- 1Pharmacy College, Chengdu University of Traditional Chinese Medicine, Chengdu, China
- 2Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing, China
- 3Kenneth P. Dietrich School of Arts and Sciences, University of Pittsburgh, Pittsburgh, PA, United States
- 4Beijing Engineering Research Center of Pediatric Surgery, Engineering and Transformation Center, Beijing Children’s Hospital, National Center for Children’s Health, Capital Medical University, Beijing, China
- 5Department of City and Regional Planning, Nanjing University, Nanjing, China
Polygonum is a generalized genus of the Polygonaceae family that includes various herbaceous plants. In order to provide aid in understanding the evolutionary and phylogenetic relationship in Polygonum at the chloroplast (cp) genome-scale level, we sequenced and annotated the complete chloroplast genomes of four Polygonum species using next-generation sequencing technology and CpGAVAS. Then, repeat sequences, IR contractions, and expansion and transformation sites of chloroplast genomes of four Polygonum species were studied, and a phylogenetic tree was built using the chloroplast genomes of Polygonum. The results indicated that the chloroplast genome construction of Polygonum also displayed characteristic four types of results, comparable to the published chloroplast genome of recorded angiosperms. The chloroplast genomes of the four Polygonum plants are highly consistent in genome size (159,015 bp–163,461 bp), number of genes (112 genes, including 78 protein-coding genes, 30 tRNA genes, and 4 rRNA genes), gene types, gene order, codon usage, and repeat sequence distribution, which identifies the high preservation among the Polygonum chloroplast genomes. The Polygonum phylogenetic tree was recreated by a full sequence of the chloroplast genome, which illustrates that the P. bistorta, P. orientale, and P. perfoliatum are divided into the same branch, and P. aviculare belongs to Fallopia. The precise system site of lots base parts requires further verification, but the study would provide a basis for developing the available genetic resources and evolutionary relationships of Polygonum.
Introduction
Polygonum is an annual genus of Polygonaceae, which are broadly spread around the world, and most of them are distributed in the north temperate zone (Macêdo et al., 2021; Mohtashami et al., 2021; Song et al., 2020). Moreover, there are about 113 species in China. Some species of Polygonaceae have been used as traditional Chinese medicine due to their remarkable effects on the treatment of edema and sore poison. Polygonum aviculare, Polygonum bistorta, Polygonum cuspidatum, and Polygonum perfoliatum have displayed medical values as diuretics in clinical practice, and it has effects of dehumidification of the wind, heat detoxification, and live blood (Lin et al., 2015). Phytochemical research studies showed that flavonoids, quinones, phenylpropanoids, and terpenoids are contained among Polygonum. Also, the pure compounds from Polygonum might have extensive bioactivities, such as anticancer, antitumor, antioxidative, anti-inflammatory, analgesic, antimicrobial, and insecticidal activities. The subordinate division of Polygonaceae is not clear and has been controversial. Since Linnaeus established the genus Polygonum in 1753, Meissner conducted an in-depth research on the genus Polygonum in the world in 1826 and established 10 groups under the genus of Polygonum (Fan et al., 2013). With the further development of research work, most groups have been promoted to the genus level by some scholars, and the genus is divided into 15 genera (Costea and Tardif 2005). In the different genera of Polygonum, the medicinal chemical components and the curative effects on diseases are different. Therefore, accurate identification of the species is the key to ensuring the clinical efficacy and safety of medicinal plants of this genus.
Chloroplast is one of the plastids and a vital organelle for transforming the energy and performing photosynthesis among the plants, which can be generally found in land plants, algae, and some protists (Asaf, Khan, Aaqil Khan, et al., 2017). Chloroplasts are composed of membranes, with thylakoids and stromata inside. The membrane of the thylakoid contains a large number of pigment molecules in photosynthesis, which are used to capture and transfer energy during photosynthesis, while the stroma contains various enzymes, inorganic salts, and DNA. Chloroplasts can not only synthesize sugars into photosynthesis but also participate in the synthesis of complex organic substances such as amino acids and fatty acids in organisms (Yao et al., 2015). The chloroplast genomes are conversed into many plants, presenting a covalently closed circular DNA, and few are linear or other shapes (Asaf et al., 2017). For example, in Acetabularia, the chloroplasts are found to be a rare linear structure rather than a conventional closed circular double-stranded DNA, and in another alga, a polycyclic structure exists because several independent micro-circular linked to each other in dinoflagellate (Cheng et al., 2017; Cheon et al., 2019; Yu, Sun, et al., 2020). The size of chloroplast genomes is usually between 100 and 200 kb in plants of different families and genera of plants (Chen et al., 2019). The chloroplast DNA size of most angiosperms is generally between 110 and 160 kb, and the chloroplast DNA size of ferns is about 140–150 kb.
The double-stranded closed circle of the chloroplast genome is generally classified into four regions: large single-copy region (LSC), small single-copy region (SSC), inverted repeat region A (IRa), and inverted repeat region B (IRb). Two IR regions are separated by LSC and SSC, and they have the same length in opposite directions (Jansen et al., 2011). Studies (Jansen, Saski, Lee, Hansen and Daniell 2011; Cheon, Kim, Kwak, Lee and Yoo 2019) have shown that changes in the IR region are the main reason for chloroplast genome changes. In the earlier reported cp genome in Polygonaceae, Yu and Ye (Yu et al., 2020; Ye et al., 2021) studied the length of the cp genome of LSC, SSC, and IR region and classified the species of Polygonum chinense and P. cuspidatum. The result would be a valuable genetic resource for studying the genetics and evolutionary relationships between the Polygonaceae species. There are about 110–130 genes encoded by chloroplast DNA, which consist of rRNA-coding genes, protein-coding genes, and tRNA-coding genes (Cheng, Li, Zhang, Cai, Gao, Qiao and Mi 2017; Tan et al., 2020). In general, gene replication occurs in all rRNA genes, along with some protein-coding and tRNA genes. Based on the functions of chloroplast DNA-encoding genes, it can be divided into three classes: genes for the photosynthetic system, such as petB, genes for the genetic system related to transcription and translation, such as tRNA-UGC, genes for biosynthesis related to the synthesis of amino acids, and open reading frames (ORF), such as accD, matK, and ycf1 (Yang et al., 2013).
The chloroplast genome is a valuable multi-level taxonomic resource with rich genetic information and has been broadly used in the aspect of plant phylogeny and evolution, species identification, and taxonomy. One reason for the fast development of chloroplast genomes is the advent of high-throughput sequencing technologies. After the complete chloroplast genomes of Nicotiana tabacum (Shinozaki et al., 1986) and Marchantia polymorpha (Kohchi et al., 1988) were established in 1986, researchers (Daniell et al., 2016) have paid more attention to chloroplast genomes of plants. After that, 3,721 cp DNA in different plants has been described, including green algae and aquatic life which can be found in National Center for Biotechnology Information (NCBI) database (Tonti-Filippini et al., 2017). First-generation sequencing technologies (Pareek et al., 2011; Slatko et al., 2011; Liao et al., 2021), including the traditional “dideoxy” sequencing technique, chemical degradation method, and the improved fluorescence automatic sequencing technology developed based on them (Dobrogojski et al., 2020). The next-generation sequencing technology that does not require DNA amplification and cloning and the third-generation sequencing technology that uses single-molecule real-time (SMRT) sequencing is now being widely used in chloroplast genome sequencing, which could facilitate de novo genome assembly (Tonti-Filippini, Nevill, Dixon and Small 2017).
Most of the Polygonum species have high financial and therapeutic values. Here, four Polygonum chloroplast genomes (P. aviculare, P. bistorta, P. orientale, P. perfoliatum) were identified and assembled, compared with other published Polygonum chloroplast genomes (Yu, Liu, Liu, Lan and Qu 2020; Ye, Lin, Zhou, He, Yan and Cheng 2021), we got considerable biological evidence, with the cp genome structure, repeat sequences, and other characteristics. This work also provided an essential foundation of the Polygonum cp genome library, encouraging the progress of phylogenetics, DNA barcoding, and population (Lee et al., 2019).
Materials and Methods
DNA Extracting, Sequencing, and Genome Annotation
DNA was extracted from four species of Polygonum, such as P. aviculare, P. bistorta, P. orientale and P. perfoliatum. Fresh Polygonum leaves were collected from Chengdu (Sichuan Province, China). The specimens of Polygonum have been kept in CDCM (Traditional Chinese medicine herbarium of Chengdu University of Traditional Chinese Medicine) (Supplementary Table S1). The cetyltrimethylammonium bromide method has been used to obtain the whole genome DNA from fresh leaves. The ND-2000 spectrometer was used to quantify the DNA. A shotgun library (250 bp) was built following the constructer guidelines. The X Ten Platform (Illumina, San Diego, CA, United States) was used to sequence through the double terminal sequencing technique with pair-end 150. The total raw data from the mensuration DNA was about 3.5G, and about 12 million paired-ends scrutinizes were finished (Supplementary Table S2).
