- 1Karunya Institute of Technology and Sciences, Coimbatore, India
- 2Tamil Nadu Agricultural University, Coimbatore, India
Small millets, also known as nutri-cereals, are smart foods that are expected to dominate food industries and diets to achieve nutritional security. Nutri-cereals are climate resilient and nutritious. Small millet-based foods are becoming popular in markets and are preferred for patients with celiac and diabetes. These crops once ruled as food and fodder but were pushed out of mainstream cultivation with shifts in dietary habits to staple crops during the green revolution. Nevertheless, small millets are rich in micronutrients and essential amino acids for regulatory activities. Hence, international and national organizations have recently aimed to restore these lost crops for their desirable traits. The major goal in reviving these crops is to boost the immune system of the upcoming generations to tackle emerging pandemics and disease infestations in crops. Earlier periods of civilization consumed these crops, which had a greater significance in ethnobotanical values. Along with nutrition, these crops also possess therapeutic traits and have shown vast medicinal use in tribal communities for the treatment of diseases like cancer, cardiovascular disease, and gastrointestinal issues. This review highlights the significance of small millets, their values in cultural heritage, and their prospects. Furthermore, this review dissects the nutritional and therapeutic traits of small millets for developing sustainable diets in near future.
1 Introduction
Recent changes in food habits related to multi-grains have generated enormous interest in food and nutritional security. People have now started to focus on the nutritional uptake of small millets and growing children are provided processed foods that meet the daily nutritional requirement (Muthamilarasan and Prasad, 2021). Small millets, otherwise known as “wonder cereals”, possess numerous health benefits and thrive in harsh conditions (Barretto et al., 2021). Increasing demand for nutrition has paved the way to revive these crops, which once ruled nations. The 11 small millet species—finger millet, foxtail millet, barnyard millet, little millet, proso millet, Kodo millet, fonio millet, teff, brown top millet, Job’s tears, and guinea millet—are commonly known as the lost crops of the world. They are so-called ‘small’ owing to their smaller seed size. Small millets are rich in micronutrients, essential amino acids, and vitamin B complex, which are very rare in our staple diets. Phytochemical studies in small millets have demonstrated their higher antioxidant contents and lower glycemic indexes compared to other food crops (Vetriventhan et al., 2020).
Several programs for diet schedules are now popularizing the use and consumption of small millets in various forms. This has resulted in the food processing industries producing value-added products like flakes, noodles, biscuits, cookies, batter, flour, bread, and rice analogs. Rice analogs are one of the most popular developments that increase the palatable value of small millets by heat extrusion (Zhang et al., 2020). Hence, the demand for small millets is increasing in food industries. In addition, small millets are free of gluten, and it is highly suitable for patients with celiac disease (Deshpande et al., 2015). Despite nutrition, traditional practices have emphasized their medicinal values. These crops were previously not only foods but were also used to treat diseases including cancer and snakebites. Hence, analyzing the therapeutic traits of small millets paves a new way for smarter food in near future. Their uptake in the regular diet would provide their caloric, nutritional, medicinal, and fiber rich-properties for a sustainable life (Banerjee and Maitra, 2020).
In response to emerging pandemics in the changing climatic scenario, crop scientists have focused on boosting the human immune system with natural supplements through food diversification. These efforts have necessitated the acceleration of small millet breeding programs (Upadhyaya and Vetriventhan, 2018). Small millet genomics has recently started analyzing agronomic traits. Dissecting the genes underlying nutritional and therapeutic traits by applying advanced omics will reveal novel metabolic pathways in cereals for biofortification (Kumar, and Kumar, 2015). Next-generation and third-generation crop sequencing and breeding have demonstrated the potential of genetic databases and tools to analyze the genomes of small millets (Aggarwal et al., 2022). Hence, the future relies on developing smart crops with climate resilience, higher nutrition, and therapeutic traits. Moreover, conserving the traditional landraces of small millets, their cultural heritage, and ethnobotanical values by national and international organization programs to protect the rights of tribal farmers is needed to preserve novel alleles for the future (Saha et al., 2016). The present review focuses on the significance of small millets, breeding techniques, and advanced approaches for improving their productivity by enhancing their desirable features in a sustainable cropping system.
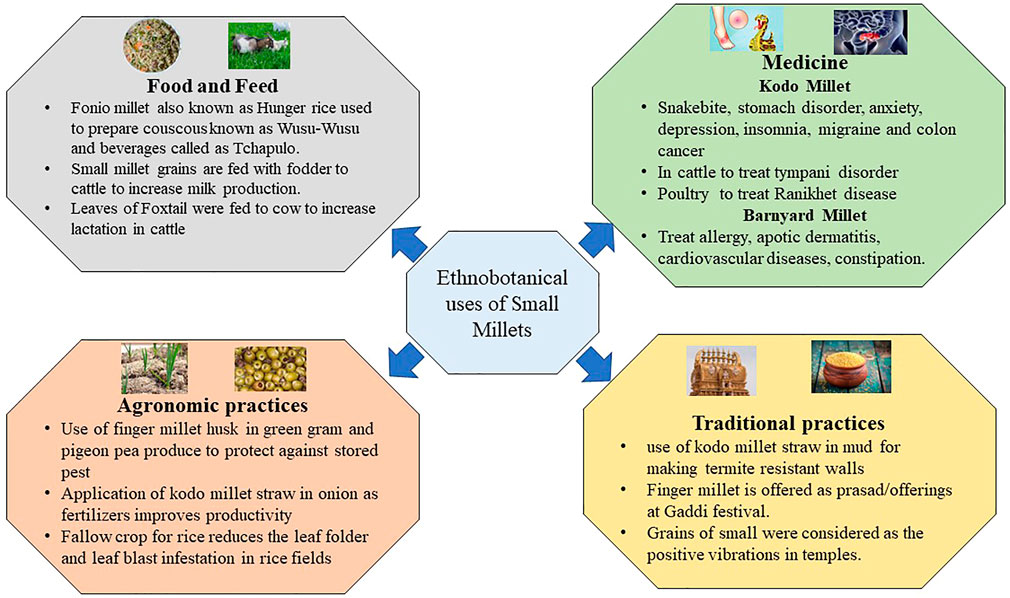
2 Ethnobotanical significance and cultural heritage of small millets
Small millets have a profound significance in our cultural heritage and until now have played significant roles in temple festivals in tribal regions. These activities are preserved as traditional knowledge in the regulations put forth by PPVFRA, 2001 (Satyarthi et al., 2018). These traditions demonstrate that these grains were recognized by our ancestors for their nutraceutical and therapeutic values (Figure 1). One common practice in small grains was their presentation as a wedding gift to the bridegroom. The amount of grains gifted was treated as a prestige; the grains were also cooked, especially during puberty and childbirth celebrations (Rawat et al., 2021). Small millets are rich in folic acid; therefore, they were treated as a special entity for women to overcome anemic disorders. In Africa, fonio millet, commonly known as “hungry rice,” has similar importance. These grains are predominantly used to prepare couscous known as wusu-wusu. Fonio is best used in the preparation of beverages called Tchapulo, which is rich in minerals. Finger millet has a similar value in the processing of beer, and its malted products are often used in African tribal communities (Hitu et al., 1997). Arake, a distilled liquor, is prepared in Ethiopia with finger millet flour. Furthermore, people residing in Sudan predominantly consume a hot porridge of finger millet with banana or sugar juice, which is a staple dish in tribal zones. A sour bread known as injera is made from teff and is used in spicy stews by well-off tribal individuals. Teff has a unique role in Africa after fonio millet. Due to its cold tolerance in higher altitudes, it is popularly known as love grass. The novel features of these lost crops are also being conserved. The major morphotypes in fonio millet are Yoro, Ipordapia, Ipordawoun, Ipoagoa, and Iporni are conserved by communities including the Hausa and pagans in west African regions (Dansi et al., 2010; National Research Council, 1996).
Regarding the conservation of cultural heritage in India, the Malayali in Eastern Ghats continue to cultivate and conserve small millet landraces (Newmaster et al., 2013). The landraces of little, foxtail, and proso millets are conserved by the Kolli Hill tribes; the characteristics of these millets suggest the presence of novel alleles for future breeding programs (Venkatesan et al., 2015; Ragupathy et al., 2016). Tracking the records of the utilization of small millets in Chhattisgarh, India has revealed the use of Kodo millet straw in mud to make termite-resistant walls. The farmers of this region also used Kodo millet straw as fertilizers in onion fields to increase productivity. Another traditional practice was the application of finger millet husk in green gram and pigeon pea products to protect these grains from pest infestation during storage. The pot makers in Northern India also are using Kodo millet straw when baking pots. Moreover, the leaves of Kodo millet possess lecithin and are used for the treatment of snakebites, stomach disorders, and joint pain. In cattle, Kodo millet straw had a significant impact on treating tympani disorder. Additionally, the older grains of Kodo millet (3–4 years) were used to cure Ranikhet disease in poultry. In Africa and India, Kodo millet was a fallow crop after rice; in other rice fields, Kodo millet straw is usually spread in the fields to protect against leaf folder and blast (Rawat et al., 2021).
The small millet grains were also previously mixed with fodder to increase milk production in cows. Several recent agro-start-ups in India for cattle feed also practice this technique to enhance milk production in rural dairy farms (Bhat et al., 2018). In traditional practice, finger millet is often a prasad/offering in the Gaddi festival. This is believed to enhance the fruiting of non-flowering mango and tamarind trees. Furthermore, thick pastes of finger millet flour are used to treat fire burns, and these grains are considered to offer positive energy in temples (Sahu and Sharma, 2013).
In addition, barnyard millet is used to treat allergies, atopic dermatitis, cardiovascular diseases, constipation, and blood-related disorders. Moreover, Kodo millet is preferred for overcoming anxiety, depression, insomnia, migraine, and colon cancer. Foxtail millet is used to treat chicken pox, heart attack, fever, cholera, and gastric problems. The leaves of foxtail millet are also fed to cattle to increase lactation. These practices underscore the nutritional and therapeutic value of small millets in our heritage. The genetics underlying these traits could be explored and used to develop sustainable diets (Satyarthi et al., 2018).
3 Nutritional importance and quality parameters of small millets
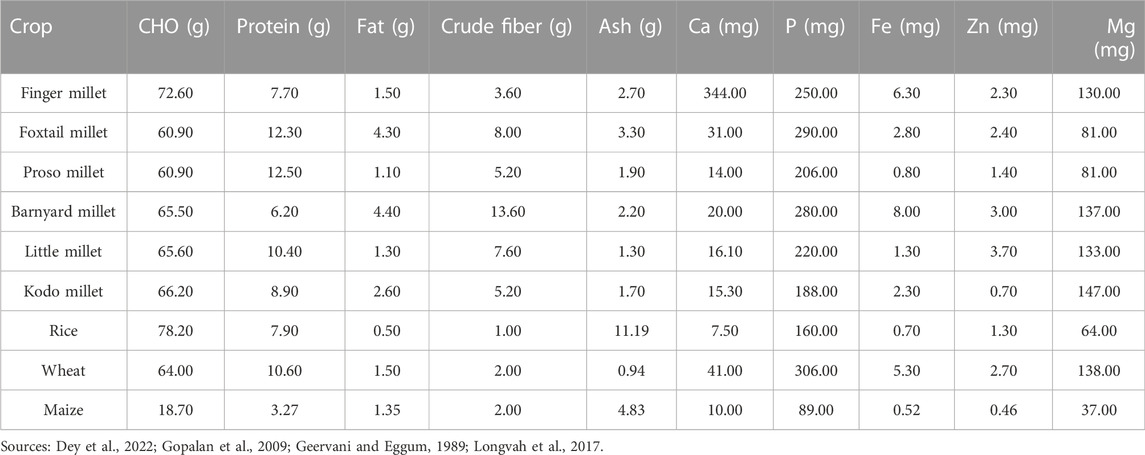
Small millets are being considered for their paramount importance in nutritional aspects not provided by other staple crops. These species have profound nutritional benefits, especially their micronutrient and protein profiles (Pramitha Lydia et al., 2021). The 11 small millets species each represent a unique beneficial feature for dietary bowls (Figure 2). Considering their acceptability for consumption, they are highly preferred for patients with diabetes and celiac disease owing to their gluten-free and higher fiber content (Table 1). Our ancestors were involved in laborious work and these foods predominantly helped them to sustain healthy lives. Due to the comfortability provided by major staples in later generations, the importance of these crops was almost forgotten, and they lost their commercial value. However, some traditional farmers still preserve this cultural heritage through festivals and traditional practices. Arising pandemics and emerging unprecedented weather have caused us to reassess the nutritional benefits of these crops (Muthamilarasan and Prasad, 2021).
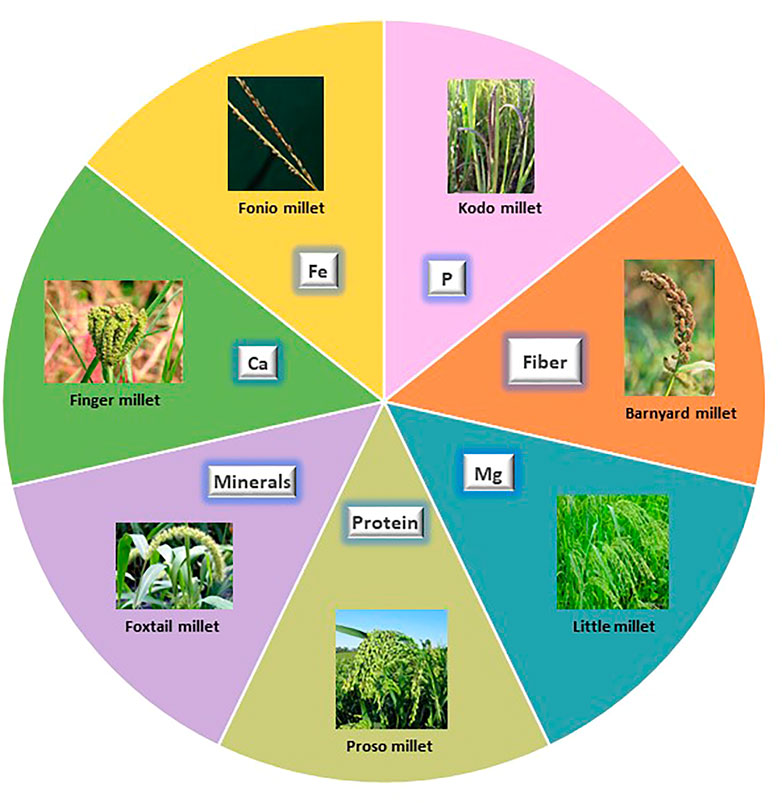
FIGURE 2. Nutritional uniqueness of small millets. Each sphere depicts the specialty of small millets in food.
