- 1Uniklinik Köln, Köln, Germany
- 2Max-Plank Institute for Evolutionary Biology, Plön, Germany
- 3Deutsches Zentrum für Neurodegenerative Erkrankungen e. V. (DZNE), Bonn, Germany
Map2k7 (synonym Mkk7) is a conserved regulatory kinase gene and a central component of the JNK signaling cascade with key functions during cellular differentiation. It shows complex transcription patterns, and different transcript isoforms are known in the mouse (Mus musculus). We have previously identified a newly evolved testis-specific transcript for the Map2k7 gene in the subspecies M. m. domesticus. Here, we identify the new promoter that drives this transcript and find that it codes for an open reading frame (ORF) of 50 amino acids. The new promoter was gained in the stem lineage of closely related mouse species but was secondarily lost in the subspecies M. m. musculus and M. m. castaneus. A single mutation can be correlated with its transcriptional activity in M. m. domesticus, and cell culture assays demonstrate the capability of this mutation to drive expression. A mouse knockout line in which the promoter region of the new transcript is deleted reveals a functional contribution of the newly evolved promoter to sperm motility and the spermatid transcriptome. Our data show that a new functional transcript (and possibly protein) can evolve within an otherwise highly conserved gene, supporting the notion of regulatory changes contributing to the emergence of evolutionary novelties.
Introduction
Mitogen activated protein kinase (MAPK) pathways are highly conserved throughout eukaryotes and trigger multistep signaling cascades mediating transcriptional response upon receiving outside stimuli (English et al., 1999; Chang and Karin, 2001). Map2k7 belongs to the JNK group of kinases and acts as its specific activator (Holland et al., 1997; Tournier et al., 1997; Fleming et al., 2000; Tournier et al., 2001; Kishimoto et al., 2003; Takekawa et al., 2005; Wang et al., 2007). Cellular stresses like UV and gamma irradiation, osmotic shock and drug treatments on the one hand and different inflammatory cytokines, such as tumor necrosis factor, interleukin-1 or interleukin-3 on the other hand, lead to JNK pathway activation (Moriguchi et al., 1997; Foltz et al., 1998; Chang and Karin, 2001; Nishina et al., 2004). Downstream targets of JNK include transcription factors (Yang et al., 2003) as well as other proteins, for example microtubule-associated proteins (Chang et al., 2003) and members of the Bcl2 family (Lei et al., 2002; Deng et al., 2003). The JNK pathway has several known functions in the immune system, apoptosis, and developmental processes (Sabapathy et al., 1999; Dong et al., 2000; Nishina et al., 2004; Wada et al., 2004; Wang et al., 2007). Knock out of both JNK1 and JNK2 (double KO) or of Mapk2k7 alone, leads to embryonic lethality in mice (Wang et al., 2007).
In their report on the first identification of Map2k7 in mice, Tournier and others (Tournier et al., 1997) showed by Northern blotting the expression of a long transcript in various organs, plus an additional shorter transcript specific to the testis. While all the further studies concentrated on the longer transcript, the origin and function of the shorter transcript was not further investigated. In a systematic study for differentially expressed genes in mouse populations, we identified Map2k7 to be differentially expressed in testis between wild populations of M. m. domesticus and M. m. musculus, with strongly elevated expression in M. m. domesticus (Harr et al., 2006). A cis-trans test via allele-specific expression analysis in F1 hybrids of both subspecies demonstrated that a cis-acting sequence causes the expression change. It turned out that the elevated expression level can be correlated with the additional testis-specific transcript in the M. m. domesticus subspecies, which is absent in M. m. musculus. This suggested that a new testis-specific promoter had evolved within the Map2k7 gene (Harr et al., 2006). Given the highly conserved nature of the Map2k7 gene, such an evolution of a strong new promoter is of particular interest. We present here comparative and functional data that allow inferences on the evolutionary history of the new promoter, which includes both a new origination event and a secondary loss event triggered by a single mutation in one of the subspecies. The knockout analysis proves that the new promoter has assumed a new function in spermatids’ maturation and the transcriptome’s regulation during this phase.
