
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 11 November 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1039677
This article is part of the Research Topic Genetic Mechanisms of Stress Tolerance in Crops View all 7 articles
 Xing Huo1
Xing Huo1 Junyi Xiao1,2
Junyi Xiao1,2 Xin Peng1
Xin Peng1 Yanhui Lin3
Yanhui Lin3 Dilin Liu1
Dilin Liu1 Wuge Liu1
Wuge Liu1 Yilong Liao1
Yilong Liao1 Jinhua Li1
Jinhua Li1 Manshan Zhu1
Manshan Zhu1 Chongyun Fu1
Chongyun Fu1 Xueqin Zeng1
Xueqin Zeng1 Xiaozhi Ma1
Xiaozhi Ma1 Le Kong1
Le Kong1 Feng Wang1*
Feng Wang1*Rice originated in tropical and subtropical regions and is distributed worldwide. Low temperature is one of the most critical abiotic stresses affecting grain yield and geographical distribution of rice. It is vital to elucidate the molecular mechanism of chilling tolerance in rice for ensuring cereals production. Previously we isolated the domestication-related gene NOG1 which affects rice grain number and yield. In this study, we specified that rice varieties harboring high-yielding NOG1 allele are more distributed in low-latitude regions. Additionally, we observed NOG1 influences the chilling tolerance of rice. Through genome-wide transcriptional analysis after cold treatment at 10°C, there were 717 differentially expressed genes (DEGs) in nog1 near-isogenic lines compared with the control Guichao 2, including 432 up-regulated DEGs and 284 down-regulated DEGs. Gene ontology annotations and KEGG enrichment analysis of DEGs showed that various biological processes and signaling pathways were related to cold stress, such as lipid metabolism and genetic information processing. These results provide new insights into the mechanism of chilling tolerance in rice and the molecular basis of environmental adaptation during rice domestication.
Rice is one of the most important food crops, feeding more than half of the world’s population (Ito and Lacerda, 2019). Coping with various biotic and abiotic stresses and maintaining stable yields during rice production is of great significance to ensuring world food security (Lesk et al., 2016). Low temperature is an essential abiotic stress factor affecting the environmental adaptability and geographical distribution of rice. Since rice originated in tropical and subtropical regions, it is extremely sensitive to low temperature. The damage caused by low temperature will reduce the growth and development of rice, and eventually lead to the reduction of yield and quality (Jaglo et al., 2001; Zhang et al., 2019). Cold stress is prevalent in the world’s main rice producing areas, and leads to huge economic losses every year. There are more than 15 million hectares of rice in the world facing the threat of low temperature (Lin et al., 2004; Zhang et al., 2014; Pradhan et al., 2019; Li et al., 2021). As an essential abiotic stress, cold stress has been a primary spot in plant science research. A series of cold tolerance-related genes were described (Saito et al., 2004; Fujino et al., 2008; Fujino and Matsuda, 2010; Su et al., 2010; Liu et al., 2013; Ma et al., 2015; Manishankar and Kudla, 2015; Shi and Gong, 2015; Mao et al., 2019). However, the molecular mechanisms of cold stress tolerance in rice are still unclear. In recent years, RNA-Seq has been widely used to perform genome-wide gene expression analysis, providing detailed and in-depth data for the molecular mechanism study of plant cold tolerance (Bashir et al., 2019; Wai et al., 2021). NOG1 is a functional gene that regulates grain number and yield in rice. The functional site of NOG1 is a 12-bp copy number variation in the promoter region. There are two haplotypes of NOG1 in cultivated rice, with two copies of 12-bp fragments in high-yielding rice and only one copy of 12-bp in low-yielding rice (Huo et al., 2017). In this study, we found that the yield-related NOG1 is associated with the geographic distribution of rice varieties and acts as a negative player in cold tolerance during rice domestication. The genome-wide transcriptional identification of NOG1 near-isogenic lines under cold stress were additionally compared, which provided new insights into the molecular mechanism of NOG1-mediated cold tolerance pathway in rice.
Geographic distribution of different NOG1 alleles were investigated using 158 rice accessions, including 84 indica and 74 japonica varieties (Supplementary Tables S1,S2). As a result, high-yielding rice varieties containing two copies of 12-bp fragment were more frequently distributed in low-latitude regions (Figure 1). Under the control of other variables, the relationship between latitude and temperature is inversely proportional, that is, the higher the latitude, the lower the temperature. The principle is that the altitude of the Sun decreases with increasing latitude, resulting in a decrease in solar radiation with increasing latitude, resulting in a decrease in temperature with increasing latitude. The geographic distribution of NOG1 alleles suggests that NOG1 might affect the response to cold stress in addition to its role in improving rice yield.
