- 1Department of Surgical Oncology, The Sir Run Run Shaw Affiliated Hospital, Zhejiang University, Hangzhou, China
- 2Key Laboratory of Cancer Prevention and Intervention, Ministry of Education, Hangzhou, China
- 3Zhejiang University School of Medicine, Hangzhou, China
- 4The First Affiliated Hospital of Zhejiang Chinese Medical University, Hangzhou, China
Systemic therapy is one of the most significant cancer treatments. However, drug resistance often appears and has become the primary cause of cancer therapy failure. Regulation of drug target, drug metabolism and drug efflux, cell death escape (apoptosis, autophagy, et al.), epigenetic changes, and many other variables are complicatedly involved in the mechanisms of drug resistance. In various types of cancers, long non-coding RNA H19 (lncRNA H19) has been shown to play critical roles in tumor development, proliferation, metastasis, and multiple drug resistance as well. The efficacy of chemotherapy, endocrine therapy, and targeted therapy are all influenced by the expression of H19, especially in breast cancer, liver cancer, lung cancer and colorectal cancer. Here, we summarize the relationship between lncRNA H19 and tumorigenesis, and illustrate the drug resistance mechanisms caused by lncRNA H19 as well. This review may provide more therapeutic potential targets for future cancer treatments.
1 Background
Cancer is a global public health epidemic and is predicted to be the leading cause of death in 2018 according to the World Health Organization (WHO). As a result, research on cancer treatment has gained growing attention (World Health Organization, 2018). Systemic therapy is an important way of treating cancer, among many treatment interventions. However, drug resistance has become a major problem in current cancer recurrence and clinical treatment failure (Holohan et al., 2013), (Chen et al., 2017a). Two forms of drug resistance (intrinsic and acquired) can significantly influence the efficacy of systemic therapy. Intrinsic resistance means that the resistance-mediating factors pre-exist in the bulk of tumor cells before systemic therapy is received (Longley and Johnston, 2005). Acquired drug resistance can be caused by mutations and other adaptive responses, such as the increased expression of therapeutic targets and the activation of alternative compensatory signaling pathways during treatment (Longley and Johnston, 2005). As tumors are increasingly recognized to be highly heterogeneous, drug resistance can occur through therapy-induced selection of a small subpopulation of resistant cells in the original tumor, and tumor cells can acquire cross-resistance to a wide variety of drugs (Kartal-Yandim et al., 2016).
Long noncoding RNAs (LncRNAs) are defined as a class of non-coding RNAs which consist of more than 200 nucleotides (Huarte, 2015). They do not encode any proteins but can be transcribed by RNA polymerase II like mRNAs (Mirzaei et al., 2021). As research deepen, more evidence has revealed the various functions of lncRNAs at chromatin, transcriptional and post-transcriptional levels (Mirzaei et al., 2022). According to the locations where lncRNAs function, they can be divided into nuclear lncRNAs and cytoplasmic lncRNAs. The nuclear lncRNAs participate in chromatin remodeling and modification, chromosomal looping, transcriptional modulation, and RNA processing; while cytoplasmic lncRNAs usually interact with mature mRNA and/or protein (Wang et al., 2017a). Based on the mechanisms above, lncRNAs have been identified to participate in a series of cellular processes including cell growth, proliferation, apoptosis, invasion, metastasis, and the regulation of gene expression, etc., Therefore, disturbances or impairment in lncRNA expression leads to emergence of pathological events, especially cancer (Ashrafizaveh et al., 2021). In different types of cancer cells, more and more lncRNAs like lncRNA H19 (here after, referred as H19), have been verified to engage in tumor development and drug resistance of systemic therapy (Qu et al., 2015).
In this review, we focus on the relation between H19 and tumorigenesis. Then we identify the drug resistant roles played by H19 in various cancers, such as breast cancers, hepatocellular carcinoma, bladder cancers, lung cancers, etc., Meanwhile, the possible association between H19 and various types of drugs is summarized. Finally, we address the functions performed by H19 in different forms of cell death and the possible directions of further research relevant to H19.
2 The mechanism of H19 in tumorigenesis
H19 was the first discovered lncRNA; it was firstly reported in 1991 by Bartolomei et al. (1991) and was shown to lack a common open reading frame (ORF). The H19 gene is a well-known imprinted oncofetal gene, which locates on human chromosome 11p15.5 and encodes for a processed 2.3 kb RNA (Pachnis et al., 1984). As an imprinting gene, H19 is maternally expressed and shares a common enhancer region with IGF2 (Insulin-like growth factor 2) gene which expresses the paternal allele (DeChiara et al., 1991). Without relevant encoding protein expression, H19 can be highly expressed in extraembryonic tissues, the embryo proper and most fetal tissues, but not expressed in most tissues postnatally (Matouk et al., 2007a). H19 has been described to be located in both cytoplasm and nucleus, although it was reported mainly in cytoplasm before (Schoenfelder et al., 2007), (Seidl et al., 2006). Recent evidence shows that the expression of H19 can be reactivated during regeneration and tumorigenesis in adult tissue, indicating that H19 is probably related to the development and progression of tumor (Gabory et al., 2010). Further study demonstrates that H19 displays a cell-dependent and/or tumor type-dependent function. However, it is found that H19 also shows a tumor suppressor function in teratocarcinomas and pituitary tumors (Yoshimizu et al., 2008), (Wu et al., 2018).
Therefore, it remains unclear whether H19’s functional role is tumor suppressive or oncogenic. The function of H19 is largely dependent on the type of cancer, the stage of tumor formation, and the level of molecular signaling pathway (Matouk et al., 2007b). There are several cancers with abnormal expression of H19: breast cancers, pancreatic cancers (Ma et al., 2014), choriocarcinomas (Arima et al., 1997), hepatocellular carcinomas (Ye et al., 2019), ovarian cancers (Tanos et al., 1999), and so on (Ariel et al., 2000a). Furthermore, it is shown that the poor prognosis of patients is correlated with overexpressed H19, especially in higher grades and invasive transitional cell carcinomas (Ariel et al., 2000b), (Ariel et al., 1995), (Gao et al., 2018).
The molecular mechanisms between H19 and tumorigenesis, as shown in Figure 1, largely depend on the partners that H19 interacts with.
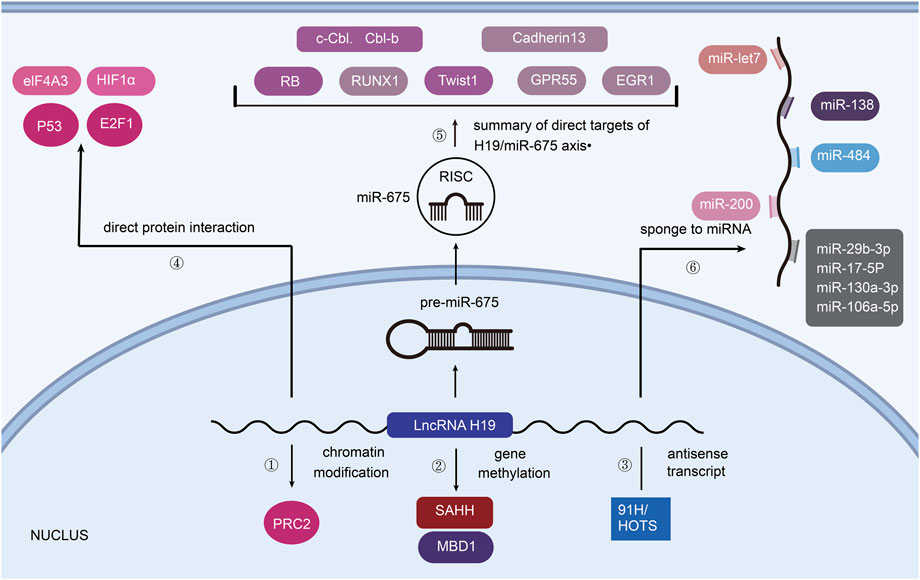
FIGURE 1. H19 associated mechanisms towards tumorigenesis. ① PRC2 consists of core components (EZH2/EZH1, EED, SUZ12). ② H19 binds SAHH or MBD1 to promote the methylation of imprinted genes. ③ 91H and HOTS are the antisense transcripts of H19. ④ p53, E2F1 and eIF4A3, HIF1α can directly interact with H19 to affect tumorigenesis. ⑤ H19 is the precursor of miR-675. ⑥ H19 acts on post-transcriptional control as ceRNA.
2.1 Chromatin modification
As reviewed by Callum Livingstone, the expression of IGF2 is associated with the development of various cancers (Livingstone, 2013). H19 and IGF2 are demonstrated to compete each other for binding enhancer. Thus, H19 could regulate the progression of cancer by changing the expression of IGF2 (Schmidt et al., 1999). What’s more, in bladder cancer, H19 has been found to interact with polycomb repressive complex 2 (PRC2) by associating with enhancer of zeste homolog 2 (EZH2), which leads to the silencing of the E-cadherin gene (Luo et al., 2013). In this way, increasing expression of H19 could downregulate E-cadherin (repressor of cell invasion and metastasis) and Nkd1 (inhibitor of Wnt/β-catenin signaling), causing the progression of cancer cells (Zhang et al., 2017) (Figure 1①).
2.2 Gene methylation
Our previous study suggested that H19 could bind to SAHH (S-Adenosylhomocysteine Hydrolase) and inhibit it, so as to catalyze SAH hydrolysis (Zhou et al., 2015) (Figure 1②). SAH affects cellular DNA methylating, which means that H19 may alter the methylation of DNA and lead to distinct tumorigenesis. (Martinez-leal et al., 2008). Besides, MBD1 (Methyl-CpG–Binding Domain Protein 1), the partner protein of H19, can induce methylation at H3K9me3 (lysine 9 of histone H3) to differentially methylated regions (DMRs) of correlated imprinted genes like IGF2, SLC38A4 (tumor suppressor in hepatocellular carcinoma) and PEG1 (Monnier et al., 2013), (Li et al., 2021).
2.3 Antisense transcript
LncRNA 91H is a novel H19 antisense RNA which was first revealed by Berteaux et al. (2008) LncRNA 91H contributes to the expression of IGF2, showing its oncogenic role in breast cancer cells. HOTS (H19 opposite tumor suppressor), an H19 antisense transcript, is confirmed to inhibit tumor growth in rhabdomyosarcoma and choriocarcinoma (Onyango and Feinberg, 2011) (Figure 1③).
2.4 Direct protein interaction
As the protein encoded by tumor suppressor gene TP53, p53 is reported to repress the expression of H19 by binding to H19 promoter (Lottin et al., 1998). E2F1 is a transcription activator of E2F family which helps to carry out cell cycle. Berteaux et al. (2005) elucidated that E2F1 could also bind to H19 promoter, resulting in G1/S transition and cell proliferation of breast cancers. Moreover, by recruiting and directly binding to eIF4A3 (an RNA-binding protein), H19 promotes the growth of colorectal cancer. Similarly, hypoxia-inducible factor 1-alpha (HIF1α) can physically interact with H19, inducing smooth muscle cell apoptosis and abdominal aortic aneurysm development (Li et al., 2018). However, the tumorigenesis pathway influenced by the interaction between H19 and different kinds of protein is still under exploration (Han et al., 2016) (Figure 1④).
2.5 H19/miR-675 axis
The regulation of miR-675 by H19 is illustrated to be responsible for limiting placental growth before birth and the progression of different Cancers (Keniry et al., 2012) (Figure 1⑤). As Matouk et al. (2015) have summarized, H19 derived miR-675 can induce epithelial mesenchymal transition (EMT) and promote tumorigenesis in many cancer types. In detail, H19 serves as the precursor of miR-675 and promotes it to directly target c-Cbl and Cbl-b mRNA so as to decrease their expression, leading to sustained activation of AKT and ERK pathways as well as enhanced cell proliferation and migration in breast cancers both in vitro and in vivo (Vennin et al., 2015). Other targets of miR-675 in tumors contain: Retinoblastoma protein (RB, a tumor suppressor) in colorectal cancer (Tsang et al., 2010), Twist 1 (a key mediator in epithelial-mesenchymal transition) in hepatocellular cancer (Hernandez et al., 2013), Runt Domain Transcription Factor1 (RUNX1, a tumor suppressor) in gastric cancer (Zhuang et al., 2014), Cadherin 13 (a member of cadherin subfamily) in glioma (Shi et al., 2014a), G protein-coupled receptor (GPR55) in non-small cell lung cancer (He et al., 2015), early growth response protein1 (EGR1) in human liver cancer (Li et al., 2015). MiR-675 is also found to modulate p53 level during bladder cancer cell growth and colorectal tumor metastasis, though p53 is not a direct target of miR-675 (Liu et al., 2016), (Cen et al., 2019).
