- 1Department of Animal Science, Texas A&M University, College Station, TX, United States
- 2Department of Animal Sciences, Washington State University, Pullman, WA, United States
In this study, actinin-3 (ACTN3) gene expression was investigated in relation to the feed efficiency phenotype in Bos indicus - Bos taurus crossbred steers. A measure of relative feed efficiency based on residual feed intake relative to predictions from the NRC beef cattle model was analyzed by the use of a mixed linear model that included sire and family nested within sire as fixed effects and age, animal type, sex, condition, and breed as random effects for 173 F2 Nellore-Angus steers. Based on these residual intake observations, individuals were ranked from most efficient to least efficient. Skeletal muscle samples were analyzed from 54 steers in three groups of 18 (high efficiency, low efficiency, and a statistically average group). ACTN3, which encodes a muscle-specific structural protein, was previously identified as a candidate gene from a microarray analysis of RNA extracted from muscle samples obtained from a subset of steers from each of these three efficiency groups. The expression of ACTN3 was evaluated by quantitative reverse transcriptase PCR analysis. The expression of ACTN3 in skeletal muscle was 1.6-fold greater in the inefficient steer group than in the efficient group (p = 0.007). In addition to expression measurements, blocks of SNP haplotypes were assessed for breed or parent of origin effects. A maternal effect was observed for ACTN3 inheritance, indicating that a maternal B. indicus block conferred improved residual feed efficiency relative to the B. taurus copy (p = 0.03). A SNP haplotype analysis was also conducted for m-calpain (CAPN2) and fibronectin 1 (FN1), and a significant breed effect was observed for both genes, with B. indicus and B. taurus alleles each conferring favorable efficiency when inherited maternally (p = 0.03 and p = 0.04). Because the ACTN3 structural protein is specific to fast-twitch (type II) muscle fibers and not present in slow-twitch muscle fibers (type I), muscle samples used for expression analysis were also assayed for fiber type ratio (type II/type I). Inefficient animals had a fast fiber type ratio 1.8-fold greater than the efficient animals (p = 0.027). Because these fiber-types exhibit different metabolic profiles, we hypothesize that animals with a greater proportion of fast-twitch muscle fibers are also less feed efficient.
Introduction
Efficient use of resources is regarded as a critical area of emphasis for livestock production (Kenny et al., 2018; Brito et al., 2021). Feed costs can contribute to 70% of total livestock production costs (Becker 2008). Environmental costs associated with beef production have become increasingly important to the consumer. Although the beef industry made substantial strides in reducing the environmental footprint of cattle production over several decades (Capper 2011), feed efficiency remains a target today in beef producers’ efforts toward sustainability. In considering nutrient security for humans, utilizing new and emerging technologies to better understand the complex physiological mechanisms of production traits will be key for enabling even greater efficiency of food animal production (Riggs et al., 2017; Riggs et al., 2018). Increased access to nutrient-dense animal source foods requires greater production volume if global population growth reaches 9.8 billion people in the coming decades (FAO, 2018). Additionally, a systematic review of literature that examined life cycle assessment in cattle (published between 2000 and 2016) contained climate change as a category, with nearly a third of those papers focusing on only that topic (McClelland et al., 2018). Understanding feed efficiency in the context of breeds adapted to specific environmental conditions—particularly hot, dry conditions found in much of the world’s grazing lands—has important economic and environmental implications.
Brunes et al. (2021) noted that feed efficiency traits in beef cattle are highly complex and affected by numerous biological and polygenic mechanisms. In their GWAS analysis of feed efficiency from 4,329 Nellore cattle, multiple genomic regions were identified that harbor sequence variation associated with greater than 0.5% of the additive genetic variance for feed efficiency in the population (Brunes et al., 2021). Genes within the identified regions contribute to the metabolism of protein, lipids, and numerous metabolic, energetic, and behavioral pathways. A study of dairy cattle in New Zealand (Puillet et al., 2021) examined gene-by-environment interactions. As might be expected, this work demonstrated that the relationships between feed efficiency and other production traits are complex and must be carefully balanced when one considers overall measures of lifetime production efficiencies.
In the present study, we focused on aspects of feed efficiency influenced by skeletal muscle by specifically examining ACTN3 within a genetic mapping herd of beef cattle that utilized Bos indicus–Bos taurus cattle suited to the subtropical environment of Texas. Resting energy consumption contributes to feed efficiency as a major component of an animal’s overall energy requirements. Skeletal muscle has lower energy requirements by weight compared to other tissue types. However, because skeletal muscle mass contributes as much as half of the live animal’s weight, a great deal of an animal’s total caloric intake may be utilized for the growth and maintenance of this tissue. Approximately 20% of an animal’s overall energy consumption is consumed by the skeletal muscle (Ortigues 1991), and this percentage can increase drastically during stress, activity, or changes in temperature.
We evaluated ACTN3 sequence haplotypes and gene expression in F2 Nellore-Angus steers that had been evaluated for efficiency. For the study we describe here, we specifically wanted to investigate whether different ACTN3 alleles in this population corresponded to feed efficiency rates. This gene was identified as one candidate contributing to a feed efficiency phenotype from a pilot gene expression network analysis, and we suspected that animals differed in muscle turnover rate. We hypothesized that we could identify genetic variation in ACTN3 responsible for differences in muscle physiology and homeostasis that are important for overall feed efficiency. In pigs, a relationship between muscle turnover and residual feed intake was previously described (Cruzen et al., 2013). In beef cattle, the negative correlation of muscle proteolysis rate and feed efficiency has been noted (McDonagh et al., 2001). Other investigators have evaluated skeletal muscle gene expression in beef cattle feed efficiency studies. For example, Lam et al., 2020 developed an RNA sequencing pipeline from liver and muscle transcriptomes (Tizioto et al., 2016) to identify single nucleotide polymorphisms (SNP) associated with feed efficiency in Nellore steers classified with high or low residual feed intake (RFI). In this study, the authors suggested relationships between feed efficiency and both immune and metabolic function in livestock. Regulation of the RAC1 gene was associated with less efficient cattle. Interestingly, the genetic pathways that include RAC1 and ACTN3 may have related functional roles associated with calcium metabolism-based regulation of immune function (Calender et al., 2020).
Also contributing to energetic requirements, different skeletal muscle fiber types have different energy requirements. Type II, or fast-twitch fibers, have greater energy demands than type I, or slow-twitch fibers. A study in horses identified ACTN3 as a potential element associated with muscle performance (Ropka-Molik et al., 2019). Welch et al., 2013 found a positive correlation between relative feed intake (RFI) and type II muscle fibers. This finding is consistent with the idea that the physiological skeletal muscle phenotype of an animal contributes to its overall efficiency phenotype. Because ACTN3 is expressed only in type II fibers, this study also examined fiber type profiles in muscle samples.
Materials and Methods
Feed Efficiency Phenotype
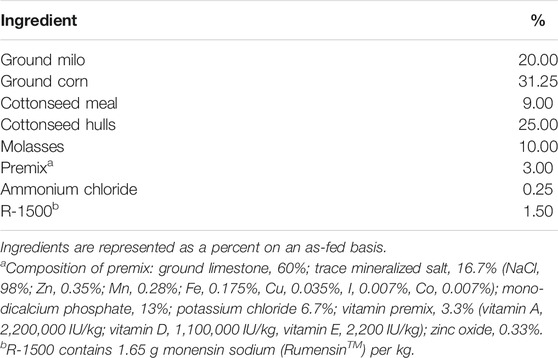
For this study, feed and carcass data, obtained from 173 Nellore-Angus F2 steers in 13 full-sibling families produced by embryo transfer from the Texas A&M McGregor Genomics research population located in central Texas (latitude: 31.3865, longitude: −97.4105), were utilized, as described by Amen (2007). Briefly, calves were produced in the spring and fall calving seasons, and this study used calves born from 2003 to 2005. After weaning (approximately 230 days of age), the animals were grass-fed for approximately 130 days until they reached 11–13 months of age. Steers were moved to a Calan Broadbent Feeding System (American Calan, Northwood, NH, USA) to measure individual feed intake. Over 28 days, the steers were adjusted to the finishing diet (Table 1). Feeding was ad libitum, and uneaten feed was removed and measured every 7 days. Animals were weighed every 28 days for ∼140 days at the same time of day and in the same order of pens at each weigh day to equalize gut fill effects across time, as much as possible.
