- 1Samsung Medical Center, Department of Laboratory Medicine and Genetics, Sungkyunkwan University School of Medicine, Seoul, South Korea
- 2Clinical Genomics Center, Samsung Medical Center, Seoul, South Korea
Titin truncating variants (TTNtvs) are the most common genetic cause of dilated cardiomyopathy (DCM). Among four regions of titin, A-band enrichment of DCM-causing TTNtvs is widely accepted but the underlying mechanism is still unknown. Meanwhile, few reports have identified exon 327 as a highly mutated A-band exon but the degree of exon 327 enrichment has not been quantitatively investigated. To find the real hotspot of DCM-causing TTNtvs, we aimed to reassess the degree of TTNtv enrichment in known titin regions and in exon 327, separately. In addition, we tried to explain exon 327 clustering in terms of nonsense-mediated mRNA decay (NMD) efficiency and a dominant negative mechanism recently proposed. Research papers focusing on TTNtvs found in patients with DCM were collected. A total of 612 patients with TTNtv-realated DCM were obtained from 10 studies. In the four regions of TTN and exon 327, the degree of TTNtvs enrichment was calculated in a way that the effect of distribution of highly expressed exons was normalized. As a result, exon 327 was the only region that showed significant enrichment for DCM-related TTNtv (p < .001). On the other hand, other A-band exons had almost the same number of TTNtv of random distribution. A review of RNAseq data revealed that the median allelic imbalance deviation of exon 327 TTNtvs was .04, indicating almost zero NMD. From these findings, we propose that the widely accepted A-band enrichment of DCM-related TTNtv is mostly attributable to exon 327 enrichment. In addition, based on the recently demonstrated dominant negative mechanism, the extremely low NMD efficiency seems to contribute to exon 327 enrichment.
Introduction
Titin-truncating variants (TTNtvs) are the most commonly observed genetic cause of dilated cardiomyopathy (DCM) and are found in up to 25% of DCM cases (Herman et al., 2012; Roberts et al., 2015; Yotti et al., 2019; Mazzarotto et al., 2020). Moreover, approximately 1% of the general population carry TTNtvs, necessitating the parameters for TTNtv pathogenicity prediction (Roberts et al., 2015; Schafer et al., 2017; Yotti et al., 2019). Percent spliced-in (PSI) has been reported as a crucial pathogenicity predictor of TTNtv in DCM. Clinical data demonstrated that TTNtvs located in exons that are constitutively expressed in the human left ventricle (PSI > .9) are significantly associated with DCM (Roberts et al., 2015; Schafer et al., 2017; Haggerty et al., 2019). In addition to PSI, the location of premature termination codon (PTC) has also been investigated as a parameter for pathogenicity prediction of TTNtv. As a result, among the four protein regions of titin, i.e., Z-disk, I-band, A-band, and M-band, A-band enrichment of TTNtv in patients with DCM has been repeatedly reported (Herman et al., 2012; Roberts et al., 2015; Akinrinade et al., 2016; Schafer et al., 2017; Rich et al., 2020; Fomin et al., 2021). TTNtvs from all titin regions can lead to DCM, and A-band TTNtvs are known to have a higher odds ratio (OR) than other regions (Akinrinade et al., 2016; Schafer et al., 2017). The mechanism of A-band enrichment remains unclear. Meanwhile, in few reports, exon 327 within the A-band has been briefly mentioned as a frequently mutated TTN exon (Akinrinade et al., 2016; Tharp et al., 2019; Fomin et al., 2021). The degree of TTNtv enrichment in exon 327 and other A-band exons has not been separately analyzed.
The A-band is the longest region of TTN, which comprises 49.8% (53,799 bp/1,07,976 bp) of the TTN coding region, and when limited to high-PSI exons (PSI > .9), the A-band accounts for 67.0% (53,799 bp/80,325 bp) of the coding region. Considering the high share A-band holds in the high-PSI exons, a high frequency of DCM-related TTNtv in the A-band is expected. In this study, we aimed to assess the degree of TTNtv enrichment in the titin regions when the effect of the distribution of high-PSI exons was removed. Additionally, we aimed to measure the degree of TTNtv enrichment in the exon 327 and other A-band exons separately.
As a disease mechanism of TTNtv-related DCM, in addition to haploinsufficiency caused by the shortage of full-length titin proteins, a dominant negative effect caused by truncated titin protein has been demonstrated in recent reports (Fomin et al., 2021; McAfee et al., 2021). In this scenario, evasion of nonsense-mediated mRNA decay (NMD) is necessary to create a truncated titin protein that acts as a poison peptide. Because the amount of truncated titin protein has been demonstrated to affect the outcome of DCM (Fomin et al., 2021), a reasonable assumption is that NMD efficiency may affect the pathogenesis of TTNtv in DCM.
