
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 07 December 2022
Sec. Livestock Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1078049
 Ran Di1†
Ran Di1† Rensen Zhang1,2†
Rensen Zhang1,2† Joram Mwashigadi Mwacharo3,4
Joram Mwashigadi Mwacharo3,4 Xiangyu Wang1
Xiangyu Wang1 Xiaoyun He1
Xiaoyun He1 Yufang Liu1
Yufang Liu1 Jinlong Zhang5
Jinlong Zhang5 Yiming Gong1
Yiming Gong1 Xiaosheng Zhang5*
Xiaosheng Zhang5* Mingxing Chu1*
Mingxing Chu1*As a novel class of small RNAs, piRNAs are highly expressed in the animal gonads and their main known role is to inhibit transposon activity for ensuring the correctness and integrity of genome. In order to explore the characteristics of piRNAs in sheep testis and their possible regulatory roles on male reproduction, deep sequencing technology was used to sequence small RNAs and identify piRNAs in testes of sheep. The length of piRNAs in sheep testes showed a unimodal distribution between 26 and 31 nt, with a peak at 29 nt. These piRNAs exhibited obvious ping-pong signature and strand specificity. In the genome, they were mainly aligned to CDS, intron, repetitive sequence regions and unannotated regions. Furthermore, in transposon analysis, piRNAs were aligned predominantly to LINE, SINE, and LTR types of retrotransposon in sheep testes, and the piRNAs derived from each type showed obvious ping-pong signature. The piRNA clusters identified in sheep testes were mainly distributed on chromosomes 3, 7, 15, 17, 18 and 20. The results combining semen determination with pathway enrichment analysis implied that differentially expressed piRNAs between the testes of rams with different fertility might participate in spermatogenesis by regulating multiple pathways closely related to stabilization of blood-testis barrier and renewal and differentiation of spermatogonial stem cell. Taken together, the study provided new insights into the characteristics, origin and expression patterns of piRNAs in sheep testes tissue, which would help us better understand the role of piRNAs in sheep reproduction.
piRNAs are a novel class of small RNAs different from microRNAs. They were firstly discovered in the testes tissue of Drosophila melanogaster (Aravin et al., 2001). They are far more numerous than microRNAs and are distinctly different from microRNAs in length, sequence characteristics and distribution. Usually, the lengths of piRNAs are between 24 and 33 nt (Czech et al., 2018; Ozata et al., 2019). They must specifically bind PIWI proteins to exert their biological functions, so they are called PIWI-interacting RNA (piRNA) (Aravin et al., 2006; Girard et al., 2006; Houwing et al., 2007). piRNAs include two forms: primary piRNAs and secondary piRNAs (Czech et al., 2018; Ozata et al., 2019). Primary piRNAs are mainly processed from precursor piRNAs which were transcript from piRNAs clusters, and usually their 5′ ends are base U (Vagin et al., 2006; Houwing et al., 2007). Then primary piRNAs bind with a kind of PIWI protein (Aub in Drosophila, Miwi in mouse) to form a complex, and they together recognize and cleave RNAs transcribed from transposon. Thus, secondary piRNAs with A preference at 10th base are formed. The secondary piRNAs can combine with another PIWI protein (Ago3 in Drosophila, Mili in mouse) to form a complex that is just enough to cleave the precursor piRNAs and generate primary piRNAs. Through this ping-pong cycle mechanism, the function of piRNAs to inhibit transposon transcription is realized (Brennecke et al., 2007; Gunawardane et al., 2007; Aravin et al., 2008; Toth et al., 2016; Madison-Villar et al., 2017).
Although the biological origin and function of piRNAs have not been fully elucidated, more and more evidence suggested that piRNAs are critical for reproduction. piRNAs are highly expressed in animal gonads (Grivna et al., 2006; Vagin et al., 2006; Houwing et al., 2007), and play important roles in maintaining integrity of the gamete genome (Vagin et al., 2006; Goriaux et al., 2014; Mohn et al., 2014; Ernst et al., 2017), gender determination (Kiuchi et al., 2014), germ cell differentiation (Gonzalez et al., 2015) and epigenetic regulation of gene expression (Teixeira et al., 2017; Czech et al., 2018). During spermatogenesis in mice, piRNAs are involved in the degradation of multiple mRNAs, whose expression is detrimental to spermatogenesis (Watanabe et al., 2011; Gou et al., 2014). Multiple studies suggested that piRNAs are essential in animal spermatogenesis (Goh et al., 2015; Zhang et al., 2015; Guo et al., 2018). Recently, we found that PIWI protein had the highest expression level in the gonad of sheep compared with other tissues (Li et al., 2020). Furthermore, the expression in testes is higher than that in ovaries (Li et al., 2020), which implies the important roles of piRNAs in male gonad.
The sequence information and visual map of piRNAs in humans and some model organisms (mouse, rat, Drosophila) can be accessed from a piRNA bank database (http://pirnabank.ibab.ac.in/) (Lakshmi and Agrawal, 2008). Recently, the database piRBase (http://bigdata.ibp.ac.cn/piRBase) collected piRNA sequence information from 44 species, and this information included gold standard piRNA sets, piRNAs expression data, regulatory relationship between piRNAs and target genes, and so on (Wang et al., 2022). However, the information about ovine piRNAs sequences can’t retrieved from these two databases. In 2019, He et al. (2019) predicted piRNAs in sheep testes tissue. However, the in-depth sequence features (including ping-pong signature, strand specificity, the genomic distribution and other characteristics of sheep piRNAs) still need to be fully revealed. Therefore in this study, solexa sequencing technology and bioinformatics methods were used to screen and identify piRNA sequences in sheep testes, and their sequence characteristics, genomic origin and chromosomal distribution were revealed. Then, differentially expressed (DE) piRNAs in the testes of rams with different fertility were analyzed and their functions were predicted by combining the results of target gene function clustering and semen index assay. These would help us better understand sequence characteristics and origin of sheep piRNAs and provide new insights into the epigenetic regulation of ram fertility.
In china, both Small-Tailed Han (STH) sheep and Sunite (SNT) sheep belong to the Chinese Mongolian sheep clade and have similar genetic backgrounds. However, their fertilities were distinct (STH with high fertility and SNT with low fertility). In this study, we selected a sheep farm where both breeds were raised in Tianjin of China, and the feed and management were the same for both breeds in this farm. Then six healthy, 3-year-old adult rams with similar weight (difference less than 1 kg) were selected in two breeds (3 rams in each breed) for semen evaluation and sample collection. At AM 9:00, semen was collected when the ram was fully climbing the back of the ewe model in order to ensure maximum semen volume in each collection. For all the rams, semen was collected three times simultaneously within 10 days. Ejaculate volume, sperm motility, viable sperm ratio, and sperm density were determined using the methods described by David et al. (2015). The pH of fresh essence was measured with a SevenExcellence™ pH meter. The semen slides were stained with 0.5% gentian violet, and 500 sperms in different view fields were viewed under a microscope to calculate the percentage of abnormal sperm. The above data were analyzed by one-way ANOVA with SPSS19.0, and p < 0.05 was considered to be statistically significant. After all measurements were completed, the above six rams were euthanized at the same time, and the testes of the rams were immediately collected and quickly stored in liquid nitrogen.
Total RNA was extracted from the testes according to the instructions of the TRIzol kit. The Agilent 2100 Bioanalyzer was used to accurately detect RNA integrity, and the RIN value of each sample was greater than 8.5 which indicates good quality of RNA for subsequent analysis. Small RNAs less than 40 nt in length were isolated from total RNA using a FlashPAGE fractiontor (Ambion, Life Technologies, Paisley, UK), and adapters (5′ seq: 5′-GUUCAGAGUUCUACAGUCCGACGAUC-3’; 3′ seq: 5′-TGGAATTCTCGGGTGCCAAGG-3′) were attached to both ends of the isolated small RNAs using the Illumina TruSeq Small RNA Sample Preparation kit. The cDNA synthesis was performed by RT-PCR and PCR products were screened using 15% polyacrylamide gels. RNA fragments with a length of 140–160 bp (the non-coding small RNA plus the 3′ and 5′ end adapter sequences) were recovered and sequenced using Illumina 2000. The construction and sequencing of the small RNA libraries were completed by Beijing Nuohezhiyuan Technology Co., Ltd. The raw sequencing data have been deposited in the Genome Sequence Archive (Chen et al., 2021) in National Genomics Data Center (Xue et al., 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA008325) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
The obtained small RNA raw data was filtered to remove the following reads: low-quality reads (reads with Qphred≤5 bases accounting for more than 50% of the entire read length), reads with N bases content greater than 10%; reads containing 5′ adapter contamination, reads without 3′ adapter sequences and inserts; reads containing poly A/T/C/G (they may be due to sequencing errors). Then the 3′ linker sequence of the remaining data was removed, and finally clean reads were obtained after removal of atypical length range (<24 nt or >33 nt) reads of piRNA (Czech et al., 2018; Ozata et al., 2019). Furthermore, the clean reads from known other types of small RNAs (rRNAs, tRNAs, snRNAs, snoRNAs, and miRNAs) were removed by aligning them to the Rfam database (http://rfam.xfam.org/) (Kalvari et al., 2018) and miRbase v21.0 (http://www.mirbase.org/) (Griffiths-Jones et al., 2008). Then the new predicted miRNA reads by Mireap 0.2 were also removed from the remaining reads. Finally, reads with 1U or 10 A feature in the remaining reads were predicted as candidate piRNAs reads. Among them, piRNAs reads with the same sequence were considered as a piRNA tag.
