
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 13 January 2023
Sec. Human and Medical Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1073851
 Yongyi Zou1,2†
Yongyi Zou1,2† Chuanxin Feng1,2†
Chuanxin Feng1,2† Jiawei Qin1,2
Jiawei Qin1,2 Xinrong Wang2
Xinrong Wang2 Tingting Huang1,2
Tingting Huang1,2 Yan Yang1,2
Yan Yang1,2 Kang Xie1,2
Kang Xie1,2 Huizhen Yuan1,2
Huizhen Yuan1,2 Shuhui Huang1,2
Shuhui Huang1,2 Bicheng Yang1,2
Bicheng Yang1,2 Wan Lu1,2*
Wan Lu1,2* Yanqiu Liu1,2*
Yanqiu Liu1,2*To evaluate the performance of expanded non-invasive prenatal testing (expanded noninvasive prenatal testing, NIPT-Plus) in screening for fetal chromosomal abnormalities includes aneuploidies and copy number variations, a total of 23,116 pregnant women with a singleton pregnancy were recruited for NIPT-Plus. Screening positive results were verified by karyotype analysis and chromosomal microarray analysis after amniocentesis. A total of 264 pregnancies (1.14%) were positive results as predicted by NIPT-Plus, including 233 aneuploidies and 31 copy number variations. Following genetic counseling, 233 (88.26%) pregnant women underwent invasive prenatal diagnosis and 136 were verified as true positives, comprising 72 common trisomies (T21, T18, T13), 47 sex chromosomal abnormalities two rare autosomal aneuploidies (RATs) and 15 copy number variations The positive predictive value for common trisomies, SCAs, RATs and CNVs were 68.57%, 68.12%, 6.67% and 51.72%, respectively. Pregnant women with screen-positive results for common trisomies have higher rates of invasive prenatal diagnosis and pregnancy termination than those with positive results for SCAs, RATs, and CNVs. NIPT-Plus showed a good performance in detecting common trisomies, SCAs and also contributed to detecting pathogenic CNVs, but higher accuracy was required in the detection of RATs. In summary, this study provides a reference for the clinical application of NIPT-Plus for screening fetal chromosomal abnormalities in this region. Therefore, we suggest that NIPT-Plus could be widely used in clinical screening for fetal chromosomal abnormalities in combination with prenatal diagnosis and genetic counseling.
Chromosomal abnormalities, which include aneuploidies and copy number variations, can result in a spectrum of diseases ranging from pregnancy loss to adults with psychiatric disorder (Yilmaz et al., 2017; Dahdouh and Kutteh, 2021). Most aneuploid embryos undergo embryonic death and spontaneous abortion in early development due to severe developmental defects. However, some trisomic aneuploid embryos can continue to develop and cause birth defects, such as trisomy 21, trisomy 18, trisomy 13 and SCAs(Hassold et al., 2007). In newborns, the prevalence of aneuploidy is approximately 0.3%, with the most common abnormalities being trisomy 21 and sex chromosome trisomy (Mikwar et al., 2020). Copy number variants are always associated with human genetic diseases and syndromes, and these changes from the genome can lead to gene dosage effects, gene breaking that affect gene expression levels and phenotypes (Grati et al., 2015). Microdeletion and microduplication syndromes (MMS) caused by pathogenic CNVs can occur in any pregnancy independent of maternal age (Chau et al., 2019). Clinically relevant CNVs have been identified in 6.0% of fetuses with structural anomalies on ultrasonography, and in 1.7% of pregnancies with standard indications for prenatal diagnosis (such as advanced maternal age and positive screening results) in previous studies (Wapner et al., 2012). Overall, chromosomal abnormalities occur in approximately one in 150 live births and are the main cause of congenital malformations and even death in infants and childhood (Carlson and Vora, 2017). Karyotyping and/or chromosomal microarray analysis after amniocentesis or chorionic villous sampling (CVS) can effectively detect fetal chromosomal abnormalities. However, the limitations of these techniques are their invasiveness and procedure-related miscarriage with a low but non-negligible risk (Alfirevic et al., 2017).
The discovery of cell-free fetal DNA (cffDNA) in maternal plasma and the rapid development of high-throughput sequencing technologies have enabled the non-invasive detection of fetal chromosomal abnormalities (Chitty and Lo, 2015), known as non-invasive prenatal testing (NIPT). NIPT has been widely used as a routine screening method for fetal chromosomal aneuploidy, and it shows high accuracy in detecting common trisomies. It was reported that the detection rate, specificity, and false positive rate of NIPT for common trisomies were 98.59%, 99.99%, and 0.02%, respectively (Yu et al., 2017). Numerous studies have shown that the PPV is in the range of 65%–94% for T21, 47%–85% for T18, 12%–62% for T13, and 45%–58% for SCAs(Quezada et al., 2015; Zhang et al., 2017; Zheng et al., 2020). The American College of Medical Genetics and Genomics (ACMG) recommended that NIPT could replace conventional screening for common trisomies in 2016 (Gregg et al., 2016).
