
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 10 February 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.756270
This article is part of the Research TopicThe Development and Application of Multi-Omics Integration Approaches to Dissecting Complex Traits in PlantsView all 25 articles
 Yujie Fan1†
Yujie Fan1† Jiyan Qi1†
Jiyan Qi1† Xiaohu Xiao2†
Xiaohu Xiao2† Heping Li1
Heping Li1 Jixian Lan1
Jixian Lan1 Yacheng Huang1
Yacheng Huang1 Jianghua Yang2
Jianghua Yang2 Yi Zhang1
Yi Zhang1 Shengmin Zhang1
Shengmin Zhang1 Jun Tao1
Jun Tao1 Chaorong Tang1*
Chaorong Tang1*Natural rubber, an important industrial raw material with wide applications, is harvested in the form of latex (cytoplasm of rubber-producing laticifers) from Hevea brasiliensis (para rubber tree) by the way of tapping. Conspicuous stimulation on latex production is observed for the first few tappings conducted on virgin (untapped before) or resting (tapped before but no tapping for a period) rubber trees. To understand the underlying mechanisms, an integrative analysis of the latex transcriptome and proteome was conducted on virgin or resting Hevea trees for the first five tappings. A total of 505 non-redundant differentially expressed (DE) transcript-derived fragments (TDFs) were identified by silver-staining cDNA-AFLP, with 217 exhibiting patterns of upregulated, 180 downregulated and 108 irregularly-regulated. Meanwhile, 117 two dimensional gel electrophoresis DE-protein spots were isolated and subjected to mass spectrometry analysis, with 89 and 57 being successfully identified by MALDI-TOF and MALDI-TOF/TOF, respectively. About 72.5% DE-TDFs and 76.1% DE-proteins were functionally annotated and categorized. Noteworthily, most of the DE-TDFs implicated in sugar transport and metabolism as well as rubber biosynthesis were upregulated by the tapping treatment. The importance of sugar metabolism in harvesting-induced latex production was reinforced by the identification of abundant relevant DE-protein spots. About 83.8% of the randomly selected DE-TDFs were validated for expression patterns by semi-quantitative RT-PCR, and an 89.7% consistency for the 29 latex regeneration-related DE-TDFs examined by quantitative RT-PCR analysis. In brief, our results reveal extensive physiological and molecular changes in Hevea laticifers incurred by the tapping treatment, and the vast number of DE genes and proteins identified here contribute to unraveling the gene regulatory network of tapping-stimulated latex production.
Natural rubber (cis-1, 4-polyisoprene, NR) is an elastomer with superior properties that cannot be completely replaced by petroleum-derived synthetic rubber and is used as an important industrial raw material. Due to the advantages of good quality, high yield, and easiness for harvesting, Hevea brasiliensis (para rubber tree, Hevea thereafter) has become the sole commercial NR source among the 2, 500 NR-bearing plant species (Van Beilen and Poirier, 2007). Hevea trees need warm and humid climate conditions for normal growth and NR production yet vulnerable to typhoon, thus confining its planting to restricted tropical areas (Huang, 2001).
Hevea rubber yield is affected mainly by three factors: duration of latex flow after tapping, the capability of latex regeneration between two consecutive tappings, and the ability of laticifer differentiation in bark cambium (d’Auzac et al., 1997; Hao et al., 2004). In the trunk bark of tapped Hevea trees, the number of laticifer rings is one to three times more than that observed in un-tapped trees, indicating the stimulating effect of tapping on laticifer differentiation (Hao and Wu, 1982). Meanwhile, both mechanical injury and jasmonic acid induce laticifer differentiation, latex regeneration and production (Hao and Wu, 2000; Sae-Lim et al., 2019). Cytologically, latex is the cytoplasm of the laticifers, 90% of the dry weight as rubber hydrocarbon (NR) (Chrestin et al., 1997). To sustain NR productivity, in regularly tapped Hevea trees, the expelled latex must be efficiently regenerated before the next tapping, usually with an interval of 2–3 days (d’Auzac et al., 1997).
After tapping, the latex flowing out of the laticifers includes cis-polyisoprene particles (rubber particles), lysosomal microvacuoles (lutoids), plastid-like Frey-Wyssling complexes, sugars, organic acids, nucleic acids, and proteins/enzymes (d'Auzac et al., 1997). A positive correlation exists between sucrose content and latex yield, and the control of sucrose metabolism has been intensively studied in relation to latex production (Tupý, 1969; Tupý, 1985; Tupý, 1989). Expression of the responsible sucrose transporter, HbSUT3, for sucrose loading into laticifers is induced by the treatments of ethylene and tapping, both bolstering the latex yield (Tang et al., 2010). In the latex, an alkaline/neutral invertase is responsible for cleaving sucrose into hexose sugars that are then exploited in subsequent latex production (Tupý, 1989). Expression and enzymatic activity of the relevant invertase gene, HbNIN2, correlate positively with the latex yield (Liu et al., 2015). Rubber particles are a kind of organelle suspended in latex where rubber biosynthesis takes place and subsequently stores, with proteins of rubber elongation factor (REF)/small rubber particle protein (SRPP) as important rubber-synthesizing participants (Cornish, 1993; Oh et al., 1999; Singh et al., 2003; Tang et al., 2016). Priya et al. (2007) observed a positive correlation between REF mRNA abundance and the latex yielding levels of different Hevea clones. After Ethrel (2-chloroethylphosphonic acid, an ethylene releaser) bark treatment, a marked increase in transtonoplast ΔpH within Hevea laticifers was observed consisting of one of the major mechanisms of ethylene stimulation on latex yield (Amalou et al., 1992). Together, these studies suggest that latex production, with NR biosynthesis as the core activity, involves a complex regulatory network of gene expression, multi-enzyme reaction and physio-biochemical processes. Regular tapping with intervals of 2–4 days significantly stimulates latex yield on both virgin (never tapped before) and resting (tapped before but left with no tapping for 3 months or above) Hevea trees in a similar way, except that the number of tappings required is larger in the former (7–10) than the latter (4–6) to yield at a relatively stable level (Pakianathan et al., 1992; Tang et al., 2010; Qi, 2011). In the latex of virgin Hevea trees, expressions of a number of genes involved in latex production are bolstered by the tapping treatment, e.g., HbSUT3 (Tang et al., 2010), HbNIN2 (Liu et al., 2015) and a farnesyl diphosphate synthase gene (Adiwilaga and Kush, 1996). Latex transcript and protein profiling of virgin or resting Hevea trees for the first few tappings could be beneficial to unraveling the mechanisms of tapping-induced latex production and to identifying the genes or proteins involved.
Since the advent of the cDNA-AFLP transcript-profiling technique in 1996 (Bachem et al., 1996), owing to its advantages of good repeatability, high sensitivity and high throughput, this technique has been successfully applied to various aspects of plant studies, such as plant’s abiotic stress response (Fatemi et al., 2019), plant-microorganism interaction (Xiao et al., 2016), hormone signaling (Akihiro et al., 2006), and plant development (Leymarie et al., 2007). In this study, the latex transcriptome and proteome were compared for the first five tappings in virgin and resting Hevea trees, respectively, using a silver staining cDNA-AFLP (Xiao et al., 2009) and two dimensional electrophoresis coupled with mass spectrometry analysis. A large number of differentially expressed (DE) transcripts and proteins were successfully isolated and functionally identified, and the results reveal the importance of sugar metabolism as well as rubber biosynthesis in tapping-activated latex production in Hevea trees.
To extensively identify the differentially expressed transcripts responding to the tapping treatment, all the 128 cDNA-AFLP selective primer pairs of Apo I / Mse I restriction system (Xiao et al., 2009) were screened in the latex RNA for the first five tappings in virgin Hevea trees. On average, about 70 transcript-derived fragments (TDFs) greater than 100 bp were discernable on the silver-stained polyacrylamide gels for each primer pair (Figure 1). Therefore, nearly 9,000 TDFs were profiled for each latex RNA sample. In total, 651 DE-TDFs were successfully cloned and sequenced.
