- Department of Clinical Laboratory, Henan Province Chest Hospital, Zhengzhou University, Zhengzhou, China
The association between polymorphisms in lncRNA H19 and cancer susceptibility remains to be inconsistent. This study aimed to provide a more precise estimation of the relationship between lncRNA H19 polymorphisms and the risk of cancer based on all available published studies. 53 studies encompassing 32,376 cases and 43,659 controls were included in our meta-analysis by searching the Pubmed, Embase, Web of Science, WanFang, and China National Knowledge Infrastructure databases. Pooled ORs and their 95% CIs were used to estimate the strength between the SNPs in H19 (rs217727, rs2839698, rs2107425, rs3024270, rs2735971, rs3741216, and rs3741219) and cancer susceptibility. The results showed that H19 rs2839698 polymorphism was associated with increased cancer risk in all participants under three genetic models. However, no significant association was identified between the other six SNPs as well as an overall cancer risk. Stratification by ethnicity showed that rs2839698 mutation indicated to be an important hazardous factor for the Asian population. While rs2107425 mutation had a protective effect on the Caucasian population. Stratification by cancer type identified that rs217727 mutation was linked to increased susceptibility to oral squamous cell carcinoma, lung cancer, and hepatocellular carcinoma; whereas rs2839698 mutation was associated with an elevated risk of hematological tumor and digestive system tumor (p < 0.05). Besides, the rs2735971 mutation was connected with the digestive system tumor. In summary, the rs217727, rs2839698, rs2107425 and rs2735971 polymorphisms in H19 have associations with cancer susceptibility.
1 Introduction
Cancer is a serious disease affecting human health, which ranks as the leading cause of mortality worldwide (Dessale et al., 2022). It was estimated that there were about 19.3 million new cases and 10 million cancer-related deaths in 2020 (Sung et al., 2021), which is an important barrier to increasing life expectancy. There are many causes of the tumor, such as individual hereditary, chronic inflammation, unhealthy lifestyles, environmental pollution, and so on (Bade and Dela Cruz, 2020; Dugué et al., 2022). Among them, the role of genetic inheritance was widely accepted (Adolf et al., 2022). Numerous genome-wide association studies (GWAS), as well as case-control studies, have been performed in the last few decades to detect the association of thousands of genes and their mutation with cancer risk and prognosis (Lin et al., 2021; Yang et al., 2021). The long non-coding RNAs (lncRNAs) gene has attracted extensive attention for its critical role in carcinogenesis.
lncRNA, larger than 200 nts, is one of the most important members of the non-coding RNA family, which regulates gene expression through various mechanisms, such as inducing chromosome refactoring, synthesizing endogenous small interfering RNA, and changing protein localization (Bridges et al., 2021). Although only more than 5,000 lncRNAs have been identified, they play important roles in cell regulation at the transcriptional and post-transcriptional levels and are involved in multiple biological processes, including cell cycle, proliferation, and apoptosis (Xia et al., 2022).
LncRNA H19, located on chromosome 11p15.5, belongs to the human imprinted gene and contains five exons as well as four introns (Chen and Sima, 2019). It has been shown to be over-expressed in multiple cancers, such as lung cancer (Zhao et al., 2021), osteosarcoma (Zhao and Ma, 2018), prostate cancer (Singh et al., 2021), breast cancer (Chen et al., 2022), and so on. Given the oncogenic roles of H19 and the function of gene variation in regulating tumor growth and prognosis, increasing studies have been performed to explore the possible role of H19 SNPs in cancer susceptibility (Qin et al., 2019; Cao et al., 2020). For instance, Wu et al. conducted a study and found that H19 polymorphism rs2839698 might influence the susceptibility to hepatocellular carcinoma (HCC) (Wu et al., 2019). However, another study conducted by Li et al. showed that H19 polymorphism rs2839698 was not associated with neuroblastoma susceptibility (Li et al., 2019). In that case, though considerable studies have been performed, consistent results have not been obtained due to inadequate statistical power, ethnic differences, different genotyping techniques, and other reasons. In addition, twenty-four new relevant studies have emerged in the past 2 years. Therefore, this updated systematic review and meta-analysis of all eligible studies aimed to explore a more precise understanding of the association between lncRNA H19 SNPs and cancer risk.
2 Materials and methods
2.1 Search strategy
We searched the Pubmed, Embase, Web of Science, WanFang, and China National Knowledge Infrastructure (CNKI) databases up to May 2022 for relevant articles using the following terms: “lncRNA H19 OR long non-coding RNA H19 OR H19”, “cancer OR carcinoma OR tumor OR malignancy”, and “SNP OR polymorphism OR variant OR mutation”. Besides, we retrieved articles by reading the abstracts, and full articles published in English or Chinese without language restrictions. The references for each retrieved article were also investigated carefully for getting related studies.
2.2 Selection criteria
The studies included in our meta-analysis should meet the following criteria: 1) to evaluate the association between the H19 polymorphism and cancer risk; 2) case-control study or cohort study; and 3) genotype frequencies of case and control groups could be obtained to calculate the odds ratio (OR) and 95% confidence interval (CI). We excluded reviews, letters, meta-analyses, conference papers, and other studies that were short of sufficient genotyping data.
