
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 10 November 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1055867
This article is part of the Research Topic Functional Genomic Approaches in Molecular Breeding for Crop Improvement View all 7 articles
 Shanshan Chu1†
Shanshan Chu1† Hui Ma1†
Hui Ma1† Ke Li1
Ke Li1 Junfeng Li1
Junfeng Li1 Hongli Liu1
Hongli Liu1 Leipo Quan1
Leipo Quan1 Xuling Zhu1
Xuling Zhu1 Meiling Chen1
Meiling Chen1 Wenyan Lu1
Wenyan Lu1 Xiaoming Chen1
Xiaoming Chen1 Xuelian Qu1
Xuelian Qu1 Jiaqi Xu1
Jiaqi Xu1 Yun Lian2
Yun Lian2 Weiguo Lu2
Weiguo Lu2 Erhui Xiong1*
Erhui Xiong1* Yongqing Jiao1*
Yongqing Jiao1*Soybean cyst nematode (SCN) is a serious damaging disease in soybean worldwide. Peking- and PI 88788-type sources of resistance are two most important germplasm used in breeding resistant soybean cultivars against this disease. However, until now, no comparisons of constitutive resistances to soybean cyst nematode between these two types of sources had been conducted, probably due to the influences of different backgrounds. In this study, we used pooled-sample analysis strategy to minimize the influence of different backgrounds and directly compared the molecular mechanisms underlying constitutive resistance to soybean cyst nematode between these two types of sources via transcriptomic and metabolomic profilings. Six resistant soybean accessions that have identical haplotypes as Peking at Rgh1 and Rhg4 loci were pooled to represent Peking-type sources. The PI88788-type and control pools were also constructed in a same way. Through transcriptomic and metabolomics anaylses, differentially expressed genes and metabolites were identified. The molecular pathways involved in the metabolism of toxic metabolites were predicted to play important roles in conferring soybean cyst nematode resistance to soybean. Functions of two resistant candidate genes were confirmed by hairy roots transformation methods in soybean. Our studies can be helpful for soybean scientists to further learn about the molecular mechanism of resistance to soybean cyst nematode in soybean.
Soybean (Glycine max) is an important oil and protein crop worldwide. However, soybean cyst nematode (SCN), a widely spread pathogen, is one of major threats to soybean production (Niblack et al., 2006). In addition to crop rotation, planting resistant cultivars is the most efficient and practical method to control this pest. To date, many sources of resistance to SCN have been found in soybean (Concibido et al., 2004). Among them, Peking, PI 88788, PI 437654 and PI 209332 are four widely used germplasm in breeding SCN-resistant cultivars.
To reveal genetic basis of resistance for these sources, quantitative trait locus (QTL) analyses were performed (Wang et al., 2001; Concibido et al., 2004; Vuong et al., 2010). Among them, two QTLs, Rhg1 and Rhg4, were found to play the most important roles in conferring SCN resistance soybean (Concibido et al., 2004). Rhg1 was located on Chromosome 18 (Chr. 18) (Concibido et al., 2004). This locus was found in almost all of reported SCN-resistant sources, except for PI 567516C (Glycine max) and one wild soybean line PI 468916 (Glycine soja) (Wang et al., 2001; Concibido et al., 2004; Vuong et al., 2010). There were two different alleles at the Rhg1 locus, Rhg1-a from Peking and Rhg1-b from PI 88788 (Concibido et al., 2004; Kim M. et al., 2010). These two alleles might have different mechanisms of resistance to SCN. Rhg1-a has to interact with Rhg4 in conferring resistance to SCN HG type 0 (race 3), while Rhg1-b is able to function independently (Meksem et al., 2001; Concibido et al., 2004). Moreover, Peking- and PI 88788-type sources showed different responses to SCN infections (Kim Y. H. et al., 2010). PI 437654, one of Peking-type sources, showed a rapid resistance reaction, which was characterized by the death of SCN at the parasitic second (J2) stage juvenile (Kim Y. H. et al., 2010). In contrast, PI 88788 has a prolonged resistance reaction where nematode die at the J3 and J4 stage (Kim Y. H. et al., 2010). Rhg1-b had been cloned from PI 88788 through map-based cloning (Cook et al., 2012). Three genes, Glyma.18g022400 (Glyma18g02580), Glyma.18g022500 (Glyma18g02590) and Glyma.18g022700 (Glyma18g02610) in together, were found to be responsible for this locus. The resistance to SCN in PI 88788 was attributed to an elevated level of constitutive expressions for these three genes, which were caused by increased copy numbers rather than sequence mutation (Cook et al., 2012). Further studies showed that Rhg1-a in Peking contained specific SNPs, around half of copies, and different levels of methylations for these three genes in comparison with Rhg1-b in PI 88788 (Cook et al., 2014; Liu et al., 2017). For Rhg4, it was identified to be a serine hydroxymethyltransferase (SHMT) that was involved in cellular one-carbon metabolism (Liu et al., 2012). In Peking, gain of function mutations that resulted in amino acid changes, altered the regulatory property of the Rhg4 protein and conferred new functions to this gene (Liu et al., 2012).
Scientists made great efforts to understand molecular mechanisms of interaction between soybean and SCN. Gene expression studies on either compatible or incompatible responses to SCN were comprehensively conducted through a serial of methods including differential display of mRNA (Hermsmeier et al., 1998), cDNA microarray (Khan et al., 2004; Alkharouf et al., 2006), affymetrix Genechip (Klink et al., 2007b; Ithal et al., 2007), and laser capture micro-dissection (LCM) (Klink et al., 2007a). And, interactions between soybean and SCN were also investigated by identifying and studying the effectors that SCN secreted during infection (Bellafiore and Briggs, 2010; Gheysen and Mitchum, 2011). All these studies suggested that the a complicated network composed of pathways of auxin, cell wall, secondary metabolites, programed cell death and defense responses, was involved in the interaction between soybean and SCN (Gheysen and Mitchum, 2011).
Currently, almost all reported SCN-resistant germplasms were either Peking- or PI 88788-type (Concibido et al., 2004; Cook et al., 2014; Jiao et al., 2015). Peking-type sources were characterized by containing both Rhg1-a and Rhg4, while PI 88788-type sources of resistance were characterized by containing Rhg1-b (Cook et al., 2014; Kadam et al., 2016). Different SCN-resistant genes as well as different reactions in response to SCN infection suggested different molecular mechanisms of resistance to SCN between these two types of sources. However, no direct comparisons between these two types of resistant sources have been reported until now. Thus, the common and different characteristics of resistance mechanism between these two resistant sources remain unknown. This might due to the influence of different backgrounds between these two germplasm and resulted difficulty to obtain accurate information.
In this studies, to investigate characteristics of constitutive resistance mechanism between Peking and PI 88788-type sources of resistance and overcome influences of different backgrounds, we used pooled-sample analysis strategy to conduct transcriptomic and metabolomic studies for these two types of resistant sources. The common and specific genes and metabolites were identified. The possible resistance mechanisms between these two sources were also put forward. Our results could be helpful for scientists to further learn about the molecular mechanisms underlying resistance to SCN in soybean.
Based on the genomic and phenotypic data (Kadam et al., 2016), three pools of soybean accessions were constructed, which included: (Pool-I) Peking-type sources containing PI 548402 (Peking), PI 90763, PI 404166, PI 89772, PI 437654 and PI 438489B, which have same haplotypes as Peking at Rhg1 and Rhg4 loci; (Pool-II) PI 88788-type sources containing PI 88788, PI 437655, PI 495017C, PI 209332, PI 438503 A and PI 467312, which have same haplotypes as PI 88788 at Rhg1 locus but do not contain Rhg4; and (Pool-III) non-resistant controls PI 518664 (Hutcheson), PI 518671 (Williams 82) and PI 595362 (Magellan), which do not contain Rhg1 and Rhg4 (Table 1). Evaluations of SCN resistances revealed that the resistant accessions in Pool-I exhibited resistance or moderately resistance to SCN HG type 2.5.7, 0 and 2.5.7, while the accessions in Pool-II showed resistant or moderately resistant to SCN HG type 0 and 1.3.5.6.7. Both Pool-I and Pool-II showed resistance to SCN HG type 0 in common (Table 1).