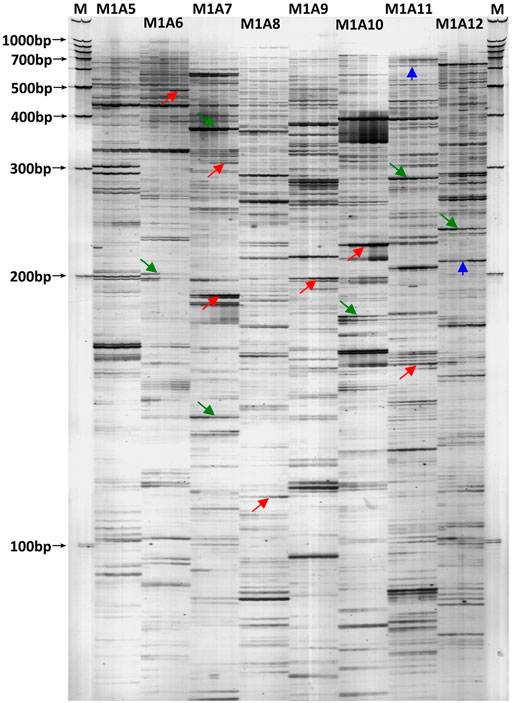
FIGURE 1. A typical cDNA-AFLP silver-stained polyacrylamide gel revealing transcript profiles in the latex of virgin Hevea trees for the first five tappings. M: 100 bp DNA ladder molecular weight standard; M1A5 to M1A12: selective primer combinations. Under each primer pair, the five lanes (from left to right) represent the latex transcript profiles for the first five tappings. The arrows marked in red indicate the DE-TDFs up-regulated by tapping, the green arrows indicate down-regulated, and the blue arrows indicate irregularly-regulated. Please note there exists subtypes for each of the three regulation types.
To identify the tapping-responsive proteins, proteomics studies were preformed on re-opened resting Hevea trees, the latex production of which is stimulated by tapping in a way similar to virgin Hevea trees (Supplementary Figure S1). The resting Hevea trees usually yield significantly higher than virgin Hevea trees for the first tapping, thus facilitating the conventional proteomics analysis that requires more latex than the virgin Hevea trees could produce (Tang et al., 2010; Qi, 2011). For each of the first five tappings in reopened resting Hevea trees, 1 mg of latex C-serum proteins were separated on two dimensional electrophoresis (2-DE) gels and stained by Coomassie brilliant blue R250. Most of the protein spots displayed superior resolution with clear background and obvious boundary on the 2-DE gels (Figure 2). On each 2-DE gel, the number of discernable protein spots ranged between 700 and 800 when pH 4–7 IPG strips were used for 1-DE (Figure 2), and between 900 and 1,000 when pH 5–8 IPG strips were used (Supplementary Figure S2). The molecular weights of latex C-serum proteins were concentrated in the range of 15–80 kDa, and the isoelectric points were in pH 4.5–7.5.
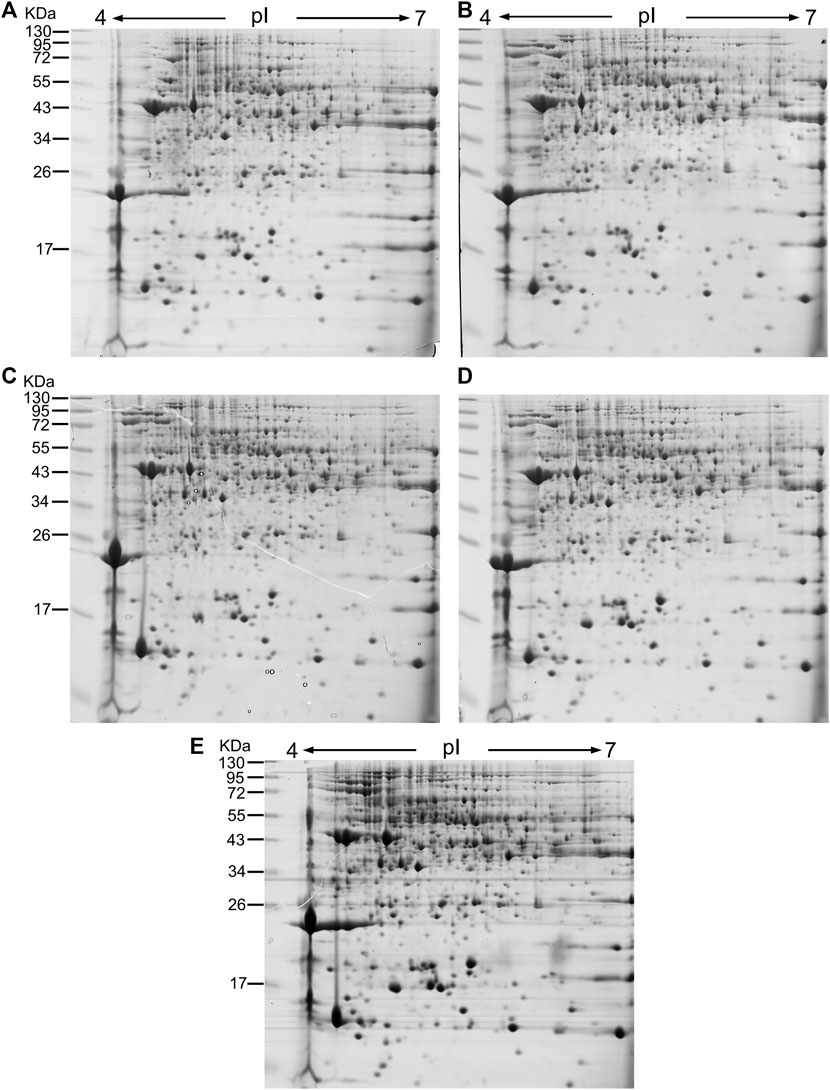
FIGURE 2. 2-DE images of latex C-serum proteins from retapped Hevea trees after a four-month resting period. IGP strips of pH4-7 were used in 2-DE, and the fresh latex C-serum samples were analyzed for the first five tappings [(A): first; (B): second; (C): third; (D): fourth; (E): fifth]. kDa: Molecular weight of protein. pI: Protein isoelectric point.
Taking the expression level at the first tapping as a reference, the DE-TDFs identified were classified into three types according to their patterns of expression across the five successive tappings: upregulation, downregulation and irregular-regulation (Figure 1). The upregulation type includes three subtypes: 1) increase successively; 2) increase first and then stabilize; 3) increase first and then decrease, but still higher than the first tapping. The downregulation type also includes three subtypes: 1) decrease successively; 2) decrease first and then stabilizes; 3) decrease first and then increase, but still lower than the first tapping. The irregular-regulation includes two subtypes: 1) increase first, reaching a high threshold, and then decrease to a level lower than the first tapping; 2) decrease first, reaching a low threshold, and then increase to a level higher than the first tapping. Accordingly, the DE-protein spots were also classified into the three types according to their expression dynamics across the first five tappings in reoppend resting Hevea trees (Figure 2). A protein spot was regarded as a DE one when its abundance varies ≥3-fold between the first tapping and any of the four other tappings. As a result, a total of 117 tapping-responsive DE-protein spots were identified, and picked out from the 2-DE gels for mass spectrometry analysis.
The DE-TDFs were made clean by removing the sequences of vector, primer, and adaptor at both ends and then annotated by Blastx online searching (http://blast.ncbi.nlm.nih.gov/Blast.cgi) against the NCBI non-redundant protein database (nr), with an E-value threshold of <10−4 and a score value of >50. According to Blastx search against the Hevea latex transcriptome database (Tang et al., 2016), the DE-TDFs belonging to the same transcript and sharing similar expression pattern in the cDNA-AFLP analysis were clustered together, and only the longest TDF was retained. As a result, a total of 505 non-redundant DE-TDFs (GenBank accession nos MZ935745—MZ936248) were obtained, including 217 (43.0%) upregulated (Supplementary Table S1), 180 (35.6%) downregulated (Supplementary Table S2) and 108 (21.4%) irregularly-regulated (Supplementary Table S3). The selected DE-protein spots were identified by a Bruker’s Ultraflex TOF/TOF mass spectrometer. The proteins identified as positive by peptide mass fingerprinting (PMF) analysis were further subjected with 2-4 matched peptides to peptide fragmentation fingerprinting (PFF) investigation. Of the 117 DE-protein spots examined, 89 were identified as positive by the PMF analysis, and 57 were further determined as positive by the PFF analysis. Of the 89 positively identified DE-proteins, 35 (39.3%) were classified as upregulated, 20 (22.5%) as downregulated and 34 (38.2%) as irregularly-regulated (Supplementary Table S4).
