- Structural Biology Lab, School of Life Sciences, Jawaharlal Nehru University, New Delhi, India
Small GTPases are the key to actin cytoskeleton signaling, which opens the lock of effector proteins to forward the signal downstream in several cellular pathways. Actin cytoskeleton assembly is associated with cell polarity, adhesion, movement and other functions in eukaryotic cells. Rho proteins, specifically Cdc42 and Rac, are the primary regulators of actin cytoskeleton dynamics in higher and lower eukaryotes. Effector proteins, present in an inactive state gets activated after binding to the GTP bound Cdc42/Rac to relay a signal downstream. Cdc42/Rac interactive binding (CRIB) motif is an essential conserved sequence found in effector proteins to interact with Cdc42 or Rac. A diverse range of Cdc42/Rac and their effector proteins have evolved from lower to higher eukaryotes. The present study has identified and further classified CRIB containing effector proteins in lower eukaryotes, focusing on parasitic protozoans causing neglected tropical diseases and taking human proteins as a reference point to the highest evolved organism in the evolutionary trait. Lower eukaryotes’ CRIB containing proteins fall into conventional effector molecules, PAKs (p21 activated kinase), Wiskoit-Aldrich Syndrome proteins family, and some have unique domain combinations unlike any known proteins. We also highlight the correlation between the effector protein isoforms and their selective specificity for Cdc42 or Rac proteins during evolution. Here, we report CRIB containing effector proteins; ten in Dictyostelium and Entamoeba, fourteen in Acanthamoeba, one in Trypanosoma and Giardia. CRIB containing effector proteins that have been studied so far in humans are potential candidates for drug targets in cancer, neurological disorders, and others. Conventional CRIB containing proteins from protozoan parasites remain largely elusive and our data provides their identification and classification for further in-depth functional validations. The tropical diseases caused by protozoan parasites lack combinatorial drug targets as effective paradigms. Targeting signaling mechanisms operative in these pathogens can provide greater molecules in combatting their infections.
Introduction
Cellular functions are a cumulative outcome of various signaling pathways that involve a downstream activation of a series of protein molecules. Cytoskeletal organization and dynamics critically regulate cell movement and migration, proliferation, adhesion, differentiation, and vesicular trafficking (Sackmann, 2015). Molecular switches are crucial proteins, which interact with their effectors to activate a signaling cascade. A varsity of small GTPase molecules is present in the cell where Rho family proteins are regulatory molecules for actin cytoskeleton dynamics (Hall, 1998).
Cdc42, Rho, and Rac are Rho family proteins’ subfamilies, a Ras superfamily subgroup (Wennerberg et al., 2005). These proteins oscillate between an active GTP-bound and inactive GDP-bound states as molecular switches in the cell (Vetter and Wittinghofer, 2001). In humans, several studies have characterized Cdc42 (filopodia formation), Rac1 (lamellipodia formation), and RhoA (establishment of stress fiber) members of the Rho family (Jordan and Canman, 2012; Ridley, 2015; Narumiya and Thumkeo, 2018). Cdc42 (Cell division control protein 42) was the first member to be reported (Shinjo et al., 1990; Marks and Kwiatkowski, 1996) with its profound functions in; cell polarity (in yeast), cell morphology regulation, motility, mammalian cell-cycle progression and induction of malignant cell transformation (summary by (Wu et al., 2000)). Rac1 modulates cytoskeleton in multiple cellular functions like phagocytosis, neural polarization and axonal growth, mesenchymal-like migration, and cellular growth and differentiation (summary by (Reijnders et al., 2017)). The signaling cascade of Cdc42 and Rac proteins recognize a consensus motif in downstream proteins for specific binding. Thus, these proteins were coined as CRIB (Cdc42/Rac interactive binding) effector proteins of the Rho family. The conserved motif is a 16 amino acid sequence “I-S-X-P-(X)2-4-F-X-H-X-X-H-V-G”, with eight core amino acids first identified by Burbelo in 1995 (Burbelo et al., 1995). Interestingly, effector proteins with one or two variations within the core sequence can still bind to Cdc42/Rac (Owen et al., 2000). Biophysical studies elucidated that the CRIB motif is essential for interaction with GTP-Cdc42/Rac, but not adequate for high-affinity binding (Rudolph et al., 1998; Thompson et al., 1998). The binding region of CRIB effector protein is thus, also called as gtpase binding domain (GBD) (Rudolph et al., 1998). Subsequently, when the motif, including a more extended sequence region was found in p21-activated kinase (PAK) CRIB effector protein, it was known as p21-binding domain (PBD) (Thompson et al., 1998).
The potential CRIB effector proteins were then categorically separated via different signaling pathways activated by Cdc42/Rac (Neudauer et al., 1998; Bishop and Hall, 2000; Phillips et al., 2008). Cdc42 and Rac activate effector proteins to signal downstream to function on actin, SRF and NF-kB (transcription factor), JNK and p38 (MAP kinase pathway), G1-cell cycle progression, cell-cell contact, and transformation (Bishop and Hall, 2000). NADPH oxidase complex (present only in professional phagocytic cells) and secretin (only in mast cells) signaling pathways get explicitly activated by binding Rac to CRIB effector protein. Similarly, the cell-polarity signaling pathway also involves the CRIB effector proteins, triggered only by Cdc42 (Neudauer et al., 1998; Bishop and Hall, 2000; Vlahou and Rivero, 2006). The effector proteins of Cdc42/Rac are diverse in domain architecture and function, which includes Ser/Thr kinase, cytosolic Tyrosine kinase, actin-associated proteins, adaptor proteins, and miscellaneous (Owen and Mott, 2018). Ser/Thr kinases include PAKs, myotonic dystrophy kinase-related Cdc42 binding kinases (MRCKs) and mixed-lineage kinases (MLKs) family. In Tyrosine kinase, only one family of activated Cdc42-associated Tyrosine kinase (ACK) contain a CRIB motif (Bishop and Hall, 2000; Pirone et al., 2000; Owen and Mott, 2018). Actin associated proteins include Wiskott Aldrich syndrome protein (WASP), WASP-like verproline-homologous protein (WAVE), IQ motif-containing GTPase-activating proteins (IQGAP) and formin families of proteins (Bishop and Hall, 2000; Owen and Mott, 2018). Partitioning defective (PAR) proteins belong to adaptor proteins of cytoskeleton assembly. Small protein effector of Cdc42 (SPEC) and Cdc42 effector protein (CEP)/Binder of Rho GTPases (Borg) (Joberty et al., 1999; Hirsch et al., 2001) family of proteins fall under the miscellaneous group because they have not been designated under any specific classification (Owen and Mott, 2018).
The diverse signaling range and protein families project the necessity of CRIB effector proteins. In humans, CRIB containing proteins have been explored structurally and functionally in depth (Pirone et al., 2001; Cotteret and Chernoff, 2002). CRIB containing effector proteins in humans belong to nine distinct families, four kinase families and five non-kinase families (Bishop and Hall, 2000). Six PAKs (PAK1-6), three MRCKs, two MLKs, three PAR-6, two WASP (WASP and N-WASP) (Kuspa and Loomis, 2006), and two SPEC (Pirone et al., 2000), which are commonly known families of Cdc42/Rac effectors. Additionally, two unique families, Gene33 (one member) (Makkinje et al., 2000) and CEP/Borg (five members) (Joberty et al., 1999; Hirsch et al., 2001) are present only in humans. In plants, eleven Rop-interactive CRIB motif-containing protein (RIC) effectors, Ric1 to Ric11, are present, involved in cell growth like CRIB effector protein in metazoans (Wu et al., 2001; Cotteret and Chernoff, 2002; Fu et al., 2005).
CRIB effector proteins present in complex and multicellular eukaryotes such as worms, flies, and frogs are mammalian homologues (Pirone et al., 2001). Nine proteins in D. melanogaster; DmMLK (Teramoto et al., 1996), DmGEK (MRCK) (Luo et al., 1997; Leung et al., 1998), DmACK (Manser et al., 1995; Clemens et al., 2000), DmPAK1-3 are kinases (Mentzel and Raabe, 2005) and DmWASP (Symons et al., 1996; Miki et al., 1998a), DmPAR-6 (Qiu et al., 2000), DmSPEC (Pirone et al., 2000) are non-kinase. All nine proteins are architecturally homologous to humans and perform alike functions. Eight proteins in C. elegans; CePAK1 and CePAK2 (Lucanic and Cheng, 2008), are PAK-like kinases, CeACK and CeMRCK are ACK (Manser et al., 1995), and MRCK (Leung et al., 1998) homologs, respectively, and other four belonging to non-kinases are CeWASP (Symons et al., 1996; Miki et al., 1998a; Zhang and Kubiseski, 2010), CePAR-6 (Qiu et al., 2000), F09F7.5 and T23G5.3 (Pirone et al., 2001; Cotteret and Chernoff, 2002). All four non-kinase Cdc42/Rac effectors share no familiar domain characteristic features except for the consensus CRIB/GBD domain. WASP and Par-6 participate in cell polarity through actin organization, while F09F7.5 and T23G5.3 are novel proteins with no homologues in humans or flies (Pirone et al., 2001). In simple eukaryotes such as yeast, there are only five such proteins, Ste20, Skm1, and Cla4, within the PAK family, and Gic1 and Gic2 (Brown et al., 1997), which are non-kinases homologous to plants, and not mammalian CRIB effector proteins. The characteristically different Gics might have a specialized role in cytoskeleton modulation during cell wall assembly.
The extended repertoire of Cdc42/Rac effector proteins in humans indicates a more complex mechanism for extracellular signals to reach Rho GTPases (Cdc42 and Rac) compared to Drosophila. Nevertheless, in worms, each group of effectors have a single protein to perform a related signaling function, while in yeast, only the PAK family is present. The trend highlights that complexity of an organism is correlated with the extension of protein members in each effector protein group. Evolutionarily, the presence of CRIB containing effector proteins in plants interestingly points an ancient origin of the CRIB motif. Possibly, the CRIB motif associated with an array of Rho signaling proteins during evolution.
The CRIB containing effector proteins from unicellular eukaryotes are not structurally characterized or classified. Here, we present the identification of effector proteins in unicellular eukaryotes, offering exciting insights into their evolutionary connection to higher eukaryotes’ signaling. Many unicellular eukaryotes organisms are pathogenic and cause dire health challenges with high prevalence. In the new world era, the treatments available for diseases like amoebiasis, sleeping sickness and protozoan parasitic diseases are not very effective and need more attention for targeted drug research (Baker et al., 2013; Diaz et al., 2014; Gonzales et al., 2019; Carrero et al., 2020). The CRIB domain effector proteins, which are barely known and investigated in protozoan parasites can be a prospective candidate for drug research as they are integral to cellular signaling cascades during parasitic pathogenesis. In humans, PAK and WASP family proteins have been thoroughly inspected, both functionally and biochemically, and they proved to be potential targets against cancer, neurodegenerative and cardiovascular diseases (Kichina et al., 2010; Li et al., 2010; Zhao and Manser, 2010; Llorens et al., 2013; Dammann et al., 2018). Rho signaling effectors are the mediators of cytoskeletal dynamics in higher organisms and are crucial for unicellular pathogens requiring a highly regulated cytoskeletal system for survival and pathogenicity. The functional and drug target-oriented research attention is needed for such proteins involved in pivotal signaling that are still undiscovered. This systematic review presents the identification and annotation of CRIB domain-containing proteins in unicellular eukaryotes, especially in pathogenic protozoans responsible for neglected tropical diseases and model organisms, from literature and database search. We have tried to present a crisp platform to select out proteins for targeted functional studies and drug development strategies.
Results
The tree of life depicts that, lower eukaryotes are simple and unicellular except Dictyostelium, an evolutionary link between unicellular and multicellular organisms. Understanding the ancient CRIB motif in signaling effector proteins in model organism-Dictyostelium, and protozoan parasite-Acanthamoeba, Entamoeba, Giardia, Trypanosoma and Leishmania to the highest evolutionary candidate will comfort to classify them suitably for further in vitro and in vivo validation. In silico studies have been conducted to identify the CRIB containing repertoire of effector proteins, further cross-referenced with available literature, and finally, a new classification has also been added.
Cdc42/Rac Interactive Binding Domain-Containing Protein in Lower Eukaryotes and Evolutionary Divergence
The complete proteome-based phylogenetic analysis of Dictyostelium, Acanthamoeba, and Entamoeba shows that animals and fungi are close to amoebozoa group (Song et al., 2005). Nevertheless, amoebozoa are distinct from early diverging unicellular eukaryotes, Leishmania, Trypanosoma, Plasmodium, Giardia, and plant as well. The more remarkable similarities despite their early divergence in amoebozoa and metazoan proteins translate into a generally higher degree of functional conservation between them. The universal domain architectures aid in delineating and organizing the proteins in their families and participating in particular cellular pathways in lower eukaryotes.
The conserved sequence-based search identifies CRIB/PBD/GBD containing proteins; ten in Dictyostelium discoideum, fourteen in Acanthamoeba castellani, nine in Entamoeba histolytica, only one protein in Trypanosoma cruzi, and Giardia lamblia. In humans, twenty-seven CRIB containing effector proteins have been identified and reported in earlier studies (Pirone et al., 2001) (Figure 1). The available functional, biophysical and biochemical characterization has been explored in detail to classify the identified CRIB containing effector proteins. Apart from the CRIB motif, the conserved structural features in identified proteins of Amoebozoa fall into PAK or PAK like kinase and actin-associated or actin assembly protein. In Acanthamoeba and Entamoeba, very few proteins have been studied earlier, on the other hand, a lot of literature is available for Dictyostelium proteins. However, in other parasite protozoans (Giardia and Trypanosoma), the single identified CRIB domain protein does not show any similarities with the conserved domains of Cdc42/Rac effector. The identified CRIB containing protein families indicated here are PAK/PAK related kinase and actin assembly protein families found conventionally during evolution, while Ser/Thr and cytosolic Tyrosine kinase, adaptor family proteins present in humans are non-conventional.
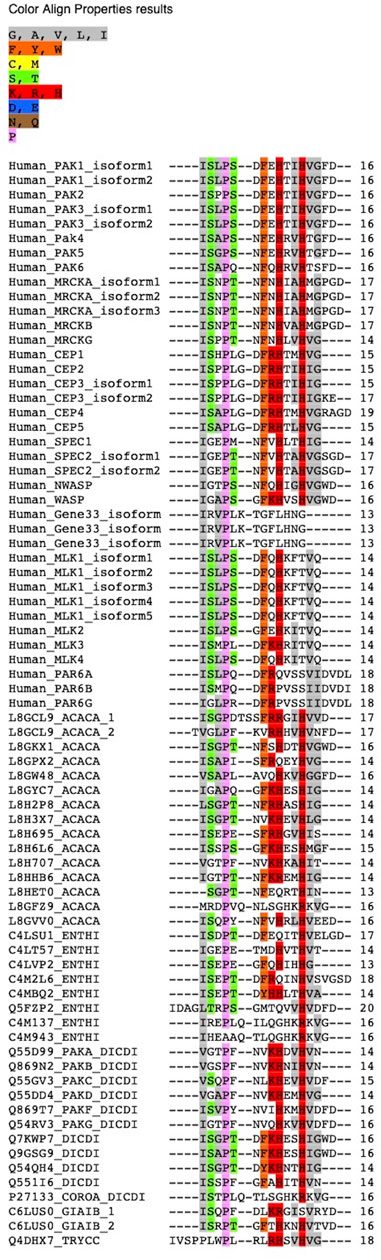
FIGURE 1. Sequence alignment of conserved CRIB motif of all the proteins of human, Dictyostelium discoideum, Acanthamoeba castellani, Entamoeba histolytica extracted from domain-based search from database.
Cdc42/Rac Interactive Binding-Containing Effector Proteins in Dictyostelium
Dictyostelium discoideum, a soil-dwelling social amoeba, is a unicellular eukaryote that forms multicellular structure fruiting bodies under limiting nutrition conditions (Gaudet et al., 2008). The D. discoideum genome (∼34 Mb) entirely encrypts about 10,300 proteins, including numerous protein families; some are involved in fundamental processes like post-translational modification, secondary metabolism, and signal transduction belong to cellular activities like cell adhesion and cytoskeleton control (Kuspa and Loomis, 2006; Loomis, 2006). Numerous Dictyostelium proteins are more similar to human orthologs than yeast, probably due to higher evolutionary changes along the fungal lineage. The small, simple genome and complex transcriptome made it an easy-going prototypical organism to dissect the signaling pathways and their elements with typical relationships throughout the metazoans (Li and Purugganan, 2011; Bozzaro, 2019).
In this study, ten CRIB domain (or PBD/GBD) proteins were found fit functionally to be Cdc42/Rac effector proteins. Conserved sequence and structural features group them into p21-activated kinase (six protein), WASP family (three protein) and a novel gelsolin-related protein (Figure 2). However, a genomic study mentions that eight PAKs (PAKa-h) are present in D. discoideum (Arasada et al., 2006), with no structural and functional shreds of evidence support. In our study we found that PAKe (Q54B33) and PAKh (Q556S2) have a kinase domain but lack a consensus CRIB domain and other accessory domains present in human homologues. Thus, these two proteins may potentially be candidates of some other subfamily of Ser/Thr kinase that is a matter of further investigation or may be pseudo PAK like E. histolytica PAK1 (Labruyere et al., 2000; Labruyere et al., 2003). The novel gelsolin homolog identified here is encoded by the gnrC gene, categorically a putative actin-binding protein (Q551I6), which suggests its regulation by a small gtpase. This is the unique protein in Dictyostelium with two CRIB domains present consecutively at its N-terminal. However, no study reports any details regarding the Rac protein, which activates it.
