- Department of Biological Sciences, University of Manitoba, Winnipeg, MB, Canada
Freshwater ecosystems and fishes are enormous resources for human uses and biodiversity worldwide. However, anthropogenic climate change and factors such as dams and environmental contaminants threaten these freshwater systems. One way that researchers can address conservation issues in freshwater fishes is via integrative non-lethal movement research. We review different methods for studying movement, such as with acoustic telemetry. Methods for connecting movement and physiology are then reviewed, by using non-lethal tissue biopsies to assay environmental contaminants, isotope composition, protein metabolism, and gene expression. Methods for connecting movement and genetics are reviewed as well, such as by using population genetics or quantitative genetics and genome-wide association studies. We present further considerations for collecting molecular data, the ethical foundations of non-lethal sampling, integrative approaches to research, and management decisions. Ultimately, we argue that non-lethal sampling is effective for conducting integrative, movement-oriented research in freshwater fishes. This research has the potential for addressing critical issues in freshwater systems in the future.
Introduction
Communities around the world rely on freshwater ecosystems for natural resources, including impoverished groups that rely on freshwater resources for survival (Sultana et al., 2003; Millenium Ecosystem Assessment Board, 2005; Vo, 2016). Fresh water is used directly by humans in numerous ways, including drinking, farming, and transportation, but human demand for fresh water is increasing because of population growth and overexploitation (Strayer and Dudgeon, 2010; Dudgeon, 2019). Freshwater fishes are critical, and often overlooked, sources of food security for hundreds of millions of people globally, providing otherwise unavailable protein via freshwater fisheries (McIntyre et al., 2016; Dudgeon, 2019), in addition to essential nutrients for human diets such as omega-3 and omega-6 fatty acids (Steffens and Wirth, 2005). Given the importance of freshwater ecosystems to biological diversity and human resource needs, conservation issues in fresh water are paramount (Dudgeon, 2019; Su et al., 2021). Aquatic species are increasingly imperiled as human needs increase, an issue exacerbated by the tendency for the tragedy of the commons to be present among communities that use freshwater resources (Strayer and Dudgeon, 2010; Dudgeon, 2019). Human activities have changed natural habitats in ways that include overexploitation, physical alterations of natural structures and ecosystem components (e.g., dams, non-native species), as well as water pollution as a result of industrial development (Strayer, 2008; Dudgeon, 2019). Anthropogenic climate change has broad, insidious effects on freshwater ecosystems that include rising temperatures, changing flow patterns, and more frequent extreme events that are all already apparent throughout many freshwater systems (Dudgeon, 2019). Conservation action is therefore necessary in many freshwater systems. Integrated terrestrial-freshwater conservation planning can increase freshwater species protection by 600% with marginal drawbacks, demonstrating the effectiveness of conservation action in freshwater systems (Leal et al., 2020).
Freshwater ecosystems contribute around 40% of global fish diversity, 25% of global vertebrate diversity, and ∼10% of all known species despite covering <1% of the earth’s surface (Dudgeon et al., 2006; Strayer and Dudgeon, 2010; Reid A. J. et al., 2019; Radinger et al., 2019; Leal et al., 2020). This diversity is reflected in species richness, phylogenetic diversity, and functional diversity, which are disproportionately high for freshwater ecosystems (Dudgeon, 2019; Su et al., 2021). For example, the Mekong, Congo, and Amazon Rivers are some of the most fish-diverse systems globally with thousands of fishes in each, and are characterized by their relative abundances of fresh water (Sverdrup-Jensen, 2002). The great diversity of life present in fresh water contributes to valuable ecosystem services, but also underscores the importance of the conservation issues that freshwater ecosystems face (Dudgeon, 2019). Physical structures such as dams can impede movement for freshwater organisms, an issue exemplified by the observation that only 37% of rivers longer than 1,000 km remain free-flowing (Dudgeon, 2019; Grill et al., 2019). Responses to global climate change often require compensatory movements by freshwater organisms, such as in finding cool water (Dudgeon, 2019). These responses are stymied by impediments to movement in freshwater systems. One reason that movement is especially important in freshwater is that these habitats are often interspersed among terrestrial habitats, in a matrix characterized by varying levels of connectivity (Griffiths, 2015; Mushet et al., 2019).
Research on freshwater fishes therefore has the potential to address critical issues in human resource needs and conservation of biodiverse ecosystems. One potential avenue of research is the study of fish movement. Movement in general is a fundamental trait for many organisms, underlying survival, reproduction, and population persistence (Nathan et al., 2008). Moreover, movement with respect to individuals is foundational for considering other mechanistic components of movement, such as the internal state of the individual or the effects of the external environment (Nathan et al., 2008). Because non-lethal sampling provides an opportunity for physiological or genetic samples to be Because non-lethal sampling provides an opportunity for physiological or genetic samples to be taken from individuals whose movement may be tracked (Prince et al., 2017; Jeffries et al., 2021), non-lethal sampling is consistent with this framework of individual-focused movement as it can provide a snapshot of the internal state of the individual at the time of sampling.
The objective of this review is to support integrative research in freshwater fishes by presenting an overview of methods for studying movement in conjunction with the physiological and genetic approaches that have been linked with movement. Beyond specific methods, various factors that we consider important to this integrative movement research are discussed. This review is not intended to provide an exhaustive survey of the relevant literature. Instead, we highlight possible approaches for linking movement with other observations, broadly classified into physiology and genetics. First, Methods for Studying Movement in the context of tagging methods and movement data analyses are addressed as a foundational step for practicing research on fish movement. In Methods for Connecting Movement and Physiology, different examples of whole-organism and molecular physiology approaches to studying movement are provided as an overview of possible research avenues. In Methods for Connecting Movement and Genetics, population genetics, quantitative genetics, and association tests are presented in the context of movement research. In Further Considerations, fish handling, collecting molecular data, the ethics of non-lethal sampling, integrating data, and applications of movement research to management are presented as ways that movement-focused research may be more effectively practiced and used. For each type of sampling method, we discuss how the sample is collected, the types of movement-relevant information it can provide, and potentially ways the sample may be used to support conservation or management decisions with case studies from the literature. While this review is focused on freshwater fish movement and non-lethal sampling, several marine fish and also non-fish studies are discussed because they illustrate concepts relevant to freshwater fishes.
Methods for Studying Movement
Tagging Methods
Animal movement can be tracked with several methods (Table 1). These methods generally fall under two approaches: mark-recapture and telemetry. In mark-recapture, organisms are observed over multiple sampling events where small tags (e.g., anchor, visible implant elastomer (VIE), coded wire, or electronic tags) are attached to fish and used to individually identify these organisms at a tagging event and subsequent recapture (Hale and Gray, 1998; Walsh and Winkelman, 2004; Hohn and Petrie-Hanson, 2013; Thorstad et al., 2013; Cayuela et al., 2018). The mark-recapture approach is useful for estimating dispersals, but it also allows the modeling of population size through rates of recapture (Nichols, 1992; Cayuela et al., 2018). A related approach is a VIE tag, where coloured tags are injected into fish in externally visible places (Walsh and Winkelman, 2004; Hohn and Petrie-Hanson, 2013). In contrast to mark-recapture, telemetry offers a different approach for tracking movement. Both radio and acoustic telemetry arrays can be built for autonomous operation, enabling the tracking of multiple tagged individuals without a researcher needing to track each individually (reviewed in Cooke et al., 2008; Donaldson et al., 2014). As multiple fish species may be tagged across fresh water and salt water (Hussey et al., 2015), acoustic telemetry arrays have the potential for increased tracking of a variety of fish species and studying interactions among entire aquatic communities (Hussey et al., 2015; Enders et al., 2019; Rudolfsen et al., 2021). Acoustic tags sensitive to predation events are used to track predators as well, for a mean of 66.5 h or 75.6 h after predation depending on the model of tag used and ambient temperatures (Halfyard et al., 2017; Schultz et al., 2017). In one test of the technology, predation transmitters accurately verified a predation event 90% of the time, although mean time from feeding event to trigger time of the tag was 59.2 h (Schultz et al., 2017).
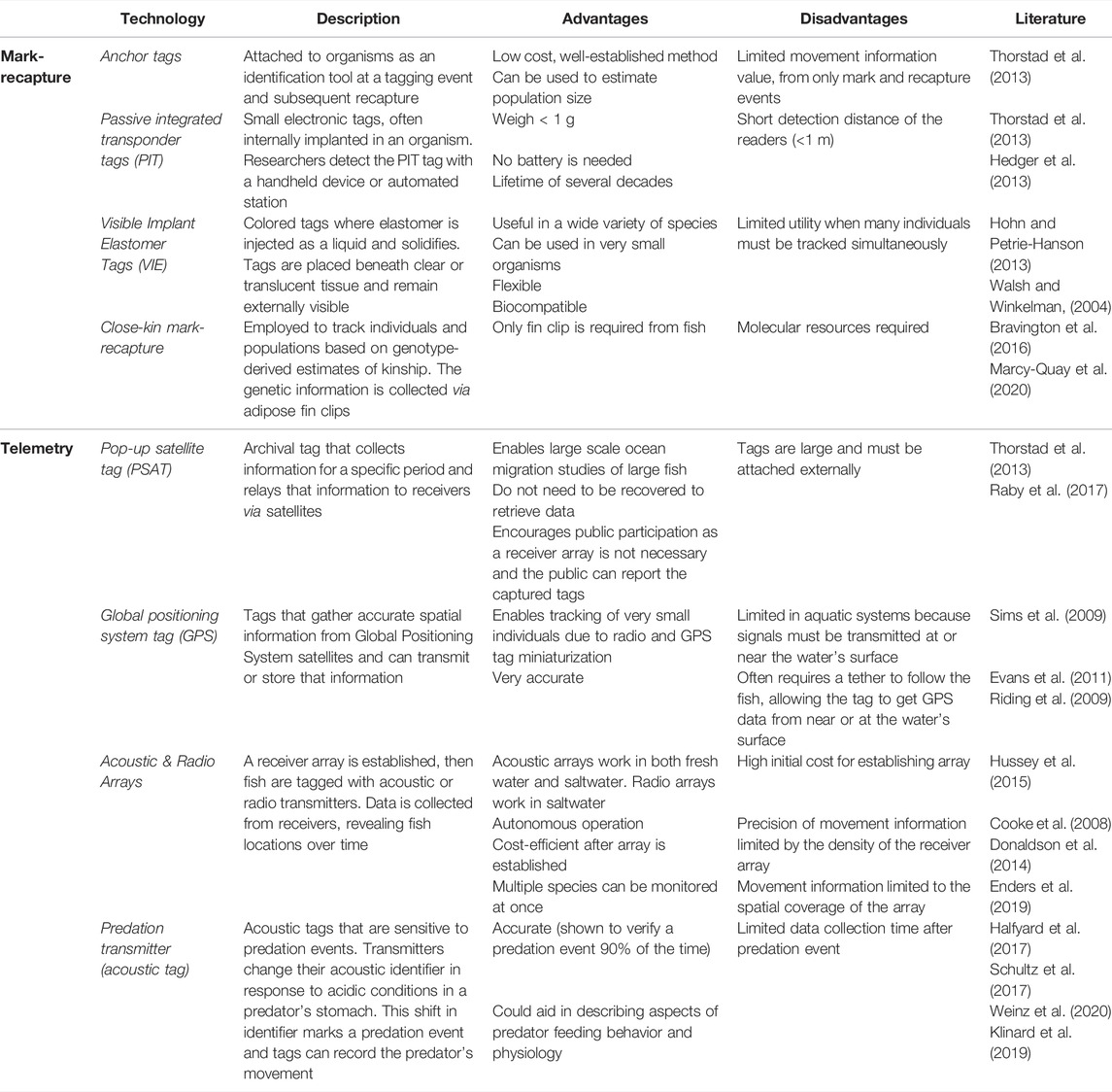
TABLE 1. Methods for studying movement in freshwater fishes. Methods are broadly classified into mark-recapture and telemetry-based approaches. Technology provides the commonly used name for each method and description provides a brief summary of how the method operates. Advantages, disadvantages, and representative literature for each method are also provided.
As capture, handling, and surgery can be acutely stressful for the animal, such procedures may induce an acute stress response resulting in physiological and behavioural perturbations (Wendelaar Bonga, 1997). This is an important consideration as tagging stress may contribute to higher rates of post-release predation/mortality and altered behavioural patterns (Mäkinen et al., 2000; Danylchuk et al., 2007; Le Pichon et al., 2015; Wilson et al., 2017), which would be deleterious to the project’s objectives. For example, Atlantic salmon (Salmo salar) caught through gill netting (i.e., slower time from capture to release, high potential for injury and mucus loss) or rod and reel angling (i.e., faster time from capture to release) exhibited distinct differences in post-release movement patterns, suggesting that capture methods do have a role in affecting acute behaviours (Mäkinen et al., 2000). To ensure that post-release behaviour/movement is marginally impacted, efforts should be made to ensure that capture and handling is minimized to reduce stress-related effects. The specific protocols used will be project-, species-, and context-dependent but should include measures that either limit handling/collection times or reduce injury to the fish. For example, rod and reel angling is used in a number of settings to collect animals for tagging (Heupel and Hueter, 2001; Gillis et al., 2010) as it has short capture durations, generally has a minor physiological impact on the fish (Brownscombe et al., 2017), and can allow for more targeted collection (c.f. gill nets or long lines). Similarly, using electro anaesthesia over chemical anaesthetics for tagging manipulations can also help to hasten tagging procedures owing to electro anaesthesia’s shorter induction and recovery times (Reid et al., 2019). In addition to reducing holding times, the tagging environment is also important in minimizing stress and injury to the animal. The fish should be held at its environmental temperature with its gills submerged to facilitate normal gas and ion exchanges. This can be achieved using a padded, water filled trough with a continual flow of fresh water, which promotes ease of access during the tagging procedure and less stress for the fish than handling without water flow (Lawrence et al., 2020). Censoring data in the days or weeks following tagging can minimize the effects of tagging stress represented in subsequent analyses of movement, as well (Wilson et al., 2017). Together, the use of proper collection methods, minimal handling, expediting tagging, and data handling can help to ensure that stress-related effects on post-release behaviours are minimized.
Movement Analyses
Movement data connects the studies in this review and its careful analysis is of fundamental importance for any project that seeks to connect movement with other non-lethally sampled variables. The field of movement ecology has developed sophisticated methods for analyzing such data. Previously, random walks with Hidden Markov Models were a common approach but more recently, connections between complex movement behaviours and landscape or geographical features have become more widely used (Schick et al., 2008; Seidel et al., 2018). Specialized statistical approaches have been developed, such as in assessing spatial autocorrelation and in specific classes of analysis techniques (reviewed in Long and Nelson, 2013). For example, just in the statistical computing environment R, 58 packages were identified as contributing to movement analyses, 11 of which had good or excellent documentation (Joo et al., 2020; R Core Team, 2021). The R package actel has been developed to reproducibly analyze acoustic telemetry array data, an increasingly common source of movement information in fishes (Flávio and Baktoft, 2021). Meanwhile, Bayesian state space models have been used to infer movement paths with telemetry data, showing a possible direction for movement analyses in the future (Munaweera Arachchilage et al., 2021). Regardless of the approach to movement analyses undertaken by researchers, careful interpretation of movement data is necessary for subsequent integrative analyses.
Methods for Connecting Movement and Physiology
Physiology and Movement
The biological impact of stressors associated with movement can be assessed through non-lethal sampling for transcriptomic (mRNA) and biochemical analyses (e.g., enzymes, hormones, osmolytes), as well as additional physiological data to investigate their roles at both the individual and population-level scales (Metcalfe et al., 2012; Connon et al., 2018; Jeffries et al., 2021). For example, salmonid (Oncorhynchus spp. & Salmo salar) migration has been studied in depth from physiological perspectives of thermal stress, cardiorespiratory performance, disease states, and stress responses using a combination of telemetry, physiological, and genomic methods (Cooke et al., 2008, 2012; Stich et al., 2015). Increased baseline levels of a commonly studied hormone, cortisol, has been linked to sooner river exit times but a lower likelihood of reaching the sea in Atlantic salmon and sea trout (S. trutta), with implications for a tradeoff between stress and migration readiness (Birnie-Gauvin et al., 2019). Elevation of cortisol and its decline during migration was consistent between landlocked kokanee salmon (Oncorhynchus nerka kennerlyi) and anadromous sockeye salmon (O. nerka nerka) sampled before, during, and after migration, providing evidence that cortisol is part of an endogenous system that is associated with migration and insights into the timing of movements in these species (Carruth et al., 2000). Physiological metrics have also been used to study Atlantic salmon, with investigation of smolt survival through telemetry and physiological assays demonstrating that individuals with the highest levels of gill Na+, K+-ATPase activity incurred 25% lower mortality than those with the lowest Na+, K+-ATPase activity, linking physiological preparedness for migration with survival success (Stich et al., 2015).
