- 1State Key Laboratory of Oral Diseases and National Clinical Research Center for Oral Diseases, Department of Cleft Lip and Palate, West China School of Stomatology, Sichuan University, Chengdu, China
- 2Division of Growth and Development and Section of Orthodontics, School of Dentistry, University of California, Los Angeles, Los Angeles, CA, United States
rs7590268 present on the 2p21 locus was identified to be associated with non-syndromic cleft lip with or without cleft palate (NSCL/P) in several populations, including the Chinese Han population, indicating that 2p21 was a susceptibility locus for NSCL/P. However, previous studies have only identified common single-nucleotide polymorphism (SNP) within the THADA gene, neglecting the rare variants and other genes in 2p21; thus, this study was designed to investigate additional variants and novel susceptibility genes in 2p21. A total of 159 NSCL/P patients and 542 controls were recruited in the discovery phase, whereas 1830 NSCL/P patients and 2,436 controls were recruited in the replication phase. After targeted region sequencing, we performed association and burden analyses for the common and rare variants, respectively. Furthermore, RNA-seq, proliferation assay and cell cycle analysis were performed to clarify the possible function of the candidate gene ZFP36L2. Association analysis showed that four SNPs were specifically associated with non-syndromic cleft lip only (NSCLO) and two SNPs were associated with both NSCLO and NSCL/P. Burden analysis indicated that ZFP36L2 was associated with NSCLO (p = .0489, OR = 2.41, 95% CI: 0.98–5.90). Moreover, SNPs in the ZFP36L2 targeted gene JUP were also associated with NSCLO. ZFP36L2 also inhibited cell proliferation and induced G2 phase arrest in the GMSM-K cell line. Therefore, we proposed that ZFP36L2 is a novel susceptibility gene of NSCLO in the 2p21 locus, which could lead to NSCLO by modulating cell proliferation and cycle.
Introduction
Non-syndromic orofacial clefts (NSOFCs), which usually occur without any other physiological abnormalities, are one of the most common birth defects. NSOFCs are commonly divided into non-syndromic cleft lip with or without cleft palate (NSCL/P) and non-syndromic cleft palate only (NSCPO), which have been historically regarded as etiologically distinct phenotypes, because they differ in epidemiology and family patterns, as well as in the developmental origin of the lip and palate (Marazita, 2012). NSCL/P, on the other hand, includes two phenotypes: non-syndromic cleft lip only (NSCLO) and non-syndromic cleft lip with palate (NSCLP), but they are usually grouped together and considered as the same defect with different severity (Mitchell et al., 2002); however, some researchers have suggested that NSCLP and NSCLO may also be etiologically distinct, and should be analyzed separately when possible (Harville et al., 2005; Sivertsen et al., 2008; Grosen et al., 2010).
Individuals with NSCL/P are usually exposed to a series of problems early in life, such as difficulties in feeding and ear infections, which impose a heavy burden on both the affected families and society. Additionally, NSCL/P impact the patients’ quality of life throughout their life span, although surgical repair, speech therapy, dental care, and psychological support are available (Wehby and Cassell, 2010). Therefore, it is of great significance to clarify the pathogenesis of NSCL/P. Currently, there is a consensus that genes, environmental factors, and their complicated interactions contribute to the occurrence of NSCL/P (Machado et al., 2016).
In the past few decades, several environmental factors have been identified to be associated with NSCL/P, including smoking, drinking, drug consumption (such as antiepileptic agents), and lack of dietary folic acid during early pregnancy (Honein et al., 2007; Stott-Miller et al., 2010). Although a variety of methods have been used to identify its susceptibility gene, progress toward its identification has been slow. Genome-wide association studies (GWASs) have been an effective tool in identifying genome variants associated with NSCL/P, aiding identification of over 40 susceptibility loci in the past few years (Birnbaum et al., 2009; Mangold et al., 2010; Ludwig et al., 2012; Sun et al., 2015).
Based on the first meta-analyses of NSCL/P, which combined the data from two large GWAS, including 666 European trios and 795 Asian trios, 2p21 was proposed to be susceptibility region for NSCL/P for the first time (Ludwig et al., 2012). Subsequent studies conducted in different populations further proved that variants in 2p21 locus were associated with NSCL/P; among them, rs7590268 was identified several times in the European population (Beaty et al., 2013; Moreno Uribe et al., 2017). Replication studies showed that rs7590268 also had a strong signal in the southern Chinese population (Pan et al., 2013).
These findings implied that 2p21, where rs7590268 was located, was a susceptibility locus for NSCL/P. However, genotyping arrays used in GWAS only capture 5% of the total SNPs occurring genome wide, potentially missing causal SNPs that are in linkage disequilibrium with SNPs captured in GWAS; and, GWAS neglects rare variants, defined here as minor allele frequency (MAF) < 0.01, which have a larger effect than common variants and could partially compensate for the missing heritability (Manolio et al., 2009; Tada et al., 2016; Sazonovs and Barrett, 2018). Additionally, the reported SNPs were mainly located in the THADA gene. Although THADA did not involve in craniofacial development according to the currently published literature, variations within it have been associated with many diseases, which may be attributable to its large size (370 kb), the effect of regulatory elements, or the other adjacent genes (Drieschner et al., 2007; Ludwig et al., 2012). Notably, a recent study proposed another gene—ZFP36L2—adjacent to THADA, as the lead risk NSCL/P gene in 2p21 for the first time (Lin-Shiao et al., 2019); however, neither common nor rare variants in the ZFP36L2 gene have been identified.
ZFP36L2 belongs to the zinc finger protein ZFP36 family and is classified as a Cys–Cys–Cys–His (CCCH)-type zinc finger tandem protein (Ramos et al., 2004; Stumpo et al., 2009). It is well-known as an RNA-binding protein, which destabilizes target mRNA and, thus, influences the targeted gene expression by binding to AU-rich elements (ARE) in the 3′ untranslated region (UTR) of labile mRNAs(Lai et al., 2000). The function of specific ARE-binding proteins could be modulated by post-translational epigenetic modifications including methylation and phosphorylation (Galloway et al., 2016). When stimulated by lipopolysaccharide, Zfp36l2 phosphorylation influences the production of inflammatory mediators by regulating Mitogen-activated protein kinase (Mkp)-1 mRNA expression (K. T. Wang et al., 2015). Additionally, ZFP36L2 also participates in the epigenetic modification process. In the absence of Zfp36l2, oocytes failed to accumulate histone methylation at H3K4 and H3K9, leading to transcriptional silence (Dumdie et al., 2018). Furthermore, Zfp36l2 could act as a safeguard against chromosomal instability and post-transcription replication stress during thymopoiesis (Vogel et al, 2016).
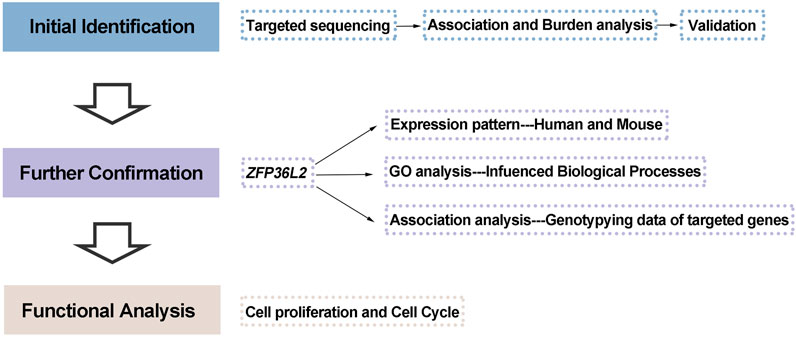
In the present study, we aimed to perform a comprehensive screening of susceptibility variants (both common and rare) in the 2p21 locus by targeted sequencing, followed by interpretation via association analysis, burden analysis, and a series of functional analyses (the study design is shown in Figure 1).
