- 1Department of Pathology, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Clinical Tuberculosis and Epidemiology Research Center, National Research Institute of Tuberculosis and Lung Diseases (NRITLD), Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 3Dental Research Center, Dentistry Research Institute, Tehran University of Medical Sciences, Tehran, Iran
- 4School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 5Chronic Respiratory Diseases Research Center, National Research Institute of Tuberculosis and Lung Diseases (NRITLD), Shahid Beheshti University of Medical Science, Tehran, Iran
- 6Department of Biology, Islamic Azad University, Damghan, Iran
- 7School of Medicine, Department of Pharmacology, Shahid Beheshti University of Medical Sciences, Tehran, Iran
The pandemic of coronavirus disease in 2019 has led to a global crisis. COVID-19 shows distinct clinical manifestations of the severity of symptoms. Numerous patients with no associated risk factors demonstrate acute respiratory distress syndrome (ARDS). The role of genetic factors in determining the severity and outcome of the disease remains unresolved. The purpose of this study was to see if a correlation exists between Angiotensin I Converting Enzyme (ACE) insertion/deletion (I/D) polymorphism and the severity of COVID-19 patients’ symptoms. 120 COVID-19 patients admitted to Masih Daneshvari Hospital in Tehran with their consent to participate entered the study. Based on the World Health Organization classification, patients were divided into moderate and severe groups, which were primarily affected by O2 saturation levels. The effects of the patients’ ACE insertion/deletion polymorphism, background disease, Angiotensin receptor blocker (ARB) drug consumption, and demographic parameters on the severity risk were calculated statistically. The ACE D allele was associated with an increased risk of disease severity (OR = 6.766, p = 0.012), but had no effect on mortality.
Introduction
COVID-19 cases show distinct clinical manifestations, most of which showing no symptoms or mild symptoms (Wu and McGoogan 2020), whereas a small proportion experience a severe form of disease resulting in severe acute respiratory syndrome. This condition is linked to high death rate (Grasselli, Zangrillo et al., 2020; Richardson, Hirsch et al., 2020). Adverse outcome is associated with some demographic characteristics, such as male gender (Zheng, Peng et al., 2020), age over 65 (Zheng, Peng et al., 2020), and being African-American (Doumas, Patoulias et al., 2020). Poor outcome is also associated with particular comorbidities, such as diabetes (Rod, Oviedo-Trespalacios et al., 2020), hypertension (Lippi, Wong et al., 2020), and body mass index (BMI) of 30 and above (Hussain, Mahawar et al., 2020), which is defined as obesity. Despite this, numerous patients without these characteristics have demonstrated acute respiratory distress syndrome (ARDS) (Zheng and Cao 2020). In the interim, the role of genetic factors in determining the severity and outcome of the disease remains unanswered (Ghafouri-Fard, Noroozi et al., 2020). Furthermore, there are many and varied factors involved in this process, such as age, environmental impacts, and the aforementioned risk factors. Therefore, further investigations are needed to assess the role of genetic susceptibility.
The novel coronavirus 2019 enters the host cells via angiotensin-converting enzyme 2 (ACE2) receptor. ACE2 is also a component of the renin-angiotensin (RAS) system and has a close interplay with ACE (Wu, Deng et al., 2020; Zhou, Yang et al., 2020). SARS-CoV epidemic began in 2002. SARS-CoV-2 and SARS-CoV have similar pathogenesis routes as they both bind to ACE2 to enter the host cells (Shi, Wang et al., 2020). In the case of SARS-CoV infection, as the virus binds to the ACE2 receptor, it can enter the cell and consequently causes down regulation of ACE2 (Ni, Yang et al., 2020). These processes are repeated in COVID-19 infection, indicating the call for a more comprehensive investigation of the role of RAS modulation in COVID-19 patients (Zheng and Cao 2020).
Angiotensin I is transformed to angiotensin II (Ang II) through ACE activity (Baudin 2002), after which Ang II is converted to Ang-(1–7) via ACE2 activity (Donoghue, Hsieh et al., 2000; Tipnis, Hooper et al., 2000). In the context of lowered ACE2 expression or raised ACE activity that happens in SARS-CoV-2 infection, an unrestricted level of Ang II is induced which, in turn, may bring about acute lung injury through several processes (Zheng and Cao 2020), (Figure 1).
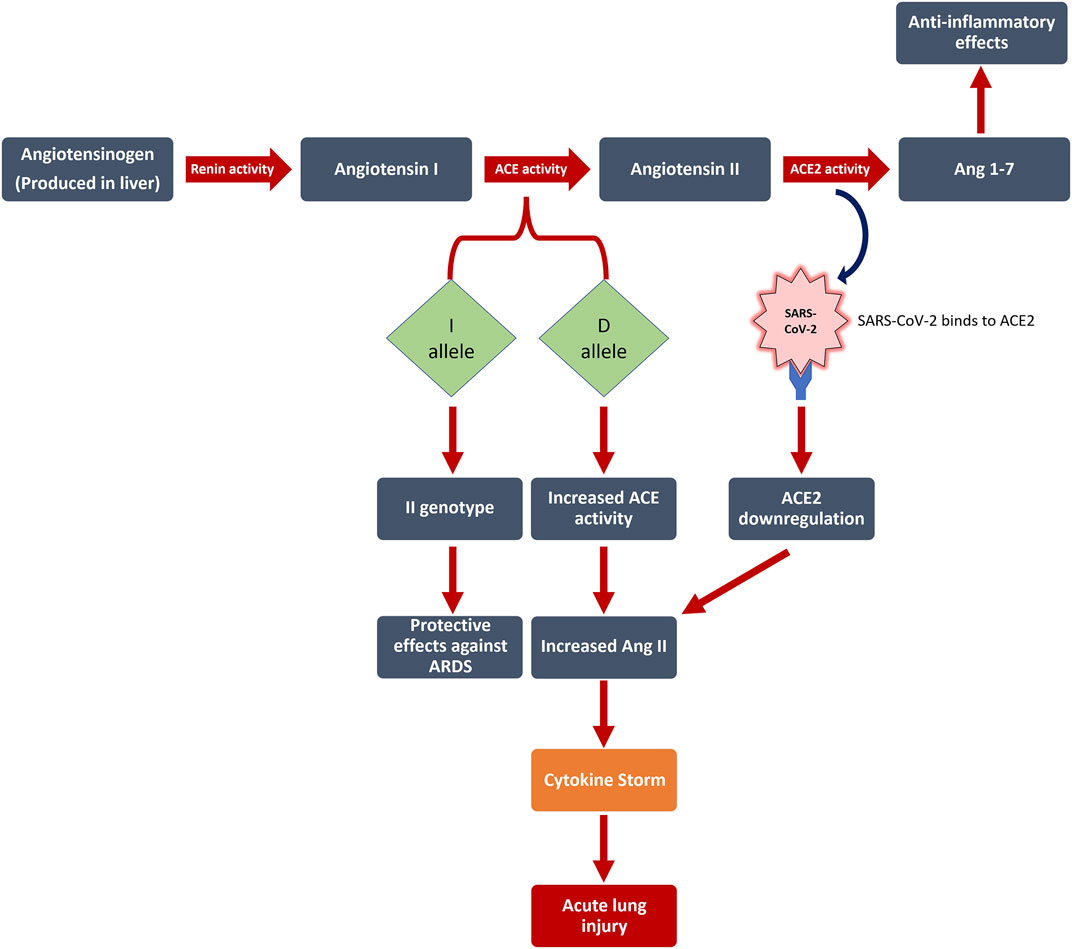
