- Zhengzhou Traditional Chinese Medicine Hospital, Zhengzhou, China
Background: Heart failure (HF) is not only a common complication in patients with end-stage renal disease (ESRD) but also a major cause of death. Although clinical studies have shown that there is a close relationship between them, the mechanism of its occurrence is unclear. The aim of this study is to explore the molecular mechanisms between HF and ESRD through comprehensive bioinformatics analysis, providing a new perspective on the crosstalk between these two diseases.
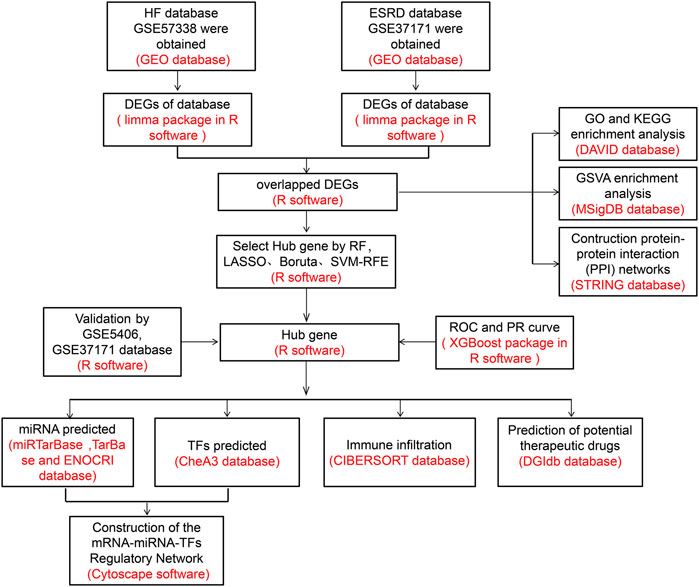
Methods: The HF and ESRD datasets were downloaded from the Gene Expression Omnibus (GEO) database; we identified and analyzed common differentially expressed genes (DEGs). First, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and gene set variation analyses (GSVA) were applied to explore the potential biological functions and construct protein−protein interaction (PPI) networks. Also, four algorithms, namely, random forest (RF), Boruta algorithm, logical regression of the selection operator (LASSO), and support vector machine-recursive feature elimination (SVM-RFE), were used to identify the candidate genes. Subsequently, the diagnostic efficacy of hub genes for HF and ESRD was evaluated using eXtreme Gradient Boosting (XGBoost) algorithm. CIBERSORT was used to analyze the infiltration of immune cells. Thereafter, we predicted target microRNAs (miRNAs) using databases (miRTarBase, TarBase, and ENOCRI), and transcription factors (TFs) were identified using the ChEA3 database. Cytoscape software was applied to construct mRNA−miRNA−TF regulatory networks. Finally, the Drug Signatures Database (DSigDB) was used to identify potential drug candidates.
Results: A total of 68 common DEGs were identified. The enrichment analysis results suggest that immune response and inflammatory factors may be common features of the pathophysiology of HF and ESRD. A total of four hub genes (BCL6, CCL5, CNN1, and PCNT) were validated using RF, LASSO, Boruta, and SVM-RFE algorithms. Their AUC values were all greater than 0.8. Immune infiltration analysis showed that immune cells such as macrophages, neutrophils, and NK cells were altered in HF myocardial tissue, while neutrophils were significantly correlated with all four hub genes. Finally, 11 target miRNAs and 10 TFs were obtained, and miRNA−mRNA−TF regulatory network construction was performed. In addition, 10 gene-targeted drugs were discovered.
Conclusion: Our study revealed important crosstalk between HF and ESRD. These common pathways and pivotal genes may provide new ideas for further clinical treatment and experimental studies.
Introduction
Heart failure (HF) is a common clinical cardiovascular disease that affects more than 26 million people worldwide (Savarese and Lund, 2017). One of the most common causes of heart failure is ischemic heart disease, which causes a loss of myocardial tissue and contractility (Severino et al., 2020). At this stage, the main goal of the pharmacological treatment of this disease is to relieve symptoms and improve residual cardiac functions. The prevalence of HF increases significantly as the renal function deteriorates (Silverberg et al., 2004), reaching 65%–70% in end-stage renal disease (ESRD). Conversely, the risk of HF is 15.8 times higher in patients with progressive chronic kidney disease (Schefold et al., 2016). On the other hand, abnormal renal function may affect the application of HF therapeutic agents, increase the risk of nephrotoxicity in HF treatment, and impair the patient’s response to diuretics. These results suggest that several susceptibility factors in kidney diseases may trigger the development of HF.
Studies have shown that the myocardium in patients with ESRD exhibits characteristic changes, often accompanied by pathological myocardial fibrosis and cardiac hypertrophy (Alhaj et al., 2013). Recent scientific work has identified several possible pathophysiological mechanisms, including hemodynamic disturbances, excessive activation of the RAAS, water and sodium retention, chronic inflammatory states, metabolic acidosis, reduced cytokine clearance, insulin resistance, oxidative stress, and post-translational modification of blood-borne molecules such as lipoproteins (Zoccali et al., 2017; Rangaswami and McCullough, 2018; Costanzo, 2020). At this stage, mechanistic studies have mostly focused on the excessive activation of the RAAS and thus cardiac remodeling (Dounousi et al., 2021). Although some studies have found that abnormalities in immune cells in ESRD patients promote cardiovascular disease, the direct effects of abnormal ratios of immune cells in the blood and alterations in inflammatory factors on the myocardial tissue in ESRD patients are often overlooked. Despite there being strong clinical and epidemiological evidence of the crosstalk between HF and ESRD, the exact mechanisms explaining the coexistence of the two diseases remain unclear. A better understanding of the relationship between them is therefore essential for the development of detection and management.
With the advancement of science and technology, bioinformatics approaches have enabled us to gain a deeper understanding of the biological processes of disease pathology at the genetic level. However, the common diagnosis and interlinked genes in HF and ESRD are unclear. Therefore, this study used bioinformatics methods to screen the biomarkers, which may serve to provide new insights into the biological mechanisms of these two diseases.
Materials and methods
Data source
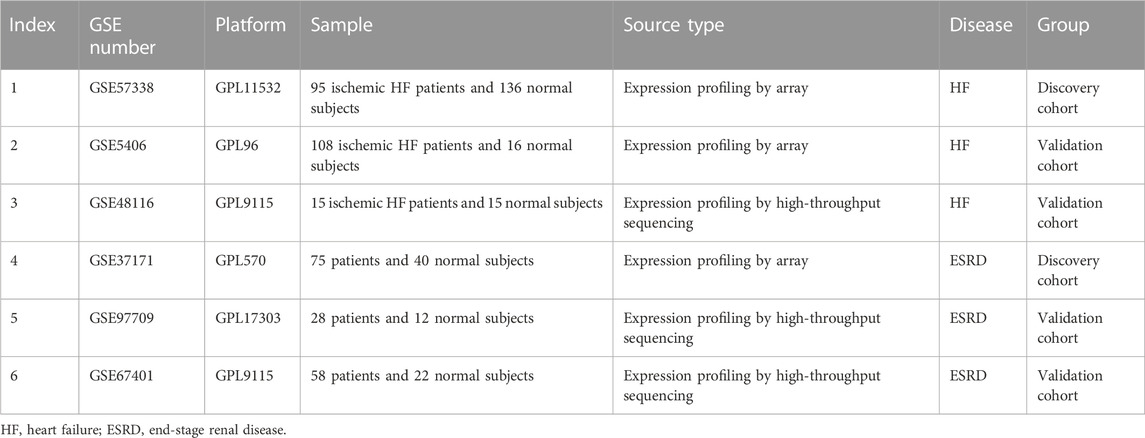
We searched the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) for gene expression datasets using the terms “heart failure” or “end stage renal disease.” The following criteria were then used to further screen the dataset: 1) studies that included cases versus healthy controls; 2) heart failure samples were obtained from ischemic HF heart tissue; 3) ESRD samples were obtained from blood samples; and 4) the sample size was greater than 30. Finally, GSE57338, GSE5604, and GSE48116 were identified as HF datasets, and GSE37171, GSE9709, and GSE67401 were identified as ESRD datasets. The information on each dataset is summarized in Table 1.