Material and Methods
Subject Characterization and Ethics Statement
In this study, we performed a two-phase case-control analysis, including an initial discovery phase and a subsequent replication phase. In the discovery phase, 159 unrelated patients with NSCL/P (80 NSCLP and 79 NSCLO patients) were selected from the Cleft Surgery Department of the West China College of Stomatology, Sichuan University. All of them self-identified as Han Chinese, and they did not have any other congenital anomalies. The whole-genome sequencing data of 542 Han Chinese normal controls (sequenced using Illumina Hiseq platform with an average coverage of 39.89) included in this phase were obtained from the Novogene internal database (http://www.novogene.com/).
The genotyping data of 1,626 patients with NSCL/P (including 579 patients with NSCLP, 1,047 patients with NSCLO) and 2,255 normal controls from two GWAS (Sun et al., 2015; Huang et al., 2019), as well as another 204 patients with NSCLO and 181 normal controls selected from the Cleft Surgery Department of West China College of Stomatology, Sichuan University, were recruited in the replication phase for inclusion in the study (Supplementary Table S1).
Human subject study protocols were approved by the Hospital Ethics Committee of West China Hospital of Stomatology, Sichuan University; these protocols conformed to the Strengthening the Reporting of Observational Studies in Epidemiology guidelines. Written informed consent was obtained from recruited individuals of consenting age and from parents on behalf of their participating children (WCHSIRB-D-2016-012R1).
DNA Extraction and Quality Control
Peripheral blood samples were collected from all the participants and their parents, from which DNA was extracted using the salting out method and then stored in Tris-EDTA buffer. The quality of the isolated genomic DNA was verified by electrophoresis on 1% agarose gel to exclude the possibility of DNA degradation or RNA/protein contamination. Furthermore, the DNA purity and concentration were detected using a NanoPhotometer® spectrophotometer (IMPLEN, CA, United States), with good quality output of ratio of optical density at 260 nm to the optical density at 280 nm (OD260/OD280) values ranging from 1.8 to 2.0.
Selection of Targeted Region and Sequencing
According to the linkage disequilibrium (LD) structure shown by the CHB/JPT HapMap, the targeted region for deep sequencing was around rs7590268, based on ranges from chr2:43,417,119 to 43,838,705 (GRCh37/hg19), including exons, introns, and the intergenic region (Supplementary Figure S1). Sequencing was efficiently carried out using 1.0 μg genomic DNA in an Agilent liquid capture system (Agilent SureSelectXT Custom Kit) according to the manufacturer’s protocol. The DNA library was sequenced on Illumina Hiseq for paired-end 150 bp reads.
Bioinformatics and Statistical Analysis
After quality control processing, including filtering of adapter-related reads, reads containing N, and low-quality reads, the clean sequence data were mapped to the GRCh37/hg19 human genome using Burrows-Wheeler Aligner (BWA) software (Li and Durbin, 2009). Then, SNVs and indels were identified by Sequence Alignment Map (SAM tools) (Li et al., 2009) and merged by VCF (Variant Call Format) tools (version 0.1.13) (Danecek et al., 2011). Later, variants were annotated by ANNOVAR (version 201707) (K. Wang et al., 2010), followed by function prediction via SIFT (Ng and Henikoff, 2003), v1.3 CADD (Rentzsch et al., 2018), Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/) (Adzhubei et al., 2013) and Mutation Taster (http://www.mutationtaster.org/) (Schwarz et al., 2010).
Variants with call rates >95% were divided into two groups: common variants (MAF ≥0.01) and rare variants (MAF <0.01). Furthermore, we performed the Hardy-Weinberg equilibrium (HWE) test at each SNP. SNPs which deviated from HWE (p < .000001) were removed from subsequent association analysis, which was conducted by PLINK (version 1.9) (Purcell et al., 2007). The p value in the replication phase was adjusted for multiple corrections. Using the R package SKAT, gene-based burden analysis was performed on rare variants in accordance with the following criteria: 1) MAF <0.01 in CHB and CHS (CHB, Han Chinese in Beijing; CHS, Southern Han Chinese) from the 1,000 Genome database and Novogene internal database (http://www.novogene.com/); 2) MAF <0.001 in the Genome Aggregation Database (GnomAD); and 3) at least two prediction tools indicate the rare variation to be damaging (PolyPhen-2, SIFT, MutationTaster, CADD), the thresholds for damaging in each tool were set as follows: SIFT scores <0.05, Polyphen2_HDIV scores ≥0.957, Mutation Taster indicates “Disease causing”, CADD scores >10. Fisher’s exact test or Pearson chi-square test was performed between the cases and controls.
Cell Culture, Transient Knockdown, and Overexpression
Considering the important role of oral epithelium in facial morphogenesis, as well as its known association with NSCL/P, a human oral epithelial cell line (GMSM-K, kindly gifted by Dr. Zhang from Peking University) was selected for functional analysis in our study (H. Liu et al., 2020; H. Liu et al., 2017). GMSM-K was cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Gibco, United States) and 1% penicillin-streptomycin solution (Gibco, United States).
Small interfering RNA (siRNA) targeting ZFP36L2 (NM_006887.4) (siRNA-ZFP36L2: 5′-GCCUUCUACGAUGUCGACUTT-3′) and negative control siRNA (siRNA-negative control: 5′-UUCUCCGAACGUGUCACGUTT-3′) were both designed and synthesized by GenePharma (Shanghai, China). Moreover, ZFP36L2 was cloned into the pcDNA3.1 overexpression plasmid (YouBio, China) (Supplementary Figure S2), and the pcDNA3.1 plasmid without additional sequence served as a negative control. Both siRNAs and plasmids were transfected into GMSM-K using the SF Cell Line 4D X Kit S (Lonza, Germany) according to the manufacturer’s instructions.
RNA-Seq, Differential Expression Analysis and Gene Ontology Analysis
GMSM-K cells were transfected with siRNA-negative control or siRNAs- ZFP36L2 for 48 h. Then, the cells were collected and RNA-seq was performed using the BGISEQ-500 platform (BGI, China). Two biological replicates were included within each group. Differential gene expression analysis was performed using DEseq2 method (|log2| ≥ 0.8, Q ≤ 0.05), and GO enrichment analysis was performed on DEGs using DAVID 6.8 (Huang da et al., 2009).
Quantitative Real-Time PCR Analysis
RNA was extracted using a Tissue/Cell RNA extraction Kit (Biobase, China) 48 h after transfection, and was then reverse-transcribed to cDNA using PrimeScript™ RT reagent Kit (Takara Biotechnology, Dalian, China). RT-qPCR was performed using TB Green® Premix Ex Taq™ (Takara Biotechnology, Dalian, China) on a LightCycler 480 System (Roche, Switzerland). All experiments were performed in triplicate at least, each with three technical replicates. The results were calculated using equation 2−ΔΔCt. The primers used are shown in Supplementary Table S2.
Proliferation Assay
Upon transfection, GMSM-K cells were seeded into 96-well plates at a density of 2 × 104 cells/100 μl. After 24, 48, and 72 h, 10 μl of Cell Counting Kit-8 (CCK-8) (APExBIO, United States) was added to each well. Then, 3 h later, OD of each well was measured at a 450 nm wavelength. Measurements at each time point were replicated three times having five replicates each. Results are shown as mean ± SEM. Statistical analysis was performed using the unpaired two-tailed t-test in GraphPad Prism eight software.