Using the NRC (2000) model application, the daily feed intake required to achieve observed ADG was predicted. Model inputs included feeding period mid-weight (used as the current reference weight for model predictions) and final weight at slaughter for each animal. Standardized inputs were used for animal type (growing/finishing), age (14 months), sex (steer), BCS (5), breed (Nellore-Angus two-way cross), management (ionophore), diet (Table 1), and quality grade (select). Environmental factors in the model were set to be thermoneutral. The model predicted intake was subtracted from the observed dry matter intake (DMI), and the difference is defined as residual feed intake based on the NRC model (RFINRC), such that those animals that consumed less than predicted (thus, were more efficient) had negative RFINRC (Liu et al., 2000). Mixed procedures of SAS were then used to analyze RFINRC with fixed factors of sire and family nested within sire (Amen 2007). This method of residual prediction parallels more typical calculations of RFI, in which individual residuals represent a deviation from a modeled mean intake at an observed rate of gain. However, the standard method is generally restricted to use within a single contemporary group fed simultaneously or with the use of the contemporary group as a model effect. Importantly, in this case, a standard model (the NRC) was utilized to construct the modeled mean intake rather than a regression on observed data, thus enabling animals in multiple contemporary groups to be evaluated against a common model.
Sample Selection
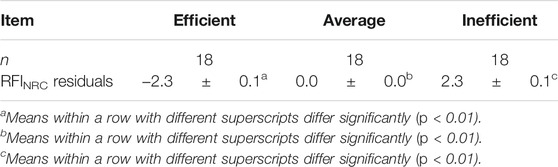
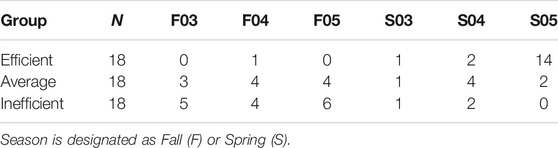
For gene expression analysis, 36 animals were identified at the tails of the efficiency distribution based on RFINRC as described above. A total of 18 animals were classified as most “efficient” for this population and had negative RFINRC residuals indicating that they had consumed less feed than would be expected based on the model. A total of 18 animals were classified as most “inefficient,” with positive RFINRC residuals indicating they had consumed more feed than would be expected based on the model. Muscle samples from these 36 animals were used for subsequent expression analysis. In addition, a statistically average group of 18 animals with an RFINRC residual clustered around zero was included for comparison purposes. Thus, a total of 54 animals from the middle, and both tails of the residual distribution were analyzed for gene expression. Means for these groups are presented in Table 2. The distribution of these groups across birth year seasons (BYS) is shown in Table 3. In addition, the frequency distribution of animals from the Amen (2007) study is presented in Table 4, with the set of most and least efficient animals shown in parentheses. Amen (2007) noted that imbalance exists among families across BYS. While this imbalance may complicate efforts to separate family effects from year-season effects, the approach provides a means to examine the combined overall impact of gene X environmental interactions on resultant phenotypes to understand the influence of gene function on phenotype despite a range of regional climate conditions.
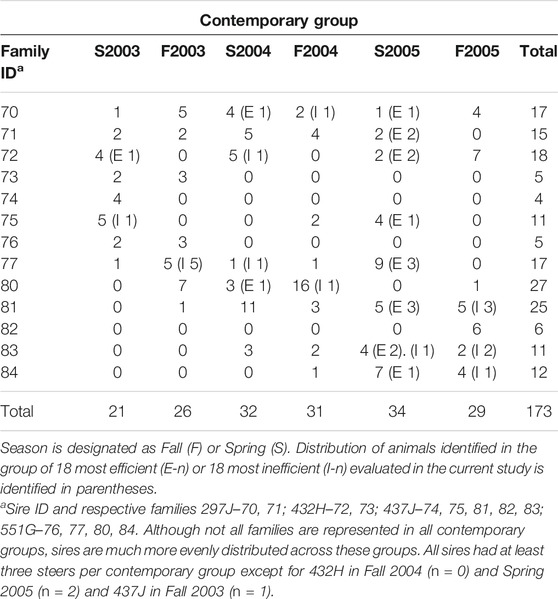
TABLE 4. Frequency table of family distribution across BYS contemporary groups from animals evaluated for feed efficiency in a previous study (Amen, 2007).
Tissue Collection and Extraction of RNA
Steers were harvested at 18 months of age at the Rosenthal Meat Center at Texas A&M University in College Station, TX using humane harvesting procedures, as described by Savell and Smith (2000). Animals were restricted from feed for approximately 12 h before harvest but had continual access to water. Animals were immobilized using a captive bolt stunning mechanism and further processed using standard industry procedures. Approximately 1 g of muscle tissue from the Longissimus cervicis (in the neck region of the carcass) was collected before electrical stimulation (ES; less than 1 h after exsanguination) of the carcass. The muscle sample was flash-frozen in liquid nitrogen to prevent mRNA degradation. Samples were stored at −80°C until RNA was extracted.
Total RNA was extracted from a portion (∼100–200 mg) of the frozen whole muscle tissue samples (L. cervicis) from each of the 54 animals with TRI Reagent® (Molecular Research Center, Cincinnati, OH, USA) and 1-bromo-3-chloropropane (Molecular Research Center). Next, RNA was precipitated with isopropanol (Sigma Aldrich, St. Louis, MO, USA), washed with 70% ethanol (Sigma Aldrich), and reconstituted in 50 µl nuclease-free water (Invitrogen, Carlsbad, CA, USA). The quality of the RNA was assessed via an Agilent 2100 series Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) according to the manufacturer’s protocol. Samples with an RNA integrity number (RIN) >8.0 and appropriate electropherogram image were treated with DNase (Invitrogen) and column-purified via the RNeasy Mini kit (Qiagen, Valencia, CA, USA). Samples were quantified by spectrophotometry (NanoDrop 1000), and identical quantities of total RNA for each sample were utilized in downstream applications. RNA extracts were stored at −80°C prior to expression analyses.
Microarray Procedures
Microarray analysis (described in Vaughn, 2013; Riggs and Vaughn, 2015) was conducted prior to this study as a pilot study with the Agilent 44k bovine array (B. taurus oligo microarray V2 Agilent 4x44k GPL11649) according to manufacturer’s recommendations. Twenty-four RNA samples (200 ng each from eight most efficient and eight least efficient animals) were labeled for two-color microarray gene expression analysis (Quick Amp Labeling Kit, Agilent Technologies) according to the manufacturer’s protocol. Following hybridization and scanning, normalization and quality control were performed using embedded quantile normalization functions in the Genespring GX v11.0.2 software. Following quality filtering, the Mann–Whitney unpaired test was utilized. A non-parametric test was used to avoid relying on the a priori assumption of a normal distribution of gene expression within these populations selected for extremes. In this experimental design, samples fail the assumption of independence, making false discovery analyses invalid. A cut-off of p ≤ 0.05 and a fold-difference of 1.4-fold or greater were set as thresholds to generate lists of probes for subsequent pathways analysis. Pathway analysis was performed using DAVID Bioinformatics Resources 6.7 (http://david.ncifcrf.gov/). Gene Ontology analysis was also performed within the DAVID software using the same data set used for pathway analysis. The GO-fat category was used with the default ease setting of 0.1 and a minimum count of 2. From these pathways analyses, as well as a separate and independent array and pathway experiments (not described here), muscle metabolic pathways we formed a hypothesis related to genes involved in skeletal muscle turnover. As a result, ACTN3 was identified as a target for further investigation by qRT-PCR and haplotype analysis. Array data are deposited in the Gene Expression Omnibus, GEO accession number GSE56705 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56705.
