
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 09 November 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1060546
This article is part of the Research TopicMolecular Dissection of the Genetic Basis Underlying Plant Complex TraitsView all 6 articles
The TEOSINTE BRANCHED1 (TBI1), CYCLOIDEA (CYC), and PROLIFERATING CELL NUCLEAR ANTIGEN FACTORS (PCF1 and PCF2) proteins truncated as TCP transcription factors carry conserved basic-helix-loop-helix (bHLH) structure, related to DNA binding functions. Evolutionary history of the TCP genes has shown their presence in early land plants. In this paper, we performed a comparative discussion on the current knowledge of the TCP Transcription Factors in lower and higher plants: their evolutionary history based on the phylogenetics of 849 TCP proteins from 37 plant species, duplication events, and biochemical roles in some of the plants species. Phylogenetics investigations confirmed the classification of TCP TFs into Class I (the PCF1/2), and Class II (the C- clade) factors; the Class II factors were further divided into the CIN- and CYC/TB1- subclade. A trace in the evolution of the TCP Factors revealed an absence of the CYC/TB1subclade in lower plants, and an independent evolution of the CYC/TB1subclade in both eudicot and monocot species. 54% of the total duplication events analyzed were biased towards the dispersed duplication, and we concluded that dispersed duplication events contributed to the expansion of the TCP gene family. Analysis in the TCP factors functional roles confirmed their involvement in various biochemical processes which mainly included promoting cell proliferation in leaves in Class I TCPs, and cell division during plant development in Class II TCP Factors. Apart from growth and development, the TCP Factors were also shown to regulate hormonal and stress response pathways. Although this paper does not exhaust the present knowledge of the TCP Transcription Factors, it provides a base for further exploration of the gene family.
Gene family size is a variable across different plant species, exhibiting essential functional roles for adaptation, growth and development, and speciation (Templeton, 2008). Protein classification and their coding genes involves two main approaches namely: i) generating varying family size and membership in profile-based protein databases such as the Hidden Markov Models (HMM), Pfam, InterPro, and SUPERFAMILY, and ii) categorizing these gene families based on parameter-based clustering of pairwise alignments. Gene duplication events (polyploidy), deletion, and the creation of new genes among other factors contribute to the expansion of gene family size (Flagel and Wendel, 2009; Magadum et al., 2013). As a result, genome size varies in individual plant species. The advent of gene transcriptomics has enabled the identification, study, characterization, and manipulation of numerous plant genomes and gene families (Chen et al., 2019; Li et al., 2022a; 2022b). Characteristic investigation of gene families contributes to the understanding of evolutionary relationship and functional differences (Li M. et al., 2022).
Among characterized gene families, the TEOSINTE BRANCHED1/CYCLOIDEA/-PROLIFERATING CELL FACTOR1 (TCP) gene family controls growth and development in plants; named from four unrelated proteins exhibiting diverse roles, and was first described in 1999, as a small group of plant genes encoding proteins sharing the TCP domain (Zhu, 2020). The cycloidea (CYC) from Antirrhinum majus, controls the floral lateral bilateral symmetry through genes differentially acting along the dorsoventral axis of the flower (Luo et al., 1996; Crawford et al., 2004; Busch et al., 2019). The Teosinte branched 1 (TB1) in Zea mays, encodes a protein with homology to the cycloidea gene of snapdragon (Doebley et al., 1997; Lukens and Doebley, 2001). Research has shown the CYC/TB1 genes to regulate apical dominance, repressing the growth of axillary organs, and enabling the formation of female inflorescences (Meshi and Iwabuchi, 1995; Yang et al., 2016). Lastly, the Proliferating Cell Factors 1 and 2 (PCF1/2) from Oryza sativa binds to the promoter region in the Proliferating Cell Nuclear Antigen (PCNA) gene (Kosugi and Ohashi, 1997). The PCF 1 and 2 are involved in the meristematic tissue-specific expression of rice PCNA gene through binding to the sites IIa and IIb, leading to the formation of either homodimer or heterodimers (Kosugi and Ohashi, 2002). The afore-mentioned genes: TB1, CYC, and PCFs are known as the TCP transcription factors (TCP TFs), characterized by the presence of a TCP domain, a 59 amino acid expanse forming a non-canonical basic-helix-loop-helix (bHLH) structure (Cubas et al., 1999). Although their ancestry remains unknown (Navaud et al., 2007), their biological roles and mode of action are conserved in plant species from the bryophytes to the angiosperms. Up to date, there have been several breakthroughs in the identification and computation of TCP TFs in numerous plant species including: A. thaliana (Aggarwal et al., 2010), G. biloba (Yu L. et al., 2022), P. edulis (Liu et al., 2018), S. lycopersicum (Parapunova et al., 2014), L. chinense (Hwarari et al., 2022), etc. However, in some plant orders, the TCP TFs have not yet been identified (Manassero et al., 2013). Research on the TCP gene expression have supported the biological functions of TB1, CYC, and PCF genes, tailoring the TCP domain to be involved in DNA binding activities, dimerization, and protein to protein interactions (Kosugi and Ohashi, 2002). Recent reports have shown their involvement in the regulation of biotic and abiotic stresses (Yu Z. et al., 2022; Hwarari et al., 2022).
Although the comprehension of the biochemical roles and evolution of the TCP TFs has improved in the past decade, there are still some gaps. In this article, we compared and discussed the current knowledge on the classification of TCP TFs in 37 plant species from lower plants to higher plants. We believe this paper will contribute valuable insights to the TCP gene family knowledge base. Additionally, we utilized available genomic data from current and previous research in TCP phylogenetic and evolution analysis to answer some important questions regarding the TCP gene family.
The TCP domain is highly conserved throughout plant species, forming a bHLH secondary structure, comprising of approximately 58–62 amino acid residues involved in: DNA binding, protein to protein interactions, and protein nuclear localization. The divergent evolution of the TCP domain from the bHLH domain was by insertion of a short stretch in the basic region thereby splitting the long helix into two (Aggarwal et al., 2010). Nonetheless, the TCP domain structure has remained intact (Kosugi and Ohashi, 1997; Cubas et al., 1999). The protein structural analysis of the bHLH TFs has shown that the TCP domain is characterized by a basic residue-rich region forming a typical fold of 3 short β -strands (β1, β2, and β3) and two consecutive α-helices (α-1 and α-2) (Sun et al., 2020). However, the TCP domain is different to the bHLH domain (Carrara and Dornelas, 2021). Recent demonstrations have exhibited that the topology of the TCP domain is different from the typical bHLH structure by comparing the ß-strand conformation of the basic region in a typical TCP domain with that of a typical bHLH protein (MyoD, PDB:1mdy), and concluded that the bHLH domain of TCP conforms to a new topology distinct from a typical bHLH structure. Interestingly, their analysis in rice OsPCF6 protein, disclosed that the TCP domain dimerize with other two TCP domains, each forming a stable conformation that adopts the ribbon-helix-helix (RHH) fold rather than the bHLH motif previously predicted. Implying that the TCP protein can also be classified into the RHH family (Liu et al., 2019; Sun et al., 2020). Although these findings are not conclusive, the homology modelling of TCP protein has also demonstrated their ability to form homodimers and/or heterodimers with other TCP proteins to bind DNA (Parapunova et al., 2014).
To confirm these findings, we compared 3D protein structures of TCP proteins from 11 plants species against a single bHLH protein representative from A. thaliana (Figure 1). Protein structure analysis confirmed the presence of 3β-strands and 2α-helices in TCP domain, and 2β-strands and 2α-helices in the bHLH protein structure. The first and the second helices of the TCP domain were amphipathic with alternating hydrophobic and hydrophilic residues. Our analysis concurred with previous TCP protein structure investigations (Pilar Cubas et al., 1999). Recently, we have also analysed the TCP protein structure of the L. chinense, and showed that most of the LcTCP proteins carry 2 to 3β-strands, and 2α-helices with alternating hydrophobic regions and are less mobile (Hwarari et al., 2022). Other researches have also exhibited that the TCP protein contain potential sites of phosphorylation and regions linked by a conserved Glycine-Aspartate-Serine residues, highly frequent in loops and Proline (Cubas et al., 1999; Tarczewska and Greb-Markiewicz, 2019; Edwards and Gorelick, 2022).This result generally confirms that the TCP bHLH domain is rather distinct from the bHLH domain.
