
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 10 November 2022
Sec. Genetics of Common and Rare Diseases
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1045738
This article is part of the Research Topic Neuronal Ceroid Lipofuscinosis: Molecular Genetics & Epigenetics View all 5 articles
Mutations in CLN5 cause a subtype of neuronal ceroid lipofuscinosis (NCL) called CLN5 disease. The NCLs, commonly referred to as Batten disease, are a family of neurodegenerative lysosomal storage diseases that affect all ages and ethnicities globally. Previous research showed that CLN5 participates in a variety of cellular processes. However, the precise function of CLN5 in the cell and the pathway(s) regulating its function are not well understood. In the model organism Dictyostelium discoideum, loss of the CLN5 homolog, cln5, impacts various cellular and developmental processes including cell proliferation, cytokinesis, aggregation, cell adhesion, and terminal differentiation. In this study, we used comparative transcriptomics to identify differentially expressed genes underlying cln5-deficiency phenotypes during growth and the early stages of multicellular development. During growth, genes associated with protein ubiquitination/deubiquitination, cell cycle progression, and proteasomal degradation were affected, while genes linked to protein and carbohydrate catabolism were affected during early development. We followed up this analysis by showing that loss of cln5 alters the intracellular and extracellular amounts of proliferation repressors during growth and increases the extracellular amount of conditioned medium factor, which regulates cAMP signalling during the early stages of development. Additionally, cln5- cells displayed increased intracellular and extracellular amounts of discoidin, which is involved in cell-substrate adhesion and migration. Previous work in mammalian models reported altered lysosomal enzyme activity due to mutation or loss of CLN5. Here, we detected altered intracellular activities of various carbohydrate enzymes and cathepsins during cln5- growth and starvation. Notably, cln5- cells displayed reduced β-hexosaminidase activity, which aligns with previous work showing that D. discoideum Cln5 and human CLN5 can cleave the substrate acted upon by β-hexosaminidase. Finally, consistent with the differential expression of genes associated with proteasomal degradation in cln5- cells, we also observed elevated amounts of a proteasome subunit and reduced proteasome 20S activity during cln5- growth and starvation. Overall, this study reveals the impact of cln5-deficiency on gene expression in D. discoideum, provides insight on the genes and proteins that play a role in regulating Cln5-dependent processes, and sheds light on the molecular mechanisms underlying CLN5 disease.
The neuronal ceroid lipofuscinoses (NCLs), also known as Batten disease, are a family of neurodegenerative diseases linked to mutations in 13 ceroid lipofuscinosis neuronal (CLN) genes (CLN1-8, CLN10-14) (Schulz et al., 2013; Mole and Cotman, 2015). Each of the 13 different subtypes of NCL are characterized by the lysosomal accumulation of autoflourescent lipid-protein aggregates called ceroid lipofuscin in neurons, as well as other cell types outside the central nervous system (Palmer et al., 1992; Mole and Cotman, 2015). The accumulation of ceroid lipofuscin has been associated with numerous clinical symptoms, including seizures, reduced motor, visual, and cognitive function, as well as a reduced lifespan (Schulz et al., 2013; Mole and Cotman, 2015). While most of the CLN genes have been studied, the precise cellular mechanisms impacted by CLN gene mutations remain elusive.
Mutations in CLN5 cause the CLN5 disease subtype of NCL (Schulz et a., 2013; Mole and Cotman, 2015). CLN5 is a soluble lysosomal and extracellular protein that is predicted to function as either a glycoside hydrolase or depalmitoylase (Isosomppi et al., 2002; Hughes et al., 2014; Jules et al., 2017; Huber and Mathavarajah, 2018a; Basak et al., 2021b; Luebben et al., 2022). CLN5 has been linked to several cellular processes including, but not limited to, endosomal sorting, biometal homeostasis, sphingolipid metabolism, and autophagy (El Haddad et al., 2012; Mamo et al., 2012; Grubman et al., 2014; Doccini et al., 2020; McLaren et al., 2021). However, like most CLN proteins, the association of CLN5 with a defined biological pathway is still under investigation.
Dictyostelium discoideum is a eukaryotic microbe that is used as a biomedical model system for studying a variety of human diseases, including the NCLs (Huber, 2016; Huber et al., 2022). The D. discoideum life cycle is comprised of single cell and multicellular phases that allow for a diversity of fundamental cellular and developmental processes to be examined in great biochemical detail (Mathavarajah et al., 2017). During the growth phase of the life cycle, haploid amoebae feed on a food source (in nature: microorganisms in the soil; in the laboratory: bacteria and nutrient-rich liquid media) and divide mitotically. When starved, amoebae initiate a 24-h multicellular developmental programme that begins with the chemotactic aggregation of cells. The multicellular mounds that form then undergo a series of morphological changes to form motile slugs. During the later stages of multicellular development, cells within slugs terminally differentiate to form fruiting bodies composed of stalk cells and viable spores that can restart the life cycle when nutrients become available.
The D. discoideum genome encodes homologs for 11 of the 13 human CLN proteins (Huber, 2016; Huber et al., 2020). Our previous work showed that the homolog of human CLN5, Cln5, has glycoside hydrolase activity and is secreted (Huber and Mathavarajah, 2018a). Our work also linked the function of Cln5 in D. discoideum to cell proliferation, cytokinesis, folic acid-mediated chemotaxis, and autophagy during growth, and since cAMP-chemotaxis, aggregation, and developmental timing during multicellular development (Huber and Mathavarajah, 2018b; McLaren et al., 2021).
In this study, we used comparative transcriptomics to identify genes that are impacted by cln5-deficiency in D. discoideum during growth and after 4 h of starvation (when cln5 expression is maximal) (Stajdohar et al., 2017). During growth, genes associated with protein ubiquitination/deubiquitination, cell cycle progression, and proteasomal degradation were affected, while genes linked to protein and carbohydrate catabolism were affected in cells starved for 4 h. We then showed that loss of cln5 affects the levels of proliferation repressors during growth and proteins required for cAMP-mediated chemotaxis and adhesion during the early stages of multicellular development. Finally, we showed that deletion of cln5 affects the activities of lysosomal enzymes and the proteasome. Together, this study examines the impact of cln5-deficiency on the D. discoideum transcriptome and provides further insight into the multifaceted role of CLN5 in the eukaryotic cell.
AX3 (hereafter referred to as WT, parental cell line of cln5-) and cln5- cells were maintained on SM/2 agar containing Klebsiella aerogenes (Fey et al., 2007). Cells used in experiments were cultured axenically in nutrient-rich medium (HL5) at 22°C and 150 RPM (Formedium, Hunstanton, Norfolk, United Kingdom). HL5 was supplemented with streptomycin sulfate (300 μg/ml) and ampicillin (100 μg/ml) to prevent bacterial growth. Blasticidin S hydrochloride (10 μg/ml) was used to select cln5- cells. For all experiments, cells were harvested during the mid-log phase of growth (1–5 x 106 cells/ml). KK2 buffer was formulated as follows: 0.7 g/L K2HPO4, 2.2 g/L KH2PO4, pH 6.5. Mouse monoclonal anti-cortexillin-I (CtxA) (241-438-1) (Faix et al., 2001), mouse monoclonal anti-cortexillin-II (CtxB) (232-238-10) (Faix et al., 2001), mouse monoclonal anti-discoidin (Dsc) (80-52-13) (Wetterauer et al., 1993), mouse monoclonal myosin heavy chain type II (MhcA) (56-396-5) (Pagh and Gerisch, 1986), mouse monoclonal anti-proteasomal subunit (PS) (159-183-10) (Schauer et al., 1993), and mouse monoclonal anti-proteasomal subunit 5 (PS5) (171-337) (Schauer et al., 1993) were purchased from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, Iowa, United States). Mouse monoclonal anti-β-actin (SC-47778) was purchased from Santa Cruz Biotechnology Incorporated (Dallas, Texas, United States). Rabbit polyclonal anti-autocrine proliferation repressor (AprA) (Brock and Gomer, 2005), rabbit polyclonal anti-counting factor-associated protein D (CfaD) (Bakthavatsalam et al., 2008), and rabbit polyclonal anti-conditioned medium factor (CmfA) (Jain et al., 1992) were generous gifts from Dr. Richard Gomer (Texas A&M University, Texas, United States). HRP-linked secondary antibodies were purchased from New England Biolabs Canada (Whitby, Ontario, Canada).
Cells grown in suspension were deposited onto Petri dishes containing HL5 medium and left to adhere to the dish for at least 1 h at 22°C (Figure 1A). The medium was replaced with fresh HL5 medium containing ampicillin (100 μg/ml) and streptomycin sulphate (300 μg/ml) and left overnight to allow for two mitotic doubling times. The following day, growth-phase cells and cells starved for 4 h in KK2 buffer were harvested and stored at −80°C for future use. A 4-h starvation timepoint was selected since this is when cln5 is maximally expressed in D. discoideum (http://dictyexpress.biolab.si/) (Stajdohar et al., 2017). Growth-phase and starved cells from −80°C storage were lysed and RNA was extracted using the Monarch Total RNA Miniprep Kit according to the manufacturer’s instructions (New England Biolabs Canada, Whitby, Ontario, Canada). A total of three biological replicates each for growth and starvation were submitted for RNA sequencing (RNA-seq) analysis.
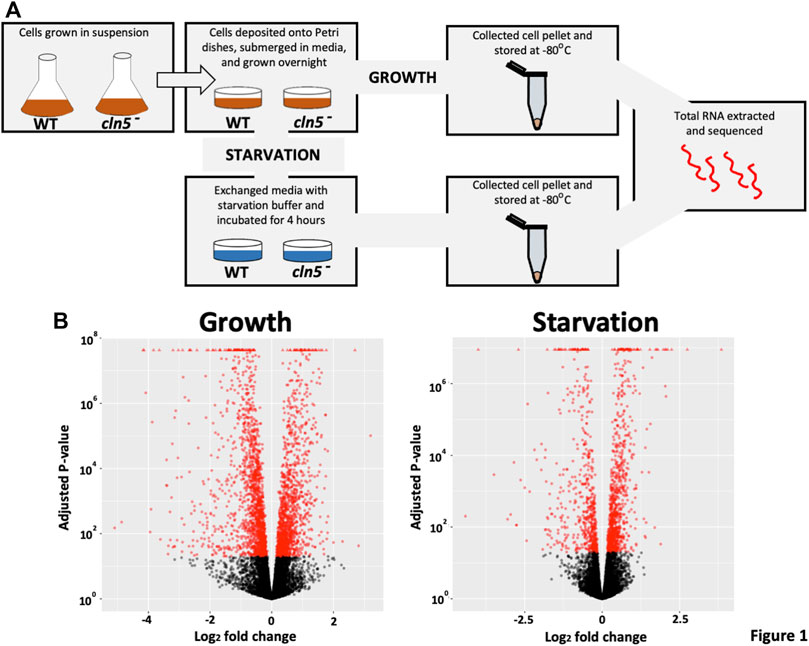
FIGURE 1. A pictorial schematic of the methods used to determine the effects of cln5 deletion on gene expression. (A) WT and cln5- cells grown in suspension and in the mid-log phase of growth were deposited onto Petri dishes and grown overnight in HL5. Growth-phase cells were harvested after the overnight incubation, while starved cells were collected after a 4-hour incubation in KK2 buffer. All cell pellets were stored at −80°C until RNA extraction was done. Extracted RNA samples were sequenced at The Hospital for Sick Children (Toronto, Ontario, Canada). (B) Following differential expression analysis via SARTools, volcano plots were generated to visualize the profile of DEGs during growth and after 4 h of starvation.