Real Time Quantitative Reverse Transcriptase Polymerase Chain Reaction
Quantitative RT-PCR analysis was conducted on the full set of 54 samples. Synthesis of cDNA from all RNA samples with the high-capacity cDNA Reverse Transcription Kit (Invitrogen) was accomplished with a starting quantity of 2 µg mRNA per 40 μl reaction. Oligo(dT)20 primers (Integrated DNA Technologies, Coralville, IA, USA), 5 μM final concentration, were used for reverse transcription. cDNA was diluted 1:2 in yeast tRNA (25 ng/μl; Invitrogen). Samples were amplified in a total volume of 20 µl containing 1X SYBR GreenER™ qPCR SuperMix (Invitrogen), 300 nM primers and 2 μl template cDNA. Real-time quantitative RT-PCR (qRT-PCR) was performed at 95°C for 10 min followed by 40 cycles of 15 s at 95°C and 60 s at 60°C, in a 7900HT thermal cycler (Applied Biosystems).
Primers for genes validated by qRT-PCR were designed within Oligo 6 Primer Analysis Software v6.71 (Molecular Biology Insights, Cascade, CO, USA). The analyzed genes included actinin-2 (ACTN2), actinin-3 (ACTN3), adipose differentiation-related protein (ADFP, also known as perilipin 2 (PLIN2)), ATP synthase H+ transporting, mitochondrial F1 complex, beta polypeptide (ATP5B), hexokinase 2 (HKII), and lactate dehydrogenase B (LDHB). Ribosomal protein S20 (RPS20) was utilized as a reference gene. Genes and primers used are described in Table 5. Primer pairs were evaluated by BLAST sequence similarity search (Altschul et al., 1990). Primer pairs were selected, which did not cross-amplify across species or between different mRNA transcripts. Additionally, primers were selected to span an exon junction to prevent genomic amplification.
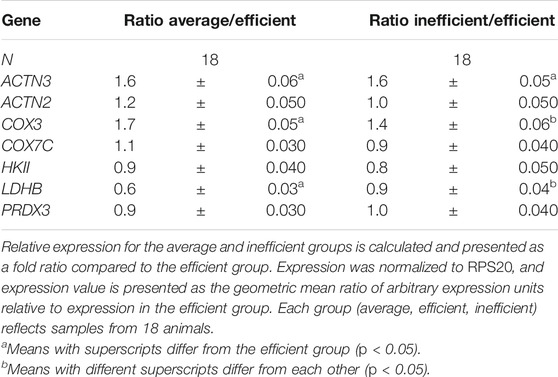
To quantify qRT-PCR results, amplification data were analyzed via Sequence Detection System v2.4 software (Applied Biosystems, Carlsbad, CA, USA). Expression was normalized to RPS20 as a reference gene (De Jonge et al., 2007), and relative expression was quantified as described by Livak and Schmittgen (2001). A threshold value of 0.20 was used to determine the Ct value. In summary, raw Ct values were normalized to RPS20 based on internal expression stability between groups (Vandesompele et al., 2002). The normalized value was subtracted from the raw Ct for each sample (Δ Ct). From the Δ Ct, the average value of the efficient group was subtracted (ΔΔ Ct). The ΔΔ Ct value was linearized by conversion to the inverse negative log2 (RQ). The efficient group in this study thus has a median expression value of 1.0. It should be noted that these are arbitrary units (AU) of expression and that no direct comparison between different genes in total expression levels can be reliably made. All values are relative and applicable directly only as a within-group comparison of relative expression.
Muscle Fiber Type Classification
Fiber type analysis was determined by gel electrophoresis. L. cervicis muscle samples (0.1 g) were homogenized in 500 μl buffer consisting of 250 mM sucrose, 100 mM KCl, 5 mM EDTA, and 20 mM Tris-HCl pH 6.8. The homogenate was filtered through nylon cloth to remove debris and centrifuged at 10,000 × g for 10 min. The pellet obtained was resuspended in 500 μl washing buffer (200 mM KCl, 5 mM EDTA, 0.5% Triton X-100, and 20 mM Tris-HCl pH 6.8). The suspension was centrifuged at 10,000 × g for 10 min. The pellet containing purified myofibrillar proteins was resuspended in 200 μl water and 300 μl of standard 2X sample loading buffer, boiled for 5 min, and then centrifuged at 12,000 × g for 5 min. The resultant supernatant was used for electrophoresis.
Stacking gels consisted of 4% acrylamide (acrylamide: bis = 50:1) and 5% (v/v) glycerol, 70 mM Tris-HCl pH 6.7, 0.4% (w/v) SDS, 4 mM EDTA, 0.1% (w/v) APS, and 0.01% (v/v) TEMED. The separation gel contained 5% (w/v) glycerol, acrylamide: bis (50:1) at a concentration ranging from 5 to 20%, 200 mM Tris pH 8.8, 4 mM EDTA, 0.4% (w/v) SDS, 0.01% (v/v) TEMED, and 0.1% (w/v) ammonium persulfate. The upper running buffer consisted of 0.1 M Tris-HCl pH 8.8, 0.1% (w/v) SDS, 150 mM glycine, and 10 mM mercaptoethanol, and the lower running buffer consisted of 50 mM Tris-HCl pH 8.8, 0.01% (w/v) SDS, and 75 mM glycine. Gels were run at 8°C in a Hoefer SE 600 (Hoefer Scientific, San Francisco, CA, USA) unit, at constant 200 V for 24 h (Bamman et al., 1999). After electrophoresis, gels were stained with Coomassie blue and scanned with a densitometer to determine the amount of each myosin isoform and the percentage of type I and type II muscle fibers was reported (Underwood et al., 2007).
Haplotype Analysis
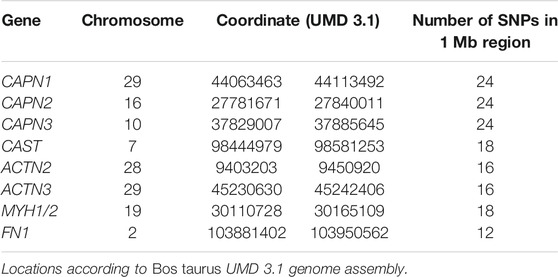
All individuals (n = 776) from the first three generations of the experimental population were previously genotyped with the Illumina BovSNP50 v1 assay (Illumina, Inc., San Diego, CA, USA), and data were available for use in this study. The haplotype analysis from these data allowed us to trace the inheritance of Nellore or Angus blocks of SNPs and identify the parent of origin for the 173 Nellore-Angus F2 steers for which RFINRC data and tissue samples were collected and used in this study. After pruning genotypes to remove animals with poor completion rate (<0.9), SNP with low minor allele frequency (<0.05), SNP with poor completion rates (<0.9), and those SNP that deviated from Hardy–Weinberg equilibrium (p < 0.0001), 39,890 genotypes per individual remained. To determine whether breed of origin or parent of origin played a role in the efficiency phenotype, SNP genotypes spanning 1 Mb intervals flanking several genes of interest (Table 6) based on expression analyses were extracted using PLINK v1.07 (Purcell et al., 2007) and formatted for phase v2.1.1 software (Stephens et al., 2001). Haplotypes were established using 100 iterations of phase v2.1.1, with a thinning interval of 2 and a burn-in of 10. Resultant phased haplotypes were ordered by generation, and breed (Nellore or Angus) and parent of origin were manually tracked through the pedigree to assign the source of each haplotype block in the F2 generation. Because gene expression analyses of μ-calpain (CAPN1), m-calpain (CAPN2), calpastatin (CAST), myosin heavy chain 1 (MYH1), and myosin heavy chain 2 (MYH2) genes were also available from another study and are relevant to pathways associated with muscle growth and proteolysis (Vaughn 2013), their relationship with feed efficiency was also examined. The ACTN2 gene was selected for analysis because it is expressed in all skeletal muscle fiber types and has conserved structural and functional similarity to ACTN3. The genes CAPN1, CAPN2, and CAST were examined because of their roles in muscle proteolysis (Goll et al., 2003), and MYH1 and MYH2 were selected for their function in muscle fibers (Wang et al., 2012).