2.3 Data extraction
The information collection was conducted by two investigators independently, and a final agreement was reached by discussing with the third person when there were divergences. The following information was collected from both cases and controls groups: the first author’s name, publication year, country, ethnicity, cancer type, source of the control group, genotyping method, and evidence of Hardy-Weinberg equilibrium (HWE) in the control group.
2.4 Statistical analysis
HWE was evaluated based on the χ2 test in control subjects. Pooled ORs and their 95%CIs were used to estimate the strength between the seven SNPs in H19 (rs217727, rs2839698, rs2107425, rs3024270, rs3741216, rs3741219, and rs2735971) and cancer susceptibility in the following genetic models: allele model, recessive model, dominant model, homozygous model, and heterozygous model. Subgroup analysis was calculated concerning ethnicity, cancer types, sources of control, HWE status, and genotyping methods. Cochran’s Q test and I2 statistic were used to calculate heterogeneity (Tsironikos et al., 2022). When no heterogeneity exists (the I2 value <50% or p > 0.05), the fixed-effect model was performed. Otherwise, a random-effect model was applied. Furthermore, sensitivity analyses were performed to examine the stability of the results. Egger’s test and Begg’s funnel plots were used to assess the publication bias (Zhang et al., 2022). p > 0.05 and symmetrical funnel plots indicate no bias exits.
3 Results
3.1 Characteristics of included studies
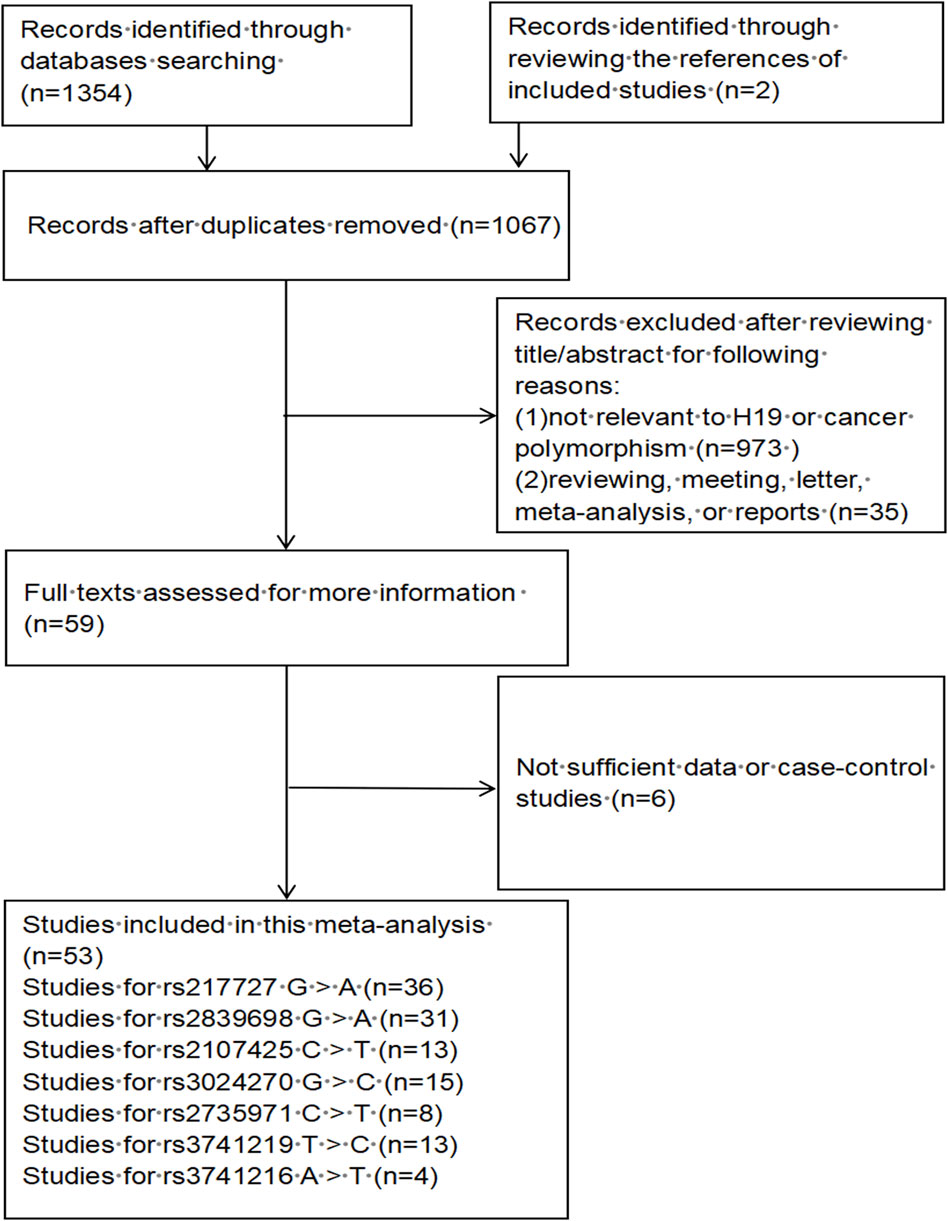
A total of 1,067 publications were preliminarily identified based on our search strategy using different search term combinations. All the abstracts and articles were scrutinized following the selection process (Figure 1). In the end, 53 studies encompassing 32,376 cases and 43,659 controls were incorporated in our meta-analysis, of which 24 studies were updated recently. The characteristics of the 53 studies are shown in Supplementary Table S1. Among the 53 studies, 45 studies were conducted in Asian populations, 6 studies in Caucasian populations, and 2 studies in African populations; 26 studies using the TaqMan genotyping method, 13 studies using PCR-RFLP, 6 studies using MassARRAY, others using KASP, Illumina and so on; the control group of 34 studies were population-based (PB), 19 studies were hospital-based (HB).
