- 1Provincial Hospital Affiliated to Anhui Medical University, Hefei, Anhui, China
- 2The First Affiliated Hospital of University of Science and Technology of China West District, Hefei, Anhui, China
- 3Wannan Medical College, Wuhu, Anhui, China
- 4Department of Medical Oncology, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, China
Background: The family with sequence similarity 83 member D (FAM83D) protein is known to play a significant role in many human diseases. However, its role in cancer remains ambiguous. This study aimed to investigate the function of FAM83D in a pan-cancer analysis, with a special focus on breast cancer.
Methods: Samples were collected from The Cancer Genome Atlas (TCGA) and used for bioinformatic analysis. Datasets from the Gene Expression Omnibus (GEO) and Genotype-Tissue Expression (GTEx) databases were also analyzed for verification. The potential value of FAM83D as a prognostic and diagnostic biomarker was visualized through R software. The “survival” and “GSVA” package were used for univariate, multivariate and pathway enrichment analyseis. We further analyzed the CancerSEA databases and TISIDB websites for single-cell and immune-related profiling. Lastly, we validated those data in vitro using quantitative reverse transcriptase-polymerase chain reaction (RT‒qPCR), cell counting kit-8 (CCK-8), transwell, flow cytometry, and tumorigenicity assays in a murine cell line model.
Results: The expression of FAM83D in tumor samples was significantly higher than in normal tissues for most cancer types in the datasets. We confirmed this finding using RT‒qPCR in a breast cancer cell line. Analysis of multiple datasets suggests that overall survival (OS) was extremely poor for breast cancer patients with high FAM83D expression. The CCK-8 assay demonstrated that MCF-7 cell proliferation was inhibited after genetic silencing of FAM83D. Transwell assay showed that knockdown of FAM83D significantly inhibited the invasion and migration ability of MCF-7 cells compared to the control. The results of flow cytometry showed that silencing FAM83D could block the G1 phase of MCF-7 cells compared with negative groups. The tumorigenicity assay in nude mice indicated that the tumorigenic ability to silence FAM83D decreased compared.
Conclusion: Results suggest that FAM83D expression can serve as a valuable biomarker and core gene across cancer types. Furthermore, FAM83D expression is significantly associated with MCF-7 cell proliferation and thus may be a prospective prognostic biomarker especially for breast cancer.
Introduction
Nearly 10 million people die annually from malignant tumors (Sung et al., 2021). In recent years, the life expectancy of cancer patients has increased in part due to ongoing improvements in treatment strategies and rapid progress in the science and technology sectors. Both the behavioral traits and treatment options of cancer are complex and variable. Certain tumors with various molecular traits and subtypes can result in a poor prognosis for some individuals, especially in breast cancer (BRCA) (Hanahan and Weinberg., 2011; Siegel et al., 2022).
The family with sequence similarity 83 (FAM83) protein belongs to a protein family where in studies have found that many members of the family have carcinogenic effects. For example, FAM83A has been identified as exhibiting a tumor-promoting role in lung cancer and BRCA and can cause epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) resistance by activating the phosphatidylinositol-3-kinase-protein kinase B (PI3K-AKT) and mitogen-activated protein kinase (MAPK) signaling pathways (Li et al., 2005; Wang et al., 2015). However, the role of FAM83D, one member of the FAM83 family, in cancer is still unclear. Previous research has revealed that FAM83D is a transactivating factor expressed in the late G1/S phase of the cell cycle and is related to cell growth and the transition from G1 to S phase (Cipriano et al., 2012; Shi et al., 2016). By adversely regulating eurogenic locus notch homolog protein 1 (Notch-1), Mu et al. demonstrated that FAM83D can enhance the invasion and metastasis of colorectal cancer cells (Mu et al., 2017). In addition, suppression of FAM83D has been shown to inhibit glioma cell proliferation, invasion, and migration by regulating the Akt- mammalian target of rapamycin (Akt-mTOR) signaling pathway (Li et al., 2022).
Despite these investigations, the pan-cancer function and prognostic value of FAM83D, especially in BRCA, remains unknown. Although some patients with BRCA achieve successful remission and increased survival from chemotherapy, targeted therapy, and immunotherapy regimens, there are still a significant number of patients with BRCA who are subject to poor outcomes, necessitating the urgent development of new, practical, and straightforward therapies. Historically, there have been some indicators for predicting prognosis in BRCA mutation carriers (Lu et al., 2020; Saikia et al., 2021; Zeng et al., 2019). However, several of these biomarkers have low sensitivity or specificity; hence, it is necessary to investigate more accurate biomarkers for predicting the prognosis of patients with BRCA. In this study, using sequencing data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO), we investigated the function and prognostic utility of FAM83D expression in breast cancer.
Materials and methods
Expression levels and diagnostic value
RNAseq data from TCGA (11,093), GSE93601 (1,110), and GSE70947 (296) datasets in the GEO database were Toil (Vivian et al., 2017) processed and converted to log2 format for analysis. In addition, 179 samples of normal breast tissue were obtained from the Genotype-Tissue Expression (GTEx) database. The Mann‒Whitney U test was used to analyze the difference in FAM83D expression levels between tumor and normal tissues. R software (× 64, 3.6.3) was used for statistical analysis and visualization. Receiver operating characteristic analysis was performed using the R software “pROC” package to calculate the diagnostic efficacy of FAM83D expression. Significance was set at p < 0.05.
Prognosis analysis
The prognostic data for TCGA came from the research archives of Liu et al. (Liu et al., 2018). The data filtering criteria used were as follows: the controlled/normal sample was removed, and the sample with clinical information was retained. Kaplan‒Meier survival analysis was applied using the R “survival” package. Subsequently, FAM83D expression data and prognostic survival information from GSE9195 and GSE12276 were validated as independent cohorts. The R “survival” package was also used to perform univariate and multivariate analyses.
Immunocyte infiltration analysis
The single sample gene set enrichment analysis (ssGSEA) algorithm built into the R “GSVA” package (Hanzelmann et al., 2013) was used to perform immunoinfiltration analysis on 33 cancers in the TCGA dataset. This algorithm uses the specific markers of each type of immune cell as a gene set to calculate the enrichment score of each type of immune cell in each sample and thus infers the infiltration of immune cells in each sample (Bindea et al., 2013). Subsequently, the TISIDB online website (http://cis.hku.hk/TISIDB/) was used to map the relationship between FAM83D expression and Immunoinhibitor, Immunostimulator, and major histocompatibility complex (MHC) molecules in different cancer types.
Differential gene expression and pathway enrichment analysis in BRCA
Single-cell sequencing data from 14 cancers in the CancerSEA dataset were used to assess the association of FAM83D expression with 14 cancer functional states. BRCA single-cell sequence data from the GSE77308 dataset were also used. In addition, the R “DESeq2” package (Love et al., 2014) was used for genetic analysis of the differences between high and low expression groups of FAM83D (LogFC = 2, p < 0.01) in BRCA. The R “clusterProfiler” (Yu et al., 2012) and “org.Hs.eg.db” packages were used for gene ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, and gene set enrichment analysis (GSEA) pathway enrichment analysis (Walter et al., 2015). The Spearman algorithm built into the R “ggplot2” package was used to visualize the correlation between FAM83D and the Polo Like Kinase 1 (PLK1) gene expression in differentdatasets.