In recent years, NIPT has expanded to detect rare autosomal aneuploidies (RATs) and copy number variations (CNVs) through deeper sequencing and higher level analyses. Several recent studies have demonstrated the possibility of expanded NIPT screening for fetal CNVs associated with MMS(Liang et al., 2019; Shi et al., 2021), including but not limited DiGeorge syndrome, Prader–Willi/Angleman syndrome, cri du chat syndrome, and 1p36 deletion syndrome. The performance of the expanded NIPT to detect MMS varied widely, with positive predictive values ranging from low (11%–18%) to moderate (29%–77%) (Shi et al., 2021). As a result, the accuracy of expanded NIPT in detecting chromosomal abnormalities other than common trisomies remains questionable (Christiaens et al., 2021). In addition, most studies of expanded NIPT originate from multiple centers in different regions. There are differences in the performance of expanded NIPT screening for chromosomal abnormalities reported by different centers. Therefore, large-scale prospective studies from a single center are necessary to guide the application of extended NIPT in local clinical practice.
In this study, we recruited a total of 23,116 singleton pregnancies who underwent expanded NIPT at a single local center to evaluate the performance of expanded NIPT in screening for aneuploidies and pathogenic CNVs. Our data have reference significance for the local clinical application of extended NIPT for screening fetal chromosomal abnormalities.
This study recruited 23,116 singleton pregnant women consecutively from October 2019 to December 2021 at the Medical Genetics Center of Jiangxi Maternal and Child Health Hospital. Base on different clinical indications, we present four common risk factors as follows: ultrasound soft index abnormalities (echocardiographic lesions, choroid plexus cysts, single umbilical artery, etc.), positive serum screening (high or intermediate risks for serum screening), advanced maternal age (age ≥35 years), adverse reproductive history (previous adverse pregnancy outcomes). Pregnant women with one of the above risk factors are classified as a high-risk group, while those with none of the above risk factors are defined as a low-risk group. Written informed consent and pretest genetic counseling were provided to all pregnant women, including test objectives, significance, and limitations.
Approximately 5 ml peripheral blood was collected in a cfDNA storage tube (Streck Cell Free DNA BCT, United States) from pregnant woman, and plasma DNA was extracted within 96 h of collection. Plasma was separated from blood samples using a two-step centrifugation protocol (Dan et al., 2012). The collected blood samples were first centrifuged at 1,600 g and 4°C for 10 min. The supernatant was then centrifuged at 16,000 g and 4°C for 10 min to remove residual blood cells. Cell-free DNA was extracted from plasma using a cfDNA isolation kit (BGI Biotechnology Co., Ltd., Wuhan) according to the reagent instructions. The extracted cfDNA was end repaired in a 10 ul end repair reaction mixture at 37°C for 10 min and 65°C for 15 min. Next, 30ul of the adapter ligation reaction mixture was added and incubated at 23°C for 20 min. Then 29ul PCR reaction mixture was added and PCR amplification was performed at 98°C 2 min, 12cycles (98°C 15 s, 56°C 15 s, 72°C 30 s) and 72°C 5 min to obtain cfDNA library. After quantifying with a QubitTM fluorometer (Thermo Fisher Scientific, United States), each cfDNA library was pooled at about 168ng. Libraries were tag sequenced with 45-cycle single-end sequencing on BGISEQ-500 platforms (BGI, China) to generate approximately 20 M Reads. Sequencing data were processed by a Non-invasive Fetal Trisomy (NIFTY) system as previously described (Jiang et al., 2012). Chromosome aneuploidies risk was evaluated using a ratio Z-score (|Z| ≥ 3 indicates a high risk of chromosomal aneuploidies, and |Z| < 3 indicates a low risk). Furthermore, algorithm based on hidden Markov models (HMM) was used to detect CNVs (Liang et al., 2019).
Genetic counseling and prenatal diagnostic testing would be offered to women receiving a high-risk screening result. T21, T18, and T13 trisomies and sex chromosome aneuploidies were confirmed using invasive amniocentesis followed by karyotyping, while CNVs high-risk results were confirmed by chromosomal microarray analysis (CMA). We then collected their invasive prenatal diagnostic results and pregnancy outcomes. Pregnant women with low-risk screening results are recommended for routine prenatal care. The postnatal follow-up was conducted by phone interview at 3 months after the expected date of delivery based on the guidelines of the National Health Commission of the People’s Republic of China (2016).
Based on the results of expanded NIPT and invasive prenatal diagnosis, the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated. Sensitivity = TP/(TP + FN), specificity = TN/(FP + TN), FNR = FN/all cases, FPR = FP/all cases, PPV = TP/(TP + FP), and negative pre-dictive value (NPV) = TN/(FN + TN). TP, FN, TN, and FP stand for true positive, false negative, true negative, and false positive, respectively. Cases without a confirmatory diagnosis or lost to follow-up were excluded from the study. All data analyses were performed using Statistical Product and Service Solutions (SPSS) software version 26.0. The Chi-square test were used to estimate the statistical significance between two categorical variables. p < 0.05 was considered statistically significant.
A total of 23,145 pregnant women with singleton pregnancy were offered NIPT-Plus at a local center during the study period. 144 cases required repeat blood sampling due to poor quality of blood or low fetal DNA fraction (<3.5%), and valid results were obtained in 115 cases. The remaining 29 cases (0.13%) still had a low fetal fraction (<3.5%) and were excluded from this study (Figure 1). We followed up these 29 pregnant women, of whom 21 gave birth to normal children and eight were lost to follow-up.
