- 1Department of Animal Sciences, College of Coastal Agricultural Sciences, Guangdong Ocean University, Zhanjiang, China
- 2College of Animal Science and Technology, Anhui Agricultural University, Hefei, China
The C4b binding protein alpha (C4BPA) chain primarily engages in critical inflammatory and coagulation processes. The previous transcriptomic analysis showed that C4BPA is a differentially expressed gene in lower and higher fat content mammary gland cell lines from Chinese Holstein. This study aimed to investigate the effects of C4BPA on the inflammation and milk fat synthesis in bMECs by C4BPA knockdown and overexpression. The results highlighted that knockdown of C4BPA in bMECs could suppress the mRNA and protein expression of IL-6, IL-8, IL-12, and the TLR-4/NF-κB pathway-related genes and promote the expression of complement and coagulation cascade pathways related genes as well as TNF-α. Moreover, knockdown of C4BPA expression in bMECs reduced the content of triglyceride (TG) and cholesterol (CHOL) in bMECs, increased NEFA content, reduced mRNA and protein expression of ACSL1 and PPARA, and increased the mRNA and protein expression of ELOVL6, FADS1, and LPL. The bMECs, with the overexpression of C4BPA, showed the enhanced expression of TLR-4/NF-κB linked genes, IL-6, IL-8, IL-12, and mRNA and protein level while reduced mRNA expression of TNF-α, compliment, and coagulation cascade related genes was observed. In bMECs, overexpression of C4BPA enhanced the content of TG and CHOL while reducing NEFA and stimulated the mRNA and protein expression of ACSL1, PPARA, and PPARG genes while inhibiting the mRNA and protein expression of FADS1 and LPL genes. Our results show that C4BPA not only regulates the lipid metabolism through the PPAR signaling pathway in bMECs but also contributes to the inflammatory response through TLR-4/NF-κB and the complement and coagulation cascade pathways. This study, for the first time, provides the primary basis for understanding the role of C4BPA in immunity and fat metabolism, which enables the researchers for innovative direction to investigate genes associated with fat metabolism and immunity. This study also advocates that the breeders must pay attention to such type of genes with multiple functions during animal breeding.
Introduction
Milk is a high fat, protein, vitamin, and other nutrient-rich liquid food secreted from the mammalian glands as a key nourishment source for infants of mammals (Winckel et al., 2011). The primary milk fat components, followed by fatty acids and cholesterol, are triglycerides (Leonard et al., 2004). Because of the enrichment of nutrients in milk, humans also consume it to meet their dietary needs. Cow milk is the largest contributor to the world’s milk output in commercial production (Gerosa and Skoet 2012).
C4BPA is one of the genes we found to function in lipid metabolism traits, and this gene is related to inflammation. Several diseases lead to inflammation of the udder, including mastitis. Mastitis is one of the most common and destructive diseases for dairy cattle. Mastitis is an inflammatory reaction in the bovine mammary tissue induced by infection with pathogens or other physical and chemical causes. Dairy cow mastitis is the consequence of issues related to genetics, environment, and food management. It can lead to a decline in milk quality and quantity, a decrease in growth and development of the animal, and a decrease in fertility, leading to significant economic losses and entailing the development of the dairy industry (Waseem Ghani et al., 2019). In order to cope with this issue, breeding of dairy cows that are resistant to mastitis is essential. Several studies have been conducted over the past few years to collect data on the genetic factors and breeding values of dairy cows with mastitis (Martin et al., 2018). Many anti-mastitis potential genes are investigated by genome-wide association studies (GWAS). Using E. coli infected cattle’s mammary tissues, Mitterhuemer identified 2,754 and 476 differential genes in the mammary tissues of the mastitis cattle. These genes included antifungal genes (S100A8, S100A9, S100A12, CXCL2, GNLY), acute-phase genes (LBP, SAA3, CP, BF, C6, C4BPA, IF), and oxidative stress genes (LBP, SAA3, GPX3, MT1A, MT2A, SOD2) (Mitterhuemer et al., 2010).
The C4BPA gene is located on chromosome 1q32 and contains 12 exons and 11 introns. The soluble alpha chain of C4BPA is a major inhibitor of the conventional complement pathway (Suankratay et al., 1999). Three types of C4BP are found: α7β1, α7β0, and α6β1 (Scharfstein, Ferreira, and Gigli 1978). In all three forms, α7β1 is the more abundant while α6β1is inconsequential (Moffat et al., 1992). C4BP has various isoforms, and the one called C4BPA is generally expressed at higher levels during inflammation. In the C4BP alpha chain, the binding sites are found for heparin (Hessing M, 1990), low-density lipoprotein receptor-related protein LRP (Westein et al., 2002), C3b, serum amyloid protein (Garcia de Frutos P and Dahlbäck B 1994), and the surface proteins of various bacteria (Blom AM, 2000; Ram S, 2001), which are the master player in lipid metabolism, inflammation, and coagulation pathways (Ridker et al., 2008; Antoniades et al., 2009).
A previous study also reported that the C4BPA possesses the binding site for heparin, C-reactive protein, and CD40, the specific key mediators during the inflammation process and the blood coagulation pathways (Antoniades et al., 2009). In human B cells, the C4BPA directly binds with CD40 at the various positions of the CD40 ligand. The vital role of C4BPA is in the induction of the proliferation of B cells and the activation of CD40, which in turn stimulates the anti-tumor response of T cells (Brodeur et al., 2003). Sogawa et al. reported that the levels of C4BPA was found to be higher in the serum of pancreatic ductal adenocarcinoma (PDAC) patients compared to the normal human serum (Sogawa et al., 2016). Due to the significant binding ability of C4BPA with CD40, the C4BPA is a key factor involved in dairy cattle mastitis. The research on mastitis development reported that the inflammatory factors including IL-6, IL-8, TNF-ɑ, and TLR4 are consistently altered (Harada et al., 1994; Strandberg et al., 2005; Fu et al., 2013).
The previous transcriptomic analysis of low- and high-fat milk-producing Chinese Holstein cattle showed that C4BPA is a differentially expressed gene with higher expression in high-fat milk cattle (Xia et al., 2021). However, the co-regulatory role of the C4BPA gene against inflammation and fat metabolism in Chinese Holstein cattle is still unknown. Therefore, this study investigates the key role of the C4BPA gene in milk fat synthesis and inflammation through its overexpression and knockdown in bMECs. In order to delineate the aforementioned co-regulatory role of C4BPA, the different regulators and indicators of inflammation and milk fat were analyzed in different experimental groups. This study provides the major basis for breeding cattle with high resistance to mastitis and good milk quality through the proper knowledge of the molecular mechanism of such type genes. According to our best knowledge, this is the first study exploring the co-regulatory role of the C4BPA in milk fat metabolism and mastitis.
Materials and Methods
Cell Source
The animal genetics and breeding laboratory at Jilin University kept BMECs from Chinese Holstein dairy cows (Lu et al., 2012). All studies were conducted under Jilin University’s rules for the care and use of experimental animals (permission number: SY201901007 (Jiang et al., 2020).
Construction of C4BPA Interference and Overexpression Plasmids
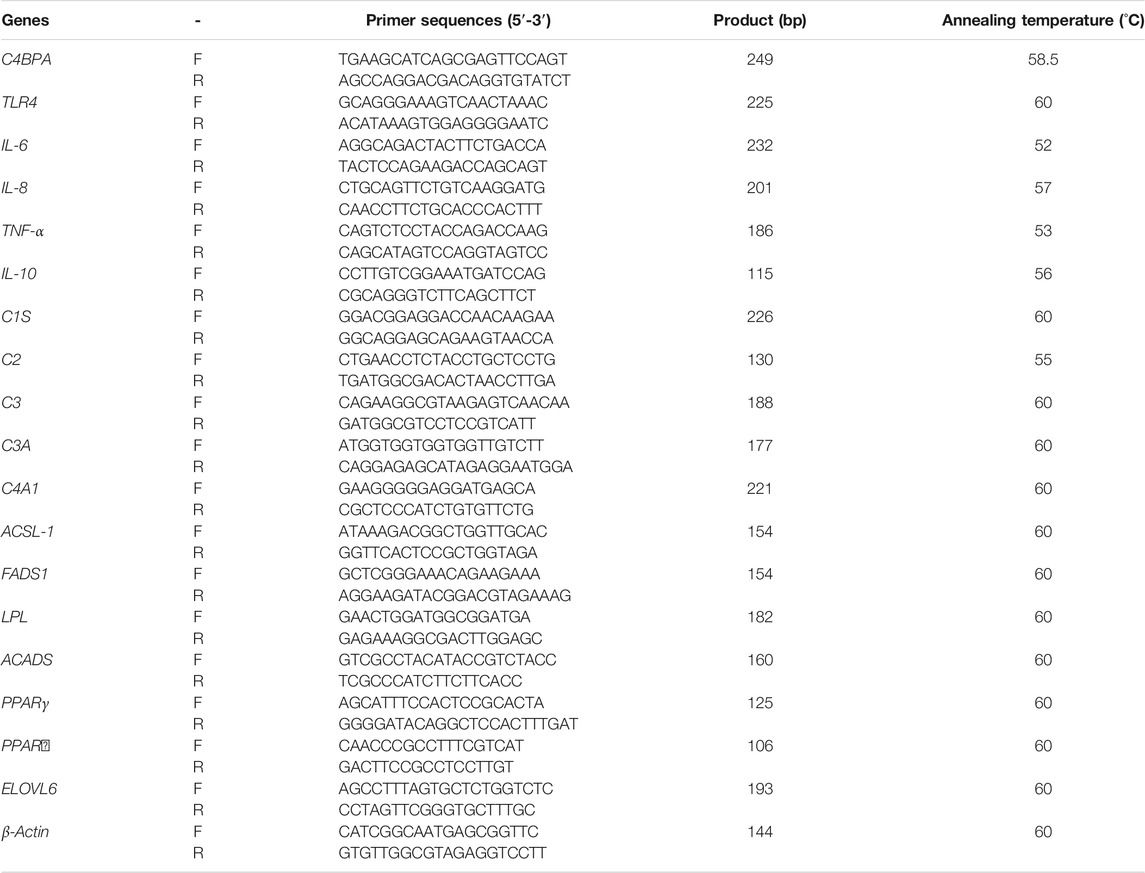
NCBI identification of a CDS-specific fragment, four C4BPA-shRNA interference sequences (pGPU6-C4BPA-shRNA1, pGPU6-C4BPA-shRNA2 pGPU6-C4BPA-shRNA3, and pGPU6-C4BPA-shRNA4), and negative control (shNC) were designed and synthesized. The sequence is shown in Table 1. Interfering RNA (shRNA) oligonucleotides were annealed and cloned into the pGPU6-GFP-Neo knockdown vector (C02007, Genepharma Corporation, Shanghai, China). Annealing system (10 µl): 1 µl each of the upper and lower primers, 10X annealing buffer 1, and 7 µl ddH2O were incubated in the PCR apparatus at 95°C for 5 min, then slowly annealed to room temperature, and stored at 4°C for later use.
