- 1Department of Obstetrics and Gynecology, Key Laboratory for Major Obstetric Diseases of Guangdong Province, The Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, China
- 2Key Laboratory of Reproduction and Genetics of Guangdong Higher Education Institutes, The Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, China
- 3Department of Clinical Medicine, The Third Clinical School of Guangzhou Medical University, Guangzhou, China
Background: Male carriers of complex chromosomal rearrangements (CCRs) may have decreased fertility and usually present with azoospermia, oligospermia or teratospermia.
Methods: High-resolution karyotype analysis using G-banding on peripheral blood lymphocytes was performed in an azoospermic male. Copy number variations (CNVs) were detected by chromosomal microarray analysis, and genetic variations were determined by long-read nanopore sequencing with Sanger sequencing for breakpoint confirmation.
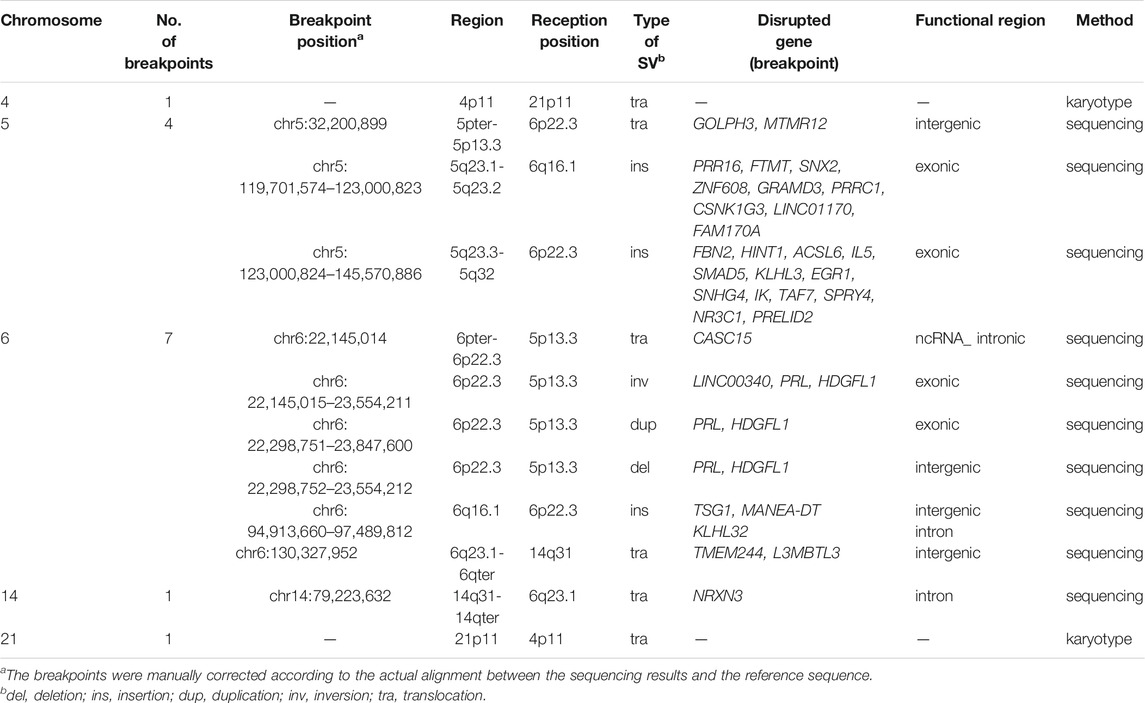
Results: The karyotype of the patient was 46,XY,t(4;21)(p11;p11),t(5;6;14)(p13q22;p22q22;q22), which did not involve CNVs with clinical significance. Twelve breakpoints in chromosomes 5, 6, and 14 were found by long-read nanopore sequencing. Reports on 17 males carrying CCRs with azoospermia were also reviewed.
Conclusion: The extent of asynaptic regions in synaptonemal complexes during pachytene and the disruption of genes involved in male gametogenesis may cause azoospermia in CCR carriers.
Introduction
Complex chromosomal rearrangements (CCRs) are chromosomal structural abnormalities that contain at least three breakpoints on two or more chromosomes, and more than 250 CCR cases have been reported (Pellestor et al., 2011; Poot and Haaf, 2015; Aristidou et al., 2018). According to the complexity and the type of rearrangement, CCRs can generally be divided into three categories: 1) three-way rearrangements, comprising three breakpoints on three chromosomes; 2) double two-way translocations, comprising two independent simple reciprocal or Robertsonian translocations; and 3) exceptional CCRs, wherein each chromosome contains multiple breakpoints (Pellestor et al., 2011).
The majority of balanced CCR carriers have normal phenotypes but have reproductive failure, including repeated miscarriages, infertility, or multiple congenital abnormalities (MCAs) (Kim et al., 2011; Madan, 2012). Most female CCR carriers bear malformed children or have recurrent miscarriages, while male CCR carriers are diagnosed with infertility problems, often presenting with azoospermia, oligozoospermia, or teratozoospermia (Coco et al., 2004).
