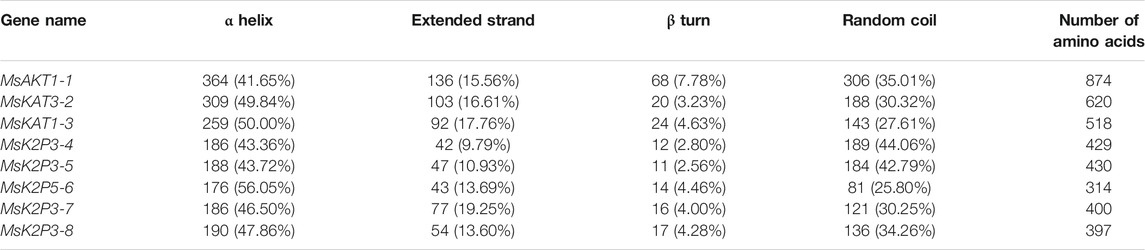
The DeepSig Server analysis showed that none of the 8 K+ channel proteins contained signal peptide sites. Secondary structure predictions (Table 5; Figures 1C–J) showed that the 8 proteins all contained α helices, extended strands, β turns, and random coils. The minimum percentage of amino acids in α helices was 41.65% (MsAKT1-1), and the maximum was 56.05% (MsK2P5-6); the percentage of amino acids in extended strands ranged from 9.79% (MsK2P3-4) to 19.25% (MsK2P3-7). The percentage of amino acids in β turns ranged from 2.56% (MsK2P3-5) to 7.78% (MsAKT1-1), and that in random coils ranged from 25.80% (MsK2P5-6) to 44.06% (MsK2P3-4). With the exception of MsK2P3-4, the amino acid percentages in the other seven proteins could be ranked α helix > random coil > extended strand > β turn. In all 8 proteins, the sum of the α helix and random coil percentages exceeded 75%.
Transmembrane structure predictions (Table 6; Figures 2A–H) showed that MsKAT1-3 had four transmembrane structures, MsKAT3-2 had six transmembrane structures, and the other six proteins had five transmembrane structures.
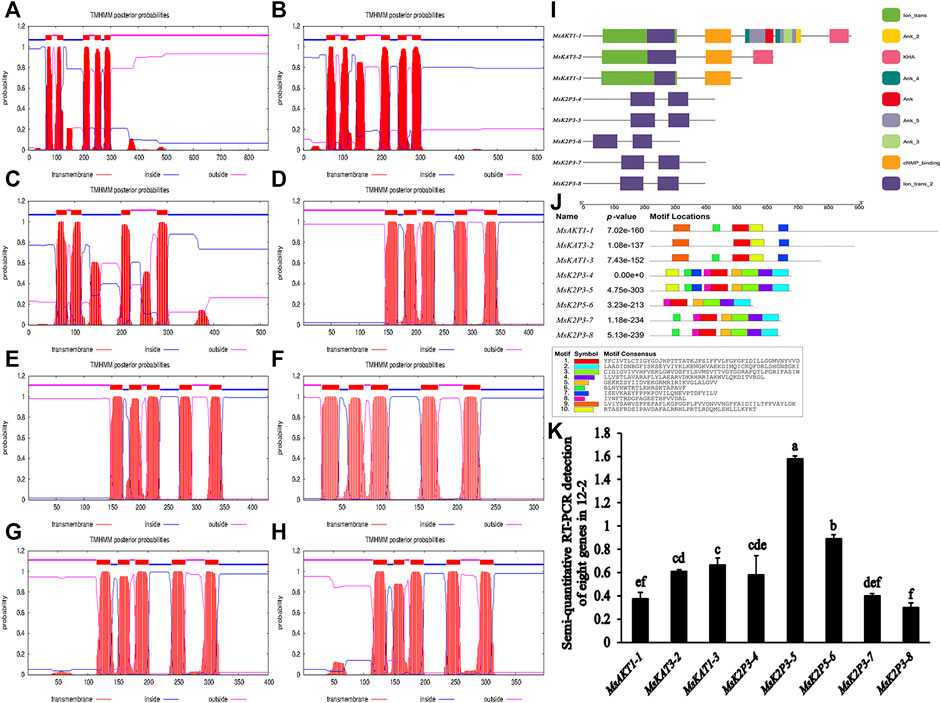
FIGURE 2. Models of transmembrane structures in K+ channel proteins, the structure of domains in K+ channel proteins, the motif distributions of K+ channel proteins, and semi-quantitative RT-PCR detection of 8 genes in roots of 12-2. (A–H): models of transmembrane structures in K+ channel proteins of 12-2. (A): MsAKT1-1. (B): MsKAT3-2. (C): MsKAT1-3. (D): MsK2P3-4. (E): MsK2P3-5. (F): MsK2P5-6. (G): MsK2P3-7. (H): MsK2P3-8. (I): the structure of domains in K+ channel proteins of 12-2. (J): motif distributions of K+ channel proteins in 12-2. (K): semi-quantitative RT-PCR detection of 8 genes in roots of 12-2.