3.2 Meta-analysis results of rs2839698 polymorphism
Thirty-one studies investigated the association of rs2839698 polymorphism with cancer susceptibility. A hazardous effect on cancer development was observed in the H19 rs2839698 polymorphism under allele model (OR = 1.09, 95% CI = 1.01–1.18, p = 0.034); dominant model (OR = 1.11, 95% CI = 1.01–1.21, p = 0.029); and homozygous model (OR = 1.25, 95% CI = 1.07–1.46, p = 0.004) in all participates (Supplementary Table S2; Figure 2A).
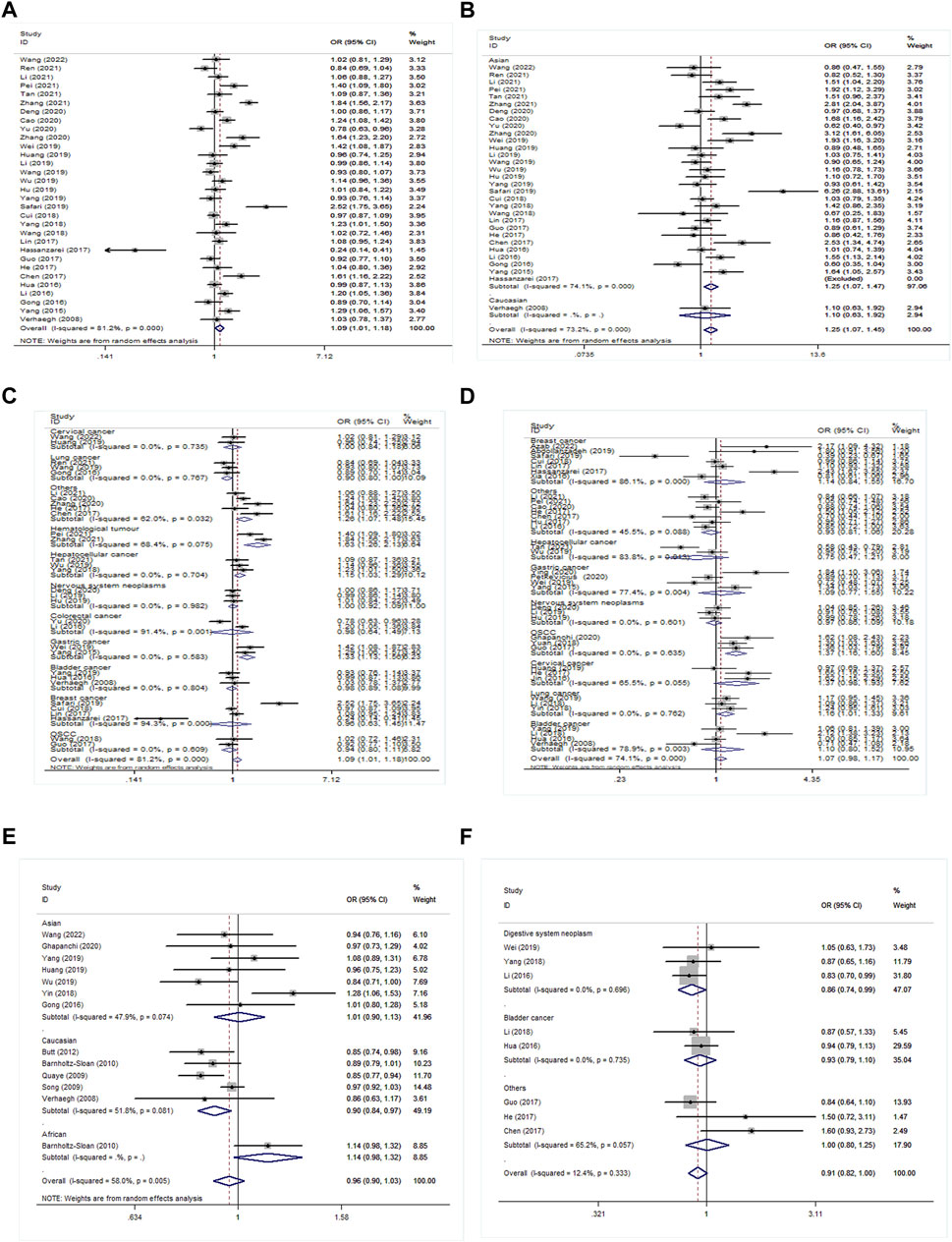
FIGURE 2. Forest plot for the association between H19 polymorphisms and cancer susceptibility. (A) Forest plot for rs2839698 polymorphism in the overall population, A vs G; (B) in ethnicity subgroup, AA vs GG; (C) and in cancer type subgroup, A vs G; (D) for rs217727 polymorphism in cancer type subgroup, AA + AG vs GG; (E) for rs2107425 polymorphism in ethnicity subgroup, T vs C; (F) for rs2735971 polymorphism in cancer type subgroup, TC vs CC.