RNA-seq was performed by The Centre for Applied Genomics at the Hospital for Sick Children (Toronto, Ontario, Canada). All RNA samples were prepared with a stranded poly(A) mRNA library preparation kit (NEBNext, New England Biolabs Canada, Whitby, Ontario, Canada) and the quality of RNA was determined using a Bioanalyzer (Agilent Technologies, Santa Cruz, California, United States). The RNA-seq analysis for all mRNA libraries were done through an Illumina NovaSeq SP flowcell PE100 sequencer (San Diego, California, United States). Adaptor sequences attached to the raw paired reads were removed with Trimmomatic (Bolger et al., 2014). The quality of the raw paired reads from the sequencer was assessed with FastQc (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The D. discoideum reference genome acquired from the Ensembl database was indexed with hierarchical indexing for spliced alignment of transcripts 2 (HISAT2) and the paired reads were mapped onto the indexed reference genome with HISAT2 (Kim et al., 2019). To obtain read counts, FeatureCounts was used on the mapped reads (Liao et al., 2014). The differential gene expression analysis was done using the DeSeq2 version of statistical analysis of RNA-seq data tools (SARTools) package (Love et al., 2014; Varet et al., 2016). In the SARTools parameters, an alpha value of 0.05 along with the Benjamin-Hochberg (BH) multiple-testing correction were used in the differential gene expression analysis. Furthermore, a log2 fold change threshold of 1.25 was applied on the list of differentially expressed genes (DEGs) (see Supplementary Table S1 for full list of DEGs). Logically accelerated gene ontology term finder (LAGO) was used for gene ontology (GO) term enrichment analyses of the DEGs (Boyle et al., 2004). A p-value of 0.05, combined with the BH correction was applied. Within each GO term enrichment analysis (e.g., biological process, molecular function, and cellular component), only annotated genes that were identified by the LAGO database were used.
Growth-phase cells and cells starved for 4 h were lysed with 0.1% NP40 in 0.05 M 2-(N-morpholino) ethanesulfonic acid (MES) (pH 6.5). All enzyme activity assays were performed using equal amounts of protein from whole cell (WC) lysates (100-200 µg). Proteins in WC lysates were quantified using a Qubit 2.0 Fluorometer (Fisher Scientific Company, Ottawa, Ontario, Canada). Each assay was performed in triplicate and measured using a Synergy HTX multi-mode plate reader (BioTek Instruments Incorporated, Winooski, Vermont, United States). All activity values were subtracted from values acquired from the blank solution. Enzyme activities in cln5- WC lysates were standardized against activities in WC lysates from WT cells. Data were statistically analyzed using the one-sample t-test and a p-value < 0.05 was considered significant.
For α-mannosidase, methods were adapted from Loomis (1970). Briefly, WC lysates were added to 5 mM acetate buffer (pH 5.0) containing a final concentration of 5 mM para-nitrophenyl-α-D-mannopyranoside substrate (N2127, Sigma Aldrich Canada, Oakville, Ontario, Canada). For β-glucosidase, methods were adapted from Coston and Loomis (1969) using a final concentration of 10 mM para-nitrophenyl-β-D-glucopyranoside substrate (487507, Sigma Aldrich Canada, Oakville, Ontario, Canada) in 50 mM acetate buffer (pH 5.0). For both assays, reaction mixtures were incubated at 35°C for 45 min, quenched with an equal volume of 1 M Na2CO3, and activity was measured at 405 nm absorbance.
Methods were adapted from Wimmer et al. (1997). Briefly, WC lysates were added to 0.1 M sodium succinate (pH 6.0) containing 2 mM p-nitrophenyl-α-D-glucopyranoside substrate (487506, Sigma Aldrich Canada, Oakville, Ontario, Canada). Reactions were incubated at 65°C for 1 h and then quenched with two equal volumes of 1 M Na2CO3. Activity was measured at 395 nm absorbance.
Methods were adapted from Kilpatrick and Stirling (1976) and Maruhn (1976). The α-galactosidase activity assay was performed using 2 mM para-nitrophenyl α-D-galactopyranoside substrate (N0877, Sigma Aldrich Canada, Oakville, Ontario, Canada) in citrate/phosphate buffer (pH 4.5), while the β-galactosidase activity assay contained a reaction solution of 5 mM ortho-nitrophenyl-β-D-galactopyranoside substrate (48712-M, Sigma Aldrich Canada, Oakville, Ontario, Canada) in 100 mM citrate buffer (pH 4.0). In both assays, the reaction solutions were incubated at 37°C for 45 min. The α-galactosidase reactions were quenched with an equal volume of 1 M sodium glycinate (pH 10.4). β-galactosidase reactions were terminated using an equal volume of 2-amino-2-methyl-propanol/HCl buffer. Activity was measured at 405 nm absorbance.
Methods were modified from van Diggelen et al. (1999). Briefly, WC lysates were added to 0.2 mM 4-methylumbelliferyl 6-thio-palmitate-β-D-glucopyranoside substrate (19524, Cedarlane Labs, Burlington, Ontario, Canada) in McIlvain’s phosphate/citrate buffer containing 15 mM dithiothreitol and 0.375% (v/v) Triton X-100 (pH 4.0). Reactions were incubated for 1 h at 37°C and then stopped by heating the solution for 2 min at 95°C. 1 U of β-glucosidase enzyme (from almonds, dissolved in distilled water containing 0.2% (w/v) bovine serum albumin) (Sigma Aldrich Canada, Oakville, Ontario, Canada) was added to the cooled reaction mixture while adjusting the pH to 5 with NaOH. The samples were left at 37°C for 1 h and then an equal volume of 0.5 M Na2CO3/NaHCO3 buffer containing 0.025% (v/v) Triton X-100 (pH 10.7) was added to terminate the reaction. Fluorescence was detected using the following filters: 360/40 nm excitation, 460/40 nm emission.
Methods were adapted from Stumpf et al. (2017). Briefly, WC lysates were combined with 120 µM Ala-Ala-Phe-7-amido-4-methylcoumarin tripeptidyl peptidase 1 (TPP1) substrate (A3401, Sigma Aldrich Canada, Oakville, Ontario, Canada) in buffer containing 100 mM sodium acetate/150 mM NaCl/0.1% (v/v) Triton X-100 (pH 4.5). Reactions were incubated at 37°C for 1 h in the dark, after which time an equal volume of quenching solution (100 mM sodium acetate, 150 mM NaCl, pH 4.3) was added to stop the reaction. Fluorescence was detected using the following filters: 360/40 nm excitation, 460/40 nm emission.
Cathepsin D (CTSD) enzymatic activity was assessed using the CTSD Activity Assay Kit according to the manufacturer’s instructions (10013-596, VWR International, Mississauga, Ontario, Canada) and fluorescence was detected using the following filters: 360/40 nm excitation, 460/40 nm emission. Cathepsin F (CTSF) activity was measured using a method adapted from Fonovič et al. (2004). Briefly, WC lysates were mixed with 5 μg/ml pepsin (10108057001, Sigma Aldrich Canada, Oakville, Ontario, Canada) (pH 4.5–5.0). Reactions were preincubated for 1 h at 37°C, after which time 1 µM Z-FR-AMC substrate (80350BP, Cedarlane Labs, Burlington, Ontario, Canada), as well as 0.1 M sodium phosphate buffer containing 1 mM EDTA and 0.1% (v/v) PEG 600 (pH 6.5) were added to the reaction mixture. The reaction was adjusted to a final DMSO content of 5% (v/v) and then incubated at 27°C for 1 h. Fluorescence was detected using the following filters: 360/40 nm excitation, 460/40 nm emission.
Methods were adapted from Barrett (1980). Briefly, samples were mixed with reaction buffer (352 mM KH2PO4, 48 mM Na2HPO4, 4 mM disodium EDTA) supplemented with fresh cysteine at a final concentration of 8 mM. Reactions were preincubated for 5 min at 40°C, after which time they were incubated for 1 h at 40°C in the presence of 1.5 mM of fluorogenic cathepsin B (CTSB) substrate (219392, Sigma Aldrich Canada, Oakville, Ontario, Canada). Reactions were stopped with two equal volumes of 100 mM sodium chloroacetate/30 mM sodium acetate/70 mM acetic acid buffer (pH 4.3) and fluorescence was detected using the following filters: 360/40 nm excitation, 460/40 nm emission.
Methods were adapted from Loomis (1969) and Huber and Mathavarajah (2018a). Briefly, WC lysates were mixed with 4.2 mM para-nitrophenyl N-acetyl-β-D-glucosaminide substrate (N9376, Sigma Aldrich Canada, Oakville, Ontario, Canada) in 100 mM acetate buffer (pH 5.0). Reactions were incubated for 5 min at 35°C. An equal volume of 1 M Na2CO3 was added to stop the reaction. Activity was measured at 405 nm absorbance.
Proteasomal activity was assessed using the Proteasome 20S Activity Assay Kit according to the manufacturer’s instructions (MAK172, Sigma Aldrich Canada, Oakville, Ontario, Canada). Briefly, growth-phase cells and cells starved for 4 h were lysed with 0.1% (v/v) NP40 in 0.05 M MES buffer (pH 6.5) and equal protein amounts (100-150 µg) from WC lysates were used in the assay. Proteins in WC lysates were quantified using a Qubit 2.0 Fluorometer. Samples were incubated for 1 h at 37°C and activity was measured using a Synergy HTX multi-mode plate reader and the following filters: 360/40 nm excitation, 460/40 nm emission. All activity values were subtracted from the blank solution and reads obtained from the cln5- line were normalized to WT activity values. Data were statistically analyzed using the one-sample t-test and a p-value of <0.05 was considered significant.
WC lysates were prepared from growth-phase cells and cells starved for 4 h. In addition, WT and cln5- conditioned media (CM) was collected during growth and conditioned buffer (CB) was collected after 4 h of starvation. CM and CB were clarified through centrifugation (4°C/1500 RPM/5 min). Protein concentrations were determined using a Qubit 2.0 Fluorometer. SDS-PAGE and western blotting were performed using standard methods (2-hour incubation at 22°C for primary and secondary antibodies in 5% (w/v) milk/TBST. The following antibody dilutions were used: anti-AprA (1:1,000), anti-CfaD (1:1,000), anti-CtxA (1:2,000), anti-CtxB (1:2,000), anti-MhcA (1:1,000), anti-CmfA (1:1,000). anti-Dsc (1:1,000), anti-PS (1:1,000), anti-PS5 (1:1,000), and anti-β-Actin (1:1,000). All primary antibodies used for western blotting detected proteins at molecular weights that were consistent with previous studies (see Section 2.1 for citations). HRP-linked secondary antibodies were used at a dilution of 1:2,000. The ChemiDoc Imaging System (Bio-Rad Laboratories Canada, Mississauga, Ontario, Canada) was used to digitally scan the immunoblots. Protein bands were quantified using Fiji/ImageJ and values obtained from WC lysates were normalized to the corresponding levels of β-Actin (Schindelin et al., 2012). Values obtained from cln5- samples were standardized against WT values and the one-sample t-test was used to assess statistical significance. A p-value of <0.05 was considered significant.
Previous work in D. discoideum linked the function of Cln5 to cell proliferation, cytokinesis, folic acid-mediated chemotaxis, and autophagy during growth, and cyclic adenosine monophosphate (cAMP)-mediated chemotaxis, adhesion, and aggregation during the early stages of multicellular development (Huber and Mathavarajah, 2018b; McLaren et al., 2021). To gain insight into the molecular mechanisms underlying cln5-deficiency phenotypes in D. discoideum and the biological pathways impacted by the loss of cln5, we performed RNA-seq analysis on growth-phase cells and cells starved for 4 h in KK2 buffer. We chose the 4-hour timepoint since this is when cln5 expression is maximal (Stajdohar et al., 2017). The list of DEGs was filtered to include only those genes with a log2 fold change of 1.25 (increase or decrease) and a p-value < 0.05. A volcano plot was generated to visualize the profile of DEGs (Figure 1B). RNA-seq revealed 2324 unique DEGs in cln5- cells during growth that included 1041 upregulated genes and 1283 downregulated genes (including cln5) (Table 1). After 4 h of starvation, 644 genes were upregulated in cln5- cells and 394 were downregulated (including cln5), totalling 1038 unique genes.