To investigate the differences of constitutive resistance between these two types of sources of resistance, we conducted transcriptomic profilings for these accessions. The root samples of Pool-I were divided into two groups, Peking and the pool of PI 90763, PI 404166, PI 89772, PI 437654, and PI 438489B (Pool-a). The root samples of Pool-II were divided into PI 88788 and the pool of PI 437655, PI 495017C, PI 209332, PI 438503 A and PI 467312 (Pool-b). And, the root samples of Pool-III were divided into Hutcheson and the pool of Williams 82 and Magellan (Pool-c). Two comparisons for Pool-I were conducted, which included Peking versus (vs.) Hutcheson and Pool-a vs. Pool-c (Figure 1; Supplementary Tables S1, 2). To minimize the influences of different backgrounds, genes with same expression patterns between these two comparisons, were chosen to represent the gene expression patterns of Peking-type sources (Pool-I) (Figure 1; Supplementary Table S3). Similarly, two comparisons including PI 88788 vs. Hutcheson and Pool-b vs. Pool-c were also conducted (Figure 1; Supplementary Tables S4, 5). And, genes with same expression patterns between these two comparisons were chosen to represent the gene expression patterns of PI 88788-type sources (Pool-II) (Figure 1; Supplementary Table S6).
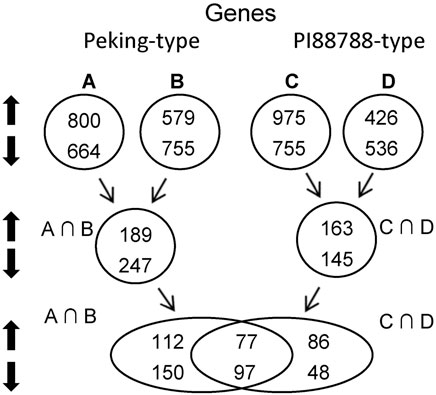
FIGURE 1. Strategy of pooled-sample analysis and the number of significant differentially expressed genes (DEGs) between Peking- and PI 88788-type sources of resistance. A, Peking versus (vs.) Hutcheson; B, Pool-a vs. Pool-c: the pool of PI 90763, PI 404166, PI 89772, PI 437654, and PI 438489B vs. the pool of Williams 82 and Magellan; C, PI 88788 vs. Hutcheson; D, Pool-b vs. Pool-c: the pool of PI 437655, PI 495017C, PI 209332, PI 438503A and PI 467312 vs. the pool of Williams 82 and Magellan; A∩B, intersection between (A) and (B); C∩D, intersection between (C) and (D).
In Peking-type sources, a total of 436 significantly differentially expressed genes (DEGs) were identified, where 189 were up-regulated and 247 were down-regulated (Figure 1; Supplementary Table S3). In PI 88788-type sources, a total of 308 DEGs were identified, where 112 were up-regulated and 150 were down-regulated (Figure 1; Supplementary Table S6). Among these DEGs, 112 up-regulated and 150 down-regulated genes were specific for Peking-type sources (Figure 1; Supplementary Table S7) while 86 up-regulated and 48 down-regulated genes were specific for PI 88788-type sources (Figure 1; Supplementary Table S8). Seventy-seven up-regulated and 97 down-regulated genes were commonly found in both of these two types of sources (Figure 1; Supplementary Table S9). Expressions of three responsible genes at Rhg1, Glyma.18g022400, Glyma.18g022500 and Glyma.18g022700 as internal standards were up-regulated in accordance with copy number variation of Rhg1 in Peking- and PI 887788- type sources (Cook et al., 2012) (Cook et al., 2014; Liu et al., 2017), which could prove the reliability of our transcriptomic profilings (Supplementary Table S9).
GO and KEGG analyses were conducted for these DEGs (Table 2). In Peking-type sources, the specifically up-regulated DEGs were significantly enriched in the biological processes including fatty acid biosynthetic process (GO:0006633), monocarboxylic acid biosynthetic process (GO:0072330), and inositol metabolic process (GO:0006020), and molecular pathways including Linoleic acid metabolism (ko00591), benzoxazinoid biosynthesis (ko00402), flavone and flavonol biosynthesis (ko00944), and plant-pathogen interaction (ko04626) (Table 2), while the specifically down-regulated DEGs were significantly enriched in the biological processes including response to stimulus (GO:0050896), oxazole or thiazole biosynthetic process (GO:0018131), oxazole or thiazole metabolic process (GO:0046484), and molecular pathways including plant-pathogen interaction (ko04626) (Table 2). In PI 88788-type sources, no significant biological processes and pathways were detected for the specific DEGs (Table 2). In both of these two sources, the common up-regulated DEGs were significantly enriched in the carotene metabolic process (GO: 0016119), while the common down-regulated DEGs were significantly enriched in the plant-pathogen interaction pathway (ko0462) (Table 2). Interestingly, the plant-pathogen interaction pathway (ko0462) was commonly detected for both up- and down-specifically regulated genes in Peking-type sources, indicating different specific pathways might be affected by these genes.
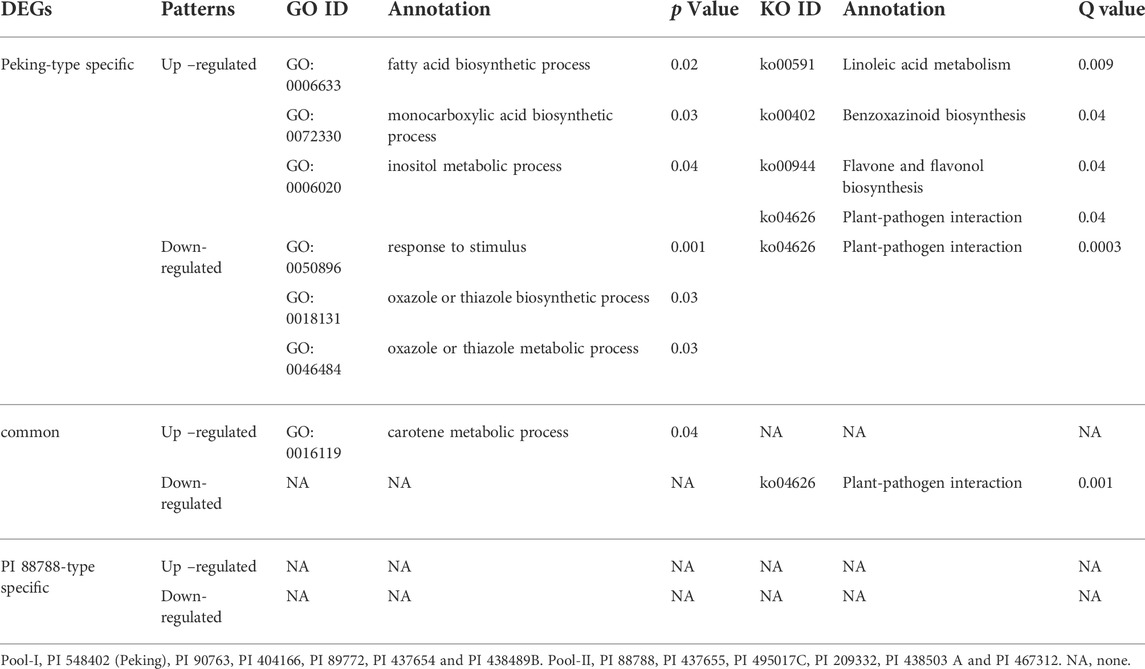
TABLE 2. The biological process ontology and pathway analysis of differentially expressed genes (DEGs) between Peking- and PI 88788- type sources of resistance (Pool-I and Pool-II).
