
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 29 November 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1077958
This article is part of the Research Topic Plant Secondary Metabolites and Their Effects on Environmental Adaptation Based on Functional Genomics View all 5 articles
B-box (BBX) which are a class of zinc finger transcription factors, play an important role in regulating of photoperiod, photomorphogenesis, and biotic and abiotic stresses in plants. However, there are few studies on the involvement of BBX transcription factors in response to abiotic stresses in sweet potato. In this paper, we cloned the DNA and promoter sequences of IbBBX28. There was one B-box conserved domain in IbBBX28, and the expression of IbBBX28 was induced under drought stress. Under drought stress, compared to wild type Arabidopsis, the protective enzyme activities (SOD, POD, and CAT) were all decreased in IbBBX28-overexpression Arabidopsis but increased in the mutant line bbx28, while the MDA content was increased in the IbBBX28-overexpression Arabidopsis and decreased in the bbx28. Moreover, the expression levels of the resistance-related genes showed the same trend as the protective enzyme activities. These results showed that IbBBX28 negatively regulates drought tolerance in transgenic Arabidopsis. Additionally, the yeast two-hybrid and BiFC assays verified that IbBBX28 interacted with IbHOX11 and IbZMAT2. The above results provide important clues for further studies on the role of IbBBX28 in regulating the stress response in sweet potato.
In nature, plants are constantly stressed by adverse abiotic environmental factors such as drought, heat, cold, nutrient deficiencies and excess salt or toxic metals in the soil. Moreover, abiotic stresses such as high salt and drought are environmental factors that affect crop yield and quality (Munns and Tester, 2008; Zhai et al., 2016; Bailey-Serres et al., 2019). Revealing the mechanisms of plant response to abiotic stress will benefit the development of resistant crops and improve agricultural sustainability. Plants will produce a series of physiological and biochemical reactions by drought stress, including stomatal closure, reduced photosynthesis and cell growth. When plants are under drought stress, the hormone ABA is also produced and the expression of stress-related genes are induced. Many transcription factors play a role in regulating drought stress signal transduction pathways, such as ARED, MYC, MYB, bZIP, NAC and BBX (Shinozaki and Yamaguchi-Shinozaki, 2007; Yang et al., 2014).
Zinc finger structure transcription factors are one of the important families in plants, and B-box (BBX) is one subgroup of zinc finger proteins. BBX transcription factor is of great interest due to its many functions in plant growth and development (Khanna et al., 2009; Crocco and Botto, 2013; Gangappa and Botto, 2014; Song et al., 2020). STO (salt tolerace)/AtBBX24 gene responses to salt stress and promotes the root growth of Arabidopsis under high salt conditions (Nagaoka and Takano, 2003). It was found that the chlorophyll content and net photosynthetic rate were lower in the overexpressed SlBBX17 tomato, and the heat tolerance of the transgenic tomato increased (Xu et al., 2022). CmBBX24 (homologous to AtBBX24) which negatively regulated the expression of genes in photoperiodic flowering pathway including GI, PRR5, CO, FT, and SOC1, delayed the flowering time in chrysanthemum. In addition, CmBBX24 is involved in regulating the response of chrysanthemum to low temperature and drought by regulating the stress response and GA synthesis-related genes (Yang et al., 2014). Overexpression of IbBBX24 and IbPRX17 in sweet potato, the salt and drought tolerance were significantly improved, and the molecular mechanisms of the IbBBX24-IbTOE3-IbPRX17 model in response to abiotic stresses were resolved (Zhang et al., 2022). BBX family members also play important roles in hormone signal transduction pathways. Studies have found that Arabidopsis BBX gene responds to plant hormones and is involved in various hormone pathways (Vaishak et al., 2019). When dealing with exogenous hormones (ABA, GA, JA and SA), the transcription levels of several BBX genes were increased. The promoters of these BBX genes contain one or more hormone-responsive cis-acting elements, such as ABRE (ABA response element), ERE (ethylene response element), CGTCA motifs, and TGACG (MeJA response element) (Gangappa and Botto, 2014; Chu et al., 2016).
The sweet potato [Ipomoea batatas (L.) Lam.] is an important root vegetable, and rank the seventh largest food crop in the world. Compared with other crops, sweet potato has many advantages, such as high adaptability, rapid growth, and high yield (O'BRIEN, 1972; Bovell-Benjamin, 2007; Kwak, 2019). The tuberous roots of sweet potato contain high levels of nutrients, such as starch, protein, fatty acids, sugars, and vitamins, and are also rich in inorganic salts such as calcium, phosphorus, and iron, as well as carotenoids and anthocyanins. It plays a vital role in human health for its antioxidant, anticancer, anti-aging, and immunomodulatory effects, and its nutritional value is gradually being recognized (Abegunde et al., 2013; Kwak, 2019; Nguyen et al., 2021).
Several genes related to abiotic stress tolerance were isolated and identified from sweet potato and have been used to improve abiotic stress resistance of sweet potato (Liu, 2017; Lyu et al., 2021). In Arabidopsis, LOW OSMOTIC STRESS 5 (LOS5)/ABA3 is an important regulator in response to cold, salt, and drought stresses, and overexpression of AtLOS5 in sweet potato improves its salt tolerance (Gao et al., 2011). The sweet potato bZIP transcription factor gene IbbZIP1 improved salt and drought tolerance in Arabidopsis (Kang et al., 2019). With the overexpression of IbP5CR gene in sweet potato, the ROS scavenging system of the transgenic sweet potato was activated, and the salt resistance was improved (Liu et al., 2014). Co-expression of the Arabidopsis Na+/H+ reverse transporter NHX1 and the DEAD-box RNA decapping enzyme elF4A1 in sweet potato enhanced its drought tolerance (Zhang et al., 2019).
It was found that AtBBX28 negatively regulates photomorphogenesis (Lin et al., 2018), and AtBBX28 negatively regulates flowering in Arabidopsis by co-acting with CO and FT (Liu et al., 2020). However, there are few studies on the response of IbBBX28 to stress in sweet potato. In this paper, we conducted a preliminary study on the biological function of IbBBX28. The IbBBX28-overexpression Arabidopsis was obtained, and the regulatory role of IbBBX28 in Arabidopsis drought stress response was explored. Furthermore, the yeast two-hybrid assay and bimolecular fluorescence assay (BiFC) were used to screen and validate the interacting proteins of IbBBX28. The mechanism of IbBBX28 in the regulation of stress response in sweet potato was initially explored.
The sweet potato variety Xuzishu-3 (XZ-3) was planted in the experimental field in Shanxi Agricultural University. Nicotiana benthamiana was used for the BiFC assay of the interaction between IbBBX28 and its interacting protein. The growth condition of N. benthamiana was as follows: light/dark for 16/8 h at 26°C. The seeds of Arabidopsis thaliana (Columbia) were kept by our laboratory. The bbx28 T-DNA insertion Arabidopsis thaliana (SALK_094193C) was obtained from AraShare Technology Service Center (https://www.arashare.cn) and analyzed by PCR. The growth condition of Arabidopsis thaliana was as follows: light/dark for 16/8 h at 22°C.
