
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 06 January 2023
Sec. Evolutionary and Population Genetics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1083578
This article is part of the Research Topic Aquatic Genomics and Transcriptomics for Evolutionary Biology - Volume II View all 6 articles
Toll-like receptors 5 (TLR5), a member of the toll-like receptors (TLRs) family, is a class of pattern recognition receptors (PRRs) that recognize pathogen-associated molecular patterns (PAMPs). It responds to vertebrate recognition of bacterial flagellin and participates in innate immune responses. However, genome-wide identification and characterization of TLR5 in fishes have not been investigated. Here, three TLR5M isotypes (TLR5Ma, TLR5Mb1, and TLR5Mb2) and a TLR5S are all extracted from fish genomes on the basis of phylogenetic and synteny analyses. We confirmed that the non-teleost fishes have one TLR5M gene, as well as additional TLR5 genes (TLR5M and TLR5S) in teleost fishes. In addition, some special teleost fishes possess two to three TLR5 genes, which have undergone the fourth whole-genome duplication (WGD). According to our results, we inferred that the diversity of TLR5 genes in fishes seems to be the result of combinations of WGD and gene loss. Furthermore, TLR5 isoforms displayed differences at the flagellin interaction sites and viral binding sites, and showed lineage-specific, which indicated that TLR5 duplicates may generate functional divergence. Bacterial experiments also supported the idea that CiTLR5Ma and CiTLR5Mb are subfunctionalized to sense bacterial flagellin. In summary, our present comparative genomic survey will benefit for further functional investigations of TLR5 genes in fish.
In the innate immune landscape, toll-like receptors (TLRs) play a major role by activating the first line of defense against invading microbial pathogens in invertebrate and vertebrate lineages (Purcell et al., 2006; Brikos and O'Neill, 2008). TLRs are antigen-recognition receptors on the surfaces or inside cells that directly recognize pathogen-associated molecular patterns (PAMPs) (Mogensen, 2009). All TLRs have a leucine-rich repeat (LRR) ectodomain for PAMP recognition, transmembrane (TM) domain for TLR dimerization and stabilization, and toll/IL-1 receptor (TIR) domain associated with signaling (Brubaker et al., 2015; Voogdt et al., 2016; Voogdt et al., 2018; Su and Yu, 2019; Gao et al., 2022; Liao et al., 2022). According to the previous data, at least 13 TLR types have been reported in mammals (Janeway and Medzhitov, 2002; Akira et al., 2006); for example, 12 (TLR1–9, TLR11–13) and 10 (TLR1–10) TLR genes have been reported in the mice and human genome, respectively. Moreover, 22 TLR genes were identified in the fish genome (Palti, 2011). Furthermore, fish-specific TLRs (soluble TLR5, TLR14, TLR18–20, and TLR22–28) and mammalian TLR orthologs (TLR1, TLR2, TLR3, TLR5, TLR7, TLR8, and TLR9) are included in the teleost fish TLRs (Matsuo et al., 2008; Palti, 2011).
As a TLR family member, TLR5 is a critical factor in initiating the innate immune response and triggering adaptive immunity (Hayashi et al., 2001; Didierlaurent et al., 2004), which can activate flagellin-mediated NF-κB via the MyD88-dependent pathway in the cellular membrane (Mizel et al., 2003). Previously, TLR5 in teleosts was divided into the membrane and soluble forms of TLR5 (TLR5M and TLR5S), respectively (Oshiumi et al., 2003; Tsujita et al., 2004), which are reported in several fishes, including orange-spotted grouper (Epinephelus coioides) (Bai et al., 2017), rainbow trout (Oncorhynchus mykiss) (Tsujita et al., 2004), fugu (Fugu rubripes) (Oshiumi et al., 2003), Japanese flounder (Paralichthys olivaceus) (Hwang et al., 2010), and golden pompano (Trachinotus ovatus) (Zhu et al., 2020). Recent studies reported that TLR5M and TLR5S play vital roles in TLR/IL-1R signaling pathways and immune response to the invasions of a broad range of pathogens in fish. Furthermore, TLR5M stimulation with Vibrio anguillarum or its flagellin possibly activated TLR5S expression. Moreover, TLR5M signaling was amplified in rainbow trout flagellin through interaction with TLR5S in positive loop feedback (Tsujita et al., 2004). In addition, studies revealed different TLR5M types in turbot and blunt snout bream, namely TLR5a and TLR5b, respectively (Liu et al., 2017; Zhan et al., 2019).
Although there were reports of TLR5 types in several bony fishes (Jiang et al., 2015; Han et al., 2017; Gao et al., 2022), the molecular evolution and a comprehensive comparative genomic survey of these gene families in different fishes have not yet been reported. Rapid advancement in sequencing technologies has enabled access to high-quality whole-genome data of many fish species, thus allowing for the extraction of fish TLR5 genes and their encoding sequences, including, tetraploid Sinocyclocheilus fishes, amphibious mudskippers, salmonids, deep-sea snailfish, cartilaginous sharks, lobe-finned fish (coelacanth), hagfish, and jawless sea lamprey. This enables the study of the presence or absence of species-specific TLR5 isotypes and differences (variations) in sequences across different species. This study investigated fish TLR5 genes and protein structures after extracting nucleotide sequences. Afterward, phylogenetic and synteny analyses were performed. Moreover, the expression patterns and primary functions of the fish TLR5 genes after bacterial infection were identified. These results are valuable for future research and lay a solid foundation for investigating the mechanisms underlying fish TLR5 genes.