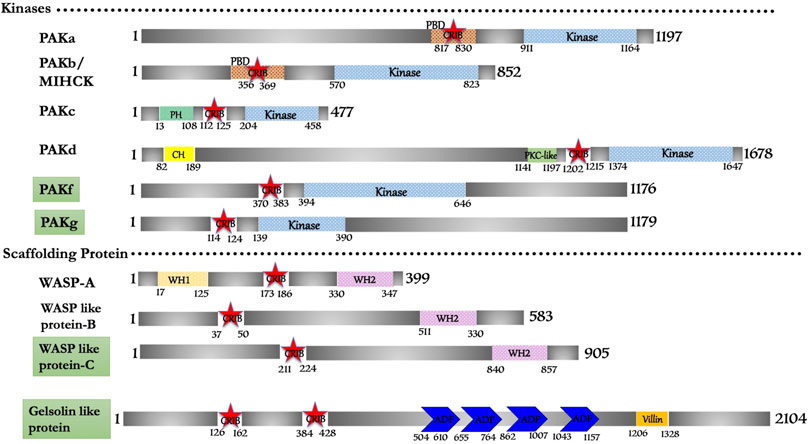
FIGURE 2. Schematic diagram of domain organization of Dictyostelium discoideum, CRIB domain containing proteins. The domain names and amino acid is labeled. The abbreviations for the domains are: CRIB, Cdc42/Rac interactive binding; CH, Calponin Homology; PBD, p21-activated binding domain; PH, pleckstrin-homology; WH1, Enabled/VASP homology 1; WH2, WASP homology 2; ADF, Actin depolymerizing factor. The proteins already known from the earlier literature, but not characterized yet are marked as green colored box. The proteins marked in orange (subtle effect) boxes are reported for the first time in this in silico study and are completely uncharacterized. Footnote: In Dictyostelium discoideum, PAKa-d can be studied for their protein structure and interaction studies with respective Rac repertoire or Cdc42. The functional role of PAKa-d in cell motility, adhesion, phagocytosis and chemotaxis represent their crucial role in unicellular protozoans, and comparative studies in parasite protozoans would be the scope of future research.
Out of six PAK family members identified in the search, PAKa, PAKb/MIHCK, PAKc, and PAKd have some similar structural domains of human Group-I PAKs. PAKf (Q869T7) and PAKg (Q556S2) possess the conserved CRIB and kinase domain; typical structural features of PAK homologs of metazoans and yeast but need to be experimentally validated. One member has identical features to human WASP protein, while, the other two (WASP like-B (Q7KWP7) and WASP like-C (Q54QH4)) have comparable domain characteristics to human N-WASP derived as WASP-related protein (Figure 2).
In Dictyostelium, PAK and WASP are the CRIB-containing effectors of Rho family GTPases, which regulate chemotaxis, phagocytosis, and cytokinesis (Wilkins and Insall, 2001; Park et al., 2004). No clear homologues of Rho and Cdc42 are present, though 15 members of Rac have been reported in the Rho GTPase family (Rivero et al., 2001; Rivero and Somesh, 2002). RacB has been proposed as a functional equivalent to Cdc42 (Rivero et al., 2001). Additionally, a component of SCAR (suppressor of cAMP-receptor)/WAVE complex, four IQGAPs, ten formins, two PCH (full form) family, several lipid kinases/phospholipase and NADPH oxidase components are also present which represent CRIB-independent effectors of Rho family (Vlahou and Rivero, 2006). Experimental evidence remarkably suggested that a CRIB motif is present in coronin protein, which interacts with GDP bound gtpase (Swaminathan et al., 2014; Swaminathan et al., 2015). In general, coronin activates the actin nucleation factor Arp2/3 complex and IQGAPs (Shina and Noegel, 2008). The presence of the CRIB motif varied amongst lower and higher eukaryotes’ coronin proteins. Nevertheless, the sequence comparison of the conserved CRIB motif and the coronin CRIB region indicated moderate similarity while displaying Cdc42/Rac binding. Reasonable similarity can possibly explain that the region not annotated as CRIB in protein domain databases can contribute to the protein-protein interaction mechanism.
WASP family consist of one WASP, two WASP related and one WASP like (SCAR) subfamilies proteins, which are the positive regulators of Arp2/3 complex in actin polymerization (Seastone et al., 2001; Myers et al., 2005). WASP controls Arp2/3 complex spatially and temporally in D. discoideum via interaction of Rac with the CRIB motif. WASP, an actin nucleation-promoting factor, also functions as a controller of cellular localization of Rac, contributing to the maintenance of front-rear polarity (Amato et al., 2019). WASP-A encoded by wasA gene has a WH1 (WASP-homology 1) domain that interacts with poly-Proline helices (Myers et al., 2005). RacC works as a connection between WASP activation and chemo-attractant stimulation in the signaling pathway regulating F-actin assembly during chemotaxis (Han et al., 2006). WASP-B regulates F-actin polymerization through attenuation that is important for regulating the dynamics of pseudopod extension and retraction (Chung et al., 2013). However, SCAR subfamily proteins possess a C-terminal VCA domain akin to human WASP and N-WASP but lack an extended N-terminal WH1 region and GBD domains (Symons et al., 1996). The human homolog of SCAR is called WAVE. SCAR/WAVE is a multi-protein complex with PIR121, Nap125, Abi2, and HSp300 components, each encoded by a single gene present in the Dictyostelium genome (Blagg et al., 2003). WASP compensates for the loss of function of SCAR/WAVE proteins in Dictyostelium (Veltman et al., 2012).
PAK family has six proteins positioned in two different clades, classified in two separate classes; PAKa-d and PAKf/g, based on phylogenetic analysis of the catalytic kinase domain. The consequence of such distinction is not clear in Dictyostelium to humans who also have two PAK groups (Arasada et al., 2006). The class I PAKs in Dictyostelium functionally established to be involved in cell polarity, actin-myosin assemble and phagocytosis (Lee et al., 2004; Yang et al., 2013; Garcia et al., 2014; Phillips and Gomer, 2014). The six PAK isoforms share high sequence identity ∼50–70% in the catalytic kinase domain and PBD regions, while the rarely display any homology outside these regions. PAKa and PAKb, both of which have CRIB/AI domains that have been linked to myosin II regulation (de la Roche and Cote, 2001).
Dictyostelium PAKa possesses a potential poly-Proline tract for SH3 domain interaction, a highly acidic N-terminal domain followed by a CRIB and a C-terminal kinase domain. The CRIB/PBD domain preferentially interacts with DdRac1B and HsCdc42 and then translocate to myosin II filament to regulate myosin heavy chain kinases (MHCKs). PAKa inhibits MHCK-B, C, and D to stabilize myosin II assembly in response to upstream cAMP response. Despite dynamic subcellular localization, PAKa co-localizes with myosin in all the cell movement process (de la Roche and Cote, 2001). PAKa was also identified at the cytokinesis cleavage furrow, and localized to rear end of the polarized migrating cell, and posterior cortex during chemotaxis (Chung and Firtel, 1999; Muller-Taubenberger et al., 2002). The signaling cascades regulated by PAKa are also dependent on coronin (Swaminathan et al., 2015).
Dictyostelium PAKb consists of an N-terminal Proline-rich region, followed by a consensus PBD, a Glutamine and Asparagine residues rich linker, and a C-terminal kinase domain for catalysis. DdPAKb, previously termed as myosin I heavy chain kinase (MIHCK), was identified (Lee et al., 1996) through its phosphorylation and regulation of a single-headed myosin I (DdMyoD) (de la Roche and Cote, 2001). PAKb phosphorylates TEDS (Thr, Glu, Asp, or Ser residue) rule site located in the MyoD motor (de la Roche and Cote, 2001). Dictyostelium Rac1a/b/c, RacA (a RhoBTB protein), RacB, RacC and RacF1 activate PAKb to regulate myosin driven motility on actin. PAKb localizes in the cytosol and enriches at the leading edge of the cells during migration, macropinocytosis, and phagocytosis, sites that show prevalence of myosin I as well (de la Roche et al., 2005). Thus PAKb plays pivotal role in myosin I activation during these events. However, loss of function mutants’ have impaired functions dependent on myosin I (Wu et al., 1996). In contrast, constitutively active PAKb mutant increases the rate of myosin I dependent processes such as pinocytosis/phagocytosis and disrupts cytokinesis. The constitutively active and C-terminal truncated active PAKb shows localization at rear-end of the migrating cell and cleavage-furrow during cell division (Yang et al., 2013). The notable fact here is the opposite localization of PAKa and PAKb in migrating cells, one at the rear end and the other at the posterior end, respectively, suggests that the two proteins do not have an overlapping function. However, both PAKa and PAKb function synergistically during phagocytosis and not pinocytosis.
The structural features of PAKc include a PH domain followed by a CRIB and a kinase domain, with a C-terminal extension Gßγ binding domain. PAKc PH domain, related to the fungal Cla4p-like PAKs, is alone responsible for the cytosolic localization (Phillips and Gomer, 2014). The PH plus CRIB domain exhibits weak membrane localization in response to chemo-attractant stimulation (Lee et al., 2004). PAKc is activated rapidly and transiently in response to chemo-attractant stimulation that enriches it at the plasma membrane. PAKc functions to inhibit lateral pseudopodia to restrict pseudopod formation to the plasma membrane facing the chemo-attractant source (Lee et al., 2004). PAKc CRIB domain preferentially binds to RacC GTP-bound form. The highly conserved Arginine 34 is required for inositol binding in the PH domain. The CRIB domain is strongly similar to human PAK1 consisting of overlapping CRIB and AID (auto-inhibitory domain). The C-terminal Gßγ binding domain shows strong conservation of the C-terminal yeast Ste20, required for transient localization to the Dictyostelium plasma membrane (Lee et al., 2004).
PAKd contains an N-terminal CH domain and additional C1 domain upstream of the CRIB domain. PAKd was implicated in F-actin aggregation during developmental processes, and actin polymerization in response to stimulation by a chemo attractant (Phillips and Gomer, 2014). Upon cell starvation, PAKd moves to cellular extensions, suggesting its presence in the Golgi apparatus (Garcia et al., 2014). PAKd kinase activity is regulated through the binding of CRIB domain to activated Cdc42/Rac molecules.
Experimental records strongly confirm that Dictyostelium coronins (Coronin A and B) have a Rac activated CRIB motif (Uetrecht and Bear, 2006). None of the previous studies on CRIB containing proteins included coronin family under Cdc42/Rac effector proteins (Clemen et al., 2008). In D. discoideum CoroninA, residues 117–133 harbor the CRIB motif highly homologous to CRIB motifs of other conventional effector proteins. Structural characterization indicates that half of the CRIB motif lies on the solvent accessible face, while the other half is embedded inside. Coronin prefers the GDP form of GTPases for binding to the CRIB motif, which is interestingly exceptional to all other CRIB containing proteins. Analysis of coronin lacking mutants reveals its role in cell motility, phagocytosis and cytokinesis (Vinet et al., 2014). Coronin functions as a Rho protein GDP dissociation inhibitor (RhoGDI) that interacts with Rac GTPases in their inactive GDP bound form, thus preventing their availability to PAKs. It also interacts with PAKs (PAKa) directly to regulate their activity (Swaminathan et al., 2015).
Cdc42/Rac Interactive Binding-Containing Effector Proteins in Acanthamoeba
Acanthamoeba is the solitary free-living soil amoeba that diverged earlier than other amoebozoans (Shabardina et al., 2018; Corsaro, 2020). The genome contains ∼15,455 coding genes and comparative genomic studies from other metazoans established many putative protein families who play an interpreting role of cytoskeleton machinery and signaling related to cell motility and cytokinesis (Clarke et al., 2013). The putative proteins involved in cytoskeleton regulation through small GTPases in downstream pathways are not explored thoroughly (Mullins and Pollard, 1999).
In silico search for the conserved CRIB domain retrieved fourteen proteins in Acanthamoeba castellanii (Figure 3). The only characterized protein in this repertoire is a Myosin I heavy chain kinase (MIHCK), belongs to the p21 activated kinase family (Brzeska et al., 1997). Including MIHCK (Q93107), seven proteins (L8GCL9, L8GPX2, L8GVV0, L8GW48, L8H707, L8HHB6, and L8HET0) have kinase domains homologous to human PAKs along with some variant domains. L8GPX2 shares a domain with myosin I heavy chain kinase, while, L8H707 and L8GW48 have substantial similarities with PAK proteins. L8HHB6 (39%) and L8GCL9 (42%) are potential Ser/Thr kinases that show moderate similarity with human PAK6 and can be categorized under the PAK family after experimental validation. L8HET0 kinase domain shows 24% similarity with HsMRCKβ but no other domains so it could be probable PAK family member. L8GVV0 shows 30% similarity with HsPAK3 can also be prospective PAK family member subjected to experimental backing. Two proteins (L8H2P8 and L8H6L6) with WH1 domain and strong homology with WASP proteins could be potential WASP proteins in Acanthamoeba. Apart from these two, another protein (L8H3X7) possesses WASP like domain characteristics/homology with WASP related proteins of metazoans. Two proteins (L8GKX1 and L8GYC7) composed of unique domains along with the CRIB domain, which does not have homology with any conventional/non-conventional proteins of higher eukaryotes, can be classified as p21 rho-binding domain-containing protein. We also found one protein (L8H695) with Leucine-rich repeats along with the CRIB domain, and have classified it as a unique group. Lastly, L8GFZ9 was with similar structural features to the coronin family, but the functional role and interactive GTPase molecule are yet to be explored. The extensive repertoire of CRIB containing proteins present in Acanthamoeba indicates and supports the ancient origin of the CRIB motif and its linkage to various effector proteins of Rho mediated signaling (see Supplementary Table S1). However, the relation with the Rho family of protein and detailed functional study will help link these proteins with cellular processes and their subcellular localization.
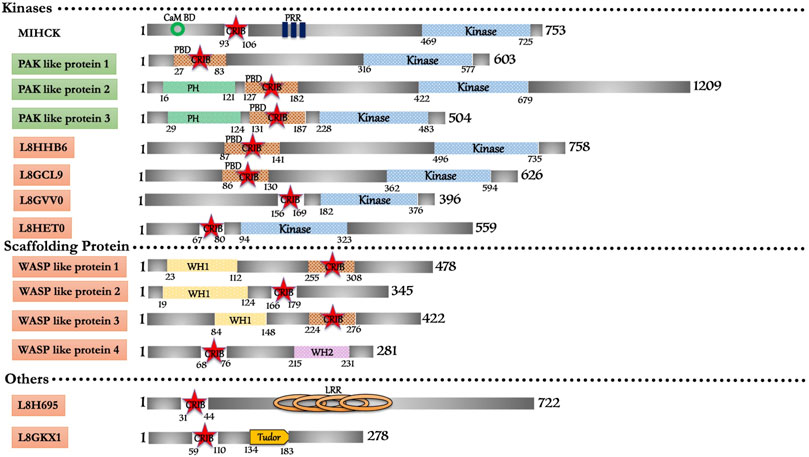
FIGURE 3. Schematic diagram of domain organization of Acanthamoeba castellani CRIB domain containing proteins. The abbreviations for the domains are: CRIB, Cdc42/Rac interactive binding; CaM BD, Calmodulin Binding Domain; LRR, Leucine Rich Repeat; PBD, p21-activated binding domain PH, pleckstrin-homology; PRR, Proline Rich Region; WH1, Enabled/VASP homology 1; WH2, WASP homology 2. The proteins already known from the earlier literature, but not characterized yet are marked as green colored box. The proteins marked in orange (subtle effect) boxes are reported for the first time in this in silico study and are completely uncharacterized. Footnote: In Acanthamoeba castellani, the biochemical characterization is only available for MIHCK, reported 2 decades ago. However, in our study, we report the highest number of CRIB domain-containing proteins from this organism. Their identification opens all kinds of structural, biochemical, biophysical and cellular study avenues for further research. The detailed study on the proteins with entirely novel domain combinations is highly relevant for further research as drug candidates.
Acanthamoeba MIHCK, member of the PAK family, phosphorylates a heavy chain of myosin IC and activates it. MIHCK, from Dictyostelium, was validated as PAKb, which performs a similar function to phosphorylate and activate myosin I. AcMIHCK has a homologous region to human PAK1, PBD (Residue 93–149), which includes the CRIB motif (Residue 93–100) and as IS domain, but lacks the kinase inhibitory (KI) domain region. It also has a putative calmodulin-binding region at its N-terminal, before PBD (Brzeska et al., 2001). The typical C-terminal Ser/Thr kinase domain has the characteristic Ser-627 phosphorylation site. The region between PBD and catalytic domain (residue 158–449) is highly Proline-rich, including multiple PXXP motifs (class I) that provide potential binding to SH3 domains (Brzeska et al., 1996). Acanthamoeba MIHCK mechanism of regulation is quite similar to mammalian PAK1. Acanhtamoeba MIHCK is fully phosphorylated (Brzeska et al., 1990a; Brzeska et al., 1999), while Dictyostelium MIHCK is partially activated in vitro by autophosphorylation in the presence of Rac and lipids (Lee et al., 1998). Human PAK1 also requires Rac or lipids for autophosphorylation (Manser et al., 1994). Calcium-dependent calmodulin inhibits phospholipids’ stimulation in Acanthamoeba MIHCK (Brzeska et al., 1992) and Dictyostelium MIHCK/PAKb (Lee et al., 1998) but is not required for mammalian PAKs. Also, the lipid that activates mammalian PAK1 (Bokoch et al., 1998) differs from those that activate Acanthamoeba and Dictyostelium proteins (Brzeska et al., 1990a; Brzeska et al., 1990b).