2.6 Sponge to miRNA
H19 can also function as ceRNA (competing endogenous RNAs) by antagonizing miRNAs (Angrand et al., 2015) (Figure 1⑥). As a molecular sponge, H19 modulates the function of let-7 family miRNA to promote the development of cancers such as pancreatic ductal adenocarcinoma (Kallen et al., 2013). Moreover, there are many other miRNAs which can be sponged by H19: 1) miR-200 family to suppress metastasis of hepatocellular carcinoma (Zhang et al., 2013), 2) miR-200a and miR-138 to promote EMT in colon cancer (Zhang et al., 2013), 3) miR-200b/c to mediate EMT and MET in breast cancer (Zhou et al., 2017a), 4) miR484 and miR29b-3p to promote cell viability and EMT in lung cancer (Zhang et al., 2018), (Liu et al., 2019), 5) miR‐130a‐3p and miR‐17‐5p to develop cardiac cancer (Jia et al., 2019), 6) miR-106a-5p to promote the growth of melanoma by upregulating E2F3 (a member of the E2F transcription factor family) expression (Luan et al., 2018).
3 H19 plays different roles in drug resistance of human cancers
3.1 Common mechanism of drug resistance
Resistance to drug therapy has always been a great barrier to overcoming cancer. Each antitumor agent interacts with cancer cells in its own specific way, and each tumor has its own specific characteristics that determine its tumor progression. Numerous drug-resisting mechanisms has arisen as the result of the interactions between different tumors and drugs (Vasan et al., 2019). Generally, by acting on the surface or entering the cells, curative drugs can function within the tumor cells and alter the micro-environment at the same time. Some tumors are intrinsically resistant to specific drug damage. As reviewed, the tumor intrinsic factors affecting drug resistance are mainly derived from the genetic, transcriptional or functional characteristics of tumor cells themselves (Kalbasi and Ribas, 2020). For example, some cancers overexpress multi-drug resistance protein1 (MDR1) without previous exposure to chemotherapeutic agents, thus possessing intrinsic drug resistance (Thomas et al., 2003). As for acquired drug resistance, the mechanisms can be split into five components at the cellular level.
Firstly, regulating drug uptake and efflux is an important way to establish drug resistance (Gottesman, 2002). ATP-binding cassette (ABC) family, including P-glycoprotein (P-gp), multi-drug resistance-associated protein1 (MRP1) and breast Cancer resistance protein (BCRP/ABCG2), is an important membrane transporter family. It can not only transport nutrients and other molecules, but also mediate the release of drugs (Fletcher et al., 2016). Secondly, compartmentalization of clinical cytotoxic agents apart from their cellular/tissue targets in lysosomes, autophagosomes, and other intercellular vesicles, will promote drug resistance in cancer (Feng et al., 2014). Thirdly, changes in drug targets and enhanced inactivation of drugs by affecting cell metabolism also play important roles in drug resistance (Bar-Zeev et al., 2017). Moreover, because the ultimate targets of many chemotherapeutic drugs are nuclear DNA, the repair of these DNA becomes one of the most well-known mechanisms of drug resistance in cancer. Nucleotide excision repair system (NER) and homologous recombination repair mechanisms (RRM) are two major DNA repair systems, which can be impaired by gene mutation and epigenetic silence (Mansoori et al., 2017). Finally, blocking cell death pathways has been found to possibly result in drug resistance (Pistritto et al., 2016). Since apoptosis is the main pathway of cell death induced by most anticancer drugs, the anti-apoptotic signaling pathways are always overactive in drug-resistant cells (Wong, 2011).
3.2 The role of H19 in therapy resistance of human cancers
H19 has been shown to be involved in and expressed in almost every form of human cancers at all stages of tumorigenesis (Raveh et al., 2015). Chemotherapy, as well as endocrine therapy and targeted therapy, is one of the most effective approaches for the treatment of human cancers. Unfortunately, once drug resistance is established, these anti-cancer drugs cannot always kill tumor cells (Szakács et al., 2006). Although there are various molecular mechanisms for MDR, as shown in Figure 2, the pathways relevant to H19 still remain unclear in the occurrence of MDR. Researchers have so far confirmed several important roles that H19 plays in drug resistance of various cancers (Ghafouri-Fard et al., 2021), (Du et al., 2020). The role of H19 in the therapeutic resistance of human cancers are summarized in Table 1.
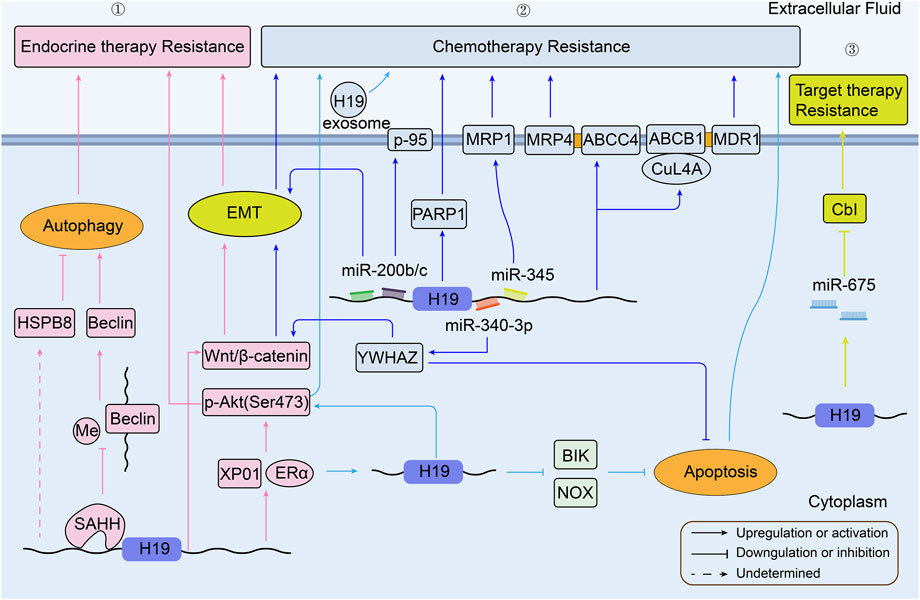
FIGURE 2. Overview of the role of H19 in modulating breast cancer therapy resistance. ①H19 related pathways in BC endocrine therapy resistance; ② H19 related pathways in BC chemotherapy resistance; ③ H19 related pathways in BC targeted therapy resistance (BC, breast cancer).
3.3 Breast cancer
Breast cancer is one of the most prominent and aggressive cancers in women (Israel et al., 2018). Female breast cancer has become the first commonly diagnosed cancer, with an estimated 2.3 million new cases (11.7%) in 2020 (Sung et al., 2021). H19 is involved in breast cancer cell growth, metastasis, and multiple drug resistance in different ways (Si et al., 2019), (Malhotra et al., 2017). A schematic illustration of the mechanisms by which H19 is involved in breast cancer therapy resistance is presented in Figure 2. In Tamoxifen-treated or Fulvestrant-treated estrogen receptor-alpha positive (ERα+) breast cancer tumors, high H19 expression is associated with increased drug resistance. H19 acts as an estrogen receptor modulator to promote the expression of ERα protein in endocrine therapy resistance (ETR) cells (Basak et al., 2018). Gao et al. (2018) found that knockdown of H19 could elevate tamoxifen sensitivity via Wnt/β-catenin pathway and EMT process in ER + breast cancers in vitro. Generally, tumors enhance autophagy activity to promote their metabolism and survival, to survive under microenvironmental stress, and to facilitate proliferation and aggressiveness (White, 2015). H19 activates autophagy via the downregulation of methylation in the promoter of Beclin1 by H19/SAHH/DNMT3B axis (SAHH and DNMT3B are two different sequences involved in tumor progression (Tan et al., 2017), (Sowińska et al., 2007). This process contributes to tamoxifen resistance (TAMR) in breast cancer (Wang et al., 2019). Moreover, N-acetyltransferase 1 (NAT1) was notably downregulated in MCF7/TAMR cell lines, but significantly elevated when knockdown H19. So it was possible that H19 conferred tamoxifen resistance via the mediation of NAT1 promoter methylation (Sun et al., 2022). Through analysis of gene functional groups, the expression of H19 is markedly higher in MCF7/TAMR cell lines (GSE26459). H19 has a positive correlation with heat shock protein family B (small) member 8 (HAPB8). And over-expression of HSPB8 may induce ETR through the regulation of autophagy (Gonzalez-Malerva et al., 2011). In another study with high H19 in BT474/TAMR (GSE112883), high exportin1 (XPO1) expression correlated with high ERα protein level, and high level of Akt signaling expression to help the tumor cell survive (Kulkoyluoglu-Cotul et al., 2019).
MiR-200 family was found to be sponged by H19 in several cancers, such as hepatocellular carcinoma (Zhang et al., 2013). Genomic analysis indicates that decreased miR-200b/c is associated with increased ZEB1 protein and the promotion of EMT in MCF7 cisplatin resistant cells (MCF7/CDDP). As reviewed above, H19 may sponge miR-345 to inhibit its expression, which upregulates MRP1 to promote cisplatin efflux in MCF7/CDDP (Pogribny et al., 2010). Apart from cisplatin, doxorubicin is another common drug tends to develop resistance to chemotherapy in breast cancer (Abe, 2005). It was reported that H19 induced 95-kilodalton membrane glycoprotein (p-95) expression to develop doxorubicin resistance in MCF-7 cells (Doyle et al., 1996). A new research revealed that H19 took part in the downregulated expression of Poly (ADP-ribose) polymerase (PARP)-1 to induce doxorubicin resistance both in vitro and in xenograft models (Wang et al., 2020a). Another study has also confirmed the H19 is over expressed in doxorubicin-resistant breast cancer cell subline compared with the matching parental cells. Additionally, H19 could be transferred from resistant cells to sensitive cells through exosomes, facilitating the chemoresistance of doxorubicin (Wang et al., 2020b). Upregulation of H19 has also enabled the chemoresistance of paclitaxel and anthracyclines analogues like doxorubicin in MCF-7 cells through H19-CUL4A-ABCB1/MDR1 (CUL4A, an ubiquitin ligase component; ABCB1, a member of the ATP-binding cassette family, which encodes MDR1) and ABCC4/MRP4 pathway (Zhu et al., 2017). In paclitaxel resistant cell line, the expression of ERα protein has a tight linkage with H19, suggesting that H19 is a downstream target of ERα. Associated with EZH2, H19 can downregulate the pro-apoptotic gene BIK and NOXA to inhibit apoptosis in ERα+ breast cancers (Si et al., 2016). Similarly, the over-expression of H19 has also been confirmed as an underlying therapeutic target in paclitaxel-resistant breast cancer cell subline. By binding with miR-340-3p, H19 subsequently regulates YWHAZ and potentiates the Wnt/β-catenin signaling. Such regulation can promote breast cancer cells’ proliferation, metastasis, and EMT features while inhibiting their apoptosis (Yan et al., 2020). Besides, H19 can also mediate Akt signaling pathway and inhibit apoptosis to make triple negative breast cancer (TNBC) resist to paclitaxel (Han et al., 2018). Another gene-expression group analysis shows high H19 in MCF7 methotrexate-resistant (MTXR) cell line and low H19 in MDA-MB-468/MTXR (GSE16080). In this study, over-expression of UGT1As was confirmed to induce methotrexate resistance in both breast cancer cell lines (Selga et al., 2009). More information about H19 expression in epirubicin-resistant breast cancer cell lines is summarized in Supplementary Table S1.
Human epidermal growth factor receptor 2 (HER2)-positive breast cancer is another common breast cancer subtype, which can be treated by targeted drugs such as trastuzumab (Cameron et al., 2017). It was hypothesized that trastuzumab resistant HER2-positive breast cancer cells might be formed by downregulating Cbl through H19-derived miR-675 (Sun et al., 2019a).
