- 1National Institute for Genomics and Advanced Biotechnology, National Agricultural Research Centre, Islamabad, Pakistan
- 2Department of Bioinformatics, College of Life Sciences, Zhejiang University, Hangzhou, China
- 3School of Biological Sciences and Technology, Beijing Forestry University, Beijing, China
- 4National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 5Department of Plant Breeding and Genetics, The University of Haripur, Haripur, Pakistan
- 6Department of Biochemistry, College of Science, King Saud University, Riyadh, Saudi Arabia
- 7Department of Biology, College of Science, Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia
- 8National Nanfan Research Institute, Chinese Academy of Agricultural Sciences, Sanya, China
- 9China National Rice Research Institute, Hangzhou, China
- 10Hainan Yazhou Bay Seed Lab, Sanya, China
Rice plants experience various biotic (such as insect and pest attack) and abiotic (such as drought, salt, heat, and cold etc.) stresses during the growing season, resulting in DNA damage and the subsequent losses in rice production. DNA Replication Helicase/Nuclease2 (DNA2) is known to be involved in DNA replication and repair. In animals and yeast DNA2 are well characterized because it has the abilities of both helicase and nuclease, it plays a crucial role in DNA replication in the nucleus and mitochondrial genomes. However; they are not fully examined in plants due to less focused on plants damage repair. To fill this research gap, the current study focused on the genome-wide identification and characterization of OsDNA2 genes, along with analyses of their transcriptional expression, duplication, and phylogeny in rice. Overall, 17 OsDNA2 members were reported to be found on eight different chromosomes (2, 3, 4, 6, 7, 9, 10, and 11). Among these chromosomes (Chr), Chr4 contained a maximum of six OsDNA2 genes. Based on phylogenetic analysis, the OsDNA2 gene members were clustered into three different groups. Furthermore, the conserved domains, gene structures, and cis-regulatory elements were systematically investigated. Gene duplication analysis revealed that OsDNA2_2 had an evolutionary relationship with OsDNA2_14, OsDNA2_5 with OsDNA2_6, and OsDNA2_1 with OsDNA2_8. Moreover, results showed that the conserved domain (AAA_11 superfamily) were present in the OsDNA2 genes, which belongs to the DEAD-like helicase superfamily. In addition, to understand the post-transcriptional modification of OsDNA2 genes, miRNAs were predicted, where 653 miRNAs were reported to target 17 OsDNA2 genes. The results indicated that at the maximum, OsDNA2_1 and OsDNA2_4 were targeted by 74 miRNAs each, and OsDNA2_9 was less targeted (20 miRNAs). The three-dimensional (3D) structures of 17 OsDNA2 proteins were also predicted. Expression of OsDNA2 members was also carried out under drought and salt stresses, and conclusively their induction indicated the possible involvement of OsDNA2 in DNA repair under stress when compared with the control. Further studies are recommended to confirm where this study will offer valuable basic data on the functioning of DNA2 genes in rice and other crop plants.
Introduction
Rice plants experience various biotic (such as insect and pest attack) and abiotic (such as drought, salt, heat, and cold etc.) stresses during the growing season, resulting in DNA damage and the subsequent losses in rice production. Homologous recombination is essential for replication, DNA repair pathways, and the exchange of genetic substances between parent chromosomes during meiosis (Li, 2008; Kuzminov, 2011). The complex reorganization of DNA structures is typically organized into several stages, and the success rate of completing these stages is entirely related to the activities of multiple helicase enzymes (Wu, 2012; Huselid and Bunting, 2020). Helicases of several families are organized to process the broken ends of DNA structures, and are also involved in the subsequent disassembly and formation of recombinant intermediate materials essential for the template-based repair of DNA structures (Raney et al., 2013; Croteau et al., 2014; Tisi et al., 2020). Therefore, the loss of recombinant-linked helicase functionality can result in genome disorder, higher risks of tumor forging, and subsequent cell death (Yousefzadeh et al., 2021). Certain helicases are associated with the anti-recombinase effects that influence the recombination efficiency, ultimately leading to other pathways directed toward repairing broken ends of DNA (Wu and Hickson, 2006; Li and Heyer, 2008). Several helicases are also responsible for adjusting the relative repair outputs for noncrossover and crossover. A typical increase in the utilization of recombination occurs during the collision of transcription material and replication forks, or when it comes across lesions in the DNA template (Nguyen et al., 2015). Amazingly, successful recombination in such situations is also regulated by helicases, permitting optimized cell growth while maintaining genome integrity (Saada et al., 2018).
During developmental processes, the unrelenting activities of apical meristems organize organ morphogenesis (Traas, 2018; Marconi and Wabnik, 2021). The development of apical meristems is maintained and promoted by cell division in meristematic areas (Murray et al., 2012; Geng et al., 2022). Plant meristems are accompanied by stem cells, and have a strong regenerative ability; they maintain plant growth mechanisms and produce new plant organs such as stems, leaves, flowers, and roots (Chang et al., 2020). The final forms of the organs and the overall plant architecture mainly depend on the temporal and spatial regulation of cell proliferation in meristems.
DNA2, typically called the DNA replication helicase/nuclease two protein, occurs in both the mitochondria and nucleus, where it performs the roles of helicase and ATPase-dependent nuclease (Zhou et al., 2015; Zheng et al., 2020). During the 1980s, the DNA2 protein was first reported in the yeast Saccharomyces cerevisiae (Budd et al., 2000). Because it has the abilities of both helicase and nuclease, it plays a crucial role in DNA replication in the nucleus and mitochondrial genomes (Dumas et al., 1982; Budd and Campbell, 1997). In humans, it is known as DNA replication ATP-dependent helicase/nuclease DNA2 (Masuda-Sasa et al., 2006). DNA2 essentially shares an important purpose in the removal of long flaps during DNA replication and DNA LP-BER (long-patch base excision) repair. Moreover, it interacts with flap endonuclease 1 (FEN1) and replication protein A (RPA) (Kleppa et al., 2012). DNA2 has the ability to promote the reactivation of the prehended replication fork along with BLM (Bloom syndrome protein) and WRN (Werner syndrome ATP-dependent helicase) (Machwe et al., 2006; Sturzenegger et al., 2014). DNA2 assists in the removal of primers during strand displacement replication in the mitochondria (Zheng et al., 2008). Additionally, DNA2 is considered to act as a key to protein-sharing activities in complex DNA damage repair. Moreover, it is accompanied by a double stranded break (DSB) and a 50 reactive adduct resulting from a chemical group attached to DNA 50 ends, produced by ionization of the radiation (Pawłowska et al., 2017). In human and animal cells, the key role of DNA2 in general cell cycle maintenance proposes its generalized function in genomic integrity. Therefore, for human and animal cells, it is essential for disease therapy.
During the repair of broken DNA ends, DNA2 intercedes the 59-end resection of DNA by splitting the 59-single-stranded DNA, ssDNA, with the assistance of RPA and Sgs1. Subsequently, it acts as helicase, whose function is mediated by RPA, and can disentangle DNA along the production of an ssDNA substrate for DNA2 (Cejka et al., 2010; Ruff et al., 2016; Bonetti et al., 2018). Homologs of RPA and Sgs1 are also preserved in plants, with essential duties in different pathways, such as DNA repair (Chatterjee and Walker, 2017; Verma et al., 2020; Wang et al., 2021). Yeast DNA2 is occupied by the compound nuclear localization signal (NLS) sequences, Pat4 and Pat7, and is decentralized to the nucleus (Jia et al., 2016; Meng et al., 2019). Yeast DNA2 mutants are sensitive to DNA damage factors, including X-ray and UV irradiation, and methyl methane sulfonate (MMS) (Bae et al., 2001; Jia et al., 2016). Moreover, yeast DNA2 exhibits DNA repair activities by assisting homologous recombination. In mammalian cells, DNA2 participates in DNA repair and replication, whereas in humans, a reduction in hDNA2 delays cell division as well as the entire cell cycle (Duxin et al., 2009; Bailey et al., 2019; Hudson and Rass, 2021).
On a broader level, DNA2 is thought to have essential roles in DNA repair and replication, along with maintaining nuclear genomic DNA and mitochondrial integrity in fungi and animals (Duxin et al., 2009; Gredilla et al., 2012). However, the role of DNA2 in plants has not yet been investigated due to less focus on plants damage repair. Therefore, this study performed with objectives of genome-wide identification and characterization of OsDNA2 genes in the rice genome along with their differential expression analysis.
Materials and methods
Evidencing the identification of OsDNA2
Phytozome database was used to obtain sequences of DNA2 proteins (Zhang et al., 2012). In the rice genome (Oryza sativa IRGSP-1.0), the Hidden Markov Model (HMM) profiles of DNA2 domain from the Pfam (protein family) database were used to scan the predicted proteins using HMMERv3 (Prince and Pickett, 2002). By using HMM model in HMMERv3, the protein sequences of rice DNA2 were aligned. For the confirmation of the presence of DNA2 conserved domain, the putative DNA2 gene core sequences were verified by searching against the SMART (http://smart.embl-heidelberg.de/) and Pfam database (https://pfam.xfam.org/). Protein sequences of Zea mays, Hordeum vulgare, Pennisetum glaucum, and Oryza sativa were obtained from previous studies (Liu and Widmer, 2014; Guo et al., 2017), and TAIR (https://www.arabidopsis.org/) source was used to download information of Arabidopsis DNA2 gene family protein sequences and annotation. ExPASy (https://www.expasy.org/) online server was used to obtain molecular weight, GRAVY, Iso-electric point information for OsDNA2 (Gasteiger et al., 2003).