The raw reads were trimmed using Skewer v0.22 (skewer -q 20 -Q 30–l 100 -t 32) (Jiang et al., 2014). BLAST was used to predict the chloroplast-like reads by cleaning the reads with the sequences of the reference P. cuspidatum (MW411186.1) (Deng et al., 2015; Ye, Lin, Zhou, He, Yan and Cheng 2021). Generally, SOAPdenovo-2. 04 (SOAPdenovo-127mer all -s config. txt -o out -K 51 -R) was used to accumulate the sequences using chloroplast reads (Gogniashvili et al., 2015). Then, those accumulated assembled sequences were extended with the help of SSPACE-3. 0 (SSPACE_standard_v3.0.pl -l library.txt -s out.config -x 1 -T 4 -b sspace.out) and GapCloser-1.12 (GapCloser -a scaffols.fa -b library.txt -o Fanal.fa)were used to fill the gaps (Boetzer et al., 2011; Acemel et al., 2016). To authenticate the accuracy of the connection splicing, a random primer was designed to check the connections of the sequence by polymerase chain reaction. The PCR primer information and amplification conditions are shown in the Supplementary Table S3. The Sanger sequencing results were compared with the assembled chloroplast genome sequence to verify the accuracy of genome linkage.
CpGAVAS (Liu et al., 2012) was used for sequence annotation. The annotation results were checked by DOGMA (http://dogma.ccbb.utexas.edu/) and BLAST (Wyman et al., 2004). In addition, the tRNA genes were classified using tRNAscanSEv1. 21 (Chan and Lowe 2019). The OGDRAWv1. 2 (Lohse et al., 2007) and MEGA5. 2 (Tamura et al., 2011) were used to plot the structural features of the chloroplast genome and define the relative utilization of synonymous codons. MEGA5. 2 (Tamura, Peterson, Peterson, Stecher, Nei and Kumar 2011) was adopted to analyze the relative synonymous codon usage (RSCU). The assembled chloroplast genome sequences of the four Polygonum species were deposited in NCBI under the Genbank accession number MZ748474–MZ748477.
Repeats and Comparative Analysis of Chloroplast Genomes
Tandem Repeats Finder (Benson 1999) and REPuter (Kurtz et al., 2001) have been used to find the tandem, forward, and palindromic repeats from the four Polygonum chloroplast genomes. The Misa.pl (Beier et al., 2017) was used to recognize the SSRs and the finding parameters of mononucleotides transfer to eight repeatable elements, dinucleotides, and trinucleotides four repeatable elements, tetranucleotides, pentanucleotides, and hexanucleotides transfer to three repeatable elements. Primer3 (Untergasser et al., 2012) was used to design the SSR primers.
Genome structures among six Polygonum chloroplast genomes, Including four polygonum species in this study and two polygonum species published by the NCBI (P. cuspidatum and P. chinense), were completed by mVISTA software (Shuffle-LAGAN mode) (Frazer et al., 2004) using the genome of P. cuspidatum as the reference. Pi values and sequence polymorphisms of six Polygonum species were analyzed using DNAsp v. 6.12.03 (Rozas et al., 2017). The step size was set to 200 bp, and the window length was set to 800 bp.
Phylogenetic Analysis
A total of 19 chloroplasts sequences (Supplementary Table S4) were used to build the phylogenetic trees. Each of the 67 protein-coding genes shared by all the genomes was compared individually and then linked end to end to form a supergene from each species. The sequences alignment was carried out using the MAFFT v7.309. The best model was determined using the modeltest-ng-0.1.6 software with default parameters; ML analysis was performed using RAxMLNG v0.9.084 based on Linux edition using default parameters. The parameters were GTR + FU + IU + G4m, noname = 1–51,039. Chrysanthemum x morifolium has been situated likewise those out-groups.
Results
Features of Polygonum Chloroplast DNA
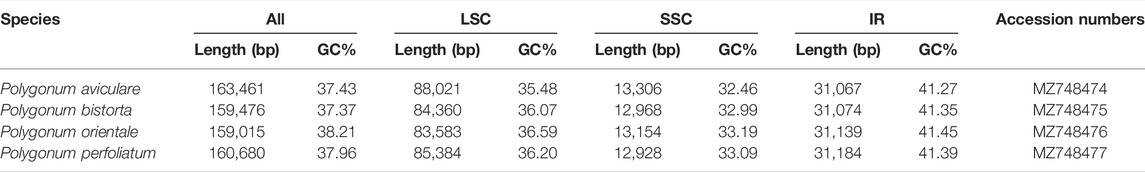
The genome sizes of the four Polygonum chloroplasts are 163,461 bp (P. aviculare), 159,476 bp (P. bistorta), 159,015 bp (P. orientale), and 160,680 bp (P. perfoliatum), respectively. The whole GC content are 37.43% (P. aviculare), 37.37% (P. bistorta), 38.21% (P. orientale), and 37.96% (P. perfoliatum), individually. The LSC region, SSC region, and a couple of inverted repeat regions (IRA/IRB) are alike in Polygonum chloroplast genomes than other plants (Guo et al., 2018). The length of the LSC region is 83,583–88,021 bp and the GC content is 35.48–36.59% in Polygonum chloroplast genomes. The distribution of length in the SSC regions is 12,928–13,306 bp and the GC content is about 32.46–33.19%. The GC content in those IR regions is 41.27–41.45% and the length is 31,067–31,184 bp (Table 1). The GC content is a significant marker to identify the genetic relationship of species; moreover, the Polygonum has comparable cpDNA GC content. The phenomenon is also common in other plants (Guo, Guo, Zhao, Xu, Li, Zhang, Shen, Wu and Hou 2018; Liang et al., 2019) that the GC content in those IR regions is more than that of other regions (LSC, SSC). The high GC content of the IR areas usually points to the rRNA and tRNA genes (He et al., 2016; Shen et al., 2017). Also, the whole chloroplast genome sequences of four Polygonum species can be checked in the National Center for Biotechnology Information (NCBI) database afterward annotation and the GenBank accession number can be found in Table1.
The annotation results (GB files) of four Polygonum chloroplast genomes which were measured in this study were submitted to the OGDraw software, and the physical map of the Polygonum chloroplast genomes were drawn. The results can be found in Figure 1. In total 112 genes were found in the chloroplast genomes, such as four rRNA genes, 30 tRNA genes, and 78 protein-coding genes (Figure 1; Table 2). The main genes of the four Polygonum chloroplasts can be coarsely divided into three classes, named chloroplast self-replication-related genes, photosynthesis-related genes, and other genes (Saski et al., 2005).
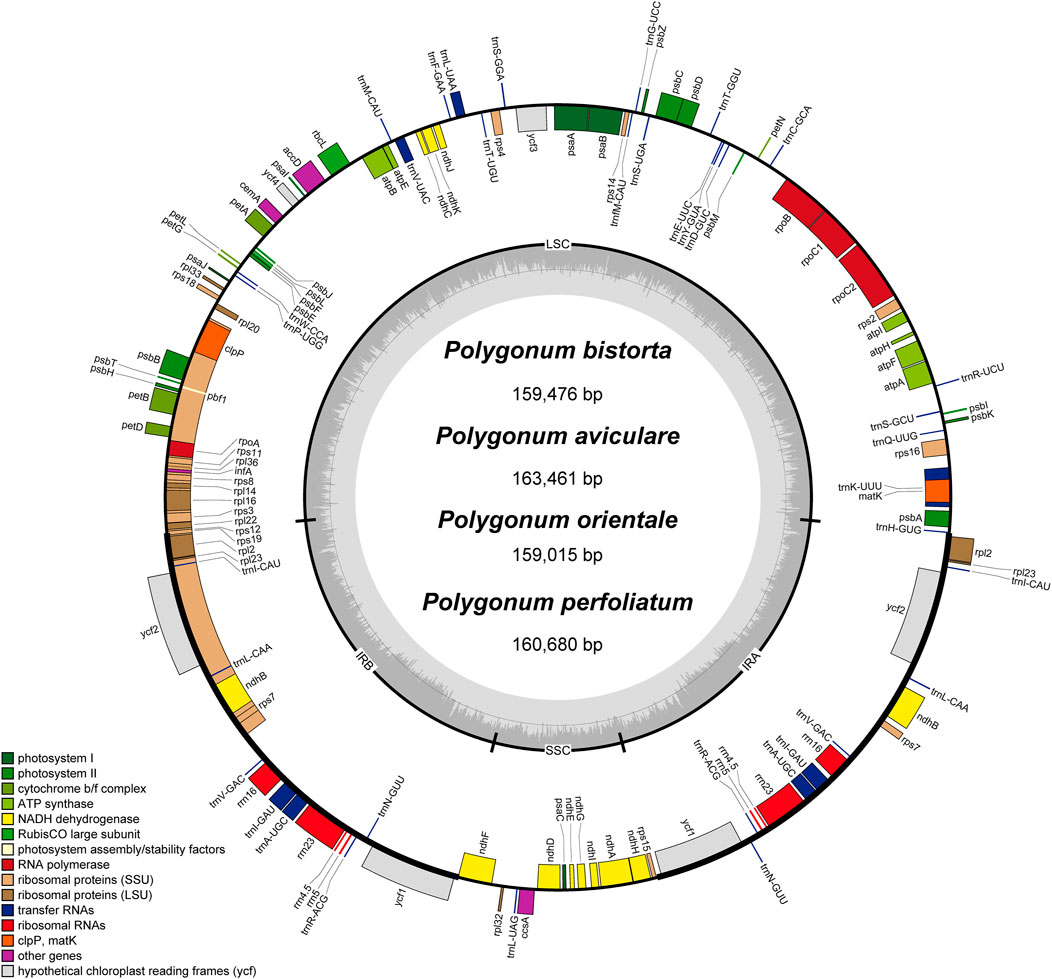
FIGURE 1. Gene map of the Polygonum chloroplast genome. Genes drawn inside the circle are transcribed clockwise, and those outside are transcribed counterclockwise. Genes belonging to different functional groups are color coded. The darker gray in the inner circle corresponds to DNA G + C content, while the lighter gray corresponds to A + T content.