3.4 Hepatocellular carcinoma
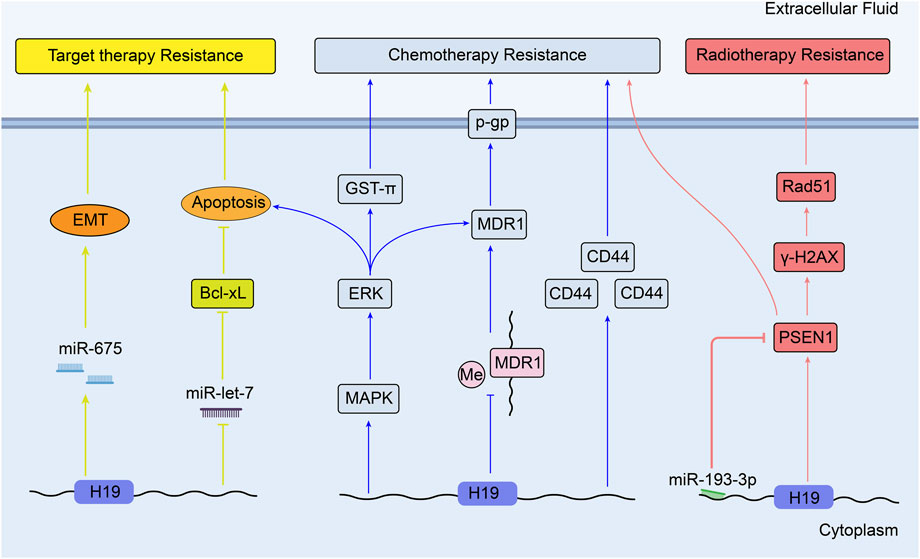
According to the Global Cancer Statistics 2020, primary liver cancer is now the sixth most commonly diagnosed cancer and the third leading cause of cancer death worldwide. HCC accounts for 75%–85% of all cases of liver cancer (Sung et al., 2021). During the progression of hepatocellular carcinoma, the expression level of H19 transcripts is found imbalanced high (Iizuka et al., 2004). It was reported that knockdown of H19 suppressed MDR1 expression and its transcript P-glycoprotein via regulating MDR1 promoter methylation. This regulation resulted in the increased doxorubicin accumulation level and sensitized doxorubicin toxicity in R-HepG2 cells (Tsang and Kwok, 2007). Similarly, another study showed that the downregulation of H19 might block MAPK/ERK signaling pathway by inhibiting drug resistance genes MDR1 and (glutathione-s-transferase-π) GST-π. H19 was shown to facilitate cell apoptosis and reduce the response of CD133+ HuH7 cells to chemotherapeutic drugs like methotrexate (MTX) (Ding et al., 2018). Moreover, in gemcitabine-resistant HepG2 cell line, H19 showed a close association with high expression of CD44, CD90, and CD133. These three proteins are HCC stem cell markers and predict worse prognosis of HCC (ZJ, 2019). Additionally, Ma et al. (2018) demonstrated that restrained expression of H19 and over-expression of miR-193a-3p enhanced the survival rate of hepatoma cell line when they were tolerant to chemotherapeutic agents [Doxorubicin, paclitaxel, vinorelbine, 5-fluorouracil (5-Fu)]. By targeting H19/miR-193a-3p axis, high expression of presenilin 1(PSEN1) increased γ- H2AX and Rad51 expression, and inducted radio-resistance to single-dose X-ray in HCC cells.
Moreover, sorafenib was confirmed to induce apoptosis in HCC, which can be inhibited by potentiating anti-apoptotic member Bcl-xL expression. In human HCC tissues and cell line, low let-7 microRNA can enhance the expression of Bcl-xL and apoptosis (Shimizu et al., 2010). This finding suggests that the effect of sorafenib may be inhibited through high expression of H19 by sponging miR-let-7. Furthermore, it was newly found that upregulated H19/miR-675 expression could elevate sorafenib resistance by promoting EMT in HCC tissue samples and cells (Xu et al., 2020). However, the role of H19 in the therapy of HCC is not completely elucidated. In contrast, in chemo-resistant cells, over-expression of H19 can reverse the drug resistance to doxorubicin, so that suppressing hepatocarcinogenesis and hepatoma cell growth (Schultheiss et al., 2017). Therefore, H19 has a dual effect on therapy resistance in HCC. Figure 3 shows the mechanisms of H19 in the therapy resistance of liver cancer.
3.5 Lung cancer
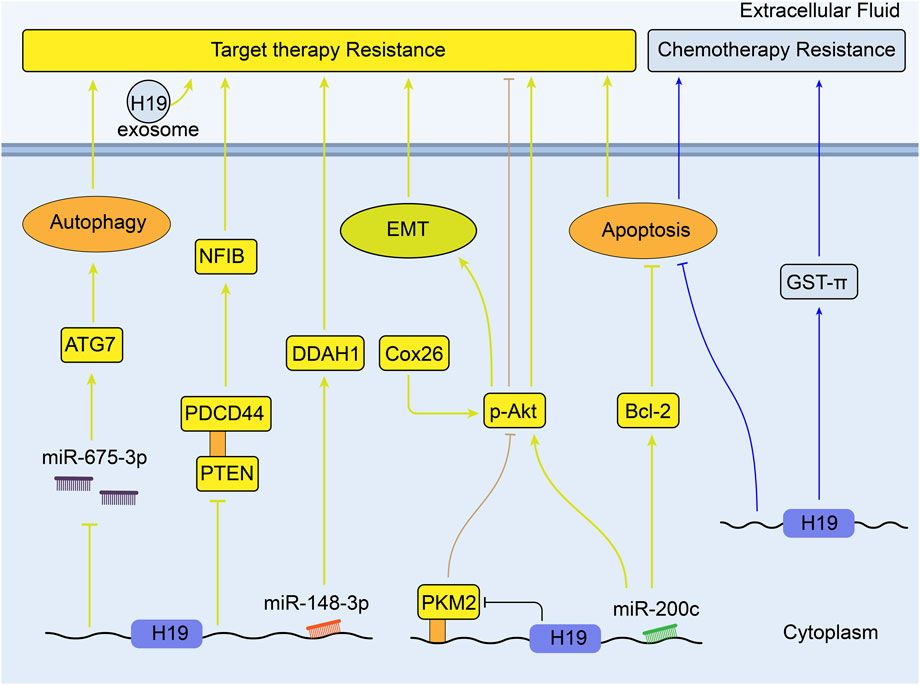
Lung cancer is the second most commonly diagnosed cancer and remains the leading cause of cancer death in 2020 (Sung et al., 2021). Currently, the impact of H19 on resistance to therapeutic option is mainly focused on non-small cell lung cancer (NSCLC). Such patients can benefit from the inhibitors of the epidermal growth factor receptor tyrosine kinase (EGFR TKIs), like gefitinib and erlotinib. (Kris et al., 2003). Lei et al. (2018) have proved that gefitinib resistance in NSCLC cells can be induced by packaging the H19 into exosomes and transferring it to these non-resistant cells. Zhou et al. (2017b) testified that miR-200c could enhance sensitivity of drug-resistant NSCLC to gefitinib by decreasing phosphorylated-Akt signaling and Bcl-2 expression. So it can be speculated that high expression of H19 induce gefitinib resistance through sponging miR-200c and inhibiting apoptosis. Similarly, it is proposed that the cooperation between PI3K/Akt pathway and connexin 26 (Cx26) can induce EMT and confer the gefitinib resistance of NSCLC cells (Yang et al., 2015). Thus, H19 silencing has been confirmed to increase the anticancer impacts of gefitinib in NSCLC through upregulation of PTEN and PDCD4 (both are tumor suppressors) and inhibition of nuclear factor I/B (NFIB) (Zhou and Zhang, 2020). Besides, H19 could also confer resistance to gefitinib via miR-148b-3p/dimethylarginine dimethylaminohydrolase-1 (DDAH1) axis in lung adenocarcinoma (Huang et al., 2019). Furthermore, unlike the usual correlation between high H19 expression and drug resistance, it is demonstrated that knockdown of H19 results in the resistance to erlotinib in vivo and in vitro by upregulating pyruvate kinase isoform muscle 2 (PKM2) expression and enhancing the phosphorylation of AKT (Chen et al., 2020). In contrast, upregulated H19 in erlotinib-resistant cells can sponge miR-615-3p to promote autophagy. Packaged exosomal H19 can also facilitate erlotinib resistance through miR-615-3p/ATG7 axis in NSCLC sensitive cells (Pan and Zhou, 2020).
H19 mediates the regulation of cisplatin resistance in human lung adenocarcinoma cells through apoptosis inhibition. Consistent with the results in vitro, over-expression of H19 is associated with worse clinical outcomes of patients who receive cisplatin-based therapy (Wang et al., 2017b). From other point, it has been discussed that H19 may promote gene GST-π expression in hepatocellular carcinoma (Ding et al., 2018). In another research, GST-π expression is reported to be positively correlated with the resistance to cisplatin in lung cancer cell lines, which means H19 may affect the lung cancer drug resistance through H19/ GST-π pathway (Wang et al., 2011). Figure 4 shows the mechanisms of H19 in the target therapy and chemotherapy resistance of lung cancer.
3.6 Colorectal cancer
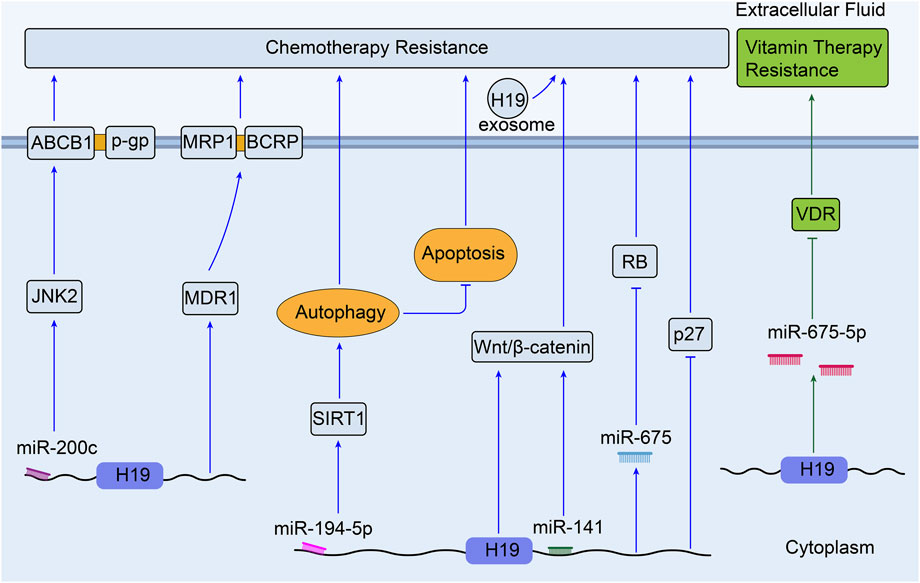
Colorectal cancer is the third most commonly diagnosed cancer and the second most common cause of cancer-associated mortality over 185 countries (Sung et al., 2021). As reviewed, 1,25(OH)2D3 (the most active form of vitamin D in the human body) and its analogs have positive anti-tumor effect in colorectal cancer (Dou et al., 2016).S. Chen et al. have found that colon cancer cells show different resistance to the treatment of 1,25 (OH) 2D3 both in vitro and in vivo when H19 is overexpressed. They also discovered that H19 is able to downregulate the expression of Vitamin D receptor (VDR) by transcribing miR-675-5p, indicating the important role of H19 underlying the development of resistance to 1,25 (OH) 2D3 treatment in advanced colon cancer cells (Chen et al., 2017b). Besides, chemotherapeutic resistance is a mainly formidable challenge in the treatment of colorectal cancer (Figure 5).
Methotrexate (MTX) is one of anti-metabolite and anti-folate chemotherapeutic agents for various cancers including CRC, and it is revealed that H19 can mediate MTX resistance by activating Wnt/β-catenin signaling in colorectal cancer cell line HT-29 (feng Wu et al., 2017). Through integrative bioinformatics analysis, H19 is observed to play key roles in the process of oxaliplatin or irinotecan resistance in colorectal cancer (Sun et al., 2019b). Meanwhile, H19 shows lower expression in oxaliplatin- and irinotecan-resistant CRC cell lines compared with the parental cells (GSE42387, Supplementary Table S1) (Jensen et al., 2015). Moreover, the exosomes derived from carcinoma-associated fibroblasts (CAFs) have been found to transfer H19 to CRC cells and induce oxaliplatin resistance in vitro and in vivo. Upregulated H19 can activate the Wnt/β-catenin pathway and promote the stemness of CRC cells through sponging miR-141 (Ren et al., 2018). What’s more, it was concluded that many lncRNAs including H19 could act as regulators of autophagy and participate in CRC drug resistance (Bermúdez et al., 2019). Wang et al. (2018a) confirmed that H19 could sponge miR-194-5p to promote autophagy via NAD-dependent deacetylase sirtuin-1(SIRT1), so that to enhance 5-Fu chemoresistance in CRC cells. H19 silencing decreased the expression of MDR1, MRP1, and BCRP, which could reverse the sensitivity to 5-Fu in CRC (Wang et al., 2018b). In 5-Fu resistant rectal cancer cells, H19 was linked with downregulation of RB and p27kip1 (p27, a tumor suppressor) (Yokoyama et al., 2019). In addition, miR-200c was found to reduce the expression of JNK2(a set of enzymes in response to a plethora of stress signals) gene and ABCB1 mediated P-gp; this sensitized the MDR colorectal cancer cells to chemotherapeutic drugs, like cisplatin, 5-FU, pirabucin (Sui et al., 2014). According to previous studies, H19 can potentially sponge miR-200c to regulate the process of MDR in CRC.