In order to further eliminate the interference of genetic background, SIL176, an introgression line harboring nog1 allele in the background of Guichao 2, was selected to backcross with Guichao 2 and then selfed. Combined with genotype identification in the BC2F2 population, a near-isogenic line with only one fragment introgressed in the genomic region of nog1 compared with Guichao 2 was screened and named NIL176. Gene sequencing validated that the NOG1 sequence in NIL176 was identical to that in SIL176. Compared with Guichao 2, NIL176 has a 12-bp deletion and 15 SNPs in the promoter region (Figure 2A), the grain number per panicle and grain yield of the near-isogenic lines were investigated, and the results showed that NIL176 exhibited a similar performance as SIL176. Compared with Guichao 2, both NIL176 and SIL176 exhibited less grains per panicle and lower yield (Figures 2B–G). Therefore, in the further study of the function of NOG1 gene and its response to cold stress, using near-isogenic lines as materials can eliminate the interference of genetic background and obtain more reliable results.
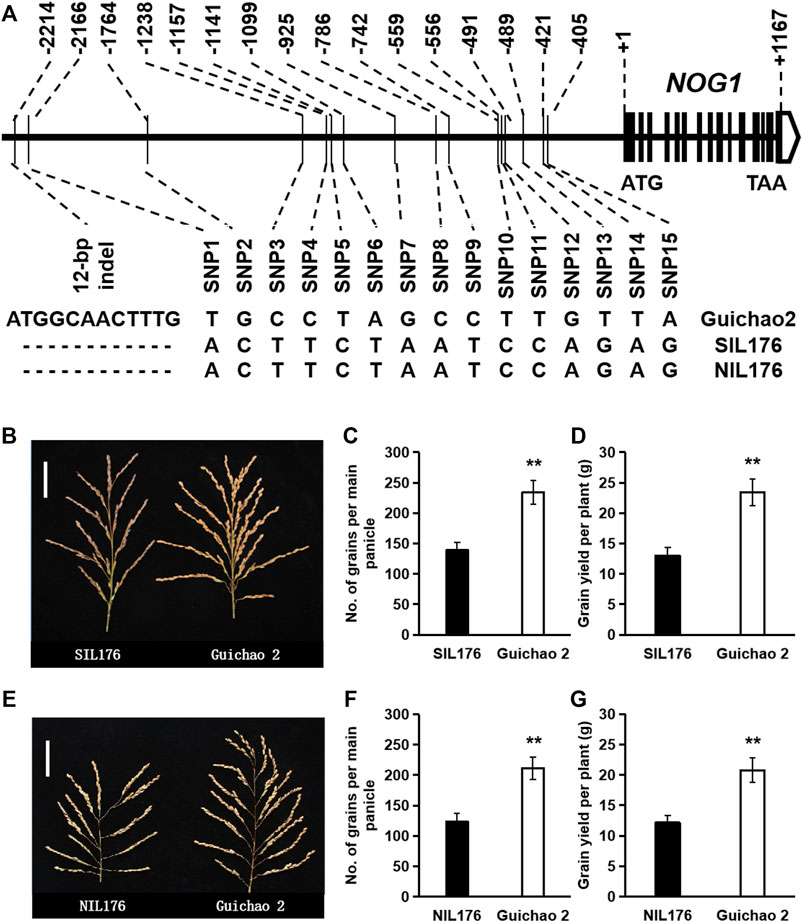
FIGURE 2. Genotypes and phenotypes of the nog1 near-isogenic line. (A): Genotypes of the nog1 near-isogenic line (NIL176), SIL176 and Guichao 2. (B–D): Phenotypic comparison between SIL176 and Guichao 2. (E–G): Phenotypic comparison between NIL176 and Guichao 2.
To verify whether NOG1 affects the response of rice to cold stress, NIL176 and Guichao 2 were used for further study. Rice seedlings were treated under low temperature stress at 10°C for 10 days and recovered at 28°C for three days to examine the phenotype. After cold stress, NIL176 still maintained a normal growth, while Guichao 2 showed obvious cold injury phenotypes including wilting and yellowing (Figure 3). The results showed that compared with Guichao 2, the tolerance of NIL176 to cold stress was significantly improved.

FIGURE 3. Phenotypes of chilling tolerance between NIL176 and Guichao 2. (A): Phenotypic comparison of NIL176 and Guichao 2 at 28°C. (B): Phenotypic comparison of NIL176 and Guichao 2 after cold stress.
RNA-seq analysis was performed for Guichao 2 and NIL176, which were subjected to low temperature treatment at 10°C for 12 h. The differentially expressed genes (DEGs) were screened with the difference of gene expression levels more than 2 folds or less than 0.5 fold. After cold treatment, NIL176 had 5,954 up-regulated DEGs and 5,612 down-regulated DEGs, while Guichao 2 had 4,499 up-regulated DEGs and 5,850 down-regulated DEGs (Figure 4A). NIL176 and Guichao 2 shared 3,463 up-regulated genes and 3,953 down-regulated genes (Figures 4B,C). Under cold stress, the number of down-regulated genes in NIL176 and Guichao 2 were highly close. However, the up-regulated genes in NIL176 were significantly more than those in Guichao 2. A total of 2,491 up-regulated genes were unique in NIL176.
