- 1Chinese Perch Research Center, College of Fisheries, Huazhong Agricultural University, Wuhan, China
- 2Engineering Research Center of Green Development for Conventional Aquatic Biological Industry in the Yangtze River Economic Belt, Ministry of Education, Wuhan, China
- 3Department of Ecophysiology and Aquaculture, Leibniz-Institute of Freshwater Ecology and Inland Fisheries, Berlin, Germany
Mandarin fish (Siniperca chuatsi) is one of the most economically important fish in China. However, it has the peculiar feeding habit that it feeds solely on live prey fish since first-feeding, while refuses dead prey fish or artificial diets. After the specific training procedure, partial individuals could accept dead prey fish and artificial diets. The genetic basis of individual difference in artificial diet feeding habit is still unknown. In the present study, the resequencing was performed between 10 individuals which could be domesticated to accept artificial diets and 10 individuals which could not. Through the selective sweep analysis based on heterozygosity (Hp) and population differentiation coefficient (Fst), 57 candidate windows were identified as the putative selected regions for feeding habit domestication of mandarin fish, involved in 149 genes. These genes were related to memory, vision and olfaction function, which could be potential targets of molecular marker assistant breeding of artificial diet feeding trait. Beside of the DNA sequence, we also explored the potential role of DNA methylation in feeding habit domestication in mandarin fish. Whole-genome bisulfite sequencing was performed between the individuals which could be domesticated to accept artificial diets and those could not. 5,976 differentially methylated regions were identified, referring to 3,522 genes, such as the genes involved in cAMP signaling pathway. The DNA methylation changes of these genes might contribute to the adaption of artificial diets in mandarin fish. In conclusion, the putative selected regions and the differentially methylated regions were identified in the whole genome, providing new insights into the feeding habit domestication from live prey fish to artificial diets in mandarin fish. And the involved genes were identified as the candidate genes for molecular breeding of artificial diet utilization in mandarin fish.
Introduction
Mandarin fish (Siniperca chuatsi) is one of the most economically important fish in China. The annual yield has been over 300,000 tonnes, and the output value has been more than three billion US dollars since 2015 (FAO). However, due to its special feeding habit of live prey fish (Liang et al., 2001; Liang et al., 2008), the stable supply of palatable live prey fish resulted in high farming cost for mandarin fish. After the specific training procedure (Liang et al., 2001), some individuals could accept dead prey fish and artificial diets, but there are still some individuals could not. The genetic basis of individual difference in artificial diet feeding habit is still unknown.
In our previous studies, we have identified several single nucleotide polymorphisms (SNPs) in functional genes through PCR product sequencing. One SNP was identified in the appetite regulatory gene npy (Sun et al., 2015) and two SNPs were identified in the digestive enzyme gene pepsinogen (pep) (Yi et al., 2013) related to dead prey fish feeding trait, two SNPs were identified in the learning and memory gene protein phosphatase 1 (pp1) related to artificial diet feeding trait (Cheng et al., 2015). Whole-genome resequencing has been applied in fish to identify candidate genes associated with traits of biological and commercial interest, such as genes associated with growth, early development and immunity traits in Nile tilapia (Cádiz et al., 2020), genes associated with body color and fin morphology in Siamese fighting fish (Kwon et al., 2022). However, the large-scale screening of selected genes and molecular markers associated with feeding habit domestication trait from the whole genome is in urgent need to research.
DNA methylation as one of the most studied epigenetic modifications, has been reported to play important roles in fish, including feeding (Cai et al., 2018; Dou et al., 2018), metabolism (Cai et al., 2020; Kumkhong et al., 2020; Zhu et al., 2020), growth (Zhong et al., 2014; Si et al., 2016; Huang et al., 2018), sex determination and gonadal differentiation (Navarro-Martín et al., 2011; Shao et al., 2014; He et al., 2020a), and environmental adaptation (Morán et al., 2013; Pierron et al., 2014; Veron et al., 2018). During the feeding habit transition from carnivory to herbivory in grass carp, the DNA methylation level of umami receptor t1r1 increased and the mRNA expression level decreased (Cai et al., 2018). In our previous study, we found that DNA methylation could regulate the mRNA expression of t1r1, further contributed to the feeding habit domestication from live prey fish to dead prey fish in mandarin fish (Dou et al., 2018). Whole-genome DNA methylation patterns have been characterized in fish, and differential methylation analyses identified key genes related to feeding, such as genes involved in diet response in Nile tilapia (Podgorniak et al., 2022), and genes involved in foraging in grass carp (Li et al., 2021). However, little is known about the whole-genome DNA methylation assay related to the feeding habit domestication in mandarin fish.
Chromosome-level reference genome assembly of mandarin fish (He et al., 2020b) allowed us to identify genome-wide variants and DNA methylation changes between mandarin fish with different performance in accepting artificial diets. In the present study, the whole-genome resequencing and bisulfite sequencing were performed between the individuals which could be domesticated to accept artificial diets and those could not. The putative selected regions and the differentially methylated regions were identified in the whole genome, and the involved genes were identified as the candidate genes for molecular breeding of artificial diet utilization in mandarin fish.
Materials and methods
Fish
Mandarin fish (Siniperca chuatsi) (Huakang No. 1) used in the present study were obtained from the Chinese Perch Research Center of Huazhong Agricultural University (Wuhan, China), and kept in tanks with continuous system of water filtration and aeration at constant temperature (25°C ± 1 C). Mandarin fish (50 ± 5 g) were trained to accept artificial diets according to our previously published training procedure (Liang et al., 2001; He et al., 2021). The composition of artificial diets was reported in our previous study (He et al., 2021). This study was approved by the Institutional Animal Care and Use Ethics Committee of Huazhong Agricultural University (Wuhan, China) (HZAUFI-2020-0020).
Whole-genome resequencing and mapping
10 individuals which could be domesticated to accept artificial diets and 10 individuals which could not were sampled. To minimize possible suffering, the fish were anesthetized with MS-222 (200 mg/l) (Redmond, WA, United States) until loss of equilibrium. Pelvic fins were dissected, and genomic DNA was extracted following the standard phenol-chloroform extraction procedure. DNA integrity was evaluated by agarose gel electrophoresis. DNA purity was checked using the NanoPhotometer® spectrophotometer (IMPLEN, CA, United States). DNA concentration was determined by Qubit® 2.0 Flurometer (Life Technologies, CA, United States) with a Qubit® DNA Assay Kit. Paired-end sequencing library was constructed for each of the 20 samples according to the manufacturer’s instructions (Illumina Inc., San Diego, CA, United States). The libraries were sequenced on Illumina HiSeq X Ten platform (PE150). Library construction and sequencing were performed by Tianjin Novogene Bioinformatic Technology Co., Ltd. (Tianjin, China).
