
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 16 September 2022
Sec. Epigenomics and Epigenetics
Volume 13 - 2022 | https://doi.org/10.3389/fgene.2022.1006582
This article is part of the Research TopicProtein Modifications in Epigenetic Dysfunctional Diseases: Mechanisms and Potential Therapeutic StrategiesView all 15 articles
B cell transposition gene 3 (BTG3) is reported to be a tumor suppressor and suppresses proliferation and cell cycle progression. This study aims to analyze the clinicopathological and prognostic significances, and signal pathways of BTG3 mRNA expression in human beings through bioinformatics analysis. We analyzed BTG3 expression using Oncomine, TCGA (the cancer genome atlas), Xiantao, UALCAN (The University of ALabama at Birmingham Cancer data analysis Portal) and Kaplan-Meier plotter databases. Down-regulated BTG3 expression was observed in lung and breast cancers, compared with normal tissues (p < 0.05), but not for gastric and ovarian cancer (p < 0.05). The methylation of BTG3 was shown to be adversely correlated with its mRNA expression (p < 0.05). BTG3 expression was higher in gastric intestinal-type than diffuse-type carcinomas, G1 than G3 carcinomas (p < 0.05), in female than male cancer patients, T1-2 than T3-4, and adenocarcinoma than squamous cell carcinoma of lung cancer (p < 0.05), in invasive ductal than lobular carcinoma, N0 than N1 and N3, TNBC (triple-negative breast cancer) than luminal and Her2+, and Her2+ than luminal cancer of breast cancer (p < 0.05), and G3 than G2 ovarian carcinoma (p < 0.05). BTG3 expression was positively related to the survival rate of gastric and ovarian cancer patients (p < 0.05), but not for breast cancer (p < 0.05). KEGG and PPI (protein-protein interaction) analysis showed that the BTG3 was involved in cell cycle and DNA replication, digestion and absorption of fat and protein, spliceosome and ribosome in cancer. BTG3 expression was positively linked to carcinogenesis, histogenesis, and aggressive behaviors, and was employed to evaluate the prognosis of cancers by regulating cell cycle, metabolism, splicing and translation of RNA.
B cell transposition gene 3 (BTG3) belongs to a member of B-cell translocation gene family, and maps to human chromosome 21q21.1 (Guéhenneux et al., 1997). Its encoding protein has been reported to be a tumor suppressor in some malignancies, including gastric cancer, breast cancer, renal cell carcinoma, esophageal adenocarcinoma, hepatocellular carcinoma, lung cancer, ovarian cancer, and prostate cancer (Yu et al., 2008; Majid et al., 2009; Lin et al., 2012; Chen et al., 2013; Deng et al., 2013; Lv et al., 2013; Du et al., 2015; Gou et al., 2015; Ren et al., 2015). In the nuclear compartment, BTG3 protein can interact with E2F1 (E2F transcription factor 1), Smad8 receptor-regulated Smad transcription factor, and CCR4 (C-C motif chemokine receptor 4) transcription factor-associated protein Caf1 to suppress finally cell proliferation and cell cycle progression (Yoshida et al., 2001; Ou et al., 2007; Miyai et al., 2009). BTG3 maintains genomic stability by promoting Lys63-linked ubiquitination and CHK1 (checkpoint kinase 1) activation, whereas BTG3 can be phosphorylated and activated via its interaction with CHK1 as a positive feedback loop (Cheng et al., 2013). In the cytosolic compartment, BTG3 binds to and suppresses src, Akt and Ras/MAP kinase signaling (Rahmani, 2006; Cheng et al., 2015). Ma et al. (2020) demonstrated that the combination of hypoxia and BTG3 expression could induce radiation resistance, indicating an important role of BTG3 in hypoxia-induced radiation resistance of colorectal cancer cells. Cucurbitacin B inhibited cell proliferation and anti-apoptosis of colorectal cancer by the reactivation of BTG3 by promoter demethylation (Mao et al., 2019).
By postnatal 21 months, BTG3-deficiency might promote bone morphogenetic protein-induced ectopic bone formation and lung adenocarcinogenesis (Yoneda et al., 2009). BTG3 deficiency triggers acute cellular senescence via the Erk-AP-1-JMJD3-p16 pathway (Lin et al., 2012). MiR-142-5p strengthens cell growth and migration in renal cell carcinoma, while miR-93 desensitizes esophageal cancer to radiotherapy by targeting BTG3 (Cui et al., 2017; Liu et al., 2017). IASPP promotes miR-20a expression that restores cell invasion and cisplatin chemoresistance of cervical cancer cells by targeting BTG3 (Xiong et al., 2017). A body of evidence identified BTG3 as a direct downstream target of miR-519c-3p and miR-20b-5p, which promoted proliferation and migration in hepatocellular carcinoma and colorectal cancer cells, respectively (Peng et al., 2019; Wang et al., 2019). Enkhnaran et al. (2022) demonstrated that miR-106b-5p promoted cell proliferation and cell cycle and increased hepatocellular carcinoma cells’ resistance to sorafenib through the BTG3/Bcl-xL/p27 signaling pathway.
In our previous work, BTG3 overexpression was demonstrated to reverse the aggressive phenotypes of gastric and colorectal cancer cells (Gou et al., 2015; Zheng et al., 2017). Here, we aimed to clarify the clinicopathological and prognostic significances, and related signal pathways of BTG3 mRNA expression in cancers by a bioinformatics analysis.
The individual gene expression level of BTG3 mRNA was analyzed using Oncomine (www.oncomine.org), a cancer microarray database and web-based data mining platform for a new discovery from genome-wide expression analyses. We compared the differences in BTG3 mRNA levels between normal tissue and cancer. All data were log-transformed, the median centered per array, and the standard deviation normalized to one per array.
The expression data (RNA-seqV2) and clinicopathological data of gastric (n = 392), lung (n = 865), breast (n = 1,093), and ovarian (n = 304) cancer patients were downloaded from the TCGA database (https://cancergenome.nih.gov/abouttcga/overview) by TCGA-assembler in R software. We integrated the raw data, analyzed BTG3 mRNA expression in the cancers, and compared it with clinicopathological and prognostic data of cancer patients. The means were compared with student t-test. Kaplan-Meier survival plots were generated with survival curves and compared by the log-rank statistic. Cox’s proportional hazards model was employed for multivariate analysis. SPSS 17.0 software was employed to analyze all data. Two-sided p < 0.05 was considered statistically significant.