FIGURE 1. The interplay between RAS, Ang II levels, and SARS-CoV-2 infection is illustrated in this figure. Numerous patients with no associated risk factors demonstrate acute respiratory distress syndrome (ARDS). The role of genetic factors in determining the severity and outcome of COVID-19 disease remains unresolved. We discovered that the Angiotensin I Converting Enzyme (ACE) insertion/deletion (I/D) polymorphism affects the severity of patients’ symptoms, with the D allele increasing the risk of disease severity by 6.8 (p = 0.012), but having no effect on mortality. ACE: Angiotensin I Converting Enzyme; ACE2: Angiotensin-Converting Enzyme two; Ang 1–7: Angiotensin 1–7; D allele: Deletion allele; I allele: Insertion allele; ARDS: acute respiratory distress syndrome; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
Unrestricted levels of Ang II may have disruptive effects related to lung injury, and yet Ang II is produced by ACE activity. Thus, it can be concluded that ACE activity levels may also have an impact.
ACE is an enzyme, which can be found profusely in neuroepithelial, epithelial, and endothelial cells (Ziai, Salehian et al., 2004). ACE has kinase activity and is a zinc-dependent metallopeptidase (Mohammadpour; Ziai et al., 2020). ACE has a main role in regulation of blood pressure as well (Kouchmeshky, Jameie et al., 2012). The ACE gene is located on the 17q23 locus as assigned by Mattei et al., in 1989 (Mattei, Hubert et al., 1989). The polymorphism of the ACE gene is defined by the deletion (D) or insertion (I) of the 287-bp-long Alu element in the ACE gene, specifically in intron 16. The D allele is associated to the elevated activity of the ACE (Zheng and Cao 2020). Altogether, it is suggested that the polymorphism of ACE gene may have a prominent role to play in determining the clinical outcomes or severity of COVID-19 cases.
In the present study, we aim to identify if ACE insertion/deletion polymorphism plays a role in mortality and severity of patients’ symptoms.
Materials and methods
This study was approved by the Ethics Committee of Shahid Beheshti University of Medical Sciences, Tehran, Iran, with the code IR. SBMU.MSP.REC.1399.519. All the performed experiments were in compliance with the relevant guidelines.
1) Study cohorts: Of the patients admitted to Masih Daneshvari Hospital in Tehran, 120 patients who tested positive for the COVID-19 polymerase chain reaction (PCR) test entered the study with their informed consent to participate. Demographic and anthropometric data and medical history of patients were obtained and recorded over the course of history taking. Recorded items included sex, age, existing comorbidities, such as diabetes mellitus, cardiovascular diseases, respiratory diseases, hypertension (systolic pressure ≥130 mm Hg or diastolic pressure ≥80 mm Hg), high BMI -in this study defined as BMI ≥30, which includes obese category according to Centers for Disease Control and Prevention (CDC), ((CDC) 2020), smoking and use of certain medications such as angiotensin II receptor blockers (ARBs), and ACE inhibitors (ACEI). Finally, the ultimate outcome of patients (recovery or death) was recorded.
According to World Health Organization (WHO) classification of COVID-19 severity, patients were divided into four groups based on the severity level of their condition. Mild condition is defined as patients meeting case definition criteria showing symptoms of COVID-19 infection, such as sore throat, cough, shortness of breath, fever, malaise, muscular pain, headache, nausea, vomiting, loss of smell and taste, diarrhea, anorexia, and nasal congestion but with no signs of hypoxia or viral pneumonia. Moderate condition is defined as having clinical signs of pneumonia, including dyspnea, tachypnea, cough, and fever, but not showing signs of severe form of pneumonia, including oxygen saturation (SpO2) ≥ 90% on room air. Severe condition is defined as manifestation of clinical signs of pneumonia, such as dyspnea, cough, fever in addition to one of the following criteria: severe respiratory distress, respiratory rate >30 per minute, or SpO2 < 90% on room air. Critical condition is defined by having the criteria for ARDS, septic shock, sepsis, or other settings that would generally necessitate providing the life-saving treatments, such as invasive or non-invasive mechanical ventilation or vasopressor administration (World Health Organization 2022). Since individuals with mild disease were managed in out-patient settings, we did not have access to them within the in-patient admissions. Patients with moderate disease were assigned to the moderate group, while the severe and critical groups were assigned to the severe group.
2) DNA extraction: In order to determine the polymorphism of the ACE gene (II/ID/DD) in patients, 5 ml of blood was collected from each participant. Blood samples were centrifuged at 2500xg for 15 min. The supernatant plasma layer was then discarded and for every 1 ml of blood, 3 ml of red blood cell (RBC) lysis solution was added to the remaining part. BioFact™ Genomic DNA Prep Kit (South Korea) was used for DNA extraction step. The purity and quantity of the extracted DNA was finally measured by the NanoDrop 1000 device and the data for each sample were recorded.