The coding region of C4BPA in the cow is amplified with forwarding primer 5′- GGATCCATGAAGCATCAGCGAGTTC-3′ (the underlined sequence indicates a BamHI) and reverse primer 5′- ATCGATCTATTCTGGATTAAAGTCACAAGTCA -3′ (the underlined sequence indicates a ClaI). Then, PCR was used to clone an 1833 bp product, followed by its purification. The C4BPA sequence, containing BamHI (#R3136L, NEB, China) and ClaI (#R0197V, NEB, China) restriction endonuclease cleavage sites, was recombined and ligated into the pBI-CMV3 expression vector (#631632, Clontech Laboratories, Mountain View, CA). Finally, it was confirmed by sequencing to obtain the C4BPA overexpression vector (pBI-CMV3-C4BPA). The Shanghai Shenggong Biotechnology Co., Ltd. constructed the plasmid, and the point mutation kits were used for site-directed mutagenesis (#C215-01, Vazyme, China).
Cell Culture and Transfection
The bMECs were cultured in culture plates (353003, Falcon, Franklin Lakes, NJ) in Dulbecco’s modified Eagle medium with nutrient mixture F12 (12-719Q, HyClone, Logan, UT) supplemented with 10% fetal bovine serum (11,011-6,123, Tian Hang, Zhejiang, China), with 1% antibiotics (SV30010, HyClone). Cells were transferred into six-well culture plates (353090, Falcon) at a concentration of 2 × 105 cells/well and cultured at 37°C and 5% CO2 in an incubator (Thermo Fisher Scientific, Waltham, MA). When cells reached 80% confluence, the cell culture medium was changed to a medium without antibiotics for transfection (Jiang et al., 2018). According to the manufacturer’s protocol, the various vectors were transfected into cells using a FuGENE HD Transfection Reagent (PRE2311, Promega, Madison, WI). Briefly, the mixture of 3 µg of plasmid DNA and 7.5 µl of FuGENE HD in 150 µl of Opti-MEM (Thermo Fisher Scientific) was incubated at room temperature for 15 min added into the culture medium in each well of bMECs. After 24 h, the expression of green fluorescent protein (GFP) in cells was observed under a fluorescence microscope (TE 2000, Nikon, Tokyo, Japan) to determine the transfection efficiency. Three replicates of the experiments were carried out by transfecting equal numbers of cells using the same vectors in the various wells.
Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction
Cells transfected with pGPU6-C4BPA-shRNA1 and the pBI-CMV3-C4BPA were harvested 24 h after transfection. Then, 1 μg of total RNA was extracted with an RNA extraction kit (Analytik Jena, Jena, Germany). The concentration and purity of the RNA were determined using a spectrophotometer, further confirmed via 1% agarose gel electrophoresis. The cDNA was obtained using a cDNA synthesis kit (RR047A, TaKaRa Biotechnology, Dalian, China). Then, real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed with SYBR Premix Ex Taq (RR420L, TaKaRa Biotechnology). The primers’ sequences are shown in Table 2. We performed PCR amplification in 20 µl reaction volume under the following conditions: an initial denaturing step of 95°C for 30 s, followed by 40 cycles of 95°C for 10 s, and then 60°C for 45 s. All samples were adjusted to the same concentration. The mRNA expression was measured via qRT-PCR with β-actin as the reference gene for internal control. All the experiments of qRT-PCR were repeated at least three times.
Triglyceride, Cholesterol, and Non-Eesterified Free Fatty Acid Detection
Cells transfected with pGPU6-C4BPA-shRNA1 and pBI-CMV3-C4BPA were harvested 24 h after transfection. A blank control group was set as the reference group using double-distilled water. Triglyceride and total cholesterol content were measured using a TG and CHOL detection kit (E1015-105 and E1013, Applygen Technologies, Beijing, China). Non-esterified free fatty acid content was measured using a NEFA detection kit (A042-2-1, Nanjing Jiancheng Bioengineering Institute, China). Each microgram of protein adjusted the cellular contents of TG, CHOL, and NEFA by the BCA (E112-01, Vazyme, China) protein quantitative analysis method. The concentrations were measured using a microplate reader (SM600, Shanghai YongChuang Medical Instrument Co. Ltd., Shanghai, China)
Western Blot
Cells transfected with pGPU6-C4BPA-shRNA1 and pBI-CMV3-C4BPA were harvested 24 h after transfection. After 24 h of transfection, trypsinize the cells and wash them three times with PBS. The cells were then resuspended in RIPA buffer with protease inhibitors (MA0151, Meilunbio). The cell lysate was removed by centrifugation at 12,000 rpm/min for 10 min at 4°C, and the supernatant was collected. The BCA protein quantitative analysis method determines the sample’s total protein concentration. For Western blot (PowerPac™ HV, Bio-Rad), separate the same amount of protein by SDS-PAGE and then transfer it to PVDF (88518, Thermo-Fisher) membrane with a membrane transfer machine. After blocking with 1×TBS buffer containing 1% BSA for 2 h, wash the PVDF membrane with TBST (B040126, Sangon Biotech, China) solution. Then, the membrane was incubated with a primary antibody at 4°C for 12 h. Incubate the membrane with a secondary antibody (RS0002, Immunoway) (diluted in 1% BSA) for 2 h. The enhanced chemiluminescence HRP substrate was used to observe protein bands. After taking a photo with a chemiluminescence instrument, grayscale analysis software (Tanon) was used to calculate the relative expression of the target protein in different samples. Primary antibodies were purchased from Bioss, China.
Statistics and Analysis
Experimental data are shown as mean ± standard error (MSE) of the mean. Statistically significant differences are shown as p < 0.05. GraphPad Prism 9 software (GraphPad Software Inc., La Jolla, CA) with a 2-tailed (unpaired) t-test used for the data analysis.
Results
Sequencing and the transfection efficiency of pBI-CMV3-C4BPA and pGPU6-C4BPA-shRNAs in bMECs: the standard sequencing results of the shRNAs and overexpression vector of C4BPA are displayed in Figure 1 (Figures BI and BII). After the successful construction and harvesting, the pBI-CMV3-C4BPA, pGPU6-C4BPA-shRNA1, pGPU6-C4BPA-shRNA2, pGPU6-C4BPA-shRNA3, and pGPU6-C4BPA-shRNA4 vectors were transferred to the cell plate when the bMECs attained 80 percent cell confluency. After 24 h of transfection, the cell morphology and the expression of green fluorescence protein were observed under an inverted fluorescent microscope (Figure 1C). The results showed that transfection of knockdown and overexpression vectors was successfully achieved, and cell morphology of the bMECs remained unchanged.
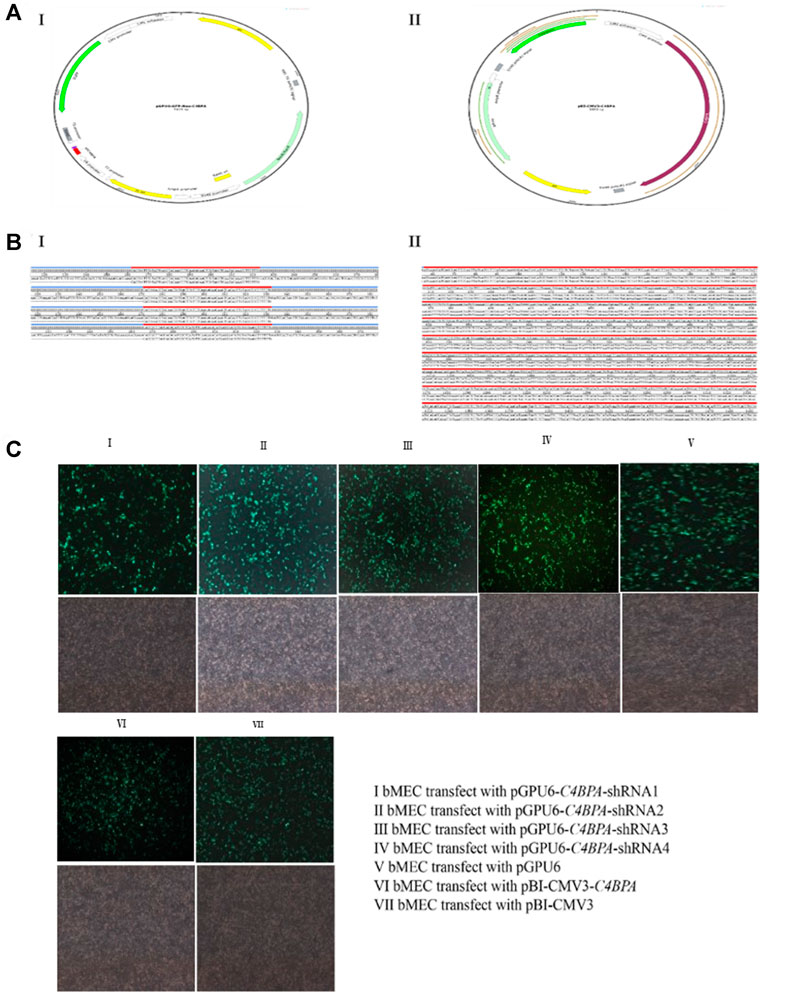
FIGURE 1. Construction of C4BPA gene interferences and overexpression vector. (AI) Construct an interference vector map. (AII) Construct C4BPA overexpression vector map. (BI) pGPU6-C4BPA-shRNA1, pGPU6-C4BPA-shRNA2, pGPU6-C4BPA-shRNA3, and pGPU6-C4BPA-shRNA4 four interferences vector sequences. (BI) pBI-CMV3-C4BPA sequencing. (CI) bMECs transfected with pGPU6-C4BPA-shRNA1. (CII) bMECs transfected with pGPU6-C4BPA-shRNA2. (CIII) bMECs transfected with pGPU6-C4BPA-shRNA3. (CIV) bMECs transfected with pGPU6-C4BPA-shRNA4. (CV) bMECs transfected with pGPU6. (CVI) bMECs transfected with pBI-CMV3-C4BPA. (CVII) bMECs transfected with pBI-CMV3.