The prognostic significance of BTG3 mRNA was also analyzed in gastric, lung, breast, and ovarian cancers using KM plotter (https://kmplot.com/analysis/).
The expression and methylation of BTG3 gene were analyzed using the UALCAN database (http://ualcan.path.uab.edu/). They were also compared with the clinicopathological and prognostic features of gastric, lung, breast, and ovarian cancers.
The expression and methylation of BTG3 gene were analyzed using the xiantao platform (https://www.xiantao.love/). Additionally, we discovered the differential and related genes using Xiantao. The differential genes were used to build the PPI (protein-protein interaction) network and identify the important hub genes. These genes were submitted to KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis in order to build signal pathways.
We used Cho’s and Chen’s datasets to perform bioinformatics analysis, and found that BTG3 expression was higher in gastric cancer than in normal tissues, even stratified into intestinal-, diffuse-, and mixed-type carcinomas (Figure 1A, p < 0.05). In TCGA (Figure 1B), Xiantao (Figure 1C) and UALCAN (Figure 1D) data, it was the same for BTG3 expression (p < 0.05). BTG3 expression was higher in G1 than G3 carcinomas by UALCAN (Figure 1E, p < 0.05). There was a negative correlation between BTG3 mRNA and methylation (cg23273752, cg12602426, cg01168851, cg04464940, cg14394939, and cg08875503) by Xiantao (Figure 1F, p < 0.05). BTG3 methylation was lower in gastric cancer than in normal tissues (Figure 1G, p < 0.05), and G1 than G3 carcinoma (Figure 1H, p < 0.05) by UALCAN. According to Kaplan-Meier plotter, we found that a higher BTG3 expression was positively correlated with overall and progression-free survival rates of all cancer patients, even stratified by gender and Her2+ expression (Figure 1I and Table 1, p < 0.05). The overall survival rate of the patients with intestinal-, diffuse-, or mixed-type carcinoma was higher in BTG3 overexpression than in underexpression groups (Table 1, p < 0.05). There appeared to be a positive relationship between BTG3 expression and the overall survival rate of the patients with 5-FU-based adjuvant (Table 1, p < 0.05). It was the same for the progression-free survival in the patients with poorly-differentiated adenocarcinoma, 5-FU-based or other adjuvant treatment (Table 1, p < 0.05).
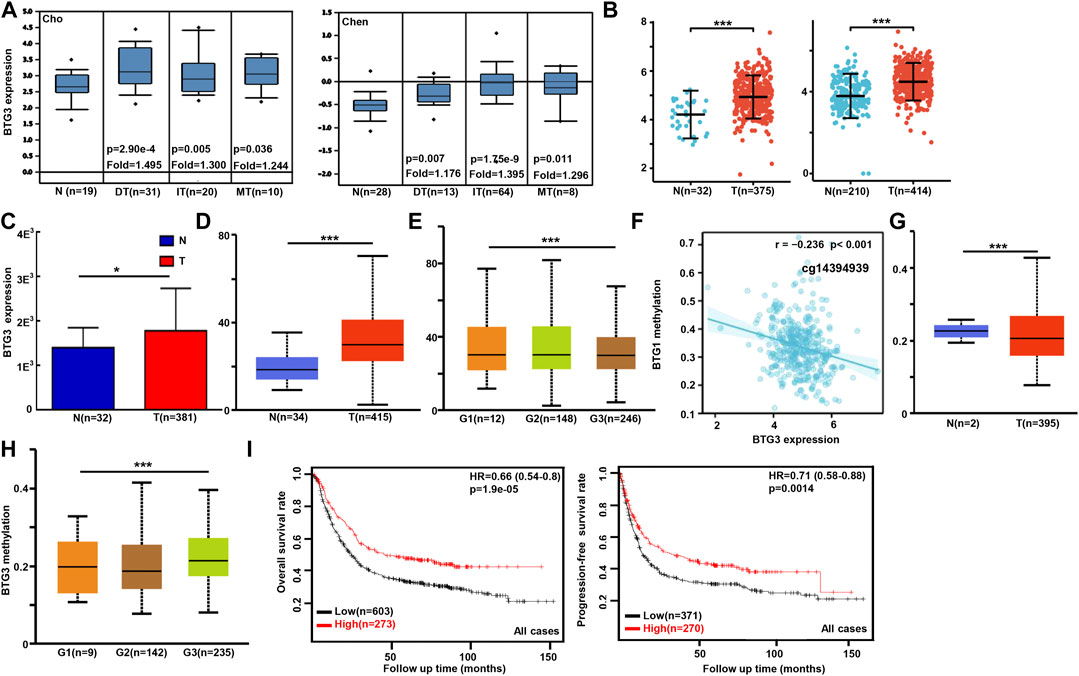
FIGURE 1. The clinicopathological and prognostic significances of BTG3 mRNA expression in gastric cancer Cho’s and Chen’s datasets were used for bioinformatics analysis to explore BTG3 expression in gastric cancer. A higher BTG3 expression was detectable in gastric cancer than that in normal mucosa, even stratified into intestinal- (IT), diffuse- (DT), and mixed-type (MT) carcinomas by Lauren’s classification [(A), p < 0.05], in line with the findings from Xiantao (B), TCGA (C) and UALCAN (D) databases (p < 0.05). UALCAN database showed that BTG3 was more expressed in G1 than G3 carcinomas [(E), p < 0.05]. The negative relationship between BTG3 mRNA expression and methylation was found in gastric cancer using Xiantao database (F). Its methylation was lower in gastric cancer than normal tissues in UALCAN (G). We also compared BTG3 methylation with histological grading of gastric cancer (H). According to the data from Kaplan-Meier plotter, BTG3 expression was positively related to both overall and progression-free survival rates of the patients with gastric cancer [(I), p < 0.05)]. Note: N, normal; T, tumor; HR, hazard ratio.