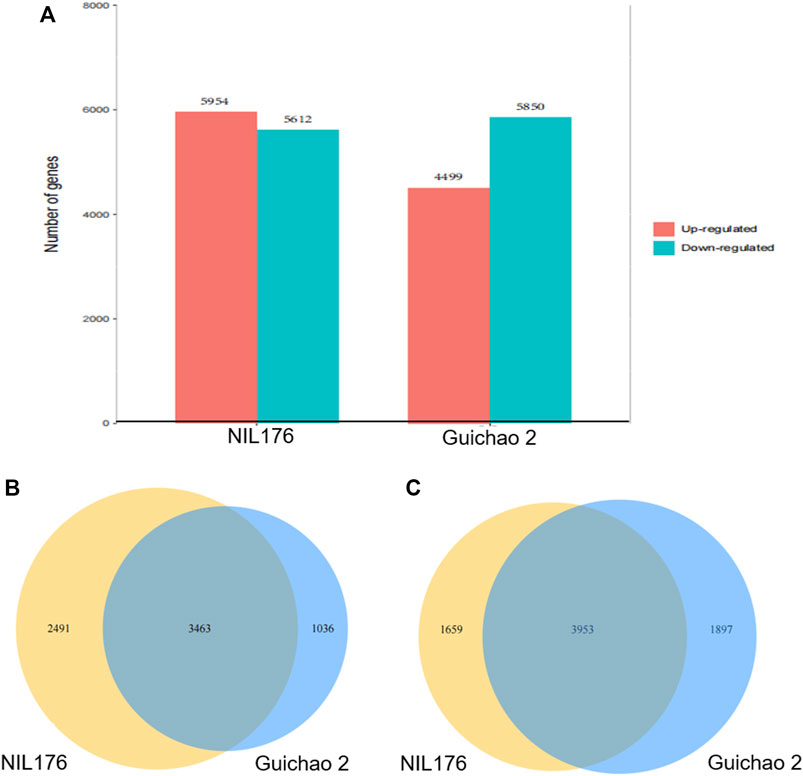
FIGURE 4. Comparisons of differentially expressed genes between NIL176 and Guichao 2 under cold stress. (A): Numbers of differentially expressed genes in NIL176 and Guichao 2. (B): Up-regulated differentially expressed genes in NIL176 and Guichao 2. (C): Down-regulated differentially expressed genes in NIL176 and Guichao 2.
After cold treatment, there are 717 DEGs when compared NIL176 with Guichao 2, of which 432 were up-regulated and 285 were down-regulated (Figure 5 and Supplementary Table S3). In order to further analyze the molecular mechanism of NOG1-mediated response to cold stress in rice, Gene ontology (GO) annotation and KEGG pathway enrichment analysis were conducted to identify cold stress-related pathways using DEGs between NIL176 and Guichao 2. The up-regulated DEGs were involved in biological processes such as cellular response to stress, response to stimulus, and defense response. It also affects cellular components including integral component of membrane and intrinsic component of membrane (Figure 6A). Down-regulation DEGs involves biological processes such as external condensing structure, response to abiotic stimulus, and response to inorganic substance (Figure 6B).
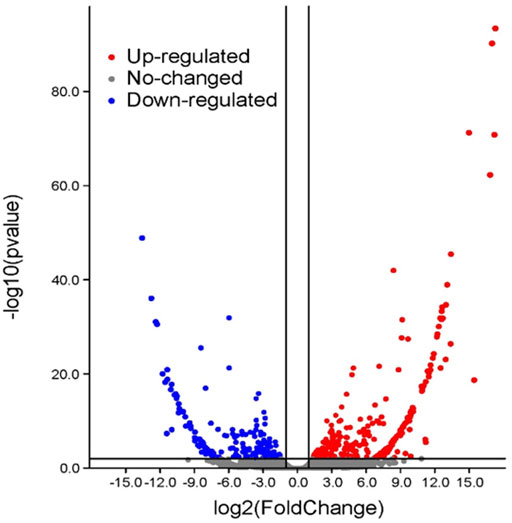
FIGURE 5. Volcano plot between NIL176 and Guichao 2. After cold-treatment, there are a total of 717 significantly differentially expressed genes in NIL176 compared with Guichao 2, including 432 up-regulated genes and 284 down-regulated genes.
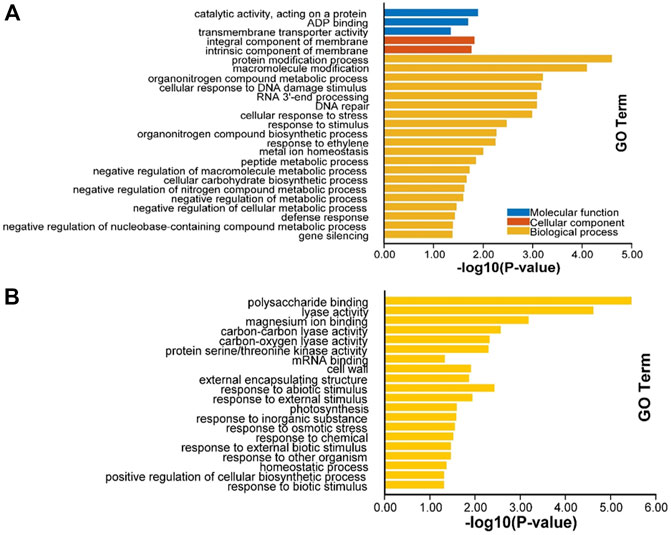
FIGURE 6. GO enrichment of differentially expressed genes between NIL176 and Guichao 2. (A): GO enrichment of up-regulated genes in NIL176 compared with Guichao 2. (B): GO enrichment of down-regulated genes in NIL176 compared with Guichao 2.