Cell Cycle Analysis
Approximately 2 × 105 transfected GMSM-K cells were plated in 6-well plates. After 48 h, cells were collected for further cell cycle analysis, in which cells were fixed in 70% ethanol at 4°C overnight, washed twice with cold phosphate buffer saline, and re-suspended in 0.5 ml PI staining reagent (25 μl propidium iodide (20X), 10 μl RNase A (50X), and 0.5 ml sodium citrate buffer) (Cell Cycle and Apoptosis Analysis Kit, Beyotime, China). The cells were then incubated in the dark at 37°C for 0.5 h. Samples were detected using an Attune NxT flow cytometer (Thermo Fisher, United States). FCS files were downloaded and analyzed using FlowJo software (version 10.4). For each group, results are shown as the mean ± SEM of three replicates. Statistical analysis was performed using the unpaired two-tailed t-test in GraphPad Prism eight software.
Results
In the discovery phase, from the 159 NSCL/P cases, we identified a total of 2,352 Single-nucleotide variants (SNVs) and 552 insertion/deletions (indels), including variants located in exons, splice sites, introns, UTRs, and intergenic regions (Supplementary Figure S3). For common and rare variants that met the inclusion criteria, we performed case-control association analysis and burden analysis among the 159 NSCL/P cases and 542 normal controls, respectively.
SNPs Within 2p21 Locus Were Significantly Associated With NSCLO
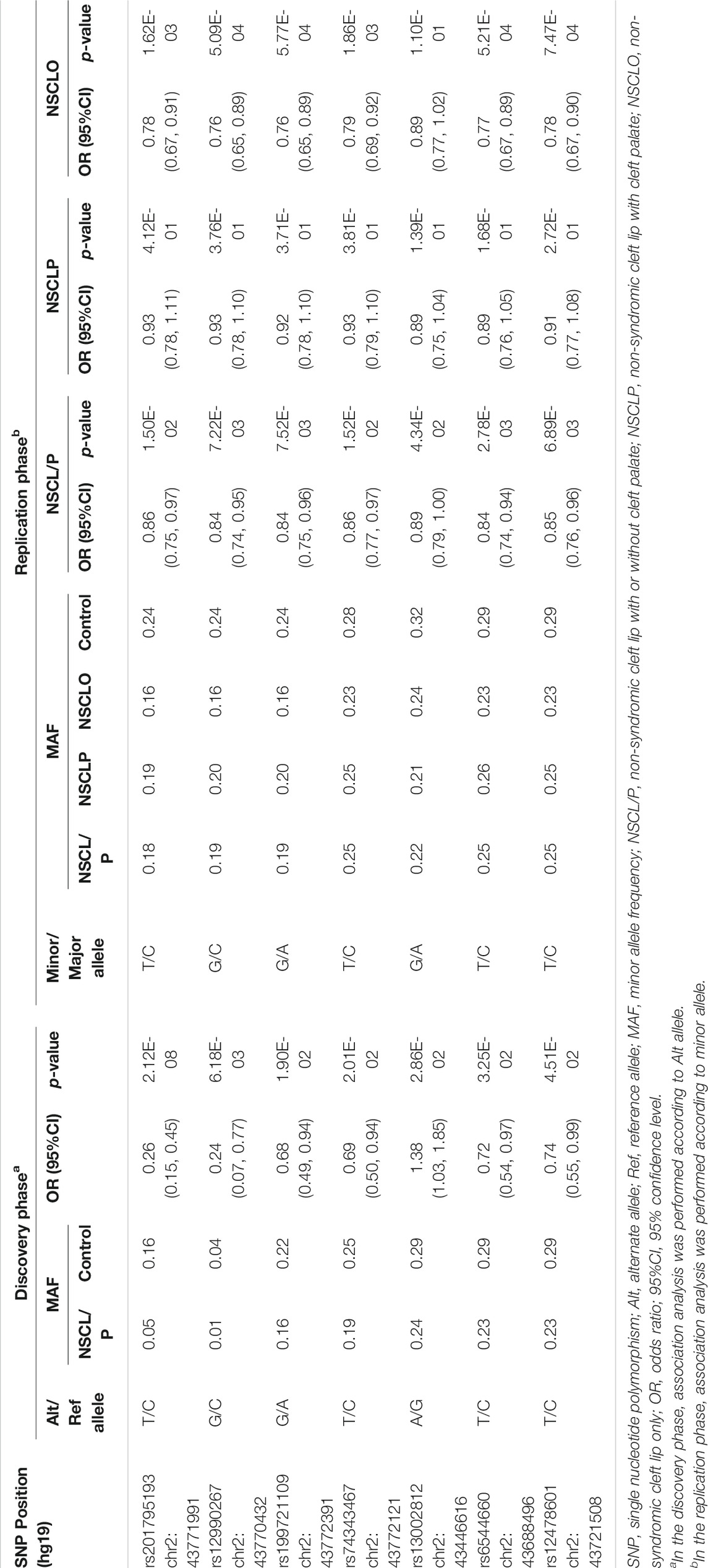
A total of 312 common SNPs (MAF ≥0.01) that passed the threshold of HWE (p > .000001) were recruited for association analysis. When compared with the p value 1.60E-04, which was adjusted for multiple corrections (p = .05/312), only rs201795193 (p = 2.12E-08, OR = 0.33, 95% CI: 0.21–0.52) was significantly associated with NSCL/P.
Seven SNPs (rs201795193, rs12990267, rs199721109, rs74343467, rs13002812, rs6544660, and rs12478601) with p value less than .05 were replicated in larger cohorts (1,626 patients with NSCL/P and 2,255 normal controls) to test their associations with NSCL/P, NSCLP, and NSCLO (Table1, Supplementary Table S3). Six out of seven SNPs were significantly associated with NSCLO after multiple corrections (p = .05/7 = 7.14E-03). Among these, four SNPs, including rs201795193 (p = 1.62E-03, OR = 0.78, 95% CI: 0.67–0.91), rs12990267 (p = 5.09E-04, OR = 0.76, 95% CI: 0.65–0.89), rs199721109 (p = 5.77E-04, OR = 0.76, 95% CI: 0.65–0.89), and rs74343467 (p = 1.86E-03, OR = 0.79, 95% CI: 0.69–0.92) were specifically associated with NSCLO; the other two SNPs, rs6544660 and rs12478601, were associated with both NSCLO (rs6544660: p = 5.21E-04, OR = 0.77, 95% CI: 0.67–0.89; rs12478601: p = 7.47E-04, OR = 0.78, 95% CI: 0.67–0.90) and NSCL/P (rs6544660: p = 2.78E-03, OR = 0.84, 95% CI: 0.74–0.94; rs12478601: p = 6.89E-03, OR = 0.85, 95% CI: 0.76–0.96), where the significant signal of NSCL/P might be driven by the NSCLO since both SNPs show lower p values in NSCLO than that in NSCL/P (Table 1). All the six significantly associated SNPs are novel, and we proposed 2p21 as a susceptibility locus for NSCLO in the Han Chinese population for the first time.