Statistical Analysis
SPSS 16.0 software (IBM, Armonk, NY) was used for all statistical analyses. To test for significance between efficiency groups, an independent samples t-test was used. Correlation analysis used a bivariate two-tailed Pearson’s correlation. All qRT-PCR expression was normalized to the RPS20 reference gene (De Jonge et al., 2007). Expression data are reported relative to the feed efficient group, which was set to a value of 1.0. The inefficient and average groups are represented as fold difference relative to the efficient group. One sample was removed from all analyses because expression values measured for the reference gene, RPS20, were not consistent with other samples.
Results
Gene Expression Analysis
Following an initial pilot microarray (GEO accession number GSE56705) and network pathway analysis, the ACTN3 gene was selected for further investigation. From the initial hypothesis generating microarray experiment, 58 genes were expressed significantly differently between efficiency groups with a fold difference between groups of 1.5-fold or greater (Supplementary Table S1). The ACTN3 gene was expressed as 2.5-fold greater in the inefficient group of steers but fell short of significance (p = 0.051). However, because different alleles of this gene have been associated with differential athletic performance in humans, and ACTN3 genotype is thought to contribute to variation in muscle phenotype (North et al., 2003; Yang et al., 2003), ACTN3 was selected for further investigation. Despite missing the initial statistical cut-off, ACTN3 also remained of interest because it is a member of skeletal muscle networks determined to be relevant. Following these early investigations, this study was developed to examine ACTN3 as a candidate gene contributing to a feed efficiency phenotype. Genes used for qRT-PCR analysis are described in Table 5. Additional genes that have some relationship to ACTN3 and the skeletal muscle pathways in which it participates were included in qRT-PCR expression and haplotype analyses. These genes were selected prior to expression and haplotype analysis as negative control genes or as genes expected to function similarly in given pathways. We thought that, by examining haplotypes according to parent of origin, we may be able to differentiate favorable Bos indicus and Bos taurus alleles that would be of use for selection. The ACTN2 gene was selected for analysis because it is expressed in all skeletal muscle fiber types and has conserved structural and functional similarity to ACTN3. Although their functions are not identical, evidence exists that ACTN2 can largely compensate for the absence of ACTN3 (Mills et al., 2001; Lek et al., 2010). The previously evaluated reference genes were tested for stability, and RPS20 was utilized as a control reference gene. Because of our hypothesis that ACTN3 was associated with feed efficiency and might reflect differences in overall metabolic activity, we also selected a set of genes that are known to play a role in other metabolic processes within muscle tissue to examine at the same time. For example, ADFP was selected because of its role in lipid metabolism and storage (Hiller et al., 2012). Genes that place a role in muscle metabolism and glucose homeostasis (ATP5B, HKII, and LDHB) were also examined because these are genes that play a role in muscle metabolism (Rajesh et al., 2011; Egan and Zierath 2013; Li et al., 2013). The genes CAPN1, CAPN2, and CAST were examined because of their roles in muscle proteolysis (Goll et al., 2003), and MYH1 and MYH2 were selected for their function in muscle fibers (Wang et al., 2012).
By qRT-PCR, expression of ACTN3 was 1.6-fold greater (p = 0.009) in the average and inefficient groups than in the efficient group (Table 7). ACTN2 expression did not vary significantly between any of the groups, nor did it correlate significantly with ACTN3 expression or fiber type ratio (as expected). Of the other genes assayed, LDHB expression was significantly lower in the average group but not the inefficient group, relative to the efficient steers. The mRNA quantity of the remaining genes examined (Table 7) did not differ significantly between groups.
Post-transcriptional modifications, other regulatory mechanisms, and differences in the timing of expression can make the correlation of mRNA expression with protein expression difficult (Greenbaum et al., 2002). To verify that the observed difference in ACTN3 gene expression translated to actual differences in muscle protein expression, 12 samples (portions of the same samples used for qRT-PCR) from each tail of the distribution (n = 24) were assayed for fiber type ratios based on gel separation of muscle fiber type specific isoforms. The ratio of fast-twitch to slow-twitch muscle fiber (type II/type I) was 1.8-fold greater (p = 0.027) in the inefficient group compared to the efficient group (Figure 1).
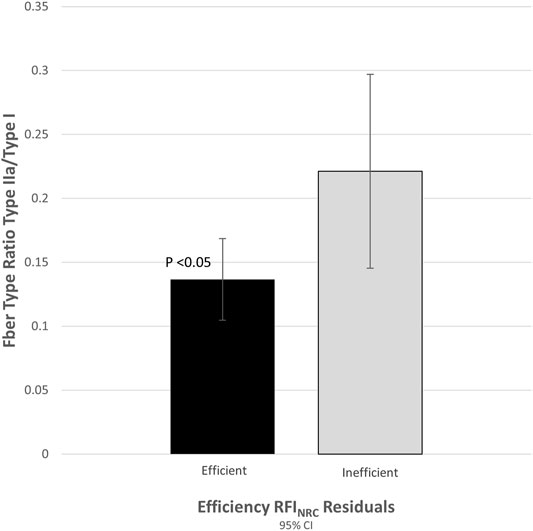
FIGURE 1. Proportion of fast-to-slow-twitch (type II/type I) fiber ratio was correlated with expression of actinin-3 (r = 0.622, p < 0.001).
Haplotype Analysis
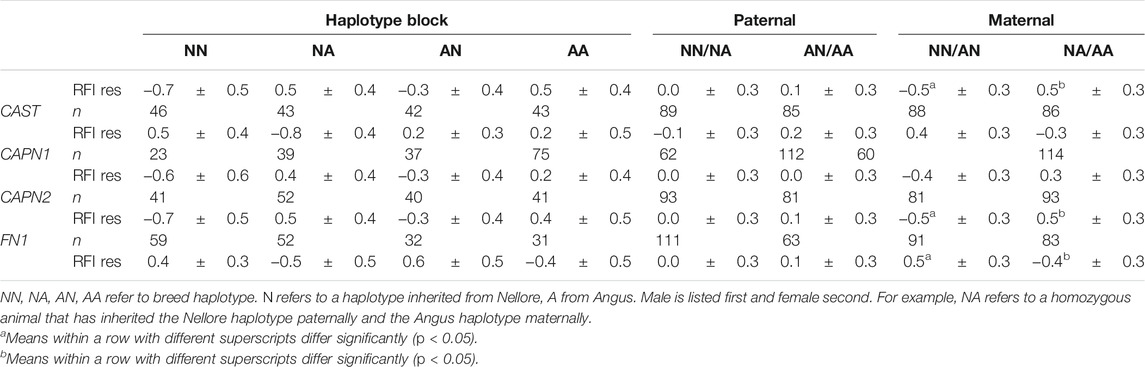
Haplotype block analysis was conducted for ACTN2, ACTN3, CAPN1, CAPN2, CAPN3, CAST, FN1, and MYH1/2. Three of the haplotype blocks were associated with significant differences in efficiency between Angus and Nellore in the larger population studied. The CAPN2 and ACTN3 Nellore haplotype blocks were associated with a superior efficiency when inherited maternally (p = 0.03). The FN1 Angus haplotype block was associated with a superior efficiency when inherited maternally (p = 0.04). Neither the CAST nor the CAPN1 haplotypes were associated with any improvement in efficiency (Table 8).