3) Genotyping: Gap-PCR method was conducted. After DNA extraction, Gap-PCR was performed to determine the genotype of I/D polymorphism of the ACE gene. Taq DNA Polymerase Master Mix (Ampliqon, Denmark) was used for carrying out the PCR. The reaction components of the PCR consisted of 3 μl of DNA sample, 10 μl of master mix (X2), 1 μl of forward and reverse primer solution, and 6 μl of DNase-RNase free water. The reaction was performed with regard to PCR thermal profile. Afterwards, agarose gel electrophoresis was used to identify the existing alleles and the results were examined under UV-transilluminators. For this step 6x loading dye-MM2121 (Sinaclon, Iran), and Ladder 50 bp (ready to use) 50-1500 bp and Ladder 100bp Plus (ready to use) 100-3000 bp (Sinaclon, Iran) were used for dyeing, and as the markers respectively. The primers’ sequence was as follows:
4) Statistical analysis: For risk estimates (odds ratio (OR)), the Pearson’s Chi-square test was used. Scale variables were tested for normal distribution using Kolmogorov–Smirnov, and independent t-tests were used to compare the mean of normal distributed variables. The Mann Whitney U test was used to compare non-parametric variables. For modeling and calculating adjusted OR, binary logistic regression (Enter method) was used. In this study, IBM SPSS software version 16 was used.
Results
In this study, 120 COVID-19 patients (61 males, 59 females) were studied, with a mean age of 52.1 ± 18.94 years (range: 14–93 years), a mean height of 1.66 ± 0.088 m (range: 1.47–1.83), and a mean BMI of 28.1 ± 6.37 (range: 15.79–49.95). 7 (5.8%) of the studied patients were smokers, 23 (19.2%) had diabetes mellitus (DM), 14 (11.7%) had cardiovascular disease (CVD), 6 (5.0%) had respiratory disease (RD), 38 (31.7%) had hypertension (HTN), and 25 (20.8%) were taking ARB drugs. Of all patients, 74 (61%) were admitted to the intensive care unit (ICU). In our study, 26, 21, and 73 patients had moderate, severe, and critical form of the disease, respectively. We genotyped all 120 patients to determine the I/D polymorphism of the ACE gene (gel electrophoresis of 19 patients is shown in Figure 2). In this study the genotypes of 120 patients were determined and 24, 82, and 14 patients were identified with DD, ID, and II genotypes respectively.

FIGURE 2. Gel electrophoresis showing genotypes of 19 patients. In this study 120 patients were genotyped and 24, 82, and 14 patients were identified with DD, ID, and II genotypes respectively. ACE polymorphism genotypes (DD/ID/II) of patients 51–69 are indicated in this figure.
Patients carrying the ACE D allele had a higher risk of disease severity (OR = 6.519, p = 0.001), and it was an independent variable in disease severity modeling using the logistic regression method (OR = 6.766, p = 0.012), (Table 1).
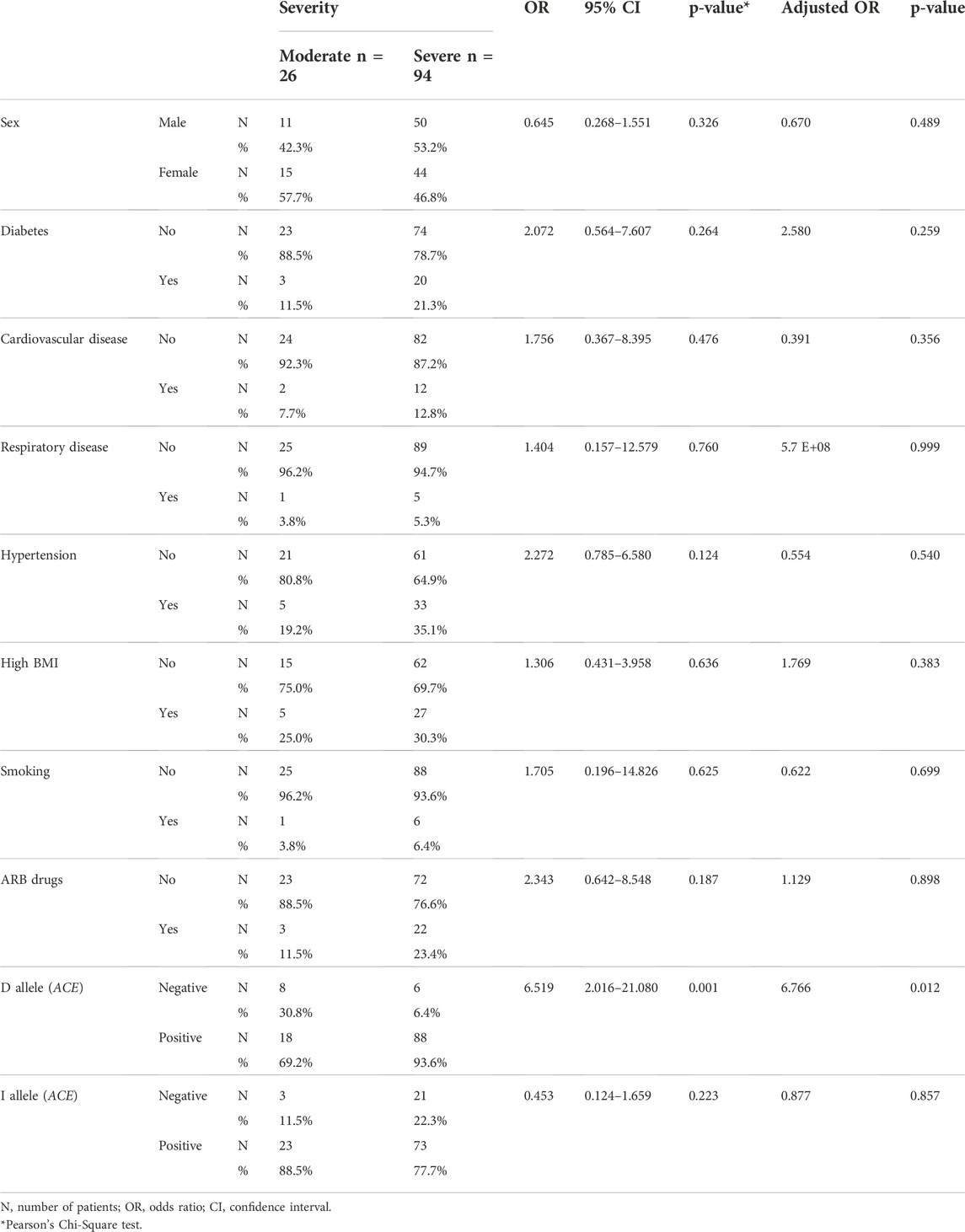
TABLE 1. Distribution of COVID-19 patients parameters based on disease severity. Adjusted odds ratio was calculated based on binary logistic regression analysis.