Protein Structures and Conserved Sites of K+ Channel Proteins
Protein domain analysis (Table 7; Figure 2I) showed that the number of domains was highest in MsAKT1-1 (nine), followed by MsKAT3-2 with four, MsKAT1-3 with three, and the other five two-pore domain potassium channels with two. MsAKT1-1, MsKAT3-2, and MsKAT1-3 were recognized as ion channel proteins, and the 8 proteins all contained Ion_trans_2 domains.
The distribution of conserved motifs (Figure 2J) showed that MsAKT1-1 and MsKAT1-3 contained five identical motifs, whereas MsKAT3-2 contained four motifs. Compared with MsAKT1-1, MsKAT3-2 lacked Motif 6. MsK2P3-4 and MsK2P3-5 both contained nine identical motifs (except Motif 9). MsK2P3-7 and MsK2P3-8 contained seven identical motifs. Compared with MsK2P3-4, they lacked Motif 7 and Motif 10. MsK2P5-6 contained six motifs. Compared with MsK2P3-7, it lacked Motif 6. All 8 K+ channel proteins contained Motif 1.
Semi-Quantitative RT-PCR Detection of 8 K+ Channel Genes in Roots of 12-2
To explore differences in expression level among the 8 K+ channel genes in roots of 12-2, we performed semi-quantitative RT-PCR detection of their expression levels by analyzing the brightness and gray levels of electrophoresis images (Figure 2K; Supplementary Figure S9). The expression levels of MsK2P3-5 and MsK2P5-6 were significantly higher than those of the other six genes, and the expression levels of the other six genes did not differ significantly.
Relative Expression of K+ Channel Genes in Roots of 12-2 After Exposure to ARD-Associated Fusarium Proliferatum MR5
qRT-PCR analysis (Figure 3) showed that the relative expression of the 8 genes differed between roots treated with MR5 spore solution and control roots. Throughout the test period, the relative expression levels of MsAKT1-1, MsK2P5-6, MsK2P3-7, and MsK2P3-8 were significantly higher in Fusarium-treated roots than in control roots. The relative expression of MsAKT1-1 and MsK2P5-6 was highest on the first day and then decreased with time. The relative expression of MsK2P3-7 and MsK2P3-8 first increased and then decreased, and their relative expression was highest on the fifth day. MsKAT3-2, MsKAT1-3, and MsK2P3-5 showed a similar expression pattern: their expression first increased and then decreased. Relative expression of MsKAT3-2 was highest on the third day and was significantly higher in Fusarium-treated roots than in control roots. Its expression was the lowest on the ninth day and was significantly lower in treated roots than in control roots. MsKAT1-3 had the highest expression on the fifth day, when its expression was significantly higher in Fusarium-treated than control roots. The relative expression of MsK2P3-5 peaked on the third day and there was no significant difference between Fusarium-treated and control roots. On the remaining days, its expression was significantly lower in Fusarium-treated than control roots. Throughout the experiment, the relative expression level of MsK2P3-4 was significantly lower in treated than in control roots, and it showed a slowly increasing trend overall. Its relative expression was higher on the first day, then decreased and stabilized from the third to the seventh day. Its relative expression was highest on the ninth day and was significantly higher than that on the seventh day.
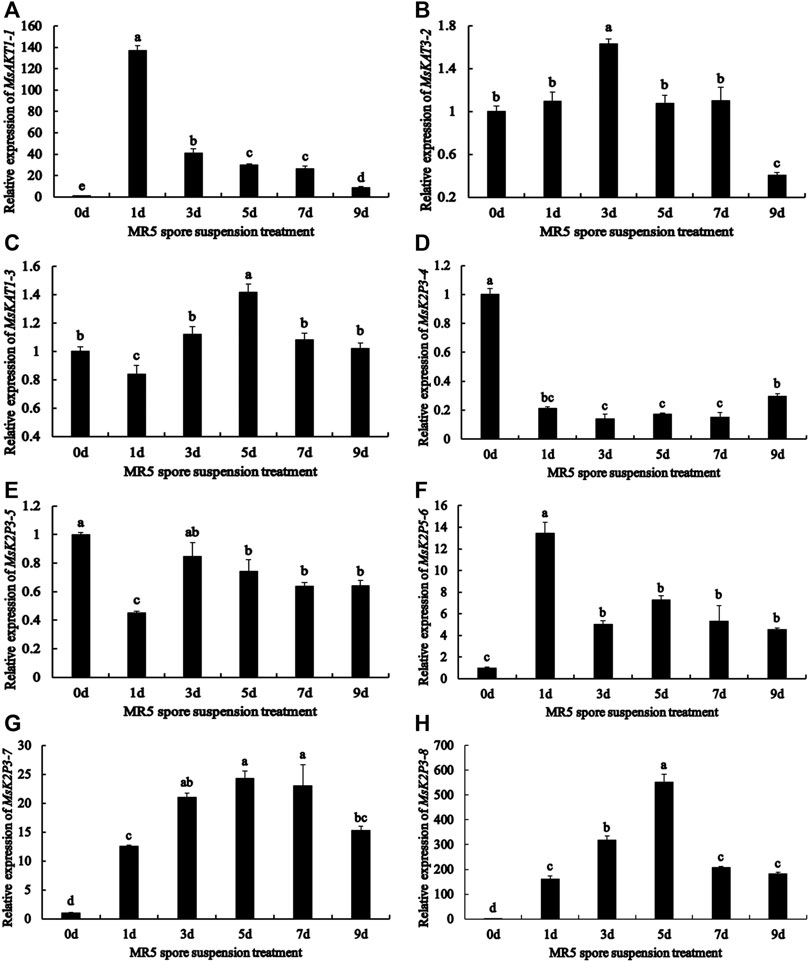
FIGURE 3. Relative expression of 8 K+ channel genes in roots at different days after spore solution treatment. (A): MsAKT1-1. (B): MsKAT3-2. (C): MsKAT1-3. (D): MsK2P3-4. (E): MsK2P3-5. (F): MsK2P5-6. (G): MsK2P3-7. (H): MsK2P3-8.