3.7 Gastric cancer
The incidence and mortality of gastric cancer (GC) have been both increasing dramatically in most countries worldwide during recent 30 years (Etemadi et al., 2020). Over-expression of H19 has been confirmed to be associated with anti-apoptotic and metastatic properties in gastric cancer, leading to multi-drug resistance of tumor (Li et al., 2014). Cisplatin-resistant gastric cancer cell line SGC7901 showed high expressions of H19/miR-675 and low expression of Fas-associated death domain (FADD), which suppressed caspase8 and caspase3 dependent apoptosis (Yan et al., 2017). What’s more, down-regulation of H19 was shown to reduce doxorubicin 50% inhibition concentration (IC50) and alleviate chemoresistance in GC cells. In this study, H19 can promote the expression of IGF2BP3(IGF2 mRNA binding protein 3) and PEG10 (Paternally Expressed 10) (Ishii et al., 2017). The miR-200 family can be divided into miR-200bc/429 cluster and miR-200a/141 cluster, and these two clusters function specifically on different cell types. In GC cell lines, miR-200bc/429 cluster could target X-linked inhibitor of apoptosis protein (XIAP) and BCL2 to modulate apoptosis, promoting the formation of vincristine (VCR) resistance (Dehghanzadeh et al., 2015), (Zhu et al., 2012).
3.8 Neuronal glioma
H19/miR-675 signaling plays a critical role in glioma progression (Shi et al., 2014b). It is the major determinant conferring oncogenic properties to the glioma cells. As reported, the over-expression of H19 can promote temozolomide (TMZ) resistance in glioma cell lines. Compared to the TMZ-sensitive tumors, the major drug resistance genes such as MDR, MRP, and ABCG2 and their expressed mRNA and protein are found to upregulate in the TMZ-resistant (TMZR) glioma cell lines (Jiang et al., 2016). Through gene expression analysis between glioblastoma LN229 cell line and LN229/TMZR, H19 shows a lower expression in resistant cell line (GSE113510). The researchers focused on increased MGMT (O6-methylguanine-DNA methyltransferase) expression regulated by lncRNA TALC (temozolomide-associated lncRNA in glioblastoma recurrence) in TMZR cells (Wu et al., 2019a). Similar study in other temozolomide resistant glioma cells shows that H19 can confer temozolomide resistance by modulating MGMT expression (Xu et al., 2017). Additionally, via integrated bioinformatics analyses, Xiao et al. (2020) have found that H19’s copy number variations could affect the infiltration level of glioma immune cells. Consequently, H19 may be future target to the immunotherapy for glioma.
3.9 Ovarian cancer
Recently, it was shown that the expression of H19 was enhanced in cisplatin-resistant ovarian cancer cells. H19 can confer cisplatin resistance to ovarian cancer cells via regulating glutathione metabolism in vitro and in vivo (Zheng et al., 2016). Sajadpoor et al. (2018) confirmed that valproic acid (VPA) could negatively regulate the H19 and EZH2 expression in ovarian cancer A2780 cisplatin-resistant cells, which subsequently lead to cell apoptosis. Therefore, H19 could increase cisplatin resistance in ovarian cells by targeting EZH2/p21/PTEN pathway. Another research revealed that EMT transcription factors snail and slug contributed to cisplatin resistance in ovarian cancer, indicating the potential new mechanism between H19 and cisplatin resistance (Haslehurst et al., 2012). Downregulation of H19 can inhibit EMT, migration and sensibility of cisplatin in these cells (Wu et al., 2019b). Another studies about gene expression difference in ovarian cancer also showed high H19 expression in cisplatin and oxaliplatin resistant ovarian cancer cells (GSE28648) (Zeller et al., 2012). The role of miR-483-3p and modulated protein kinase C α(PKCα) was focused on the occurrence of drug resistance (GSE58472) (Arrighetti et al., 2016).
3.10 Other cancers
Moreover, H19 promotes the cisplatin resistance in seminoma, resulting from the increasing expression of TDRG1 (testis developmental related gene 1) by sponging miRNA‐106b‐5p (Wei et al., 2018). H19 targeting miR‐130a‐3p and miR‐17‐5p could increase overall survival of cardiac cancer cells treated with cisplatin, doxorubicin, mitomycin, and 5‐fluorouracil (5-FU), leading to the establishment of chemoresistance for cardiac cancer (Jia et al., 2019).
H19 is also related to the drug resistance of choriocarcinoma (CC). The resistance of CC cells to MTX and 5-FU could be reduced after H19 is depressed. By knocking out gene H19, the proliferative, migratory, and invasive ability can be decreased and the apoptosis can be increased in MTX/5-FU treated CC cells (Yu et al., 2019). Besides, H19 over-expression would induce bortezomib resistance in multiple myeloma by targeting MCL-1 via miR-29b-3p (Pan et al., 2019).
Laryngeal squamous cell carcinoma (LSCC) is a highly aggressive malignancy, accounting for approximately 90% of all laryngeal cancer (Siegel et al., 2015). Notably, expression of H19 has been shown to be increased in LSCC tissues and drug-resistant cells. The resistance to cisplatin is mediated via H19/miR-107/HMGB1 axis and subsequent autophagy (Chen et al., 2021).
In nasopharyngeal carcinoma, knockdown of H19 in drug-resistant cells significantly increases their chemoresistance through apoptosis promotion. When combined with paclitaxel, silencing H19 could enhance tumor inhibition in vivo (Zhu, 2020).
Neuroendocrine prostate cancer (NEPC) is a highly lethal subtype of prostate cancer with high expression of H19. By binding to PRC2, H19 induces epigenetic changes and promotes the association of H19 with EZH2. Knockdown of H19 was testified to re-sensitize NEPC to enzalutamide (Singh et al., 2021).
4 Discussion
4.1 H19 and different kinds of anti-tumor drugs
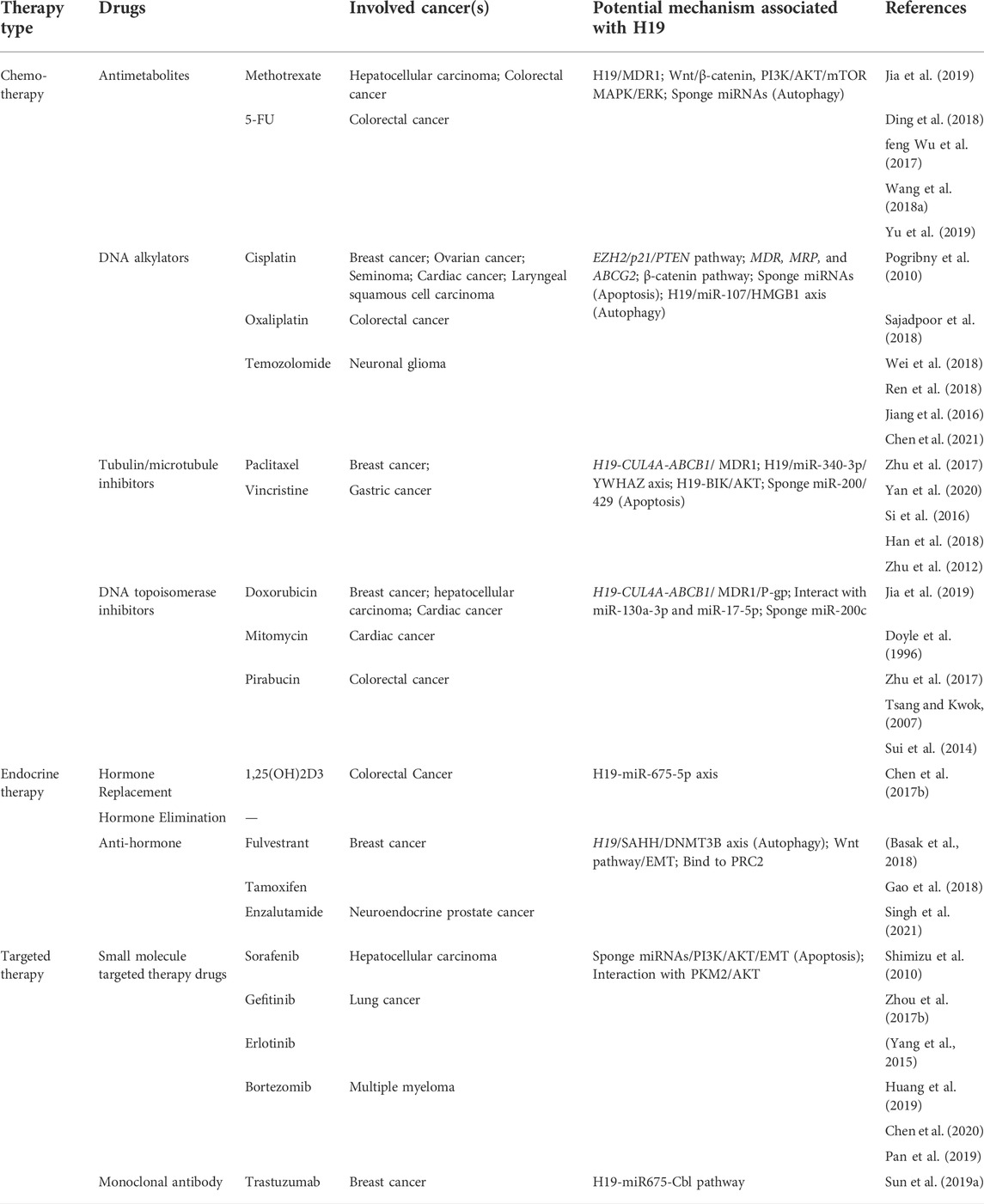
The three primary types of anti-tumor systemic treatment are chemotherapy, endocrine therapy, and targeted therapy. According to the mechanism of action on cancer cells, the chemotherapy drugs we commonly use are divided into four categories: Antimetabolites (like MTX, 5-FU), DNA alkylators (like cisplatin, oxaliplatin, temozolomide), Tubulin/microtubule inhibitors (like paclitaxel, vincristine), and DNA topoisomerase inhibitors (like doxorubicin, mitomycin, pirabucin) (Bailly et al., 2020). Endocrine therapy drugs can be divided into three types: Hormone replacement drugs (like 1,25(OH)2D3), Hormone elimination drugs, and Anti-hormone drugs (like fulvestrant, tamoxifen). Small molecule-targeted therapy drugs (such as sorafenib, gefitinib, bortezomib) and monoclonal antibody (such as trastuzumab) are typical targeted therapy drugs. Many of the above-mentioned drugs may develop tolerance when used in certain cancers (Table 2).
In chemoresistance, sponging miR-200b/c, miR-345, miR-340-3p to regulate the expression of membrane protein are important mechanisms of H19. Apoptosis intervention usually confers chemoresistance, while autophagy regulation often confers endocrine therapy resistance. H19-mediated Wnt/β-catenin and EMT signaling pathways show important roles in chemoresistance, endocrine therapy resistance and targeted therapy resistance. Particularly, encoding miR-675 shows targeted resistance phenotype in breast cancer and liver cancer. As the summary of potential mechanisms associated with H19 and different anti-tumor drugs, three common ways to promote MDR through H19 are proposed: gene methylation and nuclear epigenetic changes, miRNA control in cytoplasm, and direct association with certain protein/transcription factors (TFs) (Wang et al., 2020c).
4.2 H19 and different kinds of cell death
The research of cell death has always been closely interrelated with drug resistance research. Until now, the most widely-used classification of programmed cell death is consisted of apoptosis, necrosis, autophagy-associated cell death and ferroptosis (Tang et al., 2019). In previous studies, apoptosis is demonstrated to be the most common form of cell death in the regulation of H19. Gene methylation, miRNA regulation and direct protein interaction all play irreplaceable roles in apoptosis inhibition. Because most of the clinical therapeutic drugs induce cell death through apoptosis, silencing H19 will become a non-negligible method to increase drug efficacy or/and inhibit MDR. As another form of cell death reported frequently, autophagy can also be regulated by gene methylation and miRNA sponge. However, the researches of necrosis and ferroptosis in systemic therapy are scarce. In heart disease, necrosis is the main form of cardiomyocyte death. The miR-103/107-Fas-associated protein with death domain (FADD) pathway is demonstrated to induce necrosis in cardiac cell line H9c2. Consequently, H9c2 cells can be protected from necrosis by upregulating the expression of H19 (Wang et al., 2015). Although this finding was irrelevant to tumorigenesis and drug resistance, it revealed the possible relationship between H19 and cell necrosis.