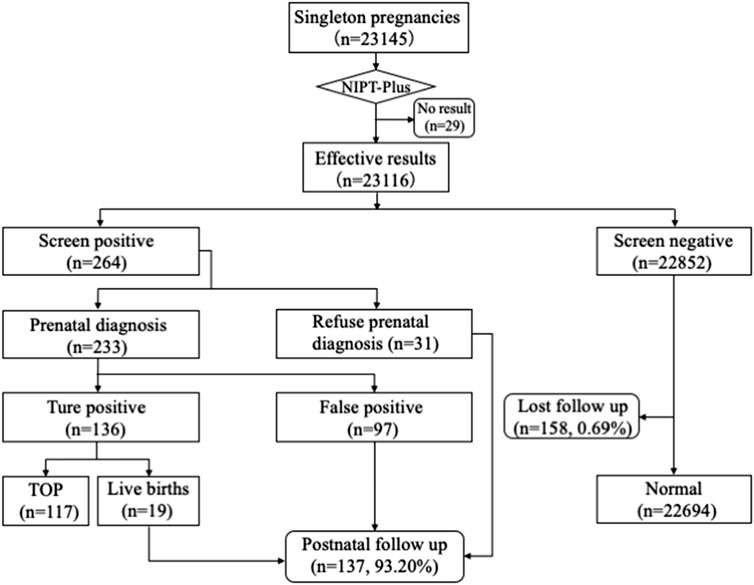
FIGURE 1. Flowchart of NIPT-Plus results and outcomes of pregnant women. TOP, termination of pregnancy.
Ultimately, a total of 23,116 samples were included in this study. The maternal age ranged from 14 to 54 years with a mean age of 29.6 years (SD, 4.9), and the gestational age ranged from 12 to 33 weeks with a mean gestational age of 15.5 weeks (SD, 2.7). The majority (83.67%) of the women were at 12–17+6 weeks when NIPT-Plus was performed. Of these 23,116 pregnant women, 8,395 pregnancies demonstrated single or multiple indications, including advanced maternal age, positive serum screening, ultrasound soft index abnormalities and adverse reproductive history (Table 1).
Among 23,116 pregnant women screened by NIPT-Plus, there were 264 (1.14%) cases reported to be positive (high-risk). There were 109 positive cases for common trisomies (60 cases of T21, 36 cases of T18 and 13 cases of T13) and 84 positive cases for SCAs (32, 12, 28 and 12 cases of 45,X, 47,XXX, 47, XXY and 47,XYY, respectively). For common trisomies, 105 cases received invasive prenatal diagnosis, and 50 T21, 19 T18 and three T13 were confirmed. Therefore, the positive predictive value (PPV) for screening T21, T18 and T13 were 86.21%, 55.88% and 23.08%, respectively. All low-risk cases of NIPT-Plus were verified to be true negative, except 158 cases, which had been lost to follow-up. Thus, the compound PPV, NPV, sensitivity and specificity for common trisomies were 68.57%, 100%, 100% and 99.86%, respectively. Meanwhile, 69 cases of SCAs received prenatal diagnosis, and 47 true positive and 22 false positive SCAs cases were confirmed, resulting in a compound PPV of 68.12%. Among four categories of SCAs, the PPV of 47, XXX (88.89%), 47, XXY (99.15%) and 47, XYY (72.73%) were higher than that of 45, X (26.09%) (Table 2). Overall, NIPT-Plus demonstrated high sensitivity and specificity for the detection of common trisomies and SCAs.
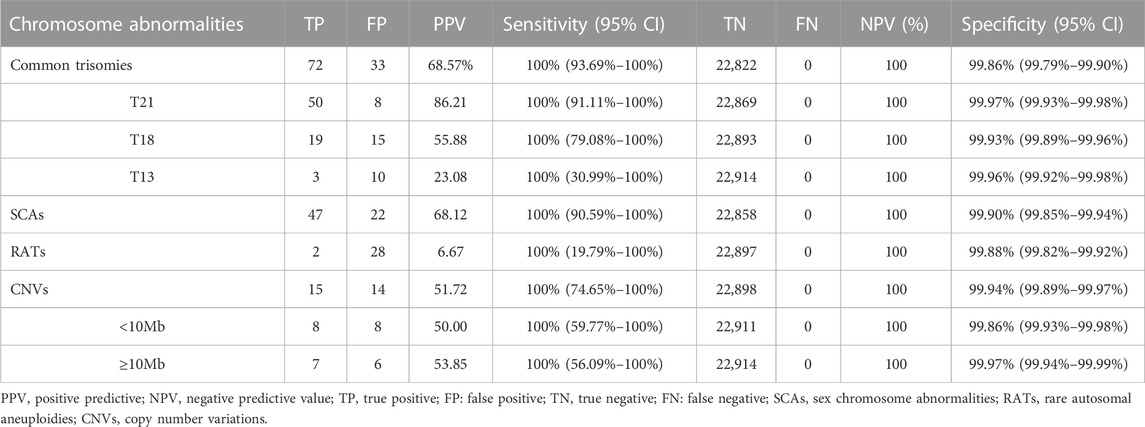
TABLE 2. The performance of NIPT-Plus in screening for chromosome aneuploidy and copy number variations.