Among the eleven species of small millets, finger millet and foxtail millet have larger areas of cultivation. Finger millet is known for its calcium, phosphorous, iron, and zinc content. It also has a higher caloric value due to its carbohydrate content. Foxtail millet is mostly preferred for its protein content, which is higher than wheat. It is also rich in minerals like phosphorous, iron, and zinc. Barnyard millet is known for its low glycemic index and high phosphorous and magnesium content. Among small millets, barnyard millet has the highest dietary fiber and niacin content (Pramitha Lydia et al., 2021). Kodo millet has the highest phosphorous content and radical scavenging activity owing to its high phenol content. Consuming Kodo millet reduces the risk of cardiovascular dysfunction (Rao Dayakar et al., 2017; Deepak Taggelli and Thakur, 2018). Little millet is known for its magnesium, phosphorous, and protein content. The unique feature of little millet is that it is rich in PUFA and flavonoids (Indirani and Devasena, 2021). Proso millet has a high protein profile following foxtail millet and also has higher magnesium and niacin content. Proso millet is also rich in essential amino acids like lysine, leucine, isoleucine, and methionine, unlike other major cereals (Pramitha Lydia et al., 2021). Fonio millet is preferably found in African regions and has high iron, dietary fiber, crude protein, flavonoid, GABA, and riboflavin content. It is highly nutritious owing to its high methionine and cysteine content. Teff is yet another wonder millet in Africa, which features the smallest grain and highest calcium and iron content. Teff is especially rich in lysine, which is deficient in all grains, and has comparatively higher protein and starch content. The other minor millets include brown top millet, Job’s tears, and guinea millet. These are also rich in phosphorous, iron, zinc, and vitamin B (Durairaj et al., 2019).
The demand for small millets is increasing in upcoming markets due to value-added products. Thus, their quality aspects should be considered for commercial production. Small millets are small-grained cereals and usually have a lower milling recovery than other grains. Therefore, uniformly sized, unbroken seeds are of concern in small millet production (Aggarwal et al., 2022). The colors vary from black to light yellow-colored grains, with consumers predominantly preferring light yellow- to brown-colored seeds for flour and cooking purposes (Durgad et al., 2021). Yellow-colored grains are also reportedly more aromatic compared to black grains (Aggarwal et al., 2022). Small millets are also a great source of resistant starch. The highest resistant starch values have been reported in Kodo millet, little millet, barnyard millet, and foxtail millet. Thus, these grains have nutraceutical value. As the grain yields of cereals deteriorate owing to harsh climates, small millets can be used to manufacture and process resistant starch in various industries (Kaimal et al., 2021). Rice analogs from small millet flours are also currently produced by heat extrusion and gelatinization. These rice granules are comparatively more desirable than normal rice varieties (Zhang et al., 2020).
Other quality parameters in small millet breeding include their fodder and forage value. Small millets are also used as fodder crops due to their higher biomass. Kodo millet, little millet, and proso millet are the most preferred animal feeds due to their higher palatability and crude protein content. Despite these factors, rancidity affects the storage of these flours in homes (Vetriventhan et al., 2020; Sruthi and Rao, 2021). Although the grains of small millets have a longer storage value, their flours are subject to rancidity; thus, efforts in advanced breeding techniques are needed to enhance the shelf life of small millet flours (He et al., 2015; Hariprasanna et al., 2014).
4 Breeding objectives and prospects in overcoming constraints
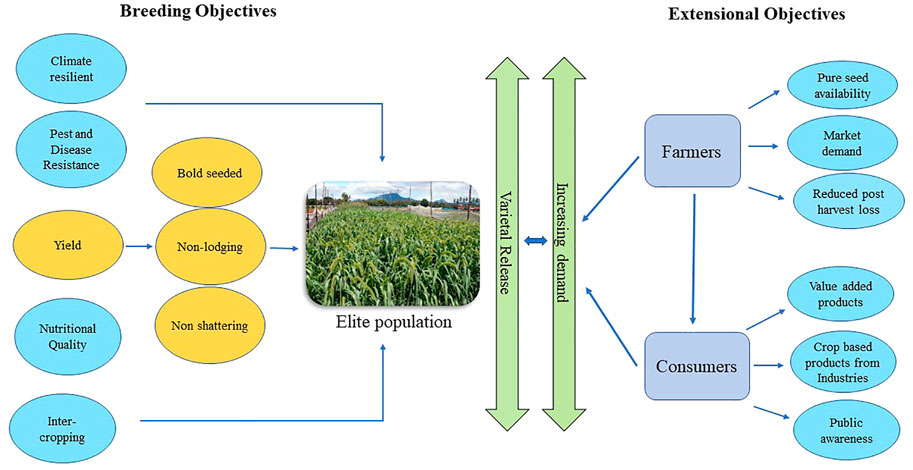
Small millets are among the less demanding crops in cultivation. They require minimum input from irrigation to fertilizers and pesticides. They can also thrive in harsh conditions, making them a reliable smart crop in the future (Durairaj et al., 2019). The major breeding objectives in small millets concern cultivation and post-harvest techniques. The initial objective predominantly focuses on higher yields. Yield is a complex trait that depends on numerous variables. The major factors causing yield loss in small millets are lodging and shattering. The increase of tillers in small millets with higher biomass at maturity results in lodging. Also, some crops are prone to shattering at harvest, which can be overcome with a stronger culm diameter while breeding for higher yield. Second, small millets have a non-preference in ideotype due to the spined hairy shoots and leaves. This causes challenges in managing field activities such as weeding and pesticide spraying. Hence, breeding for reduced bristles, spines in shoots, and leaves helps in proper crop management (Ganapathy and Patil, 2017).
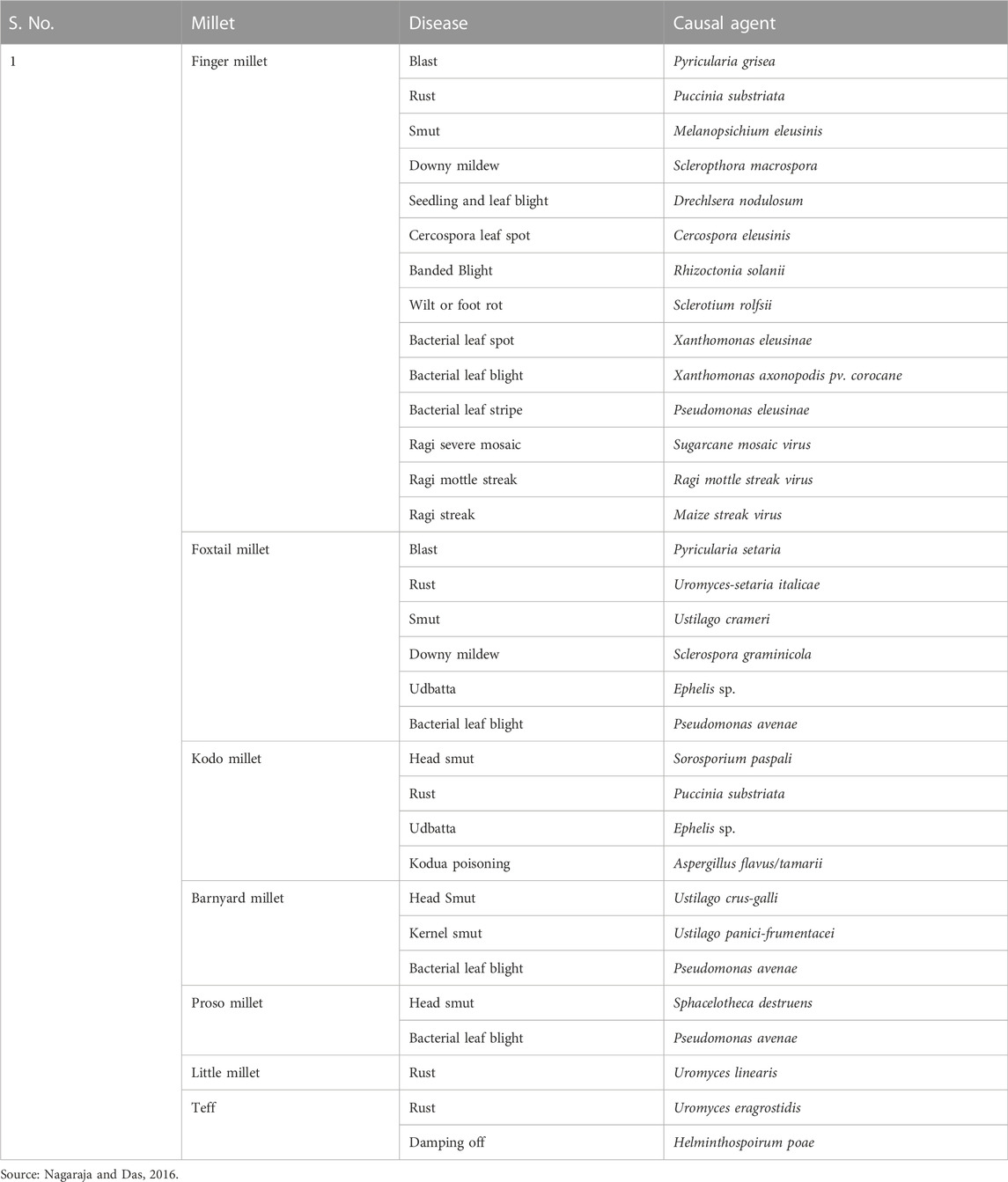
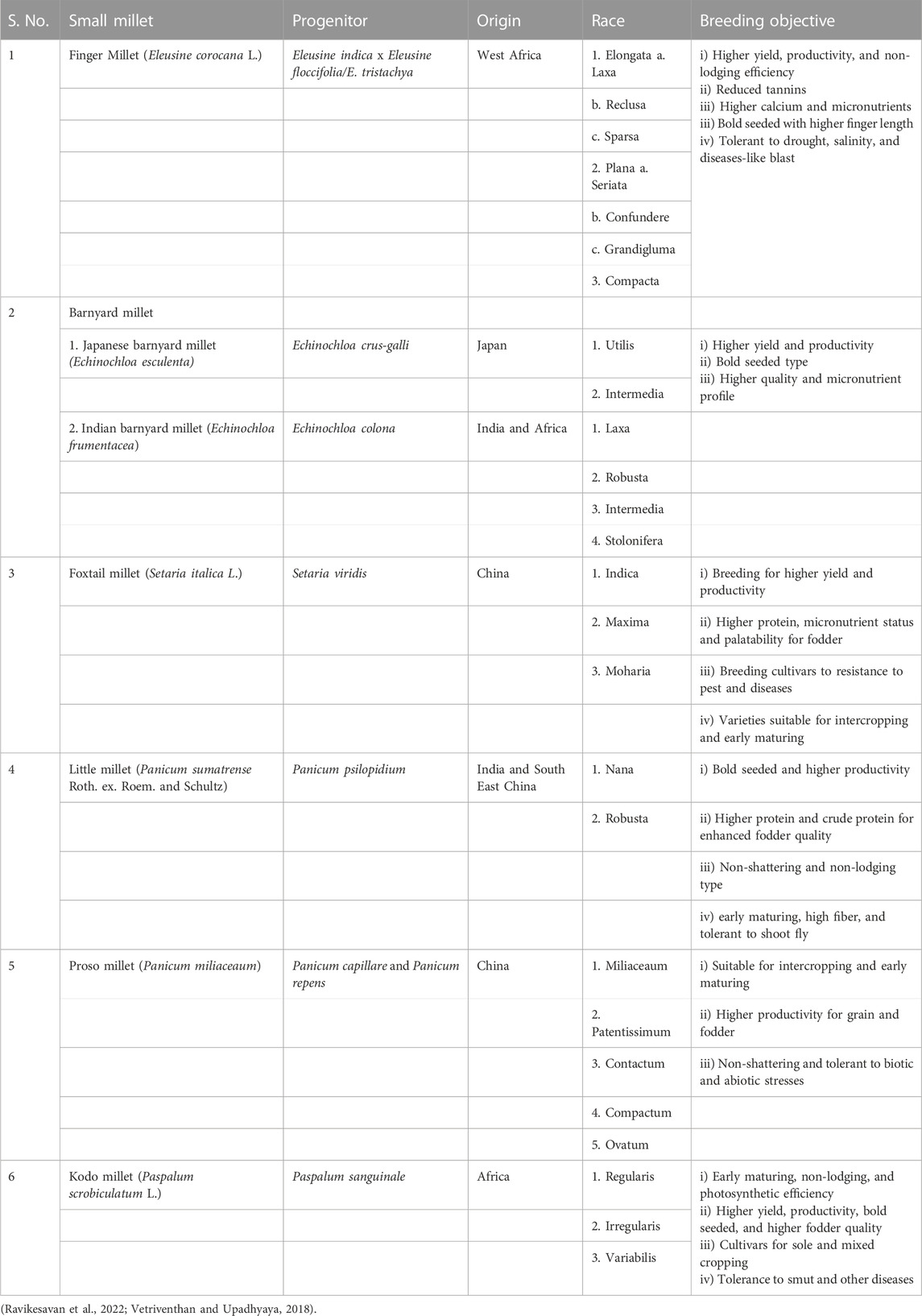
Small millets are also less prone to diseases. Some of the major diseases infecting them are shown in Table 2. These infestations are observed only in endemic regions and conducive conditions caused by poor cultivation practices (Nagaraja and Das, 2016). Therefore, the crop protection schedule and its inputs in these crops are predominantly minimal (Ravikesavan et al., 2022). Finally, as a major objective in the post-harvest technique, smaller grain size causes poor milling recovery; thus, breeding for larger seeds would help minimize post-harvest losses (Table 3). Future efforts in small millet breeding should focus on improving the color, nutritional profile, fodder yield, flour quality, and reduced antinutritional traits (Vetriventhan and Upadhyaya, 2019).