A total of 30 species of fish were included in this research. Two methods were used to obtain data; first, the unreported TLR5 sequences from 28 species of fish were used and obtained from our complete genomic data (Table 1). Second, Ensembl (https://asia.ensembl.org/index.html) and GenBank (https://www.ncbi.nlm.nih.gov/genbank/) public databases were utilized for the purpose of downloading published TLR5 sequence data. Through tBLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi), the prospective homology-based TLR5 genes were recovered from fish genomes (Pevsner, 2009), with an e-value of 10–5. The best hit for each alignment was found using the BLAST results treated with the Perl script. Finally, the TLR5 genes were predicted from the best hits using GeneWise v2.2.0 (Birney et al., 2004).
Phylogenetic analysis was conducted by utilizing nucleotide and protein sequences of all TLR5 genes. Protein sequences of TLR5 were aligned using the MAFFT software (Yamada et al., 2016), and RAxML8.0.17 was employed to conduct a maximum likelihood (ML) phylogenetic analysis (Stamatakis, 2006; Stamatakis et al., 2008). The FastTree v2.1.7 software was employed to generate the ML phylogenetic trees of the TLR5 isotypes based on their corresponding coding sequences (Price et al., 2009). In addition, a protein model of human TLR5M was downloaded from the public Protein DataBank to compare the structural differences of fish TLR5 proteins.
The conservation of TLR5 genes was assessed by observing the genes in the up and downstream regions of each TLR5M and TLR5S paralog. Moreover, associated genomic data was also obtained from GenBank and our lab. The genome of zebrafish was considered the reference standard for any down and upstream regions of TLR5.
Healthy and juvenile Ctenopharyngodon idellus (80 ± 20 g) were purchased from a farm in Guangdong Province, China. The fish were kept at 25°C–26°C for 2 weeks in a flow-through water system to ensure their acclimatization to the laboratory conditions before the experiments.
Luria-Bertani (LB) broth was used for the culturing of Aeromonas hydrophila (ZK2022061) at a temperature of 37°C and utilized for immune system challenges. After 12 h, bacteria were washed twice with sterile phosphate-buffered saline (PBS), and the bacterial concentration was adjusted to 3 × 107 colony-forming units/mL. C. idellus was divided into A. hydrophila and PBS control groups. Each fish was injected with 100 μl of the bacterial suspension or PBS (control group). At time intervals of 0, 6, 9, 12, 24, and 48 h after infection, three replicates of the fish’s important immune organs (head kidney, liver, and spleen) were collected from the groups, immediately frozen in liquid nitrogen, and stored at −80°C.
The total RNA was extracted using TRIzol (Life Technologies, California, United States), synthesized to the first-strand cDNA using HIScript Q Select RT SuperMix (Vazyme, Nanjing, China), and stored at -20°C for qRT-PCR detection. All steps were performed according to the manufacturer’s instructions. The five-fold dilution of cDNA templates was carried out, and β-actin was employed as the internal control. Using the obtained sequences of C. idellus, the primers of the gene of interest were designed using Primer Premier 6.0, and the primers of β-actin were used herein. All primers are presented in Supplementary Table S1. The total reaction volume of 20 μl was designed as follows: 4 μl diluted cDNA, 0.5 μl of each specific primer, 10 μl AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China), and 5 μl Diethyl Pyrocarbonate (DEPC) water. The amplification was performed under the following conditions: 5 min at 95°C, 10 s at 95°C (40 cycles), and 30 s at 60°C (40 cycles). The experiment was conducted in triplicates, and the relative levels of expression of the target genes were calculated by employing the 2−ΔΔCT method (Schmittgen and Livak, 2008). The discrepancy between different treatments was analyzed using the one-way analysis of variance (ANOVA), and p < 0.05 or p < 0.01 was considered statistically significant.
In total, 40 vertebrate species (Table 1) were studied to collect the TLR5 sequences, and only a single TLR5 gene was identified among the genomes of Chondrichthyes, Sarcopterygii, reptiles, amphibians, and mammals. According to previous reports, teleosts contain two TLR5 genes (Oshiumi et al., 2003); however, the present study confirmed that the teleosts contain four TLR5 genes. One or two TLR5 genes were present in the common diploid teleost, which is consistent with previous findings (Oshiumi et al., 2003; Zhu et al., 2020). The economically important Tilapia (Oreochromis mossambicus) and large yellow croaker (Larimichthys crocea) possessed two TLR5 forms, TLR5M and TLR5S (Table 1). Interestingly, in Black porgy (Acanthopagrus schlegelii) and Mariana hadal snailfish (Pseudoliparis swirei), only TLR5M was observed while TLR5S was absent. Two copies of the TLR5M gene, temporally named TLR5Ma and TLR5Mb, and one isotype, TLR5S, were observed in the typical tetraploid teleost, such as Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss), which experienced the salmonid-specific genome duplication. In tetraploid Cyprinidae, which underwent carp-specific genome duplication, two copies of TLR5 genes, TLR5Ma and TLR5Mb, were observed, with no TLR5S gene. However, the tetraploid Sinocyclocheilus fish possessed double copies of the TLR5Mb gene, temporally named TLR5Mb1 and TLR5Mb2, and one TLR5Ma gene.