KEGG enrichment analysis showed that up-regulated DEGs were enriched to lipid metabolism, genetic information processing, protein families: signaling and cellular processes, protein kinases and other pathways (Figure 7A), and down-regulated DEGs were enriched to genetic information processing, carbohydrate metabolism, energy metabolism, and other pathways (Figure 7B). Notably, pathways such as genetic information processing, lipid metabolism, protein families: signaling and cellular processes can be enriched in both up-regulated and down-regulated DEGs, indicating that these pathways may play an important role in the NOG1-mediated rice cold tolerance gene network.
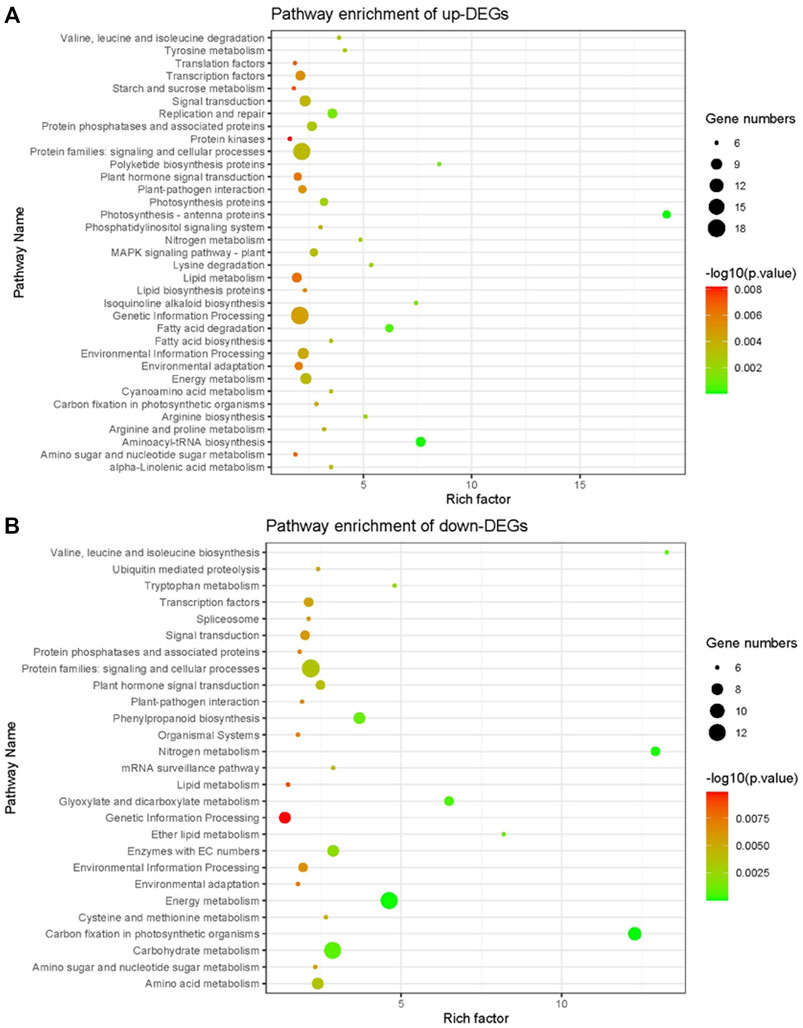
FIGURE 7. KEGG pathway enrichment of differentially expressed genes between NIL176 and Guichao 2. (A): KEGG pathway enrichment of up-regulated genes in NIL176 compared with Guichao 2. (B): KEGG pathway enrichment of down-regulated genes in NIL176 compared with Guichao 2.
Fatty acids, the major constituents of membrane glycerolipids, have important biological functions for stress response in plants. In response to a decrease in ambient temperature, plants increase the level of their unsaturated fatty acids, maintaining the appropriate fluidity of membrane lipids (Sakamoto and Murata, 2002). The related genes of fatty acid metabolism were enriched in DGEs of Guichao 2 and NIL176 (Figure 7). To investigate whether fatty acids involved in NOG1-mediated cold stress, we detected expression changes among fatty acids related genes (Figure 8). In the KEGG pathway of fatty acid biosynthesis, most of genes were down-regulated after cold treatment both in Guichao 2 and NIL176. However, seven genes are relatively high expression in NIL176 compared with Guichao 2 under normal condition, which suggests that NIL176 accumulates more fatty acids before cold stress, compared with Guichao 2. Long-chain acyl-CoA synthetase (LACS) plays a critical role in plant development and stress responses (Zhao et al., 2021). Intriguingly, the expression of LACS (K01897 [EC:6.2.1.3]) was particularly high expression in the cold-treated NIL176 plant.