Similar to patterns in more southerly salmonids, arduous migratory journeys exacerbated by changing climates and stressors can influence selective pressures and alter the physiology of subsequent generations as observed in Arctic char (Salveliunus alpinus) in northern Canada. Using a combination of non-lethal sampling for genomics and telemetry, Moore et al. (2017) demonstrated increased selection pressures (via outlier markers) on cardiac genes of char as migratory length and elevation gain increased. These genes were related to skeletal muscle differentiation and heart development, actin binding in the sarcomere Z-disc, glycogen breakdown into glucose, and the conversion of glycerol to glucose (ibid). In a related study, physiological metrics of increased thermal maxima, increased aerobic capacity, and decreased heart mass were also associated with increased population migratory difficulty (Gilbert, 2020). Together these studies apply a variety of techniques linking genetics, physiological capacity, and telemetry to investigate the migratory challenges faced by different Arctic char populations. Their endurance capabilities for reproductive migration were assayed using a combination of non-lethal sampling, movement data, and classical physiological metrics.
Anthropogenic influences of aquaculture-related pathogens, contaminants, and water usage can alter the movement of fishes (Brander, 2007; Nathan et al., 2008; Bozinovic et al., 2011; Miller et al., 2014). Salmonids making reproductive migrations may be further challenged by increased, or novel, pathogen spillover from aquaculture facilities resulting in elevated infection pressure in a critical stage of their life cycle (Cicco et al., 2018; Mordecai et al., 2021). Additionally, hormones such as 17β-estradiol can be introduced into aquatic systems from anthropogenic activities and have been demonstrated to alter migration behavior in both Atlantic and sockeye salmon (Madsen et al., 2004; Veldhoen et al., 2013a). Infectious pathogen presence, populations, and abundance have been assessed using non-lethal sampling, using transcriptomic approaches paired with telemetry, to evaluate increasing predation and limited reproductive success in stressed and infected salmonids throughout their migrations (Jeffries et al., 2014; Furey et al., 2021; Vollset et al., 2021).
In freshwater systems, anthropogenic water allocation can impact the movement and impose restrictions to swimming passage, limiting the successful progression of fishes throughout their lifecycle. Dams and other diversions can lead to habitat fragmentation or alter flow states, decreasing hydrological connectivity and restricting fishes from habitat critically important for their survival and reproduction (Yoshiyama et al., 1998; Jeffres et al., 2006; McKay et al., 2013; O’Hanley et al., 2013). In sockeye salmon monitored while crossing dam passages, using a combination of telemetry and biomonitoring, increased oxygen consumption (MO2) and anaerobic glycolysis while ascending the fishway indicated higher probability of mortality, while lower plasma glucose, earlier migration, and longer lake residency time upstream of the dam increased the probability that migrating fish reached their spawning sites (Roscoe et al., 2011; Burnett et al., 2014, 2017; Bett et al., 2020). For species like sturgeons, with poor swimming capabilities and lower success passing through dams, spillways, and diversions, these passages and the restrictions that they impose can lead to direct mortality or increased physiological stress leaving them vulnerable to predation as juveniles (Peake et al., 1997; Cheong et al., 2006; Cocherell et al., 2011; Poletto et al., 2014; Katopodis et al., 2019). By pairing movement data with non-lethal sampling and physiological metrics successfully, researchers can clarify the processes influencing, allowing, or promoting specific movement trajectories in response to anthropogenic water allocation.
Tissue Biopsies
Tissue biopsies are an effective means to collect physiological, molecular, and environmental data for use in spatial movement projects. They can non-lethally provide insight into how physiological and environmental characteristics influence fish behaviour, spatial use, and ecological interactions, thereby linking multiple levels of biological scale (Cooke et al., 2008; Donaldson et al., 2014). As well, given their minimally invasive nature, the use of non-lethal biopsies is useful in settings where lethal samples are not obtainable, as in the case of species at risk, or where obtaining unadulterated behavioural/movement-based data is the key aim of the project. The non-lethal nature of such samples also means that large sample sizes can be readily obtained without substantially diminishing a studied population (Henderson et al., 2016). Non-lethal tissue biopsies in contemporary ecological studies typically sample tissues that are easily accessible and minimize stress on the animal.
One of the most widespread biopsy approaches in spatial ecology involves the use of blood samples (Figure 1; Caputo et al., 2009; O’Toole et al., 2012; Taylor and Litvak, 2017; Thorstensen et al., 2021). While specific details on sampling methods can be found elsewhere (Houston, 1990; Lawrence et al., 2020), this approach is advantageous as it is quick, minimally invasive, and can provide insight into the fish’s physiological state (reviewed in Lawrence et al., 2020). The physiological metrics obtained from blood samples can be used to inform the proximate mechanisms driving fish movement and behavioural dynamics. For example, the use of blood sampling has been an integral component in understanding fisheries interactions and spawning migration of Pacific salmonids showing that indices of stress often correspond with poor migration success (Cooke et al., 2006, 2008; Drenner et al., 2018; Bass et al., 2019). In a recreational fishing context, blood sampling has been coupled with telemetry and monitoring to characterize the factors contributing to post-release survivorship and behaviour (Arlinghaus et al., 2009; Heberer et al., 2010; Tracey et al., 2016). Blood samples can also be used to indirectly characterize foraging and habitat use patterns through isotopic analyses when paired with telemetry datasets (Matich and Heithaus, 2014; Shipley et al., 2021). Blood sampling on live fish often involves caudal puncture where a small needle/syringe is inserted through the ventral surface of the caudal peduncle, piercing the dorsal vasculature (Lawrence et al., 2020). This method applies to a wide range of species, sizes, and settings, with smaller sized fishes requiring a slightly modified approach (e.g., lateral blood sampling in zebrafish; Zang et al., 2013, 2015). The sampling should occur in an environment that minimizes stress on the animal, restrains the fish adequately, and enhances ease of blood sampling. This is often achieved using a padded V-trough that the fish is held ventral side up to facilitate sampling access. The gills of fish can be artificially irrigated with water, or the trough filled with water to allow oxygen exchange (see Lawrence et al., 2020). As this method is minimally invasive, mortality rates are generally considered to be low making it an effective technique for non-lethal sampling (Lawrence et al., 2020).
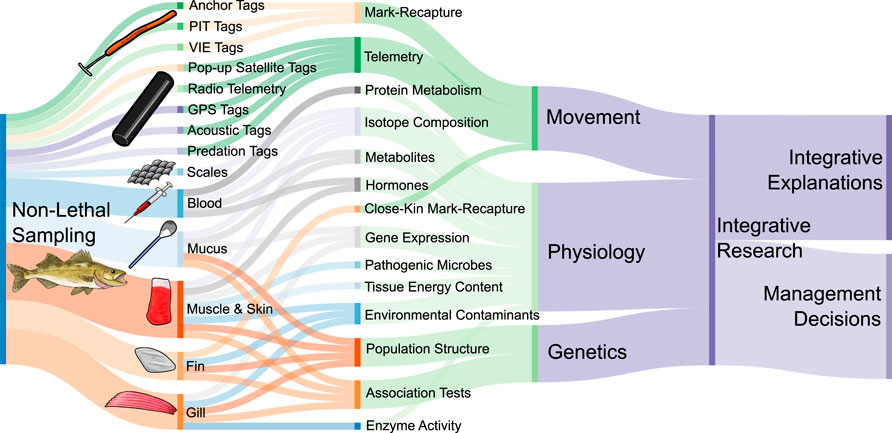
FIGURE 1. Conceptual diagram of different tagging and tissue sampling methods used for non-lethal sampling and movement studies in fishes. Specific sampling and tagging methods are linked to overall approaches in research, although for simplicity, only approaches presented in this review with examples are shown here. The different research approaches are categorized into movement, physiology, and genetics, which can be incorporated into integrative research projects. These integrative projects are useful for making informed management decisions and finding integrative explanations of natural phenomena. Therefore, movement, physiology, and genetics can all inform integrative explanations and management decisions via integrative research. Colour choices are arbitrary, and only for distinguishing elements of the diagram. Created with SankeyMATIC.
Another common non-lethal method in fish biology includes the use of gill biopsies, which involves the removal of a small portion of the distal end of several gill filaments (McCormick, 1993). This approach is a relatively non-invasive means of tissue sampling and appears to have limited consequence on the fish’s post-release survival or growth/condition indices when compared to handling controls (McCormick, 1993; Martinelli-Liedtke et al., 1999; Cooke et al., 2005; Jeffries et al., 2014). However, some work has demonstrated that biopsied fish had higher post-release mortality rates, relative to non-biopsied conspecifics (Bass et al., 2020). As with blood sampling, a large breadth of information can be obtained from gill biopsies that can be informative in movement-based projects. Gill biopsies are being used extensively to quantify pathogenic microbes and host biomarkers of disease in Pacific salmonids (Miller et al., 2014; Chapman et al., 2021). Using molecular toolsets (e.g., qPCR, RNA-seq), gene expression patterns related to disease in the gill have been coupled with telemetry datasets, with links between disease burdens and migration activities (Jeffries et al., 2014; Teffer and Miller, 2019; Chapman et al., 2020; Stevenson et al., 2020; reviewed in; Chapman et al., 2021). Because the gill is one of the most important structures in regulating ion and acid base balance, waste excretion, and oxygen uptake (Evans et al., 2005), gill biopsies can also be used to derive important physiological responses to environmental challenges. This often includes quantifying activities of relevant enzymes/transporters (e.g., Na+/K+ ATPase; McCormick, 1993; Zhao et al., 2014; Gesto et al., 2015) or by assaying gene expression patterns (Rees et al., 2005; Norman et al., 2014; Guo et al., 2018). Consequently, gill biopsies can be useful in assaying responses to a wide array of treatments and environmental challenges (reviewed in Jeffries et al., 2021). In spatial ecology specifically, gene expression analyses can be combined with telemetry data to provide a more comprehensive assessment of how fine scale transcriptomic changes can influence fish movement patterns and behaviour (Madsen et al., 2004; Crossin et al., 2009; Evans et al., 2011). In some cases, gill biopsies have been proposed as an alternative to blood sampling as a means of collecting physiological metrics such as cortisol (Gesto et al., 2015). Similar to blood biopsies, non-lethal gill biopsies may not be feasible in smaller fishes, which might require that the animal be sacrificed to obtain a sample (ibid). However, this approach has been successfully used to take a non-lethal gill biopsy from sockeye salmon smolts (Jeffries et al., 2014).
Muscle, skin, and scale biopsies could also play an important role in movement-based studies (Figure 1). These techniques employ a similar approach, where a small tissue sample is removed using either a medical-grade biopsy punch device (e.g., Crawford et al., 1977; Tyus et al., 1999; Smith et al., 2015; Henderson et al., 2016; Bøe et al., 2020) or, in the case of skin from fin samples specifically, simply clipping a portion of the fish’s fin off (Sanderson et al., 2009; Galván et al., 2015; Wang et al., 2017). Scale samples may be removed from the exterior of the fish (Cunjak et al., 2005; TinHan et al., 2018). Both skin and muscle biopsies are minimally invasive and appear to have little impact on the fish’s health and post-release mortality with wounds healing within weeks in some instances (Crawford et al., 1977; Tyus et al., 1999; Pratt and Fox, 2002; Henderson et al., 2016; Bøe et al., 2020).
Non-lethal skin and muscle samples can provide a variety of metrics that are useful in spatial ecology projects. Historically, tissues derived from muscle, scale, and skin biopsies have been used in the quantification of accumulated environmental toxicants (Aerts et al., 2015; Alves et al., 2016; Gelsleichter et al., 2020; Charette et al., 2021; Stahl et al., 2021), isotopic compositions (Cunjak et al., 2005; Kim et al., 2012; Busst et al., 2015; Peterson et al., 2017; TinHan et al., 2018), pathogenic microbes (Bowers et al., 2008; Elliott et al., 2015), DNA (John Nelson et al., 2003; Dominik et al., 2010; Smith et al., 2018; Thorstensen et al., 2019), and physiological metrics such as hormonal levels and tissue energy contents (Fagan et al., 2012; Olsen et al., 2013; Verkamp et al., 2021). Koi (Cyprinus carpio) mucus was assayed for 11-ketotestosterone (Schultz et al., 2005), while mucus was also useful for stable isotopes analysis of rainbow trout diet (Salmo gairdneri) (Church et al., 2009), and DNA in Nile tilapia (Oreochromis niloticus) (Taslima et al., 2016). While these tissues can provide a great deal of insight, it is important to ensure that values derived from such samples are interpreted within the right context. For example, when measuring levels of mercury in skin and muscle biopsies in freshwater teleosts, the degree to which these values represent whole muscle mercury concentrations can vary (Baker et al., 2004; Piraino and Taylor, 2013; Stahl et al., 2021), highlighting the need for preliminary analysis and calibration. To date though, the use of non-lethal skin and muscle biopsies in movement-based studies appears limited, but biopsies provide a wealth of context-dependent biological information. Some potential links between biopsies and movement are presented in the following sections.
Environmental Contaminants
Evaluating the effects of contaminants in fish often involves the removal of liver and/or muscle tissues during lethal sampling to examine chemical or biological indicators of toxicity (reviewed in Gray et al., 2002). These lethal samples can limit the quantity of samples one is able to collect, experimental design options, target species (i.e. species at risk), repeatability, and long-term analysis of individuals (Gray et al., 2002; Sanderson et al., 2009; Rodríguez-Jorquera et al., 2019; Jeffries et al., 2021). Because of these limitations, using non-lethal and minimally-invasive methods has become more common practice (Jeffries et al., 2021). Molecular responses to a range of contaminant exposure have been tested with many non-lethal samples, including muscle (Veldhoen et al., 2010; Somerville et al., 2020), fins (Andreasen et al., 2006; Veldhoen et al., 2013b, 2014; Baillon et al., 2016), mucus (Arukwe and Røe, 2008; Bahamonde et al., 2019; Bulloch et al., 2020), blood and plasma (Arukwe and Røe, 2008; Rodríguez-Jorquera et al., 2019), gill (Rees et al., 2005; Barriga-Vallejo et al., 2017), and scales (Aerts et al., 2015; Alves et al., 2016) (Figure 1). However, when it comes to the intersection of contaminants, non-lethal sampling, and molecular work, there are few examples that analyze aspects of movement. Contaminants can affect anadromous migrations, along with temperature changes, pathogens, and a multitude of other stressors, and therefore alter reproductive success and population survival (Veldhoen et al., 2013a). For example, Madsen et al. (2004) exposed Atlantic salmon to 17β-estradiol and 4-nonylphenol to evaluate enzyme activity, smolting, and migration success. Using non-lethal gill samples at multiple time points, Na+,K+-ATPase mRNA levels were analysed and found to be depressed in the 17β-estradiol and 4-nonylphenol treatments during smolting and migration. They also found that 17β-estradiol and 4-nonylphenol delayed smolting and migration, and increased mortality during migration compared to control fish. Exposure to contaminants has also been found to heighten sensitivity to external stressors, which may impact the long-term sustainability of migrating salmon populations (Lerner et al., 2007).
Locations of high residency time (e.g., spawning areas, areas with high food availability, migration routes) can also affect the contaminant burdens individuals face (Hoffman et al., 2020). For example, Heltsley et al. (2005) used mobile passive sampling devices (PSD) attached to tags on flathead catfish (Pylodictis olivaris) to determine location-specific exposure to polychlorinated biphenyls (PCBs) and organochlorine pesticides. To measure these concentrations, they evaluated if adipose fins could be used in replacement of lethal muscle samples. They found that the concentrations of PCBs and organochlorine pesticides in adipose fin samples correlated to lethal muscle tissue samples, providing an appropriate minimally invasive substitute for this species. They also found contaminate concentrations in the mobile PSDs correlate to the concentrations found in the stationary PSDs in several locations with varying PCB and organochlorine pesticides concentrations. The concept that the movement ecology of a species can affect their contaminant burden is significant, particularly in the context of point source pollution. Reduced movement or increased use of habitat at a point source of pollution may contribute to increased contaminant burden. By contrast, more transient movements through such an area are less likely to influence individuals’ contaminant burdens. These connections between contaminant burden and movement may have long term impacts on fish populations that can be monitored through physiological metrics.
Isotopes
Stable isotope levels derived from non-lethally collected fin clips, scales, and skin have also been used to inform habitat use and foraging patterns in fishes when coupled with movement data (Cunjak et al., 2005; TinHan et al., 2018; Eggenberger et al., 2019; Aschettino et al., 2020). For example, stable isotopes in conjunction with PIT tags were used to distinguish between movement for foraging or to seek cool water refugia in Atlantic salmon (Cunjak et al., 2005). Here, δ13C measured from muscle tissue was associated with downstream sites along the river system (Gray et al., 2004; Cunjak et al., 2005). Acoustic telemetry was paired with isotope analyses to determine that common snook (Centropomus undecimalis) of higher trophic levels moved more often into different zones than snook of lower trophic levels (Eggenberger et al., 2019). Similarly, research in seatrout (Cynoscion nebulosus), using acoustic telemetry paired with isotopic signatures from scales, revealed that fish resident to a recently restored oyster reef were larger and had narrower isotopic niches than their smaller, less reef-residential counterparts (TinHan et al., 2018). Examination of humpback whale (Megaptera novaeangliae) skin samples as well as satellite telemetry throughout the Chesapeake Bay, showed that whales utilize the area for foraging commonly in similar areas to shipping channels, demonstrating related isotopic signatures to whales feeding in the Gulf of St. Lawrence on likely similar prey items (Aschettino et al., 2020). Stable isotopes analyzed in conjunction with movement thus have wide ranging uses across a variety of species with the potential to reveal the ways diet, habitat use, and movement interact.