In Xiantao, BTG3 expression was lower in lung cancer than in normal tissues (Figure 2A, p < 0.05). In TCGA database, BTG3 expression was higher in female than male cancer patients, T1-2 than T3-4, and adenocarcinoma than squamous cell carcinoma patients (Figure 2B, p < 0.05). There was a negative correlation between BTG3 mRNA and its methylation (cg14380517, cg10696191, cg03232933, cg05762769, cg14394939, and cg03232933) by xiantao (Figure 2C, p < 0.05). BTG3 methylation was higher in lung cancer than in normal tissues (Figure 2D, p < 0.05). In TCGA, BTG3 expression was positively associated with a high overall survival rate of cancer patients (Figure 2E, p < 0.05). Cox’s risk proportional regression model indicated that T staging, lymph node status and BTG3 expression were independent prognostic factors for lung cancer patients (Table 2, p < 0.05). According to Kaplan-Meier plotter, we found that a higher BTG3 expression was negatively correlated with overall survival rates of all cancer patients, female or male patients, adenocarcinoma patients, N1, M0, Stage I and II cancer patients, smoking and non-smoking patients, or those with surgical margin negative (Figure 2F, p < 0.05). All, female T1, T2, or N1 cancer patients with high BTG3 expression showed a short progression-free survival time than those with its low expression (p < 0.05, data not shown). There appeared to be a negative relationship between BTG3 expression and the progression-free survival rate of cancer patients with surgical margin negative or smoking cancer patients (p < 0.05, data not shown).
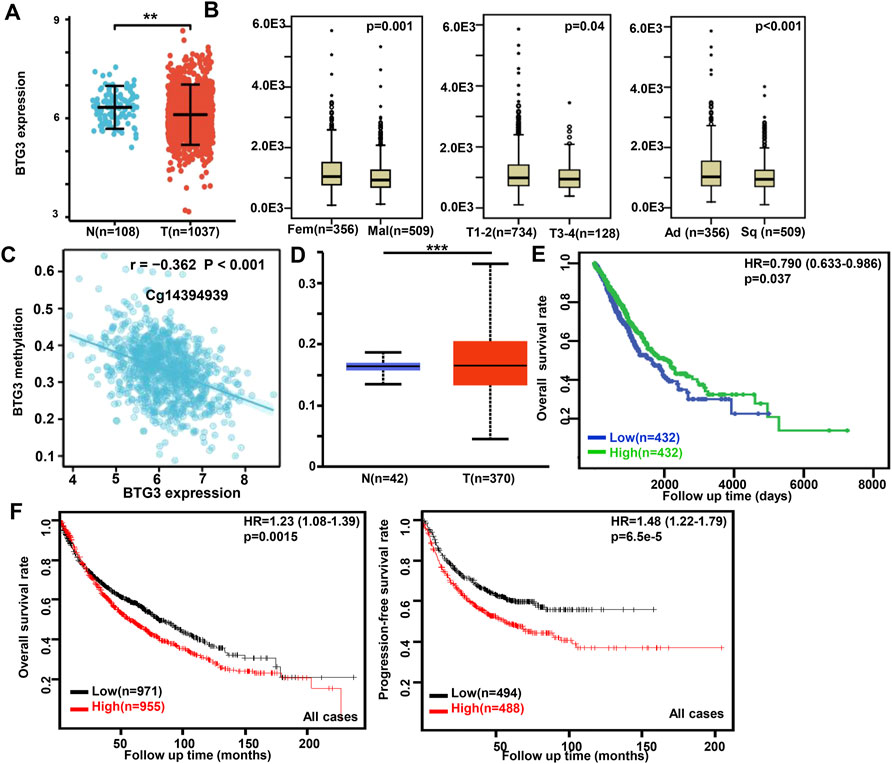
FIGURE 2. The clinicopathological and prognostic significances of BTG3 mRNA expression in lung cancer Xiantao dataset was employed for bioinformatics analysis to analyze BTG3 expression during lung carcinogenesis (A). BTG3 expression was compared with gender, histological subtyping, and T staging of the cancer patients by TCGA database (B). The negative relationship between BTG3 mRNA expression and methylation was analyzed in lung cancer using Xiantao database (C). Its methylation was higher in lung cancer than normal tissues in UALCAN (D). The correlation between BTG3 expression and overall or post-progression survival rate of the patients with lung cancer was analyzed using TCGA database (E) and Kaplan-Meier plotter (F). Note: N, normal; T, tumor; Ad, adenocarcinoma; Sq, squamous cell carcinoma; HR, hazard ratio.
According to Xiantao (Figure 3A) and UALCAN (Figure 3B) databases, we found that BTG3 expression was lower in breast cancer than in normal tissues (p < 0.05). TCGA database showed a higher BTG3 expression in invasive ductal than lobular carcinoma (Figure 3C, p < 0.05). It was higher in N0 than N1 and N3, TNBC (triple-negative breast cancer) than luminal and Her2+, and Her2+ than Luminal cancer patients by UALCAN (Figure 3D, p < 0.05). As summarized in Table 3, BTG3 mRNA expression was negatively associated with elder age, non-Asian race, non-TNBC, ER positivity, and PR positivity (p < 0.05). There was a negative correlation between BTG3 mRNA and its methylation (cg14380517, cg10696191, cg03232933, cg05762769, cg27075724, cg02652260, cg23273752, cg12602426, cg01168851, cg20227212, cg04464940, and cg14394939) by Xiantao (Figure 3E, p < 0.05). BTG3 methylation was higher in breast cancer than normal tissues (Figure 3F, p < 0.05), N3 than N0, Luminal and Her2+ than TNBC, Her2+ than Luminal cancer patients by UALCAN (Figure 3G, p < 0.05). According to Kaplan-Meier plotter, we found that a higher BTG3 expression was negatively correlated with overall, progression-free, post-progression, and distant-metastasis-free survival rates of all cancer patients (Figure 3H, p < 0.05). There appeared to be a negative relationship between BTG3 expression and the overall survival rate of patients with Luminal-B breast cancer (p < 0.05, data not shown). The relapse-free survival rate of the cancer patient with or without lymph node metastasis was lower in the groups of high BTG3 expression than its low expression (p < 0.05, data not shown). A negative association between BTG3 expression and relapse-free prognosis was observed in Luminal-B cancer patients (p < 0.05, data not shown). ER (estrogen receptor)- positive or Her2-negative cancer patients with high BTG3 expression showed a shorter overall survival time than those with its low expression (p < 0.05, data not shown).
