
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 15 September 2022
Sec. Livestock Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1008649
This article is part of the Research Topic Epigenomics Implication for Economic Traits in Domestic Animals View all 10 articles
 Yi-Fan Liu1,2†
Yi-Fan Liu1,2† Ming Zhang1,2†
Ming Zhang1,2† Yan-Ju Shan1,2
Yan-Ju Shan1,2 Li-Chuan Pang1,2
Li-Chuan Pang1,2 Gai-Ge Ji1,2
Gai-Ge Ji1,2 Xiao-Jun Ju1,2
Xiao-Jun Ju1,2 Yun-Jie Tu1,2
Yun-Jie Tu1,2 Shi-Ying Shi1,2
Shi-Ying Shi1,2 Hao Bai3
Hao Bai3 Jian-Min Zou1,2
Jian-Min Zou1,2 Jing-Ting Shu1,2*
Jing-Ting Shu1,2*MicroRNAs (miRNAs) might play critical roles in skeletal myofiber specification. In a previous study, we found that chicken miR-499-5p is specifically expressed in slow-twitch muscle and that its potential target gene is SOX6. In this study, we performed RNA sequencing to investigate the effects of SOX6 and miR-499-5p on the modulation and regulation of chicken muscle fiber type and its regulatory mechanism. The expression levels of miR-499-5p and SOX6 demonstrated opposing trends in different skeletal muscles and were associated with muscle fiber type composition. Differential expression analysis revealed that miR-499-5p overexpression led to significant changes in the expression of 297 genes in chicken primary myoblasts (CPMs). Myofiber type-related genes, including MYH7B and CSRP3, showed expression patterns similar to those in slow-twitch muscle. According to functional enrichment analysis, differentially expressed genes were mostly associated with muscle development and muscle fiber-related processes. SOX6 was identified as the target gene of miR-499-5p in CPM using target gene mining and luciferase reporter assays. SOX6 knockdown resulted in upregulation of the slow myosin genes and downregulation of fast myosin genes. Furthermore, protein-protein interaction network analysis revealed that MYH7B and RUNX2 may be the direct targets of SOX6. These results indicated that chicken miR-499-5p may promote slow-twitch muscle fiber formation by repressing SOX6 expression. Our study provides a dataset that can be used as a reference for animal meat quality and human muscle disease studies.
Meat quality is an important livestock economic characteristic and is determined by muscle color, water holding capacity, pH, tenderness, flavor substance content, and intermuscular fat. As an essential tissue accounting for 40% of an animal’s body weight, skeletal muscle contains muscle fibers that demonstrate diverse biochemical and structural features, and different fiber compositions in skeletal muscle are closely associated with muscle quality (Lee et al., 2010; Joo et al., 2013; Ismail and Joo, 2017). Myofibers within chickens are classified as white and red fibers; they indicate glycolytic (type IIB; undergo glycolytic metabolism and rapid contraction) and oxidative (type I/IIA; undergo oxidative metabolism and slow contraction) fibers, respectively (Zierath and Hawley, 2004; Hakamata et al., 2018).
The skeletal muscle fiber type is directly determined by various genetic factors. miRNAs are small (22 nucleotides) non-coding RNAs, which show negative gene regulation at the post-transcription level by combining with the 3'-untranslated region (3'UTR) in target mRNAs (Gladka et al., 2012). Several studies have suggested that miRNAs regulate skeletal muscle function, including fiber-type transition and differentiation (Zhang et al., 2014; Zhang et al., 2019; Yin et al., 2020).
miR-499-5p has been identified as a myosin-encoding miRNA, which is recognized as an essential regulatory factor for muscle fiber conversion and as a muscle disease biomarker (Liu et al., 2016; Wan et al., 2018; Shi et al., 2019). Van Rooij et al. first reported that miR-499-5p and miR-208b knockouts in mice resulted in slow-to-fast alteration of muscle fiber type (van Rooij et al., 2009). In mammals and fish, miR-499 is highly expressed within slow-twitch muscles, which possibly modulates the types of muscle fibers via target mRNA, including SOX6, THRAP1, FNIP1, ROD1, and pathways, including NFATc1/MEF2C and FNIP1/AMPK (Nachtigall et al., 2015; Liu et al., 2016; Wang et al., 2017; Xu et al., 2018). Nonetheless, the possible role of miR-499-5p and related mechanisms in chicken muscle fiber switching have not been clarified.
RNA-seq is an effective tool for gene function studies because it can obtain global gene expression levels in tissues or cells with high accuracy (Liu et al., 2017; Pu et al., 2022). In this study, RNA-seq was used to explore the biological function of miR-499-5p in chicken skeletal muscle fiber specification and to identify the target gene of miR-499-5p in chicken primary myoblasts (CPMs). Our findings may enable a better understanding of the underlying mechanisms implicated in the quality performance of chicken meat.
The Wenchang chickens used for ATPase staining and quantitative real-time PCR (qRT-PCR) assays were obtained from Tanniu Chicken Co., Ltd. (Haikou, China). Six 20-weeks-old hens of similar body weights (1,315–1,430 g) were slaughtered by electrical stunning and exsanguination. Intermediate section samples of various skeletal muscles were collected after rapid dissection, followed by freezing in liquid nitrogen and preservation at -80°C. Skeletal muscle samples included the pectoralis major (PM), sartorius (SA), lateral pectineus (LP), and medial gastrocnemius (MG) muscles.
According to our previous report, myofiber types were identified using myosin ATPase staining (Li et al., 2016). As described by Brooke and Kaiser (1970), type IIB was intensely active and stained dark brown, type IIA was weakly active and stained light brown, and type was inactive and non-stained.
We separated CPMs in E11 chicken leg muscles according to a previous description, followed by culturing in DMEM/F12 medium (Gibco, Carlsbad, CA, United States) containing 20% fetal bovine serum (FBS; Gibco) (Shan et al., 2020). When the density reached 80%, FBS was replaced with 5% horse serum (Procell, Wuhan, China) to induce myogenic differentiation. miR-499-5p mimics, NC inhibitor, siRNA targeting SOX6, or siRNA negative control were transfected into CPMs using Lipofectamine 3,000 (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s protocol. The miRNA mimics and siRNAs were prepared by Ribobio Co., Ltd (Guangzhou, China). The SOX6 siRNA sequence was 5'-GGTGAATACAAACAACTGA-3'. After 48 h of transfection, each cell group with four replicates was collected for the subsequent RNA-seq and qRT-PCR assays.
Total RNA was extracted from CPMs using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s protocol. An Agilent 2,100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States) and a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) were used to check RNA quality. High-throughput RNA-seq was performed on an Illumina HiSeq 4,000 platform (Illumina, San Diego, CA, United States) at Gene Denovo Biotechnology Co. (Guangzhou, China).