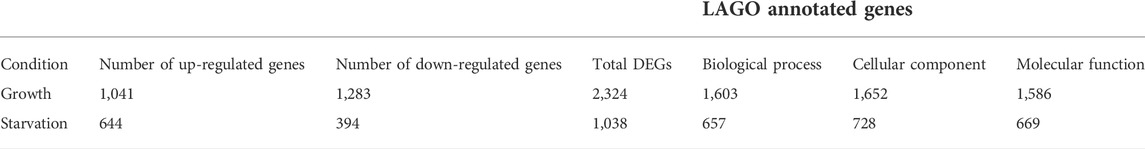
TABLE 1. Quantitative summary of DEGs and annotated genes determined by LAGO in cln5- cells during growth and after 4 h of starvation. A log2 fold threshold of 1.25 was applied.
During growth, the most significantly downregulated genes were those encoding heat shock proteins (hspE-1, hspG3, hspG4, hspG5, hspG6, hspG7, hspG8, hspG12, hspH, hspJ, hspM, dnaja1) and genes involved in protein ubiquitination (ubqG, ubqH, ubqI, ubqJ, DDB_G0285907) (Supplementary Table S1). Conversely, the most significantly upregulated genes were those associated with cell cycle progression, including but not limited to, subunits of the anaphase-promoting complex (anapc3, anapc5, anapc6, anapc7, anapc10), and genes involved in mitosis, such as kinesin-related genes (kif2, kif4, kif10, kif12, kif13) (Castro et al., 2005; Nag et al., 2008; Tikhonenko et al., 2009; Koonce, 2020). During starvation, genes related to development (cotC, cotD, cotE) were significantly downregulated in cln5- cells. In contrast, genes encoding ADP-ribosylation factors (arrJ, arrH, arrK), which modulate the trafficking of endocytic and secretory vesicles, and ponticulin-like proteins (ponC1, ponC2, ponC3, ponC4, ponC5), which are involved in actin bundling and cell-cell adhesion, were the most significantly upregulated genes in cln5-deficient cells (Ingalls et al., 1989; Shariff and Luna, 1990; Chia et al., 1993; Li and Guo, 2022).
As a first step towards analyzing DEGs during growth and starvation, we performed GO term enrichment analyses using LAGO; an online program that clusters genes based on common biological processes they have been associated with, molecular functions of the proteins encoded by the genes, and the subcellular localizations of the proteins (Boyle et al., 2004). For this analysis, we excluded those genes that were uncharacterized (e.g., annotated, but unknown function) (Table 1). Out of 2324 DEGs during growth, this resulted in a list of 1603 genes for biological process, 1586 genes for molecular function, and 1652 genes for cellular component. During growth, DEGs associated with cln5-deficiency are linked to a variety of biological processes including macroautophagy (20 genes), protein ubiquitination (60 genes) and deubiquitination (15 genes), proteasomal-mediated ubiquitin-dependent protein catabolism (56 genes), signal transduction (147 genes), and lipid metabolism (111 genes) (Supplementary Table S2). Proteins encoded by DEGs during growth have kinase activity (75 genes), hydrolase activity (330 genes), and bind to a variety of biological substrates (e.g., protein, 228 genes; carbohydrates, 296 genes; lipids, 44 genes; nucleotides, 323 genes; ions, 535 genes). In addition, the protein products of the DEGs primarily localize to the nucleus (446 genes), ubiquitin ligase complex (37 genes), cell periphery (171 genes), plasma membrane (139 genes), cytoplasm (672 genes), and cellular components associated with cell division (e.g., microtubule organizing centre, 29 genes; spindle pole, 11 genes; spindle, 20 genes; microtubule cytoskeleton, 39 genes; kinetochore, 10 genes; centromeric region of the chromosome, 15 genes; chromosome, 73 genes).
Like growth, only characterized genes (e.g., annotated with a known or predicted function) from the starvation DEG list were used in LAGO analyses, which included 657, 669, and 728 genes for biological process, molecular function, and cellular component, respectively (Table 1). DEGs during starvation are involved in several biological processes including signal transduction (83 genes), protein phosphorylation (42 genes), and catabolic processes involving carbohydrates (8 genes) and proteins (51 genes) (Supplementary Table S2). Proteins encoded by the DEGs during starvation have a variety of molecular functions including kinase (37 genes) and hydrolase (125 genes) activity and bind to a variety of substrates (carbohydrates, 113 genes; ions, 221 genes; nucleotides, 123 genes; proteins, 100 genes). Additionally, some GO terms related to molecular function were only observed in DEGs during starvation including ubiquitin conjugation (8 genes), cysteine-type endopeptidase activity (14 genes), and actin binding (19 genes). Finally, proteins encoded by DEGs during starvation are associated with lysosomes (25 genes), secretory vesicles (8 genes), the cell periphery (118 genes), the cytoplasm (320 genes), the plasma membrane (78 genes), and extracellularly (20 genes). Overall, LAGO analyses revealed that loss of cln5 influences the expression of hydrolases and genes involved in protein processing during growth and starvation. In addition, LAGO analyses revealed clusters of DEGs involved in lipid metabolism during growth and carbohydrate metabolism during starvation. Lastly, cln5-deficiency appears to impact the expression of genes that encode proteins within the secretory pathway during starvation.
In D. discoideum, cln5-deficiency reduces cell proliferation and impairs cytokinesis during growth (McLaren et al., 2021). Here, RNA-seq revealed that loss of cln5 affects the expression of several genes that encode proteins related to cell proliferation including, but not limited to, kinases (aurK and cdk1, increased; qkgA-1, LRRK family protein kinase, decreased), and proteins involved in cell division (rblA, cdc45, and cycB) (all increased) (Supplementary Table S3) (Luo et al., 1995; Sharma et al., 1999; Li et al., 2008; Phillips and Gomer, 2010; Sanchez-Pulido and Ponting, 2011; Strasser et al., 2012). Loss of cln5 also increased the expression of aprA, which encodes the secreted AprA (Brock and Gomer, 2005; Choe et al., 2009). At the protein level, we observed a decreased amount of intracellular AprA and an elevated amount extracellularly suggesting that loss of cln5 increases the secretion of AprA (Figures 2A,B). AprA exerts its effect by interacting with CfaD (Bakthavatsalam et al., 2008; Choe et al., 2009). Although cfaD was not identified as a DEG during growth, loss of cln5 increased the intracellular and extracellular amounts of CfaD (Figures 2A,C). Together, these data suggest that Cln5 regulates the levels of AprA and CfaD to modulate cellular proliferation.
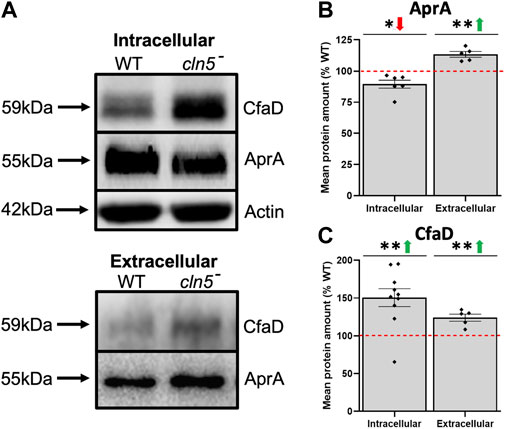
FIGURE 2. Loss of cln5 affects the intracellular and extracellular levels of AprA and CfaD during growth. Whole cell (WC) lysates (20-30 µg) and samples of conditioned media (CM) (0.1–0.375 µg) from growth-phase WT and cln5- cells were separated by SDS-PAGE and analyzed by western blotting. (A) Membranes were probed with anti-AprA, anti-CfaD, and anti-β-actin (loading control). Intracellular and extracellular amounts of (B) AprA and (C) CfaD were quantified using Fiji/ImageJ and intracellular values were standardized against the levels of β-actin. Data presented as mean protein amount (% WT) ± SEM (n ≥ 5). *p < 0.05 (one sample t-test).
Consistent with the effect of cln5-deficiency on cytokinesis, we detected several DEGs during growth that are associated with cytokinesis including vinculin A (vinA), ctxA, and mhcA, which were all decreased (Supplementary Table S4) (De Lozanne and Spudich, 1987; Faix et al., 1996; Stock et al., 1999; Weber et al., 1999; Nagasaki et al., 2009). In D. discoideum, cytokinesis can occur via two processes, one is dependent on myosin II and the other on cell adhesion (Zang et al., 1997). CtxA and MhcA participate in myosin II-dependent cytokinesis, while VinA localizes to focal adhesion regions and is thought, along with CtxB, to be involved in cell adhesion-dependent cytokinesis (Zang et al., 1997; Bukharova et al., 2005; Duran et al., 2009; Nagasaki et al., 2009). In addition, cortexillins function as actin-bundling proteins to help form the cleavage furrow during cytokinesis (Weber et al., 1999). To expand upon these findings, we assessed the amounts of CtxA, CtxB, and MhcA protein in cln5- cells. Loss of cln5 had no effect on the amount of CtxA but did decrease and increase the intracellular amounts of CtxB and MhcA, respectively (Figures 3A,B). Overall, these results support the role of Cln5 in cell proliferation and cytokinesis and provide insight into the genes and proteins affected by cln5-deficiency during growth.
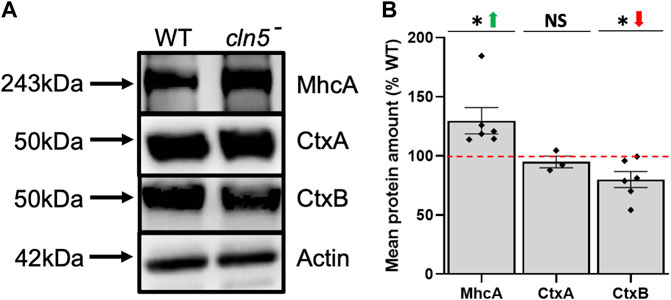
FIGURE 3. Loss of cln5 affects the abundance of proteins linked to cytokinesis during growth. (A) Whole cell (WC) lysates (20-30 µg) from growth-phase WT and cln5-deficient cells were separated by SDS-PAGE and analyzed by western blotting. Membranes were probed with anti-CtxA, anti-CtxB, anti-MhcA, and anti-β-actin (loading control). (B) Intracellular amounts of CtxA, CtxB, and MhcA were quantified using Fiji/ImageJ and standardized against the levels of β-actin. Data presented as mean protein amount (% WT) ± SEM (n ≥ 3). *p < 0.05 (one sample t-test). NS – not significant.
cln5-deficiency in D. discoideum delays aggregation, inhibits cAMP-mediated chemotaxis, and reduces adhesion during the early stages of development (Huber and Mathavarajah, 2018b; McLaren et al., 2021). Therefore, we surveyed our list of DEGs during starvation to identify dysregulated genes that may contribute to these phenotypes. When we scanned the starved DEG list for genes that have been linked to delayed aggregation, myoK (myosin-K heavy chain) and rps4 (40S ribosomal protein S4) were increased in expression, while qkgA-1 was reduced (Supplementary Table S5). RNA-seq also revealed genes that encode proteins involved in cAMP signalling (e.g., cAMP receptor D, carD, increased; cAMP-like receptor 4, crlD, decreased) and degradation (e.g., 3′,5′-cyclic-nucleotide phosphodiesterase, regA, increased) (Supplementary Table S6). The D. discoideum genome encodes four receptors that bind cAMP (CarA, CarB, CarC, and CarD) (Kim et al., 1996). carD is expressed the highest during the later stages of multicellular development, while cAMP receptor A (carA) is expressed the highest during aggregation. RNA-seq revealed that carA was not differentially expressed in cln5- cells. CarA elevates cAMP synthesis during aggregation by stimulating adenylyl cyclase activity and its expression via protein kinase A (Mann et al., 1997; Loomis, 1998; Loomis, 2014). RegA reduces intracellular cAMP levels and leads to reducing cAMP sensitivity in D. discoideum (Sun and Devreotes, 1991; Schulkes and Schaap, 1995; Loomis, 1998; Shaulsky et al., 1998; Loomis, 2014). Additionally, cAMP synthesis and binding, as well as the activation of both CarA and early developmental genes, are modulated by CmfA, which binds to the conditioned media factor receptor (CmfB) (Gomer et al., 1991; Yuen et al., 1995; van Haastert et al., 1996; Deery and Gomer, 1999). In our RNA-seq analysis, cmfA was not differentially expressed, but cmfB was elevated. Furthermore, while loss of cln5 had no effect on the amount of intracellular CmfA, cln5-deficiency elevated the amount of extracellular CmfA (Figures 4A,B). Together, these findings show that cln5-deficiency impacts the expression of genes associated with cAMP signalling during aggregation.