Stratification by ethnicity identified that H19 rs2839698 mutation significantly increased cancer risk in the Asian population under allele model (OR = 1.09, 95% CI = 1.01–1.19, p = 0.035), dominant model (OR = 1.12, 95% CI = 1.02–1.22, p = 0.020) and homozygous model (OR = 1.26, 95% CI = 1.07–1.47, p = 0.004) (Supplementary Table S2; Figure 2B). Stratification by cancer type identified that H19 rs2839698 mutation increased the risk of digestive system neoplasm under allele model (OR = 1.14, 95% CI = 1.01–1.30, p = 0.040); recessive model (OR = 1.31, 95% CI = 1.02–1.69, p = 0.03); and homozygous model (OR = 1.33, 95% CI = 1.03–1.74, p = 0.03). Disease-specific meta-analysis was performed on hepatocellular cancer (HCC), gastric cancer (GC), colorectal cancer (CRC), and other digestive system neoplasms. The analysis indicated that rs2839698 mutation increased the risk of HCC and GC rather than CRC (Supplementary Table S2; Figure 2C). Furthermore, the significant association between the rs2839698 polymorphism and hematological tumor was demonstrated in all genetic models (p < 0.05) (Supplementary Table S2; Figure 2C). In analyses according to the source of control and genotyping methods, significant associations were observed between rs2839698 G>A and cancer susceptibility in PB and TaqMan (Supplementary Table S2).
3.3 Meta-analysis results of rs217727 polymorphism
Thirty-six studies were selected to estimate the association of the rs217727 SNP with the risk of cancer. No significant association was indicated for rs217727 through the pooled risk estimation in all genetic models (Supplementary Table S3).
In the further subgroup analysis by cancer type, we identified that H19 rs217727 mutation increased oral squamous cell carcinoma (OSCC) risk under all models (allele model: OR = 1.31, 95% CI = 1.14–1.50, p < 0.001; recessive model: OR = 1.67, 95% CI = 1.04–2.67, p = 0.034; dominant model: OR = 1.37, 95% CI = 1.16–1.60, p < 0.001; homozygous model: OR = 1.89, 95% CI = 1.19–2.99, p = 0.007; heterozygote model: OR = 1.27, 95% CI = 1.07–1.51, p = 0.005). The same association with increased lung cancer (LC) risk was also observed in four models (allele model: OR = 1.16, 95% CI = 1.06–1.27, p = 0.002; recessive model: OR = 1.31, 95%CI = 1.03–1.66, p = 0.028; dominant model: OR = 1.16, 95% CI = 1.01–1.33, p = 0.031; homozygous model: OR = 1.38, 95%CI = 1.14–1.67, p = 0.001). While the variant A allele of the rs217727 polymorphism resulted in significantly decreased risk for HCC in recessive model (OR = 0.73, 95% CI = 0.54–1.00, p = 0.048) and homozygous model (OR = 0.68, 95% CI = 0.49–0.93, p = 0.017) (Supplementary Table S3; Figure 2D). In the subgroup of genotyping methods, rs217727 was linked with increased cancer risk through the PCR-RFLP method (p < 0.05, Supplementary Table S3). However, no significant results were detected in the subgroup analysis by ethnicity and source of control.
3.4 Meta-analysis results of rs2107425, rs2735971, rs3024270, rs3741219 and rs3741216 polymorphisms
The association between the H19 rs2107425, rs2735971, rs3024270, rs3741219, and rs3741216 polymorphisms and cancer risk were separately examined in 13 studies, 8 studies, 15 studies, 13 studies, and 4 studies. We did not find any significant association between these five SNPs and cancer risk in all participants. However, in subgroup analysis stratified by ethnicity, cancer type, genotyping methods, and source of control, these SNPs were related to cancer susceptibility except for rs3741216.
The analysis of rs2107425 indicated that a variant T allele decreased cancer risk in the Caucasian population under the allele model (OR = 0.90, 95% CI = 0.84–0.97, p = 0.006), dominant model (OR = 0.84, 95% CI = 0.75–0.94, p = 0.003) and heterozygote model (OR = 0.82, 95% CI = 0.72–0.94, p = 0.003) (Supplementary Table S4; Figure 2E).
For rs2735971, this locus was linked with reduced cancer risk to digestive system neoplasm under three genetic models (allele model: OR = 0.88; 95% CI = 0.78–0.98, p = 0.021; dominant model: OR = 0.85, 95% CI = 0.74–0.98, p = 0.024; and heterozygote model: OR = 0.86, 95% CI = 0.74–0.99, p = 0.034) (Supplementary Table S4; Figure 2F). Additionally, in subgroup analysis by genotyping method, feebly positive results were shown in the TaqMan group for the rs3024270 locus. While the subgroup analysis stratified by the source of the control group indicated that rs3741219 increased cancer susceptibility in the HB control population (Supplementary Table S5). No significant association existed in other subgroups.
3.5 Detection of heterogeneity, sensitivity analysis, and publication bias
There was heterogeneity in the analysis between these seven SNPs and cancer risk, except for the recessive and homozygous models of rs2107425 and rs3024270, the recessive model of rs3741219, the heterozygous model of rs2735971 as well as the dominant, homozygous and heterozygous models of rs3741216. Therefore, a random-effects model was adopted to analyze the cancer risk. Otherwise, a fixed-effects model was applied for others mentioned above. Sensitivity analyses observed no change of polled OR before and after the removal of each study in all results of all genetic models, suggesting the good stability and reliability of our study (Figure 3). Begg’s funnel plot and Egger’ test were used to evaluate publication bias. The funnel plot was symmetrical, indicating that there was no significant publication bias in the selected literature (Figure 4).