Among the 23,116 cases, 40 positive cases for RATs were reported. Confirmatory testing was performed in 30 of the 40 RATs cases. Two cases confirmed to be mosaic trisomy (mosaic T9 and mosaic T3), and the other 28 RATs cases had normal karyotype. Therefore, the combined PPV for RATs was as low as 6.67%. Thirty-one cases with positive CNVs results were reported, of which 29 cases underwent amniocentesis and CMA analysis. Fifteen cases had consistent results with NIPT-Plus, while the other 14 were discordant. Thus, the total PPV for CNVs was 51.72%. CNVs were categorized into two groups according to length (CNV<10Mb, and CNVs ≥10 Mb), and the PPV was calculated separately. There was no significant difference in PPV between CNV< 10Mb and CNV ≥10Mb positive cases (50.00% vs 53.85%, p = 0.837). The 15 true positive CNVs detected by NIPT-Plus were summarized in Table 3. 22q11 deletion syndrome (DiGeorge syndrome) was the most common (n = 3), followed by cri du chat syndrome (n = 2).
Based on different clinical indications, 8,395 (36.3%) pregnant women were classified as high-risk groups and 14,721 (63.7%) as low-risk groups. The positive rate and PPV of T21, T18, T13, SCAs, RATs and CNVs in the two groups are shown in Table 4. For T21, whose higher disease prevalence is known to associated with advanced maternal age, both positive rate and PPV were significantly higher in the high-risk groups than in the low-risk groups (PR 0.43% vs 0.16%, PPV 94.29% vs 73.91%, p < 0.05). However, there was no significant difference between the two groups in PPV for T18, T13, SCAs, RATs, and CNVs (T18 58.33% vs. 54.55%, T13 25.00% vs. 22.22%, SCAs 70.37% vs 66.67%, RTAs 16.67% vs. 4.17%, CNVs 50.00% vs 52.63%, p > 0.05). At the same time, the positive rates of these chromosomal abnormalities also showed similar characteristics between the two groups.
All the 264 cases of NIPT-Plus positive results were followed up to pregnancy outcomes (Table 5). A total of 72 positive results were confirmed as common trisomies (50 T21, 19 T18, and 3 T13), all of whom opted for termination of pregnancy (TOP). Thirty-one women carrying fetuses with SCAs terminated their pregnancy, with higher rates of termination in cases of 45,X (66.7%) and 47,XXY (88%) than in cases of 47, XXX (37.5%) and 47, XYY (25%). In addition, the termination rates for cases confirmed as RATs and CNVs were 50% (1/2) and 86.7% (13/15), respectively. For 31 cases with a NIPT-Plus positive result who refused prenatal diagnosis, 10 (32.3%) opted for termination and 21 (67.7%) continued the pregnancy. A total of 100% (4/4) of unconfirmed common trisomy positive cases terminated their pregnancy, which was higher than unverified positive cases for SCAs (20%, 3/15), RATs (20%, 2/10) and CNVs (50%, 1/2).
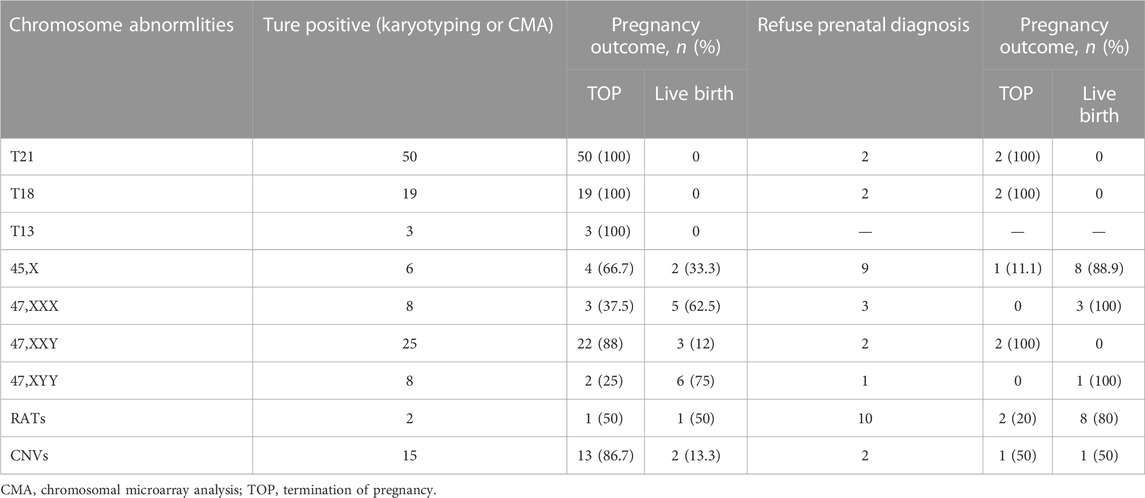
TABLE 5. Pregnancy outcome of confirmatory abnormal fetuses and positive screening cases without prenatal diagnosis.