Chromosomal location, gene structural, and phylogenetic analysis
Rice genomic database in phytozome was accessed to get the DNA2 genes genomic coordinated on rice chromosomes. All OsDNA2 were present on the eight chromosomes of rice genome. Protein sequences of DNA2 in Arabidopsis, rice, maize, barley, and millet were aligned using clustalW. Bootstrap 1000 replications were used to generate phylogenetic tree using maximum likelihood (ML) method in MEGA 10. Coding sequence was compared with the corresponding full-length sequence for the identification of intron insertion sites in the DNA2 genes. This identification was performed by using Gene Structure Display Server (Guo et al., 2007). The analysis of conserved DNA2 motifs was performed by using MEME Suit (Multiple EM for Motif Elicitation) Version 4.12.0 (http://meme-suite.org/tools/meme) (Rehman et al., 2022). Following parameters were used for this analysis: ten motifs to be found with motif width between 10 and 200; site distribution was one occurrence per sequence or set at zero (at most one occurrence for each motif was allowed for each sequence), whereas the maximum number of motifs was set to 10 (Bailey et al., 2009). TBtools (https://bio.tools/tbtools) program was used for further analysis of MEME results (Chen et al., 2020). Unipro UGENE software package (Okonechnikov et al., 2012), helped to examine the conserved domains of OsDNA2 proteins. This aligned the sequences by the ClustalW algorithm and conserved regions were displayed in the form of color patterns which differentiated each amino acid based on physiochemical properties. OsDNA2 protein sequences in SMART database containing Pfam domain search options, was used to perform protein domain analysis, and confirmation was carried out through the InterPro database (Letunic et al., 2012; Blum et al., 2021).
MicroRNA, gene ontology, cis-elements, collinearity and synteny prediction in OsDNA2
MicroRNAs (miRNAs) interacting with the DNA2 genes were predicted form the available rice miRNA reference sequences by submitting genome sequences of OsDNA2 to the psRNATARGET server (https://www.zhaolab.org/psRNATarget/). The visualization of miRNAs and OsDNA2 gene was done with Cytoscape software (https://cytoscape.org/). Online tool gProfiler (https://biit.cs.ut.ee/gprofiler/gost, accessed on 20 July 2022) was used to conduct gene ontology (GO) analysis of OsDNA2 protein sequences with default parameters. The rice genome database was downloaded from RGAP (https://cottonfgd.org/) to get the DNA2 promoter region sequence containing 2000 bp upstream of the inhibition codon (ATG). The prediction of regulatory elements in the DNA2 promoter regions was carried out by using online tool PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and visualized by TBtools. For the sequence similarity patterns evaluation, synteny analysis and sequence identity visualization were performed using the TBtools (Rahim et al., 2022). For gene duplication, homologous gene pair were calculated with the help of Ka/Ks calculator 2.0. For Synteny analysis, genome sequence (FASTA) and annotation files (gff/gtf) were used in One-step MCScanX toolkit in the TBtools.
Subcellular localization and 3D protein structure prediction of OsDNA2
CELLO v.2.5: subCELlular LOcalization predictor was used to predict the sub-cellular location of DNA2 family (Yu et al., 2014). Protein sequences were used as input and output results were further analyzed/visualized by using TBtools software. Amino acid sequences of OsDNA2 proteins were used for the prediction of 3D structures by utilizing the SWISS-MODEL database (https://www.swissmodel.expasy.org) (Waterhouse et al., 2018), while visualization of such predicted structures was carried out with the help of Pymol software (https://pymol.org/2/). Ramachandran Plot—Zlab, (https://zlab.umassmed.edu/bu/rama/), was applied for the confirmation of predicted 3D models of OsDNA2 proteins (Anderson et al., 2005).
Expression analysis of OsDNA2 under abiotic stress
RNA seq data for drought and salt stresses were assessed from online data bases which are publically available with Bio-projects GSE145869 and GSE167342 (Tarun et al., 2020; Bundó et al., 2022). In these studies, Nil-95 and Swarna genotypes were used as drought tolerant and sensitive, while IL22 and PL12 were used as a salt tolerant and sensitive rice genotype, respectively. FPKM (fragments per kilobase of transcript per million mapped reads) values were extracted and heatmaps were generated in TBtools. Furthermore, the rice genotype IR-6 was used in this study under drought and salt stresses. Plants were grown under normal conditions for 2 weeks. Then drought and salt stresses were applied to two batches of plants and one batch was kept as a control. Three biological replicates for each treatment were used. Gene specific primers of selected OsDNA2 genes along with drought and salt reported genes (OsEm1 and bZIP23) for qRT-PCR are presented in Supplementary Table S1. For this purpose, 1 g leaf tissues from control and treated samples were grinded in liquid nitrogen and used for RNA extraction. Total RNA was extracted with the help of TRIzol method. Complementary DNA (cDNA) was also synthesized from the 800 ng extracted RNA with the help of reverse transcriptase-III, first strand cDNA Synthesis Kit (K1691, Thermo Scientific Revert Aid). StepOne RT-PCR (Applied Biosystems® 7900 H T Fast RT-PCR) was used for quantification. OsActin was used as a reference gene and 2−ΔΔCT method was used for expression calculations (Uzair et al., 2021).
Results
Delineation of DNA2 gene family in rice
Wheat DNA2 protein (TraesCS2A02g301600) and Arabidopsis (AT2G03270) sequences were used as queries to identify DNA2 genes in the rice genome. From these analysis, 17 OsDNA2 members were found (Figure 1A, Supplementary Table S2). OsDNA2 was distributed on the basis of its physical position on eight chromosomes (2, 3, 4, 6, 7, 9, 10, and 11). Of these chromosomes, Chr4 had the maximum number of six OsDNA2 genes, followed by Chr3, which had three genes. Chr2 and Chr10 each had two members, whereas the rest of the chromosomes had a single member. Chr9 was the shortest one which had OsDNA2_5. This structure plays an important role in the expression of genes. For this purpose, we also checked the gene structures and found that the number of introns varied from 1 to 32 (Figure 1B). The maximum number of introns was found in OsDNA2_1, whereas the minimum was found in OsDNA2_11. The CDS length extended from 399 bp (OsDNA2_11) to 3624 bp (OsDNA2_1). Similarly, the protein length, protein molecular weight, and number of exons varied from 133 aa (OsDNA2_11) to 1208 aa (OsDNA2_1), 132.72854 KDa (OsDNA2_1) to 14.19668 KDa (OsDNA2_11), and 2 (OsDNA2_11 and OsDNA2_3) to 33 (OsDNA2_1), respectively (Table 1). A total of five genes (OsDNA2_1, OsDNA2_2, OsDNA2_4, OsDNA2_7, and OsDNA2_17) out of 17 were found on the positive strand. The isoelectric points varied between 4.2510 (OsDNA2_11) and 11.6011 (OsDNA2_3). The charge on a protein molecule depends on the ionizable groups and their pKa values. The protein becomes negatively charged when the pH becomes higher than the pI. In the present study, only four OsDNA2 genes (OsDNA2_2, OsDNA2_8, OsDNA2_10, and OsDNA2_11) were negatively charged (Table 1). All members of the OsDNA2 gene family, except OsDNA2_7, showed negative GRAVY, indicating that they are hydrophilic in nature (Table 1). Additionally, the subcellular localization of all OsDNA2 proteins was determined. The results showed that most of the genes were located in the nucleus, cytoplasm, mitochondria, and chloroplasts (Figure 1C). Meanwhile, the subcellular localization of OsDNA2_16 was predicted in the peroxisomes (Figure 1C).