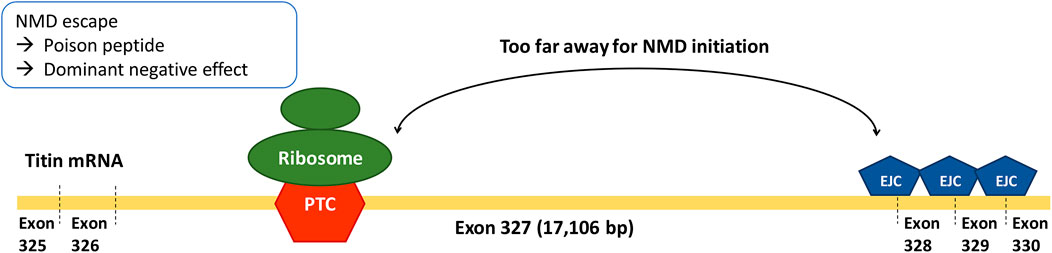
Exon 327 is the longest exon in TTN—by far—consisting of 17,106 bp (median = 261 bp, second-longest exon = 5,609 bp; Supplementary Table S1). According to reports on the emerging parameters of NMD efficiency, PTCs in long exons tend to decrease the efficiency of NMD because of the long average distance from the PTC to the downstream intron (PTC-to-intron distance). A long PTC-to-intron distance hinders the contact between PTC-bound ribosomes and the downstream exon-junction complex, which is required for NMD initiation (Lindeboom et al., 2016; Hoek et al., 2019; Supek et al., 2021). The association of a large PTC-to-intron distance and inefficient NMD was recorded in a study that utilized single-molecule imaging (Hoek et al., 2019). In addition, TTN was reported to be one of the genes enriched for pathogenic truncating variants that do not trigger NMD (Lindeboom et al., 2019). Our third aim was to identify whether the exon 327 enrichment of TTNtv in patients with DCM is the consequence of reduced NMD efficiency caused by long PTC-to-intron distance. Lastly, we also aimed to determine whether there is a correlation between NMD efficiency and clinical outcomes of TTNtv-related DCM.
Data acquisition
This study was based on a literature review and was thus exempt from any local institutional ethics board review. We searched Google Scholar and PubMed for papers in the English language from January 2010 to May 2022 for the terms “titin,” “truncating variant,” and “dilated cardiomyopathy.” A list of the included studies is presented in Table 1. Tayal et al. (2017) was excluded from the analysis because the study was performed in the same hospital as Roberts et al. (2015) and there was significant overlap in the study period and the list of variants. In addition, among the cases of Jansweijer et al. (2017), cases from the University Medical Center Groningen were excluded because Jansen et al. (2019) also retrospectively utilized DCM probands from the same hospital, and there was significant overlap in the study period and list of variants. Similarly, among the cases of Herman et al. (2012), those from Royal Brompton and Harefield Hospitals NHS Foundation Trust were excluded because of the overlap with Roberts et al. (2015) in hospital, study period, and variants.
Patients with TTN nonsense variants or frameshift variants were selected from the final included papers. Patients with splice-site variants were excluded because their exact PTC location could not be determined. Exceptionally, in RNAseq data analysis, splice-site variants could not be filtered out and all truncating variants were included because in one study (McAfee et al., 2021), the allelic imbalance (AI) deviation values were given without specific variant description. Cases with frameshift variants with incomplete descriptions for exact PTC location determination (e.g., p.Ser4228Leufs instead of p.Ser4228LeufsTer23 without cDNA description) were also excluded. The variants described according to the transcripts other than the meta-transcript (NM_001267550.2) were transformed to NM_001267550.2 using VariantValidator (Freeman et al., 2018). (https://variantvalidator.org), and the PTC location was determined based on the amino acid sequence according to NP_001254479.2. Exon number, length of PTC-containing exon, and PTC-to-intron distance were annotated using the exon boundary information downloaded from the Table Browser of the UCSC genome browser (Supplementary Table S1) (Karolchik et al., 2004). PSI and titin protein regions were annotated using the data of Roberts et al. (2015).
The gnomAD v2.1.1 data (Karczewski et al., 2020) was used as a control group, and a list of TTN variants was downloaded from the official gnomAD website (https://gnomad.broadinstitute.org/). Splice-site variants were removed; the annotation of PTC location, exon, and region information, and PSI was performed as aforementioned.
TTNtv enrichment only in exon 327 in DCM group
For known titin protein regions and exon 327, the degree of TTNtv enrichment was calculated based on the ratio of the observed and expected number of TTNtvs (expected based on the length of regions) in both the DCM and control groups. To remove the effect of PSI, a confounding variable in measuring the effect of PTC location on the pathogenesis of DCM, we utilized only high-PSI exons (PSI > .9) both for collecting TTNtvs and for calculating the coding region length (Figure 1). After removing low-PSI regions, the I-band was divided by the major splicing region, resulting in two sub-regions, the proximal I-band and distal I-band, as in other studies (Schafer et al., 2017; Fomin et al., 2021). To focus on the degree of TTNtv enrichment in exon 327, we also divided the A-band into three sub-regions: pre-exon 327 A-band, exon 327, and post-exon 327 A-band. For determining the degree of TTNtv enrichment in the DCM group compared with the control group, the ratio of the degree of TTNtv enrichment in the DCM group to that in the control group was derived.