Protein Metabolism
Biochemical and metabolic connections with movement have been observed in several fishes, such as in tradeoffs between activity and growth (in yellow perch; Rennie et al., 2005), increased reliance on protein for energy during extended swimming at elevated speeds (in Nile tilapia; Alsop et al., 1999), and protein degradation and synthesis each increased with swimming activity (in rainbow trout; Houlihan and Laurent, 1987). Related to protein metabolism, migration has been linked to increased amino acid and carbon flux in sockeye salmon (reviewed for salmon in Mommsen, 2004). Protein degradation was indirectly linked to walleye (Sander vitreus) movement across a large freshwater lake by non-lethal whole blood biopsy (Thorstensen et al., 2021). We speculate that while physiology and biochemistry are certainly affected by and affect movement, specific pathways used to address movement likely differ between species, possibly based on phylogenetic constraints and life history strategies. Protein metabolism has been assessed non-lethally through observations of urea and ammonia excretion in an experimental tank (Alsop et al., 1999), or careful selection of a metabolite panel (Thorstensen et al., 2021). However, research using non-lethal sampling applied to protein metabolism is in its infancy.
Gene Expression
Internal and external stimuli lead to cellular responses such as changes in gene expression, which reflect the physiological state of an organism (reviewed in Szymański and Barciszewski, 2003; Jeffries et al., 2021). This information on physiological state may be linked to movement in freshwater fishes (see Physiology and Movement). Gene expression can be measured by quantifying messenger RNA (mRNA) abundance, which can be sampled from tissues non-lethally (Jeffries et al., 2021). Therefore, techniques that quantify gene activity via mRNA expression have great potential for linking movement and physiological state non-lethally in animals.
Quantitative polymerase chain reaction (qPCR), microarrays, and mRNA sequencing are all techniques that have been used to study gene expression and have been paired with movement (Mantione et al., 2014). With these approaches, the abundance of individual genes in groups of fish relative to others are compared, with possible implications for functional response pathways (Miller et al., 2011; Jeffries et al., 2014; Connon et al., 2018). For example, in migrating sockeye salmon, gill gene expression was correlated with environmental changes throughout migration (Evans et al., 2011), and predictive of migration completion (Miller et al., 2011; Jeffries et al., 2014), demonstrating an application of mRNA sequencing for revealing factors influencing migration success. Similarly, hatchery reared steelhead (O. mykiss) migration survival was predicted via elevated transcript abundance of inflammatory responsive genes in the gill, via qPCR, associating gene expression responses with mortality or successful migration (Healy et al., 2018). The combination of these technologies and transcriptional markers for environmental stress, immunity, and pathogen presence provides predictive power, and enable risk assessments of predation throughout migration. These approaches further link molecular signatures of disease status and stress to interspecific predator-prey interactions, especially in wild salmonids (Jeffries et al., 2014; Miller et al., 2014, 2017; Furey et al., 2021). In the Japanese eel (Anguilla japonica), upregulation in prolactin gene expression revealed its importance during upstream migration from sea water to fresh water, and its role in relation to hyper-osmoregulation (Yada et al., 2019). Alternative migratory tactics in brown trout (Salmo trutta) were associated with gene expression in liver and brain tissue (Wynne et al., 2021). Additionally, alteration of metabolic and immune processes in the liver were identified in genes differentially expressed between migrants and residents of the same species, indicating physiological influences on movement strategies (ibid).
Integrating Gene Expression and Physiological Indices
Gene expression and movement data have often been integrated with other physiological indices. Migration success, infectious agents, and plasma variables (e.g., cortisol, lactate, potassium, sodium, leukocrit, among others) were analyzed from the gills and blood of Chinook salmon during upstream migration (O. tshawytscha) and provided additional evidence that disease status may be an important predictor of migration success in anadromous fish (Bass et al., 2019). Disease and stress state in the gills of migrating sockeye salmon were also investigated with gene expression (via microarray and qPCR) and blood plasma parameters paired with telemetry (Drenner et al., 2018). This study provided further evidence for the importance of those parameters in migration and revealed that stressed fish may enter fresh water earlier, but experience subsequently higher mortality (ibid).
Body tissues are also useful in linking gene expression with migration and overall movement. Muscle tissue can aid in linking gene expression with migration success and environmental xenobiotics. This relationship was found in sockeye and Chinook salmon including xenobiotic compounds such as organochlorine pesticides, organohalogen residues, and polybrominated diethyl ethers (Veldhoen et al., 2010). Here, sockeye salmon were found to experience greater biological stress than the Chinook salmon, in conjunction with endocrine disrupting chemicals (ibid). Brain, gill, muscle, and liver tissue from Coho salmon (O. kisutch) revealed how gene expression may change with important life history events including implications for physiological preparations for migration (Houde et al., 2019). Other work has also connected physiological and transcriptomic patterns, such as staggered increases in protein turnover, protein biosynthesis, immune responses, oxidative phosphorylation, and glycolytic potential as sockeye salmon underwent anadromous migrations (Miller et al., 2009). These gene expression and physiology-based studies may show a bias towards fish with anadromous migrations, but nevertheless demonstrate the enormous potential for describing how movement may be impacted by different environmental factors during fishes’ lifetimes.
Methods for Connecting Movement and Genetics
Population Genetics
Population genetics has been useful for describing the extent to which evolution and movement interact by measuring multigenerational population connectivity (reviewed in Ovenden et al., 2015; Cayuela et al., 2018). Here, connectivity is measured as dispersal, which is often linked to gene flow and population structure (Lowe and Allendorf, 2010; Hall and Beissinger, 2017; Cayuela et al., 2018). With population genetics, the unit of observation is genetic material ‘moving’ between places and populations over generations, while in other studies in this review, the unit of movement is an organism moving within its lifetime. Effective dispersal refers to an individual’s success at reproducing, whereas non-effective dispersal is characterized by a failure to reproduce (Lowe and Allendorf, 2010; Cayuela et al., 2018). Within effective dispersal, historical and recent gene flow are distinguishing characteristics of population connectivity (ibid). Methods for assessing dispersal using population genetics include indirect approaches that assess effective dispersal through its outcome on genotype frequencies and direct approaches that measure all dispersal by assigning individuals to their population of origin (Broquet and Petit, 2009). For example, in yellowfin tuna (Thunnus albacares), a direct approach of genetic assignment with DNA data detected asymmetric dispersal between two current stock designations, which had important implications to the management as it suggested spawning stock biomass was overestimated (Mullins et al., 2018). DNA from fin clips has been used to identify how population origin can affect migration behaviours in wild and stocked Atlantic salmon in Europe (Jepsen, 2004) and in brook trout (D’Amelio et al., 2008). Furthermore, by coupling population genetic methods with other approaches such as telemetry or mark-recapture, DNA data can be further used to describe and model species movement (Cayuela et al., 2018; DeSaix et al., 2019). For example, to describe movement in lemon sharks (Negaprion brevirostris), Kessel et al. (2014) combined population structure data with acoustic telemetry to develop a residency model for predicting aggregations.
Quantitative Genetics and Association Tests
Quantitative genetics with experimentally-manipulated pedigrees in Atlantic salmon have revealed a 50–70% heritability for migrant probability (Debes et al., 2020). Variance in migratory traits has been linked with genomics using quantitative genetic approaches in arthropods, fish, and birds, although connections with movement were often indirect (Liedvogel et al., 2011). Association testing in migration timing of Pacific salmon (Oncorhynchus spp.) has revealed a strong genomic basis for early versus late-run fish linked with the genes greb1 and rock1 (O’Malley et al., 2013; Brieuc et al., 2015; Hess et al., 2016; Prince et al., 2017; Koch and Narum, 2020; Thompson et al., 2020). A chromosomal rearrangement was associated with migration in Atlantic cod (Kess et al., 2019), showing that an association-based approach can also reveal structural genomic variants possibly underlying migration phenotypes. The age at which Atlantic salmon return from marine environments to spawn was associated with muscle development and metabolic genes, among others, and has implications for connections between life history strategies, movement, and genomics (Johnston et al., 2014). Several non-lethal association tests used in birds are presented here, for their possible relevance to conducting future research on freshwater fishes. In two warblers (Vermivora chrysoptera & V. cyanoptera), a selection-based approach was used to identify the gene vps13a as affecting wintering location choices (Toews et al., 2019). A hybrid zone of Swainson’s thrush (Catharus ustulatus) was used to identify genomic regions underlying migratory orientation, such as in endopeptidase inhibitor activity and circadian clock genes (Delmore et al., 2016). These different lines of evidence point to a genomic influence on movement in certain circumstances, with implications for potential impacts of genetic variation on the constraint or promotion of movement-based responses to changing environments. Associations between movement and genomics are in their infancy, although some consistent findings may be emerging (Harringmeyer et al., 2021). Certain genomic regions such as those containing GREB1L or circadian rhythm-related genes may be more susceptible to mutations or have an influence on migration (ibid). With more research, these genomic regions may be useful for identifying conservation units for conserving specific adaptive variation in freshwater fishes (Prince et al., 2017).
Further Considerations
Handling Fish
An important property of physiological samples is that they are a ‘snapshot’ of the fish’s physiological status at that time, however acute markers can be sensitive to handling stress during tagging/manipulations (Lower et al., 2005; Stoot et al., 2014; Dick et al., 2018; Lawrence et al., 2018), and are sometimes limited to larger fishes where samples can be obtained (Lawrence et al., 2020). Biopsies are typically taken when the fish is captured and restrained, and is often conducted in concert with tagging or other sampling events (e.g., Miller et al., 2009; Raby et al., 2012). While such biopsies are typically quick (typically a few minutes to perform; Raby et al., 2012; Henderson et al., 2016), anaesthesia can be used for longer sampling workups (Bøe et al., 2020). Importantly, the process of capture, handling, and sampling can impart stress on the animal which may affect post-release mortality and predation (Raby et al., 2014; Hoyle et al., 2015), post-release behaviours (Hoolihan et al., 2011), and alter the physiological status of the sample itself (Dick et al., 2018; Lawrence et al., 2018). Therefore, sampling should always be conducted expediently and in a manner that minimizes harm and stress to the fish itself. Proper sampling techniques and storage of tissue are important considerations that should be incorporated into study designs to ensure that sample parameters are reflective of the treatment of interest and not a methodological artefact (Clark et al., 2011; Schwieterman et al., 2019; Lawrence et al., 2020).
Collecting Molecular Data
Molecular experiments require that tissue sampling be conducted in a standardized manner that ensures sample quality is maintained. In most cases, ecologically-based studies in the context of this review generally employ non-lethal biopsies such that movement patterns can be linked to physiological state or environmental conditions (Cooke et al., 2005). Consequently, sampling must ensure that enough tissue is collected to fulfil analytical requirements while also minimizing adverse impacts on the animal (Corthals et al., 2015; Lawrence et al., 2020; Jeffries et al., 2021). As some of these analyses (i.e., gene expression) can be quite sensitive to stress or handling effects over even brief stressor durations (Krasnov et al., 2005; Milla et al., 2010; Vehniäinen et al., 2019), it is also important to ensure that sampling is done in an environment which minimizes these effects and allows for a timely collection of tissues (See tagging considerations). Granted, sampling of tissues in ecological studies are unlikely to occur in a completely stress-free environment (e.g., fish caught straight from a river and biopsied on-shore), which makes standardisation of the sampling protocol a key requisite in ensuring that handling-associated impacts remain uniform across all replicates in the study. As for the removal of the tissue biopsy, we recommend that gloves be worn at all times and that sampling tools (i.e., scissors, scalpels, punches, etc.) and frequently handled surfaces be sterilized in 70–95% ethanol in between each sampling event (e.g., Andruszkiewicz et al., 2017; Turner and Bucking, 2019). More specialized cleaners may also be employed such as RNase Away™ (Molecular BioProducts Inc. San Diego, California, United States) in RNA-based experiments, which neutralizes RNases from contacted surfaces (Peterson and Freeman, 2009; Carpenter et al., 2014; Andruszkiewicz et al., 2017) thereby reducing the likelihood of degraded RNA samples. Proper sterilization ensures that residual RNA/DNA/bacteria/degradation enzymes are removed from the surface and reduces the risk of sample contamination. Once removed, sampled tissues need to be quickly stored, which commonly includes either the use of a preserving agent (e.g., RNAlater; Invitrogen™, Carlsbad, California, United States) or by snap-freezing the tissue with liquid nitrogen. The latter method is thought to be a better means of sample preservation but may not be possible in all field applications because of transportation or safety concerns (Corthals et al., 2015; Passow et al., 2019). See also a study of sample handling best practices to preserve RNA integrity in Vehniäinen et al. (2019). Due to its ease of use and the ability to store samples in the field, RNAlater is often used in RNA and DNA-based field studies in fish biology (Garseth et al., 2013; Teffer and Miller, 2019; Vehniäinen et al., 2019; Kazyak et al., 2021). By contrast, preserving tissues for DNA sampling can be done with a variety of field-ready methods due to its stability (reviewed in Nagy, 2010). Tissue preservation for DNA extraction is commonly done with 95% ethanol, but freezing, desiccation, and even preservation in vodka or toilet paper are possible, as well (Oakenfull, 1994; Nagy, 2010; Baptista and Goodwin, 2017).
Ethics and Non-Lethal Sampling
Removal of a large number of fish for research purposes can have unintended consequences, especially in small rivers or lakes (Use of Fishes in Research Committee, 2014). For example, removal of many fish at one trophic level can lead to changes in species abundance at other trophic levels and can even lead to trophic collapse in severe situations in sensitive ecosystems (Dobson et al., 2006; Henderson et al., 2016). In addition, a practical benefit of non-lethal sampling is that it enables data collection after an organism is handled via data loggers or tags. In laboratory holding studies, non-lethal sampling allows for monitoring growth and survival post-sampling. For movement studies, this post-handling information is generally where movement data is derived. Other methods, such as isotope-based analyses, only provide information about the organism prior to sampling. Therefore, non-lethal sampling may be both favourable and required for conducting research in practice depending on the question being addressed (Use of Fishes in Research Committee, 2014).
However, a discussion of the ethical considerations underlying non-lethal sampling is useful for considering why non-lethal sampling may be important, and in what contexts it may be used. The Principles of Humane Experimental Technique (Russell and Burch, 1959) is a widely influential application of ethics to animal research, and guides policy to the present day (Tannenbaum and Bennett, 2015). It describes principles of “reduction”, “refinement”, and “replacement” (the three R’s) for humane animal use in research. In this framework, non-lethal sampling may be considered in the context of refinement, but non-lethal sampling may still cause harm to an animal. The fact the animal survives may actually increase distress by introducing negative mental states (e.g., pain, fear, or isolation). While the question of whether fish can feel pain is discussed widely (Rose et al., 2014; Chatigny, 2019), evidence that tagging and sampling affect post-release behaviours (e.g., Wilson et al., 2017) shows that non-lethal sampling does affect sampled individuals. However, underlying the principles of the three R’s is the idea of wellbeing. Definitions of wellbeing range from an absence of distress to the presence of positive mental states and experiences (Tannenbaum and Bennett, 2015). If wellbeing was interpreted as an absence of distress, then non-lethal sampling can contribute to distress and would not be consistent with wellbeing. However, if wellbeing was considered as positive experiences or mental states (Tannenbaum and Bennett, 2015), non-lethal sampling contributes to wellbeing by allowing future positive experiences for a research organism. While the principles of the three R’s of humane experimental technique are useful heuristics for many research approaches, they are sometimes insufficient for considering why non-lethal sampling is important.
Another idea relevant to non-lethal sampling is intrinsic value, which is the value of organisms beyond what they can do, acknowledging a value in and of themselves (Vucetich et al., 2015). While rarely discussed explicitly, intrinsic value is widely accepted among surveyed people and underlies many wildlife-oriented policies (Vucetich et al., 2015). Intrinsic value is significant because it explains why non-lethal sampling is more ethical than lethal sampling as non-lethal sampling respects the value inherent to individual research organisms. The value of a wild research organism’s life may be considered under other value systems, such as the ecosystem services and contributions to biodiversity it provides upon release (i.e., use value; De Groot et al., 2002; Vucetich et al., 2015). However, intrinsic value is necessary and sufficient to justify non-lethal sampling in organisms regardless of potential ecosystem services. Therefore, intrinsic value may be used along with the principles of humane experimental technique for designing ethical research projects involving wildlife.
Integrating Data
Integrating datasets is of broad interest to biologists who seek to link movement with other types of information (Figure 1). However, integration is often easier said than done. One potentially useful framework to consider integration is in distinguishing integrative explanations, methods, or data, although other points of view exist (reviewed in O’Malley and Soyer, 2012; Brigandt, 2013). Integrative explanations may be the gold standard for new knowledge but are possibly the most philosophically difficult kinds of integration to achieve. Issues with ultimate and proximate causation, scales of data collected, and diverging perspectives of collaborating researchers are all challenges for scholars that seek integrative explanations (Mayr, 1961; O’Malley and Soyer, 2012). Challenges with data and methods are addressed below, but challenges of diverging perspective and ultimate or proximate causation are project- and domain-specific.