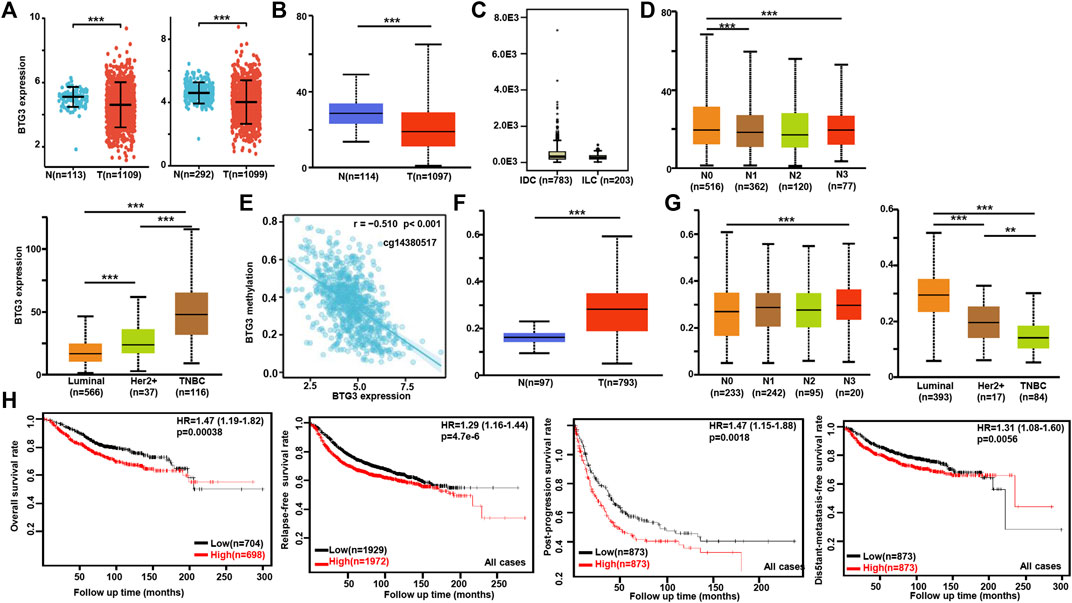
FIGURE 3. The clinicopathological and prognostic significances of BTG3 mRNA expression in breast cancer. According to Xiantao (A) and UALCAN (B) databases, BTG3 hypoexpression was detectable in breast cancer, compared with breast normal tissue (p < 0.05). TCGA database showed a higher BTG3 expression in invasive ductal (IDC) than lobular (ILC) carcinoma [(C), p < 0.05]. UALCAN database was also used to compare BTG3 expression with N stage and molecular subtyping of breast cancer (D). The negative relationship between BTG3 mRNA expression and methylation was analyzed in breast cancer using Xiantao database (E). Its methylation was higher in breast cancer than normal tissues (F), and compared with N stage and molecular subtyping (G) in UALCAN. The correlation between BTG3 expression and overall, post-progression, distant-metastasis-free or relapse-free survival rate was analyzed in the patients with breast cancer using Kaplan-Meier plotter (H). Note: N, normal tissue; T, tumor; TNBC, triple-negative breast cancer; HR, hazard ratio.
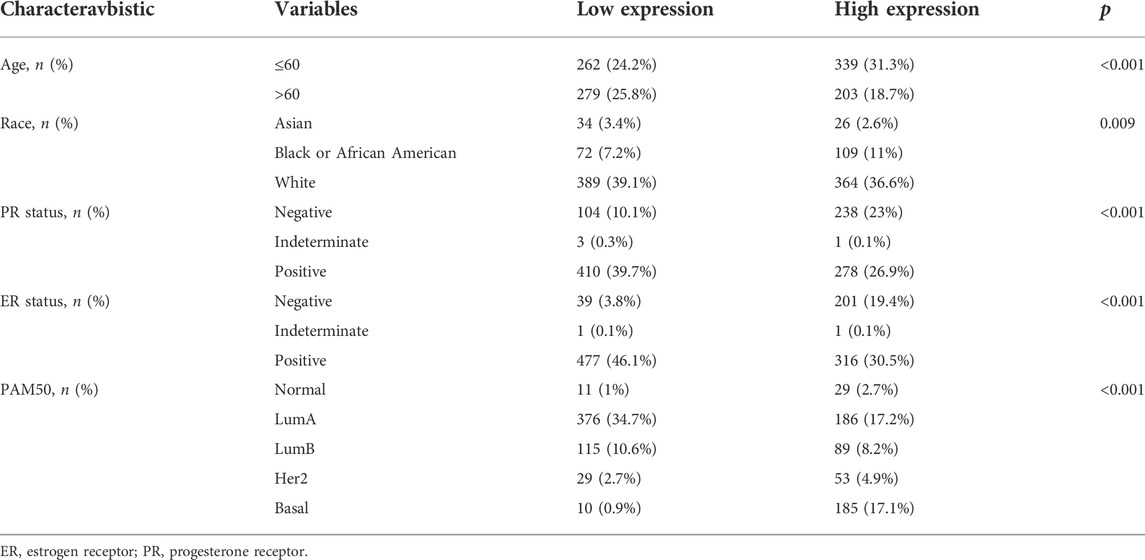
TABLE 3. The correlation between BTG3 mRNA expression and clinicopathological characteristics of breast cancer.
We performed bioinformatics analysis of BTG3 expression in ovarian cancer using Bonome’s, Hendrix’s, Lu’s, Welsh’s, and TCGA’s datasets. BTG3 expression was higher in ovarian cancer than normal mucosa regardless of histological subtyping (Figure 4A, p < 0.05). It was the same for Xiantao data (Figure 4B, p < 0.05). BTG3 expression was higher in G3 than G2 carcinoma patients by UALCAN (Figure 4C, p < 0.05). The data from Kaplan-Meier plotter showed a positive relationship between BTG3 expression and the overall survival rate of the ovarian cancer patients with paclitaxel treatment (Figure 4D, p < 0.05). A positive correlation between BTG3 expression and the post-progression survival rate was observed in all ovarian cancer patients, or G1-3, G 2-3, Grade3 or suboptimal cancer patients (Figure 4D, p < 0.05).