Season of test feeding period may have influenced efficiency. A general linear model was used to produce the least square means using RFINRC residuals as the dependent variable with birth year season (BYS) as the fixed factor described in Amen (2007). Six BYS groups were included in this study: F03 (n = 26), F04 (n = 31), F05 (n = 29), S03 (n = 21), S04 (n = 32), and S05 (n = 34), with a total of 87 spring-born animals weaned in the fall and 85 weaned in the spring. Steers weaned in the spring were more efficient than those weaned in the fall; linear contrast of fall RFINRC residuals means minus spring RFINRC residuals means was 1.09 + 0.15 (p < 0.0001). One BYS level, Spring of 2005, had a lower mean RFI than the other BYS groups (p < 0.0001). Within the subset selected for expression analysis, 14 of the 18 samples in the efficient group were from that BYS level (Table 3). The distribution of most and least efficient animals examined is also described in Table 4.
ACTN3 expression was higher in fall weaned animals than spring weaned animals by 2.1-fold (p < 0.001). No significant differences between seasons were noted in any other genes assayed. Additionally, a difference (p < 0.001) in fiber type by the season of weaning was observed. Those calves weaned in the fall (n = 11) had a fast-to-slow-twitch (type II/type I) fiber type ratio 1.6-fold greater than those weaned in the spring (n = 13). These ratios were 0.23 and 0.14, respectively.
Discussion
Multiple biological and developmental processes, driven by complex genetic networks and response to environmental conditions, each contribute to phenotypic measures of efficiency and the overall relationships of inputs (e.g., feed intake) to outputs (e.g., meat and other products). The identification of specific genetic variation contributes to better understanding and insight into mechanisms of growth and energy utilization. Such knowledge can drive further gains in productivity, reducing waste and environmental impacts. Increased feed efficiency can reduce production costs (Ho et al., 2013) and environmental footprint, both of which are important in a world with an expanding population to feed and finite resources for the production of necessary animal source foods. Waste concerns, from feed waste to methane production, have also gained emphasis in recent years (Brito et al., 2020). Simultaneously, consumers are demanding environmentally conscious options, while producers strive to maintain fiscal viability.
In the present study, a difference in ACTN3 expression between groups was confirmed by qRT-PCR analysis in skeletal muscle samples from the high and low feed efficiency groups of steers. Expression of ACTN2 did not differ between groups. Relative to the efficient group, the inefficient group overexpressed ACTN3 1.6-fold. The fiber type ratio measured by fast-twitch (type II)/slow-twitch (type I) differed between groups, with a 1.8-fold increase in the inefficient group relative to the efficient group. Season of weaning may have influenced the results since steers weaned in the fall also had a greater fast-twitch to slow-twitch fiber type ratio and greater expression of ACTN3 compared with those weaned during spring. However, the model predicted RFINRC does consider birth year season. Feed intake and efficiency have been previously shown to be affected by seasonal effects (Mujibi et al., 2010).
Although not all families are represented in all contemporary groups for which feed efficiency data were collected (Table 4), sires are much more evenly distributed across these groups. All sires had at least three steers per contemporary group except for 432H in Fall 2004 (n = 0) and Spring 2005 (n = 2) and 437J in Fall 2003 (n = 1). Because RFINRC is a population-based calculation rather than a cohort-based calculation, its use may produce unequal numbers of efficient/inefficient observations per contemporary group. Although only 1 or 0 animals are evaluated from a single contemporary group, this should not bias the results compared to if all animals were included in the analyses.
Our use of RFINRC enabled animals to be compared across BYS groups. However, as evidenced by the imbalance of animals deemed “most efficient” across BYS, we are likely observing the impact of G X E interaction on the efficiency phenotype. However, despite a potential environmental influence that also affected efficiency, we identified the most efficient animals across all BYS. We are interested in understanding genes and gene networks that contribute to multigenic traits such as feed efficiency in the context of variably subtropical conditions that are typically experienced in beef cattle-producing regions such as Texas.
As ACTN3 is expressed only in type II fibers, the fiber type data are consistent with our expression data. The inefficient group also had a larger standard error than the efficient group. A possible explanation for this variability in expression could be that an animal may be inefficient, or just simply average, due to a wide array of combined genotypic, environmental, and other factors. In contrast, an efficient animal might be expected to possess only specific genotype combinations that, in conjunction with certain environmental conditions, return an efficient phenotype. Fiber type ratio is also just one variable among many that could possibly reduce overall efficiency.
A role for ACTN3 variation in growth and metabolism is supported by studies in other species, including humans. A relationship between ACTN3 expression and metabolic rates has been reported in rodents (MacArthur et al., 2007; MacArthur et al., 2008; Ogura et al., 2009) and humans (North and Beggs 1996; North et al., 2003; Yang et al., 2003). Additionally, the relationship between the ACTN3 R577X (loss of function) polymorphism and elite athletic performance has been described in humans (Roth et al., 2008; Ahmetov et al., 2010; Fiuza-Luces et al., 2011). Interestingly, loss of function has been shown to be associated with the reduced cross-sectional area and thigh muscle volume in older women (Min et al., 2016; Kiuchi et al., 2021).
Among all steers with records, across all seasons in the study (n = 173), from which a subset was used for gene expression assays, parent and breed of origin of the ACTN3 haplotype block had a significant impact on efficiency as measured by RFI residuals. The Nellore (Bos indicus) haplotype block was associated with greater efficiency when inherited maternally (RFI residual −0.47 for maternal Nellore inheritance compared to 0.49) regardless of the breed of origin of the paternal haplotype block. If efficiency is represented in terms of feed efficiency during a 180-day feedlot finishing period in kg/d, at current US prices of $0.352/kg, this difference will result in about $63 savings per animal for the most efficient animals. Additionally, the Nellore haplotype blocks of CAPN2 and FN1 were associated with differences in efficiency when inherited maternally. For CAPN2, a maternally inherited Nellore haplotype was linked to improved efficiency (RFI residual −0.50 compared to 0.50). The FN1 Angus haplotype block was associated with improved efficiency relative to the Nellore (−0.40 compared to 0.50). No paternal haplotype blocks were implicated to play a role in this trait. These results suggest that parent of origin effects or other epigenetic effects may play an important role in the efficiency phenotype and should be investigated further to optimize management practices. Reciprocal differences in birthweight between B. indicus X B. taurus and B. taurus X B. indicus offspring are well known (Dillon et al., 2015). Epigenetic differences in regulation of skeletal muscle growth have also been described, notably in callipyge sheep, where certain transcripts detected in skeletal muscle are transcribed from only a single parental allele (Bidwell et al., 2004). Although feed efficiency data are available only for the F2 steers utilized in this study and not the parent/grandparent generations nor sibling heifers, these findings are relevant for beef cattle production strategies in subtropical regions. It will be necessary to understand the impact of these same regions on heifers, replacement cows, and future sires. Additionally, these data begin to contribute to the understanding of parent-of-origin effects on the inheritance of chromosomal regions—particularly important for regions where Bos taurus–Bos indicus cross cattle are utilized.
Regarding fiber types, fast- and slow-twitch muscle fibers rely primarily on different metabolic pathways. Fast-twitch fibers (type II) rely on the anaerobic degradation of glucose for energy (glycolysis). In contrast, slow fibers (type I) utilize the aerobic citric acid cycle as a primary energy source. Due to the differences in metabolic pathways utilized, a shift in one direction can lead to overall differences in energy utilization during the lifespan of the animal. A reduction in ACTN3 can result in differences in glycogen phosphorylase activity in muscle and changes in calcium metabolism (MacArthur et al., 2007; Quinlan et al., 2009; Quinlan et al., 2010). Mice entirely deficient in Actn3 show an increase in expression of enzymes relating to the glycolytic pathway and a decrease in expression of enzymes of the aerobic cellular respiration pathway (MacArthur et al., 2008). Aerobic cellular respiration in total produces 38 molecules of ATP per molecule of glucose input compared to only two molecules of ATP produced by glycolysis per molecule of glucose, making aerobic metabolism 19 times more efficient than glycolysis. Therefore, any shift towards one over the other may affect the overall efficiency of energy metabolism. A greater abundance of ACTN3 and Type II fibers may contribute mechanistically to seasonal differences.
