
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 24 October 2022
Sec. Cancer Genetics and Oncogenomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1050402
This article is part of the Research Topic The Genetics of Head and Neck Squamous Cell Carcinoma View all 20 articles
Objectives: Tight junction-associated marvel proteins (TAMP) is a transmembrane protein whose members are associated with tight junctions between cells and epithelial remodeling. MARVEL domain containing 3 (MARVELD3) is one of the members of the TAMP. MARVELD3, as a novel tight junction protein involved in bicellular tight junction assembly, has attracted growing attention in the field of oncology. This study aimed to investigate the prognostic role of MARVELD3 and to determine how it functions in tumorigenesis in oral squamous cell carcinoma (OSCC), thus providing additional data to help the guidance of clinical practice.
Materials and Methods: RNA-seq data and relevant clinical information were obtained from TCGA. Bioinformatics means used in this study included differential gene expression analysis, KM survival curve analysis, univariate and multivariate Cox regression analyses, nomogram analysis, ROC curve analysis, methylation level analysis, gene function enrichment analysis, and immune cell infiltration analysis.
Results: MARVELD3 was significantly higher expressed in OSCC tissue than in normal tissue, and the overall survival of the high expression group was significantly lower than that of the normal group. Univariate and multivariate Cox regression analyses showed that MARVELD3 could serve as an independent contributing factor to poor OSCC prognosis. The nomograms and ROC curves supported the results above. Its expression was negatively correlated with DNA methylation sites. Analysis of PPI networking and gene functional enrichment showed that MARVELD3 was involved in the functional activities of DNA and RNA and was associated with immune cell infiltration.
Conclusion: The high expression of MARVELD3 is associated with poor prognosis in OSCC, and MARVELD3 could be recognized as a novel independent prognostic factor for OSCC.
Oral squamous cell carcinoma (OSCC) is the most prevalent oral malignant tumor worldwide, with a five-year survival rate of only 50% (Manzano-Moreno et al., 2021). At present, the main treatment methods for OSCC are mainly surgery, chemotherapy, and radiotherapy. The major causes of high mortality are high tumor invasion, lymph node involvement, poor response to therapy, and early local recurrence (Cierpikowski et al., 2021). To be able to effectively improve the survival rate and improve prognosis in patients with OSCC, there is an urgent need to look for a potential biomarker. This marker can be used as a diagnostic indicator as well as a prognostic indicator. The Tight junction Associated Marvel Proteins (TAMP) are transmembrane proteins. The TAMP family consists of occludin, tricellulin (also called MARVELD2), and MARVEL domain containing 3 (MARVELD3) (Heymans et al., 2021). Its members are associated with tight junctions between cells and epithelial remodeling. Most studies focus on the connection function between epithelial cells and the role of epithelial mesenchymal cells in transformation (Kojima et al., 2011). Studies have confirmed that tricellulin is identified as the first marker of tricellular tight junction in epithelial cells. Its loss affects the tight binding of the tricellular tight junction as well as the barrier function of the epithelial cells (Ikenouchi et al., 2005; Ikenouchi et al., 2008). At the same time, Steed et al. found that normal expression of MARVELD3 is not necessary for the formation of functionally tight junctions, but it is a decisive factor in epithelial paracellular permeability properties (Steed et al., 2009). Some literature has also found that MARVELD3 is involved in the process of promoting cell migration by EMT in hepatocellular carcinoma cells, and it inhibits the occurrence and progression of this process through the NF-κB signaling pathway (Li et al., 2021). Some scholars have shown that MARVELD2 and MARVELD3 are included in the genes that are highly correlated with the close connection of epithelial tumor cells (Kohn et al., 2014). In addition to this, studies have shown that MARVELD3 has two isoforms and has a broad tissue distribution. MARVELD3 functions as a regulator of epithelial cell proliferation, migration, and survival in human colon and pancreatic cancer cells, but the role it plays in OSCC is unclear (Li et al., 2021). In addition to that, previous studies have found that MARVEL domain-containing 1 (MARVELD1) could inhibit tumor cell proliferation and enhance the sensitivity to chemotherapeutic drugs in hepatocellular carcinoma (Zhang et al., 2019). So, we suspect that MARVELDs may have a different expression in OSCC than in normal tissue. With the development of bioinformatics analysis technology and the establishment and improvement of various databases, we have studied this problem. This study conducted in-depth analysis of MARVELDs expression in OSCC and assessed their potential value as prognostic biomarkers, providing a new method for guiding clinical work and effectively and accurately assessing the long-term prognosis of OSCC patients.
The Cancer Genome Atlas (TCGA) database was utilized to collect the data on gene expression profiles of 329 samples with OSCC and 32 non-OSCC normal tissue samples. The RNA-seq data and the corresponding clinical information were downloaded from TCGA. Transcripts per million reads (TPM) format of RNA-seq data (from TCGA), through Toil process standardization, were downloaded from UCSC XENA (https://xenabrowser.net/datapages/). Log2 fold change (log2FC) was calculated to further compare mRNA expression levels between tumor and normal samples.