A phylogenetic analysis was performed using protein sequences of all TLR5 genes (Figure 1). These TLR5 proteins were divided into three main clades with high support values (L1, L2, and L3). In the ancient spotted garfish (Lepisosteus oculatus), TLR5M was located out of the whole teleost TLR5 clade, whereas a single TLR5 gene in ancient coelacanth fish (Latimeria chalumnae) was located within the mammalian clade. Furthermore, diploid teleost TLR5 genes were classified into TLR5M and TLR5S subgroups, consistent with the teleost experiencing the third teleost-specific WGD. TLR5Ma, TLR5Mb, and TLR5S in Protacanthopterygii, including rainbow trout, and Atlantic salmon diverged into three different clades. In Cyprinidae, such as zebrafish, grass carp, common carp, and Sinocyclocheilus, the TLR5 genes comprised the L3 clade, which was also divided into TLR5Ma and TLR5Mb/b1/b2 subgroups.
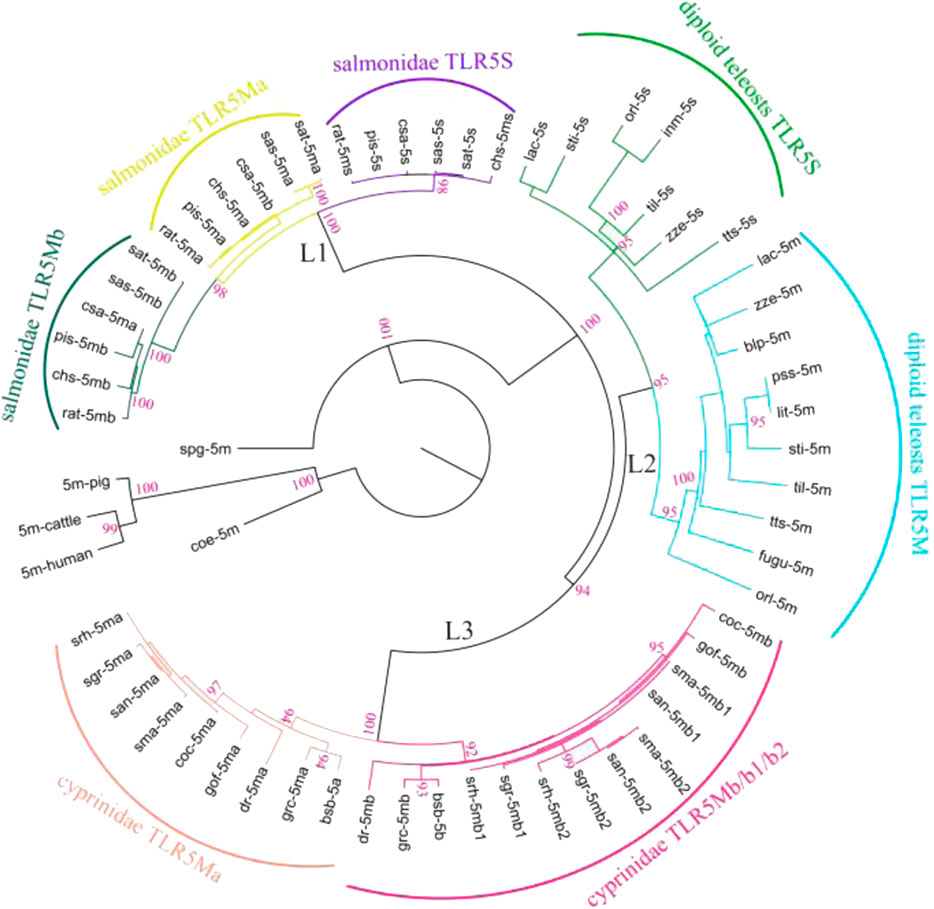
FIGURE 1. Phylogenetic relationships among various fish TLR5s. Representative TLR5 genes were categorized into three main clades (L1, L2, and L3). Furthermore, diploid teleost TLR5 genes were classified into two subgroups (diploid teleost TLR5S and TLR5M). TLR5 genes in Salmonidae were clustered into three different clades (salmonidae TLR5S, TLR5Mb, and TLR5Mb). TLR5 genes in Cyprinidae were divided into two subgroups (cyprinidae TLR5Ma and TLR5Mb/b1/b2). Detailed abbreviation of each species is available in Table 1.
For the assessment of the chromosomal or scaffold synteny, in silico searches were carried out for conserved genes downstream and upstream of the TLR5 isotypes in various fishes. It was observed that all TLR5 genes shared a conserved suite of genes binding them on their side (Figure 2); however, several species displayed gene loss. Interestingly, TLR5Ma and TLR5Mb/b1 of Cyprinidae, such as zebrafish, grass carp, common carp, and Sinocyclocheilus, were closely located within the same chromosome, whereas TLR5Mb2 of Sinocyclocheilus was located on another chromosome. Moreover, TLR5S and TLR5M reside on the same chromosome or scaffold with minor variations in the location in some diploid teleost, including medaka and tilapia. Furthermore, the collinear analysis also established the reliability of the extracted TLR5 genes.