Identification of DEGs
In GSE57338 and GSE37171 datasets, gene expression was analyzed for subsequent analyses, and the “limma” package (Ritchie et al., 2015) was used to calculate the differentially expressed genes (DEGs), with p-value <.05 and |log2 fold-change (LogFC)> 0.5 considered statistically significant. Subsequently, the batch effects were removed using the “ComBat” package for the next stage of the analysis. The “ggplot2” (Ito and Murphy, 2013) and “pheatmap” packages were used to plot DEGs for visualizing different datasets.
Enrichment analyses
To reveal the potential functions of common DEGs in HF and ESRD, Gene Ontology (GO) (Carbon et al., 2009) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000) analyses of common DEGs were performed to explore the functions of DEGs. In the present study, a protein−protein interaction (PPI) network of DEGs was constructed using the STRING database (Szklarczyk et al., 2019).
Gene set variation analysis enrichment analysis
We performed GSVA (Hänzelmann et al., 2013) enrichment analysis using the “GSVA” R package to explore the potential biological functions involved in HF and ESRD. The gene list of pathways was collected by integrating the Molecular Signatures Database (MSigDB, v7.0) (Subramanian et al., 2005). p-values <.05 indicate the statistically significant differences.
Hub gene screening and validation by multiple machine learning methods
We jointly used four algorithms, namely, random forest (RF) (Alakwaa et al., 2018), Boruta algorithm (Kursa, 2014), logical regression of the selection operator (LASSO) (Alhamzawi and Ali, 2018), and support vector machine-recursive feature elimination (SVM-RFE) (Lin et al., 2012), to screen potential marker genes in HF and ESRD datasets. RF algorithm is a supervised machine learning algorithm that classifies features based on their processing and variables. The Boruta package uses the Boruta algorithm for significant feature gene extraction. The glmnet package is used for LASSO regression, where the best λ is determined by the least binomial deviation with tenfold cross-validation. SVM-RFE is a supervised machine learning algorithm that ranks different features based on differences in predictive power (Sanz et al., 2018). The crossover genes obtained by four machine learning methods have high accuracy in determining diagnostic gene signals (Gao et al., 2021). Meanwhile, HF (GSE5406) and ESRD (GSE97709) validation datasets were used to draw the expression of hub genes.
Evaluation of the hub gene diagnostic value
eXtreme Gradient Boosting (XGBoost) (Ogunleye and Wang, 2020) is a commonly used supervised integrative learning algorithm that can use the expression values of pivotal genes as the feature values for XGBoost model training. First, we select the HF dataset (GSE57338) and ESRD dataset (GSE37171) as the training sets, and the evaluation is performed on the datasets of HF (GSE48116) and ESRD (GSE67401). The prognostic efficiency was evaluated by the receiver operating characteristic (ROC) curve, precision–recall (PR) curve, and area under the curve (AUC).
Analysis of immune cell infiltration
We used the CIBERSORT package (Newman et al., 2015) to quantify the level of immune cell infiltration in each sample and to assess the effect of hub genes on immune infiltration in the HF myocardial tissue. The Wilcoxon test was used to analyze the immune scores, the “vioplotR” package was employed to visualize the results, and p <.05 was considered significant.
Construction of the miRNA−mRNA−TF regulatory network
The miRTarBase database (Chou et al., 2018), TarBase database (Karagkouni et al., 2018), and ENOCRI database (Li et al., 2014) were used to discover potential miRNAs. Furthermore, we overlapped predicted target miRNAs and constructed a miRNA–target gene regulatory network. The ChIP-X Enrichment Analysis 3 (ChEA3) (Keenan et al., 2019) verified the targets of TFs, and the top 10 TFs were selected as target TFs. Cytoscape (Shannon et al., 2003) was used to visualize the miRNA−mRNA−TF regulatory network.
Evaluation of candidate drugs
The Enrichers platform was used to identify the relationship between drug molecules and hub genes, and the data were obtained from the Drug Signatures Database (DSigDB, http://tanlab.ucdenver.edu/DSigDB). We retrieved potential drugs for the hub genes.
Results
Identification of DEGs
The flow chart of the study design is shown in Figure 1. A total of 447 DEGs were found in the screened HF and healthy subjects in the dataset GSE57338, including 250 upregulated and 197 downregulated genes (Figures 2A, B). There were 5,299 DEGs in ESRD patients compared to healthy controls, including 4,013 upregulated and 1,286 downregulated genes (Figures 2C, D). A total of 68 common DEGs were identified after taking the intersection of the Venn diagrams (Figure 2E).
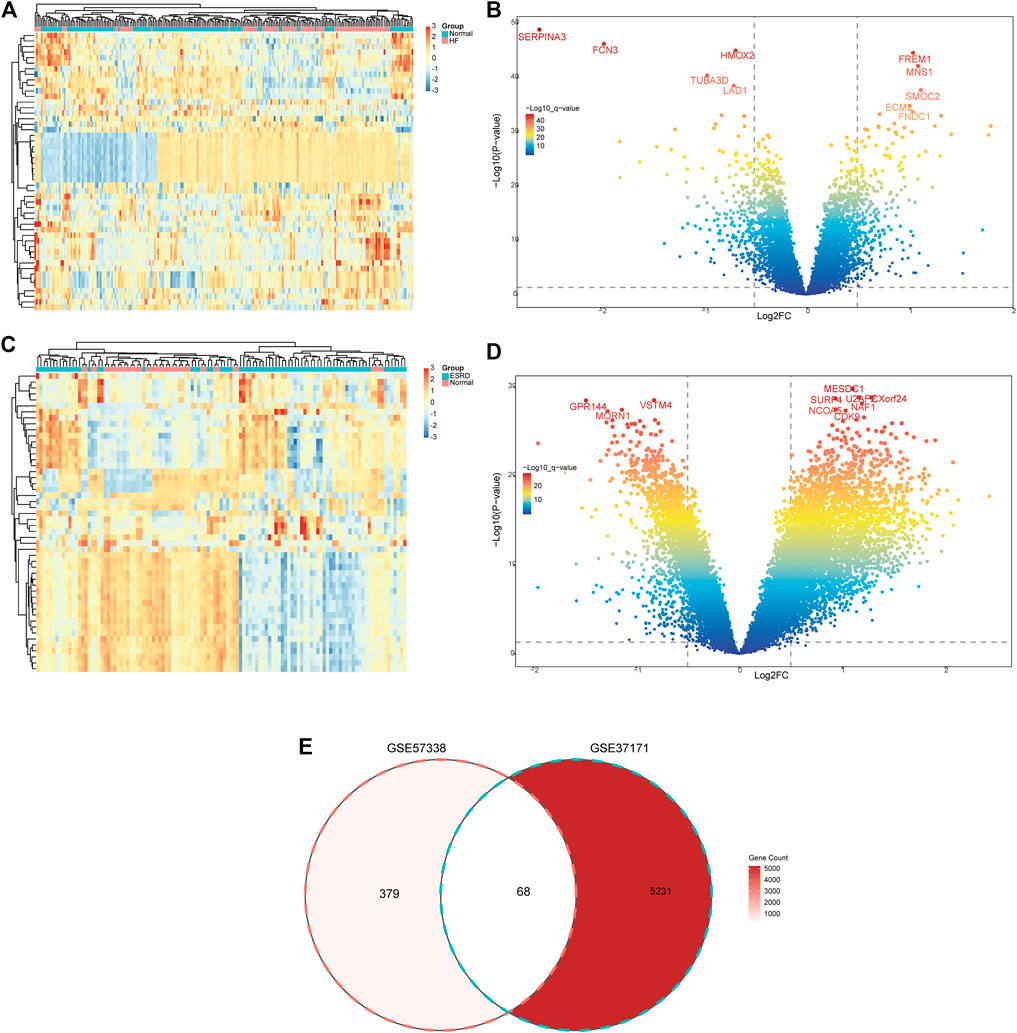
FIGURE 2. Identification of common DEGs. (A) Heatmap of the top 50 DEGs in GSE57338. (B) Volcanic plots of GSE57338. (C) Heatmap of the top 50 DEGs in GSE37171. (D) Volcanic plots of GSE37171. (E) Venn diagram of common DEGs in HF and ESRD. HF, heart failure; ESRD, end-stage renal disease.