Materials and Methods
In situ Hybridization and Northern Blotting
In situ detection of Map2k7 RNA was performed by hybridization with a digoxigenin (DIG) labeled probe (Tautz and Pfeifle, 1989). For probe generation, a fragment spanning 1197bp of Map2k7 exons 5-10 was amplified from testis C57Bl6 cDNA with primers P49 and P50 and cloned into a PCR cloning vector. The DNA fragment was reamplified from a pure plasmid clone. Reverse transcription to generate a DIG labeled probe was set up by adding 200 ng of purified PCR product to 2 μl DIG RNA Labeling Mix, 2 μl transcription buffer, 2 μl T7 polymerase, 0.5 μl RNase inhibitor (Roche, Basel). Pure water was added to the reaction mix to obtain a final volume of 20 μl. The reaction mix was incubated for 2 h at 37°C followed by a treatment with 1 μl Turbo DNAse for 15 min at 37°C to remove the DNA template. The probe was precipitated with salt and alcohol, washed and re-suspended in 40 μl of 50% formamide diluted in nuclease free water (Applied Biosystems/Ambion, Austin).
All buffers and tools that were used for the following procedure were kept RNAse free. Paraffinized sections were dewaxed in xylene for 2 × 10 min, washed for 5 min in ethanol, rehydrated in a series of decreasing ethanol concentration (95%, 90%, 70%, 30%; 3 min each) and washed for 5 min in PBS before postfixing them for 1 h in 4% PFA. After postfixation the tissue was washed in PBS for 2 × 5 min and partially digested with 10 μg/ml proteinase K in 100 mM Tris-HCl pH 7.5 for 10 min at 37°C.
Digestion was stopped with 0.2% glycine in PBS. 2 × 5 min washing in PBS was followed by 15 min incubation in 0.1 N HCl and another 2 × 5 min washing in PBS was performed previous to blocking of positively charged amino acids by 0.25% acetic anhydride in 0.1 M triethanolamine pH 8.0 for 10 min. Afterwards slides were washed for 5 min in PBS and for 5 min in pure water before prehybridization for 2 h at 65°C (50% formamide, 5x SSC, 1x Denhardt’s, 0.1% Tween-20). 1 μl of DIG labeled probe was diluted in 100 μl prehybridization buffer containing 400 ng tRNA (Sigma-Aldrich, St. Louis) and denatured at 70°C for 5 min. The hybridization mix was applied to the sections and covered with coverslips. Slides were incubated overnight at 65°C in a moist chamber. The next day, the sections were washed in 50% formamide containing 5x SSC and 1% SDS at 70°C for 30 min and subsequently with 50% formamide containing 2x SSC and 0.2% SDS for another 30 min at 65°C. Afterwards the sections were washed for 3 × 5 min in MABS (100 mM maleic acid, 150 mM NaCl, 0.1% Tween-20, and 2 mM levamisole; adjusted to pH 7.5 with NaOH). Samples were blocked with 1% blocking reagent (Roche, Basel) in MABS. Anti-DIG-AP antibody was applied in 1% blocking reagent in MABS by overnight incubation at 4°C. The next day, the sections were first washed 3 × 10 min and then 3 × 30 min in MABS. Subsequently, pH was adjusted by incubating for 3 × 10 min in NTMLT buffer (100 mM Tris-HCl pH 9.5, 50 mM MgCl2, 100 mM NaCl, 100 mM levamisole, 0.1% Tween-20). BM purple solution (Roche, Basel) was applied as a substrate for the alkaline phosphatase coupled with the anti-DIG antibody. The tissue was stained until the desired degree of the signal was observed. Slides were washed for 1 min in water and mounted with Kaisers glycerin-gelatine.
Detection of RNA in Northern Blotting (Alwine et al., 1977) was performed with radioactively labeled probes generated from the same clone used for in situ hybridization. Probes were labeled with 32P-dCTP (Hartmann Analytic, Braunschweig) using the Rediprime II DNA Labeling Kit (GE Healthcare Life Science, Little Chalfont) according to the manufacturer’s manual. Labeled probes were cleaned up with MicoSpin S-200 HR columns (GE Healthcare Life Science, Little Chalfont) according to the manufacturer’s manual.