NIPT for prenatal screening of T21, T18, and T13 has been widely accepted by clinicians and patients in recent years. However, whether this approach can be extended to detect other chromosomal abnormalities remains controversial (Evans et al., 2016; Chitty et al., 2018). Several recent studies have demonstrated the application of expanded NIPT (NIPT-Plus) in screening for multiple chromosomal abnormalities, including SCAs, RATs, and CNVs. However, the PPV varies widely across studies, ranging from 33% to 59% for SCAs, 5%–29% for RATs, and 29%–61% for CNVs(Fiorentino et al., 2017; Chen et al., 2019; 2021; Ge et al., 2021). In present study, a cohort of 23,116 singleton pregnancies from a single center were enrolled to investigate the performance of NIPT-Plus. Our study showed that NIPT-Plus had a comparable PPV for T21 (86.21%) and T18 (55.88%) but a relatively low PPV for T13 (23.08%) compared to previous studies (Chen et al., 2022). The low PPV for T13 may be related to the small number of T13 positive cases in this study, which would increase the fluctuation of PPV. Another possible reason could be related to the size of chromosome 13 or the GC ratio on chromosome 13 (Hu et al., 2019). Furthermore, our data suggested that the sensitivity and specificity of NIPT-Plus for screening common trisomies were comparable to standard NIPT (Lu, 2020). In our study, the combined PPV for SCAs was 61.82%, which was similar to that reported by expanded NIPT (Ge et al., 2021). Overall, the PPV for XXX, XXY, and XYY were 88.89%, 96.15%, and 72.73%, respectively, which was obviously higher than the 26.09% of monosomy X. The poor accuracy of screening for monosomy X may be due to confined placental mosaicism, a vanishing twin affected by monosomy X or maternal monosomy X mosaicism (Zhao et al., 2022).
In our study, we also explored the applicability of NIPT-Plus in screening for other chromosomal abnormalities, such as fetal RATs and CNVs. We screened 40 positive cases for RATs with a low PPV of 6.67%, which was similar to that reported in a recent study (Ge et al., 2021). Despite the lack of investigation into the cause of false positive RATs, fetoplacental mosaicism has been identified as a potential explanation in previous case studies (Grati et al., 2014). Since most of RATs belong to non-viable trisomy and spontaneous abortion occurs in the first trimester, it is recommended to conduct genetic counseling and prenatal diagnosis (FISH or CMA) based on the results of NIPT-Plus and ultrasonography to confirm the presence of chromosomal trisomy mosaicism in the fetus. We also evaluated the efficiency of NIPT-Plus to screen for CNVs, with a total of 31 positive cases detected and 15 confirmed as true-positive cases, resulting in a PPV of 51.72%. The most common of the 15 true positive cases identified by CMA was DiGeorge syndrome, followed by cri du chat syndrome. Meanwhile, the PPV for DiGeorge syndrome and cri du chat syndrome was also relatively high in this study, which was consistent with Liang’s study (Liang et al., 2019). In addition, there was no significant difference in PPV for CNVs <10 Mb and CNVs ≥10 Mb (50.00% vs 53.85%, p = 0.837) in present study. This indicated the effectiveness of NIPT-Plus and validated algorithms for identifying CNVs, even for small CNVs.
The American College of Obstetricians and Gynecologists (ACOG) recommends screening for aneuploidy in all pregnancies (American College of Obstetricians and Gynecologists’ Committee on Practice Bulletins—Obstetrics, 2020). Therefore, it is necessary to evaluate the performance of NIPT plus for screening chromosomal abnormalities including aneuploidy and CNV in pregnancies with different risk factors. According to our results, 8,395 (36.32%) cases with high-risk factors chose NIPT-Plus screening, from which, a total of 109 positive cases were screened. Meanwhile, 155 positive cases were screened in 14,721 (63.7%) pregnant women without any risk factors. We further analyzed the performance of NIPT-Plus in the high-risk and low-risk groups. The results showed that the positive rate and PPV for T21 in the high-risk group were significantly higher than those in the low-risk group. However, the positive rate and PPV for other chromosomal abnormalities were not significantly different. Thus, it is suggested that NIPT-Plus could be widely used in all pregnant women as a routine prenatal screening.
Among the positive cases screened, 31 cases (11.74%) refused prenatal diagnosis, of which 10 cases chose to terminate pregnancy and 21 cases continued pregnancy. Compared with RATs and CNVs, pregnant women with positive results of common trisomies were more likely to choose for prenatal diagnosis. Furthermore, because of the severity of the three diseases, all cases diagnosed as common trisomy choose to terminate pregnancy, which was similar to Zhou’s study (Zhou et al., 2019). Pregnancy termination was chosen in 66% (31/47) of confirmed SCA cases, which was comparable to other studies in the same period (61.1%–81%) (Scibetta et al., 2017; Lai et al., 2021). The pregnancy termination rate was 37.5% (3/8) for 47, XXX syndrome cases and 25% (2/8) for 47, XYY syndrome cases, which was significantly lower than other sex chromosome syndromes. This suggests that there is a growing acceptance of children with 47, XXX and 47, XYY syndrome, whose IQs are mostly in the normal range despite occasional physical abnormalities observed (Berglund et al., 2019). However, 80% (12/15) of the positive cases of SCAs without prenatal diagnosis opted for continuing pregnancies, which was significantly higher than that with confirmation of SCAs. Prenatal genetic counseling and the cognition level of pregnant women may have contributed to these differences. We also noted that pregnant women with confirmed larger CNVs or CNVs associated with MMS chose to terminate their pregnancy. There was one case chose to continue the pregnancy after it was confirmed to be inherited from the parents, and succeeded in live birth. This suggests that the origin of CNVs must be identified if necessary, which is important for genetic counseling. In addition, we also followed up 21 cases of continued pregnancy 3 months after delivery. Among them, one case was preterm at 31 weeks and had difficulty feeding at birth but no other structural abnormalities were found. No obvious abnormality was found in the other 20 patients during follow-up. We will continue to follow up for 1 and 3 years after birth to track the development.