Integration of methods involves either sequential or simultaneous application of different methods on the same biological system (O’Malley and Soyer, 2012). This form of integration is often motivated in part by the techniques available to researchers, as issues of scale and data availability may preclude direct connections between data. Integration of statistical methods can then provide a way to find integrative explanations. For example, in Thorstensen et al. (2021), different scales of time precluded direct inclusion of length-at-age, length-mass relationship, and metabolite concentrations in the same models of walleye in a large lake. Instead, different models were developed within each scale of time, yielding results that showed consistent spatial patterns across the different scales of time with indirect connections to movement (ibid). A similar conceptual approach was used in modeling walleye movements in relation to temperature (Raby et al., 2018). Separate models of outmigration timing from the western basin of a large lake, the extent of eastward migration, and of summer temperatures in the fish’s habitat did not individually integrate all datasets analyzed in the study. But together, the analyses provided an integrative explanation for walleye post-spawning emigration from the western basin as partially driven by temperature (ibid). Direct integration of data with specialized statistical tests, such as in genotype-environment associations (Forester et al., 2018) or in genome-wide associations (Choi et al., 2020) is sometimes possible. Gene expression data can be modeled in an integrative way as well, such as by using weighted gene correlation network analysis to relate correlated modules of genes with external sample traits (e.g., mass, length, experimental treatment) in RNA-seq data (Stuart et al., 2003; Langfelder and Horvath, 2008). These methods have been developed for integrating specific kinds of data and provide appropriate tools to address statistical issues. However, specific methods are not available for many of the possible connections between datasets in biology, and developing these methods is complex and requires deep statistical knowledge. One common approach is to apply multiple statistical tests on a single dataset to find overlapping patterns of significance as a way to integrate methods. In doing so, however, overall significance is limited to the weakest statistical method used (Forester et al., 2018). Nevertheless, the success of integrative methods for specific kinds of data, and the success of integrative explanations from analyzing different statistical methods concurrently, shows that this approach is challenging but rewarding.
Integration of data is the foundational step toward achieving integrative methods and explanations. Here, data must be standardized and made available beyond the original researchers that collected it such as by deposition in repositories (O’Malley and Soyer, 2012). Data standardization is an overlooked challenge as data structures may differ substantially between scientific fields or types of data (Wickham, 2014). As an illustration of challenges in integrating data, observations of temperature at different sites along a river may be recorded as three estimates per site per researcher visit, while observations of movement from an acoustic receiver array may range from data at 10-minute intervals for some fish staying near receivers to weekly observations for fish who happen to use space primarily outside the receiver array. Meaningfully combining these observations across space and time would be a non-trivial data science task, and integrative research often presents data challenges of similar scope. We recommend three basic practices in data formatting to facilitate integration: each variable forms a column, each observation forms a row, and each observational unit forms a table (i.e., tidy data; Wickham, 2014). In the above example for temperature and movement data, one table would contain information on temperature, and another would contain information on fish movement via acoustic telemetry. Data repositories can hold overlooked complexity, as well. Out of 1,379 repositories for research data, 832 were discipline-specific (Kindling et al., 2017). Repositories themselves are faced with different issues such as data formatting, documentation, validation, and accessibility (Assante et al., 2016). As such, collating data from across repositories or making data available to a community of researchers within a repository may be an unexpected challenge in integrative research.
Management
Given the enormous importance of movement to animal survival and reproduction (Nathan et al., 2008), understanding when, where, and why fish move is important to effective long-term management of fisheries resources (Cooke et al., 2016). Collecting and analyzing movement data is a large task though, one that has been addressed by large collaborations such as the Ocean Tracking Network and Great Lakes Acoustic Telemetry Observation System (Cooke et al., 2011; Krueger et al., 2018). With managers involved throughout the research process, relevancy of movement analyses can be maintained (Krueger et al., 2018). Manager participation can include direct involvement in research (e.g., co-authorship), face-to-face communication (e.g., annual meetings), and early participation in researcher networks at the start of the collaboration (ibid). The integrative research reviewed here also has potential applications for management, such as in using physiological thresholds in conservation policy by identifying transcriptomic responses in the context of habitat requirements and movement ecology (Connon et al., 2018). In addition, the genomics of premature migration can be used to potentially define conservation units as a supplement to management frameworks that conserve adaptive variation that maintains the potential for different movement strategies in conserved populations (Prince et al., 2017). As effective management has been guided by the individual components of movement, physiology, and genetics described in this review, research that integrates those approaches has great potential to address future management needs (Figure 1).
Specific examples of movement-based research being applied to management questions exist, as well. Dams and water diversion structures provide important goods and services to humans, but can alter freshwater systems and affect species movement (Grill et al., 2019). River regulation can change flow regimes by altering timing, duration, and frequency of high or low flow conditions, alter water quality, dilute chemical cues, change water temperature, and can even completely impede migration (Hatry et al., 2013; Bennett and Kozak, 2016; Enders et al., 2019; Bett et al., 2020). Not only can non-lethal sampling provide valuable information to inform management decisions while reducing the effects of the research on population size or dynamics, but integration of non-lethal sampling techniques can lead to important management decisions. For example in a long-term study by Bett et al. (2020) monitoring migrating Pacific salmon (Oncorhynchus spp.) in Seton River (a tributary of the Fraser River, British Columbia, Canada), researchers integrated multiple techniques of telemetry, behavioural tests, molecular analyses, and hydraulic monitoring to characterize the effects of a dam fishway on the survival and reproductive success of returning adults. Their findings directly influenced new operational guidelines for dam flow releases (ibid), demonstrating that non-lethal sampling with movement information can be a powerful management tool.
Discussion
Freshwater systems are faced with some of the most pressing contemporary conservation issues and are important for numerous human and ecosystem-based uses. At the landscape scale, these freshwater ecosystems are characterized by waterbodies of varying connectivity, together creating mosaics of fresh water. Movement research on freshwater systems has the potential to explain key factors underlying organism movements within and between waterbodies in these mosaics of connectivity. Non-lethal sampling has enormous potential for addressing integrative movement-oriented questions. Tagging methods require an initial interaction with a research organism before release, a situation consistent with non-lethal practice. During these brief interactions, an opportunity exists for researchers to non-lethally sample a small amount of biological tissue useful for analysis in diverse approaches across environmental toxicology, stress physiology, gene expression, and genetics. Preserving a research organism’s life is practical for species of conservation concern, such as those in small populations that may be diminished by large lethal sample sizes, and ethical within frameworks of wellbeing (which underlies the three R’s) and intrinsic value. Using movement information in conjunction with other data requires careful domain-dependent interpretations of results, and careful but creative applications of integrative methods. Given the importance of movement in freshwater, the conservation issues facing freshwater systems, and the practical and ethical advantages of non-lethal sampling, resource managers would benefit from using non-lethal sampling and movement-centered questions to guide decisions. With the array of biological sampling and tracking methods becoming available to researchers, integrative non-lethal movement studies will provide deep insights into freshwater ecosystems in the future.
Author Contributions
Other than first and last author, author order was chosen by random number generation. MJT and KMJ conceived of the review. WSB, SM, LV, TEM, MJL, and MJT contributed to physiological sections. MJT and CAV contributed to sections covering genetics. TEM, MJL, and MJT contributed to the sections directly reviewing movement analyses methods. LV, KMJ, and MJT wrote the introduction. MJL, CAV, and MJT contributed to further considerations. All authors participated in drafting and editing the manuscript.
Funding
KMJ is supported by a Natural Sciences and Engineering Research Council of Canada Discovery Grant (#05479) and a Genome Canada Large-Scale Applied Research Project grant.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Evelien de Greef for illustrating the conceptual figures in this review.
References
Aerts, J., Metz, J. R., Ampe, B., Decostere, A., Flik, G., and De Saeger, S. (2015). Scales Tell a Story on the Stress History of Fish. PLoS One 10, e0123411. doi:10.1371/journal.pone.0123411
A. H. Houston (Editor) (1990). “Blood and Circulation,” Methods for Fish Biology (Bethesda, MD: American Fisheries Society). doi:10.47886/9780913235584.ch9
Alsop, D. H., Kieffer, J. D., and Wood, C. M. (1999). The Effects of Temperature and Swimming Speed on Instantaneous Fuel Use and Nitrogenous Waste Excretion of the Nile tilapia. Physiol. Biochem. Zool. 72, 474–483. doi:10.1086/316686
Alves, R. M. S., Pereira, B. F., Ribeiro, R. G. L. G., Pitol, D. L., Ciamarro, C. M., Valim, J. R. T., et al. (2016). The Scale Epithelium as a Novel, Non-invasive Tool for Environmental Assessment in Fish: Testing Exposure to Linear Alkylbenzene Sulfonate. Ecotoxicology Environ. Saf. 129, 43–50. doi:10.1016/j.ecoenv.2016.03.010
Andreasen, E. A., Mathew, L. K., and Tanguay, R. L. (2006). Regenerative Growth Is Impacted by TCDD: Gene Expression Analysis Reveals Extracellular Matrix Modulation. Toxicol. Sci. 92, 254–269. doi:10.1093/toxsci/kfj118
Andruszkiewicz, E. A., Starks, H. A., Chavez, F. P., Sassoubre, L. M., Block, B. A., and Boehm, A. B. (2017). Biomonitoring of marine Vertebrates in Monterey Bay Using eDNA Metabarcoding. PLoS One 12, e0176343. doi:10.1371/journal.pone.0176343
Arlinghaus, R., Klefoth, T., Cooke, S. J., Gingerich, A., and Suski, C. (2009). Physiological and Behavioural Consequences of Catch-And-Release Angling on Northern pike (Esox lucius L.). Fish. Res. 97, 223–233. doi:10.1016/j.fishres.2009.02.005
Arukwe, A., and Røe, K. (2008). Molecular and Cellular Detection of Expression of Vitellogenin and Zona Radiata Protein in Liver and Skin of Juvenile salmon (Salmo salar) Exposed to Nonylphenol. Cell Tissue Res 331, 701–712. doi:10.1007/s00441-007-0543-y
Aschettino, J. M., Engelhaupt, D. T., Engelhaupt, A. G., DiMatteo, A., Pusser, T., Richlen, M. F., et al. (2020). Satellite Telemetry Reveals Spatial Overlap between Vessel High-Traffic Areas and Humpback Whales (Megaptera Novaeangliae) Near the Mouth of the Chesapeake Bay. Front. Mar. Sci. 7. doi:10.3389/fmars.2020.00121
Assante, M., Candela, L., Castelli, D., and Tani, A. (2016). Are Scientific Data Repositories Coping with Research Data Publishing? Data Sci. J. 15, 1–24. doi:10.5334/dsj-2016-006
Bahamonde, P., Berrocal, C., Barra, R., Mcmaster, M. E., Munkittrick, K. R., and Chiang, G. (2019). Mucus Phosphoproteins as an Indirect Measure of Endocrine Disruption in Native Small-Bodied Freshwater Fish, Exposed to Wastewater Treatment Plant and Pulp and Paper Mill Effluents. Gayana (Concepc.) 83, 10–20. doi:10.4067/S0717-65382019000100010
Baillon, L., Pierron, F., Oses, J., Pannetier, P., Normandeau, E., Couture, P., et al. (2016). Detecting the Exposure to Cd and PCBs by Means of a Non-invasive Transcriptomic Approach in Laboratory and Wild Contaminated European Eels (Anguilla anguilla). Environ. Sci. Pollut. Res. 23, 5431–5441. doi:10.1007/s11356-015-5754-2
Baker, R. F., Blanchfield, P. J., Paterson, M. J., Flett, R. J., and Wesson, L. (2004). Evaluation of Nonlethal Methods for the Analysis of Mercury in Fish Tissue. Trans. Am. Fish. Soc. 133, 568–576. doi:10.1577/T03-012.1
Baptista, L. V., and Goodwin, W. (2017). DNA Persistence in Soft Tissue Comparing Vodka and Absolute Ethanol. Forensic Sci. Int. Genet. Suppl. Ser. 6, e46–e48. doi:10.1016/j.fsigss.2017.09.008
Barriga-Vallejo, C., Aguilera, C., Cruz, J., Banda-Leal, J., Lazcano, D., and Mendoza, R. (2017). Ecotoxicological Biomarkers in Multiple Tissues of the Neotenic Ambystoma Spp. For a Non-lethal Monitoring of Contaminant Exposure in Wildlife and Captive Populations. Water Air Soil Pollut. 228, 415. doi:10.1007/s11270-017-3590-3
Bass, A. L., Hinch, S. G., Teffer, A. K., Patterson, D. A., and Miller, K. M. (2019). Fisheries Capture and Infectious Agents Are Associated with Travel Rate and Survival of Chinook salmon during Spawning Migration. Fish. Res. 209, 156–166. doi:10.1016/j.fishres.2018.09.009
Bass, A. L., Stevenson, C. F., Porter, A. D., Rechisky, E. L., Furey, N. B., Healy, S. J., et al. (2020). In Situ experimental Evaluation of Tag burden and Gill Biopsy Reveals Survival Impacts on Migrating Juvenile Sockeye salmon. Can. J. Fish. Aquat. Sci. 77, 1865–1869. doi:10.1139/cjfas-2020-0134
Bennett, M. G., and Kozak, J. P. (2016). Spatial and Temporal Patterns in Fish Community Structure and Abundance in the Largest U.S. River Swamp, the Atchafalaya River Floodplain, L Ouisiana. Ecol. Freshw. Fish. 25, 577–589. doi:10.1111/eff.12235
Bett, N. N., Hinch, S. G., Bass, A. L., Braun, D. C., Burnett, N. J., Casselman, M. T., et al. (2020). Using an Integrative Research Approach to Improve Fish Migrations in Regulated Rivers: a Case Study on Pacific Salmon in the Seton River, Canada. Hydrobiologia 849, 385–405. doi:10.1007/s10750-020-04371-2
Birnie-Gauvin, K., Flávio, H., Kristensen, M. L., Walton-Rabideau, S., Cooke, S. J., Willmore, W. G., et al. (2019). Cortisol Predicts Migration Timing and success in Both Atlantic salmon and Sea trout Kelts. Sci. Rep. 9, 1–9. doi:10.1038/s41598-019-39153-x
Bøe, K., Robertson, M. J., Fleming, I. A., and Power, M. (2020). Evaluating the Effect of Dorsal Muscle Biopsies on Adult Atlantic salmon Growth and marine Return Rates. Conserv. Physiol. 8. doi:10.1093/conphys/coz099
Bowers, R. M., Lapatra, S. E., and Dhar, A. K. (2008). Detection and Quantitation of Infectious Pancreatic Necrosis Virus by Real-Time Reverse Transcriptase-Polymerase Chain Reaction Using Lethal and Non-lethal Tissue Sampling. J. Virol. Methods 147, 226–234. doi:10.1016/j.jviromet.2007.09.003