In addition to general analyses, we also investigated the functions of these DEGs in detail. Because a large number of DEGs were involved in plant secondary metabolism, we selected some interesting genes that might be involved in producing toxic or antioxidant secondary metabolites and conferring constitutive SCN resistance to soybean (Table 3). Four predicted O-methyltransferase encoding genes that participate in the conversion of TRIBOA-glucoside to DIMBOA-glucoside in the last step of benzoxazinoid biosynthesis pathway (ko00402) were selected, among which Glyma.06G286600, Glyma.09G094600 and Glyma.06G286200 were specifically up-regulated in Peking-type sources and Glyma.09G094400 was commonly up-regulated in both of two resources (Table 3; Supplementary Figure S1). Two genes, Glyma.09G144900 and Glyma.09G145000, were specifically up-regulated in PI 88788-type sources, which were predicted to encode a 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase respectively and might be involved in conversion of DIBOA-glucoside to TRIBOA-glucoside, the penultimate step of benzoxazinoid biosynthesis (Table 3; Supplementary Figure S1). The natural benzoxazinoids could function as pesticides and insecticides (Barnes and Putnam, 1987) (Grombacer AW and Guthrie, 1989) (Bravo and Lazo, 1993) (Sicker et al., 2000). The DIBOA biosynthesis is regarded as the core pathway for benzoxazinoids. DIMBOA is produced by the modification of DIBOA and has more reactive activity than DIBOA. In plants, benzoxazinoids are mainly stored in vacuole as glucosides (Sicker et al., 2000). Upon cell damage, the benzoxazinone glucosides is released from the vacuole and produced the toxic aglucone by a specific glucosidase (Esen, 1992). Up-regulation of these genes might indicate the enhancement of benzoxazinoid biosynthesis pathway and DIMBOA-glucosite content in these resistance sources, especially in Peking-type sources. Except for genes in the benzoxazinoid pathway, several genes in cyanoamino acid metabolism were also selected, which included Glyma.15G031400, Glyma.18G080400, Glyma.08G150100 and Glyma.15G270900 (Table 3; Supplementary Figure S2). Cyanohydrins in plants are found usually in the form of cyanogydrin glycosides, which was relatively stable and stored in the plant cell vacuole (Flematti et al., 2013). Cyanohydrins can be hydrolyzed by a hydroxynitrile lyase to release cyanide, which was the toxic metabolites and confer defense against herbivores and pathogens (Flematti et al., 2013). Three up-regulated DEGs, Glyma.15G031400, Glyma.08G150100, and Glyma.15G270900 were Peking-type specific, common, and PI 88788-type specific, respectively. All of these three genes were predicted to encode β-glucosidases and participated in conversion of cyanoglycoside to cyanohydrin (ko00460) (Table 3; Supplementary Figure S2). The Glyma.18G080400 was a Peking-type specific down-regulated DEG, which was predicted to be a cytochrome P450 and involved in conversion of aldoxime to cyanohydrin (ko00460) (Table 3; Supplementary Figure S2). These results indicated that the branch from amygdalin to cyanohydrin in cyanoamino acid metabolism might be enhanced in both Peking-type and PI 88788-type sources, while the branch from l-amino acid to cyanohydrin might be specifically reduced in Peking-type resistant sources (ko00460).
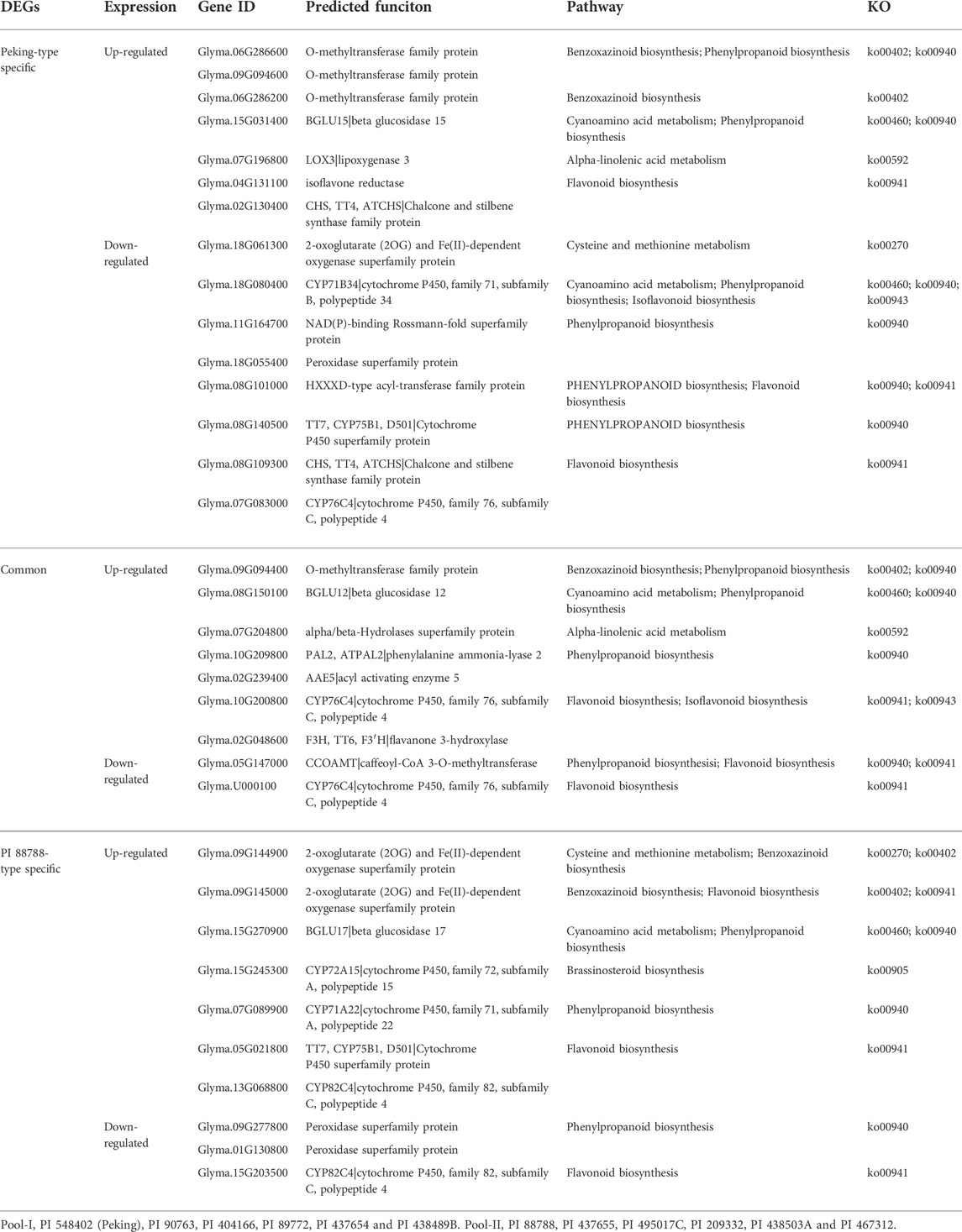
TABLE 3. The selected differentially expressed genes (DEGs) related to metabolites involved in constitutive resistances of Peking- and PI 8878- type sources of resistance (Pool-I and Pool-II).
To reveal the differences of metabolism between Peking-type and PI 88788-type sources, we conducted and compared metabolomic profilings by use of LC-MS for the same samples that were used for transcriptomic profilings.
To minimize the influences of different backgrounds, the same strategy of pooled-sample analysis was adopted for the analyses of metabolism as that in the transcriptome analysis (Figure 2; Supplementary Table S10). In comparison with the controls (Pool-III), five up-regulated metabolites including HOME, jasmonic acid, kamlolenic acid, S-adenosylmethionine and traumatic acid, and six down-regulated ones including cGMP, aminooctanoic acid, glycitein-O-glucuronide, HpOTrE, hydroxy-octadecadiynoic acid and linolenic acid, were significantly detected for Peking-type sources (Table 4). KEGG pathway analyses showed that the up-regulated metabolites were mainly involved in the pathway of biosynthesis of plant hormones (ko01070), and α-linolenic acid metabolism (ko00592) (Table 4). In comparison, the down-regulated metabolites were mainly involved in pathways of purine metabolism (ko00230), arginine and proline metabolism (ko00943),α-Linolenic acid metabolism (ko00592), and linoleic acid metabolism (ko00591) (Table 4). Two pathways of arginine and proline metabolism (ko00943) and α-Linolenic acid metabolism (ko00592) were detected for both of up- and down-regulated metabolites (Table 4). For PI 88788-type sources, four up-regulated metabolites including HOME, jasmonic acid, quercetin and traumatic acid, and three down-regulated ones including aminooctanoic acid, phenylalanine and TriHOME, were significantly identified (Table 4). KEGG Pathway analyses showed that the up-regulated metabolites were mainly involved in the pathways of α-linolenic acid metabolism (ko00592) and flavonoid biosynthesis (ko00941) (Table 4). In comparison, the down-regulated metabolites were mainly involved in the pathway of phenylalanine, tyrosine and tryptophan biosynthesis (ko00400), and linoleic acid metabolism (ko00591) (Table 4). No common pathways were detected for both up- and down-regulated genes (Table 4). By comparing the changes of metabolites between Peking-type and PI 88788-type sources, we found that three up-regulated metabolites including HOME, jasmonic acid and traumatic acid, and one down-regulated metabolite, aminooctanoic acid, were commonly detected for both of these two comparisons (Table 4). Jasmonic acid and traumatic acid were both involved in the pathways of α-linolenic acid metabolism (ko00592) (Table 4).