Approximately 50% of infertile cases can be attributed to male factors, and 10–20% of infertile men are diagnosed with azoospermia (Lee et al., 2011; Rogawski and Khafagy, 2011; Regent et al., 2020). Obstructive azoospermia (OA), caused by a blocked vas deferens with preserved spermatogenesis in the testis, occurs in 40% of men with azoospermia, whereas spermatogenetic malfunctions (nonobstructive azoospermia, NOA) occur in nearly 60% of men with azoospermia (Matzuk and Lamb, 2008; Lee et al., 2011; Ghieh et al., 2019). Genetic abnormalities, including chromosomal or gene defects, are found in approximately 30% of azoospermic men (Krausz and Cioppi, 2021). The frequency of chromosomal aberrations increases to 10–15% in azoospermic men (Matzuk and Lamb, 2008; Kuroda et al., 2020).
Molecular cytogenetic techniques such as fluorescence in situ hybridization (FISH) and comparative genomic hybridization (CGH) arrays, have been used to study CCRs to date, but how different CCRs cause azoospermia remains an important research question. In this study, CCR was identified in an azoospermic male using traditional cytogenetic and molecular genetic methods. The relationship between CCR and azoospermia has been discussed in combination with related literature.
Materials and Methods
Case Report
The patient was a 29-year-old man seeking genetic counselling with his wife due to infertility for the past 3 years. The patient was 167 cm tall and weighed 70 kg. His phenotype and intelligence were normal. No spermatozoa were found in routine semen analysis, and no microdeletions were identified in the AZF-a, AZF-b, and AZF-c genes on the Y chromosome, which are noted in the recommendations of the European Academy of Andrology (EAA) and the European Quality Monitoring Network Group (EMQN) using the Y Chromosome Deletion Detection System (Promega, Madison, WI, United States). Serum testosterone was normal at 10.94 nmol/L (normative values 4.94–33.01 nmol/L), and prolactin was 10.10 ng/ml (normative values 3.46–19.4 ng/m). Serum luteinizing hormone (LH) and follicle-stimulating hormone (FSH) were 3.01 μ/L (normative values 0.57–12.07 μ/L) and 7.24 μ/L (normative values 0.7–11.1 μ/L), respectively. The hormones and menstrual cycle of his wife were entirely normal.
The couple were not consanguineous and had no adverse family history. The patient’s family members (except his sister) underwent genetic tests after detecting his chromosomal abnormality. Written informed consent was obtained for all the tests conducted in this study.
Chromosomal Karyotype Analysis
Peripheral blood was extracted from the patient and his family members (including his wife and his parents). Peripheral blood was cultured at 37°C for 72 h in lymphocyte culture medium (Guangzhou Dahui Biotechnology). Harvested cells were exposed to a hypotonic solution (0.075 M potassium chloride solution) and then immobilized twice with methanol/acetic acid (3:1). Chromosomes were obtained after droplet, trypsin digestion and Giemsa staining. A G-band resolution of approximately 400–550 bands was routinely analysed using MetaSystems (ZEISS, Jena, Germany) on at least 20 metaphase plates per subject. Karyotype description was based on the recommendations of the International Human Cytogenetic Nomenclature (McGowan-Jordan et al., 2020).
Chromosomal Microarray Analysis (CMA)
Genomic DNA was extracted using the QIAamp DNA Blood Mini kit (Qiagen, Dusseldorf, Germany). DNA purity, integrity and concentration were detected by Nanodrop, agarose gel electrophoresis and Qubit, respectively. Next, the genomic DNA was hybridized with Affymetrix’s Cytoscan HD Array (Affymetrix, Santa Clara, CA, United States) following the manufacturer’s procedures. Afymetrix Chromosome Analysis Suite Software 4.2 was used for result analysis. The reported copy number variation (CNV) threshold was 50 kb, and the number of markers was 25. Public databases, such as the Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources (DECIPHER; https://decipher.sanger.ac.uk/), database of genomic variants (DGV; http://dgv.tcag.ca/dgv/app/home), University of California Santa Cruz Genome Browser (UCSC; http://genome.ucsc.edu/) and Online Mendelian Inheritance in Man (OMIM; http://www.omim.org), were used to interpret the data.
Long-Read Nanopore Sequencing and Breakpoint Verification
A sequencing library was prepared using the SQK-LSK109 kit (Oxford Nanopore, Oxford, United Kingdom) according to the manufacturer’s procedures. The library was sequenced on PromethION (Oxford Nanopore, Oxford, United Kingdom). FASTQ files with an average quality less than 7 were filtered. Sequencing reads were aligned to the human reference genome sequence GRCh37/hg19 using minimap2. The structural variations (SVs), including deletion, insertion, duplication, inversion and translocation, were detected using Sniffles. SVs with a frequency of variation greater than 30% and a length greater than 50 bp were retained. Functional annotation of the detected and retained SVs and related genes around the breakpoint was performed in ANNOVAR according to databases such as the 1,000 Genome SV, gnomAD, DGV, dbVar, and Decipher databases. The flow chart of the bioinformatics analysis is shown in Figure 1. The translocation breakpoints were verified using Sanger sequencing on an ABI3730XL sequencer (Applied Biosystems, Foster City, CA, United States). Primer information is available in Supplementary Table S1.