Cell culture, transfection, and reagents
MCF-7 cells were purchased from the American Type Culture Collection (ATCC; Manassas, VA, United States). We used 10% fetal bovine serum and 1% penicillin/streptomycin (Solarbio, Beijing, China) in RPMI 1640 medium (Gibco, Gland Island, NY, United States) to culture the cells. The cells were kept at 37°C in an incubator with 5% CO2. Jierui Biological Engineering Co., Ltd. (Shanghai, China) built the FAM83D knockdown vector, mixed it with the pPACKH1 packaging plasmid, and transfected it into cells. In accordance with the packaging plan described by SBI, virus particles were collected 3 days after the solution for letinous edodes concentrated virus precipitation was prepared. Infection of the cells was performed using TUNDUX viral transducers, and puromycin screening was performed to identify cells that were infected. The target sequences used were as follows: sh1-FAM83D: GATCTGAAAGTTCATCCTGAA and sh2-FAM83D: CCTGACTTTGTCACCTTTGTT.
Quantitative reverse transcriptase-polymerase chain reaction
Using TRIzol Reagent (Invitrogen, Carlsbad, CA, United States), total RNA was extracted from the MCF-7 cells. Using PrimeScript RT Master Mix (Takara Biotechnology, Dalian, China), total RNA was reverse transcribed into cDNA, which was then used for RT- qPCR using SYBR qPCR Master Mix (Vazyme, Nanjing, China). To quantify FAM83D gene expression, the 2−ΔΔCT was calculated for every sample and normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). For PCR, the FAM83D primers were as follows: forward: 5′-AGAGCGGCAATTCCACTTCG-3′, reverse: 5′- TGCCAGAATGAAGGCCAAGG-3′. GAPDH: forward: 5′-AGATCCCTCCAAAATCAAGTGG -3′, reverse: 5′-GGCAGAGATGATGACCCTTTT-3′.
Cell counting Kit-8 assay
The CCK-8 (Abcam, Shanghai, China) assay was used to measure the proliferation rate of MCF-7 cells following the manufacturer’s instructions. Further, 96-well plates were seeded with 2×103 cells per well. All wells were incubated for 2 h at 37°C, followed by the measurement of absorbance at 450 nm in a microplate reader at the desired time point.
Transwell and flow cytometry assay
After collecting cultured sh1-FAM83D, sh2-FAM83D, and negative control (NC) group cells with serum-free medium, they were counted. 1×104 cells were evenly inoculated into the upper layer of each compartment and a culture medium containing 10% fetal bovine serum (500 μl) was added to the lower layer of the compartment to induce cell migration to the other side. After incubation for 24 h at 37°C in a 5% CO2 incubator, the cells were fixed in 4% methanol for 30 min, stained with 0.1% crystalline violet for 20 min, and rinsed off with PBS. Finally, five representative images were randomly captured under an inverted microscope. For the detection of cell invasion, Matrigel (BD, Bioscience, Pharmingen) was spread in advance on the upper layer of the chambers and placed in the incubator for 2–4 h. The rest of the procedure was carried out following the migration assay steps above.
For cell cycle assay, MCF-7 cells were collected and fixed in 80% pre-cooled ethanol for about 18 h at 4°C in a refrigerator. The following day, cells were resuspended with propidium staining solution and incubated at 37°C for 30 min protected from light. Ensure that the cycling assay is completed by flow cytometry within 1 h and the results are analyzed for data using ModFit software.
Animal experiment
Female BALB/c mice that were 4-weeks old, naked, and reared in a sterile environment were used for FAM83D silencing experiments (Vitalriver, Beijing, China). The constructs sh-FAM83D and NC group were transfected into 3×106 MCF-7 cells and then injected into the nude mice through the tail vein (n = 5 for each treatment group). Every 3 days, the tumor sizes were measured. The mice were euthanized after 24 days, and their tumor weight were measured. This animal experiment was reviewed and approved by the University of Science and Technology of China Animal Experimentation Ethics Committee.
Results
Pan-cancer FAM83D expression
The expression of FAM83D was higher in cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), glioblastoma multiforme (GBM), kidney renal clear cell carcinoma (KIRC), rectum adenocarcinoma (READ), and uterine corpus endometrial carcinoma (UCEC) samples compared to that in normal tissue samples (p < 0.001; Figure 1A). Paired data also revealed that the expression level of FAM83D in tumor tissues exceeded that in normal tissues (Figure 1B). In addition, by comparing FAM83D expression data from TCGA datasets, combining the normal tissue data of GTEx with TCGA data, and reviewing paired data (Figures 1C,D), we discovered that the expression level of FAM83D in BRCA tissues was markedly higher than in normal tissues (p < 0.001). To further verify the dissimilarity in FAM83D expression between BRCA and normal tissues, 1,110 samples, including 602 tumor and 508 tumor-adjacent normal tissue samples, from the GSE93601 dataset in the GEO database were integrated and analyzed. These results also confirmed the conclusion that FAM83D was highly expressed in tumor tissues than in paired adjacent tissues (Figure 1F). Similarly, validating the expression data of 148 paired specimens in the external dataset GSE70947 found consistent results with the TCGA analysis (Figure 1G).