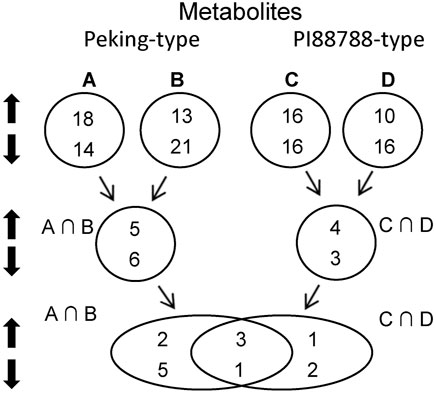
FIGURE 2. Bulk strategy and number of significant differentially expressed metabolites (DEMs) between Peking- and PI 88788-type sources of resistance. A, Peking versus (vs.) Hutcheson; B, Pool-a vs. Pool-c: the pool of PI 90763, PI 404166, PI 89772, PI 437654, and PI 438489B vs. the pool of Williams 82 and Magellan; C, PI 88788 vs. Hutcheson; D, Pool-b vs. Pool-c: the pool of PI 437655, PI 495017C, PI 209332, PI 438503A and PI 467312 vs. the pool of Williams 82 and Magellan; A∩B, intersection between (A) and (B); C∩D, intersection between (C) and (D).
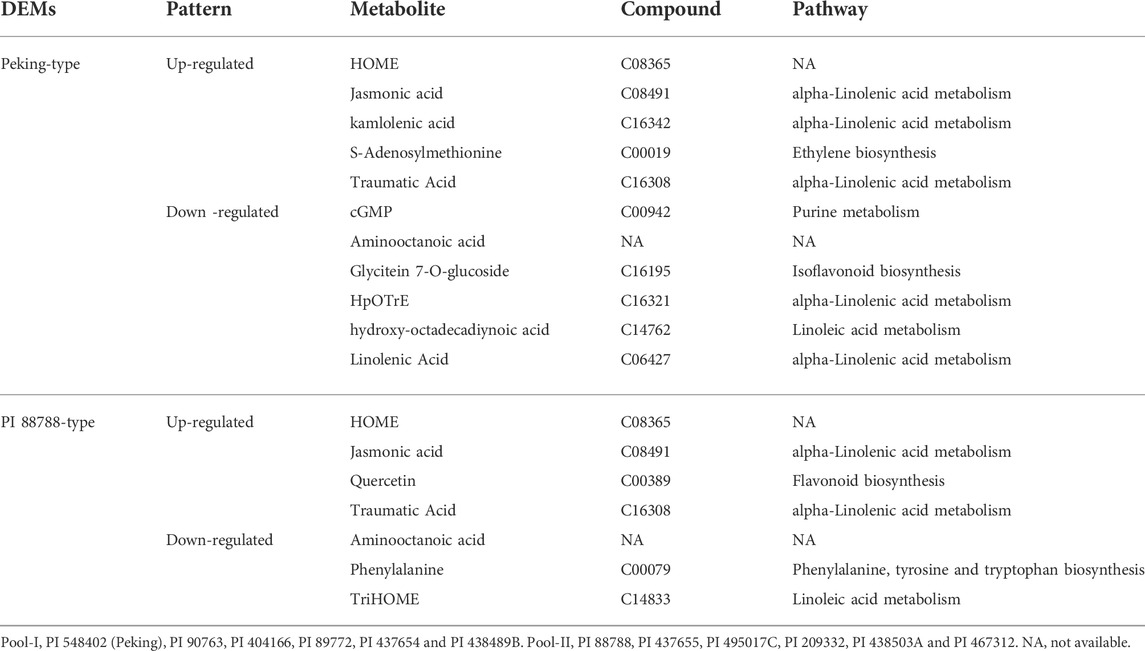
TABLE 4. Significant differentially expressed metabolites (DEMs) between Peking- and PI 88788-type sources of resistance.
Combined with transcriptome data, we found that, one gene, Glyma.07G204800, in α-Linolenic acid metabolism (KO00592) was commonly up-regulated in both of Peking-type and PI 88788-type sources (Table 3). This gene was predicted to encode an α/β -Hydrolase, which catalyzed phosphatidylcholine to α-Linolenic acid (ko00592) (Supplementary Figure S3). Up-regulation of Glyma.07G204800 might activate the production of α-Linolenic acid and enhance the content of relevant metabolites such as traumatic acid and jasmonic acid, which was proved by the metabolome data (Table 4). The metabolites analyses showed that the phenylalaninine was specifically down-regulated in PI 88788-type sources (Table 4). Accordingly, Glyma.06G235900 that encodes an aminotransferase was also specifically down-regulated in PI 88788-type sources, which was involved in conversion of phenylpyruvate to phenylalanine (ko00400) (Supplementary Figure S4).
To investigate the effectiveness of our strategy, we selected two candidate resistant genes, GmERF71 (Glyma.19G262700) and GmLOX1 (Glyma.15G026500), to confirm their functions in conferring SCN resistance to soybean through hairy-root transformation methods. The GmERF71encodes an AP2 protein and GmLOX1 encodes a lipoxygenase. Both of these two genes were up-regulated in Peking-type and PI88788-type sources of resistance (Supplementary Tables S3 and S6). We generated transgenic hairy roots by overexpressing these two genes under 35S promoter (Supplementary Figure S5). The qRT-PCR results confirmed the increased expression levels of GmERF71 and GmLOX1 (Figure 3), confirming the success of our hairy root transformation. The evaluation of SCN resistance for transgenic hairy roots showed overexpression of these two genes could significantly decrease the cyst number on hairy roots (Figure 3), confirming their functions in conferring SCN resistance to soybean.
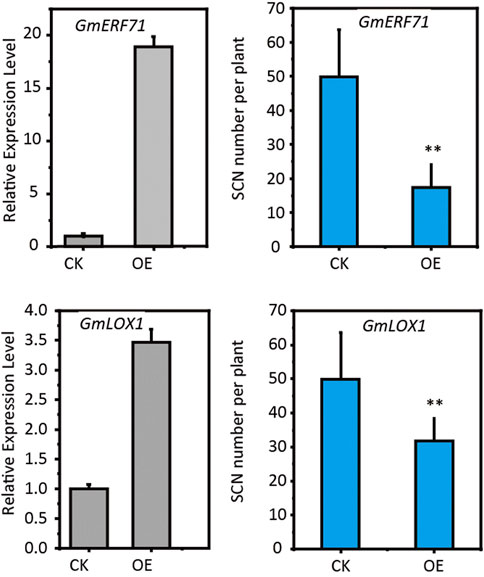
FIGURE 3. Functional confirmation of GmERF71 and GmLOX1. CK, control; OE, plants with gene overexpressed. The significance was determined by t test. n = 10.
For Rhg1-b in PI 88788, about ten copies of three responsible genes, Glyma.18g022400, Glyma.18g022500 and Glyma.18g022700 cause constitutively up-regulated expression of these three genes and as a result the changes of constitutive molecular pathways in vivo (Cook et al., 2012). In Peking, despite around four copies of these three genes were found for Rhg1-a, their expressions were also constitutively enhanced (Cook et al., 2012; Cook et al., 2014; Kadam et al., 2016). In addition, for Rhg4 in Peking, the alteration of gene function rather than loss of function could cause the alterations of constitutive metabolisms in vivo either (Liu et al., 2012). These constitutive changes were proved to play important roles in conferring SCN resistance to soybean (Liu et al., 2012; Cook et al., 2014). Our summary of phenotypic data showed that Peking-type accessions was resistant or moderately resistant to SCN HG types 2.5.7, 0 and 2.5.7, while PI 88788-type accessions were resistant or moderately resistant to SCN HG types 0 and 1.3.5.6.7 (Table 1). Both of them were resistant to SCN HG type 0. These results indicated these two types of resistance sources might have common and different characteristics of mechanisms underlying constitutive resistances. To reveal these characteristics will provide important information for scientists to elucidate the resistance mechanisms for Peking and PI88788-type resistant germplasm. Unfortunately, until now, little has been known about constitutive resistances for these two types of sources of resistance. That might be due to the influences of different backgrounds between these two sources and as a result no direct comparisons could be conducted. To overcome this problem, we developed and used the pooled-sample analysis strategy. Six resistant soybean accessions that have haplotypes identical to Peking at Rhg1 and Rhg4, were selected to constitute the Peking-type pool (Kadam et al., 2016). Similarly, the PI88788-type pool was also constructed. Theoretically, these two pools only have differences at Rhg1 and Rhg4 loci. Thus, the direct comparisons of SCN resistances between these two types of sources of resistances could be realized. The genes and metabolites associated with constitutive SCN resistances underlying these two types of sources were identified in our studies, suggesting the rationality of our strategy.