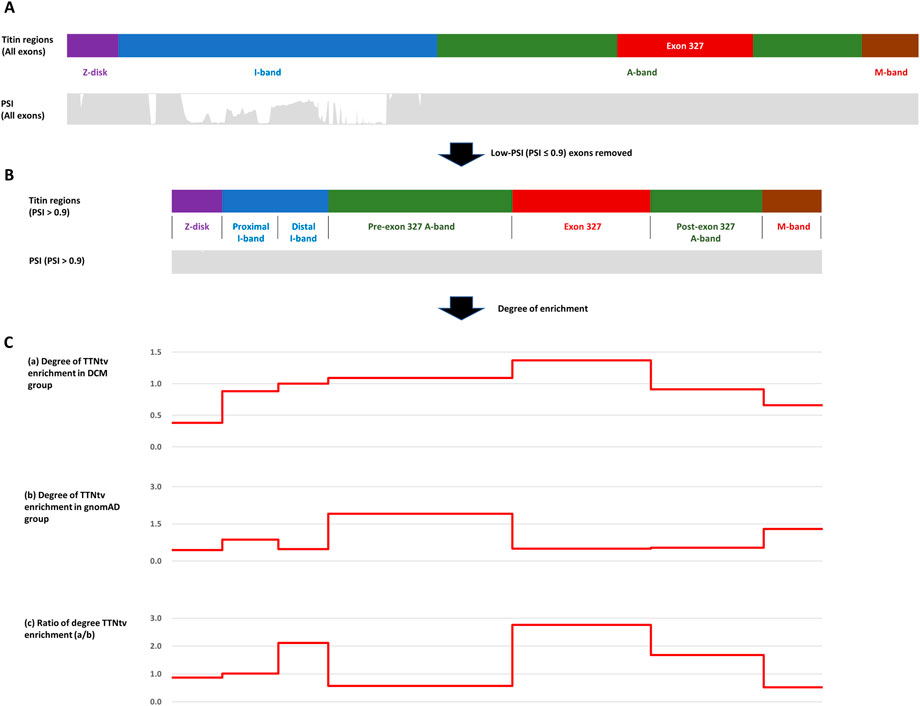
FIGURE 1. (A) Four regions of titin meta-transcript (NM_001267550.2) with PSI (Percent spliced-in) distribution. (B) After low-PSI exons were removed, I-band was divided into two sub-regions (Proximal I-band: exon 29–50, Distal I-band: exon 220–252), and A-band was divided into three sub-regions (Pre-exon 327: exon 253–326, exon 327, post-exon 327: exon 328–358). (C) Degree of Titin-truncating variant (TTNtv) enrichment calculated based on the ratio of observed and expected (based on the length of the region) number of TTNtv. (A) DCM group (B) gnomAD group (C) DCM group/gnomAD group.
From 10 cohort studies, 612 cases of TTNtv-related DCM were found (Table 1; Supplementary Table S2) (Herman et al., 2012; Pugh et al., 2014; Roberts et al., 2015; Franaszczyk et al., 2017; Jansweijer et al., 2017; Jansen et al., 2019; Akhtar et al., 2020; Rich et al., 2020; Fomin et al., 2021; McAfee et al., 2021). Among these cases, 606 had variants in the high-PSI (>.9) exons. Except for the 22 cases in which full variant descriptions were not available (McAfee et al., 2021) and 14 cases excluded because of redundancy (Herman et al., 2012), 570 cases were used in the regional enrichment analysis constituting the DCM group. The number of cases utilized in the RNAseq data analysis and phenotype analysis were 42 and 76, respectively (Table 1; Supplementary Tables S3, S4). From gnomAD v2.1.1, which consisted of 141,456 cases, 1,749 cases (1.24%) with TTNtv were found (Supplementary Tables S5, S6). After removing variants from low-PSI exons and terminal exon of novex-3 (ENST00000360870.5) that cannot be converted to the meta-transcript (Roberts et al., 2015), 957 cases (0.68%) remained and were used in the regional enrichment analysis as a control group (Supplementary Tables S6, S7).
Within the DCM group, the degree of TTNtv enrichment was the highest in exon 327 (1.37, p < .001) (Table 2; Figure 1). The enrichment of total A-band TTNtvs was statistically significant (p < .001); however, after excluding exon 327 TTNtvs, the degree of enrichment in the A-band did not show a significant increase in either pre-exon 327 or post-exon 327 TTNtvs (p = .178 and p = .347, respectively). The degree of I-band TTNtv enrichment was also non-significant, with a value close to 1.0 (.94, p = .533). The degree of Z-disk and M-band enrichment was below 1.0 (.38 and .66, respectively), which was statistically significant (p < .001 and p = .010, respectively). When separated into individual studies, statistically significant enrichment in exon 327 was observed only in two studies (Akhtar et al., 2020; Rich et al., 2020), presumably due to the smaller sample sizes (Figure 2).
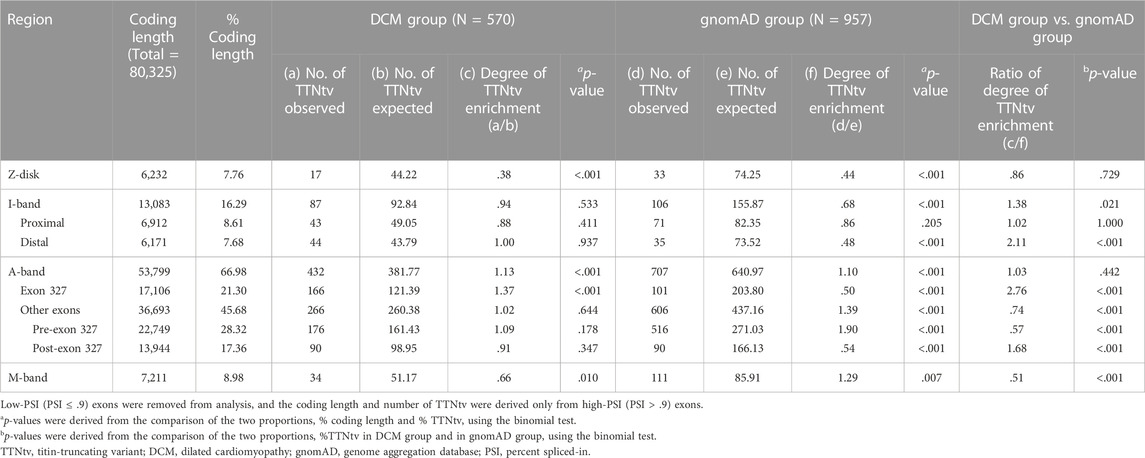
TABLE 2. Degree of TTNtv enrichment in DCM group and in gnomAD calculated based on the ratio of observed and expected number of TTNtv from exons with PSI > .9.