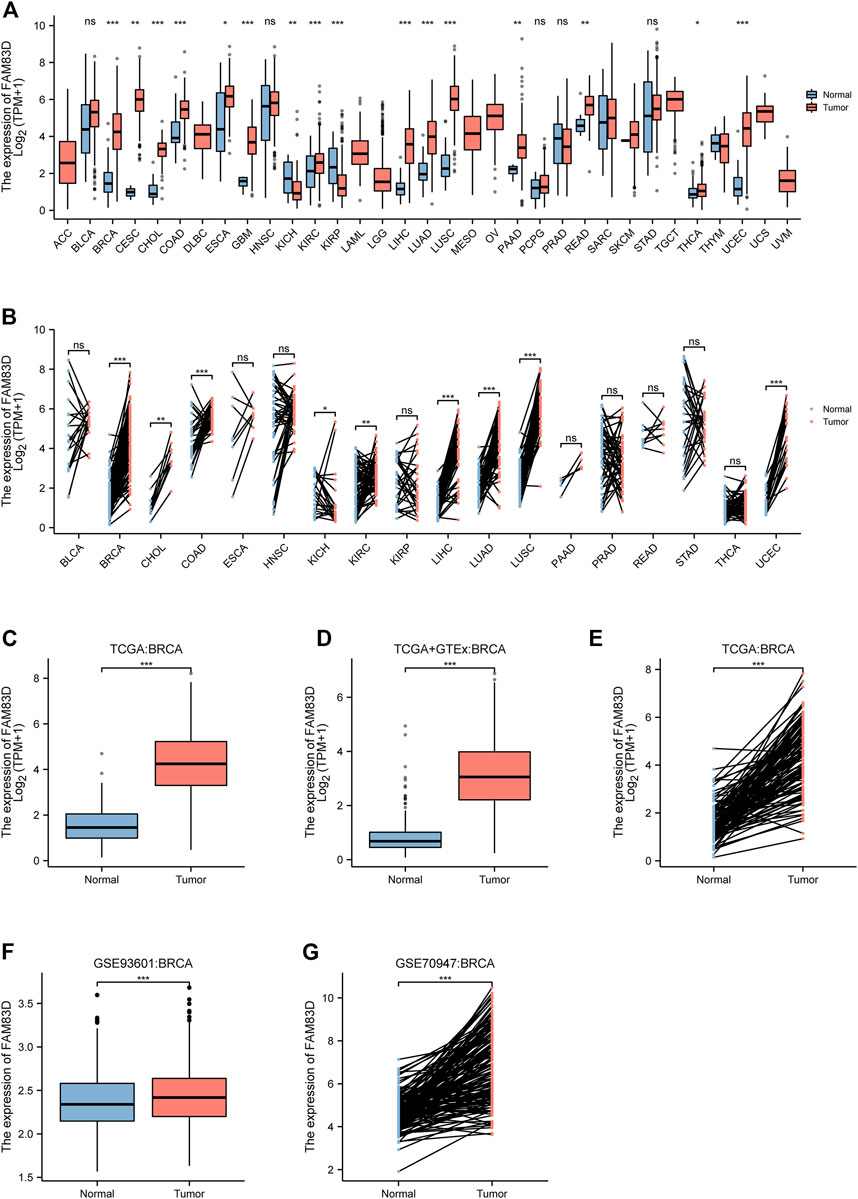
FIGURE 1. The expression level of FAM83D in human cancer (A) Pan-cancer analysis in TCGA cohort (B) Pan-cancer pair analysis in TCGA cohort (C) Expression in TCGA breast cancer tumour tissues and the normal (D) Expression in TCGA and GTEx of breast cancer (E) Expression in TCGA breast cancer paired samples (F) Expression of the GSE93601 dataset in BRCA (G) Expression of the GSE70947 dataset in BRCA (*p < 0.05, **p < 0.01, ***p < 0.001).
Pan-cancer diagnostic value of FAM83D
We next constructed a receiver operating characteristic (ROC) curve to evaluate the diagnostic value of FAM83D expression in the TCGA cohort data. Ourr analysis showed (Figures 2A–N) that FAM83D expression may act as a diagnostic marker in BRCA (area under the curve, AUC = 0.949; 95% confidence interval, CI: 0.932–0.967), cervical cancer (CESC; AUC = 0.968, 95% CI: 0.924–1.000), CHOL (AUC = 0.941, 95% CI: 0.873–1.000), COAD (AUC = 0.831, 95% CI: 0.744–0.918), GBM (AUC = 0.947, 95% CI: 0.927–0.967), kidney renal papillary cell carcinoma (KIRP; AUC = 0.732, 95% CI: 0.639–0.825), acute myeloid leukemia (LAML; AUC = 1.000, 95% CI: 1.000–1.000), liver hepatocellular carcinoma (LIHC; AUC = 0.946, 95% CI: 0.923–0.968), LUAD (AUC = 0.895, 95% CI: 0.862–0.928), LUSC (AUC = 0.990, 95% CI: 0.982–0.998), ovarian serous cystadenocarcinoma (AUC = 0.986, 95% CI: 0.967–1.000), pancreatic adenocarcinoma (PAAD; AUC = 0.984, 95% CI: 0.971–0.996), READ (AUC = 0.810, 95% CI: 0.660–0.961), UCEC (AUC = 0.961, 95% CI: 0.932–0.989). Furthermore, the ROC curve with an area of 0.876 under the subject working characteristic curve (95% CI: 0.836–0.916) indicated that FAM83D expression had a high degree of accuracy in diagnosing BRCA in the GSE70947 dataset (Figure 2O).
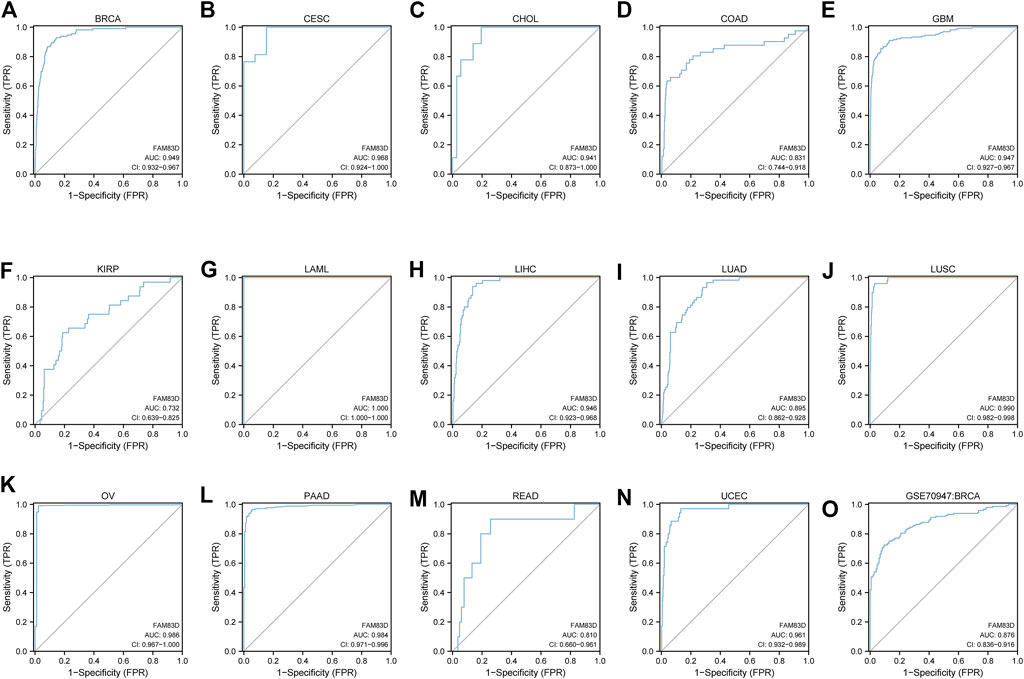
FIGURE 2. ROC curve of pan-cancer (A–N) ROC curve of BRCA, CESC, CHOL, COAD, GBM, KIRP, LAML, LIHC, LUAD, LUSC, OV, PAAD, READ, UCEC (O) ROC curve of the GSE70947 dataset in BRCA.
High FAM83D expression in patients predicts poor prognosis
As shown in Figures 3A–L, the overall survival (OS) of patients with low expression of FAM83D was better than for patients with high expression of FAM83D in adrenocortical carcinoma (ACC; p < 0.001), bladder urothelial carcinoma (BLCA; p = 0.022), BRCA (p = 0.001), kidney chromophobe (KICH; p = 0.002), KIRP (p < 0.001), brain lower grade glioma (LGG; p < 0.001), LIHC (p < 0.001), LUAD (p < 0.001), mesothelioma (MESO; p < 0.001), PAAD (p < 0.001), sarcoma (SARC; p = 0.023), skin cutaneous melanoma (SKCM; p = 0.004), and uveal melanoma (UVM; p = 0.036). To further validate this result, we sorted out the chip expression and recurrence-free survival (RFS) data from the GSE9195 dataset (p = 0.01; Figure 3N) in the GEO database. Analysis of the RFS of GSE12276 showed that patients in the high-expression FAM83D group had a worse prognosis than those in the low-expression FAM83D group (Figure 3O) (p < 0.001).