Cattle facing insufficient nutrition degrade fast-twitch muscle fibers initially to preserve slow-twitch muscle fibers (Lehnert et al., 2006), suggesting a preference for higher efficiency muscle under periods of nutritional stress. To our knowledge, a role for ACTN3 expression specifically influencing bovine metabolic efficiency has not been previously described. However, Nolte et al., 2019 examined biological networks to identify key long non-coding RNAs (lncRNA) associated with bovine metabolic efficiency. Of three key lncRNAs, one lncRNA with connectivity to low metabolic efficiency (MSTRG.10337) and expressed in muscle tissue was correlated with calcium signaling and other skeletal muscle genes, including ACTN3. It is unclear what role CAPN2 and FN1 might play in affecting efficiency, but these results are also consistent with the Nolte study of a potential role for calcium and cytoskeletal signaling. Because both CAPN2 and FN1 are associated with muscle turnover and growth, it is possible they are linked to differences in the rates of these physiological functions, which would, in turn, alter the metabolic rate of the animal. Interestingly, Karisa et al. (2013) demonstrated the association of CAST genes with efficiency. The animals in their study were crossbred Bos taurus breeds reared in Canada. Potentially, the favorable Bos indicus ACNT3, CAPN2, and FN1 haplotypes reflect the more favorable suitability of these breeds for subtropical environments.
In this experiment, we demonstrated that, in skeletal muscle of Nellore-Angus F2 steers, samples from animals classified as feed efficient (relative to RFINRC) contained fewer ACTN3 transcripts in comparison to the inefficient group. Similarly, muscle samples from the efficient group possessed fewer type II muscle fibers than the inefficient group. Maternal inheritance of the Nellore Bos indicus ACTN3 haplotype block conferred the greatest improvement in efficiency. Two additional genes involved in skeletal muscle turnover, CAPN2 and FN1, were also associated with improved efficiency when inherited as maternal Bos indicus haplotype blocks. Collectively these data demonstrate that expression of ACTN3, the gene network in which it resides, and its regulation, may represent a novel target for improving feed efficiency in cattle, particularly in subtropical environments. While ACTN3 may reflect just one element of a trait, these data, in connection with other studies, help provide a base for a better understanding of complex biological mechanisms and their interaction with fluctuating climatic conditions.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56705, GSE56705.
Ethics Statement
All procedures involving animals were approved by the Texas A&M AgriLife Institutional Animal Care and Use Committee.
Author Contributions
PR and RV designed the study. CG, DR, AH, and JS provided animal genotype and phenotype data and designed the genetic mapping herd structure. RV, KK, and AT prepared samples and conducted gene expression analyses. MD conducted fiber type analysis. RV conducted data analysis. PR, CG, and DR designed and supervised experiments and data analysis. RV and PR wrote the manuscript with input from all authors.
Funding
Financial support for this project was provided, in part, by the Beef Competitiveness Exceptional Item funding from the Texas legislature. Genotypes utilized for analyses were produced with support from The Beef Checkoff and National Research Initiative Competitive Grant no. 2008-35205-18767 from the USDA National Institute of Food and Agriculture Animal Genome Program. RV was partially supported by the Whole Systems Genomics Initiative for Human, Animal, and Environmental Wellbeing at Texas A&M University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors gratefully acknowledge numerous undergraduate and graduate students, staff, and faculty colleagues at Texas A&M who contributed to sample and data collection for this research project. This work was included as a portion of the doctoral dissertation of RV, in partial fulfillment of the degree requirements.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.796038/full#supplementary-material
References
Ahmetov, II, Druzhevskaya, A. M., Astratenkova, I. V., Popov, D. V., Vinogradova, O. L., and Rogozkin, V. A. (2010). The ACTN3 R577X Polymorphism in Russian Endurance Athletes. Br. J. Sports Med. 44, 649–652. doi:10.1136/bjsm.2008.051540
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic Local Alignment Search Tool. J. Mol. Biol. 215, 403–410. doi:10.1016/s0022-2836(05)80360-2
Amen, T. S. (2007). Feed Efficiency, Carcass and Temperament Traits in F2 Nellore-Angus Steers. Doctoral Dissertation. College Station (TX): Texas A&M University.
Bamman, M. M., Clarke, M. S. F., Talmadge, R. J., and Feeback, D. L. (1999). Enhanced Protein Electrophoresis Technique for Separating Human Skeletal Muscle Myosin Heavy Chain Isoforms. Electrophoresis 20, 466–468. doi:10.1002/(sici)1522-2683(19990301)20:3<466aid-elps466>3.0.co;2-7
Becker, G. S. 2008. Livestock Feed Costs: Concerns and Options. CRS Report for Congress. Available at: http://congressionalresearch.com/RS22908/document.php?studyDLivestockCFeedCCostsCConcernsCandCOptions (Accessed September 17, 2008)
Bidwell, C. A., Kramer, L. N., Perkins, A. C., Hadfield, T. S., Moody, D. E., and Cockett, N. E. (2004). Expression of PEG11 and PEG11AS Transcripts in Normal and Callipyge Sheep. BMC Biol. 2, 17. doi:10.1186/1741-7007-2-17
Brito, L. F., Bedere, N., Douhard, F., Oliveira, H. R., Arnal, M., Peñagaricano, F., et al. (2021). Genetic Selection of High-Yielding Dairy Cattle Toward Sustainable Farming Systems in a Rapidly Changing World. Animal 15, 100292. doi:10.1016/j.animal.2021.100292
Brito, L. F., Oliveira, H. R., Houlahan, K., Fonseca, P. A. S., Lam, S., Butty, A. M., et al. (2020). Genetic Mechanisms Underlying Feed Utilization and Implementation of Genomic Selection for Improved Feed Efficiency in Dairy Cattle. Can. J. Anim. Sci. 100, 587–604. doi:10.1139/cjas-2019-0193
Brunes, L. C., Baldi, F., Lopes, F. B., Lôbo, R. B., Espigolan, R., Costa, M. F. O., et al. (2021). Weighted Single‐step Genome‐wide Association Study and Pathway Analyses for Feed Efficiency Traits in Nellore Cattle. J. Anim. Breed. Genet. 138, 23–44. doi:10.1111/jbg.12496
Calender, A., Weichhart, T., Valeyre, D., and Pacheco, Y. (2020). Current Insights in Genetics of Sarcoidosis: Functional and Clinical Impacts. Jcm 9, 2633. doi:10.3390/jcm9082633
Capper, J. L. (2011). The Environmental Impact of Beef Production in the United States: 1977 Compared with 2007. J. Anim. Sci. 89, 4249–4261. doi:10.2527/jas.2010-3784
Cruzen, S. M., Harris, A. J., Hollinger, K., Punt, R. M., Grubbs, J. K., Selsby, J. T., et al. (2013). Evidence of Decreased Muscle Protein Turnover in Gilts Selected for Low Residual Feed Intake. J. Anim. Sci. 91, 4007–4016. doi:10.2527/jas.2013-6413
De Jonge, H. J. M., Fehrmann, R. S. N., de Bont, E. S. J. M., Hofstra, R. M. W., Gerbens, F., Kamps, W. A., et al. (2007). Evidence Based Selection of Housekeeping Genes. PLoS One 2, e898. doi:10.1371/journal.pone.0000898
Dillon, J. A., Riley, D. G., Herring, A. D., Sanders, J. O., and Thallman, R. M. (2015). Genetic Effects on Birth Weight in Reciprocal Brahman-Simmental Crossbred Calves. J. Anim. Sci. 93, 553–561. doi:10.2527/jas.2014-8525
Egan, B., and Zierath, J. R. (2013). Exercise Metabolism and the Molecular Regulation of Skeletal Muscle Adaptation. Cel Metab. 17, 162–184. doi:10.1016/j.cmet.2012.12.012
FAO (2018). “Shaping the Future of Livestock,” in 10th Global Forum for Food and Agriculture (GFFA), Berlin, January 18–20, (Food and Agriculture Organization of the United Nations). Available at: https://www.fao.org/publications/card/en/c/I8384EN/.
