- 1Department of Animal Genetics and Breeding, College of Animal Science and Technology, Key Laboratory of Animal Genetics, Breeding and Reproduction of Ministry of Agriculture and Rural Affairs, National Engineering Laboratory for Animal Breeding, China Agricultural University, Beijing, China
- 2National Dairy Innovation Center, Hohhot, China
SEC13 homolog, nuclear pore and COPII coat complex component (SEC13) is the core component of the cytoplasmic COPII complex, which mediates material transport from the endoplasmic reticulum to the Golgi complex. Our preliminary work found that SEC13 gene was differentially expressed in dairy cows during different stages of lactation, and involved in metabolic pathways of milk synthesis such as citric acid cycle, fatty acid, starch and sucrose metabolisms, so we considered that the SEC13 might be a candidate gene affecting milk production traits. In this study, we detected the polymorphisms of SEC13 gene and verified their genetic effects on milk yield and composition traits in a Chinese Holstein cow population. By sequencing the whole coding and partial flanking regions of SEC13, we found four single nucleotide polymorphisms (SNPs). Subsequent association analysis showed that these four SNPs were significantly associated with milk yield, fat yield, protein yield or protein percentage in the first and second lactations (p ≤.0351). We also found that two SNPs in SEC13 formed one haplotype block by Haploview4.2, and the block was significantly associated with milk yield, fat yield, fat percentage, protein yield or protein percentage (p ≤ .0373). In addition, we predicted the effect of SNP on 5′region on transcription factor binding sites (TFBSs), and found that the allele A of 22:g.54362761A>G could bind transcription factors (TFs) GATA5, GATA3, HOXD9, HOXA10, CDX1 and Hoxd13; and further dual-luciferase reporter assay verified that the allele A of this SNP inhibited the fluorescence activity. We speculate that the A allele of 22:g.54362761A>G might inhibit the transcriptional activity of SEC13 gene by binding the TFs, which may be a cause mutation affecting the formation of milk production traits in dairy cows. In summary, we proved that SEC13 has a significant genetic effect on milk production traits and the identified significant SNPs could be used as candidate genetic markers for GS SNP chips development; on the other hand, we verified the transcriptional regulation of 22:g.54362761A>G on SEC13 gene, providing research direction for further function validation tests.
1 Introduction
The traits that affect the production efficiency of dairy cows are calving traits, milk production traits, longevity traits and so on (Weller and Ezra 2016; Zhang H. et al., 2021), among which, milk production traits are the most important economic traits in dairy cow breeding, including milk yield, fat yield, protein yield, fat percentage and protein percentage (Spelman et al., 1996). They are quantitative traits, controlled by multiple genes and easily affected by the environment. In addition, dairy cattle breeding is faced with problems such as long generation interval and slow progress (Wiggans et al., 2017). In 2009, developed countries began to widely use genomic selection (GS) for dairy cattle breeding. GS could shorten the generation gap, accelerate the population genetic progress, reduce the breeding costs, and almost double the genetic progress rate (Wiggans et al., 2017). Since the promotion and application of GS in China in 2012, the genetic progress of Chinese Holstein cows has been significantly improved. According to studies, adding functional gene information with greater genetic effects of target traits to single nucleotide polymorphism (SNP) marker data can improve the accuracy of genomic estimated breeding value prediction (Zhang et al., 2014; Zhang et al., 2015; de Las Heras-Saldana et al., 2020). Therefore, many researchers try to find the key genes/loci that have significant impacts on milk production traits through candidate gene analysis, genome-wide association analysis and omics data analysis strategies (Schennink et al., 2009; Canovas et al., 2013; Cui et al., 2014; Liang et al., 2017; Bhat et al., 2020). Studies have shown that the milk production traits of dairy cows can be significantly affected by SNPs in the genes (Jiang et al., 2010; Dux et al., 2018; Han et al., 2019a; Han et al., 2019b; Clancey et al., 2019; Jia et al., 2021; Du et al., 2022; Fu et al., 2022; Ye et al., 2022).
Previously, we used isobaric tag for relative and absolute quantification (iTRAQ) technique to study the proteomes of nine liver tissue samples from three Holstein cows during dry period, early and peak lactations, and found that SEC13 homolog, nuclear pore and COPII coat complex component (SEC13) gene was differentially expressed among different lactations (dry period vs peak lactation: fold change = 1.37, p-value = .03134; early lactation vs. peak lactation: fold change = 1.22, p-value = .001002) and significantly enriched in the metabolic items and pathways related to milk synthesis, such as citric acid cycle, fatty acid, starch and sucrose metabolism, mTOR and PPAR signal pathways (Xu et al., 2019). SEC13, a member of the WD-Repeat protein family, is the core component of the COPII complex in the cytoplasm and nuclear pore complex on the nuclear membrane (Antonny and Schekman 2001; Stagg et al., 2006). Moreover, SEC13 shuttles between the nucleus and the cytoplasm, acting in the ER-to-Golgi vesicular transport system (Enninga et al., 2003). SEC13 interacts with SEC31 to form the outer cage of the COPII vesicle coat needed for the transport of vesicle proteins from the endoplasmic reticulum (ER) to the Golgi matrix (Stagg et al., 2006; Stagg et al., 2008). COPII coat is used to transport newly synthesized proteins, including secretory and transmembrane proteins. As part of the COPII complex, SEC13 is involved in the process of insulin stimulating glucose uptake by controlling the amount of glucose transporter 4 (GLUT4) in the plasma membrane (Martin et al., 2000; Karylowski et al., 2004; Huang and Czech 2007; Stockli et al., 2011). In addition, we found that an SNP (rs109645852; Chr.22:54428720) 67 kb away from SEC13 gene (Chr.22:54326136.54361315; Cattle Quantitative Trait Locus Database; https://www.animalgenome.org/jbrowse/q?data=db%2FbovARS&loc=Chr.22%3A54243801.54530800&tracks=Milk%20composition%20-%20fat%2CGenbank%20annotations&highlight=Chr.22%3A54428718.54428722%20(%22Milk%20fat%20yield%20(daughter%20deviation)%22) was significantly associated with milk fat yield, which was identified in a reported genome-wide association study of Holstein cows (p = .0359) (Meredith et al., 2012). Therefore, we inferred that SEC13 gene might be an important functional gene affecting milk production traits of dairy cows.