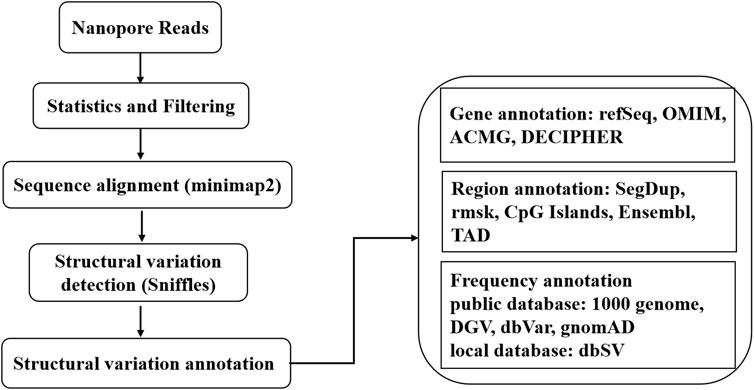
FIGURE 1. The workflow of bioinformatics analysis. The nanopore sequencer generated 3,385,816 reads containing 55.25 Gb. After filtering, 50.50 Gb with 2,964,325 reads were retained. The total number aligned to the human genome was 47.16 Gb, with 2,845,728 reads. A total of 603 SVs were annotated in ANNOVAR according to the above databases. More details are summarized in Table 1, Supplementary Table S2, S3.
Literature Review of CCRs Associated With Azoospermia
A literature search was performed on male CCR carriers with azoospermia to provide information on the relationship and mechanisms between CCRs and azoospermia.
Results
Cytogenetic Investigation
G-banding chromosomal karyotype analysis revealed complex translocation involving chromosomes 4, 5, 6, 14, and 21. The karyotype of the patient was 46,XY,t(4;21)(p11;p11),t(5;6;14)(p13q22;p22q22;q22) (Figure 2A). Both parents and his wife had normal results.
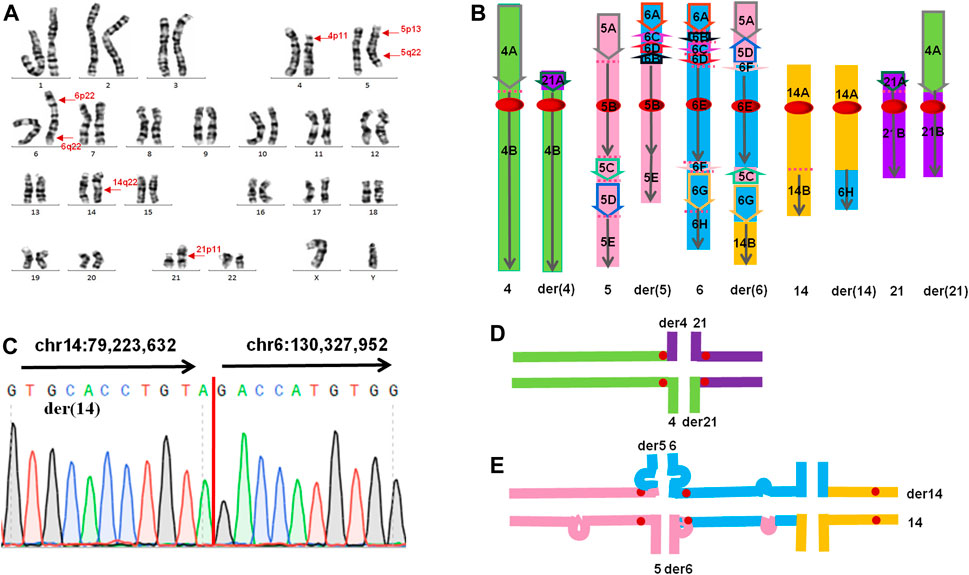
FIGURE 2. (A) The karyotype result of the patient. Arrows indicate the breakpoints. (B) The presumed pattern of variation of involved chromosomes in this CCR. The green colour represents chromosome 4, the pink colour represents chromosome 5, the blue colour represents chromosome 6, the orange colour represents chromosome 14, and the purple colour represents chromosome 21. (C) Sanger sequencing result on der(14). The translocation breakpoints (red line) detected by nanopore sequencing were confirmed by Sanger sequencing. Sequence directions from different chromosomes were indicated by arrows. (D,E) Putative chromosome pairing by complex chromosomal rearrangement during the pachytene stage of meiosis. (D) Schematic view of a quadrivalent configuration. (E) Schematic view of a hexavalent configuration. Substantial portions of the asynaptic regions in the hexavalent configuration mainly contain insertion and inversion.
Chromosomal Microarray Analysis (CMA)
CMA of the peripheral blood for this patient was arr(X,Y)×1,(1–22)×2, and no microdeletion or microduplication with clinical significance was found. The CMA result of the patient was showed in Supplementary Figure S1.