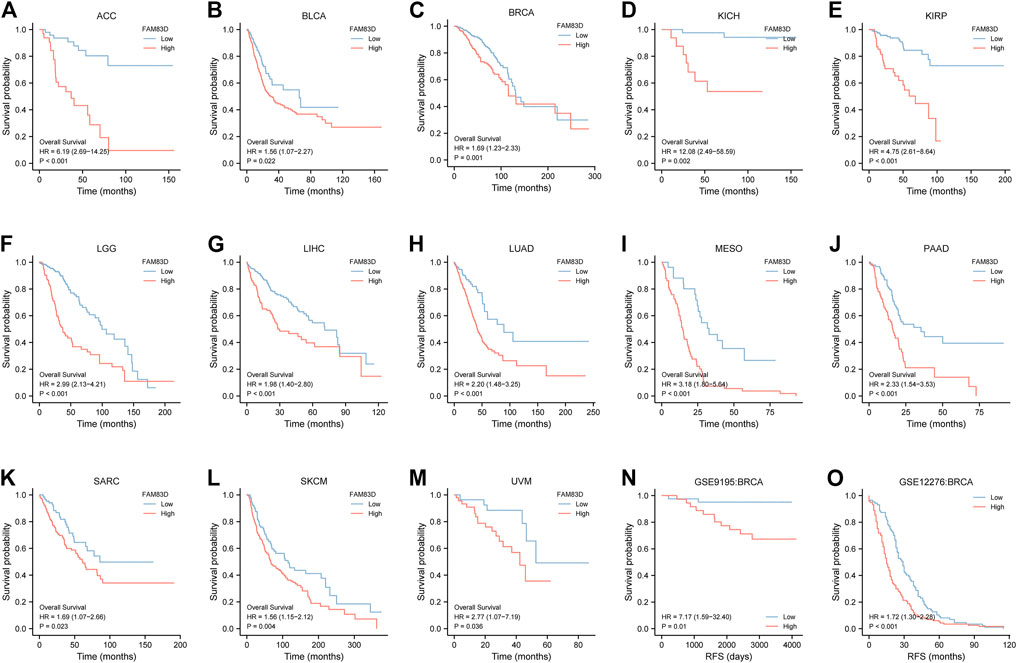
FIGURE 3. Kaplan–Meier survival curves comparing the high and low expression of FAM83D in different types of cancer in Kaplan–Meier Plotter. (A–M) Kaplan–Meier analysis of the association between FAM83D expression and OS in the TCGA cohort. (N,O) RFS analysis of RFS in GSE9195 and GSE12276 in BRCA.
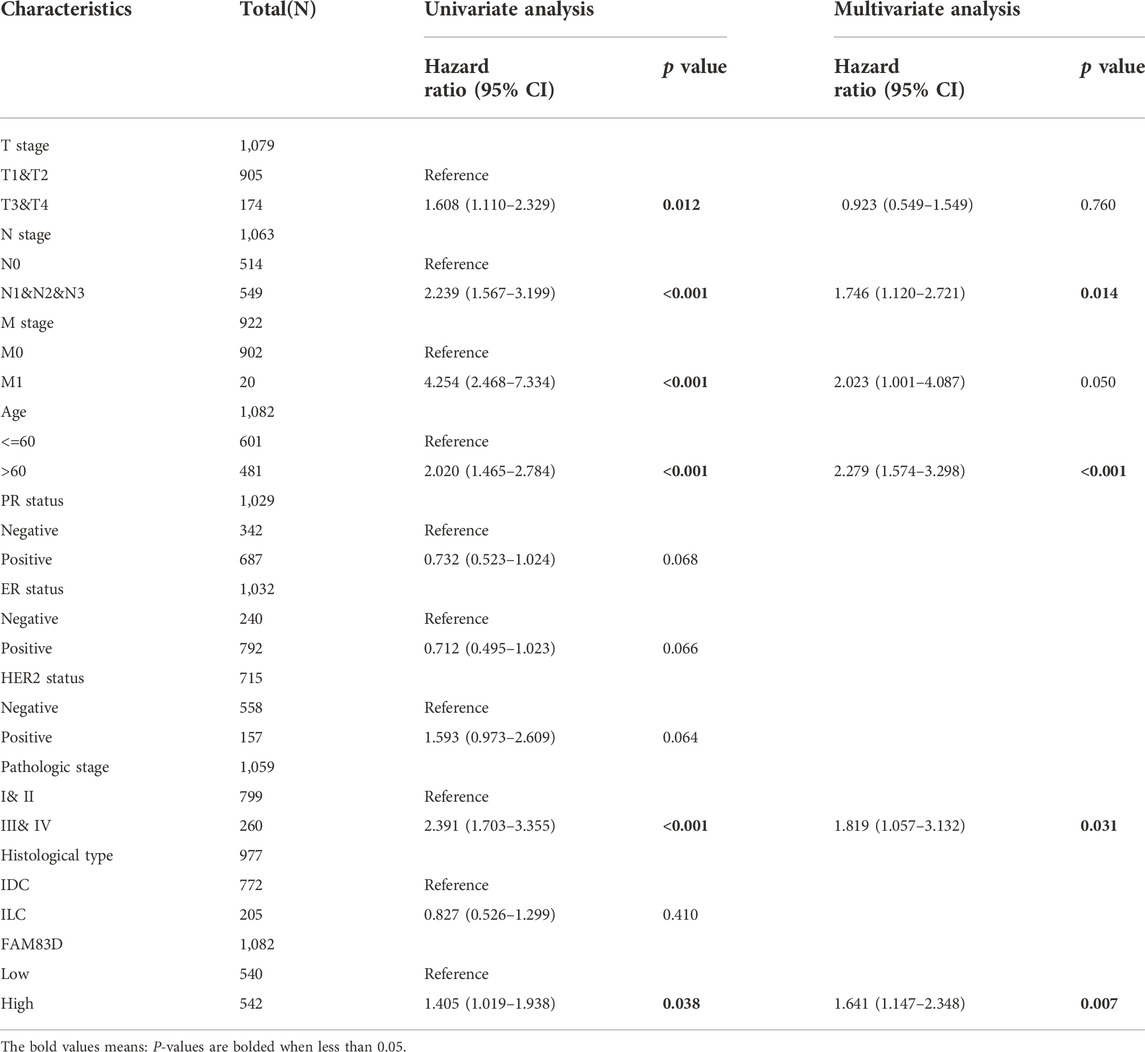
The Cox regression module of the R “survival” package was used to analyze BRCA data from the TCGA dataset with univariate and multivariate statistics. As shown in Table 1, univariate meaningful parameters were included in the multivariate analysis. Multivariate analysis showed that FAM83D expression (hazard ratio, HR = 1.641 (95% CI: 1.147–2.348)) was independently correlated with OS. Moreover, the N stage, patient age, and pathologic stage were also independent predictors of BRCA prognosis.