The Open Reading Frameb (ORF) sequence of IbBBX28 (GenBank accession number OP047916) was previously cloned in our lab, and the gene structure of the genomic DNA sequence of IbBBX28 was predicted from the database (https://121.36.193.159/blast.html). The genomic DNA was extracted from XZ-3 root sample using the CTAB method (Safeena et al., 2021). Using the genomic DNA as template, the genomic DNA sequence and promoter sequence of IbBBX28 were cloned by PCR. All the specific primers are listed in Supplementary Table S1. Based on the DNA and ORF sequences of IbBBX28, the gene structure was mapped using the online tool Gene Structure Display Server (http://gsds.gao-lab.org/). Multiple protein sequence alignments of BBX28 from different species were conducted with DNAMAN software. The conserved domain was confirmed by the Conserved Domain Database (CDD) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). A maximum-likelihood (ML, Maximum-likelihood) phylogenetic tree of the BBX proteins was constructed using MEGA 7.0 software. The bootstrap was set at 1,000 replicates (Kumar et al., 2016). The cis-acting elements of the IbBBX28 promoter were predicted at the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002).
Approximately 10 cm vines were cut from XZ-3 plants grown in the field for 30 d and placed in 1/2 Hoagland nutrient solution for hydroponics in an incubator (25°C/16 h of light and 22°C/8 h of darkness). After 2 weeks of growth, 30% PEG6000 was added to the 1/2 Hoagland nutrient solution for drought stress treatments. Samples were taken at 0, 6, 12, 24, 48, and 72 h after the stress treatment. All treatments had three biological replications. The samples were frozen in liquid nitrogen and kept in a −80°C refrigerator for further use.
Total RNA was extracted using a Quick RNA Isolation kit (Huayueyang Biotech, Beijing, China). Using 1 μg total RNA as template, a PrimeScript RT Reagent kit with gDNA Eraser (Takara Bio, Shiga, Japan) was used to synthesis cDNA by following the manufacturer’s instructions.
RT-qPCR was used to analyze the expression pattern of the IbBBX28 gene in XZ-3 leaves, stems, and tubers under different PEG treatment times (0, 6, 12, 24, 48, and 72 h). The experiments were conducted on a CFX96PCR system (Bio-Rad, United States), and the reaction system was 10 μL (1 μl cDNA, 5 μl SYBR®Premix Ex-TaqTM, 3.2 μL dd H2O, 0.4 μL PF and 0.4 μl PR). All the experiments were replicated three times, the relative expression was calculated using the 2−ΔΔCt method, and the IbActin gene (AY905538) was used as an internal reference (Gao et al., 2011).
The overexpressed vector pCAMBIA1300-IbBBX28 (pC1300-IbBBX28) was constructed and the positive transgenic plants were obtained. The seeds of wild type Arabidopsis, T3 generation of two IbBBX28-overexpression Arabidopsis lines (OE-4 and OE-5), and mutant line bbx28 were sown on 1/2 MS solid medium (Petri dishes of 90 mm diameter), respectively. After 7 d of growth on plates, the seedlings were transferred to 1/2 MS solid medium (150 mm diameter dishes) containing different concentrations (0, 200, 250, and 300 mM) of mannitol. Samples were taken after 7 d of treatment.
Seedlings (WT, OE-4, OE-5, and bbx28) which were grown on 1/2 MS solid medium for 7 d, were transplanted to pots (1:3 ratio of nutrient soil to vermiculite). After 15 d of normal growth in the incubator, the plants were rehydrated for 3 d after 10 d of drought treatment. Samples were taken after 7 d of treatment. The plant materials collected were immediately frozen in liquid nitrogen and stored in a −80°C freezer for further use. The protective enzyme activities (CAT, POD, and SOD) and MDA content were determined according to the kit instructions (Nanjing Jiancheng Biotechnology Company, Nanjing, China).
RNA from the leaves of the WT, T3 generation Arabidopsis of overexpressed IbBBX28, and bbx28 grown normally and treated with drought stress for 7 d was extracted and then reverse transcribed to cDNA, and the expression levels of the related resistance genes were analyzed using RT-qPCR. All the primers are shown in Supplementary Table S2. All the experiments were repeated three times, and the relative expression levels were calculated in the 2−ΔΔCt method. The Arabidopsis Actin gene (NM_112764) was used as the internal control (Kang et al., 2018).
The yeast two-hybrid (Y2H) assay was carried on via co-transformation method according to the protocols (OE biotech. Company). The vectors of pGADT7 and pGBKT7 were used as the prey and the bait constructs, respectively. The cDNA from sweet potato tuberous root was constructed into the prey vector pGADT7 to obtain yeast library. The coding sequence (CDS) of IbBBX28 was inserted into the empty vector pGBKT7 with EcoRI and BamHI sites to construct the bait vector pGBKT7-IbBBX28. The CDS of IbHOX11 and IbZMAT2 were insert into the empty vector pGADT7 with EcoRI and BamHI sites to obatin the prey vectors of pGADT7-IbHOX11 and pGADT7-IbZMAT2. The bait and prey vectors were co-transformed into yeast strain Y2H Gold, and positive clones were screened. The transformed yeast broth was screened on a screening plate SD/-Trp/-Leu/-His/-Ade/X-α-Gal/AbA and incubated in an incubator at 30°C for 3–5 d in an inverted position.
The CDS sequence of IbBBX28 was constructed into the pSm35s-nYFP vector and named NY-IBBBX28. The CDS sequences of the interacting protein coding genes were constructed into the pSm35s-cYFP vector and named IbHOX11-CY and IbZMAT2-CY, respectively. The primers required for the BiFC assay are shown in Supplementary Table S3. BamHⅠ and XbaⅠ were selected as the restriction sites, and the constructed vector plasmid was transformed into Agrobacterium GV3101 for the BiFC assay. Agrobacterium tumefaciens was inoculated into 10 ml LB liquid medium with spectinomycin and incubated in a shaker (200 rpm) at 28°C for 1 h. Then, it was centrifuged at 4,000 rpm for 10 min, the supernatant was discarded, and the bacterial cell was resuspended (10 mM MgCl2 and 120 μM AS). The OD600 was adjusted to about 0.6. The Agrobacterium solutions of the two genes were mixed in a ratio of 1:1, and left at room temperature for 3 h, and then injected with four to five leaf stage tobacco leaves. The injected tobacco was incubated at 26°C (16 h light/8 h dark) for 2 days; then, it was observed and photographed with a laser scanning confocal microscope (Nikon C2-ER). The excitation wavelength of yellow fluorescent protein YFP was 511 nm, and the emission wavelength was 525 nm.
Three biological replicates were performed for all experiments, and the data are presented as the mean value ±standard deviation (SD). All data were analyzed with SPSS software (Chicago, IL, United States, version 8.0) using t-test or one-way ANOVA and least significant difference (LSD) test.
In this study, a 1,022 bp genomic sequence of IbBBX28 was obtained with two exons and one intron (Figure 1A). The ORF encoding protein of IbBBX28 contained one BBX transcription factor specific domain B-box in the 3–45 region of its amino acid sequence (Figure 1B). Multiple sequence alignment of IbBBX28 and BBX28 proteins from other species showed that they all contain one B-box domain (Figure 1C). The IbBBX28 protein sequence has the highest similarity with Ipomoea trifida (Itr_sc000359.1_g00035.1) at 95.57%. The similarities of IbBBX28 with Arabidopsis thaliana (AT4G27310), Fragaria x ananassa (QOI16737), Rosa hybrid cultivar (UCU84643), and Solanum tuberosum (ARU77867) were 29.89%, 40.21%, 31.48%, and 30.74%, respectively.