For quality control, fastp v0.18 with default parameters was utilized to process the raw data, followed by alignment of paired-end clean reads against the reference genome (GRCg6a) with Hisat2 (Sirén et al., 2014; Chen et al., 2018). FeatureCounts were used to count reads mapped into diverse genes, and fragments per kilobase million (FPKM) values were determined for diverse genes (Liao et al., 2014). Differentially expressed genes (DEGs) in the treatment and control groups were identified using the DESeq2 R package (Love et al., 2014). DEGs were identified based on the threshold of false discovery rate (FDR) < 0.05 with DESeq2. Target genes of miR-499-5p were identified using the TargetScan 7.2 (http://www.targetscan.org/vert_72/) and miRDB (http://mirdb.org/) web platforms with default parameters.
Gene ontology (GO) enrichment analysis of DEGs was performed using an online tool from OMICSHARE (www.omicshare.com/tools). Significant enrichment of GO terms was indicated by p-value < 0.05. OMICSHARE was used to identify significantly enriched DEGs in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The STRING database (https://string-db.org/) was utilized for protein-protein interaction (PPI) network construction at moderate confidence. Cytoscape software v3.5.1 was used for visualization.
The Primer3 website (https://bioinfo.ut.ee/primer3-0.4.0/) and miRprimer2 software (https://sourceforge.net/projects/mirprimer) were used to design qRT-PCR primers for genes and miR-499-5p, respectively. Primer information is listed in Supplementary Table S1. For mRNA quantification, cDNA was prepared using the HiScript III 1st Strand cDNA Synthesis Kit (+gDNA wiper) (R312, Vazyme, Nanjing, China). qRT-PCR with ChamQ SYBR Color qRT-PCR Master Mix (Q411, Vazyme) was performed for cDNA quantification using a real-time PCR system (Mx3000P, Agilent Technologies). For miRNA quantification, cDNA was synthesized using the miRcute Plus miRNA First-Strand cDNA kit (KR211, Tiangen, Beijing, China), and qRT-PCR was performed using miRcute Plus miRNA qPCR kit (SYBR Green) (FP411, Tiangen). Chicken GAPDH and U6 snRNA were used as endogenous references. The 2−ΔΔCt method was used to calculate the fold-change.
DF-1 cells were used for the luciferase reporter assay. The PmiR-RB-Report vector was used to synthesize mutant (MUT) and wildtype (WT) luciferase reporter vectors at Ribobio Co., Ltd (Guangzhou, China). The information on mutant sites in the luciferase reporter vectors is presented in Supplementary Table S2. The cells were transfected with MUT or WT constructs and miR-499-5p mimics or NC mimics. After 48 h, luminescence was detected using a Dual-Glo Luciferase Assay System (E2920, Promega, Madison, WI, United States).
CPMs were seeded in six-well plates and transfected with overexpression miRNA mimics for 48 h. Cells were harvested, washed with 1× phosphate buffered saline (PBS) and lysed in RIPA lysis buffer. Immunoblotting was performed using standard procedures. The primary antibodies used were anti-SOX6 (NBP2-20458, NOVUS, Saint Louis, MI, United States) and anti-GAPDH (ERAB0003, Erwanbiotech, Shanghai, China). The goat anti-rabbit IgG-HRP (111-035-003; Jackson immune Research, Bar Harbor, ME, United States) was used as the secondary antibody.
The results of the fiber type content, miR-499-5p, and gene levels within muscles and cells are presented as mean ± SD. The expression patterns of the genes and miR-499-5p were statistically compared among the different muscle groups using the one-way analysis of variance (ANOVA) followed by Tukey’s test, as implemented in the SPSS Software Suite (SPSS version 20.0). The statistical significance of qRT-PCR analysis results and luciferase reporter assay results between the treatment and control groups was determined using t-test. Differences were considered statistically significant at p-values of < 0.05 and < 0.01.
As shown in Figures 1A,B, four different skeletal muscles—the PM, SA, LP, and MG, exhibit different muscle fiber compositions. PM is composed entirely of type ΙIB fiber, whereas the other three contain three types of muscle fibers. SA has the highest content of type ΙIA fibers, whereas MG has the most amount of type fiber.
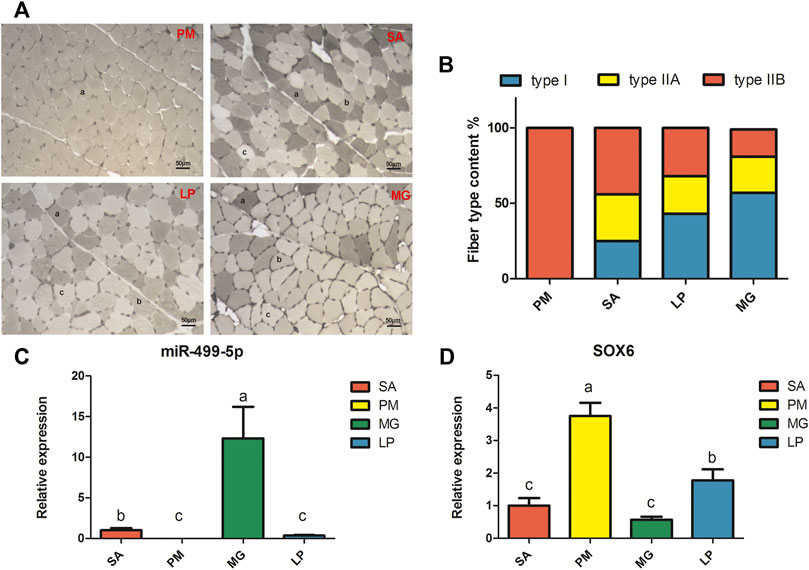
FIGURE 1. Myofiber type phenotype and gene expression levels in four different skeletal muscles. (A) Myosin ATPase staining of four different skeletal muscles. PM: pectoralis major, SA: sartorius, LP: lateral pectineus, MG: medial gastrocnemius. (A) type IIB fibers, (B) type IIA fibers, (C) type I fibers. (B) Fiber type content of four different skeletal muscles. Relative expression levels of miR-499-5p (C) and SOX6 (D) in four different skeletal muscles. All results are shown as mean ± SD. p < 0.05 or less is considered significant and indicated with different letters.
The qRT-PCR results showed that miR-499-5p and SOX6 have opposing expression patterns. As shown in Figure 1C, miR-499-5p showed the highest expression in MG, followed by SA and LP, and the lowest expression in PM. As expected, SOX6 was most significantly upregulated in PM, followed by LP, SA, and MG (Figure 1D). The expression level of miR-499-5p was found to have a significant positive correlation with type I fiber content and negative correlation with type IIB fiber content in skeletal muscles, whereas SOX6 expression showed a similar correlation with type IIB fiber content (Supplementary Table S3).