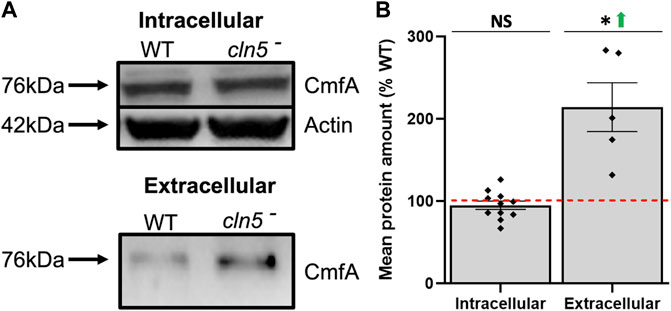
FIGURE 4. Loss of cln5 affects the extracellular amount of CmfA during starvation. (A) WT and cln5-deficient cells starved for 4 h were lysed and standard SDS-PAGE/western blotting was performed on equal protein amounts from whole cell (WC) lysates (20-40 µg) and clarified conditioned buffer (CB) (0.15–0.375 µg). Membranes were probed with anti-CmfA and anti-β-actin (loading control). (B) Intracellular and extracellular amounts of CmfA were quantified with Fiji/ImageJ and intracellular amounts of CmfA were standardized against the levels of β-actin. Data presented as mean protein amount (% WT) ± SEM (n ≥ 5). *p < 0.05 (one sample t-test). NS – not significant.
Cell-substrate and cell-cell adhesion also play roles in D. discoideum aggregation, which is delayed by the loss of cln5 (Huber and Mathavarajah, 2018b). In addition to cAMP-mediated chemotaxis, CmfA also influences the expression of discoidin I, a lectin that is secreted and involved in cell-substrate adhesion and migration (Springer et al., 1984; Barondes et al., 1985; Gomer et al., 1991). In scanning the list of DEGs during starvation, genes encoding discoidin subunits (dscB, dscC, dscD) were elevated in expression (Supplementary Table S7). In support of the differential expression, elevated amounts of intracellular and extracellular discoidin were observed with the loss of cln5 (Figures 5A,B). Similarly, D. discoideum contains a protein complex composed of counting factors (CF45-1, CF50-1, and CF60) and countin (CtnA), that collectively regulate cell adhesion and migration (Brock and Gomer, 1999; Roisin-Bouffay et al., 2000; Tang et al., 2002). Our RNA-seq analysis revealed elevated expression of cf45-1 and cf50-1 in cln5- cells during starvation. Furthermore, RNA-seq revealed no change in expression of cf60 and ctnA, which is consistent with previous findings of unaltered CtnA protein levels in cln5- cells (McLaren et al., 2021). Finally, upregulation of smlA (small aggregate formation protein) was observed, which encodes a protein that influences protein processing and secretion (Brock et al., 1996). Altogether, these observations support previous findings of Cln5 playing a role in aggregation and adhesion.
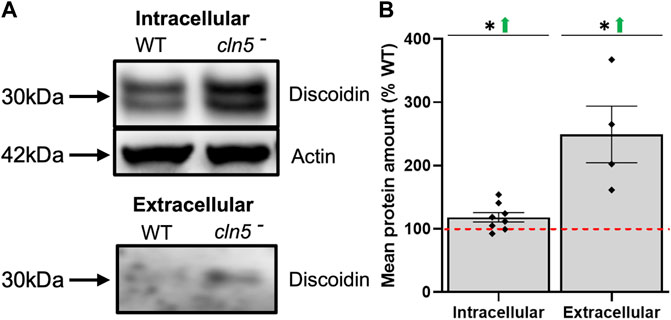
FIGURE 5. Loss of cln5 affects the intracellular and extracellular amounts of discoidin during starvation. (A) Whole cell (WC) lysates (30-50 µg) and clarified conditioned buffer (CB) (0.15–0.375 µg) were collected from WT and cln5-cells starved for 4 h. Equal protein amounts were subjected to SDS-PAGE/western blotting, and membranes were probed with anti-Dsc and anti-β-actin (loading control). (B) Protein bands were quantified using Fiji/ImageJ and the intracellular values for discoidin were standardized against the levels of β-actin. Data presented as mean protein amount (% WT) ± SEM (n ≥ 4). *p < 0.05 (one sample t-test). NS – not significant.
In previous studies, several lysosomal enzymes were identified as direct or indirect interactors of Cln5 including α-mannosidase, β-glucosidase, and NagA (Huber and Mathavarajah, 2018a). While the D. discoideum genome encodes several α-mannosidases, manA is the most highly expressed α-mannosidase gene during growth and aggregation (Stajdohar et al., 2017). The D. discoideum genome also encodes two β-galactosidases, glb1 and glb2. Two α-galactosidases are encoded by the D. discoideum genome, melA and DDB_G0291524, with the latter being expressed more than melA during the D. discoideum life cycle. Two α-glucosidases (gaa, modA) are encoded by the D. discoideum genome with modA being expressed more than gaa during the life cycle (Stajdohar et al., 2017). Finally, the D. discoideum genome encodes one β-glucosidase (gluA). In our transcriptomics dataset, we observed many carbohydrate enzymes that were differentially expressed during cln5- growth and starvation. During growth, loss of cln5 increased the expression of glb1, manA, melA, and gaa (Supplementary Table S8). When starved, cln5- cells increased the expression of gluA and decreased the expression of the putative α-galactosidase (DDB_G0291524) and α-mannosidase (DDB_G0268754) genes. Next, we assessed the intracellular activity of the aforementioned enzymes. During growth, loss of cln5 decreased β-glucosidase and NagA activity and increased the activity of α-glucosidase (Figure 6A). During starvation, cln5-deficiency decreased α-mannosidase, β-glucosidase, and NagA activity (Figure 6B). We observed no effect of cln5-deficiency on α-galactosidase and β-galactosidase activity during both growth and starvation. Collectively, these findings suggest that Cln5 regulates the activities of several carbohydrate enzymes.
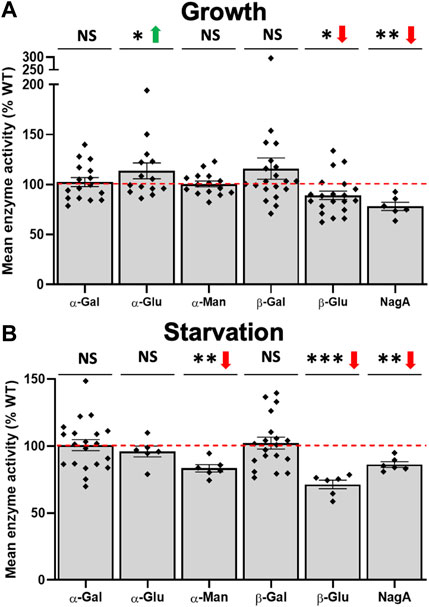
FIGURE 6. Loss of cln5 affects the activity of carbohydrate enzymes during growth and starvation. WT and cln5-deficient cells during (A) growth and (B) 4-hour starvation were lysed and the activities of various carbohydrate enzymes were assessed including α-galactosidase (α-Gal), β-galactosidase (β-Gal), α-glucosidase (α-Glu), β-glucosidase (β-Glu), α-mannosidase (α-Man), and N-acetylglucosaminidase (NagA). Raw enzymatic values were subtracted from the blank solution and activities in cln5-deficient lysates were standardized against the activities in WT samples. Data presented as mean enzyme activity (% WT) ± SEM (n ≥ 6). *p < 0.05, **p < 0.01, ***p < 0.001 (one sample t-test). NS – not significant.
Ceroid lipofuscin accumulation is a common hallmark of the NCLs and has been postulated to occur due to alterations in the autophagy pathway (Cárcel-Trullols et al., 2015; Leinonen et al., 2017; Adams et al., 2019; Mukherjee et al., 2019; McLaren et al., 2021). Dysregulated autophagy has been observed in many CLN5 disease models including D. discoideum (Best et al., 2017; Leinonen et al., 2017; Adams et al., 2019; Doccini et al., 2020; McLaren et al., 2021). By scanning the list of DEGs, we identified genes involved in autophagy initiation (atg1, atg13) and autophagosome formation and turnover (atg6A, atg8, atg18) (all decreased) during growth, but no DEGs related to autophagy during starvation (Supplementary Table S9) (Tanida et al., 2005; Mizushima, 2010; Bento et al., 2016; Mesquita et al., 2017). Since our previous work in D. discoideum reported increased numbers of ubiquitin-positive proteins in cln5- cells during growth, we also examined our growth DEG list for genes associated with protein ubiquitination (Pickart, 2001; Jin et al., 2008; Williamson et al., 2009; Shao et al., 2013; McLaren et al., 2021). Not surprisingly, we identified DEGs that encode proteins involved in ubiquitin attachment, such as polyubiquitin proteins (ubqA, ubqD, ubqF, ubqG, ubqH, ubqI, ubqJ), all of which were downregulated during growth (Supplementary Table S10). In addition, DEGs encoding proteins involved in ubiquitin conjugation (E2 complex) were reduced, including ubiquitin-conjugating enzyme E2 (ubcB) and a gene similar to human UBE2J2 (ube2j2), and genes encoding Ube2C and Ube2S (ube2c and ube2s, respectively) were elevated. Ubiquitin ligase genes (skp1B, rnf160) were also reduced in expression. However, some genes associated with the ligase activity of the E3 complex were increased such as subunits of the anaphase-promoting complex (anapc3, anapc4, anapc5, anapc6, anapc7, anapc10, cdc20, cdc26) (Supplementary Table S10) (Matyskiela et al., 2009; Wang et al., 2009). Finally, genes involved in protein deubiquitination such as ubiquitin hydrolases and thioesterases were reduced in expression (e.g., uch2, yod1) (Deol et al., 2020). Overall, these findings revealed the genetic impact of cln5-deficiency on the autophagy and protein ubiquitination pathways, which further supports the role of Cln5 in autophagy.
Interestingly, the autophagy pathway and ubiquitin-proteasome system crosstalk with each other and both use ubiquitin as a tag to signal protein degradation (Kocaturk and Gozuacik, 2018). Our RNA-seq analysis showed that loss of cln5 increased the expression of a gene predicted to be involved in proteasome assembly, psmG4, as well as reduced the expression of psmE3, which encodes a proteasome subunit that regulates the activity of the proteasome during D. discoideum growth (Supplementary Table S10) (Le Tallec et al., 2007; Masson et al., 2009). In addition, we observed elevated amounts of one proteasome subunit, PS5, during cln5- growth and starvation (Figures 7A,B), no effect on another proteasome subunit (Figure 7C), and decreased proteasome 20S activity during both growth and starvation (Figure 7D). These findings, coupled with previous work, suggest that Cln5 plays a role in proteasomal degradation and that the loss of cln5 leads to dysfunctional protein turnover via both autophagy and through the proteasome.