The Relative mRNA and Protein Expression of pGPU6-C4BPA-shRNAs in bMECs
The bMECs were cultured in six well-plates. When the cells attained 80% growth, the C4BPA knockdown vectors, including pGPU6-C4BPA-shRNA1, pGPU6-C4BPA-shRNA2, pGPU6-C4BPA-shRNA3, pGPU6-C4BPA-shRNA4, and pGPU6, were transfected into the cells. After 24 h of successful transfection, the total RNA was extracted and reverse-transcribed and qRT-PCR was performed. The results showed that the mRNA expression of the C4BPA gene was significantly downregulated (p < 0.001) in pGPU6-C4BPA-shRNA1, pGPU6-C4BPA-shRNA2, and pGPU6-C4BPA-shRNA4 as compared to the pGPU6 (control group) in bMECs, which is shown in Figure 2A. When the bMECs were transfected with the pGPU6-C4BPA-shRNA3, the mRNA expression of the C4BPA did not show any significant downregulation compared to the pGPU6 shown in Figure 2A. Moreover, the protein expression of pGPU6-C4BPA-shRNA2 through ELISA was determined. The results showed that the pGPU6-C4BPA-shRNA2 was significantly downregulated (p < 0.001) compared to the pGPU6 (Figure 2A). Both of the above-mentioned results validate that pGPU6-C4BPA-shRNA2 significantly downregulated the mRNA and protein expression of the C4BPA gene. For further experiment, we selected the pGPU6-C4BPA-shRNA2 as the shRNA for the knockdown C4BPA gene.
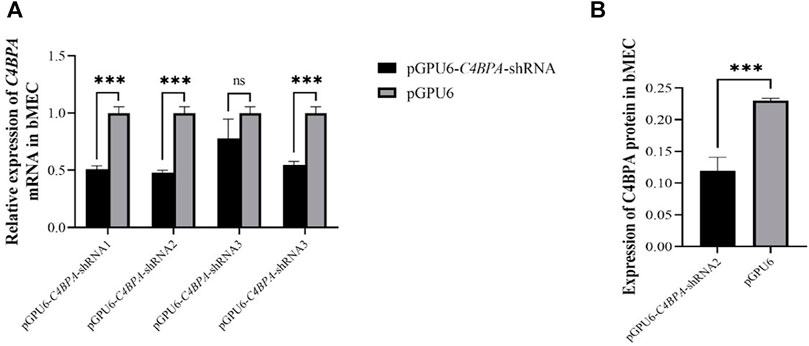
FIGURE 2. The relative expression of C4BPA at mRNA and protein level in different experimental groups of bMECs. (A) Relative mRNA expression of C4BPA gene after 24 h of transfection with pGPU6 C4BPA-shRNA1, pGPU6-C4BPA-shRNA2, pGPU6-C4BPA-shRNA3, pGPU6-C4BPA shRNA4, and pGPU6 in bMECs. (B) Relative protein expression of C4BPA after 24 h of transfection with pGPU6-C4BPA-shRNA2.
The Relative mRNA and Protein Expression of pBI-CMV3-C4BPA in bMECs
bMECs were cultured in six well-plates, and after reaching the 80% cell confluency, the shRNA vectors, including pBI-CMV3-C4BPA and pBI-CMV3, were transfected into the cells. After 24 h of successful transfection, qRT-PCR was performed. The result showed that the mRNA expression of the C4BPA gene was significantly upregulated (p < 0.001) in pBI-CMV3-C4BPA as compared to pBI-CMV3 in bMECs (Figure 3A). Moreover, the protein expression of C4BPA in bMECs transfected with pBI-CMV3-C4BPA determined through ELISA showed that pBI-CMV3-C4BPA was significantly upregulated (p < 0.001) compared to the bMECs transfected with pBI-CMV3 (Figure 3B). Taken together, these results validated that pBI-CMV3-C4BPA significantly upregulated the mRNA and protein expression of the C4BPA gene in bMECs.
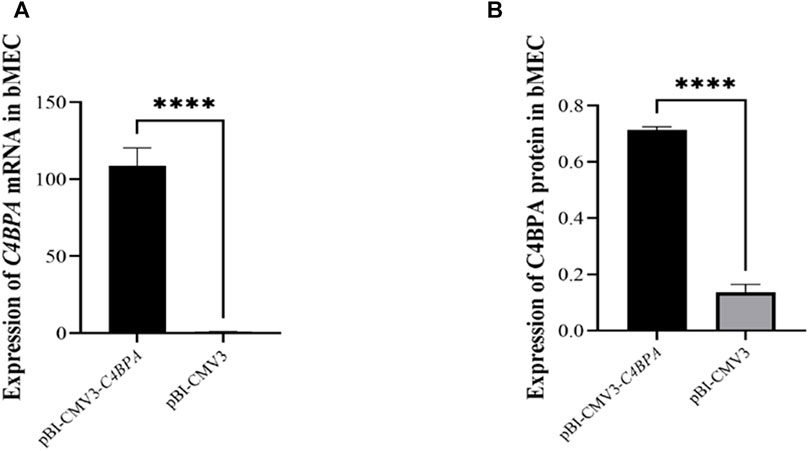
FIGURE 3. (A) Relative mRNA expression of C4BPA gene after 24 h transfection with pBI-CMV3-C4BPA and pBI-CMV3 in bMECs. (B) Relative protein expression of C4BPA gene after 24 h transfection with pBI-CMV3-C4BPA.
The mRNA Expression of C4BPA Influences the NF-κB Pathway-Related Genes in bMECs
The expression of the NF-κB pathway genes is investigated after the transfection of pGPU6-C4BPA-shRNA2 and pBI-CMV3-C4BPA vectors in bMECs. After 24 h of successful transfection, qRT-PCR was performed. The result showed that when bMECs are transfected with pGPU6-C4BPA-shRNA2, the genes that have a role in the NF-κB pathway, including the TRAF6, IKBKG-2, and NF-κB1, were significantly downregulated in the pGPU6-C4BPA-shRNA2 group as compared to pGPU6, which is shown in Figure 4A. The expression trend of NFKFBIA in the pGPU6-C4BPA-shRNA2 group had no significant downregulation trend compared to pGPU6, as shown in Figure 4A. The contrary result was obtained when bMECs were transfected with the pBI-CMV3-C4BPA. The genes involved in the NF-κB pathway, including TRAF6, IKBKG-2, NFKFBIA, and NFKB1, were all significantly upregulated in pBI-CMV3-C4BPA compared to pBI-CMV3, which is shown in Figure 4B. These results validated that C4BPA had a major influence on the NF-κB pathway genes and played a vital role in regulating these genes.
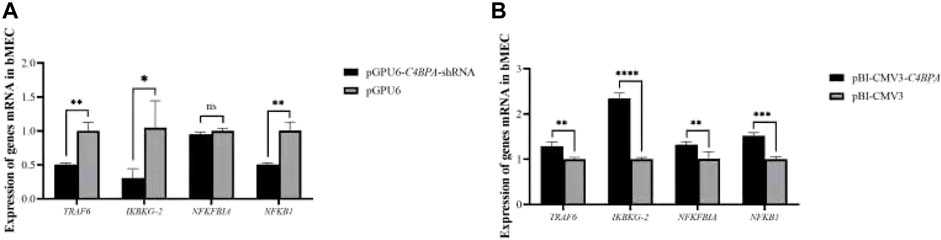
FIGURE 4. (A) The relative mRNA expression of NF-κB pathway-related genes in bMECs after transfection with pGPU6-C4BPA-shRNA2 vector. (B) The relative mRNA expression of NF-κB pathway-related genes in bMECs after transfection with pBI-CMV3-C4BPA vector.
The Relative mRNA Expression of Inflammatory Factors in bMECs With Knockdown and Overexpression of C4BPA
The mRNA expression of the inflammatory factors was investigated by the transfection of the vectors pGPU6-C4BPA-shRNA2 and pBI-CMV3-C4BPA in bMECs. After 24 h of successful transfection, qRT-PCR was performed. The result showed that when bMECs are transfected with pGPU6-C4BPA-shRNA2, the genes with a role in the inflammatory factors, including IL-8, IL-10, and IL-12, were significantly downregulated (p < 0.05) in the pGPU6-C4BPA-shRNA2 group as compared to pGPU6 (Figure 4A). The expression trend of IL-8 in the pGPU6-C4BPA-shRNA2 group had no significant downregulation trend compared to pGPU6, as shown in Figure 5A. The expression of TNF-ɑ in the pGPU6-C4BPA-shRNA2 group was significantly enhanced (p < 0.05) compared to pGPU6, as shown in Figure 4 A. The conflicting result was obtained when bMECs were transfected with pBI-CMV3-C4BPA. The genes involved in the NF-κB pathway, including inflammatory factors IL-6, IL-8, and IL-12, all were significantly upregulated in pBI-CMV3-C4BPA compared to pBI-CMV3, which is shown in Figure 5B. The expression trend of IL-10 in the pBI-CMV3-C4BPA group had no significant downregulation trend compared to pBI-CMV3, as shown in Figure 5B. The expression of TNF-ɑ in the pBI-CMV3-C4BPA group was significantly downregulated (p < 0.05) compared to pBI-CMV3, as shown in Figure 5B. Altogether, these results validated that C4BPA had a major influence on inflammatory factors and played a vital role in regulating these genes.