In this study, based on the candidate gene SEC13 for milk production traits of dairy cows, the SNPs of this gene were detected, their genetic associations with milk yield, fat yield, fat percentage, protein yield and protein percentage were analyzed, and whether they could be used in the development of GS SNP chips was evaluated. In addition, we predicted the effect of identified SNPs on transcription factor binding site (TFBS), and verify the effect of SNP at 5′flanking region on the transcriptional activity of SEC13 gene by dual-luciferase reporter experiment, thus speculating the causal mutation of milk production traits in dairy cows.
2 Materials and methods
2.1 Animal selection and phenotypic data collection
In this study, 947 cows from 45 Chinese Holstein sire families in Beijing were used as the experimental population. They were distributed in 22 farms of Beijing Shounong Animal Husbandry Development Co., Ltd. (Beijing, China), and raised under the same feeding conditions, with accurate pedigree and Dairy Herd Improvement (DHI) records. We used the phenotypic data of 947 first-lactation cows and 654 second-lactation cows (293 cows only completed the first lactation) for association analysis. The data of the entire lactation period of the parity of each cow was used as the individual milk yield phenotype. The actual total milk yield was multiplied by the corresponding estimated coefficient to get 305-day milk yield. The 305-day milk fat and protein content were calculated by multiplying the 305-day milk yield by the average percentage of milk fat and protein, respectively. The average milk fat and protein percentages were the ratio of total milk fat and protein contents to total milk yield, respectively. Results of descriptive statistics for the milk yield and composition in the two lactations were shown in Supplementary Table S1. Frozen semen and cow blood samples were provided by Beijing Dairy Center (Beijing, China).
2.2 Genomic DNA extraction
We extracted genomic DNA from frozen semen of 45 bulls by salt-out procedure, and used a TIANamp Blood DNA Kit (Tiangen, Beijing, China) to extract DNA from the blood of 947 cows. We detected the quantity and quality of extracted DNA samples by a NanoDrop2000 spectrophotometer (Thermo Science, Hudson, NH, United States) and gel electrophoresis respectively.
2.3 SNP identification and estimation of linkage disequilibrium
We designed primers with Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) to amplify all the coding regions and the 2000 bp of 5′and 3′regions of SEC13 gene. The primers were synthesized by Beijing Genomics Institute (BGI, Beijing, China). The genomic DNA of bull frozen semen was mixed with the same amount, then amplified by PCR (Supplementary Table S2). We used 2% gel electrophoresis to detect whether the PCR amplification products were qualified, and sequenced the qualified PCR amplification products by Sanger sequencing (BGI, Beijing, China). Then we compared the sequenced sequences with the reference sequences (ARS-UCD1.2) on NCBI-BLAST (https://blast.ncbi.nlm. nih. gov/Blast.cgi) to find the potential SNPs. Subsequently, we used Haploview4.2 (Broad Institute of MIT and Harvard, Cambridge, MA, United States) to estimate the degree of linkage disequilibrium (LD) between the identified SNPs. In addition, we genotyped 947 dairy cows using Genotyping by Target Sequencing (GBTS) technology in Boruidi Biotechnology Co., Ltd. (Hebei, China).
2.4 Association analyses on milk production traits
We used the SAS 9.4 software (SAS Institute Inc., Cary, NC, United States) to estimate the genetic associations of the SNPs or haplotype blocks with milk production traits, 305-day milk yield, fat yield, fat percentage, protein yield and protein percentage, on first or second lactation with the following animal model:
2.5 Prediction and verification of SNP induced changes in gene transcription activity
We used Jaspar software (http://jaspar.genereg.net/) to predict whether SNP in the 5′flanking region of SEC13 gene changed the transcription factor binding site (TFBS; relative score≥ .90).
Further, we used the dual-luciferase reporter assay to verify the effect of SNP site that affect the transcription factor binding on gene expression activity. For 22:g.54362761A>G, we synthesized the fragment with SNP site, A or G. The fragment that carried endonuclease sites KpnI and Nhel at ends, respectively, were cloned into pGL4.14 luciferase analysis vector (Promega, Madison, WI, United States). The constructed plasmid was sequenced to confirm the integrity of each insert. The Endo-free Plasmid Maxi Kit (Omega Bio-tek, Inc., Norcross, GA, United States) was used to extract plasmids needed for cell transfection. Human embryonic kidney (HEK) 293T cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco; Thermo Fisher Scientific Inc., MA, United States) supplemented with 10% fetal bovine serum (FBS; Gibco) at 5% CO2 and 37°C. The cells were seeded into 24-well plates with 2 × 105–107 cells per well before transfection. The cells were transiently transfected with liposome 2000 (Invitrogen; Thermo Fisher Scientific Inc.). For each well, 500 ng of the constructed plasmid was co-transfected along with 10 ng of pRL-TK Renilla luciferase reporter vector (Promega). 48 h after transfection, the cells were harvested and the luciferase activity was detected by a Dual-Luciferase Reporter Assay System (Promega). The relative fluorescence activity was calculated by the fluorescence activity ratio of firefly and renilla.
3 Results
3.1 SNPs identification
We totally found four SNPs in SEC13 gene, one SNP, 22:g.54362761A>G, is located in 5′flanking region, 22:g.54334911G>A in intron and two SNPs, 22:g.54326411T>C and 22:g.54326366C>T, in 3′untranslated region (UTR) (Table 1). Additionally, the genotypic and allelic frequencies of all the identified SNPs were summarized in Table 1.