First, the length distribution of piRNA reads and tags (reads with the same nucleotide sequence as a tag) in sheep testes tissue were counted, and the base frequency of each site in the 5′-3′ direction was analyzed. The putative piRNA sequences were then aligned to the sheep genome (Oar v 4.0) and the genomic composition of piRNAs was determined by bowtie2 (version 2.3.5). In the repeat region, piRNA is mainly derived from the transposable element (TE), therefore the piRNAs derived from the repeat regions were aligned with the TE sequences in Rpbase, thus the types of piRNAs in the TE were determined by RepeatMasker (version 4.1.0). The ping-pong structural feature means that there is a 10-base complementarity between the primary and secondary piRNA strands. We used the method described previously (Yan et al., 2011) to analyze the ping-pong structure of all putative piRNA sequences as a whole and piRNA sequences derived from each TE type in sheep testes, respectively.
Most piRNAs are located in piRNA clusters scattered throughout the genome. Therefore, proTRAC v2.1.2 (Rosenkranz and Zischler, 2012; Rosenkranz, 2016) was used to identity piRNA clusters in the sheep genome. Then, strand specificity within individual piRNA cluster and all clusters was analyzed. The percentage of U bases in the first bases of all piRNAs derived from individual piRNA cluster were counted respectively. In addition, the distribution of piRNA clusters across chromosomes of sheep was also analyzed.
The piRNA expression levels of each sample were normalized using TPM. The DE piRNAs were screened between testes of sheep with high fertility and low fertility by DESeq2 program (v 1.4.5). Those piRNAs with p-value <0.05 and fold change >1.5 were considered to be differentially expressed between the two groups. The programs miRanda, RNAhybrid and TargetScan were used to predict target genes of DE piRNAs, and the overlap genes among prediction results of the three programs were screened as the candidate target genes for further analysis. DAVID software (https://david.ncifcrf.gov/) (Huang et al., 2009b; Huang et al., 2009a) was used for GO and KEGG enrichment analysis of target genes. Associations between the pathways of interest were analyzed using the emapplot function in the R cluster Profiler package. In addition, the interaction network among DE genes in male reproduction-related pathways was constructed using the enrichplot function in the R cluster Profiler package.
Quantitative polymerase chain reaction (qPCR) was employed to verify the accuracy of small RNA sequencing. U6 was used as a reference gene, and five piRNAs were randomly selected. The stem-loop primer method was used to improve the specificity of detection. Reverse transcription stem-loop primers and qPCR amplification primers were designed and showed in Supplementary Table S1. Total RNA was reverse transcribed to synthesize cDNA using PrimeScript™ RT Reagent Kit (Takara, Japan). The qPCR reaction conditions were as follows: 95°C for 15 min, followed by 40 cycles of 10 s at 95°C and 30 s at 60°C, respectively. Three technical replicates were performed for each sample, and the relative expression ratios of the piRNAs were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001), and processed by one-way ANOVA with SPSS 19.0.
Sheep semen quality is an important factor affecting sheep fertility. Therefore, in order to explore whether there were significant differences in semen quality between SNT and STH rams, the volume of single ejaculate, sperm density, sperm number, pH value of semen, sperm motility, viable sperm ratio, and sperm deformity rate were detected respectively. The statistical results showed that the volume of single ejaculate, sperm number and pH value of semen in STH sheep were significantly higher than those of semen in SNT sheep (Table 1). For other parameters (including sperm motility, viable sperm ratio, sperm density and sperm deformity rate), there was no significant difference between the two breeds.
In order to reveal the sequence characteristics of piRNAs in sheep testes tissue, the small RNAs in sheep testes were firstly sequenced using solexa sequencing technology. A total of 44,388,241 and 46,551,224 small RNA raw reads were acquired in SNT and STH testes, respectively. After filtering low quality reads, we obtained 43,554,297 and 40,476,683 clean reads in SNT and STH testes (Table 2). Among them, more than 80% small RNAs can be aligned to the sheep genome. Finally, after removing the following sequences (including rRNA, tRNA, snRNA, snoRNA, known and newly predicted miRNA sequences) from these mapped reads, we obtained 4218952–6894,453 piRNAs reads and 775532–837165 piRNAs tags in testis of each ram for further analysis.
First, the length distribution of piRNAs reads and tags were analyzed in sheep testes tissue. The results in Figure 1A showed that the reads of piRNAs in sheep testes exhibit a unimodal distribution between 24 nt and 33 nt, with a peak at 29 nt. Most of piRNAs tags were concentrated between 25 nt and 30 nt. Simultaneously, we performed statistics on the base distribution frequencies of piRNAs reads and tags in the sheep testes. Their first bases in sequences had strong U preference (Figure 1), whereas the 10th base did not have base preference, implying that piRNAs in sheep testes mainly came from the primary piRNA pathway. In addition, the sequences of piRNAs in sheep testes exhibited strong ping-pong signature (Figure 2).
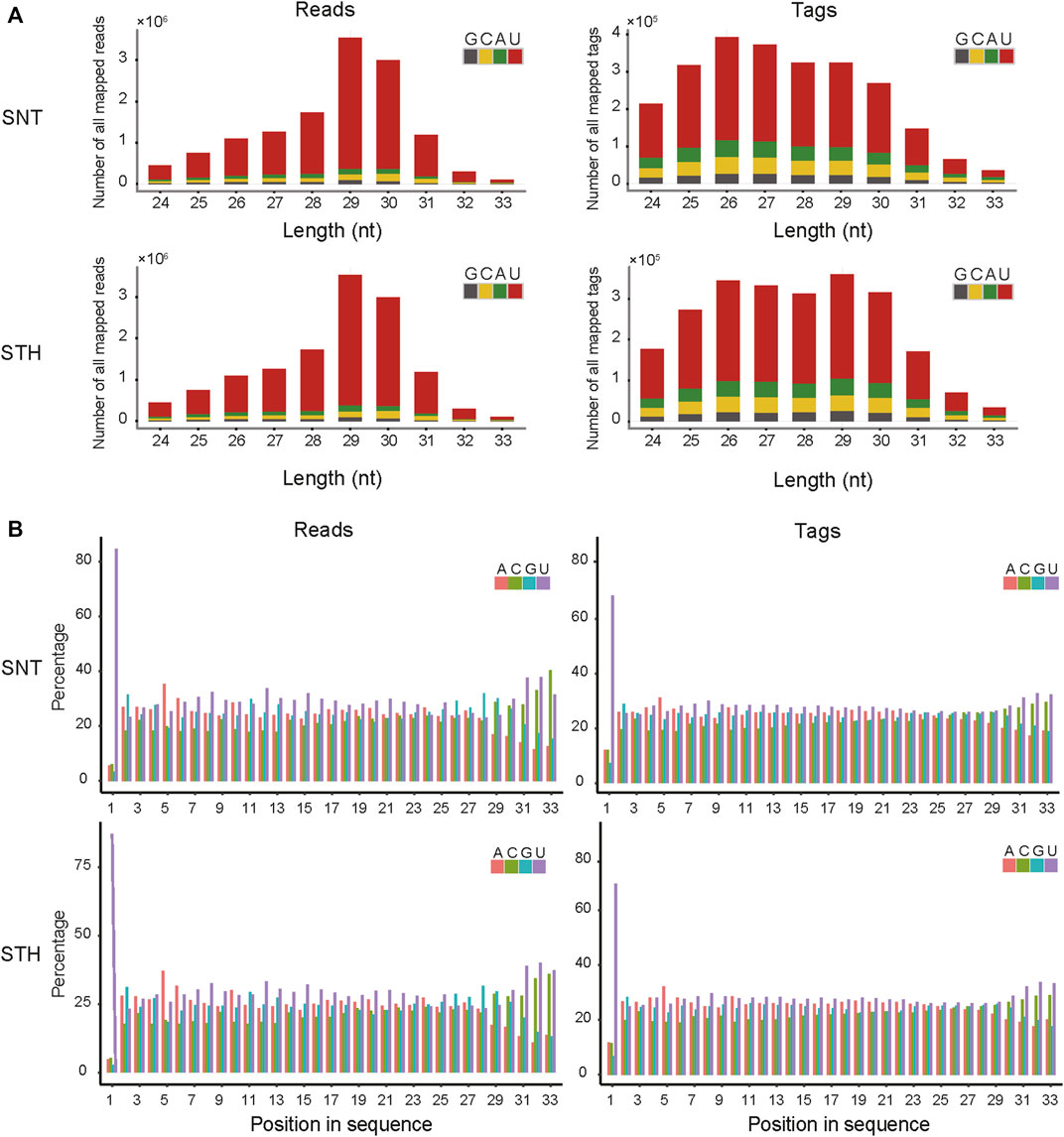
FIGURE 1. The feature of length and base composition of piRNAs in sheep testis. (A) Length distribution feature and base composition of the first nucleotide at 5′ end; (B) 5′-3′ base distribution frequencies of piRNAs reads and tags.
The piRNAs derived from different genomic regions play different biological roles. Therefore, in this study, the candidate piRNAs were aligned to the sheep genome for determining the main sources of piRNAs and their functional regions on the genome. Results showed that the alignment regions of piRNAs were similar in the testes of SNT and STH sheep, and they were mainly aligned to the CDS region, intron region, repetitive sequence region and unannotated region (Figure 3). In addition, a small part of piRNAs were aligned to 5′UTR, 3′UTR and lncRNA. All these regions can be considered as potential target regions regulated by piRNAs in sheep testis.