Patients with severe disease had a significantly higher mean age (54.32 ± 18.96 years) than those in the moderate group (44.23 ± 16.91 years) (Independent t-test, p = 0.016), Figure 3. Patients with moderate and severe disease had mean BMIs of 27.81 ± 6.09 and 28.28 ± 6.46, respectively (non-significant) which is shown in Table 2.
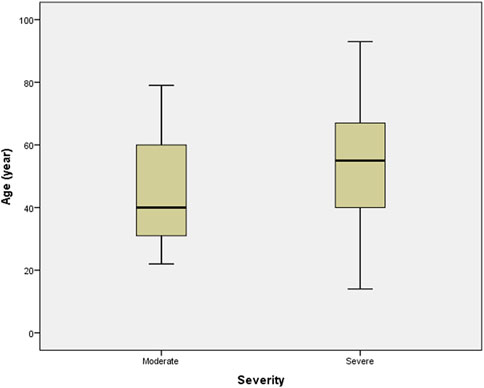
FIGURE 3. Distribution of the patients’ age among severity of the disease groups. Boxplots show the median and interquartile range, and whiskers represent the extreme cases of individual variables. The mean age of individuals in severe disease group was significantly higher than that of in the moderate disease group (p = 0.016).
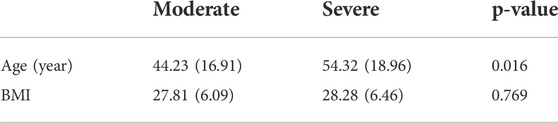
TABLE 2. Distribution of age and BMI among the disease groups (severe and moderate). Data show mean value (standard deviation) and p values were calculated based on two independent sample student t test.
Table 3 shows that while background disease (DM, HTN, CVD, and RD) did not increase the risk of COVID-19 severity, O2 saturation did. A significantly higher proportion of patients with moderate than severe disease have the ACE II polymorphism (OR = 0.153; p = 0.002).
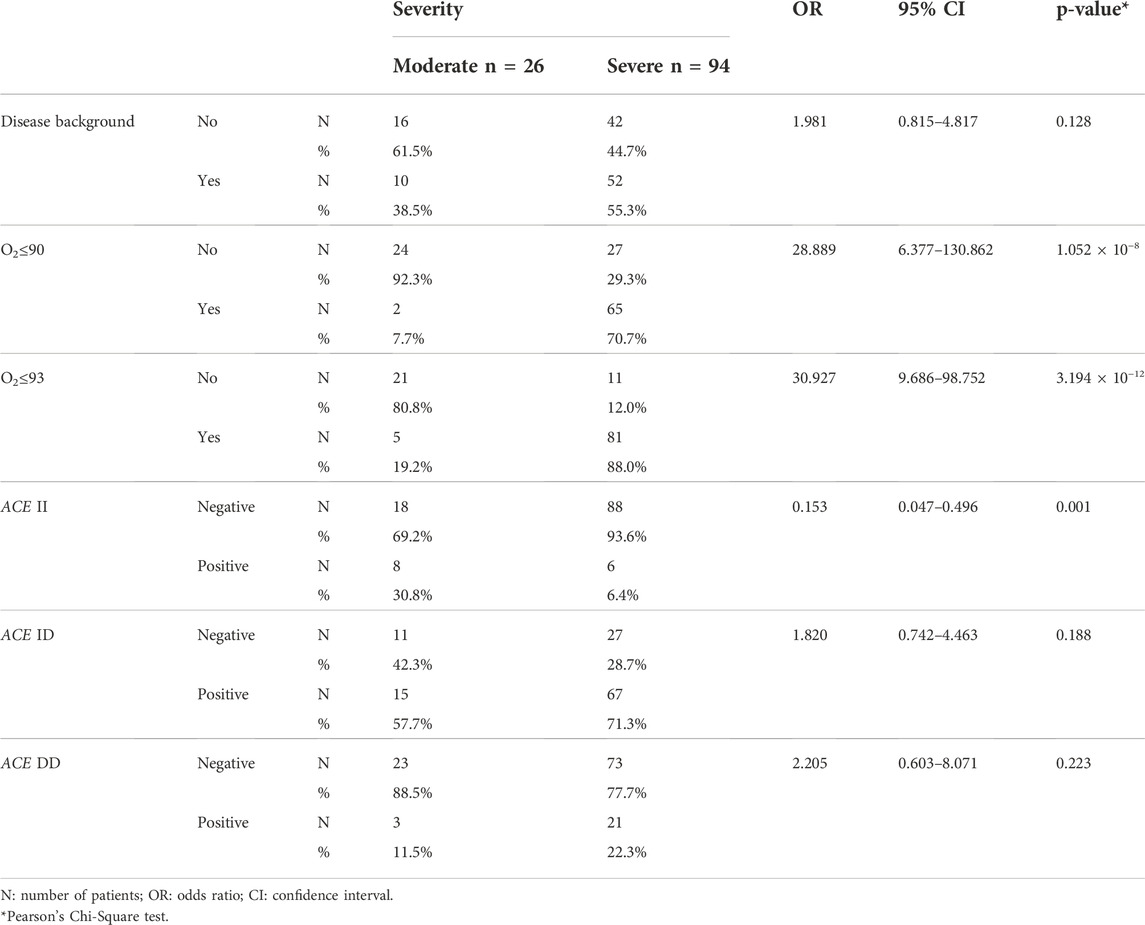
TABLE 3. Analysis of the effects of background disease (the sum of the disease mentioned in Table 1), O2 saturation, and ACE polymorphism on disease severity.
