
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 03 March 2022
Sec. RNA
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.782957
Tilapia (Oreochromis sp.) is one of the important economical fishes in the world. Streptococcosis is commonly found in tilapia, causing severe and devastating effects in tilapia cultures. Streptococcus agalactiae and Streptococcus iniae are the predominant pathogens causing tilapia streptococcosis. To understand the molecular mechanisms underlying differential streptococcal infection patterns, Nile tilapias (Oreochromis niloticus) were infected by 1 × 107 CFU/mL S. agalactiae, 1 × 107 CFU/mL S. iniae, and 1 × 107 CFU/mL S. agalactiae and S. iniae (1:1), respectively, and transcriptome analysis was conducted to the intestine samples of Nile tilapia (Oreochromis niloticus) at 6, 12, 24 h, and 7 days post-infection. A total of 6,185 genes that differentially expressed among groups were identified. Eight differentially expressed genes (DEGs) including E3 ubiquitin-protein ligase TRIM39-like, C-X-C motif chemokine 10-like(CXCL 10), C-C motif chemokine 19-like, interleukin-1 beta-like, IgM heavy chain VH region, partial, IgG Fc-binding protein, proteasome subunit beta type-8 (PSMB8), and ATP synthase F(0) complex subunit B1, mitochondrial that involved in the immune system were selected, and their expression levels in the coinfection group were significantly higher than those in either of the single infection groups. These genes were associated with four different KEGG pathways. Additionally, the differential expression of eight DEGs was validated by using the RT-qPCR approach, and their immunological importance was discussed. The results provided insights into the responses of tilapia against S. agalactiae and S. iniae at the transcriptome level, promoting our better understanding of immune responses for aquatic animal against Streptococcus.
Tilapia (Oreochromis niloticus) is an economic and important aquaculture fish species in the world. China is the world’s largest producer of tilapia. In 2020, the aquaculture production of tilapia reached 1.6417 million tons, accounting for about 40% of the world’s total tilapia production, and the export volume of tilapia ranked among the top three in aquatic product exports in China (Yearbook, 2020). However, with the rapid development of the tilapia aquaculture industry, tilapia cultures have encountered tremendous challenges caused by disease outbreaks. As a common disease caused by pathogenic bacteria, streptococcosis has devastating effects in tilapia cultures (Anshary et al., 2014). Gram-positive bacteria S. agalactiae and S. iniae are the main pathogens of streptococcosis (Maulu et al., 2021). In 2019, the economic losses for the tilapia aquaculture industry caused by S. agalactiae and S. iniae have reached around 2.5 billion RMB (Huang et al., 2014; Tavares et al., 2018; Shiry et al., 2019). S. agalactiae is also known as group B Streptococcus (Brown, 1939). Both of them are recognized as causative agents of zoonosis with a broad host range, including horse, pig, and fish (Mohan, 1947; Monteverde and Simeone, 1951; Poppert et al., 2009). Typical symptoms of the Streptococcus-infected tilapia include blackening of the fish surface, prominent or turbid eyeballs, whiteness, bleeding, abdominal spots, and bleeding on the inner side of the lid (Perera et al., 1994). Previous research studies on tilapia infected by S. agalactiae and S. iniae focused mainly on the isolation, identification and typing of strains, screening of drugs for the prevention and treatment of Streptococcus-infected tilapia, the drug resistance, and vaccine development (Suanyuk et al., 2008; Cai et al., 2020; De Sousa et al., 2021). Most research studies on the immune mechanism of tilapia mainly focused on the kidney and spleen (Zhu et al., 2015; Zhu et al., 2017). However, the knowledge of the molecular mechanisms in immune response aspects against S. agalactiae and S. iniae in the intestine of tilapia is still limited.
The outbreak of the disease is often caused by the joint action of multiple pathogens (Austin and Allen-Austin, 1985). For example, S. agalactiae and S. iniae have previous been isolated from tilapia suffering from streptococcosis (Chen et al., 2007), indicating that coinfection of the two pathogens is common in tilapia. Although the transcriptome profiling in the spleen or kidney of tilapia (Oreochromis niloticus) infected by Streptococcus agalactiae at the early stage has been studied previously (Zhang et al., 2013), investigations regarding the effect caused by coinfection are still lacking (Li et al., 2014).
With the rapid development of high-throughput sequencing, transcriptome analysis has been widely applied to investigate molecular mechanisms underlying host immune response upon infections. For example, Wang et al. carried out transcriptome sequencing of spleen samples from Oncorhynchus mykiss infected with Yersinia ruckeri and identified many KEGG pathways and differentially expressed genes associated with the immune system including CCR9, CXCL11, IL-1 beta, and CARD9 (Wang et al., 2021). Zhang et al. identified various essential genes which play roles on regulating myogenesis from the transcriptome analysis of Trichiurus lepturus (Zhang et al., 2016). Tilapia is a freshwater teleost species, and the defense action against infection is mainly achieved by the innate immune system (Kordon et al., 2018). The mucosal immune system in the intestine of tilapia is an important barrier to infectious microorganisms (Andani et al., 2012). In spite of classical immune organs, a recent study revealed that the intestinal epithelium is the main entry site of Streptococcus in tilapia, and the intestine plays an important defensive role against Streptococcus (Iregui et al., 2016). The impact of Streptococcus on the intestine of tilapia warrants further study to better understand the role of intestinal immune function in bony fish against Streptococcus. In this study, transcriptome analysis was conducted in tilapia challenged by coinfections of S. agalactiae and S. iniae, as well as by single infections of each of the two pathogens. Differentially expressed genes (DEG) and their corresponding metabolic pathways in the intestine were identified. The molecular mechanisms underlying the immune responses of tilapia following infections are discussed.
The S. agalactiae standard strain ATCC13813 and the S. iniae standard strain ATCC29178 were donated by the Pearl River Fisheries Research Institute of Chinese Academy of Fishery Sciences (Shin et al., 2006; Guo et al., 2017). The bacteria were identified and grown in BHI liquid shaken cultures (180 rpm and 28°C) for one day.
O. niloticus (24 ± 1.02 g) were obtained from the Guangdong Tilapia Fine Breeding Farm (each fish was free from Streptococcus spp.). Before experimental challenge, the fish were acclimated in the laboratory (28°C, 6.8–7.2 pH) for 2 weeks. Tilapia were randomly divided into four groups, including the single infection of S. agalactiae (AG), the single infection of S. iniae (IG), and the coinfection of S. agalactiae and S. iniae (MG) and control (CG), with 60 fish in each tank at a temperature of 28 ± 0.5°C. The fish in the AG, IG, and MG groups were injected with 0.3 ml final concentration of 1 × 107 CFU/ml S. agalactiae, 1 × 107 CFU/ml S. iniae, and 1 × 107 CFU/mL S. agalactiae and S. iniae (1:1), respectively (lethal concentration 50%). Meanwhile, the fish in the control group were injected with 0.3 ml normal saline and cultured in a tank with a continuous supply of water. At 0, 6, 12, 24 h, and 7 days following the infection, 10 fish were collected from each of the appropriate tank at each timepoint randomly. (Ten biological replicates were set up from each timepoint and treatment.) Intestine samples from each fish were collected and frozen in liquid nitrogen immediately and stored at -80 °C until RNA extraction.