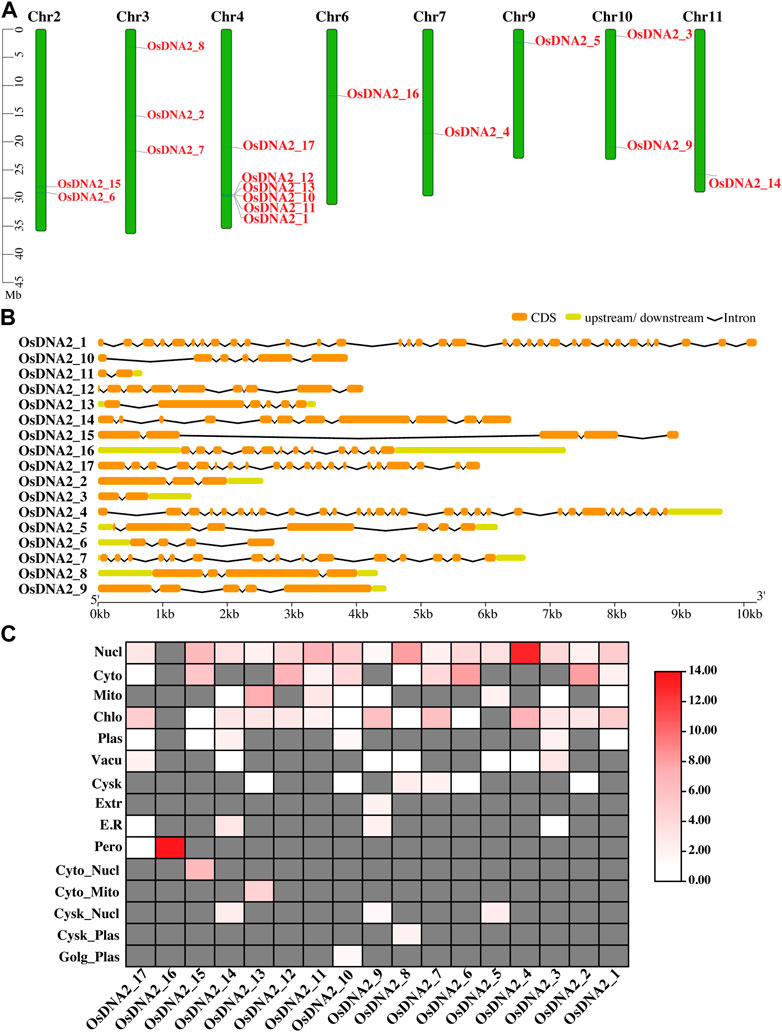
FIGURE 1. Prediction of DNA2 gene members in rice genome. (A) Distribution of OsDNA2 members on respective chromosome. Chr represent the chromosome. Left side of the figure scale was used in mega base (Mb). (B) Gene structure of OsDNA2 members. Orange color shows the CDS, light green shows upstream/downstream region, and black line represent the introns. (C) In-silico prediction of subcellular location of OsDNA2. Proteins were shown on the right side of the figure. Nucl = nucleus, Cyto = cytoplasm, Mito = mitochondria, Chlo = chloroplast, Plas = plasma-membrane, Vacu = vacuole, Cysk = cytoskeleton, Extr = extracellular, E.R = endoplasmic reticulum, Pero = peroxisome, and Golg = Golgi apparatus.
Motifs and domain analysis, and phylogenetic association among DNA2 genes
When characterizing newly identified proteins, it is very important to understand the motifs and domains of that specific protein. In the present study, motifs were predicted for all OsDNA2 genes using MEME (Bailey et al., 2009). For this purpose, we used the protein sequence of OsDNA2 and found ten conserved motifs (Figure 2A). The lengths of the predicted motifs varied between 20 and 39 amino acids (Supplementary Figure S1). Motif five was present in all genes except OsDNA2_2, OsDNA2_6, and OsDNA2_12. Motifs 1, 2, 3, and six were conserved among all members. Similarly, we also examined the domains in OsDNA2 members (Figure 2B). The conserved domain in the OsDNA2 genes of rice is the AAA_11 superfamily (Pfam: PF13086), which belongs to the DEAD-like helicase superfamily involved in the unwinding of ATP-dependent RNA or DNA.
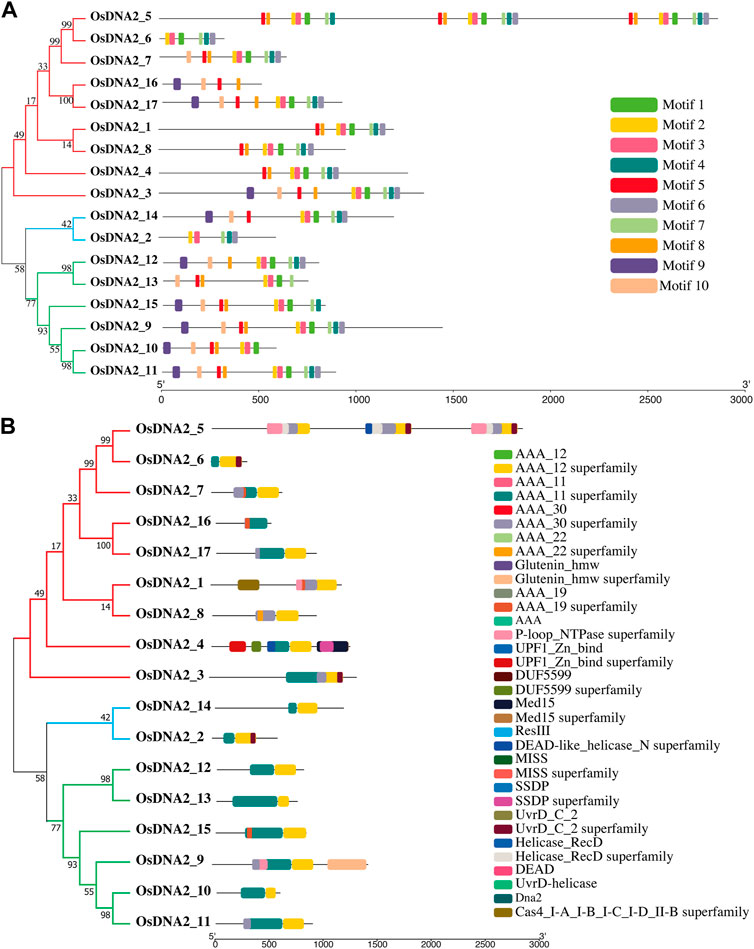
FIGURE. 2OsDNA2 members were divided into three groups. Prediction of motifs (A) and domains (B) in OsDNA2 members. Each motif and domain were represented by different colors.
In the current study, the evolutionary associations among OsDNA2s, ZmDNA2s, SbDNA2s, HvDNA2s, and AtDNA2s were assessed (Figure 3). The results revealed that 99 DNA2 molecules were clustered into six main clusters (C1 = pink, C2 = blue, C3 = yellow, C4 = green, C5 = brown, and C6 = purple). Cluster six contained a maximum of six OsDNA2 genes (OsDNA2_2 and OsDNA2_4–8). Interestingly, Cluster five contained only one, OsDNA2_1. Overall, OsDNA2 showed a closer association with ZmDNA2, HvDNA2, and SbDNA2 than with AtDNA2 in each cluster.
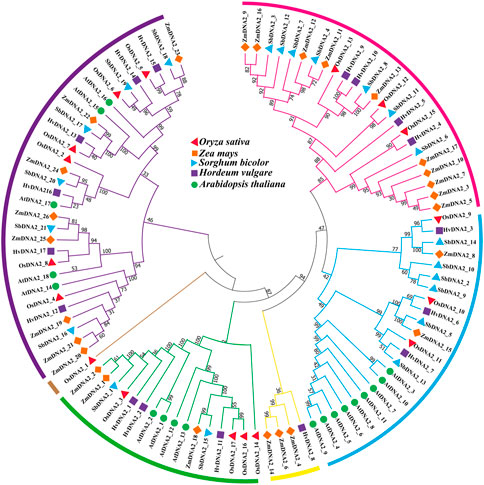
FIGURE 3. Evolutionary relationship among DNA2 members of Oryza sativa, Zea mays, Hordeum vulgare, and Arabidopsis thaliana. For the construction of phylogenetic tree, a maximum likelihood (ML) method was used with 1000 bootstrap values.
Investigating synteny and collinearity among DNA2 genes
To estimate the evolutionary relationship among DNA2 members of Oryza sativa, a synteny analysis of DNA2 protein sequences was conducted (Figure 4A and Supplementary Table S2). These analyses were performed to study gene duplication using TBtools for the 17 predicted OsDNA2 (Chen et al., 2020). For gene duplication, Ka/Ks values were calculated, and the results revealed that six gene pairs, including OsDNA2_1-OsDNA2_8, OsDNA2_2-OsDNA2_14, OsDNA2_5-OsDNA2_6, OsDNA2_10-OsDNA2_11, OsDNA2_12-OsDNA2_13, and OsDNA_16-OsDNA2_17 could be duplicated in the rice genome (Supplementary Table S3). Similarly, we also performed collinearity analysis among Oryza sativa, Sorghum bicolor, Zea mays, and Hordeum vulgare. The results revealed that rice DNA2 genes were more collinear with sorghum than with maize and barley, indicating that whole genome or segmental duplication was involved in OsDNA2 gene family progression (Figure 4B).