ZFP36L2 Might be the Susceptibility Gene for NSCLO Within 2p21 Locus
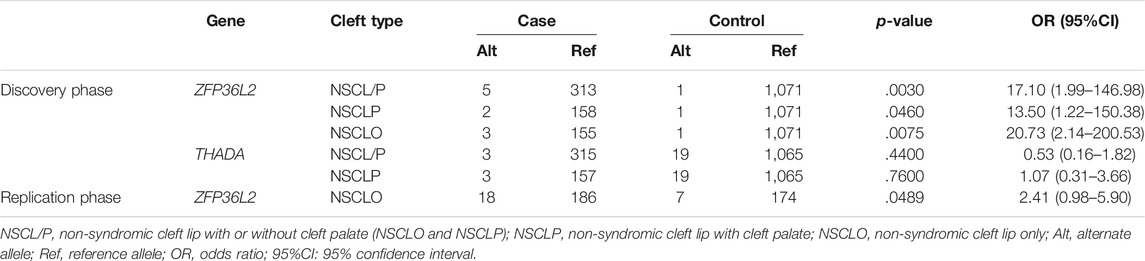
Three rare variants of THADA and four rare variants of ZFP36L2 were enrolled in the burden analysis, details about these variants were shown in the Supplementary Table S4. The collective results indicated that rare variants within ZFP36L2 at 2p21 were significantly associated with increased risk for NSCL/P (p = .003, OR = 17.10, 95% CI: 1.99–146.98), NSCLP (p = .046, OR = 13.5, 95% CI: 1.22–150.38), and NSCLO (p = .0075, OR = 20.73, 95% CI: 2.14–200.53) (Table 2). Interestingly, compared with NSCL/P and NSCLP, ZFP36L2 conveyed the highest risk to NSCLO with a lower p value and larger OR value in our study. However, rare variants within the THADA gene did not show a statistically significant association with NSCL/P or NSCLP.
Replication of the burden analysis was conducted by sequencing ZFP36L2 exons among 204 patients with NSCLO and 181 normal controls. The rare variants were filtered using the same criterion enrolled into the burden analysis, and the result indicated that ZFP36L2 was also associated with NSCLO (p = .0489, OR = 2.41, 95% CI: 0.98–5.90). Based on these findings, we further verified that the 2p21 locus was a risk factor for NSCLO and that the ZFP36L2 gene in 2p21 is a susceptibility gene for NSCLO in the Han Chinese population.
ZFP36L2 Influences Biological Processes in the Etiology of NSCLO
To investigate the potential roles of ZFP36L2 in the etiology of NSCLO, we performed RNA sequencing on human oral epithelial cell line (GMSM-K) with or without ZFP36L2 knockdown. Two biological replicates were included for each group.
Differential gene expression analysis identified 327 differentially expressed genes (DEGs) in total, among which 270 genes were upregulated and 57 genes were downregulated when ZFP36L2 was knocked down (Figure 2A). GO analysis showed that a series of biological processes were enriched, including terms of “Apoptotic signaling pathway,” “Regulation of cell proliferation” and “Positive regulation of cell migration,” which were associated with lip development (Jiang et al., 2006) (Figure 2B); genes within the two former terms relating to apoptotic and proliferation were totally upregulated, whereas genes within term of “Positive regulation of cell migration” were only partially upregulated (Figure 2C). In addition, we observed that genes involved with the term “regulation of cell proliferation” overlapped with genes involved with the terms “apoptotic signaling pathway” or “positive regulation of cell migration” (Figure 2D).
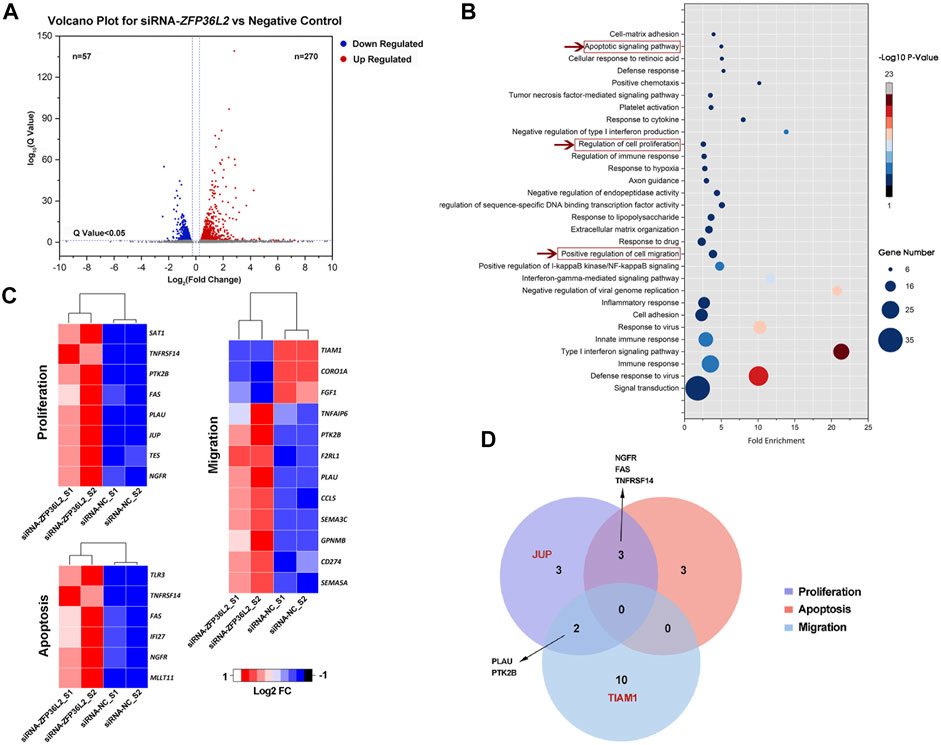
FIGURE 2. (A) Volcano plot for differential gene expression. (B) GO biological process terms enriched in DEGs. A total of 67 GO terms were significantly enriched (p < .05), the figure shows the top 30 GO terms with the most genes. (C) Fold change (Log2 FC) of genes among “Apoptotic signaling pathway,” “regulation of cell proliferation” and “positive regulation of cell migration” biological processes. (D) Venn diagram showing number of genes associated with the terms “apoptotic signaling pathway,” “regulation of cell proliferation” and “positive regulation of cell migration.” The red font is indicative of genes where the SNPs associated with NSCLO or NSCPO were located.
On the other hand, it is known that the development of the upper lip in mice begins at E10.5, where the medial and lateral nasal processes fuse; later, the maxillary and medial nasal processes grow rapidly and fuse together, marking the completion of lip development (Jiang et al., 2006). To further explore the role of ZFP36L2 in NSCLO etiology, we searched the literature and found that the Zfp36l2 gene is expressed in both the maxillary and lateral nasal processes in E10.5 mouse embryos (Supplementary Table S5) (Brunskill et al., 2014). When referring to our previous RNA-seq data of six Chinese Han patients with NSCLO, we noticed that, in the lip tissue, ZFP36L2 was expressed even higher than IRF6, a well-known susceptibility gene for NSCL/P (ZFP36L2-Average FPKM: 227.9; IRF6-Average FPKM:147.5) (Supplementary Figure S4).
ZFP36L2 Target Gene is Associated With NSCLO
Next, from two GWAS databases (Sun et al., 2015; Huang et al., 2019), we retrieved the genotyping data of SNPs at 19 genes within the terms “regulation of cell proliferation” (NGFR, TES, JUP, PLAU, FAS, PTK2B, TNFRSF14, SAT1), “apoptotic signaling pathway” (MLLT11, NGFR, IFI27, FAS, TNFRSF14, TLR3) and “positive regulation of cell migration” (SEMA5A, FGF1, CD274, CORO1A, GPNMB, SEMA3C, CCL5, PLAU, TIAM1, F2RL1, PTK2B, TNFAIP6) that were influenced by ZFP36L2. A total of 4,682 SNPs were recruited; we obtained the most SNPs in the SEMA5A gene, followed by TIAM1, SEMA3C, PTK2B, and FGF1 genes (Figure 3A). Then, we performed association analysis on these SNPs. When compared to the stringent significance threshold of 1.07E-05 derived from multiple corrections (p = .05/4,682), two SNPs within the JUP gene were significantly associated with NSCLO: rs4479305 (p = 7.42E-07, OR = 0.72, 95% CI: 0.63–0.82) and rs9913846 (p = 2.59E-06, OR = 0.73, 95% CI: 0.64–0.83) (Supplementary Table S6). Meanwhile, rs56026457 within the TIAM1 gene was significantly associated with NSCPO (p = 9.68E-07, OR = 0.70, 95% CI: 0.60–0.80), and none were identified to be associated with NSCL/P or NSCLP (Figure 3B).