As a new recognized regulated cell death first reported in 2012, ferroptosis gives rise to more and more researches, including those in anti-tumor therapy (Dixon et al., 2012). A recent study reported that inhibition of PI3K-AKT-mTOR signaling axis could sensitize breast cancer cells (BT474 and MDA-MB-453) to ferroptosis induction (Yi et al., 2020). Therefore, H19 may indirectly participate in ferroptosis by activating PI3K-AKT-mTOR signaling. Besides, the expression of iron storage protein ferritin is specifically dependent on H19/miR-675 expression levels.
Moreover, the interactive mechanism between ferritin and H19 differs in different cancer cells. It was found that the amounts of ferritin were negatively correlated with H19/miR675 levels in K562 cells (the first human established myelogenous leukemia cell line), but positively related in breast cancer cell line MCF7 cells (Di Sanzo et al., 2018). These researches develop a new level of interactive complexity between iron metabolism and H19 or some miRNAs expression. By regulating iron metabolism, ferroptosis will be broadly discussed in the cell death induction of H19.
4.3 Clinic and future prospect
Systemic therapy is one of the most important treatments for cancer patients. However, drug resistance has become the most urgent problem hampering our treatment. Along with the development of relevant studies, H19 has been testified to function in the tumorigenesis and drug resistance in human cancers via different mechanisms. Hence, increasing drug sensitivity and decreasing cancer cells drug resistance might be realized by targeting H19. Diphtheria toxin A-chain (DT-A)-H19 has shown anti-cancer effect by suppressing tumor growth in ovarian cancer (Mizrahi et al., 2009). DTA-H19 is a DNA plasmid that contains H19 gene regulatory sequences that drive the expression of an intracellular toxin. As an individualized DNA-based approach, DTA-H19 can be used in the tumors with high H19 expression. A phase 1/2a clinical trial for superficial bladder cancer has proved the therapeutic effect of intravesical DTA-19 (Sidi et al., 2008). Similar to H19, IGF2 is also highly active in various human cancers. The use of double promoter toxin vector H19-DTA-(IGF2)-P4-DTA exhibited superior inhibition towards pancreatic cancer, ovarian cancer, glioblastoma and HCC (Amit and Hochberg, 2012).
Nevertheless, more gene editing studies on H19 are still preclinical and are much needed. The regulation of cell death by H19 exerts a wide prospect of molecular research and clinical drug application. In the future, more attention needs to be paid to the additional functions and pathways related to H19, tumorigenesis and cells drug resistance. The research of H19 may provide us with a safer and more effective target to treat MDR and to enrich its function in genetics and molecular biology.
Author contributions
XZ wrote the paper. XZ, ML and JZ prepared the figure and table. BG, SS, XL, HX, SJ, JZ, YZ and LW revised the paper. All authors collected the data, read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81672729, No. 81972597, No. 81602471, and No. 81972453), Zhejiang Provincial Natural Science Foundation of China (Grant LY19H160059, LR22H160011, and LY19H160055), Zhejiang Provincial Medical and Health Science and Technology (Youth Talent Program) (Grant No. 2021RC016) and Zheng Shu Medical Elite Scholarship Fund.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1005522/full#supplementary-material
References
Abe, O. (2005). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet 365 (9472), 1687–1717. doi:10.1016/S0140-6736(05)66544-0
Amit, D., and Hochberg, A. (2012). Development of targeted therapy for a broad spectrum of cancers (pancreatic cancer, ovarian cancer, glioblastoma and HCC) mediated by a double promoter plasmid expressing diphtheria toxin under the control of H19 and IGF2-P4 regulatory sequences. Int. J. Clin. Exp. Med. 5 (4), 296–305.
Angrand, P. O., Vennin, C., Le Bourhis, X., and Adriaenssens, E. (2015). The role of long non-coding RNAs in genome formatting and expression. Front. Genet. 6, 165. doi:10.3389/fgene.2015.00165
Ariel, I., De Groot, N., and Hochberg, A. (2000). Imprinted H19 gene expression in embryogenesis and human cancer : The oncofetal connection. Am. J. Med. Genet. 91 (1), 46–50. doi:10.1002/(sici)1096-8628(20000306)91:1<46::aid-ajmg8>3.0.co;2-i
Ariel, I., Lustig, O., Schneider, T., Pizov, G., SappirM., , De-GrootN., , et al. (1995). The imprinted H19 gene as a tumor marker in bladder carcinoma. Urology 45 (2), 335–338. doi:10.1016/0090-4295(95)80030-1
Ariel, I., SughayerM., , Fellig, Y., Pizov, G., AyeSh, S., PoDeh, D., et al. (2000). The imprinted H19 gene is a marker of early recurrence in human bladder carcinoma. Mol. Pathol. 53 (6), 320–323. doi:10.1136/mp.53.6.320
Arima, T., Matsuda, T., Takagi, N., and Wake, N. (1997). Association of IGF2 and H19 imprinting with choriocarcinoma development. Cancer Genet. cytogenet. 93 (1), 39–47. doi:10.1016/s0165-4608(96)00221-x
Arrighetti, N., Cossa, G., De Cecco, L., Stucchi, S., Carenini, N., Corna, E., et al. (2016). PKC-alpha modulation by miR-483-3p in platinum-resistant ovarian carcinoma cells. Toxicol. Appl. Pharmacol. 310, 9–19. doi:10.1016/j.taap.2016.08.005
Ashrafizaveh, S., Ashrafizadeh, M., Zarrabi, A., Husmandi, K., Zabolian, A., Shahinozzaman, M., et al. (2021). Long non-coding RNAs in the doxorubicin resistance of cancer cells. Cancer Lett. 508, 104–114. doi:10.1016/j.canlet.2021.03.018
Bailly, C., Thuru, X., and Quesnel, B. (2020). Combined cytotoxic chemotherapy and immunotherapy of cancer: Modern times. Nar. Cancer 2 (1), zcaa002–20. doi:10.1093/narcan/zcaa002
Bar-Zeev, M., Livney, Y. D., and Assaraf, Y. G. (2017). Targeted nanomedicine for cancer therapeutics: Towards precision medicine overcoming drug resistance. Drug resist. updat. 31, 15–30. doi:10.1016/j.drup.2017.05.002
Bartolomei, M. S., Zemel, S., and Tilghman, S. M. (1991). Parental imprinting of the mouse H19 gene. Nature 351, 153–155. doi:10.1038/351153a0
Basak, P., Chatterjee, S., Bhat, V., Su, A., Jin, H., Lee-Wing, V., et al. (2018). Long non-coding RNA H19 acts as an estrogen receptor modulator that is required for endocrine therapy resistance in ER + breast cancer cells. Cell. Physiol. biochem. 51 (4), 1518–1532. doi:10.1159/000495643
Bermúdez, M., Aguilar-Medina, M., Lizarraga-Verdugo, E., Avendano-Felix, M., Silva-Benitez, E., Lopez-Camarillo, C., et al. (2019). LncRNAs as regulators of autophagy and drug resistance in colorectal cancer. Front. Oncol. 9 (1), 1008–1014. doi:10.3389/fonc.2019.01008
Berteaux, N., Aptel, N., Cathala, G., Genton, C., Coll, J., Daccache, A., et al. (2008). A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol. Cell. Biol. 28 (22), 6731–6745. doi:10.1128/MCB.02103-07
Berteaux, N., Lottin, S., Monte, D., Pinte, S., Quatannens, B., Coll, J., et al. (2005). H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J. Biol. Chem. 280 (33), 29625–29636. doi:10.1074/jbc.M504033200
Cameron, D., Piccart-Gebhart, M. J., Gelber, R. D., Procter, M., Goldhirsch, A., de Azambuja, E., et al. (2017). 11 years’ follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive early breast cancer: Final analysis of the HERceptin adjuvant (HERA) trial. Lancet 389, 1195–1205. doi:10.1016/S0140-6736(16)32616-2
Cen, B., Chen, J., Hu, F., Qiu, Y., Xiao, G., Zhou, J., et al. (2019). Prostaglandin E2 induces mir675-5p to promote colorectal tumor metastasis via modulation of p53 expression. Gastroenterology 158 (4), 971–984. e10. doi:10.1053/j.gastro.2019.11.013
Chen, C., Liu, W. R., Zhang, B., Zhang, L. M., Li, C. G., Liu, C., et al. (2020). LncRNA H19 downregulation confers erlotinib resistance through upregulation of PKM2 and phosphorylation of AKT in EGFR-mutant lung cancers. Cancer Lett. 486, 58–70. doi:10.1016/j.canlet.2020.05.009
Chen, L., Xu, Z., Zhao, J., Zhai, X., Li, J., Zhang, Y., et al. (2021). H19/miR-107/HMGB1 axis sensitizes laryngeal squamous cell carcinoma to cisplatin by suppressing autophagy in vitro and in vivo. Cell Biol. Int. 45 (3), 674–685. doi:10.1002/cbin.11520
Chen, Q. N., Wei, C. C., Wang, Z. X., and Sun, M. (2017). Long non-coding RNAs in anti-cancer drug resistance. Oncotarget 8 (1), 1925–1936. doi:10.18632/oncotarget.12461
Chen, S., Bu, D., Ma, Y., Zhu, J., Chen, G., Sun, L., et al. (2017). H19 overexpression induces resistance to 1, 25 (OH) 2D3 by targeting VDR through miR-675-5p in colon cancer cells. Neoplasia (United States) 19 (3), 226–236. doi:10.1016/j.neo.2016.10.007
DeChiara, T. M., Robertson, E. J., and Efstratiadis, A. (1991). Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64 (4), 849–859. doi:10.1016/0092-8674(91)90513-x
Dehghanzadeh, R., Jadidi-Niaragh, F., Gharibi, T., and Yousefi, M. (2015). MicroRNA-induced drug resistance in gastric cancer. Biomed. Pharmacother. 74, 191–199. doi:10.1016/j.biopha.2015.08.009
Di Sanzo, M., ChiRillo, R., Aversa, I., BiamonteF., , Santamaria, G., Giovannone, E. D., et al. (2018). shRNA targeting of ferritin heavy chain activates H19/miR-675 axis in K562 cells. Gene 657, 92–99. doi:10.1016/j.gene.2018.03.027
Ding, K., Liao, Y., Gong, D., Zhao, X., and Ji, W. (2018). Effect of long non-coding RNA H19 on oxidative stress and chemotherapy resistance of CD133+ cancer stem cells via the MAPK/ERK signaling pathway in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 502 (2), 194–201. doi:10.1016/j.bbrc.2018.05.143
Dixon, S. J., Lemberg, K. M., Lamprecht, M. R., Skouta, R., Zaitsev, E. M., Gleason, C. E., et al. (2012). Ferroptosis : An iron-dependent form of nonapoptotic cell death. Cell 149 (5), 1060–1072. doi:10.1016/j.cell.2012.03.042
Dou, R., Ng, K., Giovannucci, E. L., Manson, J. E., Qian, Z. R., and Ogino, S. (2016). Vitamin D and colorectal cancer: Molecular, epidemiological and clinical evidence. Br. J. Nutr. 115 (9), 1643–1660. doi:10.1017/S0007114516000696
Doyle, L. A., Yang, W., Rishi, A. K., Gao, Y., and Ross, D. D. (1996). H19 gene overexpression in atypical multidrug-resistant cells associated with expression of a 95-kilodalton membrane glycoprotein. Cancer Res. 56 (13), 2904–2907.