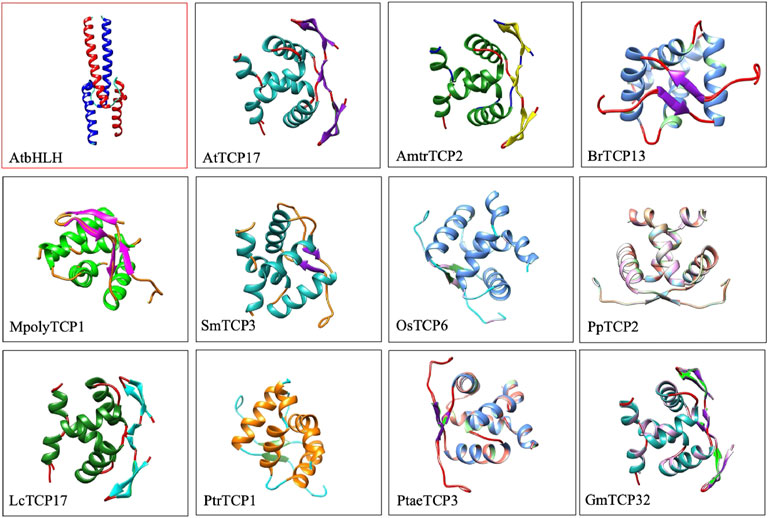
FIGURE 1. 3D protein structure comparison of Arabidopsis bHLH3 against other 11 TCP proteins. Specific names of the TCP protein are highlighted below the image. 3D protein structures were searched against the online PDB database (https://www.rcsb.org/) and viewed using the chimera software.
The Class I TCP TFs referred to as PCF1 and PCF2, are essential for DNA binding and dimerization, they also carry the TCP domain, and are characterized by two highly conserved sequences, DRHxK and RxRRxR, in the N- and C- terminal, respectively (Figure 2) (Liu et al., 2019). Although, some class II TCP proteins lack the conserved N-terminal part in the basic region, such as the S. lycopersicum TCP26 (Parapunova et al., 2014), B. rapa TCP12a/TCP1c (Du et al., 2017). The main distinction between Class I and Class II is that, Class I has a four-amino acid deletion within the TCP domain which is absent in Class II. Deeper analysis has shown a full conservation of Class I TCP amino acids within the lower plants as compared to the higher plants (Horn et al., 2015). To confirm these findings, we constructed TCP domain logo for class I and II using the protein alignments of the TCP domain (Figures 2A,B). The TCP domain comparisons evidenced the presence of 4 amino acid deletion in angiosperms which was absent in the lower plants TCP Class I. Suggesting that the lower plants are the first forms of life or rather the extend of evolution was different between the two plant clades (Qin et al., 2021). Other researchers have also shown that the Class I TCP domain is flanked by short regions recognizing a 6–10 base pair binding sequence of GGNCCC or CCNNCC, which is absent in the Class II TCP domain (Kosugi and Ohashi, 2002). In contrast, the GGNCCC has also been shown in PCF5 of rice, a Class II member (Liu et al., 2020), which has led to conclusions that these genes share a core sequence, the GGNCCC, and they have differing flanking sequences leading to either competition or cooperation (Savadel et al., 2021).
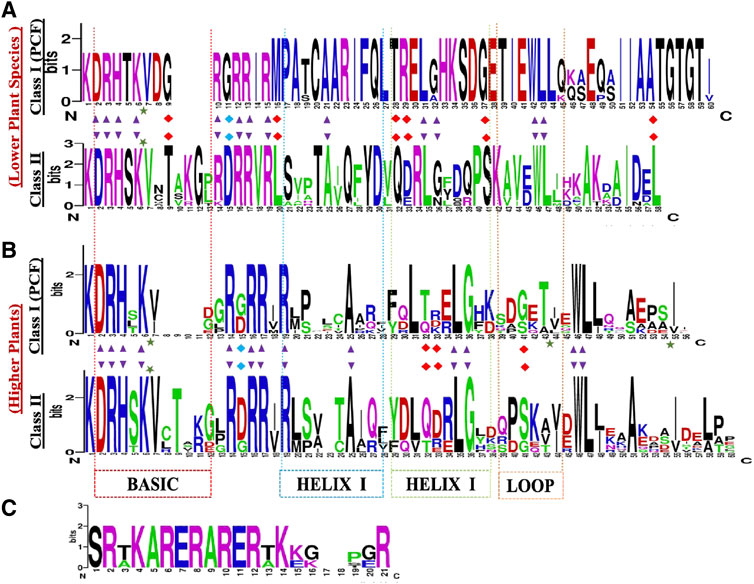
FIGURE 2. TCP protein sequence domain logos in: (A) lower plants (B) higher plants, (C) The angiosperm specific R-domain, present in the CYC/TB1 subclade. The sequence logos were generated by WebLogo online tool (http://weblogo.berkeley.edu/), based on the alignments of the TCP domains. The overall height of each stack letter indicates the sequence conservation at that position (measured in bits), whereas the height of the symbols within the stack reflects the relative frequency of the corresponding amino acid at that position. The black arrow depicts 90% conserved loci within the whole family. The red arrow depicts the key DNA binding site for the two subgroups. Top logo shows the TCP domain conserved within the lower plants. Bottom logo shows the TCP domain the higher plants.
The TCP Class II-clade, otherwise known as the CIN clade, was first isolated from snapdragon cin-mutant producing abnormal leaves and petals with rolled edges (Lan and Qin, 2020). Distinctively, the CIN protein carries conserved residues which are exclusive to the PCF proteins. In comparison, the Class II TCP domain has less conservation, studies have shown that the HLH region is 90% conserved, carrying Alanine (A)-25, Leucine (L)-35, (G)-36, Tryptophan (W)-46, and L-47. Additionally, there are notable distinctions in HELIX II, the first L residue has been replaced by Isoleucine (I) and Valine (V). The third L residue has also been replaced by an I residue (Martín-Trillo and Cubas, 2010; Liu et al., 2019). In total, our TCP domain comparisons, showed that lower plants Class II TCP domain is fully conserved, and exhibit significant differences in the protein sequence arrangements (Figure 2) in both the lower and higher plants.
The CYC/TB1 factors are a subdivision of Class II TCP proteins, and are angiosperm-specific. Protein structural studies of the CYC and TB1 genes have shown that they both have a 21 residue long basic region that includes a putative bi-partite nuclear localisation signal (NSL). In addition, they are characterized by the presence of an angiosperm conserved 18–20 amino acid Arginine-rich motif (the R-domain) (Figure 2C). Although, a few CIN-like proteins have also been shown to carry the R-domain (Wang et al., 2021). The R-domain forms an α-helix structure that coils similarly to leucine zippers which functions in protein-protein interaction (PPIs) mediation, and in evolutionary/developmental and phylogenetic studies (Busch and Sassone-Corsi, 1990). It is predicted to have originated independently in two separate clades, one of which is the ECE clade. The ECE denotes a conserved motif (Glu-Cys-Glu) between the TCP- and R-domain, found in most member of this clade (Smith et al., 2004).
System classification of the TCP gene family based on the molecular phylogeny facilitates the building of functional and genomic studies (Mondragón-Palomino and Trontin, 2011). 849 TCP proteins from 37 plant species were analyzed using the Parsimony, Maximum Likelihood (ML) and the Bayesian method. Results were consistent with corresponding values. Figure 3 shows the protein ML phylogenetic tree, our analyses concurred with previous findings that the TCP gene family can be divided into three main groups: the PCF, CIN, and the CYC/TB1 (Citerne et al., 2003). Likewise, the Bayesian inference methods have sorted TCP sequences in groups of high similarity, the class I (PCF1/2 clade) and class II (CIN and CYC/TB1) (Manassero et al., 2013; Liu et al., 2019; Yu L. et al., 2022). Nonetheless, distinctions have not been made whether the class I or class II TCP subfamily was the first to appear in plant kingdom due to the fact that lower plants believed to be first forms of plant life like Marchantia polymorpha carry both classes (Sharma et al., 2013). Some predictions have displayed the CIN-like TCP sub-clade in the Class II, to be more ancestral than the CYC/TB1-like TCPs since the Class II TCPs belong to the CIN-like TCP sub-clade in the non-vascular plants (Wang J. et al., 2022).
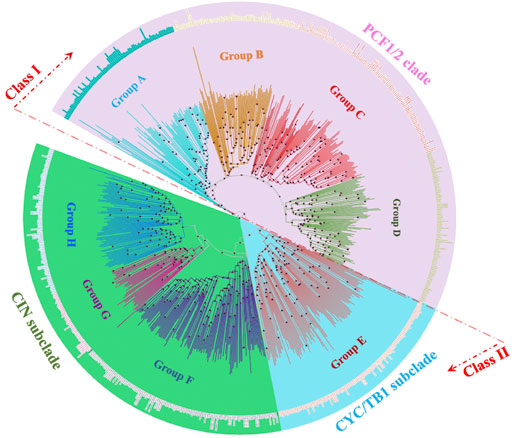
FIGURE 3. Evolutionary relationships of TCP subgroups in 28 plant species. An unrooted neighbour-joining tree was constructed with the trimmed multiple sequence alignment of MEGA 11. The phylogeny was constructed using 1,000 bootstrap replication value, bootstrap values of less than 70% are indicated by red circles on tree branches. Groups A-H were designated according to ancestral relationship of different branches and their clockwise appearance within each tree. Groups are distinguished by different branch color schemes.