10 μg of total RNA per sample were diluted in 15 μl nuclease-free pure water (Applied Biosystems/Ambion, Austin) and mixed with 10 μl sample buffer (50% formamide, 5.18% formaldehyde, 2.5x MOPS, 0.1 mg/ml ethidium bromide and 2.5x blue marker). Samples were heat-denatured for 5 min at 70°C and separated on an agarose gel (1.2% agarose, 0.666% formaldehyde, 1x MOPS). The RNA lanes were blotted through a classic upward blot onto an Amersham Hybond N+ membrane (GE Healthcare Life Science, Little Chalfont) by neutral transfer (20x SSC) overnight. Membranes were baked for 2 h at 80°C and prehybridized in ExpressHyb (Clontech, Mountain View) at 65°C for 1 h. A radioactively labeled probe was added to the prehybridized blot and hybridization took place overnight at 65°C in a rotating oven. The next day, the blots were washed 10–40 min in 2x SSC containing 0.05% SDS at room temperature and subsequently washed for 5–30 min with 0.1x SSC containing 0.1% SDS at 50°C. After washing, the blots were dipped in 2x SSC, sealed in a plastic bag, and analyzed via autoradiography using Kodak Biomax-MS films (Kodak, Rochester).
Promoter Tests in Cell Culture
The promoter tests in cell culture did require to set up an appropriate expression system. The details on the construction and testing of this system are described in (Heinen, 2008). It resulted in the construction of a “Luciflip plasmid” that contains the following elements in the given order: PGK-promoter, ATG, FRT, splice acceptor, double polyA signal, EcoRI site, Kozak, Luc-MYC, intron, polyA signal and was the basis for the Map2k7 alpha reporter assay (compare Supplementary Figure S1 for further details on this plasmid). For this, fragments spanning −487 to +43 relative to the transcription start of the Map2k7 alpha promoter (see annotation of the sequence in Supplementary Figure S2) were amplified from genomic DNA of M. m. musculus and M. m. domesticus using the primer pair P318/P319. A two-step PCR strategy was pursued to generate fragments with a deleted insulator motif. Two separate PCRs with the primer pairs P318/P320 and P321/P319 were run on top of the cloned promoter fragment. The primers P319 and P320 bind just right upstream and downstream of the insulator sequence and were tailed with a sequence stretch that is homologous to the sequence on the opposite part exactly beyond the insulator. The other primer is one of the primers that were used in the first PCR. Thus, the promoter fragment is divided into two fragments, each defined by an inner and an outer primer. The inner edges overlapped but lacked the insulator. Both PCR products were cleaned up and included in another PCR without primers. After five cycles, the outer primers P318 and P319 were added to the reaction, and PCR continued as usual. The resulting product was cloned into a PCR cloning vector and validated by Sanger sequencing with M13 primers.
The second version of all four fragments has an additional upstream CMV enhancer. For this, the CMV enhancer was amplified with the primers P322 and P323 using the phrGFPII-1 plasmid (Agilent Technologies, Santa Clara) as a template. The resulting product was cloned into a PCR cloning vector and validated by Sanger sequencing with M13 primers. Both CMV primers and the upstream primer used for promoter fragment generation (P318) were tailed with an XhoI restriction site overhang. CMV enhancer fragments were retrieved by XhoI digestion and ligated into the XhoI site of all 4 Map2k7 promoter fragments. The assembled fragments were cut out by EcoRI digestion and cloned into the EcoRI site of Luciflip. The orientation of the inserts was controlled with an XhoI digest. All 8 Luci-flip constructs were sequenced with the primers P293, P358, and P199 to control the inserts. No mutations were found.