Although small millets have significant potential, the number of research programs remains low compared to other crops. This is due to the constraints involved in the smaller inflorescence and spikelets. These features restrict the possibility of attaining desirable recombinants. Recent approaches in hot water emasculation have resulted in the successful development of recombinants. Advances in mutation breeding with MutMap+ and genotyping facilities have allowed the dissection of novel alleles in small millets. Hence, there is a need for increased focus on small millet genetics and genomics using advanced omics approaches (Muthamilarasan and Prasad, 2015).
5 Progress of crop improvement in small millets
Initiatives on small millet improvement began in the early 1950s, when conventional breeding including pureline selection and pedigree breeding were ruling the varietal releases in all crops (Paroda and Mal, 1989). All small millets were highly self-pollinated and local collections by breeders were evaluated for line development. Varieties in small millets including CO 6 and CO (7) thenai in foxtail; CO (PV) 5 in proso millet; CO (samai) 4 in little millet; and CO 9, CO 13, CO 14, and Paiyur 2 in finger millet are examples of successful lines developed from recombination breeding. These varieties were released by Tamil Nadu Agricultural University by using standardized protocols for emasculation like hot water treatment and approaches (Ravikesavan et al., 2022). The millets were dominant in earlier civilizations; however, due to the palatability of major staples like rice and wheat, the small millets lost their presence among the population. Additionally, constraints like shattering, low yield, poor milling recovery, flour rancidity, and cooking time in small millets were major factors that drove these crops out of the staple source in the pipeline. Hence, breeder efforts were lost in the middle of the century after the green revolution (Vetriventhan et al., 2020).
Although these crops lost their economic value, few farmers relied entirely on this cropping pattern due to their adaptability in arid and harsh environments. Thus, these crops were always preserved as a cultural heritage among tribal populations and traditional farmers across generations. Overcoming the difficulties in crossing, mutation breeding in small millets later began to arise for varietal development. Mutation breeding in small millets started in the 1970s based on EMS and gamma rays. The frequency spectrum of macromutations with EMS and gamma in proso millet was studied by Ganapathy et al. (2008) and Bhave et al. (2016). Eswari et al. (2014) and Ambavane et al. (2015) also evaluated the effect of mutagenic frequencies with EMS and gamma rays in the previously released cultivars of finger millet. Foxtail mutation breeding experimented with different doses of EMS, DES, and gamma which also tended to affix the LD50 for isolating desirable mutants (Anittha and Mullainathan, 2018). Ramesh et al. (2019)assess the efficiencies of EMS and gamma rays in the CO (Kv) 2 strain of barnyard millet. Sood et al. (2020) evaluated the efficacies of different concentrations of sodium azide and gamma-ray treatments in barnyard millets. Thus, the LD50 values in small millets from earlier studies provided an understanding of the frequency of mutations induced in these genomes. Dosages of 500–600 Gy for finger millet and barnyard millet; 0.3%–0.45% EMS for finger millet, barnyard millet, and Kodo millet; 0.2% EMS for teff; 0.1 M nitrous acid in fonio millet for 4 h; and 0.03% nitrosoguanidine for finger millet induced desirable variations (Jency et al., 2016; Vetriventhan et al., 2020; Francis et al., 2022). Simultaneously, the genetic resources in small millet germplasm collections were also conserved as core collections in international research institutes. Due to efforts by Upadhyaya et al. (2016), the core collections from ICRISAT for little millet (460 accessions), foxtail millet (155 accessions), barnyard millet (89 accessions), and Kodo millet (75 accessions) were preserved and utilized. Other genetic reservoirs for small millets include NBPGR, the Ethiopian Biodiversity Institute, ICARDA, USDA, the N.I. Vavilov Russian Scientific Research Institute, the Ustymivika Experimental Research Station, and the Kenya Agricultural Livestock Research Station (Upadhyaya et al., 2008; Joshi et al., 2021).
The breeding of small millets has only recently accelerated due to increases in patients with diabetes and children with nutritional absorption issues. Moreover, the publication of the genome sequence of foxtail millet brought attention to the importance of these millets. Hence, relatively fewer molecular studies are published in small millets compared to the major staples. Still, almost all kinds of markers, from RFLP to AFLP, RAPD, EST-SSR, SSR, and SNPs have been utilized for marker-assisted selection. The first linkage map in small millets was developed with RFLP in foxtail millet; later, SSR and SNPs were used to map the QTLs in a high-density linkage map (Wang et al., 1998; Wang et al., 2017). Linkage maps with SNP markers were also later developed in proso millet by Rajput et al. (2016). A high-density linkage map with SNPs for finger millet was recently developed by Pendergast et al. (2022). Subsequently, molecular markers such as AFLP, RAPD, CAP, miRNA, EST, ISSR, SRAP, DEG, and SNP in proso millet were implied for genotyping the diversity, while EST, RAPD, and AFLP were used to analyze the calcium dynamics in finger millet (Habiyaremye et al., 2017). Fukunago et al. (2002b) and Desai et al. (2021) studied chloroplast and mitochondrial diversity in foxtail millet and barnyard millet, respectively. Among all molecular markers, SSRs were predominantly utilized in small millets and were also used in comparative genomics to analyze their lineages. Hence, a separate marker database for SSRs in foxtail was developed by Bonthala et al. (2014).
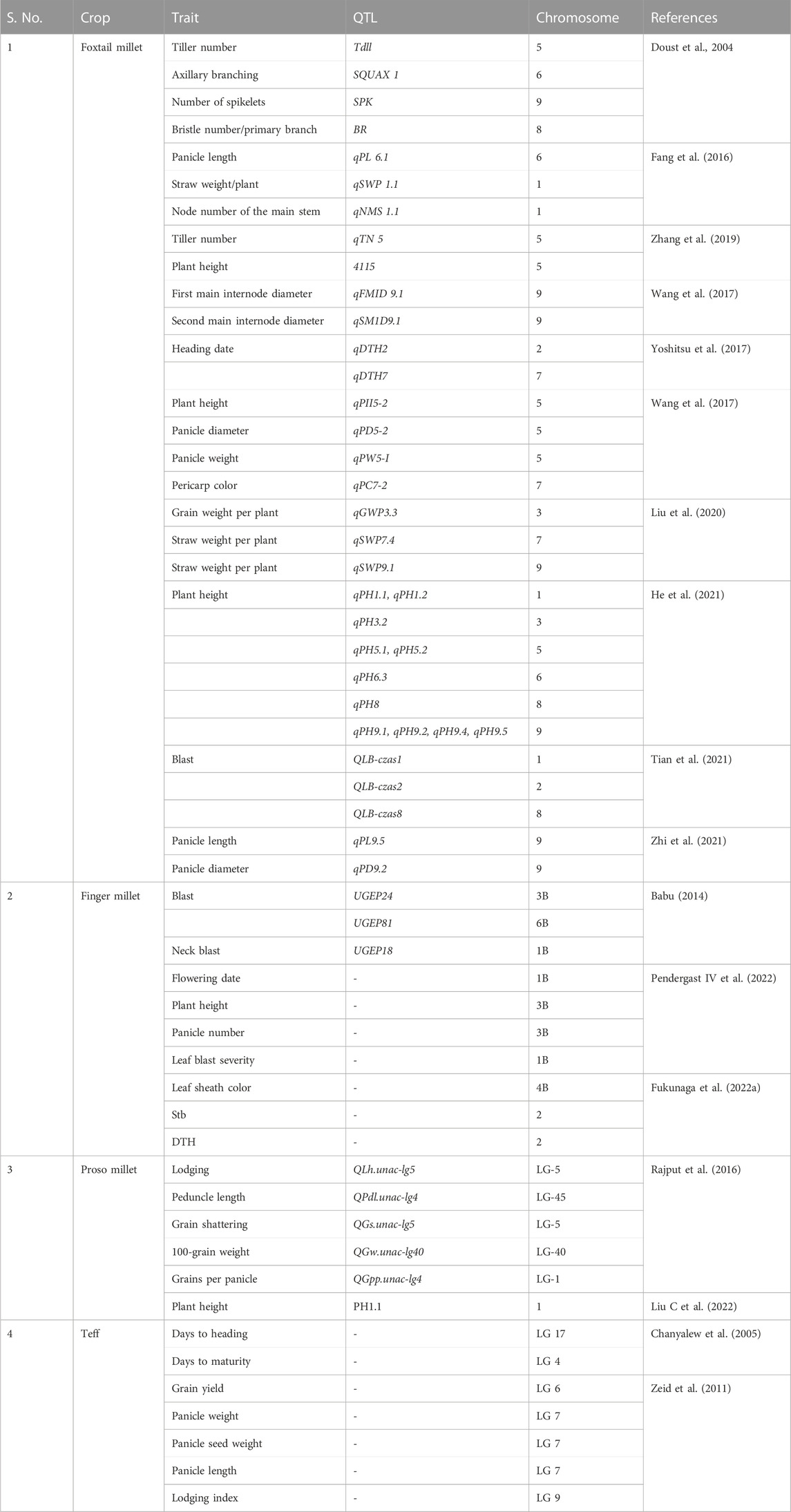
QTL mapping and trait mapping in small millets are in the initial stages; until now, QTL mapping for agronomic traits was performed (Table 4). The QTLs for traits like tiller number, branching, number of spikelets, bristles, panicle length and weight, plant height, grain weight/plant, and pericarp color have been identified in foxtail millet (Aggarwal et al., 2022). In finger millet, Babu (2014) and Ramakrishnan et al. (2016) identified QTLs for blast, neck blast, and leaf blast. Furthermore, in finger millet, the QTLs for P efficiency (Ramakrishnan et al., 2017), agronomic traits (Sharma et al., 2018), and biochemical traits like APX, CAT, GR, and SOD and POD activities (David et al., 2021) have also been studied. The populations mostly used for mapping in small millets included F2–F6 and RILs. The use of other mapping populations like NILs and double haploids remains to be explored (Vetriventhan et al., 2020; Zhang et al., 2021). Association mapping in foxtail millet and proso millet were predominantly studied with SSR and SNPs to determine the LD values for the diverse collections (Aggarwal et al., 2022; Boukail et al., 2021).
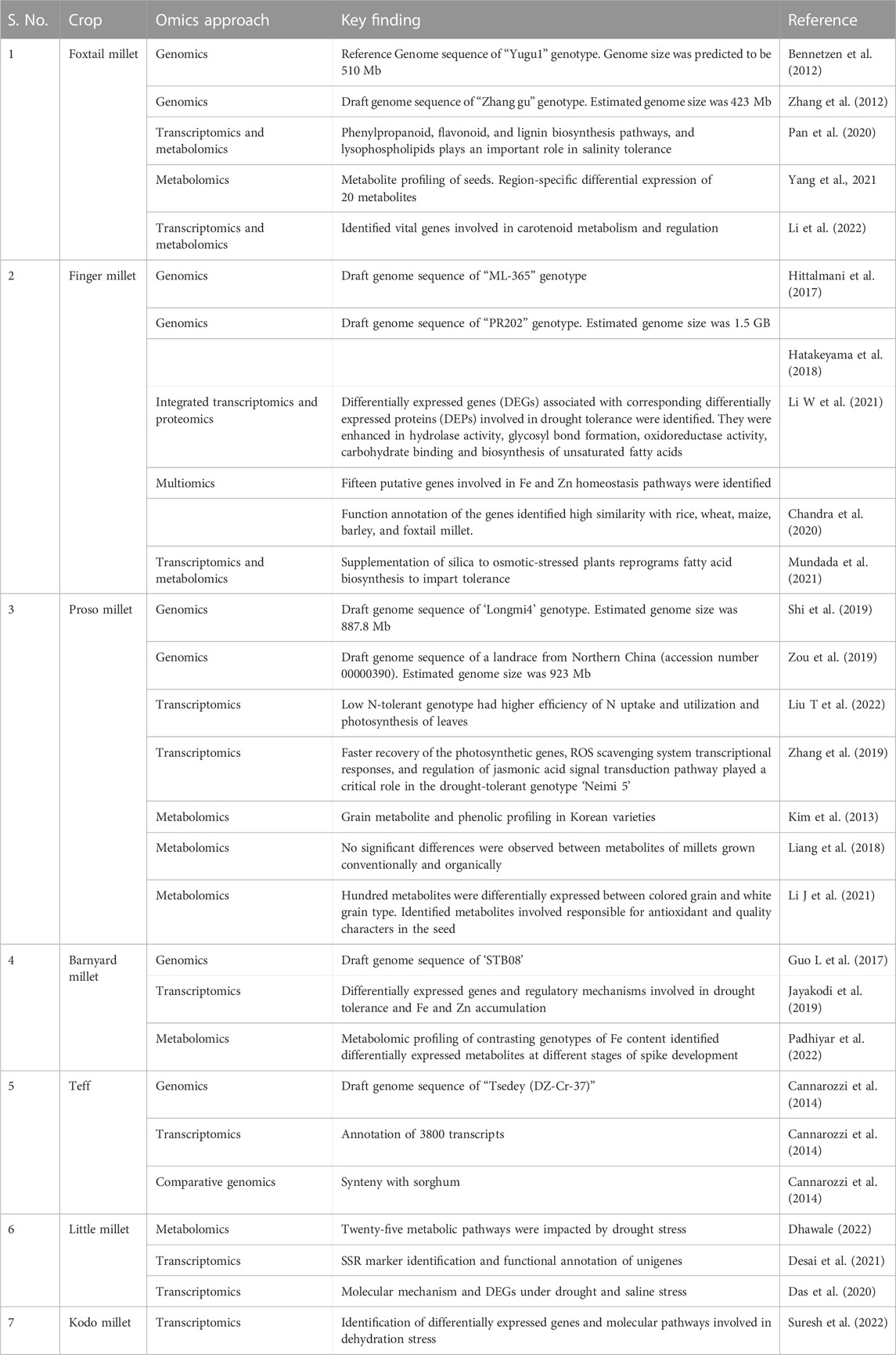
Among small millets, foxtail millet is the most exploited crop in genomics analysis (Table 5). Thus, foxtail millet is a model crop to understand genes in other crops, owing to its amenable genome and crop duration. Hence, advanced omics, genome editing, and double haploid techniques have been standardized and utilized from foxtail millet to analyze the genomics in other small millets (Aggarwal et al., 2022). The first sequence of foxtail millet was completed by Bennetzen et al. (2012) and Zhang et al. (2012) with two different cultivars. An updated sequence of this millet was later released by Ni et al. (2017). Successively, the finger millet whole genome sequence was published by Hittlamani et al. (2017) and Hatakeyama et al. (2018). The genome of sequences of barnyard millet and proso millet were also completed and published in China by Guo et al. (2017) and Zou et al. (2019), respectively. The genome sequences of teff (VanBuren et al., 2020) and fonio millet were recently completed (Abrouk et al., 2020). In addition, the protocols for genetic transformation with agrobacterium and biolistic approaches in finger millet, agrobacterium and biolistic methods in foxtail millet, biolistic methods in barnyard millet, agrobacterium-mediated approaches in teff and fonio, and agrobacterium-mediated methods in Kodo millet are now available (Vetriventhan et al., 2020; Bhatt et al., 2021). These advances are expected to enhance the prospects of understanding the underlying genomics of small millets. However, there remain research gaps in small millets regarding key nutritional and medicinal traits that could be used to revitalize our diets.