Cdc42/Rac Interactive Binding-Containing Effector Proteins in Entamoeba
Entamoeba histolytica is a primitive unicellular eukaryote and amitrochondrian protozoan parasite, which causes dysentery and liver abscess. Amoebic pathogenicity is selected coincidently in the lumen of the intestine because the parasite uses the same methods to kill bacteria or cause disease by damaging the host cells (Bosch and Siderovski, 2015). Amoebic phagocytosis and its mechanism show similarities with the action of macrophages during the phagocytosis of bacteria and unwanted cells, which supports the idea of coincidental selection (Ghosh and Samuelson, 1997; Labruyere et al., 2019). The parasite uses anterior pseudopods and posterior uroids to move inside the human intestine. The host complement system, lectin ConA (multivalent), and anti-amoeba antibodies target the invading amoebic cell, initiating the formation of cap by rearward recruitment of surface receptors and increasing the local receptor-ligand concentration (Calderon, 1980). The defense mechanism of E. histolytica against host immune response includes surface receptor capping in the uroids and membrane shedding (Espinosa-Cantellano and Martinez-Palomo, 1994). E. histolytica cytoskeleton functions actively in the capping process (Espinosa-Cantellano and Martinez-Palomo, 1994; Rath and Gourinath, 2020). The invasion and survival inside the host tissue are maintained through the phagocytosis of RBC, lumen cells and surrounding cells. It has been demonstrated the role of cytoskeleton assembly in the Entamoeba and a human macrophage (Marchat et al., 2020). Phagocytosis is a dynamic and regulated process that involved a varsity of proteins ranging from actin-binding protein, motor protein, small GTPases, kinases and phosphatases (Godbold and Mann, 1998; Marquay Markiewicz et al., 2011; Anwar and Gourinath, 2016; Gautam et al., 2017; Agarwal et al., 2019; Gautam et al., 2019). ∼20 Rho family GTPases and numerous downstream signaling effectors are present in these single-celled trophozoites that coordinate actin dynamics in pathogenesis-related processes. Hence, cell migration and chemotaxis, followed by adherence to the epithelium in the host intestine, and host cell killing and phagocytosis are all regulated by Rho family signaling toolkit (Bosch and Siderovski, 2013).
The domain-based search retrieved a total of nine CRIB domain-containing proteins (Figure 4). The homology with metazoans proteins classifies them under conventional effector proteins of Rac/Cdc42. Six (PAK2-7) and one pseudo-PAK (PAK1) belong to the p21-activated kinase with their typical PBD and kinase domain architecture. However, an earlier study shows that one isoform of PAK lacks CRIB/PBD domain in N-terminal but has C-terminal kinase domain homologous to yeast ste20 (Gangopadhyay et al., 1997). Earlier kinome study (Anamika et al., 2008) predicted 17 homologous PAKs in the E. histolytica genome. Only with the six homologues match the typical PAK features, and thus, it is now confirmed that Entamoeba possesses only six PAK isoforms, including one pseudo-PAK. Interestingly, out of six, three PAKs (PAK2, PAK3 and PAK5) have additional PH domains (Figure 4). Three proteins (C4M2L6, C4LZJ6, C4M0R3) have a domain that is homologous to C-terminal of human WASP, but no experimental studies have so far been performed to characterize its structure and function (Figure 4). The Arp2/3 complex nucleates new actin filaments, when activated by nucleation promoting factors like WASP or SCAR. No available research yet describes any WASP or SCAR protein in Entamoeba, hence, the actin nucleation activity may be regulated by other unidentified proteins. Exploring the predicted putative WASP here might reveal its role in actin nucleation and regulation of actin cytoskeleton. However, CARMIL protein binds Arp2/3 complex with an exact mechanism used by WASP via its acidic motif. CARMIL homologues have been discovered through proteomic analysis of the phagosome (Okada et al., 2005; Clark et al., 2007; Tolstrup et al., 2007) and they further provide essential clues for understanding actin nucleation. A recent in silico study on actin-binding proteins (Rath and Gourinath, 2020) from our lab shows that E. histolytica harbors three WASH (Wiskoit-Aldrich syndrome protein and SCAR homolog) proteins C4MBT4, C4LTV1 and C4M2Y0. Additionally, E. histolytica encodes six formin genes that accelerate actin filament assembly in eukaryotic cells, however, no IQGAPs are reported yet. Three proteins (C4M137, C4M943, C4M5U0), which have similar structural features to the coronin protein family, are also present in Entamoeba (see Supplementary Table S1) but the CRIB motif is not yet defined it them like Dictyostelium. Out of the seven PAK family members, the detailed account of experimental evidence is available only for five; PAK6 and PAK7 have not been characterized yet (see Supplementary Table S1).
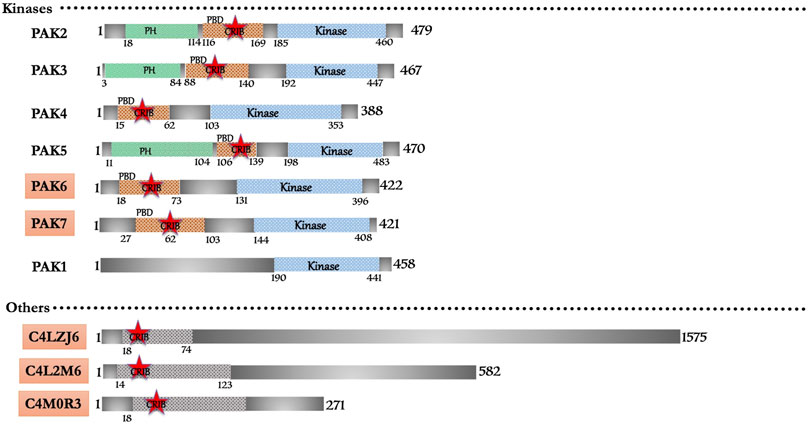
FIGURE 4. Schematic diagram of domain organization of Entamoeba histolytica CRIB domain containing proteins. The abbreviations for the domains are: CRIB, Cdc42/Rac interactive binding; PBD, p21-activated binding domain PH, pleckstrin-homology. The proteins already known from the earlier literature, but not characterized yet are marked as green colored box. The proteins marked in orange (subtle effect) boxes are reported for the first time in this in silico study and are completely uncharacterized. Footnote: In Entamoeba histolytica, PAK1 and PAK2 have been reported as crucial kinases in the survival of this pathogen and are intricately involved in the phagocytic process. PAK3 and PAK4 characterization also showed a promising role in pathogenesis. Structural and biophysical characterization is the prospective future of the already recognized proteins apart from the ones identified in this study. Experimental research on these proteins will lead to new potential drugs targets against the single drug (metronidazole) available in the market. The development of new drugs is necessary due to the reports of drug resistance found in laboratory cultured amoebic cells.
Entamoeba PAK (PAK1) is a pseudo-PAK because it lacks the consensus CRIB motif (or PBD); still, it interacts with EhRac1 via the N-terminal region, suggesting that it is a regulatory domain necessary for the maintenance of cell polarity (Labruyere et al., 2003). Migrating trophozoites display PAK1 at their leading edge, where it functions in amoeboid cellular polarity and motility, along with human red blood cell phagocytosis (Labruyere et al., 2000). The protein shares 33% identity with rat KPAK and yeast STE20 (Gangopadhyay et al., 1997). The Proline-rich N-terminal domain can potentially bind to SH3 domains of adapters like Nck (non-catalytic region of Tyrosine kinase adapter protein) or PIX (PAK Interacting eXchange factor) (Galisteo et al., 1996; Manser et al., 1998). Constitutive EhPAK expression alters the new adhesion site formation in E. histolytica (Labruyere et al., 2003).
PAK2 PBD selectively binds activated EhRacA during receptor capping and collagen matrix invasion (Arias-Romero et al., 2006). PAK2 probably controls cell movement, surface receptor capping and cytokinesis (Arias-Romero et al., 2006). The biochemical studies conducted on the C-terminal kinase domain of PAK2 described its activity towards myelin basic protein. Interestingly, the PAK and EhRacA complex homology model showed the specific interaction in PAK2 residues Met-121 and His-123 with RacA Tyr-40; and PAK2 residue Phe-145 with RacA Asp-63, Arg-66, Leu-67 and Leu-70 (Arias-Romero et al., 2006). A detailed investigation to elucidate the critical residues influencing the binding energy would guide the rationale development of small molecules that inhibit such interaction. The possible interaction of other GTPases with PAK2 can also be investigated experimentally.
Un-stimulated cells show cytoplasmic PAK3 distribution, while capping protein induction relocates it to the caps (Dutta et al., 2007). PAK3 undergoes autophosphorylation and phosphorylates histone H1 in vivo, and in vitro studies displays kinase activity in the absence of small GTPases (Dutta et al., 2007). Maximum enzymatic activity is achieved after the autophosphorylation of a critical residue present in the activation-loop of many protein kinases. It is yet to be established whether an increase in activity or change in localization is observed upon gtpase binding. PAK3 sequential feature reports a PH domain (residues 2–82), a PBD/CRIB domain (residues 84–141) and a kinase domain (residue 142–447) at its C-terminal. PAK3 shares 40% identity (50% similarity) with Dictyostelium PAK (PAKc). All the typically conserved XI subdomains from Ser/Thr kinases are visible in its kinase domain. The only variation observed, is the replacement of the conserved Leucine by Tyrosine in subdomain VII.
PAK4 and PAK5 are highly specific effectors of EhRacC (Bosch and Siderovski, 2015). EhRacCQ65LGTP and EhPAK4-PBD reveal a deviation of PBD α-helix in an otherwise conserved Rho/effector interface. The side chains of EhPAK4 PBD residues line up the EhRacC binding interface. The similar residues are well conserved in EhPAK5, hinting at a common interaction surface for the same Rho gtpase. Asp-17 of EhPAK4 (Glu-108 in EhPAK5) forms a salt bridge with Arg-30 of EhRacC, while Phe-21 of EhPAK4 (Tyr-112 of EhPAK5) contributes towards a hydrophobic RacC interface (Bosch and Siderovski, 2015). Further experiments need to be conducted to for RacC effectors PAK4 and PAK5 to elucidate their biological functions. We still lack the knowledge that correlates the autoinhibition modes of E. histolytica PAK isoforms with mammalian group-I and group-II PAKs. Here, we also acknowledge the unresolved question of the signaling specificity between Rho family gtpase and PAKs.
Entamoeba has three coronins; two belonging to short tail (Coronin1: C4M943, Coronin 2: C4M137) are 70% identical to each other, and one long tail (Coronin 3: C4M5U0). The predicted function of long coronin is crucial in the amoeboid migration and pseudopod regulation (Tolstrup et al., 2007).
Cdc42/Rac Interactive Binding-Containing Effector Proteins in Giardia
The genus Giardia comprises several species that inhabit intestinal tracts of vertebrates (fish, amphibians, reptiles, birds, rodents). It is one of the most pervasive intestinal pathogens that infects a wide range of mammals; for example, human and agricultural livestock such as cattle and sheep. However, Giardia lamblia (alternatively referred to as Giardia intestinalis and Giardia duodenalis) infects and causes giardiasis in humans, suggesting a zoonotic transmission (Ryan and Caccio, 2013). The life cycle is simple, involving two morphogenetic stages; 1) cyst form, which is environmentally resistant and infectious, and 2) vegetative trophozoite stage, which colonizes the small intestine and becomes invasive to cause disease. During encystation, the parasite relays the signals to produce, transport, and secrete the cyst wall protein (CWP). It has been demonstrated that flagella and disk structures modulate motility and host intestinal epithelial cell attachment (Di Genova and Tonelli, 2016). The molecular mechanism behind the regulation of these processes remains abstract. The sole Rho family gtpase, GlRac, regulates endomembrane organization and CWP trafficking (Krtkova et al., 2016). Subcellular localization studies indicate the association of GlRac with endoplasmic reticulum and Golgi apparatus like encystation-specific vesicles (ESV).
The CRIB domain search identifies only one protein (C6LUS0), which has the CRIB and the kinase domains homologous with other PAKs (see Supplementary Table S1). The earlier studies indicated that Giardia lamblia has only one Rac in the complete Rho family, so the identified protein in our search can be the prospective effector protein (Krtkova et al., 2016). The interaction and co-localization studies on proposed protein with known Rac will provide thoughtful insights on the signaling mechanism of cytoskeleton assembly in the parasite.
Cdc42/Rac Interactive Binding-Containing Effector Proteins in Trypanosoma
Trypanosoma cruzi causes an encumbering severe illness in humans, known as the Chagas disease, which affects millions of people globally (de Souza et al., 2010). The various species of Trypanosoma belongs to kinetoplastid protozoans in an evolutionary context (Gupta et al., 2020). The complex life cycle involves four developmental stages: 1) epimastigotes; 2) metacyclic trypomastigotes; 3) amastigotes; and 4) bloodstream trypomastigotes (Zuma et al., 2021). Trypomastigotes and extracellular amastigotes are the only infective forms that are able to invade almost any nucleated host cell (Ferri and Edreira, 2021). During the invasion, bidirectional signaling pathways are triggered in both the parasite and the host cell. The CRIB domain search identifies only one protein (Q4DHX7) in Trypanosoma cruzi (see Supplementary Table S1) which intrigued us to carryout in depth literature search for cytoskeletal regulation signaling pathways. However, we cannot categorize this CRIB domain-containing protein in any existing families of effector protein because it doesn’t any depict homologous features to other eukaryotic proteins. Further investigation is required to know more about the protein function and relation to the CRIB domain-containing protein family from other lower eukaryotes.
Interestingly, few proteins have been found that regulate cytoskeletal pathways but are only related to the invasion of the extracellular amastigote form of T. Cruzi. Extracellular amastigote engulfed by mammalian cells via phagocytic cup based on actin-dependent cytoskeleton changes (Mortara et al., 2005). Extracellular amastigotes secretes proteins like P21, mevalonate kinase (MVK) and specific-surface protein 4 (Ssp4), which mediate host cell signaling during phagocytosis. P21 is a 21 kDa secretory protein (da Silva et al., 2009), related to ERK and PI3K signaling pathways during phagocytosis and cytoskeleton remodelling (Rodrigues et al., 2012; Teixeira et al., 2015). The recombinant version of P21 (rP21) interacts with the CXCR4 chemokine receptor, inducing actin assembly to drive phagocytosis and modulate the PI3K-dependent expression of an actin-related gene (da Silva et al., 2009; Rodrigues et al., 2012). TcMVK is involved in protein glycosylation and cytoskeletal assembly through activation of p38/ERK, FAK (focal adhesion kinase) components and PAK signaling trails (Ferreira et al., 2016). Amastigote form invades the host cell through Ssp-4, another secretory molecule predicted to function as Rac1/WAVE2 and Cdc42/N-WASP signaling mediators (Florentino et al., 2018). TcSsp4 is majorly a surface GPI-anchored glycoprotein whose expression doesn’t correlate with infection, but glycosylation of protein is linked with host cell invasion. Highly infective strain’s amastigotes secrete a differentially glycosylated Ssp4 that recruits Galectin-3 (Gal3) to mediate host cell surface and parasite interaction (Florentino et al., 2018). Recent studies on actin-binding proteins from kinetoplastids, proposed a protein (Q4DEX0) as coronin because it has homologous WD repeat and coronin domains characteristic to amoebozoa and higher eukaryotes (Gupta et al., 2020).
Cdc42/Rac Interactive Binding-Containing Effector Proteins in Leishmania
Leishmania donovani also belongs to kinetoplastids, a unicellular protozoan parasite causing a fatal disease, visceral leishmaniasis, in humans. In vertebrates, it is present as invasive promastigote and amastigote, which cause infection. The best survival strategy used by promastigotes during establishment of infection in macrophages is to inhibit the fusion of phagosome and endosome (Desjardins and Descoteaux, 1997). Lipophosphoglycan (LPG), a significant surface glycoconjugate of promastigote, is crucial for intracellular survival (Handman et al., 1986). The pathogen uses the human host macrophage cell cytoskeletal assembly to sustain and prevent phagosome maturation (Scianimanico et al., 1999; Lodge and Descoteaux, 2008). However, recent report on the actin-binding protein repertoire is classifying the presence of various proteins in the pathogen itself (Gupta et al., 2020). Earlier information reveals that L. donovani recruits human Cdc42 and Rac1 to form an F-actin coat around its phagosome as protective measure from macrophage killing (Lodge and Descoteaux, 2005; Lerm et al., 2006).
The domain search retrieves one protein (Q4QEZ0), which has conserved Ser/Thr kinase domain homologous to PAK catalytic domain but no CRIB domain was present in it (see Supplementary Table S1). A coronin homolog protein (E9BGF4) is reported recently in the genome-based studies (Gupta et al., 2020). The functional characterization to link it with CRIB containing effector proteins is yet to be established.
Cdc42/Rac Interactive Binding-Containing Effector Proteins in the Human Host
Human is placed as the top ranking evolutionary evolved organism, but it is a host for several pathogens. The understanding of molecular mechanisms of host and pathogen proteins helps to prevent infections and diseases. The cellular signaling process is complex in this system due to presence of multi-layered system of tissues and organs. However, cytoskeleton signaling and its regulation are somehow similar in most of the cells and different in some cells at the same time. Investigations on complexity in regulating cytoskeleton dynamics, mediated by Rho family GTPases is fundamental to processes like motility (Murali and Rajalingam, 2014), adhesion, differentiation and development. All the twenty-seven proteins, which have been identified in the CRIB domain-based search, are effector proteins of Rac/Cdc42. The brief information of CRIB containing effectors is accounted in this study to understand the link and origin of CRIB motif and association with protein in social amoebas (Kumar et al., 2009), intestinal pathogens and kinetoplastids. In the human proteome, nine different CRIB containing protein families are present (Figure 5, see Supplementary Table S2). Additionally, six coronin isoforms are also identified in human; some of them have shown the presence of CRIB motif via experimental evidence while others need to be explored (see Supplementary Table S2).