Long-Read Nanopore Sequencing
The total number of bases aligned to the human genome was 47.16 Gb, with a read N50 of 24.36 kb and a depth of 15.72-fold (Table 1). A total of 96.0% of the reads had no less than one alignment to the hg19 reference genome (Figure 3). The sequencing data confirmed the presence of the translocation between chromosome 5 and chromosome 6 as well as the translocation between chromosome 6 and chromosome 14. It also illustrated the presence of an insertion from segments of 5q into chromosome 6. In total, 12 breakpoints in chromosomes 5, 6, and 14 were discovered, which formed three chromosomal rearrangements. Except for the failure to detect a reciprocal translocation between chromosome 4 and chromosome 21 due to method limitations, the sequencing results of the other chromosomal breakpoints were essentially consistent with those of the karyotype results. Combining the karyotype and sequencing results, 14 breakpoints in chromosomes 4, 5, 6, 14, and 21 were detected, illustrating the complex character of this rearrangement (Figure 2). Chromosome 4, chromosome 14 and chromosome 21 contained one breakpoint, chromosome 5 had four breakpoints, and chromosome 6 had seven breakpoints (Figure 2B). Detailed information about the breakpoints and influenced genes is summarized in Table 2.

FIGURE 3. The map for chromosomal coverage through reads alignment with reference sequence using long-read nanopore sequencing. The red line represents the coverage depth of different positions in the chromosome. Cytogenetic bands are inside each chromosome, and the red bands represent the centromere of each chromosome. ChrY chromosomes were not counted in male samples. More details are showed in Supplementary Figure S2.
Discussion
According to the above results, we could infer the rearrangement as follows: four breakpoints and paracentric inversions were in the short arm of chromosome 6 as well as a reciprocal translocation between chromosomes 5p13.3 and 6p22.3. The long arm of chromosome 5 broke three times and invertedly inserted segments 5q23.1 and 5q23.2-q32 into 6q16.1 and 6p22.3, respectively. Additionally, a reciprocal translocation between 4p11 and 21p11 as well as 6q23.1 and 14q31 occurred (Figure 2B). Breakpoints 4p11 and 21p11 were located in centromeric regions with highly repetitive nucleotides and had no applicable reference sequence to be aligned on hg19. According to the UCSC database, reference sequences in these regions are formed by a series of “N”. Due to the limitations of detection in the centromere region of chromosomes, no breakpoints were identified between chromosomes 4 and 21 by long-read nanopore sequencing. According to the classification of CCRs, this case is considered a double CCR.
Within more than 250 cases related to CCRs published thus far, 161 cases concerned male CCR carriers (Olszewska et al., 2020). Approximately 75% of CCRs appear de novo or are inherited maternally (70% of all familial cases) (Pellestor et al., 2011; Olszewska et al., 2020). To date, most men with CCRs have serious fertility problems due to spermatogenesis disorders (Salahshourifar et al., 2009). The source of CCRs is unclear. Ionizing radiation exposure, advanced paternal age, use of immunosuppressive drugs before or during pregnancy and maternal chromosomal instability have been identified as possible risk factors for the origin of CCRs (Farra et al., 2011). The origin of CCR in the patient reported in this study is difficult to determine because the above risk factors were excluded. In addition, it has been proposed that translocations of CCRs may be formed through chromothripsis (Kloosterman et al., 2011; Kloosterman et al., 2012; Weckselblatt et al., 2015; Weckselblatt and Rudd, 2015). Most constitutional chromothripsis events occur de novo, and those investigations thus far have been confirmed to be of patrilineal origin. Chromothripsis is produced simultaneously by multiple DNA double-strand breaks (DSBs) that are close to each other and usually occur on multiple chromosomes simultaneously (Kloosterman et al., 2011; Kloosterman et al., 2012; Weckselblatt et al., 2015; Weckselblatt and Rudd, 2015). Alternatively, mitotic errors in early embryos (Vanneste et al., 2009) or the pulverization of micronuclei (Crasta et al., 2012) could be responsible for numerous DNA breaks. Thus, we infer that this de novo CCR could be due to chromothripsis.
Previous studies have suggested that CCR-related sterility is the result of spermatogenesis stagnation caused by the complex meiosis structure during meiosis (Asia et al., 2014). In the first meiosis, homologous chromosomes pair up to form synaptic complexes (SCs) and undergo meiosis recombination. (Yakut et al., 2013). These events are critical for meiosis fidelity, and defects in these processes may lead to meiosis cessation and sterility (Ferguson et al., 2008). In cross-translocated carriers, homologous chromosome pairing can theoretically be achieved by the formation of tetravalent chromosomes (trivalent chromosomes of Robertson translocation) (Kurahashi et al., 2012). However, in tetravalent chromosomes, regions around the breakpoints are often not fully synaptic (Kurahashi et al., 2012). Defects in SCs are detected by the “pachytene checkpoint”, leading to p53-independent apoptosis (Coco et al., 2004). In this case, a tetravalent configuration (Figure 2D) and a especial hexavalent configuration (Figure 2E) will be formed in pachytene. A large number of unconjugated regions (mainly in the hexavalent configuration) can activate the “pachytene checkpoint” of spermatocytes and initiate the apoptosis process. This could explain the azoospermia observed in this carrier.