FAM83D is related to immune infiltration in pan-cancer
The ssGSEA algorithm was used to perform immune infiltration analysis on samples from the TCGA dataset. Spearman correlation analysis showed the correlation between FAM83D expression in the tumor environment and immune cells (Figure 4A). Th2 and T helper cells were positively correlated with FAM83D expression in most cancer types. In contrast, in BRCA, KIRC, and prostate adenocarcinoma (PRAD), the degree of immune cell infiltration was correlated with the expression level of FAM83D. In the ssGSEA results for BRCA (Figure 4B), the expression level of FAM83D was positively correlated with Th2 cells and aDC cells but negatively correlated with Natural Killer (NK) cells.
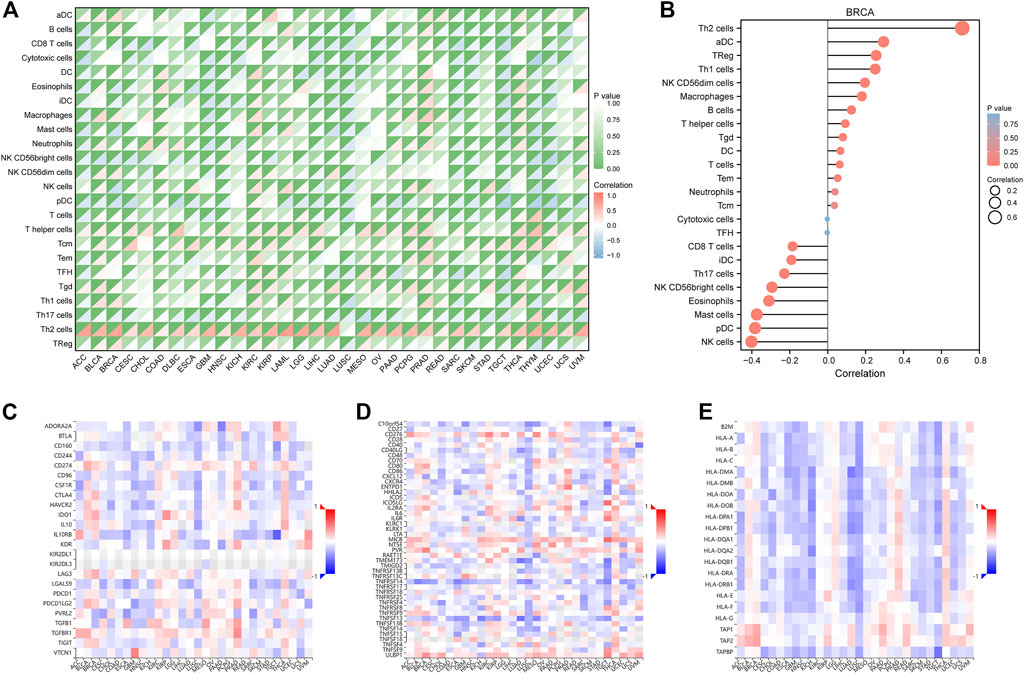
FIGURE 4. Immune-related analysis. (A) TCGA immune cell infiltration analysis in pan-cancer (B) immune cell infiltration analysis of BRCA (C–E) Correlation analysis between FAM83D and Immunoinhibitor, Immunostimulator and MHC molecule in the TISIDB database.
We further analyzed the relationship between FAM83D expression and Immunoinhibitor, Immunostimulator, and MHC molecules in the TISIDB database (Figures 4C–E). Our results revealed that FAM83D expression is negatively associated with immune-related molecules in LUSC and KICH and positively associated with these molecules in BRCA, PRAD, and thyroid carcinoma.
Single-cell sequencing analysis
Next, we analyzed the correlation between FAM83D expression and 14 cancer functional states using single-cell sequencing data from CancerSEA. FAM83D expression was positively associated with the cell cycle in most tumor types (Figure 5A), whereas it was negatively associated with hypoxia (Figure 5A). As for BRCA, there was a significant positive correlation between FAM83D expression and the cell cycle (cor = 0.54, p < 0.001), DNA damage (cor = 0.45, p < 0.001) and proliferation (cor = 0.51, p ≤ 0.001) (Figure 5B).
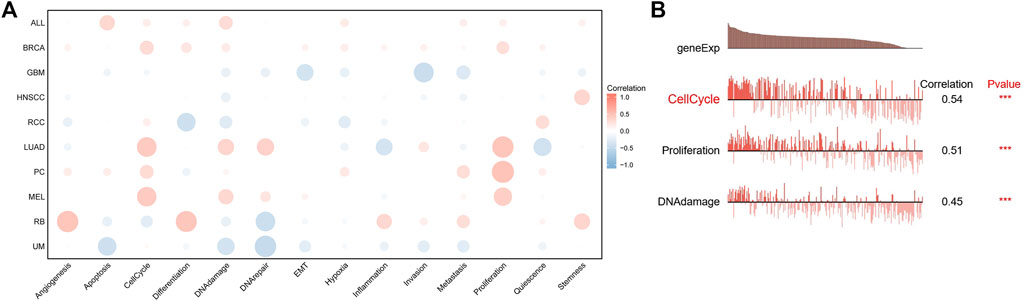
FIGURE 5. The correlation between FAM83D expression and 14 cancer functional states using single-cell sequence data from the CancerSEA database ((A) The correlation between CENPL expression and 14 cancer functional states in pan-cancer (B) The expression of CENPL is positively correlated with the DNA repai, cell cycle, DNA damage, proliferation and Hypoxia of BRCA (***p < 0.001).
Functional analysis of FAM83D in BRCA
We next used the median expression level of FAM83D in BRCA samples as a grouping cutoff value and displayed the differentially expressed genes (LogFC = 2, p < 0.01) in a volcano plot in Figure 6A. We found that the PLK1 gene was significantly elevated in the high expression group of FM83D. Subsequently, GO and KEGG enrichment analyses showed that the differentially expressed genes were primarily enriched in the cornification, antimicrobial humoral response, keratinization, keratinocyte differentiation, cell fate commitment, and intermediate filament cytoskeleton pathways (Figure 6B). GSEA pathway enrichment analysis further indicated that the cell cycle, S phase, PLK1, and DNA replication pathways are related to FAM83D expression in BRCA (Figures 6C,D). Molecular correlation analysis of BRCA data from the TCGA, GSE93601, and GSE70947 datasets showed that the expression level of PLK1 was strongly correlated with FAM83D expression (Figures 6E–G). As shown in Supplementary Figure S1, we further verify the relationship between FAM83D and PLK1 in other tumors by biogenic analysis. Consistent with BRCA results, a total of 25 cancers, FAM83D expression levels strongly correlated with PLK1.