Since both Peking- and PI 88788-type sources were resistant to SCN HG type 0, there might be a common resistant mechanism shared by these two types of resistance sources. Seventy-seven up-regulated and 97 down-regulated DEGs were commonly identified in both of these two sources. The analyses of GO and KEGG pathways revealed that the defense response (GO: 0006952), plant-pathogen interaction (ko04626), linoleic acid metabolism (ko00591), isoflavonoid biosynthesis (ko00943), phenylpropanoid biosynthesis (ko00940) etc, were significantly detected for common DEGs (Table 2). These molecular pathways were reported to be associated with resistance to abiotic and biotic stresses in plants (Hernandez et al., 2009; Suzuki et al., 2014). In addition to genes selected in Table 3, others are also worthy of being studied in the further. For example, two lipoxygenase 1-enconding genes, Glyma.15G026500 and Glyma.04G105900 were found to be up-regulated in both Peking- and PI 88788-type sources (Supplementary Table S9). Lipoxygenase is an important enzyme in the biosynthesis of jasmonic acid that is an important wounding-related phytohormone involved in resistance to biotic stresses (Dar et al., 2015; Carvalhais et al., 2017). In the process of SCN infection, the J2 SCN juveniles need enter into soybean roots and continue moving until they found the loci where the syntium can be established (Gheysen and Mitchum, 2011). During this process, the wounding to plants by SCN happened continuously. The enhancement of JA content might enable soybean to fight against this infection wounding and consequently confer SCN resistance to soybean.
Except for the common resistance to SCN HG type 0, Peking- and PI 88788- type resistant sources showed specific resistance phenotypes, where Peking-type sources are resistant or moderately resistant to SCN HG type 2.5.7 and HG type 2.5.7 while PI 88788-type sources are resistant or moderately resistant to HG type 1.3.5.6.7 (Table 1). These specific phenotypes indicated that there might be specific resistance mechanisms underlying these two types of sources. In order to gain insights into specific mechanisms, we also investigated specific DEGs between Peking- and PI 88788- type resistant sources (Supplementary Tables S7, 8). Four significant pathways including Linoleic acid metabolism (ko00591), benzoxazinoid biosynthesis (ko00402), Flavone and flavonol biosynthesis (ko00944), Plant-pathogen interaction (ko04626), were specific for Peking-type sources, which indicated that these pathways might play important roles in conferring specific SCN resistance to Peking-type sources (Table 2). In PI 88788-type sources, no significant pathways were detected (Table 2). However, a large number of DEGs that were involved in defense signal transductions were detected such as five leucine-rich repeat transmembrane protein kinase (LRR) proteins (Glyma.12G123100, Glyma.15G021400, Glyma.15G021700,Glyma.16G174100 and Glyma.16G215000) and four receptor like proteins (Glyma.20G118400, Glyma.16G176600, Glyma.16G176900 and Glyma.16G175100) (Supplementary Table S8). Based on the above, we speculated that, in Peking-type sources, defense-related metabolites might play more important roles in response to SCN. The constitutive high level of anti-pathogen/insect metabolites in Peking-type sources could lead to a rapid defense response to the infection of J2 stage nematode (Klink et al., 2011). In contrast, in PI 88788-types sources, defense signaling pathway might be more important. It might take a while for anti-pathogen/insect metabolites to reach a certain level that could influence SCN. This might be the reason why PI 88788-type sources could exhibit a delayed defense response to J3 stage nematode (Klink et al., 2011). Our results might provide clues to reveal the mechanism underlying phenotypic differences between these two resistant germplasm.
To investigate the metabolic differences between Peking- and PI 88788-type germplasm, we used the samples identical to the ones for transcriptomic analyses to conduct metabolomic analyses. For Peking-type sources, five up-regulated and six down-regulated metabolites were identified, which were involved in five different pathways including α-Linolenic acid metabolism (ko00592), biosynthesis of plant hormones (ko01070), Purine metabolism (ko00230), Isoflavonoid biosynthesis (ko00943) and Linoleic acid metabolism (ko00591). For PI 88788-type sources, four up-regulated and three down-regulated metabolites were identified, which were involved in six different pathways including α-Linolenic acid metabolism (ko00592), flavonoid biosynthesis (ko00941), phenylalanine metabolism (ko00360) and linoleic acid metabolism (ko00591). Three up-regulated metabolites including HOME, jasmonic acid and traumatic acid, and one down-regulated metabolite, aminooctanoic acid, were commonly identified in both Peking- and PI 88788-type sources (Table 4). HOME is one component of cutin, which is the polymer matrices for lipophilic cell wall barriers and play important roles in protecting plants form biotic and abiotic stresses (Pollard et al., 2008). Jasmonic acid is well known as a defense plant hormone (Dar et al., 2015). Previous studies showed JA pathway played important roles in conferring SCN resistance to soybean in both of PI 88788- and Peking-types sources of resistance (Klink et al., 2009) (Klink et al., 2010; Matsye et al., 2010). Traumatic Acid was one of major products derived from linolenic acid in the hydroperoxidelyase pathway, where a series of important signals in wounding and plant–insect communication were identified (Croft et al., 1993; Kallenbach et al., 2011). The common up-regulation of these three metabolites indicated the enhancement of molecular pathways associated with biotic stresses in both of PI 88788- and Peking-type sources, which might be the reason why both of these two types of sources displayed resistance to SCN HG type 0 in common.
The S-adenosylmethionine that takes part in ethylene and polyamine biosynthesis, were specifically up-regulated in Peking type sources (Table 4). And, glycitein-O-glucuronide that takes part in isoflavonoid biosynthesis was specifically down-regulated (Table 4). Ethylene, polyamine and isoflavonoids were reported to be important metabolites associated with plant-insect interactions (Simmonds, 2003; Hussain et al., 2011; Erb et al., 2012). The specific changes of these metabolites indicated that the ethylene, polyamine and isoflavonoids related pathways might be involved in determining specific phenotypes of Peking-type sources. In PI 88788-type sources, quercetin that takes parts in flavonoid biosynthesis was specifically up-regulated, while phenylalanine that takes parts in phenylalanine, tyrosine and tryptophan biosynthesis, was specifically down-regulated (Table 3). The flavonoids metabolites were largely involved in all aspects of plant life including plant-insect interaction (Ferreyra et al., 2012). Tryptophan is the precursor of auxin, which is an important phytohormone regulating plant development and other processes (Park, 2007) (Zhao, 2010). The changes of these specific metabolites might have important functions in determining specific phenotypes of PI 88788-type sources.
Interestingly, we noticed that some metabolites that belong to the same metabolism pathway were regulated in different patterns in same type of resistant sources. For example, jasmonic acid, kamlolenic acid, and traumatic acid, were up-regulated in Peking-type sources, while HpOTrE and linolenic Acid were down-regulated. All of these metabolites were in the same pathway of α-Linolenic acid metabolism (ko00592). We further investigated the map of α-Linolenic acid metabolism (Supplementary Figure S3) and found that jasmonic acid, kamlolenic acid, traumatic acid and HpOTrE were located in the different branches of α-Linolenic acid metabolism. These results indicated that different branches of same metabolism pathway might be regulated differently in response to SCN, which were worthy of being studied further.