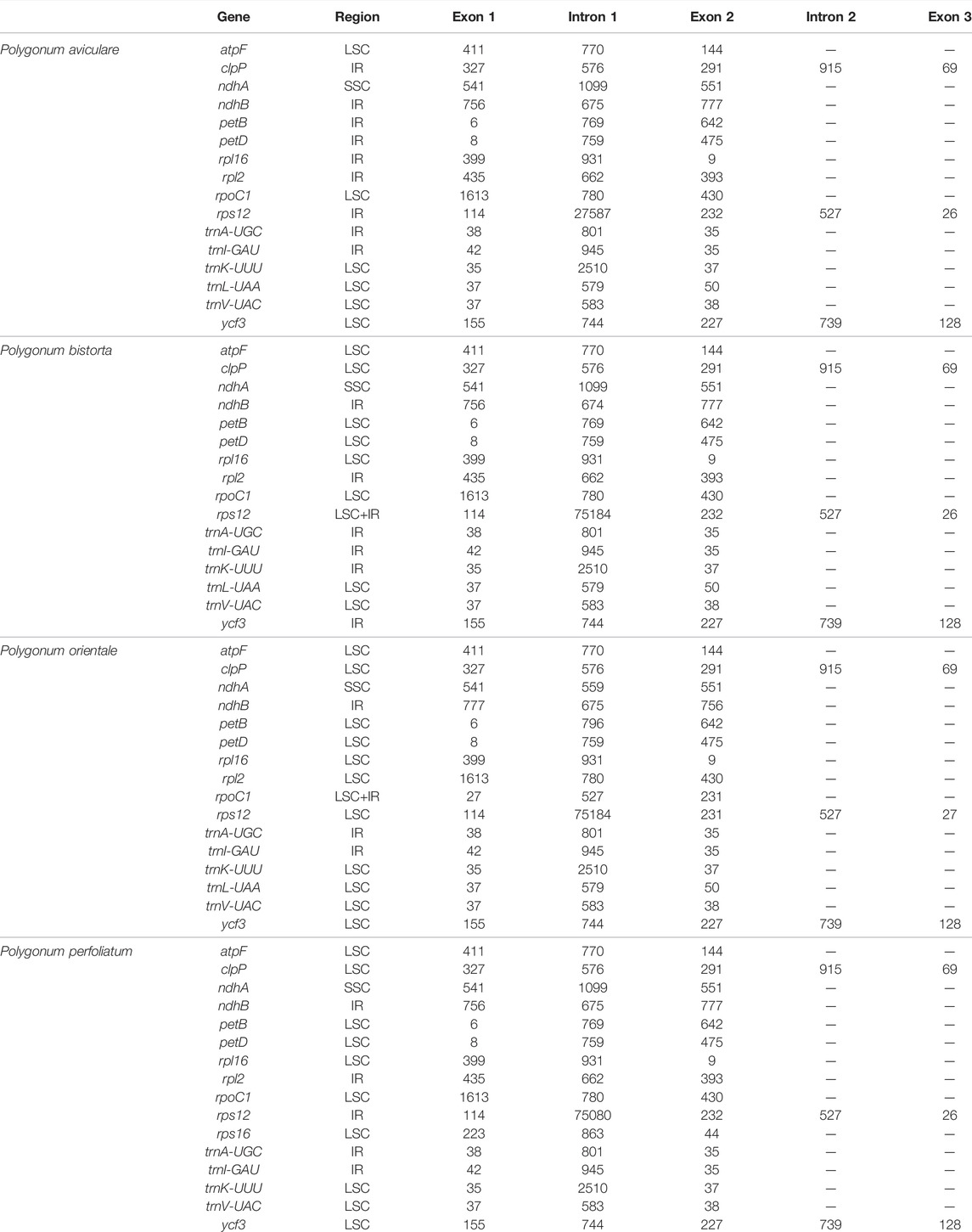
There are 16 genes with introns among the 112 genes of four Polygonum chloroplast genomes (Table 3), with 5 tRNA genes and 11 functional genes. The tRNA genes include trnI-GAU, trnL-UAA, trnV-UAC, trnK-UUU, and trnA-UGG. The 11 functional genes include ndhB, ndhA, atpF, petB, petD, rpoC1, ycf3, clpP, rps12, rpl16, and rpl2. The 5’ termination of the rps12 gene is in the LSC region of the chloroplast genome, and the 3’ end is in the IR region of the chloroplast genome. Like the other angiospermous chloroplasts trans-splicing phenomenon also occurs in the rps12 gene of the Polygonum chloroplast genome. Three of the 17 intron-containing genes cover two introns (rps12, ycf3, and clpP), and the other genes only contain one intron, of which trnK-UUU covers the main intron (2,510 bp), and this intron covers the entire gene of matK.
Relative Synonymous Codon Usage Analysis
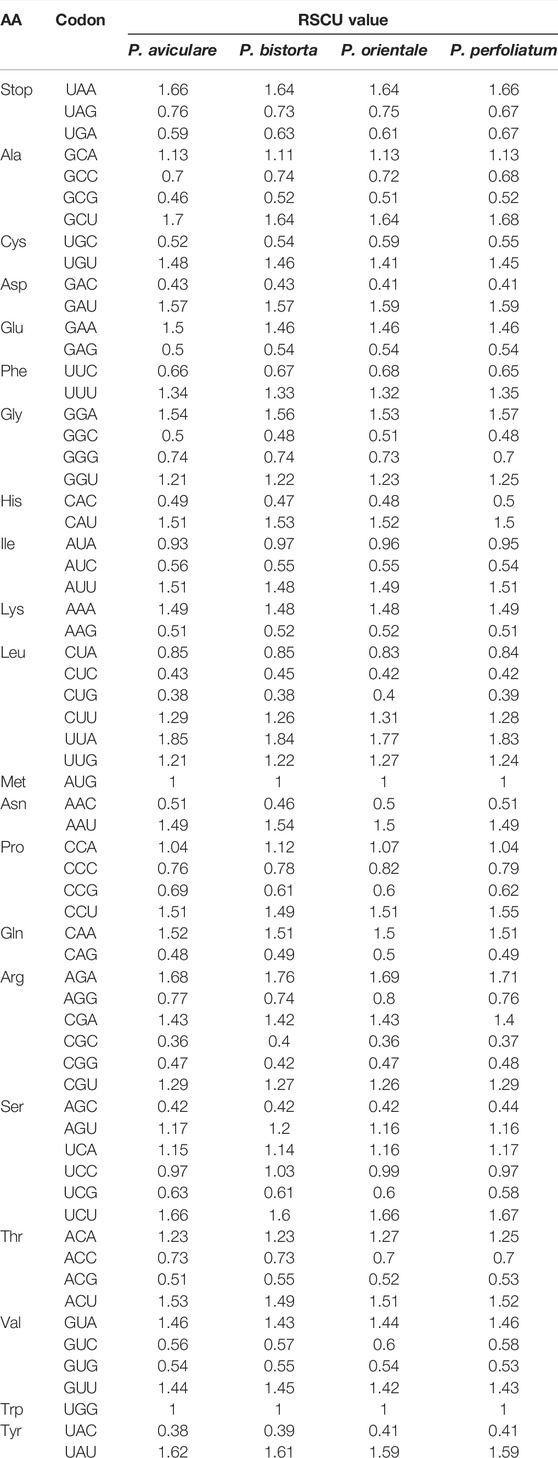
Relative synonymous codon usage (RSCU) is a synonymous codon correlative effect, which values the 64 vital synonymous codons (Wu et al., 2007). RSCU is calculated as the ratio of the actual observed value to the average usage of the synonymous codons. The value of RSCU can be divided into three types: greater than 1, less than 1, and equal to 1. If the value of RSCU is greater than 1, it indicates that the codon is used more frequently than other codons. If the value of RSCU is less than 1, it means that other synonymous codons of this codon are used more frequently than this codon. If the value of RSCU is equal to 1, it indicates that there is no bias in the use of a codon. According to the statistical study of four Polygonum chloroplast genomes, the extent of CDS is from 80,286 to 83,403 bp, and for which accounts for about 50% of the total chloroplast genome length. The number of codons is in between 26,762–27,801. As for the statistical study of RSCU, there was some certain bias in the use of other amino acids, except for Trp and Met (Tables 4).