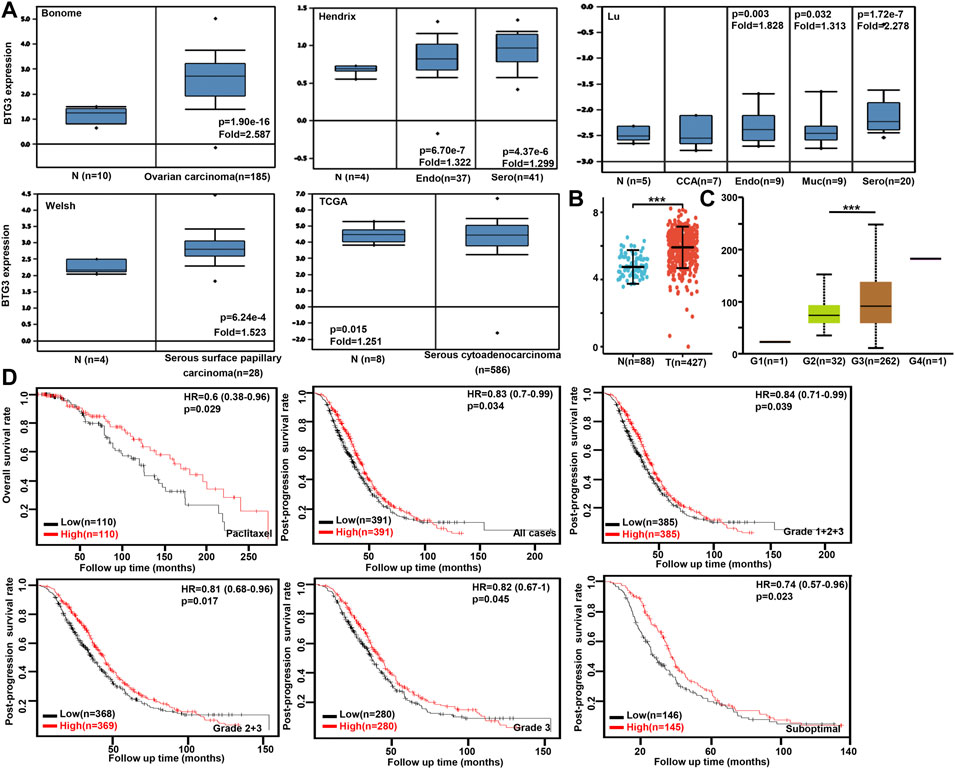
FIGURE 4. The clinicopathological and prognostic significances of BTG3 mRNA expression in ovarian cancer Oncomine (A) and Xiantao (B) datasets were employed for bioinformatics analysis to observe BTG3 expression in ovarian cancer. A lower BTG3 expression was detectable in ovary than that in ovarian carcinoma, clear cell adenocarcinoma (CCA), endometriod (Endo), mucinous (Muc) and serous (Sero) adenocarcinoma (p < 0.05). BTG3 expression was compared with histological grading of ovarian cancer (C). The correlation between BTG3 expression and overall, or post-progression survival rate was analyzed in the patients with ovarian cancer using Kaplan-Meier plotter, even stratified by different clinicopathological parameters [(D), p < 0.05]. Note: N, normal tissue; T, tumor; HR, hazard ratio.
On the Xiantao platform, we found the differential genes between low and high expression groups of BTG3 mRNA in cancers. KEGG analysis showed that the top signal pathways of the differential genes included cell cycle, calcium and p53 signal pathway, pancreatic and insulin secretion, fat digestion and absorption, DNA replication, mismatch repair and homologous recombination in gastric cancer, platelet activation and coagulation, digestion and absorption of protein and fat, metabolism of arachidonic and linoleic acids in lung cancer, cell cycle, salivary and insulin secretion, DNA replication, and ovarian steroidogenesis in breast cancer, and PI3K/Akt signal pathway, focal adhesion and ECM-receptor interaction in ovarian cancer (Figure 5A). In addition, the STRING was used to identify the PPI pairs and the cytoscape to find out the top 10 nodes ranked by degree (Figure 5B). The top hub genes mainly contained replication protein, DNA replication helicase, replication factor, WRN RecQ like helicase and exonucleases in gastric cancer, Apolipoproteins, lipase C, phospholipase A2, and cholesteryl ester transfer protein in lung cancer, minichromosome maintenance protein in breast cancer, and ribosomal proteins in ovarian cancer.
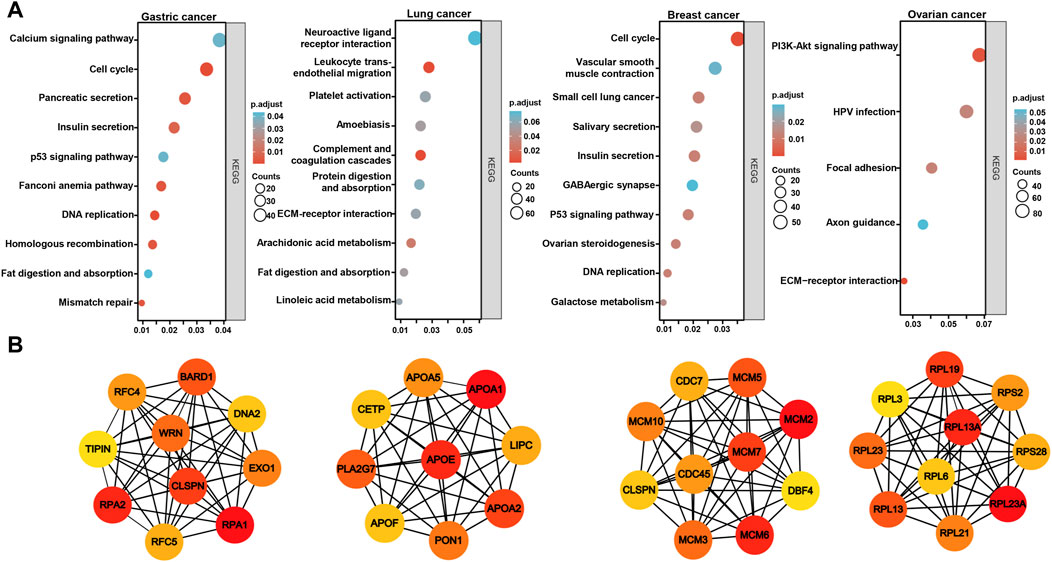
FIGURE 5. The differential genes and related signal pathways about BTG3 in cancersn The differential genes of BTG3 were subjected to the signal pathway analysis using KEGG (A). STRING was used to identify the protein-protein interaction network of differential genes about BTG3 in cancers, and Cytoscape was employed to find out the top 10 hub nodes ranked by degree (B).