In addition, the breakpoints of CCRs may interrupt the genetic structure related to male gametogenesis and interfere with spermatogenesis (Yakut et al., 2013). Gene expression is a complicated process regulated by lots of factors, including cis-acting elements and trans-acting factors at the transcription level. The cis-regulatory elements such as enhancers can act at long distances away from the transcription unit (Hu and Tee, 2017; Bylino et al., 2020). Therefore, breakpoints in the intergenic regions may disturb the interactions of the promoter and transcription unit with its cis-acting regulators by “position effects” and thus affect the expression of gene (Kleinjan and van Heyningen, 2005). All the genes affected by breakpoints in the present case are shown in Table 2. Most of these genes are protein-coding genes that are involved in intracellular transport, signal transduction, transcription regulation and tumour growth inhibition. Among them, FTMT, ACSL6 and FAM170A are mainly or highly expressed in the testis. FTMT (OMIM 608847) is a functional ferritin targeting mitochondrial gene that plays an important role in spermatogenesis. Maccarinelli et al. (Maccarinelli et al., 2017) used knockout mice to verify that FTMT contributes to spermatogenesis and thus to male fertility. ACSL6 (OMIM 604443) activates the cellular metabolism of fatty acids and is highly expressed in the testis. Hale et al. (Hale et al., 2019) demonstrated that ACSL6 activates and integrates DHA into lecithin, which is required for normal spermatogenesis. FAM170A (OMIM 618401; previously Znfd), as a new testicular-specific gene, positively regulates the expression of heat shock genes as a nuclear transcription factor (Lei et al., 2010). Devlin et al. (Devlin et al., 2020) found that FAM170A is critical to sperm head morphology, progressive sperm motility and normal spermatogenesis at the end of spermatogenesis using a knockout mouse model. Azoospermia in the present case may be also the result of disruption of these important genes.
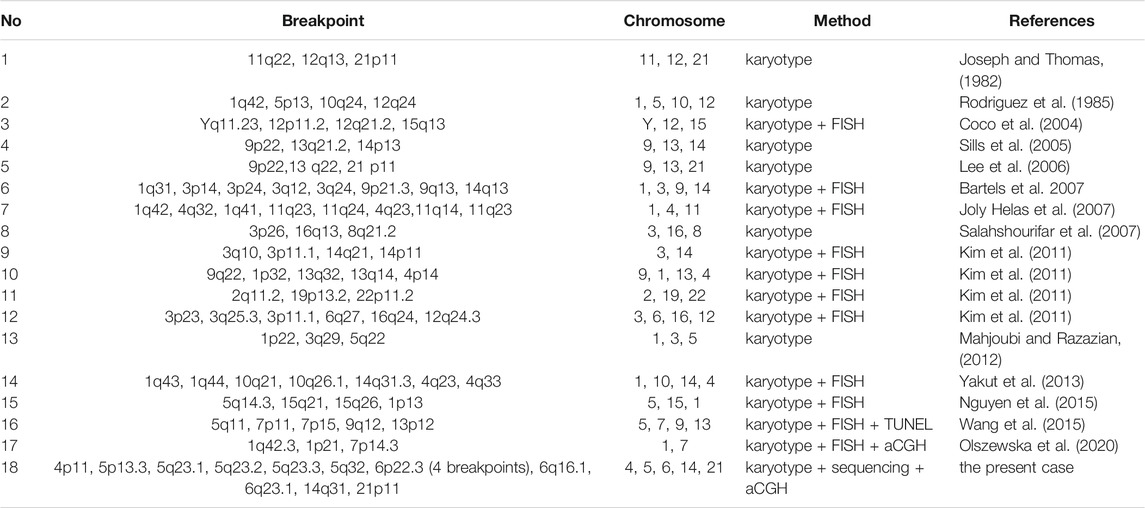
After reviewing the literature, we found reports of 18 men with azoospermia and CCRs to date, including our patient (Table 3). Most of these studies used karyotype and FISH analyses to describe the CCRs. Our study is the only one to date that uses karyotype and long-read nanopore sequencing to characterize CCR in an azoospermic male. In 18 cases, all the chromosomes except chromosomes 17, 18, 20, and X were involved in the CCRs, and chromosome 1 was the most involved. The breakpoints 5p13, 14q31.3 and 21p11 found in our case have been reported in previous studies (Joseph and Thomas, 1982; Rodriguez et al., 1985; Lee et al., 2006; Yakut et al., 2013). Notably, breakpoint 21p11 was found in three different cases (Joseph and Thomas, 1982; Lee et al., 2006), including this one, suggesting that 21p11 may be a hotspot for CCR in azoospermic men.
In conclusion, this is the first report to date to use nanopore sequencing to characterize CCR. We systematically investigated the relationship between CCR and azoospermia using conventional cytogenetic and molecular genetic methods. The occurrence of azoospermia in male CCR carriers may be related to the degree of asynaptic regions in the SC and the destruction of genes related to gametogenesis. In addition, with the development of sequencing technology, third-generation sequencing, especially nanopore sequencing, can achieve rapid sequencing and real-time base calling of long sequences at a reasonable cost. The main limitation of this study is the sample size, which needs to be increased in further studies. In addition, the specific effects of the factors identified in this study on azoospermia need to be further studied.