Prior to RNA extraction, individual samples were ground into powder in the presence of liquid nitrogen using a mortar and pestle. Total RNA was extracted according to the instructions of the RNeasy Plus Universal Mini Kit (Qiagen, China). Then the quantity and quality of RNA were determined by the BioTek microplate reader and denaturing agarose gel electrophoresis.
The NEB Next® Ultra™ RNA Library Prep Kit (NEB, United States) was used to create cDNA libraries. First Strand Synthesis Reaction Buffer, random primers, and reverse transcriptase (Invitrogen, China) were used to synthesize the first-strand cDNA. The second-strand cDNA was synthesized and purified, followed by end repair and adapter ligation. Qubit 2.0 was used to quantify the chosen products enriched by PCR amplification. Sequencing was performed using the Illumina HiSeq 4000, and the sequence data were obtained by Illumina Pipeline Software v1.6 (Biomarker Technologies Inc., Beijing, China).
Through trimming the adapter-only sequences, empty reads, poly-N stretches (>10% total N), low-quality reads, and uncertain nucleotide, we obtained clean data from raw data using the fastq filter, and the clean data were then assembled by SOAP de novo program. In addition, the Q10, Q20, Q30, GC content, and sequence duplication level of the clean data were simultaneously calculated. All following analyses were based on the high-quality clean data.
The Nile tilapia reference genome and gene model annotation file were downloaded from the NCBI (https://www.ncbi.nlm.nih.gov/genome/197), Kyoto Encyclopedia of Genes and Genomes. The index of the reference genome was built using Bowtie v2.2.3, and paired-end clean reads were aligned to the reference genome using TopHat v2.0.12.
The read numbers mapped to each gene was counted by HTSeq v0.6.1. Fragments per kilobase of transcript per millions base pairs sequenced (FPKM) was used to measure the expression levels of each sample’s transcripts or genes. Differentially expressed genes (DEGs) between each group were explored using DEseq, and we used the Benjamini–Hochberg procedure to assess the statistical significance. Genes were considered to be differentially expressed when their |fold change| ≥ 2 and the false discovery rate (FDR) < 0.01. All the DEGs were further annotated by querying against the GO and KEGG databases. The results were obtained using the GO-seq and KOBAS (2.0), respectively.
To validate the results obtained by RNA-seq, eight genes were used for quantitative real-time PCR (RT-qPCR) analysis. Primer 5.0 was used to design the gene specific primers based on the contig sequence (Table 1). The RNA samples used for RT-qPCR amplification were the same as those used to construct the RNA-seq library mentioned earlier. The RT-qPCRs were performed on the CFX96 real-time PCR detection system (BioRad) with SYBR Premix Ex Taq™ (TaKaRa). PCR cycling’s initial degeneration was at 95°C (30 s), followed by 40 cycles of degeneration at 94°C (5s) and appropriate annealing/expansion temperature (60°C, 30 s), and additional temperature-increasing step of 65–95°C was used to generate the melting curve, in response to the fluorescence intensity of three biological replicates of each gene products, with a threshold cycle (Ct) value, the non-template amplification was run as control for each experiment, then the relative quantitative were translated into fold change.
The statistical difference of results were analyzed by the independent sample t test in the SPSS statistics tool, then the data were expressed as mean ± standard deviation, and compared the change in relative gene expression between RT-qPCR and RNA-seq.
A total of 5,870 million clean data were obtained from each sample. Clean data with similar Q30 base percentage more than 92.05% were obtained from all groups. Moreover, 61.31–87.63% from each group were matched to reference genomic sequence, respectively (Supplementary Tables S1,S2).
A total of 18,972 genes from four groups at each timepoint were identified with clear annotations in intestine samples. Among them, a total of 6,185 differentially expressed genes were detected, and 3,876 and 2,309 genes were significantly upregulated and downregulated, respectively (Supplementary Table S3).
The minimum numbers of DEGs in the AG, IG, and the MG groups were 2,874 at 6 h, 2,687 at 12 h, and 2,613 at 6 h, respectively. The maximum number of DEGs in all experimental groups appeared at 24 h (3,792, 3,319, and 3,064, respectively) (Figure 1).
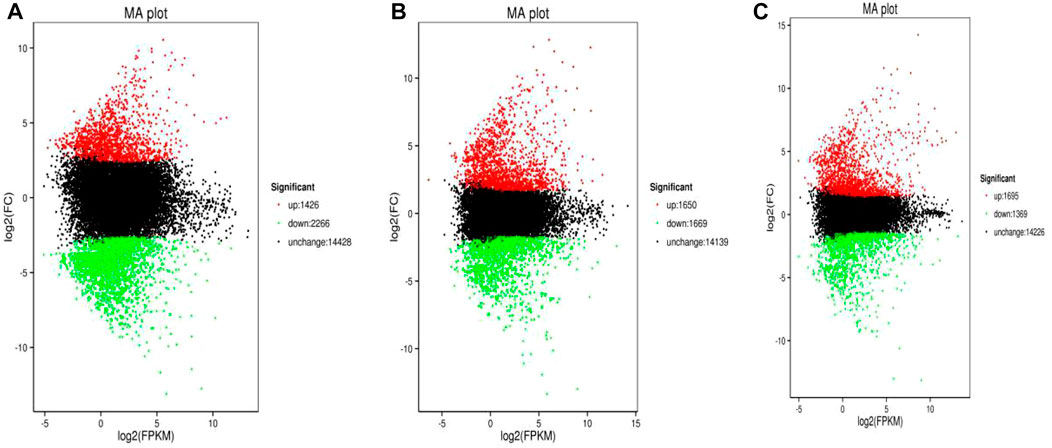
FIGURE 1. Volcano plot of Nile tilapia DEGs identified from the Streptococcus challenge. AG group 24 h vs control was shown in (A), IG group 24 h vs control was shown in (B), while MG group 24 h vs control was displayed in (C). The log2(FPKM) value represented the mean expression level of each gene, and each dot represented one gene. The up-regulated genes were shown in red dots, the down-regulated genes were shown in green dots, while genes with no differential expression were shown in black dots.
Through functional annotation and screening, we obtained 136 immunologically DEGs. Among them, we selected eight significant DEGs associated with intestinal inflammation in Nile tilapia including E3 ubiquitin-protein ligase TRIM39-like, C-X-C motif chemokine 10-like(CXCL 10), C-C motif chemokine 19-like, interleukin-1 beta-like, IgM heavy chain VH region, partial, IgG Fc-binding protein, proteasome subunit beta type-8(PSMB8), and ATP synthase F(0) complex subunit B1, mitochondrial (Figure 2; Table 2).