There were several limitations in this study that should be noted. Screening negative cases lost to follow-up and positive cases without prenatal diagnosis were excluded when assessing NIPT-Plus performance, which may have affected the accuracy of our results. In accordance with the National Health Commission guidelines of China, follow-up began 12 weeks after delivery. Although there were no further false-negative case reports, it is too early to judge the chromosomal abnormalities merely through our follow-up at 3 months after delivery, especially for SCAs and CNVs. Unlike common trisomies (T21/T18/T13) that show distinct physical signs of chromosomal disease at birth, symptoms of SCAs and pathogenic CNVs may not appear until childhood (Battaglia et al., 2013; Skuse et al., 2018). Thus, another limitation to this study is that longer follow-up will be necessary to accurately assess the performance of NIPS-Plus for detecting SCAs and pathogenic CNVs.
By increasing sequencing depth and optimizing algorithms, NIPT-Plus can not only be used in screening for common trisomies, but also for other chromosomal abnormalities, such as SCAs, RATs, and CNVs. Our data demonstrated that NIPS-Plus shows high performance in detecting common trisomes and SCAs and also contributed to detecting pathogenic CNVs. Moreover, NIPT-Plus showed comparable performance in low-risk populations as in high-risk populations. This study is the first large-scale NIPT-Plus analysis of pregnant women in Jiangxi, China, and provided sufficient data support for the clinical application of NIPT-Plus in this region. Although similar studies have been done in other regions of China, there are differences in the screening performance of NIPT-plus in different regions. The study reported in Fujian showed that the PPV of the T18 screening was as high as 78%, while the study in Guangxi was similar to ours with the PPV of only 52%. In addition, the PPV of SCAs screening in our study was 68%, which was higher than Fujian’s 59% and Guangxi’s 40% (Chen et al., 2021; Ge et al., 2021). These data have significant implications for genetic counseling by local physicians, reducing anxiety and potentially unnecessary pregnancy termination in some high-risk pregnant women. With the advancement of methodology and the accumulation of clinical experience, NIPT-Plus combined with conventional ultrasonography will be the most comprehensive and reliable method for detecting fetal chromosomal abnormalities.
The original contributions presented in the study are publicly available. This data can be found in Figshare https://doi.org/10.6084/m9.figshare.21825066.v1.
The studies involving human participants were reviewed and approved by The studies involving human participants were reviewed and approved by EC-KT-202216. The patients/participants provided their written informed consent to participate in this study.
YZ and CF were in charge of the genetic analysis, conceptualized the study, and wrote the manuscript original draft preparation. XW, TH, and YZ were responsible for the clinical evaluation. JQ, YY and KX did the investigation and data analysis. SH, BY, WL, and YL wrote, reviewed, and edited the manuscript. All authors have read and agreed to the published version of the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the Jiangxi Provincial Key Laboratory of Birth Defect for Prevention and Control (No. 20202BCD42017 to YL). Provincial Health Commission Program of Jiangxi (No. 20201095 to YZ). National Natural Science Foundation of China (Grant No. 82160318 to YZ). Key Research and Development Program of Jiangxi Province (Grant No. 20202BBG73015 to YY). Youth Science Foundation of Jiangxi Province (Grant No. 20192BAB215010 to WL).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1073851/full#supplementary-material
Alfirevic, Z., Navaratnam, K., and Mujezinovic, F. (2017). Amniocentesis and chorionic villus sampling for prenatal diagnosis. Cochrane Database Syst. Rev. 9, CD003252. doi:10.1002/14651858.CD003252.pub2
American College of Obstetricians and Gynecologists’ Committee on Practice Bulletins—Obstetrics Kaimal, A. J., Dugoff, L., Norton, M. E., et al. (2020). Committee on genetics, and society for maternal-fetal MedicineScreening for fetal chromosomal abnormalities: ACOG practice bulletin, number 226. Obstet. Gynecol. 136, e48–e69. doi:10.1097/AOG.0000000000004084
Battaglia, A., Doccini, V., Bernardini, L., Novelli, A., Loddo, S., Capalbo, A., et al. (2013). Confirmation of chromosomal microarray as a first-tier clinical diagnostic test for individuals with developmental delay, intellectual disability, autism spectrum disorders and dysmorphic features. Eur. J. Paediatr. Neurology 17, 589–599. doi:10.1016/j.ejpn.2013.04.010