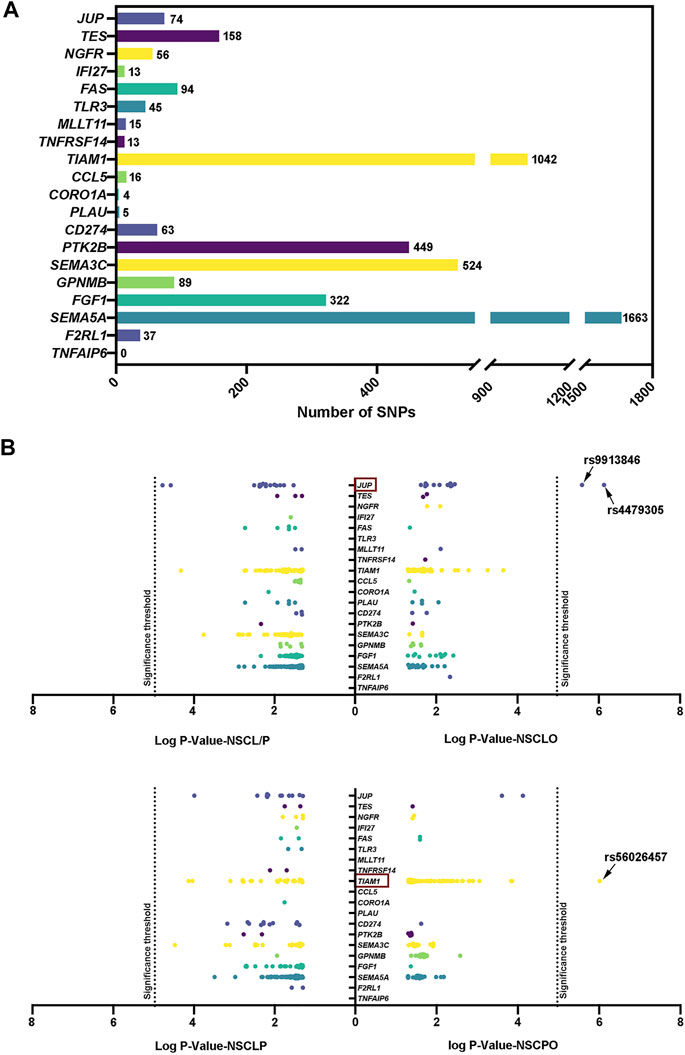
FIGURE 3. (A) Number of SNP within genes associated with the terms “apoptotic signaling pathway,” “regulation of cell proliferation,” and “positive regulation of cell migration; ” (B) Recruited SNPs with p value pass .05. Log p value with base 10 are shown in the figure. The 5.42E-05 significance threshold in the replication phase was adjusted by multiple correction.
Furthermore, we noticed that Jup was expressed in the medial nasal processes in E10.5 mouse embryos (Supplementary Table S5), whereas Tiam1 was not. In lip tissues from individuals with NSCLO, JUP showed a significantly higher expression level than TIAM1 (Supplementary Figure S4). These results indicated that ZFP36L2 might also influence genes participate in lip development.
ZFP36L2 Gene Inhibits Cell Proliferation and Induces G2 Phase Arrest
Since the JUP gene is specifically related to the term “regulation of cell proliferation” (Figure 2D), we investigated proliferation after knockdown or overexpression of ZFP36L2 in the GMSM-K cell line. RT-qPCR results indicated that the transfection of pcDNA3.1-ZFP36L2 plasmid and siRNA-ZFP36L2 could effectively overexpress or inhibit the expression level of ZFP36L2 gene (Figure 4A). Proliferation assay showed that knockdown of ZFP36L2 obviously facilitated cell proliferation; however, cell proliferation was significantly inhibited following ZFP36L2 overexpression (Figure 4B). To clarify this further, cell cycle was also investigated. When ZFP36L2 was knocked down, cells in G2 phase decreased, accompanied by a significantly increased number of cells in S phase. Conversely, we observed a significant G2 phase arrest accompanied by a reduced number of cells in the G1 phase in ZFP36L2 overexpressing cells (Figure 4C). These findings indicated that the ZFP36L2 gene could lead to abnormal lip development by affecting both cell proliferation and the cell cycle.
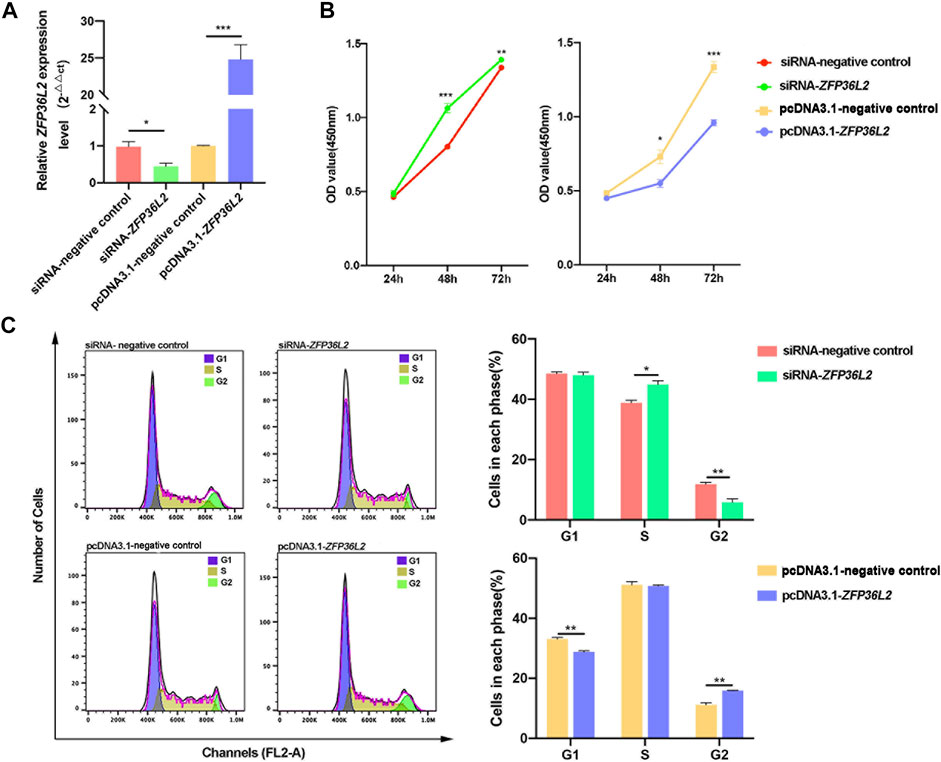
FIGURE 4. (A) RT-qPCR analysis of the expression level of ZFP36L2 after knockdown or overexpression in the GMSM-K cell line. (B) Cell proliferation after knockdown or overexpression of ZFP36L2 in the GMSM-K cell line. (C) Percentage of cells in each cell cycle after knockdown or overexpression of ZFP36L2 in the GMSM-K cell line. Data is shown as the mean ± SEM. *p < .05; **p < .01; ***p < .001.