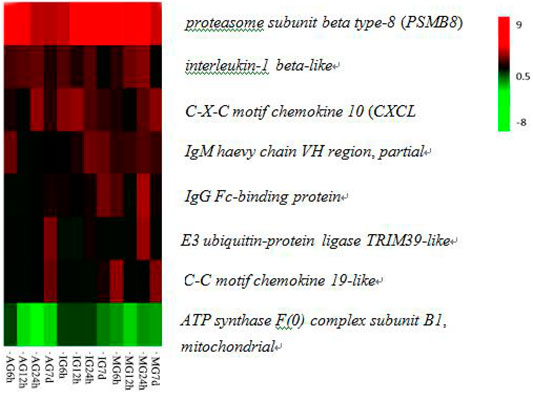
FIGURE 2. Hierarchical clustering of eight DEGs related to immune responses in the O. niloticus intestine.
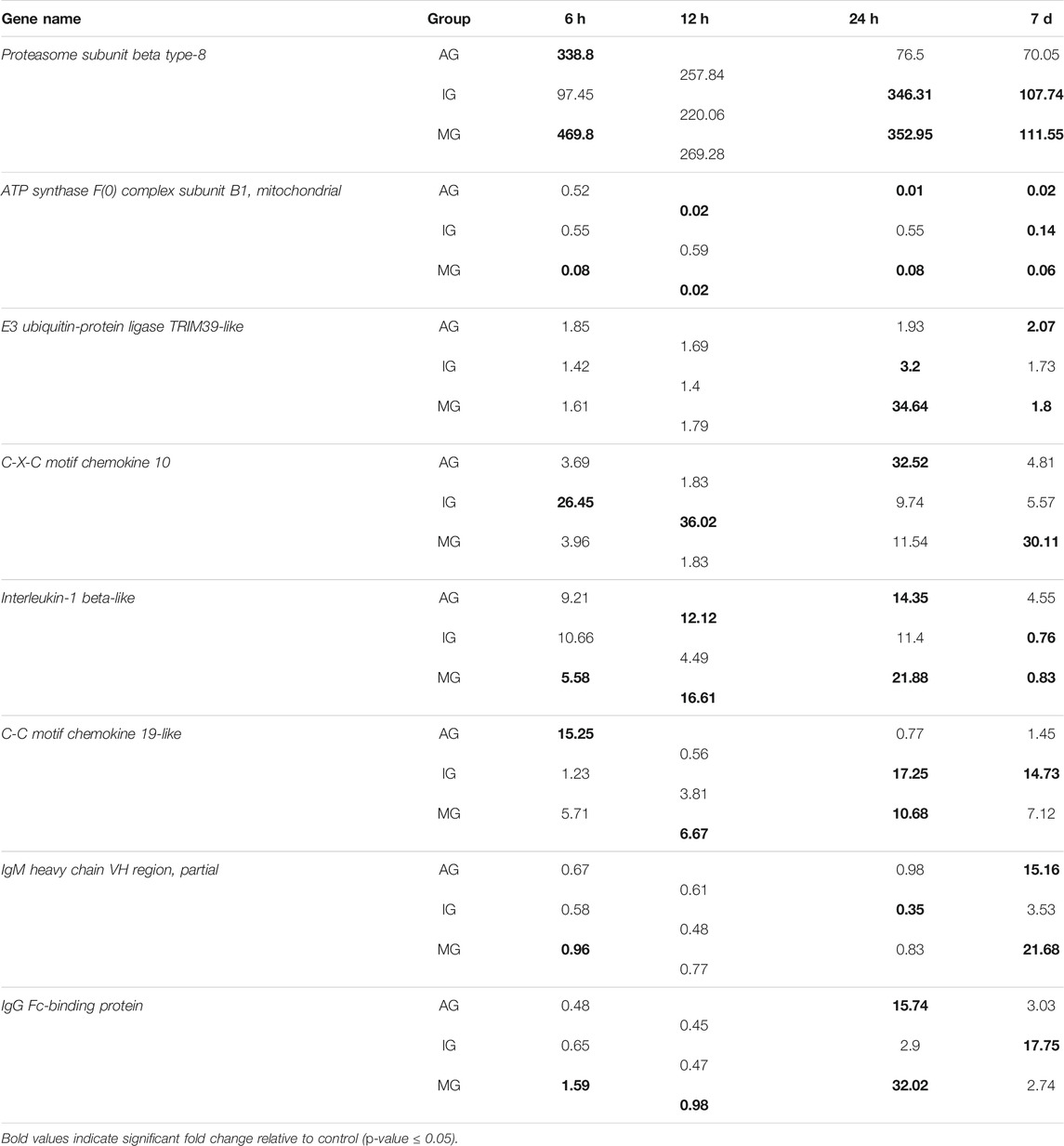
TABLE 2. Differentially expressed immune-related genes from O. niloticus following Streptococcus challenge.
The differentially expressed genes were annotated for gene ontology by Blast2GO; the gene ontology (GO) terms of different groups were classified to biological process, cellular component, and molecular function. KEGG was used to investigate gene functions. After annotation, 6,185 DEGs were grounded into 100 obviously enriched pathways. The most enriched pathways of each treatment group are shown in Figure 3. The largest enriched pathway was the cytokine–cytokine receptor interaction. In addition, many immune-related pathways also exhibited DEGs enrichment, such as phagosome, calcium signaling pathway, CAMs, proteasome, and oxidative phosphorylation. Some enriched immune-related DEGs in these pathways are shown in Supplementary Table S4.
Among the eight DEGs with significance, C-X-C motif chemokine 10-like, C-C motif chemokine 19-like, and interleukin-1 beta-like belong to the cytokine–cytokine receptor interaction pathway, IgM heavy chain VH region, partial, and IgG Fc-binding protein belonged to the calcium signaling pathway, E3 ubiquitin-protein ligase TRIM39-like and proteasome subunit beta type-8 (PSMB8) belonged to the proteasome pathway, and ATP synthase F(0) complex subunit B1 belonged to the oxidative phosphorylation pathway. These four pathways were related to intestinal inflammation in Nile tilapia (Li et al., 2020). Some key immune-related DEGs were identified from key immune pathways, including the chemokines, immunoglobulin, and ubiquitin proteasome.
To validate the DEGs identified by RNA-seq, eight differentially expressed genes associated with Nile tilapia intestinal inflammation were selected for RT-qPCR confirmation (Figure 4). Melting curve analysis revealed a single product for all tested genes, ensuring the primer specificity. The relative fold changes from RT-qPCR were compared with the RNA-seq expression analysis results. As shown in Supplementary Table S5, the RT-qPCR results revealed that these genes had the same upregulation or downregulation trend with the RNA-seq analysis, indicating the accuracy and reliability of RNA-seq expression analysis.