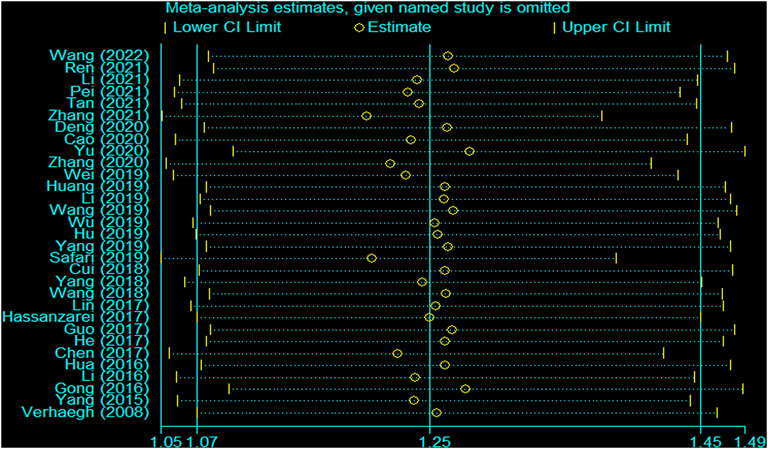
FIGURE 3. Sensitivity analysis for the H19 rs2839698 polymorphism and cancer susceptibility in the overall population (allele model: A vs G).
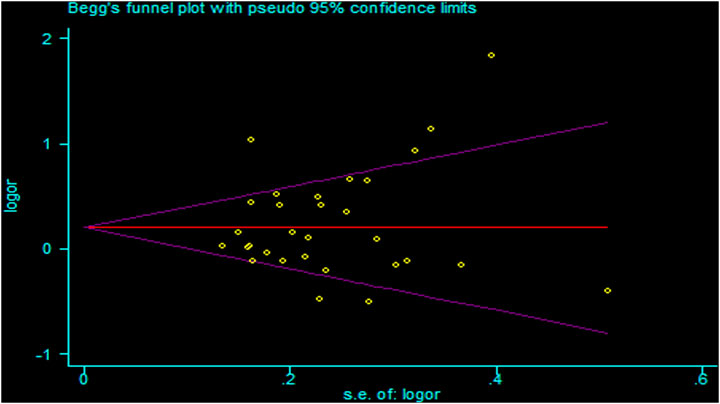
FIGURE 4. Funnel plots for publication bias of H19 rs2839698 polymorphism in the overall population (allele model: A vs G).
4 Discussion
LncRNA plays a crucial role in regulating gene expression and maintaining cell function (Rizk et al., 2022). Accumulating evidence has demonstrated that lncRNA H19 polymorphism was closely related to the initiation, development, and prognosis of cancer (Mitroi et al., 2022; Shaheen et al., 2022). Meanwhile, the study by Allemailem et al. (2021) confirmed the possibility of H19 SNPs in diagnosis and individualized treatment of cancer. A study by Liang et al. (2015) reported that lncRNA H19 can act as the miRNA sponge promoting epithelial to mesenchymal transition (EMT) in CRC. It functions as a primary miRNA precursor or competitive endogenous RNA to regulate specific mRNAs post-transcriptionally (Cai and Cullen, 2007), and the alteration of the SNPs may result in gain and loss of function of miRNA–lncRNA interactions (Correction, 2016). Similarly, alterations in the SNPs may affect the piRNA–lncRNA interactions and thus binding to PIWI proteins that involve in genome integrity, transposon silencing, and epigenetic regulation (Singh et al., 2018; Ozata et al., 2019). Besides, H19 mutation may produce antisense RNA 91H accumulating in breast cancer cells and regulate the expression of insulin growth factor 2 (IGF2) (Berteaux et al., 2008), thereby affecting oncogene and tumor suppressor gene expression. Previously, several meta-analyses have investigated the correlation between H19 polymorphisms and tumorigenesis, and no definitive conclusion has been drawn on this causal relationship (Liu et al., 2020a; Liu et al., 2020b; Wang et al., 2021). Here, we look forward to obtaining more effective evidence to precisely estimate their association and draw a more reliable conclusion. In this study, we collected all the relevant published data up to May 2022 to examine the association between lncRNA H19 rs2839698, rs217727, rs273597, rs2107425, rs3024270, rs3741216, and rs3741219 polymorphisms with cancer risk. We obtained some new conclusions.
Our combined studies demonstrated a clear increase in cancer risk associated with H19 rs2839698 polymorphism in the overall and Asian population group, which contradicts the results of Wang et al. (2021) and Liu et al. (2020b) found no association between the H19 rs2839698 polymorphism and cancer risk neither in the overall group nor in the ethnicity subgroup. The obvious alteration of estimates of effect could be due to increased sample size, given that the 13 newly included studies accounted for 27.18% of individuals. In the subgroup analysis by cancer type, rs2839698 polymorphism was associated with increased cancer risk in hematologic tumors and digestive system tumors. To our knowledge, this is the first meta-analysis to investigate the association between H19 rs2839698 and the risk of hematologic tumors. Moreover, the function of H19 in hematologic tumors is seemingly much more complex than that in other tumor types because of the highly heterogeneous pathogenesis (Pei et al., 2021; Sattarzadeh Bardsiri et al., 2022). We additionally performed a disease-specific meta-analysis on digestive tumors, and the results indicated a significant association between HCC and GC as well as H19 rs2839698 than CRC. The H19 rs2839698 polymorphism effects were tissue-specific, which may give a clue for early screening of the digestive system tumor.