The 8 expression constructs were transfected into NIH/3T3 cells. For this, NIH/3T3 fibroblast cells were grown in DMEM medium containing sodium pyruvate, non-essential amino acids, L-glutamine penicillin/streptomycin (all from Invitrogen, Carlsbad) and 10% fetal calf serum (PAN, Aidenbach) at 37°C incubation maintaining 5% CO2 concentration. One day before transfection, 3 × 103 NIH/3T3 cells were seeded with 70 μl medium into each well of 96-well plates and grown overnight. Cells were co-transfected with Luciflip plasmid and a pGL3 plasmid (Promega, Mannheim) which contains firefly luciferase under the control of SV40 promoter. Firefly luciferase was used to normalize transfection efficiency. Therefore, 0.18 μl Fugene 6 reagent (Roche, Basel) was added to 4.82 μl serum-free medium and incubated for 5 min at room temperature. Subsequently, 30 ng of Luciflip and 30 ng of pGL3 DNA were mixed and added to the medium containing Fugene 6. The mixture was incubated for 20 min and added to one well of the 96 well plate containing NIH/3T3. The transfection was performed for the Luciflip-CMV construct and for an empty Luciflip plasmid as blank control. Each transfection was performed in 8 replicates in parallel. Cells were incubated overnight. The next day, firefly and renilla luciferase substrates were applied using the Dual-Glo Luciferase Assay System (Promega, Mannheim) according to the manufacturer’s manual (the corresponding reagent was added directly to the cells and induced cell lysis). Relative light units were measured with a Mithras LB 940 Luminometer (Berthold Technologies, Bad Wildbad). The renilla luciferase signal was divided by the firefly luciferase signal to normalize transfection efficiency for every well. For every construct, median and standard deviation were calculated from the 8 individual replicates.
Construction of Knockout Mice
The general scheme for the construction of the promoter knockout is shown in Supplementary Figure S2. It consisted of two steps. In the first step, in neomycin cassette was inserted at position chr8:4,239,573 (mm10) via homologous recombination in embryonic stem cells, whereby 593 bp of the promoter region, including all possible transcription start sites were deleted (compare sequence annotation in Supplementary Figure S2). These cells were then used to generate transgenic mice via injection into blastocysts. In the second step, the neomycin cassette was removed via flp recombination at the FRT sites in the mice. The annotated wildtype sequence in this region, the neomycin cassette, and the final sequence after recombination are provided in Supplementary Figure S2. The generation of the KO mice was done by inGenious Targeting Laboratory (iTL), Stony Brook. The mice were then transferred into our facility and backcrossed against C57Bl6/J until final analysis after about 15 generations.
Sperm Analysis
Computer-assisted sperm analysis (CASA) using a CEROS Sperm Analyzer (Hamilton Thorne, Beverly MA) was used to study sperm motility, following the protocols described in (Goossens et al., 2008; Turner et al., 2012). Testis and epididymis were dissected from the right side of each mouse, and weights of the testis and the mice were determined to normalize testis weight with body weight. The cauda epididymis was excised, immediately transferred in 250 μl human tubular fluid medium (Millipore, Billerica), punctured with a needle, and placed at 37°C, 5% CO2 for 20 min. The sperm suspension was diluted to ∼1 million/ml and ∼25 μl loaded into 100 μm depth slide chambers (Leja, Nieuw-Vennep, Netherlands). The fractions of motile sperm and progressive sperm were calculated by the CEROS system [see (Turner et al., 2012) for a discussion of the different motility parameters in mice].
RNA-Seq and Data Analysis
The testis tissues of 8 WT and 8 knockout mice were carefully collected and immediately frozen in liquid nitrogen. Total RNA was purified using QIAGEN RNeasy Microarray Tissue Mini Kit (Catalog no. 73304), and prepared using Illumina TruSeq Stranded mRNA HT Library Prep Kit (Catalog no. RS-122-2103), and sequenced using Illumina NovaSeq 6000 Regaent Kit v1.5 (200 cycles) (Catalog no. 20028313). Raw reads in FASTQ format were trimmed with Trimmomatic (0.38) (Bolger et al., 2014), and only the reads left in pairs were used for further analysis. The trimmed reads were mapped to the mouse reference genome GRCm39 (Waterston et al., 2002; Howe et al., 2021) with HISAT2 (2.2.1) (Kim et al., 2015) and SAMtools (1.9) (Li et al., 2009), and the mouse gene annotation in Ensembl (Version 104) was used for indexing the genome, i.e., the options “--ss” and “--exon” were used for command “hisat2-build”. The numbers of fragments uniquely mapped to the genes annotated in Ensembl (Version 104) were calculated with featureCounts (2.0.3) (Liao et al., 2014). Principal component analysis on VST (varianceStabilizingTransformation) fragment counts and differential expression analysis on raw counts were performed with DESeq2 (1.30.1) (Love et al., 2014; Zhu et al., 2019).