The raw data was filtered to remove the adapters, reads containing more than 10% unknown bases, and reads with more than 50% low-quality bases (Phred score ≤5). Then the clean reads were aligned against our mandarin fish reference genome (version: sinChu7) (He et al., 2020b) using Burrows-Wheeler Aligner software (bwa mem -t 4 -k 32 -M) (Li and Durbin, 2009). The output SAM files were converted into BAM files, then sorted according to genome coordinates. Subsequently, PCR duplicates were removed using SAMtools software (Li et al., 2009).
Single nucleotide polymorphism calling and annotation
SNP calling was performed using SAMtools software (Li et al., 2009). SNPs were filtered by Perl script with the parameters: read depth ≥4, missing rate <0.1, minor allele frequency (MAF) ≥ 0.05, and genotype quality score ≥10. SNPs were annotated using ANNOVAR software (Wang et al., 2010) based on the GFF3 file of our mandarin fish reference genome. Synonymous and non-synonymous variants were predicted.
Selective sweep analysis
A sliding window method (the window size of 100 kb and the step size of 50 kb) was applied for the selective sweep analysis. Heterozygosity (Hp) and population differentiation coefficient (Fst) were calculated. Windows with top 10% Hp values and top 10% Fst values were considered to be candidate selected regions. To know the biological function of genes within candidate selected regions, Gene Ontology (GO) (http://geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (https://www.genome.jp/kegg/pathway.html) enrichment analyses were performed.
Whole-genome bisulfite sequencing (WGBS)
According to our previous study, the mandarin fish did not eat artificial diets during domestication processes were named Group W (n = 56), and the mandarin fish ate artificial diets were named Group X (n = 24) (He et al., 2021). Three fish were randomly selected from each group, anesthetized with MS-222 (200 mg/l) and euthanized. Genomic DNA was isolated from liver tissue with a standard phenol-chloroform method. DNA integrity and purity were evaluated by agarose gel electrophoresis. DNA concentration was determined by Qubit® 2.0 Flurometer with a Qubit® DNA Assay Kit (Life Technologies). DNA was randomly fragmented by sonication. The fragments with an average size of 200–400 bp were purified according to the manufacturer’s instructions of Agencort AMPure XP-Medium Kit (Beckman Coulter, United States), followed by DNA-end repair, 3′-dA overhang and ligation of methylated sequencing adaptors. Then bisulfite conversion was performed with ZYMO EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, United States). After desalting, size selection, PCR amplification and size selection again, qualified libraries were sequenced on Illumina HiSeq X Ten platform (PE150). Library construction and sequencing were performed by Beijing Genomics Institute (BGI, Wuhan, China).
Identification of differentially methylated regions and differentially methylated genes