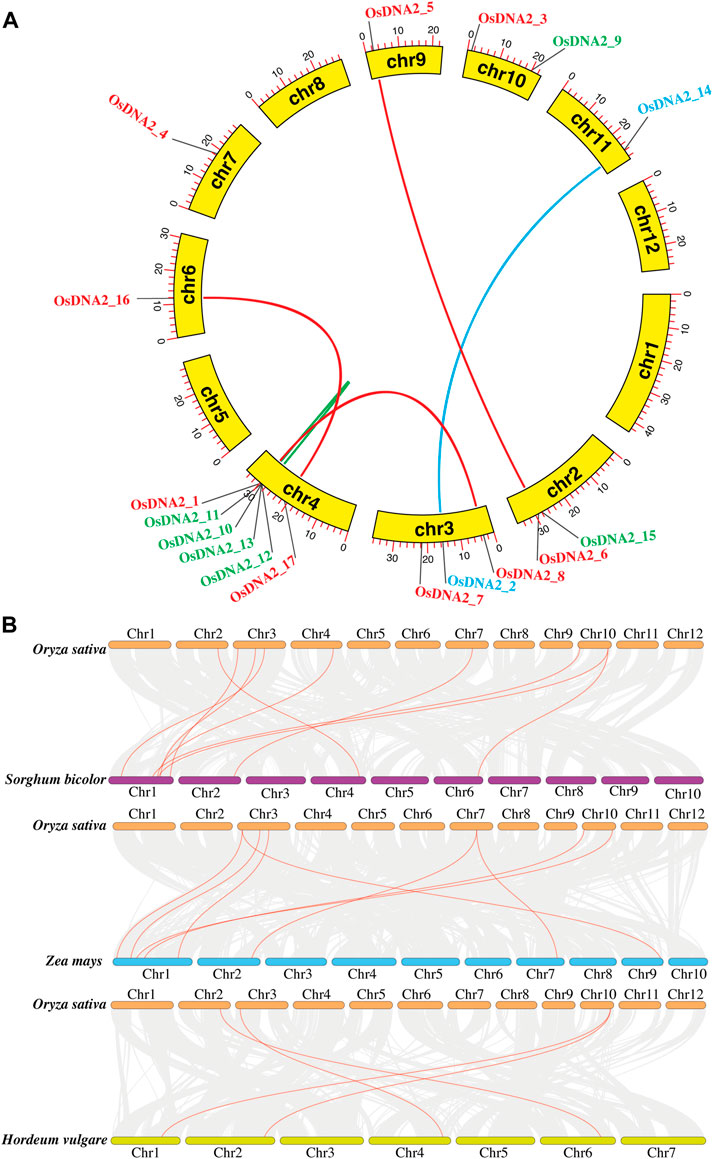
FIGURE 4. Identification of OsDNA2 gene duplication. (A) Synteny analysis and (B) Collinearity analysis of DNA2 genes amongst Oryza sativa, Sorghum bicolor, Zea mays, and Hordeum vulgare.
Promoter analysis of OsDNA2 genes
It was previously reported that the promoter region is the control center for the expression and regulation of genes (Rasool et al., 2021). Promoters are also known as cis-regulatory elements in DNA. The 2-kb upstream region of each OsDNA2 gene was subjected to the PLACE database, and the results revealed that more than 85 different types of cis-acting elements and nine unnamed types of elements were detected (Figure 5, Supplementary Table S4). CAAT-box, TATA-box, and unnamed were the most identified elements for the OsDNA2 genes. Furthermore, different stress-related regulatory elements, such as CGTCA-motif, G-box, Sp1, GATA-motif, I-box, GT1-motif, and AT-rich, were detected. Similarly, hormone-related cis-regulatory elements, such as TATC-box, ABRE, CGTCA-motif, P-box, and TGACG-motif, were also detected. ABRE was detected in all members of OsDNA2, except for OsDNA2_6 (Figure 5). The GCN4-motif, GC-motif, O2-box, and ARE were detected in the different OsDNA2 members, indicating that these genes are involved in cellular development. Notably, it has been reported that the GCN4-motif and O2-box are involved in the expression of endosperm and zein metabolism, respectively.
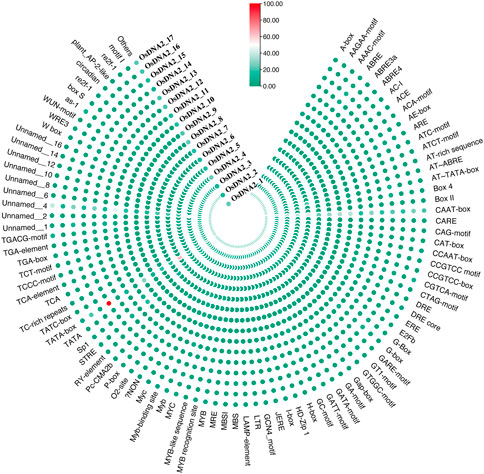
FIGURE 5. Prediction of Cis-Elements in 2 kb upstream region of OsDNA2 coding sequences. Scale bar was used to indicate the presence of numbers of specific elements in that particular DNA2 genes.
Gene ontology (GO) and MicroRNA (miRNA) targeting OsDNA2 genes
GO annotation was used for the practical investigation of OsDNA2 genes. In silico characterization based on functions was conducted, which revealed three types of biological processes (BPs), cellular components (CCs), and molecular functions (MFs) (Figure 6, Supplementary Table S5). Further analysis of the BPs’ annotations revealed that most of the terms were related to DNA replication. Similarly, the MFs also showed DNA helicase, and catalytic and ATP-dependent activities. Based on these findings, we concluded that OsDNA2 genes play an essential role in DNA replication.
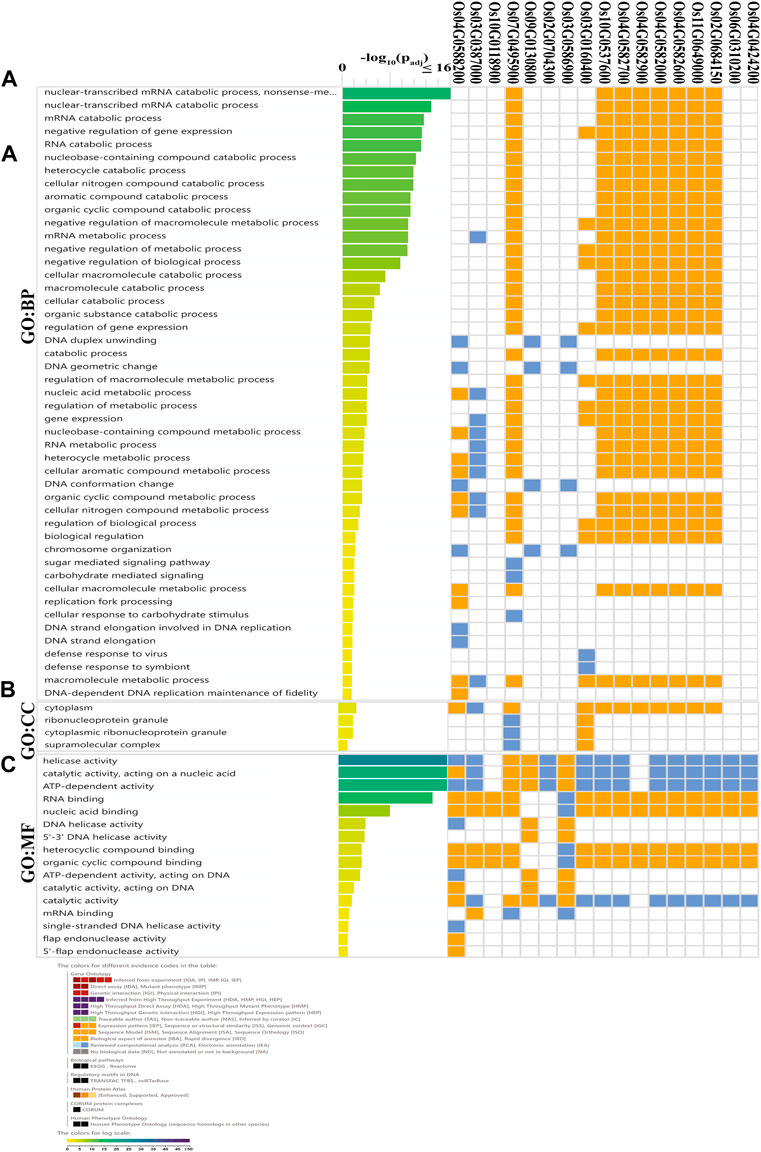
FIGURE 6. Gene ontology (GO) analysis of OsDNA2 genes was performed. OsDNA2 genes were involved in biological processes (BP, (A), cellular components (CC, (B), and molecular functions (MF, (C).
Over the past few decades, many studies have shown that miRNAs play essential roles in the regulation of genes in specific environments. Thus, to understand the post-transcriptional modification of OsDNA2 genes, we identified 653 miRNAs targeting 17 OsDNA2 genes (Figure 7, Supplementary Table S6). The results indicated that at the maximum, OsDNA2_1 and OsDNA2_4 were targeted by 74 miRNAs each, and OsDNA2_9 was less targeted (20 miRNAs). Nineteen members of the osa-miR164 family targeted four genes (OsDNA2_1, OsDNA2_4, OsDNA2_6, and OsDNA2_14). Furthermore, the single miRNAs, such as osa-miR6255, osa-miR6245, osa-miR6246, and osa-miR6248, targeted OsDNA2_10, OsDNA2_14, OsDNA2_10, and OsDNA2_4, respectively. Future studies will be required to validate the functions of these miRNAs and their target genes to understand their biological interactions in the rice genome.