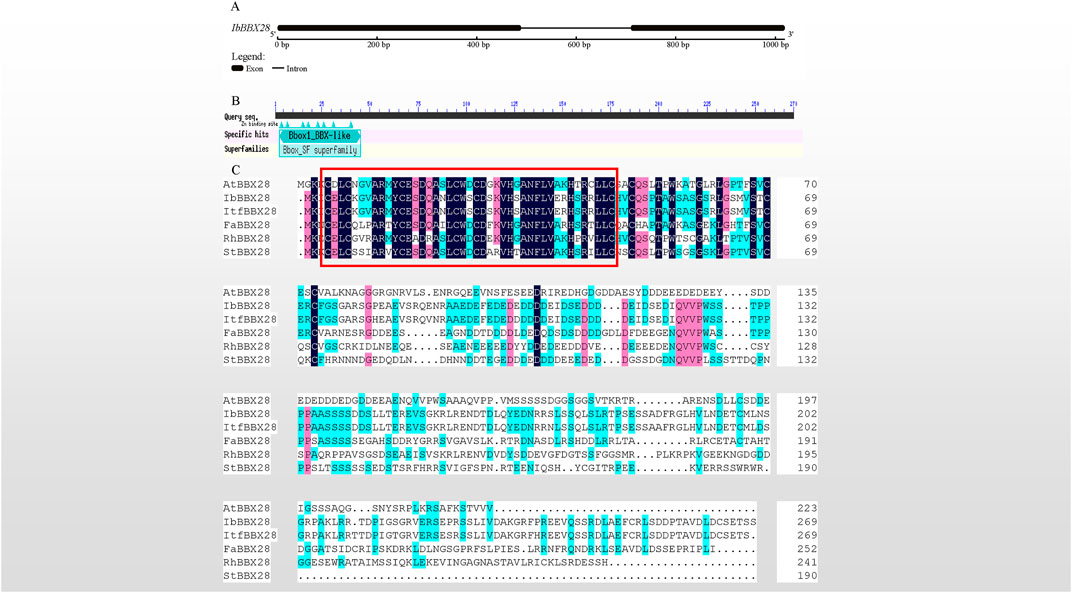
FIGURE 1. Sequence cloning of IbBBX28, gene structure, and multiple sequence alignment results. (A) The gene structure of IbBBX28. (B) The structural domain of IbBBX28 obtained by the CD-search in the NCBI. (C) The multiple sequence alignment of IbBBX28 and BBX28 proteins in other species; the sequence in the red box is the B-box domain. The amino acids sequences are: AtBBX28 (Arabidopsis thaliana, AT4G27310), ItfBBX28 (I. trifida;Itr_sc000359.1_g00035.1), FaBBX28 (Fragaria x ananassa, QOI16737), RhBBX28 (Rosa hybrid cultivar, UCU84643), and StBBX28 (Solanum tuberosum, ARU77867).
The phylogenetic tree of the above six species was constructed. It showed that IbBBX28 had the closest evolutionary relationship with ItfBBX28, a sweet potato diploid wild ancestor (Supplementary Figure S1).
The 2000 bp upstream sequence of ATG of IbBBX28 was cloned and named IbBBX28-Pro. The cis-acting element analysis showed (Supplementary Table S4) that IbBBX28-Pro contain six stress-responsive elements (in response to anaerobic, low temperature, drought, and other stresses), 10 hormone-responsive elements (GA, abscisic acid, methyl jasmonate, and salicylic acid), 19 photoresponsive elements and 34 RNA polymerase binding sites TATA box.
The sweet potato variety XZ-3 was subjected to drought stress with 30% PEG6000. Samples of the root, stem, and leaf were taken at 0, 6, 12, 24, 48, and 72 h after PEG treatment. RNA was extracted from root, stem, and leaf tissues, and these RNA were reverse transcribed into cDNA. The expression levels of the IbBBX28 gene in the three tissues was analyzed. The results showed that the expression of the IbBBX28 gene was significant induced (Figure 2) under PEG treatment. Compared with 0 h, the expression level of the IbBBX28 gene in leaf and root were peaked at 24 h with 5.5 and 2 folds, respectively. And the expression of IbBBX28 was decreased in stem. The results indicated that IbBBX28 response to drought stress.
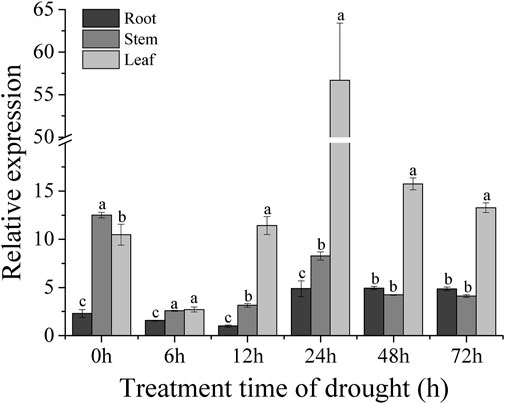
FIGURE 2. Expression analysis of IbBBX28 under PEG stress treatment. The expression of IbBBX28 at 12 h in root was set as 1-fold. Data are presented as means ± SD (n = 3). The different letters indicate significant differences in the expression level of the IbBBX28 gene in the roots, stems, and leaves under the same time treatment (p < 0.05) by one-way ANOVA and least significant difference (LSD) test.
To study the function of the IbBBX28 gene, the overexpression vector pC1300-IbBBX28 was transformed into Arabidopsis Thaliana. Seven transgenic Arabidopsis lines were obtained, and two lines (OE-4 and OE-5) in which the expression level of IbBBX28 gene was relatively high were chosen for further study.
A mannitol-simulated drought stress test was carried out on the wild type (WT), IbBBX28-overexpression Arabidopsis, and bbx28 mutant plant (Figure 3). In the medium without mannitol addition, there was no significant difference in the growth of the four lines. After adding different concentrations of mannitol to the medium, the growth of the WT, OE-4, OE-5 and bbx28 were inhibited, and the root growth was slow. Compared with the WT, the root growth of the OE-4, OE-5, and bbx28 was less (Figure 3A). On the medium supplemented with 0 mM mannitol, the root length and root elongation of the OE-4 and OE-5 plants were slightly longer than the WT after 10 days of growth. The root length of the bbx28 was not significantly different from the WT, and the root elongation was slightly larger than the WT (Figures 3B,C). On the medium supplemented with 300 mM mannitol, the root length and root elongation of the OE-4 and OE-5 plants were lower than those of the WT plants after 10 days of growth. The root length of the bbx28 was lower than that of the WT, but the root elongation was not significantly different from that of the WT (Figures 3B,C).