Discussion
It is known that NSCL/P has a far more complex genetic architecture than we originally thought, with a variety of genetic risk factors and environmental exposures contributing (Dixon et al., 2011; Boyle et al., 2017). With the advent of the GWAS era, more and more susceptibility genes/loci for NSCL/P have been identified (Birnbaum et al., 2009; Beaty et al., 2010; Mangold et al., 2010; Ludwig et al., 2012; Sun et al., 2015), however, GWASs have only explained a small portion of phenotypic variance since GWAS’ rationale is based on the hypothesis “common disease, common variants,” (Manolio et al., 2009) which results in their lack of ability to detect rare variants. Targeted sequencing, which can detect both common and rare variants, greatly solves the above problems of GWAS. At the same time, findings from GWAS provide a reasonable hypothesis for the targeted region. Therefore, in the present study, we selected the target region of the haplotype around rs7590268 for sequencing in the hope of gaining novel insights into the 2p21 locus. Notably, our results indicated that ZFP36L2 in 2p21 locus is a susceptibility gene for NSCLO, whereas the previous study only identified ZFP36L2 and SIX2 in 2p21 locus to be associated with NSCL/P and NSCPO, respectively (Lin-Shiao et al., 2019; Sweat et al., 2020).
Our evidence can be summarized into two parts, the first being bioinformatics-related. Association analysis based on a large cohort and stringent significance threshold showed that most SNPs in 2p21 locus were associated with NSCLO, from which we inferred that 2p21 was a susceptibility locus for NSCLO. We annotated the six statistically significant SNPs using HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) (Supplementary Table S7), finding that different alleles of these six SNPs resulted in altered motifs, except for rs12990267. Among these, the T allele of rs201795193 altered most motifs, including Pax7, which is critical for myogenic satellite cell specification (Seale et al., 2000). The T allele of rs74343467 altered the motif of E2F, a family of transcription factors important for life and death due to their involvement in various biological processes such as DNA replication, cell differentiation and proliferation (Helin, 1998; Polager and Ginsberg, 2008). Furthermore, rs6544660 and rs12478601 were shown to be present in regulatory elements among epidermal keratinocyte primary cells and influenced ZFP36L2 expression levels in whole blood and naive monocytes, although all six SNPs were in the intronic region of the THADA gene. These results suggested that these SNPs might be functional. Burden analysis specified our inference, that is, the ZFP36L2 gene in 2p21 locus was more significantly associated with NSCLO. ZFP36L2 conferred the highest OR of 20.73, and this association was also statistically significant in subsequent validation (p = .0489, OR = 2.41, 95% CI: .98–5.90).
Functional analysis of the ZFP36L2 was carried out to further determine its association with NSCLO. We found that Zfp36l2 gene was expressed in the facial processes related to mouse lip development (Supplementary Table S5). In human lip tissue, we also observed that ZFP36L2 gene expression level was even higher than IRF6, for which the etiology of NSCLO was quite clear since a relatively high expression level of IRF6 would lead to the occurrence of NSCLO (Huang et al., 2019). Additionally, through RNA-seq, we observed that ZFP36L2 affected three biological processes related to proliferation, migration, and apoptosis, which are critical in lip development (Jiang et al., 2006). In this section, we noticed that DEGs appeared to be most enriched in immune-related biological processes, this may be due to the involvement of ZFP36L2 in thymic development and T lymphoblastic leukemia (Hodson et al., 2010). Lastly, we conducted an association analysis of the SNPs within genes involved in these three biological processes and NSOFC, finding that two of the three statistically significant SNPs were specifically associated with NSCLO. Considering all the findings, we rationally proposed that ZFP36L2 in the 2p21 locus is a susceptibility gene for NSCLO.
We attempted to address how the ZFP36L2 gene leads to NSCLO. We noticed that four genes (Ngfr, Tes, Jup, and Sat1) associated with the term “regulation of cell proliferation” were expressed in the medial nasal, lateral nasal or maxillary processes. However, only two genes (Mllt11 and Ngfr genes) associated with the term “apoptotic signaling pathway” and three genes (Sema5a, Coro1a and Tnfaip6 gene) associated with the term “positive regulation of cell migration” were expressed in the three upper lip-related processes (Supplementary Table S5) (Brunskill et al., 2014). Furthermore, the two SNPs associated with NSCLO were both occurring within the proliferation-related gene JUP, which was influenced by ZFP36L2 (Figure 2D and Figure 3B). Therefore, we speculated that ZFP36L2 might lead to NSCLO by influencing cell proliferation, and our study proved that ZFP36L2 negatively regulates cell proliferation and induces cell arrest in the G2 phase of GMSM-K. Our results were consistent with those of previous studies, that is, ZFP36L2 inhibited cell proliferation through a cyclin D-dependent and p53-independent pathway (J. Liu et al., 2018; Suk et al., 2018). As for the effect of ZFP36L2 on the cell cycle, the results are slightly varied. Fat-Moon Suk et al. observed that ZFP36L2 led to G1 phase arrest and thus resulted in a decreased S phase (Suk et al., 2018), whereas several other studies have shown that ZFP36L2 might be a key protein in S phase progression control in the case of genome instability (Galloway et al., 2016; Vogel et al., 2016; Noguchi et al., 2018). In our study, we noticed that as long as the cells in G2 phase were influenced, the subsequent G1 or S phases would be affected more or less. Thus, it is certain that ZFP36L2 affects the cell cycle, but its effects may be slightly modified in different cell types.
In summary, our study comprehensively illustrated the important role of the ZFP36L2 gene in the etiology of NSCLO and proposed that ZFP36L2 is a novel susceptibility gene for NSCLO among the Han Chinese population. Further research is required to address limitations of this study. All samples recruited in our study were from the Han Chinese population; due to the genetic heterogeneity among different populations, further verification should be conducted in other populations. Additionally, to clarify the role of ZFP36L2 in NSCLO intuitively, follow-up experiments in animal models are necessary.
Data Availability Statement
All the datasets generated in this article were shown in the main text and the Supplementary Material. Any questions about the data, please contact to emhvbmdsaW5qaWFAc2luYS5jb20=.
Ethics Statement
The studies involving human participants were reviewed and approved by The Hospital Ethics Committee of West China Hospital of Stomatology, Sichuan University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
M-JL contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. J-YS and Q-SZ contributed to data analysis, critically revised the manuscript. BS and Z-LJ contributed to conception, design, data acquisition, analysis, and interpretation, critically revised the manuscript. All authors gave final approval and agreed to be accountable for all aspects of the work.