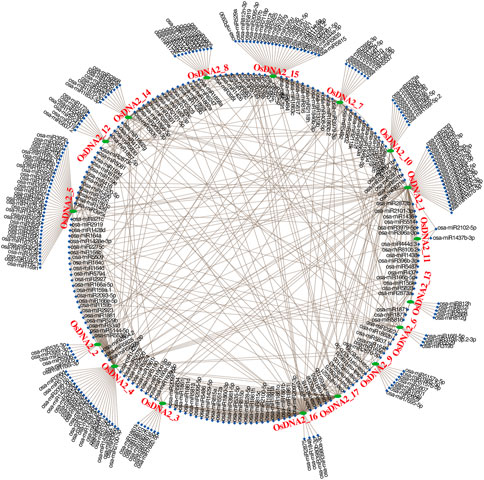
FIGURE 7. 7Prediction of MicroRNAs (miRNAs) shows how they target the respective OsDNA2 genes. Green oval shapes represent the OsDNA2 genes, blue rectangles represent the miRNAs, and brown lines represent the interaction among the miRNAs and OsDNA2 genes.
3D-protein structural analysis of OsDNA2
Understanding the structure of proteins is very difficult due to their complexity, and the fact that they contain a different number of atoms and convoluted topology. In this study, the SWISS_MODEL online server was used to predict the three-dimensional (3D) structures of 17 OsDNA2 proteins (Figure 8, Supplementary Table S7). The results revealed the successful prediction of the 17 OsDNA2 protein models. Many reports have shown that >30% identity with the template is acceptable (Xiang, 2006; Rahim et al., 2022), and our findings also showed an average of 40% similarity with the template (5ean. 1.A, 6ff7. 1.c, 5mzn. 1.A, 2wjv. 1.A, 4b3f. 1.A, 4b3g. 2.A, 2gjk. 1.A, 2wjv. 1.A, and 2xzl. 1.A). Spiral shapes represent α-helices, thick arrows depict ß-sheets, and thin lines indicate loops and turns. Most of the OsDNA2 members had similar structures (Figure 8). Furthermore, these predicted 3D models were confirmed using Ramachandran plots with the help of diahedral angles of the OsDNA2 proteins (Anderson et al., 2005). The results of the Ramachandran plots revealed that >95% of the regions of OsDNA2 proteins showed highly favorable regions. This confirmed the better quality and stability of the predicted OsDNA2 protein structures.
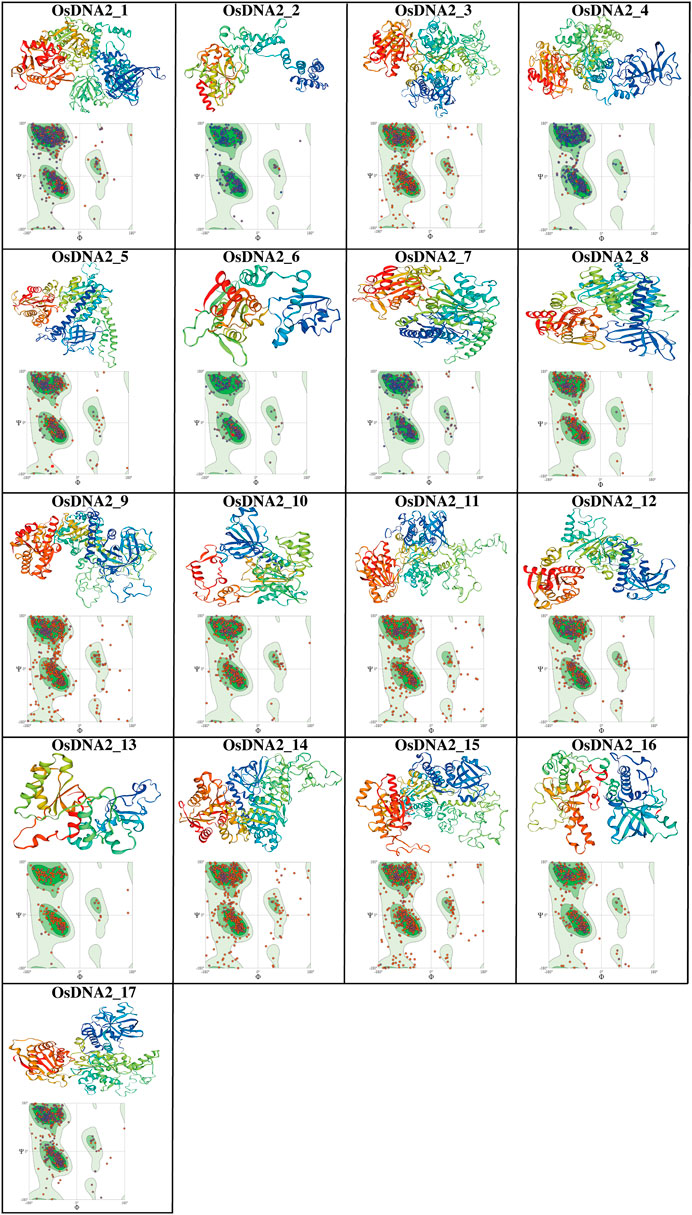
FIGURE 8. The 3D structure prediction and Ramachandran plots of OsDNA2 proteins. Different shapes in these final models represent the sheets and helicases.
Role of OsDNA2 genes in abiotic stress tolerance
RNA-seq data for drought and salt stress were retrieved from publicly available data sites. Circular heat maps for these stresses were generated from the FPKM values, showing the expression of OsDNA2 members at the seedling stage (Figures 9A,B, Supplementary Table S8). OsDNA2_11, OsDNA2_14, and OsDNA2_15 were downregulated and showed low expression under drought and salt stress conditions (Figures 9A,B). Quantitative real-time PCR (qRT-PCR) analysis of five randomly selected OsDNA2 genes was performed to study the transcription profile (Figure 9C). OsDNA2_2 and OsDNA2_5 were up-regulated under both stress conditions, whereas OsDNA2_4 and OsDNA2_7 were up-regulated under salt and drought stress, respectively. Moreover, OsDNA2_15 expression was unchanged after the treatments. These findings indicate that these genes might be involved in mitigating abiotic stresses (drought and salt), and they may provide useful information for the functional characterization of these genes in the future.
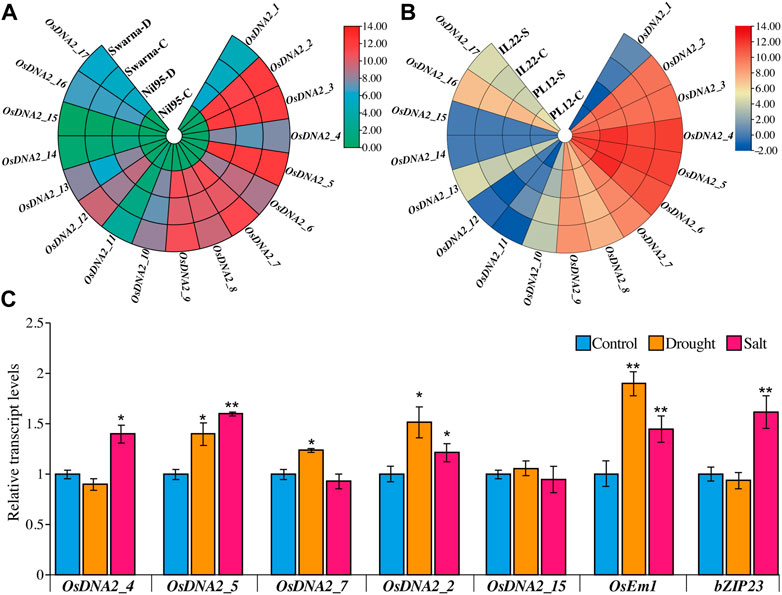
FIGURE 9. 9In-silico expression profiling of OsDNA2 genes members under drought (A) and salt (B) stress. Different capital letters C, D, and S represent the control, drought, and salt stresses, respectively. (C) Quantification of expression of OsDNA2 genes through qRT-PCR. Results are shown as fold change. Data is a mean ± SE of three biological replicates. Comparison was checked with the help of t-test (*p < 0.05, **p < 0.01).