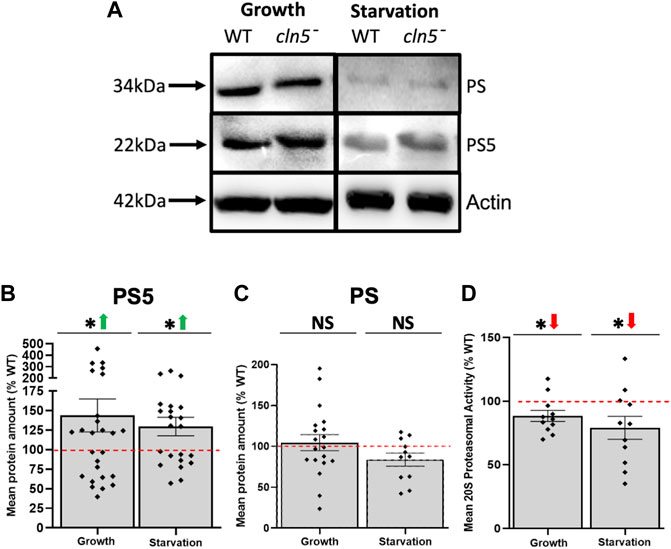
FIGURE 7. Loss of cln5 increases the abundance of a proteasomal subunit during growth and starvation but decreases proteasome 20S activity. (A) Whole cell (WC) lysates from growth-phase and 4 h starved WT and cln5-- cells were separated by SDS-PAGE and analyzed by western blotting. Membranes were probed with anti-PS, anti-PS5, and anti-β-actin (loading control). Fiji/ImageJ was used to quantify (B) PS5 and (C) PS protein bands, which were then standardized against the levels of β-actin. Data presented as mean protein amount (% WT) ± SEM (n ≥ 11). *p < 0.05 (one sample t-test). NS – not significant. (D) Proteasome 20S activity was assessed by collecting WT and cln5- cells during growth and after 4 h of starvation. Cells were then lysed and proteasome 20S activity was measured using a commercially available kit. Raw activity values were subtracted from the blank solution and activities in cln5-deficient lysates were standardized against the activities in WT samples. Data presented as mean proteasome 20S activity (% WT) ± SEM (n = 11). *p < 0.05 (one sample t-test).
It has been suggested that CLN proteins collectively participate in a shared biological pathway or pathways that converge to regulate a common cellular process (Persaud-Sawin et al., 2007; Huber, 2020). As a result, we scanned our list of DEGs and revealed several CLN-like genes that were differentially expressed in cln5- cells (Supplementary Table S11). During growth, tpp1E (similar to human TPP1/CLN2), ddj1 (similar to human DNAJC5/CLN4), and kctd9 (similar to human KCTD7/CLN14) were reduced in expression in cln5- cells relative to WT cells, while tpp1F (similar to human TPP1/CLN2) and grn (similar to human GRN/CLN11) were elevated. During starvation, the expression of tpp1B and tpp1F (similar to human TPP1/CLN2), ctsD (similar to human CTSD/CLN10), DDB_G0291191 (similar to human CTSF/CLN13), and DDB_G0269760 (similar to human KCTD7/CLN14) were increased, while cprB (similar to human CTSF/CLN13) was reduced. Based on these findings, we next assessed the intracellular activities of CLN proteins with demonstrated enzymatic activity in mammals including PPT1, TPP1, CTSD, and CTSF. While there was no effect of cln5-deficiency on ppt1 expression, we detected increased PPT1 activity in both growth and starved cells (Figures 8A,B). Despite the effects of cln5-deficiency on tpp1E and tpp1F expression during growth, and tpp1B and tpp1F expression during starvation, there was no correlated impact on TPP1 activity in cln5- cells. There was elevated CTSD and CTSF activity observed in cln5- cells during growth but no changes were observed during starvation (Figures 9A,B). Finally, elevated ctsB expression was observed in cln5- cells during growth and cells starved for 4 h, but reduced CTSB activity was observed in cln5- cells during starvation. From these findings, it is evident that Cln5 influences the expression and activity of some CLN proteins, which further supports the molecular networking of CLN genes and proteins (Huber, 2020).

FIGURE 8. Loss of cln5 increases PPT1 activity during growth and starvation but has no effect on TPP1 activity. (A) Growth-phase and (B) 4-hour starved WT and cln5- cells were lysed PPT1 and TPP1 activity in whole cell (WC) lysates were subtracted from the blank solution and activities in cln5-deficient lysates were standardized against the activities in WT samples. Data presented as mean enzyme activity (% WT) ± SEM (n ≥ 12). *p < 0.05 (one sample t-test). NS – not significant.
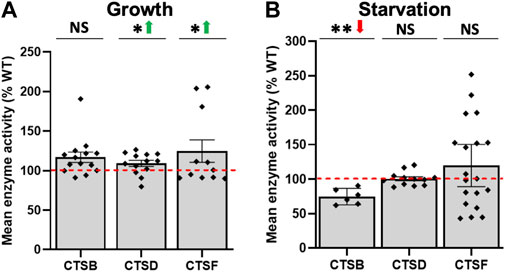
FIGURE 9. Loss of cln5 affects the activity of cathepsins during growth and starvation. The activity of cathepsins were assessed during (A) growth and (B) 4-hour starvation in WT and cln5- cells as described in the Materials and methods. Activities in whole cell (WC) lysates were subtracted from the blank solution and activities in cln5- lysates were standardized against the activities in WT samples. Data presented as mean enzyme activity (% WT) ± SEM (n ≥ 6). *p < 0.05, **p < 0.01 (one sample t-test). NS – not significant.
In D. discoideum, cln5-deficiency suppresses cell proliferation, reduces cytokinesis, increases the basal level of autophagy, delays aggregation, impairs cAMP-mediated chemotaxis, reduces cell-substrate and cell-cell adhesion, and causes precocious multicellular development after mound formation (Huber and Mathavarajah, 2018b; McLaren et al., 2021). In this study, we used comparative transcriptomics to gain insight into the molecular mechanisms underlying these phenotypes. We then used the expression data to inform follow up work that examined the effect of cln5-deficiency on protein levels and enzyme activity during growth and the early stages of development.
Loss of cln5 increases the intracellular levels of ubiquitinated proteins during growth (McLaren et al., 2021). Here, comparative transcriptomics revealed that the most significantly downregulated genes during growth were those that encode heat shock proteins and proteins associated with protein ubiquitination, which could reflect an attempt by cln5- cells to limit the expression of genes linked to protein ubiquitination. The most significantly upregulated genes were associated with cell cycle progression and mitosis. Since loss of cln5 inhibits cell proliferation (McLaren et al., 2021), these findings indicate that cln5- cells increase the expression of genes linked to cell proliferation to help restore the normal rate of proliferation. During starvation, the most significantly downregulated genes were linked to development. Conversely, the most significantly upregulated genes were involved in intracellular trafficking and cell-cell adhesion. These findings are consistent with the defects in developmental timing and adhesion observed in cln5- cells (Huber and Mathavarajah, 2018b; McLaren et al., 2021).
GO term analysis revealed an enrichment of DEGs associated with autophagy, protein homeostasis, and lipid metabolism during growth. These findings are consistent with work in D. discoideum and mammalian models of CLN5 disease that have linked the function of CLN5 to these cellular processes (El Haddad et al., 2012; Schmiedt et al., 2012; Best et al., 2017; Leinonen et al., 2017; Adams et al., 2019; Doccini et al., 2020; McLaren et al., 2021; Yasa et al., 2021; Doccini et al., 2022). Previous work suggests that CLN5 functions as either a glycoside hydrolase or depalmitoylase (Huber and Mathavarajah, 2018a; Luebben et al., 2022). Here, comparative transcriptomics showed that loss of cln5 during growth alters the expression of genes that encode enzymes. During starvation, GO term analysis identified an enrichment of DEGs that encode enzymes and proteins involved in catabolic processes. In addition, consistent with the extracellular localization of Cln5 in D. discoideum and CLN5 in mammals (Isosomppi et al., 2002; Moharir et al., 2013; Hughes et al., 2014; Huber and Mathavarajah, 2018a; Huber and Mathavarajah, 2018b; McLaren et al., 2021), GO term analyses identified an enrichment of DEGs that encode proteins that localize to secretory vesicles, the cell periphery, the plasma membrane, and extracellularly, which is consistent with previous work that indicated a role for CLN5 in protein secretion (Hersrud et al., 2016; Huber, 2021; Iwan et al., 2021). Notably, GO term enrichment analyses also showed that loss of cln5 affects the expression of genes that encode lysosomal enzymes, which aligns with the lysosomal localization of CLN5 in mammalian cells (Isosomppi et al., 2002).
LAGO analysis revealed altered expression of genes linked to cell cycle progression and mitosis in cln5- cells, which is consistent with the reduced proliferation of cln5- cells (McLaren et al., 2021). In addition, we showed that loss of cln5 significantly increases the expression of aprA, which encodes the well-established proliferation repressor AprA (Brock and Gomer, 2005). Intriguingly, cln5-deficiency decreased the intracellular amount of AprA, but increased the extracellular amount, suggesting that loss of cln5 increases AprA secretion. These findings support the reduced proliferation of cln5- cells since more AprA is present outside cells to repress proliferation. In addition, it appears that cln5- cells increase the expression of aprA to counteract the reduced intracellular amount of the protein. Finally, loss of cln5 also increased the intracellular and extracellular levels of CfaD, which participates in AprA-dependent signalling. Combined, these observations indicate that cln5- cells cannot modulate AprA-dependent signalling and the increased secretion of AprA is exacerbated by the increased expression of aprA in cln5-deficient cells, which ultimately supresses cell proliferation.
Our previous work in D. discoideum reported reduced cytokinesis in cln5-deficient cells (McLaren et al., 2021). Here, RNA-seq revealed reduced expression of genes associated with cytokinesis including ctxA, mhcA, and vinA. We also observed significantly increased amounts of MhcA protein in cln5- cells, which could explain the reduced expression of mhcA, and reflect an attempt by cln5- cells to mitigate defects in MhcA-dependent cytokinesis. Interestingly, our RNA-seq analysis revealed a cluster of DEGs that encode proteins that bind to the cytoskeleton, which is consistent with a previous study that revealed DEGs in Cln1−/− mice related to cytoskeleton organization (von Schantz et al., 2008). Moreover, altered cytokinesis has also been reported in yeast and D. discoideum knockout models of CLN3 disease (Codlin et al., 2008; Mathavarajah et al., 2018) indicating that multiple CLN genes regulate cytokinesis.
Comparative transcriptomics identified DEGs associated with aggregation, cAMP-mediated chemotaxis, and cell adhesion in cln5- cells, which is consistent with the delayed aggregation, suppressed cAMP-mediated chemotaxis, and reduced adhesion observed in cln5-deficient cells (Huber and Mathavarajah, 2018b; McLaren et al., 2021). We also found that loss of cln5 elevates the amount of secreted CmfA and expression of the gene encoding the CmfA receptor, cmfB, which together modulate cAMP-mediated chemotaxis and the expression of early developmental genes (Gomer et al., 1991; Yuen et al., 1995; van Haastert et al., 1996; Deery and Gomer, 1999). In addition, cln5-deficiency increased the expression of the gene encoding the cAMP phosphodiesterase RegA. These findings suggest that the delayed aggregation of cln5- cells can be at least partly due to altered CmfA-dependent signalling and increased degradation of cAMP due to elevated expression of regA. Finally, consistent with the role of Cln5 in cell adhesion (Huber and Mathavarajah, 2018b), loss of cln5 increased the expression of genes that encode subunits of discoidin, which resulted in a correlated increase in the intracellular and extracellular amounts of discoidin protein in starved cells. Discoidins are secreted lectins that regulate cell migration and cell-substrate adhesion in D. discoideum (Springer et al., 1984; Barondes et al., 1985). Thus, the increased expression and amount of discoidin protein likely reflected an attempt by cln5-deficient cells to restore adhesion to WT levels.
RNA-seq identified DEGs in cln5- cells associated with protein tagging, protein degradation, and autophagy, which aligns with studies from D. discoideum, mice, and humans linking the function of CLN5 to autophagy (Best et al., 2017; Leinonen et al., 2017; Huber and Mathavarajah, 2018b; Adams et al., 2019; Doccini et al., 2020; Basak et al., 2021b; McLaren et al., 2021). Intriguingly, autophagy is dysregulated in many NCL subtypes (Kim et al., 2022). Accumulated evidence suggests that autophagy and the ubiquitin-proteasome system work together to regulate protein degradation and operate in a compensatory manner when one of the pathways is inhibited (Kocaturk and Gozuacik, 2018). Our previous work showed that loss of cln5 increases the basal level of autophagy during growth (McLaren et al., 2021). Here, we reported an accumulation of one of the proteasomal subunits in cln5- cells, but a reduction in proteasome 20S activity during both growth and starvation, which is consistent with the increased abundance of ubiquitin-positive proteins in cln5- cells (McLaren et al., 2021). Together, these results indicate that Cln5 influences protein degradation via autophagy and the ubiquitin-proteasome system.