6 Genomics of climate resilience in small millets
The average global temperature is predicted to rise by 4–5°C by the end of the twenty-first century, which will adversely affect the growth of cereals crops like wheat and rice. Small millet crops, apart from being nutritionally superior to major cereal crops, are inherently tolerant to abiotic stresses like drought, high temperature, cold, poor soil fertility, and salinity (Goron and Raizada, 2015; Singh et al., 2021). They are physiologically sustainable under adverse environmental conditions owing to their excellent and efficient water and nitrogen use (Baltensperger, 2002; Saha et al., 2016). These characteristics highlight small millets as ideal smart crops for cultivation in the context of climate change. They can also aid in creating climate resilience in major cereals like wheat and rice (Bandyopadhyay et al., 2017). Hence, understanding the genetic and molecular mechanisms controlling stress tolerance in millets is critical. Modern and conventional breeding should work together to expedite the dissection and utilization of these complex mechanisms.
The advent of next-generation sequencing technologies (NGS) has revolutionized the field of genomics and created significant molecular information. Although the benefits of this technology were delayed in small millets due to limited funding and research, the genome sequences of foxtail millet, finger millet, proso millet, teff, Japanese barnyard millet, and white fonio are now available (Zou et al., 2019; Vetriventhan et al., 2020; Wang et al., 2021). Whole-genome sequencing and annotation of the ML-365 finger millet genome revealed TFs (transcription factors) and genes related to drought tolerance and the C4 photosynthetic pathway. A total of 2,866 drought-responsive genes were associated with WRKY, MYB, MYC, ZFHD, NAC, ABF, AREB, GRF, and NF-Y transcription factors (Hittalmani et al., 2017). In proso millet, 180 NAC TFs were identified and differentially expressed under various drought treatments. The expression levels of 31% of PmNAC (Proso millet NAC) genes were upregulated in the roots, indicating the critical role of root characteristics in drought tolerance (Shan et al., 2020). Numerous enzymes, including signal recognition particle receptor, farnesyl pyrophosphate synthase, calcineurin B-like interacting protein kinase 31, serine–threonine protein phosphatase 2A, and others were activated by drought stress in finger millet. Several housekeeping and basal regulatory genes were also activated by drought. Novel drought-associated genes reported in the crop included pentatricopeptide repeat proteins and tetratricopeptide repeat proteins (Parvathi et al., 2019).
Salinity-tolerant finger millet genotypes show upregulation of many genes governing cell growth and differentiation. In the salinity-tolerant strain Trichy 1, genes involved in flavonoid biosynthesis were selectively down-regulated (Rahman et al., 2014). Transcriptomic and metabolomic analyses of contrasting genotypes for salinity tolerance showed that lysophospholipids, phenylpropanoid, flavonoid, and lignin biosynthesis pathways were crucial in the salinity tolerance of foxtail millet. The tolerant Yugu2 strain showed increased antioxidant enzyme activity and non-enzymatic antioxidant content (Pan et al., 2020; Puranik et al., 2011). The genome-wide gene expression profile in foxtail millet showed that most of the crop’s drought-responsive genes were associated with photosynthesis, signal transduction, and TFs (Qin et al., 2020). Apart from the well-known stress tolerance-associated genes like peroxidases and glutamine synthetase, leaf tissue-specific expression of ricin-B lectin-like was reported in little millet under salt and drought stress. Alcohol dehydrogenase was also upregulated in both root and leaf tissues of the crop under both stresses (Das et al., 2020). Photosynthesis was a key factor in the ability of Indian barnyard millet to adapt to dry conditions, as shown in a comparative transcriptome analysis between the plant and its wild relative, barnyard grass (Echinochloa crus-galli) (Jayakodi et al., 2019). Thus, the unique expression for climate resilience in small millets suggests novel alleles for use in stress breeding programs (Saha et al., 2016).
7 Innovative breeding methods to improve nutrition and stress tolerance
Innovative breeding methods can be used to speed breeding progress and reveal genes responsible for stress tolerance and nutritional quality improvement in small millets. Additionally, this information can be used to enhance these traits in susceptible major cereal crops. Whole-genome sequencing has revolutionized the breeding and biotechnological approaches in crop improvement. The genome sequences of small millets have generated genomic resources that can be used to identify the gene position, selection, and introgression of desirable traits into other varieties, species, or crops. By considering the genome sequence of a single individual as a reference genome, we may miss many key variations present in crop genetic diversity. This reference bias can be overcome by the concept of a pangenome, wherein several individuals of the species are sequenced to better represent the crop’s diversity. This method can distinguish the diversity as core (shared among different genotypes of the species) and variable (may be absent in some individuals) genome (Tettelin et al., 2005; Morgante et al., 2007). In small millets, where genome sequence information is limited, the sequences of panicoid species can be combined using pangenome and comparative genome approaches to improve breeding efficiency (Bennetzen and Freeling, 1997; Sharma et al., 2018; Tay Fernandez et al., 2022). Moreover, trait-based pangenomes can also be devised for stress tolerance and nutritional quality (Tay Fernandez et al., 2022).
Comparative genomics between C4 and C3 grasses provides a better understanding of their evolutionary paths to accelerate the research goal of improving the photosynthetic efficiency and drought tolerance in common staple crops (Chapman et al., 2022). High-throughput sequencing platforms have made the identification of single-nucleotide polymorphisms (SNPs) easier. Currently, SNPs are the main genetic variations or markers used in marker-assisted selection (Lata and Prasad, 2013). Genome-wide association studies (GWAS) and nested association mapping (NAM) can be used to establish marker–trait associations and detect powerful QTLs. NAM uses multiple mapping populations, has higher recombination events, and can detect tightly associated QTLs compared to traditional QTL mapping. Though this approach is widely used to identify candidate genes in major cereal crops, it has not yet been applied in underutilized crops like small millets (McMullen et al., 2009; Bouchet et al., 2017).
Mutation breeding has evolved from conventional methods to MutMap populations and targeted mutagenesis. MutMap and MutMap + are genome sequence-based mutation breeding approaches that facilitate rapid gene identification and isolation. The MutMap method has been used to map the dwarfing gene D3 on chromosome 8 of the foxtail millet genome. The dwarf mutant also shows reduced drought tolerance, indicating critical trait associations (Fan et al., 2017). Mutmap-based cloning was used to establish the regulatory role of the WRKY transcription factor in panicle and seed development. MutMap + does not require crossing between mutant and wild type and is a desirable approach in millet crops in which crossing is tedious (Fekih et al., 2013). New plant breeding techniques like CRISPR-Cas9, TALENs, ZFNs, mega nucleases, oligonucleotide-directed mutagenesis (ODM), cis-genesis, transgenesis, and RNA-dependent methylation (RDM) can also be explored for small millet crop improvement (Schaart et al., 2016). These techniques can be used to decrease anti-nutritional content like phytates, polyphenols, and tannins. The higher levels of proteases and amylose inhibitors that impede millet digestibility can also be reduced using site-targeted modifications (Vinoth and Ravindhran, 2017). In foxtail millet, bioinformatic database information was used to identify target genes for waterlogging tolerance in designing guide RNA for clustered regularly interspaced short palindromic repeat (CRISPR)-aided activation of tolerance-associated transcripts (Abdulla et al., 2021).
Various approaches exist for the transgenic biofortification of finger millet and other crops. Increasing the calcium (Ca2+) storage capacity in edible parts by modifying the expression of transporter proteins, including the overexpression of channel proteins and Ca2+-binding proteins to increase calcium accumulation and mutagenesis-induced alterations of calcium content (Wyatt et al., 2002; Morris et al., 2008; Conn and Gilliham, 2010; Sharma et al., 2018). Other modern techniques like double haploid production and reverse breeding in millet crops may be useful for fixing genetically desirable cross combinations (Gis et al., 2019). Creating such platforms in the model crop foxtail millet may provide opportunities to dissect complex C4 pathways in millets and exploit their role in stress tolerance (Jacquier et al., 2020). Cheng et al. (2021) reported haploid embryo induction through the CRISPR-Cas9-mediated knockout of SiMTL in foxtail, which is an ortholog of MATRILINEAL/NOT-LIKE-DAD/PHOSPHOLIPASE A (MTL/NLD/ZmPLA) in maize used for haploid induction (Gilles et al., 2017; Kleter et al., 2019).
Transcriptomics is yet another method that has generated enormous genomic information even in crops in which genome sequence data are not available. Transcriptome sequencing (RNA-Seq) reveals genome-wide information on functional genes, differential expression of these genes, and their regulatory mechanisms. This method is widely used as it is cheaper than building a genome assembly and reveals the transcriptional activity based on the time and location of the observation. It can also provide insights into various metabolic pathways of the crop (Guo et al., 2021). Metabolomics, another promising omics method, was used to profile metabolites associated with drought stress response in little millet (Dhawale. 2022). The integrative use of transcriptomics and metabolomics was applied to identify key pathways involved in salinity tolerance in foxtail millet (Pan et al., 2020). Metabolomics analysis revealed the environmental and geographical influences on the bioactive nutrient profile of foxtail millet and the key biochemical pathways (Yang et al., 2021). Thus, combining advanced techniques to understand the genetic regulation of crops provides new gateways for the development of desirable plant types.
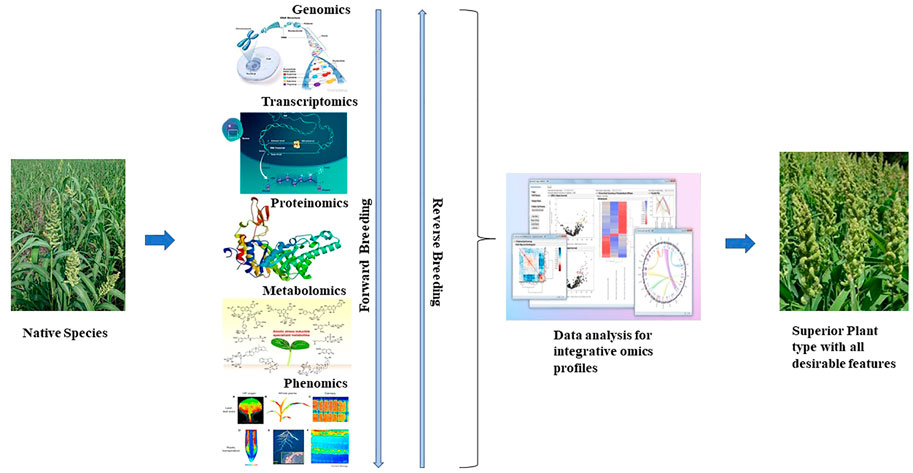
8 Advanced omics approaches in small millets for trait-specific breeding
Recent advances in next-generation sequencing have applied multiple omics. Integrated omics is a rapid platform to hasten the selection process in plant breeding programs (Figure 3). Genomics plays roles from locating QTLs to developing trait-associated markers. Structural genomics has been utilized to locate the trait of interest in genetic and physical maps. Furthermore, positional cloning of QTLs by advanced QTL-seq approach has also allowed the identification of the exact location and sequence of candidate genes, paving the way for the design of allele-specific markers (Fan et al., 2017). Inclusively, genotype by sequencing has more precisely revealed the population structure of diverse collections, which has enabled the detection of novel alleles. Allele-specific markers are now used in plant breeding programs and are more reliable than the conventional markers used previously (Scossa et al., 2021).