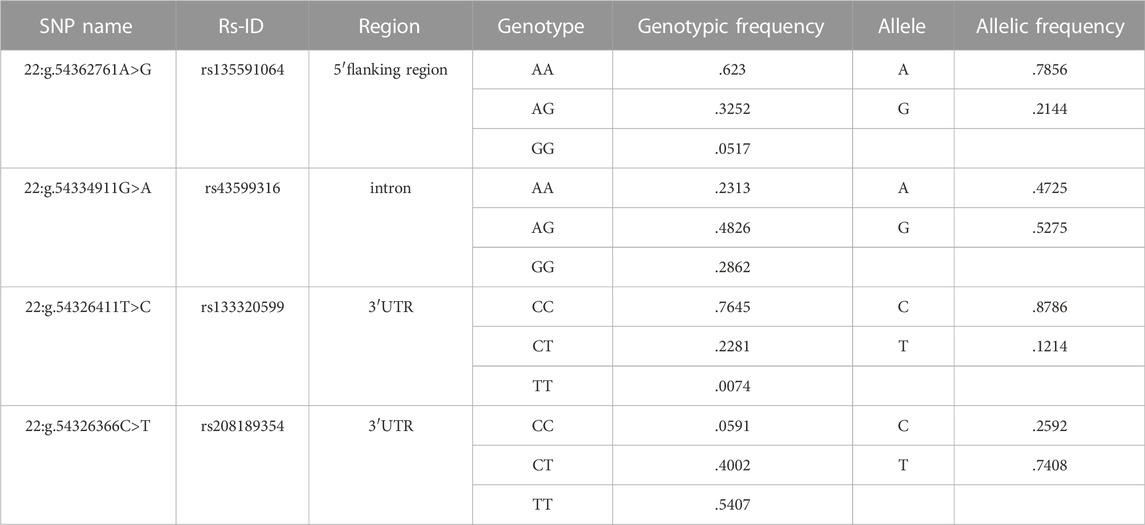
TABLE 1. Detailed information about SNPs identified in SEC13 gene and their genotypic and allelic frequencies. UTR: untranslated region.
3.2 Association analysis between SNP and the five milk production traits
We analyzed the associations between the four SNPs and five milk production traits in dairy cows, including 305-day milk yield, fat yield, protein yield, fat percentage and protein percentage (Table 2). The SNP 22:g.54362761A>G was significantly associated with protein yield (p = .036) in the first lactation, milk yield (p = .0004), fat yield (p < .0001) and protein yield (p < .0001) in the second lactation. 22:g.54334911G>A was significantly associated with fat yield (p = .0351) in the first lactation, milk yield (p = .0033), fat yield (p < .0001), protein yield (p = .0001) and protein percentage (p = .0274) in the second lactation. 22:g.54326411T>C was significantly associated with protein yield (p = .0296) in the second lactation. 22:g.54326366C>T was significantly associated with milk yield (p = .0173), fat yield (p = .0008) and protein yield (p = .0028) in the first lactation. Accordingly, the results of additive, dominant and substitution effects for the four SNPs were shown in Supplementary Table S3.
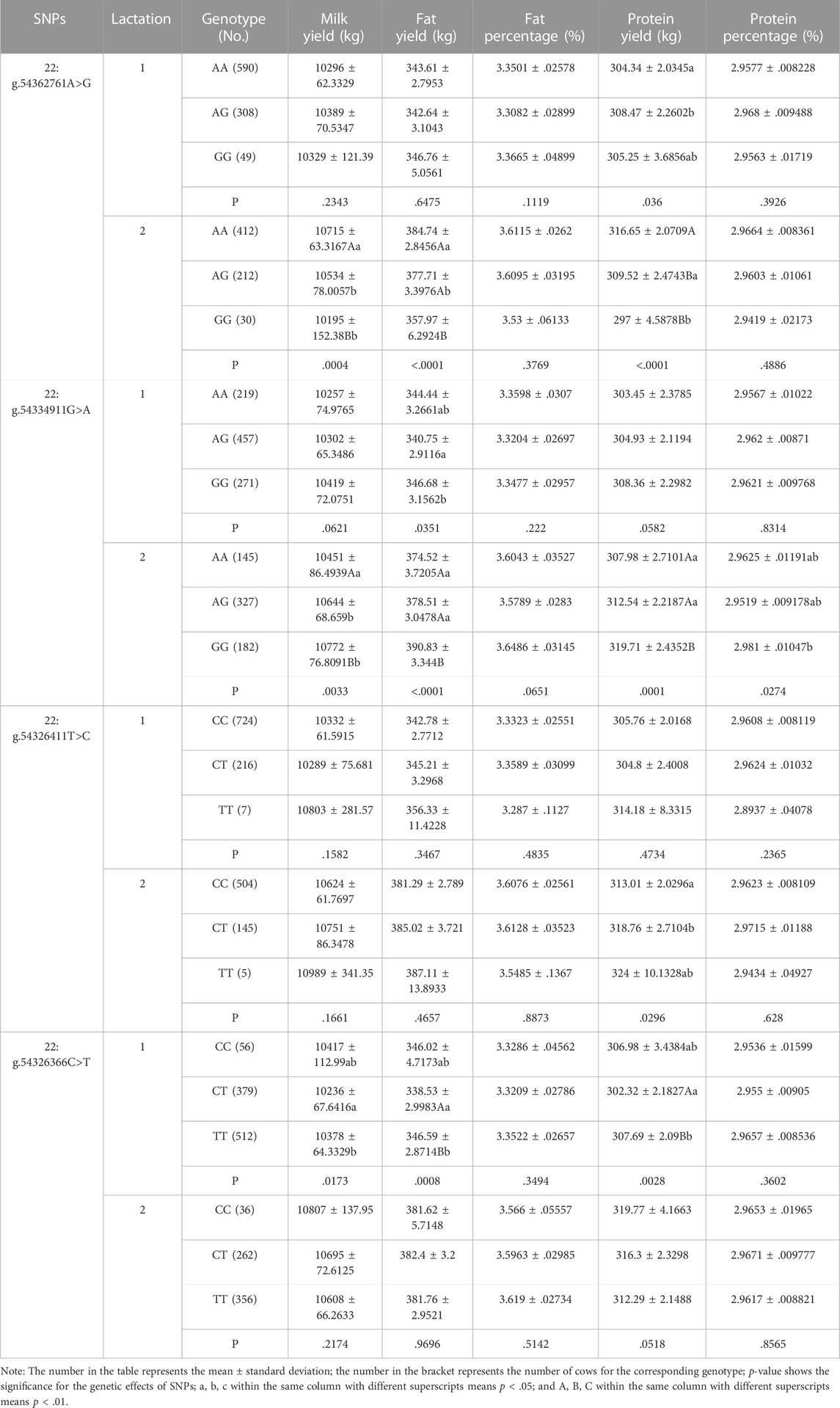
TABLE 2. Associations of four SNPs with milk production traits in Chinese Holstein cattle during two lactations.