Discussion
The DNA of a living organism contains genetic information that is important for its survival and reproduction. During cell division, DNA strands can be damaged by abiotic stresses such as exposure to radiation or chemicals. To avoid the consequent negative impacts on life, cells initiate processes that quickly fix damaged DNA using different techniques, such as homologous recombination, in which cells fix the damaged strands (Duxin et al., 2009). Different enzymes act on broken strands to form single-stranded tails, and DNA2 is one of the enzymes involved (Jia et al., 2016). In yeast, the dna2 mutant shows a lethal phenotype, indicating that the wild-type protein is necessary for cell viability (Budd et al., 2011). Similarly, the Arabidopsis dna2 mutant caused small roots at the time of germination (Diray-Arce et al., 2013; Jia et al., 2016). Another protein, Rpa, is required for the proper functioning of DNA2; however, it remains unclear how Rpa regulates it. DNA2 homologs are the same in fungi and other metazoans, indicating that they may have conserved functions (Kang et al., 2010). In plants, the alternate DNA helicase is DNA2 (Levikova and Cejka, 2015). Plants face different types of biotic and abiotic stresses that cause cell injury, DNA damage including double- and single-stranded breaks (DSBs and SSBs), and DNA lesions (Gill et al., 2015; Noctor and Foyer, 2016; Nisa et al., 2019; Rout et al., 2022). Because the function of DNA2 is conserved in DNA damage repair and Okazaki fragments, the small RNA/DNA was removed by FEN1, while the larger flaps were removed in the correct order of both FEN1 and DNA2 (Kang et al., 2010; Zheng and Shen, 2011). To date, DNA2 has mainly been studied in fungi and animals, but its role in plants has not been fully studied. Therefore, this study aimed to investigate the function of DNA2 in rice at the whole-genome level.
Rice is an important cereal used worldwide as a good source of food around the globe. Rice production is hampered by biotic and abiotic stresses. The rice genome is publicly available, which permits genome-wide identification and characterization of the DNA2 family (Table 1). In this study, we identified 17 OsDNA2 members in the rice genome (Figure 1A) that were distributed into three clusters. The expression of any gene is dependent on its structure. In this study, gene structure analysis confirm that 2 to 33 exons were present among the OsDNA2 genes, whereas the number of introns varied from 1 to 32 (Figure 1B). Conserved motifs and domain analyses were also carried out systematically. The AAA_11 superfamily (Pfam: PF13086) domain is present in the OsDNA2 gene, which belongs to the DEAD-like helicase superfamily, and is involved in ATP-dependent RNA or DNA unwinding (Figure 2). Phylogenetic association was also carried out among different plant species, such as Oryza sativa, Hordeum vulgare, Zea mays, Sorghum bicolor, and Arabidopsis thaliana (Figure 3). These results indicate that OsDNA2s were conserved with related species, and this may be due to the presence of conserved domains among them. Similarly, we performed synteny and collinearity analyses among Oryza sativa, Sorghum bicolor, Zea mays, and Hordeum vulgare. The results revealed that rice DNA2 genes were more collinear with sorghum than with maize and barley, indicating that segmental duplication is involved in OsDNA2 gene family progression (Figure 4B).
To better understand the function of OsDNA2 genes in stress tolerance, cis-regulatory elements were predicted (Figure 5). Our results showed that three main types of cis-elements (stress, hormones, and light) were detected. Among these stress-related regulatory elements, CGTCA-motif, G-box, Sp1, GATA-motif, I-box, GT1-motif, and AT-rich were detected. According to previous studies, cis-elements are involved in the stress response (Rasool et al., 2021; Su et al., 2021). The GCN4-motif, GC-motif, O2-box, and ARE were detected in the different OsDNA2 members, indicating that these genes are involved in cellular development. These results were further confirmed through gene ontology (Figure 6), in which most of the GO terms were related to DNA replication, DNA helicase, and catalytic and ATP-dependent activities. These findings further emphasized the thought that OsDNA2 genes may be involved in DNA repair and replication under environmental stress.
Recent studies have shown that most plant biological processes are controlled by microRNAs through the regulation of gene expression (Millar, 2020). In grasses, different miRNAs are expressed under drought conditions (Njaci et al., 2018; Raza et al., 2022). In another study on Arabidopsis, miRNA394 was shown to respond to cold stress. Similarly, in wheat, different miRNAs, such as tae-miR1119, tae-miR398, and tae-miR444a, were expressed in the roots under drought conditions (Rasool et al., 2021). In this study, we identified 653 miRNAs that target 17 OsDNA2 genes (Figure 7). To date, the highest number of identified miRNAs is 1077 in maize (Fu et al., 2017). These miRNAs can be upregulated or downregulated in response to environmental stress. In these findings, OsDNA2_4 was targeted by 74 miRNAs, and was upregulated under salt stress.
For further in-depth study, we predicted 3D models of the OsDNA2 proteins (Figure 8). The results showed 20–74% homology with the templates, which is widely acceptable. Our findings are in accordance with those of other studies (Su et al., 2021; Rahim et al., 2022). The Ramachandran plots also confirmed that the 17 OsDNA2 proteins had favorable regions because they showed >80% residue in the allowed regions, which indicates that the predicted 3D structures of these proteins were of good quality. Similar findings have also been reported for TATrx and TaRPK1 proteins in wheat (Bhurta et al., 2022; Rahim et al., 2022). In another study, 21 TaEIL 3D models and Ramachandran plots were used (Yi-Qin et al., 2020).
Previously, it was reported in mammals and other microorganisms that DNA2 genes are expressed differently under different circumstances. Thus, in this study, we examined the expression of OsDNA2 genes in plants under control, drought, and salt stress conditions. We found that some of the OsDNA2 genes were up- and downregulated under drought and salt stress conditions (Figures 9A,B). qRT-PCR of five randomly selected OsDNA2 genes (OsDNA2_2, OsDNA2_4, OsDNA2_5, OsDNA2_7, and OsDNA2_15), along with well-known drought- (OsEm1) and salt- (bZIP23) related genes, was performed to study the transcription profile (Figure 9C). OsDNA2_2 and OsDNA2_5 were upregulated under both stress conditions, whereas OsDNA2_4 and OsDNA2_7 were up-regulated under salt and drought stress, respectively. However, OsDNA2_15 expression was unchanged after treatment. These findings suggest that these genes are involved in abiotic stress mitigation. Moreover, this study also signifies the need for the functional characterization of these genes in the near future.
Conclusion
In animals and yeast DNA2 are well characterized because it has the abilities of both helicase and nuclease, it plays a crucial role in DNA replication in the nucleus and mitochondrial genomes. However; they are not fully examined in plants due to less focused on plants damage repair. The current study extensively examined the characteristic, properties, gene structures, chromosomal locations, cis-regulatory elements, synteny and collinearity, miRNAs, and expression of OsDNA2. Overall, 17 OsDNA2 genes were reported in the whole genome of rice, and were distributed on eight chromosomes. Phylogenetic analysis revealed that all the OsDNA2 genes were organized into three groups. We also found that the conserved domain (AAA_11 superfamily) was present in the OsDNA2 genes, which belongs to the DEAD-like helicase superfamily. In addition, 653 miRNAs targeting OsDNA2 genes were identified. Meanwhile, shifts in gene expression under abiotic stress, especially drought and salinity, were investigated using a comparative transcriptome approach to evaluate the susceptibility and tolerance to abiotic stress. These findings provide essential information for future functional characterization of OsDNA2 genes under abiotic stress to improve stress tolerance in rice as well as other crop species.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
BS, MU, and OR conceived the idea and performed the analysis related to this study. MU and AR visualized the figures. UF, MA, RK, WA, SI, MN, MC, MSF, MR, GY, and MU helped in writing of original draft. SF, KA, NK, HA, and MU helped in revision, editing and with technical expertise to further improve the article. All authors carefully read, revise and approved the article for submission.
Funding
This work was supported by the Nanfan Special Project of CAAS (YBXM06), and The Hainan Yazhou Bay Seed Laboratory (B21HJ0218).