After 48 h of miR-499-5p overexpression in the CPMs, the qRT-PCR results showed a highly significant increase in miR-499-5p expression compared with that in the control (p < 0.01, Figure 2). Four miR-499-5p overexpression and four control CPMs were collected for transcriptome sequencing. A total of 297 DEGs were detected (FDR <0.05), including 127 upregulated and 170 downregulated genes within cells overexpressing miR-499-5p compared with control cells (Supplementary Table S4). GO enrichment analysis showed that DEGs were mainly related to muscle development, muscle contraction, and muscle cell differentiation (Figure 3A). KEGG pathway analysis showed that the enriched pathways of DEGs included many muscle function-related pathways such as actin cytoskeleton regulation, focal adhesion, and cardiac muscle contraction (Figure 3B).
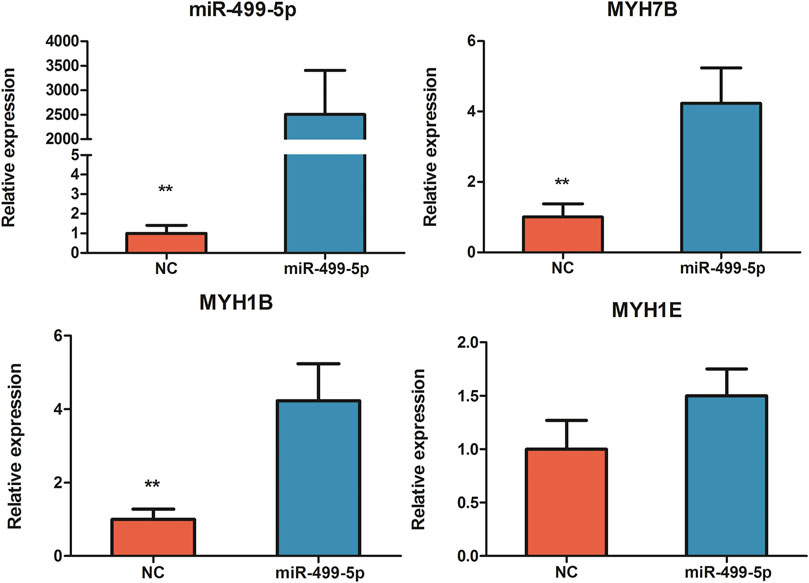
FIGURE 2. The relative expression levels of miR-499-5p, MYH7B, MYH1B, and MYH1E in miR-499-5p overexpressing CPMs and negative control by qRT-PCR. All results are shown as mean ± SD. **p < 0.01 as compared with the control.

FIGURE 3. Functional enrichment of DEGs between miR-499-5p overexpressing and NC CPMs. (A) Top 20 significantly enriched GO-BP terms; (B) Top 20 significantly enriched KEGG pathways.
Furthermore, several key genes associated with muscle fiber type were found to be differentially expressed in miR-499-5p overexpressing CPMs (Table 1). Compared with our previous transcriptome data on slow- and fast-twitch chicken muscles, most of these genes shared similar expression patterns in miR-499-5p-overexpressing CPMs and those in slow-twitch muscles (Liu et al., 2020). A series of myosin genes, including MYH1B, MYH1E, and MYH7B, were upregulated after miR-499-5p overexpression. MYH7B, which encode slow myofibrillar proteins, was highly expressed in the overexpressed cells (log2foldchange = 2.42). The MYH1B, MYH1E, and MYH7B expression patterns were confirmed using qRT-PCR (Figure 2).
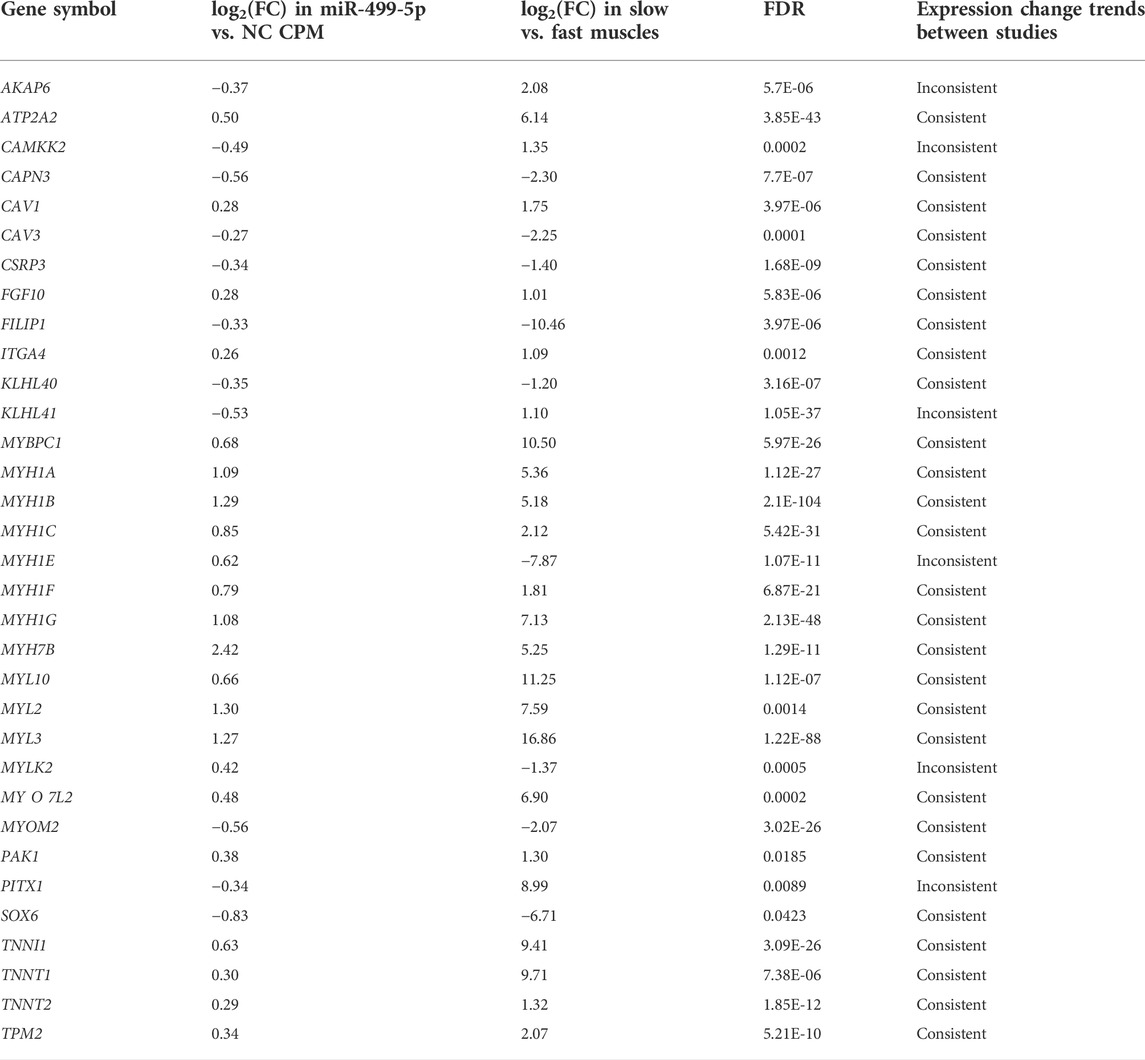
TABLE 1. Expression change of myofiber type-related DEGs between miR-499-5p overexpressing and NC CPMs. Trends in gene expression changes between the present study and our previous study on slow-twitch and fast-twitch muscles in chickens are also shown (Liu et al., 2020).