3.3 Association between haplotype block and the five milk traits
We estimated the degree of LD among four identified SNPs using Haploview4.2, and inferred one haplotype block, including two SNPs, 22:g.54326366C>T and 22:g.54326411T>C, (D' = 1; Figure 1). In the block, the frequencies of H1 (TC), H2 (CC) and H3 (CT) haplotypes were 74.1%, 13.8% and 12.1%, respectively. The haplotype block of SEC13 was significantly associated with milk yield (p = .046), fat yield (p = .0001), fat percentage (p = .0113), protein yield (p = .0056) and protein percentage (p = .0373) in the first lactation (Supplementary Table S4).

FIGURE 1. Linkage disequilibrium estimated between SNPs in SEC13 gene. The bright red boxes without numbers indicate complete LD (D′ = 1).
3.4 Effect of 22:g.54362761A>G on gene transcriptional activity
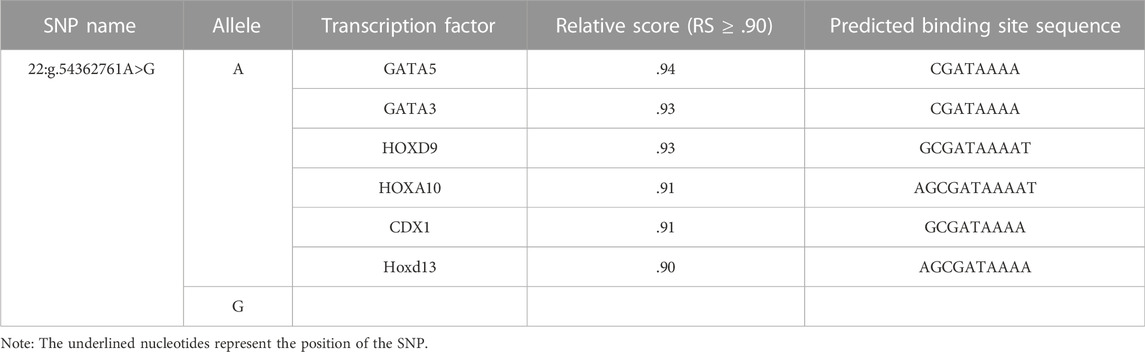
We predicted the TFBS changes for the SNP, 22:g.54362761A>G, in the 5′flanking region of SEC13 gene by Jaspar software (relative score (RS) ≥.90). The result showed that allele A of 22:g.54362761A>G created binding sites for transcription factors (TFs) GATA5 (RS = .94), GATA3 (RS = .93), HOXD9 (RS = .93), HOXA10 (RS = .91), HOXD13 (RS = .90) and CDX1 (RS = .91; Table 3).
To further determine whether this SNP changed the transcription activity of SEC13 gene, we constructed reporter plasmids containing two alleles A and G, respectively (Figure 2A). As shown in Figure 2B, the luciferase activities of the two recombinant plasmids were significantly higher than that of the empty vector (PGL4.14 + TK) and the blank cell controls (p < .01), which confirmed that the inserted fragment have transcriptional regulation function. The luciferase activity of G allele (PGL4.14(G)+TK) was significantly higher than that of A allele (PGL4.14(A)+TK; p < .01), suggesting that the 22:g.54362761A might inhibit the transcription activity of SEC13 gene.
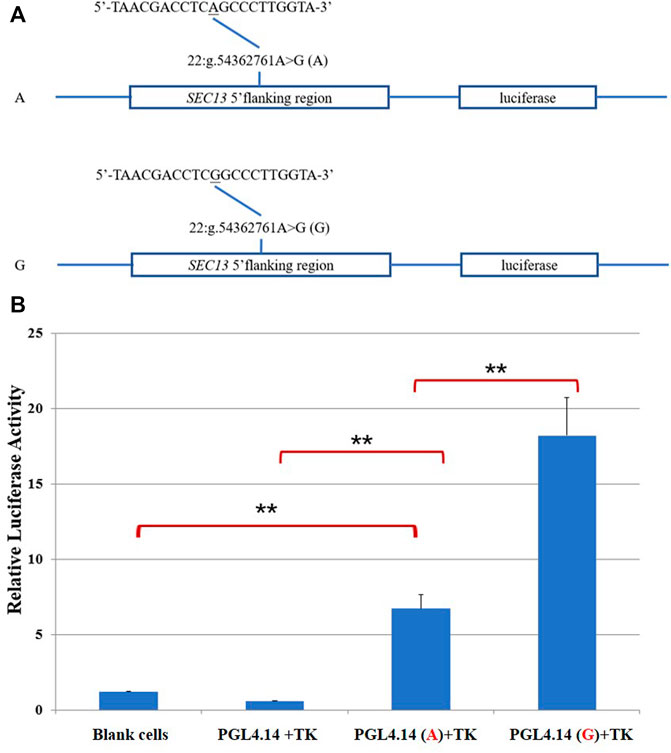
FIGURE 2. Dual-luciferase activity assay. (A)Sketches of recombinant plasmids with 22:g.54362761A>G in SEC13 gene. The underlined nucleotide was the SNP. (B) Luciferase activity analysis of the recombinant plasmids in HEK 293T cells. **p < .01.