Du, T., Shi, Y., Xu, S., Wan, X., Sun, H., and Liu, B. (2020). Long non-coding RNAs in drug resistance of breast cancer. OncoTargets Ther. 13, 7075–7087. doi:10.1016/j.drup.2022.100851
Etemadi, A., Safiri, S., Sepanlou, S., Ikuta, K., Bisignano, C., Shakeri, R., et al. (2020). The global, regional, and national burden of stomach cancer in 195 countries, 1990–2017: A systematic analysis for the global burden of disease study 2017. Lancet. Gastroenterol. Hepatol. 5 (1), 42–54. doi:10.1016/S2468-1253(19)30328-0
feng Wu, K., Liang, W. C., Feng, L., Pang, J. X., Waye, M. M. Y., Zhang, J. F., et al. (2017). H19 mediates methotrexate resistance in colorectal cancer through activating Wnt/β-catenin pathway. Exp. Cell Res. 350 (2), 312–317. doi:10.1016/j.yexcr.2016.12.003
Feng, Y., He, D., Yao, Z., and Klionsky, D. J. (2014). The machinery of macroautophagy. Cell Res. 24 (1), 24–41. doi:10.1038/cr.2013.168
Fletcher, J. I., Williams, R. T., Henderson, M. J., Norris, M. D., and Haber, M. (2016). ABC transporters as mediators of drug resistance and contributors to cancer cell biology. Drug resist. updat. 26, 1–9. doi:10.1016/j.drup.2016.03.001
Gabory, A., Jammes, H., and Dandolo, L. (2010). The H19 locus: Role of an imprinted non-coding RNA in growth and development. BioEssays 32 (6), 473–480. doi:10.1002/bies.200900170
Gao, H., Hao, G., Sun, Y., Li, L., and Wang, Y. (2018). Long noncoding RNA H19 mediated the chemosensitivity of breast cancer cells via wnt pathway and EMT process. Onco. Targets. Ther. 11, 8001–8012. doi:10.2147/OTT.S172379
Ghafouri-Fard, S., Shoorei, H., Bahroudi, Z., Abak, A., and Taheri, M. (2021). The role of H19 lncRNA in conferring chemoresistance in cancer cells. Biomed. Pharmacother. 138, 111447. doi:10.1016/j.biopha.2021.111447
Gonzalez-Malerva, L., Park, J., Zou, L., Hu, Y., Moradpour, Z., Pearlberg, J., et al. (2011). High-throughput ectopic expression screen for tamoxifen resistance identifies an atypical kinase that blocks autophagy. Proc. Natl. Acad. Sci. U. S. A. 108 (5), 2058–2063. doi:10.1073/pnas.1018157108
Gottesman, M. M. (2002). Mechanisms of cancer drug resistance. Annu. Rev. Med. 53 (1), 615–627. doi:10.1146/annurev.med.53.082901.103929
Han, D., Gao, X., Wang, M., Qiao, Y., Xu, Y., Yang, J., et al. (2016). Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget 7 (16), 22159–22173. doi:10.18632/oncotarget.8063
Han, J., Han, B., Wu, X., Hao, J., Dong, X., Shen, Q., et al. (2018). Knockdown of lncRNA H19 restores chemo-sensitivity in paclitaxel-resistant triple-negative breast cancer through triggering apoptosis and regulating Akt signaling pathway. Toxicol. Appl. Pharmacol. 359, 55–61. doi:10.1016/j.taap.2018.09.018
Haslehurst, A. M., Koti, M., Dharsee, M., Nuin, P., Evans, K., Geraci, J., et al. (2012). EMT transcription factors snail and slug directly contribute to cisplatin resistance in ovarian cancer. BMC Cancer 12, 91. doi:10.1186/1471-2407-12-91
He, D., Wang, J., Zhang, C., Shan, B., Deng, X., Li, B., et al. (2015). Down-regulation of miR-675-5p contributes to tumor progression and development by targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol. Cancer 14 (1), 73. doi:10.1186/s12943-015-0342-0
Hernandez, J. M., ElAhi, A., Clark, C. W., Wang, J., Humphries, L. A., Centeno, B., et al. (2013). MiR-675 mediates downregulation of twist1 and rb in afp-secreting hepatocellular carcinoma. Ann. Surg. Oncol. 20 (3), S625–S635. doi:10.1245/s10434-013-3106-3
Holohan, C., Van Schaeybroeck, S., Longley, D. B., and Johnston, P. G. (2013). Cancer drug resistance: An evolving paradigm. Nat. Rev. Cancer 13 (10), 714–726. doi:10.1038/nrc3599
Huang, Z., Ma, Y., Zhang, P., Si, J., Xiong, Y., and Yang, Y. (2019). Long non-coding RNA H19 confers resistance to gefitinib via miR-148b-3p/DDAH1 axis in lung adenocarcinoma. Anticancer. Drugs 1, 44–54. doi:10.1097/CAD.0000000000000831
Huarte, M. (2015). The emerging role of lncRNAs in cancer. Nat. Med. 21 (11), 1253–1261. doi:10.1038/nm.3981
Iizuka, N., Oka, M., Tamesa, T., Hamamoto, Y., and Yamada-Okabe, H. (2004). Imbalance in expression levels of insulin-like growth factor 2 and H19 transcripts linked to progression of hepatocellular carcinoma. Anticancer Res. 24 (6), 4085–4089.
Ishii, S., Yamashita, K., Harada, H., Ushiku, H., Tanaka, T., Nishizawa, N., et al. (2017). The H19-PEG10/IGF2BP3 axis promotes gastric cancer progression in patients with high lymph node ratios. Oncotarget 8 (43), 74567–74581. doi:10.18632/oncotarget.20209
Israel, B. B., Tilghman, S. L., Parker-Lemieux, K., and Payton-Stewart, F. (2018). Phytochemicals: Current strategies for treating breast cancer (review). Oncol. Lett. 15 (5), 7471–7478. doi:10.3892/ol.2018.8304
Jensen, N. F., Stenvang, J., Beck, M. K., Hanakova, B., Belling, K. C., Do, K. N., et al. (2015). Establishment and characterization of models of chemotherapy resistance in colorectal cancer: Towards a predictive signature of chemoresistance. Mol. Oncol. 9 (6), 1169–1185. doi:10.1016/j.molonc.2015.02.008
Jia, J., Zhang, X., Zhan, D., Li, J., Li, Z., Li, H., et al. (2019). LncRNA H19 interacted with miR-130a-3p and miR-17-5p to modify radio-resistance and chemo-sensitivity of cardiac carcinoma cells. Cancer Med. 8 (4), 1604–1618. doi:10.1002/cam4.1860
Jiang, P., Wang, P., Sun, X., Yuan, Z., Zhan, R., Ma, X., et al. (2016). Knockdown of long noncoding RNA H19 sensitizes human glioma cells to temozolomide therapy. Onco. Targets. Ther. 9, 3501–3509. doi:10.2147/OTT.S96278
Kalbasi, A., and Ribas, A. (2020). Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 20 (1), 25–39. doi:10.1038/s41577-019-0218-4
Kallen, A. N., Zhou, X. B., Xu, J., Qiao, C., Ma, J., Yan, L., et al. (2013). The imprinted H19 LncRNA antagonizes let-7 MicroRNAs. Mol. Cell 52 (1), 101–112. doi:10.1016/j.molcel.2013.08.027
Kartal-Yandim, M., Adan-Gokbulut, A., and Baran, Y. (2016). Molecular mechanisms of drug resistance and its reversal in cancer. Crit. Rev. Biotechnol. 36 (4), 716–726. doi:10.3109/07388551.2015.1015957
Keniry, A., Oxley, D., Monnier, P., Kyba, M., Dandolo, L., Smits, G., et al. (2012). The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 14 (7), 659–665. doi:10.1038/ncb2521
Kris, M. G., Natale, R. B., Herbst, R. S., Lynch, T. J., Prager, D., Belani, C. P., et al. (2003). Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: A randomized trial. J. Am. Med. Assoc. 290 (16), 2149–2158. doi:10.1001/jama.290.16.2149
Kulkoyluoglu-Cotul, E., Smith, B. P., Wrobel, K., Zhao, Y. C., Chen, K. L. A., Hieronymi, K., et al. (2019). Combined targeting of estrogen receptor alpha and XPO1 prevent Akt activation, Remodel metabolic pathways and induce autophagy to Overcome tamoxifen resistance. Cancers (Basel) 11 (4), E479. doi:10.3390/cancers11040479
Lei, Y., Guo, W., Chen, B., Chen, L., Gong, J., and Li, W. (2018). Tumor-released lncRNA H19 promotes gefitinib resistance via packaging into exosomes in non-small cell lung cancer. Oncol. Rep. 40 (6), 3438–3446. doi:10.3892/or.2018.6762
Li, D. Y., Busch, A., Jin, H., Chernogubova, E., Pelisek, J., Karlsson, J., et al. (2018). H19 induces abdominal aortic aneurysm development and progression. Circulation 138 (15), 1551–1568. doi:10.1161/CIRCULATIONAHA.117.032184
Li, H., Li, J., Jia, S., Wu, M., An, J., Zheng, Q., et al. (2015). miR675 upregulates long noncoding RNA H19 through activating EGR1 in human liver cancer. Oncotarget 6 (31), 31958–31984. doi:10.18632/oncotarget.5579
Li, H., Yu, B., Li, J., Su, L., Yan, M., Zhu, Z., et al. (2014). Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 5 (8), 2318–2329. doi:10.18632/oncotarget.1913
Li, Jie, Li, M. h., Wang, T. t., Liu, X. n., Zhu, X. t., Dai, Y. z., et al. (2021). SLC38A4 functions as a tumour suppressor in hepatocellular carcinoma through modulating Wnt/β-catenin/MYC/HMGCS2 axis. Br. J. Cancer 125, 865–876. doi:10.1038/s41416-021-01490-y
Liu, C., Chen, Z., Fang, J., Xu, A., Zhang, W., and Wang, Z. (2016). H19-derived miR-675 contributes to bladder cancer cell proliferation by regulating p53 activation. Tumour Biol. 37 (1), 263–270. doi:10.1007/s13277-015-3779-2
Liu, L., Liu, L., and Lu, S. (2019). lncRNA H19 promotes viability and epithelial-mesenchymal transition of lung adenocarcinoma cells by targeting miR-29b-3p and modifying STAT3. Int. J. Oncol. 54, 929–941. doi:10.3892/ijo.2019.4695
Livingstone, C. (2013). IGF2 and cancer. Endocrine-Related Cancer 20 (6), R321–R339. doi:10.1530/erc-13-0231
Longley, D. B., and Johnston, P. G. (2005). Molecular mechanisms of drug resistance. J. Pathol. 205 (2), 275–292. doi:10.1002/path.1706
Lottin, Â., Montpellier, C., AdriaEnssEns, E., Lottin, S., Dumont, L., IotsoVa, V., et al. (1998). The H19 TATA-less promoter is efficiently repressed by wild-type tumor suppressor gene product p53, ” Oncogene, vol. 16(18), no. pp. 2395–2401. doi:10.1038/sj.onc.1201742
Luan, W., Zhou, Z., Ni, X., Xia, Y., Wang, J., Yan, Y., et al. (2018). Long non-coding RNA H19 promotes glucose metabolism and cell growth in malignant melanoma via miR-106a-5p/ E2F3 axis. J. Cancer Res. Clin. Oncol. 144 (3), 531–542. doi:10.1007/s00432-018-2582-z
Luo, M., Li, Z., Wang, W., Zeng, Y., Liu, Z., and Qiu, J. (2013). Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 333 (2), 213–221. doi:10.1016/j.canlet.2013.01.033
Ma, C., Nong, K., Zhu, H., Wang, W., Huang, X., Yuan, Z., et al. (2014). H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target HMGA2-mediated EMT. Tumour Biol. 35 (9), 9163–9169. doi:10.1007/s13277-014-2185-5
Ma, H., Yuan, L., Li, W., Xu, K., and Yang, L. (2018). The LncRNA H19/miR-193a-3p axis modifies the radio-resistance and chemotherapeutic tolerance of hepatocellular carcinoma cells by targeting PSEN1. J. Cell. Biochem. 119 (10), 8325–8335. doi:10.1002/jcb.26883
Malhotra, A., Jain, M., Prakash, H., Vasquez, K. M., and Jain, A. (2017). The regulatory roles of long non-coding RNAs in the development of chemoresistance in breast cancer. Oncotarget 8 (66), 110671–110684. doi:10.18632/oncotarget.22577
Mansoori, B., Mohammadi, A., Davudian, S., Shirjang, S., and Baradaran, B. (2017). The different mechanisms of cancer drug resistance: A brief review. Adv. Pharm. Bull. 7 (3), 339–348. doi:10.15171/apb.2017.041
Martinez-leal, J. F., Ferrer, I., BlanCo-ApariCio, C., Hernandez-Losa, J., Ramon Y Cajal, S., CArnero, A., et al. (2008). S-adenosylhomocysteine hydrolase downregulation contributes to tumorigenesis. Carcinogenesis 29 (11), 2089. doi:10.1093/carcin/bgn198
Matouk, I. J., DeGroot, N., Mezan, S., Ayesh, S., Abu-lail, R., Hochberg, A., et al. (2007). The H19 non-coding RNA is essential for human tumor growth. PLoS One 2 (9), e845. doi:10.1371/journal.pone.0000845
Matouk, I. J., DeGroot, N., Mezan, S., Ayesh, S., Abu-lail, R., Hochberg, A., et al. (2007). The H19 non-coding RNA is essential for human tumor growth. PLoS One 2 (9), e845. doi:10.1371/journal.pone.0000845
Matouk, I. J., Halle, D., Gilon, M., and Hochberg, A. (2015). The non-coding RNAs of the H19-IGF2 imprinted loci: A focus on biological roles and therapeutic potential in lung cancer. J. Transl. Med. 13 (1), 113. doi:10.1186/s12967-015-0467-3
Mirzaei, S., Gholami, M. H., Hushmandi, K., Hashemi, F., Zabolian, A., Canadas, I., et al. (2022). The long and short non-coding RNAs modulating EZH2 signaling in cancer. J. Hematol. Oncol. 15 (1), 18. doi:10.1186/s13045-022-01235-1
Mirzaei, S., Zarrabi, A., Hashemi, F., Zabolian, A., Saleki, H., Ranjbar, A., et al. (2021). Regulation of Nuclear Factor-KappaB (NF-κB) signaling pathway by non-coding RNAs in cancer: Inhibiting or promoting carcinogenesis? Cancer Lett. 509, 63–80. doi:10.1016/j.canlet.2021.03.025
Mizrahi, A., Czerniak, A., Levy, T., Amiur, S., Gallula, J., Matouk, I., et al. (2009). Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J. Transl. Med. 7, 69. doi:10.1186/1479-5876-7-69
Monnier, P., Martinet, C., Pontis, J., Stancheva, I., Ait-si-ali, S., and Dandolo, L. (2013). H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1., ” Proc. Natl. Acad. Sci. U. S. A., vol. 110(51), no. pp. 20693-20698. doi:10.1073/pnas.1310201110
Onyango, P., and Feinberg, A. P. (2011). A nucleolar protein , H19 opposite tumor suppressor ( HOTS ), is a tumor growth inhibitor encoded by a human imprinted H19 antisense transcript. Proc. Natl. Acad. Sci. U. S. A. 108 (40), 16759–16764. doi:10.1073/pnas.1110904108
Pachnis, V., Belayew, A., and Tilghman, S. M. (1984). Locus unlinked to α-fetoprotein under the control of the murine raf and Rif genes. Proc. Natl. Acad. Sci. U. S. A. 81 (17), 5523–5527. doi:10.1073/pnas.81.17.5523
Pan, R., and Zhou, H. (2020). Exosomal transfer of lncRNA H19 promotes erlotinib resistance in non-small cell lung cancer via miR-615-3p/ATG7 Axis. Cancer Manag. Res. 12, 4283–4297. doi:10.2147/CMAR.S241095
Pan, Y., Zhang, Y., Liu, W., Huang, Y., Shen, X., Jing, R., et al. (2019). LncRNA H19 overexpression induces bortezomib resistance in multiple myeloma by targeting MCL-1 via miR-29b-3p. Cell Death Dis. 10 (2), 106. doi:10.1038/s41419-018-1219-0
Pistritto, G., Trisciuoglio, D., Ceci, C., Garufi, A., and D’Orazi, G. (2016). Apoptosis as anticancer mechanism: Function and dysfunction of its modulators and targeted therapeutic strategies. Aging 8 (4), 603–619. doi:10.18632/aging.100934
Pogribny, I. P., Filkowski, J. N., Tryndyak, V. P., Golubov, A., Shpyleva, S. I., and Kovalchuk, O. (2010). Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int. J. Cancer 127 (8), 1785–1794. doi:10.1002/ijc.25191
Qu, J., Li, M., Zhong, W., and Hu, C. (2015). Competing endogenous RNA in cancer: A new pattern of gene expression regulation. Int. J. Clin. Exp. Med. 8 (10), 17110–17116.