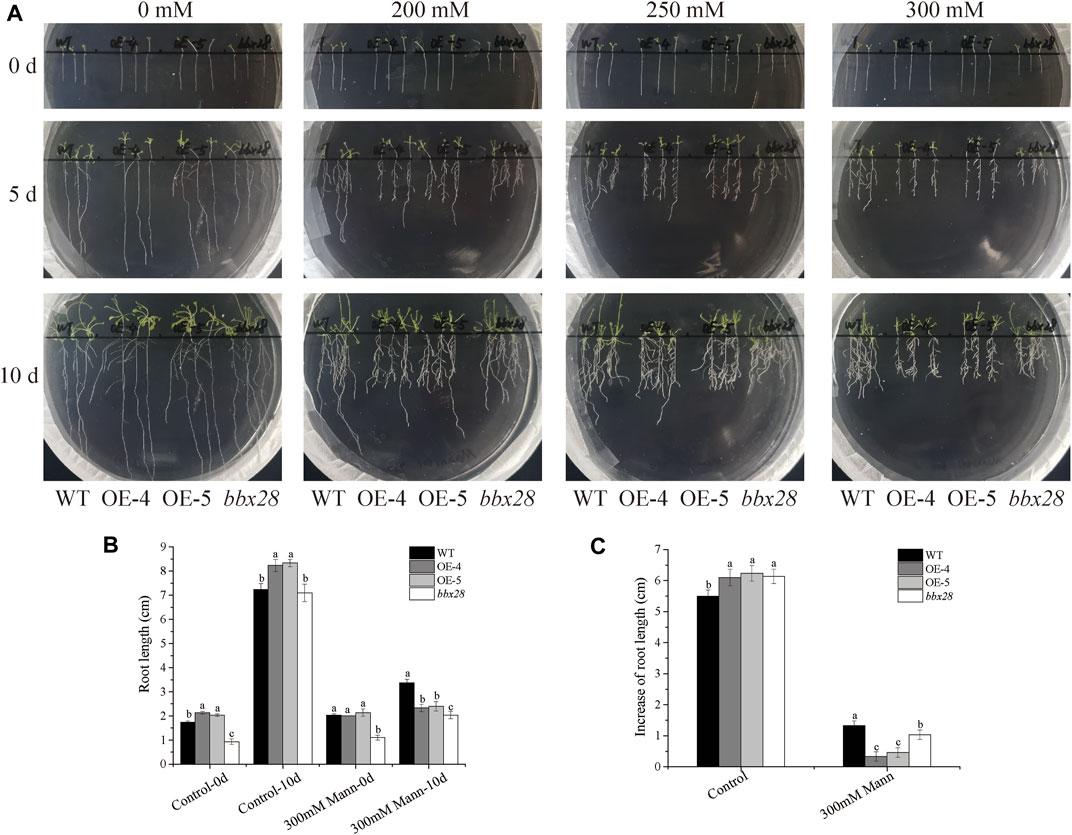
FIGURE 3. The phenotype of overexpressing IbBBX28 transgenic Arabidopsis and bbx28T-DNA insertion Arabidopsis in 1/2MS medium supplemented with different concentrations of mannitol. (A) The growth of overexpressing IbBBX28 transgenic Arabidopsis in 1/2 MS medium supplemented with 0, 200, 250, and 300 Mm mannitol. (B) The root length. (C) The root elongation. Data are presented as means ± SD (n = 3). Different lowercase letters indicate significant differences between OE-4, OE-5, bbx28, and WT (p < 0.05) by one-way ANOVA and least significant difference (LSD) test.
A soil drought stress test was conducted (Figure 4). The results showed that after 10 days of drought stress, the growth of OE-4 and OE-5 was significantly worse than that of the WT, while the growth of the mutant line bbx28 was better than that of WT. After rehydration, the growth of the WT and bbx28 recovered somewhat, however, the growth of OE-4 and OE-5 were still worse.
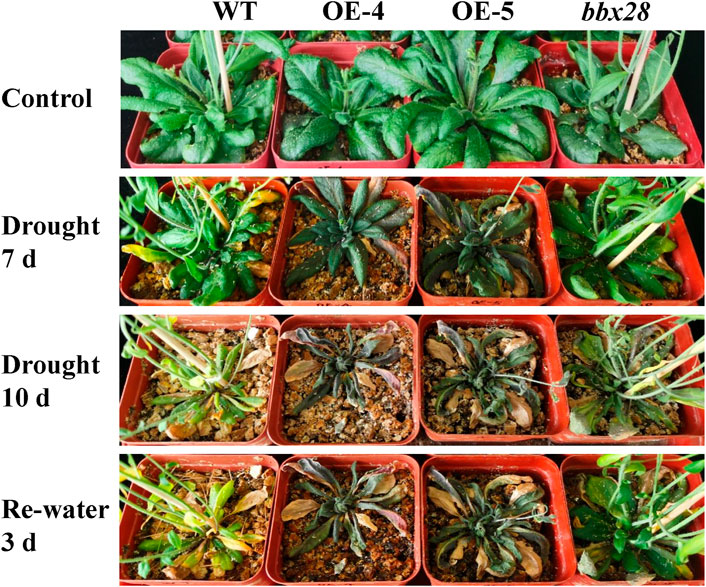
FIGURE 4. The phenotype of overexpressing IbBBX28 transgenic Arabidopsis and bbx28 T-DNA insertion Arabidopsis under drought stress. The experiments were performed three times with similar results. There were more than 10 plants in each replication. The photographs depict the results of one of the three experiments.
The activity of the protective enzymes was measured (Figures 5A–C). Under normal growth conditions (Control), the activities of superoxide dismutase (SOD), and peroxide (POD), and catalase (CAT) in the OE-4, OE-5, bbx28, and the WT showed no significant differences. Under drought stress, compared with the Control, the activities of the CAT, POD, and SOD in the WT, OE-4, OE-5, and bbx28 were increased, but the enzyme activities in the WT were significantly higher than those in OE-4 and OE-5 and lower than those in bbx28. The results of the malondialdehyde (MDA) content determination (Figure 5D) showed that under normal growth conditions, there was no significant difference in MDA content among the WT, OE-4, OE-5, and bbx28. Under drought stress, the MDA content in WT, OE-4, OE-5, and bbx28 was significantly increased compared with the Control, but the MDA content in the OE-4 and OE-5 was significantly higher than that in the WT, while the MDA content in the WT was higher than that in the bbx28. In conclusion, combined with the results of the drought stress tests in the Arabidopsis seedlings, it is speculated that IbBBX28 negatively regulates the drought tolerance of Arabidopsis.