4 Discussion
Our previous study considered SEC13 gene to be a candidate to affect milk production traits in dairy cows. In this study, we detected the polymorphisms of SEC13 gene, and found that there was a significant genetic association between the SNP/haplotype block of the gene and milk production traits. Studies have shown that in the genomic relationship matrices, SNPs can be given different weights to make more accurate and less biased prediction of traits (Tiezzi and Maltecca 2015). Therefore, when adding the SNPs that have significant effects on milk production traits in this study to the commercial SNP chip, we can give different weights to these SNPs according to their effects to improve the accuracy of GS.
Coding sequence (CDS) is a key area for creature survival, many mutations in this region can lead to death, and DNA repair in this region is more efficient than other regions. A studies had shown that the number of SNP in the exon region is less than that in other regions (Frigola et al., 2017), which may be the reason why we didn’t find SNP in the exon region. Transcription factors (TFs) can affect gene expression by combining to TFBSs to regulate the transcription of target genes (Calkhoven and Ab 1996). The SNP located at TFBS may affect the binding of TFs, resulting in differences in gene expression among individuals with different genotypes (Kasowski et al., 2010; McDaniell et al., 2010). In this study, we found that the mutation from allele A to G of 22:g.54362761A>G led to the disappearance of TFBSs for TFs GATA5, GATA3, HOXD9, HOXA10, HOXD13 and CDX1. GATA family regulates cell reprogramming to induce stem cell differentiation and normal function of cells (Shu et al., 2015). GATA5 is a member of GATA family and plays an important role in cardiovascular disease (Shi et al., 2014; Messaoudi et al., 2015). Furthermore, GATA5 is involved in suppressing expression of the reprogramming genes and stemness markers in hepatocellular carcinoma cells (Feng et al., 2020). GATA3 is estimated to be the most highly expressed transcription factor in the differentiated luminal epithelial cells lining the breast ductal structures and plays an important role in mammary gland development (Kouros-Mehr et al., 2006; Kouros-Mehr and Werb 2006). Moreover, GATA3 transcriptionally inhibits Slug expression, thereby inhibits cancer cell proliferation, migration and invasion (Zhang Z. et al., 2021). HOX is a family of many TFs that involved in embryonic development and the maintenance of normal tissues (Mallo and Alonso 2013). HOXD9 is closely related to many kinds of tumors. It can promote tumorigenesis and metastasis by increasing the expression level of other genes in gastric and breast cancer (Zhu et al., 2019; Xiong et al., 2020; Hu et al., 2022). Both HOXA10 and HOXD13 can promote the occurrence and development of various cancers by increasing the expression of genes and activating signal pathways (Chen et al., 2019; Cui et al., 2020; Yin and Guo 2021), nevertheless, both of them can inhibit the expression of genes and suppress the occurrence of prostate cancer (Hatanaka et al., 2019; Xu et al., 2021). CDX1 controls intestinal cell differentiation in the colon and has been shown to directly promote the expression of structural proteins important for epithelial differentiation, including cytokeratin 20 and villin (Chan et al., 2009; Arango et al., 2012). These TFs can activate or inhibit gene expression, and they may interact with each other to promote or inhibit the expression of SEC13 gene. It has been reported that the SEC13 gene is close to the quantitative trait locus (QTL) which has been found to have a significant effect on milk fat yield (Meredith et al., 2012). Another study showed that the QTL from the same region showed significant genetic effects on milk protein and fat traits (Bagnato et al., 2008). According to the phenotypic data of milk production traits of different genotypes, it was found that the milk, fat and protein yields of genotype AA was significantly higher than that of genotype GG of 22:g.54362761A>G. Further, the results of dual-luciferase reporter assay showed that when the A mutated to G in 22:g.54362761A>G, the transcriptional activity of SEC13 gene increased significantly. Therefore, we speculated that the change of SEC13 gene expression caused by SNP 22:g.54362761A>G may be one of the reasons for the phenotypic change of milk production traits in dairy cows. The specific mechanism of SEC13 gene on the formation of milk production traits needs further experimental verification. SEC23B, which is also the core component of COPII complex with SEC13, mediates the transport of substances from the endoplasmic reticulum to the Golgi complex and plays an important role in professional secretory tissues/cells (Tao et al., 2012). This also provides an important clue for our in-depth research. Further, we can use secretory cell models such as bovine mammary epithelial cells to verify the influence of SEC13 on the formation of milk production traits.
5 Conclusion
This study confirmed the genetic effects of SEC13 on milk production traits of Chinese Holstein cows. The SNP 22:g.54362761A>G might be a causal mutation site for milk production traits, which might regulate the transcriptional activity of SEC13 gene by binding transcription factors, the specific mechanism remains to be further verified. This study laid a foundation for the further functional verification of SEC13 for milk synthesis in dairy cattle, and the significant SNP sites could be used as genetic markers for dairy cattle GS breeding.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by The study was conducted in accordance with Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee (IACUC) at China Agricultural University (Beijing, China; permit number: DK996).
Author contributions
BH and DS: Conceptualization, methodology and funding acquisition. RJ and LX: formal analysis. RJ: Visualization. RJ: Writing–original draft preparation. RJ and BH: Writing, review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by Inner Mongolia and Hohhot Science & Technology Plan (No.2020-Ke Ji Xing Meng-National Innovation Center-12; No.2021-National Dairy Innovation Center-3), National Natural Science Foundation of China (32072716; 31872330), Key R & D project of Ningxia Hui Autonomous Region (2021BEF02018), and the Program for Changjiang Scholar and Innovation Research Team in University (IRT_15R62).