Discussion
It has been suggested that there are two different types of K+ uptake systems in plants: the high-affinity K+ absorption system for external K+ concentrations of 0.001–0.2 mM and the low-affinity K+ absorption system for external K+ concentrations of 1–10 mM (Rai and Kawabata, 2020). The high-affinity K+ absorption system mainly involves K+ transporters, whereas the low-affinity system mainly involves K+ channel proteins (Xie et al., 2020). When plants are under stress and the amount of K+ is insufficient, the high-affinity K+ system is quickly activated, and plant high-affinity K+ absorption is mediated mainly by high-affinity K+ transporters (Shin and Schachtman, 2004; Schachtman and Shin, 2007).
In this study, we cloned 8 K+ channel genes from the 12-2 rootstock through homologous cloning. Phylogenetic analysis showed that the K+ channel genes in 12-2 had high similarity to those from apple. This reflects not only the differentiation of monocots and dicots during plant evolution (Parkin, 1926) but also the fact that 12-2 and apple are members of the same genus. Research by Shealy et al. (2003) showed that the Shaker family contained the highly conserved amino acid sequence TxxTxGYGD (the T box). A TMCTIGYGD sequence was present at amino acids 191–199, 192–200, and 69–77 in MsK2P3-4, MsK2P3-5, and MsK2P5-6, respectively, and a TLCTIGYGD sequence was present at amino acids 159–167 and 156–164 in MsK2P3-7 and MsK2P3-8. The five two-pore domain potassium (K2P) channel genes therefore all contained T box structures. This may be one of the reasons why two-pore domain potassium (K2P) channel genes of 12-2 were highly similar to some AKT genes of apple, Arabidopsis, and rice in this research.
The results showed that the gene lengths and CDS lengths of the 8 K+ channel genes, and the number of translatable amino acids, unlike other species such as Arabidopsis. These results were in agreement with the study of Moulton et al. (2003), which showed that the number of amino acids differed among potassium channel proteins from distantly related species, although there was also conservation among species. The differences in gene length and CDS length also caused differences in the molecular weight of single-stranded DNA and double-stranded DNA, the base compositions of the nucleotide sequences, and the transcription factor binding sites. The numbers of amino acids also led to differences in molecular weight, isoelectric point, instability index, aliphatic index, and grand average of hydropathicity.
Studies have shown that AKT1 proteins are located on the plasma membrane (Cuéllar et al., 2013), consistent with the localization prediction for MsAKT1-1 in this study. Amino acids 260–268 of MsKAT3-2 were TLTTVGYGD, indicating that MsKAT3-2 contained a T-box and belonged to the Shaker gene family. KC1 (KAT3) family genes are mainly expressed in root hairs and endothelial cells, and they participate in the regulation of K+ absorption by the AKT1 channel (Ragel et al., 2019). Jin et al. (2021) showed that members of the Shaker family were all located on the plasma membrane and were all highly conserved. The five two-pore domain potassium (K2P) channels in 12-2 were predicted to be located on the vacuole and plasma membrane, consistent with the results of Voelker et al. (2010) on the subcellular localization of K2P channels in Arabidopsis.