Primers Used
All primers were obtained from Metabion (Martinsried). Sequences are listed 5′> 3′.
P49 AATTAACCCTCACTAAAGGGGAGCATCGAGATTGACCAGA
P50 TAATACGACTCACTATAGGGGCTCGGATGTCATAGTCAGG
(Underlined parts in P49 and P50 indicate the matches with the mouse genomic sequence).
P318 CTCGAGTGACCAACTACTTTTCACTATTGCTG
P319 CAAGCTGTGAAGGTCAGTCAGG
P320 TGGTGGACAAGCTGGATCTAGAAAGGAAGAGGAAGCACT
P321 CTCTTCCTTTCTAGATCCAGCTTGTCCACCATGACC
P322 CTCGAGCGCGTTACATAACTTACGGTAAA
P323 CTCGAGCAAAACAAACTCCCATTGACG
Results
Expression Differences Between Sub-Species
Comparison of testis expression of Map2k7 in two mouse subspecies (Mus musculus domesticus, and Mus musculus) via Northern blotting and in situ hybridization shows a major difference between the two subspecies (Figure 1). We included in our analysis testis samples from the inbred strain C57Bl6/J [derived from M. m. domesticus (Frazer et al., 2007)], as well as from wild-caught mice that were kept under outbreeding conditions (Harr et al., 2016). Northern blotting was done with a 1.2 kb probe spanning exons 5–10 shared between all RNA isoforms (see Methods for the generation of this probe). They revealed a weak 3.5 kb and a strong 1.6 kb band in M. m. domesticus (Figure 1A), but only the 3.5 kb band in M. m. musculus, with the major 1.6 kb band missing. For the inbred strain C57Bl6/J this confirms the previous observations by (Tournier et al., 1997) and for the wildtype strains the observation by (Harr et al., 2006).
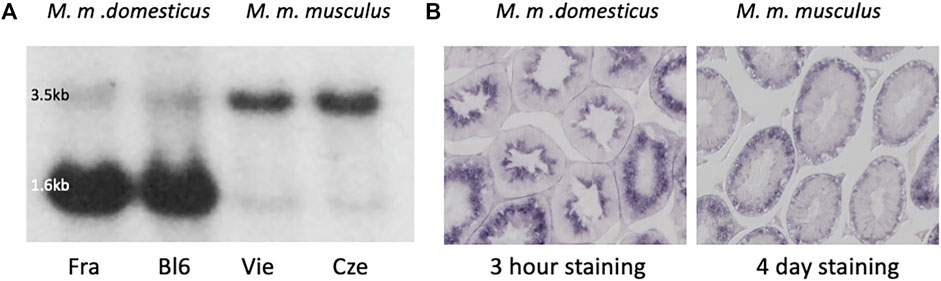
FIGURE 1. Map2k7 expression in M. m. domesticus versus M. m. musculus. (A) Northern blots with testis RNA from M. m. domesticus individuals of a wild population from France (Fra) and the laboratory inbred strain C57Bl6/J (Bl6), as well as M. m. musculus individuals from a wild-caught population from Vienna (Vie) and a wild-caught population from the Czech Republic (Cze). A fragment spanning exons 5-10 was used as a probe (see Methods for generating this fragment). The strongly expressed 1.6 kb band is only visible in the two individuals from the M. m. domesticus subspecies. The weak 1.6 kb band in M. m. musculus is derived from another transcript variant that differs with respect to an additional intron in the 3′-end of the long transcript (compare annotation in Figure 2). (B) In situ hybridization with the Map2k7 probe on cross-sections of seminiferous tubules of an individual from M. m. domesticus (left) and an individual from M. m. musculus (right) was the same fragment as used for the Northern blots. Note that the M. m. domesticus signal developed already after 3 h of color incubation, while the M. m. musculus signal developed only after 4 days of color incubation. This is compatible with the fact that the signal in M. m. domesticus comes mainly from the strong 1.6 kb transcript seen in the Northern blot, while the signal in M. m. musculus comes from the weaker 3.5 kb transcript. Microscope pictures were taken at ×100 magnification.