Long-Repeat and Simple Sequence Repeat Analysis
In this research, we also studied the repeated sequence of four Polygonum chloroplast genomes, with tandem repeats (T), forward repeats (F), reverse repeats (R), and palindromic repeats (P). The results of the repeated study of four Polygonum chloroplast genomes are in Figure 2. There are 99 repeated sequences in P. aviculare, including 49 tandem repeats, 22 forward repeats, 26 palindromic repeats, and 2 reverse repeats; 86 repeated sequences in P. bistorta, including 36 tandem repeats, 23 forward repeats, 22 palindromic repeats, and 5 reverse repeats; 72 repeated sequences in P. orientale, including 22 tandem repeats, 22 forward repeats, 22 palindromic repeats, and 6 reverse repeats; 76 repeated sequences in P. perfoliatum, including 27 tandem repeats, 20 forward repeats, 22 palindromic repeats, and 7 reverse repeats. Among all the categories of repeated sequences, the sequences of length 20–50 bp are the most (Figure 2).
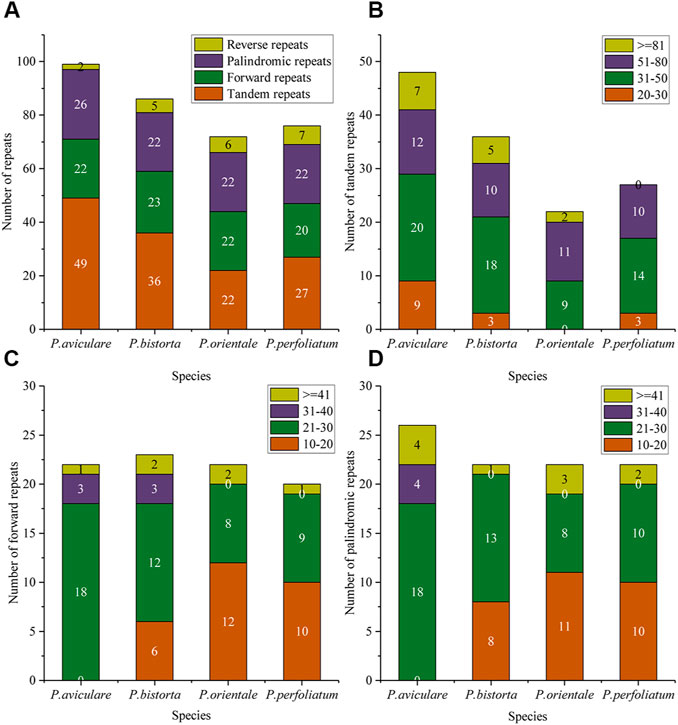
FIGURE 2. Repeat sequences analysis of the four Polygonum cp genomes. (A) Repeat types in the four cp genomes; (B) tandem repeats in the four cp genomes; (C) forward repeats in the four cp genomes; (D) palindromic repeats in the four cp genomes. In (A), different colors show different repeat types; in (B–D), different colors show different lengths. The ordinate represents the number of repeats.
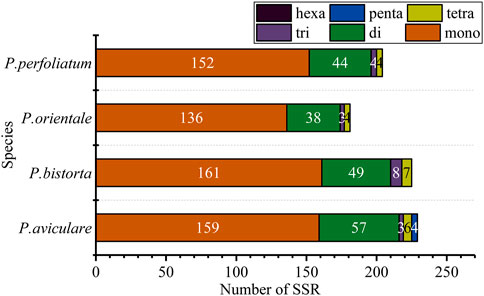
Simple sequence repeat (SSR), also known as microsatellite sequence, is a repeat sequence composed of one to six bases as repeat units in series, which is of great significance to the study of plant populations. SSRs with a length of over 10 bp are inclined on slipped-strand mispairing, which is approved to be the principal mutational mechanism of SSR polymorphisms (Asaf et al., 2017). Additionally, SSRs which are changeable at the intraspecific position in the chloroplast genome are often used as the genetic marker in the investigation of population genetics and evolution (Yang et al., 2016; Zl et al., 2016). There are 228 SSRs in P. aviculare, including, three trinucleotides, 57 dinucleotides, 159 mononucleotides, 6 tetranucleotides, and 3 pentanucleotides; 225 repeated sequences in P. bistorta, including 7 tetranucleotides, 8 trinucleotides, 161 mononucleotides and 49 dinucleotides; 181 repeated sequences in P. orientale, including 136 mononucleotides, 38 dinucleotides, 3 trinucleotides and 4 tetranucleotides; 204 repeated sequences in P. perfoliatum, including 152 mononucleotides, 44 dinucleotides, 4 trinucleotides, and 4 tetranucleotides (Figure 3). Based on these SSR results, we designed 10 pairs of primers as molecular markers. P. orientale, P. cuspidatum, and P. aviculare were used for molecular marker amplification. The results showed that four pairs of primers could distinguish well between species (Supplementary Figure S1). Although the four of them can be distinguished at the genus level, the PCR amplification products may be chloroplast DNA or nuclear DNA, which requires further study.
Comparative Chloroplast Genomic Analysis and Sequence Variation
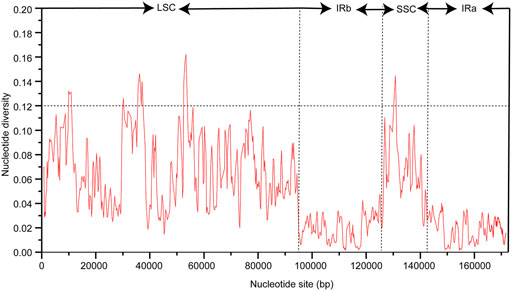
The chloroplast genome sequence of P. cuspidatum is used as a reference sequence to draw an analogy among the genomic sequences of six Polygonum chloroplast genomes (P. cupidatum, P. chinense, P. aviculare, P. bistorta, P. orientale, and P. orientale). Figure 4 displayed regions in common. The figure displayed that, aiming at the Polygonum chloroplast genomes; the rate of changes for the LSC and SSC regions is visibly greater than that of the IR region. To further clarify the variation in the coding regions, the Pi (nucleotide diversity) was also calculated (Figure 5). Six divergent loci (psbI-trnS-GCU, rpoB-trnC-GCA, trnE-UUC-trnT-GGU, trnT-GGU-psbD, trnT-UGU-trnL-UAA, and rpl32-trnL-UAG) had a Pi value greater than 0.12. All of these six divergent loci were intergenic regions and were present in the LSC region, except for rpl32-trnL-UAG, which occurred in the SSC region, with none being detected in the IR region. These highly variable regions may also resolve the interspecific relationships of Polygonum in the Polygonaceae phylogeny. With the forward progress of the chloroplast genome because of the wide usage of the chloroplast DNA fragments, some conundrums in plant species authentication, phylogenetics, and other related researche studies can be solved. Obviously, the chloroplast DNA fragments have higher resolution loci than general DNA fragments which have low distinguishability and mutation rates in some proximal related groups. Meanwhile, these high-efficiency DNA fragments from the chloroplast genome can promote the development of species identification and population diversity by comparing the differences between the chloroplast genome sequences of different groups.

FIGURE 4. Comparative analysis of chloroplast genome differences in six Polygonum cp genomes. Gray arrows and thick black lines above the alignment indicate gene orientation. Purple bars represent exons, blue bars represent untranslated regions (UTRs), pink bars represent non-coding sequences (CNS), and gray bars represent mRNA. The y-axis represents the percentage identity (shown: 50–100%).