We used TargetScan and mirDB for miR-499-5p target gene prediction and obtained the target gene information with high confidence by comparing it with transcriptome data. We first identified 444 target genes of miRNA-499-5p in CPMs that could be identified as target genes using at least one tool (Supplementary Table S5). We found that the expression of more than 65% of the target genes had reduced after miRNA overexpression, with 16 reaching significant levels (FDR <0.05, Table 2). To improve the target gene prediction accuracy, we used the intersection of the results of the two tools as the possible targets of miR-499-5p. A total of 72 target genes were identified in CPMs, four of which were significantly downregulated in the overexpressed cells (FDR <0.05).
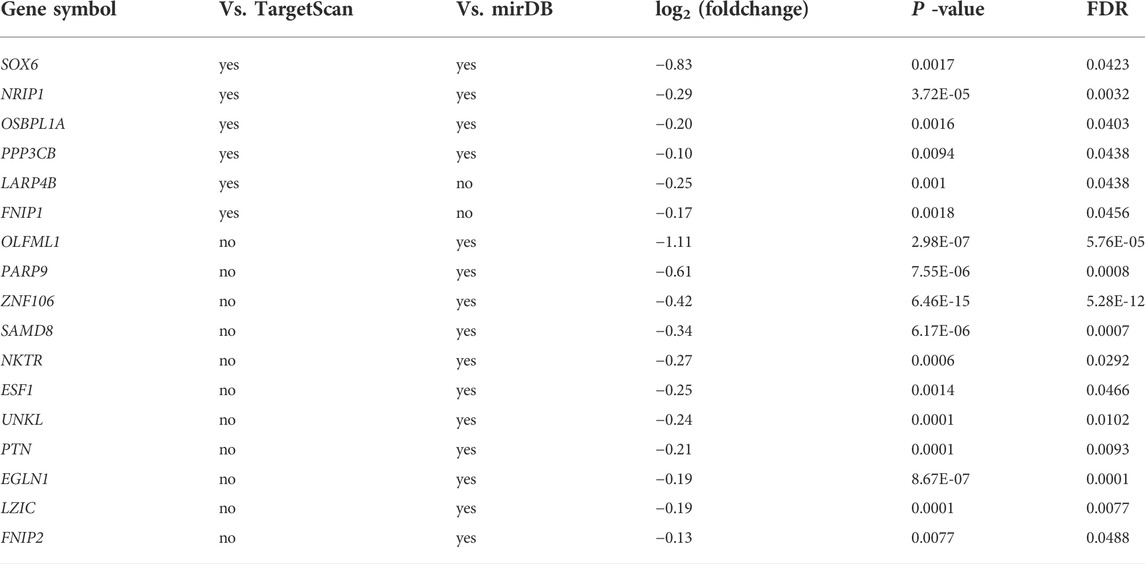
TABLE 2. Expression change of miR-499-5p target genes in miR-499-5p overexpressing CPMs compared with negative control. TargetScan and mirDB were used for miR-499-5p target gene prediction.
Among the four significantly downregulated target genes, SOX6 showed the most significant decrease in expression (log2foldchange = -0.83). In addition, target genes associated with myofiber-type switching, including PPP3CB, FNIP1, and FNIP2, were identified, but they presented only lower expression changes.
We used qRT-PCR, western blotting, and luciferase reporter assay to validate the target binding relationship between miR-499-5p and the target genes (Figure 4). The qRT-PCR results revealed that the relative SOX6, PPP3CB, and FNIP1 expression levels decreased in the overexpressed cells, with SOX6 showing the most remarkable change in expression, consistent with the RNA-seq results. Overexpression of miR-499-5p decreased the protein level of SOX6. The luciferase reporter assay showed that miR-499-5p significantly decreased luciferase activity in combination with SOX6 3'-UTR sites (p-value < 0.05). However, the PPP3CB and FNIP1 luciferase activities were not significantly altered following miR-499-5p transfection (p-value > 0.05).
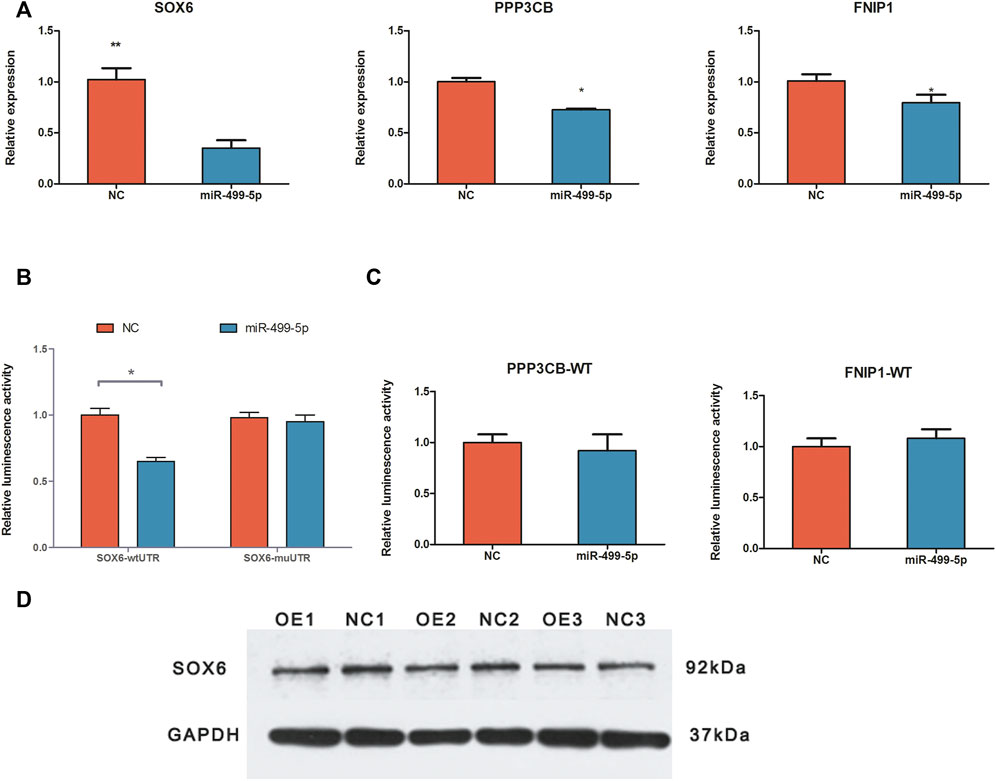
FIGURE 4. Validation of targeted binding relationship between miR-499-5p and target genes. (A) The relative expression levels of SOX6, PPP3CB, and FNIP1 in miR-499-5p overexpressing CPMs and negative control by qRT-PCR; (B) Results of luciferase reporter assay for SOX6 and miR-499-5p; miR-499-5p binding site mutant and wildtype vector of SOX6’s 3'UTR were used. (C) Results of the luciferase reporter as say for PPP3CB/miR-499-5p and FNIP1/miR-499-5p; only wildtype vectors were used. All results are shown as mean ± SD. **p < 0.01 and *p < 0.05 as compared with control. (D) The protein levels of SOX6 in miR-499-5p overexpressing CPMs and negative control by western bolting.