Functional genomics also plays a major role in identifying the functions of genes relative to the trait of interest. Mutagenomics, epigenomics, and pangenome analysis are involved in depicting gene functions. Metabolomics, proteomics, trancriptomics, ionomics, and phenomics are also used in plants to dissect functional roles in gene expression. However, to understand the complete biological process of an organism, multi-omics is essential and also plays a role in the rapid breeding of crops (Kumar et al., 2021). Previously, metabolomics and transcriptomics were associated with plant phenotypes of major traits (Table 5). Recently, owing to advances in omics technology, an integrated approach has been implemented in crops. GWAS combined with metabolite profiling has been used to detect biochemical and genetic processes in major staples (Luo, 2015; Matsuda et al., 2015). Combined HRPF and GWAS was used to inform biomass and yield in rice (Yang et al., 2015); photosynthesis and growth rates in maize were analyzed by the combination of metabolomics and ionomics (Guo R et al., 2017); metabolomics, ionomics, and genomics involving QTL mapping were applied to evaluate iron and zinc in rice (Pinson et al., 2015); integrated mutagenomics and phenomics were used to understand increased lycopene in tomato (Li et al., 2018); mQTL analysis and mGWAS have been applied in maize and rice (Li K. et al., 2019; Tiozon, 2021), and combined transcriptome and metabolome study have been used to understand defense response in wheat Trichoderma harzianum strain T22 (Coppola et al., 2019) to detect key metabolites and candidate genes involved in the pathways for grain yield, secondary metabolites, and disease resistance (Yang et al., 2021).
Finally, functional genomics plays a major role in hybrid performance. The prediction of genotype performance by multi-omics is more precise than the conventional techniques (Yang et al., 2021). Future goals of integrated omics include the development of a proteome and metabolome atlas depicting the major genes in metabolic pathways, which also ensures the development of advanced databases for integrated big data in molecular breeding (Peng et al., 2020). These future innovations could frame a new way to design crops for favorable traits by de novo domestication and allow the rapid development of crop ideotypes (Choi, 2019).
9 Therapeutic value and its incorporation through genomics-assisted breeding techniques
Small millets are known for their nutritional value; however, understanding their significance in ethnobotany reveals their therapeutic value. The wonder grains of small millets are remembered as medicines for cures; thus, studying these values offers a greater scope for feeding people with a healthier diet. Each small millet has a potential medicinal value; for example, barnyard millet is known for its fiber content and thus is highly recommended to patients with heart disease and diabetes mellitus. Barnyard millet also has a higher lysine and iso-leucine for blood formation and lipid metabolism in organisms. The components of barnyard millet have the highest linoleic acid, which is a major explanation for the plant’s antioxidant and immunological activities. The phytochemicals in barnyard millet include alkaloids, tannins, terpenoids, and flavonoids (Renganathan et al., 2020). Moreover, finger millet is rich in tryptophan; therefore, it is prescribed for decreasing appetite in weight loss programs. Finger millet also includes threonine, which obstructs the formation of fat in the liver and regulates cholesterol levels. Due to its higher calcium, potassium, and iron levels, this millet is also suggested for use in growing children, lactating women, and anemic people (Verma et al., 2018). Ragi is rich in phytates, tannins, and trypsin inhibitors. Proso millet is predominantly known for its highest protein content and is prescribed as a potential therapeutic agent for diabetes mellitus II. This is combined with folic acid, vitamin B6, phenolic acid, ferulic acid, chlorogenic acid, syringic acid, and caffeic acid. Proso millet has shown effectiveness in curing heart disease and preventing breast cancer (Goron and Raizada, 2015). However, foxtail millet has higher linoleic acid, tocopherol, phenol, and flavonoid levels, which enhance its therapeutic value. Additionally, it supplies copper for regulated metabolic reactions. Moreover, small millets contain flavonoids and folic acid and are often used to enhance immune function. Finally, fonio millet is a good source of GABA and folic acid and has the highest iron content (Satyarthi et al., 2018).
These small millet components have been associated with health. Research on their therapeutic value and their stability in the grains is needed and could be a foundation for improving the medicinal values of major staples. The biofortification of grains with micronutrients is reaching new heights and incorporating flavonoids, folates, and GABA in mainstream foods to strengthen our immune response to overcome new emerging diseases. Although rice landraces are used as sources of folate, GABA, and flavonoids, their expression in processed grains is a concern. Whole brown rice and pigmented rice are highly nutritious; however, low percentages of people have access to these landraces. Hence, small millets could be an efficient source for enhancing the medicinal value of food, and its genomics may reveal the expression of these traits in processed foods (Tiozon 2021).
Thus, studies are needed for the identification of QTLs for these traits from mapping populations; association studies in small millets to identify factors related to micronutrients, therapeutic traits, and yield; metabolite genome-wide association studies; functional characterization of key genes; dissection of stable donors; and candidate gene analysis (Xia et al., 2021). Correlation and association studies on medicinal and yield values would help improve multiple traits in small millets (Tiozon 2021). Linkage maps for agronomic traits are currently available in foxtail millet and must be further improved to include micronutrient and medicinal traits. Since small millets are climate-resilient, achieving overall improvements in yields, micronutrients, and therapeutic profiles will be rewarding for smart foods (Aggarwal et al., 2022).
10 Road map for enhancing nutritional security with small millets
The current status in crop production sheds light on enhancing small millets productivity, which is influenced by many factors. The priority is to increase the areas under small millet cultivation. Until now, the cultivation of small millets was restricted to marginal and rainfed farmers who use these as cover crops to conserve arable lands in harsh conditions (Merga, 2018). These farmers also have poor access to high-quality seeds for sowing and the availability of pure small millet varietal seeds must be meticulously formulated from seed production plots. Second, increasing the cultivation area requires accelerating the market demand. Market consumers must be familiarized with diversifying their food habits for a sustainable life (Ravikesavan et al., 2022). Food diversification results in crop diversification and the potential to reintroduce lost and underutilized crops to mainstream cultivation. Recent cropping systems have also been altered to include small millets and they are now incorporated as an intercrop with legumes for a higher profit (Maitra, 2020). This positively impacts our ecosystem for a balanced chain and may also minimize genetic erosion (Deshpande et al., 2015).
Moreover, public communities have recently focused on physical fitness, with trainers in urban regions counseling their trainees on food habits. These efforts involve nutritional supplements and popular advertisements for artificial health supplements have appeared in diet schedules. These schedules could soon be replaced with small millet-based supplements. Small millets are rich in folic acid, flavonoids, terpenoids, resistant starch, and other phytochemicals that regulate cellular metabolic activities. Hence, in-depth research on the therapeutic value of small millets and their enrichment could replace artificial supplements in fitness programs (Durairaj et al., 2019; Lloyd and Kossmann, 2021).
Another major stream in small millets that requires consideration is the incorporation of breeding strategies to yield desirable traits. Weedy features in small millets need improvement through selection. The characteristic features like shattering, lodging, small seeds, spined shoots, bristles, and awns must also be considered in the development of crop ideotypes in small millet breeding (Banerjee and Maitra, 2020). Further breeding objectives for analyzing their nutritional stability, bioavailability in processed foods, multigrain products, phytochemical expression, uniform maturity, fertilizer responsiveness, and biotic stress tolerance must be emphasized to provide higher yield and productivity (Figure 4). Thus, small millets could be an alternative source of nutritional and therapeutic traits in our regular diet (Adekunle et al., 2018; Upadhyaya and Vetriventhan, 2018). Food processing industries are also now moving toward small millets for the extraction of resistant starch, which opens a new market for small millet demand. In response, countries have increased their focus on small millets (Kaimal et al., 2021).
Small millets are arising as major cereals in countries like Ethiopia where other crops strive hard to feed the populations. The staple crop of Ethiopia is teff and Europe and the US are increasing their production of this grain owing to its nutritional value (Woldeyohannes et al., 2022). Proso millet is yet another “poor man’s food”. The cultivation of proso millet is increasing in the northern US and China. From a catch crop, it is now evolving as an alternative to the major food basket across countries (Bhat et al., 2019). Foxtail millet is well renowned for its grain and fodder. Recent ventures in foxtail yield and productivity are welcomed by farmers in Telangana and Andhra Pradesh in India. Internationally, foxtail millet is also favored for its various savories and recipes in China, Europe, and the US. Millet is also being explored as a biofuel, which has generated large industrial interest (Petti et al., 2013; Aggarwal et al., 2022). In addition, Western Africa is also now considering the fonio millet value chain in its market as a future for crop and food diversification based on small millets (Kanlindogbe et al., 2020; Ibrahim Bio Yerima and Yerima, 2021). The new-age policies from the government for introducing finger millet in Nepal and enhancing its cultivation in Ethiopia (Gebreyohannes et al., 2021) provide a new gateway for future diets. Recently, a global strategy for the intake and conservation of small millets proposed by Bramel et al. (2022) also enforces the need to enhance the cultivation of small millets to meet the continuously increasing demand. As discussed in the previous works on small millets, these crops can thrive to address future food and nutritional hunger (Muthamilarasan and Prasad, 2021; Renganathan et al., 2020).
11 Conclusion
This review highlights the overall value of small millets in our daily lives. Small millets were important food crops in ancient civilizations. Due to shifts in human dietary habits, these crops are now underutilized as food. By analyzing recent progress toward the use of small millets, we can realize support from government-aided projects and international and national collaborations to revitalize the genetic resources in small millets. Insights into the novel traits in small millets have demonstrated their paramount importance in nutritional and climate resilience. Therefore, this states the reasons behind the reverence of these crops in the cultural heritage of our ancestors. From these ethnobotanical records, we can conclude that breeding programs for improved varieties will restore the importance of on-farm conservation of wild and traditional landraces of small millets.
Small millets are climate-resilient crops that can meet the need for food and fodder and act as nutritional supplements. Hence, they are well considered as nutri-cereals and smart crops. Our detailed survey of studies in small millets revealed their great scope for applying advanced omics techniques to tap the genetic reasons for climate resilience and nutrition. Among small millets, the prospects of research in foxtail are higher and it has been utilized as a model crop system. Thus, small millets could act as a model crop for novel genes and mechanisms for manipulation by comparative genomics in mainstream cereals for increased stress tolerance and therapeutic traits. In this context, recent trends in food habits among the public have also begun to shift toward small millets, as evidenced by multigrain value-added products, processed foods, and new recipes in the markets. Therefore, small millets could be a major crop in future and a component in diversifying our food habits for a healthier life.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aggarwal, P.i, Lydia, P., Choudhary, P., Singh, R., Shukla, P., Prasad, M., et al. (2022). Multi-omics intervention in Setaria to dissect climate-resilient traits: Progress and prospects. Front. Plant Sci. 13, 892736. doi:10.3389/fpls.2022.892736
Abdullah, S. N. A., Mayes, S., and Moradpour, M. (2021). Target gene identification and sgRNA design for waterlogging tolerance in foxtail millet via CRISPR-based transcriptional activation. Curr. Chin. Sci. 1 (5), 523–533. doi:10.2174/2210298101666210709104258
Abrouk, M., Ahmed, H. I., Cubry, P., Šimoníková, D., Cauet, S., Pailles, Y., et al. (2020). Fonio millet genome unlocks African orphan crop diversity for agriculture in a changing climate. Nat. Commun. 11 (1), 1–13. doi:10.1038/s41467-020-18329-4
Adekunle, A., Darwin, L., Valérie, O., and Raghavan, V. (2018). Helping agribusinesses—small millets value chain—to grow in India. Agriculture 8 (3), 44. doi:10.3390/agriculture8030044
Ambavane, A. R., Sawardekar, S. V., Sawantdesai, S. A., and Gokhale, N. (2015). Studies on mutagenic effectiveness and efficiency of gamma rays and its effect on quantitative traits in finger millet (Eleusine coracana L. Gaertn). J. Radiat. Res. Appl. Sci. 8, 120–125. doi:10.1016/j.jrras.2014.12.004
Anittha, I., and Mullainathan, L. (2018). Mutagenic effect of EMS and DES on Tenai (Setaria italica) in M1 generation. J. Phytology, 06–08. doi:10.25081/jp.2018.v10.3406
Babu, B. K. (2014). Molecular analysis of world collection of finger millet accessions for blast disease resistance using functional SSR markers. SABRAO J. Breed. Genet. 46 (2), 202–216.
Baltensperger, D. D. (2002). “Progress with proso, pearl and other millets,” in Trends in new crops and new uses, 100–103.
Bandyopadhyay, T., Muthamilarasan, M., and Prasad, M. (2017). Millets for next generation climate-smart agriculture. Front. Plant Sci. 8, 1266. doi:10.3389/fpls.2017.01266
Banerjee, P., and Maitra, S. (2020). The role of small millets as functional food to combat malnutrition in developing countries. Indian J. Nat. Sci. 10 (60), 20412. doi:10.1073/pnas.0402892101
Barretto, R., Buenavista, R. M., Rivera, J. L., Wang, S., Prasad, P. V., and Siliveru, K. (2021). Teff (eragrostis tef) processing, utilization and future opportunities: A review. Int. J. Food Sci. Technol. 56 (7), 3125–3137. doi:10.1111/ijfs.14872
Bennetzen, J. L., and Freeling, M. (1997). The unified grass genome: Synergy in synteny. Genome Res. 7, 301–306. doi:10.1101/gr.7.4.301
Bennetzen, J. L., Schmutz, J., Wang, H., Percifield, R., Hawkins, J., Pontaroli, A. C., et al. (2012). Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 30 (6), 555–561. doi:10.1038/nbt.2196
Bhat, S., Nandini, C., Srinathareddy, S., Jayarame, G., and Prabhakar, (2019). Proso millet (Panicum miliaceum l.)-a climate resilient crop for food and nutritional security: A review. Environ. Conserv. J. 20 (3), 113–124. doi:10.36953/ECJ.2019.20315
Bhat, S., Nandini, C., and Tippeswamy, V. (2018). Significance of small millets in nutrition and health-A review. J. Dairy Foods Home Sci. 37 (1), 35–40. doi:10.18805/ajdfr.DR-1306
Bhatt, R., Asopa, P. P., Jain, R., Kothari-Chajer, A., Kothari, S. L., and Kachhwaha, S. (2021). Optimization of agrobacterium mediated genetic transformation in paspalum scrobiculatum L. (Kodo millet). Agronomy 11 (6), 1104. doi:10.3390/agronomy11061104
Bhave, K. G., Dalvi, V. V., Thaware, B. L., Mahadik, S. G., Kasture, M. C., and Desai, S. S. (2016). Mutagenesis in proso millet (Panicum miliaceum L.). Int. J. Sci. Res. 5, 1635–1638.