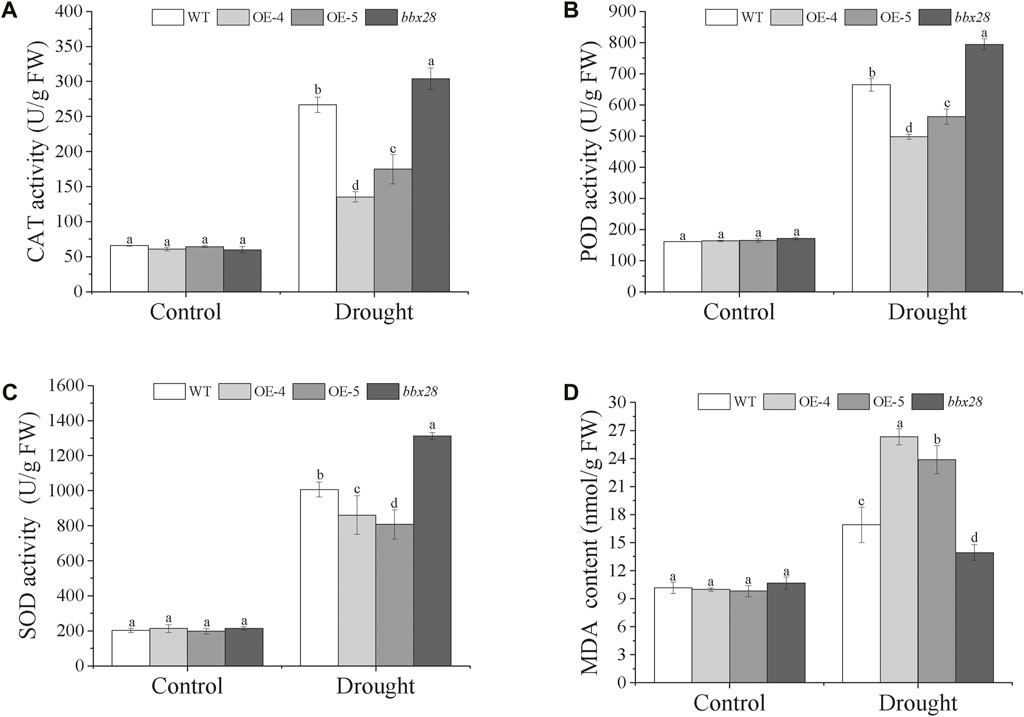
FIGURE 5. The activity of superoxide dismutase (SOD), peroxide (POD), and catalase (CAT), and the content of malondialdehyde (MDA) in the transgenic Arabidopsis and WT plants under drought stress. (A) The activity of CAT (U/g FW). (B) The activity of POD (U/g FW). (C) The activity of SOD (U/g FW). (D) The content of MDA. Data are presented as means ± SD (n = 3). The different letters indicate significant differences among OE-4, OE-5, bbx28, and WT at p < 0.05 by one-way ANOVA and least significant difference (LSD) test.
RNA was extracted from the WT, transgenic lines (OE-4, OE-5), and mutant lines (bbx28) under normal and drought treatment for 7 days. The expression of the resistance-related genes was analyzed by RT-qPCR after reverse transcription into cDNA (Figure 6). The results showed that the expression level of IbBBX28 in the OE-4 and OE-5 decreased after drought stress compared with the control group. The expression levels of the AtNCED3, AtABA3, and AtSDR genes involved in ABA biosynthesis were increased in the four lines, but the expression levels in the WT were higher than those in the OE-4 and OE-5 and lower than those in the bbx28. The expression of the stress response gene AtRD29a was significantly increased, and the expression of the AtRD29a in the WT was higher than in the OE-4 and OE-5 and lower than in the bbx28. The expression of AtKIN2 in the WT was decreased, but the expression of AtKIN2 in the WT was higher than that in the OE-4 and OE-5 and lower than that in the bbx28. After drought stress treatment, the expressions of AtAPX1, AtSOD and AtPOD were decreased, but their expressions in the WT were higher than those in the OE-4 and OE-5 and lower than those in the bbx28.
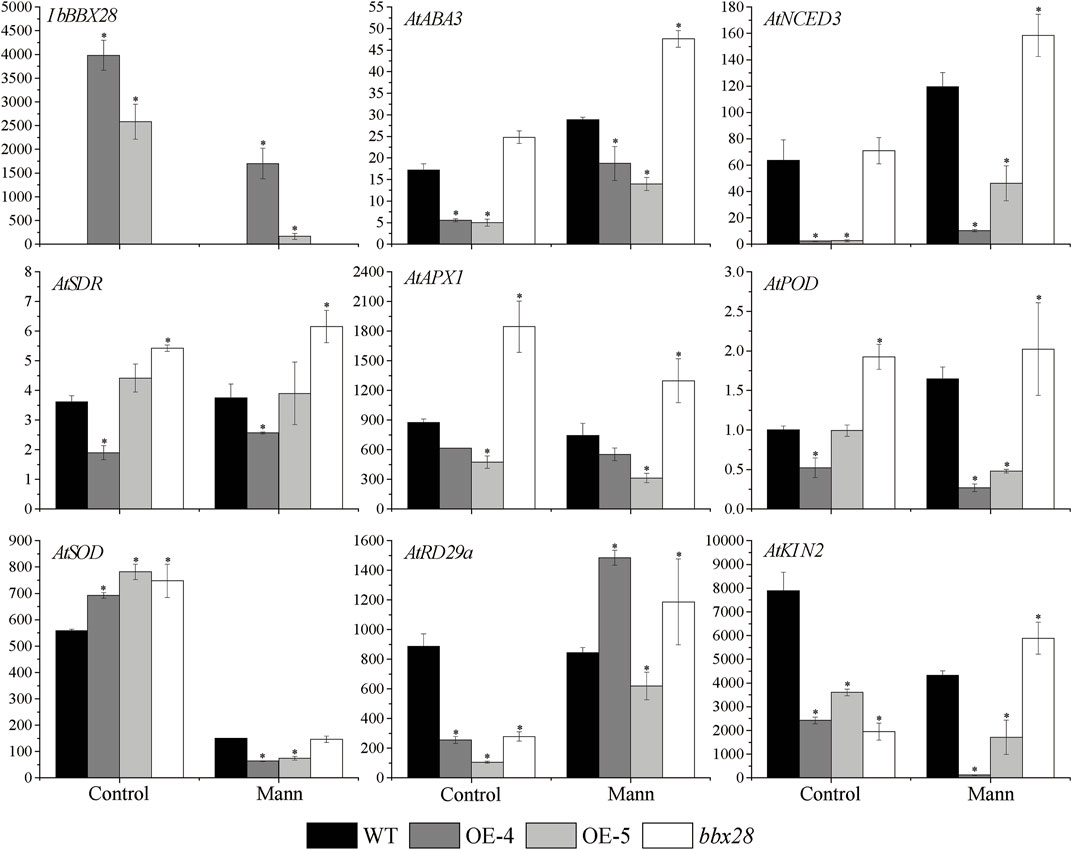
FIGURE 6. Expression analysis of overexpressing IbBBX28 transgenic Arabidopsis resistance related genes under drought stress. The expression of AtPOD in WT in control group was set as 1-fold. Data are presented as means ± SD (n = 3). * indicates a significant difference of OE-4, OE-5, and bbx28 from that of the WT at p < 0.05 by the t-test.