One main function of piRNA is to inhibit the activity of transposons. Therefore in this study, piRNAs derived from repeat sequences in sheep testes were aligned to the Repbase database in order to identify the types of transposable elements which piRNAs originate from and act in. Results showed that piRNAs in sheep testes were aligned predominantly to retrotransposon. The types of transposons were similar for piRNAs in SNT and STH testes, and they mainly included LINE, SINE, and LTR types in retrotransposons (Figure 4A). In addition, some piRNAs were also aligned to DNA-type transposons. In a finer classification, piRNAs were mainly aligned to L1 and L2 types in LINE, MIR type in SINE, ERV1, ERVK, ERVL, ERVL-MaLR types in LTR, and hAT-Charlie in DNA-type transposons. Furthermore, the piRNAs derived from each of the above TE types presented obvious ping-pong signature in sheep testes (Figure 4B), confirming that the piRNAs derived from each TE type can inhibit transposon activity through ping-pong cycle mechanism.
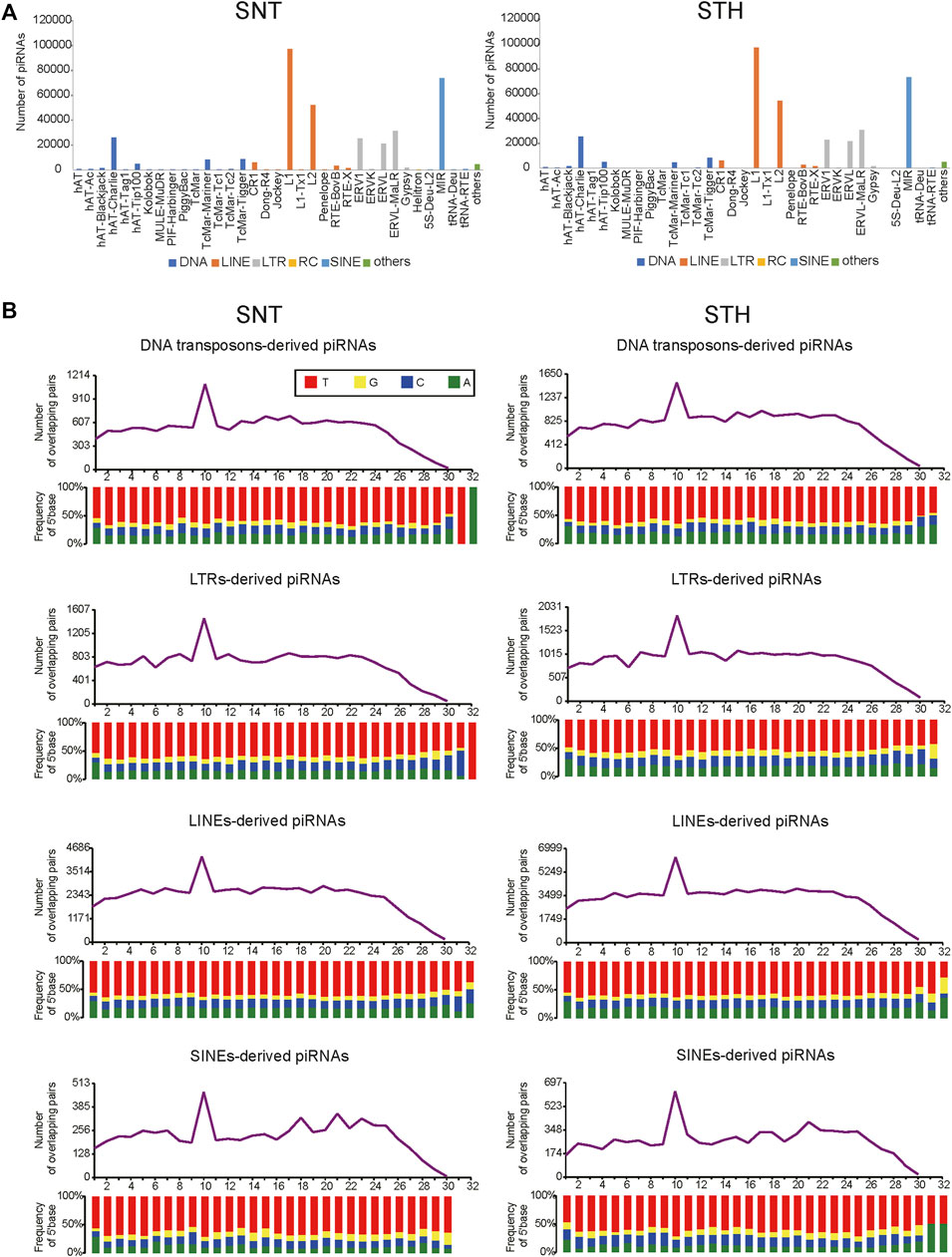
FIGURE 4. Distribution of sheep testis piRNAs among transposon elements (A) and the ping-pong signature of transposable element derived piRNAs in sheep testes (B).
In most species, piRNAs exist in piRNAs clusters on genome. In testis of each ram, a total of 284–430 piRNA clusters were identified on genome. In each piRNA cluster, piRNAs had strong strand specificity in sheep testes (Figure 5A). However when all piRNA clusters were taken into account, the ratio of piRNAs derived from the sense and antisense strands was comparable (42% and 58%), and there was no longer obvious strand specificity (Figure 5B). At the same time, the first bases of all piRNAs derived from piRNA clusters had a strong U preference, which means that most piRNAs in clusters originated from the primary piRNA pathway. In addition, results showed that the distribution of piRNA clusters was not proportional to the length of chromosomes. In sheep testes, piRNA clusters were mainly distributed on chromosomes 3, 14, 20, and 24 (Figure 5C), which implied that piRNA clusters on these chromosomes have more important roles in maintaining the stability of germ cell genome and regulating the physiological activity of testis in rams.
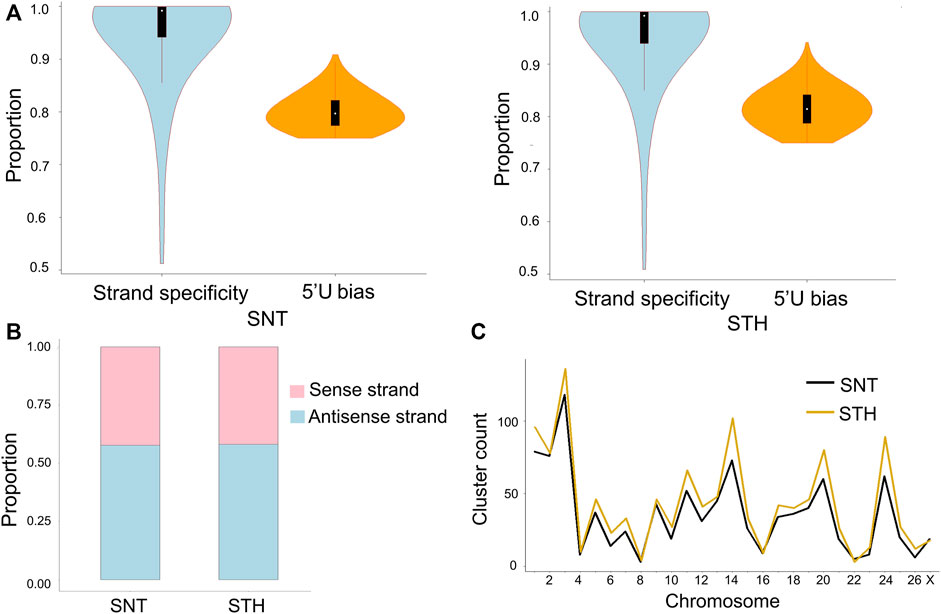
FIGURE 5. Characteristics of piRNA clusters in sheep testes. (A) Strand specificity and 5′U bias of piRNAs in clusters; (B) piRNAs distribution on the sense and antisense strands; (C) Chromosome distribution of piRNA clusters in testes of SNT and STH.
In order to verify the accuracy of sequencing data, five differentially expressed piRNA sequences were randomly selected for qPCR. The results (Supplementary Figure S1)_showed that the expression trends of five piRNAs were consistent with the sequencing results, indicating that the data obtained from small RNA sequencing were reliable for subsequent analysis. A total of 2,428 DE piRNAs were screened in the testes of rams with different fertility. The top 30 DE piRNAs were summarized in Table 3. The functions of all DE piRNAs were predicted by GO and KEGG enrichment analysis for target genes. In terms of biological processes, their target genes were mainly enriched in Biological regulation, Cellular process, Metabolic process, Response to stimulus, and Single-organism process (Figure 6). In terms of cellular components, they were mainly involved in Cell, Cell part, Membrane and Organelle. In terms of molecular functions, Binding and Catalytic activity were enriched by their target genes. KEGG enrichment analysis showed that the target genes of DE piRNAs were significantly enriched in 54 pathways, and the 25 most significantly pathways were shown in Supplementary Table S2. They included important pathways of reproduction, which were closely related to adhesion (Focal adhesion pathway, Adherens junction, Cell adhesion molecules and Rap1 signaling pathway), spermatogonial stem cell renewal and differentiation (Wnt signaling pathway, PI3K/Akt signaling pathway, MAPK signaling pathway, HIF-1 signaling pathway and Rap1 signaling pathway) and cytoskeleton (Regulation of actin cytoskeleton, Chemokine signaling pathway and Focal adhesion pathway), respectively. Among them, the pathways associated with adhesion and cytoskeleton play important roles in ensuring the stability of the blood-testicular barrier. Notably, cytoscape analysis indicated that the top 25 enriched pathways were closely interconnected (Figure 7). Moreover, genes in these pathways also formed a complex association network (Figure 8), suggesting that they would act synergistically in the regulation of ram reproduction.