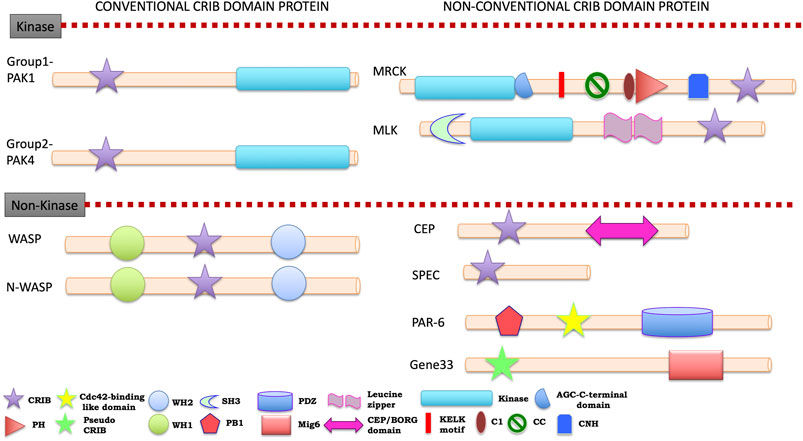
FIGURE 5. Cartoon diagram of signature domain organization of human CRIB domain containing protein families A colour key to the domain names and symbols is given below of classification. The abbreviations for the domains are: CRIB, Cdc42/Rac interactive binding; PH, pleckstrin-homology; WH1, Enabled/VASP homology 1; WH2, WASP homology 2; SH3, Src-homology 3 doamin; PB1, Phox and Bem1; PDZ, PSD-95, Drosophila discs large, and the adherens junction protein, ZO 1; CEP, CRIB effector protein; CNH, Citron homology domain. The proteins divided into conventional and unconventional classification on the basis of their evolutionary link with unicellular eukaryotes.
p21-Activated Kinases
Human PAKs are thoroughly studied, including the elucidation of their structure, function, localization and regulatory mechanism in cells. PAK isoforms have been categorized in Group-I (PAK1, 2, 3) and Group-II (PAK4, 5, 6). Most of the PAKs are ubiquitously expressed, but some are restricted to tissue specific expression (Murali and Rajalingam, 2014). PAK phosphorylates numerous substrates (membrane, cytosolic, mitochondria and nuclear) and act by remodeling cytoskeleton, employing scaffold and shuttling to specific subcellular compartment (Lian et al., 2002; Kumar et al., 2017). It has been clearly understood from evidences that their dysregulation leads to disruption of cellular homeostasis and severely impacts key cellular functions (Bokoch, 2003). Some PAKs are associated with numerous defects and disease (Chan and Manser, 2012) majorly, cancer (Kumar et al., 2006), neurological (Ma et al., 2012), and cardiac disorders (Ke et al., 2014). The in depth functional details can be referred from the recent reviews (Zhao and Manser, 2005; Rane and Minden, 2014; Kumar et al., 2017), while this study presents the classification details based of domain architecture and structural features. Both the PAK groups possess a PBD at N-terminus, an auto inhibitory domain (AID), and a kinase domain at C-terminus. The PBD domain is similar in both groups whereas AID domain of Group-I is partly similar to group-II with minor modifications. The regulatory kinase domain is structurally different and shows distinct activation mechanism in both groups (Ha et al., 2015). The kinase activity of group-I PAKs is initiated in the presence of Rac/Cdc42, while group-II doesn’t require Rac/Cdc42 stimulation for its constitutive activity. Many publications (Baker et al., 2014; Rane and Minden, 2014) highlight that group-I activity modifies through two PAK molecules acting as a dimer to exert a reciprocal auto inhibitory activity (Buchwald et al., 2001). Group-I PAKs are also stimulated by interaction of its PIX motif (PXXP motif at Proline-rich region) present between PBD and kinase domain with SH3 domain in signaling molecule, phosphorylation by 3-phospho-insositide dependent kinase1, AKT and JAK, and binding of phospholipids, exchange factor β-PIX or SH3 proteins such as NCK1 and GRB2 (summary by (Kumar et al., 2017)). The emerging role of PAK1 as potential therapeutic target in cancer was recently reviewed (Rane and Minden, 2019; Yao et al., 2020) comprehensively, elaborating interesting facts and classical evidences to understand its role in many oncogenic signaling pathways (Senapedis et al., 2016; Yao et al., 2020).
Wiskott-Aldrich Syndrome Proteins
The 2 decades of extensive evidences suggest that WASP family proteins have widened into five subfamilies in vertebrates including humans: 1) WASP and neural-WASP (N-WASP; also known as WASL), 2) three WASP family Verprolin homologue isoforms (WAVE1-3; also known as SCAR1-3 or WASF1-3), 3) WASP homolog associated with actin, membranes and microtubules (WHAMM), WASP and SCAR homologue (WASH; also known as WASHC1), and 4) junction-mediating regulatory protein (JMY) (Alekhina et al., 2017). WASP protein participates in innate and adaptive immune response through regulation of actin cytoskeleton-dependent cellular processes, including immune synapse formation, cell signaling, migration and cytokine release (Thrasher and Burns, 2010; Rivers and Thrasher, 2017). Most of the vertebrate including human possess a ubiquitous WASP-paralogue, N-WASP, which was originally described as neural-specific gene although expressed in nearly all cell types (Miki et al., 1996; Miki et al., 1998a). N-WASP commonly exists in an inactive confirmation in which Arp2/3 complex cannot interact with actin filament. Humans have four homologues of SCAR (HsSCAR1-4) (Bear et al., 1998) and one WAVE (Miki et al., 1998b) protein member representatives. Subcellular localization data of WASP family proteins underline the crucial role of these proteins in actin-based cell motility regulation (Papayannopoulos et al., 2005). The C-terminal VCA domain in all WASP family members is responsible for the activation of Arp2/3 complex to nucleate actin polymerization. On the other hand, the N-terminus contains variable domains that are considered to confer spatial and temporal regulation of Arp2/3 activating activity. The presence of Proline-rich regions, one or more WH2 (WASP homology-2) and WH1 (WASP homology-1) domain helps to interact with WIP (WASP interacting protein) and is required to stabilize WASP (Tyler et al., 2016). The GBD/CRIB domain bind to gtpase bound forms of Cdc42/Rac and alleviate auto inhibitory fold (Kim et al., 2000; Hemsath et al., 2005). The role of WASP family member as actin cytoskeleton regulators also links with the invasiveness and metastasis of cancer (review by (Kurisu and Takenawa, 2010)).
Activated Cdc42-Associated Kinase
ACK1, the only non-receptor Tyrosine kinase (or cytosolic Tyrosine kinase) is composed of diverse domains and its Tyrosine phosphorylation activates several effectors involved in cell proliferation and growth. ACK1 gets activated in response to multiple signals, majorly cell adhesion, growth factor receptors and hetero-trimeric G-protein coupled receptors (Prieto-Echague and Miller, 2011). The ACK1 possesses N-terminal catalytic kinase domain followed by a SH3 domain, C-terminal poly-Proline region (PPXY motif) and CRIB domain (Yang and Cerione, 1997). ACK1 was identified with some more domain regions which sets it apart from other non-receptor Tyrosine kinases (NRTKs) and includes at least eight distinct domains; the sterile α-motif (SAM), kinase or catalytic domain, SH3 domain, GTPase-binding domain, clathrin-interacting region, PPXY motif or WW domain-interacting region, an MIG6 homology region (MHR), also known as epidermal growth factor receptor (EGFR)-binding domain and an ubiquitin association (UBA) domain. It interacts only with Cdc42 and not Rac gtpase protein. Biochemical studies suggest that ACK1 strongly interacts with SH3 domain of Src family kinases (Src or Hck) via the C-terminal Proline-rich region (Yokoyama and Miller, 2003). Interaction of Cdc42 is required for auto phosphorylation whereas SH3 domain appears to function during auto inhibition (Galisteo et al., 2006). One of the substrates of ACK1 is WASP, which get phosphorylated and promotes its function (Yokoyama et al., 2005). The high expression of ACK1 during breast cancer makes it an appropriate marker for breast cancer detection. Apart from its role in breast cancer, ACK1 involved in stomach (Xu et al., 2015), hepatic (Xie et al., 2015; Wang et al., 2020), prostate (Furlow, 2006), ovarian, lung (Tan et al., 2014) and cervical cancer. The emergence of ACK1 as an oncogenic kinase has unraveled novel mechanisms by which dysregulated Tyrosine kinase signaling drives cancer progression through altered cellular homeostasis (Mahajan and Mahajan, 2015). Perhaps, recent data explains its function as an epigenetic regulator (Mahajan and Mahajan, 2015).
Myotonic Dystrophy-Related Cdc42-Binding Kinases Proteins
MRCK is primarily involved in actomyosin regulation by regulating Myosin II light chain (MLC) and has three isoforms (MRCKα, MRCKβ, MRCKγ) (Zhao et al., 1997; Leung et al., 1998; Ng et al., 2004) in human. MRCK, a CRIB containing effector is present in non-vertebrate and vertebrates but absent in lower eukaryotes and yeast (Pirone et al., 2001; Zhao and Manser, 2005). The Cdc42 regulated MRCK proteins are a subfamily of AGC (PKA, PAG and PKC) kinase family (Pearce et al., 2010). Apart from MRCK, Rho mediated signaling pathway for phosphorylation of MLC includes ROCK (RhoA binding coiled-coil containing kinases) (Olson, 2008) and CRIK (Citron rho interacting kinase) (201), which are functionally more established relative to MRCKs regulation mechanism. All three MRCKs have well conserved N-terminal kinase domain followed by a central linker linking to C-terminal with four domains. These four domains are protein kinase C conserved region (C1 domain) followed by PH domain, CH domain and lastly CRIB domain. C1 domain binds phorbol ester and might help in promoting kinase activation, while PH domain interacts mostly with lipid partners and leads to appropriate cellular localization of MRCKs (Loo et al., 2013). It has been expected that elevated MRCK expression might be prominent in invasiveness and metastasis of cancer, because actin myosin contractility is essential component of cell motility and is vital for cancer cell invasion and metastasis (Olson, 2008; Unbekandt and Olson, 2014; Zhao and Manser, 2015).
Mixed Lineage Kinases
MLKs are predominantly upstream kinases initiating the MAPK cascade, particularly the JNK (c-Jun N-terminal kinases). Four isoforms of MLKs (MLK1-4) are present in human and they are of immense interest because of their role in neurodegenerative conditions. The structural features of MLKs indicate N-terminal SH3 domain and kinase domain, a Leucine zipper, which connects the kinase domain to C-terminal CRIB/PBD domain (Dorow et al., 1993). Experimental evidences show that MLK3 interacts with GTP bound state of Cdc42 and Rac1 (Teramoto et al., 1996) and later on with RhoG as well (Zhang and Gallo, 2001; Wennerberg et al., 2002; Du et al., 2005). Earlier researches on gene silencing, genetically engineered mouse models and small molecule inhibitors suggest that MLKs are critical in tumor progression as well as in inflammatory processes (Handley et al., 2007; Gallo et al., 2020). Recent studies highlight the function of MLK3 in tumor cell proliferation, migration and invasion which opens the avenue for further research to investigate MLKs as potential therapeutic target for cancer treatment (Chadee, 2013; Gallo et al., 2020). Moreover, previous studies shows that the altered function/expression of MLK family of kinases leads to very wide spectrum of disorders (Handley et al., 2007; Craige et al., 2016).
Cdc42 Effector Protein
CEPs are also known as binder of Rho GTPases (Borg), with five isoforms present in humans. CEP/Borg protein family function as negative regulator of small GTPases. Borg1, Borg2, Borg4, and Borg5 (previously termed as MSE55) bind both TC10 GTPases and Cdc42, except Borg3, which only interacts with Cdc42 (Joberty et al., 1999). CEPs are structurally composed of a Cdc42 binding domain and two unique CI and CII domains (Hirsch et al., 2001), only exception is CEP5 that lacks CI domain. Alike PAK, ACK and WASP proteins, CEP family proteins also have the conserved consensus sequence at extended C-terminal CRIB domain (I-S-X-P-L-G-X-F-R-H-T-AA-H-AA-G-X-X-Gly-(X)0–2-D-AA-F-G-D-X-S-F-L, where AA represents an aliphatic amino acid) that can be involved in regulation of biological effects of CEP protein (Hirsch et al., 2001). Despite an earlier discovery, the molecular mechanism and functions CEPs or Borg family remain largely elusive. Interestingly, unlike other Cdc42 effectors, these genes are only present in vertebrates. However, recently researchers have investigated their role in tumor progression, regulation and function (Farrugia and Calvo, 2016; 2017).
Small Protein Effector of Cdc42
In humans, two members (SPEC1/Cdc42SE1 and SPEC2/Cdc42SE2) are found that contain a conserved N-terminal region and centrally located CRIB domain. Biochemical interaction studies show that it strongly interacts with Cdc42, weekly with Rac1 and not at all with RhoA (Pirone et al., 2000). One study reveals that three distinct regions (phosphoinositide-binding region within basic amino acids, N-terminal to CRIB sequence) within SPECs are likely to be involved in early contractile events in phagocytosis (Ching et al., 2007). SPECs have been shown to play an important role in Cdc42-mediated F-actin accumulation at immunological synapse (Ching et al., 2005). In disease connection, SPEC1 was down regulated during skin cancer to promote tumorigenesis, and thus proposed to be as an important marker of skin cancer progression (Kalailingam et al., 2019). This family of proteins is yet to be investigated for regulation, expression and function in details to chronology connect.
Gene33
Gene 33, also called as mitogen-inducible gene-6 (Mig-6) or ERFI1 is an immediate early gene that is induced transcriptionally by several extracellular stimuli. Physiological function of Gene-33 remains unclear but commonly occurring chronic stress stimuli (mechanical strain, vasoactive peptides and diabetic nephropathy) increase its mRNA levels in the cells (Makkinje et al., 2000; Park et al., 2017). The structure of Gene33 resembles an adaptor protein capable of binding monomeric gtpase (HsCdc42) in vivo and in vitro. Gene33/Mig-6 is a negative regulator of EGF signaling (Park et al., 2015), and Mig-6 inhibits Cdc42 signaling which is critical for Mig-6 function to suppress cell migration. The dysregulation of Cdc42 mediated Gene33 pathway may play a critical role in cancer development (Jiang et al., 2016).
PAR-6
Structurally PAR-6 have pseudo-CRIB domain in comparison to other CRIB containing effectors. PAR-6 functions mainly in determining cell polarity and are conserved throughout metazoans where it is a substrate of aPKC (atypical protein kinase C) (Izumi et al., 1998; Lin et al., 2000). The Cdc42-PAR-6-aPKC complex involved with PAR-3 localizes in epithelial cells’ apical rings and E-cadherin at adherens junction (Yamanaka et al., 2001; Atwood et al., 2007; Thompson, 2021). PAR-6 has three isoforms in humans that are involved in cell polarity regulated by Cdc42 (Joberty et al., 2000; Macara, 2004).
Discussion
Understanding the domain architecture serves as a crucial link to decipher a protein’s molecular mechanism in intracellular signaling cascades. Several protein domains and their relation to signaling components present in humans are entirely missing from the best-studied model organism or pathogens and vice versa. The signaling pathways involving Cdc42 and Rac GTPases are conserved in all eukaryotes but their interactive proteins (CRIB) are not annotated or characterized in lower eukaryotes like protozoan parasites. The current study identifies and describes the CRIB domain containing protein families and their possible role (according to domain architechture) as effector proteins for cytoskeletal regulation. The structural similarities, combination of regulatory domains, and their putative/observed functions in lower eukaryotic pathogens have been accounted here. Genome and proteome information allows us for a better understanding of pathogenic processes and consequently help improve the prevention, diagnosis, and treatment of the diseases.
The evolutionary divergence shows that owing to their ancient origin, PAK and WASP families are termed as conventional effector molecules. They contain a conserved CRIB motif with its extended region known as PBD (in PAK) and GBD (in WASP), crucial for cytoskeleton dynamics. However, coronin protein family is an exception that binds GDP-Rac/Cdc42 in Dictyostelium (Swaminathan et al., 2014). However, earlier studies on CRIB effector proteins; coronin have not been included in the list of Cdc42 effectors, despite their well-defined CRIB motif shown in the structural and functional studies (Swaminathan et al., 2014). The coronin family proteins must be include in the CRIB effectors now. The CRIB proteins in protozoan parasites is not investigated in any capacity and reported here which opens an avenue for their functional and biophysical studies as potential drug candidates against host.
The evolution of the other reported effector protein families started later on as seen in worm, flies, frog, and human, displaying a direct correlation between the increase in the complexity of the organism with the increase in several members of each family. These effector families: ACK, MRCK, MLK, SPEC, CEP, PAR-6 now classified as non-conventional Cdc42/Rac effectors. The highest evolved organism, human, consists of conventional and non-conventional Cdc42/Rac effector protein members along with some other proteins like IQGAPs (IQGAP1-3), IRSp53 etc ((Pichaud et al., 2019) Supplementary Information).