Further classifications of the TCP sequences have been shown within the phylogenetic analyses into smaller groups of high sequence similarity and possibly biological functionality. In this analysis, we divided the phylogram into eight subgroups (A-H). The PCF-clade comprised group A to D, the CYC/TB1 proteins clustered in a monophyletic group (Group E), and the CIN clade was carried in the groups F to G (Figure 4B). Group A in PCF clade had the highest number of proteins while group G in the CIN-clade had the least number of proteins. Other studies have also shown divisions of 8–10 groups depending on: the total number of sequences included in the phylogenetic research, TCP protein clustering on the same branch, and sequence structures both within and outside the TCP domain (Wang et al., 2018). We also noted that some of the plant species were fully represented in individual groups, such as G. max and P. trichocarpa, suggesting that they have undergone various gene expansion and duplications types, and also that their proteins are involved in a wide range of biological functions (Ling et al., 2020; Wang J.-L. et al., 2022). In addition, research has also shown that the TCP monocot clade can be organized into at least 20 groups, each with sequences from different species. These sequences sharing amino acid motifs extending to the TCP and carboxyl domains, and an average identity greater than 64% with the majority resembling well-supported clades of the phylogeny (Mondragón-Palomino and Trontin, 2011).
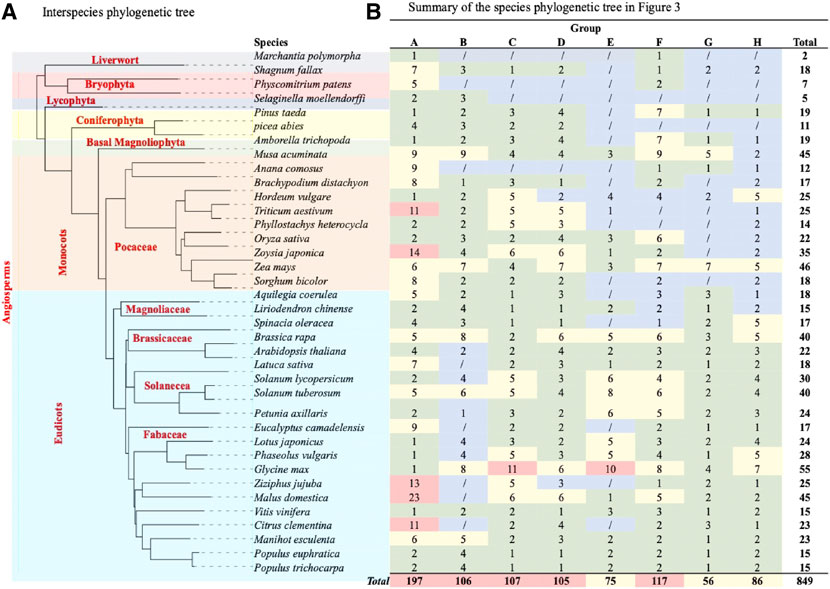
FIGURE 4. TCP phylogenetic analysis. (A) The interspecies phylogenetic tree constructed using Xshell ortholog finder, and the phylogenetic trees was constructed using online iTOL software. (B) Summary of TCP protein distribution within the phylogenetic tree shown in Figure 3. The analysed TCP proteins were grouped into eight groups (A–H) according to close ancestry and phylogeny relationships. Different color schemes represent increase in the total number of TCP proteins in specific plant species, with blue (0) to red (highest total number).
To fully understand evolution of the TCP gene family, we also analyzed the evolution of plant species in question (Figure 4A). Analysis showed that all these plants had a common ancestor, and evolutionarily events like speciation led to the formation of different plant clades and orders (Berger et al., 2016). However, monophyly of the liverwort (M.polymorpha) and bryophytes (S. fallax and P. patens) were shown to have diverged earlier, suggesting that these are amongst the earliest land plants (Fernandez-Pozo et al., 2022; Kumar et al., 2022). On the other hand, angiosperms diverged later into several different clades through speciation, these include the basal, magnoliid, eudicot and monocot angiosperms, possibly during the angiosperm evolution. These findings concurred with previous studies on the angiosperm species diversity and expansion (Qin et al., 2021; Hu et al., 2022).
Characterization of the TCP gene family in different plant species has yielded inconsistences in the total number of TCP proteins, motif arrangement and conserved domain structures. The highest and lowest total number of TCP family members are N. tabacum (61) (Chen et al., 2016) and S. officinarum (2) according to online plant transcription factor database (PTFD; http://planttfdb.gao-lab.org/) (Tian et al., 2019). TCP genes form small families in different species which have engendered larger members of angiosperms (Li X. et al., 2022). Genome-wide searches have indicated that the expansion of the TCP gene family is by independent gene or whole-genome duplication. Lower plants, P. patens, S. moellendorffii and M. polymorpha, have been branded with less total number of TCP genes and none in the unicellular algae (Chlamydomonas), this may be accounted for by the fact that angiosperms have a renowned history of WGDs driven form autopolyploid and allopolyploid events (Van de Peer et al., 2009; Li X. et al., 2022). The expansion of the monocot TCP-like genes was mainly through two rounds of whole genome duplication (WGD) (Mondragón-Palomino and Trontin, 2011). Several systematic analyses of the orthologous clades from B. distachyon, O. sativa, Z. mays and S. bicolor demonstrated that their common ancestor was formed by 21 genes. These findings were also supported by other WGD research in the angiosperm genome (Landis et al., 2018; Wang et al., 2020). Although, the TCP genes have been defined as evolutionary conserved plant transcription factors grouped according to similarity, differences among them have been related to the probability of insertion or loss of introns during evolution of the species. This phenomenon may suggest that functional diversity and expression control methods have involved more replication fragments, gene doubling, and other duplication events (Li et al., 2021).
Gene duplication events may take the form of segmental or single-gene duplications, involving: tandem, proximal, dispersed, and transposed duplications (Wang et al., 2012). Duplication event researches have shown that segmental duplications are the main driving force for expansion and evolution of the TCP gene family (Cannon et al., 2004; Wang et al., 2012). In support to these findings, research in Tartary buckwheat (Yang M. et al., 2022), and M. acuminata (Wang J. et al., 2022) have shown that the segmental duplication was responsible for the expansion of TCP gene family, and that the TCP gene family has undergone three WGDs during evolution. Therefore, to further understand the gene duplication events of TCP members in 17 different plant species, we computed for duplication event types using the plant duplicate gene database, PDGD (http://pdgd.njau.edu.cn8080) (Lee et al., 2012) (Figure 5). We observed that dispersed duplication event was the most prevalent duplication event constituting 54% of the total duplication events among the analyzed plant species, while tandem duplication events had the least prevalence of 1%. Analysis of duplicate gene pairs for each plant species showed higher percentages of dispersed duplication events in G. hirsutum. Tandem duplication events were only prevalent in M. domestica and G. hirsutum. Transposed duplication events were also noticed in all the plant species except in H. vulgare (Figure 5A). Therefore, we concluded that the dispersed duplication event contributed to a greater extend the expansion of the TCP gene family bringing about inconsistencies in the total number of TCP proteins within plant species (Figure 5B).
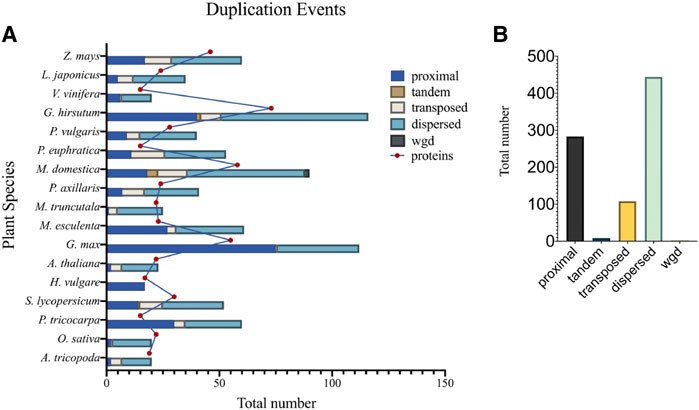
FIGURE 5. Duplication events different plant species. (A) Shows the number of duplication event types present in individual plant species, and (B) The total number of duplication events in investigated plant species. Different color backgrounds on the bar represent different gene duplication events. Also described in the key, top right corner.
In addition, the CYC/TB1clade has undergone several duplications and diversifications at the base of core eudicots, giving rise to three distinct clades: CYC1, CYC2, and CYC3 (Reeves and Olmstead, 2003; Smith et al., 2004). The CYC1 is more diverse than the other two copies, containing about 8% more sequence divergence in the TCP domain (Zhao et al., 2019), and it is sister clade to the CYC2 and CYC3 clades. While, the CYC3 clade is the only clade that does not contain additional duplications from other lineages. Notably, the CYC2 exhibits the greatest number of within-clade duplication, and contains CYCLOIDEA and its orthologs (Howarth and Donoghue, 2006). The TB1, a single copy from Zea mays is more similar to genes in the CYC1 clade, compared to CYC2 and CYC3 (Doebley et al., 1997).