In order to study the mechanism of IbBBX28 involved in stress regulation, the interacting proteins of IbBBX28 were screened in the yeast cDNA library of sweet potato tuberous roots using yeast two-hybrid method. A total of 25 proteins interacting with IbBBX28 were screened (Supplementary Table S5). According to the annotation information of each protein, the proteins of which are of significance and without frame-shifting were selected for interaction verification. Among the interaction proteins, the homeobox-leucine zipper protein HOX11 (IbHOX11) may function in stress response, and studies about zinc finger matrin-type protein 2 (IbZMAT2) in plants were few. Therefore, IbHOX11 and IbZMAT2 were selected for the next validation test. The results showed that pGBKT7-IbBBX28 + pGADT7-IbHOX11 and pGBKT7-IbBBX28 + pGADT7-IbZMAT2 grew well on SD/-Trp/-His/-Leu/-Ade/X-α-Gal deficient medium. This indicates that IbBBX28 interacts with IbHOX11 and IbZMAT2 in the yeast system (Figure 7). To further verify the interaction of IbBBX28 with IbHOX11 and IbZMAT2, a BiFC assay was performed in tobacco leaves (Figure 8). The results showed that no YFP fluorescence signal was detected in the negative controls (NY-IbBBX28 + CY、NY + IbHOX11-CY, and NY + IbZMAT2-CY) after injection into tobacco leaves. However, the YFP fluorescence signal was observed after NY-IbBBX28 + IbHOX11-CY and NY-IbBBX28 + IbZMAT2-CY were injected into tobacco leaves, respectively. This indicated that IbBBX28 interacts with IbHOX11 and IbZMAT2.
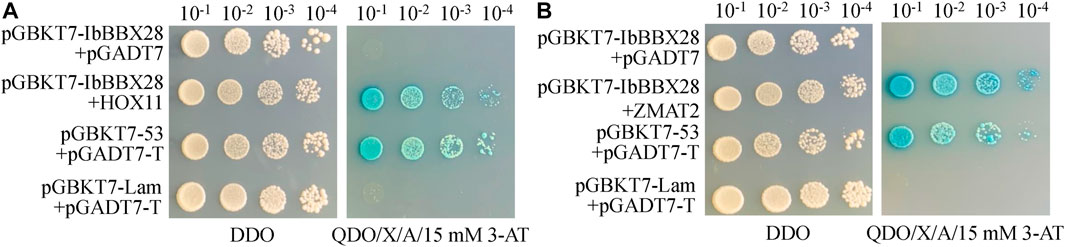
FIGURE 7. Plate validation of IbBBX28 and interacting proteins. (A) Plate validation of IbBBX28 and IbHOX11. (B) Plate validation of IbBBX28 and IbZMAT2.
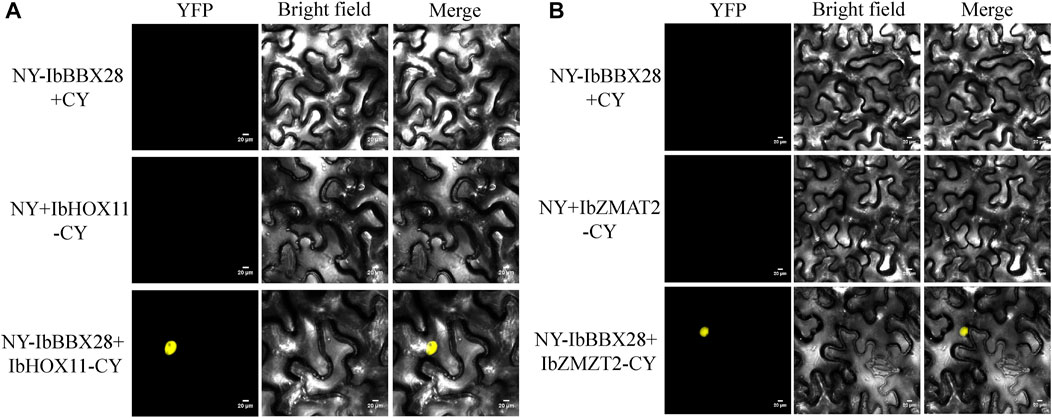
FIGURE 8. BiFC validation of the IbBBX28 interacting protein. (A) BiFC assay of IbBBX28 with IbHOX11. (B) BiFC assay of IbBBX28 with IbZMAT2.
B-box transcription factors contain characteristic B-box domains and play an important role in plant growth, development, and stress response (Gangappa and Botto, 2014). The conserved zinc finger structure of the B-box domain can bind to zinc ions to enhance protein stability and play a role in protein and protein interaction (Wang et al., 2021). Many studies have reported that BBX proteins are involved in plant signal transduction pathways and respond to various abiotic stresses, such as low temperature, high salinity, drought, and heat stress (Nagaoka and Takano, 2003; Wang et al., 2013; Yang et al., 2014). AtBBX28, a repressor of light signaling, delayed the flowering time in Arabidopsis under long day conditions (Lin et al., 2018; Liu et al., 2020). Furthermore, heterologous expression of strawberry FaBBX28C1 have been found to negatively regulate the flowering time in Arabidopsis (Ye et al., 2021). In this study, there existed drought response and hormone response cis-elements in IbBBX28-Pro, including MBS, P-box, ABRE, CGTCA-motif. And the expression of IbBBX28 in the roots, stems, and leaves of sweet potato was significantly induced under drought stress, suggesting that IbBBX28 response to drought stress.
To resist environmental stress, plants have evolved a series of regulatory pathways that allow them to respond and adapt the environment. Under adverse conditions, a large amount of ABA is accumulated in plants to enhance stress resistance (Ikegami et al., 2009). The ROS scavenging system can remove the toxic effects caused by ROS and enhance the activity of protective enzymes (Gill and Tuteja, 2010), and improving the ROS scavenging activity can also increase the stress tolerance of plants (Liu et al., 2014; Kikuchi et al., 2015). The protective enzymes of ROS scavenging in plants include CAT, SOD, POD, APX and so on. When under adversity, MDA content increases with the excessive accumulation of superoxide free radicals and hydrogen peroxide. The higher content of MDA can induce cell membrane damage, and the tolerance to stress is reduced in plants (Bao et al., 2009; Kumar et al., 2010). In this study, we found that the activities of the protective enzymes (CAT, SOD, and POD) in the transgenic Arabidopsis were lower than those in the WT, while the content of MDA was higher than that in the WT. The protective enzyme activity and MDA content in the Arabidopsis mutant line bbx28 were opposite. Moreover, the ROS scavenging enzyme related genes (AtAPX1, AtSOD, and AtPOD) and ABA synthesis-related genes (AtABA3, AtSDR, and AtNCED3) in the transgenic Arabidopsis were lower than those in the WT, while the expression level of these genes in the Arabidopsis mutant line bbx28 was higher than the WT. Therefore, we predicted that IbBBX28 negatively regulates drought tolerance in Arabidopsis thaliana.
To further explore the mechanism of IbBBX28 in regulating the stress response in sweet potato, the interacting proteins of IbBBX28 were screened. We found that IbBBX28 interacted with IbHOX11 and IbZMAT2 (Figure 7 and Figure 8). The HOX gene family is involved in several biological processes in plants, such as embryo morphology, meristem development of roots, shoots, and flowers, vascular development, and multiple stress responses, and they are key regulators of plant morphogenesis (Kikuchi et al., 2015). Lui et al. performed transcriptome sequencing of L. multiflorum under drought; they found that HOX22 and HOX24 presented high expression levels in leaf samples under drought stress, and they were identified as the enrichment of DEGs in the Intrinsic/Integral component of the Golgi membrane by GO analysis (Liu Q. et al., 2022). These results indicated that IbBBX28 may be involved in drought tolerance by interacting with IbHOX11. ZMAT2 is a matrix protein with zinc finger structure, which plays an important role in keratinocyte differentiation and RNA splicing. Studies on ZMAT mainly focus on human diseases, while there are few related studies in plants (Baral and Rotwein, 2020; Suzuki et al., 2020). The function of IbZMAT2 in the drought response of sweet potato is needs to be clarified in future studies.