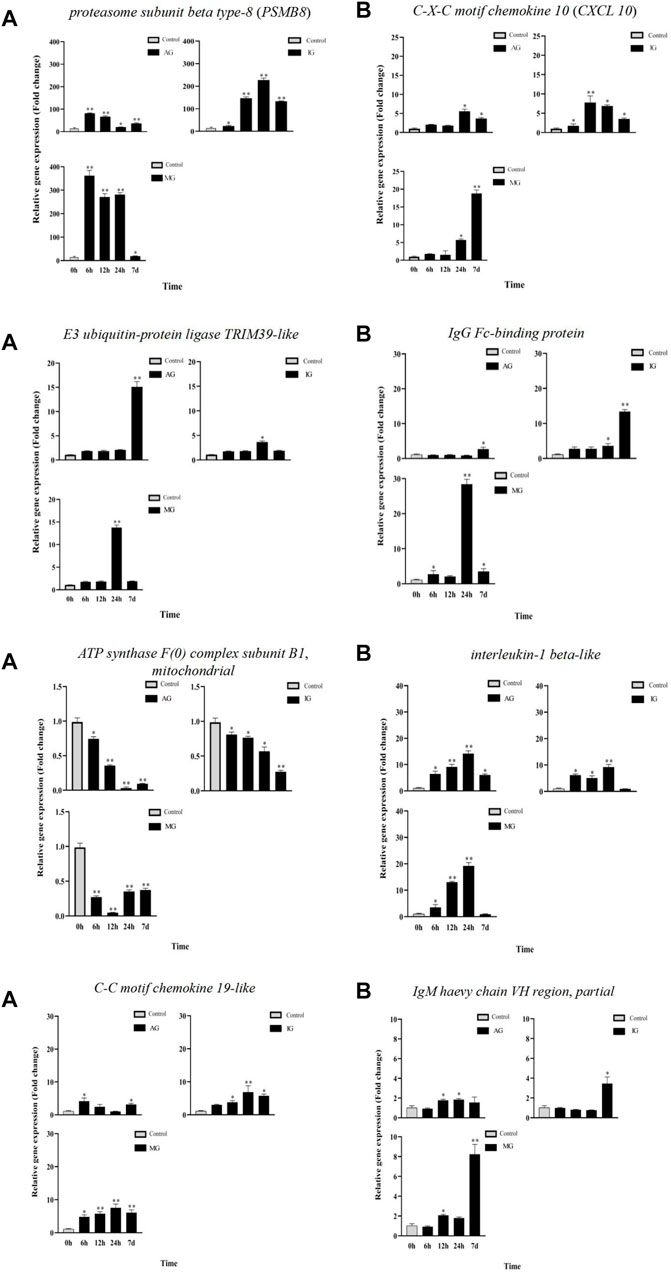
FIGURE 4. Gene expression analysis from RT-qPCR of Nile tilapia infected with Streptococcus. PSMB8, E3 ubiquitin-protein ligase TRIM39-like, ATP synthase F(0) complex subunit B1, mitochondrial and C-C motif chemokine 19-like gene expression analysisi were shown in (A). CXCL 10 , IgG Fc-binding protein, interleukin-1 beta-like and IgM haevy chain VH region, partial gene expression analysis were shown in (B).
The intestine is an important immune organ of fish, and a variety of immune cells are distributed in the epithelial layer and mucosa of the fish intestine (Salinas, 2015). Due to the significant role it plays in immune-related disorders, the intestine was chosen for RNA-seq analysis in this study. In order to understand the acute and chronic effects of Streptococcus on tilapia, fish samples were collected at 6, 12, 24 h, and 7 days, and 6,185 DEGs were identified from the intestine in tilapia, including 3,876 significantly upregulated genes and 2,309 significantly downregulated genes. Immune-related DEGs in all of the three experimental groups reached the maximum number at 24 h. Previous studies have shown that a certain concentration of bacteria had a stimulating effect on fish (Kacha et al., 2009).
According to the result of functional enrichment analyses, six immune-related pathways were highlighted, including four pathways associated with inflammatory responses (the calcium signaling pathway, the cytokine–cytokine receptor interaction pathway, the proteasome pathway, and the oxidative phosphorylation pathway). These results suggested that single infection or coinfection of Streptococcus induced an inflammatory response in the intestines of tilapia.
The intestine is one of the gut-associated lymphoid organs in the body, which can generate great amounts of IgT antibodies, serving as the first line of defense against microbial invasion (Piazzon et al., 2016). It has been reported that the immune-related genes in the calcium signaling pathway were induced in Paralichthys olivaceus with the infection of Edwardsiella tarda (Xiu et al., 2019). The results of this study revealed the significant enrichment of genes in the calcium signaling pathways, which suggested that this pathway played an important role against the invasion by Streptococcus. As part of this pathway, the IgM heavy chain VH region and IgG-Fc binding protein were significantly upregulated. In addition, the expression of these two genes in the coinfection group was significantly higher than that in the single infection group. IgG-Fc binding protein can not only bind to immunoglobulins but also stimulate the proliferation and differentiation of B lymphocytes (Fillatreau et al., 2013). After Streptococcus infection, IgG-Fc binding protein was significantly upregulated in the intestine within 7 days, implying that IgG-Fc binding protein, as an immunoglobulin binding factor, participated in the immune response to bacterial challenge. This result suggested that Streptococcus can induce the proliferation and activation of B lymphocytes. This effect was more pronounced in the coinfection group. The IgM encoded by the IgM heavy chain VH region is one of the most abundant immunoglobulin molecules in bony fish (Akula and Hellman, 2017). IgM can combine with antigen molecules to form antigen–antibody immune complexes and then be engulfed by macrophages (Gallily et al., 1982). Previous results showed that alginic acid vaccination results in a significant increase in the number of IgM in Oncorhynchus mykiss (Gioacchini et al., 2008). In this study, the expression of IgM was significantly upregulated within 7 days in the intestine, suggesting that the IgM VH region played an important role in the clearance of Streptococcus in the intestine of tilapia. As expected, the expression of the IgM VH region in the coinfection group was higher than those in the single infection group, illustrating that coinfection of S. agalactiae and S. iniae has a more significant stimulating effect on the immune system of tilapia.