The previous meta-analysis obtained inconsistent findings on the correlation between H19 rs217727 polymorphism and cancer susceptibility. Liu et al. (2020b) found that rs217727 is associated with increased cancer risk in the overall and Asian populations. However, our updated meta-analysis is more supportive of Wang’s study that no effect was detected between rs217727 polymorphism and cancer susceptibility in the general population and ethnicity subgroup (Wang et al., 2021). This difference may result from 18 new articles included in our study. Besides, in the subgroup analysis by cancer type, we found the rs217727 polymorphism may be a risk factor for the occurrence of OSCC and LC, as well as a protective factor for HCC.
In the previous meta-analysis, no association was found between the rs2735971 polymorphism and cancer risk in any genetic models (Li et al., 2020; Wang et al., 2021). However, we added five recently published articles and deleted studies using overlapping data and small sample sizes, finding that the A allele or AA genotype is a potential genetic marker for gastrointestinal tumors. Furthermore, our meta-analysis demonstrated that rs2107425 polymorphism may be a protective factor in the Caucasian population but not the Asian population, which further confirmed the results of the study by Liu et al. (2020b) and Li et al. (2020). The influence of SNP rs2107425 on tumor susceptibility was affected by ethnic factors. We also explored the role of rs3024270, rs3741219, and rs3741216 polymorphisms in cancer risk. No association was found between them.
In addition, the influence of genotypic methods and population information on cancer susceptibility was also explored in our study. When stratified by genotypic methods, rs217727 polymorphism was significantly correlated with PCR-RFLP, rs2839698, and rs3024270 polymorphisms were significantly correlated with TaqMan. Both PCR-RFLP and TaqMan have high confidence in the analysis of gene polymorphism due to their respective merits (Hubáček et al., 2015). In addition, Wang’ meta-analysis pointed out that there was a significant correlation between rs217727 polymorphism and the HB control group when subgroup analysis by a source of controls was performed (Wang et al., 2021). Nevertheless, crucial results were found concentrated in the HB control group in the rs3741219 locus in our study. Insufficient sample size and lack of representativeness may be the reason for this phenomenon.
There are some limitations to this research. Firstly, there is little published research about rs2735971 and rs3741216 polymorphisms, which may limit the statistical power and decrease the reliability of the results. Secondly, in addition to genetic factors, some characteristics of participants, such as age, alcohol intake, and tobacco smoking also influence cancer susceptibility. However, the interaction between these factors and cancer risk could not be assessed due to insufficient data. Thirdly, there exists intensity, moderate or low heterogeneity in this meta-analysis, which may reduce the reliability. Lastly, as mentioned above, because almost all the studies included focused on the Asian population, it is unreasonable to suggest that there is a significant ethnic difference between rs217727 and rs2839698 polymorphisms and cancer risk. This conclusion may not apply to all populations.
In conclusion, our results demonstrated that the H19 rs2839698 polymorphism increased cancer susceptibility, whereas the rs2107425 mutation decreased cancer susceptibility in the Caucasian population. Rs2839698 polymorphism was associated with an increased risk of hematologic tumor and digestive system cancer. Rs217727 polymorphism was associated with OSCC, LC, and HCC risk. Rs2735971 mutation significantly increased the risk of digestive system cancer. Considering the limitations mentioned above, further well-designed case-control studies with larger sample sizes and more different ethnic groups are still warranted to explore the impact of H19 genetic variants on cancer risk.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
YY and WW designed the study and finally approved the manuscript. YY, YW, XN, and YH contributed to searching the literature, analyzing the data, and interpreting the result. WL, ZL, MC, JT, and YZ participated in discussions and revised the draft.
Acknowledgments
The authors are grateful to the researchers who participated in data collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1051766/full#supplementary-material
Abbreviations
LncRNA, long non-coding RNA; GWAS, genome-wide association studies; SNP, single-nucleotide polymorphism; CNKI, China National Knowledge Infrastructure; CI, confidence interval; OR, odds ratio; HWE, Hardy–Weinberg equilibrium; PB, population-based; HB, hospital-based; EMT, epithelial to mesenchymal transition; IGF2, insulin growth factor 2.
References
Adolf, I. C., Rweyemamu, L. P., Akan, G., Mselle, T. F., Dharsee, N., Namkinga, L. A., et al. (2022). The interplay between xpg -Asp1104His polymorphism and reproductive risk factors elevates risk of breast cancer in Tanzanian women: A multiple interaction analysis. Cancer Med. doi:10.1002/cam4.4914
Allemailem, K. S., Almatroudi, A., Alrumaihi, F., Makki Almansour, N., Aldakheel, F. M., Rather, R. A., et al. (2021). Single nucleotide polymorphisms (SNPs) in prostate cancer: Its implications in diagnostics and therapeutics. Am. J. Transl. Res. 13, 3868–3889.