Through data analyses, the samples were divided into high-expression and low-expression groups according to the median expression level of MARVELDs. At the same time, Kaplan-Meier survival analysis was performed using the R package (survive, version 0.4.9 and survival, version 3.2.10). The KM curve was plotted to effectively assess the relationship between the expression level of MARVELDs and the overall survival of patients.
To investigate whether the expression level of MARVELD3, gender, age, tumor stage (T stage), lymphatic involvement grade (N stage), lymph vascular invasion, and histologic grade were independent risk factors for oral squamous cell carcinoma, univariate and multivariate Cox regression analyses were utilized. We used the R package (survival, version 3.2.10) for data processing and set Hazard ratios (HR) and 95% confidence intervals. The significance threshold was set as p < 0.05.
Constructing nomograms were allowed to predict the prognosis of patients with oral squamous cell carcinoma based on age, gender, TNM, clinical grade, and MARVELD3 expression levels. The ROC curves were plotted using the R package (pROC, version 1.17.0.1 and ggplot2, version 3.3.3) to assess the diagnostic value of MARVELD3.
For the gene methylation data, we used Illumina Human Methylation 450 array methylation chip data and the RNA-Seq data of the level 3 HTSeq-FPKM format downloaded from the Head and Neck Squamous Cell Carcinoma (HNSC) project in TCGA. Data with no clinical information were discarded. Samples that belonged to oral cancer sites (such as alveolar ridge, base of tongue, buccal mucosa, floor of mouth, hard palate, oral cavity, and oral tongue) were retained, while others (such as hypopharynx, larynx, lip, oropharynx, and tonsil) were discarded. Methylation results were visualized using the R package (ggplot2, version 3.3.3).
Statistical analysis of gene-gene correlations was performed using spearman correlation by the R package (stat, version 3.6.3). |Cor spearman|≥0.4 and p-spearman<0.05 were set as the filter criteria. Besides, the GeneMANIA (http://genemania.org) and the STRING database (https://cn.string-db.org/) (Szklarczyk et al., 2019) were used to analyze the protein-protein interaction (PPI) networks and to look for relevant signaling pathways and functional similarities. Based on the top 50 relevant genes found, the R package (org.Hs.eg.db, version 3.10.0 and clusterProfiler, version 3.14.3) (Yu et al., 2012) were used for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis to screen and evaluate potential gene functions correlated with MARVELD3.
In this study, the R package (GSVA package, version 1.34.0) (Hänzelmann et al., 2013) was used to analyze the 24 types of immune cells in OSCC.
There were 329 primary OSCC samples and 32 normal samples downloaded from TCGA. Clinical information includes tumor stage (T stage), lymphatic involvement grade (N stage), distant metastases grade (M stage), clinical stage, radiation therapy, primary therapy outcome, gender, race, age, histologic grade, anatomic tumor subdivision, smoker, alcohol history, lymphovascular invasion, lymphnode neck dissection, overall survival (OS) event, disease special survival (DSS) event, and progress free interval (PFI) event (Table 1).
Through the analyses of the data from the TCGA database, the results showed that the expression levels of MARVELD1 and MARVELD3 were significantly higher than those of normal tissues (Figures 1A,B). There was no statistical difference in the expression level of MARVELD2 compared to normal tissue. Then, through KM survival analysis, the relationships between the expression levels of MARVELDs and the overall survival of OSCC patients were analyzed. According to the KM survival curves, the high expression of MARVELD3 was significantly associated with the worse overall survival in OSCC patients (Figure 1E). There were no significant correlations between the expression levels of MARVELD1 and MARVELD2 with worse overall survival (Figures 1C,D). The results suggested that the high expression of MARVELD3 was associated with the poor prognosis of OSCC and that MARVELD3 may be considered an oncogene for OSCC.