Based on the patient outcome (dead or recovered), our findings were consistent with others in that background disease increased the risk of mortality (OR = 3.469, p = 0.008), with hypertension being a significant risk factor (OR = 3.096, p = 0.01). The ACE polymorphism, on the other hand, had no effect on mortality. Recovered patients were younger than deceased patients (49.51 ± 18.31 vs. 61.19 ± 18.55 p = 0.004).
Discussion
Two factors that play a key role in RAS modulation, are ACE and ACE2 (Crowley, Gurley et al., 2005). Since the equilibrium between ACE and ACE2 is crucial in homeostasis of RAS, it seems that ACE and ACE2 have a mutual interaction to achieve this (Zheng and Cao 2020).
Unrestricted levels of Ang II can lead to acute lung injury through several processes (Zheng and Cao 2020). These include provoking fibrosis (Marshall, Gohlke et al., 2004), promoting apoptosis of alveolar epithelial cells and endothelial cells (Dimmeler, Rippmann et al., 1997; Wang, Feng et al., 2000), increasing vasculature permeability and subsequent vasoconstriction (Zheng and Cao 2020), and finally, augmentation of pro-inflammatory cytokines, such as interleukin 6 (IL-6) and IL-8 (Suzuki, Ruiz-Ortega et al., 2003). IL-6 is a main factor of the cytokine storm and plays an important role in the severity of COVID-19 patients (Hadjadj, Yatim et al., 2020; Mahmoudi; Rezaei et al., 2020).
Our study found that in the severity outcome, the ACE D allele and older age are risk factors for disease severity, but in the mortality outcome, background disease, specifically hypertension, and older age are risk factors.
Studies on animal models and humans advocate the role of raised levels of Ang II caused by ACE and ACE2 imbalances in pathogenesis of acute lung injury (Zheng and Cao 2020).
In a cohort study of COVID-19 patients, plasma levels of Ang II were found to be meaningfully higher than the healthy control group. In addition, Ang II levels showed a linear correlation with the severity of the injury and the virus burden (Liu, Yang et al., 2020). This piece of evidence suggests that SARS-CoV-2 infection results in increased amount of ACE molecule or activity. In a study on a rat model, high-volume ventilation was employed to foster lung injury, and subsequently, aggravated lung injury which correlated with lung Ang II overproduction, was observed (Jerng, Hsu et al., 2007). As for influenza, the animal model manifested severe lung injury, seemingly correlated with decreased expression of ACE2 (Yang, Gu et al., 2014). In another mouse model, SARS-CoV binding to ACE2 in vivo showed declined expression of ACE2 and augmented acute lung injury (Kuba, Imai et al., 2005). However, some studies have demonstrated different results. Biancatelli et al. have shown that exposure of subunit one of the SARS-CoV-2 spike protein (S1SP) to in-house harvested human lung microvascular endothelial cells does not change ACE2 expression significantly (Colunga Biancatelli, Solopov et al., 2022). Another study conducted by Solopov et al. on homogenates of lung tissue of mice which were on alcohol diet versus those on control diet, showed alcohol increased ACE2 expression in alcohol-consumption group but ACE2 expression was not further affected by S1SP exposure (Solopov, Colunga Biancatelli et al., 2022).
A large body of evidence exists that supports the link between ACE genotype and severity of ARDS outcomes (Marshall, Webb et al., 2002; Pati, Mahto et al., 2020; Sarangarajan, Winn et al., 2020; Yamamoto, Ariumi et al., 2020). In individuals with DD genotype, the average levels of ACE activity were nearly twice as high as the II genotype. This insertion/deletion (I/D) polymorphism constitutes about half of the diversity of ACE tissue and serum levels observed among individuals (Zheng and Cao 2020).
Our results showed that a higher proportion of patients with severe disease had the D allele. In a meta-analysis study in 2015, it was observed that I/D polymorphism could be a risk factor if the genotype was DD, and the DD genotype showed an elevated risk in ARDS development compared to ID and II genotypes (Deng, Zhang et al., 2015). Another study by Tsantes et al. on patients with acute lung injury and ARDS demonstrated that D allele strongly correlated with higher death rates, and I/D polymorphism seemed to impact the serum ACE levels associated with prognosis in these patients (Tsantes, Kopterides et al., 2013). Another study showed the association of DD genotype with mortality in the ARDS population. In addition, patients with II polymorphism were associated with markedly better rates of survival (Ziai et al., 2004; Jerng, Yu et al., 2006; Rezaei et al., 2020). In a study conducted by Itoyama et al., in severe acute respiratory syndrome (SARS) patients, it was shown that the D allele was higher in frequency in the hypoxemic group compared to the non-hypoxemic group (Itoyama, Keicho et al., 2004). In a study by Bellone et al. ACE DD polymorphism was reported to be associated with COVID-19 mortality rate (Bellone and Calvisi 2020). Similar results have also been reported in some other studies conducted on patients with COVID-19. In the study by Pati et al. it was revealed that D allele was associated with mortality rate of COVID-19 patients in Asians (Pati, Mahto et al., 2020). The DD polymorphism has been shown to affect the outcomes and increase the severity of the disease, as reported in the study by Sarangarajan et al., (Sarangarajan, Winn et al., 2020). Furthermore, Yamamoto et al. reported that ACE II genotype correlated negatively with number of COVID-19 cases and COVID-19-related mortalities (Yamamoto, Ariumi et al., 2020). A meta-analysis also concluded that ACE I/D polymorphism can be a beneficial marker for acute lung injury/ARDS prognosis and the genotypes may be used to treat acute lung injury/ARDS in COVID-19 patients. The mentioned study also showed that significant connection existed between risk of acute lung injury/ARDS and ACE polymorphism in children and Caucasian population, in addition to in Asian population in analysis of mortality (Pabalan, Tharabenjasin et al., 2021). A similar result was stated by another study regarding the prognostic value of the ACE genotype, and its impact on results of COVID-19 treatment (Verma, Abbas et al., 2021). A recent study reported close association between ACE polymorphism and COVID-19 infection severity, in which the II genotype showed protective effects upon developing the severe form of COVID-19 (Gunal, Sezer et al., 2021).