Structural insights into the CRIB effector protein emphasizes that the C-terminal inhibitory switch (IS) domain responsible for sustaining it in an auto-inhibited basal state, is well-conserved during evolution (Hoffman and Cerione, 2000; Kim et al., 2000; Lei et al., 2000). Depending on the context of low or high-affinity binding, GBD/PBD domain adopts related but distinct folds (Rudolph et al., 1998; Kim et al., 2000). When the GBD domain is in the free state, it looks largely unstructured, while in an auto-inhibited state, it forms an β-hairpin structure following the conserved CRIB motif and a central three-helix bundle. However, during the interaction, the PBD/GBD forms high-affinity complex with their respective G-proteins, in which the unstructured region becomes structured (Hoffman and Cerione, 2000; Kim et al., 2000; Lei et al., 2000). Similarly, the Cdc42-CRIB motif interaction occurs between the β2 strand of Cdc42 through the formation of an intermolecular β-sheet (Abdul-Manan et al., 1999; Mott et al., 1999; Hoffman and Cerione, 2000; Morreale et al., 2000). The GBD domains (except CRIB motif) display divergence in their C-terminal regions with several binding mode variations, possibly determining the specificity of interaction with effector protein. Association of GBD/PBD domains with Cdc42/Rac, instigates a stagy change in the conformation that refolds the IS domain while unfolding the rest of the structure (Hoffman and Cerione, 2000). The CRIB motif interacts with switch I and II regions of GTPases (Stevens et al., 1999). It has been observed generally that two histidine at the C-terminal and one at sixth positions are well conserved, and along with the adjacent α-helix mediate sensitivity to the nucleotide switch. This establishes that the CRIB motif prefers GTP-loaded GTPases and exhibits decreased binding activity with GDP form. Apart from GTP bound state, the flanking sequence of the CRIB motif also determines Cdc42/Rac binding specificity (Pirone et al., 2001; Owen and Mott, 2018). The typical example of such specificity is an autoinhibitory domain of PAKs located C-terminally to the CRIB/PBD domain (Lei et al., 2000; Kim et al., 2016). The new concepts of the intrinsically disordered region (IDR) have also emerged, which highlight that apart from modular and defined domain, the basic rich (BR) regions like poly-Proline leads to the structure-function paradigm in CRIB containing effector proteins (Papayannopoulos et al., 2005; Pang and Zhou, 2016; Owen and Mott, 2018).
The structural component of CRIB containing effector proteins also highlights that apart from CRIB domain and signature domain of that family, various accessory domains like, Proline-rich sequences, PH domain, AID domain, and others are crucial for regulation, activation, subcellular localization and specificity of substrate to perform definite functions. The conventional or non-conventional protein family’s structures share numerous parallelisms in signaling and arrangement in course of evolution. The notable fact that lower eukaryotes represent functional equivalents/counterparts with high sequence divergence in intrinsically regions of proteins. The putative proteins identified in this study from gastric protozoan pathogens and kinetoplastids will be of great importance to study experimentally and some can be used as therapeutic targets against neglected tropical diseases cause by them in their human host. The putative proteins of occasional parasite social amoeba can also be utilized to study the function and regulation of activation of CRIB domain containing effector proteins to understand the role of comparative to the human homologues. Two unique domain combination protein of Acanthamoeba are of great importance in light of drug targets.
Taken together, we would like to summarize that CRIB motif is ancient in origin and it has conventional effector family protein members present in model organism and protozoan parasites. The primary classification and identified protein awaits experimental confirmation for being potential drug targets and key players for the survival of the pathogen.
Methods
We first tabulated the various types of CRIB-domain containing proteins from the available literature. We used the InterPro database (Blum et al., 2021) to carry out a domain-based search for all the types of CRIB domains against the selected organisms. Our data consists of proteins searched across the proteome of the following organisms: Dictyostelium discoideum, Acanthamoeba castellani, Entamoeba histolytica, Trypanosoma cruzi, Leishmania donovani, and Giardia lamblia. We also used the proteins from humans for comparative purposes using their domain architecture (Figure 6) (Page et al., 2021). The protein ID’s used in the manuscript was procured from the UniProt database for universal usage (UniProt, 2021). Once the proteins from all the organisms were fished out, we used Phyre2 software to determine their structure homology to better classify them (Kelley et al., 2015). We performed a manual sequence analysis as well to corroborate the presence of variant CRIB domains, otherwise missed out by the software, as well. Sequence alignment was performed using the ClustalW software and the final Figure was prepared with the Sequence Manipulation Suite2 website (Thompson et al., 1994; Stothard, 2000).
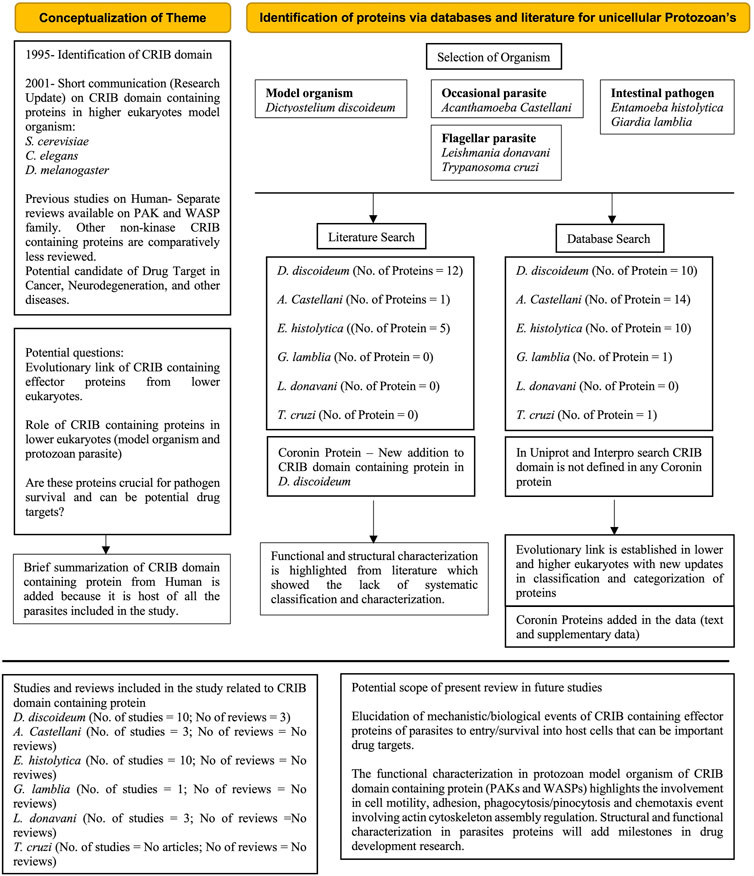
FIGURE 6. PRISMA flow diagram (Reference) explains the conceptualization, brief overview of database and literature search and outcomes of systematic review. The numbers of studies and reviews mentioned in box purely accounting CRIB containing proteins about particular organism. The future scope of systematic review explained in key bullets. Coronin proteins which are unique and proposed to be included in this review based on individual protein characterization (Protein ID’s are included in Supplementary Information).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
PU, PPR, and SG contributed to the conception and design of the study. PPR performed the bioinformatics data mining and analysis and PU did compilation of the study. PU wrote the first draft of the manuscript and PPR edited it under the supervision of SG. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
We thank Science and Engineering Research Board, Government of India for funding under grant number CRG/2020/000,177.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
PU acknowledges UGC, DBT, and British Council and PR thanks ICMR (2020–6079) for their respective fellowships.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.781885/full#supplementary-material
REFERENCES
Abdul-Manan, N., Aghazadeh, B., Liu, G. A., Majumdar, A., Ouerfelli, O., Siminovitch, K. A., et al. (1999). Structure of Cdc42 in Complex with the GTPase-Binding Domain of the 'Wiskott-Aldrich Syndrome' Protein. Nature 399, 379–383. doi:10.1038/20726
Agarwal, S., Anand, G., Sharma, S., Parimita Rath, P., Gourinath, S., and Bhattacharya, A. (2019). EhP3, a Homolog of 14-3-3 Family of Protein Participates in Actin Reorganization and Phagocytosis in Entamoeba Histolytica. PLoS Pathog. 15, e1007789. doi:10.1371/journal.ppat.1007789
Alekhina, O., Burstein, E., and Billadeau, D. D. (2017). Cellular Functions of WASP Family Proteins at a Glance. J. Cell Sci 130, 2235–2241. doi:10.1242/jcs.199570
Amato, C., Thomason, P. A., Davidson, A. J., Swaminathan, K., Ismail, S., Machesky, L. M., et al. (2019). WASP Restricts Active Rac to Maintain Cells' Front-Rear Polarization. Curr. Biol. 29, 4169–4182. doi:10.1016/j.cub.2019.10.036
Anamika, K., Bhattacharya, A., and Srinivasan, N. (2008). Analysis of the Protein Kinome ofEntamoeba Histolytica. Proteins 71, 995–1006. doi:10.1002/prot.21790
Anwar, T., and Gourinath, S. (2016). Deep Insight into the Phosphatomes of Parasitic Protozoa and a Web Resource ProtozPhosDB. PLoS One 11, e0167594. doi:10.1371/journal.pone.0167594
Arasada, R., Son, H., Ramalingam, N., Eichinger, L., Schleicher, M., and Rohlfs, M. (2006). Characterization of the Ste20-like Kinase Krs1 of Dictyostelium discoideum. Eur. J. Cell Biol. 85, 1059–1068. doi:10.1016/j.ejcb.2006.05.013
Arias-Romero, L. E., de Jesús Almáraz-Barrera, M., Díaz-Valencia, J. D., Rojo-Domínguez, A., Hernandez-Rivas, R., and Vargas, M. (2006). EhPAK2, a Novel P21-Activated Kinase, Is Required for Collagen Invasion and Capping in Entamoeba Histolytica. Mol. Biochem. Parasitol. 149, 17–26. doi:10.1016/j.molbiopara.2006.04.001
Atwood, S. X., Chabu, C., Penkert, R. R., Doe, C. Q., and Prehoda, K. E. (2007). Cdc42 Acts Downstream of Bazooka to Regulate Neuroblast Polarity through Par-6-aPKC. J. Cell Sci 120, 3200–3206. doi:10.1242/jcs.014902
Baker, N., De Koning, H. P., Mäser, P., and Horn, D. (2013). Drug Resistance in African Trypanosomiasis: the Melarsoprol and Pentamidine story. Trends Parasitol. 29, 110–118. doi:10.1016/j.pt.2012.12.005
Baker, N. M., Yee Chow, H., Chernoff, J., and Der, C. J. (2014). Molecular Pathways: Targeting RAC-P21-Activated Serine-Threonine Kinase Signaling in RAS-Driven Cancers. Clin. Cancer Res. 20, 4740–4746. doi:10.1158/1078-0432.ccr-13-1727
Bear, J. E., Rawls, J. F., and Saxe, C. L. (1998). SCAR, a WASP-Related Protein, Isolated as a Suppressor of Receptor Defects in Late Dictyostelium Development. J. Cell Biol 142, 1325–1335. doi:10.1083/jcb.142.5.1325
Bishop, A. L., and Hall, A. (2000). Rho GTPases and Their Effector Proteins. Biochem. J. 348, 241–255. doi:10.1042/bj3480241
Blagg, S. L., Stewart, M., Sambles, C., and Insall, R. H. (2003). PIR121 Regulates Pseudopod Dynamics and SCAR Activity in Dictyostelium. Curr. Biol. 13, 1480–1487. doi:10.1016/s0960-9822(03)00580-3
Blum, M., Chang, H.-Y., Chuguransky, S., Grego, T., Kandasaamy, S., Mitchell, A., et al. (2021). The InterPro Protein Families and Domains Database: 20 Years on. Nucleic Acids Res. 49, D344–D354. doi:10.1093/nar/gkaa977
Bokoch, G. M. (2003). Biology of the P21-Activated Kinases. Annu. Rev. Biochem. 72, 743–781. doi:10.1146/annurev.biochem.72.121801.161742
Bokoch, G. M., Reilly, A. M., Daniels, R. H., King, C. C., Olivera, A., Spiegel, S., et al. (1998). A GTPase-independent Mechanism of P21-Activated Kinase Activation. J. Biol. Chem. 273, 8137–8144. doi:10.1074/jbc.273.14.8137
Bosch, D. E., and Siderovski, D. P. (2015). Entamoeba Histolytica RacC Selectively Engages P21-Activated Kinase Effectors. Biochemistry 54, 404–412. doi:10.1021/bi501226f
Bosch, D. E., and Siderovski, D. P. (2013). G Protein Signaling in the Parasite Entamoeba Histolytica. Exp. Mol. Med. 45, e15. doi:10.1038/emm.2013.30
Bozzaro, S. (2019). The Past, Present and Future of Dictyostelium as a Model System. Int. J. Dev. Biol. 63, 321–331. doi:10.1387/ijdb.190128sb
Brown, J. L., Jaquenoud, M., Gulli, M.-P., Chant, J., and Peter, M. (1997). Novel Cdc42-Binding Proteins Gic1 and Gic2 Control Cell Polarity in Yeast. Genes Dev. 11, 2972–2982. doi:10.1101/gad.11.22.2972
Brzeska, H., Knaus, U. G., Wang, Z.-Y., Bokoch, G. M., and Korn, E. D. (1997). p21-activated Kinase Has Substrate Specificity Similar to Acanthamoeba Myosin I Heavy Chain Kinase and Activates Acanthamoeba Myosin I. Proc. Natl. Acad. Sci. 94, 1092–1095. doi:10.1073/pnas.94.4.1092
Brzeska, H., Kulesza-Lipka, D., and Korn, E. D. (1992). Inhibition of Acanthamoeba Myosin I Heavy Chain Kinase by Ca(2+)-Calmodulin. J. Biol. Chem. 267, 23870–23875. doi:10.1016/s0021-9258(18)35917-9
Brzeska, H., Lynch, T. J., and Korn, E. D. (1990a). Acanthamoeba Myosin I Heavy Chain Kinase Is Activated by Phosphatidylserine-Enhanced Phosphorylation. J. Biol. Chem. 265, 3591–3594. doi:10.1016/s0021-9258(19)39630-9
Brzeska, H., Lynch, T. J., Martin, B., Corigliano-Murphy, A., and Korn, E. D. (1990b). Substrate Specificity of Acanthamoeba Myosin I Heavy Chain Kinase as Determined with Synthetic Peptides. J. Biol. Chem. 265, 16138–16144. doi:10.1016/s0021-9258(17)46199-0
Brzeska, H., Szczepanowska, J., Hoey, J., and Korn, E. D. (1996). The Catalytic Domain of Acanthamoeba Myosin I Heavy Chain Kinase. J. Biol. Chem. 271, 27056–27062. doi:10.1074/jbc.271.43.27056
Brzeska, H., Young, R., Knaus, U., and Korn, E. D. (1999). Myosin I Heavy Chain Kinase: Cloning of the Full-Length Gene and Acidic Lipid-dependent Activation by Rac and Cdc42. Proc. Natl. Acad. Sci. 96, 394–399. doi:10.1073/pnas.96.2.394
Brzeska, H., Young, R., Tan, C., Szczepanowska, J., and Korn, E. D. (2001). Calmodulin-binding and Autoinhibitory Domains ofAcanthamoeba Myosin I Heavy Chain Kinase, a P21-Activated Kinase (PAK). J. Biol. Chem. 276, 47468–47473. doi:10.1074/jbc.m108957200
Buchwald, G., Hostinova, E., Rudolph, M. G., Kraemer, A., Sickmann, A., Meyer, H. E., et al. (2001). Conformational Switch and Role of Phosphorylation in PAK Activation. Mol. Cell Biol 21, 5179–5189. doi:10.1128/mcb.21.15.5179-5189.2001
Burbelo, P. D., Drechsel, D., and Hall, A. (1995). A Conserved Binding Motif Defines Numerous Candidate Target Proteins for Both Cdc42 and Rac GTPases. J. Biol. Chem. 270, 29071–29074. doi:10.1074/jbc.270.49.29071
Calderón, J. (1980). Dynamic Changes on the Surface of Entamoeba Induced by Antibodies. Arch. Invest. Med. (Mex) 11, 55–61.