FIGURE 2. Synteny analyses of the chromosome loci harboring TLR5 genes in fishes. Different colored rectangles represent different gene loci. Some gene loci near TLR5 present conserved synteny in these examined fishes. Detailed abbreviation of each species is available in Table 1.
The domain features of the TLR5 types were analyzed among various fishes (Figure 3). The domains of the representative fish TLR5M members, such as TLR5Ma and TLR5Mb/b1, contained three domains, including the LRR ectodomain, TM domain, and TIR domain. However, Sinocyclocheilus TLR5Mb2 consisted of only the LRR and TM domains. Fish TLR5S only contained the LRR domains. Moreover, the number of LRR domains varied not only across different TLR5 types of a single species but also across different fish species.

FIGURE 3. The domain features of TLR5 genes among fishes. Detailed abbreviation of each species is available in Table 1.
Representative TLR5 protein sequences were aligned to check the secondary structures of these TLR5 types in fishes (Supplementary Figure S1). The research on amino acid sites associated with the functional and structural features of TLR5 proteins was carried out. At the two disulfide bonds (583–610 and 585–629 residues), most residues were well conserved among these TLR5 sequences. Some residues around the glycosylation sites, including 37, 46, 245, 342, 422, 595, and 598, were non-conserved. Furthermore, three residues (294, 366, and 342) were responsible for the flagellin interaction; however, one of them showed differences among the fish TLR5 isoforms, such as D294-S294-N294. In the TIR domain, two significant residues, including Pro736 and Tyr798, were well conserved. Pro736 binds to the signaling adaptor molecule MyD88, and Tyr798 phosphorylates upon flagellin recognition. In addition, the amino acid difference was identified in the TLR5 residue 268 (G268S).
To determine the role of TLR5Ma and TLR5Mb in the immune response against A. hydrophila infections, we examined their expression patterns in the liver, spleen, and head kidney tissues of C. idellus (Figure 4). The transcription levels of CiTLR5a and CiTLR5b were highest in the liver, spleen, and head kidney after 9 h of A. hydrophila infection. Subsequently, the transcription levels gradually declined but were still higher than the control group. These results suggest that TLR5 plays a crucial role in the antibacterial immunity of C. idellus.
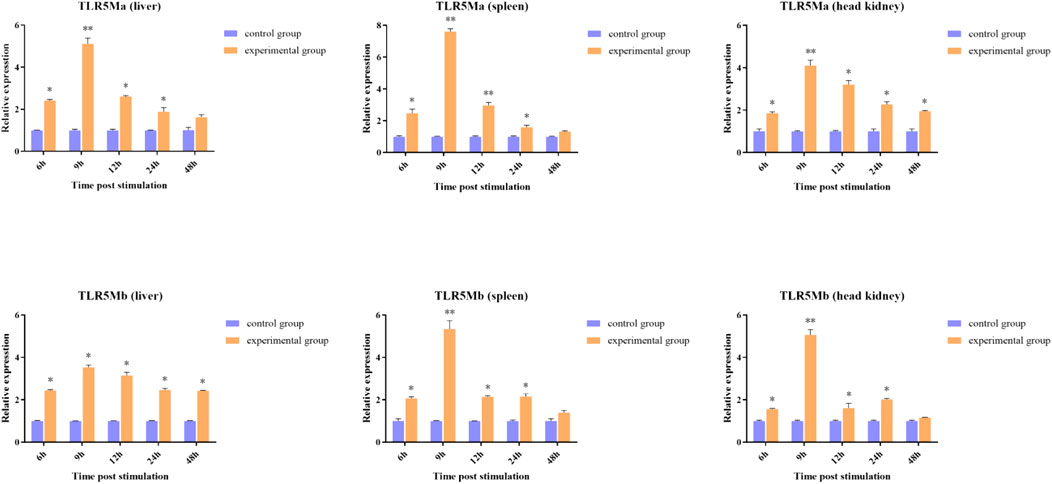
FIGURE 4. The expression patterns of CiTLR5Ma and CiTLR5Mb on transcription level in tissues of liver, spleen and head kidney, after infected with A. hydrophila. The expression level of CiTLR5Ma and CiTLR5Mb on 0 h was used as the baseline calibrator. The significance levels were obtained by comparing with corresponding 0 h (*p < 0.05, **p < 0.01).
Two rounds of WGD have been proven to occur in the common ancestry of early vertebrates before ray-finned fishes–tetrapod split (Guyomard et al., 2012; Glasauer and Neuhauss, 2014). Specifically, the first round of WGD occurred before the split of agnatha–gnathostomata, and the second round occurred before the split of chrondrichthyes–osteichthyes. While the teleost-specific WGD, i.e., the third WGD, occurred solely in teleosts (Amores et al., 1998; Meyer and Schartl, 1999; Hoegg et al., 2004; Vandepoele et al., 2004). Moreover, a fourth WGD was observed in various teleost lineages, for instance, Sinocyclocheilus fishes, goldfish, salmonids, and common carp (Mayfield-Jones et al., 2013; Lien et al., 2016; Chen et al., 2019). Genome and gene duplications have contributed to teleosts’ rich evolutionary history and genomic diversity (Kaessmann, 2010).