Functional enrichment analyses and the PPI network
The potential functions and pathways of these common DEGs were detected through the GO and KEGG clustering analyses. The GO term enrichment analysis includes biological processes (BPs), cellular components (CCs), and molecular functions (MFs). In BP, module genes were significantly enriched in immune receptor activity, cytokine binding, cytokine receptor activity, protein self-association, chemokine binding, chemokine receptor activity, and G protein-coupled chemoattractant receptor activity. In MF, module genes were significantly enriched in the cytokine-mediated signaling pathway, response to the virus, T-cell-mediated immunity, chemokine-mediated signaling pathway, dendritic cell migration, and regulation of monocyte chemotaxis. In CC, module genes were significantly enriched in the vesicle lumen, cytoplasmic vesicle lumen, and secretory granule lumen (Figure 3A). KEGG analysis showed enrichment in the cytokine−cytokine receptor interaction, viral protein interaction with cytokine and cytokine receptor, chemokine signaling pathway, human cytomegalovirus infection, ferroptosis, TNF signaling pathway, and FOXO signaling pathway (Figure 3B). KEGG analysis also showed that processes such as immune factors and chemokine and immune cell regulation were mainly enriched. We used the STRING online server to construct a PPI network (Figure 3C)//.

FIGURE 3. Enrichment analyses for common DEGs. (A) GO enrichment analysis. (B) KEGG enrichment analysis. (C) PPI network; the nodes represent proteins, and the edges represent interactions. GO, Gene Ontology; BP, biological processes; CC, cellular components; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein−protein interaction.
GSVA enrichment analysis
The results of GSVA enrichment analysis revealed that immune-related response, interferon gamma response, interferon alpha response, and KRAS signaling DN were positively correlated with HF (Figure 4A). Meanwhile, the high expression of adipogenesis, DNA repair, and KRAS signaling DN were found to be related with ESRD (Figure 4B).
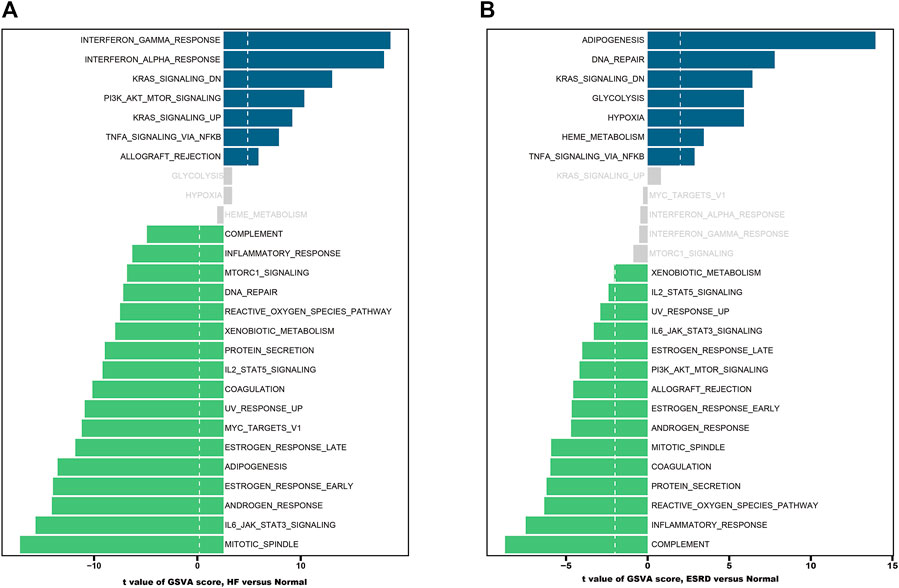
FIGURE 4. Functional and pathway analysis by GSVA enrichment analysis in HF and ESRD. (A) Significant related biological pathways in HF were obtained by GSVA. (B) Significant related biological pathways in ESRD were obtained by GSVA. The blue bars represent high expression, and the green bars represent low expression. HF, heart failure; ESRD, end-stage renal disease; GSVA, gene set variation analysis.
Identification and validation of hub genes
RF, Boruta, LASSO, and SVM-RFE algorithms were used to explore the hub genes shared in HF and ESRD. In the HF dataset, 31 genes were screened by RF algorithm (Figure 5A), 42 genes by Boruta algorithm (Figure 5B), 21 genes by LASSO algorithm (Figure 5C), and 57 genes by SVM-RFE algorithm after tenfold cross-validation (Figure 5D). Eventually, 19 overlapping genes were found in the HF dataset (Figure 5E). In the ESRD dataset, 30 genes were found by RF algorithm (Figure 5F), 34 genes were found by Boruta algorithm (Figure 5G), 19 genes were found by LASSO algorithm (Figure 5H), and 35 genes were found by SVM-RFE algorithm after tenfold cross-validation (Figure 5I). Finally, 13 overlapping genes were found in the ESRD dataset. In the end, the SCN2B, BCL6, CCL5, CNN1, and PCNT genes were found (Figure 5J).
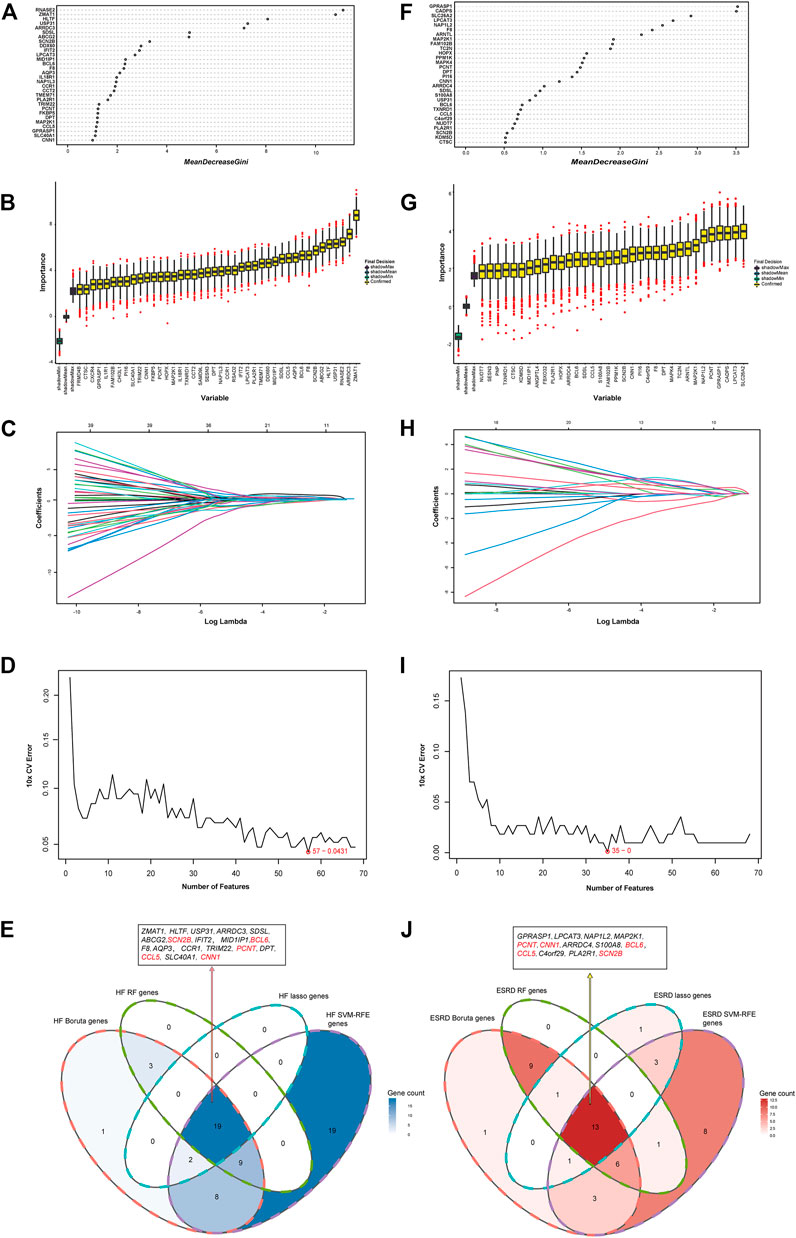
FIGURE 5. Identification of the hub gene in HF and ESRD datasets. (A) RF method identified overlapping DEGs in HF datasets. (B) Boruta method identified common DEGs in HF datasets. (C) LASSO method identified common DEGs in HF datasets. (D) SVM-RFE method identified 68 common DEGs in HF datasets. (E) Intersection of common DEGs in HF by the four analyses. (F) RF method identified common DEGs in ESRD datasets. (G) Boruta method identified common DEGs in ESRD datasets. (H) LASSO method identified common DEGs in ESRD datasets. (I) SVM-RFE method identified common DEGs in ESRD datasets. (J) Intersection of common DEGs in ESRD by the four analyses.