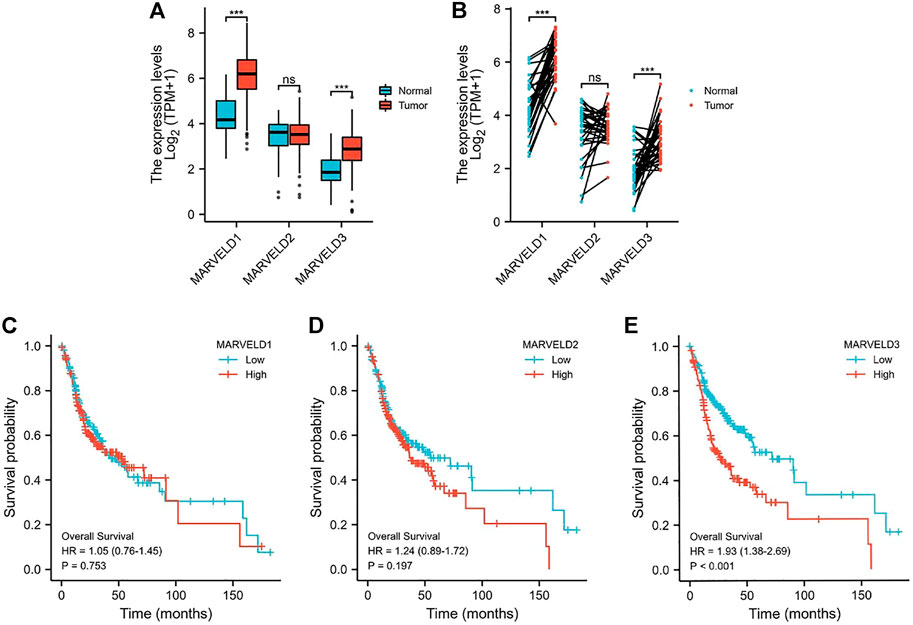
FIGURE 1. The MARVELDs expressions and survival analysis in OSCC. (A) Differential expression analysis of MARVELDs in non-paired samples of OSCC patients. (B) Differential expression analysis of MARVELDs in paired samples of OSCC patients. (C–E) K-M survival curves of the associations between MARVELDs expressions and overall survival.
Based on the results above, we selected MARVELD3 for further in-depth analysis. The results of the univariate Cox regression analysis showed that the high expression of MARVELD3 was a contributing factor to the poor prognosis of OSCC. The results of multivariate Cox regression analysis showed that the high expression of MARVELD3 was an independent prognostic factor for poor OSCC prognosis (Figure 2A). In addition, to be able to predict the survival probability of patients at 1, 3, and 5 years, we constructed nomogram analysis including factors such as age, gender, TNM stage, clinical stage, etc. (Figure 2B). At the same time, we used the ROC curves to evaluate the diagnostic value of MARVELD3 (Figure 2C). The result of the ROC curves showed that the area under the curve (AUC) of MARVELD3 was 0.808, which indicated a considerable diagnostic value. Combined with the above analyses, MARVELD3 may be an unfavorable factor for survival in OSCC patients and an independent predictor of poor prognosis for OSCC.
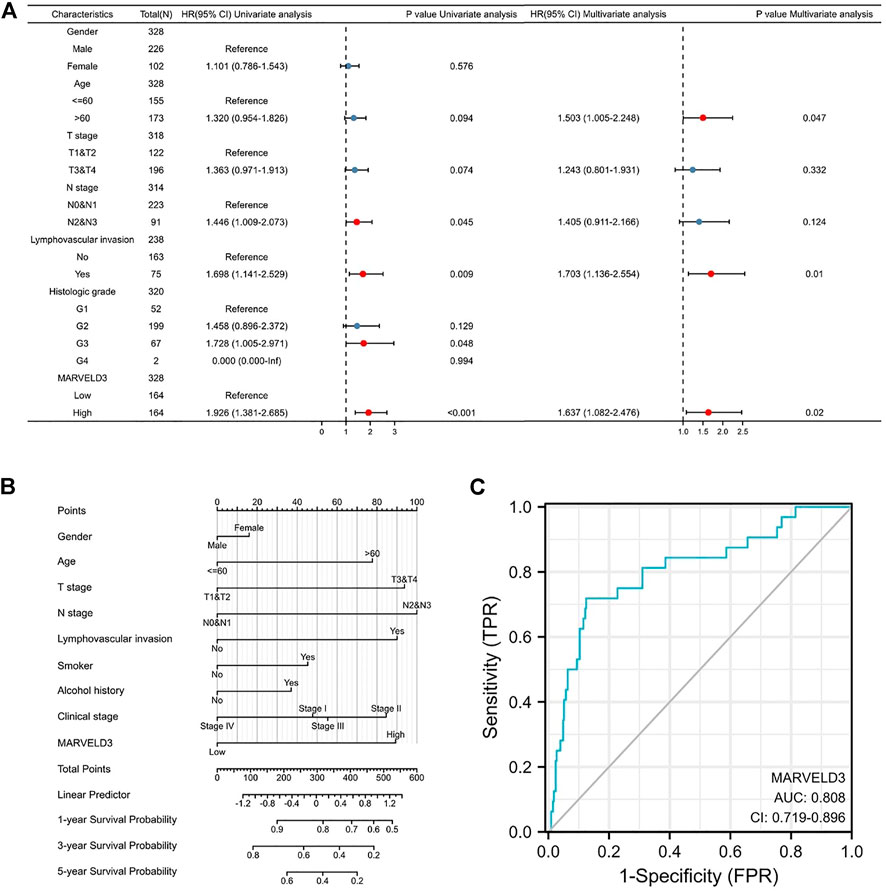
FIGURE 2. Diagnostic value of MARVELD3 in OSCC. (A) Univariate and multivariate Cox regression analyses. HR > 1 indicates disadvantageous factors, and HR < 1 indicates protective factors. Red dots are risk factors. (B) The nomograms were developed by integrating the MARVELD3 expression with key clinical characteristics. (C) The diagnostic value of MARVELD3 expression was evaluated using the ROC curve.