Berglund, A., Viuff, M. H., Skakkebæk, A., Chang, S., Stochholm, K., and Gravholt, C. H. (2019). Changes in the cohort composition of turner syndrome and severe non-diagnosis of klinefelter, 47, XXX and 47, XYY syndrome: A nationwide cohort study. Orphanet J. Rare Dis. 14, 16. doi:10.1186/s13023-018-0976-2
Carlson, L. M., and Vora, N. L. (2017). Prenatal diagnosis: Screening and diagnostic tools. Obstet. Gynecol. Clin. North Am. 44, 245–256. doi:10.1016/j.ogc.2017.02.004
Chau, M. H. K., Cao, Y., Kwok, Y. K. Y., Chan, S., Chan, Y. M., Wang, H., et al. (2019). Characteristics and mode of inheritance of pathogenic copy number variants in prenatal diagnosis. Am. J. Obstetrics Gynecol. 221, e1–e1493. e11. doi:10.1016/j.ajog.2019.06.007
Chen, Y., Lai, Y., Xu, F., Qin, H., Tang, Y., Huang, X., et al. (2021). The application of expanded noninvasive prenatal screening for genome-wide chromosomal abnormalities and genetic counseling. J. Maternal-Fetal Neonatal Med. 34, 2710–2716. doi:10.1080/14767058.2021.1907333
Chen, Y., Lu, L., Zhang, Y., Wang, F., Ni, Y., Wang, Q., et al. (2022). Clinical application of expanded noninvasive prenatal testing for fetal chromosome abnormalities in a cohort of 39, 580 pregnancies. Am. J Med Genet. Pt A 188, 1426–1434. doi:10.1002/ajmg.a.62657
Chen, Y., Yu, Q., Mao, X., Lei, W., He, M., and Lu, W. (2019). Noninvasive prenatal testing for chromosome aneuploidies and subchromosomal microdeletions/microduplications in a cohort of 42, 910 single pregnancies with different clinical features. Hum. Genomics 13, 60. doi:10.1186/s40246-019-0250-2
Chitty, L. S., Hudgins, L., and Norton, M. E. (2018). Current controversies in prenatal diagnosis 2: Cell-free DNA prenatal screening should be used to identify all chromosome abnormalities. Prenat. Diagn. 38, 160–165. doi:10.1002/pd.5216
Chitty, L. S., and Lo, Y. M. D. (2015). Noninvasive prenatal screening for genetic diseases using massively parallel sequencing of maternal plasma DNA. Cold Spring Harb. Perspect. Med. 5, a023085. doi:10.1101/cshperspect.a023085
Christiaens, L., Chitty, L. S., and Langlois, S. (2021). Current controversies in prenatal diagnosis: Expanded NIPT that includes conditions other than trisomies 13, 18, and 21 should be offered. Prenat. Diagn. 41, 1316–1323. doi:10.1002/pd.5943
Dahdouh, E. M., and Kutteh, W. H. (2021). Genetic testing of products of conception in recurrent pregnancy loss evaluation. Reprod. Biomed. Online 43, 120–126. doi:10.1016/j.rbmo.2021.03.015
Dan, S., Wang, W., Ren, J., Li, Y., Hu, H., Xu, Z., et al. (2012). Clinical application of massively parallel sequencing-based prenatal noninvasive fetal trisomy test for trisomies 21 and 18 in 11, 105 pregnancies with mixed risk factors. Prenat. Diagn 32, 1225–1232. doi:10.1002/pd.4002
Evans, M. I., Wapner, R. J., and Berkowitz, R. L. (2016). Noninvasive prenatal screening or advanced diagnostic testing: Caveat emptor. Am. J. Obstetrics Gynecol. 215, 298–305. doi:10.1016/j.ajog.2016.04.029
Fiorentino, F., Bono, S., Pizzuti, F., Duca, S., Polverari, A., Faieta, M., et al. (2017). The clinical utility of genome-wide non invasive prenatal screening. Prenat. Diagn. 10, 593–601. doi:10.1002/pd.5053
Ge, Y., Li, J., Zhuang, J., Zhang, J., Huang, Y., Tan, M., et al. (2021). Expanded noninvasive prenatal testing for fetal aneuploidy and copy number variations and parental willingness for invasive diagnosis in a cohort of 18, 516 cases. BMC Med. Genomics 14, 106. doi:10.1186/s12920-021-00955-6
Grati, F. R., Malvestiti, F., Ferreira, J. C. P. B., Bajaj, K., Gaetani, E., Agrati, C., et al. (2014). Fetoplacental mosaicism: Potential implications for false-positive and false-negative noninvasive prenatal screening results. Genet. Med. 16, 620–624. doi:10.1038/gim.2014.3
Grati, F. R., Molina Gomes, D., Ferreira, J. C. P. B., Dupont, C., Alesi, V., Gouas, L., et al. (2015). Prevalence of recurrent pathogenic microdeletions and microduplications in over 9500 pregnancies: Prevalence of recurrent pathogenic copy number in prenatal population. Prenat. Diagn 35, 801–809. doi:10.1002/pd.4613
Gregg, A. R., Skotko, B. G., Benkendorf, J. L., Monaghan, K. G., Bajaj, K., Best, R. G., et al. (2016). Noninvasive prenatal screening for fetal aneuploidy, 2016 update: A position statement of the American College of medical genetics and Genomics. Genet. Med. 18, 1056–1065. doi:10.1038/gim.2016.97
Hassold, T., Hall, H., and Hunt, P. (2007). The origin of human aneuploidy: Where we have been, where we are going. Hum. Mol. Genet. 16, R203–R208. doi:10.1093/hmg/ddm243
Hu, H., Wang, L., Wu, J., Zhou, P., Fu, J., Sun, J., et al. (2019). Noninvasive prenatal testing for chromosome aneuploidies and subchromosomal microdeletions/microduplications in a cohort of 8141 single pregnancies. Hum. Genomics 13, 14. doi:10.1186/s40246-019-0198-2