In our results, allele D was not associated with the outcome (recovery or death). In the study of Chan et al., it was reported that no association existed between ACE polymorphism and undesirable outcomes after SARS infection (Chan, Tang et al., 2005). A meta-analysis also indicated that no association exists between ACE polymorphism and susceptibility to ARDS or acute lung injury, but a connection is present between ACE polymorphism and mortality among Asian patients with ARDS (Matsuda, Kishi et al., 2012). Also, another study reported that ACE polymorphism and ARDS were not associated, however, plasma Ang II levels were higher in the ARDS group of D-allele carriers, but not in the control group, and the frequency of severe hypoxemia was less in carriers of D-allele (Cruces, Díaz et al., 2012). One study on COVID-19 showed a positive correlation between frequency ratio of I/D alleles and rate of recovery, but no difference in regard to the mortality rate (Hatami, Ahi et al., 2020).
The limitations on this study were the lack of group of patients with mild severity of the disease versus moderate and severe groups which was due to the out-patient management of individuals with mild disease and we did not have access to them within the in-patient admissions.
Conclusion
We concluded that ACE II genotype was significantly higher in proportion within the moderate disease group compared to the severe disease group, and that the D allele was associated with a higher risk of disease severity but not mortality.
Data availability statement
The datasets presented in this article are not readily available because of privacy and ethical issues. Requests to access the datasets should be directed to Seyed Ali Ziai, YWxpemlhaUBzYm11LmFjLmly.
Ethics statement
The studies involving human participants were reviewed and approved by Ethics Committee of Shahid Beheshti University of Medical Sciences, Tehran, Iran. The patients/participants provided their written informed consent to participate in this study.
Author contributions
MR contributed to conceptualization; MR and SZ contributed to methodology; SZ conducted statistical analysis; MR, HM, FN, MM, PT, and MP contributed to data collection; ME, SZ, and MR contributed in writing the original draft and editing. All authors reviewed and approved the manuscript.
Funding
Shahid Beheshti University of Medical Sciences provided financial support for this study (grant no. 23215).
Acknowledgments
This article is based on ME’s thesis, which was written in the School of Medicine at Shahid Beheshti University of Medical Sciences and has the registration number 26408 and the ethic code IR. SBMU.MSP.REC.1399.519.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Baudin, B. (2002). New aspects on angiotensin-converting enzyme: From gene to disease. Clin. Chem. Lab. Med. 40 (3), 256–265. doi:10.1515/CCLM.2002.042
Bellone, M., and Calvisi, S. L. (2020). ACE polymorphisms and COVID-19-related mortality in Europe. J. Mol. Med. 98, 1505–1509. doi:10.1007/s00109-020-01981-0
CDC, (2020). Defining adult overweight and obesity. from https://www.cdc.gov/obesity/adult/defining.html.
Chan, K. C., Tang, N. L., Hui, D. S., Chung, G. T., Wu, A. K., Chim, S. S., et al. (2005). Absence of association between angiotensin converting enzyme polymorphism and development of adult respiratory distress syndrome in patients with severe acute respiratory syndrome: A case control study. BMC Infect. Dis. 5, 26. doi:10.1186/1471-2334-5-26
Colunga Biancatelli, R. M. L., Solopov, P. A., Gregory, B., Khodour, Y., and Catravas, J. D. (2022). HSP90 inhibitors modulate SARS-CoV-2 spike protein subunit 1-induced human pulmonary microvascular endothelial activation and barrier dysfunction. Front. Physiol. 13, 812199. doi:10.3389/fphys.2022.812199
Crowley, S. D., Gurley, S. B., Oliverio, M. I., Pazmino, A. K., Griffiths, R., Flannery, P. J., et al. (2005). Distinct roles for the kidney and systemic tissues in blood pressure regulation by the renin-angiotensin system. J. Clin. Invest. 115 (4), 1092–1099. doi:10.1172/JCI23378
Cruces, P., Díaz, F., Puga, A., Erranz, B., Donoso, A., Carvajal, C., et al. (2012). Angiotensin-converting enzyme insertion/deletion polymorphism is associated with severe hypoxemia in pediatric ARDS. Intensive Care Med. 38 (1), 113–119. doi:10.1007/s00134-011-2381-3
Deng, X., Zhang, S., Jin, K., Li, L., Gu, W., Liu, M., et al. (2015). Angiotensin-converting enzyme I/D polymorphism and acute respiratory distress syndrome. J. Renin. Angiotensin. Aldosterone. Syst. 16 (4), 780–786. doi:10.1177/1470320315576255
Dimmeler, S., Rippmann, V., Weiland, U., Haendeler, J., and Zeiher, A. M. (1997). Angiotensin II induces apoptosis of human endothelial cells: Protective effect of nitric oxide. Circ. Res. 81 (6), 970–976. doi:10.1161/01.res.81.6.970
Donoghue, M., Hsieh, F., Baronas, E., Godbout, K., Gosselin, M., Stagliano, N., et al. (2000). A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 87 (5), E1–E9. doi:10.1161/01.res.87.5.e1