Since both transcriptome and metabolome were conducted in this study, we further combined these two analyses and investigated the changes of molecular pathways between Peking- and PI 88788- type sources. The results indicated the transcriptome and metabolome data exhibited good consistency. Lipoxygenase was a key enzyme in the biosynthesis of jasmonic acid, which used linoleate and α-linolenate as substrates (Schaller and Stintzi, 2009). Two putative lipoxygenase 1-encoding genes, Glyma.15G026500 and Glyma.04G105900, were commonly up-regulated in expression in both of Peking- and PI 88788- type sources (Supplementary Table S9). Up-regulation of these two lipoxygenase genes was in accordance with the enhancement of JA content in metabolite profiling (Table 4). In PI 88788-type sources, one intermediate product in flavonoid biosynthesis, quercetin, was specifically up-regulated in content (Table 4). Correspondingly, Glyma.05G021800 that encodes a putative cytochrome P450 and was involved in flavonoid biosynthesis (ko00941), was also up-regulated (Supplementary Table S8). All these information could provide us useful clues to understand molecular mechanisms underlying constitutive resistance to SCN between these two types of resistance sources. However, due to limited capability of technology for metabolomics analysis, we are not able to get a full and accurate map of metabolites in vivo in plants. Thus, changes of lots of metabolites in our comparisons were unavailable. In addition, mRNA level and metabolites are not directly related with each other. Many processes such as gene translation, protein modification, and metabolites stability can affect the consistency between mRNA and metabolite. Thus, in some cases, changes of gene expressions might not reflect actual changes of metabolites.
The APETALA2 (AP2)/ETHYLENE-RESPONSIVE FACTOR (ERF) is the second largest transcription factor family in plants and plays a important role in stress responses (Feng et al., 2020). The SCN candidate gene GmERF71 was both up-regulated in Peking-type and PI88788-type sources of resistance. The cyst number on the transgenic hairy roots overexpressing GmERF71 decreased by 59% compared with the control, confirming its resistant function (Figure 3). Previous studies showed that, in transgenic soybean plants overexpressing GmERF5 and GmERF113 the expression of defense related genes such as PR10, PR1-1 and PR10-1 were significantly induced, thereby improving the resistance to soybean phytophthora blight (Dong et al., 2015; Zhao et al., 2017). Overexpression of GmERF3 genes in transgenic tobacco also activates the expression of PR genes, enhancing its resistance to pathogens (Zhang et al., 2009). AtERF1 has been proved to act downstream of ethylene and jasmonic acid pathways, and is a key factor in the integration of these two signals to regulate defense response genes (Lorenzo et al., 2003). OsERF3 mediates the biosynthesis of salicylic acid, jasmonic acid, ethylene and hydrogen peroxide through MAPK cascade and WRKY genes, enhancing the resistance of plants to stinging or chewing insects (Lu et al., 2011). We speculate that GmERF71 participates in the regulation of resistance to SCN by activating downstream defense genes and phytohormone pathways, which need to be studied further.
The lipoxygenase is a key enzyme in the biosynthesis of jasmonic acid (Dar et al., 2015; Wasternack and Song, 2017). It has been reported that lipoxygenase genes are up-regulated in syncytium after SCN infection in Peking and PI88788 (Klink et al., 2009; Klink et al., 2010). Overexpression of three JA biosynthesis genes, AtAOS, AtAOC and AtJAR in soybeans improves the resistance to SCN (Matthews et al., 2014). The GmLOX1 (Glyma.15G026500) were commonly up-regulated in both of Peking- and PI 88788- type sources and were also identified by integrative analyses of transcriptome and metabolome, which were involved in JA biosynthesis. Our results showed that GmLOX1 overexpression could enhance resistance to SCN in transgenic hairy roots, confirming the role of JA in response to SCN infection.
In summary, we directly and systemically compared the constitutive resistances between Peking- and PI 88788- type sources of resistance in this study, results of which might be helpful for scientists to learn the resistance mechanisms for these two types of sources further.
The soybean accessions used in this study were collected from Germplasm Resources Information Network (GRIN), United States. Seeds were pre-germinated on moistened filter paper in a plant growth chamber at 27°C, 85% ambient humidity and a 16:8 (light: dark) photoperiod for 3–4 days. The seedlings were then transferred into tubes (5 cm 3 30 cm) with sandy soil at 27°C under 16 h of light in the greenhouse. When the plants had three fully expanded trifoliate leaves, roots were harvested, flash frozen in liquid nitrogen, ground to powder, and stored at −80 C until the next step. For each sample, three biological replicates were set up.
The SCN assays were performed in greenhouse according to the reported methods (Niblack et al., 2009). In brief, soybean seeds were germinated and transplanted into PVC tubes. The tubes were filled with steam-pasteurized sandy soil and packed into plastic containers prior to transplanting. Five plants of each indicator line and RIL were arranged in a randomized complete block design. Two days after transplanting, each plant was inoculated with about 2000 SCN eggs. Thirty days post-inoculation, nematode cysts were washed from the roots of each plant and counted. The SCN resistances were evaluated by SCN reaction to a set of soybean indicator lines for HG type tests (Peking, PI 88788, PI 90763, PI 437654, PI 209332, PI 89772, PI 548316, and susceptible checks (cv. Hutcheson). The female index (FI %) was estimated to evaluate the response of each plant to each HG type of SCN using the following formula:
FI (%) = (Number of female cyst nematodes on a given individual/average number of female nematodes on the susceptible check) × 100.
The root samples were grinded into two parts, one of which was used to extract total RNAs using TRIZOL reagent (Invitrogen, CA, United States) following the manufacturer’s procedure. The total RNA concentration and purity were assayed with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States). The RNA integrity was assessed on an Agilent 2,100 Bioanalyzer Lab-on-Chip system (Agilent Technologies, Palo Alto, CA, United States). The construction of cDNA libraries was according to the protocol for the mRNA-Seq sample preparation kit (Illumina, San Diego, United States). The clustering of samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia, San Diego, United States) according to the manufacturer’s instructions. And the paired-end sequencing was performed on the Illumina Hiseq2500 platform. Sequencing-received raw image data were transformed by base calling into raw sequences. The raw sequences were transformed into clean tags by removing the three adaptor sequences and low quality tags. All clean tags were mapped to the reference sequences (https://www.soybase.org/), and those tags with no more than one mismatch were considered. The differentially expressed genes (DEGs) with fold change of log2 ≧2 were deemed as significant DEGs. Gene Ontology (GO) enrichment and KEGG pathway analyses for these significant DEGs were carried out in the website of Soybase (https://www.soybase.org/) and KEGG (https://www.genome.jp/kegg/).
The other root samples were used for LC-MS analysis. Fifty mg of root powders were extracted with 800 μL methanol. After vortex-mixing for 30 s, these samples were centrifuged at 12,000 rpm for 10 min to remove proteins. The supernatant was then transferred to autosampler vials. Metabolic Profiling Analysis of Plasma by LC-Q-TOF MS. Plasma samples for metabolomics assays were thawed at room temperature for about 15 min and vortexed for 5 s. Afterwards, 300 μL methanol of HPLC grade (Merck) was added to 100 μL plasma. After shaking for 30 s and standing for 20 min at 4°C, the mixture was centrifuged at 12,000 rpm for 15 min at 4 C. The supernatant was transferred into a vial, and 4 μL sample was injected into LC-Q-TOF MS (Agilent, 1,290 Infinity LC, 6530 UHD and Accurate-Mass Q-TOF/MS). The plasma extract was injected into a C18 column (Agilent, 100 × 2.1 mm, 1.8 μm) in line with LC system and equilibrated with the mobile phase (A: water containing 0.1% formic acid; B: acetonitrile containing 0.1% formic acid). Samples were eluted by 95% A and 5% B at a flow rate of 0.4 ml/min for 16 min, and 5% A and 95% B at a flow rate of 0.4 ml/min for 3 min. The metabolites were separated by LC, analyzed, and assigned by Q-TOF-MS (Agilent). The Q-TOF was operated in ESI-positive mode.
The capillary and sampling cone voltages were set at 4 kV and 35 kV, respectively. The desolvation gas flow was set to 600 L/h at a temperature of 350°C, and the source temperature was set to 100 °C. The TOF-MS data were collected in the range of m/z 50–1,000 with a scan time of 0.03 s and inter scan time of 0.02 s. In order to insure the accuracy and repeatability, the leucine-enkephalin was applied as lock mass. The changed metabolites with p value ≦ 0.05 and FDR ≦0.05 were deemed as significantly changed metabolites. The KEGG pathway analyses for differentially changed metabolites were carried out in the website of KEGG (https://www.genome.jp/kegg/).