Data Availability Statement
The datasets for this article are not publicly available due to concerns regarding participant/patient anonymity. Requests to access the datasets should be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of The Third Affiliated Hospital of Guangzhou Medical University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
SX and XY designed the study. KS, PQ, QW, WD, LM, LS, and LZ performed the experiments. LY and XY wrote the paper. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of Guangdong Province (grant no: 2020A0505100062), the Clinical Innovation Research Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory (2018GZR0201002) and the Key Program for Health Collaborative Innovation of Guangzhou (grant no: 201803040009).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to express our gratitude for the helpful comments received from our reviews.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.792539/full#supplementary-material
Abbreviations
CCRs, Complex Chromosomal Rearrangements; CNV, Copy Number Variation; MCAs, Multiple Congenital Abnormalities; OA, Obstructive Azoospermia; NOA, Nonobstructive Azoospermia; FISH, Fluorescence In Situ Hybridization; CGH, Comparative Genomic Hybridization; AZF, Azoospermia Factor; LH, Luteinizing Hormone; FSH, Follicle-Stimulating Hormone; CMA, Chromosomal Microarray Analysis; SVs, Structural Variations; DSBs, Double-Strand Breaks; DHA, Docosahexaenoic Acid.
References
Aristidou, C., Theodosiou, A., Ketoni, A., Bak, M., Mehrjouy, M. M., Tommerup, N., et al. (2018). Cryptic Breakpoint Identified by Whole-Genome Mate-Pair Sequencing in a Rare Paternally Inherited Complex Chromosomal Rearrangement. Mol. Cytogenet. 11, 34. doi:10.1186/s13039-018-0384-2
Asia, S., Vaziri Nasab, H., Sabbaghian, M., Kalantari, H., Zari Moradi, S., Gourabi, H., et al. (2014). A Rare De Novo Complex Chromosomal Rearrangement (CCR) Involving Four Chromosomes in an Oligo-Asthenosperm Infertile Man. Cell J 16 (3), 377–382.
Bartels, I., Starke, H., Argyriou, L., Sauter, S. M., Zoll, B., and Liehr, T. (2007). An Exceptional Complex Chromosomal Rearrangement (CCR) with Eight Breakpoints Involving Four Chromosomes (1;3;9;14) in an Azoospermic Male with normal Phenotype. Eur. J. Med. Genet. 50 (2), 133–138. doi:10.1016/j.ejmg.2006.10.007
Bylino, O. V., Ibragimov, A. N., and Shidlovskii, Y. V. (2020). Evolution of Regulated Transcription. Cells 9 (7), 1675. doi:10.3390/cells9071675
Coco, R., Rahn, M. I., Estanga, P. G., Antonioli, G., and Solari, A. J. (2004). A Constitutional Complex Chromosome Rearrangement Involving Meiotic Arrest in an Azoospermic Male: Case Report. Hum. Reprod. 19 (12), 2784–2790. doi:10.1093/humrep/deh506
Crasta, K., Ganem, N. J., Dagher, R., Lantermann, A. B., Ivanova, E. V., Pan, Y., et al. (2012). DNA Breaks and Chromosome Pulverization from Errors in Mitosis. Nature 482 (7383), 53–58. doi:10.1038/nature10802
Devlin, D. J., Nozawa, K., Ikawa, M., and Matzuk, M. M. (2020). Knockout of Family with Sequence Similarity 170 Member A (Fam170a) Causes Male Subfertility, while Fam170b Is Dispensable in Mice††. Biol. Reprod. 103 (2), 205–222. doi:10.1093/biolre/ioaa082
Farra, C., Singer, S., Dufke, A., Ashkar, H., Monsef, C., and Awwad, J. (2011). De Novo exceptional Complex Chromosomal Rearrangement in a Healthy fertile Male: Case Report and Review of the Literature. Fertil. Sterility 96 (5), 1160–1164. doi:10.1016/j.fertnstert.2011.07.1114
Ferguson, K. A., Chow, V., and Ma, S. (2008). Silencing of Unpaired Meiotic Chromosomes and Altered Recombination Patterns in an Azoospermic Carrier of a T(8;13) Reciprocal Translocation. Hum. Reprod. 23 (4), 988–995. doi:10.1093/humrep/den013
Ghieh, F., Mitchell, V., Mandon-Pepin, B., and Vialard, F. (2019). Genetic Defects in Human Azoospermia. Basic Clin. Androl. 29, 4. doi:10.1186/s12610-019-0086-6
Hale, B. J., Fernandez, R. F., Kim, S. Q., Diaz, V. D., Jackson, S. N., Liu, L., et al. (2019). Acyl-CoA Synthetase 6 Enriches Seminiferous Tubules with the ω-3 Fatty Acid Docosahexaenoic Acid and Is Required for Male Fertility in the Mouse. J. Biol. Chem. 294 (39), 14394–14405. doi:10.1074/jbc.RA119.009972