The model plant Arabidopsis was often used to study gene function for its short growth cycle and mature transformation system. However, the difference in the genetic background may bring different results. The results of the heterologous gene expression in Arabidopsis may be different from those of the transformation in the background plant, which may interfere with the results. For example, the overexpression of CmBBX22 delays leaf senescence and improves drought tolerance in Arabidopsis (Liu et al., 2019). However, CmBBX22 showed reduced drought resistance in transgenic chrysanthemum, which was contrary to the results in Arabidopsis, indicating that CmBBX22 responded to stress differently in Arabidopsis and Chrysanthemum (Liu Y. et al., 2022). Therefore, based on the results in Arabidopsis, we will conduct subsequent experiments in sweet potato to further study the mechanism of IbBBX28 in response to drought stress.
In this study, the expression of the IbBBX28 was induced under drought stress in sweet potato. And IbBBX28 negatively regulated the drought tolerance in Arabidopsis. Yeast two-hybrid and BiFC verified that IbBBX28 interacted with IbHOX11 and IbZMAT2. IbBBX28 may be involved in regulating the stress response in sweet potato by interacting with IbHOX11 and IbZMAT2.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Conceptualization, JD and JZ; methodology, JD; software, CZ and YR; validation, LH and RT; resources, WW; writing—original draft preparation, JD; supervision, XJ; funding acquisition, XJ. All authors have read and agreed to the published version of the manuscript.
This work was supported by the National Key Research and Development Program of China (2018YFD1000700, 2018YFD1000705), the Graduate Research and Innovation Projects of Shanxi Province (2020BY057), the Applied Basic Research Program of Shanxi Academy of Agricultural Sci-ences (YGC2019FZ4), the High-level Foreign Experts Introduction Project (G2022004007L), the Central Guidance for Local Science and Technology Development Project, the Lvliang Key Research and Development Program of High-level Technological Talent (2021RC-2-21), the National Natural Science Foundation for Young Scientists of China (31900450) and Science and Technology Innovation Fund of Shanxi Agricultural University (2018YJ28).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1077958/full#supplementary-material
Supplementary Figure S1 | Phylogenetic analysis of IbBBX28 in different species.
Abegunde, O. K., Mu, T. H., Chen, J. W., and Deng, F. M. (2013). Physicochemical characterization of sweet potato starches popularly used in Chinese starch industry. Food Hydrocoll. 33 (2), 169–177. doi:10.1016/j.foodhyd.2013.03.005
Bailey-Serres, J., Parker, J. E., Ainsworth, E. A., Oldroyd, G. E. D., and Schroeder, J. I. (2019). Genetic strategies for improving crop yields. Nature 575 (7781), 109–118. doi:10.1038/s41586-019-1679-0
Bao, A. K., Wang, S. M., Wu, G. Q., Xi, J. J., Zhang, J. L., and Wang, C. M. (2009). Overexpression of the Arabidopsis H+-PPase enhanced resistance to salt and drought stress in transgenic alfalfa (Medicago sativa L.). Plant Sci. 176 (2), 232–240. doi:10.1016/j.plantsci.2008.10.009
Baral, K., and Rotwein, P. (2020). ZMAT2 in humans and other primates: A highly conserved and understudied gene. Evol. Bioinform. Online 16, 1176934320941500–1176934320941516. doi:10.1177/1176934320941500
Bovell-Benjamin, A. C. (2007). Sweet potato: A review of its past, present, and future role in human nutrition. Adv. Food Nutr. Res. 52, 1–59. doi:10.1016/S1043-4526(06)52001-7
Chu, Z., Wang, X., Li, Y., Yu, H., Li, J., Lu, Y., et al. (2016). Genomic organization, phylogenetic and expression analysis of the B-BOX gene family in tomato. Front. Plant Sci. 7, 1552–1566. doi:10.3389/fpls.2016.01552
Crocco, C. D., and Botto, J. F. (2013). BBX proteins in green plants: Insights into their evolution, structure, feature and functional diversification. Gene 531 (1), 44–52. doi:10.1016/j.gene.2013.08.037
Gangappa, S. N., and Botto, J. F. (2014). The BBX family of plant transcription factors. Trends Plant Sci. 19 (7), 460–470. doi:10.1016/j.tplants.2014.01.010
Gao, S., Yuan, L., Zhai, H., Liu, C., He, S., and Liu, Q. (2011). Transgenic sweetpotato plants expressing an LOS5 gene are tolerant to salt stress. Plant Cell Tissue Organ Cult. 107 (2), 205–213. doi:10.1007/s11240-011-9971-1
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. biochem. 48 (12), 909–930. doi:10.1016/j.plaphy.2010.08.016
Ikegami, K., Okamoto, M., Seo, M., and Koshiba, T. (2009). Activation of abscisic acid biosynthesis in the leaves of Arabidopsis thaliana in response to water deficit. J. Plant Res. 122 (2), 235–243. doi:10.1007/s10265-008-0201-9
Kang, C., He, S., Zhai, H., Li, R., Zhao, N., and Liu, Q. (2018). A sweetpotato auxin response factor gene (IbARF5) is involved in carotenoid biosynthesis and salt and drought tolerance in transgenic Arabidopsis. Front. Plant Sci. 9, 1307–1319. doi:10.3389/fpls.2018.01307
Kang, C., Zhai, H., He, S., Zhao, N., and Liu, Q. (2019). A novel sweetpotato bZIP transcription factor gene, IbbZIP1, is involved in salt and drought tolerance in transgenic Arabidopsis. Plant Cell Rep. 38 (11), 1373–1382. doi:10.1007/s00299-019-02441-x
Khanna, R., Kronmiller, B., Maszle, D. R., Coupland, G., Holm, M., Mizuno, T., et al. (2009). The Arabidopsis B-box zinc finger family. Plant Cell 21 (11), 3416–3420. doi:10.1105/tpc.109.069088
Kikuchi, A., Huynh, H. D., Endo, T., and Watanabe, K. (2015). Review of recent transgenic studies on abiotic stress tolerance and future molecular breeding in potato. Breed. Sci. 65 (1), 85–102. doi:10.1270/jsbbs.65.85
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi:10.1093/molbev/msw054
Kumar, V., Shriram, V., Kavi Kishor, P. B., Jawali, N., and Shitole, M. G. (2010). Enhanced proline accumulation and salt stress tolerance of transgenic indica rice by over-expressing P5CSF129A gene. Plant Biotechnol. Rep. 4 (1), 37–48. doi:10.1007/s11816-009-0118-3