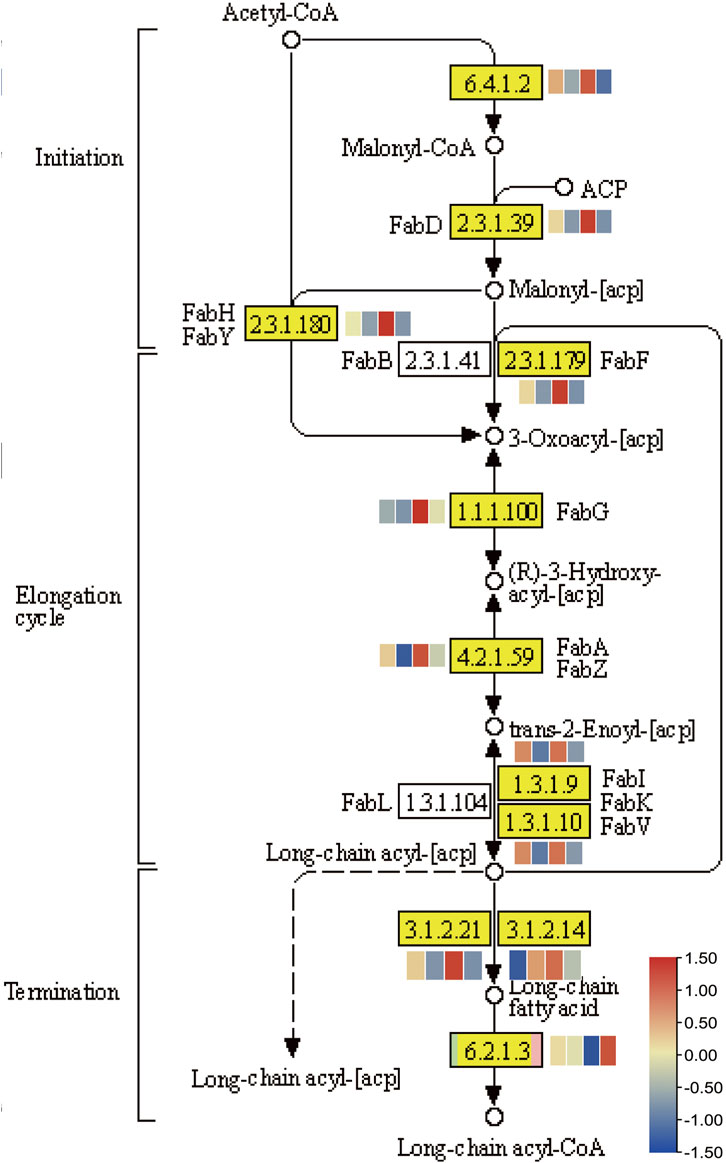
FIGURE 8. The KEGG pathway of fatty acid biosynthesis. The heatmap was generated by using the FPKM values of Guichao 2 under normal condition, Guichao 2 after cold treatment, NIL176 under normal condition, and NIL176 after cold treatment with row normalization.
Rice is one of the earliest crops domesticated by humans. A series of key domestication traits were artificially selected, such as seed shattering, plant architecture, grain color and awn development (Konishi et al., 2006; Li et al., 2006; Sweeney et al., 2006; Tan et al., 2008; Zhu et al., 2011; Zhu et al., 2013; Hua et al., 2015). At the same time, rice originated in tropical and subtropical regions. During the process of domestication, local varieties need a series of adaptive improvements to the local environment, including photoperiod, temperature tolerance, drought and other stress resistance. In this study, the domestication-related NOG1 gene affects the cold tolerance of rice, thereby affecting the latitudinal distribution of NOG1 alleles. In the future molecular breeding process for genetic improvement using domesticated genes such as NOG1, it is necessary to comprehensively consider the geographic location and temperature of the planting area.
Biofilm is the main damaged part of plants under cold stress, and the damage can lead to irreversible phase transition of the biofilm system. The fluidity and stability of biofilms are closely related to the low temperature tolerance of rice (Sakamoto and Murata, 2002). Fatty acids are important components of biofilms, and the content and composition of unsaturated fatty acids play an influential role in the cold tolerance of rice (Ariizumi et al., 2002). The NOG1 gene that regulates grain number and yield in rice encodes enoyl-CoA hydratase/isomerase (ECH), a key enzyme in fatty acid metabolism, which affects the content of endogenous fatty acids and the composition of unsaturated fatty acids in rice, especially C18:3. The fatty acid biosynthesis pathway was enriched in DEGs, which indicates that fatty acid metabolism likely played an essential role in NOG1-mediated low temperature stress. Lipid metabolism, alpha-Linolenic acid metabolism, fatty acid biosynthesis and fatty acid degradation pathways were all enriched.
RNA-seq can analyze the differential expression of genome-wide genes, which is helpful to comprehensively analyze the response pathway of rice cold tolerance. The results of GO annotation and KEGG enrichment analysis of DEGs showed that in addition to fatty acid metabolism pathways, there are multiple pathways involved in cold stress responses. GO annotation enriched in transmembrane transporter activity, integral component of membrane, intrinsic component of membrane and response to inorganic substance. Membrane transporters are important functional proteins that change biomembrane permeability, maintain osmotic pressure and ion transport inside and outside the biomembrane, and are one of the essential ways to improve plant cold tolerance (Lissarre et al., 2010; Miura and Furumoto, 2013).