Our analysis showed that the DNA methylation levels were inversely correlated with the expression of MARVELD3 in four methylation sites (cg09326345, cg19311153, cg004477917, and cg18468219) (Figure 3).
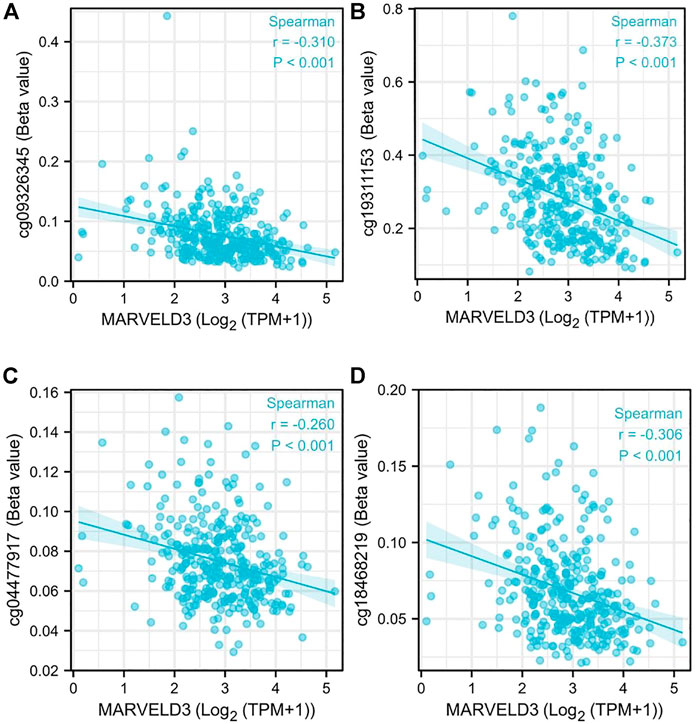
FIGURE 3. MARVELD3 methylation level analysis. (A–D) Correlation of MARVELD3 expression with methylation sites of cg09326345, cg19311153, cg004477917, and cg18468219.
Gene correlation analyses were performed and the top 50 relevant genes were presented (Figures 4A,B). In addition, PPI network analysis was plotted using the GeneMANIA database and the STRING database (Figures 4C,D). Among them, the related proteins are mainly related to the tight junction, leukocyte transendothelial migration, and cell adhesion molecules.
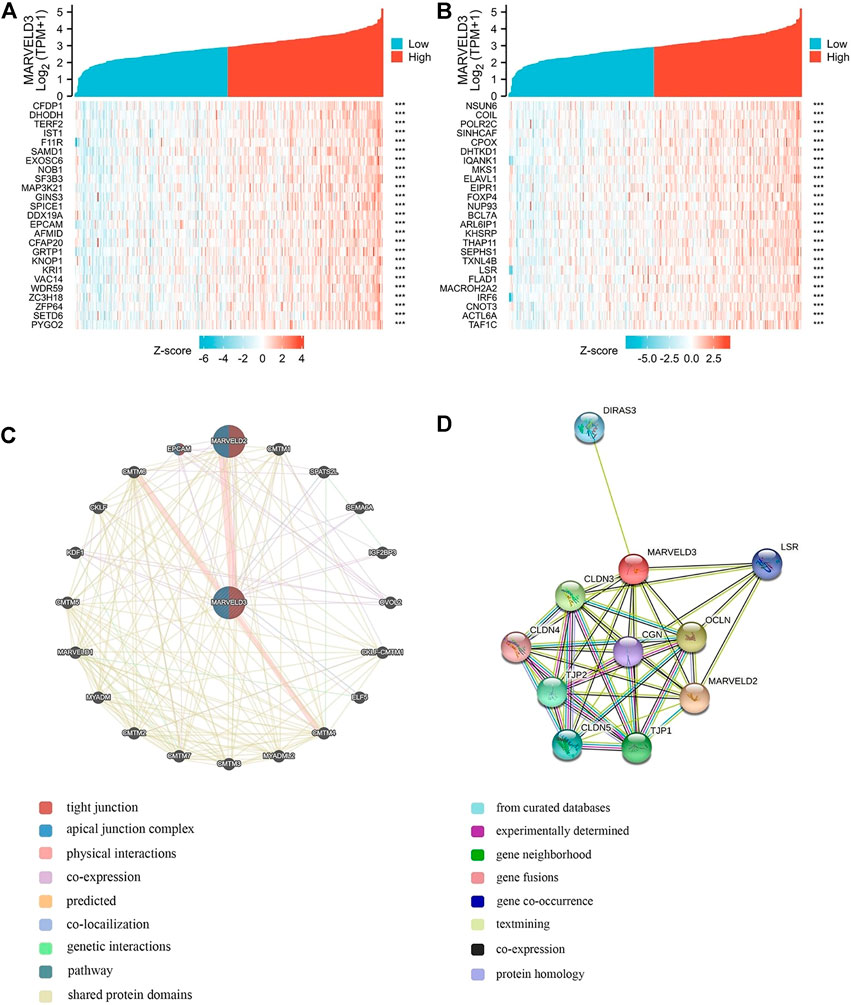
FIGURE 4. MARVELD3 correlation interactive networks. (A–B) Top 50 genes related to MARVELD3 expression. (C) Network diagram of 20 genes associated with MARVELD3. (D) Protein-protein interaction (PPI) network analysis of 11 interacting proteins correlated with MARVELD3.