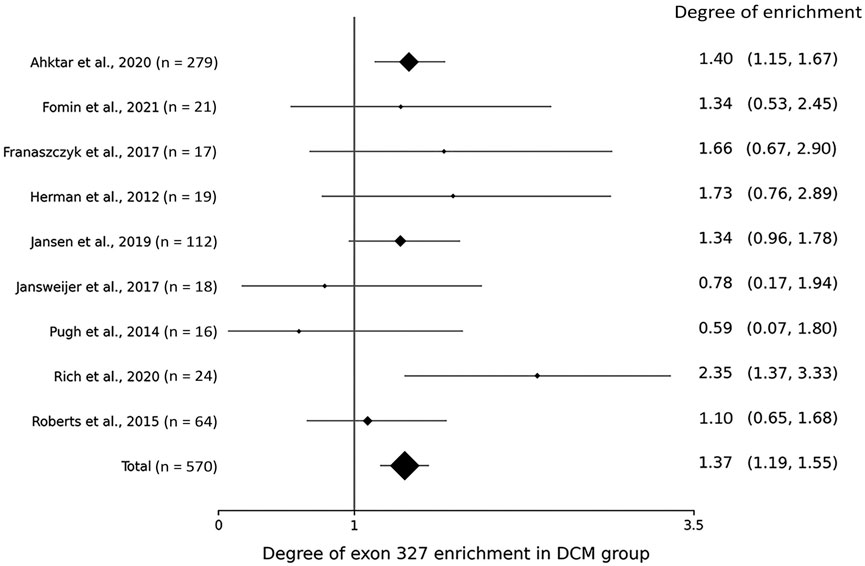
FIGURE 2. Forest plot showing the degree of titin-truncating variant (TTNtv) enrichment in exon 327 from individual studies.
From the gnomAD group, while the degree of TTNtv enrichment showed considerable fluctuation, the degree of enrichment exon 327 was low (.50, p < .001) reflecting deprivation of exon 327 TTNtv in general population (Table 2; Figure 1). When the degree of enrichment in the DCM group was divided by that of the gnomAD group, the ratio was the highest in exon 327 among all regions of TTN (2.76, p < .001).
Generally reduced NMD efficiency especially in exon 327
In the three studies in which RNAseq was performed (Roberts et al., 2015; Fomin et al., 2021; McAfee et al., 2021), AI deviation was obtained as a surrogate for the actual NMD efficiency such that 0 and .5 indicate no NMD at all and 100% NMD efficiency, respectively. AI deviation was directly provided in one study (McAfee et al., 2021), and in two other studies (Roberts et al., 2015; Fomin et al., 2021), AI deviation was derived from altered allele frequency (altered AF) simply by subtracting the altered AF from .5. A value of zero was used as the AI deviation when a negative value was obtained. From the three studies, PTC location, age at heart transplantation (HT), and left ventricular ejection fraction (LVEF) were also obtained from the available cases, and the correlations among the four variables (AI deviation, PTC location, age at HT, and LVEF) were analyzed.
The results of RNAseq data analysis and phenotype analysis are shown in Figure 3. The PTC-to-intron distance was negatively correlated with allelic imbalance (AI) deviation (r = .318, p = .0404, Figure 3A). The level of NMD measured using AI deviation was low in most DCM cases. The median AI deviation was higher in non-exon 327 TTNtvs than in exon 327 TTNtvs, but the difference was non-significant (median AI deviation of .04 and .10, respectively, p = .050, Mann–Whitney U test, Figure 3B). When limited to exon 327 TTNtvs, PTC-to-intron distance was negatively correlated with age at HT (r = −.560, p = .0467, Figure 3C). AI deviation also showed a similar effect on the age at HT, but without statistical significance, probably because of the reduced sample size (r = −.497, p = .1438, Figure 3D). Other correlation analyses among available parameters, PTC-to-intron distance, AI deviation, age at HT, and LVEF, including subgroup analysis (exon 327 and non-exon 327 TTNtvs), did not show any statistical significance (Supplementary Figure S1).