Raveh, E., Matouk, I. J., Gilon, M., and Hochberg, A. (2015). The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol. Cancer 14, 184. doi:10.1186/s12943-015-0458-2
Ren, J., Ding, L., Zhang, D., Shi, G., Xu, Q., Shen, S., et al. (2018). Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics 8 (14), 3932–3948. doi:10.7150/thno.25541
Sajadpoor, Z., Amini-Farsani, Z., Teimori, H., Shamsara, M., Sangtarash, M. H., Ghasemi-Dehkordi, P., et al. (2018). Valproic acid promotes apoptosis and cisplatin sensitivity through downregulation of H19 noncoding RNA in ovarian A2780 cells. Appl. Biochem. Biotechnol. 185 (4), 1132–1144. doi:10.1007/s12010-017-2684-0
Schmidt, J. V., Levorse, J. M., and Tilghman, S. M. (1999). Enhancer competition between H19 and Igf2 does not mediate their imprinting. Proc. Natl. Acad. Sci. U. S. A. 96 (17), 9733–9738. doi:10.1073/pnas.96.17.9733
Schoenfelder, S., Smits, G., Fraser, P., Reik, W., and Paro, R. (2007). Non-coding transcripts in the H19 imprinting control region mediate gene silencing in transgenic Drosophila. EMBO Rep. 8 (11), 1068–1073. doi:10.1038/sj.embor.7401094
Schultheiss, C. S., Laggai, S., Czepukojc, B., Hussein, U. K., List, M., Barghash, A., et al. (2017). The long non-coding RNA H19 suppresses carcinogenesis and chemoresistance in hepatocellular carcinoma. Cell Stress 1 (1), 37–54. doi:10.15698/cst2017.10.105
Seidl, C. I. M., Stricker, S. H., and Barlow, D. P. (2006). The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. EMBO J. 25 (15), 3565–3575. doi:10.1038/sj.emboj.7601245
Selga, E., Oleaga, C., Ramírez, S., de Almagro, M. C., Noé, V., and Ciudad, C. J. (2009). Networking of differentially expressed genes in human cancer cells resistant to methotrexate. Genome Med. 1 (9), 83. doi:10.1186/gm83
Shi, Y., Wang, Y., Luan, W., Wang, P., Tao, T., Zhang, J., et al. (2014). Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One 9 (1), e86295. doi:10.1371/journal.pone.0086295
Shi, Y., Wang, Y., Luan, W., Wang, P., Tao, T., Zhang, J., et al. (2014). Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One 9 (1), e86295. doi:10.1371/journal.pone.0086295
Shimizu, S., Takehara, T., Hikita, H., Kodama, T., Miyagi, T., Hosui, A., et al. (2010). The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J. Hepatol. 52 (5), 698–704. doi:10.1016/j.jhep.2009.12.024
Si, H., Chen, P., Li, H., and Wang, X. (2019). Long non-coding RNA H19 regulates cell growth and metastasis via miR-138 in breast cancer. Am. J. Transl. Res. 11 (5), 3213–3225.
Si, X., Zang, R., Zhang, E., Liu, Y., Shi, X., Zhang, E., et al. (2016). LncRNA H19 confers chemoresistance in ERα-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget 7 (49), 81452–81462. doi:10.18632/oncotarget.13263
Sidi, A. A., Ohana, P., Benjamin, S., Shalev, M., Ransom, J. H., Lamm, D., et al. (2008). Phase I/II marker lesion study of intravesical BC-819 DNA plasmid in H19 over expressing superficial bladder cancer refractory to bacillus Calmette-Guerin. J. Urol. 180 (6), 2379–2383. doi:10.1016/j.juro.2008.08.006
Siegel, R. L., Miller, K. D., Jemal, A., Goding-Sauer, A., Pinheiro, P. S., Martinez-Tyson, D., et al. (2015). Cancer statistics for hispanics/latinos, 2015. Ca. Cancer J. Clin. 65 (1), 457–480. doi:10.3322/caac.21314
Singh, N., Ramnarine, V. R., Song, J. H., Pandey, R., Padi, S. K. R., Nouri, M., et al. (2021). The long noncoding RNA H19 regulates tumor plasticity in neuroendocrine prostate cancer. Nat. Commun. 12 (1), 7349. doi:10.1038/s41467-021-26901-9
Sowińska, A., Jagodzinski, P. P., and SowinskA, A. (2007). RNA interference-mediated knockdown of DNMT1 and DNMT3B induces CXCL12 expression in MCF-7 breast cancer and AsPC1 pancreatic carcinoma cell lines. Cancer Lett. 255 (1), 153–159. doi:10.1016/j.canlet.2007.04.004
Sui, H., Cai, G. X., Pan, S. F., Deng, W. L., Wang, Y. W., Chen, Z. S., et al. (2014). miR200c attenuates P-gp-mediated MDR and metastasis by targeting JNK2/c-Jun signaling pathway in colorectal cancer. Mol. Cancer Ther. 13 (12), 3137–3151. doi:10.1158/1535-7163.MCT-14-0167
Sun, F., Liang, W., and Qian, J. (2019). The identification of CRNDE, H19, UCA1 and HOTAIR as the key lncRNAs involved in oxaliplatin or irinotecan resistance in the chemotherapy of colorectal cancer based on integrative bioinformatics analysis. Mol. Med. Rep. 20 (4), 3583–3596. doi:10.3892/mmr.2019.10588
Sun, H., Wang, G., Cai, J., Wei, X., Zeng, Y., Peng, Y., et al. (2022). Long non-coding RNA H19 mediates N-acetyltransferase 1 gene methylation in the development of tamoxifen resistance in breast cancer. Exp. Ther. Med. 23 (1), 12. doi:10.3892/etm.2021.10934
Sun, Z., Zhang, C., Wang, T., Shi, P., Tian, X., and Guo, Y. (2019). Correlation between long non-coding RNAs (lncRNAs) H19 expression and trastuzumab resistance in breast cancer. J. Cancer Res. Ther. 15, 933–940. doi:10.4103/jcrt.JCRT_208_19
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. A Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Szakács, G., Paterson, J. K., Ludwig, J. A., Booth-Genthe, C., and Gottesman, M. M. (2006). Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 5 (3), 219–234. doi:10.1038/nrd1984
Tan, D., Wu, Y., Hu, L., He, P., Xiong, G., Bai, Y., et al. (2017). Long noncoding RNA H19 is up-regulated in esophageal squamous cell carcinoma and promotes cell proliferation and metastasis. Dis. Esophagus 30 (1), 1–9. doi:10.1111/dote.12481
Tang, D., Kang, R., Vanden Berghe, T., and Vandenabeele, P. (2019)., 0. January, 1–18.03 the molecular machinery of regulated cell deathCell Res.
Tanos, V., Prus, D., AyeSh, S., Weinstein, D., Tykocinski, M. L., De-GrootN., , et al. (1999). Expression of the imprinted H19 oncofetal RNA in epithelial ovarian cancer. Eur. J. Obstet. Gynecol. Reprod. Biol. 85 (1), 7–11. doi:10.1016/s0301-2115(98)00275-9
Thomas, H., Coley, H. M., Parker Kodiak, J., Pastel, A., and Ellefson, J. (2003). Overcoming multidrug resistance in cancer: An update on the clinical strategy of inhibiting p-glycoprotein. Cancer control. 10 (2), 159–165. doi:10.1177/107327480301000207
Tsang, W. P., and Kwok, T. T. (2007). Riboregulator H19 induction of MDR1-associated drug resistance in human hepatocellular carcinoma cells. Oncogene 26 (33), 4877–4881. doi:10.1038/sj.onc.1210266
Tsang, W. P., Ng, E. K. O., Ng, S. S. M., Jin, H., Yu, J., Sung, J. J. Y., et al. (2010). Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 31 (3), 350–358. doi:10.1093/carcin/bgp181
Vasan, N., Baselga, J., and Hyman, D. M. (2019). A view on drug resistance in cancer. Nature 575 (7782), 299–309. doi:10.1038/s41586-019-1730-1
Vennin, C., Spruyt, N., Dahmani, F., Julien, S., Bertucci, F., Finetti, P., et al. (2015). H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget 6 (30), 29209–29223. doi:10.18632/oncotarget.4976
Wang, J., Sun, J., and Yang, F. (2020). The role of long non-coding RNA H19 in breast cancer. Oncol. Lett. 19 (1), 7–16. doi:10.3892/ol.2019.11093
Wang, J., Xie, S., Yang, J., Xiong, H., Jia, Y., Zhou, Y., et al. (2019). The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J. Hematol. Oncol. 12 (1), 81–14. doi:10.1186/s13045-019-0747-0
Wang, J. X., Zhang, X. J., Li, Q., Wang, K., Wang, Y., Jiao, J. Q., et al. (2015). MicroRNA-103/107 regulate programmed necrosis and myocardial ischemia/reperfusion injury through targeting FADD. Circ. Res. 117 (4), 352–363. doi:10.1161/CIRCRESAHA.117.305781
Wang, J., Ye, C., Xiong, H., Shen, Y., Lu, Y., Zhou, J., et al. (2017). Dysregulation of long non-coding RNA in breast cancer: An overview of mechanism and clinical implication. Oncotarget 8 (3), 5508–5522. doi:10.18632/oncotarget.12537
Wang, J., Zhang, J., Zhang, L., Zhao, L., Fan, S., Yang, Z., et al. (2011). Expression of P-gp, MRP, LRP, GST-π and TopoIIα and intrinsic resistance in human lung cancer cell lines. Oncol. Rep. 26 (5), 1081–1089. doi:10.3892/or.2011.1405
Wang, M., Han, D., Yuan, Z., Hu, H., Zhao, Z., Yang, R., et al. (2018). Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 9 (12), 1149. doi:10.1038/s41419-018-1187-4
Wang, Q., Cheng, N., Li, X., Pan, H., Li, C., Ren, S., et al. (2017). Correlation of long non-coding RNA H19 expression with cisplatin-resistance and clinical outcome in lung adenocarcinoma. Oncotarget 8 (2), 2558–2567. doi:10.18632/oncotarget.13708
Wang, X., Pei, X., Guo, G., Qian, X., Dou, D., Zhang, Z., et al. (2020). Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J. Cell. Physiol. 235 (10), 6896–6904. doi:10.1002/jcp.29585
Wang, X., Zhao, H., Qin, Q., Liu, Y., Zhang, B., and Wu, Y. (2018). Role of long noncoding RNA H19 in chemosensitivity of colorectal cancer. Chin. J. Color. Dis. Electron. Ed. 07 (05), 437–441.