Detailed comparison revealed that the DE-protein spots of three small rubber particle proteins (nos. 163, 723 and 730), one tubulin alpha-3 chain-like protein (no. 552) and two latex abundant family proteins (nos. 554 and 628) had their DE-TDF counterparts (M1-A10-2, M12-A6-4, M6-A10-4, M6-A7-1 and M8-A7-2) (Supplementary Tables S1, S2, S4, S5), of which only one pair (protein spot 163 and TDF #M1-A10-2) revealed a consistency of upregulation. The other nine DE-TDFs overlapped in functional annotations with a number of DE- protein spots, belonging to distinct genes within the same gene families (Supplementary Table S5). As reported in studies covering various types of organisms (Taniguchi et al., 2010; Vogel and Marcotte, 2012; Walley et al., 2013), low overlap and poor correlation have been frequently observed between transcriptomic and proteomic data due to several possible explanations. One more plausible explanation for the low overlap revealed here is the detection limit of the proteome technique that is capable of profiling only medium to high expressed proteins (Humphery-Smith et al., 1997) in comparison with the high transcript-detecting sensitivity of the cDNA-AFLP technique (Vuylsteke et al., 2007; Xiao et al., 2009).
According to the results of Blastx search, the 505 non-redundant DE-TDFs were divided into four categories: 1) Proteins with clear functional annotation; 2) Unclassified proteins, the functional annotation of the proteins being multiple; 3) Predicted protein, showing high homology with a predicted protein in the database; 4) No hit, no homologous sequence in the database. Most (366, 72.5%) of these DE-TDFs were homologous to genes with known functions, whereas 30 (5.9%) and 59 (11.7%), respectively, belonged to unclassified proteins and predicted proteins and the remaining 50 (9.9%) with no hit (Table 1). Meanwhile, the 89 positive DE-protein spots were divided into two categories: 1) Proteins with clear functional annotation; 2) Unknown proteins. Most (78, 87.6%) of these DE-protein spots were functionally annotated, whereas the remaining 11 (12.4%) belonged to predicted or hypothetical proteins (Table 1).
With reference to the functional categories of plant genes defined by Bevan et al. (1998), the 366 DE-TDFs and 78 DE-proteins with known functions were classified, respectively, into 11 and 7 functional categories, among which a new category, rubber biosynthesis, was singled out from “secondary metabolism” (Figure 3; Table 1). Of these DE-TDFs, five categories including cell growth and division, protein degradation and storage, cellular structure, secondary metabolism, and rubber biosynthesis had a higher portion of upregulated DE-TDFs than that of the down- and irregularly-regulated DE-TDFs together (Table 1). Noteworthily, nine of the ten DE-TDFs implicated in rubber biosynthesis were upregulated, and the remaining one is irregularly-regulated. Of the 89 DE-protein spots, the number of upregulated (35) was significantly larger than the downregulated (20), but similar to the irregular-regulated (34). Expression patterns of the DE-protein spots varied in most functional categories, whereas all four in the signal transduction category were upregulated. It is worth noting that there were many cases that more than one protein spots corresponded to an identical gene, e.g., six spots (nos. 33, 229, 516, 534, 539, and 591) for an enolase, five (nos. 149, 163, 723, 730, and 858) for a small rubber particle protein, and five (nos. 688, 778, 779, 789, 797, and 803) for a heat shock protein (Supplementary Table S4), indicating extensive posttranslational modifications in Hevea latex proteins.
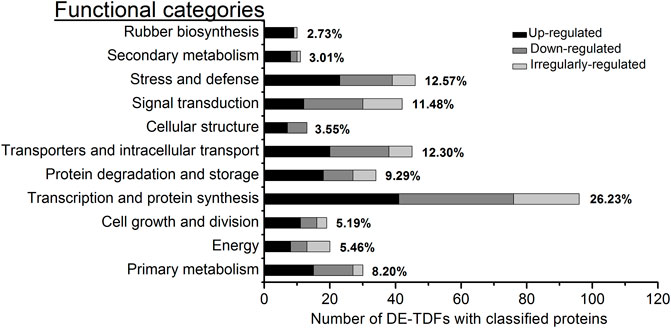
FIGURE 3. Functional category and percentage of the DE-TDFs with known function. The 366 non-redundant DE-TDFs of annotation proteins with known function were identified in the latex for the first five tappings in rubber trees. These DE-TDFs were classified into 11 functional categories. In each category, the percentage covering the total know functional DE-TDFs was placed at its right whereas the three types of expression were shown in differing gray bars.
To determine the reliability of the cDNA-AFLP results, 80 DE-TDFs, covering >10% of the DE-TDFs we identified were randomly selected from each functional category and subjected to semi-quantitative reverse transcription PCR (sqRT-PCR) analysis using specific primers for TDFs with 18S rRNA as the reference gene. The ImageJ software was used to quantify the sqRT-PCR amplicons fractionated in agarose gel electrophoresis, with the value in the first tapping sample taken as 1.0 (Table 2). About 84% (67 TDFs) showed expression patterns consistent with their cDNA-AFLP gel profiles, indicating the high reliability of the cDNA-AFLP screening.
A total of 29 DE-TDFs implicated in latex-regeneration were investigated by quantitative RT-PCR (qRT-PCR) analysis for their expression patterns across the five successive tappings. About 90% of the qRT-PCR results were consistent with their original cDNA-AFLP expression profiles (Figure 4; Table 3). The genes of these DE-TDFs are putatively involved in the pathways of primary metabolism, rubber biosynthesis and regulation, transporters, and intracellular transport. Of the ten rubber biosynthesis pathway DE-TDFs, nine revealed qRT-PCR patterns similar to their cDNA-AFLP results (Figure 4; Table 3).
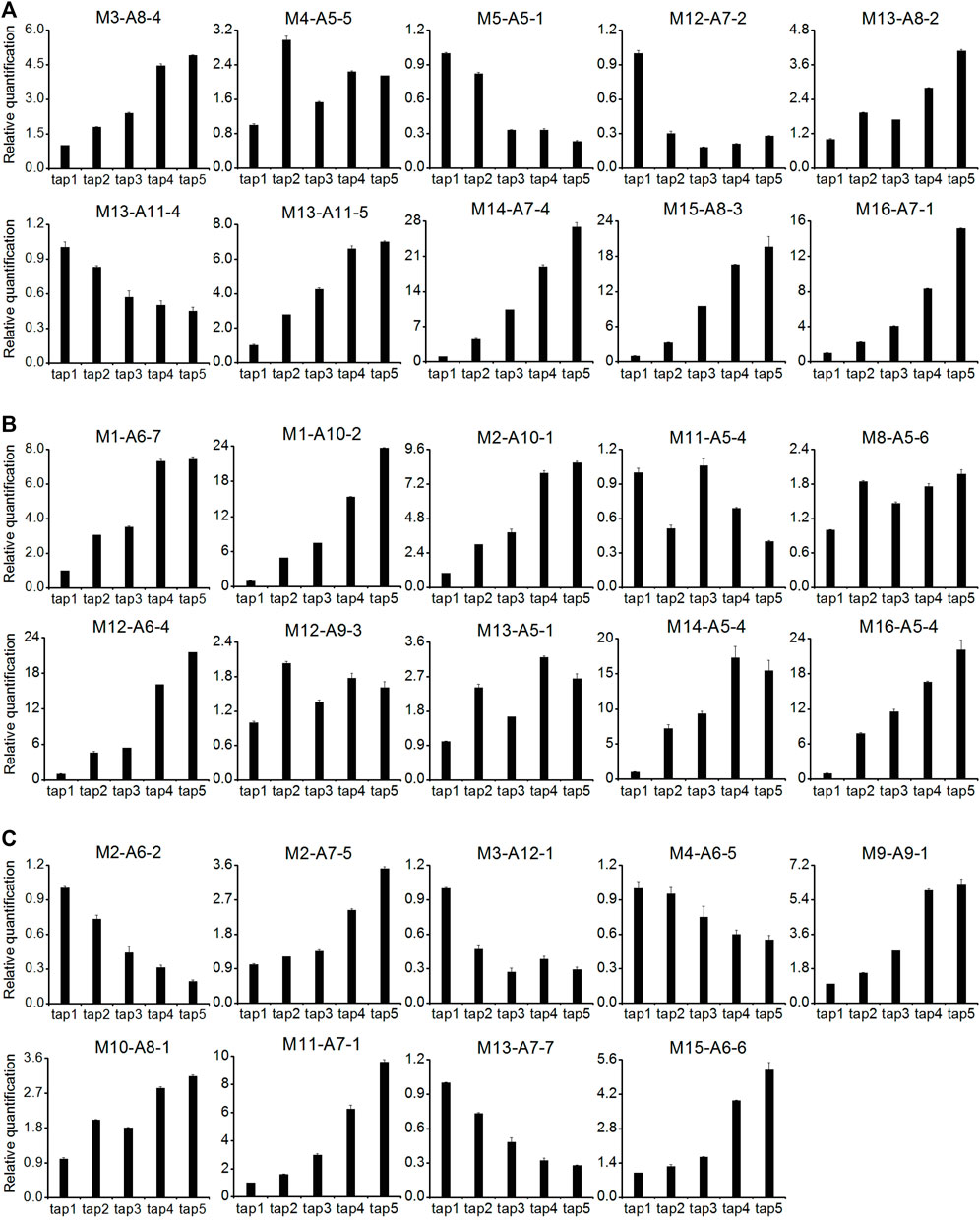
FIGURE 4. qRT-PCR analysis for expression of latex regeneration genes in the latex of the first five tappings. (A): primary metabolism pathway (ten DE-TDFs); (B): rubber biosynthesis and regulation pathway (ten DE-TDFs); (C): transporters and intracellular transport pathway (nine DE-TDFs). Except for M3-A8-4, M11-A5-4 and M16-A7-1, 26 of the 29 DE-TDFs shows the results consistent with their original cDNA-AFLP expression patterns and were presented here. All data were normalized to the expression level of HbYLS8 gene. Values are averages ± STDEV of three technical replicates. The latex samples of three individual trees were analyzed with similar patterns of expression, and one representative was shown.