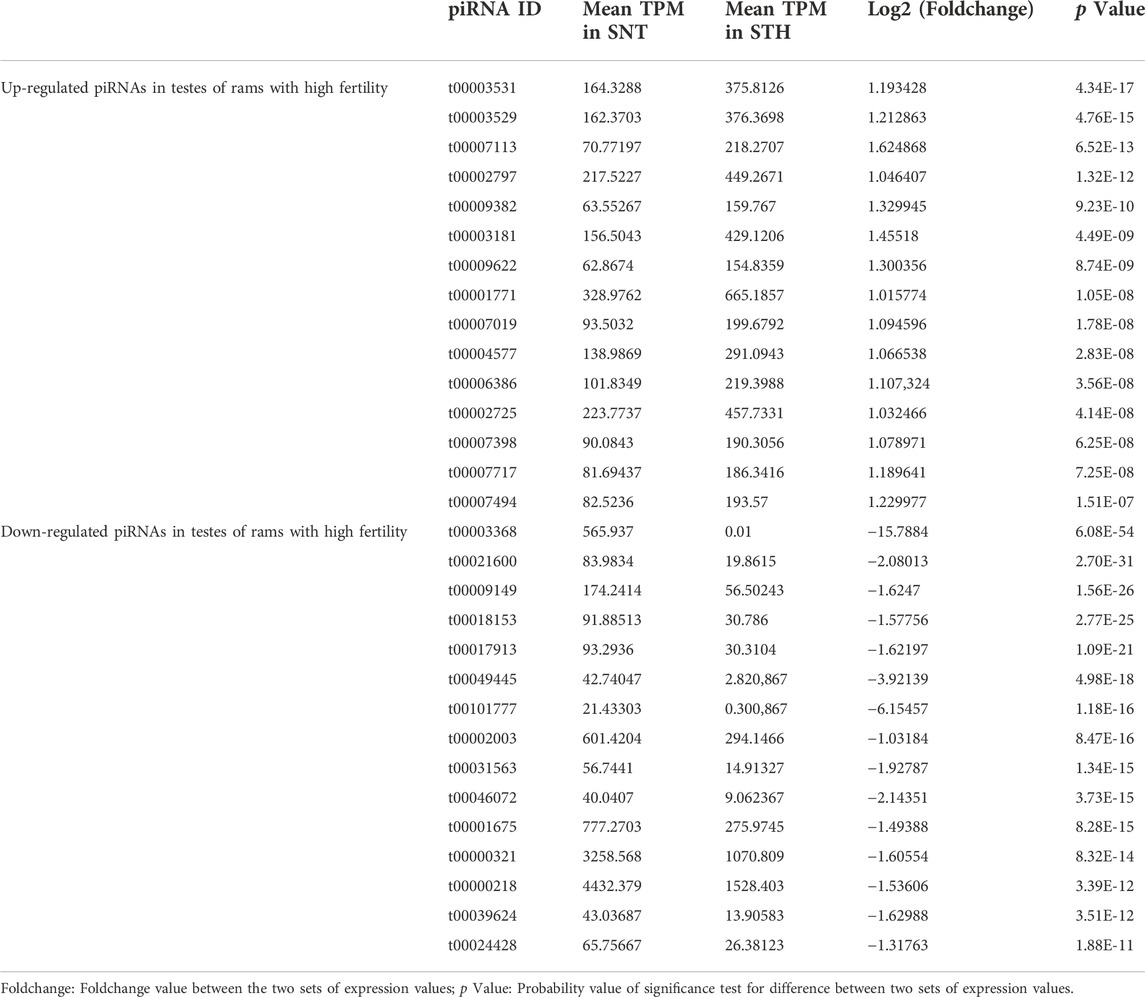
TABLE 3. Top 30 significantly differentially expressed piRNAs in testes of rams with different fertility.
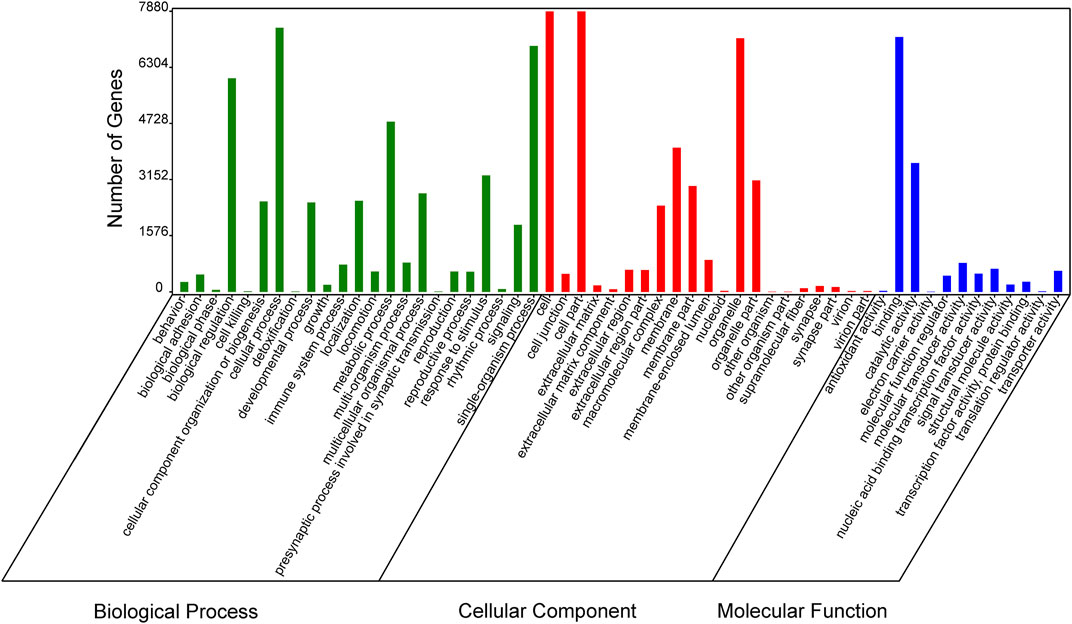
FIGURE 6. Enriched Gene Ontology (GO) terms of DE-piRNAs targeted gene between the testes of SNT and STH sheep.
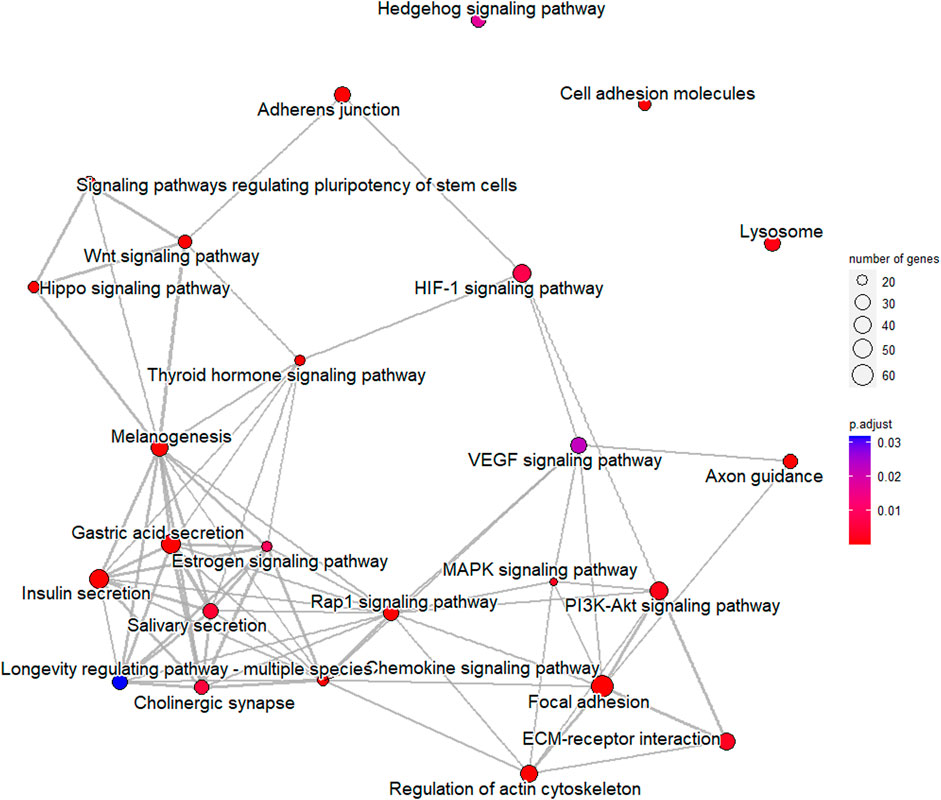
FIGURE 7. Associations among top 25 significantly enriched pathways for target genes of differentially expressed piRNAs.