The miRNAs control gene expression by binding to the target messenger RNA (mRNA), studies have shown the miR319 among other mRNAs to bind the TCP genes for effective regulation of their biological functions (Fang et al., 2021; Gao et al., 2022). Accumulating evidence has revealed the role of miR319-regulated TCPs (MRTCPs) in various biological pathways controlling growth and development and abiotic stress regulations (Fang et al., 2021). In A. thaliana the CIN-like TCP clade comprise eight members, divided into two clades based on the presence of microRNA (miRNA) binding sites outside the TCP domain. miRNA binding sites exist in TCP2, TCP3, TCP4, TCP10, and TCP24 and are post-transcriptionally regulated by miR319. On the other hand, a small clade called the TCP5-like CIN-TCPS is formed by TCP5, TCP13, and TCP17, and it is critical in plant thermophogenesis (Han et al., 2019). Nonetheless, other miRNAs have been shown to bind TCP genes, a total of nine microRNAs have been shown to regulate twenty TCP genes in three Apiaceae species with miR319 having most target genes targeting 11 TCP genes, miR172 and miR181 targeting 3 TCP genes each. Thereby, evidencing that the miRNAs target TCP genes to execute their biological functions (Pei et al., 2021). Studies in sweet potato have identified 4 IbTCP genes containing miR319-bindibg sites, further investigation have confirmed that IbmiR319 plays a crucial role in leaf anatomical morphology, and inhibits the expression levels of IbTCP11/17 (Ren et al., 2021). The miR319 is also involved in modulating leaf morphogenesis and flowering, and the positive regulation of leaf senescence in Arabidopsis through the overexpressed ApTCP2 influencing the JA biosynthesis (Zhu et al., 2022). The miRNA319 target 3 TCP genes in C. nankingense (CnTCP2/4/14). Expression analysis in Arabidopsis transgenic confirmed that the CnTCP4 negatively regulates the cold stress by downregulating the cold-induced genes such as AtCBF1/2/3, AtCOR15A, and AtKIN1(Tian et al., 2022).
Similar to other transcription factors, the TCP factors have undergone considerable evolutionary measures and rearrangements that created novel protein biological functions (Bornberg-Bauer and Albà, 2013). Thus, they regulate several aspects of plant development including; whole plant stature, leaf morphogenesis and maturation, inflorescence stem growth and floral organ development (Chai et al., 2017). To have an insight of the potential biological roles of the TCP genes, we performed Gene ontology analysis using A. thaliana TCP protein sequences (Figure 6). We observed that many processes were assigned to the biological processes (BP), mainly involved in growth and development, concurring with previous findings (İlhan et al., 2018; Kiseleva et al., 2022). A fewer processes in molecular function (MF), also had significant fold changes, and most of them were involved in DNA binding, supporting previous findings that TCPs are involved in DNA binding (Kosugi and Ohashi, 2002; He et al., 2021). The least number of processes were assigned to the cellular component (CC) category, although significant fold changes were observed. We assumed that fewer TCPs were involved with cell and cytoplasm processes. In accordance with studies in Tartary buckwheat (Yang Q. et al., 2022).
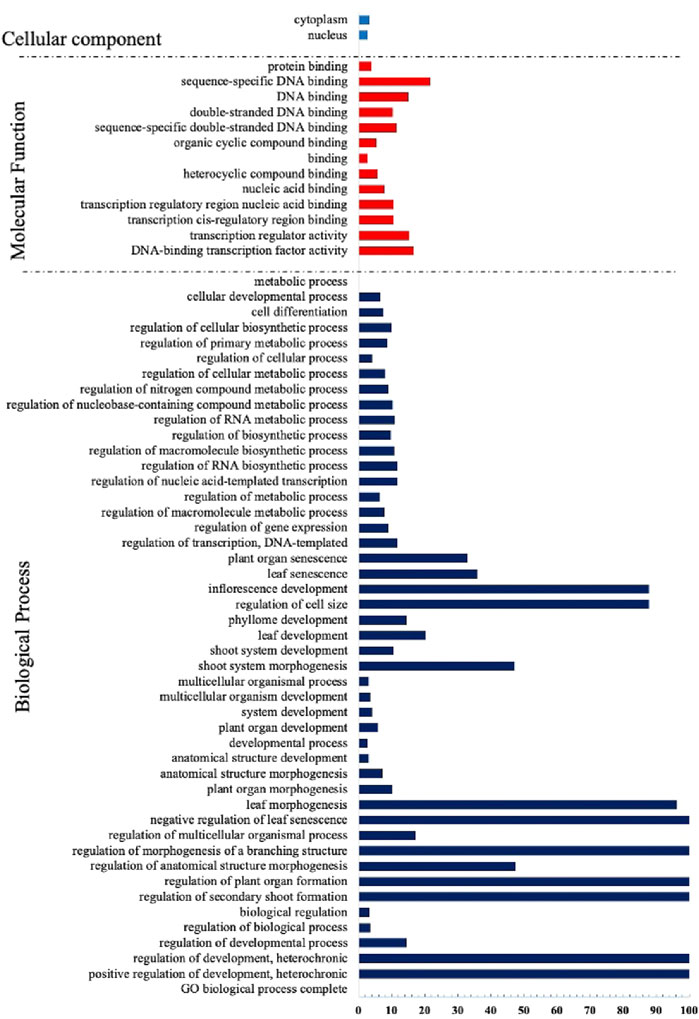
FIGURE 6. Gene Ontology Analysis of the TCP proteins in A. thaliana. GO terms were assigned using the online tool Gene Ontology Resources (http://geneontology.org/) from A. thaliana gene IDs.
Expression analysis of the Class I TCP genes in A. thaliana (AtTCP15/14/8/22) has demonstrated that the plants that lack functionality in Class I TCPs display poor stamen elongation. Suggesting that the Class I TCPs are involved in stamen elongation. For instance, AtTCP15 regulates the expression of the Small Auxin Up RNA 63 (SAUR63) gene family, which is involved in both petal and stamen development suggesting a AtTCP15- SAUR63 mode of action in the regulation of stamen development. Studies have also shown that AtTCP14 and AtTCP15 are required for optimal petiole and hypocotyl elongation during heat stress (Gastaldi et al., 2020; Ferrero et al., 2021). Overexpression analysis of Class I GbTCP, a homolog of AtTCP15 yielded fewer trichomes on the stem, smaller flowers, longer pedicel, and more buds. Interestingly, the overexpression of GbTCP in A. thaliana promoted root hair initiation and elongation. Further analysis demonstrated that GbTCP regulates the biosynthesis of jasmonates (LOX4, AOS, AOC3, and OPCL1) and other phytohormones including Ethylene and Auxin Response genes (Hao et al., 2012).
The Class I TCP transcription factors are also involved in abiotic stress regulation. The PeTCP10 in P. edulis was recently shown to alleviate salt stress, and reduce cellular damage in A. thaliana. Additional biochemical analysis exhibited that overexpressed PeTCP10 increases chlorophyll content, improves antioxidant capacity, and reduces water loss. Implying that the PeTCP10 might regulate salt stress through the CBL/SCaBP-CIPK/PKS pathway. Additionally, PeTCP10 was also shown to repress seedling growth and seed germination under high saline conditions (Pandey et al., 2015; Xu et al., 2021). Previous research has also heralded similar pathway in salt stress regulation by AtMYB49 (Zhang et al., 2020), and OsTCP19 in the AB14-mediated pathway (Mukhopadhyay and Tyagi, 2015). Recently, the CmTCP9 from C. morifolium expressed in A. thaliana has enhanced the enlargement of leaves and petals, and shown to interact with multiple hormone pathway protein, such as the GA3ox4, a key enzyme in the biosynthesis of gibberellin (Yu et al.).Furthermore, the class I TCPs are involved in plant biotic stress defence. The expression patterns of StTCP23 from Potato were analysed in susceptible variety inoculated with pathogen Streptomyces turgidiscabies. Their results prompted a suggestion that StTCP23 decrease the pathogenicity of bacterial scab in Potato (Bao et al., 2019). The PIF4 transcription factors bind to the GA20ox1 and the growth regulator genes, HB1 and PRE6, for gibberellin biosynthesis (Filo et al., 2015). AtTCP14 and AtTCP15 were shown to significantly play a role in hormonal biosynthesis, by directly participating in the induction of HB1 and PRE6 and targeting the same genes targeted by PIF4, enhancing PIF4 binding affinity to growth regulator genes (Ferrero et al., 2019).