Tapping can stimulate latex production, especially in virgin and reopened resting Hevea trees (Pakianathan et al., 1992; Tang et al., 2010; Qi, 2011). A number of early studies have shown that the first few tappings greatly stimulate the metabolism of laticifers, accompanied by the enhanced expression of several specific genes involved in latex production (Dennis et al., 1989; Adiwilaga and Kush, 1996; Gohet, 1996; Priya et al., 2007). The latex flows out of laticifers after tapping, and in order to compensate for the loss of cytoplasm (latex) and maintain the balance of intracellular metabolism, the laticifers require large amounts of RNA and proteins to be synthesized before the next tapping. Of the 366 DE-TDFs identified in the latex with known functions (Table 1), 26.2% were classified into the functional category of transcription and protein synthesis (Figure 3), representing the largest category, 42.7% of which were upregulated by the tapping treatment. These results indicated that tapping significantly affects the ways of laticifers to synthesize RNA and proteins, providing a prerequisite for multiple subsequent biological responses to the tapping treatment. Of the 78 DE-protein spots identified in the latex with known functions, 29.5% were classified into the functional category of stress and defense (Table 1) and represented the largest category, corresponding well to the defense functions of Hevea laticifers (Sharples, 1918; Chow et al., 2007). Laticifers are believed to be a defense system for Hevea trees to cope with biotic and abiotic stresses, and the latex exuded after bark wounding has been found to play roles in resisting pathogen infection, insect feeding and abiotic stress (Chow et al., 2007). The tapping itself is a kind of abiotic stress upon Hevea trees. Consistent with the proteome study, “stress and defense” also accounted for a large portion of the functional DE-TDFs, ranking the second place among the DE-TDF functional categories (Figure 3; Table 1; Supplementary Table S4). The harvesting stress response has been suggested to be one of the key factors affecting latex production and rubber productivity in Hevea trees (Pirrello et al., 2014).
The category of transporters and intracellular transport was the third largest among the 11 functional categories, accounting for 12.3% of the total functional DE-TDFs (Figure 3). This agrees well with the sink effect caused by the large loss of latex after tapping. The process of regenerating the expelled latex involves the synthesis, transport, loading and subcellular localization of a large number of organelles, proteins, nucleic acids, sugars, etc., all of which require the active involvement of transporters and intracellular transport-related proteins (d’Auzac et al., 1997; Tungngoen et al., 2011). DE-TDFs involved in signal transduction were also highly represented, accounting for a proportion of 8.3% for the total DE-TDFs (Table 1), and all four DE-proteins involved in signal transduction were upregulated (Table 1; Supplementary Table S4). A variety of signaling pathways within Hevea laticifers, including ethylene, jasmonic acid and wound signaling, have been reported to be extensively participate in latex regeneration and regulation (Dennis et al., 1989; Hao et al., 2004; Duan et al., 2010; Lacote et al., 2010; Pirrello et al., 2014; Tang et al., 2016). The proportions for the two categories, protein degradation and storage and primary metabolism were also high, covering, respectively, 9.3% and 8.2% of the total functional DE-TDFs (Figure 3; Table 1). Their high representation suggested that with the progress of tapping, in order to meet the balance of supply and demand of all substances in latex regeneration, protein turnover rate becomes faster and primary metabolism gets active. In a word, these results indicated that the latex production induced by tapping involves a complex multi-gene regulatory network, as well as multiple physiological and biochemical response processes.
In regularly tapped Hevea trees, the main metabolic activity of the laticifers is latex regeneration, which centers on the biosynthesis of NR that consists of about 90% of the latex dry weight (d’Auzac et al., 1997). Sucrose is the precursor material for rubber biosynthesis in laticifers, and provides the carbon skeleton and energy required for latex regeneration (Tupý, 1989; Duangngam et al., 2020). In Hevea trees that are tapped at two-four days of intervals, a tree produces dozens to hundreds of milliliters of fresh latex, and the removed latex could be effectively recovered before the next tapping (d’Auzac et al., 1997; Tang et al., 2010). Therefore, the laticifers represent an active carbon sink, and the effective supply of sucrose is a key factor determining latex production (Tupý, 1985; Chantuma et al., 2009). In this study, the genes of a sucrose transporter (M10-A9-1) and a sugar transporter (M10-A8-1) were among the DE-TDFs identified, both of which were significantly upregulated with the increase of tappings (Supplementary Table S1). Interestingly, the sucrose transporter (M10-A9-1) identified here is the HbSUT3 we previously reported to be critical in sucrose uptake into laticifers and Hevea rubber production (Tang et al., 2010). The upregulation of these two transporters indicated an active involvement of sucrose and sugar transport in tapping-stimulated latex production. Sucrose catabolism and the following pathways including glycolysis, tricarboxylic acid cycle and pentose phosphate pathway provide essential components, i.e., the carbon skeleton (acetyl CoA), the reducing power (NADPH) and the energy (ATP) for the final rubber biosynthesis pathway (Tupý, 1989; d’Auzac et al., 1997). Therefore, sugar metabolism becomes one of the core metabolic pathways contributing to latex production in Hevea (Tupý, 1989; d’Auzac et al., 1997; Silpi et al., 2007). This study identified multiple DE-TDFs involved in sucrose cleavage and the three above mentioned sugar metabolism pathways (Table 3; Supplementary Tables S1–S3), including neutral/alkaline invertase (M16-A7-1), fructokinase (M14-A7-4), phosphofructokinase (M12-A7-2), glyceraldehyde 3-phosphate dehydrogenase (M15-A8-3), pyruvate kinase (M13-A11-5), pyruvate dehydrogenase (M13-A8-2), and glucose-6-phosphate dehydrogenase (M5-A5-1), etc. Most of these DE-TDFs were upregulated in the latex for the first few tappings (Figure 3; Supplementary Table S1). It is worth noting that the upregulated DE-TDF (M16-A7-1) as identified by both cDNA-AFLP (Supplementary Table S1) and qRT-PCR (Figure 4) turned out to be HbNIN2, the neutral/alkaline invertase that is responsible for sucrose catabolism in Hevea laticifers (Liu et al., 2015). Here, the proteomic research also backed up the importance of sugar metabolism in tapping-stimulated latex production in reopened resting Hevea trees although a low overlap was observed among the DE-genes and DE-proteins we identified this study. Among the 18 DE-protein spots identified in category of primary metabolism and energy, 13 were involved in sugar metabolism (Supplementary Table S4). Notably, nine spots (nos. 356, 372, 593, 33, 229, 516, 535, 539, and 591) were implicated in the glycolytic pathway. By contrast, no DE-protein spots were in tricarboxylic acid (TCA) cycle, collaborating the proposition of the relative importance of glycolysis versus the TCA cycle in sugar degradation in hypoxic Hevea latex (d’Auzac and Jacob, 1969; Tupý and Resing, 1969) and thus in tapping-stimulated latex production.