According to the present study, WGD and gene loss may lead to copy number variations of the TLR5 gene in various fishes. Tetrapods and ancient fishes, including Actinistia coelacanth and Neopterygii spotted-gar, possess only one TLR5 gene, i.e., TLR5M. The TLR5 gene formed two copies, TLR5M and TLR5S, after undergoing teleost-specific WGD; therefore, two copies of the gene might be present in the teleost genomes compared to their tetrapod counterparts. Therefore, the teleost TLR5 gene splits into two forms, including TLR5M and TLR5S. Furthermore, the existence of two TLR5 genes is due to tandem duplication and not genome duplication (Figure 5).
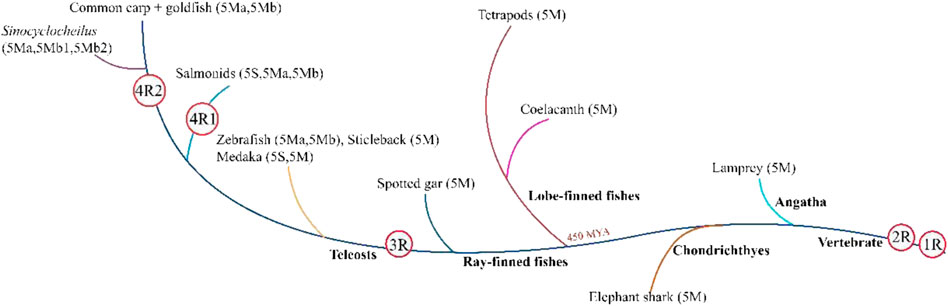
FIGURE 5. All teleost fishes undergone at least three whole-genome duplication (WGD) events, the two earlier rounds of vertebrate genome duplication (1R and 2R) that occurred before the di-vergence of lamprey and hagfish from the jawed vertebrates, and the third teleost-specific WGD (3R) at the base of the teleosts 320 million years ago (MYA). Atlantic salmon and river trout share the salmonid-specific WGD (4R 1) that occurred in the common ancestor of salmonids 80 MYA. Common carp and goldfish share the fourth WGD (4R) that occurred approximately 14 MYA in their common ancestor. Atlantic salmon and river trout share the salmonid-specific WGD (4R 1) that occurred in the common ancestor of salmonids 80 MYA. Common and goldfish share the fourth WGD (4R 2) that occurred approximately 14 MYA in their common ancestor. The TLR5 isoform is given in parentheses.
In tetraploid teleosts, such as rainbow trout, the salmonid-specific genome duplication develops three copies of the TLR5 genes, also studied in the Atlantic salmon (Lien et al., 2016). TLR5S was lost in other tetraploid carps, generating two TLR5M genes in its genome. However, in tetraploid golden-line fishes, three TLR5M isoforms are present, including TLR5Ma, TLR5Mb1, and TLR5Mb2. Additionally, as per the synteny analysis, the generation of three TLR5M genes is due to tandem (TLR5Ma and TLR5Mb1) and genome duplications (TLR5Mb2). Moreover, copy numbers are not always in the one-to-two correspondence between tetraploid and diploid species, presumably due to the selective loss of some copies.
Generally, WGD duplicates escape gene loss due to subfunctionalization or neofunctionalization (Jaillon et al., 2004). Teleost retained multiple isoforms, and Chondrichthyes only had one isoform of TLR5. The study revealed that fish TLR5 genes went through the primitive M (spotted gar), duplicated M/M (zebrafish), M/S (medaka and tilapia), tetraploid fish M/M/S (salmonids), and M/M/M (Sinocyclocheilus fish) evolution types. Membrane and soluble members of the M/S type play a synergistic role in sensing flagellin (Tsujita et al., 2004) possibly implying that soluble TLR5 is redundant in function and disappears in some fish and endothermic vertebrates. Cyprinid-specific duplicated membrane TLR5 (M/M type) demonstrated neofunctionalization to sense viral dsRNA as functional homodimeric receptors in antagonistic effect (Liao et al., 2022). Domain number variation was identified in the fish TLR5 isotypes, generating functional diversity and complexity (Asami and Shimizu, 2021). Furthermore, the TLR5 genes vary in the number of the extracellular LRR domain involved in the recognition of pathogens, contributing to TLR5 specificity (Asami and Shimizu, 2021; Shan et al., 2021). These findings suggest that the duplicated membrane TLR5 was sequence and species-specific for dsRNA binding. Many residues around the putative flagellin binding site were highly conserved among the TLR5 types, with only a few variations, such as residues 268 and 294. A study conducted previously indicates that these differences might influence the affinity of TLR5 for flagellins from different bacterial species (Andersen-Nissen et al., 2007). The fish TLR5 isoforms exhibited variations at the dsRNA binding sites (98 residues), possibly affecting viral immune response. Therefore, the newly predicated TLR5 duplicates in teleost may generate functional divergence and play a part in the adaption of these fishes during the evolutionary process.