Acknowledgments
We appreciate Beijing Dairy Cattle Center for providing the semen and blood samples and phenotypic data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1065096/full#supplementary-material
SUPPLEMENTARY TABLE S1 | Descriptive statistics of phenotypic values for milk production in two lactations.
SUPPLEMENTARY TABLE S2 | Primers and procedures for PCR used in SNPs identification of SEC13 gene.
SUPPLEMENTARY TABLE S3 | Additive, dominant and allele substitution effects of four SNPs in SEC13 gene on milk yield and composition traits in Chinese Holstein cows during two lactations.
SUPPLEMENTARY TABLE S4 | Haplotype analysis for SEC13 gene.
References
Antonny, B., and Schekman, R. (2001). ER export: Public transportation by the COPII coach. Curr. Opin. Cell. Biol. 13, 438–443.
Arango, D., Al-Obaidi, S., Williams, D. S., Dopeso, H., Mazzolini, R., Corner, G., et al. (2012). Villin expression is frequently lost in poorly differentiated colon cancer. Am. J. Pathol. 180, 1509–1521.
Bagnato, A., Schiavini, F., Rossoni, A., Maltecca, C., Dolezal, M., Medugorac, I., et al. (2008). Quantitative trait loci affecting milk yield and protein percentage in a three-country Brown Swiss population. J. Dairy Sci. 91, 767–783. doi:10.3168/jds.2007-0507
Bhat, S. A., Ahmad, S. M., Ibeagha-Awemu, E. M., Mobashir, M., Dar, M. A., Mumtaz, P. T., et al. (2020). Comparative milk proteome analysis of Kashmiri and Jersey cattle identifies differential expression of key proteins involved in immune system regulation and milk quality. BMC Genomics 21, 161. doi:10.1186/s12864-020-6574-4
Calkhoven, C. F., and Ab, G. (1996). Multiple steps in the regulation of transcription-factor level and activity. Biochem. J. 317 (2), 329–342. doi:10.1042/bj3170329
Canovas, A., Rincon, G., Islas-Trejo, A., Jimenez-Flores, R., Laubscher, A., Medrano, J. F., et al. (2013). RNA sequencing to study gene expression and single nucleotide polymorphism variation associated with citrate content in cow milk. J. Dairy Sci. 96, 2637–2648. doi:10.3168/jds.2012-6213
Chan, C. W., Wong, N. A., Liu, Y., Bicknell, D., Turley, H., Hollins, L., et al. (2009). Gastrointestinal differentiation marker Cytokeratin 20 is regulated by homeobox gene CDX1. Proc. Natl. Acad. Sci. U. S. A. 106, 1936–1941. doi:10.1073/pnas.0812904106
Chen, W., Wu, G., Zhu, Y., Zhang, W., Zhang, H., Zhou, Y., et al. (2019). HOXA10 deteriorates gastric cancer through activating JAK1/STAT3 signaling pathway. Cancer Manag. Res. 11, 6625–6635. doi:10.2147/CMAR.S201342
Clancey, E., Kiser, J. N., Moraes, J. G. N., Dalton, J. C., Spencer, T. E., and Neibergs, H. L. (2019). Genome-wide association analysis and gene set enrichment analysis with SNP data identify genes associated with 305-day milk yield in Holstein dairy cows. Anim. Genet. 50, 254–258. doi:10.1111/age.12792
Cui, X. G., Hou, Y. L., Yang, S. H., Xie, Y., Zhang, S. L., Zhang, Y., et al. (2014). Transcriptional profiling of mammary gland in Holstein cows with extremely different milk protein and fat percentage using RNA sequencing. BMC Genomics 15, 226. doi:10.1186/1471-2164-15-226
Cui, Y. P., Xie, M., Pan, W. X., Zhang, Z. Y., and Li, W. F. (2020). HOXA10 promotes the development of bladder cancer through regulating FOSL1. Eur. Rev. Med. Pharmacol. Sci. 24, 2945–2954. doi:10.26355/eurrev_202003_20659
de Las Heras-Saldana, S., Lopez, B. I., Moghaddar, N., Park, W., Park, J. E., Chung, K. Y., et al. (2020). Use of gene expression and whole-genome sequence information to improve the accuracy of genomic prediction for carcass traits in Hanwoo cattle. Genet. Sel. Evol. 52, 54. doi:10.1186/s12711-020-00574-2
Du, A., Zhao, F., Liu, Y., Xu, L., Chen, K., Sun, D., et al. (2022). Genetic polymorphisms of PKLR gene and their associations with milk production traits in Chinese Holstein cows. Front. Genet. 13, 1002706. doi:10.3389/fgene.2022.1002706
Dux, M., Muranowicz, M., Siadkowska, E., Robakowska-Hyzorek, D., Flisikowski, K., Bagnicka, E., et al. (2018). Association of SNP and STR polymorphisms of insulin-like growth factor 2 receptor (IGF2R) gene with milk traits in Holstein-Friesian cows. J. Dairy Res. 85, 138–141. doi:10.1017/S0022029918000110
Enninga, J., Levay, A., and Fontoura, B. M. (2003). Sec13 shuttles between the nucleus and the cytoplasm and stably interacts with Nup96 at the nuclear pore complex. Mol. Cell. Biol. 23, 7271–7284. doi:10.1128/mcb.23.20.7271-7284.2003
Feng, H., Lin, B., Zheng, Y., Xu, J., Zhou, Y., Liu, K., et al. (2020). Overexpression of GATA5 stimulates paclitaxel to inhibit malignant behaviors of hepatocellular carcinoma cells. Cell. J. 22, 89–100. doi:10.22074/cellj.2020.6894