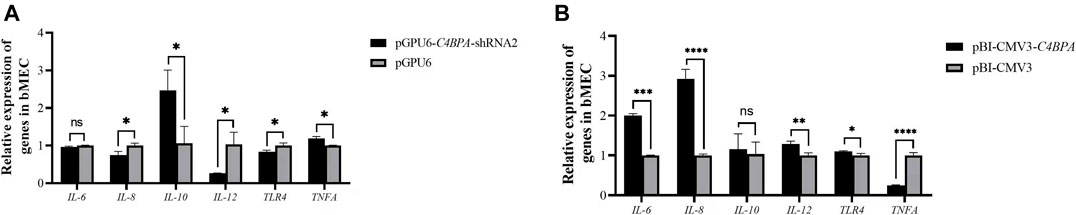
FIGURE 5. (A) The relative mRNA expression of inflammatory factors in bMECs after transfection with pGPU6-C4BPA-shRNA2 vector. (B) The relative mRNA expression of inflammatory factors in bMECs after transfection with pBI-CMV3-C4BPA vector.
The Relative Protein Expression of C4BPA on Inflammatory Factors in bMECs
The protein expression of the inflammatory factors was explored by transfecting the vectors pGPU6-C4BPA-shRNA2 and pBI-CMV3-C4BPA in bMECs after 24 h of successful transfection, and the western blot was performed. The result showed that when bMECs were transfected with pGPU6-C4BPA-shRNA2, the genes that played a role in the inflammatory process, including the IL-6 and TLR4, were significantly downregulated (p < 0.05) in the pGPU6-C4BPA-shRNA2 group compared to pGPU6, which is shown in Figure 6A. The expression trend of IL-8 in the pGPU6-C4BPA-shRNA2 group had no significant downregulation trend compared to pGPU6, which is shown in Figure 5A. The expression of TNF-ɑ in the pGPU6-C4BPA-shRNA2 group was significantly enhanced (p < 0.001) compared to pGPU6, as shown in Figure 6A. The differing result was obtained when bMECs were transfected with pBI-CMV3-C4BPA. The genes involved in the NF-κB pathway, including inflammatory factors IL-6, IL-8, and TLR4, were all significantly upregulated (p < 0.05) in pBI-CMV3-C4BPA compared to pBI-CMV3, which is shown in Figure 5B. The expression of TNF-ɑ in the pBI-CMV3-C4BPA group was significantly downregulated (p < 0.05) compared to pBI-CMV3, which is shown in Figure 6B. These western blot results also validated that the expression of C4BPA had a major impact on the relative protein expression of inflammatory process related genes.
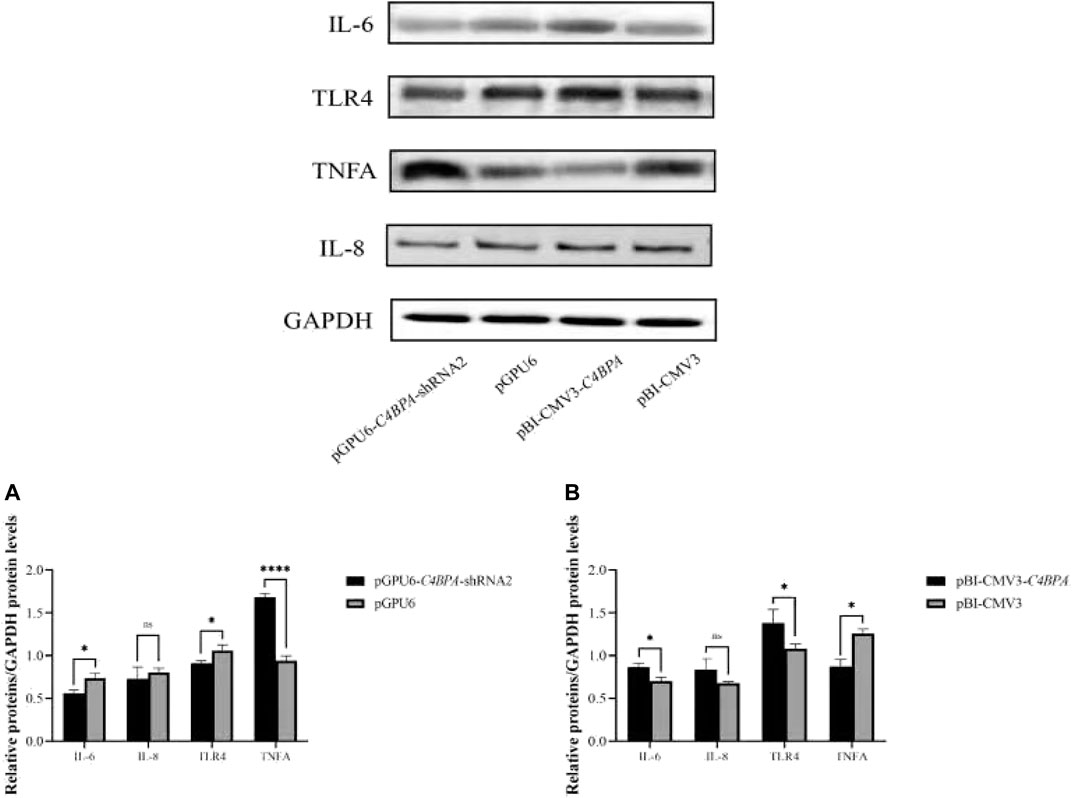
FIGURE 6. Relative protein expression of the immunity related factors in bMECs. (A) Relative protein expression of inflammatory factors in bMECs after transfection with pGPU6 C4BPA-shRNA2 vector. (B) Relative protein expression of inflammatory factors in bMECs after transfection with pBI-CMV3-C4BPA vector.
The Relative mRNA Expression of C4BPA on Complement and Coagulation Cascades Genes in bMECs
The mRNA expression of the genes related to the complement and the coagulation cascade pathways is investigated after 24 h of successful transfection with the vector pGPU6-C4BPA-shRNA2 and pBI-CMV3-C4BPA in bMECs. The results showed that when bMECs were transfected with pGPU6-C4BPA-shRNA2, the genes that played a role in the complement and coagulation cascade pathways, including C1S, C2, C3, and C3A, were significantly upregulated in the pGPU6-C4BPA-shRNA2 group, compared to pGPU6 as given in Figure 7A. The expression trend of the C4A1 gene in the pGPU6-C4BPA-shRNA2 group had no significant upregulation trend compared to pGPU6, as shown in Figure 7A. The contrary result was obtained when bMECs were transfected with pBI-CMV3-C4BPA. The genes involved in the complement and coagulation cascade pathways, including C1S, C2, C3, and C3A, were significantly downregulated in pBI-CMV3-C4BPA compared to the pBI-CMV3, as given in Figure 7B. The expression trend of C4A1 in the pBI-CMV3-C4BPA group had no significant downregulation trend compared to pBI-CMV3, as shown in Figure 7B. This result validates that C4BPA has a major effect on the complement and coagulation cascade pathways and plays a vital role in regulating these genes.
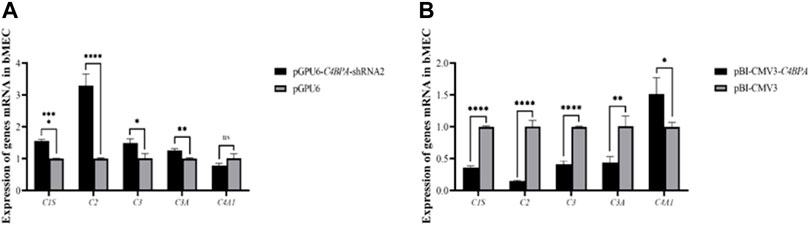
FIGURE 7. (A) The mRNA expression of complement and coagulation cascade pathways related genes in bMECs after transfection with pGPU6-C4BPA-shRNA2. (B) The mRNA expression of complements and coagulation cascade pathways related genes in bMECs after transfection with pBI-CMV3-C4BPA.
The Effect of C4BPA on the Content of Triglyceride (TG) in bMECs
The role of C4BPA in triglycerides was investigated by the transfection of the vectors pGPU6-C4BPA-shRNA2 and pBI-CMV3-C4BPA in bMECs. The TG extraction was performed after 24 h of successful transfection. The result showed that when bMECs were transfected with pGPU6-C4BPA-shRNA2, the TG content was significantly downregulated (p < 0.001) in the pGPU6-C4BPA-shRNA2 group compared to pGPU6, which is shown in Figure 8A. The contrary result was obtained when the bMECs were transfected with pBI-CMV3-C4BPA. The TG content was significantly upregulated (p < 0.001) in pBI-CMV3-C4BPA compared to pBI-CMV3, as given in Figure 8B. This result validates that C4BPA has a significant role in the triglycerides metabolism of bMECs.
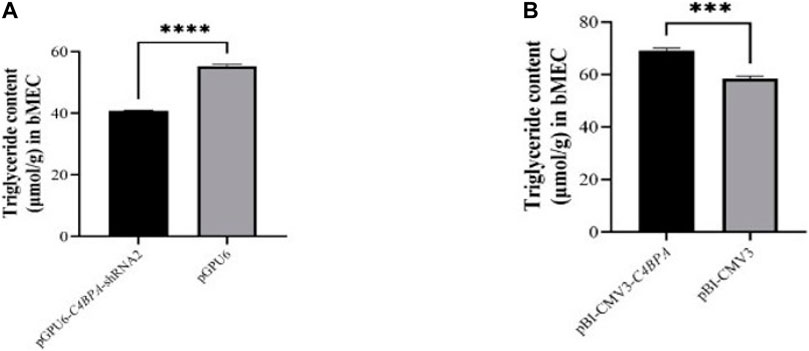
FIGURE 8. (A) The relative content of triglycerides in bMECs after transfection with pGPU6-C4BPA-shRNA2. (B) The relative content of triglycerides in bMECs after transfection with pBI-CMV3-C4BPA.