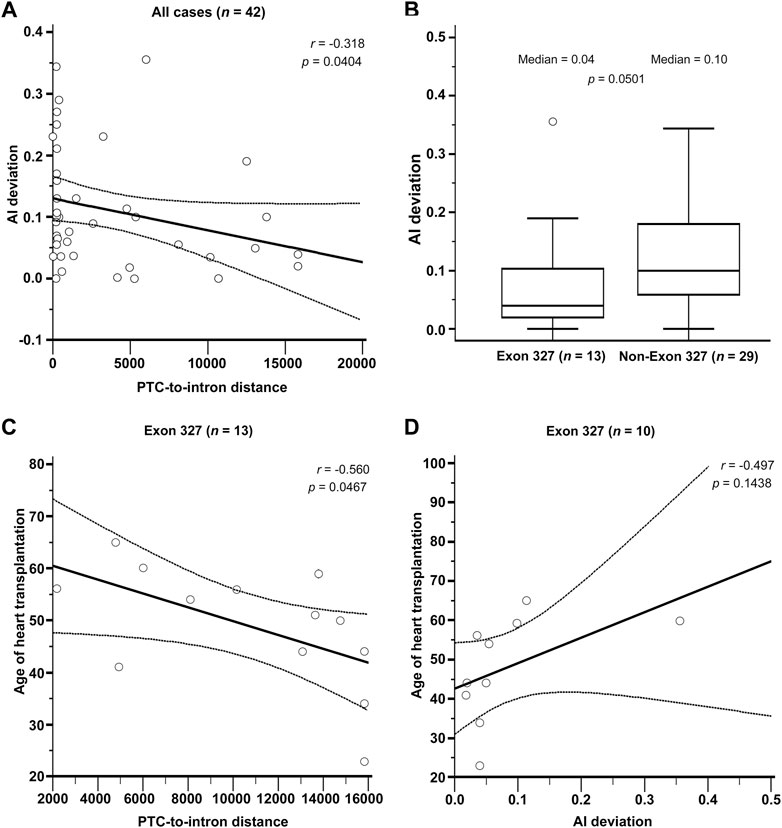
FIGURE 3. (A) Relationship between Premature termination codon (PTC)-to-intron distance and allelic imbalance deviation (AI deviation). (B) Difference in nonsense-mediated mRNA decay efficiency measured by AI deviation level between exon 327 and non-exon 327 titin-truncating variants (TTNtvs). Median PTC-to-intron distance was 10,693 and 223 in exon 327 group and non-exon 327 TTNtvs, respectively (p < .001). (C) Relationship between PTC-to-intron distance and age of heart transplantation in carriers of exon 327 TTNtv. (D) Relationship between AI deviation and age of heart transplantation in carriers of exon 327 TTNtvs. p-values are from Pearson’s correlation test and the Mann–Whitney test. PTC, premature termination codon; AI, allelic imbalance; TTNtv, titin-truncating variant.
Phenotype analysis
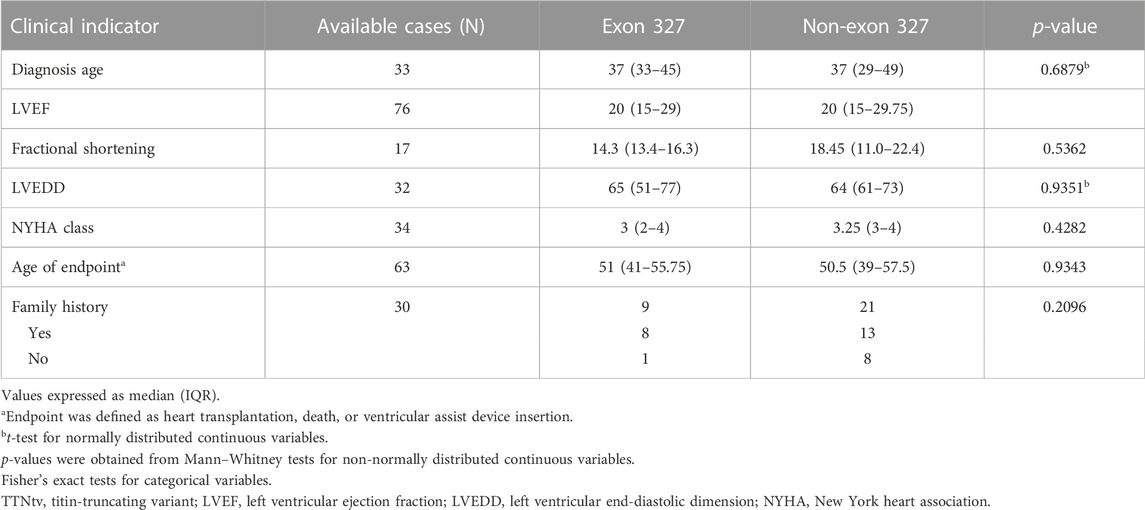
Utilizing studies in which clinical outcome data were provided and the data could be linked to specific TTNtvs (Herman et al., 2012; Fomin et al., 2021; McAfee et al., 2021), clinical outcomes between carriers of exon 327 TTNtv and non-exon 327 TTNtv were compared (Table 3). Seven clinical indicators, including diagnosis age, LVEF, and family history, were used; however, statistical significance was not observed for all parameters.
Statistical analysis
The Shapiro test was used to test the normality of the data. For the comparison of the two groups, the t-test was used when the normality assumption was satisfied; otherwise, the Mann–Whitney test was used. Pearson’s correlation was used to analyze the correlation between two numerical variables when the normality assumption was satisfied, and Spearman’s correlation analysis was used otherwise. The Fisher’s exact test was used to compare categorical variables between groups and binomial test was used for the comparison of proportions. Shapiro test and binomial test were performed using R (version 4.0.3) software and all other tests were performed using MedCalc Statistical Software version 19.2.6 (MedCalc Software Ltd., Ostend, Belgium; https://www.medcalc.org;2020).