We validated the expression of candidate hub genes in the HF dataset (GSE5406) and the ESRD dataset (GSE97709), as shown in Figures 6A, B. However, in the HF dataset, there was no significant difference between the two groups in the expression of SCN2B. Therefore, BCL6, CCL5, CNN1, and PCNT were defined as hub genes in HF and ESRD. Surprisingly, CCL5 expression was increased in HF tissues but decreased in ESRD blood samples. To assess the diagnostic efficacy of the hub genes, we selected one HF dataset (GSE57338) for training and another (GSE48166) for validation. The performance of the training set showed that the values of ROC and PR curves were 0.976 and 0.958 (Figure 7A), respectively, and that of AUC was 0.924 and 0.92 for the validation set (Figure 7B), which illustrated the diagnostic efficacy of the model. To verify its ability to identify ESRD patients, we used the ESRD dataset (GSE37171) for training (ROC = 0.996; PR = 0.998; Figure 7C) and the ESRD dataset (GSE67401) for validation (ROC = 0.886; PR = 0.724; Figure 7D), and the results are also applicable and practical.
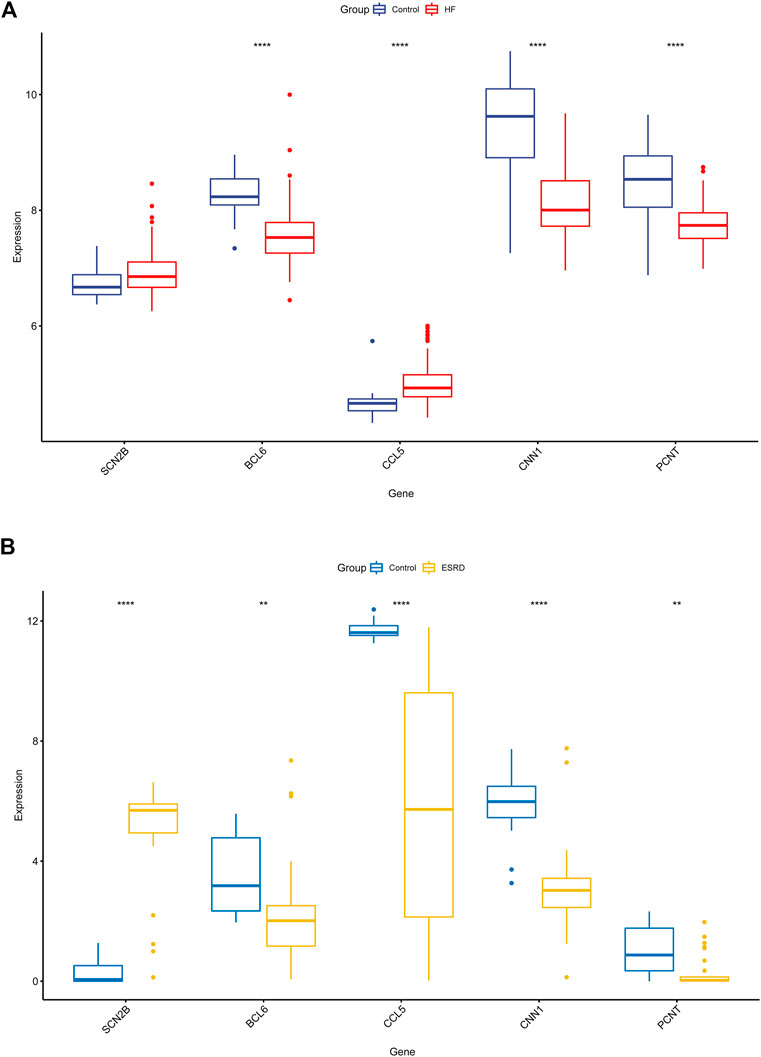
FIGURE 6. Validation of the hub genes in GSE5406 and GSE97709 datasets. (A) Expression levels in GSE5406 datasets. (B) Expression levels of the hub gene in GSE97709 datasets. HF, heart failure; ESRD, end-stage renal disease; p-values are shown as *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
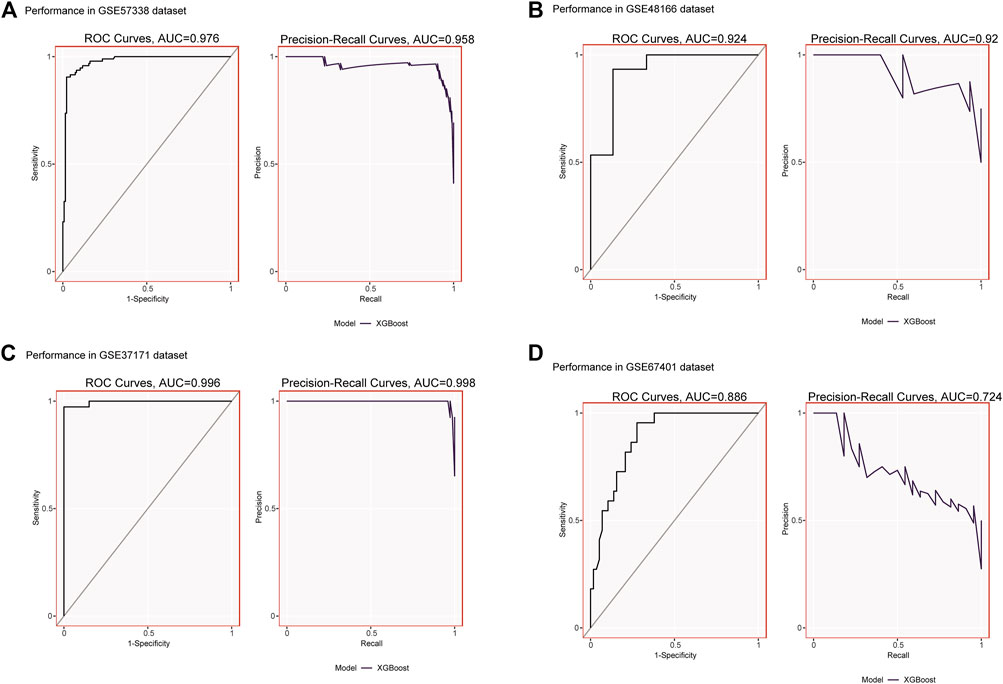
FIGURE 7. (A) XGBoost modeling in the HF training set (GSE57338). (B) Validation through the HF validation set (GSE48166). (C) XGBoost modeling in the ESRD training set (GSE37171). (D) Validation through the ESRD validation set (GSE67401).
Analysis of immune infiltration in the HF and normal groups
We used CIBERSORT to quantify the enrichment scores of 22 immune cell species and related functions in HF patients and the normal population to explore the differences between myocardial tissue and normal tissue immune cells in HF. The first bar graph clearly shows the proportion of different immune cell subpopulations in each sample (Figure 8A). Regarding the correlation between immune cell subtypes, activated mast cells and M1 macrophages (r = 0.49) have the most significant positive correlation. In contrast, B memory (r = −0.57) cells have the most significant negative correlation (Figure 8B). We find significant differences between HF and normal tissues in the 21 immune cell subpopulations. Therefore, the occurrence of HF is closely related to abnormal immune cell infiltration, and changes in the correlation between immune cells may be related to the occurrence and development of HF (Figure 8C). Specifically, all hub genes showed a remarkable correlation with neutrophil cells (Figure 8D).