To investigate the effect of SOX6 on chicken muscle fiber specification, RNA interference and RNA-seq techniques were used to obtain the expression patterns of muscle fiber-related genes and to construct a regulatory network for muscle fiber specification. After RNA interference, SOX6 expression was 0.6- and 2.6-fold downregulated in CPMs, as determined using qRT-PCR and RNA-seq, respectively (Figure 5 and Table 3). We identified 1,448 DEGs in si-SOX6 and control CPMs using FDR <0.05 as the screening criterion. Among these, 107 genes overlapped with the results of the miR-499-5p overexpression experiment (Supplementary Table S6). The foldchange of MYH7B, MYH1B, CSRP3, TNNI1, MYL2 and MYL10 expression were confirmed using qRT-PCR (Figure 6).
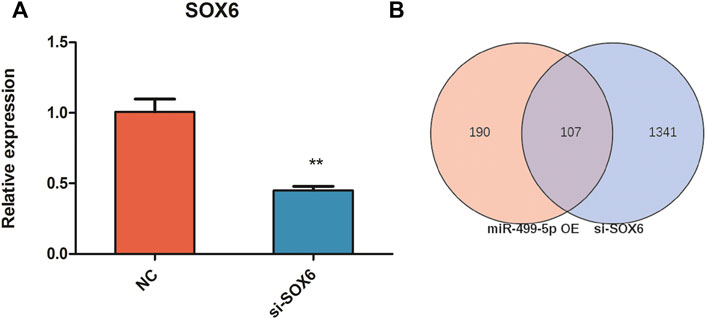
FIGURE 5. The inhibition efficiency of si-SOX6. (A) The relative expression levels of SOX6 in SOX6 knockdown CPMs and negative control by qRT-PCR. (B) The Venn diagram of differential genes in miR-499 overexpressing and SOX6 knockdown assays. All results are shown as mean ± SD. **p < 0.01 as compared with the control.

TABLE 3. Expression change of SOX6 and myosin genes in SOX6 knockdown CPMs compared with negative control.
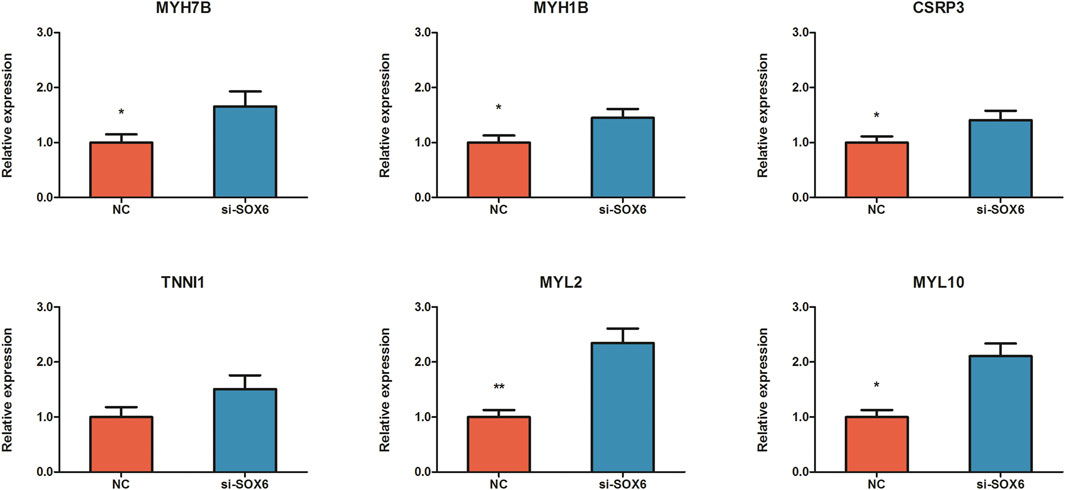
FIGURE 6. Validaton of expression change of MYH7B, MYH1B, CSRP3, TNNI1, MYL2 and MYL10 between SOX6 knockdown and NC CPMs using qRT-PCR. All results are shown as mean ± SD. **p < 0.01 and *p < 0.05 as compared with control.
The myosin gene expression pattern revealed that the knockdown of SOX6 resulted in significant upregulation of slow-twitch muscle-associated MYH7, MYH7B, and MYH1B and significant downregulation of fast-twitch muscle-associated MYH1D and MYH1F (Table 3). A total of 24 differentially expressed genes, known for myofiber type switching, were selected to build a protein-protein interaction network based on the STRING database. Among them, a total of six genes, MYH7B, MYH1B, CSRP3, TNNI1, MYL2 and MYL10, were down-regulated after SOX6 interference, while the remaining genes were up-regulated. According to the network, SOX6 may have a direct regulatory relationship with RUNX2, PRDM1, HDAC4, and MYH7B (Figure 7).
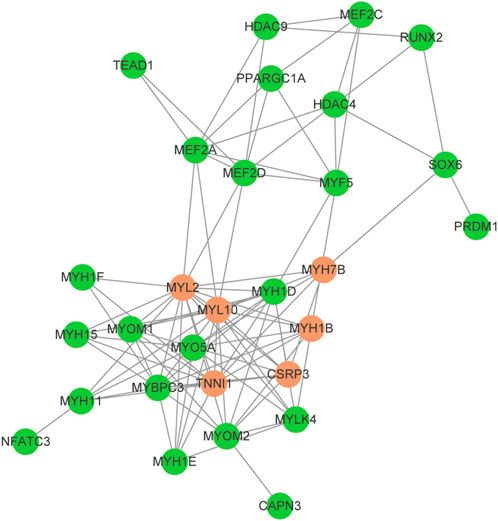
FIGURE 7. Protein-protein interaction network for the selected DEGs between SOX6 knockdown and NC CPMs. Upregulated genes are shown in red and downregulated genes are shown in blue.