Previous studies on Cyprinus carpio have revealed that TLR5M responds to flagellin from A. hydrophila (Duan et al., 2013). TLR5M transcription was subjected to remarkable upregulation after stimulation with lipopolysaccharide (LPS) in immune-related tissues in Siniperca chuatsi (Wang et al., 2021). The transcription of TLR5S and TLR5M was clearly altered after stimulation by polyinosinic:polycytidylic acid (poly (I:C)), LPS, and flagellin in immune-related organs of Golden Pompano, Trachinotus ovatus (Zhu et al., 2020). Moreover, the expression level of TLR5 was remarkably upregulated in all tissues tested of Nile tilapia in response to Streptococcus agalactiae infection (Gao et al., 2022). These studies indicated that TLR5 plays important roles in the immune response to pathogen invasion. For the investigation of the expression of genes in response to A. hydrophila infection, to determine the role of TLR5M duplicates in the immune response, we employed Quantitative real-time Polymerase Chain Reaction (qRT-PCR). After A. hydrophila infection, overexpression of CiTLR5Ma and CiTLR5Mb genes were observed in immune system tissues and vital organs, including the liver, spleen, and head kidney, suggestive of their important roles in the immune response to the invasion of pathogens. Moreover, CiTLR5Ma and CiTLR5Mb showed similar expression patterns across tissues, which supports the finding that CiTLR5Ma and CiTLR5Mb are subfunctionalized to sense bacterial flagellin as a heterodimer (Liao et al., 2022).
In summary, the present study surveyed many facets of the fish TLR5 genes, offering a worldwide genomic view of the diversity of the TLR5 gene family of the fish. The existence of three TLR5M genes, TLR5Ma, TLR5Mb1, and TLR5Mb2, and one TLR5S in fish was confirmed in the study. Furthermore, the copy number variation in TLR5 genes in fishes likely resulted from the integration of WGD and gene loss. In addition, differences between the flagellin interaction sites and viral binding sites were identified among TLR5 isoforms, speculating that the fish TLR5 genes possibly have different functions. The present study indicated that TLR5 duplicates are expressed in several tissues with similar transcription levels after a bacterial infection. These findings supported the view that TLR5 duplicates obtain sub-functionalization or neofunctionalization to sense pathogens.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by All animal experiments were conducted after the formal approval of the Animal Ethics Committee of Zhongkai University of Agriculture and Engineering.
KZ analysed the data and wrote the manuscript. MC conducted the experiments. KZ and MC prepared the figures and tables. HH and HK collected the samples and discussed the experimental results. RL, LL, and KZ conceived and designed the experiments, contributed reagents/materials/analysis tools, supervised the work, and reviewed drafts of the paper. All authors have read and agreed to the published version of the manuscript.
This research was funded by the “Innovation and Strong Universities” special fund from the Department of Education of Guangdong Province (KA2001960), the Science and Technology Plan Project of Qingyuan City (2022KJJH064), and the National Natural Science Foundation of China (31802300).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1083578/full#supplementary-material
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell. 124, 783–801. doi:10.1016/j.cell.2006.02.015
Amores, A., Force, A., Yan, Y. L., Joly, L., Amemiya, C., Fritz, A., et al. (1998). Zebrafish hox clusters and vertebrate genome evolution. Science 282, 1711–1714. doi:10.1126/science.282.5394.1711
Andersen-Nissen, E., Smith, K. D., Bonneau, R., Strong, R. K., and Aderem, A. (2007). A conserved surface on Toll-like receptor 5 recognizes bacterial flagellin. J. Exp. Med. 204, 393–403. doi:10.1084/jem.20061400
Asami, J., and Shimizu, T. (2021). Structural and functional understanding of the toll-like receptors. Protein Sci. 30, 761–772. doi:10.1002/pro.4043
Bai, J. S., Li, Y. W., Deng, Y., Huang, Y. Q., He, S. H., Dai, J., et al. (2017). Molecular identification and expression analysis of TLR5M and TLR5S from orange-spotted grouper (Epinepheluscoioides). Fish. Shellfish Immunol. 63, 97–102. doi:10.1016/j.fsi.2017.01.037
Birney, E., Clamp, M., and Durbin, R. (2004). GeneWise and genomewise. Genome Res. 14, 988–995. doi:10.1101/gr.1865504
Brikos, C., and O'neill, L. A. (2008). Signalling of toll-like receptors. Handb. Exp. Pharmacol. 2008, 21–50. doi:10.1007/978-3-540-72167-3_2
Brubaker, S. W., Bonham, K. S., Zanoni, I., and Kagan, J. C. (2015). Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 33, 257–290. doi:10.1146/annurev-immunol-032414-112240
Chen, Z., Omori, Y., Koren, S., Shirokiya, T., Kuroda, T., Miyamoto, A., et al. (2019). De novo assembly of the goldfish (Carassius auratus) genome and the evolution of genes after whole-genome duplication. Sci. Adv. 5, eaav0547. doi:10.1126/sciadv.aav0547
Didierlaurent, A., Ferrero, I., Otten, L. A., Dubois, B., Reinhardt, M., Carlsen, H., et al. (2004). Flagellin promotes myeloid differentiation factor 88-dependent development of Th2-type response. J. Immunol. 172, 6922–6930. doi:10.4049/jimmunol.172.11.6922