The results of secondary structure in this research were in agreement with those of Gao et al. (2021), and the 8 K+ channel proteins all contained multiple transmembrane structures. MsAKT1-1 channel proteins had the five transmembrane structures typical of Shaker proteins (Yang et al., 2019), and amino acids 63–299 were the K+ transport region. This was in contrast to a study of AKT1 channel proteins in soybean by Wang Y. Q. et al. (2015). The number of transmembrane structures of the other seven K+ channel genes were also different. These results might also be related to the diversity of expanded membrane structures in the Shaker family (Jin et al., 2021). The results of protein domain analysis further verified their identity as potassium channel proteins. Conserved motif analysis showed that the number of conserved amino acid sites was positively correlated with the number of nucleotide transcription factor binding sites (Yura and Go, 2008). The 8 K+ channel proteins all contained Motif 1, and Motif 1 contained a T box, providing further evidence that the K2P channel genes of 12-2 were highly similar to some AKT genes from apple, Arabidopsis, and rice.
Semi-quantitative RT-PCR detection and qRT-PCR analysis showed that the 8 potassium channel genes may participate in rhizosphere K+ homeostasis of 12-2 by responding to ARD stress. These results also help to explain our previous findings on ion currents in 12-2 roots (Mao et al., 2021b). A study by Fuchs et al. (2005) showed that the K+ channel gene OsAKT1 regulated the absorption of K+ to improve rice salt tolerance, and a study by Jin et al. (2017) showed that ion channels could increase plant drought resistance through K+ efflux. KAT1 genes encode plant inward K+ channels (Philippar et al., 2003) and are closely related to plant abiotic stress tolerance (Hasanuzzaman et al., 2018). Guo et al. (2021) found that KAT genes were related to drought resistance. K2P channel proteins have been found in humans and mammals and are located on membranes (Goldstein et al., 2005). Five genes encoding K2Ps have been found in the Arabidopsis genome and are involved in a variety of physiological functions, such as cell volume regulation and apoptosis (Nematian-Ardestani et al., 2020). Although no studies have shown a direct relationship between K2P genes and resistance, these genes have a marked effect on vacuole homeostasis (Kintzer and Stroud, 2016). Feng et al. (2021) showed that a vacuole gene was closely related to stress resistance in cotton.
Conclusion
8 K+ channel genes were cloned from the superior apple rootstock line 12-2 with improved ARD resistance by homologous cloning. Basic biochemical characterization, semi-quantitative RT-PCR, and qRT-PCR showed that the 8 potassium channel genes were associated with ARD resistance and played an important role in maintaining the rhizosphere K+ homeostasis of 12-2. The above results provide a theoretical basis for further research on the functions of potassium channel genes in 12-2 and also provide a scientific basis for the breeding of ARD-resistant rootstock.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author, and have been submitted to GenBank (GenBank accession numbers for 8 genes are OL742694, OL742695, OL742696, OL742697, OL742698, OL742699, OL742700, and OL742701). Nucleotide alignments from this article are listed in Additional file 1 (Supplementary Figures S1–S8).
Author Contributions
YM and XS planned and designed the research. YM, YY, XC, HW, XFS, XQ, and YL performed experiments, conducted fieldwork, analyzed data. YM, YY, YH, and XS wrote the manuscript. Every author contributed equally.
Funding
This research was funded by Project supported by the National Natural Science Foundation of China, grant number 32072520; Fruit innovation team project of Shandong Province (CN), grant number SDAIT-06-07; Natural Science Foundation of Shandong Province (CN), grant number ZR2020MC132 and Industrialization project of improved varieties in Shandong Province (CN), grant number 2019LZGC007.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.803160/full#supplementary-material
Abbreviations
12-2, superior apple rootstock line named 12-2; ARD, apple replant disease; MR5, ARD-associated Fusarium proliferatum MR5; K2P, two-pore domain potassium.