Frigola, J., Sabarinathan, R., Mularoni, L., Muinos, F., Gonzalez-Perez, A., and Lopez-Bigas, N. (2017). Reduced mutation rate in exons due to differential mismatch repair. Nat. Genet. 49, 1684–1692. doi:10.1038/ng.3991
Fu, Y., Jia, R., Xu, L., Su, D., Li, Y., Liu, L., et al. (2022). Fatty acid desaturase 2 affects the milk-production traits in Chinese Holsteins. Anim. Genet. 53, 422–426. doi:10.1111/age.13192
Han, B., Yuan, Y., Liang, R., Li, Y., Liu, L., and Sun, D. (2019a). Genetic effects of LPIN1 polymorphisms on milk production traits in dairy cattle. Genes (Basel) 10, 265. doi:10.3390/genes10040265
Han, B., Yuan, Y., Shi, L., Li, Y., Liu, L., and Sun, D. (2019b). Identification of single nucleotide polymorphisms of PIK3R1 and DUSP1 genes and their genetic associations with milk production traits in dairy cows. J. Anim. Sci. Biotechnol. 10, 81. doi:10.1186/s40104-019-0392-z
Hatanaka, Y., de Velasco, M. A., Oki, T., Shimizu, N., Nozawa, M., Yoshimura, K., et al. (2019). HOXA10 expression profiling in prostate cancer. Prostate 79, 554–563. doi:10.1002/pros.23761
Hu, X. C., Chu, J., Zhou, Y., Li, C. C., Zhou, G. J., and Jiang, G. Q. (2022). HOXD9 transcriptionally induced UXT facilitate breast cancer progression via epigenetic modification of RND3. Cell. Signal 90, 110188. doi:10.1016/j.cellsig.2021.110188
Huang, S., and Czech, M. P. (2007). The GLUT4 glucose transporter. Cell. Metab. 5, 237–252. doi:10.1016/j.cmet.2007.03.006
Jia, R., Fu, Y., Xu, L., Li, H., Li, Y., Liu, L., et al. (2021). Associations between polymorphisms of SLC22A7, NGFR, ARNTL and PPP2R2B genes and Milk production traits in Chinese Holstein. BMC Genom Data 22, 47. doi:10.1186/s12863-021-01002-0
Jiang, L., Liu, J., Sun, D., Ma, P., Ding, X., Yu, Y., et al. (2010). Genome wide association studies for milk production traits in Chinese Holstein population. PLoS One 5, e13661. doi:10.1371/journal.pone.0013661
Karylowski, O., Zeigerer, A., Cohen, A., and McGraw, T. E. (2004). GLUT4 is retained by an intracellular cycle of vesicle formation and fusion with endosomes. Mol. Biol. Cell. 15, 870–882. doi:10.1091/mbc.e03-07-0517
Kasowski, M., Grubert, F., Heffelfinger, C., Hariharan, M., Asabere, A., Waszak, S. M., et al. (2010). Variation in transcription factor binding among humans. Science 328, 232–235. doi:10.1126/science.1183621
Kouros-Mehr, H., Slorach, E. M., Sternlicht, M. D., and Werb, Z. (2006). GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 127, 1041–1055. doi:10.1016/j.cell.2006.09.048
Kouros-Mehr, H., and Werb, Z. (2006). Candidate regulators of mammary branching morphogenesis identified by genome-wide transcript analysis. Dev. Dyn. 235, 3404–3412. doi:10.1002/dvdy.20978
Liang, R., Han, B., Li, Q., Yuan, Y., Li, J., and Sun, D. (2017). Using RNA sequencing to identify putative competing endogenous RNAs (ceRNAs) potentially regulating fat metabolism in bovine liver. Sci. Rep. 7, 6396. doi:10.1038/s41598-017-06634-w
Mallo, M., and Alonso, C. R. (2013). The regulation of Hox gene expression during animal development. Development 140, 3951–3963. doi:10.1242/dev.068346
Martin, S., Millar, C. A., Lyttle, C. T., Meerloo, T., Marsh, B. J., Gould, G. W., et al. (2000). Effects of insulin on intracellular GLUT4 vesicles in adipocytes: Evidence for a secretory mode of regulation. J. Cell. Sci. 113 Pt 19, 3427–3438. doi:10.1242/jcs.113.19.3427
McDaniell, R., Lee, B. K., Song, L., Liu, Z., Boyle, A. P., Erdos, M. R., et al. (2010). Heritable individual-specific and allele-specific chromatin signatures in humans. Science 328, 235–239. doi:10.1126/science.1184655
Meredith, B. K., Kearney, F. J., Finlay, E. K., Bradley, D. G., Fahey, A. G., Berry, D. P., et al. (2012). Genome-wide associations for milk production and somatic cell score in Holstein-Friesian cattle in Ireland. BMC Genet. 13, 21. doi:10.1186/1471-2156-13-21
Messaoudi, S., He, Y., Gutsol, A., Wight, A., Hebert, R. L., Vilmundarson, R. O., et al. (2015). Endothelial Gata5 transcription factor regulates blood pressure. Nat. Commun. 6, 8835. doi:10.1038/ncomms9835
Schennink, A., Stoop, W. M., Visker, M. H., van der Poel, J. J., Bovenhuis, H., and van Arendonk, J. A. (2009). Short communication: Genome-wide scan for bovine milk-fat composition. II. Quantitative trait loci for long-chain fatty acids. J. Dairy Sci. 92, 4676–4682. doi:10.3168/jds.2008-1965
Shi, L. M., Tao, J. W., Qiu, X. B., Wang, J., Yuan, F., Xu, L., et al. (2014). GATA5 loss-of-function mutations associated with congenital bicuspid aortic valve. Int. J. Mol. Med. 33, 1219–1226. doi:10.3892/ijmm.2014.1700