Doumas, M., Patoulias, D., Katsimardou, A., Stavropoulos, K., Imprialos, K., and Karagiannis, A. (2020). COVID19 and increased mortality in african Americans: Socioeconomic differences or does the renin angiotensin system also contribute? J. Hum. Hypertens. 34, 764–767. doi:10.1038/s41371-020-0380-y
Ghafouri-Fard, S., Noroozi, R., Vafaee, R., Branicki, W., Poṡpiech, E., Pyrc, K., et al. (2020). Effects of host genetic variations on response to, susceptibility and severity of respiratory infections. Biomed. Pharmacother. 128, 110296. doi:10.1016/j.biopha.2020.110296
Grasselli, G., Zangrillo, A., Zanella, A., Antonelli, M., Cabrini, L., Castelli, A., et al. (2020). Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the lombardy region, Italy. JAMA 323 (16), 1574–1581. doi:10.1001/jama.2020.5394
Gunal, O., Sezer, O., Ustun, G. U., Ozturk, C. E., Sen, A., Yigit, S., et al. (2021). Angiotensin-converting enzyme-1 gene insertion/deletion polymorphism may be associated with COVID-19 clinical severity: A prospective cohort study. Ann. Saudi Med. 41 (3), 141–146. doi:10.5144/0256-4947.2021.141
Hadjadj, J., Yatim, N., Barnabei, L., Corneau, A., Boussier, J., Smith, N., et al. (2020). Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Sci. (New York, N.Y.) 369 (6504), 718–724. doi:10.1126/science.abc6027
Hatami, N., Ahi, S., Sadeghinikoo, A., Foroughian, M., Javdani, F., Kalani, N., et al. (2020). Worldwide ACE (I/D) polymorphism may affect COVID-19 recovery rate: An ecological meta-regression. Endocrine 68 (3), 479–484. doi:10.1007/s12020-020-02381-7
Hussain, A., Mahawar, K., Xia, Z., Yang, W., and El-Hasani, S. (2020). Obesity and mortality of COVID-19. Meta-analysis. Obes. Res. Clin. Pract. 14 (4), 295–300. doi:10.1016/j.orcp.2020.07.002
Itoyama, S., Keicho, N., Quy, T., Phi, N. C., Long, H. T., Ha, L. D., et al. (2004). ACE1 polymorphism and progression of SARS. Biochem. Biophys. Res. Commun. 323 (3), 1124–1129. doi:10.1016/j.bbrc.2004.08.208
Jerng, J.-S., Hsu, Y.-C., Wu, H.-D., Pan, H.-Z., Wang, H.-C., Shun, C.-T., et al. (2007). Role of the renin-angiotensin system in ventilator-induced lung injury: An in vivo study in a rat model. Thorax 62 (6), 527–535. doi:10.1136/thx.2006.061945
Jerng, J. S., Yu, C. J., Wang, H. C., Chen, K. Y., Cheng, S. L., and Yang, P. C. (2006). Polymorphism of the angiotensin-converting enzyme gene affects the outcome of acute respiratory distress syndrome. Crit. Care Med. 34 (4), 1001–1006. doi:10.1097/01.CCM.0000206107.92476.39
Kouchmeshky, A., Jameie, S., Amin, G., and Ziai, S. (2012). Investigation of angiotensin-convertings enzyme inhibitory effects of medicinal plants used in traditional Persian medicine for treatment of hypertension: Screening study. Thrita 1, 13–23. doi:10.5812/thrita.4264
Kuba, K., Imai, Y., Rao, S., Gao, H., Guo, F., Guan, B., et al. (2005). A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 11 (8), 875–879. doi:10.1038/nm1267
Lippi, G., Wong, J., and Henry, B. M. (2020). Hypertension in patients with coronavirus disease 2019 (COVID-19): A pooled analysis. Pol. Arch. Intern. Med. 130 (4), 304–309. doi:10.20452/pamw.15272
Liu, Y., Yang, Y., Zhang, C., Huang, F., Wang, F., Yuan, J., et al. (2020). Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China. Life Sci. 63 (3), 364–374. doi:10.1007/s11427-020-1643-8
Mahmoudi, S., Rezaei, M., Mansouri, N., Marjani, M., and Mansouri, D. (2020). Immunologic features in coronavirus disease 2019: Functional exhaustion of T cells and cytokine storm. J. Clin. Immunol. 40 (7), 974–976. doi:10.1007/s10875-020-00824-4
Marshall, R. P., Gohlke, P., Chambers, R. C., Howell, D. C., Bottoms, S. E., Unger, T., et al. (2004). Angiotensin II and the fibroproliferative response to acute lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 286 (1), L156–L164. doi:10.1152/ajplung.00313.2002
Marshall, R. P., Webb, S., Bellingan, G. J., Montgomery, H. E., Chaudhari, B., McAnulty, R. J., et al. (2002). Angiotensin converting enzyme insertion/deletion polymorphism is associated with susceptibility and outcome in acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 166 (5), 646–650. doi:10.1164/rccm.2108086
Matsuda, A., Kishi, T., Jacob, A., Aziz, M., and Wang, P. (2012). Association between insertion/deletion polymorphism in angiotensin-converting enzyme gene and acute lung injury/acute respiratory distress syndrome: A meta-analysis. BMC Med. Genet. 13, 76. doi:10.1186/1471-2350-13-76
Mattei, M., Hubert, C., AlhencGelas, F., Roeckel, N., Corvol, P., and Soubrier, F. (1989). Angiotensin i converting enzyme gene is on chromosome 17. Cytogenetics and cell genetics. Karger Allschwillerstrasse 10, CH-4009 Basel 266 (23), 15377.
Mohammadpour, H., Ziai, A., Sadr, M., Rezaei, M., Marjani, M., and Tabarsi, P. (2020). A novel coronavirus disease (COVID-19): A review of host cell signaling pathways. Tanaffos 19 (2), 108–111.