Hu, Z., and Tee, W.-W. (2017). Enhancers and Chromatin Structures: Regulatory Hubs in Gene Expression and Diseases. Biosci. Rep. 37 (2), BSR20160183. doi:10.1042/BSR20160183
Joly-Helas, G., de La Rochebrochard, C., Mousset-Siméon, N., Moirot, H., Tiercin, C., Romana, S. P., et al. (2007). Complex Chromosomal Rearrangement and Intracytoplasmic Sperm Injection: a Case Report. Hum. Reprod. 22 (5), 1292–1297. doi:10.1093/humrep/del507
Joseph, A., and Thomas, I. M. (1982). A Complex Rearrangement Involving Three Autosomes in a Phenotypically normal Male Presenting with Sterility. J. Med. Genet. 19 (5), 375–377. doi:10.1136/jmg.19.5.375
Kim, J. W., Chang, E. M., Song, S.-H., Park, S. H., Yoon, T. K., and Shim, S. H. (2011). Complex Chromosomal Rearrangements in Infertile Males: Complexity of Rearrangement Affects Spermatogenesis. Fertil. Sterility 95 (1), 349352e341–352345. doi:10.1016/j.fertnstert.2010.08.014
Kleinjan, D. A., and van Heyningen, V. (2005). Long-range Control of Gene Expression: Emerging Mechanisms and Disruption in Disease. Am. J. Hum. Genet. 76 (1), 8–32. doi:10.1086/426833
Kloosterman, W. P., Guryev, V., van Roosmalen, M., Duran, K. J., de Bruijn, E., Bakker, S. C. M., et al. (2011). Chromothripsis as a Mechanism Driving Complex De Novo Structural Rearrangements in the Germline†. Hum. Mol. Genet. 20 (10), 1916–1924. doi:10.1093/hmg/ddr073
Kloosterman, W. P., Tavakoli-Yaraki, M., van Roosmalen, M. J., van Binsbergen, E., Renkens, I., Duran, K., et al. (2012). Constitutional Chromothripsis Rearrangements Involve Clustered Double-Stranded DNA Breaks and Nonhomologous Repair Mechanisms. Cel Rep. 1 (6), 648–655. doi:10.1016/j.celrep.2012.05.009
Krausz, C., and Cioppi, F. (2021). Genetic Factors of Non-obstructive Azoospermia: Consequences on Patients' and Offspring Health. Jcm 10 (17), 4009. doi:10.3390/jcm10174009
Kurahashi, H., Kogo, H., Tsutsumi, M., Inagaki, H., and Ohye, T. (2012). Failure of Homologous Synapsis and Sex-specific Reproduction Problems. Front. Gene 3, 112. doi:10.3389/fgene.2012.00112
Kuroda, S., Usui, K., Sanjo, H., Takeshima, T., Kawahara, T., Uemura, H., et al. (2020). Genetic Disorders and Male Infertility. Reprod. Med. Biol. 19 (4), 314–322. doi:10.1002/rmb2.12336
Lee, I. W., Su, M. T., Hsu, C. C., Lin, Y. H., Chen, P. Y., and Kuo, P. L. (2006). Constitutional Complex Chromosomal Rearrangements in Azoospermic Men-Ccase Report and Literature Review. Urology 68 (6), 1343e1345–8. doi:10.1016/j.urology.2006.09.007
Lee, J. Y., Dada, R., Sabanegh, E., Carpi, A., and Agarwal, A. (2011). Role of Genetics in Azoospermia. Urology 77 (3), 598–601. doi:10.1016/j.urology.2010.10.001
Lei, C., Liu, Q., Wang, W., Li, J., Xu, F., Liu, Y., et al. (2010). Isolation and Characterization of a Novel Zinc finger Gene, ZNFD, Activating AP1(PMA) Transcriptional Activities. Mol. Cel Biochem 340 (1-2), 63–71. doi:10.1007/s11010-010-0401-1
Maccarinelli, F., Regoni, M., Carmona, F., Poli, M., Meyron-Holtz, E. G., and Arosio, P. (2017). Mitochondrial Ferritin Deficiency Reduces Male Fertility in Mice. Reprod. Fertil. Dev. 29 (10), 2005–2010. doi:10.1071/rd16348
Madan, K. (2012). Balanced Complex Chromosome Rearrangements: Reproductive Aspects. A Review. Am. J. Med. Genet. 158a (4), 947–963. doi:10.1002/ajmg.a.35220
Mahjoubi, F., and Razazian, F. (2012). Constitutional Complex Chromosomal Rearrangements in a Klinefelter Patient: Case Report and Review of Literature. J. Assist. Reprod. Genet. 29 (5), 437–441. doi:10.1007/s10815-012-9725-y
Matzuk, M. M., and Lamb, D. J. (2008). The Biology of Infertility: Research Advances and Clinical Challenges. Nat. Med. 14 (11), 1197–1213. doi:10.1038/nm.f.1895
McGowan-Jordan, J., Hastings, R. J., and Moore, S. (2020). ISCN 2020: An International System for Human Cytogenomic Nomenclature. Basel: Karger. doi:10.1159/isbn.978-3-318-06867-2
Nguyen, M. H., Morel, F., Pennamen, P., Parent, P., Douet-Guilbert, N., Le Bris, M. J., et al. (2015). Balanced Complex Chromosome Rearrangement in Male Infertility: Case Report and Literature Review. Andrologia 47 (2), 178–185. doi:10.1111/and.12245