According to the Xiantao database, the BTG3-correlated genes in cancers were analyzed and subjected to the KEGG analysis (Figure 6). The BTG3-correlated genes were involved in RNA transport, splicing and degradation, DNA replication and cell cycle, proteasomal degradation for gastric cancer, cell cycle, DNA replication, and mismatch repair, TNF and NF-κB signal pathways for lung cancer, ribosome and spliceosome, and metabolism of amino acids for breast cancer, neural diseases, viral infection, oxidative phosphorylation for ovarian cancer.

FIGURE 6. The BTG3-related signal pathways in cancers The top BTG3-related genes were screened and classified into the signal pathways of KEGG using Xiantao database.
BTG3 overexpression suppressed the proliferation and invasion of epithelial ovarian and colorectal cancer cells by weakening Akt/GSK3β/β-catenin signaling (Mao et al., 2016; An et al., 2017). Lv et al. (2018) found that BTG3 knockdown promoted cell proliferation, migration, invasion, relieved G2 arrest, and inhibited apoptosis in colorectal cancer cells with PAK2 (p21 activated kinase 2), RPS6KA5 (ribosomal protein S6 kinase A5), YWHAB (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta), and STAT3 up-regulated and RAP1A (ras-related protein rap-1A), DUSP6 (dual specificity phosphatase 6), and STAT (signal transducer and activator of transcription) 1 down-regulated. Our group demonstrated that BTG3 expression inhibited proliferation, tumor growth, migration and invasion, and induced autophagy, apoptosis, and chemosensitivity to cisplatin, MG132 (proteasome inhibitor), paclitaxel, and SAHA (histone deacetylase inhibitor) in gastric and colorectal cancer cells (Gou et al., 2015; Zheng et al., 2017). In esophageal adenocarcinoma cells, BTG3 upregulation suppressed the proliferation and invasion (Du et al., 2015). Lv et al. (2013) found that BTG3 suppressed proliferation, invasion and induced G1/S cycle arrest of hepatocellular carcinoma cells. Reportedly, iASPP promoted epithelial-mesenchymal transition, and conferred cisplatin resistance in cervical cancer via miR-20a- FBXL5/BTG3 signaling (Xiong et al., 2017). Yanagida et al. (2013) demonstrated that the antisense transcript of BTG3 gene (ASBEL) down-regulated BTG3 protein and promoted proliferation and tumorigenicity of ovarian clear cell carcinoma. ASBEL knockdown functioned as a tumor suppressive role in breast cancer cells by up-regulating BTG3 (Xia et al., 2017). BTG3 anti-sense transcript mediated the down-regulation of ATF3 expression, which was essential for the proliferation and tumorigenicity of colon cancer cells (Taniue et al., 2016). In combination with these findings, it was suggested that BTG3 might be employed as a molecular target of cancer gene therapy because of its inhibitory effects on aggressive phenotypes of cancer cells.
A body of evidence indicates that BTG3 expression is down-regulated in gastric cancer (Gou et al., 2015; Ren et al., 2015), esophageal adenocarcinoma (Du et al., 2015), hepatocellular carcinoma (Lv et al., 2013), lung cancer (Chen et al., 2013), ovarian cancer (Deng et al., 2013), renal cell carcinoma (Majid et al., 2009), and colorectal cancer (Xiong et al., 2017) due to its promoter methylation (Yu et al., 2008; Majid et al., 2009; Lv et al., 2013; Gou et al., 2015), in line with our findings about lung and breast cancers. Additionally, BTG3 methylation was negatively correlated with its mRNA expression, and was higher in cancer than in normal tissues of the stomach, lung, and breast. In breast and gastric cancer, the correlation between BTG3 mRNA and aggressive behaviors was the opposite to that between BTG3 methylation and them. These findings suggested that BTG3 hypoexpression might be due to its promoter methylation. In contrast, our results showed BTG3 mRNA overexpression in gastric and ovarian cancers. The controversial findings might be explained by different approaches: previous reports from immunohistochemistry, Western blot or RT-PCR, but the present study from the cDNA chip or transcriptomics.
In addition, BTG3 mRNA expression was positively linked to the differentiation of gastric cancer, in line with our previous report about gastric cancer tissues (Gou et al., 2015). BTG3 was also reported to induce the differentiation of gastric and colorectal cancer cells, evidenced by a higher level of alkaline phosphatase (Gou et al., 2015; Zheng et al., 2017). These findings suggested that BTG3 mRNA expression might underlie the molecular mechanisms of gastric cancer differentiation. In addition, BTG3 expression was found to be negatively associated with the tumor size of lung cancer. Pulmonary adenocarcinoma patients had a higher BTG3 mRNA expression than squamous cell carcinoma patients. A higher BTG3 mRNA expression was seen in invasive ductal than lobular carcinomas and negatively correlated with N staging and favorable molecular subtypes (Luminal-type), suggesting that BTG3 might be involved in the progression of breast cancer, and underlay the mechanisms of molecular subtyping. These results suggested that BTG3 mRNA was employed to indicate the aggressive behaviors of lung cancer, and histogenesis of lung and breast cancers.
The prognostic significance of BTG3 expression was analyzed, but controversial. Ren et al. (2015) found that BTG3 expression was positively correlated with distant metastasis of gastric cancer, and the cancer patients with lower BTG3 expression had a shorter overall survival time. Deng et al. (2013) demonstrated that BTG3 expression was negatively associated with a higher incidence of metastasis, a better differentiation, longer disease-free time, and overall survival time of epithelial ovarian cancer as an independent factor of prognosis. However, there was no relationship between BTG3 protein expression and overall survival rates of the patients with gastric (Gou et al., 2015) or colorectal (Zheng et al., 2017) cancer. In the present study, the positive correlation between BTG3 mRNA expression and survival rate was seen in gastric and breast cancer patients, but not for lung and ovarian cancers according to Kaplan-Meier plotter. TCGA database showed that BTG3 mRNA expression was an independent factor for favorable overall survival of lung cancer patients. BTG3 mRNA was documented to indicate the adverse prognosis for pediatric T-cell acute lymphoblastic leukemia (Gottardo et al., 2007). Therefore, we concluded that the prognostic significance of BTG3 expression was dependent on cancer type, pathological grouping, distinct methodologies, and different databases. Therefore, it should be careful to employ BTG3 mRNA as a prognostic marker in clinicopathological practice.