There are 20 gene families directly involved in the NR biosynthesis and termed as rubber biosynthesis (RB) genes (Tang et al., 2016; Chow et al., 2007). In this study, a total of nine DE-TDFs involving six such families were identified, including cis-prenyltransferase (M1-A6-7), hydroxymethylglutaryl coenzyme A synthase (M16-A5-4), 3-hydroxy-3-methylglutaryl-coenzyme A reductase (M12-A9-3), farnesyl diphosphate synthase (M11-A5-4), REF/SRPP proteins (M2-A10-1, M13-A5-1, M14-A5-4, M1-A10-2, M12-A6-4) (Table 3). Among the nine DE-TDFs, eight were demonstrated by qRT-PCR to be upregulated with the tappings (Figure 4). In addition, a DE-TDF (M8-A5-6) annotated as inorganic pyrophosphatase, a vacuolar type of which being located on rubber particles and essential for IPP incorporation into elongating rubber hydrocarbon molecules (Zeng et al., 2009), was also bolstered by the tapping treatment (Figure 4). The tapping treatment also changes the expression of RB genes at the protein levels in reopened resting Hevea latex. A total of eight relevant DE-protein spots were identified, corresponding to REF/SRPP proteins and an acetyl-CoA C-acetyltransferase (Supplementary Table S4).
The cDNA-AFLP technique has been widely applied in various eukaryotes including the Hevea tree for transcript profiling due to its advantages of stringency, reproducibility, cost-effectiveness, genome-wide coverage and the ability to distinguish among highly homologous genes (Ko et al., 2003; Vuylsteke et al., 2007; Ganeshan et al., 2011; Tang et al., 2013; Xiao et al., 2016). In this study, as determined by sqRT-PCR and qRT-PCR, about 84% and 90%, respectively, of the selected DE-TDFs were verified for their cDNA-AFLP profiles (Figure 4; Tables 2, 3), reflecting a high reliability of this technique in screening tapping-responsive DE-TDFs in Hevea latex. According to a previous in-silico estimation (Xiao et al., 2009), about 84% of the genes expressed in Hevea latex could be visualized using the silver-staining cDNA-AFLP technique with the restriction enzyme pair of Apo I and Mse I exploited here. The sucrose transporter HbSUT3 (Tang et al., 2010) and the neutral/alkaline invertase HbNIN2 (Liu et al., 2015) that have been reported to be upregulated in the latex of virgin Hevea trees by the tapping treatment were among the DE-TDFs identified in this study (Figure 4; Table 3; Supplementary Table S1), demonstrating a high transcript coverage of this technique. However, compared with the currently popularly used next generation RNA-sequencing technique that relies on expensive DNA sequencers and specialized bioinformatics, the cDNA-AFLP is labor-intensive. Nevertheless, the cDNA-AFLP technique still have its niche among the various transcript profiling techniques, and can be readily established in a mediocrely equipped and stringently funded lab to fulfill its customized transcript profiling task.
An integrative transcriptome and proteome analysis was conducted to identify important regulators and pathways participated in tapping stimulated latex production in Hevea trees. A total of 505 tapping-responsive DE-TDFs and 89 DE-proteins were identified in the rubber-producing laticifers of Hevea trees (virgin or reopened). The low overlap between the DE-TDFs and DE-protein spots identified is indicative of posttranscriptional regulation and the strong complementarities of transcriptome and proteome analysis. According to the 366 DE-TDFs and 78 DE-proteins with functional annotations, the tapping treatment brought about extensive physiological and molecular changes in laticifers. The integration of these changes, especially those of sugar metabolism and rubber biosynthesis, upgraded the mediocre level of laticifer metabolism in virgin or reopened trees to a high dynamic equilibrium of latex regeneration in regularly tapped trees. Further integrative studies will benefit a deeper insight into the exact relationships (synergy or antagonism) among the vast number of biological pathways implicated in tapping-stimulated latex production.
Hevea trees of Reyan7-33-97 clone for cDNA-ALFP transcript profiling (virgin trees) and 2-DE protein profiling (resting trees) analysis, were all planted in the experimental field of Chinese Academy of Tropical Agricultural Sciences (Danzhou, Hainan). i.e., removing a slice of trunk bark by a special knife, with a half spiral tapping system, every 3 days, and with no ethylene stimulation (Tang et al., 2010). The virgin trees were planted for eight years and first subjected to the tapping. The resting trees (after four months of no tapping) were planted for 10 years, and re-tapped for the third year. Because the virgin rubber trees produced little latex in the first tapping that could not satisfy the requirement of C-serum preparation for 2-DE analysis as described below, re-tapped resting trees were therefore exploited. Such Hevea trees produce much more latex than virgin trees in the first tapping and in the subsequent several tappings the latex production increases in a pattern similar to that observed in virgin trees albeit not striking.
Three batches of five rubber trees attaining the tapping standard (trunk girth >= 50 cm at 1 m above the ground) were selected for latex collection and RNA extraction. Twenty seconds after tapping, about 5 ml of latex was allowed to flow into a centrifuge tube containing 5 ml 2×RNA extraction buffer (.3 M LiCl, 10 mM EDTA, 10% SDS, 100 mM Tris-HCl, pH8.0). The collected latex was placed in ice box and brought to laboratory for RNA extraction as described in Tang et al. (2007). Electrophoresis on a 1.5% formaldehyde denaturing agarose gel was used to detect the integrity of RNA samples. RNA samples from each batch of rubber trees for the five tappings were used as one biological replicate.
A total of 50 μg latex total RNA taken from each of the five tapping samples was subjected to cDNA-AFLP analysis. The detailed manipulations were conducted according to the silver-staining cDNA-AFLP procedure which we previously established suitable for Hevea latex transcriptome profiling (Xiao et al., 2009). Briefly, the synthesized double-stranded cDNA was cut by the restriction enzymes of Apo I and Mse I (Thermo Fisher Scientific, Vilnius, Lithuania), and ligated with adaptors. The ligation product, termed primary template, was used directly for pre-amplification. The pre-amplification product, termed secondary template, was then used for selective amplification. All the 128 possible selective primer combinations with 8 Apo I primers and 16 Mse I primers as reported previously (Xiao et al., 2009) were applied in screening DE-TDFs affected by the tapping treatment in mature virgin Hevea trees.
With the expression level of the TDFs at the first tapping as a reference, the DE-TDFs were identified for the tapping treatment and isolated from the polyacrylamide gel. The DE-TDFs was scraped from the polyacrylamide gel with a surgical blade, put into a sterile PCR tube containing 30 μl .1× TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH8.0), heated at 95°C for 15 min, kept overnight at 4°C, and centrifuged at 10,000 g for 5 min to collect the dissolved DNA solution for PCR amplification. A total of 25 μl PCR reaction mixture includes: 2 μl DNA solution, 2.5 μl 10× PCR buffer (plus Mg2+), 1.2 μl 2.5 mM dNTPs Mix, .5 μl 50 ng/μl MseⅠselective primer, .5 μl 50 ng/μl ApoⅠselective primer, .2 μl 5U/μl Taq DNA Polymerase (Takara, Dalian, China) and 18.1 μl ddH2O. The PCR amplification procedure is the same as the pre-amplification in cDNA-AFLP analysis (Xiao et al., 2009). PCR products were fractionated by 1.2% agarose gel electrophoresis, and the target band was sliced and purified using AxyPrepTM DNA Gel Extraction Kit (AxyGen, shanghai, China).
The purified DE-TDFs were ligated with the T-vectors using the pMD18-T Vector Kit (Takara, Dalian, China) in accordance with the manufacture’s manual. The ligation mixture was used to transform E. coli JM109 competent cells and the transformants were sent to BGI Genomics Co., Ltd. (Shenzhen, China) for sequencing.