Additionally, cold stress is related to plant stress resistance (Saijo and Loo, 2020). In this study, GO analysis showed that stress resistance-related pathways such as external encapsulating structure, response to abiotic stimulus, and response to inorganic substance were activated. The results of KEGG also showed that environmental adaptation, environmental adaptation pathways such as Information processing and plant-pathogen interaction are enriched. Hence, NOG1 may also be related to other abiotic stresses and disease resistance in rice in addition to its role in low temperature stress. These results enrich the understanding of cold resistance pathways in rice and provide new evidence for analyzing the molecular mechanism of rice cold tolerance.
The NOG1 gene, which regulates grain number and yield in rice, encodes a key enzyme in fatty acid metabolism. Allele identification and geographic origin of 158 rice varieties including 84 indica varieties and 74 japonica varieties indicated a strong latitude adaptation of the NOG1 alleles. Through the cold treatment of Guichao 2 and NIL176 (the near-isogenic line of nog1) at the seedling stage, it was found that NOG1 regulates the cold tolerance in rice. Using whole transcriptome analysis of near-isogenic lines, 5,954 up-regulated DEGs and 5,612 down-regulated DEGs were observed in NIL176, whereas 4,499 up-regulated DEGs and 5,850 down-regulated DEGs were found in Guichao 2 after cold treatment. The number of up-regulated genes in NIL176 was significantly higher than that in Guichao 2, among which 2,491 genes were only up-regulated in NIL176. GO annotation and KEGG enrichment analysis of DEGs showed that fatty acid metabolism, transmembrane transporter activity, integral component of membrane and plant-pathogen interaction pathways DEGs were enriched.
The sources of SIL176 and Guichao 2 were introduced in previous studies (Huo et al., 2017). The near-isogenic line NIL176 was selected from BC2F2, using SIL176 as a donor and Guichao 2 as the recipient. A collection of 158 rice materials, including 84 indica (O. sativa L. ssp. indica) varieties and 74 japonica (O. sativa L. ssp. japonica) varieties, were used to verify the alleles of NOG1 and to investigate the geographic distribution of material origins, as listed in Supplementary Table S1.
The primers used in this study for NOG1 sequencing are listed in Supplementary Table S2.
The seeds of the test materials were treated at 45°C for 48 h to break dormancy. Soak seeds with ddH20 at room temperature for 24 h, keep moist for 24–48 h to promote germination. Seeds with uniform whiteness were selected and sown in bottomless 96-well PCR plates. Plant 3 plates of each material, hydroponically in an artificial climate chamber. The temperature of the artificial climate chamber is 28°C, with a light/darkness regime of 12/12 h and a relative humidity of 85%. The tested materials were grown to the 3-leaf stage and subjected to low temperature treatment in the artificial climate chamber. The treatment temperature was 10°C, 12 h of light, 12 h of darkness, and a relative humidity of 85%. After 10 days of treatment, growth was recovered at 28°C for 3 days and the phenotype was investigated.
Total RNA was extracted from the leaves of the plant with cold-treatment for 12 h using the QIAGEN plant RNA kit (Hilden, Germany). The concentration and quality of RNA were evaluated using NanoDrop 2000 UV-VIS spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States). Paired-end reads were generated on a HiSeq2000 platform following the manufacturer’s instructions (Illumina, United States). The analysis of RNA-sequencing (RNA-seq) data and differentially expressed genes (DEGs) were carried out as previously reported (Peng et al., 2021). Briefly, RNA-seq data were mapped on the reference genome with HISAT2 2.1.0 (Kim et al., 2019). FeatureCounts 1.6.2 was used to count the number of reads mapped on exons (Liao et al., 2014). DEGs were evaluated by edgeR 3.32.0 (Robinson et al., 2010). Genes with p < 0.05 and log2 fold-changes > 1 were considered as DEGs. Further screening among the initial DEGs was performed based on fragments per kilo-base per million fragments mapped (FPKM) values. Afterward, the GO and KEGG annotations of DEGs were conducted by pannzer2 (Toronen et al., 2018) and KEGG Automatic Annotation Server (Moriya et al., 2007).
The raw sequence data reported in this study have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA008173) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa (Chen et al., 2021).
FW and WL designed and supervised this work. XH, DL, and XP analyzed the data and wrote the article. JX constructed near-isogenic lines and performed cold stress. XH, JX, YLL, and JL. investigated the distribution and genotype of 158 rice varieties. XH, MZ, LK, and XZ. performed RNA extraction and detection. XH, YHL, XZ, ZM, and CF. participated in the revision of the manuscript. All authors reviewed and approved the final manuscript.