The TCP CIN-clade is involved in cell elongation in the hypocotyl (Rath et al., 2022). In A. thaliana TCP4 and TCP24 suppress photomorphogenesis as compensatory measures to promote hypocotyl elongation. Analysis in the Chirita heterotricha CYC homologous genes, ChCYC1C and ChCYC1D has led to the conclusion that the ChRAD genes contain promoter binding sites for the CYC genes. Directly targeting the ChCYC1 genes, and enhancing the floral dorsoventral asymmetry (zygomorphy) in C. heterotricha and other core eudicots (Yang et al., 2010; Rath et al., 2022). In T.fournieri, TfTCP8 and TfTCP13 genes were demonstrated to reduce the leaf breadth, affirming previous research that ectopic overexpression of AtTCP15 a homologous gene to TfTCP8 narrows leaf size (Huang and Irish, 2015; Zhang et al., 2021). The OsPCF7 in rice has been shown to have a great relationship with rice tiller and heading. Comparative expression analysis between transgenic rice carrying the OsPCF7 gene with the wild seedlings evidenced its functional roles in increasing shoot height, root length, and grain yield. Deeper analysis in the mode of function demonstrated that the OsPCF7 increased the expression of downstream genes, the Class I KNOX genes such as: STM, KNAT2, KNAT6, LOX2, AS1 and indoleacetic acid-induced protein3 (IAA3) (Li et al., 2020).
Comparative expression analysis of the CIN-clade in Antirrhinum majus between wild type and the CIN-mutant genes, has shown that the CIN-clade inhibits excess cell proliferation, maintaining the leaf surface flatness by regulating its phytohormonal pathways. Molecular analysis revealed that CIN-clade TCPs directly bind to genomic regions promoting the transcription of cytokinin receptor homolog HISTININE KINASE 4 (AmHK4) and INDOLE-3-ACETIC ACID INDUCIBLE3/SHORT HYPOCOTYL 2 (IAA3/SHY2) (Das Gupta et al., 2014). Ectopic expression of G. raimondii TCP11 in A. thaliana has shown that GrTCp11 suppresses JA and Ethylene biosynthesis pathways thereby, reducing root hair elongation; through targeting genes that are directly associated with AtLOX4, AtAOC3, AtJAZ1, AtJAZ2, and AtMYC2 (Hao et al., 2021).
The TB1 subfamily regulates branching in various plant species (Aguilar-Martínez et al., 2007). Expression analysis of G. hirsutum TCP62 have proved these findings, and showed a high enrichment in the auxiliary buds and phyllophores. Nonetheless, overexpression of GhTCP62 in A. thaliana has shown a negative regulation of total number of shoots and reduced growth vigor (Liu J. et al., 2021). The Branched and Indeterminate spikelet 1 (BDI1), which encodes a TCP transcription factor, and highly conserved in both wild and cultivated barley has been shown to play a crucial role in determining barley inflorescence architecture and spikelet development; through regulating the gene transcription of cell wall modification and known Trehalose-6-phosphate homeostasis (Shang et al., 2020). The COMPUSITUM1 (COM1) in Class II CYC/TB1 subclade, working independent of the COM2 has been shown to inhibit spike-branching through boundary defined signals linked to the SM identity pathway, VRS4(HRA2)—COM1- HvLG1 (Poursarebani et al., 2020). Ranunculales, a sister order to three eudicots, P. somniferum, E. californica, and C. vesicaria, has been shown to play a crucial role with a wide diversity in developmental traits through the expression of CYL genes (Zhao et al., 2018). Although, the mode of action still requires further research. The overexpression of V. vaccinium TCP18 has been demonstrated to significantly decrease seed germination which can be alleviated by stratification and low temperature regulation, through a negative feedback loop. VvTCP18 is downregulated by low temperatures further preventing its binding to the FT, thereby retaining the normal function of the FT (Maurya et al., 2020; Li et al., 2021). Additional regulatory roles of theTCP gene members from various plant species are summarized in Table 1.
Characterization and expression research of the TCP TFs has progressed quite well in the past decades, and has improved the understanding of the TCP gene family. In this article, we summarized recent findings and answered a few questions in regard to the TCP gene family through phylogenetic and duplication analysis. TCP factors have been classified into the PCF-, CIN-, and CYC/TB1 clades. This diversity has brought a wide range of biological functionality in hormonal, growth and development, biotic and abiotic, and other numerous biological processes. Implying a crucial role within the TCP gene family. Biomolecular studies have also revealed the basis of functionality of the TCPs, which is the bHLH domain responsible for DNA-binding and protein to protein interaction. Suggesting that TCPs can bind to other proteins or DNA to effectively perform their biological roles. Although, the TCP TFs may carry similar TCP domain in different plant species, studies have also revealed inconsistencies in the TCP gene family size amongst plant species. This phenomenon can be related to duplication and deletion events of plant genome that contribute to the expansion of gene families. Several duplication events have been discussed and shown to have contributed to the overall expansion of the gene family notably, here we concluded that the dispersed duplication event contributed to a greater extend in the investigated plant species. Evolution can also be accounted for in plant diversity, we investigated the evolution of TCP genes, basically we noted that the PCF-clade was fully conserved in bryophytes, lycophytes and liverworts as compared to the angiosperms, and that lower plants lacks the R-domain in the CYC/TB1 subclade. This we related to the fact that the angiosperm evolution brought about increased speciation and probably deletions within the TCP conserved motifs and domain. Nonetheless, TCP genes in other plants are still yet to be characterized and their substantial functional roles elucidated.
Future characterizations of TCP gene family should provide resources for plant genetic improvements, offer directions for practical use, and fully disclose the regulatory mechanism by which the TCP genes control abiotic stress response, and growth and development through genetic transformation or gene editing (Min et al., 2022). In addition, future TCP protein to protein network studies should map different pathways that interact with the other proteins. In conclusion, cumulative knowledge gained from these summarized studies will generate novel morphologies of agronomic interests and help bio-engineer enhanced resistant plants to environmental stress and pathogens.
HZ and DH took part in confirming the topic and goals of this study, searching and screening the literatures, extracting the data, assessing the quality of literatures and writing the manuscript. HM and HX reviewed and edited the manuscript, and provided software for analyses. LY and YL sourced funding and supervised the progress of this manuscript.
The work was funded by Open Project of Jiangsu Key Laboratory for Eco-Agricultural Biotechnology around Hongze Lake (HZHLAB2001), and the Natural Science Foundation of Jiangsu Provincial Department of Education (17KJA180002 and 19KJB180011).