Shu, J., Zhang, K., Zhang, M., Yao, A., Shao, S., Du, F., et al. (2015). GATA family members as inducers for cellular reprogramming to pluripotency. Cell. Res. 25, 169–180. doi:10.1038/cr.2015.6
Spelman, R. J., Coppieters, W., Karim, L., van Arendonk, J. A., and Bovenhuis, H. (1996). Quantitative trait loci analysis for five milk production traits on chromosome six in the Dutch Holstein-Friesian population. Genetics 144, 1799–1808. doi:10.1093/genetics/144.4.1799
Stagg, S. M., Gurkan, C., Fowler, D. M., LaPointe, P., Foss, T. R., Potter, C. S., et al. (2006). Structure of the Sec13/31 COPII coat cage. Nature 439, 234–238. doi:10.1038/nature04339
Stagg, S. M., LaPointe, P., Razvi, A., Gurkan, C., Potter, C. S., Carragher, B., et al. (2008). Structural basis for cargo regulation of COPII coat assembly. Cell. 134, 474–484. doi:10.1016/j.cell.2008.06.024
Stockli, J., Fazakerley, D. J., and James, D. E. (2011). GLUT4 exocytosis. J. Cell. Sci. 124, 4147–4159. doi:10.1242/jcs.097063
Tao, J., Zhu, M., Wang, H., Afelik, S., Vasievich, M. P., Chen, X. W., et al. (2012). SEC23B is required for the maintenance of murine professional secretory tissues. Proc. Natl. Acad. Sci. U. S. A. 109, E2001–E2009. doi:10.1073/pnas.1209207109
Tiezzi, F., and Maltecca, C. (2015). Accounting for trait architecture in genomic predictions of US Holstein cattle using a weighted realized relationship matrix. Genet. Sel. Evol. 47, 24. doi:10.1186/s12711-015-0100-1
Weller, J. I., and Ezra, E. (2016). Genetic analysis of calving traits by the multi-trait individual animal model. J. Dairy Sci. 99, 427–442. doi:10.3168/jds.2015-9768
Wiggans, G. R., Cole, J. B., Hubbard, S. M., and Sonstegard, T. S. (2017). Genomic selection in dairy cattle: The USDA experience. Annu. Rev. Anim. Biosci. 5, 309–327. doi:10.1146/annurev-animal-021815-111422
Xiong, R., Yin, T., Gao, J. L., and Yuan, Y. F. (2020). HOXD9 activates the TGF-β/smad signaling pathway to promote gastric cancer. Onco Targets Ther. 13, 2163–2172. doi:10.2147/OTT.S234829
Xu, F., Shangguan, X., Pan, J., Yue, Z., Shen, K., Ji, Y., et al. (2021). HOXD13 suppresses prostate cancer metastasis and BMP4-induced epithelial-mesenchymal transition by inhibiting SMAD1. Int. J. Cancer 148, 3060–3070. doi:10.1002/ijc.33494
Xu, L., Shi, L., Liu, L., Liang, R., Li, Q., Li, J., et al. (2019). Analysis of liver proteome and identification of critical proteins affecting milk fat, protein, and lactose metabolism in dariy cattle with iTRAQ. Proteomics 19, e1800387. doi:10.1002/pmic.201800387
Ye, W., Xu, L., Li, Y., Liu, L., Ma, Z., Sun, D., et al. (2022). Single nucleotide polymorphisms of ALDH18A1 and MAT2A genes and their genetic associations with milk production traits of Chinese Holstein cows. Genes (Basel) 13, 1437. doi:10.3390/genes13081437
Yin, J., and Guo, Y. (2021). HOXD13 promotes the malignant progression of colon cancer by upregulating PTPRN2. Cancer Med. 10, 5524–5533. doi:10.1002/cam4.4078
Zhang, H., Liu, A., Wang, Y., Luo, H., Yan, X., Guo, X., et al. (2021a). Genetic parameters and genome-wide association studies of eight longevity traits representing either full or partial lifespan in Chinese holsteins. Front. Genet. 12, 634986. doi:10.3389/fgene.2021.634986
Zhang, Z., Erbe, M., He, J., Ober, U., Gao, N., Zhang, H., et al. (2015). Accuracy of whole-genome prediction using a genetic architecture-enhanced variance-covariance matrix. G3 (Bethesda) 5, 615–627. doi:10.1534/g3.114.016261
Zhang, Z., Fang, X., Xie, G., and Zhu, J. (2021b). Erratum: GATA3 is downregulated in HCC and accelerates HCC aggressiveness by transcriptionally inhibiting slug expression. Oncol. Lett. 21, 575. doi:10.3892/ol.2021.12836
Zhang, Z., Ober, U., Erbe, M., Zhang, H., Gao, N., He, J., et al. (2014). Improving the accuracy of whole genome prediction for complex traits using the results of genome wide association studies. PLoS One 9, e93017. doi:10.1371/journal.pone.0093017
Keywords: genetic marker, SNP, milk yield and composition, dairy cattle, dual-luciferase reporter assay
Citation: Jia R, Xu L, Sun D and Han B (2023) Genetic marker identification of SEC13 gene for milk production traits in Chinese holstein. Front. Genet. 13:1065096. doi: 10.3389/fgene.2022.1065096
Received: 09 October 2022; Accepted: 15 December 2022;
Published: 04 January 2023.
Edited by:
Ibrar Muhammad Khan, Fuyang Normal University, ChinaReviewed by:
Xiaoxue Zhang, Gansu Agricultural University, ChinaCai Yafei, Nanjing Agricultural University, China
Rajwali Khan, University of Agriculture, Peshawar, Pakistan
Copyright © 2023 Jia, Xu, Sun and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Han, Ym9oYW5AY2F1LmVkdS5jbg==
 Ruike Jia1
Ruike Jia1 Dongxiao Sun
Dongxiao Sun Bo Han
Bo Han