Discussion
Studies have reported that TTNtvs that affect high-PSI (>.9) exons have a 93% probability of being pathogenic, and for A-band TTNtvs, the probability is higher, at 97% (Roberts et al., 2015; Akinrinade et al., 2016). From case control studies, A-band TTNtv showed an OR of 49.8–70.4, and TTNtv from other regions showed an OR of 5.3 (Z-disk), 3.7 (M-band), 19.0 (proximal I-band), and 19.5–32.0 (distal I-band), when limited to constitutive exons (Akinrinade et al., 2016; Schafer et al., 2017). In our study, the total number of patients with DCM (with or without TTNtv) was not available in three studies (Table 1) (Jansen et al., 2019; Akhtar et al., 2020; Rich et al., 2020), making it impossible to derive OR. Instead, we analyzed the degree of enrichment based on the ratio of the observed and expected numbers of TTNtv (expected based on the length of the high-PSI coding region). The degree of TTNtv enrichment among titin regions and exon 327 could be quantitatively compared.
Although exon 327 has been designated as a potential hotspot in several reports (Akinrinade et al., 2016; Tharp et al., 2019; Fomin et al., 2021), the degree of enrichment of exon 327 and other A-band exons has not been separately analyzed. In the DCM cohorts in our study, the number of TTNtv observed in exon 327 was 37% higher than the expected number based on uniform distribution (p < .001), and no other regions showed a significant increment or increment of >10%. The non-exon 327 A-band showed a minimally (2%) higher number of TTNtv than that with the uniform distribution, which was statistically non-significant (p = 0.644). Overall, except for exon 327, the number of DCM-related TTNtv was almost the same as the expected number of uniform distributions in the A-band and I-band, whereas that of the Z-disk and M-band was significantly lower. Based on this result, the widely accepted A-band enrichment appears to be mostly attributable to exon 327.
In contrast with the DCM cohorts, in which the fluctuation in the degree of TTNtv enrichment was not substantial and consistent with the existing knowledge except for the exon 327 vs. other A-band exons, in the gnomAD cohort, the degree of TTNtv enrichment showed considerable fluctuation in an unpredicted manner. Although the allele frequency (AF) of a variant in the general population is known to reflect the severity of the phenotype by the purifying selection pressure (Gorlov et al., 2011), this might not work in a precise, quantitative way for late-onset, incompletely-penetrant phenotypes, such as TTN-related DCM. A high proportion of Clinvar (likely) pathogenic variants (21.6% among high-PSI variants, 12.1% among all variants, Supplementary Table S8) among the TTNtv included in gnomAD cohort indicates that the control group may not consist of individuals with a purely normal cardiac phenotype. Although exon 327 remained the most significantly enriched area when both DCM and gnomAD cohorts were considered, we posit that the results from DCM cohorts are a more direct reflection of the status of TTNtv distribution in patients with DCM.
The pathogenic mechanism of TTNtv leading to DCM is not well understood. It was once proposed that Cronos titin isoform, which encodes the most distal region of the I-band, the A-band and the M-band, can rescue the phenotype explaining the A-band enrichment (Zou et al., 2015; Schafer et al., 2017; Santiago et al., 2021). However, TTNtvs located in pre-Cronos regions also showed high OR (Schafer et al., 2017) and the absence of compensatory changes in Cronos isoform was demonstrated (Fomin et al., 2021), contradicting the hypothesis. The debate between haploinsufficiency and a dominant negative mechanism is ongoing (Santiago et al., 2021) and despite the clues for dominant negative mechanisms, such as the tendency of higher pathogenicity of more distally located TTNtv (Roberts et al., 2015), direct evidence had not been observed until the existence of truncated titin proteins was proved in affected human heart tissue in two recent studies (Fomin et al., 2021; McAfee et al., 2021). In these two reports, truncated titin protein was quantitatively detected by methods, such as Western blotting. Truncated titin proteins were present in intracellular aggregates rather than sarcomeres (Fomin et al., 2021), and the sizes of truncated proteins corresponded to that predicted by the location of TTNtvs (McAfee et al., 2021). Disease severity did not correlate with TTNtv location, but the level of truncated titin protein negatively correlated with patient age at HT (Fomin et al., 2021). These two reports suggested a multifactorial mechanism of combined dominant-negative and haploinsufficiency.
In the RNAseq data analysis, a negative correlation between PTC-to-intron distance and AI deviation was observed, confirming the negative effect of long PTC-to-intron distance on NMD efficiency in the literature (Hoek et al., 2019; Supek et al., 2021). DCM-related TTNtv showed generally reduced NMD efficiency (median AI deviation = .10), and exon 327 TTNtv showed almost no NMD (median AI deviation = .04). Although the difference between exon 327 TTNtv and other DCM-related TTNtv did not reach statistical significance (p = .0501), the low AI deviation values generally observed for DCM-related TTNtvs demonstrated the importance of NMD evasion in the pathogenesis of TTNtv-related DCM. This result is in concordance with a report in which TTN was included in the list of genes enriched for pathogenic truncating variants that do not trigger NMD (Lindeboom et al., 2019). The extremely low NMD efficiency of exon 327 TTNtvs, presumably caused by the long PTC-to-intron distance, seems to be a cause of the high frequency of exon 327 TTNtvs observed in patients with DCM (Figure 4).