Autophagy is dependent on lysosomal function. Not surprisingly, our RNA-seq dataset contained several DEGs in cln5- cells that encode carbohydrate enzymes including β-glucosidase and α-mannosidase, which both interact with Cln5 (Huber and Mathavarajah, 2018a). We followed up this analysis by showing that cln5-deficiency affects the activity of various carbohydrate enzymes. Notably, cln5- cells displayed reduced NagA activity during growth and starvation, which is consistent with previous work that reported that D. discoideum Cln5 and human CLN5 can also cleave the NagA substrate and that Cln5 and NagA may participate in a common pathway in D. discoideum that regulates autophagy (Huber and Mathavarajah, 2018a; McLaren et al., 2021). We also showed that loss of cln5 alters the expression and activities of other CLN-like proteins in D. discoideum. This aligns with previous work that reported an interaction between CLN5 and PPT1 in COS-1 cells (Lyly et al., 2009). In addition, D. discoideum Cln5 interacts with CtsD, various homologs of human CTSF (CprA, CprD, CprE, and CprG), NagA, α-mannosidase, and β-glucosidase, all of which may have displayed altered enzymatic activity due to the absence, and hence, lack of interaction with Cln5, in cln5- cells (Huber and Mathavarajah, 2018a). While our previous work showed that loss of cln5 decreases the intracellular amount of CtsD during starvation (McLaren et al., 2021), in this study, comparative transcriptomics revealed increased expression of ctsD in starved cln5- cells. These findings indicate that cln5- cells upregulate the expression of ctsD to restore the intracellular amount of the protein. Also, our work showed that loss of cln5 elevates ctsB expression but reduces CTSB activity. Previous work also reported reduced CTSB activity in CLN5-deficient cells and it was suggested that the loss of CLN5 impairs intracellular trafficking and the movement of lysosomes (Mamo et al., 2012; Basak et al., 2021a; Yasa et al., 2021). Consistent with these findings, LAGO analyses revealed an enrichment of GO terms associated with intracellular vesicles including lysosomes, endocytic vesicles, and phagocytic vesicles. In CLN5-depleted HeLa cells, endosomal sorting is perturbed due to abolished retromer interaction and recruitment to the endosome, as well as poor fusion between the lysosome and autophagosome (Mamo et al., 2012; Yasa et al., 2021). Combined, these findings indicate that loss or mutation of CLN5 affects the expression, synthesis, and/or delivery of enzymes to lysosomes.
In conclusion, this study sheds light on the pathways perturbed in cln5- cells, highlights the multifaceted role of CLN5 in eukaryotic cells, and contributes knowledge to the mechanisms that may be disrupted in CLN5 disease patients.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA888040.
WK: data acquisition and analysis, writing—original draft, review and editing; RH: conceptualization, writing—review and editing, funding acquisition, supervision. All authors read and approved the final draft of the manuscript.
This work was supported by a Project Grant from the Canadian Institutes of Health Research (PJT165873 to RH). WK was supported by a Canada Graduate Scholarship from the Natural Sciences and Engineering Council of Canada and a Queen Elizabeth II Graduate Scholarship in Science and Technology.
The authors would like to thank Richard Gomer (Texas A&M University) for providing antibodies against AprA, CfaD, and CmfA, Compute Canada for allowing the authors to use the SHARCNET cluster for the bioinformatics pipeline, and Aaron Shafer and Michael Donaldson (both from Trent University, Peterborough, Ontario, Canada) for their advice in the bioinformatics analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1045738/full#supplementary-material
Adams, J., Feuerborn, M., Molina, J. A., Wilden, A. R., Adhikari, B., Budden, T., et al. (2019). Autophagy–lysosome pathway alterations and alpha-synuclein up-regulation in the subtype of neuronal ceroid lipofuscinosis, CLN5 disease. Sci. Rep. 9, 151. doi:10.1038/s41598-018-36379-z
Bakthavatsalam, D., Brock, D. A., Nikravan, N. N., Houston, K. D., Hatton, R. D., and Gomer, R. H. (2008). The secreted Dictyostelium protein CfaD is a chalone. J. Cell Sci. 121, 2473–2480. doi:10.1242/jcs.026682
Barondes, S. H., Haywood-Reid, P. L., and Cooper, D. N. W. (1985). Discoidin I, an endogenous lectin, is externalized from Dictyostelium discoideum in multilamellar bodies. J. Cell Biol. 100, 1825–1833. doi:10.1083/jcb.100.6.1825
Barrett, A. (1980). Fluorimetric assays for cathepsin B and cathepsin H with methylcoumarylamide substrates. Biochem. J. 187, 909–912. doi:10.1042/bj1870909
Basak, I., Hansen, R. A., Ward, M. E., and Hughes, S. M. (2021a). Deficiency of the lysosomal protein cln5 alters lysosomal function and movement. Biomolecules 11, 1412. doi:10.3390/biom11101412
Basak, I., Wicky, H. E., McDonald, K. O., Xu, J. B., Palmer, J. E., Best, H. L., et al. (2021b). A lysosomal enigma CLN5 and its significance in understanding neuronal ceroid lipofuscinosis. Cell. Mol. Life Sci. 78, 4735–4763. doi:10.1007/s00018-021-03813-x
Bento, C. F., Renna, M., Ghislat, G., Puri, C., Ashkenazi, A., Vicinanza, M., et al. (2016). Mammalian autophagy: How does it work? Annu. Rev. Biochem. 85, 685–713529. doi:10.1146/annurev-biochem-060815-014556
Best, H. L., Neverman, N. J., Wicky, H. E., Mitchell, N. L., Leitch, B., and Hughes, S. M. (2017). Characterisation of early changes in ovine CLN5 and CLN6 Batten disease neural cultures for the rapid screening of therapeutics. Neurobiol. Dis. 100, 62–74. doi:10.1016/j.nbd.2017.01.001
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi:10.1093/bioinformatics/btu170
Boyle, E. I., Weng, S., Gollub, J., Jin, H., Botstein, D., Cherry, J. M., et al. (2004). GO::TermFinder – open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20, 3710–3715. doi:10.1093/bioinformatics/bth456
Brock, D. A., and Gomer, R. H. (1999). A cell-counting factor regulating structure size in Dictyostelium. Genes Dev. 13, 1960–1969. doi:10.1101/gad.13.15.1960
Brock, D. A., and Gomer, R. H. (2005). A secreted factor represses cell proliferation in Dictyostelium. Development 132, 4553–4562. doi:10.1242/dev.02032
Brock, D. A., Buczynski, G., Spann, T. P., Wood, S. A., Cardelli, J., and Gomer, R. H. (1996). A Dictyostelium mutant with defective aggregate size determination. Development 122, 2569–2578. doi:10.1242/dev.122.9.2569
Bukharova, T., Weijer, G., Bosgraaf, L., Dormann, D., van Haastert, P. J., Weijer, C. J., et al. (2005). Paxillin is required for cell-substrate adhesion, cell sorting and slug migration during Dictyostelium development. J. Cell Sci. 118, 4295–4310. doi:10.1242/jcs.02557
Cárcel-Trullols, J., Kovács, A. D., and Pearce, D. A. (2015). Cell biology of the NCL proteins: What they do and don’t do. Biochim. Biophys. Acta 1852, 2242–2255. doi:10.1016/j.bbadis.2015.04.027
Castro, A., Bernis, C., Vigneron, S., Labbé, J. C., and Lorca, T. (2005). The anaphase-promoting complex: A key factor in the regulation of cell cycle. Oncogene 24, 314–325. doi:10.1038/sj.onc.1207973
Chia, C. P., Shariff, A., Savage, S. A., and Luna, E. J. (1993). The integral membrane protein, ponticulin, acts as a monomer in nucleating actin assembly. J. Cell Biol. 120, 909–922. doi:10.1083/jcb.120.4.909
Choe, J. M., Bakthavatsalam, D., Phillips, J. E., and Gomer, R. H. (2009). Dictyostelium cells bind a secreted autocrine factor that represses cell proliferation. BMC Biochem. 10, 4. doi:10.1186/1471-2091-10-4
Codlin, S., Haines, R. L., Burden, J. J. E., and Mole, S. E. (2008). Btn1 affects cytokinesis and cell-wall deposition by independent mechanisms, one of which is linked to dysregulation of vacuole pH. J. Cell Sci. 121, 2860–2870. doi:10.1242/jcs.030122
Coston, M. B., and Loomis, W. F. (1969). Isozymes of beta-glucosidase in Dictyostelium discoideum. J. Bacteriol. 100, 1208–1217. doi:10.1128/jb.100.3.1208-1217.1969
De Lozanne, A., and Spudich, J. A. (1987). Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science 236, 1086–1091. doi:10.1126/science.3576222
Deery, W. J., and Gomer, R. H. (1999). A putative receptor mediating cell-density sensing in Dictyostelium. J. Biol. Chem. 274, 34476–34482. doi:10.1074/jbc.274.48.34476
Deol, K. K., Crowe, S. O., Du, J., Bisbee, H. A., Guenette, R. G., and Strieter, E. R. (2020). Proteasome-bound UCH37/UCHL5 debranches ubiquitin chains to promote degradation. Mol. Cell 80, 796–809. doi:10.1016/j.molcel.2020.10.017
Doccini, S., Morani, F., Nesti, C., Pezzini, F., Calza, G., Soliymani, R., et al. (2020). Proteomic and functional analyses in disease models reveal CLN5 protein involvement in mitochondrial dysfunction. Cell Death Discov. 6, 18. doi:10.1038/s41420-020-0250-y
Doccini, S., Marchese, M., Morani, F., Gammaldi, N., Mero, S., Pezzini, F., et al. (2022). Lysosomal proteomics links disturbances in lipid homeostasis and sphingolipid metabolism to CLN5 disease. Cells 11, 1840. doi:10.3390/cells11111840
Duran, M. B., Rahman, A., Colten, M., and Brazill, D. (2009). Dictyostelium discoideum paxillin regulates actin-based processes. Protist 160, 221–232. doi:10.1016/j.protis.2008.09.005
El Haddad, S., Khoury, M., Daoud, M., Kantar, R., Harati, H., Mousallem, T., et al. (2012). CLN5 and CLN8 protein association with ceramide synthase: Biochemical and proteomic approaches. Electrophoresis 33, 3798–3809. doi:10.1002/elps.201200472
Faix, J., Steinmetz, M., Boves, H., Kammerer, R. A., Lottspeich, F., Mintert, U., et al. (1996). Cortexillins, major determinants of cell shape and size, are actin-bundling proteins with a parallel coiled-coil tail. Cell 86, 631–642. doi:10.1016/S0092-8674(00)80136-1
Faix, J., Weber, I., Mintert, U., Köhler, J., Lottspeich, F., and Marriott, G. (2001). Recruitment of cortexillin into the cleavage furrow is controlled by Rac1 and IQGAP-related proteins. EMBO J. 20, 3705–3715. doi:10.1093/emboj/20.14.3705
Fey, P., Kowal, A. S., Gaudet, P., Pilcher, K. E., and Chisholm, R. L. (2007). Protocols for growth and development of Dictyostelium discoideum. Nat. Protoc. 2, 1307–1316. doi:10.1038/nprot.2007.178