Bozinovic, F., Calosi, P., and Spicer, J. I. (2011). Physiological Correlates of Geographic Range in Animals. Annu. Rev. Ecol. Evol. Syst. 42, 155–179. doi:10.1146/annurev-ecolsys-102710-145055
Brander, K. M. (2007). Global Fish Production and Climate Change. Proc. Natl. Acad. Sci. U.S.A. 104, 19709–19714. doi:10.1073/pnas.0702059104
Brieuc, M. S. O., Ono, K., Drinan, D. P., and Naish, K. A. (2015). Integration of Random Forest with Population-Based Outlier Analyses Provides Insight on the Genomic Basis and Evolution of Run Timing in Chinook salmon (Oncorhynchus tshawytscha). Mol. Ecol. 24, 2729–2746. doi:10.1111/mec.13211
Brigandt, I. (2013). Integration in Biology: Philosophical Perspectives on the Dynamics of Interdisciplinarity. Stud. Hist. Philos. Sci. C: Stud. Hist. Philos. Biol. Biomed. Sci. 44, 461–465. doi:10.1016/j.shpsc.2013.09.009
Broquet, T., and Petit, E. J. (2009). Molecular Estimation of Dispersal for Ecology and Population Genetics. Annu. Rev. Ecol. Evol. Syst. 40, 193–216. doi:10.1146/annurev.ecolsys.110308.120324
Brownscombe, J. W., Danylchuk, A. J., Chapman, J. M., Gutowsky, L. F. G., and Cooke, S. J. (2017). Best Practices for Catch-And-Release Recreational Fisheries - Angling Tools and Tactics. Fish. Res. 186, 693–705. doi:10.1016/j.fishres.2016.04.018
Bulloch, P., Schur, S., Muthumuni, D., Xia, Z., Johnson, W., Chu, M., et al. (2020). F2-isoprostanes in Fish Mucus: A New, Non-invasive Method for Analyzing a Biomarker of Oxidative Stress. Chemosphere 239, 124797. doi:10.1016/j.chemosphere.2019.124797
Burnett, N. J., Hinch, S. G., Bett, N. N., Braun, D. C., Casselman, M. T., Cooke, S. J., et al. (2017). Reducing Carryover Effects on the Migration and Spawning Success of Sockeye Salmon through a Management Experiment of Dam Flows. River Res. Applic. 33, 3–15. doi:10.1002/rra.3051
Burnett, N. J., Hinch, S. G., Braun, D. C., Casselman, M. T., Middleton, C. T., Wilson, S. M., et al. (2014). Burst Swimming in Areas of High Flow: Delayed Consequences of Anaerobiosis in Wild Adult Sockeye Salmon. Physiol. Biochem. Zool. 87, 587–598. doi:10.1086/677219
Busst, G. M. A., Bašić, T., and Britton, J. R. (2015). Stable Isotope Signatures and Trophic-step Fractionation Factors of Fish Tissues Collected as Non-lethal Surrogates of Dorsal Muscle. Rapid Commun. Mass. Spectrom. 29, 1535–1544. doi:10.1002/rcm.7247
Caputo, M., O’Connor, C., Hasler, C., Hanson, K., and Cooke, S. (2009). Long-term Effects of Surgically Implanted Telemetry Tags on the Nutritional Physiology and Condition of Wild Freshwater Fish. Dis. Aquat. Org. 84, 35–41. doi:10.3354/dao02025
Carpenter, R. E., Maruska, K. P., Becker, L., and Fernald, R. D. (2014). Social Opportunity Rapidly Regulates Expression of CRF and CRF Receptors in the Brain during Social Ascent of a Teleost Fish, Astatotilapia Burtoni. PLoS One 9, e96632. doi:10.1371/journal.pone.0096632
Carruth, L. L., Dores, R. M., Maldonado, T. A., Norris, D. O., Ruth, T., and Jones, R. E. (2000). Elevation of Plasma Cortisol during the Spawning Migration of Landlocked Kokanee salmon (Oncorhynchus nerka Kennerlyi). Comp. Biochem. Physiol. C: Pharmacol. Toxicol. Endocrinol. 127, 123–131. doi:10.1016/S0742-8413(00)00140-7
Cayuela, H., Rougemont, Q., Prunier, J. G., Moore, J.-S., Clobert, J., Besnard, A., et al. (2018). Demographic and Genetic Approaches to Study Dispersal in Wild Animal Populations: A Methodological Review. Mol. Ecol. 27, 3976–4010. doi:10.1111/mec.14848
Chapman, J. M., Kelly, L. A., Teffer, A. K., Miller, K. M., and Cooke, S. J. (2021). Disease Ecology of Wild Fish: Opportunities and Challenges for Linking Infection Metrics with Behaviour, Condition, and Survival. Can. J. Fish. Aquat. Sci. 78, 995–1007. doi:10.1139/cjfas-2020-0315
Chapman, J. M., Teffer, A. K., Bass, A. L., Hinch, S. G., Patterson, D. A., Miller, K. M., et al. (2020). Handling, Infectious Agents and Physiological Condition Influence Survival and post-release Behaviour in Migratory Adult Coho salmon after Experimental Displacement. Conserv. Physiol. 8, coaa033. doi:10.1093/conphys/coaa033
Charette, T., Rosabal, M., and Amyot, M. (2021). Mapping Metal (Hg, as, Se), Lipid and Protein Levels within Fish Muscular System in Two Fish Species (Striped Bass and Northern Pike). Chemosphere 265, 129036. doi:10.1016/j.chemosphere.2020.129036
Chatigny, F. (2019). The Controversy on Fish Pain: A Veterinarian's Perspective. J. Appl. Anim. Welfare Sci. 22, 400–410. doi:10.1080/10888705.2018.1530596
Cheong, T. S., Kavvas, M. L., and Anderson, E. K. (2006). Evaluation of Adult White Sturgeon Swimming Capabilities and Applications to Fishway Design. Environ. Biol. Fish. 77, 197–208. doi:10.1007/s10641-006-9071-y
Choi, S. W., Mak, T. S.-H., and O’Reilly, P. F. (2020). Tutorial: a Guide to Performing Polygenic Risk Score Analyses. Nat. Protoc. 15, 2759–2772. doi:10.1038/s41596-020-0353-1
Church, M. R., Ebersole, J. L., Rensmeyer, K. M., Couture, R. B., Barrows, F. T., and Noakes, D. L. G. (2009). Mucus: A New Tissue Fraction for Rapid Determination of Fish Diet Switching Using Stable Isotope Analysis. Can. J. Fish. Aquat. Sci. 66, 1–5. doi:10.1139/F08-206
Clark, T. D., Donaldson, M. R., Drenner, S. M., Hinch, S. G., Patterson, D. A., Hills, J., et al. (2011). The Efficacy of Field Techniques for Obtaining and Storing Blood Samples from Fishes. J. Fish. Biol. 79, 1322–1333. doi:10.1111/j.1095-8649.2011.03118.x
Cocherell, D. E., Kawabata, A., Kratville, D. W., Cocherell, S. A., Kaufman, R. C., Anderson, E. K., et al. (2011). Passage Performance and Physiological Stress Response of Adult white sturgeon Ascending a Laboratory Fishway. J. Appl. Ichthyol. 27, 327–334. doi:10.1111/j.1439-0426.2010.01650.x
Connon, R. E., Jeffries, K. M., Komoroske, L. M., Todgham, A. E., and Fangue, N. A. (2018). The Utility of Transcriptomics in Fish Conservation. J. Exp. Biol. 221, jeb148833. doi:10.1242/jeb.148833
Cooke, S. J., Crossin, G. T., Patterson, D. A., English, K. K., Hinch, S. G., Young, J. L., et al. (2005). Coupling Non-invasive Physiological Assessments with Telemetry to Understand Inter-individual Variation in Behaviour and Survivorship of Sockeye salmon: Development and Validation of a Technique. J. Fish. Biol. 67, 1342–1358. doi:10.1111/j.1095-8649.2005.00830.x
Cooke, S. J., Hinch, S. G., Crossin, G. T., Patterson, D. A., English, K. K., Healey, M. C., et al. (2006). Mechanistic Basis of Individual Mortality in Pacific salmon during Spawning Migrations. Ecology 87, 1575–1586. doi:10.1890/0012-9658(2006)87[1575:mboimi]2.0.co;2
Cooke, S. J., Hinch, S. G., Donaldson, M. R., Clark, T. D., Eliason, E. J., Crossin, G. T., et al. (2012). Conservation Physiology in Practice: How Physiological Knowledge Has Improved Our Ability to Sustainably Manage Pacific salmon during Up-River Migration. Phil. Trans. R. Soc. B 367, 1757–1769. doi:10.1098/rstb.2012.0022
Cooke, S. J., Hinch, S. G., Farrell, A. P., Patterson, D. A., Miller-Saunders, K., Welch, D. W., et al. (2008). Developing a Mechanistic Understanding of Fish Migrations by Linking Telemetry with Physiology, Behavior, Genomics and Experimental Biology: An Interdisciplinary Case Study on Adult Fraser River Sockeye Salmon. Fisheries 33, 321–339. doi:10.1577/1548-8446-33.7.321
Cooke, S. J., Iverson, S. J., Stokesbury, M. J. W., Hinch, S. G., Fisk, A. T., VanderZwaag, D. L., et al. (2011). Ocean Tracking Network Canada: A Network Approach to Addressing Critical Issues in Fisheries and Resource Management with Implications for Ocean Governance. Fisheries 36, 583–592. doi:10.1080/03632415.2011.633464
Cooke, S. J., Martins, E. G., Struthers, D. P., Gutowsky, L. F. G., Power, M., Doka, S. E., et al. (2016). A Moving Target-Incorporating Knowledge of the Spatial Ecology of Fish into the Assessment and Management of Freshwater Fish Populations. Environ. Monit. Assess. 188, 239. doi:10.1007/s10661-016-5228-0
Corthals, A., Martin, A., Warsi, O. M., Woller-Skar, M., Lancaster, W., Russell, A., et al. (2015). From the Field to the Lab: Best Practices for Field Preservation of Bat Specimens for Molecular Analyses. PLoS One 10, e0118994. doi:10.1371/journal.pone.0118994
Crawford, B. A., Leider, S. A., and Tipping, J. M. (1977). Technique for Rapidly Taking Samples of Skeletal Muscle from Live Adult Steelhead Trout. The Progressive Fish-CulturistFish-Culturist 39, 125. doi:10.1577/1548-8659(1977)39[125:tfrtso]2.0.co;2
Crossin, G. T., Hinch, S. G., Cooke, S. J., Cooperman, M. S., Patterson, D. A., Welch, D. W., et al. (2009). Mechanisms Influencing the Timing and Success of Reproductive Migration in a Capital Breeding Semelparous Fish Species, the Sockeye Salmon. Physiol. Biochem. Zool. 82, 635–652. doi:10.1086/605878
Cunjak, R. A., Roussel, J.-M., Gray, M. A., Dietrich, J. P., Cartwright, D. F., Munkittrick, K. R., et al. (2005). Using Stable Isotope Analysis with Telemetry or Mark-Recapture Data to Identify Fish Movement and Foraging. Oecologia 144, 636–646. doi:10.1007/s00442-005-0101-9
D'Amelio, S., Mucha, J., Mackereth, R., and Wilson, C. C. (2008). Tracking Coaster Brook Trout to Their Sources: Combining Telemetry and Genetic Profiles to Determine Source Populations. North Am. J. Fish. Manage. 28, 1343–1349. doi:10.1577/M05-193.1
Danylchuk, A. J., Danylchuk, S. E., Cooke, S. J., Goldberg, T. L., Koppelman, J. B., and Philipp, D. P. (2007). Post-release Mortality of Bonefish, Albula vulpes, Exposed to Different Handling Practices during Catch-And-Release Angling in Eleuthera, the Bahamas. Fish. Manage 14, 149–154. doi:10.1111/j.1365-2400.2007.00535.x
De Groot, R. S., Wilson, M. A., and Boumans, R. M. J. (2002). A Typology for the Classification, Description and Valuation of Ecosystem Functions, Goods and Services. Ecol. Econ. 41, 393–408. doi:10.1016/S0921-8009(02)00089-7
Debes, P. V., Piavchenko, N., Erkinaro, J., and Primmer, C. R. (2020). Genetic Growth Potential, rather Than Phenotypic Size, Predicts Migration Phenotype in Atlantic salmon. Proc. Biol. Sci. 287, 20200867. doi:10.1098/rspb.2020.0867rspb20200867
Delmore, K. E., Toews, D. P. L., Germain, R. R., Owens, G. L., and Irwin, D. E. (2016). The Genetics of Seasonal Migration and Plumage Color. Curr. Biol. 26, 2167–2173. doi:10.1016/j.cub.2016.06.015
DeSaix, M. G., Bulluck, L. P., Eckert, A. J., Viverette, C. B., Boves, T. J., Reese, J. A., et al. (2019). Population Assignment Reveals Low Migratory Connectivity in a Weakly Structured Songbird. Mol. Ecol. 28, 2122–2135. doi:10.1111/mec.15083
Di Cicco, E., Ferguson, H. W., Kaukinen, K. H., Schulze, A. D., Li, S., Tabata, A., et al. (2018). The Same Strain of Piscine Orthoreovirus (PRV-1) Is Involved in the Development of Different, but Related, Diseases in Atlantic and Pacific Salmon in British Columbia. Facets 3, 599–641. doi:10.1139/facets-2018-0008
Dick, M., Eliason, E. J., Patterson, D. A., Robinson, K. A., Hinch, S. G., and Cooke, S. J. (2018). Short-term Physiological Response Profiles of Tagged Migrating Adult Sockeye Salmon: A Comparison of Gastric Insertion and External Tagging Methods. Trans. Am. Fish. Soc. 147, 300–315. doi:10.1002/tafs.10027
Dobson, A., Lodge, D., Alder, J., Cumming, G. S., Keymer, J., McGlade, J., et al. (2006). Habitat Loss, Trophic Collapse, and the Decline of Ecosystem Services. Ecology 87, 1915–1924. doi:10.1890/0012-9658(2006)87[1915:hltcat]2.0.co;2
Dominik, S., Henshall, J. M., Kube, P. D., King, H., Lien, S., Kent, M. P., et al. (2010). Evaluation of an Atlantic salmon SNP Chip as a Genomic Tool for the Application in a Tasmanian Atlantic salmon (Salmo salar) Breeding Population. Aquaculture 308, S56–S61. doi:10.1016/j.aquaculture.2010.05.038
Donaldson, M. R., Hinch, S. G., Suski, C. D., Fisk, A. T., Heupel, M. R., and Cooke, S. J. (2014). Making Connections in Aquatic Ecosystems with Acoustic Telemetry Monitoring. Front. Ecol. Environ. 12, 565–573. doi:10.1890/130283
Drenner, S. M., Hinch, S. G., Furey, N. B., Clark, T. D., Li, S., Ming, T., et al. (2018). Transcriptome Patterns and Blood Physiology Associated with Homing success of Sockeye salmon during Their Final Stage of marine Migration. Can. J. Fish. Aquat. Sci. 75, 1511–1524. doi:10.1139/cjfas-2017-0391
Dudgeon, D., Arthington, A. H., Gessner, M. O., Kawabata, Z.-I., Knowler, D. J., Lévêque, C., et al. (2006). Freshwater Biodiversity: Importance, Threats, Status and Conservation Challenges. Biol. Rev. 81, 163. doi:10.1017/S1464793105006950
Dudgeon, D. (2019). Multiple Threats Imperil Freshwater Biodiversity in the Anthropocene. Curr. Biol. 29, R960–R967. doi:10.1016/j.cub.2019.08.002
Eggenberger, C. W., Santos, R. O., Frankovich, T. A., James, W. R., Madden, C. J., Nelson, J. A., et al. (2019). Coupling Telemetry and Stable Isotope Techniques to Unravel Movement: Snook Habitat Use across Variable Nutrient Environments. Fish. Res. 218, 35–47. doi:10.1016/j.fishres.2019.04.008
Elliott, D., McKibben, C., Conway, C., Purcell, M., Chase, D., and Applegate, L. (2015). Testing of Candidate Non-lethal Sampling Methods for Detection of Renibacterium Salmoninarum in Juvenile Chinook salmon Oncorhynchus tshawytscha. Dis. Aquat. Org. 114, 21–43. doi:10.3354/dao02846
Enders, E. C., Charles, C., Watkinson, D. A., Kovachik, C., Leroux, D. R., Hansen, H., et al. (2019). Analysing Habitat Connectivity and home Ranges of Bigmouth Buffalo and Channel Catfish Using a Large-Scale Acoustic Receiver Network. Sustainability 11, 3051. doi:10.3390/su11113051
Evans, D. H., Piermarini, P. M., and Choe, K. P. (2005). The Multifunctional Fish Gill: Dominant Site of Gas Exchange, Osmoregulation, Acid-Base Regulation, and Excretion of Nitrogenous Waste. Physiol. Rev. 85, 97–177. doi:10.1152/physrev.00050.2003
Evans, T. G., Hammill, E., Kaukinen, K., Schulze, A. D., Patterson, D. A., English, K. K., et al. (2011). Transcriptomics of Environmental Acclimatization and Survival in Wild Adult Pacific Sockeye salmon (Oncorhynchus nerka) during Spawning Migration. Mol. Ecol. 20, 4472–4489. doi:10.1111/j.1365-294X.2011.05276.x
Fagan, K.-A., Koops, M. A., Arts, M. T., Sutton, T. M., and Power, M. (2012). Lake Whitefish Feeding Habits and Condition in Lake Michigan. adv_limnology 63, 399–415. doi:10.1127/advlim/63/2012/399
Flávio, H., and Baktoft, H. (2021). Actel: Standardised Analysis of Acoustic Telemetry Data from Animals Moving through Receiver Arrays. Methods Ecol. Evol. 12, 196–203. doi:10.1111/2041-210X.13503
Forester, B. R., Lasky, J. R., Wagner, H. H., and Urban, D. L. (2018). Comparing Methods for Detecting Multilocus Adaptation with Multivariate Genotype-Environment Associations. Mol. Ecol. 27, 2215–2233. doi:10.1111/mec.14584
Furey, N. B., Bass, A. L., Miller, K. M., Li, S., Lotto, A. G., Healy, S. J., et al. (2021). Infected Juvenile salmon Can Experience Increased Predation during Freshwater Migration. R. Soc. Open Sci. 8, rsos.201522. doi:10.1098/rsos.201522
Galván, D. E., Funes, M., Liberoff, A. L., Botto, F., and Iribarne, O. O. (2015). Important Sources of Variation to Be Considered when Using Fin Clips as a Surrogate for Muscle in Trophic Studies Using Stable Isotopes. Mar. Freshw. Res. 66, 730. doi:10.1071/MF13346
Garseth, Å. H., Biering, E., and Aunsmo, A. (2013). Associations between Piscine Reovirus Infection and Life History Traits in Wild-Caught Atlantic salmon Salmo salar L. In Norway. Prev. Vet. Med. 112, 138–146. doi:10.1016/j.prevetmed.2013.06.007
Gelsleichter, J., Sparkman, G., Howey, L., Brooks, E., and Shipley, O. (2020). Elevated Accumulation of the Toxic Metal Mercury in the Critically Endangered Oceanic Whitetip Shark Carcharhinus longimanus from the Northwestern Atlantic Ocean. Endang. Species Res. 43, 267–279. doi:10.3354/esr01068
Gesto, M., Hernández, J., López-Patiño, M. A., Soengas, J. L., and Míguez, J. M. (2015). Is Gill Cortisol Concentration a Good Acute Stress Indicator in Fish? A Study in Rainbow trout and Zebrafish. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 188, 65–69. doi:10.1016/j.cbpa.2015.06.020
Gilbert, M. J. H. (2020). Thermal Limits to the Cardiorespiratory Performance of Arctic Char (Salvelinus alpinus) in a Rapidly Warming north.