The first strand of cDNA was synthesized by reverse transcriptase kit (RevertAidTM First Strand cDNA Synthesis Kit-K1622, Thermo Fisher Scientific, Vilnius, Lithuania), and then diluted ten times as the template for sqRT-PCR with 18S rRNA used as the reference (Xiao et al., 2009). The annealing temperatures for the PCR amplification were set according to the primers designed for each specific gene. The PCR cycles were controlled to keep the amplification under the plateau phase. All of the amplified fragments were cloned, and were sequenced for target confirmation. The amount of cDNA samples used in sqRT-PCR was adjusted to be the same for the five tappings based on the level of 18S rRNA expression, because it is conserved for sqRT-PCR in most of the plant species (Morris and Davila, 1996).
The expression pattern of candidate genes was detected by qRT-PCR. The first strand of cDNA was diluted 20 times as the template for qRT-PCR with HbYLS8 as the reference gene which was recommended in our previous study as a suitable internal standard gene in qRT-PCR analysis for the tapping treatment in rubber tree (Li et al., 2011). The PCR reaction mixture includes: 2 μl template, .3 μl each for 10 μM forward and reverse primers, 10 μl 2×SYBR® Premix Ex TaqTMⅡ (Takara, Dalian, China) and 7.4 μl ddH2O. Roche’s LightCycler 2.0 system was used for qRT-PCR analysis with the program as follows: 95°C 30 s; 94°C 5 s, 60°C 20 s, 72°C 20 s, 45cycles. Three technical replicates were analyzed for each of the three biological samples. All the cycle threshold (Ct) values from one gene were determined at the same threshold fluorescence value of .2 using the 2−ΔΔCT method (Livak and Schmittgen, 2001). The primers of target and reference genes were listed in Supplementary Table S6. Student’s t-test was performed using Statistical analysis was performed using Student’s t-test.
Three batches of five rubber trees were selected for latex collection and protein extraction. After tapping, sixty drops of latex were discarded and ∼30 ml latex per tree was then collected in a 50 ml centrifuge tube placed in an ice bath. The collected latex was immediately taken back to the laboratory, and centrifuged at 22, 000 g 4°C for 2 h to acquire the middle aqueous layer, latex cytosol (C-serum). The C-serum proteins were extracted by TCA/acetone-diethyl method (Saravanan and Rose, 2004), freeze-dried, and stored at −80°C. Protein dissolution was performed by incubating for 1 h at 4°C with ultrasonic treatment for 5 min (2s on, 3s off, 15W), with a proportion of 1 mg freeze-dried protein powder added into 10 μl lysis buffer (7M urea, 2M thiourea, 4% CHAPS, 2 mM TBP, 65 mM DTT, .2% w/v IPG buffer). The insoluble precipitate was removed by centrifugation at 20°C 15, 000 g for 10 min, and the supernatant was used for protein separation by two-dimensional electrophoresis (2-DE).
The protein content was determined according to Bradford (1976). For each of the first five tappings conducted on re-tapped resting Hevea trees, 1 mg of latex C-serum proteins were dissolved in hydration loading buffer [7M urea, 4% CHAPS, 65 mM DTT, .2% Bio-Lyte pH3/10 or pH5/8 ampholyte, .002% (W/V) bromophenol blue] to a final volume of 300 μl. The protein mixture was centrifuged at 20°C 15, 000 g for 20 min to remove bubbles and undissolved precipitates, and then loaded into the IPG strips (Bio-Rad, USA) as described in Sanchez et al. (1997). The strips were then subjected to isoelectric focusing and SDS-PAGE, in an Ettan IPGphor and DALT system according to the manufacturer’s 2-DE manual (GE Healthcare). Three biological replicates for each tapping were conducted.
The gels were stained by a modified CBB R250 staining protocol (Wang et al., 2007), and images were scanned with a Bio-Rad GS-800TM calibrated optical density scanner at 600 dpi. Images were then analyzed with Image Master 2D Platinum 5.0 (GE Healthcare) to integrate three replicates of the protein spots for each tapping. With the expression of the protein spot at the first tapping as a reference, the DE-protein spots (|ratio| ≥3.0, T value ≤.05)) were isolated from the SDS-PAGE gel, and sent to BPI Co., Ltd. (Beijing, China) for mass spectrometry identification on an ultraflex MALDI-TOF/TOF mass spectrometer instrument (Bruker Daltonics, Billerica, MA, United States).
For DE-TDFs analysis, sequences of vectors and adaptors were first trimmed off by using the VecScreen program on the NCBI website (https://www.ncbi.nlm.nih.gov/tools/VecScreen). Then, the clean DE-TDF sequences were subjected for homology analysis to publicly available GenBank non-redundant sequences databases (http://www.ncbi.nlm.nih.gov) using the BLASTX program. Also, the Gene Ontology (http://amigo1.geneontology.org/cgi-bin/amigo/go.cgi) database was used to investigate the molecular function of each DE-TDF in the cell, which was used as the basis for functional classification.
At least three individual virgin or resting Hevea trees, with one tree serving as one biological replicate, were exploited. Data were analyzed by one-way ANOVA (SAS6.11) for comparison of TDF expressions across the five tappings. Student’s t-test was performed using the software embedded in Excel 2007 for comparison of TDF or protein spot abundance between the first tapping and any of the other four tappings. Differences were accepted as significant at p < .05 or .01.
The original contributions presented in the study are publicly available in NCBI under accession numbers MZ935745-MZ936248.
CT designed the research; YF, JQ, XX, HL, JL, and YH performed the research; YF, XX, JQ, CT, JY, YZ, SZ, and JT analyzed the data; YF and CT wrote the manuscript. The authors have read and approved the manuscript.
The research was supported partially by the National Natural Science Foundation of China (Nos. 31825007; 31770711) and the Starting Research Fund of Hainan University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.756270/full#supplementary-material
2-DE, Two dimensional electrophoresis; AFLP, Amplified fragment length polymorphism; ATP, Adenosine triphosphate; CoA, Coenzyme A; DE-proteins, Differentially expressed proteins; DE-TDF, Differentially expressed transcript derived fragment; DNA, Deoxyribonucleic acid; dNTP, Deoxy-ribonucleoside triphosphate; EDTA, Ethylene diamine tetraacetic acid; IPG, Immobilized pH gradient; NADPH, Reduced form of nicotinamide-adenine dinucleotide phosphate; NIN, Alkaline/neutral invertase; NR, Natural rubber; PCR, Polymerase chain reaction; qRT-PCR, Quantitative reverse transcription PCR; RB, Rubber biosynthesis; REF, Rubber elongation factor; sqRT-PCR, Semi-quantitative reverse transcription PCR; SRPP, Small rubber particle protein; SUT, Sucrose transporter; TDF, Transcript derived fragment.
Adiwilaga, K., and Kush, A. (1996). Cloning and Characterization of cDNA Encoding Farnesyl Diphosphate Synthase from Rubber Tree (Hevea Brasiliensis). Plant Mol. Biol. 30, 935–946. doi:10.1007/bf00020805
Akihiro, T., Umezawa, T., Ueki, C., Lobna, B. M., Mizuno, K., Ohta, M., et al. (2006). Genome Wide cDNA-AFLP Analysis of Genes Rapidly Induced by Combined Sucrose and ABA Treatment in rice Cultured Cells. FEBS Lett. 580, 5947–5952. doi:10.1016/j.febslet.2006.09.065
Amalou, Z., Bangratz, J., and Chrestin, H. (1992). Ethrel (Ethylene Releaser)-Induced Increases in the Adenylate Pool and Transtonoplast ΔpH within Hevea Latex Cells. Plant Physiol. 98, 1270–1276. doi:10.1104/pp.98.4.1270
Bachem, C. W. B., van der Hoeven, R. S., de Bruijn, S. M., Vreugdenhil, D., Zabeau, M., and Visser, R. G. F. (1996). Visualization of Differential Gene Expression Using a Novel Method of RNA Fingerprinting Based on AFLP: Analysis of Gene Expression during Potato Tuber Development. Plant J. 9, 745–753. doi:10.1046/j.1365-313x.1996.9050745.x
Bevan, M., Bancroft, I., Bent, E., Love, K., Goodman, H., Dean, C., et al. (1998). Analysis of 1.9 Mb of Contiguous Sequence from Chromosome 4 of Arabidopsis thaliana. Nature 391, 485–488. doi:10.1038/35140
Bradford, M. M. (1976). A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 72, 248–254. doi:10.1016/0003-2697(76)90527-3
Chantuma, P., Lacointe, A., Kasemsap, P., Thanisawanyangkura, S., Gohet, E., Clément, A., et al. (2009). Carbohydrate Storage in wood and Bark of Rubber Trees Submitted to Different Level of C Demand Induced by Latex Tapping. Tree Physiol. 29, 1021–1031. doi:10.1093/treephys/tpp043
Chow, K. S., Wan, K. L., Isa, M. N. M., Bahari, A., Tan, S. H., Harikrishna, K., et al. (2007). Insights into Rubber Biosynthesis from Transcriptome Analysis of Hevea Brasiliensis Latex. J. Exp. Bot. 58, 2429–2440. doi:10.1093/jxb/erm093
Chrestin, H., Gidrol, X., and Kush, A. (1997). Towards a Latex Molecular Diagnostic of Yield Potential and the Genetic Engineering of the Rubber Tree. Euphytica 96, 77–82. doi:10.1023/a:1002950300536
Cornish, K. (1993). The Separate Roles of Plant Cis and Trans Prenyl Transferases in Cis-1,4-Polyisoprene Biosynthesis. Eur. J. Biochem. 218, 267–271. doi:10.1111/j.1432-1033.1993.tb18374.x
d’Auzac, J., and Jacob, J. L. (1969). Regulation of Glycolysis in the Latex of Hevea Brasiliensis. J. Rubber Res. Inst. Malaya. 21, 417.