Carrero, J. C., Reyes-López, M., Serrano-Luna, J., Shibayama, M., Unzueta, J., León-Sicairos, N., et al. (2020). Intestinal Amoebiasis: 160 Years of its First Detection and Still Remains as a Health Problem in Developing Countries. Int. J. Med. Microbiol. 310, 151358. doi:10.1016/j.ijmm.2019.151358
Chadee, D. N. (2013). Involvement of Mixed Lineage Kinase 3 in Cancer. Can. J. Physiol. Pharmacol. 91, 268–274. doi:10.1139/cjpp-2012-0258
Chan, P. M., and Manser, E. (2012). PAKs in Human Disease. Prog. Mol. Biol. Transl Sci. 106, 171–187. doi:10.1016/b978-0-12-396456-4.00011-0
Ching, K. H., Kisailus, A. E., and Burbelo, P. D. (2007). Biochemical Characterization of Distinct Regions of SPEC Molecules and Their Role in Phagocytosis. Exp. Cell Res. 313, 10–21. doi:10.1016/j.yexcr.2006.09.011
Ching, K. H., Kisailus, A. E., and Burbelo, P. D. (2005). The Role of SPECs, Small Cdc42-Binding Proteins, in F-Actin Accumulation at the Immunological Synapse. J. Biol. Chem. 280, 23660–23667. doi:10.1074/jbc.m500128200
Chung, C. Y., Feoktistov, A., Hollingsworth, R. J., Rivero, F., and Mandel, N. S. (2013). An Attenuating Role of a WASP-Related Protein, WASP-B, in the Regulation of F-Actin Polymerization and Pseudopod Formation via the Regulation of RacC during Dictyostelium Chemotaxis. Biochem. Biophysical Res. Commun. 436, 719–724. doi:10.1016/j.bbrc.2013.06.022
Chung, C. Y., and Firtel, R. A. (1999). PAKa, a Putative PAK Family Member, Is Required for Cytokinesis and the Regulation of the Cytoskeleton in Dictyostelium discoideum Cells during Chemotaxis. J. Cell Biol 147, 559–576. doi:10.1083/jcb.147.3.559
Clark, C. G., Alsmark, U. C. M., Tazreiter, M., Saito‐Nakano, Y., Ali, V., Marion, S., et al. (2007). Structure and Content of the Entamoeba Histolytica Genome. Adv. Parasitol. 65, 51–190. doi:10.1016/s0065-308x(07)65002-7
Clarke, M., Lohan, A. J., Liu, B., Lagkouvardos, I., Roy, S., Zafar, N., et al. (2013). Genome of Acanthamoeba Castellanii Highlights Extensive Lateral Gene Transfer and Early Evolution of Tyrosine Kinase Signaling. Genome Biol. 14, R11. doi:10.1186/gb-2013-14-2-r11
Clemen, C. S., Rybakin, V., and Eichinger, L. (2008). The Coronin Family of Proteins. Subcell Biochem. 48, 1–5. doi:10.1007/978-0-387-09595-0_1
Clemens, J. C., Worby, C. A., Simonson-Leff, N., Muda, M., Maehama, T., Hemmings, B. A., et al. (2000). Use of Double-Stranded RNA Interference in Drosophila Cell Lines to Dissect Signal Transduction Pathways. Proc. Natl. Acad. Sci. 97, 6499–6503. doi:10.1073/pnas.110149597
Corsaro, D. (2020). Update on Acanthamoeba Phylogeny. Parasitol. Res. 119, 3327–3338. doi:10.1007/s00436-020-06843-9
Cotteret, S., and Chernoff, J. (2002). The Evolutionary History of Effectors Downstream of Cdc42 and Rac. Genome Biol. 3, REVIEWS0002. doi:10.1186/gb-2002-3-2-reviews0002
Craige, S. M., Reif, M. M., and Kant, S. (2016). Mixed - Lineage Protein Kinases (MLKs) in Inflammation, Metabolism, and Other Disease States. Biochim. Biophys. Acta (Bba) - Mol. Basis Dis. 1862, 1581–1586. doi:10.1016/j.bbadis.2016.05.022
Da Silva, C. V., Kawashita, S. Y., Probst, C. M., Dallagiovanna, B., Cruz, M. C., Da Silva, E. A., et al. (2009). Characterization of a 21 kDa Protein from Trypanosoma Cruzi Associated with Mammalian Cell Invasion. Microbes Infect. 11, 563–570. doi:10.1016/j.micinf.2009.03.007
Dammann, K., Khare, V., Coleman, C., Berdel, H., and Gasche, C. (2018). p-21 Activated Kinase as a Molecular Target for Chemoprevention in Diabetes. Geriatrics (Basel) 3, 73. doi:10.3390/geriatrics3040073
De La Roche, M. A., and Côté, G. P. (2001). Regulation of Dictyostelium Myosin I and II. Biochim. Biophys. Acta (Bba) - Gen. Subjects 1525, 245–261. doi:10.1016/s0304-4165(01)00110-6
De La Roche, M., Mahasneh, A., Lee, S.-F., Rivero, F., and Côté, G. P. (2005). Cellular Distribution and Functions of Wild-type and Constitutively ActivatedDictyosteliumPakB. MBoC 16, 238–247. doi:10.1091/mbc.e04-06-0534
De Souza, W., De Carvalho, T. M., and Barrias, E. S. (2010). Review on Trypanosoma Cruzi: Host Cell Interaction. Int. J. Cell Biol 2010, 295394. doi:10.1155/2010/295394
Desjardins, M., and Descoteaux, A. (1997). Inhibition of Phagolysosomal Biogenesis by the Leishmania Lipophosphoglycan. J. Exp. Med. 185, 2061–2068. doi:10.1084/jem.185.12.2061
Di Genova, B. M., and Tonelli, R. R. (2016). Infection Strategies of Intestinal Parasite Pathogens and Host Cell Responses. Front. Microbiol. 7, 256. doi:10.3389/fmicb.2016.00256
Díaz, M. V., Miranda, M. R., Campos-Estrada, C., Reigada, C., Maya, J. D., Pereira, C. A., et al. (2014). Pentamidine Exerts In Vitro and In Vivo Anti Trypanosoma Cruzi Activity and Inhibits the Polyamine Transport in Trypanosoma Cruzi. Acta Tropica 134, 1–9. doi:10.1016/j.actatropica.2014.02.012
Dorow, D. S., Devereux, L., Dietzsch, E., and De Kretser, T. (1993). Identification of a New Family of Human Epithelial Protein Kinases Containing Two Leucine/isoleucine-Zipper Domains. Eur. J. Biochem. 213, 701–710. doi:10.1111/j.1432-1033.1993.tb17810.x
Du, Y., Böck, B. C., Schachter, K. A., Chao, M., and Gallo, K. A. (2005). Cdc42 Induces Activation Loop Phosphorylation and Membrane Targeting of Mixed Lineage Kinase 3. J. Biol. Chem. 280, 42984–42993. doi:10.1074/jbc.m502671200
Dutta, S., Sardar, A., Ray, D., and Raha, S. (2007). Molecular and Functional Characterization of EhPAK3, a P21 Activated Kinase from Entamoeba Histolytica. Gene 402, 57–67. doi:10.1016/j.gene.2007.07.022
Espinosacantellano, M., and Martinezpalomo, A. (1994). Entamoeba Histolytica: Mechanism of Surface Receptor Capping. Exp. Parasitol. 79, 424–435. doi:10.1006/expr.1994.1104
Farrugia, A. J., and Calvo, F. (2017). Cdc42 Regulates Cdc42EP3 Function in Cancer-Associated Fibroblasts. Small GTPases 8, 49–57. doi:10.1080/21541248.2016.1194952
Farrugia, A. J., and Calvo, F. (2016). The Borg Family of Cdc42 Effector Proteins Cdc42EP1-5. Biochem. Soc. Trans. 44, 1709–1716. doi:10.1042/bst20160219
Ferreira, É. R., Horjales, E., Bonfim-Melo, A., Cortez, C., Da Silva, C. V., De Groote, M., et al. (2016). Unique Behavior of Trypanosoma Cruzi Mevalonate Kinase: A Conserved Glycosomal Enzyme Involved in Host Cell Invasion and Signaling. Sci. Rep. 6, 24610. doi:10.1038/srep24610
Ferri, G., and Edreira, M. M. (2021). All Roads Lead to Cytosol: Trypanosoma Cruzi Multi-Strategic Approach to Invasion. Front. Cell. Infect. Microbiol. 11, 634793. doi:10.3389/fcimb.2021.634793
Florentino, P. T. V., Real, F., Orikaza, C. M., Da Cunha, J. P. C., Vitorino, F. N. L., Cordero, E. M., et al. (2018). A Carbohydrate Moiety of Secreted Stage-specific Glycoprotein 4 Participates in Host Cell Invasion by Trypanosoma Cruzi Extracellular Amastigotes. Front. Microbiol. 9, 693. doi:10.3389/fmicb.2018.00693
Fu, Y., Gu, Y., Zheng, Z., Wasteneys, G., and Yang, Z. (2005). Arabidopsis Interdigitating Cell Growth Requires Two Antagonistic Pathways with Opposing Action on Cell Morphogenesis. Cell 120, 687–700. doi:10.1016/j.cell.2004.12.026
Furlow, B. (2006). Tyrosine Kinase ACK1 Promotes Prostate Tumorigenesis. Lancet Oncol. 7, 17. doi:10.1016/s1470-2045(05)70525-8
Galisteo, M. L., Chernoff, J., Su, Y. C., Skolnik, E. Y., and Schlessinger, J. (1996). The Adaptor Protein Nck Links Receptor Tyrosine Kinases with the Serine-Threonine Kinase Pak1. J. Biol. Chem. 271, 20997–21000. doi:10.1074/jbc.271.35.20997
Galisteo, M. L., Yang, Y., Urena, J., and Schlessinger, J. (2006). Activation of the Nonreceptor Protein Tyrosine Kinase Ack by Multiple Extracellular Stimuli. Proc. Natl. Acad. Sci. 103, 9796–9801. doi:10.1073/pnas.0603714103
Gallo, K. A., Ellsworth, E., Stoub, H., and Conrad, S. E. (2020). Therapeutic Potential of Targeting Mixed Lineage Kinases in Cancer and Inflammation. Pharmacol. Ther. 207, 107457. doi:10.1016/j.pharmthera.2019.107457
Gangopadhyay, S. S., Sankar Ray, S., Sinha, P., and Lohia, A. (1997). Unusual Genome Organisation in Entamoeba Histolytica Leads to Two Overlapping Transcripts. Mol. Biochem. Parasitol. 89, 73–83. doi:10.1016/s0166-6851(97)00110-2
Garcia, M., Ray, S., Brown, I., Irom, J., and Brazill, D. (2014). PakD, a Putative P21-Activated Protein Kinase in Dictyostelium discoideum, Regulates Actin. Eukaryot. Cell 13, 119–126. doi:10.1128/ec.00216-13
Gaudet, P., Fey, P., and Chisholm, R. (2008). Dictyostelium discoideum: The Social Ameba. CSH Protoc. 2008, pdb.emo109. doi:10.1101/pdb.emo109
Gautam, G., Ali, M. S., Bhattacharya, A., and Gourinath, S. (2019). EhFP10: A FYVE Family GEF Interacts with Myosin IB to Regulate Cytoskeletal Dynamics during Endocytosis in Entamoeba Histolytica. PLoS Pathog. 15, e1007573. doi:10.1371/journal.ppat.1007573
Gautam, G., Rehman, S. A. A., Pandey, P., and Gourinath, S. (2017). Crystal Structure of the PEG-Bound SH3 Domain of Myosin IB fromEntamoeba Histolyticareveals its Mode of Ligand Recognition. Acta Cryst. Sect D Struct. Biol. 73, 672–682. doi:10.1107/s2059798317009639
Ghosh, S. K., and Samuelson, J. (1997). Involvement of p21racA, Phosphoinositide 3-kinase, and Vacuolar ATPase in Phagocytosis of Bacteria and Erythrocytes by Entamoeba Histolytica: Suggestive Evidence for Coincidental Evolution of Amebic Invasiveness. Infect. Immun. 65, 4243–4249. doi:10.1128/iai.65.10.4243-4249.1997
Godbold, G. D., and Mann, B. J. (1998). Involvement of the Actin Cytoskeleton and P21rho-Family GTPases in the Pathogenesis of the Human Protozoan Parasite Entamoeba Histolytica. Braz. J. Med. Biol. Res. 31, 1049–1058. doi:10.1590/s0100-879x1998000800004
Gonzales, M. L. M., Dans, L. F., and Sio-Aguilar, J. (2019). Antiamoebic Drugs for Treating Amoebic Colitis. Cochrane Database Syst. Rev. 1, CD006085. doi:10.1002/14651858.CD006085.pub3
Gupta, C. M., Ambaru, B., and Bajaj, R. (2020). Emerging Functions of Actins and Actin Binding Proteins in Trypanosomatids. Front. Cell Dev. Biol. 8, 587685. doi:10.3389/fcell.2020.587685
Ha, B. H., Morse, E. M., Turk, B. E., and Boggon, T. J. (2015). Signaling, Regulation, and Specificity of the Type II P21-Activated Kinases. J. Biol. Chem. 290, 12975–12983. doi:10.1074/jbc.r115.650416
Hall, A. (1998). Rho GTPases and the Actin Cytoskeleton. Science 279, 509–514. doi:10.1126/science.279.5350.509
Han, J. W., Leeper, L., Rivero, F., and Chung, C. Y. (2006). Role of RacC for the Regulation of WASP and Phosphatidylinositol 3-kinase during Chemotaxis of Dictyostelium. J. Biol. Chem. 281, 35224–35234. doi:10.1074/jbc.m605997200
Handley, M. E., Rasaiyaah, J., Chain, B. M., and Katz, D. R. (2007). Mixed Lineage Kinases (MLKs): a Role in Dendritic Cells, Inflammation and Immunity? Int. J. Exp. Pathol. 88, 111–126. doi:10.1111/j.1365-2613.2007.00531.x
Handman, E., Schnur, L. F., Spithill, T. W., and Mitchell, G. F. (1986). Passive Transfer of Leishmania Lipopolysaccharide Confers Parasite Survival in Macrophages. J. Immunol. 137, 3608–3613.