WGBS reads mapping to our mandarin fish reference genome (version: sinChu7) (He et al., 2020b) (bismark --bowtie2), deduplication (deduplicate_bismark) and methylation calling (bismark_methylation_extractor --ignore 10 --ignore_r2 10) were performed with Bismark (Krueger and Andrews, 2011). Data were filtered so that only sites with read depth ≥10 were retained. Sites that were in the 99.9th percentile of coverage were also removed from the analysis to account for potential PCR bias. Differentially methylated regions were identified by methylKit (tiling windows, win. size = 200 bp, step. size = 200 bp) (Akalin et al., 2012), with the methylation difference larger than 25% and q-value <0.01. To know the biological function of differentially methylated genes, GO and KEGG pathway enrichment analyses were performed.
Results
Whole-genome resequencing and mapping
Resequencing was performed between 10 individuals which could be domesticated to accept artificial diets and 10 individuals which could not. A total of 169.45 Gb raw data was generated. After filtering, 169.29 Gb clean data was obtained. The effective rates (the ratio of clean data to raw data) were 99.81%–99.94%. The Q20 (sequencing error rate <1%) and Q30 (sequencing error rate <0.1%) were 97.03%–97.84% and 91.78%–93.64%, respectively. The GC contents were 40.64%–41.77% (Supplementary Table S1). The average sequencing depth was 8.57–10.19×. Of the clean reads, 99.65%–99.79% were mapped to our mandarin fish reference genome (version: sinChu7). 97.75% reference genome bases had at least 1 × coverage, and 92.69% had at least 4 × coverage (Supplementary Table S2).
Variation discovery
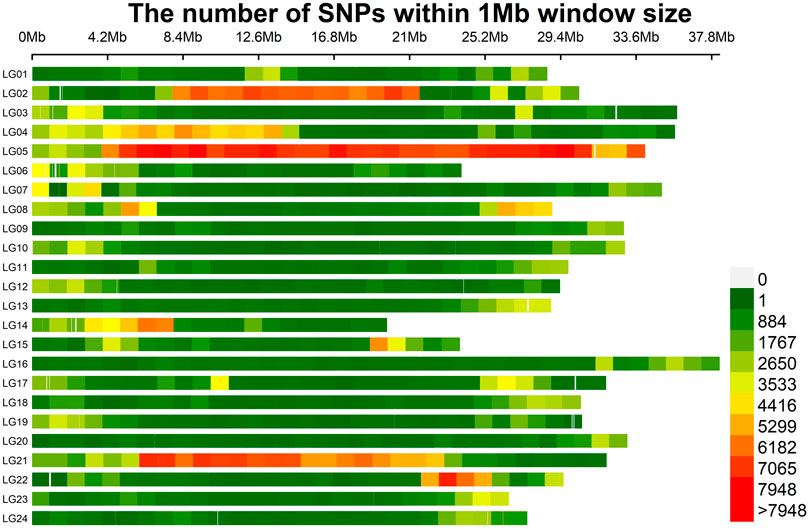
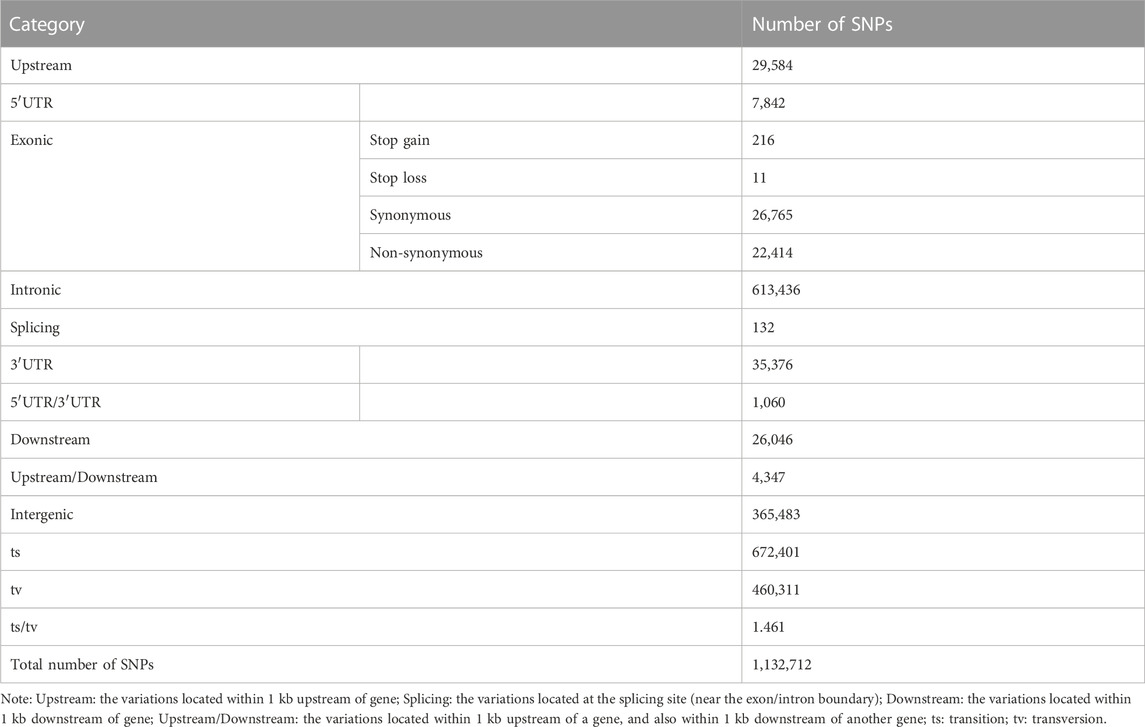
A total of 1,132,712 SNPs were identified, of which 1,119,169 (98.80%) were located on 24 assembled chromosomes of mandarin fish, with the highest density on LG05 and LG21 (Figure 1). 93,684 (8.27%) SNPs were in exonic regions, 613,568 (54.17%) SNPs were in intronic regions, and 425,460 (37.56%) SNPs were in intergenic regions (Table 1). In exonic regions, 216 stop-gain SNPs, 11 stop-loss SNPs, 26,765 synonymous SNPs and 22,414 non-synonymous SNPs were identified, respectively. The ratio of transition to transversion (ts/tv) was 1.461.
Selective sweep analysis
To identify selected regions for feeding habit domestication, selective sweep analysis based on heterozygosity (Hp) and population differentiation coefficient (Fst) was performed. A total of 15,023 windows were analyzed. The Fst values of 751 windows were above the threshold of 0.220615 (top 5% of Fst values) (Figure 2). 57 candidate windows were identified as putative selective regions for feeding habit domestication, involved in 149 genes (Figure 3). GO enrichment analysis revealed that these genes were significantly enriched in 162 GO items, including 88 GO terms in biological process, 11 GO terms in cellular component, and 63 GO terms in molecular function (Figure 4A, Supplementary Table S3). KEGG enrichment analysis revealed that these genes were significantly enriched in three KEGG pathways, including metabolic pathways, MAPK signaling pathway, and fatty acid metabolism (Figure 4B, Supplementary Table S4).
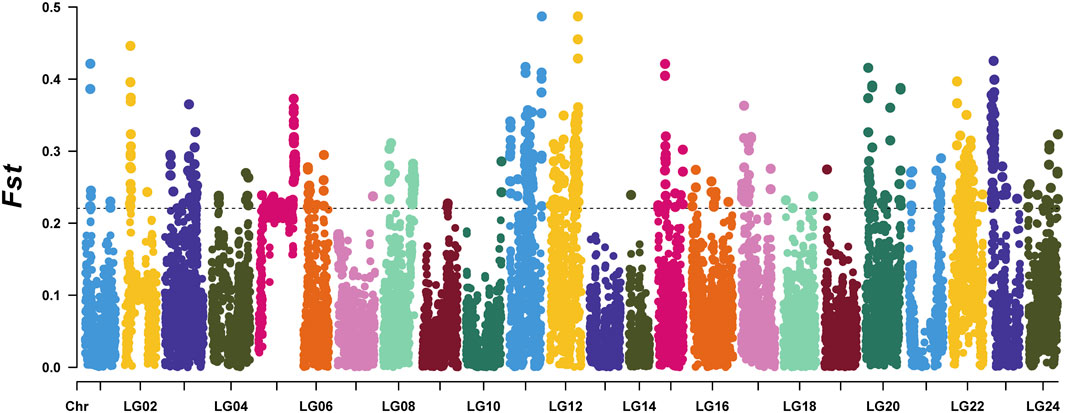
FIGURE 2. The distribution of Fst values between the mandarin fish which could be domesticated to accept artificial diets and those could not.
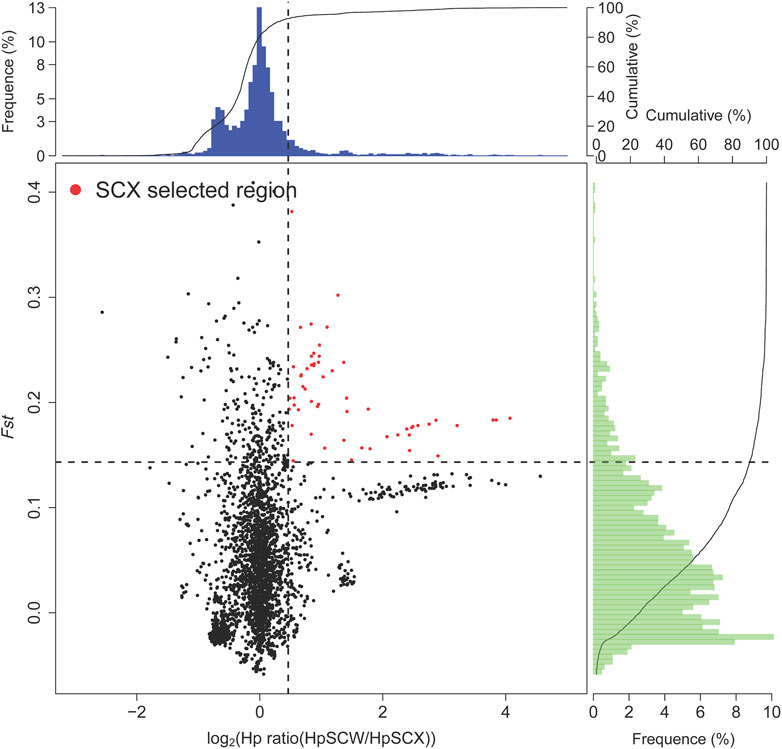
FIGURE 3. Selective sweep analysis. The regions were considered to be selected when both conditions are met: top 10% heterozygosity (Hp) and top 10% population differentiation coefficient (Fst).