There was no significant difference in the clinical outcome parameters between patients with exon 327 TTNtvs and those with other TTNtvs. However, when limited to exon 327 TTNtvs, the age at HT showed a negative correlation with PTC-to-intron distance, indicating that extremely reduced NMD efficiency may affect the clinical outcome. However, whether predicting the severity of DCM based on the PTC-to-intron distance of TTNtv is possible remains unclear.
Although NMD efficiency seems to explain certain levels of the pathogenicity of TTNtv, especially in exon 327, other factors might contribute to its pathogenesis. The level of truncated titin protein should have a more direct effect on penetrance and pathogenicity than the mRNA level. Considering the observation that decreased NMD efficiency did not lead to a proportionally increased production of truncated proteins (Fomin et al., 2021), factors other than NMD might regulate the production and degradation of truncated proteins. Because of the small sample size used in the study that negated the direct relationship between NMD efficiency and protein levels (n = 11) (Fomin et al., 2021), further investigation of the regulation of mRNA and protein levels and its effect on clinical outcomes is warranted.
In conclusion, the accepted A-band enrichment of DCM-related TTNtv was mostly attributable to exon 327 enrichment. The extremely low NMD efficiency of exon 327 TTNtvs, presumably caused by the long PTC-to-intron distance, might cause the high frequency of exon 327 TTNtvs observed in patients with DCM. Reduced NMD efficiency may play an important role in the pathogenesis of TTNtv-related DCM, particularly in exon 327.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent from the participants’ legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author contributions
Y-gK conceived the study and drafted the manuscript. CH carried out data acquisition and data analysis. SS, J-hP, and J-HJ contributed to data interpretation and manuscript revision. J-WK supervised the study and performed the final manuscript revision.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (NRF-2022R1A2C2091571, to J-WK).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1087359/full#supplementary-material
References
Akhtar, M. M., Lorenzini, M., Cicerchia, M., Ochoa, J. P., Hey, T. M., Sabater Molina, M., et al. (2020). Clinical phenotypes and prognosis of dilated cardiomyopathy caused by truncating variants in the TTN gene. Circ. Heart Fail. 13, e006832. doi:10.1161/CIRCHEARTFAILURE.119.006832
Akinrinade, O., Alastalo, T. P., and Koskenvuo, J. W. (2016). Relevance of truncating titin mutations in dilated cardiomyopathy. Clin. Genet. 90, 49–54. doi:10.1111/cge.12741
Fomin, A., Gartner, A., Cyganek, L., Tiburcy, M., Tuleta, I., Wellers, L., et al. (2021). Truncated titin proteins and titin haploinsufficiency are targets for functional recovery in human cardiomyopathy due to TTN mutations. Sci. Transl. Med. 13, eabd3079. doi:10.1126/scitranslmed.abd3079
Franaszczyk, M., Chmielewski, P., Truszkowska, G., Stawinski, P., Michalak, E., Rydzanicz, M., et al. (2017). Titin truncating variants in dilated cardiomyopathy - prevalence and genotype-phenotype correlations. PLoS One 12, e0169007. doi:10.1371/journal.pone.0169007
Freeman, P. J., Hart, R. K., Gretton, L. J., Brookes, A. J., and Dalgleish, R. (2018). VariantValidator: Accurate validation, mapping, and formatting of sequence variation descriptions. Hum. Mutat. 39, 61–68. doi:10.1002/humu.23348
Gorlov, I. P., Gorlova, O. Y., Frazier, M. L., Spitz, M. R., and Amos, C. I. (2011). Evolutionary evidence of the effect of rare variants on disease etiology. Clin. Genet. 79, 199–206. doi:10.1111/j.1399-0004.2010.01535.x
Haggerty, C. M., Damrauer, S. M., Levin, M. G., Birtwell, D., Carey, D. J., Golden, A. M., et al. (2019). Genomics-first evaluation of heart disease associated with titin-truncating variants. Circulation 140, 42–54. doi:10.1161/CIRCULATIONAHA.119.039573
Herman, D. S., Lam, L., Taylor, M. R., Wang, L., Teekakirikul, P., Christodoulou, D., et al. (2012). Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 366, 619–628. doi:10.1056/NEJMoa1110186
Hoek, T. A., Khuperkar, D., Lindeboom, R. G. H., Sonneveld, S., Verhagen, B. M. P., Boersma, S., et al. (2019). Single-molecule imaging uncovers rules governing nonsense-mediated mRNA decay. Mol. Cell. 75, 324–339. doi:10.1016/j.molcel.2019.05.008
Jansen, M., Baas, A. F., van Spaendonck-Zwarts, K. Y., Ummels, A. S., van den Wijngaard, A., Jongbloed, J. D. H., et al. (2019). Mortality risk associated with truncating founder mutations in titin. Circ. Genom. Precis. Med. 12, e002436. doi:10.1161/CIRCGEN.118.002436