To assess the stage of spermatid development at which Map2k7 is expressed, we used in situ hybridization on testis sections from M. m. domesticus and M. m. musculus. Testis tissue mainly consists of seminiferous tubules, which are the location of spermatogenesis. Spermatogonial stem cells adjacent to the inner tubule wall divide and form spermatocytes which undergo meiosis. After meiosis, the spermatocytes develop into spermatids and change morphologically from round to elongated spermatids before the generation of mature spermatozoa is completed. The three main stages, spermatogonia, spermatocytes, and spermatids, are classified into further substages (Russell et al., 1990). Sperm precursor cells are embedded in Sertoli cells, which define the shape of the spermatogenic epithelium and support the germ cells. Through the influence of Sertoli cells, developing sperm precursor cells proceed towards the lumen of seminiferous tubules according to their degree of maturation. Terminal spermiation releases the sperm cells into the luminar fluid of the tubules that transfers them to the epididymis. Hence, ring-shaped zones representing different cell stages can be distinguished in a transverse section of seminiferous tubules.
In situ hybridization results on testis sections using the same probe as used for the Northern blotting are shown in Figure 1B. In M. m. domesticus, we found a strong signal in post-meiotic spermatid stages. In contrast, the Map2k7 expression pattern in M. m. musculus is very weak and becomes only visible after several days of color development. This signal is restricted to pre-meiotic stages, and we interpret it as the expression of the long transcript. This expression would be expected to be present also in M. m. domesticus, but the sections are over-stained after several days of incubation, making it impossible to visualize this directly.
Transcript Structures
A nomenclature for the different isoforms of Map2k7 was introduced by (Tournier et al., 1999) based on the analysis of various cDNA clones. They distinguish α, β, and γ isoforms that have different 5′-exons, which can be alternatively combined with two 3′-exon variants (named 1 and 2 isoforms). However, with the availability of more genome data, their analysis needed to be further updated (for example, in their γ isoform, the exons 2 and 3 should be in reverse order). In the current annotation of the UCSC genome browser, annotators have introduced a new nomenclature by just numbering the isoforms as “variants 1–6” (see Figure 2—top panel). They fall into two major length classes, variants 2 and 3 [nominally γ in (Tournier et al., 1999)] are about 3.5 kb in length and differ only by the presence/absence of a small exon. Variants 1 and 4 [nominally β in (Tournier et al., 1999)] also differ by the presence/absence of this small exon, but have an additional intron in the 3′-exon, which shortens them to about 1.6 kb. Variants 5 and 6 [nominally α in (Tournier et al., 1999)] also have a length of about 1.6 kb, but start with a different 5′-exon and differ at the 3′-end (see Figure 2—top panel).
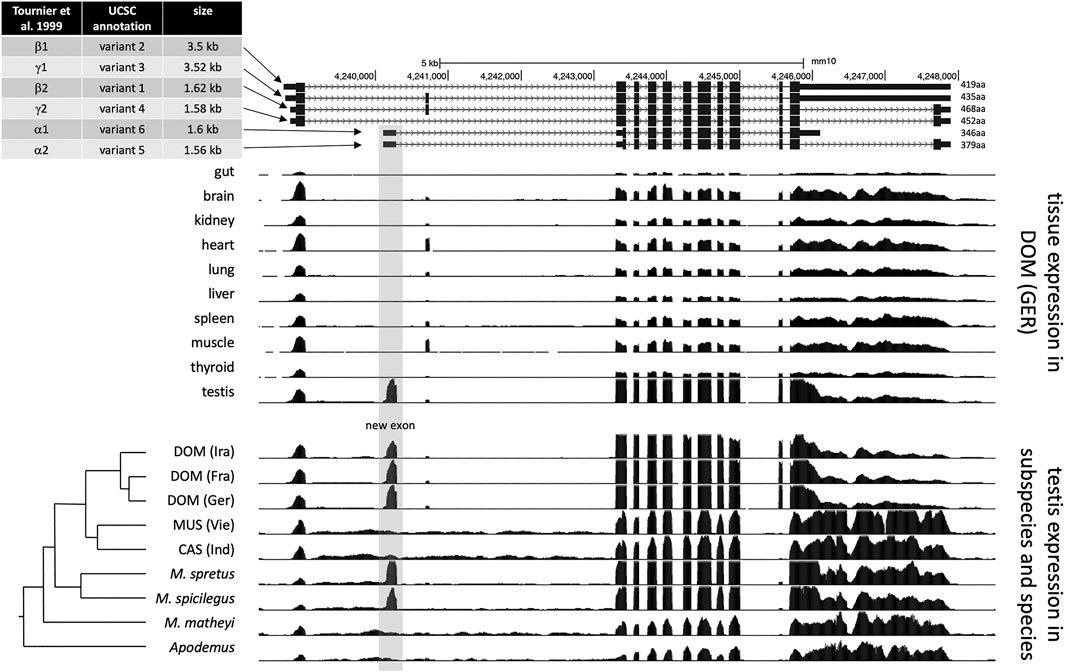