This work was supported by National Natural Science Foundation of China (32071992), Guangdong Basic and Applied Basic Research Foundation (2019A1515011903), Special fund for scientific innovation strategy-construction of high level Academy of Agriculture Science (R2018YJ-YB1001), The Discipline Team of Agricultural Competitive Industries in Guangdong Academy of Agricultural Sciences (202101TD), and Guangdong Key Laboratory of New Technology in Rice Breeding (2020B1212060047).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1039677/full#supplementary-material
Ariizumi, T., Kishitani, S., Inatsugi, R., Nishida, I., Murata, N., and Toriyama, K. (2002). An increase in unsaturation of fatty acids in phosphatidylglycerol from leaves improves the rates of photosynthesis and growth at low temperatures in transgenic rice seedlings. Plant Cell Physiol. 43, 751–758. doi:10.1093/pcp/pcf087
Bashir, K., Matsui, A., Rasheed, S., and Seki, M. (2019). Recent advances in the characterization of plant transcriptomes in response to drought, salinity, heat, and cold stress. F1000Res 8, 658. doi:10.12688/f1000research.18424.1
Chen, T., Chen, X., Zhang, S., Zhu, J., Tang, B., Wang, A., et al. (2021). The genome sequence archive family: toward explosive data growth and diverse data types. Genomics Proteomics Bioinforma. 19, 578–583. doi:10.1016/j.gpb.2021.08.001
Fujino, K., and Matsuda, Y. (2010). Genome-wide analysis of genes targeted by qltg3-1 controlling low-temperature germinability in rice. Plant Mol. Biol. 72, 137–152. doi:10.1007/s11103-009-9559-x
Fujino, K., Sekiguchi, H., Matsuda, Y., Sugimoto, K., Ono, K., and Yano, M. (2008). Molecular identification of a major quantitative trait locus, qltg3-1, controlling low-temperature germinability in rice. Proc. Natl. Acad. Sci. U. S. A. 105, 12623–12628. doi:10.1073/pnas.0805303105
Hua, L., Wang, D. R., Tan, L., Fu, Y., Liu, F., Xiao, L., et al. (2015). Laba1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell 27, 1875–1888. doi:10.1105/tpc.15.00260
Huo, X., Wu, S., Zhu, Z., Liu, F., Fu, Y., Cai, H., et al. (2017). Nog1 increases grain production in rice. Nat. Commun. 8, 1497. doi:10.1038/s41467-017-01501-8
Ito, V. C., and Lacerda, L. G. (2019). Black rice (oryza sativa l.): A review of its historical aspects, chemical composition, nutritional and functional properties, and applications and processing technologies. Food Chem. 301, 125304. doi:10.1016/j.foodchem.2019.125304
Jaglo, K. R., Kleff, S., Amundsen, K. L., Zhang, X., Haake, V., Zhang, J. Z., et al. (2001). Components of the arabidopsis c-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in brassica napus and other plant species. Plant Physiol. 127, 910–917. doi:10.1104/pp.010548
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with hisat2 and hisat-genotype. Nat. Biotechnol. 37, 907–915. doi:10.1038/s41587-019-0201-4
Konishi, S., Izawa, T., Lin, S. Y., Ebana, K., Fukuta, Y., Sasaki, T., et al. (2006). An snp caused loss of seed shattering during rice domestication. Science 312, 1392–1396. doi:10.1126/science.1126410
Lesk, C., Rowhani, P., and Ramankutty, N. (2016). Influence of extreme weather disasters on global crop production. Nature 529, 84–87. doi:10.1038/nature16467
Li, C., Zhou, A., and Sang, T. (2006). Rice domestication by reducing shattering. Science 311, 1936–1939. doi:10.1126/science.1123604
Li, C., Liu, J., Bian, J., Jin, T., Zou, B., Liu, S., et al. (2021). Identification of cold tolerance qtls at the bud burst stage in 211 rice landraces by gwas. BMC Plant Biol. 21, 542. doi:10.1186/s12870-021-03317-7
Liao, Y., Smyth, G. K., and Shi, W. (2014). Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi:10.1093/bioinformatics/btt656
Lin, S. Z., Zhang, Z. Y., Lin, Y. Z., Zhang, Q., and Guo, H. (2004). The role of calcium and calmodulin in freezing-induced freezing resistance of populus tomentosa cuttings. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 30, 59–68.