Acknowledgments
The authors would like to thank Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R292), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. Furthermore, authors would like to acknowledge the research facilities provided by Bioinformatics Lab, NIGAB, NARC, Pakistan.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1039548/full#supplementary-material
References
Anderson, R. J., Weng, Z., Campbell, R. K., and Jiang, X. (2005). Main-chain conformational tendencies of amino acids. Proteins 60, 679–689. doi:10.1002/prot.20530
Bae, S.-H., Kim, J.-A., Choi, E., Lee, K.-H., Kang, H.-Y., Kim, H.-D., et al. (2001). Tripartite structure of Saccharomyces cerevisiae DNA2 helicase/endonuclease. Nucleic Acids Res. 29, 3069–3079. doi:10.1093/nar/29.14.3069
Bailey, L. J., Bianchi, J., and Doherty, A. J. (2019). PrimPol is required for the maintenance of efficient nuclear and mitochondrial DNA replication in human cells. Nucleic Acids Res. 47, 4026–4038. doi:10.1093/nar/gkz056
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). Meme suite: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi:10.1093/nar/gkp335
Bhurta, R., Hurali, D. T., Tyagi, S., Sathee, L., Adavi, B., Singh, D., et al. (2022). Genome-wide identification and expression analysis of the thioredoxin (trx) gene family reveals its role in leaf rust resistance in wheat (Triticum aestivum L.). Front. Genet. 563, 836030. doi:10.3389/fgene.2022.836030
Blum, M., Chang, H.-Y., Chuguransky, S., Grego, T., Kandasaamy, S., Mitchell, A., et al. (2021). The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49, D344–D354. doi:10.1093/nar/gkaa977
Bonetti, D., Colombo, C. V., Clerici, M., and Longhese, M. P. (2018). Processing of DNA ends in the maintenance of genome stability. Front. Genet. 9, 390. doi:10.3389/fgene.2018.00390
Budd, M. E., Antoshechkin, I. A., Reis, C., Wold, B. J., and Campbell, J. L. (2011). Inviability of a DNA2 deletion mutant is due to the DNA damage checkpoint. Cell Cycle 10, 1690–1698. doi:10.4161/cc.10.10.15643
Budd, M. E., and Campbell, J. L. (1997). A yeast replicative helicase, DNA2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 17, 2136–2142. doi:10.1128/mcb.17.4.2136
Budd, M. E., Choe, W.-C., and Campbell, J. L. (2000). The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 275, 16518–16529. doi:10.1074/jbc.M909511199
Bundó, M., Martín-Cardoso, H., Gómez-Ariza, J., Pesenti, M., Castillo, L., Frouin, J., et al. (2022). Integrative approach for precise genotyping and transcriptomics of salt tolerant introgression rice lines. Front. Plant Sci. 12, 3217. doi:10.3389/fpls.2021.797141
Cejka, P., Cannavo, E., Polaczek, P., Masuda-Sasa, T., Pokharel, S., Campbell, J. L., et al. (2010). DNA end resection by DNA2–sgs1–RPA and its stimulation by top3–rmi1 and mre11–rad50–xrs2. Nature 467, 112–116. doi:10.1038/nature09355
Chang, W., Guo, Y., Zhang, H., Liu, X., and Guo, L. (2020). Same actor in different stages: Genes in shoot apical meristem maintenance and floral meristem determinacy in Arabidopsis. Front. Ecol. Evol. 8, 89. doi:10.3389/fevo.2020.00089
Chatterjee, N., and Walker, G. C. (2017). Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 58, 235–263. doi:10.1002/em.22087
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Croteau, D. L., Popuri, V., Opresko, P. L., and Bohr, V. A. (2014). Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 83, 519–552. doi:10.1146/annurev-biochem-060713-035428
Diray-Arce, J., Liu, B., Cupp, J. D., Hunt, T., and Nielsen, B. L. (2013). The Arabidopsis At1g30680 gene encodes a homologue to the phage T7 gp4 protein that has both DNA primase and DNA helicase activities. BMC Plant Biol. 13, 36–11. doi:10.1186/1471-2229-13-36
Dumas, L. B., Lussky, J. P., Mcfarland, E. J., and Shampay, J. (1982). New temperature-sensitive mutants of Saccharomyces cerevisiae affecting DNA replication. Mol. Gen. Genet. 187, 42–46. doi:10.1007/BF00384381
Duxin, J. P., Dao, B., Martinsson, P., Rajala, N., Guittat, L., Campbell, J. L., et al. (2009). Human DNA2 is a nuclear and mitochondrial DNA maintenance protein. Mol. Cell. Biol. 29, 4274–4282. doi:10.1128/MCB.01834-08
Fu, R., Zhang, M., Zhao, Y., He, X., Ding, C., Wang, S., et al. (2017). Identification of salt tolerance-related microRNAs and their targets in maize (Zea mays L.) using high-throughput sequencing and degradome analysis. Front. Plant Sci. 8, 864. doi:10.3389/fpls.2017.00864
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi:10.1093/nar/gkg563
Geng, Y., Yan, A., and Zhou, Y. (2022). Positional cues and cell division dynamics drive meristem development and archegonium formation in Ceratopteris gametophytes. Commun. Biol. 5, 650. doi:10.1038/s42003-022-03627-y
Gill, S. S., Anjum, N. A., Gill, R., Jha, M., and Tuteja, N. (2015). DNA damage and repair in plants under ultraviolet and ionizing radiations. ScientificWorldJournal. 2015, 250158. doi:10.1155/2015/250158
Gredilla, R., Garm, C., and Stevnsner, T. (2012). Nuclear and mitochondrial DNA repair in selected eukaryotic aging model systems. Oxid. Med. Cell. Longev. 2012, 282438. doi:10.1155/2012/282438
Guo, A.-Y., Zhu, Q.-H., Chen, X., and Luo, J.-C. (2007). Gsds: A gene structure display server. Yi chuan= Hered. 29, 1023–1026. doi:10.1360/yc-007-1023
Guo, Y., Wu, H., Li, X., Li, Q., Zhao, X., Duan, X., et al. (2017). Identification and expression of GRAS family genes in maize (Zea mays L.). PLoS One 12, e0185418. doi:10.1371/journal.pone.0185418
Hudson, J. J., and Rass, U. (2021). DNA2 in chromosome stability and cell survival—is it all about replication forks? Int. J. Mol. Sci. 22, 3984. doi:10.3390/ijms22083984
Huselid, E., and Bunting, S. F. (2020). The regulation of homologous recombination by helicases. Genes 11, 498. doi:10.3390/genes11050498
Jia, N., Liu, X., and Gao, H. (2016). A DNA2 homolog is required for DNA damage repair, cell cycle regulation, and meristem maintenance in plants. Plant Physiol. 171, 318–333. doi:10.1104/pp.16.00312
Kang, Y.-H., Lee, C.-H., and Seo, Y.-S. (2010). DNA2 on the road to Okazaki fragment processing and genome stability in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 45, 71–96. doi:10.3109/10409230903578593
Kleppa, L., Mari, P.-O., Larsen, E., Lien, G. F., Godon, C., Theil, A. F., et al. (2012). Kinetics of endogenous mouse FEN1 in base excision repair. Nucleic Acids Res. 40, 9044–9059. doi:10.1093/nar/gks673
Kuzminov, A. (2011). Homologous recombination experimental systems, analysis, and significance. EcoSal Plus 4. doi:10.1128/ecosalplus.7.2.6
Letunic, I., Doerks, T., and Bork, P. (2012). Smart 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. doi:10.1093/nar/gkr931
Levikova, M., and Cejka, P. (2015). The Saccharomyces cerevisiae Dna2 can function as a sole nuclease in the processing of Okazaki fragments in DNA replication. Nucleic Acids Res. 43, 7888–7897. doi:10.1093/nar/gkv710
Li, G.-M. (2008). Mechanisms and functions of DNA mismatch repair. Cell Res. 18, 85–98. doi:10.1038/cr.2007.115
Li, X., and Heyer, W.-D. (2008). Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 18, 99–113. doi:10.1038/cr.2008.1
Liu, X., and Widmer, A. (2014). Genome-wide comparative analysis of the GRAS gene family in Populus, Arabidopsis and rice. Plant Mol. Biol. Rep. 32, 1129–1145. doi:10.1007/s11105-014-0721-5
Machwe, A., Xiao, L., Groden, J., and Orren, D. K. (2006). The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry 45, 13939–13946. doi:10.1021/bi0615487
Marconi, M., and Wabnik, K. (2021). Shaping the organ: A biologist guide to quantitative models of plant morphogenesis. Front. Plant Sci. 12, 746183. doi:10.3389/fpls.2021.746183
Masuda-Sasa, T., Imamura, O., and Campbell, J. L. (2006). Biochemical analysis of human DNA2. Nucleic Acids Res. 34, 1865–1875. doi:10.1093/nar/gkl070
Meng, Y., Liu, C., Shen, L., Zhou, M., Liu, W., Kowolik, C., et al. (2019). TRAF6 mediates human DNA2 polyubiquitination and nuclear localization to maintain nuclear genome integrity. Nucleic Acids Res. 47, 7564–7579. doi:10.1093/nar/gkz537
Murray, J. A., Jones, A., Godin, C., and Traas, J. (2012). Systems analysis of shoot apical meristem growth and development: Integrating hormonal and mechanical signaling. Plant Cell 24, 3907–3919. doi:10.1105/tpc.112.102194
Nguyen, M. O., Jalan, M., Morrow, C. A., Osman, F., and Whitby, M. C. (2015). Recombination occurs within minutes of replication blockage by RTS1 producing restarted forks that are prone to collapse. Elife 4, e04539. doi:10.7554/eLife.04539
Nisa, M.-U., Huang, Y., Benhamed, M., and Raynaud, C. (2019). The plant DNA damage response: Signaling pathways leading to growth inhibition and putative role in response to stress conditions. Front. Plant Sci. 10, 653. doi:10.3389/fpls.2019.00653
Njaci, I., Williams, B., Castillo-González, C., Dickman, M. B., Zhang, X., and Mundree, S. (2018). Genome-wide investigation of the role of microRNAs in desiccation tolerance in the resurrection grass Tripogon loliiformis. Plants 7, 68. doi:10.3390/plants7030068
Noctor, G., and Foyer, C. H. (2016). Intracellular redox compartmentation and ROS-related communication in regulation and signaling. Plant Physiol. 171, 1581–1592. doi:10.1104/pp.16.00346
Okonechnikov, K., Golosova, O., Fursov, M., and Team, U. (2012). Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 28, 1166–1167. doi:10.1093/bioinformatics/bts091
Pawłowska, E., Szczepanska, J., and Blasiak, J. (2017). DNA2—An important player in DNA damage response or just another DNA maintenance protein? Int. J. Mol. Sci. 18, 1562. doi:10.3390/ijms18071562
Prince, V. E., and Pickett, F. B. (2002). Splitting pairs: The diverging fates of duplicated genes. Nat. Rev. Genet. 3, 827–837. doi:10.1038/nrg928
Rahim, A. A., Uzair, M., Rehman, N., Rehman, O. U., Zahra, N., and Khan, M. R. (2022). Genome-wide identification and characterization of receptor-like protein kinase 1 (RPK1) gene family in Triticum aestivum under drought stress. Front. Genet. 13, 912251. doi:10.3389/fgene.2022.912251
Raney, K. D., Byrd, A. K., and Aarattuthodiyil, S. (2013). “Structure and mechanisms of SF1 DNA helicases,” in DNA helicases and DNA motor proteins (Berlin, Germany: Springer), 17–46.