Fonovič, M., Brömme, D., Turk, V., and Turk, B. (2004). Human cathepsin F: Expression in baculovirus system, characterization and inhibition by protein inhibitors. Biol. Chem. 385, 505–509. doi:10.1515/BC.2004.059
Gomer, R. H., Yuen, I. S., and Firtel, R. A. (1991). A secreted 80 x 103 Mr protein mediates sensing of cell density and the onset of development of Dictyostelium. Development 112, 269–278. doi:10.1242/dev.112.1.269
Grubman, A., Pollari, E., Duncan, C., Caragounis, A., Blom, T., Volitakis, I., et al. (2014). Deregulation of biometal homeostasis: The missing link for neuronal ceroid lipofuscinoses? Metallomics 6, 932–943. doi:10.1039/c4mt00032c
Hersrud, S. L., Geraets, R. D., Weber, K. L., Chan, C. H., and Pearce, D. A. (2016). Plasma biomarkers for neuronal ceroid lipofuscinosis. FEBS J. 283, 459–471. doi:10.1111/febs.13593
Huber, R. J., and Mathavarajah, S. (2018a). Cln5 is secreted and functions as a glycoside hydrolase in Dictyostelium. Cell. Signal. 42, 236–248. doi:10.1016/j.cellsig.2017.11.001
Huber, R. J., and Mathavarajah, S. (2018b). Secretion and function of Cln5 during the early stages of Dictyostelium development. Biochim. Biophys. Acta. Mol. Cell Res. 1865, 1437–1450. doi:10.1016/j.bbamcr.2018.07.017
Huber, R. J., Hughes, S. M., Liu, W., Morgan, A., Tuxworth, R. I., and Russell, C. (2020). The contribution of multicellular model organisms to neuronal ceroid lipofuscinosis research. Biochim. Biophys. Acta. Mol. Basis Dis. 1866, 165614. doi:10.1016/j.bbadis.2019.165614
Huber, R. J., Williams, R. S. B., and Müller-Taubenberger, A. (2022). Editorial: Dictyostelium: A tractable cell and developmental model in biomedical research. Front. Cell Dev. Biol. 10, 909619. doi:10.3389/fcell.2022.909619
Huber, R. J. (2016). Using the social amoeba Dictyostelium to study the functions of proteins linked to neuronal ceroid lipofuscinosis. J. Biomed. Sci. 23, 83. doi:10.1186/s12929-016-0301-0
Huber, R. J. (2020). Molecular networking in the neuronal ceroid lipofuscinoses: Insights from mammalian models and the social amoeba Dictyostelium discoideum. J. Biomed. Sci. 27, 64. doi:10.1186/s12929-020-00653-y
Huber, R. J. (2021). Altered protein secretion in Batten disease. Dis. Model. Mech. 14, dmm049152. doi:10.1242/dmm.049152
Hughes, S. M., Hope, K. M., Xu, J. B., Mitchell, N. L., and Palmer, D. N. (2014). Inhibition of storage pathology in prenatal CLN5-deficient sheep neural cultures by lentiviral gene therapy. Neurobiol. Dis. 62, 543–550. doi:10.1016/j.nbd.2013.11.011
Ingalls, H. M., Barcelo, G., Wuestehube, L. J., and Luna, E. J. (1989). Developmental changes in protein composition and the actin-binding protein ponticulin in Dictyostelium discoideum plasma membranes purified by an improved method. Differentiation. 41, 87–98. doi:10.1111/j.1432-0436.1989.tb00736.x
Isosomppi, J., Vesa, J., Jalanko, A., and Peltonen, L. (2002). Lysosomal localization of the neuronal ceroid lipofuscinosis CLN5 protein. Hum. Mol. Genet. 11, 885–891. doi:10.1093/hmg/11.8.885
Iwan, K., Clayton, R., Mills, P., Csanyi, B., Gissen, P., Mole, S. E., et al. (2021). Urine proteomics analysis of patients with neuronal ceroid lipofuscinoses. iScience 24, 102020. doi:10.1016/j.isci.2020.102020
Jain, R., Yuen, I. S., Taphouse, C. R., and Gomer, R. H. (1992). A density-sensing factor controls development in Dictyostelium. Genes Dev. 6, 390–400. doi:10.1101/gad.6.3.390
Jin, L., Williamson, A., Banerjee, S., Philipp, I., and Rape, M. (2008). Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133, 653–665. doi:10.1016/j.cell.2008.04.012
Jules, F., Sauvageau, E., Dumaresq-Doiron, K., Mazzaferri, J., Haug-Kröper, M., Fluhrer, R., et al. (2017). CLN5 is cleaved by members of the SPP/SPPL family to produce a mature soluble protein. Exp. Cell Res. 357, 40–50. doi:10.1016/j.yexcr.2017.04.024
Kilpatrick, D. C., and Stirling, J. L. (1976). Properties and developmental regulation of an alpha-d-galactosidase from Dictyostelium discoideum. Biochem. J. 158, 409–417. doi:10.1042/bj1580409
Kim, J. Y., Van Haastert, P., and Devreotes, P. N. (1996). Social senses: G-protein-coupled receptor signaling pathways in Dictyostelium discoideum. Chem. Biol. 3, 239–243. doi:10.1016/S1074-5521(96)90103-9
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi:10.1038/s41587-019-0201-4
Kim, W. D., Wilson-Smillie, M. L. D. M., Thanabalasingam, A., Lefrancois, S., Cotman, S. L., and Huber, R. J. (2022). Autophagy in the neuronal ceroid lipofuscinoses (batten disease). Front. Cell Dev. Biol. 10, 812728. doi:10.3389/fcell.2022.812728
Kocaturk, N. M., and Gozuacik, D. (2018). Crosstalk between mammalian autophagy and the ubiquitin-proteasome system. Front. Cell Dev. Biol. 6, 128. doi:10.3389/fcell.2018.00128
Koonce, M. P. (2020). 13 plus 1: A 30-year perspective on microtubule-based motility in Dictyostelium. Cells 9, 528. doi:10.3390/cells9030528
Le Tallec, B., Barrault, M. B., Courbeyrette, R., Guérois, R., Marsolier-Kergoat, M.-C., and Peyroche, A. (2007). 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol. Cell 27, 660–674. doi:10.1016/j.molcel.2007.06.025
Leinonen, H., Keksa-Goldsteine, V., Ragauskas, S., Kohlmann, P., Sing, Y., Savchenko, E., et al. (2017). Retinal degeneration in A mouse model of CLN5 disease is associated with compromised autophagy. Sci. Rep. 7, 1597. doi:10.1038/s41598-017-01716-1
Li, T., and Guo, Y. (2022). ADP-ribosylation factor family of small GTP-binding proteins: Their membrane recruitment, activation, crosstalk and functions. Front. Cell Dev. Biol. 10, 813353–813359. doi:10.3389/fcell.2022.813353
Li, H., Chen, Q., Kaller, M., Nellen, W., Gräf, R., and De Lozanne, A. (2008). Dictyostelium Aurora kinase has properties of both Aurora A and Aurora B kinases. Eukaryot. Cell 7, 894–905. doi:10.1128/EC.00422-07
Liao, Y., Smyth, G. K., and Shi, W. (2014). FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi:10.1093/bioinformatics/btt656
Loomis, W. F. (1969). Acetylglucosaminidase, an early enzyme in the development of Dictyostelium discoideum. J. Bacteriol. 97, 1149–1154. doi:10.1128/JB.97.3.1149-1154.1969
Loomis, W. F. (1970). Developmental regulation of alpha-mannosidase in Dictyostelium discoideum. J. Bacteriol. 103, 375–381. doi:10.1128/JB.103.2.375-381.1970
Loomis, W. F. (1998). Role of PKA in the timing of developmental events in Dictyostelium cells. Microbiol. Mol. Biol. Rev. 62, 684–694. doi:10.1128/mmbr.62.3.684-694.1998
Loomis, W. F. (2014). Cell signalling during development of Dictyostelium. Dev. Biol. 391, 1–16. doi:10.1016/j.ydbio.2014.04.001
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi:10.1186/s13059-014-0550-8
Luebben, A. V., Bender, D., Becker, S., Crowther, L. M., Erven, I., Hofmann, K., et al. (2022). Cln5 represents a new type of cysteine-based S-depalmitoylase linked to neurodegeneration. Sci. Adv. 8, eabj8633–10. doi:10.1126/sciadv.abj8633
Luo, Q., Michaelis, C., and Weeks, G. (1995). Cyclin B and Cdc2 expression and Cd2 kinase activity during Dictyostelium differentiation. DNA Cell Biol. 14, 901–908. doi:10.1089/dna.1995.14.901
Lyly, A., Von Schantz, C., Heine, C., Schmiedt, M., Sipilä, T., Jalanko, A., et al. (2009). Novel interactions of CLN5 support molecular networking between Neuronal Ceroid Lipofuscinosis proteins. BMC Cell Biol. 10, 83. doi:10.1186/1471-2121-10-83
Mamo, A., Jules, F., Dumaresq-Doiron, K., Costantino, S., and Lefrancois, S. (2012). The role of ceroid lipofuscinosis neuronal protein 5 (CLN5) in endosomal sorting. Mol. Cell. Biol. 32, 1855–1866. doi:10.1128/mcb.06726-11
Mann, S. K. O., Brown, J. M., Briscoe, C., Parent, C., Pitt, G., Devreotes, P. N., et al. (1997). Role of cAMP-dependent protein kinase in controlling aggregation and postaggregative development in Dictyostelium. Dev. Biol. 183, 208–221. doi:10.1006/dbio.1996.8499
Maruhn, D. (1976). Rapid colorimetric assay of β-galactosidase and N-acetyl-β-glucosaminidase in human urine. Clin. Chim. Acta. 73, 453–461. doi:10.1016/0009-8981(76)90147-9
Masson, P., Lundin, D., Söderbom, F., and Young, P. (2009). Characterization of a REG/PA28 proteasome activator homolog in Dictyostelium discoideum indicates that the ubiquitin- and ATP-independent REGγ proteasome Is an ancient nuclear protease. Eukaryot. Cell 8, 844–851. doi:10.1128/EC.00165-08
Mathavarajah, S., Flores, A., and Huber, R. J. (2017). Dictyostelium discoideum: A model system for cell and developmental biology. Curr. Protoc. Essent. Lab. Tech. 14, 1.1–14.1.19. doi:10.1002/cpet.15
Mathavarajah, S., McLaren, M. D., and Huber, R. J. (2018). Cln3 function is linked to osmoregulation in a Dictyostelium model of Batten disease. Biochim. Biophys. Acta. Mol. Basis Dis. 1864, 3559–3573. doi:10.1016/j.bbadis.2018.08.013
Matyskiela, M. E., Rodrigo-Brenni, M. C., and Morgan, D. O. (2009). Mechanisms of ubiquitin transfer by the anaphase-promoting complex. J. Biol. 8, 92. doi:10.1186/jbiol184
McLaren, M. D., Mathavarajah, S., Kim, W. D., Yap, S. Q., and Huber, R. J. (2021). Aberrant autophagy impacts growth and multicellular development in a Dictyostelium knockout model of CLN5 disease. Front. Cell Dev. Biol. 9, 657406. doi:10.3389/fcell.2021.657406
Mesquita, A., Cardenal-Muñoz, E., Dominguez, E., Muñoz-Braceras, S., Nuñez-Corcuera, B., Phillips, B. A., et al. (2017). Autophagy in Dictyostelium: Mechanisms, regulation and disease in a simple biomedical model. Autophagy 13, 24–40. doi:10.1080/15548627.2016.1226737
Mizushima, N. (2010). The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22, 132–139. doi:10.1016/j.ceb.2009.12.004
Moharir, A., Peck, S. H., Budden, T., and Lee, S. Y. (2013). The role of N-glycosylation in folding, trafficking, and functionality of lysosomal protein CLN5. PloS One 8, e74299. doi:10.1371/journal.pone.0074299
Mole, S. E., and Cotman, S. L. (2015). Genetics of the neuronal ceroid lipofuscinoses (Batten disease). Biochim. Biophys. Acta 1852, 2237–2241. doi:10.1016/j.bbadis.2015.05.011
Mukherjee, A. B., Appu, A. P., Sadhukhan, T., Casey, S., Mondal, A., Zhang, Z., et al. (2019). Emerging new roles of the lysosome and neuronal ceroid lipofuscinoses. Mol. Neurodegener. 14, 4. doi:10.1186/s13024-018-0300-6