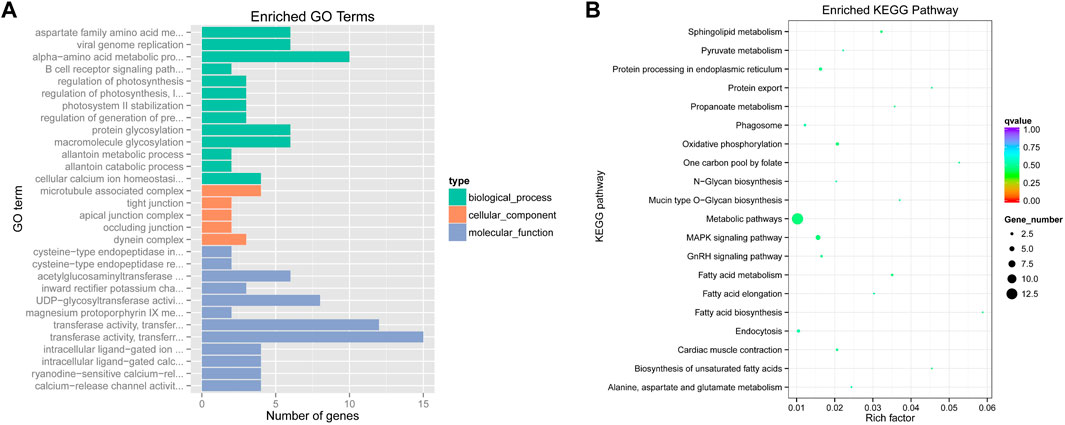
FIGURE 4. Functional enrichment analyses of potential selected genes. (A). GO enrichment terms; (B). KEGG enrichment pathways.
Whole-genome bisulfite sequencing and mapping
In the present study, the hepatic DNA methylation of mandarin fish with different performance in accepting artificial diets were sequenced with whole-genome bisulfite sequencing (WGBS). A total of 225.34 Gb clean data were obtained. The Q20 (sequencing error rate <1%) were 97.75%–97.97% (Supplementary Table S5). 80.74%–91.22% read pairs could be mapped to our mandarin fish reference genome (version: sinChu7), and 76.93%–87.55% read pairs were uniquely mapped (Supplementary Table S6).
DNA methylation patterns
The percentage of methylated cytosines to reference genomic cytosines were 6.96% and 6.44% in mandarin fish which could be domesticated to accept artificial diets and those could not. Methylated sites were counted in CG, CHG, and CHH contexts. Most methylated cytosines were in the CG dinucleotide context. The methylation levels of cytosines in CG context were 76.95% and 77.10% in mandarin fish which could be domesticated to accept artificial diets and those could not, respectively. The methylation levels of cytosines in CHG context were 0.60% and 0.55%, respectively. The methylation levels of cytosines in CHH context were 0.60% and 0.55%, respectively (Supplementary Table S7).
Identification of differentially methylated regions and differentially methylated genes
5,976 differentially methylated regions (DMRs) were identified between mandarin fish with different performance in accepting artificial diets, of which 2,941 were hypermethylated and 3,035 were hypomethylated in mandarin fish which could be domesticated to accept artificial diets. A total of 3,522 differentially methylated genes were identified. GO enrichment analysis revealed that these genes were significantly enriched in 63 GO items, including 16 GO terms in biological process, 41 GO terms in cellular component, and 6 GO terms in molecular function (Supplementary Table 8). The representative KEGG pathway was cAMP signaling pathway, involved differentially methylated genes including cAMP-dependent protein kinase catalytic subunit beta (prkacba), cyclic AMP-responsive element-binding protein 5 (creb5b) and brain-derived neurotrophic factor (bdnf) (Figure 5). The region located within 2 kb upstream of prkacba gene, and the regions located in the 5′UTR introns of creb5b and bdnf genes were hypomethylated in mandarin fish which could be domesticated to accept artificial diets. In addition, the region located within 2 kb upstream of lipoprotein lipase gene (lpl) was hypomethylated in mandarin fish which could be domesticated to accept artificial diets.
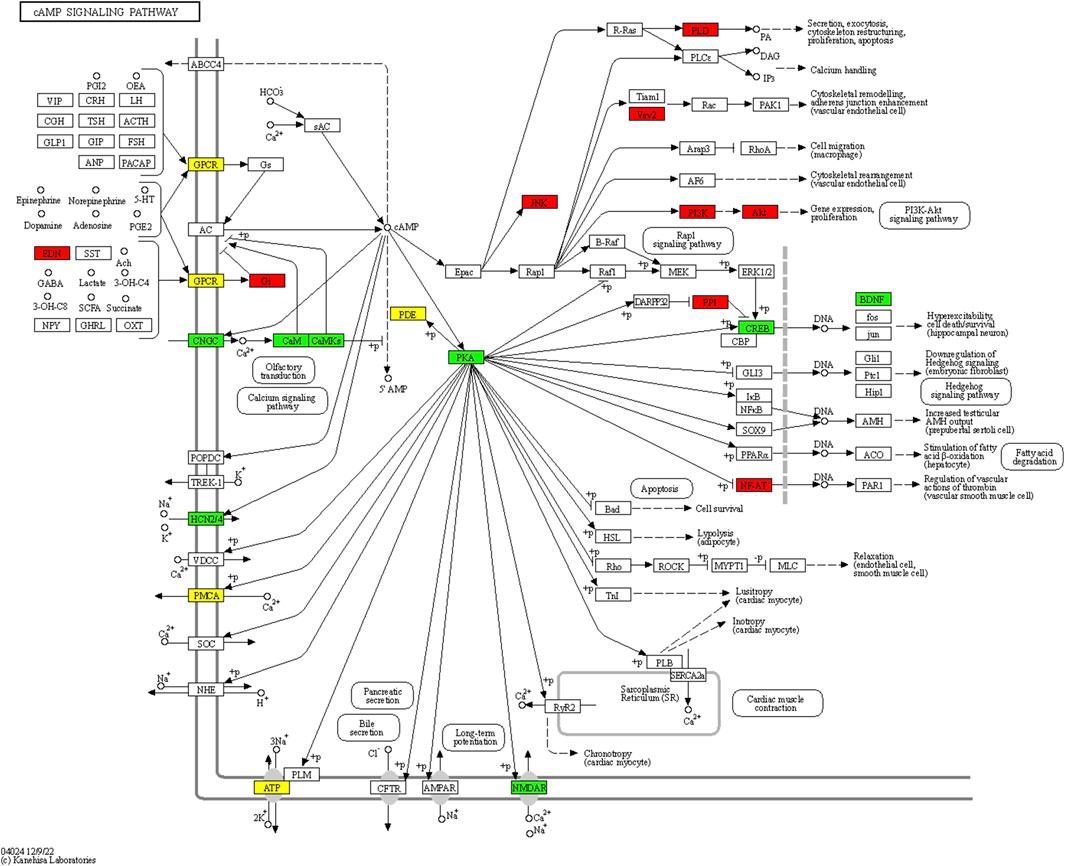
FIGURE 5. Differentially methylated genes in cAMP signaling pathway. Red: genes were hypermethylated; green: genes were hypomethylated; yellow: genes were both hypermethylated and hypomethylated.