Duan, D., Sun, Z., Jia, S., Chen, Y., Feng, X., and Lu, Q. (2013). Characterization and expression analysis of common carp Cyprinus carpio TLR5M. DNA Cell. Biol. 32, 611–620. doi:10.1089/dna.2013.2051
Gao, F., Pang, J., Lu, M., Liu, Z., Wang, M., Ke, X., et al. (2022). TLR5 recognizes Aeromonas hydrophila flagellin and interacts with MyD88 in Nile tilapia. Dev. Comp. Immunol. 133, 104409. doi:10.1016/j.dci.2022.104409
Glasauer, S. M., and Neuhauss, S. C. (2014). Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genomics 289, 1045–1060. doi:10.1007/s00438-014-0889-2
Guyomard, R., Boussaha, M., Krieg, F., Hervet, C., and Quillet, E. (2012). A synthetic rainbow trout linkage map provides new insights into the salmonid whole genome duplication and the conservation of synteny among teleosts. BMC Genet. 13, 15. doi:10.1186/1471-2156-13-15
Han, C., Li, Q., Zhang, Z., and Huang, J. (2017). Characterization, expression, and evolutionary analysis of new TLR3 and TLR5M genes cloned from the spiny eel Mastacembelus armatus. Dev. Comp. Immunol. 77, 174–187. doi:10.1016/j.dci.2017.08.007
Hayashi, F., Smith, K. D., Ozinsky, A., Hawn, T. R., Yi, E. C., Goodlett, D. R., et al. (2001). The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103. doi:10.1038/35074106
Hoegg, S., Brinkmann, H., Taylor, J. S., and Meyer, A. (2004). Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J. Mol. Evol. 59, 190–203. doi:10.1007/s00239-004-2613-z
Hwang, S. D., Asahi, T., Kondo, H., Hirono, I., and Aoki, T. (2010). Molecular cloning and expression study on Toll-like receptor 5 paralogs in Japanese flounder, Paralichthys olivaceus. Fish. Shellfish Immunol. 29, 630–638. doi:10.1016/j.fsi.2010.06.011
Ivison, S. M., Khan, M. A., Graham, N. R., Bernales, C. Q., Kaleem, A., Tirling, C. O., et al. (2007). A phosphorylation site in the Toll-like receptor 5 TIR domain is required for inflammatory signalling in response to flagellin. Biochem. Biophys. Res. Commun. 352, 936–941. doi:10.1016/j.bbrc.2006.11.132
Jaillon, O., Aury, J. M., Brunet, F., Petit, J. L., Stange-Thomann, N., Mauceli, E., et al. (2004). Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431, 946–957. doi:10.1038/nature03025
Janeway, C. A., and Medzhitov, R. (2002). Innate immune recognition. Annu. Rev. Immunol. 20, 197–216. doi:10.1146/annurev.immunol.20.083001.084359
Jiang, Y., He, L., Ju, C., Pei, Y., Ji, M., Li, Y., et al. (2015). Isolation and expression of grass carp toll-like receptor 5a (CiTLR5a) and 5b (CiTLR5b) gene involved in the response to flagellin stimulation and grass carp reovirus infection. Fish. Shellfish Immunol. 44, 88–99. doi:10.1016/j.fsi.2015.01.024
Kaessmann, H. (2010). Origins, evolution, and phenotypic impact of new genes. Genome Res. 20, 1313–1326. doi:10.1101/gr.101386.109
Keestra, A. M., De Zoete, M. R., Van Aubel, R. A., and Van Putten, J. P. (2008). Functional characterization of chicken TLR5 reveals species-specific recognition of flagellin. Mol. Immunol. 45, 1298–1307. doi:10.1016/j.molimm.2007.09.013
Liao, Z., Yang, C., Jiang, R., Zhu, W., Zhang, Y., and Su, J. (2022). Cyprinid-specific duplicated membrane TLR5 senses dsRNA as functional homodimeric receptors. EMBO Rep. 23, e54281. doi:10.15252/embr.202154281
Lien, S., Koop, B. F., Sandve, S. R., Miller, J. R., Kent, M. P., Nome, T., et al. (2016). The Atlantic salmon genome provides insights into rediploidization. Nature 533, 200–205. doi:10.1038/nature17164
Liu, F., Su, B., Fu, Q., Shang, M., Gao, C., Tan, F., et al. (2017). Identification, characterization and expression analysis of TLR5 in the mucosal tissues of turbot (Scophthalmus maximus L.) following bacterial challenge. Fish. Shellfish Immunol. 68, 272–279. doi:10.1016/j.fsi.2017.07.021
Matsuo, A., Oshiumi, H., Tsujita, T., Mitani, H., Kasai, H., Yoshimizu, M., et al. (2008). Teleost TLR22 recognizes RNA duplex to induce IFN and protect cells from birnaviruses. J. Immunol. 181, 3474–3485. doi:10.4049/jimmunol.181.5.3474
Mayfield-Jones, D., Washburn, J. D., Arias, T., Edger, P. P., Pires, J. C., and Conant, G. C. (2013). Watching the grin fade: Tracing the effects of polyploidy on different evolutionary time scales. Semin. Cell. Dev. Biol. 24, 320–331. doi:10.1016/j.semcdb.2013.02.002
Meyer, A., and Schartl, M. (1999). Gene and genome duplications in vertebrates: The one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell. Biol. 11, 699–704. doi:10.1016/s0955-0674(99)00039-3
Mizel, S. B., Honko, A. N., Moors, M. A., Smith, P. S., and West, A. P. (2003). Induction of macrophage nitric oxide production by Gram-negative flagellin involves signaling via heteromeric Toll-like receptor 5/Toll-like receptor 4 complexes. J. Immunol. 170, 6217–6223. doi:10.4049/jimmunol.170.12.6217
Mogensen, T. H. (2009). Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22, 240–273. Table of Contents. doi:10.1128/CMR.00046-08
Oshiumi, H., Tsujita, T., Shida, K., Matsumoto, M., Ikeo, K., and Seya, T. (2003). Prediction of the prototype of the human Toll-like receptor gene family from the pufferfish, Fugu rubripes, genome. Immunogenetics 54, 791–800. doi:10.1007/s00251-002-0519-8
Palti, Y. (2011). Toll-like receptors in bony fish: From genomics to function. Dev. Comp. Immunol. 35, 1263–1272. doi:10.1016/j.dci.2011.03.006
Pevsner, J. (2009). “Basic local alignment search tool (BLAST),” in Bioinformatics and functional genomics. 2nd ed. (Hoboken, NJ, USA: Wiley).