Soybean seeds were sterilized with chlorine gas for 12–14 h and then germinated in Hogland medium for 4 days under 16 h/8 h light/dark conditions in a growth chamber at 25–26 C. Germinating seedlings were used for hairy root transformation as described previously with Agrobacterium rhizogenes strain K599 (Kereszt et al., 2007). Transgenic composite plants and controls (cv. Tianlong 2) were transplanted to pots (10 × 10 cm) containing sterilized sands and grown for 3 days. About 2000 SCN eggs were inoculated for each container. Thirty days after inoculation, the SCN female numbers were counted. For each gene, at least ten plants with transgenic hairy roots were obtained and analyzed.
The data presented in the study are deposited in the BioStudies database, accession number E-MTAB-12363.
YJ designed the research and wrote the article. SC, HM, KL, JL, HL, LQ, XZ, MC, WL, XC, XQ, JX, YL, WL, EX. Performed the data analysis and experiments. YJ coordinated the study. All authors read and approved the final manuscript.
This research was funded by National Natural Science Foundation of China (31971971, 32172418, and 31901895), Joint Fund for Science and Technology Research and Development Plan of Henan (222301420028) and Henan Agricultural University (30500689).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1055867/full#supplementary-material
Alkharouf, N. W., Klink, V. P., Chouikha, I. B., Beard, H. S., MacDonald, M. H., Meyer, S., et al. (2006). Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode). Planta 224 (4), 838–852. doi:10.1007/s00425-006-0270-8
Barnes, J. P., and Putnam, A. R. (1987). Role of benzoxazinones in allelopathy by rye (Secale cereale L.). J. Chem. Ecol. 13 (4), 889–906. doi:10.1007/BF01020168
Bellafiore, S., and Briggs, S. P. (2010). Nematode effectors and plant responses to infection. Curr. Opin. Plant Biol. 13 (4), 442–448. doi:10.1016/j.pbi.2010.05.006
Bravo, H. R., and Lazo, W. (1993). Antimicrobial activity of cereal hydroxamic acids and related compounds. Phytochemistry 33, 569–571. doi:10.1016/0031-9422(93)85450-6
Carvalhais, L. C., Schenk, P. M., and Dennis, P. G. (2017). Jasmonic acid signalling and the plant holobiont. Curr. Opin. Microbiol. 37, 42–47. doi:10.1016/j.mib.2017.03.009
Concibido, V. C., Diers, B. W., and Arelli, P. R. (2004). A decade of QTL mapping for cyst nematode resistance in soybean. Crop Sci. 44 (4), 1121–1131. doi:10.2135/cropsci2004.1121
Cook, D. E., Bayless, A. M., Wang, K., Guo, X. L., Song, Q. J., Jiang, J. M., et al. (2014). Distinct copy number, coding sequence, and locus methylation patterns underlie rhg1-mediated soybean resistance to soybean cyst nematode. Plant Physiol. 165 (2), 630–647. doi:10.1104/pp.114.235952
Cook, D. E., Lee, T. G., Guo, X. L., Melito, S., Wang, K., Bayless, A. M., et al. (2012). Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338 (6111), 1206–1209. doi:10.1126/science.1228746
Croft, K., Juttner, F., and Slusarenko, A. J. (1993). Volatile products of the lipoxygenase pathway evolved from Phaseolus vulgaris (L.) leaves inoculated with Pseudomonas syringae pv phaseolicola. Plant Physiol. 101 (1), 13–24. doi:10.1104/pp.101.1.13
Dar, T. A., Uddin, M., Khan, M. M. A., Hakeem, K. R., and Jaleel, H. (2015). Jasmonates counter plant stress: A review. Environ. Exp. Bot. 115, 49–57. doi:10.1016/j.envexpbot.2015.02.010
Dong, L., Cheng, Y., Wu, J., Cheng, Q., Li, W., Fan, S., et al. (2015). Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J. Exp. Bot. 66 (9), 2635–2647. doi:10.1093/jxb/erv078
Erb, M., Meldau, S., and Howe, G. A. (2012). Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 17 (5), 250–259. doi:10.1016/j.tplants.2012.01.003
Esen, A. (1992). Purification and partial characterization of maize (Zea mays L.) beta-glucosidase. Plant Physiol. 98 (1), 174–182. doi:10.1104/pp.98.1.174
Feng, K., Hou, X. L., Xing, G. M., Liu, J. X., Duan, A. Q., Xu, Z. S., et al. (2020). Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 40 (6), 750–776. doi:10.1080/07388551.2020.1768509
Ferreyra, M. L. F., Rius, S. P., and Casati, P. (2012). Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3, 222. Artn 222. doi:10.3389/fpls.2012.00222
Flematti, G. R., Waters, M. T., Scaffidi, A., Merritt, D. J., Ghisalberti, E. L., Dixon, K. W., et al. (2013). Karrikin and cyanohydrin smoke signals provide clues to new endogenous plant signaling compounds. Mol. Plant 6 (1), 29–37. doi:10.1093/mp/sss132
Gheysen, G., and Mitchum, M. G. (2011). How nematodes manipulate plant development pathways for infection. Curr. Opin. Plant Biol. 14 (4), 415–421. doi:10.1016/j.pbi.2011.03.012
Grombacer Aw, R. W., and Guthrie, W. D. (1989). Resistance to firstgerneration European corn borer (Lepidoptera: Pyralidae) and DIMBOA concentration in midwhorl leaves of the BS9 maize synthetic. J. Kans Entomol. Soc. 62, 103–107.
Hermsmeier, D., Mazarei, M., and Baum, T. J. (1998). Differential display analysis of the early compatible interaction between soybean and the soybean cyst nematode. Mol. Plant. Microbe. Interact. 11(12), 1258–1263. doi:10.1094/Mpmi.1998.11.12.1258
Hernandez, I., Alegre, L., Van Breusegem, F., and Munne-Bosch, S. (2009). How relevant are flavonoids as antioxidants in plants? Trends Plant Sci. 14 (3), 125–132. doi:10.1016/j.tplants.2008.12.003
Hussain, S. S., Ali, M., Ahmad, M., and Siddique, K. H. (2011). Polyamines: Natural and engineered abiotic and biotic stress tolerance in plants. Biotechnol. Adv. 29 (3), 300–311. doi:10.1016/j.biotechadv.2011.01.003
Ithal, N., Recknor, J., Nettleton, D., Maier, T., Baum, T. J., and Mitchum, M. G. (2007). Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Mol. Plant. Microbe. Interact. 20 (5), 510–525. doi:10.1094/Mpmi-20-5-0510
Jiao, Y., Vuong, T. D., Liu, Y., Meinhardt, C., Liu, Y., Joshi, T., et al. (2015). Identification and evaluation of quantitative trait loci underlying resistance to multiple HG types of soybean cyst nematode in soybean PI 437655. Theor. Appl. Genet. 128 (1), 15–23. doi:10.1007/s00122-014-2409-5
Kadam, S., Vuong, T. D., Qiu, D., Meinhardt, C. G., Song, L., Deshmukh, R., et al. (2016). Genomic-assisted phylogenetic analysis and marker development for next generation soybean cyst nematode resistance breeding. Plant Sci. 242, 342–350. doi:10.1016/j.plantsci.2015.08.015
Kallenbach, M., Gilardoni, P. A., Allmann, S., Baldwin, I. T., and Bonaventure, G. (2011). C12 derivatives of the hydroperoxide lyase pathway are produced by product recycling through lipoxygenase-2 in Nicotiana attenuata leaves. New Phytol. 191 (4), 1054–1068. doi:10.1111/j.1469-8137.2011.03767.x
Kereszt, A., Li, D., Indrasumunar, A., Nguyen, C. D., Nontachaiyapoom, S., Kinkema, M., et al. (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2 (4), 948–952. doi:10.1038/nprot.2007.141
Khan, R., Alkharouf, N., Beard, H., MacDonald, M., Chouikha, I., Meyer, S., et al. (2004). Microarray analysis of gene expression in soybean roots susceptible to the soybean cyst nematode two days post invasion. J. Nematol. 36 (3), 241–248.