The Effect of C4BPA on the Content of Total Cholesterol (CHOL) in bMECs
The role of C4BPA on total cholesterol was investigated by the transfection of the vectors pGPU6-C4BPA-shRNA2 and pBI-CMV3-C4BPA in bMECs. After 24 h of successful transfection, the CHOL extraction was performed. The result showed that when bMECs are transfected with pGPU6-C4BPA-shRNA2, the CHOL content was significantly downregulated (p < 0.001) in the pGPU6-C4BPA-shRNA2 group compared to pGPU6, which is shown in Figure 9A. The contrary result was obtained when bMECs were transfected with pBI-CMV3-C4BPA. The CHOL content is significantly upregulated (p < 0.001) in pBI-CMV3-C4BPA compared to the pBI-CMV3 shown in Figure 9B. This result validates that C4BPA has a significant role in total cholesterol metabolism in bMECs in addition to the TG metabolism and, therefore, it regulates fat milk metabolism.
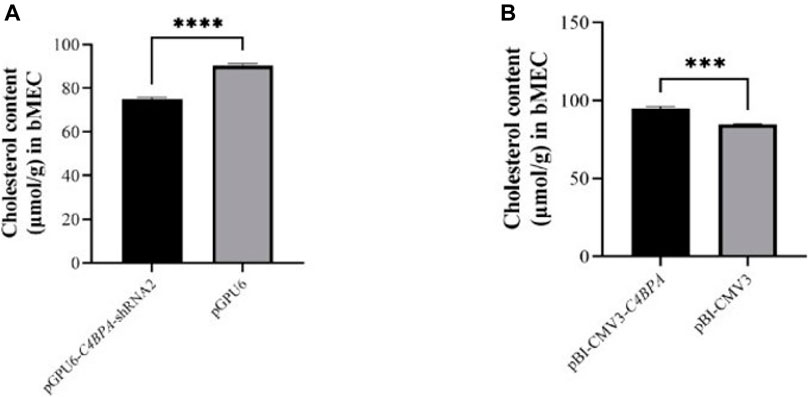
FIGURE 9. (A) The relative content of cholesterol in bMECs after transfection with pGPU6-C4BPA-shRNA2. (B) The relative content of cholesterol in bMECs after transfection with pBI-CMV3-C4BPA.
The Effect of C4BPA on the Content of Free Fatty Acids/NEFA in bMECs
The role of C4BPA in NEFA metabolism was investigated by the transfection of the pGPU6-C4BPA-shRNA2 and pBI-CMV3 C4BPA vectors in bMECs. After 24 h of successful transfection, the NEFA was analyzed. The results showed that when bMECs were transfected with pGPU6-C4BPA-shRNA2, the NEFA content was significantly upregulated (p < 0.05) in the pGPU6-C4BPA-shRNA2 group compared to pGPU6, which is shown in Figure 10A. The contrary result was obtained when bMECs were transfected with pBI-CMV3-C4BPA. In contrast, the NEFA content was significantly downregulated (p < 0.05) in pBI-CMV3-C4BPA compared to pBI-CMV3, as shown in Figure 10B. This result validated that C4BPA played a significant role in NEFA metabolism in bMECs.
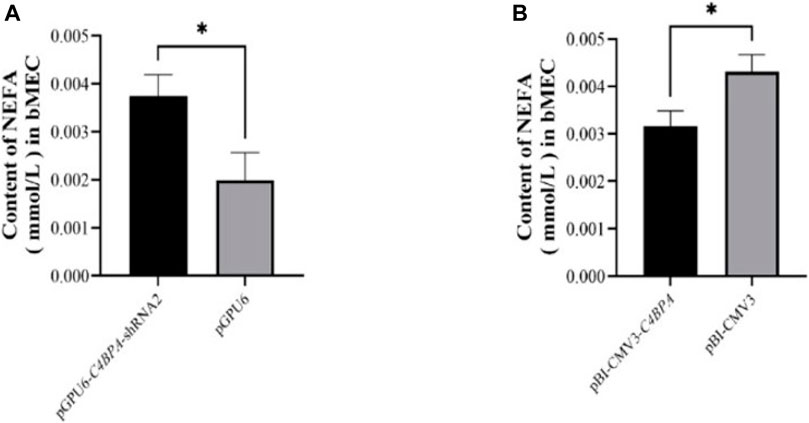
FIGURE 10. (A) The relative content of NEFA in bMECs after transfection with pGPU6-C4BPA-shRNA2. (B) The relative content of NEFA in bMECs after transfection with pBI-CMV3-C4BPA.
The Effect of C4BPA on the Relative mRNA Expression of Lipid Metabolism-Related Genes in bMECs
The mRNA expression of the lipid metabolism-related genes was investigated after 24 h of transfecting pGPU6-C4BPA-shRNA2 and pBI-CMV3-C4BPA of the vector in bMECs. The results showed that when bMECs were transfected with pGPU6-C4BPA-shRNA2, the genes that played a role in the lipid metabolism, including ELOVL6, FADS1, and LPL genes, were significantly upregulated in the pGPU6-C4BPA-shRNA2 group compared to pGPU6, as given in Figure 11A. The expression trend of ACADS in the pGPU6-C4BPA-shRNA2 group had no significant downregulation trend compared to pGPU6, which is shown in Figure 11A. The expression of ACSL1, PPARA, and FADS1 in the pGPU6-C4BPA-shRNA2 group was significantly downregulated compared to pGPU6, which is shown in Figure 11A. The contrary result was obtained when bMECs were transfected with pBI-CMV3-C4BPA. The genes involved in lipid metabolism, including the ACSL1, PPARA, and PPARG, were significantly upregulated in pBI-CMV3-C4BPA compared to pBI-CMV3 (Figure 11B). The expression trend of ACADS and ELOVL6 genes in the pBI-CMV3-C4BPA group had no significant upregulation trend compared to pBI-CMV3, which is shown in Figure 11B. Further, the expression of FADS1 and LPL in the pBI-CMV3-C4BPA group was significantly downregulated as compared to pBI-CMV3, as shown in Figure 11B. Overall, these results authenticated that C4BPA has a major impact on lipid metabolism-related genes and played a vital role in regulating these genes.
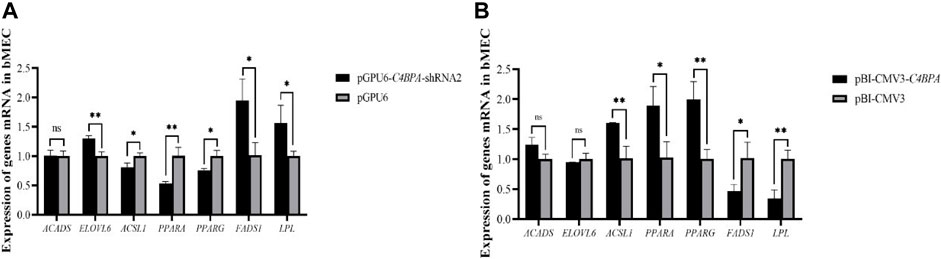
FIGURE 11. (A) Relative mRNA expression of lipid metabolism genes in bMECs after transfection with pGPU6-C4BPA-shRNA2. (B) Relative mRNA expression of lipid metabolism genes in bMECs after transfection with pBI-CMV3-C4BPA.
The Effect of C4BPA on Relative Protein Expression of Lipid Metabolism-Related Genes in bMECs
The protein expression of the lipid metabolism-related genes was investigated by the transfection of the vectors pGPU6-C4BPA-shRNA2 and pBI-CMV3-C4BPA in bMECs. After 24 h of successful transfection, the western blot was performed. The results showed that when bMECs were transfected with pGPU6-C4BPA-shRNA2, the genes that played a role in the lipid metabolism, including ELOVL6, FADS1, and LPL genes, were significantly upregulated in the pGPU6-C4BPA-shRNA2 group compared to pGPU6, which is shown in Figure 12A. The protein expression of ACSL1, PPARA, and PPARG in the pGPU6-C4BPA-shRNA2 group was significantly downregulated compared to pGPU6, as shown in Figure 12A. The contrary result was obtained when the bMECs were transfected with pBI-CMV3-C4BPA. The protein expression of genes involved in the lipid metabolism, including ACSL1 and PPARA, was significantly upregulated in pBI-CMV3-C4BPA compared to the pBI-CMV3, as shown in Figure 12B. In contrast, the expression trend of ELOVL6 and PPARG genes in the pBI-CMV3-C4BPA group had no significant upregulation trend compared to pBI-CMV3, as shown in Figure 12B. Contrarily, the FADS1 and LPL protein expression in the pBI-CMV3-C4BPA group was significantly downregulated compared to pBI-CMV3, which is shown in Figure 12B. Together, these results validated that the expression of C4BPA regulated the expression of the lipid metabolism-related genes.