Discussion
To identify candidate genomic regions and genes for feeding habit domestication from live prey fish to artificial diets, the whole-genome resequencing was performed between mandarin fish which could be domesticated to accept artificial diets and those could not. Through the selective sweep analysis based on heterozygosity (Hp) and population differentiation coefficient (Fst), 57 candidate windows were identified as the putative selective regions for feeding habit domestication of mandarin fish, involved in 149 genes. These genes were related to memory, vision and olfaction function. Asparagine synthetase (ASNS) catalyses the synthesis of asparagine which is essential for brain development and function (Lomelino et al., 2017). Mutations in human ASNS gene caused a severe neurological condition, and the deficiency of Asns leads to brain structural abnormalities and memory deficits in mice (Ruzzo et al., 2013). In our previous study, comparative genomics analysis for four species of mandarin fish (Sinipercidae) indicated that asns gene was positively selected in S. scherzeri, which is the easiest to wean onto dead prey fish or artificial diets (He et al., 2020b), and the RNA-seq data also showed the lower mRNA expression level of asns in the mandarin fish which could accept dead prey fish than those could not (He et al., 2013). In the present study, asns gene was identified in the selected regions of mandarin fish which could be domesticated to accept artificial diets. And SNPs were identified within 2 kb upstream transcription start sites, 5′UTR, Exon 4 and introns of asns gene related to the artificial diet feeding habit. These results suggested that the asns gene might play an important role in the feeding habit domestication of mandarin fish. Microfibrillar associated protein 4 (MFAP4) is an extracellular matrix protein, contributing to innate immune defense in teleosts (Mohammadi et al., 2022), and mfap4 was reported to be significantly up-regulated in the stomach of artificial diet domesticated mandarin fish (Shen et al., 2021). In the present study, mfap4 gene was identified in the selected regions of mandarin fish which could be domesticated to accept artificial diets, suggesting that the mfap4 gene might play an important role in the feeding habit domestication of mandarin fish through improving innate immunity.
Vision and olfaction are the most important sensory modalities, which are essential for the recognition of food in many fish species (DeBose and Nevitt, 2008; Konishi et al., 2022). Our previous study found that vision was the major sensory modality for mandarin fish to detect and catch prey (Liang et al., 2001). T-box transcription factor 2a (tbx2a) plays important roles in determination of photoreceptor fate (Angueyra et al., 2022) and expression regulation of opsins (Sandkam et al., 2020). TBC1 domain family member 20 (TBC1D20) gene encodes a key regulator of autophagosome maturation (Sidjanin et al., 2016). Mutations in human TBC1D20 gene cause Warburg Micro syndrome, and one of the main symptoms is visual impairment (Liegel et al., 2013). tbc1d20 was identified as a candidate adaptive allele in Caribbean pupfishes (Richards et al., 2021), and a candidate gene for domestication in rainbow trout (Oncorhynchus mykiss) (Cádiz et al., 2021). In the present study, tbx2a and tbc1d20 were identified in the selected regions of mandarin fish which could be domesticated to accept artificial diets, suggesting that potential adaptive changes of vision might contribute to the feeding habit domestication from live prey fish to artificial diets in mandarin fish. Trace amine-associated receptor 13c (taar13c) is a member of olfactory receptor family, the encoded protein product was reported to have high-affinity for cadaverine (Hussain et al., 2013; Dieris et al., 2017). Cadaverine which is produced by decarboxylation of basic amino acids, is considered to be death-associated odor (Hussain et al., 2013). Cadaverine was detected during fishmeal processing, including raw materials and fishmeal (Nguyen et al., 2022). In the present study, taar13c was identified in the selected regions of mandarin fish which could be domesticated to accept artificial diets, indicating that they might have higher tolerance to cadaverine in the fishmeal, thus displaying higher acceptance in artificial diets.