Bade, B. C., and Dela Cruz, C. S. (2020). Lung cancer 2020: Epidemiology, etiology, and prevention. Clin. Chest Med. 41, 1–24. doi:10.1016/j.ccm.2019.10.001
Berteaux, N., Aptel, N., Cathala, G., Genton, C., Coll, J., Daccache, A., et al. (2008). A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol. Cell. Biol. 28, 6731–6745. doi:10.1128/MCB.02103-07
Bridges, M. C., Daulagala, A. C., and Kourtidis, A. (2021). LNCcation: lncRNA localization and function. J. Cell. Biol. 220, e202009045. doi:10.1083/jcb.202009045
Cai, X., and Cullen, B. R. (2007). The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA (New York, N.Y.) 13, 313–316. doi:10.1261/rna.351707
Cao, Q., Li, P., Cao, P., Qian, J., Du, M., Li, L., et al. (2020). Genetic variant in long non-coding RNA H19 modulates its expression and predicts renal cell carcinoma susceptibility and mortality. Front. Oncol. 10, 785. doi:10.3389/fonc.2020.00785
Chen, J., Qin, C., Zhou, Y., Chen, Y., Mao, M., and Yang, J. (2022). Metformin may induce ferroptosis by inhibiting autophagy via lncRNA H19 in breast cancer. FEBS open bio 12, 146–153. doi:10.1002/2211-5463.13314
Chen, Y., and Sima, X. (2019). Replication of GWAS loci revealed an increased risk of BET1L and H19 polymorphisms with intracranial aneurysm. Dis. Markers 2019, 9490639. doi:10.1155/2019/9490639
Correction, (2016). fMiRNA-192 and miRNA-204 directly suppress lncRNA HOTTIP and interrupt GLS1-mediated glutaminolysis in hepatocellular carcinoma. PLoS Genet. 12, e1005825.
Dessale, M., Mengistu, G., and Mengist, H. M. (2022). Nanotechnology: A promising approach for cancer diagnosis, therapeutics and theragnosis. Int. J. Nanomedicine 17, 3735–3749. doi:10.2147/IJN.S378074
Dugué, P. A., Bodelon, C., Chung, F. F., Brewer, H. R., Ambatipudi, S., Sampson, J. N., et al. (2022). Blood DNA methylation and breast cancer risk: A meta-analysis of four prospective cohort studies. Breast Cancer Res. 24, 62. doi:10.1186/s13058-019-1145-9
Hubáček, J. A., Pikhart, H., Peasey, A., Kubínová, R., and Bobák, M. (2015). Nobody is perfect: Comparison of the accuracy of PCR-RFLP and KASP™ method for genotyping. ADH1B and FTO polymorphisms as examples. Folia Biol. 61, 156–160.
Li, W., Jiang, X., Jin, X., Yan, W., Liu, Y., Li, D., et al. (2020). Significant association between long non-coding RNA H19 polymorphisms and cancer susceptibility: A PRISMA-compliant meta-analysis and bioinformatics prediction. Medicine 99, e19322. doi:10.1097/MD.0000000000019322
Li, Y., Zhuo, Z. J., Zhou, H., Liu, J., Zhang, J., Cheng, J., et al. (2019). H19 gene polymorphisms and neuroblastoma susceptibility in Chinese children: A six-center case-control study. J. Cancer 10, 6358–6363. doi:10.7150/jca.37564
Liang, W. C., Fu, W. M., Wong, C. W., Wang, Y., Wang, W. M., Hu, G. X., et al. (2015). The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 6, 22513–22525. doi:10.18632/oncotarget.4154
Lin, X., Wu, Y., Liu, F., Na, R., Huang, D., Xu, D., et al. (2021). A germline variant at 8q24 contributes to the serum p2PSA level in a Chinese prostate biopsy cohort. Front. Oncol. 11, 753920. doi:10.3389/fonc.2021.753920
Liu, C., Chen, L., You, Z., Wu, Y., Wang, C., Zhang, G., et al. (2020). Association between lncRNA H19 polymorphisms and cancer susceptibility based on a meta-analysis from 25 studies. Gene 729, 144317. doi:10.1016/j.gene.2019.144317
Liu, X., Zhao, Y., Li, Y., and Zhang, J. (2020). Quantitative assessment of lncRNA H19 polymorphisms and cancer risk: A meta-analysis based on 48, 166 subjects. Artif. Cells Nanomed. Biotechnol. 48, 15–27. doi:10.1080/21691401.2019.1699804
Mitroi, A. F., Leopa, N., Dumitru, E., Brînzan, C., Tocia, C., Dumitru, A., et al. (2022). Association of TCF7L2, CASC8 and GREM1 polymorphisms in patients with colorectal cancer and type II diabetes mellitus. Genes. 13, 1297. doi:10.3390/genes13081297
Ozata, D. M., Gainetdinov, I., Zoch, A., O'Carroll, D., and Zamore, P. D. (2019). PIWI-Interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 20, 89–108. doi:10.1038/s41576-018-0073-3
Pei, J. S., Chen, C. C., Chang, W. S., Wang, Y. C., Chen, J. C., Hsiau, Y. C., et al. (2021). Significant associations of lncRNA H19 genotypes with susceptibility to childhood leukemia in taiwan. Pharm. (Basel, Switz. 14, 235. doi:10.3390/ph14030235