Kim, M., Hyten, D. L., Bent, A. F., and Diers, B. W. (2010a). Fine mapping of the SCN resistance locus rhg1-b from PI 88788. Plant Genome 3 (2), plantgenome2010.02.0001–89. doi:10.3835/plantgenome2010.02.0001
Kim, Y. H., Kim, K. S., and Riggs, R. D. (2010b). Differential subcellular responses in resistance soybeans infected with soybean cyst nematode races. Plant Pathol. J. 26(2), 154–158. doi:10.5423/Ppj.2010.26.2.154
Klink, V. P., Hosseini, P., Matsye, P., Alkharouf, N. W., and Matthews, B. F. (2009). A gene expression analysis of syncytia laser microdissected from the roots of the Glycine max (soybean) genotype PI 548402 (Peking) undergoing a resistant reaction after infection by Heterodera glycines (soybean cyst nematode). Plant Mol. Biol. 71 (6), 525–567. doi:10.1007/s11103-009-9539-1
Klink, V. P., Hosseini, P., Matsye, P. D., Alkharouf, N. W., and Matthews, B. F. (2011). Differences in gene expression amplitude overlie a conserved transcriptomic program occurring between the rapid and potent localized resistant reaction at the syncytium of the Glycine max genotype Peking (PI 548402) as compared to the prolonged and potent resistant reaction of PI 88788. Plant Mol. Biol. 75 (1-2), 141–165. doi:10.1007/s11103-010-9715-3
Klink, V. P., Hosseini, P., Matsye, P. D., Alkharouf, N. W., and Matthews, B. F. (2010). Syncytium gene expression in Glycine max([PI 88788]) roots undergoing a resistant reaction to the parasitic nematode Heterodera glycines. Plant Physiol. biochem. 48 (2-3), 176–193. doi:10.1016/j.plaphy.2009.12.003
Klink, V. P., Overall, C. C., Alkharouf, N. W., MacDonald, M. H., and Matthews, B. F. (2007b). A time-course comparative microarray analysis of an incompatible and compatible response by Glycine max (soybean) to Heterodera glycines (soybean cyst nematode) infection. Planta 226 (6), 1423–1447. doi:10.1007/s00425-007-0581-4
Klink, V. P., Overall, C. C., Alkharouf, N. W., MacDonald, M. H., and Matthews, B. F. (2007a). Laser capture microdissection (LCM) and comparative microarray expression analysis of syncytial cells isolated from incompatible and compatible soybean (Glycine max) roots infected by the soybean cyst nematode (Heterodera glycines). Planta 226 (6), 1389–1409. doi:10.1007/s00425-007-0578-z
Liu, S., Kandoth, P. K., Lakhssassi, N., Kang, J., Colantonio, V., Heinz, R., et al. (2017). The soybean GmSNAP18 gene underlies two types of resistance to soybean cyst nematode. Nat. Commun. 8, 14822. doi:10.1038/ncomms14822
Liu, S. M., Kandoth, P. K., Warren, S. D., Yeckel, G., Heinz, R., Alden, J., et al. (2012). A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature 492(7428), 256–260. doi:10.1038/nature11651
Lorenzo, O., Piqueras, R., Sanchez-Serrano, J. J., and Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 15 (1), 165–178. doi:10.1105/tpc.007468
Lu, J., Ju, H., Zhou, G., Zhu, C., Erb, M., Wang, X., et al. (2011). An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant J. 68 (4), 583–596. doi:10.1111/j.1365-313X.2011.04709.x
Matsye, P. D., Showmaker, K. C., Hosseini, P., Alkharouf, N. W., Lawrence, G. W., Matthews, B. F., et al. (2010). Syncytium gene expression in Glycine max (pi 88788) roots undergoing a resistant reaction to the parasitic nematode Heterodera Glycines. J. Nematology 42 (3), 255–256.
Matthews, B. F., Beard, H., Brewer, E., Kabir, S., MacDonald, M. H., and Youssef, R. M. (2014). Arabidopsis genes, AtNPR1, AtTGA2 and AtPR-5, confer partial resistance to soybean cyst nematode (Heterodera glycines) when overexpressed in transgenic soybean roots. BMC Plant Biol. 14, 96. doi:10.1186/1471-2229-14-96
Meksem, K., Pantazopoulos, P., Njiti, V. N., Hyten, L. D., Arelli, P. R., and Lightfoot, D. A. (2001). 'Forrest' resistance to the soybean cyst nematode is bigenic: Saturation mapping of the Rhg1 and Rhg4 loci. Theor. Appl. Genet. 103(5), 710–717. doi:10.1007/s001220100597
Niblack, T. L., Lambert, K. N., and Tylka, G. L. (2006). A model plant pathogen from the kingdom animalia: Heterodera glycines, the soybean cyst nematode. Annu. Rev. Phytopathol. 44, 283–303. doi:10.1146/annurev.phyto.43.040204.140218
Niblack, T. L., Tylka, G. L., Arelli, P., Bond, J., Diers, B., Donald, P., et al. (2009). Proposal for a standard greenhouse method of assessing soybean cyst nematode resistance in soybean: Sce08 (standardized cyst evaluation 2008). J. Nematology 41 (4), 364.
Park, C. M. (2007). Auxin homeostasis in plant stress adaptation response. Plant Signal. Behav. 2 (4), 306–307. doi:10.4161/psb.2.4.4069
Pollard, M., Beisson, F., Li, Y., and Ohlrogge, J. B. (2008). Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plant Sci. 13 (5), 236–246. doi:10.1016/j.tplants.2008.03.003
Schaller, A., and Stintzi, A. (2009). Enzymes in jasmonate biosynthesis - structure, function, regulation. Phytochemistry 70 (13-14), 1532–1538. doi:10.1016/j.phytochem.2009.07.032
Sicker, D., Frey, M., Schulz, M., and Gierl, A. (2000). Role of natural benzoxazinones in the survival strategy of plants. Int. Rev. Cytol. 198, 319–346. doi:10.1016/s0074-7696(00)98008-2
Simmonds, M. S. J. (2003). Flavonoid-insect interactions: Recent advances in our knowledge. Phytochemistry 64 (1), 21–30. doi:10.1016/S0031-9422(03)00293-0
Suzuki, N., Rivero, R. M., Shulaev, V., Blumwald, E., and Mittler, R. (2014). Abiotic and biotic stress combinations. New Phytol. 203 (1), 32–43. doi:10.1111/nph.12797
Vuong, T. D., Sleper, D. A., Shannon, J. G., and Nguyen, H. T. (2010). Novel quantitative trait loci for broad-based resistance to soybean cyst nematode (Heterodera glycines Ichinohe) in soybean PI 567516C. Theor. Appl. Genet. 121 (7), 1253–1266. doi:10.1007/s00122-010-1385-7
Wang, D., Arelli, P. R., Shoemaker, R. C., and Diers, B. W. (2001). Loci underlying resistance to Race 3 of soybean cyst nematode in Glycine soja plant introduction 468916. Theor. Appl. Genet. 103(4), 561–566. doi:10.1007/Pl00002910
Wasternack, C., and Song, S. (2017). Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 68 (6), 1303–1321. doi:10.1093/jxb/erw443
Zhang, G., Chen, M., Li, L., Xu, Z., Chen, X., Guo, J., et al. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 60 (13), 3781–3796. doi:10.1093/jxb/erp214
Zhao, Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61, 49–64. doi:10.1146/annurev-arplant-042809-112308
Keywords: soybean, soybean cyst nematode, Peking, PI 88788, transcriptome, metabolome
Citation: Chu S, Ma H, Li K, Li J, Liu H, Quan L, Zhu X, Chen M, Lu W, Chen X, Qu X, Xu J, Lian Y, Lu W, Xiong E and Jiao Y (2022) Comparisons of constitutive resistances to soybean cyst nematode between PI 88788- and Peking-type sources of resistance in soybean by transcriptomic and metabolomic profilings. Front. Genet. 13:1055867. doi: 10.3389/fgene.2022.1055867
Received: 28 September 2022; Accepted: 24 October 2022;
Published: 10 November 2022.
Edited by:
Yuanhu Xuan, Shenyang Agricultural University, ChinaReviewed by:
Xiaofeng Zhu, Shenyang Agricultural University, ChinaCopyright © 2022 Chu, Ma, Li, Li, Liu, Quan, Zhu, Chen, Lu, Chen, Qu, Xu, Lian, Lu, Xiong and Jiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongqing Jiao, amlhb3lvbmdxaW5nQGhlbmF1LmVkdS5jbg==; Erhui Xiong, eGlvbmdlcmh1aUBoZW5hdS5lZHUuY24=
†These authors contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.