Wang, Y., Zhou, P., Li, P., Yang, F., and qiang Gao, X. (2020). Long non-coding RNA H19 regulates proliferation and doxorubicin resistance in MCF-7 cells by targeting PARP1. Bioengineered 11 (1), 536–546. doi:10.1080/21655979.2020.1761512
Wei, J., Gan, Y., Peng, D., Jiang, X., Kitazawa, R., Xiang, Y., et al. (2018). Long non-coding RNA H19 promotes TDRG1 expression and cisplatin resistance by sequestering miRNA-106b-5p in seminoma. Cancer Med. 7 (12), 6247–6257. doi:10.1002/cam4.1871
White, E. (2015). The role for autophagy in cancer. J. Clin. Invest. 125 (1), 42–46. doi:10.1172/jci73941
Wong, R. S. Y. (2011). Apoptosis in cancer: From pathogenesis to treatment1. J. Exp. Clin. Cancer Res. 30, 87. doi:10.1186/1756-9966-30-87
World Health Organization, (2018). Global health observatory. Geneva: World Heal. Organ. Available at: who.int/gho/database/en/ (Accessed June 21, 2018).
Wu, P., Cai, J., Chen, Q., Han, B., Meng, X., Li, Y., et al. (2019). Lnc-TALC promotes O6-methylguanine-DNA methyltransferase expression via regulating the c-Met pathway by competitively binding with miR-20b-3p. Nat. Commun. 10 (1), 2045. doi:10.1038/s41467-019-10025-2
Wu, Y., Zhou, Y., He, J., Sun, H., and Jin, Z. (2019). Long non-coding RNA H19 mediates ovarian cancer cell cisplatin-resistance and migration during EMT. Int. J. Clin. Exp. Pathol. 12 (7), 2506–2515.
Wu, Z. R., Yan, L., Liu, Y. T., Cao, L., Guo, Y. H., Zhang, Y., et al. (2018). Inhibition of mTORC1 by lncRNA H19 via disrupting 4E-BP1/Raptor interaction in pituitary tumours. Nat. Commun. 9 (1), 4624. doi:10.1038/s41467-018-06853-3
Xiao, Y., Zhu, Z., Li, J., Yao, J., Jiang, H., Ran, R., et al. (2020). Expression and prognostic value of long non-coding RNA H19 in glioma via integrated bioinformatics analyses. Aging (Albany NY) 12 (4), 3407–3430. doi:10.18632/aging.102819
Xu, P.-X., Li, Q., Xu, Q.-G., and Zhang, C.-C. (2017). A study on the relationship between long non-coding RNA H19 and high-grade glioma temozolomide resistance and their related mechanism. J. Hainan Med. Univ. 23 (12), 95–100.
Xu, Y., Liu, Y., Li, Z., Li, H., Li, X., Yan, L., et al. (2020). Long non-coding RNA H19 is involved in sorafenib resistance in hepatocellular carcinoma by upregulating miR-675. Oncol. Rep. 44 (1), 165–173. doi:10.3892/or.2020.7608
Yan, J., Zhang, Y., She, Q., Li, X., Peng, L., Wang, X., et al. (2017). Long noncoding RNA H19/miR-675 Axis promotes gastric cancer via FADD/caspase 8/caspase 3 signaling pathway. Cell. Physiol. biochem. 42 (6), 2364–2376. doi:10.1159/000480028
Yan, L., Yang, S., Yue, C. X., Wei, X. Y., Peng, W., Dong, Z. Y., et al. (2020). Long noncoding RNA H19 acts as a miR-340-3p sponge to promote epithelial-mesenchymal transition by regulating YWHAZ expression in paclitaxel-resistant breast cancer cells. Environ. Toxicol. 35 (9), 1015–1028. doi:10.1002/tox.22938
Yang, J., Qin, G., LuoM., , Chen, J., Zhang, Q., Li, L., et al. (2015). Reciprocal positive regulation between Cx26 and PI3K/Akt pathway confers acquired gefitinib resistance in NSCLC cells via GJIC-independent induction of EMT. Cell Death Dis. 6 (7), e1829. doi:10.1038/cddis.2015.197
Ye, Y., Shen, A., and Liu, A. (2019). Long non-coding RNA H19 and cancer: A competing endogenous RNA. Bull. Cancer 106 (12), 1152–1159. doi:10.1016/j.bulcan.2019.08.011
Yi, J., Zhu, J., Wu, J., Thompson, C. B., and Jiang, X. (2020). Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc. Natl. Acad. Sci. U. S. A. 117, 31189–31197. doi:10.1073/pnas.2017152117
Yokoyama, Y., Sakatani, T., Wada, R., Ishino, K., Kudo, M., Koizumi, M., et al. (2019). In vitro and in vivo studies on the association of long non-coding RNAs H19 and urothelial cancer associated 1 with the susceptibility to 5-fluorouracil in rectal cancer. Int. J. Oncol. 55 (6), 1361–1371. doi:10.3892/ijo.2019.4895
Yoshimizu, T., Miroglio, A., Ripoche, M. A., Gabory, A., Vernucci, M., Riccio, A., et al. (2008). The H19 locus acts in vivo as a tumor suppressor. Proc. Natl. Acad. Sci. U. S. A. 105 (34), 12417–12422. doi:10.1073/pnas.0801540105
Yu, S., Wu, C., Tan, Q., and Liu, H. (2019). Long noncoding RNA H19 promotes chemotherapy resistance in choriocarcinoma cells. J. Cell. Biochem. 120 (9), 15131–15144. doi:10.1002/jcb.28775
Zeller, C., Dai, W., Steele, N. L., Siddiq, A., Walley, A. J., Wilhelm-Benartzi, C. S. M., et al. (2012). Candidate DNA methylation drivers of acquired cisplatin resistance in ovarian cancer identified by methylome and expression profiling. Oncogene 31 (42), 4567–4576. doi:10.1038/onc.2011.611
Zhang, L., Yang, F., Yuan, J. h., Yuan, S. x., Zhou, W. p., Huo, X. s., et al. (2013). Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis 34 (3), 577–586. doi:10.1093/carcin/bgs381
Zhang, L., Zhou, Y., Huang, T., Cheng, A. S. L., Yu, J., Kang, W., et al. (2017). The interplay of LncRNA-H19 and its binding partners in physiological process and gastric carcinogenesis. Int. J. Mol. Sci. 18 (2), E450. doi:10.3390/ijms18020450
Zhang, Q., Li, X., Li, X., Li, X., and Chen, Z. (2018). LncRNA H19 promotes epithelial-mesenchymal transition ( EMT ) by targeting miR-484 in human lung cancer cells. J. Cell. Biochem. 1196, 4447–4457. doi:10.1002/jcb.26537
Zheng, Z. G., Xu, H., Suo, S. S., Xu, X. L., Ni, M. W., Gu, L. H., et al. (2016). The essential role of H19 contributing to cisplatin resistance by regulating glutathione metabolism in high-grade serous ovarian cancer. Sci. Rep. 6 (1), 26093. doi:10.1038/srep26093
Zhou, G., Zhang, F., Guo, Y., Huang, J., Xie, Y., Yue, S., et al. (2017). miR-200c enhances sensitivity of drug-resistant non-small cell lung cancer to gefitinib by suppression of PI3K/Akt signaling pathway and inhibites cell migration via targeting ZEB1. Biomed. Pharmacother. 85, 113–119. doi:10.1016/j.biopha.2016.11.100
Zhou, J., Yang, L., Zhong, T., Mueller, M., Men, Y., Zhang, N., et al. (2015). H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat. Commun. 6, 10221. doi:10.1038/ncomms10221
Zhou, W., Ye, X. L., Xu, J., Cao, M. G., Fang, Z. Y., Li, L. Y., et al. (2017). The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci. Signal. 10 (483), eaak9557. doi:10.1126/scisignal.aak9557
Zhou, Y., and Zhang, Y. (2020). Inhibition of LncRNAH19 has the effect of anti-tumour and enhancing sensitivity to Gefitinib and Chemotherapy in Non-small-cell lung cancer in vivo. J. Cell. Mol. Med. 24 (10), 5811–5816. doi:10.1111/jcmm.15245
Zhu, H. (2020). Silencing long non-coding RNA H19 combined with paclitaxel inhibits nasopharyngeal carcinoma progression. Int. J. Pediatr. Otorhinolaryngol. 138 (1), 110249. doi:10.1016/j.ijporl.2020.110249
Zhu, Q. N., Wang, G., Guo, Y., Peng, Y., Zhang, R., Deng, J. L., et al. (2017). LncRNA H19 is a major mediator of doxorubicin chemoresistance in breast cancer cells through a cullin4A-MDR1 pathway. Oncotarget 8 (54), 91990–92003. doi:10.18632/oncotarget.21121
Zhu, W., Xu, H., Zhu, D., Zhi, H., Wang, T., Wang, J., et al. (2012). miR-200bc/429 cluster modulates multidrug resistance of human cancer cell lines by targeting BCL2 and XIAP. Cancer Chemother. Pharmacol. 69 (3), 723–731. doi:10.1007/s00280-011-1752-3
Zhuang, M., Gao, W., Xu, J., Wang, P., and Shu, Y. (2014). The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem. Biophys. Res. Commun. 448 (3), 315–322. doi:10.1016/j.bbrc.2013.12.126
Keywords: lncRNA H19, drug resistance, tumorigenesis, miRNA, chemotherapy, endocrine therapy, targeted therapy
Citation: Zhang X, Luo M, Zhang J, Guo B, Singh S, Lin X, Xiong H, Ju S, Wang L, Zhou Y and Zhou J (2022) The role of lncRNA H19 in tumorigenesis and drug resistance of human Cancers. Front. Genet. 13:1005522. doi: 10.3389/fgene.2022.1005522
Received: 28 July 2022; Accepted: 22 August 2022;
Published: 27 September 2022.
Edited by:
William C. Cho, QEH, Hong Kong SAR, ChinaReviewed by:
Linjiang Song, Chengdu University of Traditional Chinese Medicine, ChinaAli Zarrabi, University of Istinye, Turkey
Shantanu Gupta, University of São Paulo, Brazil
Copyright © 2022 Zhang, Luo, Zhang, Guo, Singh, Lin, Xiong, Ju, Wang, Zhou and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linbo Wang, bGluYm93YW5nQHpqdS5lZHUuY24=; Yulu Zhou, MTE2MTgzOTBAemp1LmVkdS5jbg==; Jichun Zhou, amljaHVuLXpob3VAemp1LmVkdS5jbg==
 Xun Zhang
Xun Zhang Mingpeng Luo
Mingpeng Luo Jiahang Zhang1,2,3
Jiahang Zhang1,2,3 Bize Guo
Bize Guo Shreya Singh
Shreya Singh Xixi Lin
Xixi Lin Hanchu Xiong
Hanchu Xiong Siwei Ju
Siwei Ju Linbo Wang
Linbo Wang Yulu Zhou
Yulu Zhou Jichun Zhou
Jichun Zhou