The biological functions of the genes associated with the expression of MARVELD3 were analyzed by GO and KEGG. The results showed that there was a range of functions related to the expression of MARVELD3, such as RNA splicing via transesterification reactions, RNA splicing via transesterification reactions with bulged adenosine as nucleophile, DNA recombination, RNA localization, nuclear chromatin, and catalytic activity acting on RNA, etc. (Figures 5A,B).
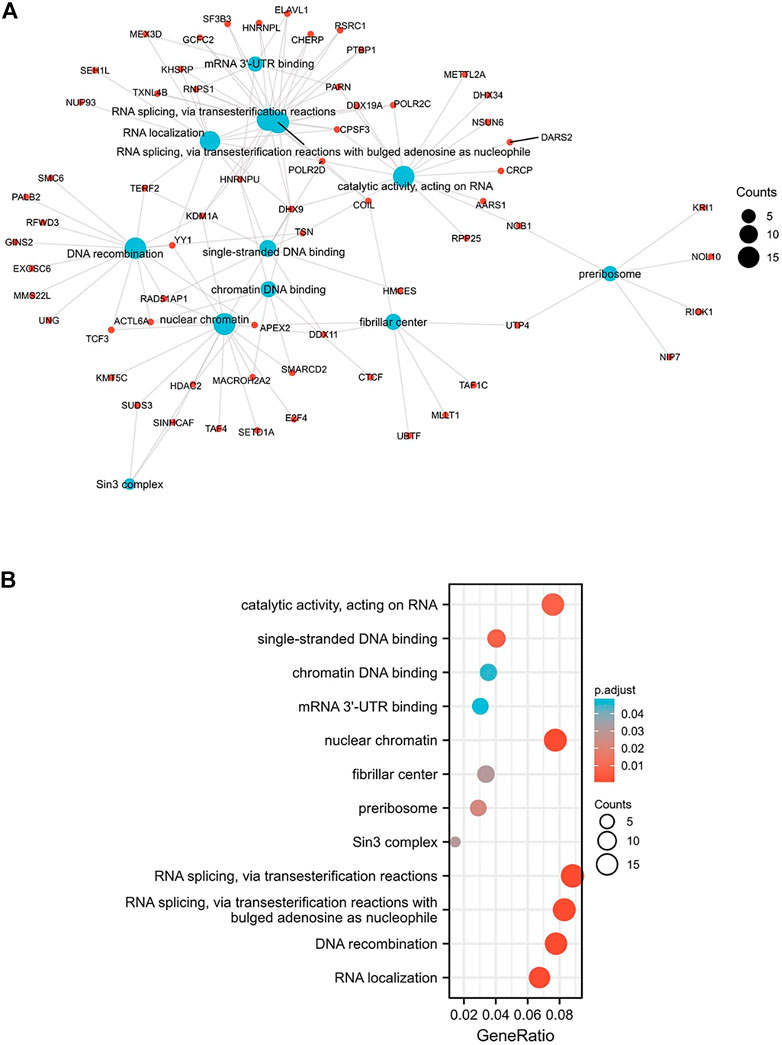
FIGURE 5. GO/KEGG enrichment analysis. (A) Network visualization of GO/KEGG enrichment analysis. (B) Bubble plot of GO/KEGG enrichment analysis.
Based on the above results, we further explored the relationship between the MARVELD3 expression and immune cell infiltration. Data downloaded from TCGAs related to 24 types of immune cells. The results showed that the expression level of MARVELD3 was inversely correlated with the infiltration level of various immune cells such as B cells, T cells, and DC cells (Figure 6).