IR Contraction and Expansion in Polygonum Chloroplast Genome
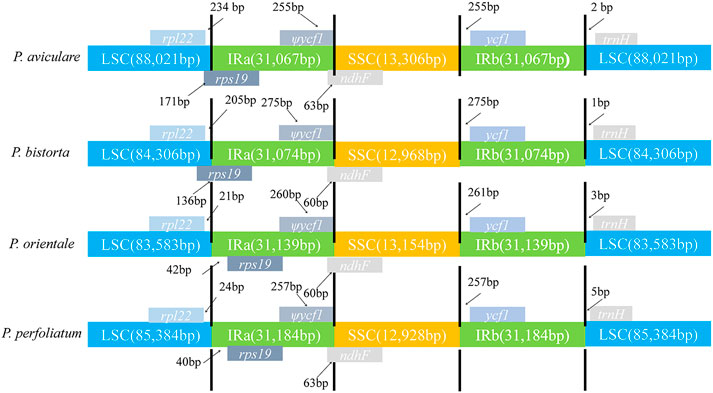
In this research, the analysis of IR-LSC and IR-SSC border structure and location of four Polygonum species was finished (Figure 6). The results found out that the SSC/IRa assembly was located in the ndhF region in the four species of Polygonum chloroplast genome, and spread a length of 60–63 bp into the IRa region in the four species. Also, the rps19 gene was located in the IRa region and the length is about (P. orientale, 21 bp; P. perfoliatum, 40 bp) into the LSC region. The ycf1 gene of four Polygonum chloroplast genomes completely exists in the IRb region, with a terminal 225 bp (P. aviculare), 275 bp (P. bistorta), 261 bp (P. orientale), and 257 bp (P. perfoliatum) from the SSC/IRa border. For now, the trnH gene existed in the LSC region, and it had a length of 2, 1, 3, and 5 bp from the LSC/IRb border in the chloroplast genome of P. aviculare, P. bistorta, P. orientale, and P. perfoliatum, individually.
Until now, the expansion mechanism of the IR region has been debated; moreover, the double-strand break repair (DCBR) theory is written off as the prime mechanism for the expansion of the IR region (Machour and Ayoub 2020). There is little probability of the IR region, which has a large shrink. Furthermore, it is believed that the DCBR model is not only the core mechanism of IR region expansion but also the mechanism of IR region contraction.
Phylogenetic Analysis
Phylogenetic analysis was accomplished on an alliance of concatenated nucleotide sequences of all genes from 19 angiosperm species. We used ML to construct a phylogenetic tree ground based on these gene data, while C. morifolium was agreed as the outgroup. The P. bistorta, P. orientale, P. perfoliatum, and P. chinense are divided into a branch, namely, Polygonum. And in the branch of Polygonum, it is further divided into two small sub-branches. The upper sub-branch includes P. orientale, P. perfoliatum, and P. chinense. Moreover, P. orientale and P. perfoliatum clustered on the small branch. The P. aviculare is closer to F. sachalinensis, F. aubertii, and Polygonum cuspidatum, which is far away from the branch of Polygonum. Therefore, we put P. aviculare and P. cuspidatum into the branch of Fallopia (Figure 7).
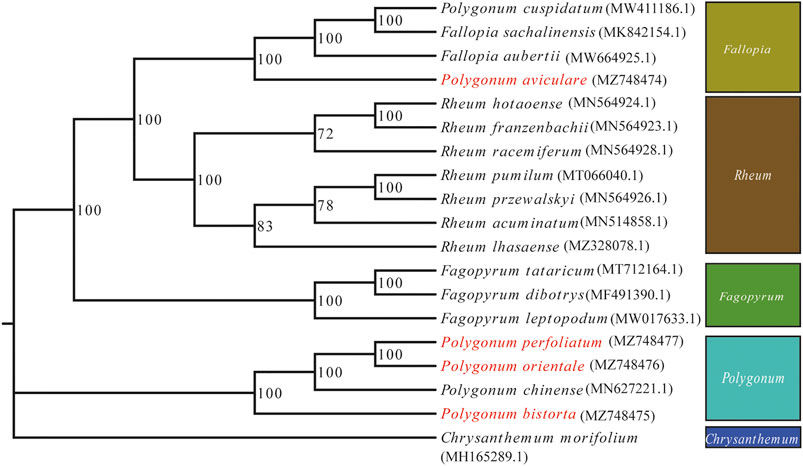
FIGURE 7. ML phylogenetic tree reconstruction containing the cp genomes of 19 plants. Chrysanthemum x morifolium was set as the out-group.
Discussion
Overall, we examined the four species of Polygonum chloroplast genomes, and then the results inferred that the four Polygonum species were similar in the angiospermous features both in structure and content. Also this shows that the characteristics of the chloroplast genome in other medicinal angiosperms (He et al., 2017) would be reliable with the characteristic quadripartite structure of the Polygonum chloroplast genome. The phenomenon is general among other angiospermous chloroplast genomes (Raubeson et al., 2007; Yang et al., 2010; Asaf, Khan, Khan, Waqas, Kang, Yun and Lee 2017) that the AT content was higher than the GC content in the chloroplast genome among all four Polygonum species, then entirely these presented that there were no significant variances in chloroplast genomes among those four Polygonum species. The consequences also confirmed that the GC content was much higher, probably because of the presence of the huge quantity of rRNA in the IR regions. However, the precise details are still poorly unknown. The consequences turned up in coding and extremely different regions among Polygonum chloroplast genomes were also exposed among other floristic chloroplast genomes (Ni et al., 2016; Chen et al., 2017).
The length of introns and exons was important among various plant chloroplast genomes. Here, the results indicated that only one gene (rps12) included three exons, meanwhile two genes (ycf3 and clpP) had two introns among the four Polygonum chloroplast genomes. The rps12 genes’ were located at the 5’ end on the LSC region and meanwhile the duplicated 3’ ends was set in the IRs regions due to the phenomenon it has been called trans-spliced gene (Guo, Guo, Zhao, Xu, Li, Zhang, Shen, Wu and Hou 2018). Furthermore, ycf3 is known as a photosynthesis-related gene as reported before (Naver et al., 2001). Therefore, the attendance of the ycf3 gene might result in an extra study of Polygonum chloroplast. The ycf1 gene also expected a basic part in the chloroplast genome, there were some related studies focused on gene capacity around ycf1, and these reports exposed ycf1 as a paramount pseudogene for those varieties of chloroplast genome and similarly encodes for Tic214 in plants (de Cambiaire et al., 2006; Nakai 2015). It has been testified in an earlier study that the introns play a significant part in regulating the expression of genes (Xu et al., 2017), which might control the gene expression level in different spatiotemporal (Le Hir et al., 2003; Niu and Yang 2011). Additionally, we reached a status that the attendance of intron or the loss of genes can be discovered in the chloroplast genomes (Graveley 2001; Ueda et al., 2007), and the regulating function of intron similarly need to be exposed through the research among a large number floristic chloroplast genomes (Emami et al., 2013). However, currently, there was no related research on the intron regulation mechanisms among those Polygonums. So, we will obtain more appropriate information through further studies to find out the functions of introns in the chloroplast genomes. And, these data around chloroplast genome will entitle significant theoretical basis for plant identification, especially medicinal plants.
It is important to identify the resources of plant germplasm and molecular markers by the finding the long repeat sequences and the SSRs of the chloroplast genomes. The results of the study demonstrated that the genes which have long repeats may be produced as a genetic marker for identifying the related species, but the exact use of it still needs to be proved by further studies. SSRs play a crucial status role in the chloroplast genomes. Because of its extreme variability, it was always used in genetic research (Ebert and Peakall 2009; Dong et al., 2013). The previous studies presented that SSR is widely distributed in the genome, and because of its special parental inheritance features, SSR is usually analyzed for genetic population structure and maternal analysis. Former researchers have studied that A. formosae has the most plentiful repeats on mononucleotides, and the phenomenon among the four Polygonum chloroplast genomes was in common. Consequently, the development found in SSRs of chloroplast genomes will significantly inspire the learning of identification among plenty species, genetic variety, and evolutionary development in Polygonum (Provan 2000; Flannery et al., 2006).
The method of DNA barcoding, which was put forward by Hebert (Hebert et al., 2003; Hu et al., 2019), can be utilized to identify the species through DNA sequences, ITS2, matK, psbA-trnH, and rbcL. However, the identification of related species—and predominantly the morphologically confusing species in the same genus—still exist in various problems. For that reason, discovering a proper DNA marker for such species is indispensable. The cp genomes have habitually been utilized for phylogenetic studies and species identification as a result of they have slower evolution than nuclear genomes (Song et al., 2017). In the current study, an analysis of five Polygonum cp genomic alignments has shown an enlarged figure of mutable sites in the intergenic spacer of the atpI-rps2, atpB-rbcL, psbD-rps14, ycf4-cemA, etc. Thus, these regions may be utilized as different nominee fragments to identify Polygonum. Moreover, ycf1a or ycf1b is the most mutable plastid genome region and can be used as a principal barcode for land plants (Dong et al., 2015). On the other hand, more Polygonum cp sequence data is necessary to be tested and it should be handled in future studies.