The cytokine–cytokine receptor interaction pathway, including soluble extracellular proteins and glycoproteins, played a bridge role between the immune signal and the immune effect (Cui and Ma, 2020). It has been reported that the expression of IL-1β and other pro-inflammatory factors in the cytokine–cytokine receptor interaction pathway was upregulated in the intestine of Ctenopharyngodon idella following Aeromonas hydrophila infection (Bai et al., 2018). Three significantly differentially expressed genes in the cytokine–cytokine receptor interaction pathway were altered after infection with S. agalactiae and S. iniae. They were involved in the inflammatory response including CXCL 10, C-C motif chemokine 19-like, and interleukin-1 beta-like. As members of the chemokine family, CXCL 10 and C-C motif chemokine play an important role in the inflammatory response. They mediated and attracted cytokines to the inflammation site to exert immune effects not only in humans but also in teleost fish (Hasni et al., 2017; Ting et al., 2018; Kim et al., 2019). In this study, the expression of CXCL 10 and C-C motif chemokine 19-like were both increased significantly within 24 h in the intestine and then decreased at 7 days. It has reported that CXCL was significantly upregulated in the spleen and kidney of tilapia infected by S. agalactiae after 6 h (Zhang et al., 2013). Combined our results, we supposed that CXCL played an important defensive role in different tissues of tilapia against Streptococcus at the early stage. Studies have shown that CXCL exerted significant roles in regulating immune cells migration and activation such as B lymphocytes and macrophages (Schwenteit et al., 2013; Mu et al., 2019). This result indicated that after being infected by Streptococcus, macrophages and B lymphocytes reached the inflammation area under the mediation and activation of CXCL 10 and C-C chemokine 19 in the intestine. Interleukin 1β can activate the proliferation and differentiation of immune cells. In this study, the expression of IL-1β was upregulated significantly within 24 h in the intestine and then decreased between 24 h and 7 days. Similar to our results, the expression of IL-1β significantly increased at 12 h and then decreased between 12 and 24 h following a bacterial infection in Piaractus mesopotamicus (Carriero et al., 2020). Our results suggested that after streptococcal infection, B cells and other immune cells were activated by IL-1β to protect the organisms from pathogenic stress, exerting a role in modulating the cellular immune response. It is worth mentioning that the expression of these three DEGs in the cytokine–cytokine receptor interaction pathway was significantly higher in the coinfection group than in the single infection group, suggesting that coinfection of S. agalactiae and S. iniae has a more significant effect on the interaction between immune cells in the intestine.
The ubiquitin-proteasome in the proteasome pathway played a significant role in mediating the degradation of intracellular proteins, presenting antigens, and mediating inflammation (Zheng et al., 2020). In the present study, two genes in this pathway were altered after infection with Streptococcus, including ubiquitin-protein ligase TRIM39-like, and proteasome subunit beta type-8 (PSMB8). They were involved in the formation of the ubiquitin proteasome system. In Paralichthys olivaceus, the expression of ubiquitin proteasome was significantly increased in some immune organs, including gill, heart, muscle, brain, and especially intestine (Liu et al., 2020). The immune catalytic subunit β5i in the ubiquitin-proteasome system was encoded by PSMB8, which assisted the activation of antigen-presenting cells to a certain extent (Takezaki et al., 2002). The E3 ubiquitin ligase encoded by E3 ubiquitin-protein ligase TRIM39-like catalyzed the ligation of ubiquitin factors to form ubiquitin bodies (Huang et al., 2017). According to our results, the maximum expression of PSMB8 occurred at 12 h post-injection and then began to decrease from 24 h onward. In the three experimental groups, the expression of E3 ubiquitin-protein ligase TRIM39-like also upregulated. In addition, in the coinfection group, the relative expression of PSMD8 was more than 250 times higher than that of the single infection group. Our results revealed that more ubiquitin proteasomes were formed in the tilapia to participate in the immune response against the coinfection of S. agalactiae and S. iniae.
The oxidative phosphorylation pathway played an important role in the synthesis of ATP and the release of reactive oxygen species (ROS) (Nolfi-Donegan et al., 2020). As an active molecule, ROS oxidized metabolic wastes and mediated and regulated a variety of signal transduction pathways (Gauron et al., 2013). In this study, the oxidative phosphorylation pathway was significantly enriched. Furthermore, the expression of ATP synthase F(0) complex subunit B1 significantly decreased. The core catalytic subunit γ chain subunit B of ATPase encoded by ATP synthase F (0) complex subunit B1 can catalyze the synthesis of ATP (Schredelseker and Pelster, 2004). Taken all these information together, we speculated that Streptococcus inhibited the synthesis of ATP in the intestine. This inhibition trend was more obvious in the coinfection group.
Immune pathways of eight selected immune DEGs in the O. niloticus intestine are shown in Figures 5,6.
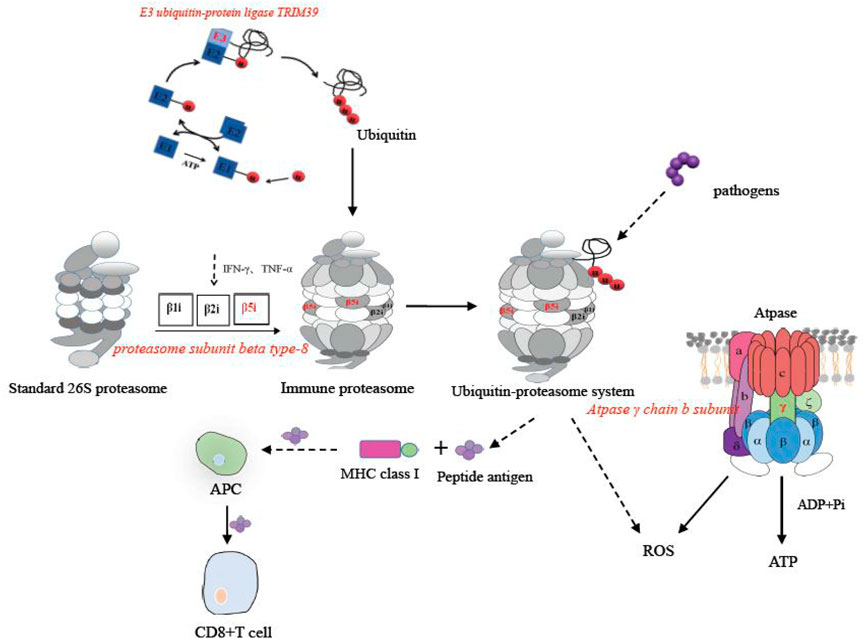
FIGURE 5. Immune pathways of PSMB8, E3 ubiquitin-protein ligase TRIM39-like, and ATP synthase F(0) complex subunit B1 in the O. niloticus intestine.
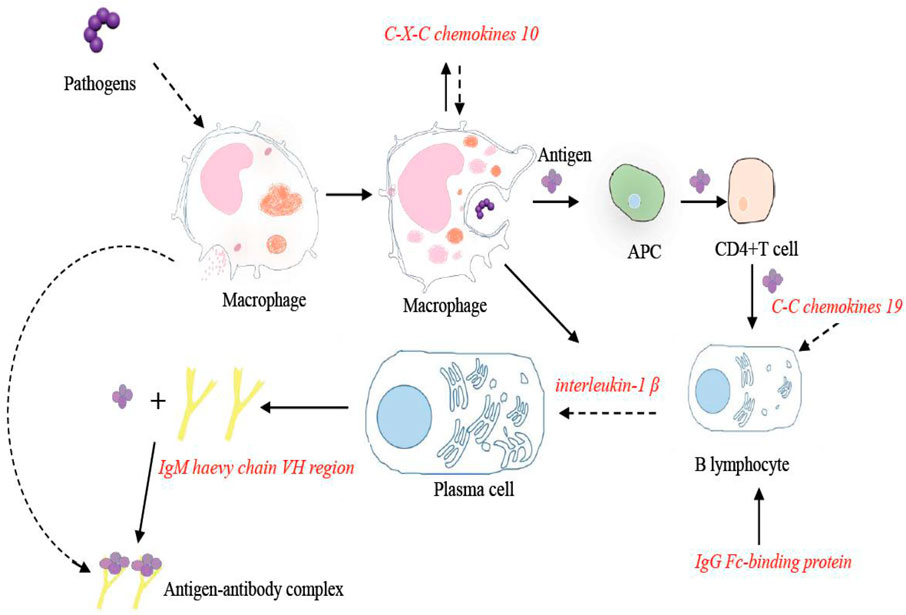
FIGURE 6. Immune pathways of CXCL 10, C-C motif chemokine 19-like, interleukin-1 beta-like, IgM heavy chain VH region, partial, and IgG Fc-binding protein in the O. niloticus intestine.