References
Adams, E., and Shin, R. (2014). Transport, Signaling, and Homeostasis of Potassium and Sodium in Plants. J. Integr. Plant Biol. 56, 231–249. doi:10.1111/jipb.12159
Anderson, J. A., Huprikar, S. S., Kochian, L. V., Lucas, W. J., and Gaber, R. F. (1992). Functional Expression of a Probable Arabidopsis thaliana Potassium Channel in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 89, 3736–3740. doi:10.1073/pnas.89.9.3736
Assaha, D. V. M., Ueda, A., Saneoka, H., Al-Yahyai, R., and Yaish, M. W. (2017). The Role of Na+ and K+ Transporters in Salt Stress Adaptation in Glycophytes. Front. Physiol. 8, 509. doi:10.3389/fphys.2017.00509
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME Suite. Nucleic Acids Res. 43, W39–W49. –W49. doi:10.1093/nar/gkv416
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen, P., Wang, Y.-z., Liu, Q.-z., Zhang, Y.-t., Li, X.-y., Li, H.-q., et al. (2020). Phase Changes of Continuous Cropping Obstacles in Strawberry (Fragaria × Ananassa Duch.) Production. Appl. Soil Ecol. 155, 103626. doi:10.1016/j.apsoil.2020.103626
Chou, K.-C., and Shen, H.-B. (2010). Cell-PLoc 2.0: an Improved Package of Web-Servers for Predicting Subcellular Localization of Proteins in Various Organisms. Ns 02, 1090–1103. doi:10.4236/ns.2010.210136
Cuéllar, T., Azeem, F., Andrianteranagna, M., Pascaud, F., Verdeil, J.-L., Sentenac, H., et al. (2013). Potassium Transport in Developing Fleshy Fruits: the grapevine Inward K+channel VvK1.2 Is Activated by CIPK-CBL Complexes and Induced in Ripening berry Flesh Cells. Plant J. 73, 1006–1018. doi:10.1111/tpj.12092
Czempinski, K., Gaedeke, N., Zimmexmann, S., and Muller-Rober, B. (1999). Molecular Mechanisms and Regulation of Plant Ion Channels. J. Exp. Bot. 50, 955–966. doi:10.1093/jxb/50.Special_Issue.95510.1093/jexbot/50.suppl_1.955
Czempinski, K., Zimmermann, S., Ehrhardt, T., and Muller-Rober, B. (1997). New Structure and Function in Plant K+ Channels: KCO1, an Outward Rectifier with a Steep Ca2+ Dependency. EMBO J. 16, 2565–2575. doi:10.1093/emboj/16.10.2565
Dreyer, I., Antunes, S., Hoshi, T., Müller-Röber, B., Palme, K., Pongs, O., et al. (1997). Plant K+ Channel Alpha-Subunits Assemble Indiscriminately. Biophysical J. 72, 2143–2150. doi:10.1016/s0006-3495(97)78857-x
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam Protein Families Database in 2019. Nucleic Acids Res. 47, D427–D432. doi:10.1093/nar/gky995
Enyedi, P., and Czirják, G. (2010). Molecular Background of Leak K+Currents: Two-Pore Domain Potassium Channels. Physiol. Rev. 90, 559–605. doi:10.1152/physrev.00029.2009
Feng, J., Ma, W., Ma, Z., Ren, Z., Zhou, Y., Zhao, J., et al. (2021). GhNHX3D, a Vacuolar-Localized Na+/H+ Antiporter, Positively Regulates Salt Response in Upland Cotton. Ijms 22, 4047. doi:10.3390/ijms22084047
Fuchs, I., Stölzle, S., Ivashikina, N., and Hedrich, R. (2005). Rice K+ Uptake Channel OsAKT1 Is Sensitive to Salt Stress. Planta 221, 212–221. doi:10.1007/s00425-004-1437-9
Gallin, W. J., and Boutet, P. A. (2011). VKCDB: Voltage-Gated K+ Channel Database Updated and Upgraded. Nucleic Acids Res. 39, D362–D366. doi:10.1093/nar/gkq1000
Gambale, F., and Uozumi, N. (2006). Properties of Shaker-type Potassium Channels in Higher Plants. J. Membr. Biol. 210, 1–19. doi:10.1007/s00232-006-0856-x
Gao, P. H., Li, Y., and Yan, B. (2021). Cloning and Expression Analysis of QtKAT1 Gene in Qiongzhuea Tumidinoda. Mol. Plant Breed. 19, 1113–1120. (In Chinese). doi:10.13271/j.mpb.019.001113
Garg, V. K., Avashthi, H., Avashthi, H., Tiwari, A., Jain, P. A., Ramkete, P. W. R., et al. (2016). MFPPI - Multi FASTA ProtParam Interface. Bioinformation 12, 74–77. doi:10.6026/97320630012074
Gaymard, F., Pilot, G., Lacombe, B., Bouchez, D., Bruneau, D., Boucherez, J., et al. (1998). Identification and Disruption of a Plant Shaker-like Outward Channel Involved in K+ Release into the Xylem Sap. Cell 94, 647–655. doi:10.1016/s0092-8674(00)81606-2
Gierth, M., and Mäser, P. (2007). Potassium Transporters in Plants - Involvement in K+acquisition, Redistribution and Homeostasis. FEBS Lett. 581, 2348–2356. doi:10.1016/j.febslet.2007.03.035
Goldstein, S. A. N., Bayliss, D. A., Kim, D., Lesage, F., Plant, L. D., and Rajan, S. (2005). International union of Pharmacology. LV. Nomenclature and Molecular Relationships of Two-P Potassium Channels. Pharmacol. Rev. 57, 527–540. doi:10.1124/pr.57.4.12
González, W., Valdebenito, B., Caballero, J., Riadi, G., Riedelsberger, J., Martínez, G., et al. (2015). K2P Channels in Plants and Animals. Pflugers Arch. - Eur. J. Physiol. 467, 1091–1104. doi:10.1007/s00424-014-1638-4