Improving meat quality is the ultimate goal of broiler breeding, especially in native Chinese breeds, and the composition of muscle fibers is a critical factor affecting meat quality. For instance, high oxidative fiber level facilitates meat flavor and juiciness (Sibut et al., 2011). Muscle fiber characteristics are affected by muscle type, location, and function within an animal (Shu et al., 2017). To reveal the regulatory mechanisms affecting myofiber type in chicken skeletal muscle, we analyzed miRNA expression profiles in oxidative and glycolytic muscles in our previous study (Liu et al., 2020). miR-499-5p showed specific expression in oxidative muscles, consistent with the results observed in fish and pigs, suggesting that this miRNA is a crucial factor in regulating chicken myofiber type conversion (Duran et al., 2015; Jiang et al., 2021). In this study, a comparison of different muscles revealed a positive correlation between miR-499-5p and type I muscle fiber content and a negative correlation between miR-499-5p and type IIB muscle fiber content, implying that miR-499-5p is associated with meat quality traits.
miR-499-5p belongs to MyomiRs, which are mainly expressed in the heart and skeletal muscles and can be encoded via specific introns in slow muscle-specific myosin MYH7B (McCarthy, 2011). miR-499-5p positively regulates the formation of slow-twitch muscle fibers and negatively regulates the formation of fast-twitch muscle fibers in mouse and pig models (van Rooij et al., 2009; Wang et al., 2017). Although miRNAs are highly conserved among species, there is a lack of evidence regarding the effect of miR-499-5p on chicken myofiber type specification (Lee et al., 2007). In this study, RNA-seq was used to investigate the effect of miR-499-5p on the CPM transcriptome. Therefore, miR-499-5p overexpression markedly enhanced MYH7B and MYH1B mRNA expression, demonstrating its ability to promote slow-twitch muscle fiber formation in chickens. Many myofiber type-related genes were differentially expressed after overexpression and showed a similar expression pattern to that of the slow-twitch muscle. For example, cysteine and glycine-rich protein 3 (CSRP3), also known as muscle LIM protein (MLP), which promotes fast-twitch muscle fiber formation in chickens, showed significantly decreased mRNA expression post miR-499-5p overexpression (Shan et al., 2022). Calpain 3 (CAPN3) is the only muscle-specific calpain that has impotant role in muscle function such as muscle formation and remodeling (Chen et al., 2021). Studies have indicated that mutations in CAPN3 are associated with meat quality traits including muscle fiber composion in chicken and pig (Gandolfi et al., 2011; Felício et al., 2013). In the present study, CAPN3 was found to be downregulated following miR-499-5p overexpression, suggesting their roles in the regulation of muscle fiber types. Functional enrichment analysis revealed that DEGs are involved in several muscle development-related processes and signaling pathways associated with myofiber-type switching, including focal adhesion and regulation of the actin cytoskeleton (Ohnuki et al., 2013; Graham et al., 2015). Collectively, miR-499-5p promotes slow-twitch muscle fiber formation through diverse regulatory factors and signaling pathways.
miRNAs are involved in regulating biological processes by binding to the 3'UTR of target mRNA to repress gene expression (Bartel, 2009). Transcriptome data and bioinformatic tools were used to identify miR-499-5p target genes. More than 60% of the predicted target genes showed decreased expression in the CPM, indicating that the miRNA transfection in this study was effective. Under stringent prediction criteria, SOX6 was identified as the target gene with the highest decrease in expression. SOX6 is a transcription factor that has a critical effect on embryonic muscle development and myofiber maintenance (Hagiwara et al., 2005; Hagiwara et al., 2007; Quiat et al., 2011; Jackson et al., 2015). In chickens, copy number variation in SOX6 is associated with muscle development and body weight (Lin et al., 2018; Fernandes et al., 2021). Previous studies have reported that SOX6 is a target of miR-499-5p in skeletal and cardiac muscles (Jia et al., 2016; Wang et al., 2017). Our results revealed opposing expression trends of miR-499-5p and SOX6 in different chicken skeletal muscles, consistent with the results observed in Nile tilapia and pigs (Nachtigall et al., 2015; Wang et al., 2017). The luciferase reporter assay showed that SOX6 is the direct target of miR-499-5p, which may regulate chicken myofiber specification by controlling SOX6 expression. Other genes, including FNIP1 and THRAP1, have also been reported to be important targets of miR-499-5p related to muscle fiber-type specification (Liu et al., 2016; Xu et al., 2018). THRAP1 did not show a significant decrease in expression after miR-499-5p overexpression, and FNIP1 was not significantly associated with miR-499-5p. This study revealed that SOX6 is the primary target of miR-499-5p in chicken myofiber-type conversion.
To further understand the effect of SOX6 on chicken myofiber type specification and the regulatory network involved, we performed a transcriptome analysis of SOX6-knockdown CPMs. As expected, the SOX6 knockdown resulted in the formation of slow-twitch muscle fibers and a decrease in fast-twitch muscle fibers. Protein-protein interaction network analysis revealed that SOX6 could act directly on myosin and functional genes related to muscle fiber type. An et al. found that SOX6 could directly target and bind MYH7B and inhibit its expression using CHIP-seq analysis, and the same result was obtained in the present study (An et al., 2011). Although fast-twitch muscle myosin genes, such as MYH1D and MYH1F, were significantly reduced after SOX6 knockdown, there was no evidence that SOX6 could act directly on them. RUNX2 is a critical transcription factor, and SOX6 and RUNX2 interact during osteogenesis (Zhang et al., 2015). RUNX2 has not been determined to play a role in the specification of muscle fiber types; combined with our findings, we hypothesize that SOX6 and RUNX2 can initiate muscle fiber type-related signaling pathways such as the PGC1α/MEF2C pathway (Shan et al., 2020).
In conclusion, we first investigated the role of miR-499-5p and SOX6 in regulating myofiber type in chicken skeletal muscle using RNA-seq. The results revealed that both miR-499-5p and SOX6 enhanced slow-twitch muscle fiber formation and that SOX6 is the primary target of miR-499-5p in this process. This study provides detailed transcriptome sequencing results and serves as a reference for related studies on chicken meat quality and other species.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://ngdc.cncb.ac.cn/gsa, CRA007272.
The animal study was reviewed and approved by the Animal Care and Use Committee at the Jiangsu Institute of Poultry Science.
Y-FL, MZ, and J-TS contributed to conception and design of the study. Y-FL, Y-J-TS, L-CP, G-GJ, and X-JJ performed the experiments and participated in data collection. MZ, Y-JT, HB, and S-YS analyzed the data. Y-FL and MZ drafted the manuscript. J-MZ and J-TS revised the manuscript critically. All authors contributed to manuscript revision, read, and approved the submitted version.