Fiuza-Luces, C., Ruiz, J. R., Rodríguez-Romo, G., Santiago, C., Gómez-Gallego, F., Yvert, T., et al. (2011). Are 'Endurance' Alleles 'Survival' Alleles? Insights from the ACTN3 R577X Polymorphism. PLoS One 6, e17558. doi:10.1371/journal.pone.0017558
Goll, D. E., Thompson, V. F., Li, H., Wei, W., and Cong, J. (2003). The Calpain System. Physiol. Rev. 83, 731–801. doi:10.1152/physrev.00029.2002
Greenbaum, D., Jansen, R., and Gerstein, M. (2002). Analysis of mRNA Expression and Protein Abundance Data: An Approach for the Comparison of the Enrichment of Features in the Cellular Population of Proteins and Transcripts. Bioinformatics 18, 585–596. doi:10.1093/bioinformatics/18.4.585
Hiller, B., Hocquette, J.-F., Cassar-Malek, I., Nuernberg, G., and Nuernberg, K. (2012). Dietary N-3 PUFA Affect Lipid Metabolism and Tissue Function-Related Genes in Bovine Muscle. Br. J. Nutr. 108, 858–863. doi:10.1017/s0007114511006179
Ho, C. K. M., Malcolm, B., and Doyle, P. T. (2013). Potential Impacts of Negative Associative Effects between Concentrate Supplements, Pasture and Conserved Forage for Milk Production and Dairy Farm Profit. Anim. Prod. Sci. 53, 437–452. doi:10.1071/an12140
Karisa, B. K., Thomson, J., Wang, Z., Stothard, P., Moore, S. S., and Plastow, G. S. (2013). Candidate Genes and Single Nucleotide Polymorphisms Associated with Variation in Residual Feed Intake in Beef Cattle. J. Anim. Sci. 91, 3502–3513. doi:10.2527/jas.2012-6170
Kenny, D. A., Fitzsimons, C., Waters, S. M., and McGee, M. (2018). Invited Review: Improving Feed Efficiency of Beef Cattle - The Current State of the Art and Future Challenges. Animal 12, 1815–1826. doi:10.1017/s1751731118000976
Kiuchi, Y., Makizako, H., Nakai, Y., Taniguchi, Y., Tomioka, K., Sato, N., et al. (2021). Associations of Alpha-Actinin-3 Genotype with Thigh Muscle Volume and Physical Performance in Older Adults with Sarcopenia or Pre-sarcopenia. Exp. Gerontol. 154, 111525. doi:10.1016/j.exger.2021.111525
Kochan, K. J., Vaughn, R. N., Amen, T. S., Abbey, C. A., Sanders, J. O., Lunt, D. K., et al. (2009). Expression of Mitochondrial Respiratory Complex Genes in Liver Tissue of Cattle with Different Feed Efficiency Phenotypes. Proc. Assoc. Advmt Anim. Breed. Genet. 18, 175–178.
Lam, S., Zeidan, J., Miglior, F., Suárez-Vega, A., Gómez-Redondo, I., Fonseca, P. A. S., et al. (2020). Development and Comparison of RNA-Sequencing Pipelines for More Accurate SNP Identification: Practical Example of Functional SNP Detection Associated with Feed Efficiency in Nellore Beef Cattle. BMC Genomics 21, 703. doi:10.1186/s12864-020-07107-7
Lehnert, S. A., Byrne, K. A., Reverter, A., Nattrass, G. S., Greenwood, P. L., Wang, Y. H., et al. (2006). Gene Expression Profiling of Bovine Skeletal Muscle in Response to and During Recovery from Chronic and Severe Undernutrition. J. Anim. Sci. 84, 3239–3250. doi:10.2527/jas.2006-192
Lek, M., Quinlan, K. G. R., and North, K. N. (2010). The Evolution of Skeletal Muscle Performance: Gene Duplication and Divergence of Human Sarcomeric α-Actinins. Bioessays 32, 17–25. doi:10.1002/bies.200900110
Li, A., Mo, D., Zhao, X., Jiang, W., Cong, P., He, Z., et al. (2013). Comparison of the Longissimus Muscle Proteome Between Obese and Lean Pigs at 180 Days. Mamm. Genome 24, 72–79. doi:10.1007/s00335-012-9440-0
Liu, M. F., Goonewardene, L. A., Bailey, D. R. C., Basarab, J. A., Kemp, R. A., Arthur, P. F., et al. (2000). A Study on the Variation of Feed Efficiency in Station Tested Beef Bulls. Can. J. Anim. Sci. 80, 435–441. doi:10.4141/a99-030
MacArthur, D. G., Seto, J. T., Chan, S., Quinlan, K. G. R., Raftery, J. M., Turner, N., et al. (2008). An Actn3 Knockout Mouse Provides Mechanistic Insights into the Association between -actinin-3 Deficiency and Human Athletic Performance. Hum. Mol. Genet. 17, 1076–1086. doi:10.1093/hmg/ddm380
MacArthur, D. G., Seto, J. T., Raftery, J. M., Quinlan, K. G., Huttley, G. A., Hook, J. W., et al. (2007). Loss of ACTN3 Gene Function Alters Mouse Muscle Metabolism and Shows Evidence of Positive Selection in Humans. Nat. Genet. 39, 1261–1265. doi:10.1038/ng2122
McClelland, S. C., Arndt, C., Gordon, D. R., and Thoma, G. (2018). Type and Number of Environmental Impact Categories Used in Livestock Life Cycle Assessment: A Systematic Review. Livestock Sci. 209, 39–45. doi:10.1016/j.livsci.2018.01.008
McDonagh, M. B., Herd, R. M., Richardson, E. C., Oddy, V. H., Archer, J. A., and Arthur, P. F. (2001). Meat Quality and the Calpain System of Feedlot Steers Following a Single Generation of Divergent Selection for Residual Feed Intake. Aust. J. Exp. Agric. 41, 1013–1021. doi:10.1071/ea00024
Mills, M., Yang, N., Weinberger, R., Vander Woude, D. L., Beggs, A. H., Easteal, S., et al. (2001). Differential Expression of the Actin-Binding Proteins, Alpha-Actinin-2 and -3, in Different Species: Implications for the Evolution of Functional Redundancy. Hum. Mol. Genet. 10, 1335–1346. doi:10.1093/hmg/10.13.1335
Min, S.-K., Lim, S.-T., and Kim, C.-S. (2016). Association of ACTN3 Polymorphisms with BMD, and Physical Fitness of Elderly Women. J. Phys. Ther. Sci. 28, 2731–2736. doi:10.1589/jpts.28.2731
Mujibi, F. D. N., Moore, S. S., Nkrumah, D. J., Wang, Z., and Basarab, J. A. (2010). Season of Testing and its Effect on Feed Intake and Efficiency in Growing Beef Cattle1. J. Anim. Sci. 88, 3789–3799. doi:10.2527/jas.2009-2407
Nolte, W., Weikard, R., Brunner, R. M., Albrecht, E., Hammon, H. M., Reverter, A., et al. (2019). Biological Network Approach for the Identification of Regulatory Long Non-coding RNAs Associated with Metabolic Efficiency in Cattle. Front. Genet. 10, 1130. doi:10.3389/fgene.2019.01130
North, K. N., Yang, N., Macarthur, D., Gulbin, J., and Easteal, S. (2003). “A Common Polymorphism in the Skeletal Muscle Gene ACTN3 Influences Human Athletic Performance,” in Meeting8th International Congress of the World-Muscle-Society, Sneged, Hungary, September 3–6, 2003 (Sponsor World Muscle Soc).