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi:10.1093/molbev/msp077
Purcell, M. K., Smith, K. D., Hood, L., Winton, J. R., and Roach, J. C. (2006). Conservation of toll-like receptor signaling pathways in teleost fish. Comp. Biochem. Physiol. Part D. Genomics Proteomics 1, 77–88. doi:10.1016/j.cbd.2005.07.003
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi:10.1038/nprot.2008.73
Shan, S., Liu, R., Feng, H., Meng, F., Aizaz, M., and Yang, G. (2021). Identification and functional characterization of a fish-specific tlr19 in common carp (Cyprinus carpio L.) that recruits TRIF as an adaptor and induces ifn expression during the immune response. Vet. Res. 52, 88. doi:10.1186/s13567-021-00957-3
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57, 758–771. doi:10.1080/10635150802429642
Stamatakis, A. (2006). RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi:10.1093/bioinformatics/btl446
Su, J., and Yu, X. (2019). Editorial: Ligands, adaptors and pathways of TLRs in non-mammals. Front. Immunol. 10, 2439. doi:10.3389/fimmu.2019.02439
Tsujita, T., Tsukada, H., Nakao, M., Oshiumi, H., Matsumoto, M., and Seya, T. (2004). Sensing bacterial flagellin by membrane and soluble orthologs of Toll-like receptor 5 in rainbow trout (Onchorhynchus mikiss). J. Biol. Chem. 279, 48588–48597. doi:10.1074/jbc.M407634200
Vandepoele, K., De Vos, W., Taylor, J. S., Meyer, A., and Van De Peer, Y. (2004). Major events in the genome evolution of vertebrates: Paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc. Natl. Acad. Sci. U. S. A. 101, 1638–1643. doi:10.1073/pnas.0307968100
Voogdt, C. G., Bouwman, L. I., Kik, M. J., Wagenaar, J. A., and Van Putten, J. P. (2016). Reptile Toll-like receptor 5 unveils adaptive evolution of bacterial flagellin recognition. Sci. Rep. 6, 19046. doi:10.1038/srep19046
Voogdt, C. G. P., Wagenaar, J. A., and Van Putten, J. P. M. (2018). Duplicated TLR5 of zebrafish functions as a heterodimeric receptor. Proc. Natl. Acad. Sci. U. S. A. 115, E3221–e3229. doi:10.1073/pnas.1719245115
Wang, K. L., Chen, S. N., Huo, H. J., and Nie, P. (2021). Identification and expression analysis of sixteen Toll-like receptor genes, TLR1, TLR2a, TLR2b, TLR3, TLR5M, TLR5S, TLR7-9, TLR13a-c, TLR14, TLR21-23 in Mandarin fish Siniperca chuatsi. Dev. Comp. Immunol. 121, 104100. doi:10.1016/j.dci.2021.104100
Yamada, K. D., Tomii, K., and Katoh, K. (2016). Application of the MAFFT sequence alignment program to large data-reexamination of the usefulness of chained guide trees. Bioinformatics 32, 3246–3251. doi:10.1093/bioinformatics/btw412
Zhan, F. B., Tan, K., Song, X., Yu, J., and Wang, W. M. (2019). Isolation and expression of four Megalobrama amblycephala toll-like receptor genes in response to a bacterial infection. Fish. Shellfish Immunol. 93, 1028–1040. doi:10.1016/j.fsi.2019.08.051
Zhou, K., Kanai, R., Lee, P., Wang, H. W., and Modis, Y. (2012). Toll-like receptor 5 forms asymmetric dimers in the absence of flagellin. J. Struct. Biol. 177, 402–409. doi:10.1016/j.jsb.2011.12.002
Keywords: TLR5, fish, whole-genome duplications, gene loss, adaptive evolution
Citation: Zhang K, Chen M, He H, Kou H, Lin L and Liang R (2023) Genome-wide identification and characterization of toll-like receptor 5 (TLR5) in fishes. Front. Genet. 13:1083578. doi: 10.3389/fgene.2022.1083578
Received: 29 October 2022; Accepted: 05 December 2022;
Published: 06 January 2023.
Edited by:
Zheng-Yong Wen, Neijiang Normal University, ChinaReviewed by:
Erlong Wang, Northwest A & F University, ChinaCopyright © 2023 Zhang, Chen, He, Kou, Lin and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rishen Liang, Y2hlZXRhaGxpYW5nQDE2My5jb20=; Li Lin, bGlubGlAemhrdS5lZHUuY24=
†These authors contributed equally to this work and share first authorship
‡These authors contributed equally to this work and share last authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.