The authors would also like to thank reviewers and editors for their valuable effort in reading and commenting on this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aggarwal, P., Das Gupta, M., Joseph, A. P., Chatterjee, N., Srinivasan, N., and Nath, U. (2010). Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22 (4), 1174–1189. doi:10.1105/tpc.109.066647
Aguilar-Martínez, J. A., Poza-Carrión, C., and Cubas, P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19 (2), 458–472. doi:10.1105/tpc.106.048934
Bao, S., Owens, R. A., Sun, Q., Song, H., Liu, Y., Eamens, A. L., et al. (2019). Silencing of transcription factor encoding gene StTCP23 by small RNAs derived from the virulence modulating region of potato spindle tuber viroid is associated with symptom development in potato. PLoS Pathog. 15 (12), e1008110. doi:10.1371/journal.ppat.1008110
Berger, B. A., Kriebel, R., Spalink, D., and Sytsma, K. J. (2016). Divergence times, historical biogeography, and shifts in speciation rates of Myrtales. Mol. Phylogenet. Evol. 95, 116–136. doi:10.1016/j.ympev.2015.10.001
Bornberg-Bauer, E., and Albà, M. M. (2013). Dynamics and adaptive benefits of modular protein evolution. Curr. Opin. Struct. Biol. 23 (3), 459–466. doi:10.1016/j.sbi.2013.02.012
Busch, A., Deckena, M., Almeida-Trapp, M., Kopischke, S., Kock, C., Schüssler, E., et al. (2019). Mp TCP 1 controls cell proliferation and redox processes in Marchantia polymorpha. New Phytol. 224 (4), 1627–1641. doi:10.1111/nph.16132
Busch, S. J., and Sassone-Corsi, P. (1990). Dimers, leucine zippers and DNA-binding domains. Trends Genet. 6, 36–40. doi:10.1016/0168-9525(90)90071-d
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi:10.1186/1471-2229-4-10
Carrara, S., and Dornelas, M. C. (2021). TCP genes and the orchestration of plant architecture. Trop. Plant Biol. 14 (1), 1–10. doi:10.1007/s12042-020-09274-z
Chai, W., Jiang, P., Huang, G., Jiang, H., and Li, X. (2017). Identification and expression profiling analysis of TCP family genes involved in growth and development in maize. Physiol. Mol. Biol. Plants 23 (4), 779–791. doi:10.1007/s12298-017-0476-1
Chen, J., Hao, Z., Guang, X., Zhao, C., Wang, P., Xue, L., et al. (2019). Author Correction: Liriodendron genome sheds light on angiosperm phylogeny and species-pair differentiation. Nat. Plants 5 (3), 328. doi:10.1038/s41477-019-0368-1
Chen, L., Chen, Y., Ding, A., Chen, H., Xia, F., Wang, W., et al. (2016). Genome-wide analysis of TCP family in tobacco. Genet. Mol. Res. 15 (7728), 10–4238. doi:10.4238/gmr.15027728
Citerne, H. L., Luo, D., Pennington, R. T., Coen, E., and Cronk, Q. C. (2003). A phylogenomic investigation of CYCLOIDEA-like TCP genes in the Leguminosae. Plant Physiol. 131 (3), 1042–1053. doi:10.1104/pp.102.016311
Crawford, B. C., Nath, U., Carpenter, R., and Coen, E. S. (2004). CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol. 135 (1), 244–253. doi:10.1104/pp.103.036368
Cubas, P., Lauter, N., Doebley, J., and Coen, E. (1999). The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 18 (2), 215–222. doi:10.1046/j.1365-313x.1999.00444.x
Das Gupta, M., Aggarwal, P., and Nath, U. (2014). CINCINNATA in Antirrhinum majus directly modulates genes involved in cytokinin and auxin signaling. New Phytol. 204 (4), 901–912. doi:10.1111/nph.12963
Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386 (6624), 485–488. doi:10.1038/386485a0
Du, J., Hu, S., Yu, Q., Wang, C., Yang, Y., Sun, H., et al. (2017). Genome-wide identification and characterization of BrrTCP transcription factors in Brassica rapa ssp. rapa. Front. Plant Sci. 8, 1588. doi:10.3389/fpls.2017.01588
Edwards, H. E., and Gorelick, D. A. (2022). The evolution and structure/function of bHLH–PAS transcription factor family. Biochem. Soc. Trans. 50 (3), 1227–1243. doi:10.1042/BST20211225
Fang, Y., Zheng, Y., Lu, W., Li, J., Duan, Y., Zhang, S., et al. (2021). Roles of miR319-regulated TCPs in plant development and response to abiotic stress. Crop J. 9 (1), 17–28. doi:10.1016/j.cj.2020.07.007
Fernandez-Pozo, N., Haas, F. B., Gould, S. B., and Rensing, S. A. (2022). An overview of bioinformatics, genomics, and transcriptomics resources for bryophytes. J. Exp. Bot. 73 (13), 4291–4305. doi:10.1093/jxb/erac052
Ferrero, L. V., Gastaldi, V., Ariel, F. D., Viola, I. L., and Gonzalez, D. H. (2021). Class I TCP proteins TCP14 and TCP15 are required for elongation and gene expression responses to auxin. Plant Mol. Biol. 105 (1-2), 147–159. doi:10.1007/s11103-020-01075-y
Ferrero, V., Viola, I. L., Ariel, F. D., and Gonzalez, D. H. (2019). Class I TCP transcription factors target the gibberellin biosynthesis gene GA20ox1 and the growth-promoting genes HBI1 and PRE6 during thermomorphogenic growth in Arabidopsis. Plant Cell Physiol. 60 (8), 1633–1645. doi:10.1093/pcp/pcz137
Filo, J., Wu, A., Eliason, E., Richardson, T., Thines, B. C., and Harmon, F. G. (2015). Gibberellin driven growth in elf3 mutants requires PIF4 and PIF5. Plant Signal. Behav. 10 (3), e992707. doi:10.4161/15592324.2014.992707
Flagel, L. E., and Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183 (3), 557–564. doi:10.1111/j.1469-8137.2009.02923.x
Gao, Z., Ma, C., Zheng, C., Yao, Y., and Du, Y. (2022). Advances in the regulation of plant salt-stress tolerance by miRNA. Mol. Biol. Rep. 1-15, 5041–5055. doi:10.1007/s11033-022-07179-6
Gastaldi, V., Lucero, L. E., Ferrero, L. V., Ariel, F. D., and Gonzalez, D. H. (2020). Class-I TCP transcription factors activate the SAUR63 gene subfamily in gibberellin-dependent stamen filament elongation. Plant Physiol. 182 (4), 2096–2110. doi:10.1104/pp.19.01501
Han, X., Yu, H., Yuan, R., Yang, Y., An, F., and Qin, G. (2019). Arabidopsis transcription factor TCP5 controls plant thermomorphogenesis by positively regulating PIF4 activity. Iscience 15, 611–622. doi:10.1016/j.isci.2019.04.005
Hao, J., Lou, P., Han, Y., Chen, Z., Chen, J., Ni, J., et al. (2021). GrTCP11, a cotton TCP transcription factor, inhibits root hair elongation by down-regulating jasmonic acid pathway in Arabidopsis thaliana. Front. Plant Sci. 12, 769675. doi:10.3389/fpls.2021.769675
Hao, J., Tu, L., Hu, H., Tan, J., Deng, F., Tang, W., et al. (2012). GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J. Exp. Bot. 63 (17), 6267–6281. doi:10.1093/jxb/ers278
He, Z., Zhou, X., Chen, J., Yin, L., Zeng, Z., Xiang, J., et al. (2021). Identification of a consensus DNA-binding site for the TCP domain transcription factor TCP2 and its important roles in the growth and development of Arabidopsis. Mol. Biol. Rep. 48 (3), 2223–2233. doi:10.1007/s11033-021-06233-z
Horn, S., Pabón-Mora, N., Theuß, V. S., Busch, A., and Zachgo, S. (2015). Analysis of the CYC/TB 1 class of TCP transcription factors in basal angiosperms and magnoliids. Plant J. 81 (4), 559–571. doi:10.1111/tpj.12750
Howarth, D. G., and Donoghue, M. J. (2006). Phylogenetic analysis of the "ECE" (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. U. S. A. 103 (24), 9101–9106. doi:10.1073/pnas.0602827103
Hu, H., Ye, J., Liu, B., Mao, L., Smith, S. A., Barrett, R. L., et al. (2022). Temporal and spatial comparisons of angiosperm diversity between eastern Asia and North America. Natl. Sci. Rev. 9 (6), nwab199. doi:10.1093/nsr/nwab199
Huang, T., and Irish, V. F. (2015). Temporal control of plant organ growth by TCP transcription factors. Curr. Biol. 25 (13), 1765–1770. doi:10.1016/j.cub.2015.05.024
Hwarari, D., Guan, Y., Li, R., Movahedi, A., Chen, J., and Yang, L. (2022). Comprehensive bioinformatics and expression analysis of TCP transcription factors in liriodendron chinense reveals putative abiotic stress regulatory roles. Forests 13 (9), 1401. doi:10.3390/f13091401
İlhan, E., Büyük, İ., and İnal, B. (2018). Transcriptome–Scale characterization of salt responsive bean TCP transcription factors. Gene 642, 64–73. doi:10.1016/j.gene.2017.11.021
Jin, K., Wang, Y., Zhuo, R., Xu, J., Lu, Z., Fan, H., et al. (2022). TCP transcription factors involved in shoot development of ma bamboo (dendrocalamus latiflorus munro). Front. Plant Sci. 13, 884443. doi:10.3389/fpls.2022.884443