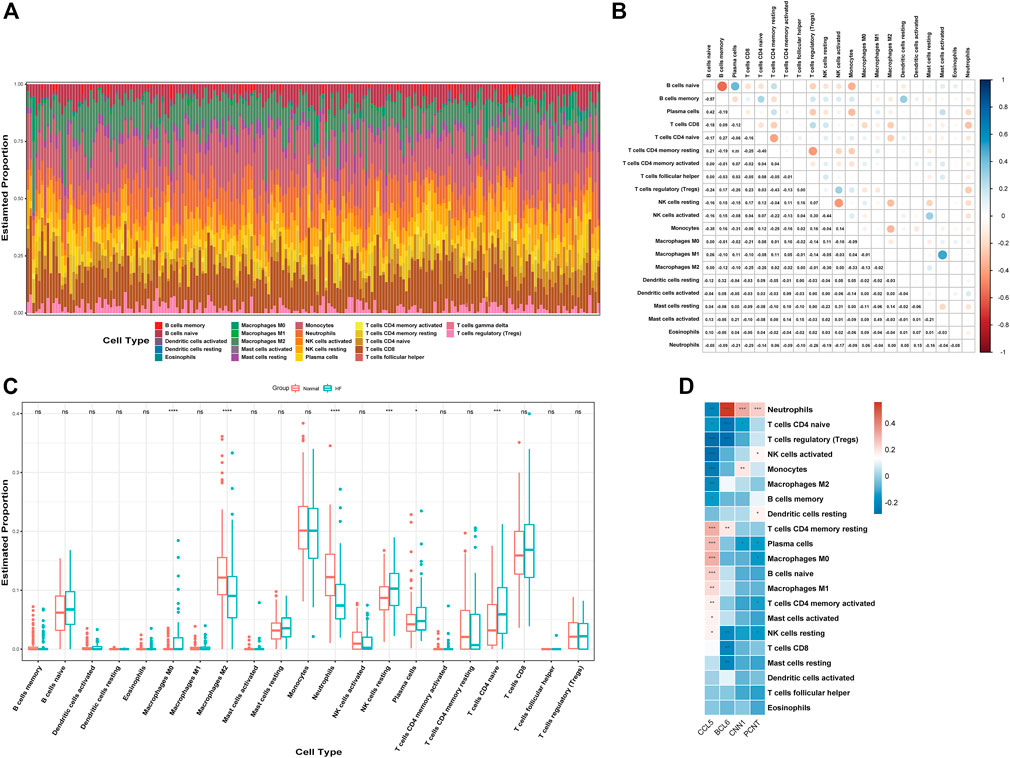
FIGURE 8. Analysis of immune cell infiltration. (A) Relative percentage of 22 immune cell subtypes. (B) Correlation heatmap of 21 immune cells. (C) Immune cells in HF and normal samples. (D) Relationship between hub genes and immune cells related to HF. HF, heart failure; p-values are shown as *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
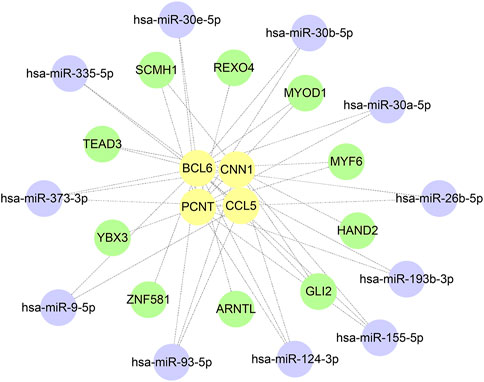
miRNA−mRNA−TF regulatory network construction
A total of 385 miRNA gene pairs were obtained after mapping by three experimentally verified miRNA target prediction databases (Supplementary Tables S1–S3). Finally, 11 overlapping target miRNAs were regarded as key miRNAs. The ChEA3 database was used to enrich TF targets of hub genes to explore their distribution further. The results of CHEA3 prediction showed that SCMH1 ranked the highest among TFs of hub genes (Supplementary Table S4). The top 10 TFs are SCMH1, TEAD3, YBX3, HAND2, MYF6, ARNTL, ZNF581, MYOD1, GLI2, and REXO4. Subsequently, we merged the mRNA−miRNA−TF regulatory network with Cytoscape software (Figure 9).
Screening for potential pharmacological targets
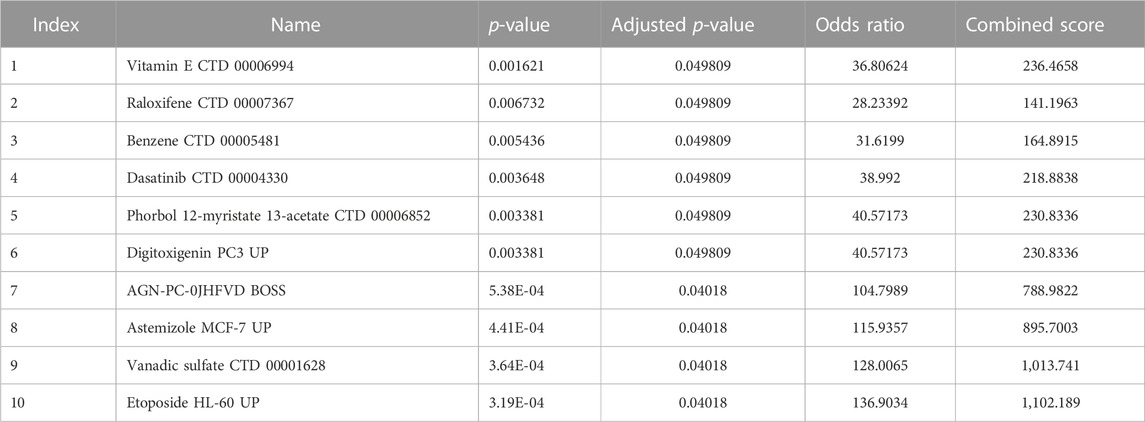
We used the DSigDB database built on the Enrichr website to search for target drugs for the hub genes. The target drugs for these genes were predicted, and the potential pharmacological target screening was downloaded. The top 10 drug candidates associated with hub genes were selected based on p-values and adjusted p-values (Table 2).
Discussion
Patients with HF have benefited from many trials, but patients with ESRD are often excluded from these trials. However, there is little evidence that the benefit of treatment for patients with heart failure combined with advanced kidney disease is altered by the presence or absence of kidney disease. Therefore, more direct evidence is needed to clarify the link between heart failure and end-stage renal disease. In this study, common DEGs and associated pathways between HF and ESRD were explored by bioinformatics analysis. Enrichment analysis revealed that immune and inflammatory responses might play an essential role in HF and ESRD. These findings highlight that immune mechanisms may play a key role in linking HF and ESRD (Rangaswami et al., 2019). Immune cell activation is usually accompanied by the production of inflammatory mediators, such as cytokines, chemokines, and cellular immune receptors, which directly or indirectly affect cardiomyocyte metabolism and promote the development of cardiomyocyte hypertrophy and myocardial fibrosis. GSVA showed that KRAS signaling and TNF-ɑ signaling via NF-κB were positively correlated with both HF and ESRD. The KRAS belongs to the Ras gene family, and the role of KRAS in cardiovascular disease was less studied. However, studies have shown a potential role in its association with cardiac cell proliferation and pathological cardiac hypertrophy (Ramos-Kuri et al., 2021). The NF-κB is a transcription factor that has crucial roles in inflammation, cell proliferation, and immunity, and its activation contributes to the pathogenic processes of various inflammatory diseases (Gordon et al., 2011; Song et al., 2019).