Ni, W., Yang, X., Yang, D., Bao, J., Li, R., Xiao, Y., et al. (2020). Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care 24 (1), 422. doi:10.1186/s13054-020-03120-0
Pabalan, N., Tharabenjasin, P., Suntornsaratoon, P., Jarjanazi, H., and Muanprasat, C. (2021). Ethnic and age-specific acute lung injury/acute respiratory distress syndrome risk associated with angiotensin-converting enzyme insertion/deletion polymorphisms, implications for COVID-19: A meta-analysis. Infect. Genet. Evol. 88, 104682. doi:10.1016/j.meegid.2020.104682
Pati, A., Mahto, H., Padhi, S., and Panda, A. K. (2020). ACE deletion allele is associated with susceptibility to SARS-CoV-2 infection and mortality rate: An epidemiological study in the Asian population. Clin. Chim. Acta. 510, 455–458. doi:10.1016/j.cca.2020.08.008
Rezaei, M., Ziai, S. A., Fakhri, S., and Pouriran, R. (2020). ACE2: Its potential role and regulation in severe acute respiratory syndrome and COVID-19. J. Cell. Physiol. 236 (3), 2430–2442. doi:10.1002/jcp.30041
Richardson, S., Hirsch, J. S., Narasimhan, M., Crawford, J. M., McGinn, T., Davidson, K. W., et al. (2020). Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York city area. JAMA 323 (20), 2052–2059. doi:10.1001/jama.2020.6775
Rod, J. E., Oviedo-Trespalacios, O., and Cortes-Ramirez, J. (2020). A brief-review of the risk factors for Covid-19 severity. Rev. Saude Publica 54, 60. doi:10.11606/s1518-8787.2020054002481
Sarangarajan, R., Winn, R., Kiebish, M. A., Bountra, C., Granger, E., and Narain, N. R. (2020). Ethnic prevalence of angiotensin-converting enzyme deletion (D) polymorphism and COVID-19 risk: Rationale for use of angiotensin-converting enzyme inhibitors/angiotensin receptor blockers. J. Racial Ethn. Health Disparities 8 (4), 973–980. doi:10.1007/s40615-020-00853-0
Shi, Y., Wang, G., Cai, X.-P., Deng, J.-W., Zheng, L., Zhu, H.-H., et al. (2020). An overview of COVID-19. J. Zhejiang Univ. Sci. B 21 (5), 343–360. doi:10.1631/jzus.B2000083
Solopov, P. A., Colunga Biancatelli, R. M. L., and Catravas, J. D. (2022). Alcohol increases lung angiotensin-converting enzyme 2 expression and exacerbates severe acute respiratory syndrome coronavirus 2 spike protein subunit 1-induced acute lung injury in K18-hACE2 transgenic mice. Am. J. Pathol. 192 (7), 990–1000. doi:10.1016/j.ajpath.2022.03.012
Suzuki, Y., Ruiz-Ortega, M., Lorenzo, O., Ruperez, M., Esteban, V., and Egido, J. (2003). Inflammation and angiotensin II. Int. J. Biochem. Cell Biol. 35 (6), 881–900. doi:10.1016/s1357-2725(02)00271-6
Tipnis, S. R., Hooper, N. M., Hyde, R., Karran, E., Christie, G., and Turner, A. J. (2000). A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 275 (43), 33238–33243. doi:10.1074/jbc.M002615200
Tsantes, A. E., Kopterides, P., Bonovas, S., Bagos, P., Antonakos, G., Nikolopoulos, G. K., et al. (2013). Effect of angiotensin converting enzyme gene I/D polymorphism and its expression on clinical outcome in acute respiratory distress syndrome. Minerva Anestesiol. 79 (8), 861–870.
Verma, S., Abbas, M., Verma, S., Khan, F. H., Raza, S. T., Siddiqi, Z., et al. (2021). Impact of I/D polymorphism of angiotensin-converting enzyme 1 (ACE1) gene on the severity of COVID-19 patients. Infect. Genet. Evol. 91, 104801. doi:10.1016/j.meegid.2021.104801
Wang, R.-M., Feng, H.-X., He, Y.-F., Xia, C.-G., Ji-Shuan, S., and Wang, Y.-P. (2000). Preparation and catalysis of NaY-encapsulated Mn(III) Schiff-base complex in presence of molecular oxygen. J. Mol. Catal. A Chem. 151 (1), 253–259. doi:10.1016/s1381-1169(99)00256-3
World Health Organization, (2022). Clinical management of COVID-19: Living guideline. Geneva, Switzerland: Cataloguing-in-Publication.
Wu, J., Deng, W., Li, S., and Yang, X. (2020). Advances in research on ACE2 as a receptor for 2019-nCoV. Cell. Mol. Life Sci. 78 (2), 531–544. doi:10.1007/s00018-020-03611-x
Wu, Z., and McGoogan, J. M. (2020). Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA 323 (13), 1239–1242. doi:10.1001/jama.2020.2648
Yamamoto, N., Ariumi, Y., Nishida, N., Yamamoto, R., Bauer, G., Gojobori, T., et al. (2020). SARS-CoV-2 infections and COVID-19 mortalities strongly correlate with ACE1 I/D genotype. Gene 758, 144944. doi:10.1016/j.gene.2020.144944
Yang, P., Gu, H., Zhongpeng, Z., Wang, W., Cao, B., Lai, C., et al. (2014). Angiotensin-converting enzyme 2 (ACE2) mediates influenza H7N9 virus-induced acute lung injury. Sci. Rep. 4, 7027. doi:10.1038/srep07027
Zheng, H., and Cao, J. J. (2020). Angiotensin-converting enzyme gene polymorphism and severe lung injury in patients with coronavirus disease 2019. Am. J. Pathol. 190 (10), 2013–2017. doi:10.1016/j.ajpath.2020.07.009
Zheng, Z., Peng, F., Xu, B., Zhao, J., Liu, H., Peng, J., et al. (2020). Risk factors of critical & mortal COVID-19 cases: A systematic literature review and meta-analysis. J. Infect. 81 (2), e16–e25. doi:10.1016/j.jinf.2020.04.021
Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579 (7798), 270–273. doi:10.1038/s41586-020-2012-7
Keywords: COVID-19, angiotensin I converting enzyme, insertion/deletion polymorphism, SARS-CoV-2, peptidyl dipeptidase A, ACE, angiotensin-converting enzyme, polymorphism
Citation: Rezaei M, Mohammadpour H, Eftekhari M, Pourabdollah M, Nasr Azadani F, Tabarsi P, Marjani M and Ziai SA (2022) The role of angiotensin I converting enzyme insertion/deletion polymorphism in the severity and outcomes of COVID-19 patients. Front. Genet. 13:1035796. doi: 10.3389/fgene.2022.1035796
Received: 03 September 2022; Accepted: 07 November 2022;
Published: 29 November 2022.
Edited by:
Stephen J. Bush, University of Oxford, United KingdomReviewed by:
Ana Cristina Simões E. Silva, Federal University of Minas Gerais, BrazilPavel Solopov, Old Dominion University, United States
Lilong Jia, The University of Hong Kong, Hong Kong SAR, China
Copyright © 2022 Rezaei, Mohammadpour, Eftekhari, Pourabdollah, Nasr Azadani, Tabarsi, Marjani and Ziai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seyed Ali Ziai, YWxpemlhaUBzYm11LmFjLmly
†ORCID: Seyed Ali Ziai, orcid.org/0000-0002-1102-8119
 Mitra Rezaei
Mitra Rezaei Hadiseh Mohammadpour3
Hadiseh Mohammadpour3 Mahya Eftekhari
Mahya Eftekhari Payam Tabarsi
Payam Tabarsi Majid Marjani
Majid Marjani Seyed Ali Ziai
Seyed Ali Ziai