Kiseleva, A. A., Bragina, M. K., Muterko, A. F., and Salina, E. A. (2022). Functional characterization of genes with daily expression patterns in common wheat. Plant Mol. Biol. 109 (1), 135–146. doi:10.1007/s11103-022-01262-z
Kosugi, S., and Ohashi, Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30 (3), 337–348. doi:10.1046/j.1365-313x.2002.01294.x
Kosugi, S., and Ohashi, Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9 (9), 1607–1619. doi:10.1105/tpc.9.9.1607
Kumar, A., Giri, P., and Uniyal, P. L. (2022). Plastogenomics provides a tool to study phylogenetic interrelationships of monilophytes: A review. Ferns, 59–98. doi:10.1007/978-981-16-6170-9_4
Lan, J., and Qin, G. (2020). The regulation of CIN-like TCP transcription factors. Int. J. Mol. Sci. 21 (12), 4498. doi:10.3390/ijms21124498
Landis, J. B., Soltis, D. E., Li, Z., Marx, H. E., Barker, M. S., Tank, D. C., et al. (2018). Impact of whole-genome duplication events on diversification rates in angiosperms. Am. J. Bot. 105 (3), 348–363. doi:10.1002/ajb2.1060
Lee, T.-H., Tang, H., Wang, X., and Paterson, A. H. (2012). Pgdd: A database of gene and genome duplication in plants. Nucleic Acids Res. 41 (D1), D1152–D1158. doi:10.1093/nar/gks1104
Li, M., Hwarari, D., Li, Y., Ahmad, B., Min, T., Zhang, W., et al. (2022c). The bZIP transcription factors in liriodendron chinense: Genome-wide recognition, characteristics and cold stress response. Front. Plant Sci. 441. doi:10.3389/fpls.2022.1035627
Li, R., Ahmad, B., Hwarari, D., Li, D. A., Lu, Y., Gao, M., et al. (2022a). Genomic survey and cold-induced expression patterns of bHLH transcription factors in liriodendron chinense (hemsl) sarg. Forests 13 (4), 518. doi:10.3390/f13040518
Li, R., Radani, Y., Ahmad, B., Movahedi, A., and Yang, L. (2022b). Identification and characteristics of SnRK genes and cold stress-induced expression profiles in Liriodendron chinense. BMC genomics 23 (1), 708–718. doi:10.1186/s12864-022-08902-0
Li, W., Chen, G., Xiao, G., Zhu, S., Zhou, N., Zhu, P., et al. (2020). Overexpression of TCP transcription factor OsPCF7 improves agronomic trait in rice. Mol. Breed. 40 (5), 48–13. doi:10.1007/s11032-020-01129-5
Li, X., Cai, K., Han, Z., Zhang, S., Sun, A., Xie, Y., et al. (2022d). Chromosome-level genome assembly for acer pseudosieboldianum and highlights to mechanisms for leaf color and shape change. Front. Plant Sci. 13, 850054. doi:10.3389/fpls.2022.850054
Li, Y., An, S., Cheng, Q., Zong, Y., Chen, W., Guo, W., et al. (2021). Analysis of evolution, expression and genetic transformation of TCP transcription factors in blueberry reveal that VcTCP18 negatively regulates the release of flower bud dormancy. Front. Plant Sci. 1382, 697609. doi:10.3389/fpls.2021.697609
Ling, L., Zhang, W., An, Y., Du, B., Wang, D., and Guo, C. (2020). Genome-wide analysis of the TCP transcription factor genes in five legume genomes and their response to salt and drought stresses. Funct. Integr. Genomics 20 (4), 537–550. doi:10.1007/s10142-020-00733-0
Liu, H.-L., Wu, M., Li, F., Gao, Y.-M., Chen, F., and Xiang, Y. (2018). TCP transcription factors in moso bamboo (phyllostachys edulis): Genome-wide identification and expression analysis. Front. Plant Sci. 9, 1263. doi:10.3389/fpls.2018.01263
Liu, H., Gao, Y., Wu, M., Shi, Y., Wang, H., Wu, L., et al. (2020). TCP10, a TCP transcription factor in moso bamboo (Phyllostachys edulis), confers drought tolerance to transgenic plants. Environ. Exp. Bot. 172, 104002. doi:10.1016/j.envexpbot.2020.104002
Liu, J., Wang, J.-J., Wu, J., Wang, Y., Liu, Q., Liu, F.-P., et al. (2021a). An optimized transformation system and functional test of CYC-Like TCP gene CpCYC in Chirita pumila (Gesneriaceae). Int. J. Mol. Sci. 22 (9), 4544. doi:10.3390/ijms22094544
Liu, M. M., Wang, M. M., Yang, J., Wen, J., Guo, P. C., Wu, Y. W., et al. (2019). Evolutionary and comparative expression analyses of TCP transcription factor gene family in land plants. Int. J. Mol. Sci. 20 (14), E3591. doi:10.3390/ijms20143591
Liu, Z., Yang, J., Li, S., Liu, L., Qanmber, G., Chen, G., et al. (2021b). Systematic characterization of TCP gene family in four cotton species revealed that GhTCP62 regulates branching in Arabidopsis. Biology 10 (11), 1104. doi:10.3390/biology10111104
Lukens, L., and Doebley, J. (2001). Molecular evolution of the teosinte branched gene among maize and related grasses. Mol. Biol. Evol. 18 (4), 627–638. doi:10.1093/oxfordjournals.molbev.a003843
Luo, D., Carpenter, R., Vincent, C., Copsey, L., and Coen, E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383 (6603), 794–799. doi:10.1038/383794a0
Magadum, S., Banerjee, U., Murugan, P., Gangapur, D., and Ravikesavan, R. (2013). Gene duplication as a major force in evolution. J. Genet. 92 (1), 155–161. doi:10.1007/s12041-013-0212-8
Manassero, N. G. U., Viola, I. L., Welchen, E., and Gonzalez, D. H. (2013). TCP transcription factors: Architectures of plant form. Biomol. Concepts 4 (2), 111–127. doi:10.1515/bmc-2012-0051
Martín-Trillo, M., and Cubas, P. (2010). TCP genes: A family snapshot ten years later. Trends Plant Sci. 15 (1), 31–39. doi:10.1016/j.tplants.2009.11.003
Maurya, J. P., Singh, R. K., Miskolczi, P. C., Prasad, A. N., Jonsson, K., Wu, F., et al. (2020). Branching regulator BRC1 mediates photoperiodic control of seasonal growth in hybrid aspen. Curr. Biol. 30 (1), 122–126. e122. doi:10.1016/j.cub.2019.11.001
Meshi, T., and Iwabuchi, M. (1995). Plant transcription factors. Plant Cell Physiol. 36 (8), 1405–1420.
Min, T., Hwarari, D., Li, D. A., Movahedi, A., and Yang, L. (2022). CRISPR-based genome editing and its applications in woody plants. Int. J. Mol. Sci. 23 (17), 10175. doi:10.3390/ijms231710175
Mondragón-Palomino, M., and Trontin, C. (2011). High time for a roll call: Gene duplication and phylogenetic relationships of TCP-like genes in monocots. Ann. Bot. 107 (9), 1533–1544. doi:10.1093/aob/mcr059
Mukhopadhyay, P., and Tyagi, A. K. (2015). OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci. Rep. 5, 9998. doi:10.1038/srep09998
Navaud, O., Dabos, P., Carnus, E., Tremousaygue, D., and Hervé, C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65 (1), 23–33. doi:10.1007/s00239-006-0174-z
Pandey, G. K., Kanwar, P., Singh, A., Steinhorst, L., Pandey, A., Yadav, A. K., et al. (2015). Calcineurin B-like protein-interacting protein kinase CIPK21 regulates osmotic and salt stress responses in Arabidopsis. Plant Physiol. 169 (1), 780–792. doi:10.1104/pp.15.00623
Parapunova, V., Busscher, M., Busscher-Lange, J., Lammers, M., Karlova, R., Bovy, A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 14, 157. doi:10.1186/1471-2229-14-157
Pei, Q., Li, N., Bai, Y., Wu, T., Yang, Q., Yu, T., et al. (2021). Comparative analysis of the <i>TCP</i> gene family in celery, coriander and carrot (family Apiaceae). Veg. Res. 1 (1), 1–12. doi:10.48130/vr-2021-0005
Poursarebani, N., Trautewig, C., Melzer, M., Nussbaumer, T., Lundqvist, U., Rutten, T., et al. (2020). COMPOSITUM 1 contributes to the architectural simplification of barley inflorescence via meristem identity signals. Nat. Commun. 11 (1), 1–6. doi:10.1038/s41467-020-18890-y
Qin, L., Hu, Y., Wang, J., Wang, X., Zhao, R., Shan, H., et al. (2021). Insights into angiosperm evolution, floral development and chemical biosynthesis from the Aristolochia fimbriata genome. Nat. Plants 7 (9), 1239–1253. doi:10.1038/s41477-021-00990-2
Rath, M., Challa, K. R., Sarvepalli, K., and Nath, U. (2022). CINCINNATA-like TCP transcription factors in cell growth - an expanding portfolio. Front. Plant Sci. 13, 825341. doi:10.3389/fpls.2022.825341
Reeves, P. A., and Olmstead, R. G. (2003). Evolution of the TCP gene family in asteridae: Cladistic and network approaches to understanding regulatory gene family diversification and its impact on morphological evolution. Mol. Biol. Evol. 20 (12), 1997–2009. doi:10.1093/molbev/msg211
Ren, L., Wu, H., Zhang, T., Ge, X., Wang, T., Zhou, W., et al. (2021). Genome-wide identification of TCP transcription factors family in sweet potato reveals significant roles of miR319-targeted TCPs in leaf anatomical morphology. Front. Plant Sci. 12, 686698. doi:10.3389/fpls.2021.686698