Lissarre, M., Ohta, M., Sato, A., and Miura, K. (2010). Cold-responsive gene regulation during cold acclimation in plants. Plant Signal. Behav. 5, 948–952. doi:10.4161/psb.5.8.12135
Liu, F., Xu, W., Song, Q., Tan, L., Liu, J., Zhu, Z., et al. (2013). Microarray-assisted fine-mapping of quantitative trait loci for cold tolerance in rice. Mol. Plant 6, 757–767. doi:10.1093/mp/sss161
Ma, Y., Dai, X., Xu, Y., Luo, W., Zheng, X., Zeng, D., et al. (2015). Cold1 confers chilling tolerance in rice. Cell 160, 1209–1221. doi:10.1016/j.cell.2015.01.046
Manishankar, P., and Kudla, J. (2015). Cold tolerance encoded in one snp. Cell 160, 1045–1046. doi:10.1016/j.cell.2015.02.037
Mao, D., Xin, Y., Tan, Y., Hu, X., Bai, J., Liu, Z. Y., et al. (2019). Natural variation in the han1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. Proc. Natl. Acad. Sci. U. S. A. 116, 3494–3501. doi:10.1073/pnas.1819769116
Miura, K., and Furumoto, T. (2013). Cold signaling and cold response in plants. Int. J. Mol. Sci. 14, 5312–5337. doi:10.3390/ijms14035312
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M. (2007). Kaas: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. doi:10.1093/nar/gkm321
Peng, X., Tun, W., Dai, S. F., Li, J. Y., Zhang, Q. J., Yin, G. Y., et al. (2021). Genome-wide analysis of cct transcript factors to identify genes contributing to photoperiodic flowering in oryza rufipogon. Front. Plant Sci. 12, 736419. doi:10.3389/fpls.2021.736419
Pradhan, S. K., Pandit, E., Nayak, D. K., Behera, L., and Mohapatra, T. (2019). Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through rna-seq analysis. BMC Plant Biol. 19, 352. doi:10.1186/s12870-019-1922-8
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi:10.1093/bioinformatics/btp616
Saijo, Y., and Loo, E. P. (2020). Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 225, 87–104. doi:10.1111/nph.15989
Saito, K., Hayano-Saito, Y., Maruyama-Funatsuki, W., Sato, Y., and Kato, A. (2004). Physical mapping and putative candidate gene identification of a quantitative trait locus ctb1 for cold tolerance at the booting stage of rice. Theor. Appl. Genet. 109, 515–522. doi:10.1007/s00122-004-1667-z
Sakamoto, T., and Murata, N. (2002). Regulation of the desaturation of fatty acids and its role in tolerance to cold and salt stress. Curr. Opin. Microbiol. 5, 208–210. doi:10.1016/s1369-5274(02)00306-5
Shi, Y., and Gong, Z. (2015). One snp in cold1 determines cold tolerance during rice domestication. J. Genet. Genomics. 42, 133–134. doi:10.1016/j.jgg.2015.03.007
Su, C. F., Wang, Y. C., Hsieh, T. H., Lu, C. A., Tseng, T. H., and Yu, S. M. (2010). A novel mybs3-dependent pathway confers cold tolerance in rice. Plant Physiol. 153, 145–158. doi:10.1104/pp.110.153015
Sweeney, M. T., Thomson, M. J., Pfeil, B. E., and McCouch, S. (2006). Caught red-handed: rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18, 283–294. doi:10.1105/tpc.105.038430
Tan, L., Li, X., Liu, F., Sun, X., Li, C., Zhu, Z., et al. (2008). Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40, 1360–1364. doi:10.1038/ng.197
Toronen, P., Medlar, A., and Holm, L. (2018). Pannzer2: a rapid functional annotation web server. Nucleic Acids Res. 46, W84–W88. doi:10.1093/nar/gky350
Wai, A. H., Cho, L. H., Peng, X., Waseem, M., Lee, D. J., Lee, J. M., et al. (2021). Genome-wide identification and expression profiling of alba gene family members in response to abiotic stress in tomato (solanum lycopersicum l.). BMC Plant Biol. 21, 530. doi:10.1186/s12870-021-03310-0
Zhang, Q., Chen, Q., Wang, S., Hong, Y., and Wang, Z. (2014). Rice and cold stress: methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice 7, 24. doi:10.1186/s12284-014-0024-3
Zhang, J., Li, X. M., Lin, H. X., and Chong, K. (2019). Crop improvement through temperature resilience. Annu. Rev. Plant Biol. 70, 753–780. doi:10.1146/annurev-arplant-050718-100016
Zhao, H., Kosma, D. K., and Lu, S. (2021). Functional role of long-chain acyl-coa synthetases in plant development and stress responses. Front. Plant Sci. 12, 640996. doi:10.3389/fpls.2021.640996
Zhu, B. F., Si, L., Wang, Z., Zhou, Y., Zhu, J., Shangguan, Y., et al. (2011). Genetic control of a transition from black to straw-white seed hull in rice domestication. Plant Physiol. 155, 1301–1311. doi:10.1104/pp.110.168500
Keywords: NOG1, cold stress, domestication, differentially expressed genes, RNA-seq
Citation: Huo X, Xiao J, Peng X, Lin Y, Liu D, Liu W, Liao Y, Li J, Zhu M, Fu C, Zeng X, Ma X, Kong L and Wang F (2022) The grain yield regulator NOG1 plays a dual role in latitudinal adaptation and cold tolerance during rice domestication. Front. Genet. 13:1039677. doi: 10.3389/fgene.2022.1039677
Received: 08 September 2022; Accepted: 26 October 2022;
Published: 11 November 2022.
Edited by:
Dezhi Wu, Hunan Agricultural University, ChinaReviewed by:
Bohan Liu, Hunan Agricultural University, ChinaCopyright © 2022 Huo, Xiao, Peng, Lin, Liu, Liu, Liao, Li, Zhu, Fu, Zeng, Ma, Kong and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Wang, ZndhbmcxNjMxQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.