Rasool, F., Uzair, M., Naeem, M. K., Rehman, N., Afroz, A., Shah, H., et al. (2021). Phenylalanine ammonia-lyase (PAL) genes family in wheat (Triticum aestivum L.): Genome-wide characterization and expression profiling. Agronomy 11, 2511. doi:10.3390/agronomy11122511
Raza, A., Tabassum, J., Fakhar, A. Z., Sharif, R., Chen, H., Zhang, C., et al. (2022). Smart reprograming of plants against salinity stress using modern biotechnological tools. Crit. Rev. Biotechnol., 1–28. doi:10.1080/07388551.2022.2093695
Rehman, O. U., Uzair, M., Chao, H., Fiaz, S., Khan, M. R., and Chen, M. (2022). Role of the type-B authentic response regulator gene family in fragrant rice under alkaline-salt stress. Physiol. Plant. 174, e13696. doi:10.1111/ppl.13696
Rout, S. S., Rout, P., Uzair, M., Kumar, G., and Nanda, S. (2022). Genome-wide identification and expression analysis of CRK gene family in chili pepper (Capsicum annuum L.) in response to Colletotrichum truncatum infection. J. Hortic. Sci. Biotechnol., 1–13. doi:10.1080/14620316.2022.2117654
Ruff, P., Donnianni, R. A., Glancy, E., Oh, J., and Symington, L. S. (2016). RPA stabilization of single-stranded DNA is critical for break-induced replication. Cell Rep. 17, 3359–3368. doi:10.1016/j.celrep.2016.12.003
Saada, A. A., Lambert, S. A., and Carr, A. M. (2018). Preserving replication fork integrity and competence via the homologous recombination pathway. DNA repair 71, 135–147. doi:10.1016/j.dnarep.2018.08.017
Sturzenegger, A., Burdova, K., Kanagaraj, R., Levikova, M., Pinto, C., Cejka, P., et al. (2014). DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J. Biol. Chem. 289, 27314–27326. doi:10.1074/jbc.M114.578823
Su, W., Raza, A., Gao, A., Jia, Z., Zhang, Y., Hussain, M. A., et al. (2021). Genome-wide analysis and expression profile of superoxide dismutase (SOD) gene family in rapeseed (Brassica napus L.) under different hormones and abiotic stress conditions. Antioxidants 10, 1182. doi:10.3390/antiox10081182
Tarun, J. A., Mauleon, R., Arbelaez, J. D., Catausan, S., Dixit, S., Kumar, A., et al. (2020). Comparative transcriptomics and co-expression networks reveal tissue-and genotype-specific responses of qDTYs to reproductive-stage drought stress in rice (Oryza sativa L.). Genes 11, 1124. doi:10.3390/genes11101124
Tisi, R., Vertemara, J., Zampella, G., and Longhese, M. P. (2020). Functional and structural insights into the MRX/MRN complex, a key player in recognition and repair of DNA double-strand breaks. Comput. Struct. Biotechnol. J. 18, 1137–1152. doi:10.1016/j.csbj.2020.05.013
Traas, J. (2018). Organogenesis at the shoot apical meristem. Plants 8, 6. doi:10.3390/plants8010006
Uzair, M., Long, H., Zafar, S. A., Patil, S. B., Chun, Y., Li, L., et al. (2021). Narrow Leaf21, encoding ribosomal protein RPS3A, controls leaf development in rice. Plant Physiol. 186, 497–518. doi:10.1093/plphys/kiab075
Verma, P., Tandon, R., Yadav, G., and Gaur, V. (2020). Structural aspects of DNA repair and recombination in crop improvement. Front. Genet. 11, 574549. doi:10.3389/fgene.2020.574549
Wang, X., Dong, Y., Zhao, X., Li, J., Lee, J., Yan, Z., et al. (2021). Rtt105 promotes high-fidelity DNA replication and repair by regulating the single-stranded DNA-binding factor RPA. Proc. Natl. Acad. Sci. U. S. A. 118, e2106393118. doi:10.1073/pnas.2106393118
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi:10.1093/nar/gky427
Wu, L., and Hickson, I. D. (2006). DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 40, 279–306. doi:10.1146/annurev.genet.40.110405.090636
Wu, Y. (2012). Unwinding and rewinding: Double faces of helicase? J. Nucleic Acids 2012, 140601. doi:10.1155/2012/140601
Xiang, Z. (2006). Advances in homology protein structure modeling. Curr. Protein Pept. Sci. 7, 217–227. doi:10.2174/138920306777452312
Yi-Qin, H., Wen-Di, H., Lei, Y., Yi-Ting, L., Chen, L., Yong-Xing, Z., et al. (2020). Genome-wide analysis of ethylene-insensitive3 (EIN3/EIL) in Triticum aestivum. Crop Sci. 60, 2019–2037. doi:10.1002/csc2.20115
Yousefzadeh, M., Henpita, C., Vyas, R., Soto-Palma, C., Robbins, P., and Niedernhofer, L. (2021). DNA damage—How and why we age? Elife 10, e62852. doi:10.7554/eLife.62852
Yu, C.-S., Cheng, C.-W., Su, W.-C., Chang, K.-C., Huang, S.-W., Hwang, J.-K., et al. (2014). CELLO2GO: A web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PloS one 9, e99368. doi:10.1371/journal.pone.0099368
Zhang, D., Iyer, L. M., and Aravind, L. (2012). Bacterial GRAS domain proteins throw new light on gibberellic acid response mechanisms. Bioinformatics 28, 2407–2411. doi:10.1093/bioinformatics/bts464
Zheng, L., Meng, Y., Campbell, J. L., and Shen, B. (2020). Multiple roles of DNA2 nuclease/helicase in DNA metabolism, genome stability and human diseases. Nucleic Acids Res. 48, 16–35. doi:10.1093/nar/gkz1101
Zheng, L., and Shen, B. (2011). Okazaki fragment maturation: Nucleases take centre stage. J. Mol. Cell Biol. 3, 23–30. doi:10.1093/jmcb/mjq048
Zheng, L., Zhou, M., Guo, Z., Lu, H., Qian, L., Dai, H., et al. (2008). Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell 32, 325–336. doi:10.1016/j.molcel.2008.09.024
Keywords: rice, abiotic stress, DNA2, DNA damage-repair, gene expression
Citation: Saleem B, Farooq U, Rehman OU, Aqeel M, Farooq MS, Naeem MK, Inam S, Ajmal W, Rahim AA, Chen M, Kalsoom R, Uzair M, Fiaz S, Attia K, Alafari HA, Khan MR and Yu G (2022) Genome-wide and molecular characterization of the DNA replication helicase 2 (DNA2) gene family in rice under drought and salt stress. Front. Genet. 13:1039548. doi: 10.3389/fgene.2022.1039548
Received: 08 September 2022; Accepted: 31 October 2022;
Published: 22 November 2022.
Edited by:
Karansher Singh Sandhu, Bayer Crop Science (United States), United StatesReviewed by:
Muhammad Noman, University of Florida, United StatesMuhammad Riaz, Shanghai Jiao Tong University, China
Zaid Khan, Huazhong Agricultural University, China
Copyright © 2022 Saleem, Farooq, Rehman, Aqeel, Farooq, Naeem, Inam, Ajmal, Rahim, Chen, Kalsoom, Uzair, Fiaz, Attia, Alafari, Khan and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Uzair, dXphaXJicmVlZGVyQGdtYWlsLmNvbQ==; Muhammad Ramzan Khan, bXJraGFuQHBhcmMuZ292LnBr; Guoping Yu, eXVndW9waW5nQGNhYXMuY24=
†These authors have contributed equally to this work
 Bilal Saleem
Bilal Saleem Umer Farooq1†
Umer Farooq1† Obaid Ur Rehman
Obaid Ur Rehman Muhammad Aqeel
Muhammad Aqeel Muhammad Shahbaz Farooq
Muhammad Shahbaz Farooq Muhammad Kashif Naeem
Muhammad Kashif Naeem Safeena Inam
Safeena Inam Ming Chen
Ming Chen Rabia Kalsoom
Rabia Kalsoom Muhammad Uzair
Muhammad Uzair Sajid Fiaz
Sajid Fiaz Kotb Attia
Kotb Attia Muhammad Ramzan Khan
Muhammad Ramzan Khan