Guo, R., Zhao, L., Zhang, K., Lu, H., Bhanbhro, N., and Yang, C. (2021). Comparative Genomics and Transcriptomics of the Extreme Halophyte Puccinellia Tenuiflora Provides Insights into Salinity Tolerance Differentiation between Halophytes and Glycophytes. Front. Plant Sci. 12, 649001. doi:10.3389/fpls.2021.649001
Guo, X. J., Han, T. T., Wang, R., Zhang, Z., Ge, R., Mao, Z. Q., et al. (2015). Differences of Growth and Root Absorption of Different Apple Rootstock Seedlings in Replant Stress. J. Plant Nutr. Fertilizers 21, 1312–1319. (In Chinese). doi:10.11674/zwyf.2015.0526
Hasanuzzaman, M., Bhuyan, M., Nahar, K., Hossain, M., Mahmud, J., Hossen, M., et al. (2018). Potassium: A Vital Regulator of Plant Responses and Tolerance to Abiotic Stresses. Agronomy 8, 31. doi:10.3390/agronomy8030031
Jin, R., Zhang, A., Sun, J., Chen, X., Liu, M., Zhao, P., et al. (2021). Identification of Shaker K+ Channel Family Members in Sweetpotato and Functional Exploration of IbAKT1. Gene 768, 145311. doi:10.1016/j.gene.2020.145311
Jin, Z., Wang, Z., Ma, Q., Sun, L., Zhang, L., Liu, Z., et al. (2017). Hydrogen Sulfide Mediates Ion Fluxes Inducing Stomatal Closure in Response to Drought Stress in Arabidopsis thaliana. Plant Soil 419, 141–152. doi:10.1007/s11104-017-3335-5
Kintzer, A. F., and Stroud, R. M. (2016). Structure, Inhibition and Regulation of Two-Pore Channel TPC1 from Arabidopsis thaliana. Nature 531, 258–264. doi:10.1038/nature17194
Kobayashi, Y., Sadhukhan, A., Tazib, T., Nakano, Y., Kusunoki, K., Kamara, M., et al. (2016). Joint Genetic and Network Analyses Identify Loci Associated with Root Growth under NaCl Stress in Arabidopsis thaliana. Plant Cel Environ. 39, 918–934. doi:10.1111/pce.12691
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. L. (2001). Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete genomes11Edited by F. Cohen. J. Mol. Biol. 305, 567–580. doi:10.1006/jmbi.2000.4315
Lainé, M., Papazian, D. M., and Roux, B. (2004). Critical Assessment of a Proposed Model of Shaker. FEBS Lett. 564, 257–263. doi:10.1016/S0014-5793(04)00273-X
Leinfelder, M. M., and Merwin, I. A. (2006). Rootstock Selection, Preplant Soil Treatments, and Tree Planting Positions as Factors in Managing Apple Replant Disease. HortSci 41, 394–401. doi:10.21273/HORTSCI.41.2.394
Li, Q., Qin, Y., Hu, X., Li, G., Ding, H., Xiong, X., et al. (2020). Transcriptome Analysis Uncovers the Gene Expression Profile of Salt-Stressed Potato (Solanum tuberosum L.). Sci. Rep. 10, 5411. doi:10.1038/s41598-020-62057-0
Manici, L. M., Kelderer, M., Franke-Whittle, I. H., Rühmer, T., Baab, G., Nicoletti, F., et al. (2013). Relationship between Root-Endophytic Microbial Communities and Replant Disease in Specialized Apple Growing Areas in Europe. Appl. Soil Ecol. 72, 207–214. doi:10.1016/j.apsoil.2013.07.011
Mao, Y., Yin, Y., Cui, X., Wang, H., Su, X., Qin, X., et al. (2021a). Detection of Above-Ground Physiological Indices of an Apple Rootstock Superior Line 12-2 with Improved Apple Replant Disease (ARD) Resistance. Horticulturae 7, 337. doi:10.3390/horticulturae7100337
Mao, Y., Yin, Y., Cui, X., Wang, H., Su, X., Qin, X., et al. (2021b). Detection of Root Physiological Parameters and Potassium and Calcium Currents in the Rhizoplane of the Apple Rootstock Superior Line 12-2 with Improved Apple Replant Disease Resistance. Front. Plant Sci. 12, 734430. doi:10.3389/fpls.2021.734430
Mao, Y., Zhang, L., Wang, Y., Yang, L., Yin, Y., Su, X., et al. (2021c). Effects of Polycyclic Aromatic Hydrocarbons (PAHs) from Different Sources on Soil Enzymes and Microorganisms of Malus Prunifolia Var. Ringo. Arch. Agron. Soil Sci. 67, 2048–2062. doi:10.1080/03650340.2020.1820488
Mazzola, M., and Manici, L. M. (2012). Apple Replant Disease: Role of Microbial Ecology in Cause and Control. Annu. Rev. Phytopathol. 50, 45–65. doi:10.1146/annurev-phyto-081211-173005
Mazzola, M., and Zhao, X. (2010). Brassica Juncea Seed Meal Particle Size Influences Chemistry but Not Soil Biology-Based Suppression of Individual Agents Inciting Apple Replant Disease. Plant Soil 337, 313–324. doi:10.1007/s11104-010-0529-5
Moulton, G., Attwood, T. K., Parry-Smith, D. J., and Packer, J. C. L. (2003). Phylogenomic Analysis and Evolution of the Potassium Channel Gene Family. Receptors and Channels 9, 363–377. doi:10.1080/1060682039025292510.3109/714041017
Narwal, S. S. (2010). Allelopathy in Ecological Sustainable Organic Agriculture. Allelopathy J. 25, 51–72.