North, K. N., and Beggs, A. H. (1996). Deficiency of a Skeletal Muscle Isoform of α-actinin (α-Actinin-3) in Merosin-Positive Congenital Muscular Dystrophy. Neuromuscul. Disord. 6, 229–235. doi:10.1016/0960-8966(96)00361-6
NRC (2000). Nutrient Requirements of Beef Cattle. 7th Revised Edition. Washington, DC: The National Academies Press.
Ogura, Y., Naito, H., Kakigi, R., Akema, T., Sugiura, T., Katamoto, S., et al. (2009). Different Adaptations of Alpha-Actinin Isoforms to Exercise Training in Rat Skeletal Muscles. Acta Physiol. 196, 341–349. doi:10.1111/j.1748-1716.2008.01945.x
Ortigues, I. (1991). Adaptation du métabolisme énergétique des ruminants à la sous-alimentation. Quantification au niveau de l'animal entier et de tissus corporels. Reprod. Nutr. Dev. 31, 593–616. doi:10.1051/rnd:19910601
Puillet, L., Ducrocq, V., Friggens, N. C., and Amer, P. R. (2021). Exploring Underlying Drivers of Genotype by Environment Interactions in Feed Efficiency Traits for Dairy Cattle with a Mechanistic Model Involving Energy Acquisition and Allocation. J. Dairy Sci. 104, 5805–5816. doi:10.3168/jds.2020-19610
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 81, 559–575. doi:10.1086/519795
Quinlan, K. G. R., Seto, J. T., Turner, N., Floetenmeyer, M., Macarthur, D. G., Raftery, J. M., et al. (2009). G.O.4 α-Actinin-3 Regulates Muscle Glycogen Phosphorylase: A Potential Mechanism for the Metabolic Consequences of the Common Human Null Allele of ACTN3. Neuromuscul. Disord. 19, 545–546. doi:10.1016/j.nmd.2009.06.011
Quinlan, K. G. R., Seto, J. T., Turner, N., Vandebrouck, A., Floetenmeyer, M., Macarthur, D. G., et al. (2010). α-Actinin-3 Deficiency Results in Reduced Glycogen Phosphorylase Activity and Altered Calcium Handling in Skeletal Muscle. Hum. Mol. Genet. 19, 1335–1346. doi:10.1093/hmg/ddq010
Rajesh, R. V., Jang, E. J., Choi, I., Heo, K.-N., Yoon, D., Kim, T.-H., et al. (2011). Proteomic Analysis of Bovine Muscle Satellite Cells During Myogenic Differentiation. Asian Australas. J. Anim. Sci. 24, 1288–1302. doi:10.5713/ajas.2011.10344
Riggs, P. K., Tedeschi, L. O., Turner, N. D., Braga-Neto, U., and Jayaraman, A. (2017). The Role of 'omics Technologies for Livestock Sustainability. Archivos Latinoamerocanos de Prodccion Anim. 25, 149–155.
Riggs, P. K., and Vaughn, R. N. (2015). DNA Markers for Feed Efficiency in Cattle. United States Letters Patent, Application No. 14/724,696.
Riggs, P. K., Fields, M. J., and Cross, H. R. (2018). Food and Nutrient Security for a Growing Population. Anim. Front. 8, 3–4. doi:10.1093/af/vfy014
Ropka-Molik, K., Stefaniuk-Szmukier, M., Musiał, A. D., Piórkowska, K., and Szmatoła, T. (2019). Sequence Analysis and Expression Profiling of the Equine ACTN3 Gene During Exercise in Arabian Horses. Gene 685, 149–155. doi:10.1016/j.gene.2018.10.079
Roth, S. M., Walsh, S., Liu, D., Metter, E. J., Ferrucci, L., and Hurley, B. F. (2008). The ACTN3 R577X Nonsense Allele Is Under-represented in Elite-Level Strength Athletes. Eur. J. Hum. Genet. 16, 391–394. doi:10.1038/sj.ejhg.5201964
Savell, J. W., and Smith, G. C. (2000). Meat Science Laboratory Manual. 7th Edn. Boston, MA: American Press.
Stephens, M., Smith, N. J., and Donnelly, P. (2001). A New Statistical Method for Haplotype Reconstruction from Population Data. Am. J. Hum. Genet. 68, 978–989. doi:10.1086/319501
Tizioto, P. C., Coutinho, L. L., Oliveira, P. S. N., Cesar, A. S. M., Diniz, W. J. S., Lima, A. O., et al. (2016). Gene Expression Differences in Longissimus Muscle of Nelore Steers Genetically Divergent for Residual Feed Intake. Sci. Rep. 6, 39493. doi:10.1038/srep39493
Underwood, K. R., Tong, J., Zhu, M. J., Shen, Q. W., Means, W. J., Ford, S. P., et al. (2007). Relationship Between Kinase Phosphorylation, Muscle Fiber Typing, and Glycogen Accumulation in Longissimus Muscle of Beef Cattle with High and Low Intramuscular Fat. J. Agric. Food Chem. 55, 9698–9703. doi:10.1021/jf071573z
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 3, RESEARCH0034. doi:10.1186/gb-2002-3-7-research0034
Vaughn, R. N. (2013). Genetic Mechanisms that Contribute to Differences in Beef Tenderness Following Electrical Stimulation. Doctoral Dissertation. College Station (TX): Texas A&M University.
Wang, M., Yu, H., Kim, Y. S., Bidwell, C. A., and Kuang, S. (2012). Myostatin Facilitates Slow and Inhibits Fast Myosin Heavy Chain Expression During Myogenic Differentiation. Biochem. Biophysical Res. Commun. 426, 83–88. doi:10.1016/j.bbrc.2012.08.040
Welch, C. M., Thornton, K. J., Murdoch, G. K., Chapalamadugu, K. C., Schneider, C. S., Ahola, J. K., et al. (2013). An Examination of the Association of Serum IGF-I Concentration, Potential Candidate Genes, and Fiber Type Composition with Variation in Residual Feed Intake in Progeny of Red Angus Sires Divergent for Maintenance Energy EPD1. J. Anim. Sci. 91, 5626–5636. doi:10.2527/jas.2013-6609
Keywords: bovine, feed efficiency, fiber type, metabolism, gene network, SNP, ACTN3, sustainability
Citation: Vaughn RN, Kochan KJ, Torres AK, Du M, Riley DG, Gill CA, Herring AD, Sanders JO and Riggs PK (2022) Skeletal Muscle Expression of Actinin-3 (ACTN3) in Relation to Feed Efficiency Phenotype of F2 Bos indicus - Bos taurus Steers. Front. Genet. 13:796038. doi: 10.3389/fgene.2022.796038
Received: 15 October 2021; Accepted: 10 January 2022;
Published: 03 February 2022.
Edited by:
Marcos De Donato, Instituto de Tecnología y Educación Superior de Monterrey (ITESM), MexicoCopyright © 2022 Vaughn, Kochan, Torres, Du, Riley, Gill, Herring, Sanders and Riggs. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Penny K. Riggs, cmlnZ3NAdGFtdS5lZHU=
 Robert N. Vaughn
Robert N. Vaughn Kelli J. Kochan
Kelli J. Kochan Aline K. Torres
Aline K. Torres Min Du
Min Du David G. Riley1
David G. Riley1 Clare A. Gill
Clare A. Gill Penny K. Riggs
Penny K. Riggs