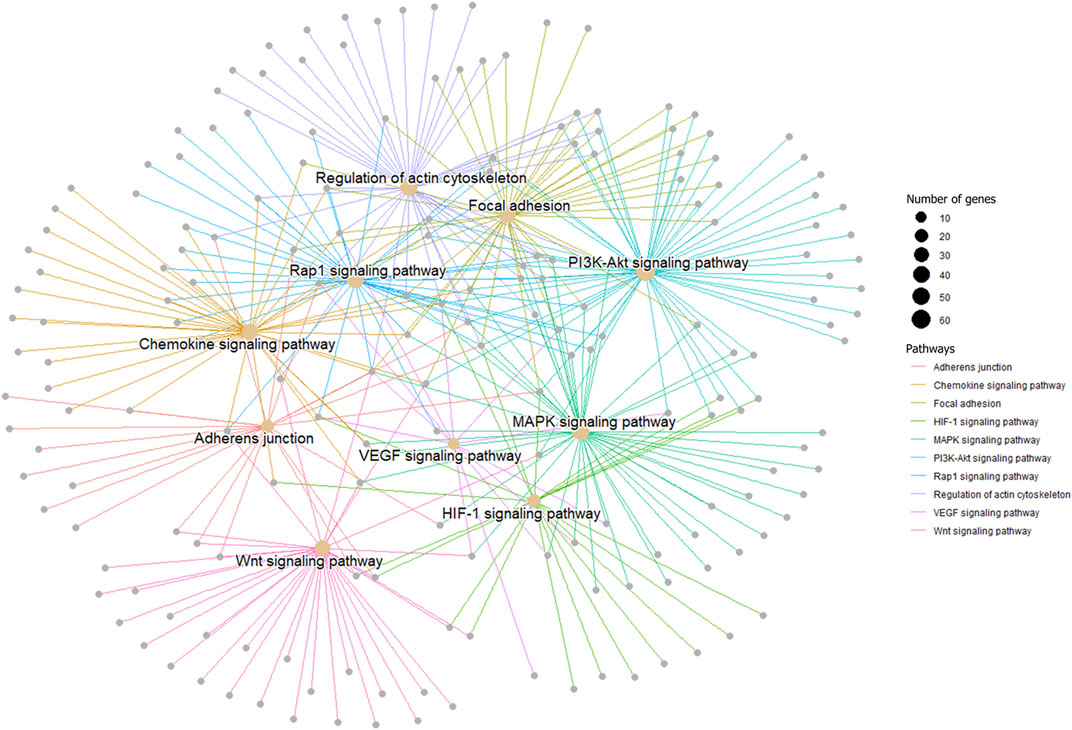
FIGURE 8. A gene network among male reproduction-related signal pathways. Genes in each pathway are represented by one color.
In this study, the characteristics of piRNA including length distribution, ping-pong signature, strand specificity, genomic origin and chromosomal distribution were revealed in sheep testes. The length distribution of microRNAs (miRNAs) ranges from 18–24 nt (Loganantharaj et al., 2017; Singh and Storey, 2021). However, as a new class of small RNAs, the length range of piRNAs is different from that of microRNAs. In this study, the length of piRNAs reads showed a unimodal distribution between 24 and 33 nt in sheep testes. The length range of piRNAs was similar to those piRNAs in testes of mammals such as mouse (Grivna et al., 2006), horse (Li et al., 2019), yak (Gong et al., 2018) and human (Ha et al., 2014), whereas it was longer than that of piRNAs sequences in reproductive tissues of Drosophila (Aravin et al., 2003) and C. elegans (Ruby et al., 2006). The piRNA reads and tags in testes and ovaries of mouse also showed a single peak distribution, with the peak appearing at 26 and 27 nt (Yan et al., 2011). The piRNA reads and tags in testes of rhesus macaque also exhibit a single peak distribution, moreover the peak was also consistent with that of sheep (29 nt) (Yan et al., 2011). Our previous study on piRNAs in ovaries showed that peak values of reads number appeared at 24 nt and 27–29 nt, and the number of tags showed a decreasing distribution between 24 and 33 nt (Li et al., 2022). This hinted that sequence distribution of piRNAs has its own characteristics in different tissues of sheep. Studies indicated that different PIWI proteins tend to bind piRNAs with different lengths. For example, Piwi protein tends to bind 25 nt piRNAs while Aub and Ago3 tend to bind 23–24 nt piRNAs in Drosophila; MILI tends to bind 28 nt piRNAs while MIWI and MIWI2 tend to bind 30 nt piRNAs in mouse (Aravin et al., 2006; Grivna et al., 2006; Aravin et al., 2008). Our previous study found that four PIWI proteins (MIWI, MILI, MIWI2 and AGO3) were expressed in sheep testes tissue (Li et al., 2020), which was similar with PIWI proteins in mouse. Referring to the study results in mouse, the first three PIWI proteins just bind to 28–30 nt piRNA, which was in good agreement with the peak (28–30 nt) of piRNA reads in this study. Thus, it also implied that the piRNA length and the synergistic PIWI proteins have specificity between mammal and non-mammal.
piRNAs derived from different genomic regions play different biological roles (Aravin et al., 2003; Girard et al., 2006; Yan et al., 2011). Therefore, in this study, alignment regions of the sheep testes-derived piRNA in genome were analyzed. Results showed that the alignment regions of piRNAs were similar in the testes of sheep with different fertility, and they were mainly aligned to the CDS region, repetitive sequence region, intron region and unannotated region. In addition, there were a small number of piRNAs aligned to 5′UTR, 3′UTR and lncRNA. This means that piRNAs in sheep testes also take part in the regulation of gene expression besides inhibiting the activity of transposons. The target region of microRNA includes 3′ UTR, CDS region and 5′ UTR of target mRNA (Liu et al., 2013). Compared with other species, we found that the proportion of piRNAs aligned to repetitive regions (14.98%–15.27%) in sheep testes was similar to that in testes of adult mouse (∼17%) (Grivna et al., 2006) and human (∼22%) (Ha et al., 2014). However, the proportion of piRNA aligned to CDS regions (26.81%–27.64%) in sheep testes was higher than that in mice (∼1.3%) (Grivna et al., 2006) and humans (∼10%) (Ha et al., 2014). This suggested that a larger proportion of piRNAs were involved in gene expression regulation in sheep testes. In addition, the proportion of piRNAs aligned to intronic regions (17.39%–21.35%) was also higher than that of mice (∼9.3%) (Grivna et al., 2006) and pigs (7.6%) (Gebert et al., 2015). In summary, these differences of piRNA distribution in different species suggest that the biological roles of piRNAs were diverse in different species. In addition, it is worth noting that some piRNAs in sheep testes were also aligned to the lncRNA region. Kelley et al. found that lncRNA sequences in human testes tissue contain many retrotransposon sequences (Kelley and Rinn, 2012). It was reported that piRNAs in mouse testes can mediate the degradation of lncRNAs derived from Rasgrf1-marked sites (Watanabe et al., 2011). These results suggest that piRNAs may be also involved in regulating the expression of lncRNAs.
In order to further understand the inhibition of piRNAs for transposons in sheep testes, piRNAs derived from repeat sequences were aligned to the Repbase database to identify the types of transposons. Results showed that piRNAs in sheep testes were aligned predominantly to retrotransposon, which was consistent with the type in human (Ha et al., 2014), pig (Gebert et al., 2015) and yak (Gong et al., 2018). In a finer classification of retrotransposon, piRNAs in humans and pigs mainly aligned to LTR (Ha et al., 2014) and SINE (Gebert et al., 2015) respectively, while sheep and yak (Gong et al., 2018) mainly aligned to LINE type. It suggested that transposon components with high activity, i.e. action targets of piRNA, differs in these species. Furthermore, piRNAs from each TE type (LINE, SINE, and LTR) showed obvious ping-pong cycle characteristics, which implied that ping-pong cycle mechanism was adopted in sheep testes to cleave mRNA produced by transposons in order to inhibit the activity of transposons and maintain the stability of genome.
piRNAs exist in piRNA clusters on chromosomes (Toth et al., 2016; Ozata et al., 2019), which are considered to be the graves of transposons (Brennecke et al., 2007). We identified 284–430 piRNA clusters in each sample of sheep testis, and the numbers are similar with those identified in model animals (Lakshmi and Agrawal, 2008) and bovids (Russell et al., 2017). The first bases of all piRNAs derived from piRNA clusters had a strong U preference, which means that most piRNAs in clusters originated from the primary piRNA pathway. However, this feature does not exist in microRNA sequences. It was previously reported that primary piRNAs in gonadal tissue were generally derived from the antisense strand of transposons and have strong strand specificity (Brennecke et al., 2007; Aravin et al., 2008; Lin and Yin, 2008; Ozata et al., 2019). Therefore, we performed an analysis on the strand specificity of piRNA clusters, and found that within a single piRNA cluster, piRNAs have strong strand specificity. Interestingly, the statistic analysis for all piRNA clusters revealed that the number of piRNAs derived from the sense and antisense strands was comparable and the strand-specificity was no longer obvious. In fact, this phenomenon also exists in pigs (Gebert et al., 2015; Russell et al., 2017), but its specific mechanism need to be further explored.