Nematian-Ardestani, E., Abd-Wahab, F., Chatelain, F. C., Sun, H., Schewe, M., Baukrowitz, T., et al. (2020). Selectivity Filter Instability Dominates the Low Intrinsic Activity of the TWIK-1 K2P K+ Channel. J. Biol. Chem. 295, 610–618. doi:10.1074/jbc.RA119.010612
Philippar, K., Ivashikina, N., Ache, P., Christian, M., Lüthen, H., Palme, K., et al. (2004). Auxin activatesKAT1andKAT2, Two K+-channel Genes Expressed in Seedlings ofArabidopsis Thaliana. Plant J. 37, 815–827. doi:10.1111/j.1365-313x.2003.02006.x
Ragel, P., Raddatz, N., Leidi, E. O., Quintero, F. J., and Pardo, J. M. (2019). Regulation of K+ Nutrition in Plants. Front. Plant Sci. 10, 281. doi:10.3389/fpls.2019.00281
Rai, H., and Kawabata, M. (2020). The Dynamics of Radio-Cesium in Soils and Mechanism of Cesium Uptake into Higher Plants: Newly Elucidated Mechanism of Cesium Uptake into Rice Plants. Front. Plant Sci. 11, 528. doi:10.3389/fpls.2020.00528
Rumberger, A., Yao, S., Merwin, I. A., Nelson, E. B., and Thies, J. E. (2004). Rootstock Genotype and Orchard Replant Position rather Than Soil Fumigation or Compost Amendment Determine Tree Growth and Rhizosphere Bacterial Community Composition in an Apple Replant Soil. Plant and Soil 264, 247–260. doi:10.1023/b:plso.0000047760.13004.94
Savojardo, C., Martelli, P. L., Fariselli, P., and Casadio, R. (2018). DeepSig: Deep Learning Improves Signal Peptide Detection in Proteins. Bioinformatics 34, 1690–1696. doi:10.1093/bioinformatics/btx818
Schachtman, D. P., and Shin, R. (2007). Nutrient Sensing and Signaling: NPKS. Annu. Rev. Plant Biol. 58, 47–69. doi:10.1146/annurev.arplant.58.032806.103750
Sentenac, H., Bonneaud, N., Minet, M., Lacroute, F., Salmon, J.-M., Gaymard, F., et al. (1992). Cloning and Expression in Yeast of a Plant Potassium Ion Transport System. Science 256, 663–665. doi:10.1126/science.1585180
Shealy, R. T., Murphy, A. D., Ramarathnam, R., Jakobsson, E., and Subramaniam, S. (2003). Sequence-Function Analysis of the K+-Selective Family of Ion Channels Using a Comprehensive Alignment and the KcsA Channel Structure. Biophysical J. 84, 2929–2942. doi:10.1016/S0006-3495(03)70020-4
Shen, X., Mao, Z. Q., Ni, W. R., Wang, R., Hu, Y. L., Chen, X. S., et al. (2015). Situ Breeding Method for Three-Stage Selection of Apple Rootstock Tolerant to ARD. China Patent No. CN104488645A. (In Chinese).