The results in this study demonstrated two potential immune pathways in the intestine of tilapia infected by S. agalactiae or S. iniae. These two immune pathways medicate inflammatory responses mainly by activating the expression of E3 ubiquitin-protein ligase TRIM39-like, CXCL 10, C-C motif chemokine 19-like, interleukin-1 beta-like, IgM heavy chain VH region, partial, IgG Fc-binding protein, PSMB8, and ATP synthase F(0) complex subunit B1, mitochondrial. In addition, the expression of these eight genes in the MG group was significantly higher than that in the AG and IG groups, which suggested that the coinfection of S. agalactiae and S. iniae is more significant than the single infection of each of the two pathogens on the stimulation of inflammatory response in tilapia intestines at equivalent concentrations of bacteria. Our study provided a theoretical basis for analyzing the complex molecular mechanism of tilapia Streptococcus infection response and a reference for the further research of other fish against streptococcal disease.
The original contributions presented in the study are publicly available. This data can be found here: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA766813.
The animal study was reviewed and approved, and all of the experiments were conducted in accordance with the guidelines and regulations of the Management and Use of Laboratory Animals of Guangdong Province and complied with China’s existing laws and regulations for biological research. This study did not involve endangered or protected species.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was supported by the Guangdong Basic and Applied Basic Research Foundation (Nos. 2019A1515012112 and 2019A1515011791), the Science and Technology Planning Project of Guangdong Province (No. (2019)170), the Natural Science Foundation of Guangdong Province (No. 2018A030313578), and the Guangzhou Science and Technology Project (No. GZKTP202032).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.782957/full#supplementary-material
Akula, S., and Hellman, L. (2017). “The Appearance and Diversification of Receptors for IgM during Vertebrate Evolution,” in Igm and its Receptors and Binding Proteins. Editors H. Kubagawa, and P. D. Burrows (Berlin: Springer), 1–23. doi:10.1007/82_2017_22
Andani, H. R. R., Tukmechi, A., Meshkini, S., and Sheikhzadeh, N. (2012). Antagonistic Activity of Two Potential Probiotic Bacteria from Fish Intestines and Investigation of Their Effects on Growth Performance and Immune Response in Rainbow trout (Oncorhynchus mykiss). J. Appl. Ichthyology 28 (5), 728–734. doi:10.1111/j.1439-0426.2012.01974.x
Anshary, H., Kurniawan, R. A., Sriwulan, S., Ramli, R., and Baxa, D. V. (2014). Isolation and Molecular Identification of the Etiological Agents of Streptococcosis in Nile tilapia (Oreochromis niloticus) Cultured in Net Cages in Lake Sentani, Papua, Indonesia. SpringerPlus 3 (1), 627. doi:10.1186/2193-1801-3-627
Austin, B., and Allen-Austin, D. (1985). A Review. J. Appl. Bacteriol. 58 (5), 483–506. doi:10.1111/j.1365-2672.1985.tb01490.x
Bai, Y., Shen, Y., Xu, X.-y., Bai, Y., Fang, Y., Zhang, M., et al. (2018). Growth Arrest and DNA Damage Inducible 45-beta Activates Pro-inflammatory Cytokines and Phagocytosis in the Grass Carp (Ctenopharyngodon Idella) after Aeromonas Hydrophila Infection. Develop. Comp. Immunol. 87, 176–181. doi:10.1016/j.dci.2018.06.010
Brown, J. H. (1939). Double-Zone Beta-Hemolytic Streptococci. J. Bacteriol. 37 (2), 133–144. doi:10.1128/jb.37.2.133-144.1939
Cai, Y.-z., Liu, Z.-g., Lu, M.-x., Ke, X.-l., Zhang, D.-f., Gao, F.-y., et al. (2020). Oral Immunization with Surface Immunogenic Protein from Streptococcus Agalactiae Expressed in Lactococcus Lactis Induces Protective Immune Responses of tilapia (Oreochromis niloticus). Aquacult. Rep. 18, 100538. doi:10.1016/j.aqrep.2020.100538
Carriero, M. M., Henrique-Silva, F., Meira, C. M., Gato, I. M. Q., Caetano, A. R., Lobo, F. P., et al. (2020). Molecular Characterization and Gene Expression Analysis of the Pro-inflammatory Cytokines IL-1β and IL-8 in the South American Fish Piaractus Mesopotamicus Challenged with Aeromonas Dhakensis. Genet. Mol. Biol. 43 (4), 15. doi:10.1590/1678-4685-gmb-2020-0006
Chen, C. Y., Cha, C. B., and Bowser, P. R. (2007). Comparative Histopathology of Streptococcus Iniae and Streptococcus Agalactiae-Infected tilapia. Bull. Eur. Assoc. Fish Pathol. 27 (1), 2–9.