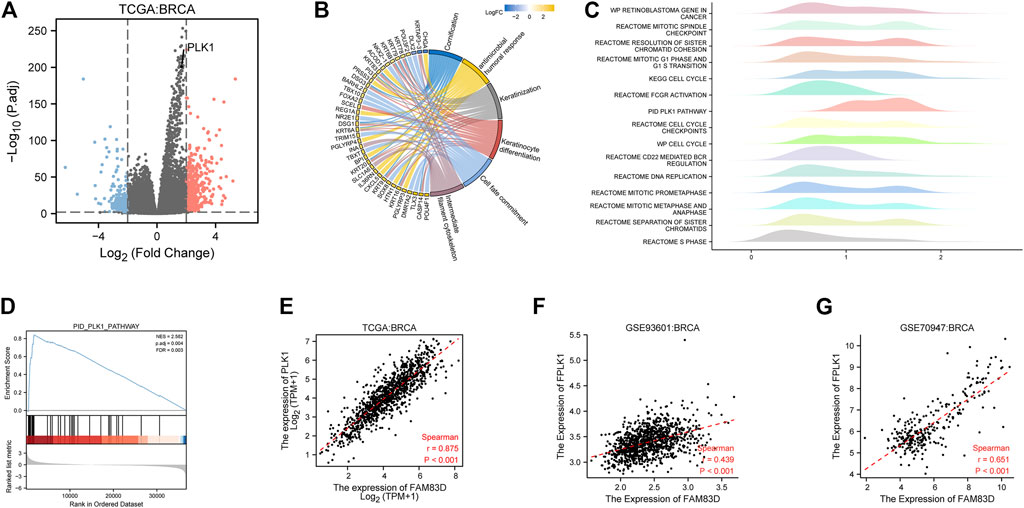
FIGURE 6. Pathway enrichment and correlation analysis in BRCA. (A) Differential genes in the TCGA cohort. (B) KEGG and GO analyses of the top ranked pathways. (C) Pathway enrichment by GSEA. (D) PLK1 pathway of GSEA. (E–G) Correlation between the FAM83D and PLK1 of TCGA, GSE93601 and GSE70947.
Verification of FAM83D results using RT‒qPCR and CCK-8 assay
RT‒qPCR results showed that FAM83D mRNA expression level in 14 pairs of BRCA tissue samples was higher than in adjacent normal tissue samples (p < 0.01, Figure 7A). This confirmed the results of our bioinformatics analysis of the TCGA, GSE93601, and GSE70947 datasets. Additionally, the CCK-8 assay revealed a significant difference between the knockdown FAM83D and NC groups in terms of cell proliferation (Figure 7B).
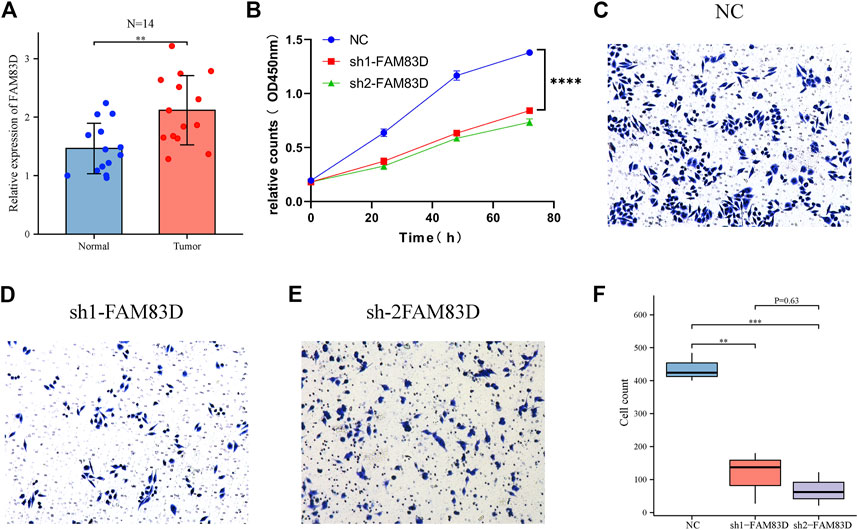
FIGURE 7. FAM83D was verified by RT‒qPCR, CCK-8 and transwell assay in MCF-7 cells. (A) RT‒qPCR analysis of tumor FAM83D expression in comparison to normal cells. (B) CCK-8 assay demonstrated that FAM83D knockdown prevented the growth of MCF-7 cells. (C–F) Transwell assay showed that knockdown of FAM83D significantly inhibited the invasion and migration ability (**p < 0.01, ***p < 0.001, ****p < 0.0001).
Verification of FAM83D results using transwell and the flow cytometric assay
Transwell assay showed that knockdown of FAM83D significantly inhibited the invasion and migration ability of BRCA cells compared to control (Figures 7C–F). Moreover, Fflow cytometric assays revealed that knockdown of FAM83D can alter the cell cycle by arresting the G1 phase (Figures 8A–D). This is consistent with previous pathway enrichment results, suggesting that FAM83D may play a role in affecting the proliferation of MCF-7 cells by regulating the cell cycle.
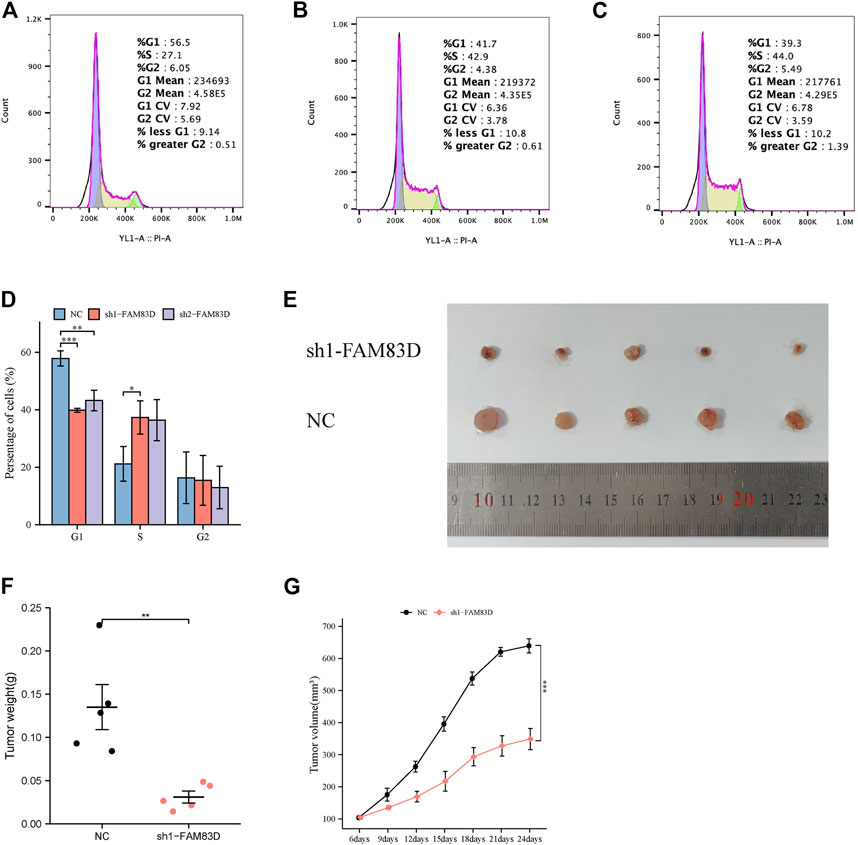
FIGURE 8. Flow cytometry assay and animal experiment. (A–D) Flow cytometric assays revealed that knockdown of FAM83D blocked cell cycle. (E–G) Knockdown of FAM83D inhibits tumor growth in vivo (*p < 0.05, **p < 0.01, ***p < 0.001).