Shin, R., and Schachtman, D. P. (2004). Hydrogen Peroxide Mediates Plant Root Cell Response to Nutrient Deprivation. Proc. Natl. Acad. Sci. 101, 8827–8832. doi:10.1073/pnas.0401707101
Voelker, C., Gomez-Porras, J. L., Becker, D., Hamamoto, S., Uozumi, N., Gambale, F., et al. (2010). Roles of Tandem-Pore K+ Channels in Plants - a Puzzle Still to Be Solved*. Plant Biol. 12, 56–63. doi:10.1111/j.1438-8677.2010.00353.x
Wang, G., Yin, C., Pan, F., Wang, X., Xiang, L., Wang, Y., et al. (2018). Analysis of the Fungal Community in Apple Replanted Soil Around Bohai Gulf. Hortic. Plant J. 4, 175–181. doi:10.1016/j.hpj.2018.05.003
Wang, N., Zheng, Y., Duan, N., Zhang, Z., Ji, X., Jiang, S., et al. (2015). Comparative Transcriptomes Analysis of Red- and White-Fleshed Apples in an F1 Population of Malus Sieversii F. Niedzwetzkyana Crossed with M. domestica 'Fuji'. PLoS ONE 10, e0133468. doi:10.1371/journal.pone.0133468
Wang, Y.-H., Liu, X.-H., Zhang, R.-R., Yan, Z.-M., Xiong, A.-S., and Su, X.-J. (2020). Sequencing, Assembly, Annotation, and Gene Expression: Novel Insights into browning-resistant Luffa Cylindrica. PeerJ 8, e9661. doi:10.7717/peerj.9661
Wang, Y. Q., Li, Y. G., Wang, T., Zhang, Y. H., Sun, M. Y., and Li, W. B. (2015). Cloning of Gene GmAKT1 from Soybean and Construction of its Sense and Antisense Plant Expression Vector. Genomics Appl. Biol. 34, 143–148. (In Chinese). doi:10.13417/j.gab.034.000143
Xiang, L., Wang, M., Pan, F., Wang, G., Jiang, W., Wang, Y., et al. (2021). Transcriptome Analysis Malus Domestica 'M9T337' Root Molecular Responses to Fusarium Solani Infection. Physiol. Mol. Plant Pathol. 113, 101567. doi:10.1016/j.pmpp.2020.101567
Xie, Q., Ma, L., Tan, P., Deng, W., Huang, C., Liu, D., et al. (2020). Multiple High-Affinity K+ Transporters and ABC Transporters Involved in K+ Uptake/Transport in the Potassium-Hyperaccumulator Plant Phytolacca Acinosa Roxb. Plants 9, 470. doi:10.3390/plants9040470
Xu, J., Tian, X., Egrinya Eneji, A., and Li, Z. (2014). Functional Characterization of GhAKT1, a Novel Shaker-like K+ Channel Gene Involved in K+ Uptake from Cotton (Gossypium Hirsutum). Gene 545, 61–71. doi:10.1016/j.gene.2014.05.006
Xu, X., Du, X., Wang, F., Sha, J., Chen, Q., Tian, G., et al. (2020). Effects of Potassium Levels on Plant Growth, Accumulation and Distribution of Carbon, and Nitrate Metabolism in Apple Dwarf Rootstock Seedlings. Front. Plant Sci. 11, 904. doi:10.3389/fpls.2020.00904
Yang, H., Li, Y., Shen, C. W., Jin, Y. M., Shi, X. Q., Xie, C. Y., et al. (2019). Cloning and Expression Analysis of Potassium Channel Gene PbAKT1 in Pear (Pyrus Betulifolia). J. Agric. Biotechnol. 27, 1314–1350. (In Chinese). doi:10.3969/j.issn.1674-7968.2019.08.002
Yura, K., and Go, M. (2008). Correlation between Amino Acid Residues Converted by RNA Editing and Functional Residues in Protein Three-Dimensional Structures in Plant Organelles. BMC Plant Biol. 8, 79. doi:10.1186/1471-2229-8-79
Zhang, J., Xu, H., Wang, N., Jiang, S., Fang, H., Zhang, Z., et al. (2018). The Ethylene Response Factor MdERF1B Regulates Anthocyanin and Proanthocyanidin Biosynthesis in Apple. Plant Mol. Biol. 98, 205–218. doi:10.1007/s11103-018-0770-5
Zhou, Z., Zhu, Y., Tian, Y., Yao, J.-L., Bian, S., Zhang, H., et al. (2021). MdPR4, a Pathogenesis-Related Protein in Apple, Is Involved in Chitin Recognition and Resistance Response to Apple Replant Disease Pathogens. J. Plant Physiol. 260, 153390. doi:10.1016/j.jplph.2021.153390
Keywords: K+ channel genes, homologous molecular cloning, biochemical information, apple replant disease, ARD-resistant rootstock
Citation: Mao Y, Yin Y, Cui X, Wang H, Su X, Qin X, Liu Y, Hu Y and Shen X (2022) Homologous Cloning of Potassium Channel Genes From the Superior Apple Rootstock Line 12-2, Which is Tolerant to Apple Replant Disease. Front. Genet. 13:803160. doi: 10.3389/fgene.2022.803160
Received: 27 October 2021; Accepted: 10 January 2022;
Published: 26 January 2022.
Edited by:
Nidhi Rawat, University of Maryland, United StatesReviewed by:
Muhammad Sajjad, COMSATS University, PakistanSantosh Kumar Upadhyay, Panjab University, India
Copyright © 2022 Mao, Yin, Cui, Wang, Su, Qin, Liu, Hu and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang Shen, c2hlbnhAc2RhdS5lZHUuY24=
 Yunfei Mao
Yunfei Mao Yijun Yin
Yijun Yin Xueli Cui
Xueli Cui Haiyan Wang
Haiyan Wang XiaFei Su
XiaFei Su Xin Qin
Xin Qin Yangbo Liu
Yangbo Liu Yanli Hu
Yanli Hu Xiang Shen
Xiang Shen