Earlier research studies had shown that IRs regions were the most conserved regions in the chloroplast genomes (Daniell et al., 2016). Its shrinkage and expansion at the borders are a common evolutionary occasion, and characterize the governing cause for the size variation and rearrangement of the chloroplast genome. There were a lot of reports that showed that the chloroplast gene had been conserved in most land plants but there were also reports that many sequences were rearranged in the chloroplast genomes of most plant species, then the IR shrinkage and extensions with inversions, the inversions in the LSC region, and the re-inversion in the SSC region were involved, and some reports showed that the wide rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes (Raubeson and Jansen 2005; Haberle et al., 2008). The sequence rearrangements caused by the alteration of chloroplast genome structure in related species may be connected with the plant genetic multiplicity information, so it can be utilized for molecular identification and evolutionary study (Chumley et al., 2006).
At present, it has been known that the chloroplast genome can serve as a super barcode to identify the plant species (Hernández-León et al., 2013). By phylogenetic analysis of the chloroplast genome of four Polygonum species, we suggested that the chloroplast genome of Polygonum might be a key marker for species identification. Furthermore, research is necessary to study and identify this assumption. The results are of great value to the genetic diversity and phylogenetic research of Polygonum at some point. Nevertheless, our research did not completely figure out the relationship between genera. Furthermore, our phylogenetic study is grounded on the chloroplast genome. If we want to completely figure out the phylogeny of the species in Polygonum, we may need to study the nuclear genes of plants, and more genera should be involved in the future. Nonetheless, our phylogeny research provided treasured resources for the classification, phylogeny, and evolutionary history of Polygonum.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, MZ748474 https://www.ncbi.nlm.nih.gov/genbank/, MZ748475 https://www.ncbi.nlm.nih.gov/genbank/, MZ748476 https://www.ncbi.nlm.nih.gov/genbank/, MZ748477 https://www.ncbi.nlm.nih.gov/, SRR15604831 https://www.ncbi.nlm.nih.gov/, SRR15604830 https://www.ncbi.nlm.nih.gov/, SRR15604829 https://www.ncbi.nlm.nih.gov/, and SRR15604828.
Author Contributions
Design and supervision: JX and SnC. Experiment: SG, XL, SuC, BL, RC, SX, HH, JC, JP, and YC. Data analysis and visualization: SG. Writing: SG, XL, SuC, and YG.
Funding
This work was supported by the National Major Science and Technology Projects (No. 2019ZX09201005), the National Key R&D Program of China (No. 2019YFC1711100), and the Fundamental Research Funds for the Central public welfare research institutes (No. ZZ13-YQ-047).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank all the peer reviewers and editors for their opinions and suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.764534/full#supplementary-material
References
Acemel, R. D., Tena, J. J., Irastorza-Azcarate, I., Marlétaz, F., Gómez-Marín, C., de la Calle-Mustienes, E., et al. (2016). A Single Three-Dimensional Chromatin Compartment in Amphioxus Indicates a Stepwise Evolution of Vertebrate Hox Bimodal Regulation. Nat. Genet. 48, 336–341. doi:10.1038/ng.3497
Asaf, S., Khan, A. L., Khan, M. A., Waqas, M., Kang, S. M., Yun, B. W., et al. (2017). Chloroplast Genomes of Arabidopsis Halleri Ssp. Gemmifera and Arabidopsis Lyrata Ssp. Petraea: Structures and Comparative Analysis. Sci. Rep. 7, 7556–7615. doi:10.1038/s41598-017-07891-5
Asaf, S., Khan, A. L., Aaqil Khan, M., Muhammad Imran, Q., Kang, S.-M., Al-Hosni, K., et al. (2017). Comparative Analysis of Complete Plastid Genomes from Wild Soybean (Glycine Soja) and Nine Other Glycine Species. PloS one 12, 1–27. doi:10.1371/journal.pone.0182281
Beier, S., Thiel, T., Münch, T., Scholz, U., and Mascher, M. (2017). MISA-web: a Web Server for Microsatellite Prediction. Bioinformatics 33, 2583–2585. doi:10.1093/bioinformatics/btx198
Benson, G. (1999). Tandem Repeats Finder: a Program to Analyze DNA Sequences. Nucleic Acids Res. 27, 573–580. doi:10.1093/nar/27.2.573
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D., and Pirovano, W. (2011). Scaffolding Pre-assembled Contigs Using SSPACE. Bioinformatics 27, 578–579. doi:10.1093/bioinformatics/btq683
Chan, P. P., and Lowe, T. M. (2019). tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol., 1–14. doi:10.1007/978-1-4939-9173-0_1
Chen, C., Zheng, Y., Liu, S., Zhong, Y., Wu, Y., Li, J., et al. (2017). The Complete Chloroplast Genome of Cinnamomum Camphoraand its Comparison with relatedLauraceaespecies. PeerJ 5, e3820. doi:10.7717/peerj.3820
Chen, Q., Wu, X., and Zhang, D. (2019). Phylogenetic Analysis of Fritillaria Cirrhosa D. Don and its Closely Related Species Based on Complete Chloroplast Genomes. PeerJ 7, e7480. doi:10.7717/peerj.7480
Cheng, H., Li, J., Zhang, H., Cai, B., Gao, Z., Qiao, Y., et al. (2017). The Complete Chloroplast Genome Sequence of Strawberry (Fragaria × ananassaDuch.) and Comparison with Related Species of Rosaceae. PeerJ 5, e3919. doi:10.7717/peerj.3919
Cheon, K.-S., Kim, K.-A., Kwak, M., Lee, B., and Yoo, K.-O. (2019). The Complete Chloroplast Genome Sequences of Four Viola Species (Violaceae) and Comparative Analyses with its Congeneric Species. PloS one 14, e0214162. doi:10.1371/journal.pone.0214162
Chumley, T. W., Palmer, J. D., Mower, J. P., Fourcade, H. M., Calie, P. J., Boore, J. L., et al. (2006). The Complete Chloroplast Genome Sequence of Pelargonium × Hortorum: Organization and Evolution of the Largest and Most Highly Rearranged Chloroplast Genome of Land Plants. Mol. Biol. Evol. 23, 2175–2190. doi:10.1093/molbev/msl089
Costea, M., and Tardif, F. J. (2005). Taxonomy of the Polygonum Douglasii (Polygonaceae) Complex with a New Species from Oregon. Brittonia 57, 1–27. doi:10.1663/0007-196x(2005)057[0001:totpdp]2.0.co;2
Daniell, H., Lin, C. S., Yu, M., and Chang, W. J. (2016). Chloroplast Genomes: Diversity, Evolution, and Applications in Genetic Engineering. Genome Biol. 17 (1), 1–29. doi:10.1186/s13059-016-1004-2
de Cambiaire, J. C., Otis, C., Lemieux, C., and Turmel, M. (2006). The Complete Chloroplast Genome Sequence of the Chlorophycean green Alga Scenedesmus Obliquus Reveals a Compact Gene Organization and a Biased Distribution of Genes on the Two DNA Strands. BMC Evol. Biol. 6, 37–15. doi:10.1186/1471-2148-6-37
Deng, P., Wang, L., Cui, L., Feng, K., Liu, F., Du, X., et al. (2015). Global Identification of microRNAs and Their Targets in Barley under Salinity Stress. PLoS One 10, e0137990. doi:10.1371/journal.pone.0137990
Dobrogojski, J., Adamiec, M., and Luciński, R. (2020). The Chloroplast Genome: A Review. Acta Physiologiae Plantarum 42, 1–13. doi:10.1007/s11738-020-03089-x
Dong, W., Xu, C., Li, C., Sun, J., Zuo, Y., Shi, S., et al. (2015). ycf1, the Most Promising Plastid DNA Barcode of Land Plants. Sci. Rep. 5 (1), 1–5. doi:10.1038/srep08348