Bonthala, V. S., Muthamilarasan, M., Roy, R., and Prasad, M. (2014). FmTFDb: A foxtail millet transcription factors database for expediting functional genomics in millets. Mol. Biol. Rep. 41 (10), 6343–6348. doi:10.1007/s11033-014-3574-y
Bouchet, S., Marcus, O., Olatoye, S. R., Marla, S. R., Ramasamy, P., Tesfaye, T., et al. (2017). Increased power to dissect adaptive traits in global sorghum diversity using a nested association mapping population. Genetics 206 (2), 573–585. doi:10.1534/genetics.116.198499
Boukail, S., Macharia, M., Miculan, M., Masoni, A., Calamai, A., Palchetti, E., et al. (2021). Genome wide association study of agronomic and seed traits in a world collection of proso millet (Panicum miliaceum L.). BMC Plant Biol. 21 (1), 1–12. doi:10.1186/s12870-021-03111-5
Bramel, P., Giovannini, P., and Eshan Dulloo, M. (2022). Global strategy for the conservation and use of genetic resources of selected millets. Bonn, Germany: Global Crop Diversity Trust.
Cannarozzi, G., Plaza-wüthrich, S., Esfeld, K., Larti, S., Wilson, Y. S., Girma, D., et al. (2014). Genome and transcriptome sequencing identifies breeding targets in the orphan crop tef (Eragrostis tef). BMC Genomics 15, 581. doi:10.1186/1471-2164-15-581
Chandra, A. K., Pandey, D., Tiwari, A., Sharma, D., Agarwal, A., Sood, S., et al. (2020). An omics study of iron and zinc homeostasis in finger millet: Biofortified foods for micronutrient deficiency in an era of climate change? OMICS A J. Integr. Biol. 24 (12), 688–705. doi:10.1089/omi.2020.0095
Chanyalew, S., Singh, H., Tefera, H., and Sorrels, M. (2005). Molecular genetic map and QTL analysis of agronomic traits based on a Eragrostis tef x E. pilosa recombinant inbred population. J. Genet. Breed. 59 (1), 53.
Chapman, M. A., He, Y., and Zhou, M. (2022). Beyond a reference genome: Pangenomes and population genomics of underutilized and orphan crops for future food and nutrition security. New Phytol. 234 (5), 1583–1597. doi:10.1111/nph.18021
Cheng, Z., Sun, Y., Yang, S., Zhi, H., Yin, T., Ma, X., and Sui, Y. (2021). Establishing in planta haploid inducer line by edited SiMTL in foxtail millet (Setaria italica). Plant Biotechnology Journal, 19(6), 1089. doi:10.1111/pbi.13584
Choi, H-K (2019). Translational genomics and multi-omics integrated approaches as a useful strategy for crop breeding. Genes. Genomics 41 (2), 133–146. doi:10.1007/s13258-018-0751-8
Conn, S., and Gilliham, M. (2010). Comparative physiology of elemental distributions in plants. Ann. Bot. 105, 1081–1102. doi:10.1093/aob/mcq027
Coppola, M., Diretto, G., Cristina Digilio, M., Woo, S., Giuliano, G., Molisso, D., et al. (2019). Transcriptome and metabolome reprogramming in tomato plants by Trichoderma harzianum strain T22 primes and enhances defense responses against aphids. Front. Physiol. 10, 745. doi:10.3389/fphys.2019.00745
Dansi, A., Adoukonou-Sagbadja, H., and Vodouhe, R. (2010). Diversity, conservation and related wild species of Fonio millet (Digitaria spp.) in the northwest of Benin. Genet. Resour. Crop Evol. 57 (6), 827–839. doi:10.1007/s10722-009-9522-3
Das, R. R., Pradhan, S., and Parida, A. (2020). De-novo transcriptome analysis unveils differentially expressed genes regulating drought and salt stress response in Panicum sumatrense. Sci. Rep. 10 (1), 1–14. doi:10.1038/s41598-020-78118-3
David, R. H. A., Ramakrishnan, M., Maharajan, T., BarathiKannan, K., Babu, G. A., Daniel, M. A., et al. (2021). Mining QTL and genes for root traits and biochemical parameters under vegetative drought in South Indian genotypes of finger millet (Eleusine coracana (L.) Gaertn) by association mapping and in silico comparative genomics. Biocatal. Agric. Biotechnol. 32, 101935. doi:10.1016/j.bcab.2021.101935
Deepak Taggelli, R. G., and Thakur, V. (2018). Minor millets-their potential health benefits and medicinal properties: A review. Int. J. Pure Appl. Sci. 6 (1), 1677.
Desai, H., Hamid, R., Ghorbanzadeh, Z., Bhut, N., Padhiyar, S. M., Kheni, J., et al. (2021). Genic microsatellite marker characterization and development in little millet (Panicum sumatrense) using transcriptome sequencing. Sci. Rep. 11 (1), 1–14. doi:10.1038/s41598-021-00100-4
Deshpande, S. S., Mohapatra, D., Tripathi, M. K., and Sadvatha, R. H. (2015). Kodo millet-nutritional value and utilization in Indian foods. J. grain Process. storage 2 (2), 16.
Dey, S., Saxena, A., Kumar, Y., Maity, T., and Tarafdar, A. (2022). Understanding the antinutritional factors and bioactive compounds of kodo millet (paspalum scrobiculatum) and little millet (Panicum sumatrense). J. Food Qual. 2022, 1–19. doi:10.1155/2022/1578448
Dhawale, R. N. (2022). Metabolomic profiling of drought-tolerant little millet (Panicum sumatrense L.) genotype in response to drought stress. 11(4), 1754.
Doust, A. N., Devos, K. M., Gadberry, M. D., Gale, M. D., and Kellogg, E. A. (2004). Genetic control of branching in foxtail millet. Proc. Natl. Acad. Sci. U.S.A. 101 (24), 9045–9050.
Durairaj, M., Gurumurthy, G., Nachimuthu, V., Karthikeyan, M., and Balasubramanian, S. (2019). Dehulled small millets: The promising nutricereals for improving the nutrition of children. Matern. Child. Nutr. 15, e12791. doi:10.1111/mcn.12791
Durgad, A. G., AmruthaJoshi, T., and Hiremath, G. M. (2021). Consumer preference for foxtail and little millets in north eastern region of Karnataka. Econ. Aff. 66 (1), 101–108. doi:10.46852/0424-2513.1.2021.13
Eswari, K., Gogulan, G., Hari Prathab, K. A., and Sakila, M. (2014). Development of early maturing mutants in finger millet. Res. J. Agric. For. Sci. , ISSN 2320, 6063.
Fan, X., Tang, S., Zhi, H., He, M., Ma, W., Jia, Y., et al. (2017). Identification and fine mapping of SiDWARF3 (D3), a pleiotropic locus controlling environment-independent dwarfism in foxtail millet. Crop Sci. 57 (5), 2431–2442. doi:10.2135/cropsci2016.11.0952
Fang, X., Dong, K., Wang, X., Liu, T., Jihong, H., Ren, R., et al. (2016). A high-density genetic map and QTL for agronomic and yield traits in Foxtail millet [Setaria italica (L.) P. Beauv.]. BMC genomics 17 (1), 1–12. doi:10.1186/s12864-016-2628-z
Fekih, R., Takagi, H., Tamiru, M., Abe, A., Natsume, S., Yaegashi, H., et al. (2013). MutMap+: Genetic mapping and mutant identification without crossing in rice. PloS one 8 (7), pe68529. doi:10.1371/journal.pone.0068529
Francis, N., Rajasekaran, R., Krishnamoorthy, I., Muthurajan, R., Thiyagarajan, C., and Alagarswamy, S. (2022). Gamma irradiation to induce beneficial mutants in proso millet (Panicum miliaceum L.): An underutilized food crop. Int. J. Radiat. Biol. 98, 1277–1288. doi:10.1080/09553002.2022.2024292
Fukunaga, K., Abe, A., Mukainari, Y., Komori, K., Tanaka, K., Fujihara, A., et al. (2022a). Recombinant inbred lines and next-generation sequencing enable rapid identification of candidate genes involved in morphological and agronomic traits in foxtail millet. Sci. Rep. 12 (1), 1–12. doi:10.1038/s41598-021-04012-1
Fukunaga, K., Wang, Z., Kato, K., and Kawase, M. (2002b). Geographical variation of nuclear genome RFLPs and genetic differentiation in foxtail millet, Setaria italica (L.) P. Beauv. Genet. Resour. Crop Evol. 49 (1), 95–101. doi:10.1023/a:1013852007770
Ganapathy, K. N., and Patil, J. V. (2017). Improvement in finger millet: Status and future prospects. Millets sorghum Biol. Genet. Improv., 87–111. doi:10.1002/9781119130765.ch3
Ganapathy, S. A., Nirmalakumari, N., Senthil, J., Souframanien, , , and Raveendran, T. S. (2008). Isolation of macromutations and mutagenic effectiveness and efficiency in little millet varieties. World J. Agric. Sci., 4, 483. ISSN 1817- 3047.
Gebreyohannes, A., Shimelis, H., Laing, M., Mathew, I., Odeny, D. A., and Ojulong, H. (2021). Finger millet production in Ethiopia: Opportunities, problem diagnosis, key challenges and recommendations for breeding. Sustainability 13 (23), 13463. doi:10.3390/su132313463
Geervani, P., and Eggum, O. (1989). Nutrient composition and protein quality of minor millets. Plant Foods Hum. Nutr. 39, 201–208. doi:10.1007/BF01091900
Gilles, L. M., Khaled, A., Laffaire, J. B., Chaignon, S., Gendrot, G., Laplaige, J., et al. (2017). Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. Embo J. 36, 707–717. doi:10.15252/embj.201796603
Gopalan, C., Rama Sastri, B. V., and Balasubramanian, S. C. (2009). Nutritive value of Indian foods. Hyderabad, India: National Institute of Nutrition, Indian Council of Medical Research.
Goron, T. L., and Raizada, M. (2015). Genetic diversity and genomic resources available for the small millet crops to accelerate a New Green Revolution. Front. Plant Sci. 6, 157. doi:10.3389/fpls.2015.00157
Guo, J., Huang, Z., Sun, J., Cui, X., and Liu, Y. (2021). Research progress and future development trends in medicinal plant transcriptomics. Front. Plant Sci. 12, 691838. doi:10.3389/fpls.2021.691838
Guo, L, L., Qiu, J., Ye, C., Jin, G., Mao, L., Zhang, H., et al. (2017). Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat. Commun. 8 (1), 1–10. doi:10.1038/s41467-017-01067-5
Guo, R, R., Shi, L., Yan, C., Zhong, X., Gu, F., Liu, Q., et al. (2017). Ionomic and metabolic responses to neutral salt or alkaline salt stresses in maize (Zea mays L.) seedlings. BMC Plant Biol. 17, 41. doi:10.1186/s12870-017-0994-6
Habiyaremye, C., Matanguihan, J. B., D’Alpoim Guedes, J., Ganjyal, G. M., Whiteman, M. R., Kidwell, K. K., et al. (2017). Proso millet (Panicum miliaceum L.) and its potential for cultivation in the pacific northwest, US: A review. Front. Plant Sci. 7, 1961. doi:10.3389/fpls.2016.01961
Hariprasanna, K., Agte, V., Elangovan, M., and Patil, J. V. (2014). Genetic variability for grain iron and zinc content in cultivars, breeding lines and selected germplasm accessions of sorghum [Sorghum bicolor (L.) Moench]. Ind. Jrnl. Gen. Plnt. Bree. 74, 42–49. doi:10.5958/j.0975-6906.74.1.006
Hatakeyama, M., Aluri, S., Balachadran, M. T., Sivarajan, S. R., Patrignani, A., Grüter, S., et al. (2018). Multiple hybrid de novo genome assembly of finger millet, an orphan allotetraploid crop. DNA Res. 25 (1), 39–47. doi:10.1093/dnares/dsx036
He, L., Zhang, B., Wang, X., Li, H., and Han, Y. (2015). Foxtail millet: Nutritional and eating quality, and prospects for genetic improvement. Front. Agric. Sci. Eng. 2, 124–133. doi:10.15302/J-FASE-2015054
He, Q., Zhi, H., Tang, S., Xing, L., Wang, S., Wang, H., et al. (2021). QTL mapping for foxtail millet plant height in multi-environment using an ultra-high density bin map. Theor. Appl. Genet. 134 (2), 557–572. doi:10.1007/s00122-020-03714-w
Hittalmani, S., Mahesh, H. B., Shirke, M. D., Biradar, H., Uday, G., Aruna, Y. R., et al. (2017). Genome and Transcriptome sequence of Finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genomics 18 (1), 1–16. doi:10.1186/s12864-017-3850-z
Hitu, K. W., M’ribu, K., Liang, H., and Mandelbaum, C. (1997). Fonio millets: Ethnobotany, genetic diversity and evolution. South Afr. J. Bot. 63 (4), 185–190. doi:10.1016/S0254-6299(15)30742-0
Ibrahim Bio Yerima, A. R., and Achigan-Dako, E. G. (2021). A review of the orphan small grain cereals improvement with a comprehensive plan for genomics-assisted breeding of fonio millet in West Africa. Plant Breed. 140 (4), 561–574. doi:10.1111/pbr.12930
Indirani, K., and Devasena, M. (2021). Review on nutritional profiles and health benefits of little millets –India. Int. J. Res. Eng. Sci. (IJRES) 9, 7.