Funding
This work was supported by the National Science Funds of China (Nos. 82170919 and 81600849), and the Major frontier issues of the application foundation project of Sichuan Science and Technology Department (2020YJ0211).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank all the participants who donated samples in this study. Thanks to Dr. Zhang from Peking University for kindly providing GMSM-K cell line.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.802229/full#supplementary-material
References
Adzhubei, I., Jordan, D. M., and Sunyaev, S. R. (2013). Predicting Functional Effect of Human Missense Mutations Using PolyPhen‐2. Curr. Protoc. Hum. Genet. 76. Chapter 7: Unit7.20. doi:10.1002/0471142905.hg0720s76
Beaty, T. H., Murray, J. C., Marazita, M. L., Munger, R. G., Ruczinski, I., Hetmanski, J. B., et al. (2010). A Genome-wide Association Study of Cleft Lip with and without Cleft Palate Identifies Risk Variants Near MAFB and ABCA4. Nat. Genet. 42, 525–529. doi:10.1038/ng.580
Beaty, T. H., Taub, M. A., Scott, A. F., Murray, J. C., Marazita, M. L., Schwender, H., et al. (2013). Confirming Genes Influencing Risk to Cleft Lip With/without Cleft Palate in a Case-Parent Trio Study. Hum. Genet. 132, 771–781. doi:10.1007/s00439-013-1283-6
Birnbaum, S., Ludwig, K. U., Reutter, H., Herms, S., Steffens, M., Rubini, M., et al. (2009). Key Susceptibility Locus for Nonsyndromic Cleft Lip with or without Cleft Palate on Chromosome 8q24. Nat. Genet. 41, 473–477. doi:10.1038/ng.333
Boyle, E. A., Li, Y. I., and Pritchard, J. K. (2017). An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 169, 1177–1186. doi:10.1016/j.cell.2017.05.038
Brunskill, E. W., Potter, A. S., Distasio, A., Dexheimer, P., Plassard, A., Aronow, B. J., et al. (2014). A Gene Expression Atlas of Early Craniofacial Development. Dev. Biol. 391, 133–146. doi:10.1016/j.ydbio.2014.04.016
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The Variant Call Format and VCFtools. Bioinformatics 27, 2156–2158. doi:10.1093/bioinformatics/btr330
Dixon, M. J., Marazita, M. L., Beaty, T. H., and Murray, J. C. (2011). Cleft Lip and Palate: Understanding Genetic and Environmental Influences. Nat. Rev. Genet. 12, 167–178. doi:10.1038/nrg2933
Drieschner, N., Kerschling, S., Soller, J. T., Rippe, V., Belge, G., Bullerdiek, J., et al. (2007). A Domain of the Thyroid Adenoma Associated Gene (THADA) Conserved in Vertebrates Becomes Destroyed by Chromosomal Rearrangements Observed in Thyroid Adenomas. Gene 403, 110–117. doi:10.1016/j.gene.2007.06.029
Dumdie, J. N., Cho, K., Ramaiah, M., Skarbrevik, D., Mora-Castilla, S., Stumpo, D. J., et al. (2018). Chromatin Modification and Global Transcriptional Silencing in the Oocyte Mediated by the mRNA Decay Activator ZFP36L2. Dev. Cel 44, 392–402. doi:10.1016/j.devcel.2018.01.006
Galloway, A., Saveliev, A., Łukasiak, S., Hodson, D. J., Bolland, D., Balmanno, K., et al. (2016). RNA-binding Proteins ZFP36L1 and ZFP36L2 Promote Cell Quiescence. Science 352, 453–459. doi:10.1126/science.aad5978
Grosen, D., Chevrier, C., Skytthe, A., Bille, C., Molsted, K., Sivertsen, A., et al. (2010). A Cohort Study of Recurrence Patterns Among More Than 54 000 Relatives of Oral Cleft Cases in Denmark: Support for the Multifactorial Threshold Model of Inheritance. J. Med. Genet. 47, 162–168. doi:10.1136/jmg.2009.069385
Harville, E. W., Wilcox, A. J., Lie, R. T., Vindenes, H., and Åbyholm, F. (2005). Cleft Lip and Palate versus Cleft Lip Only: Are They Distinct Defects? Am. J. Epidemiol. 162, 448–453. doi:10.1093/aje/kwi214
Helin, K. (1998). Regulation of Cell Proliferation by the E2F Transcription Factors. Curr. Opin. Genet. Dev. 8, 28–35. doi:10.1016/s0959-437x(98)80058-0
Hodson, D. J., Janas, M. L., Galloway, A., Bell, S. E., Andrews, S., Li, C. M., et al. (2010). Deletion of the RNA-Binding Proteins ZFP36L1 and ZFP36L2 Leads to Perturbed Thymic Development and T Lymphoblastic Leukemia. Nat. Immunol. 11, 717–724. doi:10.1038/ni.1901
Honein, M. A., Rasmussen, S. A., Reefhuis, J., Romitti, P. A., Lammer, E. J., Sun, L., et al. (2007). Maternal Smoking and Environmental Tobacco Smoke Exposure and the Risk of Orofacial Clefts. Epidemiology 18, 226–233. doi:10.1097/01.ede.0000254430.61294.c0
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 4, 44–57. doi:10.1038/nprot.2008.211
Huang, L., Jia, Z., Shi, Y., Du, Q., Shi, J., Wang, Z., et al. (2019). Genetic Factors Define CPO and CLO Subtypes of Nonsyndromicorofacial Cleft. Plos Genet. 15, e1008357. doi:10.1371/journal.pgen.1008357
Jiang, R., Bush, J. O., and Lidral, A. C. (2006). Development of the Upper Lip: Morphogenetic and Molecular Mechanisms. Dev. Dyn. 235, 1152–1166. doi:10.1002/dvdy.20646
Lai, W. S., Carballo, E., Thorn, J. M., Kennington, E. A., and Blackshear, P. J. (2000). Interactions of CCCH Zinc Finger Proteins with mRNA. J. Biol. Chem. 275, 17827–17837. doi:10.1074/jbc.M001696200
Li, H., and Durbin, R. (2009). Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 25, 1754–1760. doi:10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map Format and SAMtools. Bioinformatics 25, 2078–2079. doi:10.1093/bioinformatics/btp352
Lin-Shiao, E., Lan, Y., Welzenbach, J., Alexander, K. A., Zhang, Z., Knapp, M., et al. (2019). p63 Establishes Epithelial Enhancers at Critical Craniofacial Development Genes. Sci. Adv. 5. doi:10.1126/sciadv.aaw0946
Liu, H., Duncan, K., Helverson, A., Kumari, P., Mumm, C., Xiao, Y., et al. (2020). Analysis of Zebrafish Periderm Enhancers Facilitates Identification of a Regulatory Variant Near Human KRT8/18. Elife 9, 9. doi:10.7554/eLife.51325
Liu, H., Leslie, E. J., Carlson, J. C., Beaty, T. H., Marazita, M. L., Lidral, A. C., et al. (2017). Identification of Common Non-coding Variants at 1p22 that Are Functional for Non-syndromic Orofacial Clefting. Nat. Commun. 8, 14759. doi:10.1038/ncomms14759
Liu, J., Lu, W., Liu, S., Wang, Y., Li, S., Xu, Y., et al. (2018). ZFP36L2, a Novel AML1 Target Gene, Induces AML Cells Apoptosis and Inhibits Cell Proliferation. Leuk. Res. 68, 15–21. doi:10.1016/j.leukres.2018.02.017
Ludwig, K. U., Mangold, E., Herms, S., Nowak, S., Reutter, H., Paul, A., et al. (2012). Genome-wide Meta-Analyses of Nonsyndromic Cleft Lip with or without Cleft Palate Identify Six New Risk Loci. Nat. Genet. 44, 968–971. doi:10.1038/ng.2360
Machado, R. A., Moreira, H. S. B., de Aquino, S. N., Martelli-Junior, H., de Almeida Reis, S. R., Persuhn, D. C., et al. (2016). Interactions betweenRAD51rs1801321 and Maternal Cigarette Smoking as Risk Factor for Nonsyndromic Cleft Lip with or without Cleft Palate. Am. J. Med. Genet. 170, 536–539. doi:10.1002/ajmg.a.37281