Savadel, S. D., Hartwig, T., Turpin, Z. M., Vera, D. L., Lung, P.-Y., Sui, X., et al. (2021). The native cistrome and sequence motif families of the maize ear. PLoS Genet. 17 (8), e1009689. doi:10.1371/journal.pgen.1009689
Shang, Y., Yuan, L., Di, Z., Jia, Y., Zhang, Z., Li, S., et al. (2020). A CYC/TB1-type TCP transcription factor controls spikelet meristem identity in barley. J. Exp. Bot. 71 (22), 7118–7131. doi:10.1093/jxb/eraa416
Sharma, N., Bhalla, P. L., and Singh, M. B. (2013). Transcriptome-wide profiling and expression analysis of transcription factor families in a liverwort, Marchantia polymorpha. BMC Genomics 14 (1), 915. doi:10.1186/1471-2164-14-915
Smith, J. F., Hileman, L. C., Powell, M. P., and Baum, D. A. (2004). Evolution of GCYC, a gesneriaceae homolog of CYCLOIDEA, within gesnerioideae (gesneriaceae). Mol. Phylogenet. Evol. 31 (2), 765–779. doi:10.1016/j.ympev.2003.09.012
Sun, L., Zou, X., Jiang, M., Wu, X., Chen, Y., Wang, Q., et al. (2020). The crystal structure of the TCP domain of PCF6 in Oryza sativa L. reveals an RHH-like fold. FEBS Lett. 594 (8), 1296–1306. doi:10.1002/1873-3468.13727
Tarczewska, A., and Greb-Markiewicz, B. (2019). The significance of the intrinsically disordered regions for the functions of the bHLH transcription factors. Int. J. Mol. Sci. 20 (21), 5306. doi:10.3390/ijms20215306
Templeton, A. R. (2008). The reality and importance of founder speciation in evolution. BioEssays 30 (5), 470–479. doi:10.1002/bies.20745
Tian, C., Zhai, L., Zhu, W., Qi, X., Yu, Z., Wang, H., et al. (2022). Characterization of the TCP gene family in Chrysanthemum nankingense and the role of CnTCP4 in cold tolerance. Plants 11 (7), 936. doi:10.3390/plants11070936
Tian, F., Yang, D.-C., Meng, Y.-Q., Jin, J., and Gao, G. (2019). PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 48 (D1), D1104–D1113. doi:10.1093/nar/gkz1020
Van de Peer, Y., Fawcett, J. A., Proost, S., Sterck, L., and Vandepoele, K. (2009). The flowering world: A tale of duplications. Trends Plant Sci. 14 (12), 680–688. doi:10.1016/j.tplants.2009.09.001
Wang, C., Hao, X., Wang, Y., Shi, M., Zhou, Z. G., and Kai, G. (2021). Genome-wide identification and comparative analysis of the teosinte branched 1/cycloidea/proliferating cell factors 1/2 transcription factors related to anti-cancer drug camptothecin biosynthesis in ophiorrhiza pumila. Front. Plant Sci. 12, 746648. doi:10.3389/fpls.2021.746648
Wang, H., Wang, H., Liu, R., Xu, Y., Lu, Z., and Zhou, C. (2018). Genome-wide identification of TCP family transcription factors in medicago truncatula reveals significant roles of miR319-targeted TCPs in nodule development. Front. Plant Sci. 9, 774. doi:10.3389/fpls.2018.00774
Wang, J.-L., Wang, H.-W., Cao, Y.-N., Kan, S.-L., and Liu, Y.-Y. (2022b). Comprehensive evolutionary analysis of the TCP gene family: Further insights for its origin, expansion, and diversification. Front. Plant Sci. 13, 994567. doi:10.3389/fpls.2022.994567
Wang, J., Wang, Z., Jia, C., Miao, H., Zhang, J., Liu, J., et al. (2022a). Genome-wide identification and transcript analysis of TCP gene family in banana (musa acuminata L.). Biochem. Genet. 60 (1), 204–222. doi:10.1007/s10528-021-10100-8
Wang, L. B., Ma, H., and Lin, J. (2020). Preferential retention of the slowly evolving gene in pairs of duplicates in angiosperm genomes. J. Syst. Evol.
Wang, S., Shen, Y., Guo, L., Tan, L., Ye, X., Yang, Y., et al. (2022c). Innovation and emerging roles of Populus trichocarpa TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR transcription factors in abiotic stresses by whole-genome duplication. Front. Plant Sci. 13, 850064. doi:10.3389/fpls.2022.850064
Wang, Y., Wang, X., and Paterson, A. H. (2012). Genome and gene duplications and gene expression divergence: A view from plants. Ann. N. Y. Acad. Sci. 1256, 1–14. doi:10.1111/j.1749-6632.2011.06384.x
Xu, Y., Liu, H., Gao, Y., Xiong, R., Wu, M., Zhang, K., et al. (2021). The TCP transcription factor PeTCP10 modulates salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 40 (10), 1971–1987. doi:10.1007/s00299-021-02765-7
Yang, C. J., Kursel, L. E., Studer, A. J., Bartlett, M. E., Whipple, C. J., and Doebley, J. F. (2016). A gene for genetic background in Zea mays: Fine-mapping enhancer of teosinte branched1. 2 to a YABBY class transcription factor. Genetics 204 (4), 1573–1585. doi:10.1534/genetics.116.194928
Yang, M., He, G., Hou, Q., Fan, Y., Duan, L., Li, K., et al. (2022a). Systematic analysis and expression profiles of TCP gene family in Tartary buckwheat (Fagopyrum tataricum (L.) Gaertn.) revealed the potential function of FtTCP15 and FtTCP18 in response to abiotic stress. BMC Genomics 23 (1), 415. doi:10.1186/s12864-022-08618-1
Yang, Q., Li, Q., Gu, L., Chen, P., Zhang, Y., Li, Y., et al. (2022b). The jujube TCP transcription factor ZjTCP16 regulates plant growth and cell size by affecting the expression of genes involved in plant morphogenesis. Forests 13 (5), 723. doi:10.3390/f13050723
Yang, X., Cui, H., Yuan, Z. L., and Wang, Y. Z. (2010). Significance of consensus CYC-binding sites found in the promoters of both ChCYC and ChRAD genes in Chirita heterotricha (Gesneriaceae). J. Syst. Evol. 48 (4), 249–256. doi:10.1111/j.1759-6831.2010.00086.x
Yu, L., Chen, Q., Zheng, J., Xu, F., Ye, J., Zhang, W., et al. (2022a). Genome-wide identification and expression pattern analysis of the TCP transcription factor family in Ginkgo biloba. Plant Signal. Behav. 17 (1), 1994248. doi:10.1080/15592324.2021.1994248
Yu, Z., Tian, C., Guan, Y., He, J., Wang, Z., Wang, L., et al. (2022b). Expression analysis of TCP transcription factor family in autopolyploids of Chrysanthemum nankingense. Front. Plant Sci. 13, 860956. doi:10.3389/fpls.2022.860956
Zhang, L., Zhou, L., Yung, W. S., Su, W., and Huang, M. (2021). Ectopic expression of Torenia fournieri TCP8 and TCP13 alters the leaf and petal phenotypes in Arabidopsis thaliana. Physiol. Plant. 173 (3), 856–866. doi:10.1111/ppl.13479
Zhang, P., Wang, R., Yang, X., Ju, Q., Li, W., Lü, S., et al. (2020). The R2R3-MYB transcription factor AtMYB49 modulates salt tolerance in Arabidopsis by modulating the cuticle formation and antioxidant defence. Plant Cell Environ. 43 (8), 1925–1943. doi:10.1111/pce.13784
Zhao, M., Peng, X., Chen, N., and Shen, S. (2020). Genome-wide identification of the TCP gene family in broussonetia papyrifera and functional analysis of BpTCP8, 14 and 19 in shoot branching. Plants 9 (10), 1301. doi:10.3390/plants9101301
Zhao, Y., Pfannebecker, K., Dommes, A. B., Hidalgo, O., Becker, A., and Elomaa, P. (2018). Evolutionary diversification of CYC/TB1-like TCP homologs and their recruitment for the control of branching and floral morphology in Papaveraceae (basal eudicots). New Phytol. 220 (1), 317–331. doi:10.1111/nph.15289
Zhao, Z., Hu, J., Chen, S., Luo, Z., Luo, D., Wen, J., et al. (2019). Evolution of CYCLOIDEA-like genes in Fabales: Insights into duplication patterns and the control of floral symmetry. Mol. Phylogenet. Evol. 132, 81–89. doi:10.1016/j.ympev.2018.11.007
Zhu, L., Li, S., Ma, Q., Wen, J., Yan, K., and Li, Q. (2022). The acer palmatum TCP transcription factor ApTCP2 controls leaf morphogenesis, accelerates senescence, and affects flowering via miR319 in Arabidopsis thaliana. J. Plant Growth Regul. 41 (1), 244–256. doi:10.1007/s00344-021-10299-1
Keywords: TCP transcription factors, evolutionary relationship, TCP domain, basic-helix-loop-helix structure, phylogenetic analysis
Citation: Zhou H, Hwarari D, Ma H, Xu H, Yang L and Luo Y (2022) Genomic survey of TCP transcription factors in plants: Phylogenomics, evolution and their biology. Front. Genet. 13:1060546. doi: 10.3389/fgene.2022.1060546
Received: 03 October 2022; Accepted: 27 October 2022;
Published: 09 November 2022.
Edited by:
Xingwang Yu, North Carolina State University, United StatesReviewed by:
Assar Ali Shah, Jiangsu University, ChinaCopyright © 2022 Zhou, Hwarari, Ma, Xu, Yang and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liming Yang, eWFuZ2xpbWluZ0BuamZ1LmVkdS5jbg==; Yuming Luo, eXVtaW5nbHVvQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.