Four machine learning algorithms, namely, RF, Boruta, LASSO, and SVM-RFE, were used to screen potential hub genes, and a total of four hub genes were identified to elucidate the crosstalk between HF and ESRD. The XGBoost machine learning model analysis shows that they have a good prediction effect. BCL6 is a transcriptional repressor with anti-apoptotic and pro-oncogenic properties that was initially identified as an oncogene in non-Hodgkin’s B-cell lymphoma. BCL6 is the most strongly characterized inflammatory marker of cardiac aging and predicts the decline in the LV filling rate from early to late stages (Ma et al., 2015). BCL6 is also a regulator of immune cells, and Treg cells lacking BCL6 are uniquely deficient in their ability to suppress Th2 inflammation (Sawant et al., 1950). However, BCL6 may play a role in protecting mature cardiomyocytes from eosinophilic inflammation in myocardial tissue (Yoshida et al., 1999). In cardiac fibroblasts, BCL6 may inhibit cardiac fibroblast activation and function through direct binding to SMAD4 (Ni et al., 2019).
CCL5/RANTES is a T-cell chemotactic agent that is essential for the recruitment of leukocytes to the sites of inflammation, and CCL5 serves as one of the natural ligands for CCR5, which binds to form a family of secreted proteins involved in immune regulation and inflammatory processes (Zeng et al., 2022). CCL5 is thought to drive immune cell migration to the heart tissue of patients with HF, and studies have also revealed an essential role of CCL5 in ventricular remodeling. Stevenson et al. (2019) found significantly increased levels of CCL5 in human hearts with ischemic cardiomyopathy compared to non-failing hearts. Batista et al. (2018) found that in chasmic cardiomyopathy, CCL5 drives the migration of immune cells to myocardial tissue. The protective role of CCL5 in kidney injury has been demonstrated in other models of kidney disease (Krensky and Ahn, 2007). We found that CCL5 expression was increased in the HF dataset but decreased in the ESRD dataset. First, CCL5 in the blood of ESRD patients is influenced by several factors, such as hemodialysis, drug application, and other diseases (Naumnik et al., 2013; Elmoselhi et al., 2016). Second, impaired immune cells in the blood of patients with ESRD may lead to reduced expression of CCL5 (Betjes, 2013). However, this still needs to be confirmed by further studies.
CNN1 encodes a protein that plays a role in regulating and modulating smooth muscle contraction (Takahashi et al., 1986). It inhibits actin-activated myosin ATPase and Ca2+-dependent migration of actin. It has therefore been identified as a critical player in the stabilization of actin stress fibers (Liu and Jin, 2016) and its role in cardiomyopathy, placental vascular development, and development of tissue morphology has been revealed. A study of human heart failure gene expression has shown downregulation of CNN1 expression in heart failure, which is consistent with our findings (Tan et al., 2002). In contrast, basic studies found that CNN1 plays an essential role in DCM ventricular remodeling and can inhibit the progression of dilated cardiomyopathy in mice through εPKC signaling (Lu et al., 2014). PCTN is a multifunctional scaffolding protein that binds to various centrosomal proteins (Kim et al., 2019). PCTN regulates many centrosomal functions, such as controlling cell cycle progression, mitotic spindle organization and orientation, and directed cell division. Some studies have shown that it is associated with the development of congenital heart disease (Liang et al., 2020). However, the specific mechanism of the role of CNN1 and PCNT in renal diseases is not well understood and still to be revealed. While our study showed that CNN1 and PCNT are associated with neutrophils and plasma cells, we speculate that it may be related to the differentiation and activation of immune cells in the HF myocardial tissue.
Abnormal immune cells are an important basis for immune dysfunction. Although the quantitative changes and pathway activation/inhibition of these subtypes are not well studied in HF, our study further clarifies the relevant directions. The expression of macrophages, CD4+ T cells, NK cells, plasma cells, and neutrophils in myocardial tissue is significantly different from the normal tissue (Li et al., 2021). Consistent with previous findings, M2 macrophages are reduced in heart failure myocardial tissue, which in turn increases cardiac apoptosis and myocardial CD4+ T-cell accumulation (Stevens et al., 2022). In contrast, neutrophils exert a deleterious function in an experimental model of heart failure induced by too much pressure (Liu et al., 2021). This finding is consistent with the underlying mechanism of action of neutrophils, which can be involved in the development of multiple cardiovascular diseases through the release of degranulation and recruitment of microvesicles. Therefore, changes in these cell subtypes play an important role in the process of immune response in HF and have significant prognostic and therapeutic value.
It is well known that miRNAs mainly control gene expression, while TFs are involved in target gene transcription. We identified 11 target miRNAs and 10 associated transcription factors to further understand the hub gene associations. Some miRNAs, such as miR-30b-5p (Ren et al., 2021), miR-30a-5p (Qian et al., 2022), and miR-155-5p (Wang et al., 2022), can play a role in the pathological process of HF. The expression of BCL6 can be regulated by the transcription factors TEAD3, MYF6, ARNTL, ZNF581, MYOD1 and GLI2, and TEAD3 (Han et al., 2020). GLI2 (Voronova et al., 2012) plays an essential role in embryonic myocardial development and cardiomyogenesis. Meanwhile, to predict potentially effective therapeutic agents, we applied the DSigDB to identify 10 possible therapeutic agents. Among them, vitamin E can reduce the activation of inflammatory factor NF-қB, which leads to cytokine/chemokine and mast cell activation, and has been confirmed in related studies (Tettamanti et al., 2018). However, the molecular pathways and potential therapeutic compounds screened by our bioinformatics approach still need further validation by cellular experiments and clinical samples.
Conclusion
To conclude, we identified BCL6, CCL5, CNN1, and PCNT as hub genes between HF and ESRD. This finding contributes to understanding the close interrelationship in the development of ESRD and HF. This study also provides some theoretical basis for the possible search of new drug targets and developing new therapeutic approaches.
Data availability statement
Publicly available datasets were analyzed in this study. These data can be found at: GEO database (https://www.ncbi.nlm.nih.gov/geo/), accession numbers GSE57338, GSE5406, GSE135055, GSE37171, and GSE97709.
Author contributions
RB and XX conceived and designed the study. RB performed data analysis and data interpretation. RB and WL conducted bioinformatics and statistical analyses. RB and WL wrote the first draft, and XX conceptualized and revised the manuscript. All authors have read, edited, and approved the final manuscript.
Funding
This study was funded by the Henan Province Medical Science and Technology Research Program (LHGJ20220896) and Henan Province Traditional Chinese Medicine Scientific Research Special Program (2022ZYZD022).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1037520/full#supplementary-material
Abbreviations
AUC, area under the curve; Boruta, Boruta algorithm; CHEA3, ChIP-X Enrichment Analysis 3; DEG, differentially expressed gene; DSigDB, Drug Signatures Database; ESRD, end-stage renal disease; GEO, Gene Expression Omnibus; GSVA, gene set variation analysis; GO, Gene Ontology; HF, heart failure; KEGG, Kyoto Encyclopedia of Genes and Genomes; LASSO, logical regression of the selection operator; PPI, protein−protein interaction; PR, precision–recall; RF, random forest; ROC, receiver operating characteristic; SVM-RFE, support vector machine-recursive feature elimination; XGBoost, eXtreme Gradient Boosting.