Statistical results for indicators related to sperm quality (the ejaculate volume, pH value of semen, sperm viability, sperm motility, sperm density and sperm deformity rate) in SNT and STH rams showed that the single ejaculate volume and sperm number of STH sheep were significantly higher than that of SNT and the pH value of semen was closer to seven in STH sheep. Higher ejaculate volume and more motile sperm should be related to the higher fertility of STH sheep. Bovine sperm motility is highest when the pH of the semen is between 7 and 7.5 (Contri et al., 2013). In this study, the semen pH of STH was closer to seven compared with SNT, implying that sperm from STH had higher motility. All of the above factors contribute to increased fertility in STH rams.
In a succinct summary for signaling pathways enriched by DE piRNA target genes in the testes of rams with different fertility, they were closely related to adhesion, cytoskeleton, spermatogonial stem cell renewal and differentiation, which play crucial roles during spermatogenesis. Studies demonstrated that multiple adhesion proteins could act synergistically to safeguard the function of blood-testis barrier (BTB) (Cheng and Mruk, 2012), that is an important ultrastructure to support spermatogenesis. In this study, four adhesion-related pathways (Focal adhesion pathway, Adherens junction, Cell adhesion molecules and Rap1 signaling pathway) were noted. These pathways affect the normal physiological function of tight junctions of BTB during spermatogenesis (Siu et al., 2009). Loss of tight junctions could result in disruption of the integrity of BTB (Aivatiadou et al., 2007; Le Borgne et al., 2014; Okada et al., 2014), fibrosis of seminiferous tubule epithelial cells (Mazaud-Guittot et al., 2010), apoptosis of germ cell in the meiotic stage, decrease in sperm number and even azoospermia (Wu et al., 2012; Chakraborty et al., 2020). In this study, five pathways related to spermatogonial stem cell renewal and differentiation (Wnt signaling, PI3K/Akt pathway, MAPK signaling pathway, HIF-1 signaling pathway, Rap1 signaling pathway) were also significantly enriched. Previous studies showed that these pathways were involved in the regulation of testicular growth and development (Chambard et al., 2007), specifically playing important roles in proliferation, mitosis, and meiosis of germ cells (Godet et al., 2008; Nicol and Guiguen, 2011; Jiang et al., 2013; Kerr et al., 2014) and proliferation and lactate production of Sertoli cells (Riera et al., 2003; Lucas et al., 2012; Galardo et al., 2013; Yao et al., 2015; Ni et al., 2019) during both early and late spermatogenesis. When the above pathways were disturbed, the proliferation ability of SSCs (Lee et al., 2007; Yao et al., 2020) and Sertoli cell (Niu et al., 2015; Wang et al., 2015; Wang et al., 2017) would be inhibited. In addition, a study on asthenozoospermia population indicated that differentially expressed miRNAs in seminal plasma samples between patients with asthenospermia and healthy men were significantly associated with PI3K-Akt signaling pathway, MAPK signaling pathway and HIF-1 signaling pathway (Liang et al., 2022), which were consistent with our findings. Apart from that, the target genes of DE piRNAs were also significantly enriched in three pathways related to the cytoskeleton: Regulation of actin cytoskeleton, Chemokine signaling pathway and Focal adhesion pathway. Cytoskeletal components in the BTB are critical to development of germ cells along the Sertoli cells and successful spermatogenesis (Li et al., 2018; Wen et al., 2018).
Spermatogenesis is a complex process that is coordinated by multiple pathways. Notably, some of the pathways mentioned above were interrelated. We mapped the network relationships among the top 25 significantly enriched pathways and found that the complex associations exist among pathways (Figure 7). Furthermore, genes in pathways related to male reproduction also formed an associative network with each other (Figure 8). In fact, many previous studies suggested links among these pathways. For example, after VEGF activates the PI3K/Akt signaling pathway, expression of translation regulators (p70S6K and 4EBP1) will raise, and the synthesis of HIF-1α will be promoted (Dery et al., 2005; Marhold et al., 2015; Park et al., 2015; Zhu et al., 2018), which revealed the upstream-downstream relationship among VEGF signaling pathway, PI3K/Akt signaling pathway and HIF-1 signaling pathway. In this study, combined with the results of semen characterization and functional cluster analysis for DE piRNAs, it was implied that the differentially expressed piRNAs in the testes of rams with different fertility might be involved in regulation of BTB stability, proliferation of germ cells and accuracy of the genome through the above-mentioned pathways, which work synergistically to ultimately affect spermatogenesis and sperm number.
In this study, the sequence characteristics, origin and expression patterns of piRNAs were revealed in sheep testes. The length of piRNAs showed a unimodal distribution between 26 and 31 nt in sheep testes, with a peak at 29 nt. These piRNAs exhibited obvious ping-pong signature and strand specificity. In the genome, they were mainly aligned to CDS, intron, repetitive sequence regions and unannotated regions. Furthermore, in transposon analysis, piRNAs were aligned predominantly to LINE, SINE, and LTR types of retrotransposon in sheep testes, and the piRNAs derived from each type showed obvious ping-pong signature. The piRNA clusters identified in sheep testes were mainly distributed on chromosomes 3, 7, 15, 17, 18 and 20. The results combining semen determination with pathway enrichment analysis implied that differentially expressed piRNAs between the testes of rams with different fertility may participate in spermatogenesis by regulating multiple pathways closely related to stabilization of blood-testis barrier and renewal and differentiation of spermatogonial stem cell. Together, the study provide new insights into the characteristics and expression patterns of piRNAs in sheep testes tissue, which would help us better understand the role of piRNAs in sheep reproduction.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material
The animal study was reviewed and approved by the Science Research Department of the Institute of Animal Sciences of CAAS.
RD and RZ performed the experiment, analyzed data and wrote the manuscript. JM provided good suggestions and revised the manuscript. XH provided help in sample collection. XW, XH, YL, and YG provided help in data analysis. XZ and JZ took part in detecting semen indicators. XZ provided good suggestions and revised the manuscript. MC participated in design experiments and revision of the final manuscript. All authors read and approved the final version.
This study was supported by the National Natural Science Foundation of China (31861143012, 31572371), Earmarked Fund for China Agriculture Research System of MOF and MARA (CARS-38), and the Agricultural Science and Technology Innovation Program of China (CAASZDRW202106 and ASTIP-IAS13).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1078049/full#supplementary-material
Supplementary Figure S1 | Results of RNA-seq and qPCR for five random selected piRNAs in testes of SNT and STH sheep.
Aivatiadou, E., Mattei, E., Ceriani, M., Tilia, L., and Berruti, G. (2007). Impaired fertility and spermiogenetic disorders with loss of cell adhesion in male mice expressing an interfering Rap1 mutant. Mol. Biol. Cell. 18 (4), 1530–1542. doi:10.1091/mbc.E06-10-0902
Aravin, A. A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., et al. (2003). The small RNA profile during Drosophila melanogaster development. Dev. Cell. 5 (2), 337–350. doi:10.1016/s1534-5807(03)00228-4
Aravin, A. A., Naumova, N. M., Tulin, A. V., Vagin, V. V., Rozovsky, Y. M., and Gvozdev, V. A. (2001). Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11 (13), 1017–1027. doi:10.1016/s0960-9822(01)00299-8
Aravin, A. A., Sachidanandam, R., Bourc'his, D., Schaefer, C., Pezic, D., Toth, K. F., et al. (2008). A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 31 (6), 785–799. doi:10.1016/j.molcel.2008.09.003
Aravin, A., Gaidatzis, D., Pfeffer, S., Lagos-Quintana, M., Landgraf, P., Iovino, N., et al. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442 (7099), 203–207. doi:10.1038/nature04916