OSCC is a common malignancy in the human oral cavity, and its prognosis is mostly poor, which significantly affects the overall survival rate of patients. The incidence tends to increase year by year. The clinical outcome and prognosis of OSCC remains dismal; more than 50% of patients die of this disease or complications within 5 years (Lo et al., 2003). Lip, oral cavity, and oropharynx combined were responsible for about 447,751 new cancer cases with an estimated 228,389 deaths in 2018, which accounts for 2.4% of all cancer deaths. In addition to this, head and neck cancer is the fourteenth in terms of incidence but the thirteenth in terms of mortality (WHO, 2020). It is important to look for a potential biomarker that can diagnose, predict the prognosis of OSCC, and serve as a therapeutic target.
In this study, we found that the expressions of MARVELD1 and MARVELD3 were significantly higher in OSCC than in normal tissues. Further studies found that high expression of MARVELD3 correlated significantly with poor overall survival in patients. In addition, univariate and multivariate Cox regression analyses showed that MARVELD3 could be served as an independent predictor of poor prognosis for OSCC. The clinical predictive and diagnostic value of MARVELD3 were further evaluated by the nomograms and the ROC curve, and the results also confirmed the potential value of MARVELD3. Moreover, MARVELD3 was inversely correlated with methylation sites, which was consistent with the characteristics of the oncogene.
Members of the TAMP family are predominantly transmembrane proteins. The functions of transmembrane proteins are ensuring the interaction of tight junctions (TJs) strands between adjacent cells (Heymans et al., 2021). The major TJ proteins are classified according to their physiological role in enabling or preventing paracellular transport. MARVELD3 is linked to a multitude of TJ-associated regulatory and scaffolding proteins (Günzel and Fromm, 2012). By exploring the associated gene functions of MARVELD3 in OSCC, the results suggested that its underlying biological function may be related to RNA splicing via transesterification reactions, RNA splicing via transesterification reactions with bulged adenosine as nucleophile, DNA recombination, RNA localization, nuclear chromatin, and catalytic activity acting on RNA. Through PPI network analysis, we found that MARVELD3 and the claudins (CLDNs) family were closely related. CLDNs are a family of at least 27 transmembrane proteins (Krause et al., 2008; Bhat et al., 2020). Structurally and functionally, CLDNs are commonly used for intercellular adhesion, maintaining cell polarity, and playing a role in barrier function (Hashimoto and Oshima, 2022). Alternatively, overexpression of CLDNs has also been reported to increase aberrant localization and function in gastric, lung, prostate, ovarian, colorectal, and breast cancers, promoting metastasis and progression (Tabariès and Siegel, 2017). These evidences further validated the research value of MARVELD3 high expression in tumor tissues. Coincidentally, in the squamous epithelium of the oral cavity, cancer may occur when the dynamic structure of TJs localized in its tissues changes. At the same time, scholars have studied that the loss of claudin-7 (CLDN7) expression is associated closely with invasion and lymph metastasis. It is an unfavorable prognostic factor in patients with OSCC (Yoshizawa et al., 2013). This finding further illustrates the abnormality in the expression of genes associated with cell tight junctions in OSCC, thus providing evidence for our research.
The results of immune cell infiltration analysis showed that MARVELD3 was inversely correlated with a variety of immune cells including DCs, T cells, neutrophils, and B cells. Studies have reported similar situations (Li et al., 2016). We speculate that this phenomenon may be explained by the inability of the immune system in OSCC patients to recognize MARVELD3 enough. It cannot be recognized as a reliable antigen and aggregated to it. At the same time, this also meant that with the upregulation of MARVELD3 expression, OSCC would not have a strong host immune response. Interestingly, according to our findings, the expression of MARVELD3 was positively correlated with NK CD56bright cells. Natural cytotoxicity, mediated by natural killer (NK) cells plays an important role in the inhibition and elimination of malignant tumor cells (Koo et al., 2013). The phenotype of NK cells is defined by their CD56 expression and lack of CD3 expression, of which CD56bright and CD56dim subpopulations can be divided according to the membrane density of CD56. CD56bright cells mediate low cytotoxicity, CD56dim mainly exerts strong cytotoxicity, and CD56bright may be a precursor to CD56dim (Cooper et al., 2001; Bauernhofer et al., 2003; Watanabe et al., 2010; Dowell et al., 2012). We speculated that this may be that the high expression of MARVELD3 stimulates the conversion of CD56bright to CD56dim, resulting in the phenomenon that MARVELD3 is positively correlated with it. But still, the mechanism by which MARVELD3 is negatively correlated with immune cell infiltration needs to be further studied. Anyway, the relationship between the high expression of MARVELD3 and the clinical characteristics of OSCC obtained in this study may provide evidence for such studies. For OSCC patients in the process of postoperative recovery, doctors can detect the expression of prognostic markers such as MARVELD3 to timely grasp their situation and make correct and reasonable treatment measures, so as to effectively improve the survival status of OSCC patients. However, there are limitations to our study. The underlying molecular mechanism of MARVELD3 in OSCC immune infiltration has not been thoroughly studied. Meanwhile, the reason and mechanism of MARVELD3 overexpression in OSCC have not been thoroughly studied, and the reason for the differential expression between MARVELD1,2,3 is not clear. Therefore, more research needs to be included to explore the underlying molecular mechanism of MARVELD3 in OSCC. Therefore, more researches are needed in the future and our research group will continue to focus on this topic.