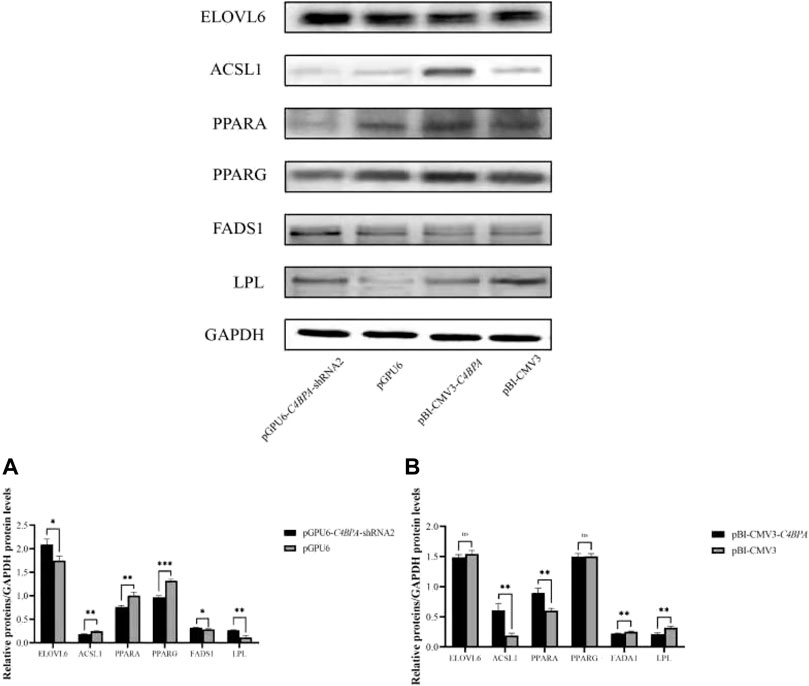
FIGURE 12. Relative protein expression of the lipid metabolism-related genes in bMECs. (A) Relative protein expression of lipid metabolism-related genes in bMECs after transfection with pGPU6-C4BPA-shRNA2. (B) Relative protein expression of lipid metabolism-related genes in bMECs after transfection with pBI-CMV3-C4BPA.
Discussion
The rapid rate of increasing human population demands an ample supply of good quality milk. One way to achieve that is to breed the dairy cows based on selecting those genes that increase milk fat and inhibit mastitis. Mastitis confreres the massive shortage of milk production and the economic loss to the dairy sector, which hinders the development of the dairy industry. Thus, cows with high-fat milk production and higher resistance to mastitis should be selected based on the molecular marker genes that play dual roles in the immunity and fat metabolism in bMECs (Waseem Ghani et al., 2019). Our previous transcriptome analysis between the high- and low-fat Chinese Holstein cattle showed that the C4BPA gene is differentially expressed with its higher expression in high-fat cattle. Therefore, in this study, we thoroughly explored the mechanism of the C4BPA gene in milk fat metabolism and mastitis in bMECs. The mammary epithelial cells can synthesize and secrete the milk components after the in vitro culture, so this study uses bMECs as a model (Jiang et al., 2020). The previous study reported that bMECs played a role in bacterial infection and acted as a key player in the anti-inflammatory response (He et al., 2017).
Role of C4BPA Gene in Inflammatory Factors and Immunity in bMECs
C4BPA is a part of the extracellular complement regulator C4B binding protein (C4BP). C4BPA is a candidate gene for anti-mastitis and high-fat milk. The complement system is the first-line defense system that plays a key role in functioning the innate immune effectors and coordinates against the foreign invaders (Olcina et al., 2018). In contrast, the previous work demonstrated that the role of complement is mostly in tumor progression and treatment. During mastitis caused by E. coli, approximately within 10–12 h, the strong inflammatory response began as a consequence of chemotactic neutrophil infiltration, complement components, tumor necrosis factor, and large amount of interleukin are produced (Shuster et al., 1997). For the further progression of the inflammation toll-like receptors (TLR), NF-κB signaling plays a vital role (Khan et al., 2020). The role of the complement system in mastitis is still not explored deeply. Therefore, to validate the role of the C4BPA gene in the inflammatory process, the overexpression and interfering vector of the C4BPA gene were transfected into bMECs. This showed that when pGPU6-C4BPA-shRNA2 was transferred into bMECs, the expression of TLR4 and the NF-κB pathway-related genes was significantly reduced. The contrary results were obtained when bMECs were treated with pBI-CMV3-C4BPA, in which the expression of TLR4 and the NF-κB pathway-related genes was significantly enhanced. In LPS-induced mastitis, increased levels of TNF-ɑ and IL-6 in the mammary gland were observed in a previous study (Ershun et al., 2014). This study reported that when the wild-type mice were treated with the lipopolysaccharides, TLR4, TRAF6, and NF-κB increased significantly. However, when TLR4 knockout mice were treated with lipopolysaccharides, the mRNA and protein expression of all the genes responsible for the activation of the NF-κB pathway remained the same as the control group (Ershun et al., 2014). With the advancement of technology, it becomes evident that the protein expression of TLR4 is responsible for the secretion of the inflammatory cytokines that mediate the NF-κB signaling pathway (Tian et al., 2019). The other study through HCT116 colorectal cancer cell immunoprecipitation validated the association between C4BPA and NF-κB pathway-related genes, which showed that C4BPA played a significant role in the regulation of NF-κB dependent apoptosis (Olcina et al., 2020). These studies validated that the C4BPA gene played a vital role in the regulation of NF-κB signaling pathway-related genes. Our results also showed that when bMECs were treated with pGPU6-C4BPA-shRNA2, the expression of IL-8 and IL-12 decreased.
Moreover, the reverse trend was found in the expression of TNF-ɑ and IL-10 when bMECs were transfected with pGPU6-C4BPA-shRNA2. However, the expression of the IL-6 gene was not changed when bMECs were transfected with pGPU6-C4BPA-shRNA2. On the contrary, when bMECs were treated with pBI-CMV3-C4BPA, the increased expression of IL-6, IL-8, and IL-12 was observed. In comparison, the expression of the TNF-ɑ was decreased in pBI-CMV3-C4BPA. In the early stages of infection, IL-6 is mainly categorized as the pro-inflammatory cytokine (Tilg, Dinarello, and Mier 1997). Other researchers also reported that IL-6 could inhibit the production of TNF-ɑ (Fiers 1991; Matthys et al., 1995). Similar to IL-6, the other inflammatory factors, IL-10 and IL-12, are the critical anti-inflammatory cytokines secreted by various cells (Oliveira et al., 2017; Zhu et al., 2017). Overall, the results of this study showed that the C4BPA gene through TLR4/NF-κB played a significant role in regulating the expression of all the major genes of immunity and inflammation factors associated with mastitis in dairy cattle.
Role of C4BPA Gene in Complement System in bMECs
The micro-organism is phagocytosed by the neutrophils in infected animals (Grewal, Rouse, and Babiuk 1978). This phagocytosis activity is regulated through the immunoglobulin G (IgG) and complement component 3 (C3). Despite playing an important role in phagocytosis, the complement system is also known as a non-specific humoral immune system that plays an essential role in initiating and controlling inflammation. A previous study reported that activating the alternative pathway (AP) causes more attraction and accumulation of the C3 during mastitis (Rainard and Poutrel 1995). We hypothesized that the C4BPA is a potential member of the cascade of complement and coagulation reactions. Our results indicated that when bMECs were transfected with pGPU6-C4BPA-shRNA2, the expression of the complement and coagulation factors C1S, C2, C3, and C3A was significantly upregulated. While the reverse result was obtained in the case of C4A1, in which the expression of C4A1 was significantly downregulated in pGPU6-C4BPA-shRNA2 treated bMECs. In contrast, when bMECs were treated with pBI-CMV3-C4BPA, the expression of the C1S, C2, C3, and C3A was significantly downregulated. Contrary results were obtained in the case of the C4A1 gene that was significantly upregulated in pBI-CMV3-C4BPA transfected bMECs. Therefore, our results highlight that C4BPA not only involves the regulation of the inflammatory factors through TLR4/NF-κB but also functions as a critical player in regulating the complement system.
Role of C4BPA Gene in Lipid Metabolism in bMECs
Development of the mammary glands and the onset and maintenance of lactation are incredibly complicated processes tightly regulated at multiple levels, including endocrine and molecular mechanisms (Sun et al., 2016). The lipid metabolism in bMECs is governed by many genes. Herein, we validate the role of the C4BPA gene in lipid metabolism. Our results indicated that when bMECs were transfected with pGPU6-C4BPA-shRNA2, the expression of ACSL1 was significantly downregulated while when bMECs were treated with pBI-CMV3-C4BPA, the expression of ACSL1 was significantly upregulated. A previous study reported that the overexpression of ACSL1 was related to the upregulation of the PPARG gene, which enhanced the synthesis of fatty acids and lipid droplets in bovine adipocytes (Zhao et al., 2020). Our results indicated that when bMECs were transfected with pGPU6-C4BPA-shRNA2, the expression of ACADS was not changed while when bMECs were treated with pBI-CMV3-C4BPA, the expression of ACADS was significantly upregulated. ACADS is a member of the acyl-CoA dehydrogenase family and catalyzes the mitochondrial fatty acid β-oxidation pathway (Dessein et al., 2017).
Further, our results indicated that when bMECs were transfected with pGPU6-C4BPA-shRNA2, the expression of FADS1 was significantly upregulated while in pBI-CMV3-C4BPA transfected with bMECs, the expression of FADS1 was significantly downregulated. The analysis of human plasma lipid profile suggested that the fatty acid desaturase 1 (FADS1) has a strong association with the regulation of the polyunsaturated fatty acids (PUFA) and long-chain polyunsaturated fatty acids (LC PUFA) in human blood (Xie and Innis 2008; Glaser, Heinrich, and Koletzko 2010). Our results indicated that when bMECs were transfected with pGPU6-C4BPA-shRNA2, the expression of ELOVL6 was significantly upregulated. When bMECs were treated with pBI-CMV3-C4BPA, the expression of ELOVL6 was significantly downregulated. The previous study reported that the elongation of very long-chain fatty acids protein 6 (ELOVL6) plays a pivotal role in the energy metabolism, insulin sensitivity, and elongation of the fatty acids (Matsuzaka and Shimano 2009).