Qin, W., Wang, X., Wang, Y., Li, Y., Chen, Q., Hu, X., et al. (2019). Functional polymorphisms of the lncRNA H19 promoter region contribute to the cancer risk and clinical outcomes in advanced colorectal cancer. Cancer Cell. Int. 19, 215. doi:10.1186/s12935-019-0895-x
Rizk, N. I., Abulsoud, A. I., Kamal, M. M., Kassem, D. H., and Hamdy, N. M. (2022). Exosomal-long non-coding RNAs journey in colorectal cancer: Evil and goodness faces of key players. Life Sci. 292, 120325. doi:10.1016/j.lfs.2022.120325
Sattarzadeh Bardsiri, M., Zehtab, S., Karami, N., Farsinejad, A., Ehsan, M., and Fatemi, A. (2022). Association of IKZF1 and CDKN2A gene polymorphisms with childhood acute lymphoblastic leukemia: A high-resolution melting analysis. BMC Med. Genomics 15, 171. doi:10.1186/s12920-022-01325-6
Shaheen, S., Alshammari, E. M., Mokhtar, S. H., Alshanwani, A. R., Toraih, E. A., Ibrahiem, A. T., et al. (2022). PUNISHER rs12318065 C>A transversion: A putative somatic driver mutation for poor prognosis in colon cancer. Biosci. Rep. 42, BSR20220465. doi:10.1042/BSR20220465
Singh, G., Roy, J., Rout, P., and Mallick, B. (2018). Genome-wide profiling of the PIWI-interacting RNA-mRNA regulatory networks in epithelial ovarian cancers. PloS one 13, e0190485. doi:10.1371/journal.pone.0190485
Singh, N., Ramnarine, V. R., Song, J. H., Pandey, R., Padi, S. K. R., Nouri, M., et al. (2021). The long noncoding RNA H19 regulates tumor plasticity in neuroendocrine prostate cancer. Nat. Commun. 12, 7349. doi:10.1038/s41467-021-26901-9
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Tsironikos, G. I., Perivoliotis, K., Bargiota, A., Zintzaras, E., Doxani, C., and Tatsioni, A. (2022). Effectiveness of exercise intervention during pregnancy on high-risk women for gestational diabetes mellitus prevention: A meta-analysis of published RCTs. PloS one 17, e0272711. doi:10.1371/journal.pone.0272711
Wang, K., Zhu, Z., Wang, Y., Zong, D., Xue, P., Gu, J., et al. (2021). The influence of LncRNA H19 polymorphic variants on susceptibility to cancer: A systematic review and updated meta-analysis of 28 case-control studies. PloS one 16, e0254943. doi:10.1371/journal.pone.0254943
Wu, E. R., Chou, Y. E., Liu, Y. F., Hsueh, K. C., Lee, H. L., Yang, S. F., et al. (2019). Association of lncRNA H19 gene polymorphisms with the occurrence of hepatocellular carcinoma. Genes. 10, 506. doi:10.3390/genes10070506
Xia, L., Chen, J., Huang, M., Mei, J., and Lin, M. (2022). The functions of long noncoding RNAs on regulation of F-box proteins in tumorigenesis and progression. Front. Oncol. 12, 963617. doi:10.3389/fonc.2022.963617
Yang, S., Wang, Y. L., Lyu, Y., Jiang, Y., Xiang, J., Ji, S., et al. (2021). mGWAS identification of six novel single nucleotide polymorphism loci with strong correlation to gastric cancer. Cancer Metab. 9, 34. doi:10.1186/s40170-021-00269-2
Zhang, M. Y., Zhao, P., Zhang, Y., and Wang, J. S. (2022). Efficacy and safety of ruxolitinib for steroid-refractory graft-versus-host disease: Systematic review and meta-analysis of randomised and non-randomised studies. PloS one 17, e0271979. doi:10.1371/journal.pone.0271979
Zhao, J., and Ma, S. T. (2018). Downregulation of lncRNA H19 inhibits migration and invasion of human osteosarcoma through the NF-κB pathway. Mol. Med. Rep. 17, 7388–7394. doi:10.3892/mmr.2018.8746
Keywords: lncRNA, H19, cancer susceptibility, polymorphism, meta-analysis
Citation: Yuan Y, Wang Y, Niu X, Han Y, Li W, Cheng M, Li Z, Tan J, Zhao Y and Wang W (2022) Association of lncRNA H19 polymorphisms with cancer susceptibility: An updated meta-analysis based on 53 studies. Front. Genet. 13:1051766. doi: 10.3389/fgene.2022.1051766
Received: 23 September 2022; Accepted: 28 November 2022;
Published: 14 December 2022.
Edited by:
Anshuman Mishra, Shobhit University, IndiaReviewed by:
Santanu Patra, Khon Kaen University, ThailandVinoth Kumar Lakshmanan, Sri Ramachandra Institute of Higher Education and Research, India
Copyright © 2022 Yuan, Wang, Niu, Han, Li, Cheng, Li, Tan, Zhao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Wang, anlrMjc4NUAxNjMuY29t
 Yingying Yuan
Yingying Yuan Yachun Wang
Yachun Wang Xiaodong Niu
Xiaodong Niu Wei Wang
Wei Wang