Hemsath, L., Dvorsky, R., Fiegen, D., Carlier, M.-F., and Ahmadian, M. R. (2005). An Electrostatic Steering Mechanism of Cdc42 Recognition by Wiskott-Aldrich Syndrome Proteins. Mol. Cell 20, 313–324. doi:10.1016/j.molcel.2005.08.036
Hirsch, D. S., Pirone, D. M., and Burbelo, P. D. (2001). A New Family of Cdc42 Effector Proteins, CEPs, Function in Fibroblast and Epithelial Cell Shape Changes. J. Biol. Chem. 276, 875–883. doi:10.1074/jbc.m007039200
Hoffman, G. R., and Cerione, R. A. (2000). Flipping the Switch. Cell 102, 403–406. doi:10.1016/s0092-8674(00)00045-3
Izumi, Y., Hirose, T., Tamai, Y., Hirai, S.-i., Nagashima, Y., Fujimoto, T., et al. (1998). An Atypical PKC Directly Associates and Colocalizes at the Epithelial Tight junction with ASIP, a Mammalian Homologue of Caenorhabditis elegans Polarity Protein PAR-3. J. Cell Biol 143, 95–106. doi:10.1083/jcb.143.1.95
Jiang, X., Niu, M., Chen, D., Chen, J., Cao, Y., Li, X., et al. (2016). Inhibition of Cdc42 Is Essential for Mig-6 Suppression of Cell Migration Induced by EGF. Oncotarget 7, 49180–49193. doi:10.18632/oncotarget.10205
Joberty, G., Perlungher, R. R., and Macara, I. G. (1999). The Borgs, a New Family of Cdc42 and TC10 GTPase-Interacting Proteins. Mol. Cell Biol 19, 6585–6597. doi:10.1128/mcb.19.10.6585
Joberty, G., Petersen, C., Gao, L., and Macara, I. G. (2000). The Cell-Polarity Protein Par6 Links Par3 and Atypical Protein Kinase C to Cdc42. Nat. Cell Biol 2, 531–539. doi:10.1038/35019573
Jordan, S. N., and Canman, J. C. (2012). Rho GTPases in Animal Cell Cytokinesis: an Occupation by the One Percent. Cytoskeleton 69, 919–930. doi:10.1002/cm.21071
Kalailingam, P., Tan, H. B., Pan, J. Y., Tan, S. H., and Thanabalu, T. (2019). Overexpression of CDC42SE1 in A431 Cells Reduced Cell Proliferation by Inhibiting the Akt Pathway. Cells 8, 117. doi:10.3390/cells8020117
Ke, Y., Wang, X., Jin, X. Y., Solaro, R. J., and Lei, M. (2014). PAK1 Is a Novel Cardiac Protective Signaling Molecule. Front. Med. 8, 399–403. doi:10.1007/s11684-014-0380-9
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. E. (2015). The Phyre2 Web portal for Protein Modeling, Prediction and Analysis. Nat. Protoc. 10, 845–858. doi:10.1038/nprot.2015.053
Kichina, J. V., Goc, A., Al-Husein, B., Somanath, P. R., and Kandel, E. S. (2010). PAK1 as a Therapeutic Target. Expert Opin. Ther. Targets 14, 703–725. doi:10.1517/14728222.2010.492779
Kim, A. S., Kakalis, L. T., Abdul-Manan, N., Liu, G. A., and Rosen, M. K. (2000). Autoinhibition and Activation Mechanisms of the Wiskott-Aldrich Syndrome Protein. Nature 404, 151–158. doi:10.1038/35004513
Kim, D.-J., Choi, C.-K., Lee, C.-S., Park, M.-H., Tian, X., Kim, N. D., et al. (2016). Small Molecules that Allosterically Inhibit P21-Activated Kinase Activity by Binding to the Regulatory P21-Binding Domain. Exp. Mol. Med. 48, e229. doi:10.1038/emm.2016.13
Krtková, J., Thomas, E. B., Alas, G. C., Schraner, E. M., Behjatnia, H. R., Hehl, A. B., et al. (2016). Rac Regulates Giardia Lamblia Encystation by Coordinating Cyst Wall Protein Trafficking and Secretion. mBio 7. doi:10.1128/mBio.01003-16
Kumar, A., Molli, P. R., Pakala, S. B., Nguyen, T. M. B., Rayala, S. K., and Kumar, R. (2009). PAK Thread from Amoeba to Mammals. J. Cell. Biochem. 107, 579–585. doi:10.1002/jcb.22159
Kumar, R., Gururaj, A. E., and Barnes, C. J. (2006). p21-activated Kinases in Cancer. Nat. Rev. Cancer 6, 459–471. doi:10.1038/nrc1892
Kumar, R., Sanawar, R., Li, X., and Li, F. (2017). Structure, Biochemistry, and Biology of PAK Kinases. Gene 605, 20–31. doi:10.1016/j.gene.2016.12.014
Kurisu, S., and Takenawa, T. (2010). WASP and WAVE Family Proteins: Friends or Foes in Cancer Invasion? Cancer Sci. 101, 2093–2104. doi:10.1111/j.1349-7006.2010.01654.x
Kuspa, A., and Loomis, W. F. (2006). The Genome of Dictyostelium discoideum. Methods Mol. Biol. 346, 15–30. doi:10.1385/1-59745-144-4:15
Labruyère, E., Zimmer, C., Galy, V., Olivo-Marin, J. C., and Guillén, N. (2003). EhPAK, a Member of the P21-Activated Kinase Family, Is Involved in the Control of Entamoeba Histolytica Migration and Phagocytosis. J. Cell Sci 116, 61–71. doi:10.1242/jcs.00190
Labruyere, E., Galy, V., Sansonetti, P., and Guillén, N. (2000). Distribution of a Potential P21-Activated Serine/threonine Kinase (PAK) in Entamoeba Histolytica. Arch. Med. Res. 31, S128–S130. doi:10.1016/s0188-4409(00)00143-0
Labruyère, E., Thibeaux, R., Olivo-Marin, J.-C., and Guillén, N. (2019). Crosstalk between Entamoeba Histolytica and the Human Intestinal Tract during Amoebiasis. Parasitology 146, 1140–1149. doi:10.1017/s0031182017002190
Lee, S.-F., Egelhoff, T. T., Mahasneh, A., and Côté, G. P. (1996). Cloning and Characterization of a Dictyostelium Myosin I Heavy Chain Kinase Activated by Cdc42 and Rac. J. Biol. Chem. 271, 27044–27048. doi:10.1074/jbc.271.43.27044
Lee, S.-F., Mahasneh, A., De La Roche, M., and Côté, G. P. (1998). Regulation of the P21-Activated Kinase-relatedDictyostelium Myosin I Heavy Chain Kinase by Autophosphorylation, Acidic Phospholipids, and Ca2+-Calmodulin. J. Biol. Chem. 273, 27911–27917. doi:10.1074/jbc.273.43.27911
Lee, S., Rivero, F., Park, K. C., Huang, E., Funamoto, S., and Firtel, R. A. (2004). DictyosteliumPAKc Is Required for Proper Chemotaxis. MBoC 15, 5456–5469. doi:10.1091/mbc.e04-04-0323
Lei, M., Lu, W., Meng, W., Parrini, M.-C., Eck, M. J., Mayer, B. J., et al. (2000). Structure of PAK1 in an Autoinhibited Conformation Reveals a Multistage Activation Switch. Cell 102, 387–397. doi:10.1016/s0092-8674(00)00043-x
Lerm, M., Holm, A., Seiron, A., Särndahl, E., Magnusson, K.-E., and Rasmusson, B. (2006). Leishmania Donovani Requires Functional Cdc42 and Rac1 to Prevent Phagosomal Maturation. Infect. Immun. 74, 2613–2618. doi:10.1128/iai.74.5.2613-2618.2006
Leung, T., Chen, X.-Q., Tan, I., Manser, E., and Lim, L. (1998). Myotonic Dystrophy Kinase-Related Cdc42-Binding Kinase Acts as a Cdc42 Effector in Promoting Cytoskeletal Reorganization. Mol. Cell Biol 18, 130–140. doi:10.1128/mcb.18.1.130
Li, S. I., and Purugganan, M. D. (2011). The Cooperative Amoeba: Dictyostelium as a Model for Social Evolution. Trends Genet. 27, 48–54. doi:10.1016/j.tig.2010.11.003
Li, X., Liu, F., and Li, F. (2010). PAK as a Therapeutic Target in Gastric Cancer. Expert Opin. Ther. Targets 14, 419–433. doi:10.1517/14728221003642019
Lian, J. P., Crossley, L., Zhan, Q., Huang, R., Robinson, D., and Badwey, J. A. (2002). The P21-Activated Protein Kinases (Paks) Receive and Integrate Messages from a Variety of Signaling Pathways. Adv. Exp. Med. Biol. 507, 497–502. doi:10.1007/978-1-4615-0193-0_76
Lin, D., Edwards, A. S., Fawcett, J. P., Mbamalu, G., Scott, J. D., and Pawson, T. (2000). A Mammalian PAR-3-PAR-6 Complex Implicated in Cdc42/Rac1 and aPKC Signalling and Cell Polarity. Nat. Cell Biol 2, 540–547. doi:10.1038/35019582
Llorens, F., Carulla, P., Villa, A., Torres, J. M., Fortes, P., Ferrer, I., et al. (2013). PrPC Regulates Epidermal Growth Factor Receptor Function and Cell Shape Dynamics in Neuro2a Cells. J. Neurochem. 127, 124–138. doi:10.1111/jnc.12283
Lodge, R., and Descoteaux, A. (2008). Leishmania Invasion and Phagosome Biogenesis. Subcell Biochem. 47, 174–181. doi:10.1007/978-0-387-78267-6_14
Lodge, R., and Descoteaux, A. (2005). Leishmania Donovani Promastigotes Induce Periphagosomal F-Actin Accumulation through Retention of the GTPase Cdc42. Cell Microbiol 7, 1647–1658. doi:10.1111/j.1462-5822.2005.00582.x
Loo, C.-S., Chen, C.-W., Wang, P.-J., Chen, P.-Y., Lin, S.-Y., Khoo, K.-H., et al. (2013). Quantitative Apical Membrane Proteomics Reveals Vasopressin-Induced Actin Dynamics in Collecting Duct Cells. Proc. Natl. Acad. Sci. 110, 17119–17124. doi:10.1073/pnas.1309219110
Lucanic, M., and Cheng, H.-J. (2008). A RAC/CDC-42-independent GIT/PIX/PAK Signaling Pathway Mediates Cell Migration in C. elegans. PLoS Genet. 4, e1000269. doi:10.1371/journal.pgen.1000269
Luo, L., Lee, T., Tsai, L., Tang, G., Jan, L. Y., and Jan, Y. N. (1997). Genghis Khan (Gek) as a Putative Effector for Drosophila Cdc42 and Regulator of Actin Polymerization. Proc. Natl. Acad. Sci. 94, 12963–12968. doi:10.1073/pnas.94.24.12963
Ma, Q.-L., Yang, F., Frautschy, S. A., and Cole, G. M. (2012). PAK in Alzheimer Disease, Huntington Disease and X-Linked Mental Retardation. Cell Logistics 2, 117–125. doi:10.4161/cl.21602
Macara, I. G. (2004). Par Proteins: Partners in Polarization. Curr. Biol. 14, R160–R162. doi:10.1016/j.cub.2004.01.048
Mahajan, K., and Mahajan, N. P. (2015). ACK1/TNK2 Tyrosine Kinase: Molecular Signaling and Evolving Role in Cancers. Oncogene 34, 4162–4167. doi:10.1038/onc.2014.350
Makkinje, A., Quinn, D. A., Chen, A., Cadilla, C. L., Force, T., Bonventre, J. V., et al. (2000). Gene 33/Mig-6, a Transcriptionally Inducible Adapter Protein that Binds GTP-Cdc42 and Activates SAPK/JNK. J. Biol. Chem. 275, 17838–17847. doi:10.1074/jbc.m909735199
Manser, E., Chong, C., Zhao, Z.-S., Leung, T., Michael, G., Hall, C., et al. (1995). Molecular Cloning of a New Member of the P21-Cdc42/Rac-Activated Kinase (PAK) Family. J. Biol. Chem. 270, 25070–25078. doi:10.1074/jbc.270.42.25070
Manser, E., Leung, T., Salihuddin, H., Zhao, Z.-s., and Lim, L. (1994). A Brain Serine/threonine Protein Kinase Activated by Cdc42 and Rac1. Nature 367, 40–46. doi:10.1038/367040a0
Manser, E., Loo, T.-H., Koh, C.-G., Zhao, Z.-S., Chen, X.-Q., Tan, L., et al. (1998). PAK Kinases Are Directly Coupled to the PIX Family of Nucleotide Exchange Factors. Mol. Cell 1, 183–192. doi:10.1016/s1097-2765(00)80019-2
Marchat, L. A., Hernández-de la Cruz, O. N., Ramírez-Moreno, E., Silva-Cázares, M. B., and López-Camarillo, C. (2020). Proteomics Approaches to Understand Cell Biology and Virulence of Entamoeba Histolytica Protozoan Parasite. J. Proteomics 226, 103897. doi:10.1016/j.jprot.2020.103897
Marks, P. W., and Kwiatkowski, D. J. (1996). Genomic Organization and Chromosomal Location of MurineCdc42. Genomics 38, 13–18. doi:10.1006/geno.1996.0586
Marquay Markiewicz, J., Syan, S., Hon, C.-C., Weber, C., Faust, D., and Guillen, N. (2011). A Proteomic and Cellular Analysis of Uropods in the Pathogen Entamoeba Histolytica. PLoS Negl. Trop. Dis. 5, e1002. doi:10.1371/journal.pntd.0001002
Mentzel, B., and Raabe, T. (2005). Phylogenetic and Structural Analysis of the Drosophila melanogaster P21-Activated Kinase DmPAK3. Gene 349, 25–33. doi:10.1016/j.gene.2004.12.030
Miki, H., Miura, K., and Takenawa, T. (1996). N-WASP, a Novel Actin-Depolymerizing Protein, Regulates the Cortical Cytoskeletal Rearrangement in a PIP2-dependent Manner Downstream of Tyrosine Kinases. EMBO J. 15, 5326–5335. doi:10.1002/j.1460-2075.1996.tb00917.x
Miki, H., Sasaki, T., Takai, Y., and Takenawa, T. (1998a). Induction of Filopodium Formation by a WASP-Related Actin-Depolymerizing Protein N-WASP. Nature 391, 93–96. doi:10.1038/34208
Miki, H., Suetsugu, S., and Takenawa, T. (1998b). WAVE, a Novel WASP-Family Protein Involved in Actin Reorganization Induced by Rac. EMBO J. 17, 6932–6941. doi:10.1093/emboj/17.23.6932
Morreale, A., Venkatesan, M., Mott, H. R., Owen, D., Nietlispach, D., Lowe, P. N., et al. (2000). Structure of Cdc42 Bound to the GTPase Binding Domain of PAK. Nat. Struct. Biol. 7, 384–388. doi:10.1038/75158
Mortara, R. A., Andreoli, W. K., Taniwaki, N. N., Fernandes, A. B., Silva, C. V. d., Fernandes, M. C. D. C., et al. (2005). Mammalian Cell Invasion and Intracellular Trafficking by Trypanosoma Cruzi Infective Forms. Acad. Bras. Ciênc. 77, 77–94. doi:10.1590/s0001-37652005000100006
Mott, H. R., Owen, D., Nietlispach, D., Lowe, P. N., Manser, E., Lim, L., et al. (1999). Structure of the Small G Protein Cdc42 Bound to the GTPase-Binding Domain of ACK. Nature 399, 384–388. doi:10.1038/20732
Müller-Taubenberger, A., Bretschneider, T., Faix, J., Konzok, A., Simmeth, E., and Weber, I. (2002). Differential Localization of the Dictyostelium Kinase DPAKa during Cytokinesis and Cell Migration. J. Muscle Res. Cell Motil 23, 751–763. doi:10.1023/a:1024475628061
Mullins, R. D., and Pollard, T. D. (1999). Rho-family GTPases Require the Arp2/3 Complex to Stimulate Actin Polymerizationin Acanthamoeba Extracts. Curr. Biol. 9, 405–415. doi:10.1016/s0960-9822(99)80187-0
Murali, A., and Rajalingam, K. (2014). Small Rho GTPases in the Control of Cell Shape and Mobility. Cell. Mol. Life Sci. 71, 1703–1721. doi:10.1007/s00018-013-1519-6
Myers, S. A., Han, J. W., Lee, Y., Firtel, R. A., and Chung, C. Y. (2005). ADictyosteliumHomologue of WASP Is Required for Polarized F-Actin Assembly during Chemotaxis. MBoC 16, 2191–2206. doi:10.1091/mbc.e04-09-0844
Narumiya, S., and Thumkeo, D. (2018). Rho Signaling Research: History, Current Status and Future Directions. FEBS Lett. 592, 1763–1776. doi:10.1002/1873-3468.13087
Neudauer, C. L., Joberty, G., Tatsis, N., and Macara, I. G. (1998). Distinct Cellular Effects and Interactions of the Rho-Family GTPase TC10. Curr. Biol. 8, 1151–1161. doi:10.1016/s0960-9822(07)00486-1
Ng, Y., Tan, I., Lim, L., and Leung, T. (2004). Expression of the Human Myotonic Dystrophy Kinase-Related Cdc42-Binding Kinase γ Is Regulated by Promoter DNA Methylation and Sp1 Binding. J. Biol. Chem. 279, 34156–34164. doi:10.1074/jbc.m405252200
Okada, M., Huston, C. D., Mann, B. J., Petri, W. A., Kita, K., and Nozaki, T. (2005). Proteomic Analysis of Phagocytosis in the Enteric Protozoan Parasite Entamoeba Histolytica. Eukaryot. Cell 4, 827–831. doi:10.1128/ec.4.4.827-831.2005
Olson, M. F. (2008). Applications for ROCK Kinase Inhibition. Curr. Opin. Cell Biol. 20, 242–248. doi:10.1016/j.ceb.2008.01.002
Owen, D., and Mott, H. R. (2018). CRIB Effector Disorder: Exquisite Function from Chaos. Biochem. Soc. Trans. 46, 1289–1302. doi:10.1042/bst20170570
Owen, D., Mott, H. R., Laue, E. D., and Lowe, P. N. (2000). Residues in Cdc42 that Specify Binding to Individual CRIB Effector Proteins. Biochemistry 39, 1243–1250. doi:10.1021/bi991567z
Page, M. J., Mckenzie, J. E., Bossuyt, P. M., Boutron, I., Hoffmann, T. C., Mulrow, C. D., et al. (2021). The PRISMA 2020 Statement: an Updated Guideline for Reporting Systematic Reviews. BMJ 372, n71. doi:10.1136/bmj.n71
Pang, X., and Zhou, H.-X. (2016). Mechanism and Rate Constants of the Cdc42 GTPase Binding with Intrinsically Disordered Effectors. Proteins 84, 674–685. doi:10.1002/prot.25018
Papayannopoulos, V., Co, C., Prehoda, K. E., Snapper, S., Taunton, J., and Lim, W. A. (2005). A Polybasic Motif Allows N-WASP to Act as a Sensor of PIP2 Density. Mol. Cell 17, 181–191. doi:10.1016/j.molcel.2004.11.054
Park, E., Kim, N., Ficarro, S. B., Zhang, Y., Lee, B. I., Cho, A., et al. (2015). Structure and Mechanism of Activity-Based Inhibition of the EGF Receptor by Mig6. Nat. Struct. Mol. Biol. 22, 703–711. doi:10.1038/nsmb.3074
Park, K. C., Rivero, F., Meili, R., Lee, S., Apone, F., and Firtel, R. A. (2004). Rac Regulation of Chemotaxis and Morphogenesis in Dictyostelium. EMBO J. 23, 4177–4189. doi:10.1038/sj.emboj.7600368