In comparison, the previous work on the bovine adipocytes suggested that the overexpression of ELOVL6 had various effects on different fatty acid contents. The myristic acid (C14:0) and palmitic acid (C16:0) fatty acids in adipocytes decreased in the bovine adipocytes with the overexpression of ELOVL6. At the same time, the opposite result was obtained when the bovine adipocytes were treated with the overexpression of ELOVL6, and the ratio of the stearic acid (C18:0) and arachidonic acid (C20:4n6) fatty acids increased. Therefore, we speculate that C4BPA might regulate the fatty acid content in bMECs by regulating the gene expression of FADS1 and ELOVL6. In order to authenticate the role of ELOVL6, our results indicated that when bMECs were transfected with pGPU6-C4BPA-shRNA2, the expression of ELOVL6 was significantly upregulated. In contrast, when bMECs were treated with pBI-CMV3-C4BPA, the expression of ELOVL6 was significantly downregulated. Our results also indicated that when bMECs were transfected with pGPU6-C4BPA-shRNA2, LPL was significantly upregulated while when bMECs were treated with pBI-CMV3-C4BPA, the LPL expression was significantly downregulated. Lipoprotein lipase (LPL) played an essential role in controlling plasma triglyceride levels. The LPL, through irretrievable hydrolysis of triglycerides, enhanced the removal of the triglyceride-rich lipoprotein in the plasma (Cheung et al., 2003). Another study reported that the overexpression of ANGPTL4 in mice severely reduced the LPL-dependent plasma TG and cholesterol clearance and enhanced the cholesterol synthesis rate (Lichtenstein et al., 2007). Our results are in line with this study, and thus, we report that C4BPA, through regulating the expression of LPL, along with other lipid metabolism-related genes, manipulated the cholesterol content in bMECs.
The peroxisome proliferator-activated receptor (PPAR) family consisted of peroxisome proliferator-activated receptor alpha (PPARA), peroxisome proliferator-activated receptor beta (PPARB), and peroxisome proliferator-activated receptor gamma (PPARG). The peroxisome proliferator-activated receptor members had different distributions and functions in different tissues (Giorgino et al., 2009). The PPARG is a marker gene that played a significant role in early fat differentiation (Gavrilova et al., 2003) (Raza et al., 2021). Our results indicated that when bMECs were transfected with pGPU6-C4BPA-shRNA2, the expression of PPARA and PPARG was significantly downregulated while when bMECs were treated with pBI-CMV3-C4BPA, the expressions of PPARA and PPARG were significantly upregulated.
Triglycerides, cholesterol, and fatty acids are the main components of milk fat, which act as important nutrients in the organism for the synthesis of cellular components and providing essential energy (Lock and Bauman 2004). Our results indicated that the knockdown of C4BPA in bMECs resulted in significantly lower contents of triglycerides, and cholesterol while the content of the NEFA was up-regulated. In contrast, the overexpression of C4BPA in bMECs culminated with significantly higher contents of triglycerides, cholesterol and lower content of NEFA. Altogether, these results showed that the C4BPA gene is a major player in regulating the lipid metabolism of bMECs. This study provides the primary basis for the role of the C4BPA gene in lipid metabolism and immunity, but an in-depth analysis is needed for properly understanding the mechanism. This study aims to attract the researchers’ attention to focus on such type of genes that can cross-talk and affect the more important trait together. This work raises many questions, which should be considered for further extrapolation. For example, can we increase the fat milk metabolism for producing milk with high quantity of good quality fat without compromising udder health or even increasing the resistance to mastitis? Which type of association between milk fat synthesis and mastitis is present?
Conclusion
The C4BPA gene involves the synthesis of triglycerides, total cholesterol, and free fatty acids acid in bMECs through regulating the expression of lipid metabolism-related genes. Additionally, the C4BPA gene is a candidate gene that plays a significant role in immunity by targeting TLR4/NF-κB and the factors involved in the complement and coagulation cascade pathways. This study reveals the dual functions of the C4BPA gene in bMECs in lipid metabolism and immunity. Our results provide the primary basis for understanding the role of C4BPA in mastitis and fat metabolism, which enables the researchers to follow the innovative direction of investigating genes associated with fat metabolism and mastitis. This research work provides crucial information for breeding cows with supreme milk quality and significant resistance to mastitis.
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors without undue reservation.
Author Contributions
AI conceived the idea, experimented and analyzed the data, and drafted the manuscript. PZ and HY helped with data collection and analysis. LJ, WH, and FJ participated in the cell transfection and plasmid extraction. ZZ and JP helped to procure the materials and supervised and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (nos. 32072717, 32002165, and 31772562) and the Key Platform Project of Innovative Strong School Engineering by the Department of Education of Guangdong Province (2018302).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Antoniades, C., Bakogiannis, C., Tousoulis, D., Antonopoulos, A. S., and Stefanadis, C. (2009). The CD40/CD40 Ligand System. J. Am. Coll. Cardiol. 54 (8), 669–677. doi:10.1016/j.jacc.2009.03.076
Brodeur, S. R., AngeliniBlom, F., Bacharier, L. B., Blom, A. M., Mizoguchi, E., Fujiwara, H., et al. (2003). C4b-Binding Protein (C4BP) Activates B Cells through the CD40 Receptor. Immunity 18 (6), 837–848. doi:10.1016/S1074-7613(03)00149-3
Cheung, M. C., Sibley, S. D., Palmer, J. P., Oram, J. F., and Brunzell, J. D. (2003). Lipoprotein Lipase and Hepatic Lipase. J. Lipid Res. 44 (8), 1552–1558. doi:10.1194/jlr.M300091-JLR200
Dessein, A.-F., Fontaine, M., Joncquel-Chevalier Curt, M., Briand, G., Sechter, C., Mention-Mulliez, K., et al. (2017). Fluxomic Evidence for Impaired Contribution of Short-Chain Acyl-CoA Dehydrogenase to Mitochondrial Palmitate β-oxidation in Symptomatic Patients with ACADS Gene Susceptibility Variants. Clinica Chim. Acta 471 (5), 101–106. doi:10.1016/j.cca.2017.05.026
Ershun, Z., Yunhe, F., Zhengkai, W., Yongguo, C., Naisheng, Z., and Zhengtao, Y. (2014). Cepharanthine Attenuates Lipopolysaccharide-Induced Mice Mastitis by Suppressing the NF-Κb Signaling Pathway. Inflammation 37 (2), 331–337. doi:10.1007/s10753-013-9744-6
Fiers, W. (1991). Tumor Necrosis Factor Characterization at the Molecular, Cellular and In Vivo Level. FEBS Lett. 285 (2), 199–212. doi:10.1016/0014-5793(91)80803-B
Fu, Y., Zhou, E., Liu, Z., Li, F., Liang, D., Liu, B., et al. (2013). Staphylococcus Aureus and Escherichia Coli Elicit Different Innate Immune Responses from Bovine Mammary Epithelial Cells. Vet. Immunol. Immunopathology 155 (4), 245–252. doi:10.1016/j.vetimm.2013.08.003
Gavrilova, O., Haluzik, M., Matsusue, K., Cutson, J. J., Johnson, L., Dietz, K. R., et al. (2003). Liver Peroxisome Proliferator-Activated Receptor γ Contributes to Hepatic Steatosis, Triglyceride Clearance, and Regulation of Body Fat Mass. J. Biol. Chem. 278 (36), 34268–34276. doi:10.1074/jbc.M300043200
Gerosa, S., and Skoet, J. (2012). Milk Availability: Trends in Production and Demand and Medium-Term Outlook, 12, 1–40. ESA, FAO, Rome.
Glaser, C., Heinrich, J., and Koletzko, B. (2010). Role of FADS1 and FADS2 Polymorphisms in Polyunsaturated Fatty Acid Metabolism. Metabolism 59 (7), 993–999. doi:10.1016/j.metabol.2009.10.022
Grewal, A. S., Rouse, B. T., and Babiuk, L. A. (1978). Characterization of Surface Receptors on Bovine Leukocytes. Int. Arch. Allergy Immunol. 56 (4), 289–300. doi:10.1159/000232034
Harada, A., Sekido, N., Akahoshi, T., Wada, T., Mukaida, N., and Matsushima, K. (1994). Essential Involvement of Interleukin-8 (IL-8) in Acute Inflammation. J. Leukoc. Biol. 56 (5), 559–564. doi:10.1002/jlb.56.5.559
He, X., Liu, W., Shi, M., Yang, Z., Zhang, X., and Gong, P. (2017). Docosahexaenoic Acid Attenuates LPS-Stimulated Inflammatory Response by Regulating the PPARγ/NF-Κb Pathways in Primary Bovine Mammary Epithelial Cells. Res. Vet. Sci. 112 (12), 7–12. doi:10.1016/j.rvsc.2016.12.011
Jiang, P., Fang, X., Zhao, Z., Yu, X., Sun, B., Yu, H., et al. (2018). The Effect of Short/Branched Chain Acyl-Coenzyme A Dehydrogenase Gene on Triglyceride Synthesis of Bovine Mammary Epithelial Cells. Arch. Anim. Breed. 61 (1), 115–122. doi:10.5194/aab-61-115-2018