Dong, W., Xu, C., Cheng, T., Lin, K., and Zhou, S. (2013). Sequencing Angiosperm Plastid Genomes Made Easy: a Complete Set of Universal Primers and a Case Study on the Phylogeny of Saxifragales. Genome Biol. Evol. 5, 989–997. doi:10.1093/gbe/evt063
Ebert, D., and Peakall, R. (2009). Chloroplast Simple Sequence Repeats (cpSSRs): Technical Resources and Recommendations for Expanding cpSSR Discovery and Applications to a Wide Array of Plant Species. Mol. Ecol. Resour. 9, 673–690. doi:10.1111/j.1755-0998.2008.02319.x
Emami, S., Arumainayagam, D., Korf, I., and Rose, A. B. (2013). The Effects of a Stimulating Intron on the Expression of Heterologous Genes inArabidopsis Thaliana. Plant Biotechnol. J. 11, 555–563. doi:10.1111/pbi.12043
Fan, D.-M., Chen, J.-H., Meng, Y., Wen, J., Huang, J.-L., and Yang, Y.-P. (2013). Molecular Phylogeny of Koenigia L. (Polygonaceae: Persicarieae): Implications for Classification, Character Evolution and Biogeography. Mol. Phylogenet. Evol. 69, 1093–1100. doi:10.1016/j.ympev.2013.08.018
Flannery, M. L., Mitchell, F. J. G., Coyne, S., Kavanagh, T. A., Burke, J. I., Salamin, N., et al. (2006). Plastid Genome Characterisation in Brassica and Brassicaceae Using a New Set of Nine SSRs. Theor. Appl. Genet. 113, 1221–1231. doi:10.1007/s00122-006-0377-0
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004). VISTA: Computational Tools for Comparative Genomics. Nucleic Acids Res. 32, W273–W279. doi:10.1093/nar/gkh458
Gogniashvili, M., Naskidashvili, P., Bedoshvili, D., Kotorashvili, A., Kotaria, N., and Beridze, T. (2015). Complete Chloroplast DNA Sequences of Zanduri Wheat (Triticum spp.). Genet. Resour. Crop Evol. 62, 1269–1277. doi:10.1007/s10722-015-0230-x
Graveley, B. R. (2001). Alternative Splicing: Increasing Diversity in the Proteomic World. Trends. Genet. 17, 100–107. doi:10.1016/s0168-9525(00)02176-4
Guo, S., Guo, L., Zhao, W., Xu, J., Li, Y., Zhang, X., et al. (2018). Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Paeonia Ostii. Molecules 23, 246. doi:10.3390/molecules23020246
Haberle, R. C., Fourcade, H. M., Boore, J. L., and Jansen, R. K. (2008). Extensive Rearrangements in the Chloroplast Genome of Trachelium Caeruleum Are Associated with Repeats and tRNA Genes. J. Mol. Evol. 66, 350–361. doi:10.1007/s00239-008-9086-4
He, L., Qian, J., Li, X., Sun, Z., Xu, X., and Chen, S. (2017). Complete Chloroplast Genome of Medicinal Plant Lonicera japonica: Genome Rearrangement, Intron Gain and Loss, and Implications for Phylogenetic Studies. Molecules 22, 249. doi:10.3390/molecules22020249
He, Y., Xiao, H., Deng, C., Xiong, L., Yang, J., and Peng, C. (2016). The Complete Chloroplast Genome Sequences of the Medicinal Plant Pogostemon Cablin. Ijms 17, 820. doi:10.3390/ijms17060820
Hebert, P. D. N., Cywinska, A., Ball, S. L., and DeWaard, J. R. (2003). Biological Identifications through DNA Barcodes. Proc. R. Soc. Lond. B 270, 313–321. doi:10.1098/rspb.2002.2218
Hernández-León, S., Gernandt, D. S., Pérez de la Rosa, J. A., and Jardón-Barbolla, L. (2013). Phylogenetic Relationships and Species Delimitation in Pinus Section Trifoliae Inferrred from Plastid DNA. PloS one 8, e70501. doi:10.1371/journal.pone.0070501
Hu, H., Shen, X., Liao, B., Luo, L., Xu, J., and Chen, S. (1902/01). Herbgenomics: A Stepping Stone for Research into Herbal Medicine. Sci. China Life Sci. 62, 913–920. doi:10.1007/s11427-018-9472-y
Jansen, R. K., Saski, C., Lee, S.-B., Hansen, A. K., and Daniell, H. (2011). Complete Plastid Genome Sequences of Three Rosids (Castanea, Prunus, Theobroma): Evidence for at Least Two Independent Transfers of Rpl22 to the Nucleus. Mol. Biol. Evol. 28, 835–847. doi:10.1093/molbev/msq261
Jiang, H., Lei, R., Ding, S. W., and Zhu, S. (2014). Skewer: a Fast and Accurate Adapter Trimmer for Next-Generation Sequencing Paired-End Reads. BMC bioinformatics 15 (1), 1–12. doi:10.1186/1471-2105-15-182
Kohchi, T., Umesono, K., Ogura, Y., Komine, Y., Nakahigashi, K., Komano, T., et al. (1988). A Nicked Group II Intron Andtrans-Splicing in liverwort,Marchantia Polymorpha, Chloroplasts. Nucl. Acids Res. 16, 10025–10036. doi:10.1093/nar/16.21.10025
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., and Giegerich, R. (2001). REPuter: the Manifold Applications of Repeat Analysis on a Genomic Scale. Nucleic Acids Res. 29, 4633–4642. doi:10.1093/nar/29.22.4633
Le Hir, H., Nott, A., and Moore, M. J. (2003). How Introns Influence and Enhance Eukaryotic Gene Expression. Trends Biochemical Sciences 28, 215–220. doi:10.1016/s0968-0004(03)00052-5
Lee, H. O., Joh, H. J., Kim, K., Lee, S. C., Kim, N. H., Park, J. Y., et al. (2019). Dynamic Chloroplast Genome Rearrangement and DNA Barcoding for Three Apiaceae Species Known as the Medicinal Herb "Bang-Poong". Int. J. Mol. Sci. 20. doi:10.3390/ijms20092196
Liang, C., Wang, L., Lei, J., Duan, B., Ma, W., Xiao, S., et al. (2019). A Comparative Analysis of the Chloroplast Genomes of Four Salvia Medicinal Plants. Engineering 08/01, 5. doi:10.1016/j.eng.2019.01.017
Liao, B., Hu, H., Xiao, S., Zhou, G., Sun, W., Chu, Y., et al. (2021). Global Pharmacopoeia Genome Database is an Integrated and Mineable Genomic Database for Traditional Medicines Derived from Eight International Pharmacopoeias. Sci. China Life Sci. [Online ahead of print], 1–9. doi:10.1007/s11427-021-1968-7
Lin, L., Ni, B., Lin, H., Zhang, M., Li, X., Yin, X., et al. (2015). Traditional Usages, Botany, Phytochemistry, Pharmacology and Toxicology of Polygonum Multiflorum Thunb. a Review. J. ethnopharmacology 159, 158–183. doi:10.1016/j.jep.2014.11.009
Liu, C., Shi, L., Zhu, Y., Chen, H., Zhang, J., Lin, X., et al. (2012). CpGAVAS, an Integrated Web Server for the Annotation, Visualization, Analysis, and GenBank Submission of Completely Sequenced Chloroplast Genome Sequences. BMC genomics 13, 715–717. doi:10.1186/1471-2164-13-715
Lohse, M., Drechsel, O., and Bock, R. (2007). OrganellarGenomeDRAW (OGDRAW): a Tool for the Easy Generation of High-Quality Custom Graphical Maps of Plastid and Mitochondrial Genomes. Curr. Genet. 52, 267–274. doi:10.1007/s00294-007-0161-y
Macêdo, S. K. S., Lima, K. S. B., Silva, N. D. d. S., Campos, S. S. G., Araújo, B. R., Almeida, J. R. G. d. S., et al. (2021). Genus Triplaris (Polygonaceae): A Review on Traditional Medicinal Use, Phytochemistry and Biological Activities. J. ethnopharmacology 277, 114188. doi:10.1016/j.jep.2021.114188
Machour, F. E., and Ayoub, N. (2020). Transcriptional Regulation at DSBs: Mechanisms and Consequences. Trends Genet. 36, 981–997. doi:10.1016/j.tig.2020.01.001
Mohtashami, L., Amiri, M. S., Ayati, Z., Ramezani, M., Jamialahmadi, T., Emami, S. A., et al. (2021). Ethnobotanical Uses, Phytochemistry and Pharmacology of Different Rheum Species (Polygonaceae): A Review. Adv. Exp. Med. Biol. 1308, 309–352. doi:10.1007/978-3-030-64872-5_22
Nakai, M. (2015). The TIC Complex Uncovered: the Alternative View on the Molecular Mechanism of Protein Translocation across the Inner Envelope Membrane of Chloroplasts. Biochim. Biophys. Acta (Bba) - Bioenerg. 1847, 957–967. doi:10.1016/j.bbabio.2015.02.011
Naver, H., Boudreau, E., and Rochaix, J.-D. (2001). Functional Studies of Ycf3. Plant Cell 13, 2731–2745. doi:10.1105/tpc.010253