Our previous studies indicated that DNA methylation might contribute to the feeding habit transformation in grass carp and mandarin fish (Cai et al., 2018; Dou et al., 2018). In the present study, the whole-genome bisulfite sequencing was performed between mandarin fish with different performance in accepting artificial diets. 5,976 differentially methylated regions were identified, involved in 3,522 genes. The representative KEGG pathway was cAMP signaling pathway, which could facilitate lipid metabolism in liver (Ravnskjaer et al., 2016). cAMP is an intracellular second messenger, and cAMP-dependent protein kinase (PKA) is essential for intracellular signal transduction (London and Stratakis, 2022). Cellular cAMP accumulation triggers the activation of PKA, then induces the phosphorylation of cAMP-response element binding protein (CREB), further activates the expression of BDNF (Wahlang et al., 2018). Brain-derived neurotrophic factor (BDNF) is a highly conserved member of neurotrophin family, playing important roles in synaptic plasticity and cognitive function (Lu et al., 2014). Except for the functions in nervous system, BDNF was reported to improve lipid and glucose metabolism in type-2-diabetic mice (Tsuchida et al., 2002). Impair of BDNF signaling might lead to metabolic syndrome (Marosi and Mattson, 2014). In the present study, differentially methylated regions were identified in genes involved in cAMP signaling pathway, including cAMP-dependent protein kinase catalytic subunit beta (prkacba), cyclic AMP-responsive element-binding protein 5 (creb5) and bdnf. The region located within 2 kb upstream of prkacba gene, and the regions located in the 5′UTR introns of creb5b and bdnf genes were hypomethylated in mandarin fish which could be domesticated to accept artificial diets. The hypomethylation of the regions located in upstream or 5′UTR intron of genes might increase the mRNA expression level of prkacba, creb5 and bdnf, activating PKA-CREB-BDNF signaling, further improving lipid metabolism of mandarin fish which could accept artificial diets. This would be beneficial for mandarin fish to adapt to the artificial diets. In addition, the region located within 2 kb upstream of lpl gene was hypomethylated in mandarin fish which could be domesticated to accept artificial diets. Lipoprotein lipase (LPL) catalyzes the hydrolysis of triglycerides from circulating chylomicrons and very low density lipoproteins (Mead et al., 2002), playing important roles in lipid utilization and storage (Olivecrona, 2016). Loss-of-function mutations in LPL caused type I hyperlipoproteinemia in human, characterized by very severe hypertriglyceridemia (Caddeo et al., 2018). The DNA methylation change of placental LPL gene influenced fetal growth and fat accretion in childhood, which might further influence the metabolism in later life (Gagné-Ouellet et al., 2017). In the present study, due to the higher fat content of artificial diets than that of the live prey fish, the hypomethylation of the region located within 2 kb upstream of lpl gene might increase the mRNA expression of lpl gene, improving the lipid utilization. And our previous study found the differential metabolites related to lipid metabolism between mandarin fish which could be domesticated to accept artificial diets and those could not (He et al., 2021). These results suggested that lipid metabolism might play an important role in the adaption of artificial diets in mandarin fish.
In conclusion, we identified the selective genomic regions and genes, and the differentially methylated regions and genes for feeding habit domestication by whole-genome resequencing and bisulfite sequencing, respectively. These candidate genes could be critical for understanding the feeding habit domestication from live prey fish to artificial diets in mandarin fish, and further research is needed to clarify the roles of these candidate genes in feeding habit domestication.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. The resequencing data and WGBS data have been deposited into the NCBI Sequence Read Archive (SRA) database (BioProject: PRJNA895188; PRJNA896929, respectively).
Ethics statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committees of Huazhong Agricultural University (Wuhan, China) (HZAUFI-2020-0020).
Author contributions
LL analyzed the data and wrote the manuscript. M-HL and Y-PZ contributed to the sample collection and preparation. X-FL, HK and SH gave technical advice and contributed to the study design. SH revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the China Postdoctoral Science Foundation (2021M701352), the National Natural Science Foundation of China (32202903 and 31972809), and the German Research Foundation (grant KU 3596/1-1; project number: 324050651).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1088081/full#supplementary-material
References
Akalin, A., Kormaksson, M., Li, S., Garrett-Bakelman, F. E., Figueroa, M. E., Melnick, A., et al. (2012). methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87. doi:10.1186/gb-2012-13-10-r87
Angueyra, J., Kunze, V. P., Patak, L. K., Kim, H., Kindt, K., and Li, W. (2022). Transcription factors involved in determination of photoreceptor fate. Invest. Ophthalmol. Vis. Sci. 63 (7), 4277.
Caddeo, A., Mancina, R. M., Pirazzi, C., Russo, C., Sasidharan, K., Sandstedt, J., et al. (2018). Molecular analysis of three known and one novel LPL variants in patients with type I hyperlipoproteinemia. Nutr. Metab. Cardiovasc. Dis. 28 (2), 158–164. doi:10.1016/j.numecd.2017.11.003
Cádiz, M. I., López, M. E., Díaz-Domínguez, D., Cáceres, G., Marin-Nahuelpi, R., Gomez-Uchida, D., et al. (2021). Detection of selection signatures in the genome of a farmed population of anadromous rainbow trout (Oncorhynchus mykiss). Genomics 113 (5), 3395–3404. doi:10.1016/j.ygeno.2021.07.027
Cádiz, M. I., López, M. E., Díaz-Domínguez, D., Cáceres, G., Yoshida, G. M., Gomez-Uchida, D., et al. (2020). Whole genome re-sequencing reveals recent signatures of selection in three strains of farmed Nile tilapia (Oreochromis niloticus). Sci. Rep. 10, 11514. doi:10.1038/s41598-020-68064-5