In conclusion, our preliminary findings revealed that the high expression of MARVELD3 was strongly and positively correlated with the poorer prognosis in OSCC, and MARVELD3 could serve as a novel independent prognostic factor in OSCC.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Data curation: KH, YM, JL, and LX; Formal analysis: KH, YM, JL, and LX; Investigation: KH and YM; Software: KH; Writing—original draft: KH and YM; Visualization: SW; Funding acquisition: HW and ZX; Writing—review and editing: HW and ZX; Supervision: HW and ZX. All authors have read and agreed to the published version of the manuscript.
This study was funded by Open Subject Foundation of Key Laboratory of Dental Maxillofacial Reconstruction and Biological Intelligence Manufacturing (20JR10RA653-ZDKF20210401 to ZX), School of Stomatology, Lanzhou University, Gansu Province, Lanzhou 730000, Foundation of School/Hospital of Stomatology, Lanzhou University (No. lzukqky-2021-q04 to HW), the Gansu Provincial Natural Science Funds (grant no. 18JR3RA341). And we sincerely thank the experimental guidance from HK_Potions_Lab: (https://mp.weixin.qq.com/s/SkjneqFslhkoKrTKs7QA3Q, Blog, WeChat Official Accounts).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bauernhofer, T., Kuss, I., Henderson, B., Baum, A. S., and Whiteside, T. L. (2003). Preferential apoptosis of CD56dim natural killer cell subset in patients with cancer. Eur. J. Immunol 33, 119–124. doi:10.1002/immu.200390014
Bhat, A. A., Syed, N., Therachiyil, L., Nisar, S., Hashem, S., Macha, M. A., et al. (2020). Claudin-1, A double-edged sword in cancer. Int. J. Mol. Sci 21, 569. doi:10.3390/ijms21020569
Cierpikowski, P., Lis-Nawara, A., and Bar, J. (2021). Sonic Hedgehog is a novel prognostic biomarker in patients with oral squamous cell carcinoma. Neoplasma 68 (4), 867–874. doi:10.4149/neo_2021_201204N1304
Cooper, M. A., Fehniger, T. A., and Caligiuri, M. A. (2001). The biology of human natural killer-cell subsets. Trends Immunol 22, 633–640. doi:10.1016/s1471-4906(01)02060-9
Dowell, A. C., Oldham, K. A., Bhatt, R. I., Lee, S. P., and Searle, P. F. (2012). Long-term proliferation of functional human NK cells, with conversion of CD56dim NK cells to a CD56bright phenotype, induced by carcinoma cells co-expressing 4-1BBL and IL-12. Cancer Immunol. Immunother 61, 615–628. doi:10.1007/s00262-011-1122-3
Günzel, D., and Fromm, M. (2012). Claudins and other tight junction proteins. Compr. Physiol 2 (3), 1819–1852. doi:10.1002/cphy.c110045
Hänzelmann, S., Castelo, R., and Guinney, J. (2013). Gsva: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinforma 14, 7. doi:10.1186/1471-2105-14-7
Hashimoto, I., and Oshima, T. (2022). Claudins and gastric cancer: An overview. Cancers (Basel) 14 (2), 290. doi:10.3390/cancers14020290
Heymans, C., Delcorte, O., Spourquet, C., Villacorte-Tabelin, M., Dupasquier, S., Achouri, Y., et al. (2021). Spatio-temporal expression pattern and role of the tight junction protein MarvelD3 in pancreas development and function. Sci. Rep. 11 (1), 14519. doi:10.1038/s41598-0.21-93654-2
Ikenouchi, J., Furuse, M., Furuse, K., Sasaki, H., Tsukita, S., and Tsukita, S. (2005). Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J. Cell Biol 171 (6), 939–945. doi:10.1083/jcb.200510043
Ikenouchi, J., Sasaki, H., Tsukita, S., Furuse, M., and Tsukita, S. (2008). Loss of occludin affects tricellular localization of tricellulin. Mol. Biol. Cell 19 (11), 4687–4693. doi:10.1091/mbc.e08-05-0530
Kohn, K. W., Zeeberg, B. M., Reinhold, W. C., and Pommier, Y. (2014). Gene expression correlations in human cancer cell lines define molecular interaction networks for epithelial phenotype. PLoS One 9 (6), e99269. doi:10.1371/journal.pone.0099269
Kojima, T., Takasawa, A., Kyuno, D., Ito, T., Yamaguchi, H., Hirata, K., et al. (2011). Downregulation of tight junction-associated MARVEL protein marvelD3 during epithelial-mesenchymal transition in human pancreatic cancer cells. Exp. Cell Res 317 (16), 2288–2298. doi:10.1016/j.yexcr.2011.06.020