Kwak, S. S. (2019). Biotechnology of the sweetpotato: Ensuring global food and nutrition security in the face of climate change. Plant Cell Rep. 38 (11), 1361–1363. doi:10.1007/s00299-019-02468-0
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi:10.1093/nar/30.1.325
Lin, F., Jiang, Y., Li, J., Yan, T., Fan, L., Liang, J., et al. (2018). B-BOX DOMAIN PROTEIN28 negatively regulates photomorphogenesis by repressing the activity of transcription factor HY5 and undergoes COP1-mediated degradation. Plant Cell 30 (9), 2006–2019. doi:10.1105/tpc.18.00226
Liu, D., He, S., Zhai, H., Wang, L., Zhao, Y., Wang, B., et al. (2014). Overexpression of IbP5CR enhances salt tolerance in transgenic sweetpotato. Plant Cell Tissue Organ Cult. 117 (1), 1–16. doi:10.1007/s11240-013-0415-y
Liu, Q. (2017). Improvement for agronomically important traits by gene engineering in sweetpotato. Breed. Sci. 67 (1), 15–26. doi:10.1270/jsbbs.16126
Liu, Q., Wang, F., Shuai, Y., Huang, L., and Zhang, X. (2022a). Integrated analysis of single-molecule real-time sequencing and next-generation sequencing eveals insights into drought tolerance mechanism of Lolium multiflorum. Int. J. Mol. Sci. 23 (14), 7921–7940. doi:10.3390/ijms23147921
Liu, Y., Chen, H., Ping, Q., Zhang, Z., Guan, Z., Fang, W., et al. (2019). The heterologous expression of CmBBX22 delays leaf senescence and improves drought tolerance in Arabidopsis. Plant Cell Rep. 38 (1), 15–24. doi:10.1007/s00299-018-2345-y
Liu, Y., Cheng, H., Cheng, P., Wang, C., Li, J., Liu, Y., et al. (2022b). The BBX gene CmBBX22 negatively regulates drought stress tolerance in chrysanthemum. Hortic. Res. 9, uhac181. doi:10.1093/hr/uhac181
Liu, Y., Lin, G., Yin, C., and Fang, Y. (2020). B-box transcription factor 28 regulates flowering by interacting with constans. Sci. Rep. 10 (1), 17789–17801. doi:10.1038/s41598-020-74445-7
Lyu, R., Ahmed, S., Fan, W., Yang, J., Wu, X., Zhou, W., et al. (2021). Engineering properties of sweet potato starch for industrial applications by biotechnological techniques including genome editing. Int. J. Mol. Sci. 22 (17), 9533–9552. doi:10.3390/ijms22179533
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi:10.1146/annurev.arplant.59.032607.092911
Nagaoka, S., and Takano, T. (2003). Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J. Exp. Bot. 54 (391), 2231–2237. doi:10.1093/jxb/erg241
Nguyen, H. C., Chen, C. C., Lin, K. H., Chao, P. Y., Lin, H. H., and Huang, M. Y. (2021). Bioactive compounds, antioxidants, and health benefits of sweet potato leaves. Molecules 26 (7), 1820–1832. doi:10.3390/molecules26071820
O'Brien, P. J. (1972). The sweet potato: Its origin and dispersal. Am. Anthropol. 74 (3), 342–365. doi:10.1525/aa.1972.74.3.02a00070
Safeena, M. I. S., Dissanayake, Y., Zakeel, M. C. M., Warnakula, L., Cooray, R., and Dayarathna, D. (2021). An improved method for efficient recovery of high quality DNA from date palm (Phoenix dactylifera L; Arecaceae. MethodsX 8, 101384. doi:10.1016/j.mex.2021.101384
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58 (2), 221–227. doi:10.1093/jxb/erl164
Song, Z., Bian, Y., Liu, J., Sun, Y., and Xu, D. (2020). B-box proteins: Pivotal players in light-mediated development in plants. J. Integr. Plant Biol. 62 (9), 1293–1309. doi:10.1111/jipb.12935
Suzuki, T., Nakano, M., Komatsu, M., Takahashi, J., Kato, H. i., and Nakamura, Y. (2020). ZMAT2, a newly-identified potential disease-causing gene in congenital radioulnar synostosis, modulates BMP signaling. Bone 136, 115349. doi:10.1016/j.bone.2020.115349
Vaishak, K. P., Yadukrishnan, P., Bakshi, S., Kushwaha, A. K., Ramachandran, H., Job, N., et al. (2019). The B-box bridge between light and hormones in plants. J. Photochem. Photobiol. B 191, 164–174. doi:10.1016/j.jphotobiol.2018.12.021
Wang, M. J., Ding, L., Liu, X. H., and Liu, J. X. (2021). Two B-box domain proteins, BBX28 and BBX29, regulate flowering time at low ambient temperature in Arabidopsis. Plant Mol. Biol. 106 (1-2), 21–32. doi:10.1007/s11103-021-01123-1
Wang, Q., Tu, X., Zhang, J., Chen, X., and Rao, L. (2013). Heat stress-induced BBX18 negatively regulates the thermotolerance in Arabidopsis. Mol. Biol. Rep. 40 (3), 2679–2688. doi:10.1007/s11033-012-2354-9
Xu, X., Wang, Q., Li, W., Hu, T., Wang, Q., Yin, Y., et al. (2022). Overexpression of SlBBX17 affects plant growth and enhances heat tolerance in tomato. Int. J. Biol. Macromol. 206, 799–811. doi:10.1016/j.ijbiomac.2022.03.080
Yang, Y., Ma, C., Xu, Y., Wei, Q., Imtiaz, M., Lan, H., et al. (2014). A zinc finger protein regulates flowering time and abiotic stress tolerance in chrysanthemum by modulating gibberellin biosynthesis. Plant Cell 26 (5), 2038–2054. doi:10.1105/tpc.114.124867
Ye, Y., Liu, Y., Li, X., Wang, G. D., Zhou, Q., Chen, Q. J., et al. (2021). An evolutionary analysis of B-Box transcription factors in strawberry reveals the role of FaBBx28c1 in the regulation of flowering time. Int. J. Mol. Sci. 22 (21), 11766–11788. doi:10.3390/ijms222111766
Zhai, H., Wang, F., Si, Z., Huo, J., Xing, L., An, Y., et al. (2016). A myo-inositol-1-phosphate synthase gene, IbMIPS 1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweet potato. Plant Biotechnol. J. 14 (2), 592–602. doi:10.1111/pbi.12402
Zhang, H., Wang, Z., Li, X., Gao, X., Dai, Z., Cui, Y., et al. (2022). The IbBBX24-IbTOE3-IbPRX17 module enhances abiotic stress tolerance by scavenging reactive oxygen species in sweet potato. New Phytol. 233 (3), 1133–1152. doi:10.1111/nph.17860
Keywords: sweet potato, IbBBX28, Arabidopsis, drought tolerance, yeast two-hybrid
Citation: Dong J, Zhao C, Zhang J, Ren Y, He L, Tang R, Wang W and Jia X (2022) The sweet potato B-box transcription factor gene IbBBX28 negatively regulates drought tolerance in transgenic Arabidopsis. Front. Genet. 13:1077958. doi: 10.3389/fgene.2022.1077958
Received: 23 October 2022; Accepted: 14 November 2022;
Published: 29 November 2022.
Edited by:
Ling Xu, Zhejiang Sci-Tech University, ChinaReviewed by:
Xinmin An, Beijing Forestry University, ChinaCopyright © 2022 Dong, Zhao, Zhang, Ren, He, Tang, Wang and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyun Jia, amlheGlhb3l1bkBzeGF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.