d'Auzac, J., Jacob, J., Prévôt, J., Clément, A., Gallois, R., Crestin, H., et al. (1997). “The Regulation of Cis-Polyisoprene Production (Natural Rubber) from Hevea Brasiliensis,” in Recent Research Developments in Plant Physiology. Editor S. G. Pandalai (Kerala India: Research Signpost), 1, 273–332.
Dennis, M. S., Henzel, W. J., Bell, J., Kohr, W., and Light, D. R. (1989). Amino Acid Sequence of Rubber Elongation Factor Protein Associated with Rubber Particles in Hevea Latex. J. Biol. Chem. 264, 18618–18626. doi:10.1016/s0021-9258(18)51511-8
Duan, C., Rio, M., Leclercq, J., Bonnot, F., Oliver, G., and Montoro, P. (2010). Gene Expression Pattern in Response to Wounding, Methyl Jasmonate and Ethylene in the Bark of Hevea Brasiliensis. Tree Physiol. 30, 1349–1359. doi:10.1093/treephys/tpq066
Duangngam, O., Desalme, D., Thaler, P., Kasemsap, P., Sathornkich, J., Satakhun, D., et al. (2020). In Situ 13CO2 Labelling of Rubber Trees Reveals a Seasonal Shift in the Contribution of the Carbon Sources Involved in Latex Regeneration. J. Exp. Bot. 71, 2028–2039. doi:10.1093/jxb/erz551
Fatemi, F., Hashemi-Petroudi, S. H., Nematzadeh, G., Askari, H., and Abdollahi, M. R. (2019). Exploiting Differential Gene Expression to Discover Ionic and Osmotic-Associated Transcripts in the Halophyte Grass Aeluropus Littoralis. Biol. Proced. Online 21, 14. doi:10.1186/s12575-019-0103-3
Ganeshan, S., Sharma, P., Young, L., Kumar, A., Fowler, D. B., and Chibbar, R. N. (2011). Contrasting cDNA-AFLP Profiles between crown and Leaf Tissues of Cold-Acclimated Wheat Plants Indicate Differing Regulatory Circuitries for Low Temperature Tolerance. Plant Mol. Biol. 75, 379–398. doi:10.1007/s11103-011-9734-8
Gohet, E. (1996). La production de latex par Hevea brasiliensis, relations avec la croissance. Influence de différents facteurs: origine clonale, stimulation hormonale, réserves hydrocarbonées. PhD Thesis. Montpellier: Université Montpellier II.
Hao, B. Z., and Wu, J. L. (1982). Effects of Wound (Tapping) on Laticifer Differentiation in the Hevea Brasiliensis. Acta Bot. Sin. 24, 388–391.
Hao, B., and Wu, J. L. (2000). Laticifer Differentiation in Hevea Brasiliensis: Induction by Exogenous Jasmonic Acid and Linolenic Acid. Ann. Bot. 85, 37–43. doi:10.1006/anbo.1999.0995
Hao, B. Z., Wu, J. L., Meng, C. X., Gao, Z. Q., and Tan, H. Y. (2004). Laticifer Wound Plugging in Hevea Brasiliensis: the Role of a Protein-Network with Rubber Particle Aggregations in Stopping Latex Flow and Protecting Wounded Laticifers. J. Rubber Res. 7, 281–299.
Huang, Z. D. (2001). Challenges and Development Strategies of China's Natural Rubber. Chin. J. Eng. Sci. 3, 28–32.
Humphery-Smith, I., Cordwell, S. J., and Blackstock, W. P. (1997). Proteome Research: Complementarity and Limitations with Respect to the RNA and DNA Worlds. Electrophoresis 18 (8), 1217–1242. doi:10.1002/elps.1150180804
Ko, J. H., Chow, K. S., and Han, K. H. (2003). Transcriptome analysis reveals novel features of the molecular events occurring in the laticifers of Hevea brasiliensis (para rubber tree). Plant Mol. Biol. 53, 479–492. doi:10.1023/b:plan.0000019119.66643.5d
Lacote, R., Gabla, O., Obouayeba, S., Eschbach, J. M., Rivano, F., Dian, K., et al. (2010). Long-term Effect of Ethylene Stimulation on the Yield of Rubber Trees Is Linked to Latex Cell Biochemistry. Field Crops Res. 115, 94–98. doi:10.1016/j.fcr.2009.10.007
Leymarie, J., Bruneaux, E., Gibot-Leclerc, S., and Corbineau, F. (2007). Identification of Transcripts Potentially Involved in Barley Seed Germination and Dormancy Using cDNA-AFLP. J. Exp. Bot. 58, 425–437. doi:10.1093/jxb/erl211
Li, H., Qin, Y., Xiao, X., and Tang, C. (2011). Screening of Valid Reference Genes for Real-Time RT-PCR Data Normalization in Hevea Brasiliensis and Expression Validation of a Sucrose Transporter Gene HbSUT3. Plant Sci. 181, 132–139. doi:10.1016/j.plantsci.2011.04.014
Liu, S., Lan, J., Zhou, B., Qin, Y., Zhou, Y., Xiao, X., et al. (2015). H b NIN 2, a cytosolic alkaline/neutral‐invertase, is responsible for sucrose catabolism in rubber‐producing laticifers of H evea brasiliensis (para rubber tree). New Phytol. 206, 709–725. doi:10.1111/nph.13257
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Morris, D. L., and Davila, J. C. (1996). Analysis of Rat Cytochrome P450 Isoenzyme Expression Using Semi-quantitative Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR). Biochem. Pharmacol. 52, 781–792. doi:10.1016/0006-2952(96)00390-5
Oh, S. K., Kang, H., Shin, D. H., Yang, J., Chow, K. S., Yeang, H. Y., et al. (1999). Isolation, Characterization, and Functional Analysis of a Novel cDNA Clone Encoding a Small Rubber Particle Protein from Hevea Brasiliensis. J. Biol. Chem. 274, 17132–17138. doi:10.1074/jbc.274.24.17132
Pakianathan, S. W., Tata, S. J., Chon, L. F., and Sethuraj, M. R. (1992). Certain Aspects of Physiology and Biochemistry of Latex Production. Dev. Crop Sci. 23, 298–323. doi:10.1016/b978-0-444-88329-2.50020-9
Pirrello, J., Leclercq, J., Dessailly, F., Rio, M., Piyatrakul, P., Kuswanhadi, K., et al. (2014). Transcriptional and post-transcriptional Regulation of the Jasmonate Signalling Pathway in Response to Abiotic and Harvesting Stress in Hevea Brasiliensis. BMC Plant Biol. 14, 341. doi:10.1186/s12870-014-0341-0
Priya, P., Venkatachalam, P., and Thulaseedharan, A. (2007). Differential Expression Pattern of Rubber Elongation Factor (REF) mRNA Transcripts from High and Low Yielding Clones of Rubber Tree (Hevea Brasiliensis Muell. Arg.). Plant Cel Rep. 26, 1833–1838. doi:10.1007/s00299-007-0402-z
Qi, J. Y. (2011). Large-scale screening by proteomic approaches of candidate latex-regeneration-related proteins in para rubber trees (Hevea brasiliensis). Master's Thesis. China: Hainan University.