Jacquier, N. M. A., Gilles, L. M., Pyott, D. E., Martinant, J. P., Rogowsky, P. M., and Widiez, T. (2020). Puzzling out plant reproduction by haploid induction for innovations in plant breeding. Nat. Plants 6, 610–619. doi:10.1038/s41477-020-0664-9
Jayakodi, M., Madheswaran, M., Adhimoolam, K., Perumal, S., Manickam, D., Kandasamy, T., et al. (2019). Transcriptomes of Indian barnyard millet and barnyardgrass reveal putative genes involved in drought adaptation and micronutrient accumulation. Acta Physiol. Plant. 41 (5), 66–11. doi:10.1007/s11738-019-2855-4
Jency, J. P., Ravikesavan, R., Sumathi, P., and Raveendran, M. (2016). Determination of lethal dose and effect of physical mutagen on germination percentage and seedling parameters in kodo millet variety CO 3. Electron. Journ. Plan. Breed. 7 (4), 1122–1126. doi:10.5958/0975-928x.2016.00155.1
Joshi, D. C., Meena, R. P., and Chandora, R. (2021). “Genetic resources: Collection, characterization, conservation, and documentation,” in Millets and Pseudo Cereals (India: Woodhead Publishing), 19–31. doi:10.1016/B978-0-12-820089-6.00003-3
Kaimal, A. M., Mujumdar, A. S., and Thorat, B. N. (2021). Resistant starch from millets: Recent developments and applications in food industries. Trends Food Sci. Technol. 111, 563–580. doi:10.1016/j.tifs.2021.02.074
Kanlindogbe, C., Sekloka, E., and Kwon-Ndung, E. H. (2020). Genetic resources and varietal environment of grown fonio millets in West Africa: Challenges and perspectives. Plant Breed. Biotech. 8 (2), 77–88. doi:10.9787/PBB.2020.8.2.77
Kim, J. K., Park, S. Y., Yeo, Y., Cho, H. S., Kim, Y. B., Bae, H., et al. (2013). Metabolic profiling of millet (Panicum miliaceum) using gas chromatography-time-of-flight mass spectrometry (GC-TOFMS) for quality assessment. Plant Omics 6 (1), 73–78.
Kleter, G. A., Kuiper, H. A., and Kok, E. J. (2019). Gene-edited crops: Towards a harmonized safety assessment. Trends Biotechnol. 37 (5), 443–447. doi:10.1016/j.tibtech.2018.11.014
Kumar, A., Kumar, S., and Kumar, S. (2015). Promoting minor millets for food and nutritional security. Acta Hortic. 1241, 571–576. doi:10.17660/actahortic.2019.1241.84
Kumar, R., Sharma, V., Srinivas, S., Ramrao, D., Akash, V., Kumar, S., et al. (2021). Understanding omics driven plant improvement and de novo crop domestication: Some examples. Front. Genet. 415, 637141. doi:10.3389/fgene.2021.637141
Lata, C., and Prasad, M. (2013). Validation of an allele-specific marker associated with dehydration stress tolerance in a core set of foxtail millet accessions. Plant Breed. 132, 496–499.
Li, H., Han, S., Huo, Y., Ma, G., Sun, Z., Li, H., et al. (2022). Comparative metabolomic and transcriptomic analysis reveals a coexpression network of the carotenoid metabolism pathway in the panicle of Setaria italica. BMC Plant Biol. 22 (1), 105–116. doi:10.1186/s12870-022-03467-2
Li, K., Wen, W., Alseekh, S., Yang, X., Guo, H., Li, W., et al. (2019). Largescale metabolite quantitative trait locus analysis provides new insights for high-quality maize improvement. Plant J. 99, 216–230. doi:10.1111/tpj.14317
Li, T., Yang, X., Yu, Y., Si, X., Zhai, X., Zhang, H., et al. (2018). Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36, 1160–1163. doi:10.1038/nbt.4273
Li, J, J., Wang, Y., Wang, L., Zhu, J., Deng, J., Tang, R., et al. (2021). Integration of transcriptomic and proteomic analyses for finger millet [Eleusine coracana (L.) Gaertn.] in response to drought stress. Plos one 16 (2), e0247181. doi:10.1371/journal.pone.0247181
Li, W, W., Wen, L., Chen, Z., Zhang, Z., Pang, X., Deng, Z., et al. (2021). Study on metabolic variation in whole grains of four proso millet varieties reveals metabolites important for antioxidant properties and quality traits. Food Chem. 357, 129791. doi:10.1016/j.foodchem.2021.129791
Liang, K., Liang, S., Lu, L., Zhu, D., Zhu, H., Liu, P., et al. (2018). Metabolic variation and cooking qualities of millet cultivars grown both organically and conventionally. Food Res. Int. 106, 825–833. doi:10.1016/j.foodres.2018.01.023
Liu, T., He, J., Dong, K., Wang, X., Wang, W., Yang, P., et al. (2020). QTL mapping of yield component traits on bin map generated from resequencing a RIL population of foxtail millet (Setaria italica). BMC genomics 21 (1), 1–13. doi:10.1186/s12864-020-6553-9
Liu, C, C., Yuan, Y., Liu, J., Wang, H., Ma, Q., Zhou, Y., et al. (2022). Comparative transcriptome and physiological analysis unravel proso millet (Panicum miliaceum L.) source leaf adaptation to nitrogen deficiency with high nitrogen use efficiency. Environ. Exp. Bot. 199, 104891. doi:10.1016/j.envexpbot.2022.104891
Liu, T, T., Liu, X., He, J., Dong, K., Pan, W., Zhang, L., et al. (2022). Identification and fine-mapping of a major QTL (PH1. 1) conferring plant height in broomcorn millet (Panicum miliaceum). Front. Plant Sci. 13, 1010057. doi:10.3389/fpls.2022.1010057
Lloyd, J. R., and Kossmann, J. (2021). Improving crops for a changing world. Front. Plant Sci. 12, 728328. doi:10.3389/fpls.2021.728328
Longvah, T., Ananthan, R., Bhaskarachary, K., and Venkaiah, K. (2017). Indian food composition tables. Hyderabad, India: National Institute of Nutrition.
Luo, J. (2015). Metabolite-based genome-wide association studies in plants. Curr. Opin. Plant Biol. 24, 31–38. doi:10.1016/j.pbi.2015.01.006
Maitra, S. (2020). Intercropping of small millets for agricultural sustainability in drylands: A review. Odisha, India: Crop Research, 55.
Matsuda, F., Nakabayashi, R., Yang, Z., Okazaki, Y., Yonemaru, J., Ebana, K., et al. (2015). Metabolome-genome-wide association study dissects genetic architecture for generating natural variation in rice secondary metabolism. Plant J. 81, 13–23. doi:10.1111/tpj.12681
McMullen, M. D., Stephen, K., Sanchez Villeda., H., Peter, B., Huihui, L., Qi, S., et al. (2009). Genetic properties of the maize nested association mapping population. Science 325, 737–740. doi:10.1126/science.1174320
Merga, M. (2018). Progress, achievements and challenges of tef breeding in Ethiopia. J. Agric. Res. 9 (1), 1–8.
Morgante, M., De Paoli, E., and Radovic, S. (2007). Transposable elements and the plant pan-genomes. Curr. Opin. Plant Biol. 10, 149–155. doi:10.1016/j.pbi.2007.02.001
Morris, J., Hawthorne, K. M., Hotze, T., Abrams, S. A., and Hirschi, K. D. (2008). Nutritional impact of elevated calcium transport activity in carrots. Proc. Natl. Acad. Sci. U. S. A. 105, 1431–1435. doi:10.1073/pnas.0709005105
Mundada, P. S., Barvkar, V. T., Umdale, S. D., Kumar, S. A., Nikam, T. D., and Ahire, M. L. (2021). An insight into the role of silicon on retaliation to osmotic stress in finger millet (Eleusine coracana (L.) Gaertn). J. Hazard. Mat. 403, 124078. doi:10.1016/j.jhazmat.2020.124078
Muthamilarasan, M., and Prasad, M. (2015). Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theor. Appl. Genet. 128 (1), 1–14. doi:10.1007/s00122-014-2399-3
Muthamilarasan, M., and Prasad, M. (2021). Small millets for enduring food security amidst pandemics. Trends Plant Sci. 26 (1), 33–40. doi:10.1016/j.tplants.2020.08.008
Nagaraja, A., and Das, I. K. (2016). “Disease resistance in pearl millet and small millets,” in Biotic stress resistance in millets (Academic Press).
National Research Council (1996). Lost crops of Africa: Volume I: Grains. Washington, DC: National Academies Press. doi:10.17226/2305
Newmaster, S. G., Ragupathy, S., Dhivya, S., Jijo, C. J., Sathishkumar, R., and Patel, K. (2013). Chitilappilly Joseph Jijo., Ramalingam Sathishkumar., and Kirit PatelGenomic valorization of the fine scale classification of small millet landraces in southern India. Genome 56 (2), 123–127. doi:10.1139/gen-2012-0183
Ni, X., Xia, Q., Zhang, H., Cheng, S., Li, H., Fan, G., et al. (2017). Updated foxtail millet genome assembly and gene mapping of nine key agronomic traits by resequencing a RIL population. GigaScience 6 (2), 1–8. doi:10.1093/gigascience/giw005
Padhiyar, S. M., Kheni, J., and Singh, H. V. D. R. (2022). Metabolomic characterization of barnyard millet (Echinochloa frumentacea L.) involved in different stages of spike development. Pharma innov. 11 (6), 604–611.
Pan, J., Li, Z., Dai, S., Ding, H., Wang, Q., Li, X., et al. (2020). Integrative analyses of transcriptomics and metabolomics upon seed germination of foxtail millet in response to salinity. Sci. Rep. 10 (1), 1–16. Available from. doi:10.1038/s41598-020-70520-1
Paroda, R. S., and Mal, B. (1989). New plant sources for food and industry in. New crops food industry.
Parvathi, M. S., Nataraja, K. N., Reddy, Y. A. N., Naika, M. B. N., and Gowda, M. V. C. (2019). Transcriptome analysis of finger millet (Eleusine coracana (L.) Gaertn.) reveals unique drought responsive genes. J. Genet. 98 (2), 46–12. doi:10.1007/s12041-019-1087-0
Pendergast IV, T. H., Qi, P., Odeny, D. A., Dida, M. M., and Devos, K. M. (2022). A high-density linkage map of finger millet provides QTL for blast resistance and other agronomic traits. Plant Genome 15 (1), e20175. doi:10.1002/tpg2.20175
Peng, H., Wang, K., Chen, Z., Cao, Y., Gao, Q., Yan, L., et al. (2020). MBKbase for rice: An integrated omics knowledgebase for molecular breeding in rice. Nucleic Acids Res. 48 (1), D1085-D1092–D1092. doi:10.1093/nar/gkz921
Petti, C., Shearer, A., Tateno, M., Ruwaya, M., Nokes, S., Brutnell, T., et al. (2013). Comparative feedstock analysis in Setaria viridis L. as a model for C4 bioenergy grasses and Panicoid crop species. Front. Plant Sci. 4, 181. doi:10.3389/fpls.2013.00181
Pinson, S. R. M., Tarpley, L., Yan, W., Yeater, K., Lahner, B., Yakubova, E., et al. (2015). Worldwide genetic diversity for mineral element concentrations in rice grain. Crop Sci. 55, 294–311. doi:10.2135/cropsci2013.10.0656
Pramitha Lydia, J., Subhalakshmi, V. K. I., Jeeva, G., and Surendar, A. (2021). Small millets exploring the hidden potential. North West Delhi, India: Scripown Publications.
Puranik, S., Jha, S., Srivastava, P. S., Sreenivasulu, N., and Prasad, M. (2011). Comparative transcriptome analysis of contrasting foxtail millet cultivars in response to short-term salinity stress. J. Plant Physiol. 168 (3), 280–287. doi:10.1016/j.jplph.2010.07.005
Qin, L., Chen, E., Li, F., Yu, X., Liu, Z., Yang, Y., et al. (2020). Genome-wide gene expression profiles analysis reveal novel insights into drought stress in foxtail millet (Setaria italica L.). Int. J. Mol. Sci. 21 (22), 8520–8521. doi:10.3390/ijms21228520
Ragupathy, S., Dhivya, S., Patel., Kirit, Abiran, S., Sambandan, K., Gartaula, H., et al. (2016). DNA record of some traditional small millet landraces in India and Nepal. 3 Biotech. 6 (2), 1–19. doi:10.1007/s13205-016-0450-6
Rahman, H., Jagadeesh, s. N., Valarmathi, R., Sachin, B., Sasikala, R., Senthil, N., et al. (2014). Transcriptome analysis of salinity responsiveness in contrasting genotypes of finger millet (Eleusine coracana L.) through RNA-sequencing. Plant Mol. Biol. 85 (4), 485–503. doi:10.1007/s11103-014-0199-4
Rajput, S. G., Santra, D. K., and Schnable, J. (2016). Mapping QTLs for morpho-agronomic traits in proso millet (Panicum miliaceum L.). Mol. Breed. 36 (4), 37–18. doi:10.1007/s11032-016-0460-4
Ramakrishnan, M., Antony Ceasar, S., Duraipandiyan, V., Vinod, K. K., Kalpana, K., Al-Dhabi, N. A., et al. (2016). Tracing QTLs for leaf blast resistance and agronomic performance of finger millet (Eleusine coracana (L.) Gaertn.) genotypes through association mapping and in silico comparative genomics analyses. PLoS One 11 (7), pe0159264. doi:10.1371/journal.pone.0159264
Ramakrishnan, M., Ceasar, S. A., Vinod, K. K., Duraipandiyan, V., Ajeesh Krishna, T. P., Upadhyaya, H. D., et al. (2017). Identification of putative QTLs for seedling stage phosphorus starvation response in finger millet (Eleusine coracana L. Gaertn.) by association mapping and cross species synteny analysis. PloS one 12 (8), e0183261. doi:10.1371/journal.pone.0183261
Ramesh, M., Vanniarajan, C., Ravikesavan, R., Aiyan, K. E. A., and Mahendran, P. P. (2019). Determination of lethal dose and effect of EMS and gamma ray on germination percentage and seedling parameters in barnyard millet variety Co Kv 2. Electron. Journ. Plan. Breed. 10 (2), 957–962. doi:10.5958/0975-928x.2019.00123.6
Rao Dayakar, B., Bhaskarachary, K., Arlene Christina, G. D., Sudha Devi, G., Vilas, A. T., and Tonapi, A. (2017). Nutritional and health benefits of millets. Rajendranagar, Hyderabad: ICAR, Indian Institute of Millets Research IIMR, 112.