Jiang, P., Xia, L., Jin, Z., Ali, S., Wang, M., Li, X., et al. (2020). New Function of the CD44 Gene: Lipid Metabolism Regulation in Bovine Mammary Epithelial Cells. J. Dairy Sci. 103 (7), 6661–6671. doi:10.3168/jds.2019-17415
Khan, M. Z., Khan, A., Xiao, J., Ma, J., Ma, Y., Chen, T., et al. (2020). Overview of Research Development on the Role of NF-Κb Signaling in Mastitis. Animals 10 (9), 1625. doi:10.3390/ani10091625
Leonard, A. E., Pereira, S. L., Sprecher, H., and Huang, Y. S. (2004). Elongation of Long-Chain Fatty Acids. Prog. Lipid Res. 43, 36–54. doi:10.1016/S0163-7827(03)00040-7
Leonardini, A., Laviola, L., Perrini, S., Natalicchio, A., and Giorgino, F. (2009). Cross-Talk between PPARγand Insulin Signaling and Modulation of Insulin Sensitivity. PPAR Res. 2009 (10), 1–12. doi:10.1155/2009/818945
Lichtenstein, L., Berbée, J. F. P., van Dijk, S. J., van Dijk, K. W., Bensadoun, A., Kema, I. P., et al. (2007). Angptl4 Upregulates Cholesterol Synthesis in Liver via Inhibition of LPL- and HL-dependent Hepatic Cholesterol Uptake. Atvb 27 (11), 2420–2427. doi:10.1161/ATVBAHA.107.151894
Lock, A. L., and Bauman, D. E. (2004). Modifying Milk Fat Composition of Dairy Cows to Enhance Fatty Acids Beneficial to Human Health. Lipids 39 (12), 1197–1206. doi:10.1007/s11745-004-1348-6
Lu, C., Yang., R., Liu, B., Li, Z., Shen, B., Yan, S., et al. (2012). Establishment of Two Types of Mammary Epithelial Cell Lines from Chinese Holstein Dairy Cow. J. Anim. Vet. Adv. 11 (8), 1166–1172. doi:10.3923/javaa.2012.1166.1172
Martin, P., Barkema, H. W., Brito, L. F., Narayana, S. G., and Miglior, F. (2018). Symposium Review: Novel Strategies to Genetically Improve Mastitis Resistance in Dairy Cattle. J. Dairy Sci. 101 (3), 2724–2736. doi:10.3168/jds.2017-13554
Matsuzaka, T., and Shimano, H. (2009). Elovl6: A New Player in Fatty Acid Metabolism and Insulin Sensitivity. J. Mol. Med. 87 (4), 379–384. doi:10.1007/s00109-009-0449-0
Matthys, P., Mitera, T., Heremans, H., Van Damme, J., and Billiau, A. (1995). Anti-Gamma Interferon and Anti-interleukin-6 Antibodies Affect Staphylococcal Enterotoxin B-Induced Weight Loss, Hypoglycemia, and Cytokine Release in D-Galactosamine-Sensitized and Unsensitized Mice. Infect. Immun. 63 (4), 1158–1164. doi:10.1128/iai.63.4.1158-1164.1995
Mitterhuemer, S., Petzl, W., Krebs, S., Mehne, D., Klanner, A., Wolf, E., et al. (2010). Escherichia Coli Infection Induces Distinct Local and Systemic Transcriptome Responses in the Mammary Gland. BMC Genomics 11 (1), 1471–2164. doi:10.1186/1471-2164-11-138
Moffat, G. J., Vik, D. P., Noack, D., and Tack, B. F. (1992). Complete Structure of the Murine C4b-Binding Protein Gene and Regulation of its Expression by Dexamethasone. J. Biol. Chem. 267 (28), 20400–20406. doi:10.1016/s0021-9258(19)88715-x
Olcina, M. M., Balanis, N. G., Kim, R. K., Aksoy, B. A., Kodysh, J., Thompson, M. J., et al. (2018). Mutations in an Innate Immunity Pathway Are Associated with Poor Overall Survival Outcomes and Hypoxic Signaling in Cancer. Cel Rep. 25 (13), 3721–3732. e6. doi:10.1016/j.celrep.2018.11.093
Olcina, M. M., Kim, R. K., Balanis, N. G., Li, C. G., von Eyben, R., Graeber, T. G., et al. (2020). Intracellular C4BPA Levels Regulate NF-κb-dependent Apoptosis. IScience 23 (10), 101594. doi:10.1016/j.isci.2020.101594
Oliveira, A. B., Bachi, A. L. L., Ribeiro, R. T., Mello, M. T., Tufik, S., and Peres, M. F. P. (2017). Unbalanced Plasma TNF-α and IL-12/IL-10 Profile in Women with Migraine Is Associated with Psychological and Physiological Outcomes. J. Neuroimmunology 313 (May), 138–144. doi:10.1016/j.jneuroim.2017.09.008
Rainard, P., and Poutrel, B. (1995). Deposition of Complement Components on Streptococcus Agalactiae in Bovine Milk in the Absence of Inflammation. Infect. Immun. 63 (9), 3422–3427. doi:10.1128/iai.63.9.3422-3427.1995
Raza, S. H. A., Khan, R., Cheng, G., Long, F., Bing, S., Easa, A. A., et al. (2022). RNA-seq Reveals the Potential Molecular Mechanisms of Bovine KLF6 Gene in the Regulation of Adipogenesis. Int. J. Biol. Macromolecules 195 (22), 198–206. doi:10.1016/j.ijbiomac.2021.11.202
Ridker, P. M., Silvertown, J. D., and JoshSilvertown, D. (2008). Inflammation, C-Reactive Protein, and Atherothrombosis. J. Periodontol. 79 (8s), 1544–1551. doi:10.1902/jop.2008.080249
Scharfstein, J., Ferreira, A., Gigli, I., and Nussenzweig, V. (1978). Human C4-Binding Protein. I. Isolation and Characterization. J. Exp. Med. 148, 207–222. doi:10.1084/jem.148.1.207
Shuster, D. E., Kehrli, M. E., Rainard, P., and Paape, M. (1997). Complement Fragment C5a and Inflammatory Cytokines in Neutrophil Recruitment during Intramammary Infection with Escherichia Coli. Infect. Immun. 65 (8), 3286–3292. doi:10.1128/iai.65.8.3286-3292.1997
Sogawa, K., Takano, S., Iida, F., Satoh, M., Tsuchida, S., Kawashima, Y., et al. (2016). Identification of a Novel Serum Biomarker for Pancreatic Cancer, C4b-Binding Protein α-chain (C4BPA) by Quantitative Proteomic Analysis Using Tandem Mass Tags. Br. J. Cancer 115 (8), 949–956. doi:10.1038/bjc.2016.295
Strandberg, Y., Gray, C., Vuocolo, T., Donaldson, L., Broadway, M., and Tellam, R. (2005). Lipopolysaccharide and Lipoteichoic Acid Induce Different Innate Immune Responses in Bovine Mammary Epithelial Cells. Cytokine 31 (1), 72–86. doi:10.1016/j.cyto.2005.02.010
Suankratay, C., Mold, C., Zhang, Y., Lint, T. F., and Gewurz, Gewurz. (1999). Mechanism of Complement-dependent Haemolysis via the Lectin Pathway: Role of the Complement Regulatory Proteins. Clin. Exp. Immunol. 117 (3), 442–448. doi:10.1046/j.1365-2249.1999.00998.x
Sun, X., Lin, J., Zhang, Y., Kang, S., Belkin, N., Wara, A. K., et al. (2016). MicroRNA-181b Improves Glucose Homeostasis and Insulin Sensitivity by Regulating Endothelial Function in White Adipose Tissue. Circ. Res. 118 (5), 810–821. doi:10.1161/CIRCRESAHA.115.308166
Tian, H., Liu, Z., Pu, Y., and Bao, Y. (2019). Immunomodulatory Effects Exerted by Poria Cocos Polysaccharides via TLR4/TRAF6/NF-Κb Signaling In Vitro and In Vivo. Biomed. Pharmacother. 112 (76), 108709. doi:10.1016/j.biopha.2019.108709
Tilg, H., Dinarello, C. A., and Mier, J. W. (1997). IL-6 and APPs: Anti-inflammatory and Immunosuppressive Mediators. Immunol. Today 18 (9), 428–432. doi:10.1016/S0167-5699(97)01103-1
Van Winckel, M., Vande Velde, S., De Bruyne, R., and Van Biervliet, S. (2011). Clinical Practice. Eur. J. Pediatr. 170 (12), 1489–1494. doi:10.1007/s00431-011-1547-x
Waseem Ghani, M., Bin, L., Waseem Birmani, M., Nawab, A., Guan Cun, L. Cun., Ye, L., et al. (2019). Impact of Subclinical Mastitis on Reproductive Performance of Dairy Animals. Int. J. Vet. Sci. Res. 5 (2), 48–57. doi:10.18488/journal.110.2019.52.48.57
Westein, E., Denis, C. V., Bouma, B. N., and Lenting, P. J. (2002). The α-Chains of C4b-Binding Protein Mediate Complex Formation with Low Density Lipoprotein Receptor-Related Protein. J. Biol. Chem. 277 (4), 2511–2516. doi:10.1074/jbc.M102293200
Xia, L., Zhao, R. Yang. Z., Fang, X., and Lu, C. (2021). Conjoint Analysis of Transcriptome and MicroRNA of Mammary Epithelial Cells of Chinese Holstein Cow. J. Anim. Sci. 96 (Suppl. l_3), 134. doi:10.1093/jas/sky404.294
Xie, L., and Innis, S. M. (2008). Genetic Variants of the FADS1 FADS2 Gene Cluster Are Associated with Altered (N-6) and (N-3) Essential Fatty Acids in Plasma and Erythrocyte Phospholipids in Women during Pregnancy and in Breast Milk during Lactation. J. Nutr. 138 (11), 2222–2228. doi:10.3945/jn.108.096156
Zhao, Z., Abbas Raza, S. H., Tian, H., Shi, B., Luo, Y., Wang, J., et al. (2020). Effects of Overexpression of ACSL1 Gene on the Synthesis of Unsaturated Fatty Acids in Adipocytes of Bovine. Arch. Biochem. Biophys. 695 (October), 108648. doi:10.1016/j.abb.2020.108648
Keywords: C4BPA, bMECs, TLR-4/NF-κB, complement, fat metabolism
Citation: Iqbal A, Ziyi P, Yu H, Jialing L, Haochen W, Jing F, Ping J and Zhihui Z (2022) C4BPA: A Novel Co-Regulator of Immunity and Fat Metabolism in the Bovine Mammary Epithelial Cells. Front. Genet. 12:830566. doi: 10.3389/fgene.2021.830566
Received: 07 December 2021; Accepted: 23 December 2021;
Published: 31 January 2022.
Edited by:
Zhi Chen, Yangzhou University, ChinaReviewed by:
Xianzhong Yu, Clemson University, United StatesSayed Haidar Abbas Raza, Northwest A&F University, China
Copyright © 2022 Iqbal, Ziyi, Yu, Jialing, Haochen, Jing, Ping and Zhihui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiang Ping, amlhbmdwQGdkb3UuZWR1LmNu; Zhao Zhihui, emh6aGFvQGdkb3UuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Ambreen Iqbal
Ambreen Iqbal Pan Ziyi2†
Pan Ziyi2† Haibin Yu
Haibin Yu Jiang Ping
Jiang Ping