Brennecke, J., Aravin, A. A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R., et al. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 128 (6), 1089–1103. doi:10.1016/j.cell.2007.01.043
Chakraborty, A., Singh, V., Singh, K., and Rajender, S. (2020). Excess iodine impairs spermatogenesis by inducing oxidative stress and perturbing the blood testis barrier. Reprod. Toxicol. 96, 128–140. doi:10.1016/j.reprotox.2020.06.012
Chambard, J. C., Lefloch, R., Pouyssegur, J., and Lenormand, P. (2007). ERK implication in cell cycle regulation. Biochim. Biophys. Acta 1773 (8), 1299–1310. doi:10.1016/j.bbamcr.2006.11.010
Chen, T. T., Chen, X., Zhang, S. S., Zhu, J. W., Tang, B. X., Wang, A. K., et al. (2021). The genome sequence archive family: Toward explosive data growth and diverse data types. Genomics Proteomics Bioinforma. 19 (4), 578–583. doi:10.1016/j.gpb.2021.08.001
Cheng, C. Y., and Mruk, D. D. (2012). The blood-testis barrier and its implications for male contraception. Phrmacol. Rev. 64 (1), 16–64. doi:10.1124/pr.110.002790
Contri, A., Gloria, A., Robbe, D., Valorz, C., Wegher, L., and Carluccio, A. (2013). Kinematic study on the effect of pH on bull sperm function. Anim. Reprod. Sci. 136 (4), 252–259. doi:10.1016/j.anireprosci.2012.11.008
Czech, B., Munafo, M., Ciabrelli, F., Eastwood, E. L., Fabry, M. H., Kneuss, E., et al. (2018). piRNA-guided genome defense: From biogenesis to silencing. Annu. Rev. Genet. 52, 131–157. doi:10.1146/annurev-genet-120417-031441
David, I., Kohnke, P., Lagriffoule, G., Praud, O., Plouarboue, F., Degond, P., et al. (2015). Mass sperm motility is associated with fertility in sheep. Anim. Reprod. Sci. 161, 75–81. doi:10.1016/j.anireprosci.2015.08.006
Dery, M. A., Michaud, M. D., and Richard, D. E. (2005). Hypoxia-inducible factor 1: Regulation by hypoxic and non-hypoxic activators. Int. J. Biochem. Cell. Biol. 37 (3), 535–540. doi:10.1016/j.biocel.2004.08.012
Ernst, C., Odom, D. T., and Kutter, C. (2017). The emergence of piRNAs against transposon invasion to preserve mammalian genome integrity. Nat. Commun. 8, 1411. doi:10.1038/s41467-017-01049-7
Galardo, M. N., Riera, M. F., Regueira, M., Pellizzari, E. H., Cigorraga, S. B., and Meroni, S. B. (2013). Different signal transduction pathways elicited by basic fibroblast growth factor and interleukin 1β regulate CREB phosphorylation in Sertoli cells. J. Endocrinol. Investig. 36 (5), 331–338. doi:10.3275/8582
Gebert, D., Ketting, R. F., Zischler, H., and Rosenkranz, D. (2015). piRNAs from pig testis provide evidence for a conserved role of the Piwi pathway in post-transcriptional gene regulation in mammals. PLoS One 10 (5), e0124860. doi:10.1371/journal.pone.0124860
Girard, A., Sachidanandam, R., Hannon, G. J., and Carmell, M. A. (2006). A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442 (7099), 199–202. doi:10.1038/nature04917
Godet, M., Sabido, O., Gilleron, J., and Durand, P. (2008). Meiotic progression of rat spermatocytes requires mitogen-activated protein kinases of Sertoli cells and close contacts between the germ cells and the Sertoli cells. Dev. Biol. 315 (1), 173–188. doi:10.1016/j.ydbio.2007.12.019
Goh, W. S., Falciatori, I., Tam, O. H., Burgess, R., Meikar, O., Kotaja, N., et al. (2015). piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes. Dev. 29 (10), 1032–1044. doi:10.1101/gad.260455.115
Gong, J. S., Zhang, Q. W., Wang, Q., Ma, Y. J., Du, J. X., Zhang, Y., et al. (2018). Identification and verification of potential piRNAs from domesticated yak testis. Reproduction 155 (2), 117–127. doi:10.1530/Rep-17-0592
Gonzalez, J., Qi, H. Y., Liu, N., and Lin, H. F. (2015). Piwi is a key regulator of both somatic and germline stem cells in the Drosophila testis. Cell. Rep. 12 (1), 150–161. doi:10.1016/j.celrep.2015.06.004
Goriaux, C., Desset, S., Renaud, Y., Vaury, C., and Brasset, E. (2014). Transcriptional properties and splicing of the flamenco piRNA cluster. EMBO Rep. 15 (4), 411–418. doi:10.1002/embr.201337898
Gou, L. T., Dai, P., Yang, J. H., Xue, Y., Hu, Y. P., Zhou, Y., et al. (2014). Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell. Res. 24 (6), 680–700. doi:10.1038/cr.2014.41
Griffiths-Jones, S., Saini, H. K., van Dongen, S., and Enright, A. J. (2008). miRBase: tools for microRNA genomics. Nucleic Acids Res. 36, D154–D158. doi:10.1093/nar/gkm952
Grivna, S. T., Beyret, E., Wang, Z., and Lin, H. (2006). A novel class of small RNAs in mouse spermatogenic cells. Genes. Dev. 20 (13), 1709–1714. doi:10.1101/gad.1434406
Gunawardane, L. S., Saito, K., Nishida, K. M., Miyoshi, K., Kawamura, Y., Nagami, T., et al. (2007). A slicer-mediated mechanism for repeat-associated siRNA 5 ' end formation in Drosophila. Science 315 (5818), 1587–1590. doi:10.1126/science.1140494
Guo, Q., Xu, L., Bi, Y., Qiu, L., Chen, Y., Kong, L., et al. (2018). piRNA-19128 regulates spermatogenesis by silencing of KIT in chicken. J. Cell. Biochem. 119 (10), 7998–8010. doi:10.1002/jcb.26695
Ha, H., Song, J., Wang, S., Kapusta, A., Feschotte, C., Chen, K. C., et al. (2014). A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genomics 15 (1), 545. doi:10.1186/1471-2164-15-545
He, X., Li, B., Fu, S., Wang, B., Qi, Y., Da, L., et al. (2019). Identification of piRNAs in the testes of sunite and small-tailed han sheep. Anim. Biotechnol. 32, 13–20. doi:10.1080/10495398.2019.1640717
Houwing, S., Kamminga, L. M., Berezikov, E., Cronembold, D., Girard, A., van den Elst, H., et al. (2007). A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 129 (1), 69–82. doi:10.1016/j.cell.2007.03.026
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009a). Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37 (1), 1–13. doi:10.1093/nar/gkn923
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4 (1), 44–57. doi:10.1038/nprot.2008.211
Jiang, X., Skibba, M., Zhang, C., Tan, Y., Xin, Y., and Qu, Y. (2013). The roles of fibroblast growth factors in the testicular development and tumor. J. Diabetes Res. 2013, 489095. doi:10.1155/2013/489095
Kalvari, I., Argasinska, J., Quinones-Olvera, N., Nawrocki, E. P., Rivas, E., Eddy, S. R., et al. (2018). Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 46 (D1), D335–D342. doi:10.1093/nar/gkx1038
Kelley, D., and Rinn, J. (2012). Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 13 (11), R107. doi:10.1186/gb-2012-13-11-r107
Kerr, G. E., Young, J. C., Horvay, K., Abud, H. E., and Loveland, K. L. (2014). Regulated wnt/beta-catenin signaling sustains adult spermatogenesis in mice. Biol. Reprod. 90 (1), 3. doi:10.1095/biolreprod.112.105809
Kiuchi, T., Koga, H., Kawamoto, M., Shoji, K., Sakai, H., Arai, Y., et al. (2014). A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 509 (7502), 633–636. doi:10.1038/nature13315
Lakshmi, S. S., and Agrawal, S. (2008). piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 36, D173–D177. doi:10.1093/nar/gkm696
Le Borgne, M., Chartier, N., Buchet-Poyau, K., Destaing, O., Faurobert, E., Thibert, C., et al. (2014). The RNA-binding protein Mex3b regulates the spatial organization of the Rap1 pathway. Development 141 (10), 2096–2107. doi:10.1242/dev.108514
Lee, J., Kanatsu-Shinohara, M., Inoue, K., Ogonuki, N., Miki, H., Toyokuni, S., et al. (2007). Akt mediates self-renewal division of mouse spermatogonial stem cells. Development 134 (10), 1853–1859. doi:10.1242/dev.003004
Li, B., He, X., Zhao, Y. P., Bai, D. Y., Bou, G., Zhang, X. Z., et al. (2019). Identification of piRNAs and piRNA clusters in the testes of the Mongolian horse. Sci. Rep. 9, 5022. doi:10.1038/s41598-019-41475-9
Li, C. Y., Liu, Q. Y., Wang, X. Y., Hu, W. P., Han, D. P., Mwacharo, J. M., et al. (2020). Expression and localization of PIWI proteins in testis and ovary of domestic sheep. Czech J. Anim. Sci. 65 (3), 86–96. doi:10.17221/7/2020-Cjas
Li, C. Y., Zhang, R. S., Zhang, Z. J., Ren, C. H., Wang, X. Y., He, X. Y., et al. (2022). Expression characteristics of piRNAs in ovine luteal phase and follicular phase ovaries. Front. Vet. Sci. 9, 921868. doi:10.3389/fvets.2022.921868
Li, X. Y., Zhang, Y., Wang, X. X., Jin, C., Wang, Y. Q., Sun, T. C., et al. (2018). Regulation of blood-testis barrier assembly in vivo by germ cells. FASEB J. official Publ. Fed. Am. Soc. Exp. Biol. 32 (3), 1653–1664. doi:10.1096/fj.201700681R
Liang, G., Wang, Q., Zhang, G., Li, Z., and Wang, Q. (2022). Differentially expressed miRNAs and potential therapeutic targets for asthenospermia. Andrologia 54 (1), e14265. doi:10.1111/and.14265