References
Alakwaa, F. M., Chaudhary, K., and Garmire, L. X. (2018). Deep learning accurately predicts estrogen receptor status in breast cancer metabolomics data. J. proteome Res. 17 (1), 337–347. doi:10.1021/acs.jproteome.7b00595
Alhaj, E., Alhaj, N., Rahman, I., Niazi, T. O., Berkowitz, R., and Klapholz, M. (2013). Uremic cardiomyopathy: An underdiagnosed disease. Congest. heart Fail. (Greenwich, Conn) 19 (4), E40–E45. doi:10.1111/chf.12030
Alhamzawi, R., and Ali, H. T. M. (2018). The bayesian adaptive Lasso regression. Math. Biosci. 303, 75–82. doi:10.1016/j.mbs.2018.06.004
Batista, A. M., Alvarado-Arnez, L. E., Alves, S. M., Melo, G., Pereira, I. R., Ruivo, L. A. S., et al. (2018). Genetic polymorphism at Ccl5 is associated with protection in chagas' heart disease: Antagonistic participation of Ccr1(+) and Ccr5(+) cells in chronic chagasic cardiomyopathy. Front. Immunol. 9, 615. doi:10.3389/fimmu.2018.00615
Betjes, M. G. (2013). Immune cell dysfunction and inflammation in end-stage renal disease. Nat. Rev. Nephrol. 9 (5), 255–265. doi:10.1038/nrneph.2013.44
Carbon, S., Ireland, A., Mungall, C. J., Shu, S., Marshall, B., Lewis, S., et al. (2009). Amigo: Online access to Ontology and annotation data. Bioinforma. Oxf. Engl. 25 (2), 288–289. doi:10.1093/bioinformatics/btn615
Chou, C. H., Shrestha, S., Yang, C. D., Chang, N. W., Lin, Y. L., Liao, K. W., et al. (2018). Mirtarbase update 2018: A resource for experimentally validated microrna-target interactions. Nucleic acids Res. 46 (D1), D296-D302–d302. doi:10.1093/nar/gkx1067
Costanzo, M. R. (2020). The cardiorenal syndrome in heart failure. Heart Fail. Clin. 16 (1), 81–97. doi:10.1016/j.hfc.2019.08.010
Dounousi, E., Duni, A., Naka, K. K., Vartholomatos, G., and Zoccali, C. (2021). The innate immune system and cardiovascular disease in eskd: Monocytes and natural killer cells. Curr. Vasc. Pharmacol. 19 (1), 63–76. doi:10.2174/1570161118666200628024027
Elmoselhi, H., Mansell, H., Soliman, M., and Shoker, A. (2016). Circulating chemokine ligand levels before and after successful kidney transplantation. J. Inflamm. Lond. Engl. 13, 32. doi:10.1186/s12950-016-0141-4
Gao, C. H., Yu, G., and Cai, P. (2021). Ggvenndiagram: An intuitive, easy-to-use, and highly customizable R package to generate Venn diagram. Front. Genet. 12, 706907. doi:10.3389/fgene.2021.706907
Gordon, J. W., Shaw, J. A., and Kirshenbaum, L. A. (2011). Multiple facets of nf-?b in the heart: To Be or not to nf-?b. Circulation Res. 108 (9), 1122–1132. Epub 2011/04/30. doi:10.1161/circresaha.110.226928
Han, Z., Yu, Y., Cai, B., Xu, Z., Bao, Z., Zhang, Y., et al. (2020). Yap/Tead3 signal mediates cardiac lineage commitment of human-induced pluripotent stem cells. J. Cell. physiology 235 (3), 2753–2760. Epub 2019/09/22. doi:10.1002/jcp.29179
Hänzelmann, S., Castelo, R., and Guinney, J. (2013). Gsva: Gene set variation analysis for microarray and rna-seq data. BMC Bioinforma. 14, 7. Epub 2013/01/18. doi:10.1186/1471-2105-14-7
Ito, K., and Murphy, D. (2013). Application of Ggplot2 to pharmacometric graphics. CPT pharmacometrics Syst. Pharmacol. 2 (10), e79. doi:10.1038/psp.2013.56
Kanehisa, M., and Goto, S. (2000). Kegg: Kyoto Encyclopedia of genes and Genomes. Nucleic acids Res. 28 (1), 27–30. doi:10.1093/nar/28.1.27
Karagkouni, D., Paraskevopoulou, M. D., Chatzopoulos, S., Vlachos, I. S., Tastsoglou, S., Kanellos, I., et al. (2018). Diana-tarbase V8: A decade-long collection of experimentally supported mirna-gene interactions. Nucleic acids Res. 46 (D1), D239-D245–d45. Epub 2017/11/21. doi:10.1093/nar/gkx1141
Keenan, A. B., Torre, D., Lachmann, A., Leong, A. K., Wojciechowicz, M. L., Utti, V., et al. (2019). Chea3: Transcription factor enrichment analysis by orthogonal omics integration. Nucleic acids Res. 47 (W1), W212-W224–w24. Epub 2019/05/23. doi:10.1093/nar/gkz446
Kim, J., Kim, J., and Rhee, K. (2019). Pcnt is critical for the association and conversion of centrioles to centrosomes during mitosis. J. Cell. Sci. 132 (6), jcs225789. Epub 2019/03/01. doi:10.1242/jcs.225789
Krensky, A. M., and Ahn, Y. T. (2007). Mechanisms of disease: Regulation of rantes (Ccl5) in renal disease. Nat. Clin. Pract. Nephrol. 3 (3), 164–170. doi:10.1038/ncpneph0418
Kursa, M. B. (2014). Robustness of random forest-based gene selection methods. BMC Bioinforma. 15, 8. doi:10.1186/1471-2105-15-8
Li, H., Chen, C., and Wang, D. W. (2021). Inflammatory cytokines, immune cells, and organ interactions in heart failure. Front. physiology 12, 695047. Epub 2021/07/20. doi:10.3389/fphys.2021.695047
Li, J. H., Liu, S., Zhou, H., Qu, L. H., and Yang, J. H. (2014). Starbase V2.0: Decoding mirna-cerna, mirna-ncrna and protein-rna interaction networks from large-scale clip-seq data. Nucleic acids Res. 42, D92–D97. doi:10.1093/nar/gkt1248
Liang, S., Shi, X., Yu, C., Shao, X., Zhou, H., Li, X., et al. (2020). Identification of novel candidate genes in heterotaxy syndrome patients with congenital heart diseases by whole exome sequencing. Biochimica biophysica acta Mol. basis Dis. 1866 (12), 165906. doi:10.1016/j.bbadis.2020.165906
Lin, X., Yang, F., Zhou, L., Yin, P., Kong, H., Xing, W., et al. (2012). A support vector machine-recursive feature elimination feature selection method based on artificial contrast variables and mutual information. J. Chromatogr. B, Anal. Technol. Biomed. life Sci. 910, 149–155. doi:10.1016/j.jchromb.2012.05.020
Liu, R., and Jin, J. P. (2016). Calponin isoforms Cnn1, Cnn2 and Cnn3: Regulators for actin cytoskeleton functions in smooth muscle and non-muscle cells. Gene 585 (1), 143–153. doi:10.1016/j.gene.2016.02.040
Liu, X., Shi, G. P., and Guo, J. (2021). Innate immune cells in pressure overload-induced cardiac hypertrophy and remodeling. Front. Cell. Dev. Biol. 9, 659666. doi:10.3389/fcell.2021.659666
Lu, D., Zhang, L., Bao, D., Lu, Y., Zhang, X., Liu, N., et al. (2014). Calponin1 inhibits dilated cardiomyopathy development in mice through the epkc pathway. Int. J. Cardiol. 173 (2), 146–153. doi:10.1016/j.ijcard.2014.02.032
Ma, Y., Chiao, Y. A., Clark, R., Flynn, E. R., Yabluchanskiy, A., Ghasemi, O., et al. (2015). Deriving a cardiac ageing signature to reveal mmp-9-dependent inflammatory signalling in senescence. Cardiovasc. Res. 106 (3), 421–431. Epub 2015/04/18. doi:10.1093/cvr/cvv128
Naumnik, B., Koc-Żórawska, E., and Myśliwiec, M. (2013). Over-dialysis plasma rantes increase depends on heparin dose and cardiovascular disease status. Adv. Med. Sci. 58 (2), 311–319. Epub 2013/08/21. doi:10.2478/ams-2013-0008
Newman, A. M., Liu, C. L., Green, M. R., Gentles, A. J., Feng, W., Xu, Y., et al. (2015). Robust enumeration of cell subsets from tissue expression profiles. Nat. methods 12 (5), 453–457. Epub 2015/03/31. doi:10.1038/nmeth.3337