This research was funded by the Natural Science Foundation of Jiangsu Province (BK20211121 and BK20210955), the ′JBGS′ Project of Seed Industry Revitalization in Jiangsu Province (JBGS [2021]107), the Special Fund for Independent Innovation of Agricultural Science and Technology in Jiangsu Province of China (CZ(20)3001), the Open Project Program of Joint International Research Laboratory of Agriculture and Agri-Product Safety, the Ministry of Education of China, Yangzhou University (JI-LARKF202020).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1008649/full#supplementary-material
An, C. I., Dong, Y., and Hagiwara, N. (2011). Genome-wide mapping of SOX6 binding sites in skeletal muscle reveals both direct and indirect regulation of muscle terminal differentiation by SOX6. BMC Dev. Biol. 11, 59. doi:10.1186/1471-213X-11-59
Bartel, D. P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell. 136, 215–233. doi:10.1016/j.cell.2009.01.002
Brooke, M. H., and Kaiser, K. K. (1970). Muscle fiber types: How many and what kind? Arch. Neurol. 23, 369–379. doi:10.1001/archneur.1970.00480280083010
Chen, L., Tang, F., Gao, H., Zhang, X., Li, X., and Xiao, D. (2021). CAPN3: A muscle-specific calpain with an important role in the pathogenesis of diseases (review). Int. J. Mol. Med. 48, 203. doi:10.3892/ijmm.2021.5036
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34, i884-i890. doi:10.1093/bioinformatics/bty560
Duran, B. O. d. S., Fernandez, G. J., Mareco, E. A., Moraes, L. N., Salomão, R. A. S., Gutierrez de Paula, T., et al. (2015). Differential microRNA expression in fast- and slow-twitch skeletal muscle of Piaractus mesopotamicus during growth. PLOS ONE. 10, e0141967. doi:10.1371/journal.pone.0141967
Felício, A. M., Boschiero, C., Balieiro, J. C., Ledur, M. C., Ferraz, J. B., Michelan Filho, T., et al. (2013). Identification and association of polymorphisms in CAPN1 and CAPN3 candidate genes related to performance and meat quality traits in chickens. Genet. Mol. Res. 12, 472–482. doi:10.4238/2013.February.8.12
Fernandes, A. C., da Silva, V. H., Goes, C. P., Moreira, G. C. M., Godoy, T. F., Ibelli, A. M. G., et al. (2021). Genome-wide detection of CNVs and their association with performance traits in broilers. BMC Genomics. 22, 354. doi:10.1186/s12864-021-07676-1
Gandolfi, G., Pomponio, L., Ertbjerg, P., Karlsson, A. H., Nanni Costa, L., Lametsch, R., et al. (2011). Investigation on CAST, CAPN1 and CAPN3 porcine gene polymorphisms and expression in relation to post-mortem calpain activity in muscle and meat quality. Meat Sci. 88, 694–700. doi:10.1016/j.meatsci.2011.02.031
Gladka, M. M., da Costa Martins, P. A., and De Windt, L. J. (2012). Small changes can make a big difference—microRNA regulation of cardiac hypertrophy. J. Mol. Cell. Cardiol. 52, 74–82. doi:10.1016/j.yjmcc.2011.09.015
Graham, Z. A., Gallagher, P. M., and Cardozo, C. P. (2015). Focal adhesion kinase and its role in skeletal muscle. J. Muscle Res. Cell. Motil. 36, 305–315. doi:10.1007/s10974-015-9415-3
Hagiwara, N., Ma, B., and Ly, A. (2005). Slow and fast fiber isoform gene expression is systematically altered in skeletal muscle of the SOX6 mutant, p100H. Dev. Dyn. 234, 301–311. doi:10.1002/dvdy.20535
Hagiwara, N., Yeh, M., and Liu, A. (2007). SOX6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Dev. Dyn. 236, 2062–2076. doi:10.1002/dvdy.21223
Hakamata, Y., Watanabe, K., Amo, T., Toyomizu, M., and Kikusato, M. (2018). Characterization of mitochondrial content and respiratory capacities of broiler chicken skeletal muscles with different muscle fiber compositions. J. Poult. Sci. 55, 210–216. doi:10.2141/jpsa.0170141
Ismail, I., and Joo, S. T. (2017). Poultry meat quality in relation to muscle growth and muscle fiber characteristics. Korean J. Food Sci. Anim. Resour. 37, 873–883. doi:10.5851/kosfa.2017.37.6.87
Jackson, H. E., Ono, Y., Wang, X., Elworthy, S., Cunliffe, V. T., and Ingham, P. W. (2015). The role of SOX6 in zebrafish muscle fiber type specification. Skelet. Muscle 5, 2. doi:10.1186/s13395-014-0026-2
Jia, Z., Wang, J., Shi, Q., Liu, S., Wang, W., Tian, Y., et al. (2016). SOX6 and PDCD4 enhance cardiomyocyte apoptosis through LPS-induced miR-499 inhibition. Apoptosis. 21, 174–183. doi:10.1007/s10495-015-1201-6
Jiang, A., Yin, D., Zhang, L., Li, B., Li, R., Zhang, X., et al. (2021). Parsing the microRNA genetics basis regulating skeletal muscle fiber types and meat quality traits in pigs. Anim. Genet. 52, 292–303. doi:10.1111/age.13064
Joo, S. T., Kim, G. D., Hwang, Y. H., and Ryu, Y. C. (2013). Control of fresh meat quality through manipulation of muscle fiber characteristics. Meat Sci. 95, 828–836. doi:10.1016/j.meatsci.2013.04.044
Lee, C. T., Risom, T., and Strauss, W. M. (2007). Evolutionary conservation of microRNA regulatory circuits: An examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell. Biol. 26, 209–218. doi:10.1089/dna.2006.0545
Lee, S. H., Joo, S. T., and Ryu, Y. C. (2010). Skeletal muscle fiber type and myofibrillar proteins in relation to meat quality. Meat Sci. 86, 166–170. doi:10.1016/j.meatsci.2010.04.040
Li, H., Shu, J., Shan, Y., Chen, W., Song, C., and Xu, W. (2016). Myofiber development during embryonic to neonatal development in duck breeds differing in muscle growth rates. J. Integr. Agric. 15, 403–413. doi:10.1016/S2095-3119(14)60949-7
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30, 923–930. doi:10.1093/bioinformatics/btt656
Lin, S., Lin, X., Zhang, Z., Jiang, M., Rao, Y., Nie, Q., et al. (2018). Copy number variation in SOX6 contributes to chicken muscle development. Genes. 9, 42. doi:10.3390/genes9010042
Liu, J., Liang, X., Zhou, D., Lai, L., Xiao, L., Liu, L., et al. (2016). Coupling of mitochondrial function and skeletal muscle fiber type by a miR-499/FNIP1/AMPK circuit. EMBO Mol. Med. 8, 1212–1228. doi:10.15252/emmm.201606372