Knockdown ofFAM83D inhibits tumor growth in vivo
To further validate the function of FAM83D, we divided nude mice into sh1-FAM83D and NC groups. Tumor volume was measured every 3 days starting at approximately 1 week after MCF-7 cell injection. Tumor volume in the sh1-FAM83D group was much lower than that of the NC group, as shown in Figure 8E. Mice were then euthanized and their tumors were 24 days after cell injection. These tumorigenicity experiments revealed that silencing FAM83D had a lower tumorigenic potential than was shown for the control mice. The size, weight, and volume of the tumors in the treatment group were considerably lower than those in the NC group (Figures 8F,G).
Discussion
It has previously been reported that FAM83D is involved in various complex biological processes, including cell proliferation, apoptosis, and invasion (Cooper et al., 2020; Yin et al., 2020; Yu et al., 2020). In addition, emerging studies have suggested a functional link between FAM83D and tumor development (Meng et al., 2021; Zhao et al., 2022). However, there is no unified conclusion on how FAM83D plays a role in different tumor types or the underlying mechanisms. To our knowledge, there is currently no pan-cancer analysis of FAM83D. Therefore, we comprehensively analyzed the role of FAM83D expression in various tumor types, with a special focus on BRCA, from the perspectives of gene expression, prognosis, immune infiltration, and related pathways using multiple cohort datasets.
To analyze FAM83D expression among different cancer types, we examined pan-cancer differential FAM83D expression between cancer tissue samples and their matched normal tissue samples from the TCGA dataset. First, we investigated the expression level of FAM83D. We found that in most cancer types, such as COAD, LUAD, LUSC, GBM, and BRCA, FAM83D expression in tumor tissue was significantly higher than in normal tissue. Subsequently, based on survival data from the TCGA, we found a highly consistent trend between poor patient prognosis and high FAM83D expression in multiple tumor types, including BRCA. These findings were further verified in the independent dataset of BRCA in the GEO database.
Next, we focused on exploring the role of FAM83D in BRCA. GSEA enrichment analysis of BRCA data in TCGA showed that genes from proliferation and cell cycle-related pathways were more likely to be enriched in the high FAM83D expression BRCA cohort. In addition, single-cell analysis demonstrated that FAM83D expression is strongly associated with cell proliferation and cell cycle in BRCA. CCK-8 assay also demonstrated this functionality. It is worth mentioning that the PLK1 pathway is closely related to the expression level of FAM83D. In addition, in the molecular correlation analysis of TCGA, GSE93601, and GSE70947, BRCA samples showed a strong positive correlation between the expression of PLK1 and FAM83D. This suggests that FAM83D may play a role in promoting tumorigenesis by affecting the PLK1 pathway. The PLK1 pathway has previously been shown in a large number of studies to be closely related to a variety of cancers, including BRCA (Mao et al., 2021; Shi et al., 2016), and plays a role in multiple biological processes such as BRCA cell proliferation, drug resistance, and metastasis (Chiappa et al., 2022; Qian et al., 2011; Wang et al., 2021).
In the treatment of patients with breast cancer, immunotherapy plays a critical role, and the continuous emergence of new biomarkers adds to the early and successful treatment of BRCA. The expression of FAM83D was firmly related to immune cells such as Th2 cells through our pathway enrichment and immune infiltration analyses. There is evidence that Th2 cells can promote tumorigenesis and progression, thus leading to poor tumor immunotherapy efficacy. This may one reason for the poor survival outcomes seen in patients with high expression of FAM83D (Espinoza et al., 2016). In contrast, there was a negative correlation between the expression level of FAM8D and NK cells. As powerful innate immune cells with multiple mechanisms to kill cancer cells, NK cells play a crucial role in the immune response to cancer. Therefore, further studies are needed to reveal the complex relationship between FAM83D and NK cells in BRCA (Lee et al., 2021).
Despite our comprehensive and detailed analysis of FAM83D, this study mainly focuses on the pan-cancer expression of FAM83D and its prognostic significance. There is a need for further research on FAM83D protein levels and the specific molecular mechanisms of FAM83D function in BRCA.
Conclusion
In sum, our study demonstrates the important pan-cancer role of FAM83D gene expression, especially in BRCA. Thus, it may serve as a key prospective prognostic biomarker of BRCA.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by University of Science and Technology of China Animal Experimentation Ethics Committee.
Author contributions
HY, QC planned and designed this study. HY, QC conducted the statistical analysis. HY, QC wrote the first draft. ZW, XQ assisted with molecular experiments. HY, QC, ZW, XQ, YP revised the manuscript. YP, XQ conducted arbitration in case of disagreement and ensured that there was no error. Each author reviewed and approved the final version of the manuscript.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the National Natural Science Funds of China (No. 82172775), National Key Clinical Specialty Construction Project 2021 Oncology(No. 2021GJLC01), Anhui Province Cancer Bioimmunotherapy Clinical Medical Research Center (No. 202101B10202005) and Construction Project of Provincial Key Medical and Health Specialties (No. 2021sjlczdzk). Funders only provide financial support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1009325/full#supplementary-material
References
Bindea, G., Mlecnik, B., Tosolini, M., Kirilovsky, A., Waldner, M., Obenauf, A. C., et al. (2013). Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39 (4), 782–795. doi:10.1016/j.immuni.2013.10.003
Chiappa, M., Petrella, S., Damia, G., Broggini, M., Guffanti, F., and Ricci, F. (2022). Present and future perspective on plk1 inhibition in cancer treatment. Front. Oncol. 12, 903016. doi:10.3389/fonc.2022.903016
Cipriano, R., Graham, J., Miskimen, K. L. S., Bryson, B. L., Bruntz, R. C., Scott, S. A., et al. (2012). Fam83b mediates egfr- and ras-driven oncogenic transformation. J. Clin. Invest. 122 (9), 3197–3210. doi:10.1172/JCI60517