Koo, K. C., Shim, D. H., Yang, C. M., Lee, S. B., Kim, S. M., Shin, T. Y., et al. (2013). Reduction of the CD16(-)CD56bright NK cell subset precedes NK cell dysfunction in prostate cancer. PLoS One 8 (11), e78049. doi:10.1371/journal.pone.0078049
Krause, G., Winkler, L., Mueller, S. L., Haseloff, R. F., Piontek, J., and Blasig, I. E. (2008). Structure and function of claudins. Biochim. Biophys. Acta 1778, 631–645. doi:10.1016/j.bbamem.2007.10.018
Li, B., Severson, E., Pignon, J. C., Zhao, H., Li, T., Novak, J., et al. (2016). Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol 17 (1), 174. doi:10.1186/s13059-016-1028-7
Li, Y., Li, T., Zhou, D., Wei, J., Li, Z., Li, X., et al. (2021). Role of tight junction-associated MARVEL protein marvelD3 in migration and epithelial-mesenchymal transition of hepatocellular carcinoma. Cell adh. Migr 15 (1), 249–260. doi:10.1080/19336918.2021.1958441
Lo, W. L., Kao, S. Y., Chi, L. Y., Wong, Y. K., and Chang, R. C. (2003). Outcomes of oral squamous cell carcinoma in taiwan after surgical therapy: Factors affecting survival. J. Oral Maxillofac. Surg 61 (7), 751–758. doi:10.1016/s0278-2391(03)00149-6
Manzano-Moreno, F. J., Costela-Ruiz, V. J., García-Recio, E., Olmedo-Gaya, M. V., Ruiz, C., and Reyes-Botella, C. (2021). Role of salivary MicroRNA and cytokines in the diagnosis and prognosis of oral squamous cell carcinoma. Int. J. Mol. Sci 22 (22), 12215. doi:10.3390/ijms222212215
Steed, E., Rodrigues, N. T., Balda, M. S., and Matter, K. (2009). Identification of MarvelD3 as a tight junction-associated transmembrane protein of the occludin family. BMC Cell Biol 10, 95. doi:10.1186/1471-2121-10-95
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). String V11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47, D607–D613. doi:10.1093/nar/gky1131
Tabariès, S., and Siegel, P. M. (2017). The role of claudins in cancer metastasis. Oncogene 36, 1176–1190. doi:10.1038/onc.2016.289
Watanabe, M., Kono, K., Kawaguchi, Y., Mizukami, Y., Mimura, K., Maruyama, T., et al. (2010). NK cell dysfunction with down-regulated CD16 and up-regulated CD56 molecules in patients with esophageal squamous cell carcinoma. Dis. Esophagus 23, 675–681. doi:10.1111/j.1442-2050.2010.01073.x
Yoshizawa, K., Nozaki, S., Kato, A., Hirai, M., Yanase, M., Yoshimoto, T., et al. (2013). Loss of claudin-7 is a negative prognostic factor for invasion and metastasis in oral squamous cell carcinoma. Oncol. Rep 29 (2), 445–450. doi:10.3892/or.2012.2161
Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012). ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol 16, 284–287. doi:10.1089/omi.2011.0118
Zhang, C., Han, F., Shi, M., Sun, H., Li, Y., Ci, Y., et al. (2019). MARVELD1 interacting with catalase regulates reactive oxygen species metabolism and mediates the sensitivity to chemotherapeutic drugs in epithelial tumors of the reproductive system. Mol. Carcinog 58 (8), 1410–1426. doi:10.1002/mc.23024
WHO (2020). Saudi Arabia Source: Globocan, Switzerland: WHO, Availble at: https://gco.iarc.fr/today/data/factsheets/populations/682-saudi-arabia-fact-sheets.pdf [Accessed 2 March 2020].
Keywords: MARVELD3, TAMP, prognostic biomarker, oral squamous cell carcinoma, immune infiltration
Citation: Huang K, Meng Y, Lu J, Xu L, Wang S, Wang H and Xu Z (2022) High expression of MARVELD3 as a potential prognostic biomarker for oral squamous cell carcinoma. Front. Genet. 13:1050402. doi: 10.3389/fgene.2022.1050402
Received: 21 September 2022; Accepted: 11 October 2022;
Published: 24 October 2022.
Edited by:
Juan Wang, Guilin Medical University, ChinaReviewed by:
Monica Charlotte Solomon, Manipal College of Dental Sciences, Manipal, IndiaCopyright © 2022 Huang, Meng, Lu, Xu, Wang, Wang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaoqing Xu, enF4dUBsenUuZWR1LmNu; Huihui Wang, bHp1X3dhbmdodWlodWlAbHp1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.