Liu, Y., Sun, Y., Li, Y., Bai, H., Xue, F., Xu, S., et al. (2017). Analyses of Long non-coding RNA and mRNA profiling using RNA sequencing in chicken testis with extreme sperm motility. Sci. Rep. 7, 9055. doi:10.1038/s41598-017-08738-9
Liu, Y., Zhang, M., Shan, Y., Ji, G., Ju, X., Tu, Y., et al. (2020). miRNA–mRNA network regulation in the skeletal muscle fiber phenotype of chickens revealed by integrated analysis of miRNAome and transcriptome. Sci. Rep. 10, 10619. doi:10.1038/s41598-020-67482-9
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi:10.1186/s13059-014-0550-8
McCarthy, J. J. (2011). The MyomiR network in skeletal muscle plasticity. Exerc. Sport Sci. Rev. 39, 150–154. doi:10.1097/JES.0b013e31821c01e1
Nachtigall, P. G., Dias, M. C., Carvalho, R. F., Martins, C., and Pinhal, D. (2015). MicroRNA-499 expression distinctively correlates to target genes SOX6 and rod1 profiles to resolve the skeletal muscle phenotype in Nile tilapia. PLOS ONE. 10, e0119804. doi:10.1371/journal.pone.0119804
Ohnuki, Y., Umeki, D., Cai, W., Kawai, N., Mototani, Y., Shiozawa, K., et al. (2013). Role of masseter muscle β2-adrenergic signaling in regulation of muscle activity, myosin heavy chain transition, and hypertrophy. J. Pharmacol. Sci. 123, 36–46. doi:10.1254/jphs.12271fp
Pu, F., Xiong, X., Li, Y., Xi, Y., Ma, S., Bai, L., et al. (2022). Transcriptome analysis of oviduct in laying ducks under different stocking densities. Br. Poult. Sci. 63, 283–290. doi:10.1080/00071668.2021.1983917
Quiat, D., Voelker, K. A., Pei, J., Grishin, N. V., Grange, R. W., Bassel-Duby, R., et al. (2011). Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor SOX6. Proc. Natl. Acad. Sci. U. S. A. 108, 10196–10201. doi:10.1073/pnas.1107413108
Shan, Y., Ji, G., Zhang, M., Liu, Y., Tu, Y., Ju, X., et al. (2022). Use of transcriptome sequencing to explore the effect of CSRP3 on chicken myoblasts. J. Integr. Agric.
Shan, Y., Ji, G., Zou, J., Zhang, M., Tu, Y., Liu, Y., et al. (2020). PGC-1α differentially regulates the mRNA expression profiles of genes related to myofiber type specificity in chicken. J. Integr. Agric. 19, 2083–2094. doi:10.1016/S2095-3119(20)63177-X
Shi, Y., Han, Y., Niu, L., Li, J., and Chen, Y. (2019). MiR-499 inhibited hypoxia/reoxygenation induced cardiomyocytes injury by targeting SOX6. Biotechnol. Lett. 41, 837–847. doi:10.1007/s10529-019-02685-3
Shu, J., Xiao, Q., Shan, Y., Zhang, M., Tu, Y., Ji, G., et al. (2017). Oxidative and glycolytic skeletal muscles show marked differences in gene expression profile in Chinese Qingyuan partridge chickens. PLOS ONE. 12, e0183118. doi:10.1371/journal.pone.0183118
Sibut, V., Hennequet-Antier, C., Le Bihan-Duval, E., Marthey, S., Duclos, M. J., and Berri, C. (2011). Identification of differentially expressed genes in chickens differing in muscle glycogen content and meat quality. BMC Genomics. 12, 112. doi:10.1186/1471-2164-12-112
Sirén, J., Välimäki, N., and Mäkinen, V. (2014). Indexing graphs for path queries with applications in genome research. IEEE/ACM Trans. Comput. Biol. Bioinform. 11, 375–388. doi:10.1109/TCBB.2013.2297101
van Rooij, E., Quiat, D., Johnson, B. A., Sutherland, L. B., Qi, X., Richardson, J. A., et al. (2009). A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell. 17, 662–673. doi:10.1016/j.devcel.2009.10.013
Wan, Q., Xu, T., Ding, W., Zhang, X., Ji, X., Yu, T., et al. (2018). miR-499-5p attenuates mitochondrial fission and cell apoptosis via p21 in doxorubicin cardiotoxicity. Front. Genet. 9, 734. doi:10.3389/fgene.2018.00734
Wang, X. Y., Chen, X. L., Huang, Z. Q., Chen, D. W., Yu, B., He, J., et al. (2017). MicroRNA-499-5p regulates porcine myofiber specification by controlling SOX6 expression. Animal. 11, 2268–2274. doi:10.1017/S1751731117001008
Xu, M., Chen, X., Chen, D., Yu, B., Li, M., He, J., et al. (2018). MicroRNA-499-5p regulates skeletal myofiber specification via NFATc1/MEF2C pathway and THRAP1/MEF2C axis. Life Sci. 215, 236–245. doi:10.1016/j.lfs.2018.11.020
Yin, H., Zhao, J., He, H., Chen, Y., Wang, Y., Li, D., et al. (2020). Gga-miR-3525 targets PDLIM3 through the MAPK signaling pathway to regulate the proliferation and differentiation of skeletal muscle satellite cells. Int. J. Mol. Sci. 21, 5573. doi:10.3390/ijms21155573
Zhang, D., Wang, X., Li, Y., Zhao, L., Lu, M., Yao, X., et al. (2014). Thyroid hormone regulates muscle fiber type conversion via miR-133a1. J. Cell. Biol. 207, 753–766. doi:10.1083/jcb.201406068
Zhang, Y., Yan, H., Zhou, P., Zhang, Z., Liu, J., and Zhang, H. (2019). MicroRNA-152 promotes slow-twitch myofiber formation via targeting uncoupling protein-3 gene. Animals. 9, 669. doi:10.3390/ani9090669
Zhang, Y., Yang, T. L., Li, X., and Guo, Y. (2015). Functional analyses reveal the essential role of SOX6 and RUNX2 in the communication of chondrocyte and osteoblast. Osteoporos. Int. 26, 553–561. doi:10.1007/s00198-014-2882-3
Keywords: chicken, meat quality, muscle fiber, RNA sequencing, miR-499-5p, Sox6
Citation: Liu Y-F, Zhang M, Shan Y-J, Pang L-C, Ji G-G, Ju X-J, Tu Y-J, Shi S-Y, Bai H, Zou J-M and Shu J-T (2022) Transcriptome sequencing analysis of the role of miR-499-5p and SOX6 in chicken skeletal myofiber specification. Front. Genet. 13:1008649. doi: 10.3389/fgene.2022.1008649
Received: 01 August 2022; Accepted: 01 September 2022;
Published: 15 September 2022.
Edited by:
Chunfang Zhao, Anhui Science and Technology University, ChinaReviewed by:
Ran Di, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2022 Liu, Zhang, Shan, Pang, Ji, Ju, Tu, Shi, Bai, Zou and Shu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing-Ting Shu, c2h1amluZ3RpbmdAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.