Ni, L., Zhao, Z., Xu, H., Chen, S., and Dorje, G. (2016). The Complete Chloroplast Genome of Gentiana Straminea (Gentianaceae), an Endemic Species to the Sino-Himalayan Subregion. Gene 577, 281–288. doi:10.1016/j.gene.2015.12.005
Niu, D. K., and Yang, Y. F. (2011). Why Eukaryotic Cells Use Introns to Enhance Gene Expression: Splicing Reduces Transcription-Associated Mutagenesis by Inhibiting Topoisomerase I Cutting Activity. Biol. Direct 6, 24–10. doi:10.1186/1745-6150-6-24
Pareek, C. S., Smoczynski, R., and Tretyn, A. (2011). Sequencing Technologies and Genome Sequencing. J. Appl. Genet. 52, 413–435. doi:10.1007/s13353-011-0057-x
Provan, J. (2000). Novel Chloroplast Microsatellites Reveal Cytoplasmic Variation in Arabidopsis thaliana. Mol. Ecol. 9, 2183–2185. doi:10.1046/j.1365-294x.2000.105316.x
Raubeson, L. A., Peery, R., Chumley, T. W., Dziubek, C., Fourcade, H. M., Boore, J. L., et al. (2007). Comparative Chloroplast Genomics: Analyses Including New Sequences from the Angiosperms Nuphar Advena and Ranunculus Macranthus. BMC genomics 8 (1), 1–27. doi:10.1186/1471-2164-8-174
Raubeson, L. A., and Jansen, R. K. (2005). “Chloroplast Genomes of Plants,” in Plant Diversity and Evolution: Genotypic and Phenotypic Variation in Higher Plants, 45–68. doi:10.1079/9780851999043.0045
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 34, 3299–3302. doi:10.1093/molbev/msx248
Saski, C., Lee, S.-B., Daniell, H., Wood, T. C., Tomkins, J., Kim, H.-G., et al. (2005). Complete Chloroplast Genome Sequence of Glycine max and Comparative Analyses with Other Legume Genomes. Plant Mol. Biol. 59, 309–322. doi:10.1007/s11103-005-8882-0
Shen, X., Wu, M., Liao, B., Liu, Z., Bai, R., Xiao, S., et al. (2017). Complete Chloroplast Genome Sequence and Phylogenetic Analysis of the Medicinal Plant Artemisia Annua. Molecules 22, 1330. doi:10.3390/molecules22081330
Shinozaki, K., Ohme, M., Tanaka, M., Wakasugi, T., Hayashida, N., Matsubayashi, T., et al. (1986). The Complete Nucleotide Sequence of the Tobacco Chloroplast Genome: its Gene Organization and Expression. EMBO J. 5, 2043–2049. doi:10.1002/j.1460-2075.1986.tb04464.x
Slatko, B., Kieleczawa, J., Ju, J., Gardner, A., Hendrickson, C., and Ausubel, F. (2011). First Generation" Automated DNA Sequencing Technology. Current Protocols in Molecular Biology. Unit7 2. doi:10.1002/0471142727.mb0702s96
Song, F., Li, T., Burgess, K. S., Feng, Y., and Ge, X.-J. (2020). Complete Plastome Sequencing Resolves Taxonomic Relationships Among Species of Calligonum L. (Polygonaceae) in China. BMC Plant Biol. 20, 261. doi:10.1186/s12870-020-02466-5
Song, Y., Chen, Y., Lv, J., Xu, J., Zhu, S., Li, M., et al. (2017). Development of Chloroplast Genomic Resources for Oryza Species Discrimination. Front. Plant Sci. 8, 1854. doi:10.3389/fpls.2017.01854
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28, 2731–2739. doi:10.1093/molbev/msr121
Tan, W., Gao, H., Jiang, W., Zhang, H., Yu, X., Liu, E., et al. (2020). The Complete Chloroplast Genome of Gleditsia Sinensis and Gleditsia Japonica: Genome Organization, Comparative Analysis, and Development of Taxon Specific DNA Mini-Barcodes. Sci. Rep. 10, 16309. doi:10.1038/s41598-020-73392-7
Tonti‐Filippini, J., Nevill, P. G., Dixon, K., and Small, I. (2017). What Can We Do with 1000 Plastid Genomes? Wiley Online Library.
Ueda, M., Fujimoto, M., Arimura, S.-i., Murata, J., Tsutsumi, N., and Kadowaki, K.-i. (2007). Loss of the Rpl32 Gene from the Chloroplast Genome and Subsequent Acquisition of a Preexisting Transit Peptide within the Nuclear Gene in Populus. Gene 402, 51–56. doi:10.1016/j.gene.2007.07.019
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3-new Capabilities and Interfaces. Nucleic Acids Res. 40, e115. doi:10.1093/nar/gks596
Wu, X.-M., Wu, S-F., Ren, D-M., Zhu, Y-P., and He, F-C. (2007). The Analysis Method and Progress in the Study of Codon Bias. Hereditas 29, 420–426. doi:10.1360/yc-007-0420
Wyman, S. K., Jansen, R. K., and Boore, J. L. (2004). Automatic Annotation of Organellar Genomes with DOGMA. Bioinformatics 20, 3252–3255. doi:10.1093/bioinformatics/bth352
Xu, J., Chu, Y., Liao, B., Xiao, S., Yin, Q., Bai, R., et al. (2017). Panax Ginseng Genome Examination for Ginsenoside Biosynthesis. Gigascience 6, 1–15. doi:10.1093/gigascience/gix093
Yang, J.-B., Tang, M., Li, H.-T., Zhang, Z.-R., and Li, D.-Z. (2013). Complete Chloroplast Genome of the Genus Cymbidium: Lights into the Species Identification, Phylogenetic Implications and Population Genetic Analyses. BMC Evol. Biol. 13, 84. doi:10.1186/1471-2148-13-84
Yang, M., Zhang, X., Liu, G., Yin, Y., Chen, K., Yun, Q., et al. (2010). The Complete Chloroplast Genome Sequence of Date palm (Phoenix Dactylifera L.). PloS one 5, e12762. doi:10.1371/journal.pone.0012762
Yang, Y., Zhou, T., Duan, D., Yang, J., Feng, L., and Zhao, G. (2016). Comparative Analysis of the Complete Chloroplast Genomes of Five Quercus Species. Front. Plant Sci. 07, 959. doi:10.3389/fpls.2016.00959
Yao, X., Tang, P., Li, Z., Li, D., Liu, Y., and Huang, H. (2015). The First Complete Chloroplast Genome Sequences in Actinidiaceae: Genome Structure and Comparative Analysis. PloS one 10, e0129347. doi:10.1371/journal.pone.0129347
Ye, X., Lin, J., Zhou, M., He, X., Yan, M., and Cheng, R. (2021). The Complete Chloroplast Genome of the Medicinal Plant Polygonum Cuspidatum (Polygonaceae) and its Phylogenetic Implications within the Subfamily Polygonoideae. Mitochondrial DNA B 6, 1563–1565. doi:10.1080/23802359.2021.1917313
Yu, B., Liu, J., Liu, X., Lan, X., and Qu, X. (2020). The Complete Chloroplast Genome Sequence of Polygonum Chinense L. Mitochondrial DNA Part B 5, 2139–2140. doi:10.1080/23802359.2019.1693931
Yu, B., Sun, Y.-b., Liu, X.-f., Huang, L.-l., Xu, Y.-c., and Zhao, C.-y. (2020). Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Camellia Fraterna. Mitochondrial DNA Part B 5, 3840–3842. doi:10.1080/23802359.2020.1841576
Keywords: Polygonum, comparative analysis, phylogenetic analysis, repeats analysis, complete chloroplast genome
Citation: Guo S, Liao X, Chen S, Liao B, Guo Y, Cheng R, Xiao S, Hu H, Chen J, Pei J, Chen Y, Xu J and Chen S (2022) A Comparative Analysis of the Chloroplast Genomes of Four Polygonum Medicinal Plants. Front. Genet. 13:764534. doi: 10.3389/fgene.2022.764534
Received: 25 August 2021; Accepted: 11 March 2022;
Published: 25 April 2022.
Edited by:
Mohan Lal, North East Institute of Science and Technology (CSIR), IndiaReviewed by:
Xiaojun Nie, Northwest A&F University, ChinaYong Qi Zheng, Chinese Academy of Forestry, China
Xiaoxuan Tian, Tianjin University of Traditional Chinese Medicine, China
Copyright © 2022 Guo, Liao, Chen, Liao, Guo, Cheng, Xiao, Hu, Chen, Pei, Chen, Xu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiang Xu, anh1QGljbW0uYWMuY24=; Shilin Chen, c2xjaGVuQGljbW0uYWMuY24=
†These authors have contributed equally to this work
 Shuai Guo
Shuai Guo Xuejiao Liao
Xuejiao Liao Shiyu Chen1,2†
Shiyu Chen1,2† Baosheng Liao
Baosheng Liao Shuiming Xiao
Shuiming Xiao Haoyu Hu
Haoyu Hu Jun Chen
Jun Chen Jin Pei
Jin Pei Jiang Xu
Jiang Xu Shilin Chen
Shilin Chen