Nag, D. K., Tikhonenko, I., Soga, I., and Koonce, M. P. (2008). Disruption of four kinesin genes in Dictyostelium. BMC Cell Biol. 9, 21. doi:10.1186/1471-2121-9-21
Nagasaki, A., Kanada, M., and Uyeda, T. Q. (2009). Cell adhesion molecules regulate contractile ring-independent cytokinesis in Dictyostelium discoideum. Cell Res. 19, 236–246. doi:10.1038/cr.2008.318
Pagh, K., and Gerisch, G. (1986). Monoclonal antibodies binding to the tail of Dictyostelium discoideum myosin: Their effects on antiparallel and parallel assembly and actin-activated ATPase activity. J. Cell Biol. 103, 1527–1538. doi:10.1083/jcb.103.4.1527
Palmer, D. N., Fearnley, I. M., Walker, J. E., Hall, N. A., Lake, B. D., Wolfe, L. S., et al. (1992). Mitochondrial ATP synthase subunit c storage in the ceroid-lipofuscinoses (Batten disease). Am. J. Med. Genet. 42, 561–567. doi:10.1002/ajmg.1320420428
Persaud-Sawin, D.-A., Mousallem, T., Wang, C., Zucker, A., Kominami, E., and Boustany, R.-M. N. (2007). Neuronal ceroid lipofuscinosis: A common pathway? Pediatr. Res. 61, 146–152. doi:10.1203/pdr.0b013e31802d8a4a
Phillips, J. E., and Gomer, R. H. (2010). The ROCO kinase QkgA is necessary for proliferation inhibition by autocrine signals in Dictyostelium discoideum. Eukaryot. Cell 9, 1557–1565. doi:10.1128/EC.00121-10
Pickart, C. M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533. doi:10.1146/annurev.biochem.70.1.503
Roisin-Bouffay, C., Jang, W., Caprette, D. R., and Gomer, R. H. (2000). A precise group size in Dictyostelium is generated by a cell-counting factor modulating cell-cell adhesion. Mol. Cell 6, 953–959. doi:10.1016/S1097-2765(05)00082-1
Sanchez-Pulido, L., and Ponting, C. P. (2011). Cdc45: The missing recj ortholog in eukaryotes? Bioinformatics 27, 1885–1888. doi:10.1093/bioinformatics/btr332
Schauer, T. M., Nesper, M., Kehl, M., Lottspeich, F., Müller-Taubenberger, A., Gerisch, G., et al. (1993). Proteasomes from Dictyostelium discoideum: Characterization of structure and function. J. Struct. Biol. 111, 135–147. doi:10.1006/jsbi.1993.1044
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi:10.1038/nmeth.2019
Schmiedt, M. L., Blom, T., Blom, T., Kopra, O., Wong, A., von Schantz-Fant, C., et al. (2012). Cln5-deficiency in mice leads to microglial activation, defective myelination and changes in lipid metabolism. Neurobiol. Dis. 46, 19–29. doi:10.1016/j.nbd.2011.12.009
Schulkes, C., and Schaap, P. (1995). cAMP-dependent protein kinase activity is essential for preaggregative gene expression in Dictyostelium. FEBS Lett. 368, 381–384. doi:10.1016/0014-5793(95)00676-Z
Schulz, A., Kohlschütter, A., Mink, J., Simonati, A., and Williams, R. (2013). NCL diseases — Clinical perspectives. Biochim. Biophys. Acta 1832, 1801–1806. doi:10.1016/j.bbadis.2013.04.008
Shao, S., Von der Malsburg, K., and Hegde, R. S. (2013). Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell 50, 637–648. doi:10.1016/j.molcel.2013.04.015
Shariff, A., and Luna, E. J. (1990). Dictyostelium discoideum plasma membranes contain an actin-nucleating activity that requires ponticulin, an integral membrane glycoprotein. J. Cell Biol. 110, 681–692. doi:10.1083/jcb.110.3.681
Sharma, S. K., Michaelis, C., Lee, K. Y., Wang, J. H., and Weeks, G. (1999). Binding and catalytic properties of the Cdc2 and Crp proteins of Dictyostelium. Eur. J. Biochem. 260, 603–608. doi:10.1046/j.1432-1327.1999.00236.x
Shaulsky, G., Fuller, D., and Loomis, W. F. (1998). A cAMP-phosphodiesterase controls PKA-dependent differentiation. Development 125, 691–699. doi:10.1242/dev.125.4.691
Springer, W. R., Cooper, D. N. W., and Barondes, S. H. (1984). Discoidin I is implicated in cell-substratum attachment and ordered cell migration of Dictyostelium discoideum and resembles fibronectin. Cell 39, 557–564. doi:10.1016/0092-8674(84)90462-8
Stajdohar, M., Rosengarten, R. D., Kokosar, J., Jeran, L., Blenkus, D., Shaulsky, G., et al. (2017). dictyExpress: A web-based platform for sequence data management and analytics in Dictyostelium and beyond. BMC Bioinforma. 18, 291. doi:10.1186/s12859-017-1706-9
Stock, A., Steinmetz, M. O., Janmey, P. A., Aebi, U., Gerisch, G., Kammerer, R. A., et al. (1999). Domain analysis of cortexillin I: Actin-bundling, PIP2-binding and the rescue of cytokinesis. EMBO J. 18, 5274–5284. doi:10.1093/emboj/18.19.5274
Strasser, K., Bloomfield, G., MacWilliams, A., Ceccarelli, A., MacWilliams, H., and Tsang, A. (2012). A retinoblastoma orthologue is a major regulator of S-phase, mitotic, and developmental gene expression in Dictyostelium. PloS One 7, e39914. doi:10.1371/journal.pone.0039914
Stumpf, M., Müller, R., Gaßen, B., Wehrstedt, R., Fey, P., Karow, M. A., et al. (2017). A tripeptidyl peptidase 1 is a binding partner of the Golgi pH regulator (GPHR) in Dictyostelium. Dis. Model. Mech. 10, 897–907. doi:10.1242/dmm.029280
Sun, T. J., and Devreotes, P. N. (1991). Gene targeting of the aggregation stage cAMP receptor cAR1 in Dictyostelium. Genes Dev. 5, 572–582. doi:10.1101/gad.5.4.572
Tang, L., Gao, T., McCollum, C., Jang, W., Vicker, M. G., Ammann, R. R., et al. (2002). A cell number-counting factor regulates the cytoskeleton and cell motility in Dictyostelium. Proc. Natl. Acad. Sci. U. S. A. 99, 1371–1376. doi:10.1073/pnas.022516099
Tanida, I., Minematsu-Ikeguchi, N., Ueno, T., and Kominami, E. (2005). Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 1, 84–91. doi:10.4161/auto.1.2.1697
Tikhonenko, I., Nag, D. K., Robinson, D. N., and Koonce, M. P. (2009). Microtubule-nucleus interactions in Dictyostelium discoideum mediated by central motor kinesins. Eukaryot. Cell 8, 723–731. doi:10.1128/EC.00018-09
Van Diggelen, O. P., Keulemans, J. L. M., Winchester, B., Hofman, I. L., Vanhanen, S. L., Santavuori, P., et al. (1999). A rapid fluorogenic palmitoyl-protein thioesterase assay: Pre- and postnatal diagnosis of INCL. Mol. Genet. Metab. 66, 240–244. doi:10.1006/mgme.1999.2809
Van Haastert, P. J. M., Bishop, J. D., and Gomer, R. H. (1996). The cell density factor CMF regulates the chemoattractant receptor cAR1 in Dictyostelium. J. Cell Biol. 134, 1543–1549. doi:10.1083/jcb.134.6.1543
Varet, H., Brillet-Guéguen, L., Coppée, J. Y., and Dillies, M. A. (2016). SARTools: A DESeq2- and edgeR-based R pipeline for comprehensive differential analysis of RNA-seq data. PloS One 11, e0157022. doi:10.1371/journal.pone.0157022
von Schantz, C., Saharinen, J., Kopra, O., Cooper, J. D., Gentile, M., Hovatta, I., et al. (2008). Brain gene expression profiles of Cln1 and Cln5 deficient mice unravels common molecular pathways underlying neuronal degeneration in NCL diseases. BMC Genomics 9, 146. doi:10.1186/1471-2164-9-146
Wang, J., Dye, B. T., Rajashankar, K. R., Kurinov, I., and Schulman, B. A. (2009). Insights into anaphase promoting complex TPR subdomain assembly from a CDC26-APC6 structure. Nat. Struct. Mol. Biol. 16, 987–989. doi:10.1038/nsmb.1645
Weber, I., Gerisch, G., Heizer, C., Murphy, J., Badelt, K., Stock, A., et al. (1999). Cytokinesis mediated through the recruitment of cortexillins into the cleavage furrow. EMBO J. 18, 586–594. doi:10.1093/emboj/18.3.586
Wetterauer, B., Jacobsen, G., Morandini, P., and MacWilliams, H. (1993). Mutants of Dictyostelium discoideum with defects in the regulation of discoidin I expression. Dev. Biol. 159, 184–195. doi:10.1006/dbio.1993.1232
Williamson, A., Wickliffe, K. E., Mellone, B. G., Song, L., Karpen, G. H., and Rape, M. (2009). Identification of a physiological E2 module for the human anaphase-promoting complex. Proc. Natl. Acad. Sci. U. S. A. 106, 18213–18218. doi:10.1073/pnas.0907887106
Wimmer, B., Lottspeich, F., Ritter, J., and Bronnenmeier, K. (1997). A novel type of thermostable alpha-D-glucosidase from Thermoanaerobacter thermohydrosulfuricus exhibiting maltodextrinohydrolase activity. Biochem. J. 328, 581–586. doi:10.1042/bj3280581
Yasa, S., Sauvageau, E., Modica, G., and Lefrancois, S. (2021). CLN5 and CLN3 function as a complex to regulate endolysosome function. Biochem. J. 478, 2339–2357. doi:10.1042/BCJ20210171
Yuen, I. S., Jain, R., Bishop, J. D., Lindsey, D. F., Deery, W. J., Van Haastert, P. J. M., et al. (1995). A density-sensing factor regulates signal transduction in Dictyostelium. J. Cell Biol. 129, 1251–1262. doi:10.1083/jcb.129.5.1251
Zang, J.-H., Cavet, G., Sabry, J. H., Wagner, P., Moores, S. L., and Spudich, J. A. (1997). On the role of myosin-II in cytokinesis: Division of Dictyostelium cells under adhesive and nonadhesive conditions. Mol. Biol. Cell 8, 2617–2629. doi:10.1091/mbc.8.12.2617
AprA autocrine proliferation repressor
BH Benjamin-Hochberg
CadA calcium-dependent cell adhesion molecule
cAMP cyclic adenosine monophosphate
carD cAMP receptor D
CB conditioned buffer
CfaD counting factor-associated protein D
CLN ceroid lipofuscinosis neuronal
CM conditioned media
CmfA conditioned medium factor
CmfB conditioned medium factor receptor
CtnA countin
CTSB cathepsin B
CTSD cathepsin D
CTSF cathepsin F
CtxA cortexillin I
CtxB cortexillin II
DEG differentially expressed gene
Dsc discoidin
GO gene ontology
HISAT2 hierarchical indexing for spliced alignment of transcripts 2
LAGO logically accelerated gene ontology
MES 2-(N-morpholino)ethanesulfonic acid
MhcA myosin heavy chain A type II
NagA N-acetylglucosaminidase A
NCL neuronal ceroid lipofuscinosis
PPT1 palmitoyl-protein thioesterase 1
PS proteasomal subunit
RegA 3′,5′-cyclic-nucleotide phosphodiesterase
RNA-seq RNA sequencing
SARTools statistical analysis of RNA-seq data tools
TPP1 tripeptidyl peptidase 1
VinA vinculin A
WC whole cell
WT wild-type
Keywords: Batten disease, CLN5, Dictyostelium discoideum, enzyme activity, neuronal ceroid lipofucinosis, proteasome, RNA-seq
Citation: Kim WD and Huber RJ (2022) An altered transcriptome underlies cln5-deficiency phenotypes in Dictyostelium discoideum. Front. Genet. 13:1045738. doi: 10.3389/fgene.2022.1045738
Received: 16 September 2022; Accepted: 27 October 2022;
Published: 10 November 2022.
Edited by:
Paschalis Nicolaou, The Cyprus Institute of Neurology and Genetics, CyprusReviewed by:
Matt Scaglione, Duke University, United StatesCopyright © 2022 Kim and Huber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert J. Huber, cm9iZXJ0aHViZXJAdHJlbnR1LmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.