KEGG analysis demonstrated that the BTG3-related pathways included cell cycle, pancreatic and insulin secretion, fat digestion and absorption, DNA replication, mismatch repair and homologous recombination in gastric cancer, platelet activation and coagulation, digestion and absorption of protein and fat, metabolism of arachidonic and linoleic acids in lung cancer, cell cycle, salivary and insulin secretion, and DNA replication in breast cancer. The top hub genes mainly contained replication protein and related enzymes in gastric cancer, the key enzymes and proteins for fat metabolism in lung cancer, minichromosome maintenance protein in breast cancer, and ribosomal proteins in ovarian cancer. Therefore, we speculate that BTG3 might play important role in the cell cycle, fat digestion and metabolism, and protein biosynthesis, which be deeply investigated in the future.
In summary, BTG3 mRNA might underlie the molecular mechanisms of the histogenesis of gastric, lung, and breast cancers. The paradoxical results about the prognostic significances of BTG3 mRNA might result from tissue specificity, distinct grouping, and different data sources.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Conception and design: H-CZ. Collection and assembly of data: HX and C-YZ. Data analysis and interpretation: K-HS and RZ. Manuscript writing: H-CZ. All authors contributed to the article and approved the submitted version.
This study was supported by Award for Liaoning Distinguished Professor, Natural Science Foundation of Hebei Province (21377772D), and National Natural Scientific Foundation of China (81672700).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
An, Q., Zhou, Y., Han, C., Zhou, Y., Li, F., and Li, D. (2017). BTG3 overexpression suppresses the proliferation and invasion in epithelial ovarian cancer cell by regulating AKT/GSK3β/β-catenin signaling. Reprod. Sci. 24 (10), 1462–1468. doi:10.1177/1933719117691143
Chen, X., Chen, G., Cao, X., Zhou, Y., Yang, T., and Wei, S. (2013). Downregulation of BTG3 in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 437 (1), 173–178. doi:10.1016/j.bbrc.2013.06.062
Cheng, Y. C., Chen, P. H., Chiang, H. Y., Suen, C. S., Hwang, M. J., Lin, T. Y., et al. (2015). Candidate tumor suppressor B-cell translocation gene 3 impedes neoplastic progression by suppression of AKT. Cell Death Dis. 6 (1), e1584. doi:10.1038/cddis.2014.550
Cheng, Y. C., Lin, T. Y., and Shieh, S. Y. (2013). Candidate tumor suppressor BTG3 maintains genomic stability by promoting Lys63-linked ubiquitination and activation of the checkpoint kinase CHK1. Proc. Natl. Acad. Sci. U. S. A. 110 (15), 5993–5998. doi:10.1073/pnas.1220635110
Cui, H., Zhang, S., Zhou, H., and Guo, L. (2017). Direct downregulation of B-cell translocation gene 3 by microRNA-93 is required for desensitizing esophageal cancer to radiotherapy. Dig. Dis. Sci. 62 (8), 1995–2003. doi:10.1007/s10620-017-4579-x
Deng, B., Zhao, Y., Gou, W., Chen, S., Mao, X., Takano, Y., et al. (2013). Decreased expression of BTG3 was linked to carcinogenesis, aggressiveness, and prognosis of ovarian carcinoma. Tumour Biol. 34 (5), 2617–2624. doi:10.1007/s13277-013-0811-2
Du, Y., Liu, P., Zang, W., Wang, Y., Chen, X., Li, M., et al. (2015). BTG3 upregulation induces cell apoptosis and suppresses invasion in esophageal adenocarcinoma. Mol. Cell. Biochem. 404 (1-2), 31–38. doi:10.1007/s11010-015-2363-9
Enkhnaran, B., Zhang, G. C., Zhang, N. P., Liu, H. N., Wu, H., Xuan, S., et al. (2022). microRNA-106b-5p promotes cell growth and sensitizes chemosensitivity to sorafenib by targeting the BTG3/Bcl-xL/p27 signaling pathway in hepatocellular carcinoma. J. Oncol. 2022, 1971559. doi:10.1155/2022/1971559
Gottardo, N. G., Hoffmann, K., Beesley, A. H., Freitas, J. R., Firth, M. J., Perera, K. U., et al. (2007). Identification of novel molecular prognostic markers for paediatric T-cell acute lymphoblastic leukaemia. Br. J. Haematol. 137 (4), 319–328. doi:10.1111/j.1365-2141.2007.06576.x
Gou, W. F., Yang, X. F., Shen, D. F., Zhao, S., Liu, Y. P., Sun, H. Z., et al. (2015). The roles of BTG3 expression in gastric cancer: A potential marker for carcinogenesis and a target molecule for gene therapy. Oncotarget 6 (23), 19841–19867. doi:10.18632/oncotarget.3734
Guéhenneux, F., Duret, L., Callanan, M. B., Bouhas, R., Hayette, S., Berthet, C., et al. (1997). Cloning of the mouse BTG3gene and definition of a new gene family (the BTG family) involved in the negative control of the cell cycle. Leukemia 11 (3), 370–375. doi:10.1038/sj.leu.2400599
Lin, T. Y., Cheng, Y. C., Yang, H. C., Lin, W. C., Wang, C. C., Lai, P. L., et al. (2012). Loss of the candidate tumor suppressor BTG3 triggers acute cellular senescence via the ERK- JMJD3-p16 (INK4a) signaling axis. Oncogene 31, 3287–3297. doi:10.1038/onc.2011.491
Liu, L., Liu, S., Duan, Q., Chen, L., Wu, T., Qian, H., et al. (2017). MicroRNA- 142-5p promotes cell growth and migration in renal cell carcinoma by targeting BTG3. Am. J. Transl. Res. 9 (5), 2394–2402.