Cai, W., He, S., Liang, X. F., and Yuan, X. (2018). DNA methylation of T1R1 gene in the vegetarian adaptation of grass carp Ctenopharyngodon idella. Sci. Rep. 8, 6934. doi:10.1038/s41598-018-25121-4
Cai, W. J., Liang, X. F., Yuan, X. C., Li, A. X., and He, S. (2020). Changes of DNA methylation pattern in metabolic pathways induced by high-carbohydrate diet contribute to hyperglycemia and fat deposition in grass carp (Ctenopharyngodon idellus). Front. Endocrinol. 11, 398. doi:10.3389/fendo.2020.00398
Cheng, X. Y., He, S., Liang, X. F., Song, Y., Yuan, X. C., Li, L., et al. (2015). Molecular cloning, expression and single nucleotide polymorphisms of protein phosphatase 1 (PP1) in Mandarin fish (Siniperca chuatsi). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 189, 69–79. doi:10.1016/j.cbpb.2015.08.001
DeBose, J. L., and Nevitt, G. A. (2008). The use of odors at different spatial scales: Comparing birds with fish. J. Chem. Ecol. 34, 867–881. doi:10.1007/s10886-008-9493-4
Dieris, M., Ahuja, G., Krishna, V., and Korsching, S. I. (2017). A single identified glomerulus in the zebrafish olfactory bulb carries the high-affinity response to death-associated odor cadaverine. Sci. Rep. 7, 40892. doi:10.1038/srep40892
Dou, Y., He, S., Liang, X. F., Cai, W., Wang, J., Shi, L., et al. (2018). Memory function in feeding habit transformation of Mandarin fish (Siniperca chuatsi). Int. J. Mol. Sci. 19 (4), 1254. doi:10.3390/ijms19041254
Gagné-Ouellet, V., Houde, A. A., Guay, S. P., Perron, P., Gaudet, D., Guérin, R., et al. (2017). Placental lipoprotein lipase DNA methylation alterations are associated with gestational diabetes and body composition at 5 years of age. Epigenetics 12 (8), 616–625. doi:10.1080/15592294.2017.1322254
He, A., Li, W., Wang, W., Ye, K., Gong, S., and Wang, Z. (2020a). Analysis of DNA methylation differences in gonads of the large yellow croaker. Gene 749, 144754. doi:10.1016/j.gene.2020.144754
He, S., Li, L., Lv, L. Y., Cai, W. J., Dou, Y. Q., Li, J., et al. (2020b). Mandarin fish (Sinipercidae) genomes provide insights into innate predatory feeding. Commun. Biol. 3, 361. doi:10.1038/s42003-020-1094-y
He, S., Liang, X. F., Sun, J., Li, L., Yu, Y., Huang, W., et al. (2013). Insights into food preference in hybrid F1 of Siniperca chuatsi (♀) × Siniperca scherzeri (♂) Mandarin fish through transcriptome analysis. BMC Genomics 14, 601. doi:10.1186/1471-2164-14-601
He, S., You, J. J., Liang, X. F., Zhang, Z. L., and Zhang, Y. P. (2021). Transcriptome sequencing and metabolome analysis of food habits domestication from live prey fish to artificial diets in Mandarin fish (Siniperca chuatsi). BMC Genomics 22, 129. doi:10.1186/s12864-021-07403-w
Huang, Y., Wen, H., Zhang, M., Hu, N., Si, Y., Li, S., et al. (2018). The DNA methylation status of MyoD and IGF-I genes are correlated with muscle growth during different developmental stages of Japanese flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 219-220, 33–43. doi:10.1016/j.cbpb.2018.02.005
Hussain, A., Saraiva, L. R., Ferrero, D. M., Ahuja, G., Krishna, V. S., Liberles, S. D., et al. (2013). High-affinity olfactory receptor for the death-associated odor cadaverine. Proc. Natl. Acad. Sci. U.S.A. 110 (48), 19579–19584. doi:10.1073/pnas.1318596110
Konishi, J., Abe, T., Ogihara, A., Adachi, D., Denboh, T., and Kudo, H. (2022). Olfactory behavioural and neural responses of planktivorous lacustrine sockeye salmon (Oncorhynchus nerka) to prey odours. J. Fish. Biol. 101 (1), 269–275. doi:10.1111/jfb.15110
Krueger, F., and Andrews, S. R. (2011). Bismark: A flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27 (11), 1571–1572. doi:10.1093/bioinformatics/btr167
Kumkhong, S., Marandel, L., Plagnes-Juan, E., Veron, V., Boonanuntanasarn, S., and Panserat, S. (2020). Glucose injection into yolk positively modulates intermediary metabolism and growth performance in juvenile Nile tilapia (Oreochromis niloticus). Front. Physiol. 11, 286. doi:10.3389/fphys.2020.00286
Kwon, Y. M., Vranken, N., Hoge, C., Lichak, M. R., Norovich, A. L., Francis, K. X., et al. (2022). Genomic consequences of domestication of the Siamese fighting fish. Sci. Adv. 8 (10), eabm4950. doi:10.1126/sciadv.abm4950
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 (14), 1754–1760. doi:10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25 (16), 2078–2079. doi:10.1093/bioinformatics/btp352
Li, Y., Fu, B., Zhang, J., Xie, J., Wang, G., Jiang, P., et al. (2021). Genome-wide analysis of DNA methylation reveals selection signatures of grass carp during the domestication. bioRxiv. doi:10.1101/2021.11.04.467282
Liang, X. F., Lin, X., Li, S., and Liu, J. K. (2008). Impact of environmental and innate factors on the food habit of Chinese perch Siniperca chuatsi (Basilewsky) (Percichthyidae). Aquac. Res. 39 (2), 150–157. doi:10.1111/j.1365-2109.2007.01870.x
Liang, X. F., Oku, H., Ogata, H. Y., Liu, J., and He, X. (2001). Weaning Chinese perch Siniperca chuatsi (Basilewsky) onto artificial diets based upon its specific sensory modality in feeding. Aquac. Res. 32 (1), 76–82. doi:10.1046/j.1355-557x.2001.00006.x
Liegel, R. P., Handley, M. T., Ronchetti, A., Brown, S., Langemeyer, L., Linford, A., et al. (2013). Loss-of-function mutations in TBC1D20 cause cataracts and male infertility in blind sterile mice and Warburg micro syndrome in humans. Am. J. Hum. Genet. 93 (6), 1001–1014. doi:10.1016/j.ajhg.2013.10.011
Lomelino, C. L., Andring, J. T., McKenna, R., and Kilberg, M. S. (2017). Asparagine synthetase: Function, structure, and role in disease. J. Biol. Chem. 292 (49), 19952–19958. doi:10.1074/jbc.R117.819060
London, E., and Stratakis, C. A. (2022). The regulation of PKA signaling in obesity and in the maintenance of metabolic health. Pharmacol. Ther. 237, 108113. doi:10.1016/j.pharmthera.2022.108113
Lu, B., Nagappan, G., and Lu, Y. (2014). BDNF and synaptic plasticity, cognitive function, and dysfunction. Handb. Exp. Pharmacol. 220, 223–250. doi:10.1007/978-3-642-45106-5_9
Marosi, K., and Mattson, M. P. (2014). BDNF mediates adaptive brain and body responses to energetic challenges. Trends Endocrinol. Metab. 25 (2), 89–98. doi:10.1016/j.tem.2013.10.006
Mead, J. R., Irvine, S. A., and Ramji, D. P. (2002). Lipoprotein lipase: Structure, function, regulation, and role in disease. J. Mol. Med. 80, 753–769. doi:10.1007/s00109-002-0384-9