Sae-Lim, P., Naktang, C., Yoocha, T., Nirapathpongporn, K., Viboonjun, U., Kongsawadworakul, P., et al. (2019). Unraveling Vascular Development-Related Genes in Laticifer-Containing Tissue of Rubber Tree by High-Throughput Transcriptome Sequencing. Curr. Plant Biol. 19, 100112. doi:10.1016/j.cpb.2019.100112
Sanchez, J. C., Rouge, V., Pisteur, M., Ravier, F., Tonella, L., Moosmayer, M., et al. (1997). Improved and Simplified In-Gel Sample Application Using Reswelling of Dry Immobilized pH Gradients. Electrophoresis 18, 324–327. doi:10.1002/elps.1150180305
Saravanan, R. S., and Rose, J. K. C. (2004). A Critical Evaluation of Sample Extraction Techniques for Enhanced Proteomic Analysis of Recalcitrant Plant Tissues. Proteomics 4 (9), 2522–2532. doi:10.1002/pmic.200300789
Sharples, A. (1918). The Laticiferous System of Hevea Brasiliensis and its Protective Function. Ann. Bot. os-32, 247–251. doi:10.1093/oxfordjournals.aob.a089674
Silpi, U., Lacointe, A., Kasempsap, P., Thanysawanyangkura, S., Chantuma, P., Gohet, E., et al. (2007). Carbohydrate Reserves as a Competing Sink: Evidence from Tapping Rubber Trees. Tree Physiol. 27, 881–889. doi:10.1093/treephys/27.6.881
Singh, A. P., Wi, S. G., Chung, G. C., Kim, Y. S., and Kang, H. (2003). The Micromorphology and Protein Characterization of Rubber Particles in Ficus Carica, Ficus Benghalensis and Hevea Brasiliensis. J. Exp. Bot. 54, 985–992. doi:10.1093/jxb/erg107
Tang, C., Qi, J., Li, H., Zhang, C., and Wang, Y. (2007). A convenient and efficient protocol for isolating high-quality RNA from latex of Hevea brasiliensis (para rubber tree). J. Biochem. Biophys. Methods 70, 749–754. doi:10.1016/j.jbbm.2007.04.002
Tang, C., Huang, D., Yang, J., Liu, S., Sakr, S., Li, H., et al. (2010). The sucrose transporter HbSUT3 plays an active role in sucrose loading to laticifer and rubber productivity in exploited trees of Hevea brasiliensis (para rubber tree). Plant Cel Environ. 33, 1708–1720. doi:10.1111/j.1365-3040.2010.02175.x
Tang, C., Xiao, X., Li, H., Fan, Y., Yang, J., Qi, J., et al. (2013). Comparative Analysis of Latex Transcriptome Reveals Putative Molecular Mechanisms Underlying Super Productivity of Hevea Brasiliensis. PLoS One 8, e75307. doi:10.1371/journal.pone.0075307
Tang, C., Yang, M., Fang, Y., Luo, Y., Gao, S., Xiao, X., et al. (2016). The Rubber Tree Genome Reveals New Insights into Rubber Production and Species Adaptation. Nat. Plants 2, 16073. doi:10.1038/nplants.2016.73
Taniguchi, Y., Choi, P. J., Li, G. W., Chen, H., Babu, M., Hearn, J., et al. (2010). Quantifying E. coli Proteome and Transcriptome with Single-Molecule Sensitivity in Single Cells. Science 329, 533–538. doi:10.1126/science.1188308
Tungngoen, K., Viboonjun, U., Kongsawadworakul, P., Katsuhara, M., Julien, J. L., Sakr, S., et al. (2011). Hormonal Treatment of the Bark of Rubber Trees (Hevea Brasiliensis) Increases Latex Yield through Latex Dilution in Relation with the Differential Expression of Two Aquaporin Genes. J. Plant Physiol. 168, 253–262. doi:10.1016/j.jplph.2010.06.009
Tupý, J. (1969). Stimulatory Effects of 2,4-dichlorophenoxyacetic Acid and of 1-naphthylacetic Acid on Sucrose Level, Invertase Activity and Sucrose Utilization in the Latex ofHevea Brasiliensis. Planta 88, 144–153. doi:10.1007/BF01391120
Tupý, J. (1985). Some Aspects of Sucrose Transport and Utilization in Latex Producing Bark ofHevea Brasiliensis Muel. Arg. Biol. Plant 27, 51–64. doi:10.1007/bf02894634
Tupý, J. (1989). “Sucrose Supply and Utilization for Latex Production,” in Physiology of Rubber Tree Latex. Editors J. d'Auzac, J. L. Jacob, and H. Chrestin (Boca Raton, FL, USA: CRC Press), 179–199.
Tupý, J., and Resing, W. L. (1969). Substrate and Metabolism of Carbon Dioxide Formation in Hevea Latex, In Vitro. J. Rubber Res. Inst. Malaya. 21, 456.
Van Beilen, J. B., and Poirier, Y. (2007). Establishment of New Crops for the Production of Natural Rubber. Trends Biotechnol. 25, 522–529. doi:10.1016/j.tibtech.2007.08.009
Vogel, C., and Marcotte, E. M. (2012). Insights into the Regulation of Protein Abundance from Proteomic and Transcriptomic Analyses. Nat. Rev. Genet. 13, 227–232. doi:10.1038/nrg3185
Vuylsteke, M., Peleman, J. D., and van Eijk, M. J. (2007). AFLP-based Transcript Profiling (cDNA-AFLP) for Genome-wide Expression Analysis. Nat. Protoc. 2, 1399–1413. doi:10.1038/nprot.2007.174
Walley, J. W., Shen, Z., Sartor, R., Wu, K. J., Osborn, J., Smith, L. G., et al. (2013). Reconstruction of Protein Networks from an Atlas of maize Seed Proteotypes. Proc. Natl. Acad. Sci. U S A. 110, E4808–E4817. doi:10.1073/pnas.1319113110
Wang, X., Li, X., and Li, Y. (2007). A Modified Coomassie Brilliant Blue Staining Method at Nanogram Sensitivity Compatible with Proteomic Analysis. Biotechnol. Lett. 29, 1599–1603. doi:10.1007/s10529-007-9425-3
Xiao, X., Li, H., and Tang, C. (2009). A silver-staining cDNA-AFLP protocol suitable for transcript profiling in the latex of Hevea brasiliensis (para rubber tree). Mol. Biotechnol. 42, 91–99. doi:10.1007/s12033-008-9139-3
Xiao, D., Liu, S. T., Wei, Y. P., Zhou, D. Y., Hou, X. L., Li, Y., et al. (2016). cDNA-AFLP Analysis Reveals Differential Gene Expression in Incompatible Interaction between Infected Non-heading Chinese Cabbage and Hyaloperonospora Parasitica. Hortic. Res. 3, 16034. doi:10.1038/hortres.2016.34
Zeng, R. Z., Duan, C. F., Li, X. Y., Tian, W. M., and Nie, Z. Y. (2009). Vacuolar-type Inorganic Pyrophosphatase Located on the Rubber Particle in the Latex Is an Essential Enzyme in Regulation of the Rubber Biosynthesis in Hevea Brasiliensis. Plant Sci. 176, 602–607. doi:10.1016/j.plantsci.2009.01.009
Keywords: Hevea brasiliensis, cDNA-AFLP, 2-DE, gene and protein expression, harvesting, latex production
Citation: Fan Y, Qi J, Xiao X, Li H, Lan J, Huang Y, Yang J, Zhang Y, Zhang S, Tao J and Tang C (2022) Transcript and Protein Profiling Provides Insights Into the Molecular Mechanisms of Harvesting-Induced Latex Production in Rubber Tree. Front. Genet. 13:756270. doi: 10.3389/fgene.2022.756270
Received: 10 August 2021; Accepted: 17 January 2022;
Published: 10 February 2022.
Edited by:
Lidan Sun, Beijing Forestry University, ChinaReviewed by:
Ertugrul Filiz, Duzce University, TurkeyCopyright © 2022 Fan, Qi, Xiao, Li, Lan, Huang, Yang, Zhang, Zhang, Tao and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chaorong Tang, Y2hhb3Jvbmd0YW5nQDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.