Cui, W., and Ma, A. (2020). Transcriptome Analysis Provides Insights into the Effects of Myo-Inositol on the Turbot Scophthalmus maximus. Fish Shellfish Immunol. 106, 691–704. doi:10.1016/j.fsi.2020.07.019
de Sousa, E. L., Assane, I. M., Santos-Filho, N. A., Cilli, E. M., de Jesus, R. B., and Pilarski, F. (2021). Haematological, Biochemical and Immunological Biomarkers, Antibacterial Activity, and Survival in Nile tilapia Oreochromis niloticus after Treatment Using Antimicrobial Peptide LL-37 against Streptococcus Agalactiae. Aquaculture 533, 736181. doi:10.1016/j.aquaculture.2020.736181
Fillatreau, S., Six, A., Magadan, S., Castro, R., Sunyer, J. O., and Boudinot, P. (2013). The Astonishing Diversity of Ig Classes and B Cell Repertoires in Teleost Fish. Front. Immun. 4, 14. doi:10.3389/fimmu.2013.00028
Gallily, R., Yamin, A., Caspi, R. R., and Avtalion, R. R. (1982). Non-Immunological Recognition and Killing of Xenogeneic Cells by Macrophages. Develop. Comp. Immunol. 6 (3), 569–578. doi:10.1016/s0145-305x(82)80043-8
Gauron, C., Rampon, C., Bouzaffour, M., Ipendey, E., Teillon, J., Volovitch, M., et al. (2013). Sustained Production of ROS Triggers Compensatory Proliferation and Is Required for Regeneration to Proceed. Sci. Rep. 3, 2084. doi:10.1038/srep02084
Gioacchini, G., Smith, P., and Carnevali, O. (2008). Effects of Ergosan on the Expression of Cytokine Genes in the Liver of Juvenile Rainbow trout (Oncorhynchus mykiss) Exposed to Enteric Red Mouth Vaccine. Vet. Immunol. Immunopathology 123 (3-4), 215–222. doi:10.1016/j.vetimm.2008.01.037
Guo, C., Yuan, C., Zhu, S., Liu, G., Lu, C., and Liu, Y. (2017). Quantitative Identification of Differential Proteins in Streptococcus Agalactiae Piscine Strain and Bovine Strain Using iTRAQ. Jiangsu J. Agric. Sci. 33 (4), 868–873. doi:10.11964/jfc.20170110690
Hasni, D. S. M., Lim, S. M., Chin, A. V., Tan, M. P., Poi, P. J. H., Kamaruzzaman, S. B., et al. (2017). Peripheral Cytokines, C-X-C Motif Ligand10 and Interleukin-13, Are Associated with Malaysian Alzheimer's Disease. Geriatr. Gerontol. Int. 17 (5), 839–846. doi:10.1111/ggi.12783
Huang, R., Zhang, J., Zhu, G., He, J., and Xie, J. (2017). The Core Ubiquitin System of Mandarin Fish, Siniperca chuatsi, Can Be Utilized by Infectious Spleen and Kidney Necrosis Virus. Fish Shellfish Immunol. 70, 293–301. doi:10.1016/j.fsi.2017.09.017
Huang, T., Li, L., Wang, R., Zhang, B., Gan, X., Luo, H., et al. (2014). Pathogenic Bacteria Streptococcus Agalactiae and S.Iniae in Diseased Ovate Pompano Trachinotus Ovatus. J. Dalian Ocean Univ. 29 (2), 161–166. doi:10.3969/J.ISSN.2095-1388.2014.02.012
Iregui, C. A., Comas, J., Vásquez, G. M., and Verján, N. (2016). Experimental Early Pathogenesis of Streptococcus Agalactiae Infection in Red tilapia Oreochromis Spp. J. Fish. Dis. 39 (2), 205–215. doi:10.1111/jfd.12347
Kacha, A., Hbid, M. H., and Bravo de la Parra, R. (2009). Mathematical Study of a Bacteria-Fish Model with Level of Infection Structure. Nonlinear Anal. Real World Appl. 10 (3), 1662–1678. doi:10.1016/j.nonrwa.2008.02.021
Kim, J.-H., Macqueen, D. J., Winton, J. R., Hansen, J. D., Park, H., and Devlin, R. H. (2019). Effect of Growth Rate on Transcriptomic Responses to Immune Stimulation in Wild-type, Domesticated, and GH-Transgenic Coho salmon. Bmc Genomics 20 (1), 1024. doi:10.1186/s12864-019-6408-4
Kordon, A. O., Karsi, A., and Pinchuk, L. (2018). Innate Immune Responses in Fish: Antigen Presenting Cells and Professional Phagocytes. Turkish J. Fish. Aquat. Sci. 18 (9), 1123–1139. doi:10.4194/1303-2712-v18_9_11
Li, M., Hu, F.-C., Qiao, F., Du, Z.-Y., and Zhang, M.-L. (2020). Sodium Acetate Alleviated High-Carbohydrate Induced Intestinal Inflammation by Suppressing MAPK and NF-Κb Signaling Pathways in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol., 98, 758–765. doi:10.1016/j.fsi.2019.11.024
Li, Y., Luo, F., Zuo, Y., Yi, Y., Wu, S., and Yang, J. (2014). Isolation and Identification of Gamma-Hemolytic streptococcus Agalactiae from tilapia. Freshw. Fish. 44 (4), 63. doi:10.13721/j.cnki.dsyy.2014.04.010
Liu, J., Pan, M., Huang, D., Guo, Y., Yang, M., Zhang, W., et al. (2020). Myostatin-1 Inhibits Cell Proliferation by Inhibiting the mTOR Signal Pathway and MRFs, and Activating the Ubiquitin-Proteasomal System in Skeletal Muscle Cells of Japanese Flounder Paralichthys olivaceus. Cells 9 (11), 2376. doi:10.3390/cells9112376
Maulu, S., Hasimuna, O. J., Mphande, J., and Munang’andu, H. M. (2021). Prevention and Control of Streptococcosis in Tilapia Culture: A Systematic Review. J. Aquat. Anim. Health 33, 162–177. doi:10.1002/aah.10132
Mohan, R. N. (1947). Bacteriology of Bovine Mastitis in India with Special Reference to the Incidence of Streptococcus Agalactiae. Indian Vet. J. 24 (3), 169–178.
Monteverde, J. J., and Simeone, D. H. (1951). Non Hemolytic Streptococcus of the Lancefield Group, Natural Pathogen of guinea Pig. Prensa Med. Argent 38 (19), 1124–1125.