Jansweijer, J. A., Nieuwhof, K., Russo, F., Hoorntje, E. T., Jongbloed, J. D., Lekanne Deprez, R. H., et al. (2017). Truncating titin mutations are associated with a mild and treatable form of dilated cardiomyopathy. Eur. J. Heart Fail. 19, 512–521. doi:10.1002/ejhf.673
Karczewski, K. J., Francioli, L. C., Tiao, G., Cummings, B. B., Alfoldi, J., Wang, Q., et al. (2020). The mutational constraint spectrum quantified from variation in 141, 456 humans. Nature 581, 434–443. doi:10.1038/s41586-020-2308-7
Karolchik, D., Hinrichs, A. S., Furey, T. S., Roskin, K. M., Sugnet, C. W., Haussler, D., et al. (2004). The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496. doi:10.1093/nar/gkh103
Lindeboom, R. G. H., Vermeulen, M., Lehner, B., and Supek, F. (2019). The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy. Nat. Genet. 51, 1645–1651. doi:10.1038/s41588-019-0517-5
Lindeboom, R. G., Supek, F., and Lehner, B. (2016). The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat. Genet. 48, 1112–1118. doi:10.1038/ng.3664
Mazzarotto, F., Tayal, U., Buchan, R. J., Midwinter, W., Wilk, A., Whiffin, N., et al. (2020). Reevaluating the genetic contribution of monogenic dilated cardiomyopathy. Circulation 141, 387–398. doi:10.1161/CIRCULATIONAHA.119.037661
McAfee, Q., Chen, C. Y., Yang, Y., Caporizzo, M. A., Morley, M., Babu, A., et al. (2021). Truncated titin proteins in dilated cardiomyopathy. Sci. Transl. Med. 13, eabd7287. doi:10.1126/scitranslmed.abd7287
Pugh, T. J., Kelly, M. A., Gowrisankar, S., Hynes, E., Seidman, M. A., Baxter, S. M., et al. (2014). The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet. Med. 16, 601–608. doi:10.1038/gim.2013.204
Rich, K. A., Moscarello, T., Siskind, C., Brock, G., Tan, C. A., Vatta, M., et al. (2020). Novel heterozygous truncating titin variants affecting the A-band are associated with cardiomyopathy and myopathy/muscular dystrophy. Mol. Genet. Genomic Med. 8, e1460. doi:10.1002/mgg3.1460
Roberts, A. M., Ware, J. S., Herman, D. S., Schafer, S., Baksi, J., Bick, A. G., et al. (2015). Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci. Transl. Med. 7, 270ra6. doi:10.1126/scitranslmed.3010134
Santiago, C. F., Huttner, I. G., and Fatkin, D. (2021). Mechanisms of TTNtv-related dilated cardiomyopathy: Insights from zebrafish models. J. Cardiovasc. Dev. Dis. 8, 10. doi:10.3390/jcdd8020010
Schafer, S., de Marvao, A., Adami, E., Fiedler, L. R., Ng, B., Khin, E., et al. (2017). Titin-truncating variants affect heart function in disease cohorts and the general population. Nat. Genet. 49, 46–53. doi:10.1038/ng.3719
Supek, F., Lehner, B., and Lindeboom, R. G. H. (2021). To NMD or not to NMD: Nonsense-mediated mRNA decay in cancer and other genetic diseases. Trends Genet. 37, 657–668. doi:10.1016/j.tig.2020.11.002
Tayal, U., Newsome, S., Buchan, R., Whiffin, N., Halliday, B., Lota, A., et al. (2017). Phenotype and clinical outcomes of titin cardiomyopathy. J. Am. Coll. Cardiol. 70, 2264–2274. doi:10.1016/j.jacc.2017.08.063
Tharp, C. A., Haywood, M. E., Sbaizero, O., Taylor, M. R. G., and Mestroni, L. (2019). The giant protein titin's role in cardiomyopathy: Genetic, transcriptional, and post-translational modifications of TTN and their contribution to cardiac disease. Front. Physiol. 10, 1436. doi:10.3389/fphys.2019.01436
Yotti, R., Seidman, C. E., and Seidman, J. G. (2019). Advances in the genetic basis and pathogenesis of sarcomere cardiomyopathies. Annu. Rev. Genomics Hum. Genet. 20, 129–153. doi:10.1146/annurev-genom-083118-015306
Keywords: TTN, truncating variant, exon 327, dilated cardiomyopathy, nonsense-mediated mRNA decay
Citation: Kim Y-g, Ha C, Shin S, Park J-h, Jang J-H and Kim J-W (2023) Enrichment of titin-truncating variants in exon 327 in dilated cardiomyopathy and its relevance to reduced nonsense-mediated mRNA decay efficiency. Front. Genet. 13:1087359. doi: 10.3389/fgene.2022.1087359
Received: 02 November 2022; Accepted: 07 December 2022;
Published: 04 January 2023.
Edited by:
Andrea Ciolfi, Bambino Gesù Children’s Hospital (IRCCS), ItalyReviewed by:
Bhagyalaxmi Mohapatra, Banaras Hindu University, IndiaPer Harald Jonson, Faculty of Medicine, University of Helsinki, Finland
Copyright © 2023 Kim, Ha, Shin, Park, Jang and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jong-Won Kim, Y3VsdHVyZS5qa2ltQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Young-gon Kim
Young-gon Kim Changhee Ha1†
Changhee Ha1† Sunghwan Shin
Sunghwan Shin Jong-ho Park
Jong-ho Park Jong-Won Kim
Jong-Won Kim