Gillis, N. C., Rapp, T., Hasler, C. T., Wachelka, H., and Cooke, S. J. (2010). Spatial Ecology of Adult Muskellunge (Esox masquinongy) in the Urban Ottawa Reach of the Historic Rideau Canal, Canada. Aquat. Living Resour. 23, 225–230. doi:10.1051/alr/2010019
Gray, M. A., Cunjak, R. A., and Munkittrick, K. R. (2004). Site Fidelity of Slimy Sculpin (Cottus Cognatus): Insights from Stable Carbon and Nitrogen Analysis. Can. J. Fish. Aquat. Sci. 61, 1717–1722. doi:10.1139/f04-108
Gray, M. A., Curry, A. R., and Munkittrick, K. R. (2002). Non-Lethal Sampling Methods for Assessing Environmental Impacts Using a Small-Bodied Sentinel Fish Species. Water Qual. Res. J. 37, 195–211. doi:10.2166/wqrj.2002.012
Griffiths, D. (2015). Connectivity and Vagility Determine Spatial Richness Gradients and Diversification of Freshwater Fish in North America and Europe. Biol. J. Linn. Soc. 116, 773–786. doi:10.1111/bij.12638
Grill, G., Lehner, B., Thieme, M., Geenen, B., Tickner, D., Antonelli, F., et al. (2019). Mapping the World's Free-Flowing Rivers. Nature 569, 215–221. doi:10.1038/s41586-019-1111-9
Guo, B., Tang, Z., Wu, C., Xu, K., and Qi, P. (2018). Transcriptomic Analysis Reveal an Efficient Osmoregulatory System in Siberian sturgeon Acipenser Baeri in Response to Salinity Stress. Sci. Rep. 8, 14353. doi:10.1038/s41598-018-32771-x
Hale, R. S., and Gray, J. H. (1998). Retention and Detection of Coded Wire Tags and Elastomer Tags in Trout. North Am. J. Fish. Manage. 18, 197–201. doi:10.1577/1548-8675(1998)018<0197:radocw>2.0.co;2
Halfyard, E. A., Webber, D., Del Papa, J., Leadley, T., Kessel, S. T., Colborne, S. F., et al. (2017). Evaluation of an Acoustic Telemetry Transmitter Designed to Identify Predation Events. Methods Ecol. Evol. 8, 1063–1071. doi:10.1111/2041-210X.12726
Hall, L. A., and Beissinger, S. R. (2017). Inferring the Timing of Long-Distance Dispersal between Rail Metapopulations Using Genetic and Isotopic Assignments. Ecol. Appl. 27, 208–218. doi:10.1002/eap.1432
Harringmeyer, O. S., Woolfolk, M. L., and Hoekstra, H. E. (2021). Fishing for the Genetic Basis of Migratory Behavior. Cell 184, 303–305. doi:10.1016/j.cell.2020.12.037
Hatry, C., Binder, T. R., Thiem, J. D., Hasler, C. T., Smokorowski, K. E., Clarke, K. D., et al. (2013). The Status of Fishways in Canada: Trends Identified Using the National CanFishPass Database. Rev. Fish. Biol. Fish. 23, 271–281. doi:10.1007/s11160-012-9293-3
Healy, S. J., Hinch, S. G., Bass, A. L., Furey, N. B., Welch, D. W., Rechisky, E. L., et al. (2018). Transcriptome Profiles Relate to Migration Fate in Hatchery Steelhead (Oncorhynchus mykiss) Smolts. Can. J. Fish. Aquat. Sci. 75, 2053–2068. doi:10.1139/cjfas-2017-0424
Heberer, C., Aalbers, S. A., Bernal, D., Kohin, S., DiFiore, B., and Sepulveda, C. A. (2010). Insights into Catch-And-Release Survivorship and Stress-Induced Blood Biochemistry of Common Thresher Sharks (Alopias Vulpinus) Captured in the Southern California Recreational Fishery. Fish. Res. 106, 495–500. doi:10.1016/j.fishres.2010.09.024
Hedger, R. D., Næsje, T. F., Fiske, P., Ugedal, O., Finstad, A. G., and Thorstad, E. B. (2013). Ice-dependent winter survival of juvenile Atlantic salmon. Ecol. Evol. 3, 523–535. doi:10.1002/ece3.481
Heltsley, R. M., Cope, W. G., Shea, D., Bringolf, R. B., Kwak, T. J., and Malindzak, E. G. (2005). Assessing Organic Contaminants in Fish: Comparison of a Nonlethal Tissue Sampling Technique to Mobile and Stationary Passive Sampling Devices. Environ. Sci. Technol. 39, 7601–7608. doi:10.1021/es051037s
Henderson, C. J., Stevens, T. F., and Lee, S. Y. (2016). Assessing the Suitability of a Non-lethal Biopsy Punch for Sampling Fish Muscle Tissue. Fish. Physiol. Biochem. 42, 1521–1526. doi:10.1007/s10695-016-0237-z
Hess, J. E., Zendt, J. S., Matala, A. R., and Narum, S. R. (2016). Genetic Basis of Adult Migration Timing in Anadromous Steelhead Discovered through Multivariate Association Testing. Proc. R. Soc. B. 283, 20153064. doi:10.1098/rspb.2015.3064
Heupel, M. R., and Hueter, R. E. (2001). “Use of an Automated Acoustic Telemetry System to Passively Track Juvenile Blacktip Shark Movements,” in Electronic Tagging and Tracking in Marine Fisheries: Proceedings of the Symposium on Tagging and Tracking Marine Fish with Electronic Devices, February 7--11, 2000, East-West Center, University of Hawaii. Editors J. R. Sibert, and J. L. Nielsen (Dordrecht: Springer Netherlands), 217–236. doi:10.1007/978-94-017-1402-0_10
Hoffman, J. C., Blazer, V. S., Walsh, H. H., Shaw, C. H., Braham, R., and Mazik, P. M. (2020). Influence of Demographics, Exposure, and Habitat Use in an Urban, Coastal River on Tumor Prevalence in a Demersal Fish. Sci. Total Environ. 712, 136512. doi:10.1016/j.scitotenv.2020.136512
Hohn, C., and Petrie-Hanson, L. (2013). Evaluation of Visible Implant Elastomer Tags in Zebrafish (Danio rerio). Biol. Open 2, 1397–1401. doi:10.1242/bio.20136460
Hoolihan, J. P., Luo, J., Abascal, F. J., Campana, S. E., De Metrio, G., Dewar, H., et al. (2011). Evaluating post-release Behaviour Modification in Large Pelagic Fish Deployed with Pop-Up Satellite Archival Tags. ICES J. Mar. Sci. 68, 880–889. doi:10.1093/icesjms/fsr024
Houde, A. L. S., Schulze, A. D., Kaukinen, K. H., Strohm, J., Patterson, D. A., Beacham, T. D., et al. (2019). Transcriptional Shifts during Juvenile Coho salmon (Oncorhynchus kisutch) Life Stage Changes in Freshwater and Early marine Environments. Comp. Biochem. Physiol. D: Genomics Proteomics 29, 32–42. doi:10.1016/j.cbd.2018.10.002
Houlihan, D. F., and Laurent, P. (1987). Effects of Exercise Training on the Performance, Growth, and Protein Turnover of Rainbow Trout (Salmo Gairdneri). Can. J. Fish. Aquat. Sci. 44, 1614–1621. doi:10.1139/f87-195
Hoyle, S. D., Leroy, B. M., Nicol, S. J., and Hampton, W. J. (2015). Covariates of Release Mortality and Tag Loss in Large-Scale Tuna Tagging Experiments. Fish. Res. 163, 106–118. doi:10.1016/j.fishres.2014.02.023
Hussey, N. E., Kessel, S. T., Aarestrup, K., Cooke, S. J., Cowley, P. D., Fisk, A. T., et al. (2015). Aquatic Animal Telemetry: A Panoramic Window into the Underwater World. Science 348, 1255642. doi:10.1126/science.1255642
Use of Fishes in Research Committee (2014). in Guidelines for the Use of Fishes in Research. Editors J. A. Jenkins, H. L. Bart, J. D. Bowker, P. R. Powser, J. R. MacMillan, and J. G. Nickum (Bethesda, MD: American Fisheries Society).
Jeffres, C. A., Klimley, A. P., Merz, J. E., and Cech, J. J. (2006). Movement of Sacramento Sucker, Catostomus Occidentalis, and Hitch, Lavinia exilicauda, during a spring Release of Water from Camanche Dam in the Mokelumne River, California. Environ. Biol. Fish. 75, 365–373. doi:10.1007/s10641-005-2924-y
Jeffries, K. M., Hinch, S. G., Gale, M. K., Clark, T. D., Lotto, A. G., Casselman, M. T., et al. (2014). Immune Response Genes and Pathogen Presence Predict Migration Survival in Wild salmon Smolts. Mol. Ecol. 23, 5803–5815. doi:10.1111/mec.12980
Jeffries, K. M., Teffer, A., Michaleski, S., Bernier, N. J., Heath, D. D., and Miller, K. M. (2021). The Use of Non-lethal Sampling for Transcriptomics to Assess the Physiological Status of Wild Fishes. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 256, 110629. doi:10.1016/j.cbpb.2021.110629
Jepsen, N. (2004). “Linking Individual Behaviour to Genetic Origin,” in Aquatic Telemetry: Advances and Applications: Proceedings of the Fifth Conference on Fish Telemetry Held in Europe. Rome: Food and Agriculture Organization of the United Nations, 45–51.
John Nelson, R., Wood, C. C., Cooper, G., Smith, C., and Koop, B. (2003). Population Structure of Sockeye Salmon of the Central Coast of British Columbia: Implications for Recovery Planning. North Am. J. Fish. Manage. 23, 703–720. doi:10.1577/M01-194
Johnston, S. E., Orell, P., Pritchard, V. L., Kent, M. P., Lien, S., Niemelä, E., et al. (2014). Genome-wide SNP Analysis Reveals a Genetic Basis for Sea-Age Variation in a Wild Population of Atlantic salmon (Salmo salar). Mol. Ecol. 23, 3452–3468. doi:10.1111/mec.12832
Joo, R., Boone, M. E., Clay, T. A., Patrick, S. C., Clusella-Trullas, S., and Basille, M. (2020). Navigating through the R Packages for Movement. J. Anim. Ecol. 89, 248–267. doi:10.1111/1365-2656.13116
Katopodis, C., Cai, L., and Johnson, D. (2019). Sturgeon Survival: The Role of Swimming Performance and Fish Passage Research. Fish. Res. 212, 162–171. doi:10.1016/j.fishres.2018.12.027
Kazyak, D. C., White, S. L., Lubinski, B. A., Johnson, R., and Eackles, M. (2021). Stock Composition of Atlantic sturgeon (Acipenser Oxyrinchus Oxyrinchus) Encountered in marine and Estuarine Environments on the U.S. Atlantic Coast. Conserv. Genet. 22, 767–781. doi:10.1007/s10592-021-01361-2
Kess, T., Bentzen, P., Lehnert, S. J., Sylvester, E. V. A., Lien, S., Kent, M. P., et al. (2019). A Migration-Associated Supergene Reveals Loss of Biocomplexity in Atlantic Cod. Sci. Adv. 5, eaav2461. doi:10.1126/sciadv.aav2461
Kessel, S., Chapman, D., Franks, B., Gedamke, T., Gruber, S., Newman, J., et al. (2014). Predictable Temperature-Regulated Residency, Movement and Migration in a Large, Highly mobile marine Predator (Negaprion brevirostris). Mar. Ecol. Prog. Ser. 514, 175–190. doi:10.3354/meps10966
Kim, S. L., del Rio, C. M., Casper, D., and Koch, P. L. (2012). Isotopic Incorporation Rates for Shark Tissues from a Long-Term Captive Feeding Study. J. Exp. Biol. 215, 2495–2500. doi:10.1242/jeb.070656
Kindling, M., Pampel, H., van de Sandt, S., Rücknagel, J., Vierkant, P., Kloska, G., et al. (2017). The Landscape of Research Data Repositories in 2015: A Re3data Analysis. D-Lib Mag. 23, 0–3. doi:10.1045/march2017-kindling
Klinard, N. V., Matley, J. K., Fisk, A. T., and Johnson, T. B. (2019). Long-term Retention of Acoustic Telemetry Transmitters in Temperate Predators Revealed by Predation Tags Implanted in Wild Prey Fish. J. Fish Biol. 95, 1512–1516. doi:10.1111/jfb.14156
Koch, I. J., and Narum, S. R. (2020). Validation and Association of Candidate Markers for Adult Migration Timing and Fitness in Chinook Salmon. Evol. Appl. 13, 2316–2332. doi:10.1111/eva.13026
Krasnov, A., Koskinen, H., Pehkonen, P., Rexroad, C. E., Afanasyev, S., and Mölsä, H. (2005). Gene Expression in the Brain and Kidney of Rainbow trout in Response to Handling Stress. BMC Genomics 6, 3. doi:10.1186/1471-2164-6-3
Krueger, C. C., Holbrook, C. M., Binder, T. R., Vandergoot, C. S., Hayden, T. A., Hondorp, D. W., et al. (2018). Acoustic Telemetry Observation Systems: Challenges Encountered and Overcome in the Laurentian great lakes. Can. J. Fish. Aquat. Sci. 75, 1755–1763. doi:10.1139/cjfas-2017-0406
Langfelder, P., and Horvath, S. (2008). WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinformatics 9, 559. doi:10.1186/1471-2105-9-559
Lawrence, M. J., Jain-Schlaepfer, S., Zolderdo, A. J., Algera, D. A., Gilmour, K. M., Gallagher, A. J., et al. (2018). Are 3 minutes Good Enough for Obtaining Baseline Physiological Samples from Teleost Fish? Can. J. Zool. 96, 774–786. doi:10.1139/cjz-2017-0093
Lawrence, M. J., Raby, G. D., Teffer, A. K., Jeffries, K. M., Danylchuk, A. J., Eliason, E. J., Hasler, C. T., Clark, T. D., and Cooke, S. J. (2020). Best Practices for non- l, ethal Blood Sampling of Fish via the Caudal Vasculature. J. Fish. Biol. 97, 4–15. doi:10.1111/jfb.14339
Le Pichon, C., Coustillas, J., and Rochard, E. (2015). Using a Multi-Criteria Approach to Assess post-release Recovery Periods in Behavioural Studies: Study of a Fish Telemetry Project in the Seine Estuary. Anim. Biotelemetry 3, 30. doi:10.1186/s40317-015-0062-7
Leal, C. G., Lennox, G. D., Ferraz, S. F. B., Ferreira, J., Gardner, T. A., Thomson, J. R., et al. (2020). Integrated Terrestrial-Freshwater Planning Doubles Conservation of Tropical Aquatic Species. Science 370, 117–121. doi:10.1126/science.aba7580
Lerner, D. T., Björnsson, B. T., and McCormick, S. D. (2007). Aqueous Exposure to 4-Nonylphenol and 17β-Estradiol Increases Stress Sensitivity and Disrupts Ion Regulatory Ability of Juvenile Atlantic Salmon. Environ. Toxicol. Chem. 26, 1433–1440. doi:10.1897/06-451r1.1
Liedvogel, M., Åkesson, S., and Bensch, S. (2011). The Genetics of Migration on the Move. Trends Ecol. Evol. 26, 561–569. doi:10.1016/j.tree.2011.07.009
Linh, V. L. G. (2016). Describing Urban Artisanal Fisheries in Vietnam's Lower Mekong River: A Student's Ventures into Fisheries Science. Fisheries 41, 519–523. doi:10.1080/03632415.2016.1210966
Long, J. A., and Nelson, T. A. (2013). A Review of Quantitative Methods for Movement Data. Int. J. Geographical Inf. Sci. 27, 292–318. doi:10.1080/13658816.2012.682578
Lowe, W. H., and Allendorf, F. W. (2010). What Can Genetics Tell Us about Population Connectivity? Mol. Ecol. 19, 3038–3051. doi:10.1111/j.1365-294X.2010.04688.x
Lower, N., Moore, A., Scott, A. P., Ellis, T., James, J. D., and Russell, I. C. (2005). A Non-invasive Method to Assess the Impact of Electronic Tag Insertion on Stress Levels in Fishes. J. Fish. Biol. 67, 1202–1212. doi:10.1111/j.1095-8649.2005.00815.x
Madsen, S. S., Skovbølling, S., Nielsen, C., and Korsgaard, B. (2004). 17-β Estradiol and 4-nonylphenol Delay Smolt Development and Downstream Migration in Atlantic salmon, Salmo salar. Aquat. Toxicol. 68, 109–120. doi:10.1016/j.aquatox.2004.03.008
Marcy-Quay, B., Sethi, S. A., Therkildsen, N. O., and Kraft, C. E. (2020). Expanding the feasibility of fish and wildlife assessments with close-kin mark–recapture. Ecosphere 11. doi:10.1002/ecs2.3259
Mäkinen, T. S., Niemelä, E., Moen, K., and Lindström, R. (2000). Behaviour of Gill-Net and Rod-Captured Atlantic salmon (Salmo salar L.) during Upstream Migration and Following Radio Tagging. Fish. Res. 45, 117–127. doi:10.1016/S0165-7836(99)00107-1
Martinelli-Liedtke, T. L., Shively, R. S., Holmberg, G. S., Sheer, M. B., and Schrock, R. M. (1999). Nonlethal Gill Biopsy Does Not Affect Juvenile Chinook Salmon Implanted with Radio Transmitters. North Am. J. Fish. Manage. 19, 856–859. doi:10.1577/1548-8675(1999)019<0856:ngbdna>2.0.co;2
Matich, P., and Heithaus, M. R. (2014). Multi-tissue Stable Isotope Analysis and Acoustic Telemetry Reveal Seasonal Variability in the Trophic Interactions of Juvenile Bull Sharks in a Coastal Estuary. J. Anim. Ecol. 83, 199–213. doi:10.1111/1365-2656.12106
Mayr, E. (1961). Cause and Effect in Biology. Science 134, 1501–1506. doi:10.1126/science.134.3489.1501
McCormick, S. D. (1993). Methods for Nonlethal Gill Biopsy and Measurement of Na+, K+-ATPase Activity. Can. J. Fish. Aquat. Sci. 50, 656–658. doi:10.1139/f93-075
McIntyre, P. B., Reidy Liermann, C. A., and Revenga, C. (2016). Linking Freshwater Fishery Management to Global Food Security and Biodiversity Conservation. Proc. Natl. Acad. Sci. U.S.A. 113, 12880–12885. doi:10.1073/pnas.1521540113
McKay, S. K., Schramski, J. R., Conyngham, J. N., and Fischenich, J. C. (2013). Assessing Upstream Fish Passage Connectivity with Network Analysis. Ecol. Appl. 23, 1396–1409. doi:10.1890/12-1564.1
Metcalfe, J. D., Le Quesne, W. J. F., Cheung, W. W. L., and Righton, D. A. (2012). Conservation Physiology for Applied Management of marine Fish: an Overview with Perspectives on the Role and Value of Telemetry. Phil. Trans. R. Soc. B 367, 1746–1756. doi:10.1098/rstb.2012.0017
Milla, S., Mathieu, C., Wang, N., Lambert, S., Nadzialek, S., Massart, S., et al. (2010). Spleen Immune Status Is Affected after Acute Handling Stress but Not Regulated by Cortisol in Eurasian Perch, Perca fluviatilis. Fish Shellfish Immunol. 28, 931–941. doi:10.1016/j.fsi.2010.02.012
Millenium Ecosystem Assessment Board (2005). Ecosystem and Human Well-Being: Wetlands and Water Synthesis. Washington, DC: World Resources Institute.