Jiang, F., Ren, J., Chen, F., Zhou, Y., Xie, J., Dan, S., et al. (2012). Noninvasive fetal trisomy (NIFTY) test: An advanced noninvasive prenatal diagnosis methodology for fetal autosomal and sex chromosomal aneuploidies. BMC Med. Genomics 5, 57. doi:10.1186/1755-8794-5-57
Lai, Y., Zhu, X., He, S., Dong, Z., Tang, Y., Xu, F., et al. (2021). Performance of cell-free DNA screening for fetal common aneuploidies and sex chromosomal abnormalities: A prospective study from a less developed autonomous region in mainland China. Genes 12, 478. doi:10.3390/genes12040478
Liang, D., Cram, D. S., Tan, H., Linpeng, S., Liu, Y., Sun, H., et al. (2019). Clinical utility of noninvasive prenatal screening for expanded chromosome disease syndromes. Genet. Med. 21, 1998–2006. doi:10.1038/s41436-019-0467-4
Lu, W., Huang, T., Wang, X. R., Zhou, J. H., Yuan, H. Z., Yang, Y., et al. (2020). Next-generation sequencing: A follow-up of 36, 913 singleton pregnancies with noninvasive prenatal testing in central China. J. Assist. Reprod. Genet. 8, 3143–3150. doi:10.1007/s10815-020-01977-2
Mikwar, M., MacFarlane, A. J., and Marchetti, F. (2020). Mechanisms of oocyte aneuploidy associated with advanced maternal age. Mutat. Res. Rev. Mutat. Res. 785, 108320. doi:10.1016/j.mrrev.2020.108320
Quezada, M. S., Gil, M. M., Francisco, C., Oròsz, G., and Nicolaides, K. H. (2015). Screening for trisomies 21, 18 and 13 by cell-free DNA analysis of maternal blood at 10-11 weeks’ gestation and the combined test at 11-13 weeks. Ultrasound Obstet. Gynecol. 45, 36–41. doi:10.1002/uog.14664
Scibetta, E. W., Gaw, S. L., Rao, R. R., Silverman, N. S., Han, C. S., and Platt, L. D. (2017). Clinical accuracy of abnormal cell-free fetal DNA results for the sex chromosomes. Prenat. Diagn. 37, 1291–1297. doi:10.1002/pd.5146
Shi, P., Wang, Y., Liang, H., Hou, Y., Chen, D., Zhao, G., et al. (2021). The potential of expanded noninvasive prenatal screening for detection of microdeletion and microduplication syndromes. Prenat. Diagn. 41, 1332–1342. doi:10.1002/pd.6002
Skuse, D., Printzlau, F., and Wolstencroft, J. (2018). Sex chromosome aneuploidies. Handb. Clin. Neurology 147, 355–376. doi:10.1016/B978-0-444-63233-3.00024-5
Wapner, R. J., Martin, C. L., Levy, B., Ballif, B. C., Eng, C. M., Zachary, J. M., et al. (2012). Chromosomal microarray versus karyotyping for prenatal diagnosis. N. Engl. J. Med. 367, 2175–2184. doi:10.1056/NEJMoa1203382
Yilmaz, Z., Szatkiewicz, J. P., Crowley, J. J., Ancalade, N., Brandys, M. K., van Elburg, A., et al. (2017). Exploration of large, rare copy number variants associated with psychiatric and neurodevelopmental disorders in individuals with anorexia nervosa. Psychiatr. Genet. 27, 152–158. doi:10.1097/YPG.0000000000000172
Yu, B., Lu, B.-Y., Zhang, B., Zhang, X.-Q., Chen, Y.-P., Zhou, Q., et al. (2017). Overall evaluation of the clinical value of prenatal screening for fetal-free DNA in maternal blood. Medicine 96, e7114. doi:10.1097/MD.0000000000007114
Zhang, B., Lu, B.-Y., Yu, B., Zheng, F.-X., Zhou, Q., Chen, Y.-P., et al. (2017). Noninvasive prenatal screening for fetal common sex chromosome aneuploidies from maternal blood. J. Int. Med. Res. 45, 621–630. doi:10.1177/0300060517695008
Zhao, G., Dai, P., Wang, C., Liu, L., Zhao, X., and Kong, X. (2022). Clinical application of noninvasive prenatal testing for sex chromosome aneuploidies in central China. Front. Med. 8, 672211. doi:10.3389/fmed.2021.672211
Zheng, Y., Wan, S., Dang, Y., Song, T., Chen, B., and Zhang, J. (2020). Clinical experience regarding the accuracy of NIPT in the detection of sex chromosome abnormality. J. Gene Med. 22, e3199. doi:10.1002/jgm.3199
Keywords: expanded non-invasive prenatal testing (NIPT-plus), chromosome aneuploidies, copy number variations, positive predictive value, Jiangxi province
Citation: Zou Y, Feng C, Qin J, Wang X, Huang T, Yang Y, Xie K, Yuan H, Huang S, Yang B, Lu W and Liu Y (2023) Performance of expanded non-invasive prenatal testing for fetal aneuploidies and copy number variations: A prospective study from a single center in Jiangxi province, China. Front. Genet. 13:1073851. doi: 10.3389/fgene.2022.1073851
Received: 19 October 2022; Accepted: 27 December 2022;
Published: 13 January 2023.
Edited by:
José María Frade, Spanish National Research Council (CSIC), SpainReviewed by:
Cristina Montagna, The State University of New Jersey, United StatesCopyright © 2023 Zou, Feng, Qin, Wang, Huang, Yang, Xie, Yuan, Huang, Yang, Lu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wan Lu, Mjg1MDI4NzAyQHFxLmNvbQ==; Yanqiu Liu, bHlxMDkxNEAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.