Olszewska, M., Stokowy, T., Pollock, N., Huleyuk, N., Georgiadis, A., Yatsenko, S., et al. (2020). Familial Infertility (Azoospermia and Cryptozoospermia) in Two Brothers-Carriers of T(1;7) Complex Chromosomal Rearrangement (CCR): Molecular Cytogenetic Analysis. Int. J. Mol. Sci. 21 (12). doi:10.3390/ijms21124559
Pellestor, F., Anahory, T., Lefort, G., Puechberty, J., Liehr, T., Hédon, B., et al. (2011). Complex Chromosomal Rearrangements: Origin and Meiotic Behavior. Hum. Reprod. Update 17 (4), 476–494. doi:10.1093/humupd/dmr010
Poot, M., and Haaf, T. (2015). Mechanisms of Origin, Phenotypic Effects and Diagnostic Implications of Complex Chromosome Rearrangements. Mol. Syndromol 6 (3), 110–134. doi:10.1159/000438812
Regent, B., Skrobisz, K., Skrobisz, K., Kozak, O., Matuszewski, M., and Studniarek, M. (2020). MRI in the Evaluation of the Azoospermic Male. Diagn. Interv. Radiol. 26 (4), 271–276. doi:10.5152/dir.2019.19189
Rodriguez, M. T., Martin, M. J., and Abrisqueta, J. A. (1985). A Complex Balanced Rearrangement Involving Four Chromosomes in an Azoospermic Man. J. Med. Genet. 22 (1), 66–67. doi:10.1136/jmg.22.1.66
Rogawski, K. M., and Khafagy, R. (2011). Re: Lee et al.: Role of genetics in azoospermia (Urology 2011;77:598-601). Urology 78 (6), 1445–1446. ; author reply 1445. doi:10.1016/j.urology.2011.08.035
Salahshourifar, I., Gilani, M. A. S., Vosough, A., Tavakolzadeh, T., Tahsili, M., Mansori, Z., et al. (2007). De Novo complex Chromosomal Rearrangement of 46, XY, T (3; 16; 8) (P26; Q13; q21.2) in a Non-obstructive Azoospermic Male. J. Appl. Genet. 48 (1), 93–94. doi:10.1007/bf03194664
Salahshourifar, I., Shahrokhshahi, N., Tavakolzadeh, T., Beheshti, Z., and Gourabi, H. (2009). Complex Chromosomal Rearrangement Involving Chromosomes 1, 4 and 22 in an Infertile Male: Case Report and Literature Review. J. Appl. Genet. 50 (1), 69–72. doi:10.1007/bf03195655
Sills, E. S., Kim, J. J., Witt, M. A., and Palermo, G. D. (2005). Non-obstructive Azoospermia and Maturation Arrest with Complex Translocation 46,XY t(9;13;14)(p22;q21.2;p13) Is Consistent with the Luciani-Guo Hypothesis of Latent Aberrant Autosomal Regions and Infertility. Cell Chromosome 4, 2. doi:10.1186/1475-9268-4-2
Vanneste, E., Voet, T., Le Caignec, C., Ampe, M., Konings, P., Melotte, C., et al. (2009). Chromosome Instability Is Common in Human Cleavage-Stage Embryos. Nat. Med. 15 (5), 577–583. doi:10.1038/nm.1924
Wang, L., Iqbal, F., Li, G., Jiang, X., Bukhari, I., Jiang, H., et al. (2015). Abnormal Meiotic Recombination with Complex Chromosomal Rearrangement in an Azoospermic Man. Reprod. BioMedicine Online 30 (6), 651–658. doi:10.1016/j.rbmo.2015.02.015
Weckselblatt, B., Hermetz, K. E., and Rudd, M. K. (2015). Unbalanced Translocations Arise from Diverse Mutational Mechanisms Including Chromothripsis. Genome Res. 25 (7), 937–947. doi:10.1101/gr.191247.115
Weckselblatt, B., and Rudd, M. K. (2015). Human Structural Variation: Mechanisms of Chromosome Rearrangements. Trends Genet. 31 (10), 587–599. doi:10.1016/j.tig.2015.05.010
Keywords: complex chromosomal rearrangement (CCR), azoospermia, karyotype, whole-genome sequencing, gametogenesis
Citation: Liang Y, Xie Y, Kong S, Pan Q, Qiu W, Wang D, Li M, Lin S, Liu Z and Sun X (2022) Complex Chromosomal Rearrangement Causes Male Azoospermia: A Case Report and Literature Review. Front. Genet. 13:792539. doi: 10.3389/fgene.2022.792539
Received: 10 October 2021; Accepted: 25 January 2022;
Published: 24 February 2022.
Edited by:
Laia Rodriguez-Revenga, Hospital Clínic de Barcelona, SpainReviewed by:
Meritxell Jodar, Hospital Clínic de Barcelona, SpainMarta Olszewska, Institute of Human Genetics (PAN), Poland
Copyright © 2022 Liang, Xie, Kong, Pan, Qiu, Wang, Li, Lin, Liu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofang Sun, eGlhb2ZhbmdzdW5AZ3pobXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Yi Liang1,2†
Yi Liang1,2† Yingjun Xie
Yingjun Xie Ding Wang
Ding Wang Xiaofang Sun
Xiaofang Sun