Cooper, L. M., Hanson, A., Kavanagh, J. A., and Waddell, D. S. (2020). Fam83d modulates map kinase and akt signaling and is induced during neurogenic skeletal muscle atrophy. Cell. Signal. 70, 109576. doi:10.1016/j.cellsig.2020.109576
Espinoza, J. A., Jabeen, S., Batra, R., Papaleo, E., Haakensen, V., Wielenga, V. T., et al. (2016). Cytokine profiling of tumor interstitial fluid of the breast and its relationship with lymphocyte infiltration and clinicopathological characteristics. OncoImmunology 5 (12), e1248015. doi:10.1080/2162402X.2016.1248015
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: The next generation. Cell 144 (5), 646–674. doi:10.1016/j.cell.2011.02.013
Hanzelmann, S., Castelo, R., and Guinney, J. (2013). Gsva: Gene set variation analysis for microarray and rna-seq data. BMC Bioinforma. 14, 7. doi:10.1186/1471-2105-14-7
Lee, E. H. C., Wong, D. C. P., and Ding, J. L. (2021). Nk cells in a tug-of-war with cancer: The roles of transcription factors and cytoskeleton. Front. Immunol. 12, 734551. doi:10.3389/fimmu.2021.734551
Li, X., Sun, C., Chen, J., Ma, J., and Pan, Y. (2022). Suppression of fam83d inhibits glioma proliferation, invasion and migration by regulating the akt/mtor signaling pathway. Transl. Oncol. 22, 101454. doi:10.1016/j.tranon.2022.101454
Li, Y., Dong, X., Yin, Y., Su, Y., Xu, Q., Zhang, Y., et al. (2005). Bj-tsa-9, a novel human tumor-specific gene, has potential as a biomarker of lung cancer. Neoplasia 7 (12), 1073–1080. doi:10.1593/neo.05406
Liu, J., Lichtenberg, T., Hoadley, K. A., Poisson, L. M., Lazar, A. J., Cherniack, A. D., et al. (2018). An integrated tcga pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173 (2), 400–416. doi:10.1016/j.cell.2018.02.052
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol. 15 (12), 550. doi:10.1186/s13059-014-0550-8
Lu, H., Shi, C., Wang, S., Yang, C., Wan, X., Luo, Y., et al. (2020). Identification of ncaph as a biomarker for prognosis of breast cancer. Mol. Biol. Rep. 47 (10), 7831–7842. doi:10.1007/s11033-020-05859-9
Mao, S., Tian, S., Luo, X., Zhou, M., Cao, Z., and Li, J. (2021). Overexpression of plk1 relieved the myocardial ischemia-reperfusion injury of rats through inducing the mitophagy and regulating the p-ampk/fundc1 axis. Bioengineered 12 (1), 2676–2687. doi:10.1080/21655979.2021.1938500
Meng, T., Tong, Z., Yang, M. Y., Zhang, Y., Liu, Y., Wang, Z. Z., et al. (2021). Immune implication of fam83d gene in hepatocellular carcinoma. J. Artic. 12 (1), 3578–3592. doi:10.1080/21655979.2021.1950260
Mu, Y., Zou, H., Chen, B., Fan, Y., and Luo, S. (2017). Fam83d knockdown regulates proliferation, migration and invasion of colorectal cancer through inhibiting fbxw7/notch-1 signalling pathway. Biomed. Pharmacother. 90, 548–554. doi:10.1016/j.biopha.2017.03.073
Qian, Y., Hua, E., Bisht, K., Woditschka, S., Skordos, K. W., Liewehr, D. J., et al. (2011). Inhibition of polo-like kinase 1 prevents the growth of metastatic breast cancer cells in the brain. Clin. Exp. Metastasis 28 (8), 899–908. doi:10.1007/s10585-011-9421-9
Saikia, S., Pal, U., Kalita, D. J., Rai, A. K., Sarma, A., Kataki, A. C., et al. (2021). Runx1t1, a potential prognostic marker in breast cancer, is co-ordinately expressed with erα, and regulated by estrogen receptor signalling in breast cancer cells. Mol. Biol. Rep. 48 (7), 5399–5409. doi:10.1007/s11033-021-06542-3
Shi, R., Sun, J., Sun, Q., Zhang, Q. L., Xia, W. J., Dong, G. C., et al. (2016). Upregulation of fam83d promotes malignant phenotypes of lung adenocarcinoma by regulating cell cycle. Am. J. Cancer Res. 6 (11), 2587–2598.
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer statistics, 2022. Ca. Cancer J. Clin. 72 (1), 7–33. doi:10.3322/caac.21708
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Vivian, J., Rao, A. A., Nothaft, F. A., Ketchum, C., Armstrong, J., Novak, A., et al. (2017). Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 35 (4), 314–316. doi:10.1038/nbt.3772
Walter, W., Sanchez-Cabo, F., and Ricote, M. (2015). Goplot: An r package for visually combining expression data with functional analysis. Bioinformatics 31 (17), 2912–2914. doi:10.1093/bioinformatics/btv300
Wang, B., Huang, X., Liang, H., Yang, H., Guo, Z., Ai, M., et al. (2021). Plk1 inhibition sensitizes breast cancer cells to radiation via suppressing autophagy. Int. J. Radiat. Oncol. Biol. Phys. 110 (4), 1234–1247. doi:10.1016/j.ijrobp.2021.02.025
Wang, D., Han, S., Peng, R., Wang, X., Yang, X., Yang, R., et al. (2015). Fam83d activates the mek/erk signaling pathway and promotes cell proliferation in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 458 (2), 313–320. doi:10.1016/j.bbrc.2015.01.108
Yin, C., Lin, X., Wang, Y., Liu, X., Xiao, Y., Liu, J., et al. (2020). Fam83d promotes epithelial-mesenchymal transition, invasion and cisplatin resistance through regulating the akt/mtor pathway in non-small-cell lung cancer. Cell. Oncol. 43 (3), 395–407. doi:10.1007/s13402-020-00494-9
Yu, C., Cheng, Z., Cui, S., Mao, X., Li, B., Fu, Y., et al. (2020). Circfoxm1 promotes proliferation of non-small cell lung carcinoma cells by acting as a cerna to upregulate fam83d. J. Exp. Clin. Cancer Res. 39 (1), 55. doi:10.1186/s13046-020-01555-5
Yu, G., Wang, L., Han, Y., and He, Q. (2012). Clusterprofiler: An r package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 16 (5), 284–287. doi:10.1089/omi.2011.0118
Zeng, C. X., Fu, S. B., Feng, W. S., Zhao, J. Y., Li, F. X., and Gao, P. (2019). Tcf 19 enhances cell proliferation in hepatocellular carcinoma by activating the atk/foxo1 signaling pathway. Neoplasma 66 (1), 46–53. doi:10.4149/neo_2018_171227N845
Keywords: pan-cancer, breast cancer, prognosis, FAM83D, plk1, biomarker
Citation: Yu H, Chen Q, Wang Z, Qian X and Pan Y (2022) Pan-cancer and single-cell analysis reveals FAM83D expression as a cancer prognostic biomarker. Front. Genet. 13:1009325. doi: 10.3389/fgene.2022.1009325
Received: 01 August 2022; Accepted: 24 November 2022;
Published: 09 December 2022.
Edited by:
Shihai Liu, The Affiliated Hospital of Qingdao University, ChinaReviewed by:
Nousheen Bibi, Shaheed Benazir Bhutto Women University, PakistanJianping Bi, Hubei Cancer Hospital, China
Copyright © 2022 Yu, Chen, Wang, Qian and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojun Qian, cWlhbnhqQHVzdGMuZWR1LmNu; Yueyin Pan, cGFueXVleWluQHVzdGMuZWR1LmNu
†These authors have contributed equally to this work
 Haiyang Yu
Haiyang Yu Qinhao Chen
Qinhao Chen Ziming Wang
Ziming Wang Xiaojun Qian
Xiaojun Qian Yueyin Pan
Yueyin Pan