Miller, K. M., Günther, O. P., Li, S., Kaukinen, K. H., and Ming, T. J. (2017). Molecular Indices of Viral Disease Development in Wild Migrating salmon†. Conserv. Physiol. 5. doi:10.1093/conphys/cox036
Miller, K. M., Li, S., Kaukinen, K. H., Ginther, N., Hammill, E., Curtis, J. M. R., et al. (2011). Genomic Signatures Predict Migration and Spawning Failure in Wild Canadian salmon. Science 331, 214–217. doi:10.1126/science.1196901
Miller, K. M., Schulze, A. D., Ginther, N., Li, S., Patterson, D. A., Farrell, A. P., et al. (2009). Salmon Spawning Migration: Metabolic Shifts and Environmental Triggers. Comp. Biochem. Physiol. Part D: Genomics Proteomics 4, 75–89. doi:10.1016/j.cbd.2008.11.002
Miller, K. M., Teffer, A., Tucker, S., Li, S., Schulze, A. D., Trudel, M., et al. (2014). Infectious Disease, Shifting Climates, and Opportunistic Predators: Cumulative Factors Potentially Impacting Wild salmon Declines. Evol. Appl. 7, 812–855. doi:10.1111/eva.12164
Mommsen, T. P. (2004). Salmon Spawning Migration and Muscle Protein Metabolism: The August Krogh Principle at Work. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 139, 383–400. doi:10.1016/j.cbpc.2004.09.018
Moore, J.-S., Harris, L. N., Le Luyer, J., Sutherland, B. J. G., Rougemont, Q., Tallman, R. F., et al. (2017). Genomics and Telemetry Suggest a Role for Migration Harshness in Determining Overwintering Habitat Choice, but Not Gene Flow, in Anadromous Arctic Char. Mol. Ecol. 26, 6784–6800. doi:10.1111/mec.14393
Mordecai, G. J., Miller, K. M., Bass, A. L., Bateman, A. W., Teffer, A. K., Caleta, J. M., et al. (2021). Aquaculture Mediates Global Transmission of a Viral Pathogen to Wild salmon. Sci. Adv. 7, 1–11. doi:10.1126/sciadv.abe2592
Mullins, R. B., McKeown, N. J., Sauer, W. H. H., and Shaw, P. W. (2018). Genomic Analysis Reveals Multiple Mismatches between Biological and Management Units in Yellowfin Tuna (Thunnus albacares). ICES J. Mar. Sci. 75, 2145–2152. doi:10.1093/icesjms/fsy102
Munaweera, I., Muthukumarana, S., Gillis, D. M., Watkinson, D. A., Charles, C., and Enders, E. C. (2021). Assessing Movement Patterns Using Bayesian State Space Models on Lake Winnipeg Walleye. Can. J. Fish. Aquat. Sci. 78, 1407–1421. doi:10.1139/cjfas-2020-0262
Mushet, D. M., Alexander, L. C., Bennett, M., Schofield, K., Christensen, J. R., Ali, G., et al. (2019). Differing Modes of Biotic Connectivity within Freshwater Ecosystem Mosaics. J. Am. Water Resour. Assoc. 55, 307–317. doi:10.1111/1752-1688.12683
Nagy, Z. T. (2010). A Hands-On Overview of Tissue Preservation Methods for Molecular Genetic Analyses. Org. Divers. Evol. 10, 91–105. doi:10.1007/s13127-010-0012-4
Nathan, R., Getz, W. M., Revilla, E., Holyoak, M., Kadmon, R., Saltz, D., et al. (2008). A Movement Ecology Paradigm for Unifying Organismal Movement Research. Proc. Natl. Acad. Sci. U.S.A. 105, 19052–19059. doi:10.1073/pnas.0800375105
Norman, J. D., Ferguson, M. M., and Danzmann, R. G. (2014). Transcriptomics of Salinity Tolerance Capacity in Arctic Charr (Salvelinus alpinus): a Comparison of Gene Expression Profiles between Divergent QTL Genotypes. Physiol. Genomics 46, 123–137. doi:10.1152/physiolgenomics.00105.2013
O'Hanley, J. R., Wright, J., Diebel, M., Fedora, M. A., and Soucy, C. L. (2013). Restoring Stream Habitat Connectivity: A Proposed Method for Prioritizing the Removal of Resident Fish Passage Barriers. J. Environ. Manage. 125, 19–27. doi:10.1016/j.jenvman.2013.02.055
O'Malley, K. G., Jacobson, D. P., Kurth, R., Dill, A. J., and Banks, M. A. (2013). Adaptive Genetic Markers Discriminate Migratory Runs of C Hinook salmon ( O Ncorhynchus Tshawytscha ) amid Continued Gene Flow. Evol. Appl. 6, 1184–1194. doi:10.1111/eva.12095
O'Toole, A. C., Dechraoui Bottein, M.-Y., Danylchuk, A. J., Ramsdell, J. S., and Cooke, S. J. (2012). Linking Ciguatera Poisoning to Spatial Ecology of Fish: A Novel Approach to Examining the Distribution of Biotoxin Levels in the Great Barracuda by Combining Non-lethal Blood Sampling and Biotelemetry. Sci. Total Environ. 427-428, 98–105. doi:10.1016/j.scitotenv.2011.11.053
Oakenfull, E. A. (1994). Vodka, Meths and DNA. Trends Ecol. Evol. 9, 26. doi:10.1016/0169-5347(94)90230-5
Olsen, R., Taranger, G., Svåsand, T., and Skilbrei, O. (2013). Improved Method for Triacylglycerol-Derived Fatty Acid Profiling by Various Non-lethal and Lethal Sampling Techniques in Atlantic salmon. Aquacult. Environ. Interact. 4, 251–261. doi:10.3354/aei00087
O’Malley, M. A., and Soyer, O. S. (2012). The Roles of Integration in Molecular Systems Biology. Stud. Hist. Philos. Sci. Part C: Stud. Hist. Philos. Biol. Biomed. Sci. 43, 58–68. doi:10.1016/j.shpsc.2011.10.006
Ovenden, J. R., Berry, O., Welch, D. J., Buckworth, R. C., and Dichmont, C. M. (2015). Ocean's Eleven: a Critical Evaluation of the Role of Population, Evolutionary and Molecular Genetics in the Management of Wild Fisheries. Fish Fish 16, 125–159. doi:10.1111/faf.12052
Passow, C. N., Kono, T. J. Y., Stahl, B. A., Jaggard, J. B., Keene, A. C., and McGaugh, S. E. (2019). Nonrandom RNAseq Gene Expression Associated with RNAlater and Flash Freezing Storage Methods. Mol. Ecol. Resour. 19, 456–464. doi:10.1111/1755-0998.12965
Peake, S., Beamish, F. W., McKinley, R. S., Scruton, D. A., and Katopodis, C. (1997). Relating Swimming Performance of lake sturgeon, Acipenser fulvescens, to Fishway Design. Can. J. Fish. Aquat. Sci. 54, 1361–1366. doi:10.1139/cjfas-54-6-136110.1139/f97-039
Peterson, C. T., Grubbs, R. D., and Mickle, A. (2017). An Investigation of Effects of the Deepwater Horizon Oil Spill on Coastal Fishes in the Florida Big Bend Using Fishery-independent Surveys and Stable Isotope Analysis. Southeast. Naturalist 16, G93–G108. doi:10.1656/058.016.0101
Peterson, S. M., and Freeman, J. L. (2009). RNA Isolation from Embryonic Zebrafish and cDNA Synthesis for Gene Expression Analysis. J. Vis. Exp. (30), e1470. doi:10.3791/1470
Piraino, M. N., and Taylor, D. L. (2013). Assessment of Nonlethal Methods for Predicting Muscle Tissue Mercury Concentrations in Coastal Marine Fishes. Arch. Environ. Contam. Toxicol. 65, 715–723. doi:10.1007/s00244-013-9946-9
Poletto, J. B., Cocherell, D. E., Mussen, T. D., Ercan, A., Bandeh, H., Levent Kavvas, M., et al. (2014). Efficacy of a Sensory Deterrent and Pipe Modifications in Decreasing Entrainment of Juvenile green sturgeon (Acipenser medirostris) at Unscreened Water Diversions. Conservation Physiol. 2, cou056. doi:10.1093/conphys/cou056
Pratt, T. C., and Fox, M. G. (2002). Effect of Fin Clipping on Overwinter Growth and Survival of Age-0 Walleyes. North Am. J. Fish. Manage. 22, 1290–1294. doi:10.1577/1548-8675(2002)022<1290:eofcoo>2.0.co;2
Prince, D. J., O’Rourke, S. M., Thompson, T. Q., Ali, O. A., Lyman, H. S., Saglam, I. K., et al. (2017). The Evolutionary Basis of Premature Migration in Pacific salmon Highlights the Utility of Genomics for Informing Conservation. Sci. Adv. 3, e1603198. doi:10.1126/sciadv.1603198
R Core Team (2021). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. Available at: https://www.r-project.org/.
Raby, G. D., Donaldson, M. R., Hinch, S. G., Patterson, D. A., Lotto, A. G., Robichaud, D., et al. (2012). Validation of Reflex Indicators for Measuring Vitality and Predicting the Delayed Mortality of Wild Coho salmon Bycatch Released from Fishing Gears. J. Appl. Ecol. 49, 90–98. doi:10.1111/j.1365-2664.2011.02073.x
Raby, G. D., Packer, J. R., Danylchuk, A. J., and Cooke, S. J. (2014). The Understudied and Underappreciated Role of Predation in the Mortality of Fish Released from Fishing Gears. Fish Fish 15, 489–505. doi:10.1111/faf.12033
Raby, G. D., Johnson, T. B., Kessel, S. T., Stewart, T. J., and Fisk, A. T. (2017). A Field Test of the use of Pop-off Data Storage Tags in Freshwater Fishes. J. Fish Biol. 91, 1623–1641. doi:10.1111/jfb.13476
Raby, G. D., Vandergoot, C. S., Hayden, T. A., Faust, M. D., Kraus, R. T., Dettmers, J. M., et al. (2018). Does Behavioural Thermoregulation Underlie Seasonal Movements in Lake Erie Walleye? Can. J. Fish. Aquat. Sci. 75, 488–496. doi:10.1139/cjfas-2017-0145
Radinger, J., Britton, J. R., Carlson, S. M., Magurran, A. E., Alcaraz-Hernández, J. D., Almodóvar, A., et al. (2019). Effective Monitoring of Freshwater Fish. Fish Fish 20, 729–747. doi:10.1111/faf.12373
Rees, C. B., McCormick, S. D., and Li, W. (2005). A Non-lethal Method to Estimate CYP1A Expression in Laboratory and Wild Atlantic salmon (Salmo salar). Comp. Biochem. Physiol. C: Toxicol. Pharmacol. 141, 217–224. doi:10.1016/j.cca.2005.04.010
Reid, A. J., Carlson, A. K., Creed, I. F., Eliason, E. J., Gell, P. A., Johnson, P. T. J., et al. (2019). Emerging Threats and Persistent Conservation Challenges for Freshwater Biodiversity. Biol. Rev. 94, 849–873. doi:10.1111/brv.12480
Reid, C. H., Vandergoot, C. S., Midwood, J. D., Stevens, E. D., Bowker, J., and Cooke, S. J. (2019). On the Electroimmobilization of Fishes for Research and Practice: Opportunities, Challenges, and Research Needs. Fisheries 44, 576–585. doi:10.1002/fsh.10307
Rennie, M. D., Collins, N. C., Shuter, B. J., Rajotte, J. W., and Couture, P. (2005). A Comparison of Methods for Estimating Activity Costs of Wild Fish Populations: More Active Fish Observed to Grow Slower. Can. J. Fish. Aquat. Sci. 62, 767–780. doi:10.1139/f05-052
Riding, T. A. C., Dennis, T. E., Stewart, C. L., Walker, M. M., and Montgomery, J. C. (2009). Tracking Fish Using “Buoy-based” GPS Telemetry. Mar. Ecol. Prog. Ser. 377, 255–262. doi:10.3354/meps07809
Rodríguez-Jorquera, I. A., Colli-Dula, R. C., Kroll, K., Jayasinghe, B. S., Parachu Marco, M. V., Silva-Sanchez, C., et al. (2019). Blood Transcriptomics Analysis of Fish Exposed to Perfluoro Alkyls Substances: Assessment of a Non-lethal Sampling Technique for Advancing Aquatic Toxicology Research. Environ. Sci. Technol. 53, 1441–1452. doi:10.1021/acs.est.8b03603
Roscoe, D. W., Hinch, S. G., Cooke, S. J., and Patterson, D. A. (2011). Fishway Passage and post-passage Mortality of Up-River Migrating Sockeye salmon in the Seton River, British Columbia. River Res. Applic. 27, 693–705. doi:10.1002/rra.1384
Rose, J. D., Arlinghaus, R., Cooke, S. J., Diggles, B. K., Sawynok, W., Stevens, E. D., et al. (2014). Can Fish Really Feel Pain? Fish Fish 15, 97–133. doi:10.1111/faf.12010
Rudolfsen, T. A., Watkinson, D. A., Charles, C., Kovachik, C., and Enders, E. C. (2021). Developing Habitat Associations for Fishes in Lake Winnipeg by Linking Large Scale Bathymetric and Substrate Data with Fish Telemetry Detections. J. Great Lakes Res. 47, 635–647. doi:10.1016/j.jglr.2021.02.002
Russell, W. M. S., and Burch, R. L. (1959). The Principles of Humane Experimental Technique. London: Methuen.