Lv, C., Wang, H., Tong, Y., Yin, H., Wang, D., Yan, Z., et al. (2018). The function of BTG3 in colorectal cancer cells and its possible signaling pathway. J. Cancer Res. Clin. Oncol. 144 (2), 295–308. doi:10.1007/s00432-017-2561-9
Lv, Z., Zou, H., Peng, K., Wang, J., Ding, Y., Li, Y., et al. (2013). The suppressive role and aberrent promoter methylation of BTG3 in the progression of hepatocellular carcinoma. PLoS One 8 (10), e77473. doi:10.1371/journal.pone.0077473
Ma, D., Gao, X., Tao, J., Yu, H., and Chai, Z. (2020). Hypoxia-induced downregulation of B-cell translocation gene 3 confers resistance to radiation therapy of colorectal cancer. J. Cancer Res. Clin. Oncol. 146 (10), 2509–2517. doi:10.1007/s00432-020-03307-6
Majid, S., Dar, A. A., Ahmad, A. E., Hirata, H., Kawakami, K., Shahryari, V., et al. (2009). BTG3 tumor suppressor gene promoter demethylation, histone modification and cell cycle arrest by genistein in renal cancer. Carcinogenesis 30 (4), 662–670. doi:10.1093/carcin/bgp042
Mao, D., Liu, A. H., Wang, Z. P., Zhang, X. W., and Lu, H. (2019). Cucurbitacin B inhibits cell proliferation and induces cell apoptosis in colorectal cancer by modulating methylation status of BTG3. Neoplasma 66 (4), 593–602. doi:10.4149/neo_2018_180929N729
Mao, D., Qiao, L., Lu, H., and Feng, Y. (2016). B-cell translocation gene 3 overexpression inhibits proliferation and invasion of colorectal cancer SW480 cells via Wnt/β-catenin signaling pathway. Neoplasma 63 (5), 705–716. doi:10.4149/neo_2016_507
Miyai, K., Yoneda, M., Hasegawa, U., Toita, S., Izu, Y., Hemmi, H., et al. (2009). ANA deficiency enhances bone morphogenetic protein-induced ectopic bone formation via transcriptional events. J. Biol. Chem. 284 (16), 10593–10600. doi:10.1074/jbc.M807677200
Ou, Y. H., Chung, P. H., Hsu, F. F., Sun, T. P., Chang, W. Y., and Shieh, S. Y. (2007). The candidate tumor suppressor BTG3 is a transcriptional target of p53 that inhibits E2F1. EMBO J. 26 (27), 3968–3980. doi:10.1038/sj.emboj.7601825
Peng, L., Li, S., Li, Y., Wan, M., Fang, X., Zhao, Y., et al. (2019). Regulation of BTG3 by microRNA-20b-5p in non-small cell lung cancer. Oncol. Lett. 18 (1), 137–144. doi:10.3892/ol.2019.10333
Rahmani, Z. (2006). APRO4 negatively regulates Src tyrosine kinase activity in PC12 cells. J. Cell Sci. 119 (4), 646–658. doi:10.1242/jcs.02778
Ren, X. L., Zhu, X. H., Li, X. M., Li, Y. L., Wang, J. M., Wu, P. X., et al. (2015). Down-regulation of BTG3 promotes cell proliferation, migration and invasion and predicts survival in gastric cancer. J. Cancer Res. Clin. Oncol. 141 (3), 397–405. doi:10.1007/s00432-014-1826-9
Taniue, K., Kurimoto, A., Takeda, Y., Nagashima, T., Okada-Hatakeyama, M., Katou, Y., et al. (2016). ASBEL-TCF3 complex is required for the tumorigenicity of colorectal cancer cells. Proc. Natl. Acad. Sci. U. S. A. 113 (45), 12739–12744. doi:10.1073/pnas.1605938113
Wang, L., Mo, H., Jiang, Y., Wang, Y., Sun, L., Yao, B., et al. (2019). MicroRNA-519c-3p promotes tumor growth and metastasis of hepatocellular carcinoma by targeting BTG3. Biomed. Pharmacother. 118, 109267. doi:10.1016/j.biopha.2019.109267
Xia, Y., Xiao, X., Deng, X., Zhang, F., Zhang, X., Hu, Q., et al. (2017). Targeting long non-coding RNA ASBEL with oligonucleotide antagonist for breast cancer therapy. Biochem. Biophys. Res. Commun. 489 (4), 386–392. doi:10.1016/j.bbrc.2017.05.136
Xiong, Y., Sun, F., Dong, P., Watari, H., Yue, J., Yu, M. F., et al. (2017). iASPP induces EMT and cisplatin resistance in human cervical cancer through miR-20a- FBXL5/BTG3 signaling. J. Exp. Clin. Cancer Res. 36 (1), 48. doi:10.1186/s13046-017-0520-6
Yanagida, S., Taniue, K., Sugimasa, H., Nasu, E., Takeda, Y., Kobayashi, M., et al. (2013). ASBEL, an ANA/BTG3 antisense transcript required for tumorigenicity of ovarian carcinoma. Sci. Rep. 3, 1305. doi:10.1038/srep01305
Yoneda, M., Suzuki, T., Nakamura, T., Ajima, R., Yoshida, Y., Kakuta, S., et al. (2009). Deficiency of antiproliferative family protein Ana correlates with development of lung adenocarcinoma. Cancer Sci. 100 (2), 225–232. doi:10.1111/j.1349-7006.2008.01030.x
Yoshida, Y., Hosoda, E., Nakamura, T., and Yamamoto, T. (2001). Association of ANA, a member of the antiproliferative Tob family proteins, with a Caf1 component of the CCR4 transcriptional regulatory complex. Jpn. J. Cancer Res. 92 (6), 592–596. doi:10.1111/j.1349-7006.2001.tb01135.x
Yu, J., Zhang, Y., Qi, Z., Kurtycz, D., Vacano, G., and Patterson, D. (2008). Methylation-mediated downregulation of the B-cell translocation gene 3 (BTG3) in breast cancer cells. Gene Expr. 14 (3), 173–182.
Keywords: BTG3, bioinformatics analysis, carcinogenesis, aggressive behavior, prognosis
Citation: Zheng H-C, Xue H, Zhang C-Y, Shi K-H and Zhang R (2022) The clinicopathological significances and related signal pathways of BTG3 mRNA expression in cancers: A bioinformatics analysis. Front. Genet. 13:1006582. doi: 10.3389/fgene.2022.1006582
Received: 29 July 2022; Accepted: 11 August 2022;
Published: 16 September 2022.
Edited by:
Bin Liu, Jiangsu Ocean Universiity, ChinaReviewed by:
Weiqiang Zhou, Shenyang Medical College, ChinaCopyright © 2022 Zheng, Xue, Zhang, Shi and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua-Chuan Zheng, SHVhLUNodWFuLlpoZW5nQGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.