Ni, J., Wu, Q. Q., Liao, H. H., Fan, D., and Tang, Q. Z. (2019). Bcl6 suppresses cardiac fibroblast activation and function via directly binding to Smad4. Curr. Med. Sci. 39 (4), 534–540. Epub 2019/07/28. doi:10.1007/s11596-019-2070-y
Ogunleye, A., and Wang, Q. G. (2020). Xgboost model for chronic kidney disease diagnosis. IEEE/ACM Trans. Comput. Biol. Bioinforma. 17 (6), 2131–2140. Epub 2019/04/19. doi:10.1109/tcbb.2019.2911071
Qian, L., Zhao, Q., Yu, P., Lü, J., Guo, Y., Gong, X., et al. (2022). Diagnostic potential of a circulating mirna model associated with therapeutic effect in heart failure. J. Transl. Med. 20 (1), 267. Epub 2022/06/12. doi:10.1186/s12967-022-03465-w
Ramos-Kuri, M., Meka, S. H., Salamanca-Buentello, F., Hajjar, R. J., Lipskaia, L., and Chemaly, E. R. (2021). Molecules linked to Ras signaling as therapeutic targets in cardiac pathologies. Biol. Res. 54 (1), 23. Epub 2021/08/05. doi:10.1186/s40659-021-00342-6
Rangaswami, J., Bhalla, V., Blair, J. E. A., Chang, T. I., Costa, S., Lentine, K. L., et al. (2019). Cardiorenal syndrome: Classification, pathophysiology, diagnosis, and treatment strategies: A scientific statement from the American heart association. Circulation 139 (16), e840–e878. doi:10.1161/cir.0000000000000664
Rangaswami, J., and McCullough, P. A. (2018). Heart failure in end-stage kidney disease: Pathophysiology, diagnosis, and therapeutic strategies. Seminars Nephrol. 38 (6), 600–617. Epub 2018/11/11. doi:10.1016/j.semnephrol.2018.08.005
Ren, Y., Wang, X., Liang, H., He, W., and Zhao, X. (2021). Mechanism of mir-30b-5p-loaded peg-plga nanoparticles for targeted treatment of heart failure. Front. Pharmacol. 12, 745429. Epub 2021/10/19. doi:10.3389/fphar.2021.745429
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). Limma powers differential expression analyses for rna-sequencing and microarray studies. Nucleic acids Res. 43 (7), e47. Epub 2015/01/22. doi:10.1093/nar/gkv007
Sanz, H., Valim, C., Vegas, E., Oller, J. M., and Reverter, F. (2018). Svm-Rfe: Selection and visualization of the most relevant features through non-linear kernels. BMC Bioinforma. 19 (1), 432. Epub 2018/11/21. doi:10.1186/s12859-018-2451-4
Savarese, G., and Lund, L. H. (2017). Global public health burden of heart failure. Card. Fail. Rev. 3 (1), 7–11. Epub 2017/08/09. doi:10.15420/cfr.2016:25:2
Sawant, D. V., Sehra, S., Nguyen, E. T., Jadhav, R., Englert, K., Shinnakasu, R., et al. (1950)., 189. Baltimore, Md, 4759–4769. Epub 2012/10/12. doi:10.4049/jimmunol.1201794Bcl6 controls the Th2 inflammatory activity of regulatory T cells by repressing Gata3 functionJ. Immunol.10
Schefold, J. C., Filippatos, G., Hasenfuss, G., Anker, S. D., and von Haehling, S. (2016). Heart failure and kidney dysfunction: Epidemiology, mechanisms and management. Nat. Rev. Nephrol. 12 (10), 610–623. Epub 2016/08/31. doi:10.1038/nrneph.2016.113
Severino, P., D'Amato, A., Pucci, M., Infusino, F., Birtolo, L. I., Mariani, M. V., et al. (2020). Ischemic heart disease and heart failure: Role of coronary ion channels. Int. J. Mol. Sci. 21 (9), 3167. Epub 2020/05/06. doi:10.3390/ijms21093167
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. Epub 2003/11/05. doi:10.1101/gr.1239303
Silverberg, D., Wexler, D., Blum, M., Schwartz, D., and Iaina, A. (2004). The association between congestive heart failure and chronic renal disease. Curr. Opin. Nephrol. Hypertens. 13 (2), 163–170. Epub 2004/06/19. doi:10.1097/00041552-200403000-00004
Song, N., Thaiss, F., and Guo, L. (2019). Nfκb and kidney injury. Front. Immunol. 10, 815. Epub 2019/05/02. doi:10.3389/fimmu.2019.00815
Stevens, T. W., Khalaf, F. K., Soehnlen, S., Hegde, P., Storm, K., Meenakshisundaram, C., et al. (2022). Dirty jobs: Macrophages at the heart of cardiovascular disease. Biomedicines 10 (7), 1579. Epub 2022/07/28. doi:10.3390/biomedicines10071579
Stevenson, M. D., Canugovi, C., Vendrov, A. E., Hayami, T., Bowles, D. E., Krause, K. H., et al. (2019). Nadph oxidase 4 regulates inflammation in ischemic heart failure: Role of soluble epoxide hydrolase. Antioxidants redox Signal. 31 (1), 39–58. Epub 2018/11/20. doi:10.1089/ars.2018.7548
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102 (43), 15545–15550. doi:10.1073/pnas.0506580102
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). String V11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic acids Res. 47 (D1), D607-D613–d13. Epub 2018/11/27. doi:10.1093/nar/gky1131
Takahashi, K., Hiwada, K., and Kokubu, T. (1986). Isolation and characterization of a 34, 000-dalton calmodulin- and F-Actin-Binding protein from chicken gizzard smooth muscle. Biochem. biophysical Res. Commun. 141 (1), 20–26. Epub 1986/11/26. doi:10.1016/s0006-291x(86)80328-x
Tan, F. L., Moravec, C. S., Li, J., Apperson-Hansen, C., McCarthy, P. M., Young, J. B., et al. (2002). The gene expression fingerprint of human heart failure. Proc. Natl. Acad. Sci. U. S. A. 99 (17), 11387–11392. Epub 2002/08/15. doi:10.1073/pnas.162370099
Tettamanti, L., Caraffa, A., Mastrangelo, F., Ronconi, G., Kritas, S., Frydas, I., et al. (2018). Different signals induce mast cell inflammatory activity: Inhibitory effect of Vitamin E. J. Biol. Regul. Homeost. agents 32 (1), 13–19. Epub 2018/03/06.
Voronova, A., Al Madhoun, A., Fischer, A., Shelton, M., Karamboulas, C., and Skerjanc, I. S. (2012). Gli2 and Mef2c activate each other's expression and function synergistically during cardiomyogenesis in vitro. Nucleic acids Res. 40 (8), 3329–3347. Epub 2011/12/27. doi:10.1093/nar/gkr1232
Wang, F., Zhang, J., Niu, G., Weng, J., Zhang, Q., Xie, M., et al. (2022). Apigenin inhibits isoproterenol-induced myocardial fibrosis and smad pathway in mice by regulating oxidative stress and mir-122-5p/155-5p expressions. Drug Dev. Res. 83 (4), 1003–1015. Epub 2022/03/13. doi:10.1002/ddr.21928
Yoshida, T., Fukuda, T., Hatano, M., Koseki, H., Okabe, S., Ishibashi, K., et al. (1999). The role of Bcl6 in mature cardiac myocytes. Cardiovasc. Res. 42 (3), 670–679. Epub 1999/10/26. doi:10.1016/s0008-6363(99)00007-3
Zeng, Z., Lan, T., Wei, Y., and Wei, X. (2022). Ccl5/Ccr5 Axis in human diseases and related treatments. Genes. & Dis. 9 (1), 12–27. Epub 2021/09/14. doi:10.1016/j.gendis.2021.08.004
Keywords: heart failure, end-stage renal disease, hub gene, bioinformatics analysis, immune infiltration
Citation: Bian R, Xu X and Li W (2023) Uncovering the molecular mechanisms between heart failure and end-stage renal disease via a bioinformatics study. Front. Genet. 13:1037520. doi: 10.3389/fgene.2022.1037520
Received: 06 September 2022; Accepted: 20 December 2022;
Published: 10 January 2023.
Edited by:
Ahmed Rebai, Centre of Biotechnology of Sfax, TunisiaReviewed by:
Najla Kharrat, Centre of Biotechnology of Sfax, TunisiaQing Qin, Fudan University, China
Copyright © 2023 Bian, Xu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuegong Xu, Mjg2NDMwNTE3QHFxLmNvbQ==
 Rutao Bian
Rutao Bian Xuegong Xu*
Xuegong Xu*