Sanderson, B. L., Tran, C. D., Coe, H. J., Pelekis, V., Steel, E. A., and Reichert, W. L. (2009). Nonlethal Sampling of Fish Caudal Fins Yields Valuable Stable Isotope Data for Threatened and Endangered Fishes. Trans. Am. Fish. Soc. 138, 1166–1177. doi:10.1577/T08-086.1
Schick, R. S., Loarie, S. R., Colchero, F., Best, B. D., Boustany, A., Conde, D. A., et al. (2008). Understanding Movement Data and Movement Processes: Current and Emerging Directions. Ecol. Lett. 11, 1338–1350. doi:10.1111/j.1461-0248.2008.01249.x
Schultz, A. A., Afentoulis, V. B., Yip, C. J., and Johnson, M. N. (2017). Efficacy of an Acoustic Tag with Predation Detection Technology. North Am. J. Fish. Manage. 37, 574–581. doi:10.1080/02755947.2017.1290720
Schultz, D. R., Perez, N., Tan, C.-K., Mendez, A. J., Capo, T. R., Snodgrass, D., et al. (2005). Concurrent Levels of 11-ketotestosterone in Fish Surface Mucus, Muscle Tissue and Blood. J. Appl. Ichthyol. 21, 394–398. doi:10.1111/j.1439-0426.2005.00650.x
Schwieterman, G. D., Bouyoucos, I. A., Potgieter, K., Simpfendorfer, C. A., Brill, R. W., and Rummer, J. L. (2019). Analysing Tropical Elasmobranch Blood Samples in the Field: Blood Stability during Storage and Validation of the HemoCue Haemoglobin Analyser. Conserv. Physiol. 7, coz081. doi:10.1093/conphys/coz081
Seidel, D. P., Dougherty, E., Carlson, C., and Getz, W. M. (2018). Ecological Metrics and Methods for GPS Movement Data. Int. J. Geographical Inf. Sci. 32, 2272–2293. doi:10.1080/13658816.2018.1498097
Shipley, O. N., Newton, A. L., Frisk, M. G., Henkes, G. A., LaBelle, J. S., Camhi, M. D., et al. (2021). Telemetry-validated Nitrogen Stable Isotope Clocks Identify Ocean-To-Estuarine Habitat Shifts in mobile Organisms. Methods Ecol. Evol. 12, 897–908. doi:10.1111/2041-210X.13567
Sims, D. W., Queiroz, N., Humphries, N. E., Lima, F. P., and Hays, G. C. (2009). Long-term GPS Tracking of Ocean Sunfish Mola Mola Offers a new Direction in Fish Monitoring. PLoS One 4, 1–6. doi:10.1371/journal.pone.0007351
Smith, A., Marty, J., and Power, M. (2015). Non-lethal Sampling of lake sturgeon for Stable Isotope Analysis: Comparing Pectoral Fin-Clip and Dorsal Muscle for Use in Trophic Studies. J. Great Lakes Res. 41, 292–297. doi:10.1016/j.jglr.2014.11.014
Smith, J., Malone, D., Baetscher, D., and Carr, M. (2018). Evaluation and Application of BIOPOLE, a Biopsy Device for In Situ Non-lethal Tissue Extraction for Fishes. Mar. Ecol. Prog. Ser. 595, 149–156. doi:10.3354/meps12575
Somerville, R., Fisher, M., Persson, L., Ehnert-Russo, S., Gelsleichter, J., and Bielmyer-Fraser, G. (2020). Analysis of Trace Element Concentrations and Antioxidant Enzyme Activity in Muscle Tissue of the Atlantic Sharpnose Shark, Rhizoprionodon terraenovae. Arch. Environ. Contam. Toxicol. 79, 371–390. doi:10.1007/s00244-020-00753-8
Stahl, L. L., Snyder, B. D., McCarty, H. B., Cohen, T. R., Miller, K. M., Fernandez, M. B., et al. (2021). An Evaluation of Fish Tissue Monitoring Alternatives for Mercury and Selenium: Fish Muscle Biopsy Samples versus Homogenized Whole Fillets. Arch. Environ. Contam. Toxicol. 81, 236–254. doi:10.1007/s00244-021-00872-w
Stefano, G. B., Kream, R. M., Kuzelova, H., Ptacek, R., Raboch, J., Samuel, J. M., et al. (2014). Comparing Bioinformatic Gene Expression Profiling Methods: Microarray and RNA-Seq. Med. Sci. Monit. Basic Res. 20, 138–142. doi:10.12659/MSMBR.892101
Steffens, W., and Wirth, M. (2005). Freshwater Fish - an Important Source of N-3 Polyunsaturated Fatty Acids: a Review. Arch. Polish Fish. 13, 5–16.
Stevenson, C. F., Bass, A. L., Furey, N. B., Miller, K. M., Li, S., Rechisky, E. L., et al. (2020). Infectious Agents and Gene Expression Differ between Sockeye salmon (Oncorhynchus nerka) Smolt Age Classes but Do Not Predict Migration Survival. Can. J. Fish. Aquat. Sci. 77, 484–495. doi:10.1139/cjfas-2019-0113
Stich, D. S., Zydlewski, G. B., Kocik, J. F., and Zydlewski, J. D. (2015). Linking Behavior, Physiology, and Survival of Atlantic Salmon Smolts during Estuary Migration. Mar. Coastal Fish. 7, 68–86. doi:10.1080/19425120.2015.1007185
Stoot, L. J., Cairns, N. A., Cull, F., Taylor, J. J., Jeffrey, J. D., Morin, F., et al. (2014). Use of Portable Blood Physiology point-of-care Devices for Basic and Applied Research on Vertebrates: a Review. Conservation Physiol. 2, cou011. doi:10.1093/conphys/cou011
Strayer, D. L., and Dudgeon, D. (2010). Freshwater Biodiversity Conservation: Recent Progress and Future Challenges. J. North Am. Benthological Soc. 29, 344–358. doi:10.1899/08-171.1
Strayer, D. L. (2008). Freshwater Mussel Ecology. Berkeley: University of California Press. doi:10.1525/9780520942523
Stuart, J. M., Segal, E., Koller, D., and Kim, S. K. (2003). A Gene-Coexpression Network for Global Discovery of Conserved Genetic Modules. Science 302, 249–255. doi:10.1126/science.1087447
Su, G., Logez, M., Xu, J., Tao, S., Villéger, S., and Brosse, S. (2021). Human Impacts on Global Freshwater Fish Biodiversity. Science 371, 835–838. doi:10.1126/science.abd3369
Sultana, P., tong Anh, V., and Chiem, N. N. (2003). Understanding Livelihoods Dependent on Inland Fisheries in Bangladesh and Southeast Asia: Vietnam Summary Report. Penang: The WorldFish Center.
Sverdrup-Jensen, S. (2002). in Fisheries in the Lower Mekong Basin: Status and Perspectives. Editors A. Bishop, T. Clayton, and C. Barlow (Mekong River Commission: Phnom Penh).
Szymański, M., and Barciszewski, J. (2003). Regulation by RNA. New York: Academic Press, 197–258. doi:10.1016/S0074-7696(03)31005-8
Tannenbaum, J., and Bennett, B. T. (2015). Russell and Burch's 3Rs Then and Now: the Need for Clarity in Definition and Purpose. J. Am. Assoc. Lab. Anim. Sci. 54, 120–132.
Taslima, K., Davie, A., McAndrew, B. J., and Penman, D. J. (2016). DNA Sampling from Mucus in the Nile tilapia,Oreochromis niloticus: Minimally Invasive Sampling for Aquaculture-Related Genetics Research. Aquac. Res. 47, 4032–4037. doi:10.1111/are.12809
Taylor, A. D., and Litvak, M. K. (2017). Timing and Location of Spawning Based on Larval Capture and Ultrasonic Telemetry of Atlantic Sturgeon in the Saint John River, New Brunswick. Trans. Am. Fish. Soc. 146, 283–290. doi:10.1080/00028487.2016.1264471
Teffer, A. K., and Miller, K. M. (2019). A Comparison of Nonlethal and Destructive Methods for Broad-Based Infectious Agent Screening of Chinook Salmon Using High-Throughput qPCR. J. Aquat. Anim. Health 31, 274–289. doi:10.1002/aah.10079
Thompson, N. F., Anderson, E. C., Clemento, A. J., Campbell, M. A., Pearse, D. E., Hearsey, J. W., et al. (2020). A Complex Phenotype in salmon Controlled by a Simple Change in Migratory Timing. Science 370, 609–613. doi:10.1126/SCIENCE.ABA9059
Thorstad, E. B., Rikardsen, A. H., Alp, A., and Okland, F. (2013). The Use of Electronic Tags in Fish Research - an Overview of Fish Telemetry Methods. Turkish J. Fish. Aquat. Sci. 13, 881–896. doi:10.4194/1303-2712-v13
Thorstensen, M., Bates, P., Lepla, K., and Schreier, A. (2019). To Breed or Not to Breed? Maintaining Genetic Diversity in white sturgeon Supplementation Programs. Conserv. Genet. 20, 997–1007. doi:10.1007/s10592-019-01190-4
Thorstensen, M. J., Wiens, L. M., Jeffrey, J. D., Klein, G. M., Jeffries, K. M., and Treberg, J. R. (2021). Morphology and Blood Metabolites Reflect Recent Spatial and Temporal Differences Among Lake Winnipeg Walleye, Sander vitreus. J. Great Lakes Res. 47, 603–613. doi:10.1016/j.jglr.2020.06.015
TinHan, T. C., Mohan, J. A., Dumesnil, M., DeAngelis, B. M., and Wells, R. J. D. (2018). Linking Habitat Use and Trophic Ecology of Spotted Seatrout (Cynoscion Nebulosus) on a Restored Oyster Reef in a Subtropical Estuary. Estuaries and Coasts 41, 1793–1805. doi:10.1007/s12237-018-0391-x
Toews, D. P. L., Taylor, S. A., Streby, H. M., Kramer, G. R., and Lovette, I. J. (2019). Selection on VPS13A Linked to Migration in a Songbird. Proc. Natl. Acad. Sci. U.S.A. 116, 18272–18274. doi:10.1073/pnas.1909186116
Tracey, S. R., Hartmann, K., Leef, M., and McAllister, J. (2016). Capture-induced Physiological Stress and Postrelease Mortality for Southern Bluefin Tuna (Thunnus Maccoyii) from a Recreational Fishery. Can. J. Fish. Aquat. Sci. 73, 1547–1556. doi:10.1139/cjfas-2015-0516
Turner, L. A., and Bucking, C. (2019). The Role of Intestinal Bacteria in Ammonia Detoxification Ability of Teleost Fish. J. Exp. Biol. 222, jeb209882. doi:10.1242/jeb.209882
Tyus, H. M., Starnes, W. C., Karp, C. A., and Saunders, J. F. (1999). Effects of Invasive Tissue Collection on Rainbow Trout, Razorback Sucker, and Bonytail Chub. North Am. J. Fish. Manage. 19, 848–855. doi:10.1577/1548-8675(1999)019<0848:eoitco>2.0.co;2
Vehniäinen, E.-R., Ruusunen, M., Vuorinen, P. J., Keinänen, M., Oikari, A. O. J., and Kukkonen, J. V. K. (2019). How to Preserve and Handle Fish Liver Samples to Conserve RNA Integrity. Environ. Sci. Pollut. Res. 26, 17204–17213. doi:10.1007/s11356-019-05033-0
Veldhoen, N., Beckerton, J. E., Mackenzie-Grieve, J., Stevenson, M. R., Truelson, R. L., and Helbing, C. C. (2014). Development of a Non-lethal Method for Evaluating Transcriptomic Endpoints in Arctic grayling (Thymallus arcticus). Ecotoxicology Environ. Saf. 105, 43–50. doi:10.1016/j.ecoenv.2014.03.030
Veldhoen, N., Ikonomou, M. G., Dubetz, C., MacPherson, N., Sampson, T., Kelly, B. C., et al. (2010). Gene Expression Profiling and Environmental Contaminant Assessment of Migrating Pacific salmon in the Fraser River Watershed of British Columbia. Aquat. Toxicol. 97, 212–225. doi:10.1016/j.aquatox.2009.09.009
Veldhoen, N., Ikonomou, M. G., Rehaume, V., Dubetz, C., Patterson, D. A., and Helbing, C. C. (2013a). Evidence of Disruption in Estrogen-Associated Signaling in the Liver Transcriptome of In-Migrating Sockeye salmon of British Columbia, Canada. Comp. Biochem. Physiol. Part C: Toxicol. Pharmacol. 157, 150–161. doi:10.1016/j.cbpc.2012.10.007
Veldhoen, N., Stevenson, M. R., Skirrow, R. C., Rieberger, K. J., van Aggelen, G., Meays, C. L., et al. (2013b). Minimally Invasive Transcriptome Profiling in salmon: Detection of Biological Response in Rainbow trout Caudal Fin Following Exposure to Environmental Chemical Contaminants. Aquat. Toxicol. 142-143, 239–247. doi:10.1016/j.aquatox.2013.08.016
Verkamp, H., Skomal, G., Winton, M., and Sulikowski, J. (2021). Using Reproductive Hormone Concentrations from the Muscle of white Sharks Carcharodon carcharias to Evaluate Reproductive Status in the Northwest Atlantic Ocean. Endang. Species Res. 44, 231–236. doi:10.3354/esr01109
Vollset, K. W., Lennox, R. J., Davidsen, J. G., Eldøy, S. H., Isaksen, T. E., Madhun, A., et al. (2021). Wild Salmonids Are Running the Gauntlet of Pathogens and Climate as Fish Farms Expand Northwards. ICES J. Mar. Sci. 78, 388–401. doi:10.1093/icesjms/fsaa138
Vucetich, J. A., Bruskotter, J. T., and Nelson, M. P. (2015). Evaluating whether Nature's Intrinsic Value Is an Axiom of or Anathema to Conservation. Conservation Biol. 29, 321–332. doi:10.1111/cobi.12464
Walsh, M. G., and Winkelman, D. L. (2004). Anchor and Visible Implant Elastomer Tag Retention by Hatchery Rainbow Trout Stocked into an Ozark Stream. North Am. J. Fish. Manage. 24, 1435–1439. doi:10.1577/M03-246.1
Wang, S. K., Xu, C., Wang, Y., Yang, G., Zhang, T. T., Zhao, F., et al. (2017). Nonlethal Sampling for Stable Isotope Analysis of Juvenile Chinese sturgeon (Acipenser Sinensis ): Comparing δ13 C and δ15 N Signatures in Muscle and Fin Tissues. J. Appl. Ichthyol. 33, 877–884. doi:10.1111/jai.13433
Weinz, A. A., Matley, J. K., Klinard, N. V., Fisk, A. T., and Colborne, S. F. (2020). Identification of Predation Events in Wild Fish Using Novel Acoustic Transmitters. Anim. Biotelemetry 8, 1–14. doi:10.1186/s40317-020-00215-x
Wendelaar Bonga, S. E. (1997). The Stress Response in Fish. Physiol. Rev. 77, 591–625. doi:10.1152/physrev.1997.77.3.591
Wilson, A. D. M., Hayden, T. A., Vandergoot, C. S., Kraus, R. T., Dettmers, J. M., Cooke, S. J., et al. (2017). Do intracoelomic Telemetry Transmitters Alter the post-release Behaviour of Migratory Fish? Ecol. Freshw. Fish. 26, 292–300. doi:10.1111/eff.12275
Wynne, R., Archer, L. C., Hutton, S. A., Harman, L., Gargan, P., Moran, P. A., et al. (2021). Alternative Migratory Tactics in Brown trout ( Salmo trutta ) Are Underpinned by Divergent Regulation of Metabolic but Not Neurological Genes. Ecol. Evol. 11, 8347–8362. doi:10.1002/ece3.7664
Yada, T., Fukuda, N., Abe, M., and Tsukamoto, K. (2019). Changes in PRL Gene Expression during Upstream Movement of the Japanese Eel, Anguilla Japonica. Zoolog. Sci. 36, 521. doi:10.2108/zs190012
Yoshiyama, R. M., Fisher, F. W., and Moyle, P. B. (1998). Historical Abundance and Decline of Chinook Salmon in the Central Valley Region of California. North Am. J. Fish. Manage. 18, 487–521. doi:10.1577/1548-8675(1998)018<0487:haadoc>2.0.co;2
Zang, L., Shimada, Y., Nishimura, Y., Tanaka, T., and Nishimura, N. (2013). A Novel, Reliable Method for Repeated Blood Collection from Aquarium Fish. Zebrafish 10, 425–432. doi:10.1089/zeb.2012.0862
Zang, L., Shimada, Y., Nishimura, Y., Tanaka, T., and Nishimura, N. (2015). Repeated Blood Collection for Blood Tests in Adult Zebrafish. J. Vis. Exp. (102), e53272. doi:10.3791/53272
Keywords: sublethal, aquatic, genomic, RNA-seq, transcriptomic, molecular, biopsy, interdisciplinary
Citation: Thorstensen MJ, Vandervelde CA, Bugg WS, Michaleski S, Vo L, Mackey TE, Lawrence MJ and Jeffries KM (2022) Non-Lethal Sampling Supports Integrative Movement Research in Freshwater Fish. Front. Genet. 13:795355. doi: 10.3389/fgene.2022.795355
Received: 14 October 2021; Accepted: 17 March 2022;
Published: 25 April 2022.
Edited by:
Denina Bobbie Dawn Simmons, Ontario Tech University, CanadaReviewed by:
Julián Torres-Dowdall, University of Konstanz, GermanyJon Midwood, Department of Fisheries and Oceans, Canada
Copyright © 2022 Thorstensen, Vandervelde, Bugg, Michaleski, Vo, Mackey, Lawrence and Jeffries. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matt J. Thorstensen, bWF0dC50aG9yc3RlbnNlbkBnbWFpbC5jb20=
 Matt J. Thorstensen
Matt J. Thorstensen Carolyn A. Vandervelde
Carolyn A. Vandervelde William S. Bugg
William S. Bugg Ken M. Jeffries
Ken M. Jeffries