Lin, H., and Yin, H. (2008). A novel epigenetic mechanism in Drosophila somatic cells mediated by PIWI and piRNAs. Cold Spring Harb. Symp. Quant. Biol. 73, 273–281. doi:10.1101/sqb.2008.73.056
Liu, C. C., Mallick, B., Long, D., Rennie, W. A., Wolenc, A., Carmack, C. S., et al. (2013). CLIP-based prediction of mammalian microRNA binding sites. Nucleic Acids Res. 41 (14), e138. ARTN e13810. doi:10.1093/nar/gkt435
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Loganantharaj, R., Randall, T. A., Lucas, T. F., Amaral, L. S., Porto, C. S., and Quintas, L. E. (2017). Na+/K+-ATPase α1 isoform mediates ouabain-induced expression of cyclin D1 and proliferation of rat sertoli cells.Na+/K+-ATPase alpha1 isoform mediates ouabain-induced expression of cyclin D1 and proliferation of rat sertoli cells. Bioinforma. Microrna Res. 1617144 (6), 133737–158745. doi:10.1007/978-1-4939-7046-9_1010.1530/REP-12-0232
Madison-Villar, M. J., Sun, C., Lau, N. C., Settles, M. L., and Mueller, R. L. (2017). Erratum to: Small RNAs from a big genome: The piRNA pathway and transposable elements in the salamander species Desmognathus fuscus. J. Mol. Evol. 84 (1), 67. doi:10.1007/s00239-016-9769-1
Marhold, M., Tomasich, E., El-Gazzar, A., Heller, G., Spittler, A., Horvat, R., et al. (2015). HIF1α regulates mTOR signaling and viability of prostate cancer stem cells. Mol. Cancer Res. 13 (3), 556–564. doi:10.1158/1541-7786.Mcr-14-0153-t
Mazaud-Guittot, S., Meugnier, E., Pesenti, S., Wu, X., Vidal, H., Gow, A., et al. (2010). Claudin 11 deficiency in mice results in loss of the Sertoli cell epithelial phenotype in the testis. Biol. Reprod. 82 (1), 202–213. doi:10.1095/biolreprod.109.078907
Mohn, F., Sienski, G., Handler, D., and Brennecke, J. (2014). The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 157 (6), 1364–1379. doi:10.1016/j.cell.2014.04.031
Ni, F. D., Hao, S. L., and Yang, W. X. (2019). Multiple signaling pathways in sertoli cells: Recent findings in spermatogenesis. Cell. Death Dis. 10 (8), 541. doi:10.1038/s41419-019-1782-z
Nicol, B., and Guiguen, Y. (2011). Expression profiling of wnt signaling genes during gonadal differentiation and gametogenesis in rainbow trout. Sex. Dev. 5 (6), 318–329. doi:10.1159/000334515
Niu, Z., Zheng, L., Wu, S., Mu, H., Ma, F., Song, W., et al. (2015). Ras/ERK1/2 pathway regulates the self-renewal of dairy goat spermatogonia stem cells. Reproduction 149 (5), 445–452. doi:10.1530/REP-14-0506
Okada, K., Miyake, H., Yamaguchi, K., Chiba, K., Maeta, K., Bilasy, S. E., et al. (2014). Critical function of RA-GEF-2/Rapgef6, a guanine nucleotide exchange factor for Rap1, in mouse spermatogenesis. Biochem. Biophys. Res. Commun. 445 (1), 89–94. doi:10.1016/j.bbrc.2014.01.149
Ozata, D. M., Gainetdinov, I., Zoch, A., O'Carroll, D., and Zamore, P. D. (2019). PIWI-Interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 20 (2), 89–108. doi:10.1038/s41576-018-0073-3
Park, S. J., Ryu, J., Kim, I. H., Choi, Y. H., and Nam, T. J. (2015). Activation of the mTOR signaling pathway in breast cancer MCF-7 cells by a peptide derived from Porphyra yezoensis. Oncol. Rep. 33 (1), 19–24. doi:10.3892/or.2014.3557
Riera, M. F., Meroni, S. B., Pellizzari, E. H., and Cigorraga, S. B. (2003). Assessment of the roles of mitogen-activated protein kinase and phosphatidyl inositol 3-kinase/protein kinase B pathways in the basic fibroblast growth factor regulation of Sertoli cell function. J. Mol. Endocrinol. 31 (2), 279–289. doi:10.1677/jme.0.0310279
Rosenkranz, D. (2016). piRNA cluster database: a web resource for piRNA producing loci. Nucleic Acids Res. 44 (D1), D223–D230. doi:10.1093/nar/gkv1265
Rosenkranz, D., and Zischler, H. (2012). proTRAC - a software for probabilistic piRNA cluster detection, visualization and analysis. Bmc Bioinforma. 13, 5. doi:10.1186/1471-2105-13-5
Ruby, J. G., Jan, C., Player, C., Axtell, M. J., Lee, W., Nusbaum, C., et al. (2006). Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C-elegans. Cell. 127 (6), 1193–1207. doi:10.1016/j.cell.2006.10.040
Russell, S., Patel, M., Gilchrist, G., Stalker, L., Gillis, D., Rosenkranz, D., et al. (2017). Bovine piRNA-like RNAs are associated with both transposable elements and mRNAs. Reproduction 153 (3), 305–318. doi:10.1530/Rep-16-0620
Singh, G., and Storey, K. B. (2021). MicroRNA cues from nature: A roadmap to decipher and combat challenges in human health and disease? Cells 10 (12), 3374. doi:10.3390/cells10123374
Siu, E. R., Wong, E. W. P., Mruk, D. D., Porto, C. S., and Cheng, C. Y. (2009). Focal adhesion kinase is a blood-testis barrier regulator. Proc. Natl. Acad. Sci. U. S. A. 106 (23), 9298–9303. doi:10.1073/pnas.0813113106
Teixeira, F. K., Okuniewska, M., Malone, C. D., Coux, R. X., Rio, D. C., and Lehmann, R. (2017). piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature 552 (7684), 268–272. doi:10.1038/nature25018
Toth, K. F., Pezic, D., Stuwe, E., and Webster, A. (2016). The piRNA pathway guards the germline genome against transposable elements. Adv. Exp. Med. Biol. 886, 51–77. doi:10.1007/978-94-017-7417-8_4
Vagin, V. V., Sigova, A., Li, C., Seitz, H., Gvozdev, V., and Zamore, P. D. (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313 (5785), 320–324. doi:10.1126/science.1129333
Wang, C., Fu, W., Quan, C., Yan, M., Liu, C., Qi, S., et al. (2015). The role of Pten/Akt signaling pathway involved in BPA-induced apoptosis of rat Sertoli cells. Environ. Toxicol. 30 (7), 793–802. doi:10.1002/tox.21958
Wang, H., Wang, J., Zhang, J., Jin, S., and Li, H. (2017). Role of PI3K/AKT/mTOR signaling pathway in DBP-induced apoptosis of testicular sertoli cells in vitro. Environ. Toxicol. Pharmacol. 53, 145–150. doi:10.1016/j.etap.2017.05.013
Wang, J. J., Shi, Y. R., Zhou, H. H., Zhang, P., Song, T. R., Ying, Z. Y., et al. (2022). piRBase: integrating piRNA annotation in all aspects. Nucleic Acids Res. 50 (D1), D265–D272. doi:10.1093/nar/gkab1012
Watanabe, T., Tomizawa, S., Mitsuya, K., Totoki, Y., Yamamoto, Y., Kuramochi-Miyagawa, S., et al. (2011). Role for piRNAs and noncoding RNA in de Novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332 (6031), 848–852. doi:10.1126/science.1203919
Wen, Q., Mruk, D., Tang, E. I., Wong, C., Lui, W. Y., Lee, W. M., et al. (2018). Cell polarity and cytoskeletons-Lesson from the testis. Semin. Cell. Dev. Biol. 81, 21–32. doi:10.1016/j.semcdb.2017.09.037
Wu, X., Peppi, M., Vengalil, M. J., Maheras, K. J., Southwood, C. M., Bradley, M., et al. (2012). Transgene-mediated rescue of spermatogenesis in Cldn11-null mice. Biol. Reprod. 86 (5), 139,1–11. doi:10.1095/biolreprod.111.096230
Xue, Y. B., Bao, Y. M., Zhang, Z., Zhao, W. M., Xiao, J. F., He, S. M., et al. (2022). Database resources of the national genomics data center, China national center for bioinformation in 2022. Nucleic Acids Res. 50 (D1), D27–D38. doi:10.1093/nar/gkab951
Yan, Z., Hu, H. Y., Jiang, X., Maierhofer, V., Neb, E., He, L., et al. (2011). Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 39 (15), 6596–6607. doi:10.1093/nar/gkr298
Yao, L., Peng, H., Xu, Z., Shi, L., Li, Y., and Dai, Y. (2020). The effect of regulating the Wnt signaling pathway on the proliferation and differentiation of spermatogonial stem cells. Ann. Transl. Med. 8 (16), 1003. doi:10.21037/atm-20-5321
Yao, P. L., Chen, L., Hess, R. A., Müller, R., Gonzalez, F. J., and Peters, J. M. (2015). Peroxisome proliferator-activated receptor-D (PPARD) coordinates mouse spermatogenesis by modulating extracellular signal-regulated kinase (ERK)-dependent signaling. J. Biol. Chem. 290 (38), 23416–23431. doi:10.1074/jbc.M115.664508
Zhang, P., Kang, J. Y., Gou, L. T., Wang, J., Xue, Y., Skogerboe, G., et al. (2015). MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell. Res. 25 (2), 193–207. doi:10.1038/cr.2015.4
Keywords: piRNAs, sheep, testis, reproduction, ping-pong signature
Citation: Di R, Zhang R, Mwacharo JM, Wang X, He X, Liu Y, Zhang J, Gong Y, Zhang X and Chu M (2022) Characteristics of piRNAs and their comparative profiling in testes of sheep with different fertility. Front. Genet. 13:1078049. doi: 10.3389/fgene.2022.1078049
Received: 24 October 2022; Accepted: 23 November 2022;
Published: 07 December 2022.
Edited by:
Qinghua Nie, South China Agricultural University, ChinaReviewed by:
Yinghui Ling, Anhui Agricultural University, ChinaCopyright © 2022 Di, Zhang, Mwacharo, Wang, He, Liu, Zhang, Gong, Zhang and Chu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaosheng Zhang, emhhbmd4czAyMjFAMTI2LmNvbQ==; Mingxing Chu, Y2h1bWluZ3hpbmdAY2Fhcy5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.