Mangold, E., Ludwig, K. U., Birnbaum, S., Baluardo, C., Ferrian, M., Herms, S., et al. (2010). Genome-wide Association Study Identifies Two Susceptibility Loci for Nonsyndromic Cleft Lip with or without Cleft Palate. Nat. Genet. 42, 24–26. doi:10.1038/ng.506
Manolio, T. A., Collins, F. S., Cox, N. J., Goldstein, D. B., Hindorff, L. A., Hunter, D. J., et al. (2009). Finding the Missing Heritability of Complex Diseases. Nature 461, 747–753. doi:10.1038/nature08494
Marazita, M. L. (2012). The Evolution of Human Genetic Studies of Cleft Lip and Cleft Palate. Annu. Rev. Genom. Hum. Genet. 13, 263–283. doi:10.1146/annurev-genom-090711-163729
Mitchell, L. E., Beaty, T. H., Lidral, A. C., Munger, R. G., Murray, J. C., Saal, H. M., et al. (2002). Guidelines for the Design and Analysis of Studies on Nonsyndromic Cleft Lip and Cleft Palate in Humans: Summary Report from a Workshop of the International Consortium for Oral Clefts Genetics. Cleft Palate-Craniofacial J. 39, 93–100. doi:10.1597/1545-1569_2002_039_0093_gftdaa_2.0.co_2
Moreno Uribe, L. M., Fomina, T., Munger, R. G., Romitti, P. A., Jenkins, M. M., Gjessing, H. K., et al. (2017). A Population-Based Study of Effects of Genetic Loci on Orofacial Clefts. J. Dent Res. 96, 1322–1329. doi:10.1177/0022034517716914
Ng, P. C., and Henikoff, S. (2003). SIFT: Predicting Amino Acid Changes that Affect Protein Function. Nucleic Acids Res. 31, 812–3814. doi:10.1093/nar/gkg509
Noguchi, A., Adachi, S., Yokota, N., Hatta, T., Natsume, T., and Kawahara, H. (2018). ZFP36L2 Is a Cell Cycle-Regulated CCCH Protein Necessary for DNA Lesion-Induced S-phase Arrest. Biol. Open 7. doi:10.1242/bio.031575
Pan, Y., Han, Y., Zhang, H., Zhou, L., Li, D., Cai, Q., et al. (2013). Association and Cumulative Effects of GWAS-Identified Genetic Variants for Nonsyndromic Orofacial Clefts in a Chinese Population. Environ. Mol. Mutagen. 54, 261–267. doi:10.1002/em.21773
Polager, S., and Ginsberg, D. (2008). E2F - at the Crossroads of Life and Death. Trends Cel Biol 18, 528–535. doi:10.1016/j.tcb.2008.08.003
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: a Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 81, 559–575. doi:10.1086/519795
Ramos, S. B. V., Stumpo, D. J., Kennington, E. A., Phillips, R. S., Bock, C. B., Ribeiro-Neto, F., et al. (2004). The CCCH Tandem Zinc-finger Protein Zfp36l2 Is Crucial for Female Fertility and Early Embryonic Development. Development, 131, 4883–4893. doi:10.1242/dev.01336
Rentzsch, P., Witten, D., Cooper, G. M., Shendure, J., and Kircher, M. (2018). CADD: Predicting the Deleteriousness of Variants throughout the Human Genome. Nucleic Acids Res. 47, D886–D894. doi:10.1093/nar/gky1016
Sazonovs, A., and Barrett, J. C. (2018). Rare-Variant Studies to Complement Genome-wide Association Studies. Annu. Rev. Genom. Hum. Genet. 19, 97–112. doi:10.1146/annurev-genom-083117-021641
Schwarz, J. M., Rödelsperger, C., Schuelke, M., and Seelow, D. (2010). MutationTaster Evaluates Disease-Causing Potential of Sequence Alterations. Nat. Methods 7, 575–576. doi:10.1038/nmeth0810-575
Seale, P., Sabourin, L. A., Girgis-Gabardo, A., Mansouri, A., Gruss, P., and Rudnicki, M. A. (2000). Pax7 Is Required for the Specification of Myogenic Satellite Cells. Cell 102, 777–786. doi:10.1016/s0092-8674(00)00066-0
Sivertsen, A., Wilcox, A. J., Skjaerven, R., Vindenes, H. A., Abyholm, F., Harville, E., et al. (2008).Familial Risk of Oral Clefts by Morphological Type and Severity: Population Based Cohort Study of First Degree Relatives. Bmj 336, 432–434. doi:10.1136/bmj.39458.56361110.1136/bmj.39458.563611.AE
Stott-Miller, M., Heike, C. L., Kratz, M., and Starr, J. R. (2010). Increased Risk of Orofacial Clefts Associated with Maternal Obesity: Case-Control Study and Monte Carlo-Based Bias Analysis. Paediatr. Perinat Epidemiol. 24, 502–512. doi:10.1111/j.1365-3016.2010.01142.x
Stumpo, D. J., Broxmeyer, H. E., Ward, T., Cooper, S., Hangoc, G., Chung, Y. J., et al. (2009). Targeted Disruption of Zfp36l2, Encoding a CCCH Tandem Zinc finger RNA-Binding Protein, Results in Defective Hematopoiesis. Blood 114, 2401–2410. doi:10.1182/blood-2009-04-214619
Suk, F.-M., Chang, C.-C., Lin, R.-J., Lin, S.-Y., Liu, S.-C., Jau, C.-F., et al. (2018). ZFP36L1 and ZFP36L2 Inhibit Cell Proliferation in a Cyclin D-dependent and P53-independent Manner. Sci. Rep. 8. doi:10.1038/s41598-018-21160-z
Sun, Y., Huang, Y., Yin, A., Pan, Y., Wang, Y., Wang, C., et al. (2015). Genome-wide Association Study Identifies a New Susceptibility Locus for Cleft Lip with or without a Cleft Palate. Nat. Commun. 6, 6414. doi:10.1038/ncomms7414
Sweat, Y. Y., Sweat, M., Mansaray, M., Cao, H., Eliason, S., Adeyemo, W. L., et al. (2020). Six2 Regulates Pax9 Expression, Palatogenesis and Craniofacial Bone Formation. Dev. Biol. 458, 246–256. doi:10.1016/j.ydbio.2019.11.010
Tada, H., Kawashiri, M.-a., Konno, T., Yamagishi, M., and Hayashi, K. (2016). Common and Rare Variant Association Study for Plasma Lipids and Coronary Artery Disease. Jat 23, 241–256. doi:10.5551/jat.31393
Vogel, K. U., Bell, L. S., Galloway, A., Ahlfors, H., and Turner, M. (2016). The RNA-Binding Proteins Zfp36l1 and Zfp36l2 Enforce the Thymic β-Selection Checkpoint by Limiting DNA Damage Response Signaling and Cell Cycle Progression. J.I. 197, 2673–2685. doi:10.4049/jimmunol.1600854
Wang, K.-T., Wang, H.-H., Wu, Y.-Y., Su, Y.-L., Chiang, P.-Y., Lin, N.-Y., et al. (2015). Functional Regulation of Zfp36l1 and Zfp36l2 in Response to Lipopolysaccharide in Mouse RAW264.7 Macrophages. J. Inflamm. 12, 42. doi:10.1186/s12950-015-0088-x
Wang, K., Li, M., and Hakonarson, H. (2010). ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 38, e164. doi:10.1093/nar/gkq603
Keywords: 2p21, non-syndromic cleft lip only, ZFP36L2, proliferation, cell cycle
Citation: Li M-J, Shi J-Y, Zhu Q-S, Shi B and Jia Z-L (2022) Targeted Re-Sequencing of the 2p21 Locus Identifies Non-Syndromic Cleft Lip Only Novel Susceptibility Gene ZFP36L2. Front. Genet. 13:802229. doi: 10.3389/fgene.2022.802229
Received: 10 November 2021; Accepted: 12 January 2022;
Published: 09 February 2022.
Edited by:
Gerson Shigeru Kobayashi, University of São Paulo, BrazilReviewed by:
Luciano Abreu Brito, University of São Paulo, BrazilSahin Naqvi, Stanford University, United States
Copyright © 2022 Li, Shi, Zhu, Shi and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Shi, c2hpYmluZ2NuQHZpcC5zaW5hLmNvbQ==; Zhong-Lin Jia, emhvbmdsaW5qaWFAc2luYS5jb20=
†These authors have contributed equally to this work
 Mu-Jia Li
Mu-Jia Li Jia-Yu Shi2†
Jia-Yu Shi2† Qiu-Shuang Zhu
Qiu-Shuang Zhu Zhong-Lin Jia
Zhong-Lin Jia