Ravikesavan, R., Sivamurugan, A. P., Iyanar, K., Pramitha, J. L., and Nirmalakumari, A. (2022). Millet cultivation: An overview. Handbook of millets-processing, quality, and nutrition status, 23.
Rawat, J. M., Pandey, S., Debbarma, P., and Rawat, B. (2021). Preparation of alcoholic beverages by tribal communities in the Indian himalayan region: A review on traditional and ethnic consideration. Front. Sustain. Food Syst. 5, 672411. doi:10.3389/fsufs.2021.672411
Renganathan, V. G., Vanniarajan, C., Karthikeyan, A., and Ramalingam, J. (2020). Barnyard millet for food and nutritional security: Current status and future research direction. Front. Genet. 11, 500. doi:10.3389/fgene.2020.00500
Saha, M. V., Gowda, C., Arya, L., Verma, M., and Bansal, K; C. (2016). Genetic and genomic resources of small millets. Crit. Rev. Plant Sci. 35 (1), 56–79. doi:10.1080/07352689.2016.1147907
Sahu, K. R., and Sharma, M. L. (2013). Medicinal and other uses of small millets by the tribal farmers of the Bastar Plateau Zone of Chhattisgarh. Agric. Update 4 (8), 596–599.
Satyarthi, K. S., Sharma, Y. P., Sharma, P., Vaidiya, P., and Randhawa, S. S. (2018). Traditional food grain crops of Himachal Pradesh. Shimla, Himachal Pradesh: Himachal Pradesh State Biodiversity Board. Himachal Pradesh Council for Science, Technology and Environment, Bemole.
Schaart, J. G., van de Wiel, C. C., Lotz, L. A., and Smulders, M. J. (2016). Opportunities for products of new plant breeding techniques. Trends in Plant Science, 21(5), 438–449. doi:10.1016/j.tplants.2015.11.006
Scossa, F., Saleh, A., and Fernie, A. R. (2021). Integrating multi-omics data for crop improvement. J. Plant Physiol. 257, 153352. doi:10.1016/j.jplph.2020.153352
Shan, Z., Jiang, Y., Li, H., Guo, J., Dong, M., Zhang, J., et al. (2020). Genome-wide analysis of the NAC transcription factor family in broomcorn millet (Panicum miliaceum L.) and expression analysis under drought stress. BMC Genomics 21 (1), 96–13. doi:10.1186/s12864-020-6479-2
Sharma, D., Tiwari, A., Sood, S., Jamra, G., Singh, N. K., Meher, P. K., et al. (2018). Genome wide association mapping of agro-morphological traits among a diverse collection of finger millet (Eleusine coracana L.) genotypes using SNP markers. PloS one 13 (8), e0199444. doi:10.1371/journal.pone.0199444
Shi, J., Ma, X., Zhang, J., Zhou, Y., Liu, M., Huang, L., et al. (2019). Chromosome conformation capture resolved near complete genome assembly of broomcorn millet. Nat. Commun. 10 (1), 464–469. doi:10.1038/s41467-018-07876-6
Singh, R. K., Muthamilarasan, M., and Prasad, M. (2021). Biotechnological approaches to dissect climate-resilient traits in millets and their application in crop improvement. J. Biotechnol. 327, 64–73. doi:10.1016/j.jbiotec.2021.01.002
Sood, S., Joshi, D. C., and Pattanayak, A. (2020). Breeding advancements in barnyard millet. Accel. Plant Breed. 1, 391. doi:10.1007/978-3-030-41866-3_15
Sruthi, N. U., and Rao, P. S. (2021). Effect of processing on storage stability of millet flour: A review. Trends Food Sci. Technol. 112, 58–74. doi:10.1016/j.tifs.2021.03.043
Suresh, B. V., Choudhary, P., Aggarwal, P. R., Rana, S., Singh, R. K., Ravikesavan, R., et al. (2022). De novo transcriptome analysis identifies key genes involved in dehydration stress response in kodo millet (Paspalum scrobiculatum L.). Genomics 114 (3), 110347. doi:10.1016/j.ygeno.2022.110347
Tay Fernandez, C. G., Nestor, B. J., Danilevicz, M. F., Gill, M., Petereit, J., Bayer, P. E., et al. (2022). Pangenomes as a resource to accelerate breeding of under-utilised crop species. Int. J. Mol. Sci. 23 (5), 2671. doi:10.3390/ijms23052671
Tettelin, H., Masignani, V., Cieslewicz, M. J., Donati, C., Medini, D., Ward, N. L., et al. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan- genome”. Proc. Natl. Acad. Sci. U. S. A. 102, 13950–13955. doi:10.1073/pnas.0506758102
Tian, B., Zhang, L., Liu, Y., Wu, P., Wang, W., Zhang, Y., et al. (2021). Identification of QTL for resistance to leaf blast in foxtail millet by genome re-sequencing analysis. (2), 743–754. doi:10.1007/s00122-020-03730-w
Tiozon, R., Sartagoda, K. J. D., Fernie, A. R., and Sreenivasulu, N. (2021). Kristel june D., sartagoda, alisdair R. Fernie., and nese SreenivasuluThe nutritional profile and human health benefit of pigmented rice and the impact of post-harvest processes and product development on the nutritional components: A review. Crit. Rev. Food Sci. Nutr., 1–28. doi:10.1080/10408398.2021.1995697
Upadhyaya, Hari D., Gowda, C. L. L., Gopal Reddy, V., and Singh, S. (2008). Diversity of small millets germplasm in gene bank at ICRISAT, 173.
Upadhyaya, H. D., Vetriventhan, M., Dwivedi, S. L., Pattanashetti, S. K., and Singh, S. K. (2016). “Proso, barnyard, little, and kodo millets,” in Genetic and genomic resources for grain cereals improvement (Academic Press), 321.
Upadhyaya, H. D., and Vetriventhan, M. (2018). Underutilized climate-smart nutrient rich small millets for food and nutritional security. Patancheru, India: International Crops Research Institute for the Semi-Arid Tropics (ICRISAT).
VanBuren, R., Man Wai, C., Wang, X., Pardo, J., Yocca, A. E., Wang, H., et al. (2020). Exceptional sub genome stability and functional divergence in the allotetraploid Ethiopian cereal teff. Nat. Commun. 11 (1), 884. doi:10.1038/s41467-020-14724-z
Venkatesan, P., Sundaramari, M., and Venkattakumar, R. (2015). Ethno taxonomical classification of little millet (Panicum sumatrance) by the tribal people in Tamilnadu. India.
Verma, V. C., Verma., V. C., Singh., A., and Agrawal, S. (2018). Ethnobotanical study of small millets from India: Prodigious grain for nutritional and industrial aspects. Int. J. Chem. Stud. 6 (4), 2155.
Vetriventhan, M., and Upadhyaya, H. D. (2019). Variability for productivity and nutritional traits in germplasm of kodo millet, an underutilized nutrient-rich climate smart crop. Crop Sci. 59 (3), 1095–1106. doi:10.2135/cropsci2018.07.0450
Vetriventhan, M., Azevedo, V. C., Upadhyaya, H. D., Nirmalakumari, A., Kane-Potaka, J., Anitha, S., et al. (2020). Genetic and genomic resources, and breeding for accelerating improvement of small millets: Current status and future interventions. Nucleus 63 (3), 217–239. doi:10.1007/s13237-020-00322-3
Vetriventhan, Mani., and Upadhyaya, Hari D. (2018). Diversity and trait-specific sources for productivity and nutritional traits in the global proso millet (Panicum miliaceum L.) germplasm collection. Crop J. 6 (5), 451–463. doi:10.1016/j.cj.2018.04.002
Vinoth, A., and Ravindhran, R. (2017). Biofortification in millets: A sustainable approach for nutritional security. Front. Plant Sci. 8, 29. doi:10.3389/fpls.2017.00029
Wang, J., Wang, Z. L., Du, X. F., Yang, H. Q., Han, F., Han, Y. H., et al. (2017). A high-density genetic map and QTL analysis of agronomic traits in foxtail millet [Setaria italica (L.) P. Beauv.] using RAD-seq. PLoS One 12, e0179717. doi:10.1371/journal.pone.0179717
Wang, X., Chen, S., Ma, X., Yssel, A. E., Chaluvadi, S. R., Johnson, M. S., et al. (2021). Genome sequence and genetic diversity analysis of an under-domesticated orphan crop, white fonio (Digitaria exilis). Gigascience 10 (3), giab013. doi:10.1093/gigascience/giab013
Wang, Z. M., Devos, K. M., Liu, C. J., Wang, R. Q., and Gale, M. D. (1998). Construction of RFLP-based maps of foxtail millet, Setaria italica (L.) P. Beauv. Theor. Appl. Genet. 96 (1), 31–36. doi:10.1007/s001220050705
Woldeyohannes, A. B., Desta, E. A., Fadda, C., Pè, M. E., and Dell’Acqua, M. (2022). Value of teff (eragrostis tef) genetic resources to support breeding for conventional and smallholder farming: A review. CABI Agric. Biosci. 3 (1), 27–16. doi:10.1186/s43170-022-00076-9
Wyatt, S. E., Tsou, P. L., and Robertson, D. (2002). Expression of the high-capacity calcium binding domain of calreticulin increases bioavailable calcium stores in plants. Transgenic Res. 11, 1–10. doi:10.1023/A:1013917701701
Xia, D., Zhou, H., Wang, Y., Li, P., Fu, P., Wu, B., et al. (2021). How rice organs are colored: The genetic basis of anthocyanin biosynthesis in rice. Crop J. 9 (3), 598–608. doi:10.1016/j.cj.2021.03.013
Yang, L., Li, R., Cui, Y., Qin, X., and Li, Z. (2021). Comparison of nutritional compositions of foxtail millet from the different cultivation regions by UPLC-Q-Orbitrap HRMS based metabolomics approach. J. Food Biochem. 45 (10), e13940. doi:10.1111/jfbc.13940
Yang, W., Guo, Z., Huang, C., Wang, K., Jiang, N., Fen, H., et al. (2015). Genome wide association study of rice (Oryza sativa L.) leaf traits with a high-throughput leaf scorer. J. Exp. Bot. 66, 5605–5615. doi:10.1093/jxb/erv100
Yoshitsu, Y., Takakusagi, M., Abe, A., Takagi, H., Uemura, A., Yaegashi, H., et al. (2017). QTL-seq analysis identifies two genomic regions determining the heading date of foxtail millet, Setaria italica (L.) P. Beauv. Breed. Sci. 67 (5), 518–527. doi:10.1270/jsbbs.17061
Zeid, M., Belay, G., Mulkey, S., Poland, J., and Sorrells, M. E. (2011). QTL mapping for yield and lodging resistance in an enhanced SSR-based map for tef. Theor. Appl. Genet. 122 (1), 77–93. doi:10.1007/s00122-010-1424-4
Zhang, G., Liu, X., Quan, Z., Cheng, S., Xu, X., Pan, S., et al. (2012). Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 30 (6), 549–554. doi:10.1038/nbt.2195
Zhang, H., Tang, S., Schnable, J. C., He, Q., Gao, Y., Luo, M., et al. (2021). SiSTL2 is required for cell cycle, leaf organ development, chloroplast biogenesis, and has effects on C4 photosynthesis in setaria italica (L.) P. Beauv. Front. Plant Sci. 12, 1103. doi:10.3389/fpls.2018.01103
Zhang, K., Jia, X., Zhu, Z., and Xue, W. (2020). Physicochemical properties of rice analogs based on multi-level: Influence of the interaction of extrusion parameters. Int. J. Food Prop. 23 (1), 2033–2049. doi:10.1080/10942912.2020.1840389
Zhang, Y., Gao, X., Li, J., Gong, X., Yang, P., Gao, J., et al. (2019). Comparative analysis of proso millet (Panicum miliaceum L.) leaf transcriptomes for insight into drought tolerance mechanisms. BMC Plant Biol. 19 (1), 1–17. doi:10.1186/s12870-019-2001-x
Zhi, H., He, Q., Tang, S., Yang, J., Zhang, W., Liu, H., et al. (2021). Genetic control and phenotypic characterization of panicle architecture and grain yield-related traits in foxtail millet (Setaria italica). Theor. Appl. Genet. 134 (9), 3023–3036. doi:10.1007/s00122-021-03875-2
Keywords: small millets, nutrition, therapeutic traits, ethnobotany, multi-omics
Citation: Lydia Pramitha J, Ganesan J, Francis N, Rajasekharan R and Thinakaran J (2023) Revitalization of small millets for nutritional and food security by advanced genetics and genomics approaches. Front. Genet. 13:1007552. doi: 10.3389/fgene.2022.1007552
Received: 30 July 2022; Accepted: 07 December 2022;
Published: 09 January 2023.
Edited by:
Reyazul Rouf Mir, Sher-e-Kashmir University of Agricultural Sciences and Technology, IndiaReviewed by:
Manjusha Verma, National Bureau of Plant Genetic Resources (ICAR), IndiaPaul Kiprotich Kimurto, Egerton University, Kenya
Vanisri Satturu, Professor Jayashankar Telangana State Agricultural University, India
Copyright © 2023 Lydia Pramitha, Ganesan, Francis, Rajasekharan and Thinakaran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: J. Lydia Pramitha, bHlkaWFwcmFtaXRoYUBnbWFpbC5jb20=
†These authors have contributed equally to this work
 J. Lydia Pramitha
J. Lydia Pramitha Jeeva Ganesan2†
Jeeva Ganesan2† Neethu Francis
Neethu Francis Ravikesavan Rajasekharan
Ravikesavan Rajasekharan Jenita Thinakaran
Jenita Thinakaran