Mu, Y., Zhou, S., Ding, N., Ao, J., and Chen, X. (2019). Molecular Characterization of a New Fish Specific Chemokine CXCL_F6 in Large Yellow Croaker (Larimichthys Crocea) and its Role in Inflammatory Response. Fish Shellfish Immunol. 84, 787–794. doi:10.1016/j.fsi.2018.10.068
Nolfi-Donegan, D., Braganza, A., and Shiva, S. (2020). Mitochondrial Electron Transport Chain: Oxidative Phosphorylation, Oxidant Production, and Methods of Measurement. Redox Biol. 37, 101674. doi:10.1016/j.redox.2020.101674
Perera, R. P., Johnson, S. K., Collins, M. D., and Lewis, D. H. (1994). Streptococcus iniaeAssociated with Mortality ofTilapia nilotica×T. aureaHybrids. J. Aquat. Anim. Health 6 (4), 335–340. doi:10.1577/1548-8667(1994)00610.1577/1548-8667(1994)006<0335:siawmo>2.3.co;2
Piazzon, M. C., Galindo-Villegas, J., Pereiro, P., Estensoro, I., Calduch-Giner, J. A., Gómez-Casado, E., et al. (2016). Differential Modulation of IgT and IgM upon Parasitic, Bacterial, Viral, and Dietary Challenges in a Perciform Fish. Front. Immunol. 7, 637. doi:10.3389/fimmu.2016.00637
Poppert, S., Nickel, D., Berger, A., Yildiz, T., Kaestner, N., Mauerer, S., et al. (2009). Rapid Identification of Beta-Hemolytic Streptococci by Fluorescence in situ Hybridization (FISH). Int. J. Med. Microbiol. 299 (6), 421–426. doi:10.1016/j.ijmm.2009.02.001
Salinas, I. (2015). The Mucosal Immune System of Teleost Fish. Biology 4 (3), 525–539. doi:10.3390/biology4030525
Schredelseker, J., and Pelster, B. (2004). Isoforms vatB1 and vatB2 of the Vacuolar Type ATPase Subunit B Are Differentially Expressed in Embryos of the Zebrafish (Danio rerio). Dev. Dyn. 230 (3), 569–575. doi:10.1002/dvdy.20074
Schwenteit, J. M., Breithaupt, A., Teifke, J. P., Koppang, E. O., Bornscheuer, U. T., Fischer, U., et al. (2013). Innate and Adaptive Immune Responses of Arctic Charr (Salvelinus alpinus, L.) during Infection with Aeromonas Salmonicida Subsp. Achromogenes and the Effect of the AsaP1 Toxin. Fish Shellfish Immunol. 35 (3), 866–873. doi:10.1016/j.fsi.2013.06.024
Shin, G., Palaksha, K., Yang, H., Shin, Y., Kim, Y., Lee, E., et al. (2006). Partial Two-Dimensional Gel Electrophoresis (2-DE) Maps of Streptococcus Iniae ATCC29178 and Lactococcus Garvieae KG9408. Dis. Aquat. Org. 70 (1-2), 71–79. doi:10.3354/dao070071
Shiry, N., Soltanian, S., Shomali, T., Paknejad, H., and Hoseinifar, S. H. (2019). Immunomodulatory Effects of Orally Administrated Florfenicol in Rainbow trout (Oncorhynchus mykiss) Following Experimental challenge with Streptococcosis/lactococcosis. Int. Immunopharmacology 73, 236–245. doi:10.1016/j.intimp.2019.05.007
Suanyuk, N., Kong, F., Ko, D., Gilbert, G. L., and Supamattaya, K. (2008). Occurrence of Rare Genotypes of Streptococcus Agalactiae in Cultured Red tilapia Oreochromis Sp. And Nile tilapia O. niloticus in Thailand-Relationship to Human Isolates? Aquaculture 284 (1-4), 35–40. doi:10.1016/j.aquaculture.2008.07.034
Takezaki, N., Zaleska-Rutczynska, Z., and Figueroa, F. (2002). Sequencing of Amphioxus PSMB5/8 Gene and Phylogenetic Position of Agnathan Sequences. Gene 282 (1-2), 179–187. doi:10.1016/s0378-1119(01)00834-4
Tavares, G. C., de Queiroz, G. A., Assis, G. B. N., Leibowitz, M. P., Teixeira, J. P., Figueiredo, H. C. P., et al. (2018). Disease Outbreaks in Farmed Amazon Catfish (Leiarius Marmoratus X Pseudoplatystoma Corruscans) Caused by Streptococcus Agalactiae, S. Iniae, and S. Dysgalactiae. Aquaculture 495, 384–392. doi:10.1016/j.aquaculture.2018.06.027
Ting, C.-H., Chen, Y.-C., and Chen, J.-Y. (2018). Nile tilapia Fry Fed on Antimicrobial Peptide Epinecidin-1-Expressing Artemia Cyst Exhibit Enhanced Immunity against Acute Bacterial Infection. Fish Shellfish Immunol. 81, 37–48. doi:10.1016/j.fsi.2018.07.008
Wang, D., Sun, S., Li, S., Lu, T., and Shi, D. (2021). Transcriptome Profiling of Immune Response to Yersinia ruckeri in Spleen of Rainbow trout (Oncorhynchus mykiss). Bmc Genomics 22 (1), 11. doi:10.1186/s12864-021-07611-4
Xiu, Y., Jiang, G., Zhou, S., Diao, J., Liu, H., Su, B., et al. (2019). Identification of Potential Immune-Related circRNA-miRNA-mRNA Regulatory Network in Intestine of Paralichthys olivaceus during Edwardsiella Tarda Infection. Front. Genet. 10, 16. doi:10.3389/fgene.2019.00731
Yearbook (2020). China Fishery Statistical Yearbook. 2020. Beijing, China: China Statistics Press, 24–34.
Zhang, H., Chang, C.-M., Shen, K.-N., Xian, W., and Hsiao, C.-D. (2016). Identification of Myogenic Regulatory Genes in the Muscle Transcriptome of Beltfish (Trichiurus Lepturus): A Major Commercial marine Fish Species with Robust Swimming Ability. Genomics Data 8, 81–84. doi:10.1016/j.gdata.2016.04.005
Zhang, R., Zhang, L.-l., Ye, X., Tian, Y.-y., Sun, C.-f., Lu, M.-x., et al. (2013). Transcriptome Profiling and Digital Gene Expression Analysis of Nile tilapia (Oreochromis niloticus) Infected by Streptococcus Agalactiae. Mol. Biol. Rep. 40 (10), 5657–5668. doi:10.1007/s11033-013-2667-3
Zheng, W., Yan, X., Huo, R., Zhao, X., Sun, Y., and Xu, T. (2020). IRF11 Enhances the Inhibitory Effect of IκBα on NF-Κb Activation in Miiuy Croaker. Fish Shellfish Immunol. 107, 156–162. doi:10.1016/j.fsi.2020.09.009
Zhu, J., Fu, Q., Ao, Q., Tan, Y., Luo, Y., Jiang, H., et al. (2017). Transcriptomic Profiling Analysis of tilapia (Oreochromis niloticus) Following Streptococcus Agalactiae challenge. Fish Shellfish Immunol. 62, 202–212. doi:10.1016/j.fsi.2017.01.023
Keywords: tilapia, transcriptomics, Streptococcus agalactiae, Streptococcus iniae, immune responses, intestine
Citation: Cui M, Wang Z, Yang Y, Liu R, Wu M, Li Y, Zhang Q and Xu D (2022) Comparative Transcriptomic Analysis Reveals the Regulated Expression Profiles in Oreochromis niloticus in Response to Coinfection of Streptococcus agalactiae and Streptococcus iniae. Front. Genet. 13:782957. doi: 10.3389/fgene.2022.782957
Received: 25 September 2021; Accepted: 31 January 2022;
Published: 03 March 2022.
Edited by:
Chiara Medaglia, Geneva University Hospitals (HUG), SwitzerlandReviewed by:
Sarbjeet Makkar, Washington University in St. Louis, United StatesCopyright © 2022 Cui, Wang, Yang, Liu, Wu, Li, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Miao Cui, Y3Vpc2Fuc2h1aUAxMjYuY29t; Delin Xu, eHVkZWxpbkBsaXZlLmNvbQ==
†ORCID: Miao Cui, orcid.org/0000-0001-6952-031X; Zibin Wang, orcid.org/0000-0002-3951-7520; Delin Xu, orcid.org/0000-0003-3091-7170
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.