Mohammadi, A., Sorensen, G. L., and Pilecki, B. (2022). MFAP4-mediated effects in elastic fiber homeostasis, integrin signaling and cancer, and its role in teleost fish. Cells 11 (13), 2115. doi:10.3390/cells11132115
Morán, P., Marco-Rius, F., Megías, M., Covelo-Soto, L., and Pérez-Figueroa, A. (2013). Environmental induced methylation changes associated with seawater adaptation in Brown trout. Aquaculture 392-395, 77–83. doi:10.1016/j.aquaculture.2013.02.006
Navarro-Martín, L., Viñas, J., Ribas, L., Díaz, N., Gutiérrez, A., Croce, L. D., et al. (2011). DNA methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genet. 7 (12), e1002447. doi:10.1371/journal.pgen.1002447
Nguyen, H. T., Hilmarsdóttir, G. S., Tómasson, T., Arason, S., and Gudjónsdóttir, M. (2022). Changes in protein and non-protein nitrogen compounds during fishmeal processing —Identification of unoptimized processing steps. Processes 10 (4), 621. doi:10.3390/pr10040621
Olivecrona, G. (2016). Role of lipoprotein lipase in lipid metabolism. Curr. Opin. Lipidol. 27 (3), 233–241. doi:10.1097/MOL.0000000000000297
Pierron, F., Baillon, L., Sow, M., Gotreau, S., and Gonzalez, P. (2014). Effect of low-dose cadmium exposure on DNA methylation in the endangered European eel. Environ. Sci. Technol. 48 (1), 797–803. doi:10.1021/es4048347
Podgorniak, T., Dhanasiri, A., Chen, X., Ren, X., Kuan, P. F., and Fernandes, J. (2022). Early fish domestication affects methylation of key genes involved in the rapid onset of the farmed phenotype. Epigenetics 17 (10), 1281–1298. doi:10.1080/15592294.2021.2017554
Ravnskjaer, K., Madiraju, A., and Montminy, M. (2016). Role of the cAMP pathway in glucose and lipid metabolism. Handb. Exp. Pharmacol. 233, 29–49. doi:10.1007/164_2015_32
Richards, E. J., McGirr, J. A., Wang, J. R., St John, M. E., Poelstra, J. W., Solano, M. J., et al. (2021). A vertebrate adaptive radiation is assembled from an ancient and disjunct spatiotemporal landscape. Proc. Natl. Acad. Sci. U.S.A. 118 (20), e2011811118. doi:10.1073/pnas.2011811118
Ruzzo, E. K., Capo-Chichi, J. M., Ben-Zeev, B., Chitayat, D., Mao, H., Pappas, A. L., et al. (2013). Deficiency of asparagine synthetase causes congenital microcephaly and a progressive form of encephalopathy. Neuron 80 (2), 429–441. doi:10.1016/j.neuron.2013.08.013
Sandkam, B. A., Campello, L., O'Brien, C., Nandamuri, S. P., Gammerdinger, W. J., Conte, M. A., et al. (2020). Tbx2a modulates switching of RH2 and LWS opsin gene expression. Mol. Biol. Evol. 37 (7), 2002–2014. doi:10.1093/molbev/msaa062
Shao, C., Li, Q., Chen, S., Zhang, P., Lian, J., Hu, Q., et al. (2014). Epigenetic modification and inheritance in sexual reversal of fish. Genome Res. 24, 604–615. doi:10.1101/gr.162172.113
Shen, Y., Li, H., Zhao, J., Tang, S., Zhao, Y., Bi, Y., et al. (2021). The digestive system of Mandarin fish (Siniperca chuatsi) can adapt to domestication by feeding with artificial diet. Aquaculture 538, 736546. doi:10.1016/j.aquaculture.2021.736546
Si, Y., He, F., Wen, H., Li, J., Zhao, J., Ren, Y., et al. (2016). Genetic polymorphisms and DNA methylation in exon 1 CpG-rich regions of PACAP gene and its effect on mRNA expression and growth traits in half smooth tongue sole (Cynoglossus semilaevis). Fish. Physiol. Biochem. 42, 407–421. doi:10.1007/s10695-015-0147-5
Sidjanin, D. J., Park, A. K., Ronchetti, A., Martins, J., and Jackson, W. T. (2016). TBC1D20 mediates autophagy as a key regulator of autophagosome maturation. Autophagy 12 (10), 1759–1775. doi:10.1080/15548627.2016.1199300
Sun, J., He, S., Liang, X. F., Li, L., Wen, Z., Zhu, T., et al. (2015). Identification of SNPs in NPY and LEP and the association with food habit domestication traits in Mandarin fish. J. Genet. 94, 118–122. doi:10.1007/s12041-014-0442-4
Tsuchida, A., Nonomura, T., Nakagawa, T., Itakura, Y., Ono-Kishino, M., Yamanaka, M., et al. (2002). Brain-derived neurotrophic factor ameliorates lipid metabolism in diabetic mice. Diabetes Obes. Metab. 4 (4), 262–269. doi:10.1046/j.1463-1326.2002.00206.x
Veron, V., Marandel, L., Liu, J., Vélez, E. J., Lepais, O., Panserat, S., et al. (2018). DNA methylation of the promoter region of bnip3 and bnip3l genes induced by metabolic programming. BMC Genomics 19, 677. doi:10.1186/s12864-018-5048-4
Wahlang, B., McClain, C., Barve, S., and Gobejishvili, L. (2018). Role of cAMP and phosphodiesterase signaling in liver health and disease. Cell. Signal. 49, 105–115. doi:10.1016/j.cellsig.2018.06.005
Wang, K., Li, M., and Hakonarson, H. (2010). Annovar: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38 (16), e164. doi:10.1093/nar/gkq603
Yi, T., Sun, J., Liang, X., He, S., Li, L., Wen, Z., et al. (2013). Effects of polymorphisms in pepsinogen (PEP), amylase (AMY) and trypsin (TRY) genes on food habit domestication traits in Mandarin fish. Int. J. Mol. Sci. 14 (11), 21504–21512. doi:10.3390/ijms141121504
Zhong, H., Xiao, J., Chen, W., Zhou, Y., Tang, Z., Guo, Z., et al. (2014). DNA methylation of pituitary growth hormone is involved in male growth superiority of Nile tilapia (Oreochromis niloticus). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 171, 42–48. doi:10.1016/j.cbpb.2014.03.006
Keywords: Mandarin fish, resequencing, whole-genome bisulfite sequencing, feeding habit domestication, selective sweep
Citation: Li L, He S, Lin M-H, Zhang Y-P, Kuhl H and Liang X-F (2023) Whole-genome resequencing and bisulfite sequencing provide new insights into the feeding habit domestication in mandarin fish (Siniperca chuatsi). Front. Genet. 13:1088081. doi: 10.3389/fgene.2022.1088081
Received: 03 November 2022; Accepted: 28 December 2022;
Published: 12 January 2023.
Edited by:
Zheng-Yong Wen, Neijiang Normal University, ChinaReviewed by:
Chao Bian, BGI Academy of Marine Sciences, ChinaDengyue Yuan, Southwest University, China
Copyright © 2023 Li, He, Lin, Zhang, Kuhl and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xu-Fang Liang, eHVmYW5nX2xpYW5nQGhvdG1haWwuY29t; Heiner Kuhl, a3VobEBpZ2ItYmVybGluLmRl
 Ling Li
Ling Li Shan He
Shan He Ming-Hui Lin1,2
Ming-Hui Lin1,2 Yan-Peng Zhang
Yan-Peng Zhang Heiner Kuhl
Heiner Kuhl Xu-Fang Liang
Xu-Fang Liang