Park, S., Zhang, X., Li, C., Yin, C., Li, J., Fallon, J. T., et al. (2017). Single-cell RNA Sequencing Reveals an Altered Gene Expression Pattern as a Result of CRISPR/cas9-mediated Deletion of Gene 33/Mig6 and Chronic Exposure to Hexavalent Chromium in Human Lung Epithelial Cells. Toxicol. Appl. Pharmacol. 330, 30–39. doi:10.1016/j.taap.2017.07.003
Pearce, L. R., Komander, D., and Alessi, D. R. (2010). The Nuts and Bolts of AGC Protein Kinases. Nat. Rev. Mol. Cell Biol 11, 9–22. doi:10.1038/nrm2822
Phillips, J. E., and Gomer, R. H. (2014). The P21-Activated Kinase (PAK) Family Member PakD Is Required for Chemorepulsion and Proliferation Inhibition by Autocrine Signals in Dictyostelium discoideum. PLoS One 9, e96633. doi:10.1371/journal.pone.0096633
Phillips, M. J., Calero, G., Chan, B., Ramachandran, S., and Cerione, R. A. (2008). Effector Proteins Exert an Important Influence on the Signaling-Active State of the Small GTPase Cdc42. J. Biol. Chem. 283, 14153–14164. doi:10.1074/jbc.m706271200
Pichaud, F., Walther, R. F., and Nunes De Almeida, F. (2019). Regulation of Cdc42 and its Effectors in Epithelial Morphogenesis. J. Cell Sci 132. doi:10.1242/jcs.217869
Pirone, D., Carter, D. E., and Burbelo, P. D. (2001). Evolutionary Expansion of CRIB-Containing Cdc42 Effector Proteins. Trends Genet. 17, 370–373. doi:10.1016/s0168-9525(01)02311-3
Pirone, D. M., Fukuhara, S., Gutkind, J. S., and Burbelo, P. D. (2000). SPECs, Small Binding Proteins for Cdc42. J. Biol. Chem. 275, 22650–22656. doi:10.1074/jbc.m002832200
Prieto-Echagüe, V., and Miller, W. T. (2011). Regulation of Ack-Family Nonreceptor Tyrosine Kinases. J. Signal. Transduct 2011, 742372. doi:10.1155/2011/742372
Qiu, R.-G., Abo, A., and Martin, G. S. (2000). A Human Homolog of the C. elegans Polarity Determinant Par-6 Links Rac and Cdc42 to PKCζ Signaling and Cell Transformation. Curr. Biol. 10, 697–707. doi:10.1016/s0960-9822(00)00535-2
Rane, C. K., and Minden, A. (2014). P21 Activated Kinases: Structure, Regulation, and Functions. Small GTPases 5. doi:10.4161/sgtp.28003
Rane, C. K., and Minden, A. (2019). P21 Activated Kinase Signaling in Cancer. Semin. Cancer Biol. 54, 40–49. doi:10.1016/j.semcancer.2018.01.006
Rath, P. P., and Gourinath, S. (2020). The Actin Cytoskeleton Orchestra inEntamoeba Histolytica. Proteins 88, 1361–1375. doi:10.1002/prot.25955
Reijnders, M. R. F., Ansor, N. M., Kousi, M., Yue, W. W., Tan, P. L., Clarkson, K., et al. (2017). Deciphering Developmental DisordersRAC1 Missense Mutations in Developmental Disorders with Diverse Phenotypes. Am. J. Hum. Genet. 101, 466–477. doi:10.1016/j.ajhg.2017.08.007
Ridley, A. J. (2015). Rho GTPase Signalling in Cell Migration. Curr. Opin. Cell Biol. 36, 103–112. doi:10.1016/j.ceb.2015.08.005
Rivero, F., Dislich, H., Glockner, G., and Noegel, A. A. (2001). The Dictyostelium discoideum Family of Rho-Related Proteins. Nucleic Acids Res. 29, 1068–1079. doi:10.1093/nar/29.5.1068
Rivero, F., and Somesh, B. P. (2002). Signal Transduction Pathways Regulated by Rho GTPases in Dictyostelium. J. Muscle Res. Cell Motil 23, 737–749. doi:10.1023/a:1024423611223
Rivers, E., and Thrasher, A. J. (2017). Wiskott-Aldrich Syndrome Protein: Emerging Mechanisms in Immunity. Eur. J. Immunol. 47, 1857–1866. doi:10.1002/eji.201646715
Rodrigues, A. A., Clemente, T. M., Dos Santos, M. A., Machado, F. C., Gomes, R. G. B., Moreira, H. H. T., et al. (2012). A Recombinant Protein Based on Trypanosoma Cruzi P21 Enhances Phagocytosis. PLoS One 7, e51384. doi:10.1371/journal.pone.0051384
Rudolph, M. G., Bayer, P., Abo, A., Kuhlmann, J., Vetter, I. R., and Wittinghofer, A. (1998). The Cdc42/Rac Interactive Binding Region Motif of the Wiskott Aldrich Syndrome Protein (WASP) Is Necessary but Not Sufficient for Tight Binding to Cdc42 and Structure Formation. J. Biol. Chem. 273, 18067–18076. doi:10.1074/jbc.273.29.18067
Ryan, U., and Cacciò, S. M. (2013). Zoonotic Potential of Giardia. Int. J. Parasitol. 43, 943–956. doi:10.1016/j.ijpara.2013.06.001
Sackmann, E. (2015). How Actin/myosin Crosstalks Guide the Adhesion, Locomotion and Polarization of Cells. Biochim. Biophys. Acta (Bba) - Mol. Cell Res. 1853, 3132–3142. doi:10.1016/j.bbamcr.2015.06.012
Scianimanico, S., Desrosiers, M., Dermine, J.-F., Meresse, S., Descoteaux, A., and Desjardins, M. (1999). Impaired Recruitment of the Small GTPase Rab7 Correlates with the Inhibition of Phagosome Maturation by Leishmania Donovani Promastigotes. Cell Microbiol 1, 19–32. doi:10.1046/j.1462-5822.1999.00002.x
Seastone, D. J., Harris, E., Temesvari, L. A., Bear, J. E., Saxe, C. L., and Cardelli, J. (2001). The WASp-like Protein Scar Regulates Macropinocytosis, Phagocytosis and Endosomal Membrane Flow in Dictyostelium. J. Cell Sci 114, 2673–2683. doi:10.1242/jcs.114.14.2673
Senapedis, W., Crochiere, M., Baloglu, E., and Landesman, Y. (2016). Therapeutic Potential of Targeting PAK Signaling. Anticancer Agents Med. Chem. 16, 75–88. doi:10.2174/1871520615666150617111414
Shabardina, V., Kischka, T., Kmita, H., Suzuki, Y., and Makałowski, W. (2018). Environmental Adaptation of Acanthamoeba Castellanii and Entamoeba Histolytica at Genome Level as Seen by Comparative Genomic Analysis. Int. J. Biol. Sci. 14, 306–320. doi:10.7150/ijbs.23869
Shina, M. C., and Noegel, A. A. (2008). Invertebrate Coronins. Subcell Biochem. 48, 88–97. doi:10.1007/978-0-387-09595-0_8
Shinjo, K., Koland, J. G., Hart, M. J., Narasimhan, V., Johnson, D. I., Evans, T., et al. (1990). Molecular Cloning of the Gene for the Human Placental GTP-Binding Protein Gp (G25K): Identification of This GTP-Binding Protein as the Human Homolog of the Yeast Cell-Division-Cycle Protein CDC42. Proc. Natl. Acad. Sci. 87, 9853–9857. doi:10.1073/pnas.87.24.9853
Song, J., Xu, Q., Olsen, R., Loomis, W. F., Shaulsky, G., Kuspa, A., et al. (2005). Comparing the Dictyostelium and Entamoeba Genomes Reveals an Ancient Split in the Conosa Lineage. PLoS Comput. Biol. 1, e71. doi:10.1371/journal.pcbi.0010071
Stevens, W. K., Vranken, W., Goudreau, N., Xiang, H., Xu, P., and Ni, F. (1999). Conformation of a Cdc42/Rac Interactive Binding Peptide in Complex with Cdc42 and Analysis of the Binding Interface. Biochemistry 38, 5968–5975. doi:10.1021/bi990426u
Stothard, P. (2000). The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. Biotechniques 28, 11021104–1104. doi:10.2144/00286ir01
Swaminathan, K., Müller-Taubenberger, A., Faix, J., Rivero, F., and Noegel, A. A. (2014). A Cdc42- and Rac-Interactive Binding (CRIB) Domain Mediates Functions of Coronin. Proc. Natl. Acad. Sci. USA 111, E25–E33. doi:10.1073/pnas.1315368111
Swaminathan, K., Stumpf, M., Müller, R., Horn, A.-C., Schmidbauer, J., Eichinger, L., et al. (2015). Coronin7 Regulates WASP and SCAR through CRIB Mediated Interaction with Rac Proteins. Sci. Rep. 5, 14437. doi:10.1038/srep14437
Symons, M., Derry, J. M. J., Karlak, B., Jiang, S., Lemahieu, V., Mccormick, F., et al. (1996). Wiskott-Aldrich Syndrome Protein, a Novel Effector for the GTPase CDC42Hs, Is Implicated in Actin Polymerization. Cell 84, 723–734. doi:10.1016/s0092-8674(00)81050-8
Tan, D. S., Haaland, B., Gan, J. M., Tham, S. C., Sinha, I., Tan, E. H., et al. (2014). Bosutinib Inhibits Migration and Invasion via ACK1 in KRAS Mutant Non-small Cell Lung Cancer. Mol. Cancer 13, 13. doi:10.1186/1476-4598-13-13
Teixeira, T. L., Machado, F. C., Alves Da Silva, A., Teixeira, S. C., Borges, B. C., Dos Santos, M. A., et al. (2015). Trypanosoma Cruzi P21: a Potential Novel Target for Chagasic Cardiomyopathy Therapy. Sci. Rep. 5, 16877. doi:10.1038/srep16877
Teramoto, H., Coso, O. A., Miyata, H., Igishi, T., Miki, T., and Gutkind, J. S. (1996). Signaling from the Small GTP-Binding Proteins Rac1 and Cdc42 to the C-Jun N-Terminal Kinase/Stress-Activated Protein Kinase Pathway. J. Biol. Chem. 271, 27225–27228. doi:10.1074/jbc.271.44.27225
Thompson, B. J. (2021). Par-3 Family Proteins in Cell Polarity & Adhesion. FEBS J. doi:10.1111/febs.15754
Thompson, G., Owen, D., Chalk, P. A., and Lowe, P. N. (1998). Delineation of the Cdc42/Rac-Binding Domain of P21-Activated Kinase. Biochemistry 37, 7885–7891. doi:10.1021/bi980140+
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-specific gap Penalties and Weight Matrix Choice. Nucl. Acids Res. 22, 4673–4680. doi:10.1093/nar/22.22.4673
Thrasher, A. J., and Burns, S. O. (2010). WASP: a Key Immunological Multitasker. Nat. Rev. Immunol. 10, 182–192. doi:10.1038/nri2724
Tolstrup, J., Krause, E., Tannich, E., and Bruchhaus, I. (2007). Proteomic Analysis ofEntamoeba Histolytica. Parasitology 134, 289–298. doi:10.1017/s0031182006001442
Tyler, J. J., Allwood, E. G., and Ayscough, K. R. (2016). WASP Family Proteins, More Than Arp2/3 Activators. Biochem. Soc. Trans. 44, 1339–1345. doi:10.1042/bst20160176
Uetrecht, A. C., and Bear, J. E. (2006). Coronins: the Return of the crown. Trends Cell Biol. 16, 421–426. doi:10.1016/j.tcb.2006.06.002
Unbekandt, M., and Olson, M. F. (2014). The Actin-Myosin Regulatory MRCK Kinases: Regulation, Biological Functions and Associations with Human Cancer. J. Mol. Med. 92, 217–225. doi:10.1007/s00109-014-1133-6
Uniprot, C. (2021). UniProt: the Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489. doi:10.1093/nar/gkaa1100
Veltman, D. M., King, J. S., Machesky, L. M., and Insall, R. H. (2012). SCAR Knockouts in Dictyostelium: WASP Assumes SCAR's Position and Upstream Regulators in Pseudopods. J. Cell Biol 198, 501–508. doi:10.1083/jcb.201205058
Vetter, I. R., and Wittinghofer, A. (2001). The Guanine Nucleotide-Binding Switch in Three Dimensions. Science 294, 1299–1304. doi:10.1126/science.1062023
Vinet, A. F., Fiedler, T., Studer, V., Froquet, R., Dardel, A., Cosson, P., et al. (2014). Initiation of Multicellular Differentiation inDictyostelium Discoideumis Regulated by Coronin A. MBoC 25, 688–701. doi:10.1091/mbc.e13-04-0219
Vlahou, G., and Rivero, F. (2006). Rho GTPase Signaling in Dictyostelium discoideum: Insights from the Genome. Eur. J. Cell Biol. 85, 947–959. doi:10.1016/j.ejcb.2006.04.011
Wang, B., Song, K., Chen, L., Su, H., Gao, L., Liu, J., et al. (2020). Targeted Inhibition of ACK1 Can Inhibit the Proliferation of Hepatocellular Carcinoma Cells through the PTEN/AKT/mTOR Pathway. Cell Biochem Funct 38, 642–650. doi:10.1002/cbf.3522
Wennerberg, K., Ellerbroek, S. M., Liu, R.-Y., Karnoub, A. E., Burridge, K., and Der, C. J. (2002). RhoG Signals in Parallel with Rac1 and Cdc42. J. Biol. Chem. 277, 47810–47817. doi:10.1074/jbc.m203816200
Wennerberg, K., Rossman, K. L., and Der, C. J. (2005). The Ras Superfamily at a Glance. J. Cell Sci 118, 843–846. doi:10.1242/jcs.01660
Wilkins, A., and Insall, R. H. (2001). Small GTPases in Dictyostelium: Lessons from a Social Amoeba. Trends Genet. 17, 41–48. doi:10.1016/s0168-9525(00)02181-8
Wu, C., Lee, S.-F., Furmaniak-Kazmierczak, E., Côté, G. P., Thomas, D. Y., and Leberer, E. (1996). Activation of Myosin-I by Members of the Ste20p Protein Kinase Family. J. Biol. Chem. 271, 31787–31790. doi:10.1074/jbc.271.50.31787
Wu, G., Gu, Y., Li, S., and Yang, Z. (2001). A Genome-wide Analysis of Arabidopsis Rop-Interactive CRIB Motif-Containing Proteins that Act as Rop GTPase Targets. Plant Cell 13, 2841–2856. doi:10.1105/tpc.010218
Wu, W. J., Erickson, J. W., Lin, R., and Cerione, R. A. (2000). The γ-subunit of the Coatomer Complex Binds Cdc42 to Mediate Transformation. Nature 405, 800–804. doi:10.1038/35015585
Xie, B., Zen, Q., Wang, X., He, X., Xie, Y., Zhang, Z., et al. (2015). ACK1 Promotes Hepatocellular Carcinoma Progression via Downregulating WWOX and Activating AKT Signaling. Int. J. Oncol. 46, 2057–2066. doi:10.3892/ijo.2015.2910
Xu, S.-H., Huang, J.-Z., Xu, M.-L., Yu, G., Yin, X.-F., Chen, D., et al. (2015). ACK1 Promotes Gastric Cancer Epithelial-Mesenchymal Transition and Metastasis through AKT-POU2F1-ECD Signalling. J. Pathol. 236, 175–185. doi:10.1002/path.4515
Yamanaka, T., Horikoshi, Y., Suzuki, A., Sugiyama, Y., Kitamura, K., Maniwa, R., et al. (2001). PAR-6 Regulates aPKC Activity in a Novel Way and Mediates Cell-Cell Contact-Induced Formation of the Epithelial Junctional Complex. Genes Cells 6, 721–731. doi:10.1046/j.1365-2443.2001.00453.x
Yang, W., and Cerione, R. A. (1997). Cloning and Characterization of a Novel Cdc42-Associated Tyrosine Kinase, ACK-2, from Bovine Brain. J. Biol. Chem. 272, 24819–24824. doi:10.1074/jbc.272.40.24819
Yang, Y., Roche, M. d. l., Crawley, S. W., Li, Z., Furmaniak-Kazmierczak, E., and Côté, G. P. (2013). PakB Binds to the SH3 Domain of Dictyostelium Abp1 and Regulates its Effects on Cell Polarity and Early Development. MBoC 24, 2216–2227. doi:10.1091/mbc.e12-12-0883
Yao, D., Li, C., Rajoka, M. S. R., He, Z., Huang, J., Wang, J., et al. (2020). P21-Activated Kinase 1: Emerging Biological Functions and Potential Therapeutic Targets in Cancer. Theranostics 10, 9741–9766. doi:10.7150/thno.46913
Yokoyama, N., Lougheed, J., and Miller, W. T. (2005). Phosphorylation of WASP by the Cdc42-Associated Kinase ACK1. J. Biol. Chem. 280, 42219–42226. doi:10.1074/jbc.m506996200
Yokoyama, N., and Miller, W. T. (2003). Biochemical Properties of the Cdc42-Associated Tyrosine Kinase ACK1. J. Biol. Chem. 278, 47713–47723. doi:10.1074/jbc.m306716200
Zhang, H., and Gallo, K. A. (2001). Autoinhibition of Mixed Lineage Kinase 3 through its Src Homology 3 Domain. J. Biol. Chem. 276, 45598–45603. doi:10.1074/jbc.m107176200
Zhang, Y., and Kubiseski, T. J. (2010). Caenorhabditis elegans Wsp-1 Regulation of Synaptic Function at the Neuromuscular junction. J. Biol. Chem. 285, 23040–23046. doi:10.1074/jbc.m109.096164
Zhao, Y., Loyer, P., Li, H., Valentine, V., Kidd, V., and Kraft, A. S. (1997). Cloning and Chromosomal Location of a Novel Member of the Myotonic Dystrophy Family of Protein Kinases. J. Biol. Chem. 272, 10013–10020. doi:10.1074/jbc.272.15.10013
Zhao, Z.-s., and Manser, E. (2005). PAK and Other Rho-Associated Kinases - Effectors with Surprisingly Diverse Mechanisms of Regulation. Biochem. J. 386, 201–214. doi:10.1042/bj20041638
Zhao, Z., and Manser, E. (2015). Myotonic Dystrophy Kinase-Related Cdc42-Binding Kinases (MRCK), the ROCK-like Effectors of Cdc42 and Rac1. Small GTPases 6, 81–88. doi:10.1080/21541248.2014.1000699
Zhao, Z. S., and Manser, E. (2010). Do PAKs Make Good Drug Targets? F1000 Biol. Rep. 2, 70. doi:10.3410/B2-70
Keywords: amoebozoa, CRIB, PAK, CDC42/RAC, WASP, coronin
Citation: Umarao P, Rath PP and Gourinath S (2022) Cdc42/Rac Interactive Binding Containing Effector Proteins in Unicellular Protozoans With Reference to Human Host: Locks of the Rho Signaling. Front. Genet. 13:781885. doi: 10.3389/fgene.2022.781885
Received: 23 September 2021; Accepted: 14 January 2022;
Published: 02 February 2022.
Edited by:
Anupam Nath Jha, Tezpur University, IndiaReviewed by:
Tanima Bose, Ludwig Maximilian University of Munich, GermanyPadmani Sandhu, Chandigarh University, India
Copyright © 2022 Umarao, Rath and Gourinath. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Samudrala Gourinath, c2dvdXJpbmF0aEBtYWlsLmpudS5hYy5pbg==
†These authors have contributed equally to this work and share first authorship
 Preeti Umarao†
Preeti Umarao† Pragyan Parimita Rath
Pragyan Parimita Rath Samudrala Gourinath
Samudrala Gourinath