FIGURE 2. Map2k7 transcript variants and transcriptome read coverage. The figure is based on UCSC genome browser (Kent et al., 2002) tracks for the mouse GRCm38/mm10 reference sequence (Waterston et al., 2002). The top panel shows the different transcript and splice versions from the mouse reference genome, representing M. m. domesticus and therefore includes the new promoter/exon (highlighted by grey shading). The middle panel is based on data from (Harr et al., 2016) and shows transcriptome read mapping tracks for different tissues. The lower panel is based on transcriptome data from (Harr et al., 2016) and (Neme and Tautz, 2016) and shows transcriptome read mapping tracks for different populations, subspecies, and species. The phylogenetic relationships are depicted to the left. DOM: M. m. domestics, MUS: M. m. musculus, CAS: M. m. castaneus.
To better resolve the transcript structures and assess the origin of the new exon, we used the transcriptome data described in (Harr et al., 2016). Figure 2 shows the read coverage as browser tracks aligned to the annotated versions of the Map2k7 transcripts in the UCSC browser (Figure 2—top panel). The set of browser tracks representing the transcriptomes from 10 different tissues of individuals from a M. m. domesticus population show that the new exon (highlighted in grey) is only expressed in the testis (Figure 2—middle panel). The second set of browser tracks shows testis transcriptomes from different populations of the subspecies and closely related species. The new exon is only present in M. m. domesticus populations (DOM), as well as in the sister species M. spretus and M. spicilegus, but absent in the subspecies M. m. musculus, M. m. castaneus, as well as the further distant mouse species M. matheyi and the wood mouse Apodemus (Figure 2—lower panel).
Northern blotting and qPCR using different exon-specific probes and primers (Supplementary Figure S3) lead us to conclude that the highly expressed testis-specific ∼1.6 kb fragment corresponds to variant 6 in the UCSC annotation and to Mkk7-α1 in (Tournier et al., 1999). In the following, we will call it Map2k7-α1 to account for the change in the official gene nomenclature.
Protein Coding Potential
The transcripts Map2k7-β and Map2k7-γ, include the JNK-binding site (D-domains) in the first exon (Figure 3). They are about ∼3.5 kb in size and can be found in all analyzed populations and species. The newly evolved transcription start is situated within the first intron of the conserved transcripts. Its transcript does not include the exon with the D-domains but encodes potentially a protein with the kinase and DVD domain. (Tournier et al., 1999). have shown that this truncated protein has a detectable, but very weak kinase activity when expressed from an expression vector in cell culture. However, the first AUG in the new transcript is before this long ORF and in a different reading frame. It initiates a novel 50aa ORF (Figure 3), and the nucleotides surrounding the start codon of this new ORF (UGGCCAACG AUG G) match much better to the Kozak-consensus-sequence (Kozak, 1987) than the nucleotides surrounding the start codon of the remaining reading frame of the Map2k7-α1 transcript (CCCCGCCAC AUG C). A purine at position −3 and a guanine at position +4 are the most critical sequence elements for translation initiation. It is therefore questionable whether the shortened form of Map2k7 is translated at all under natural conditions.
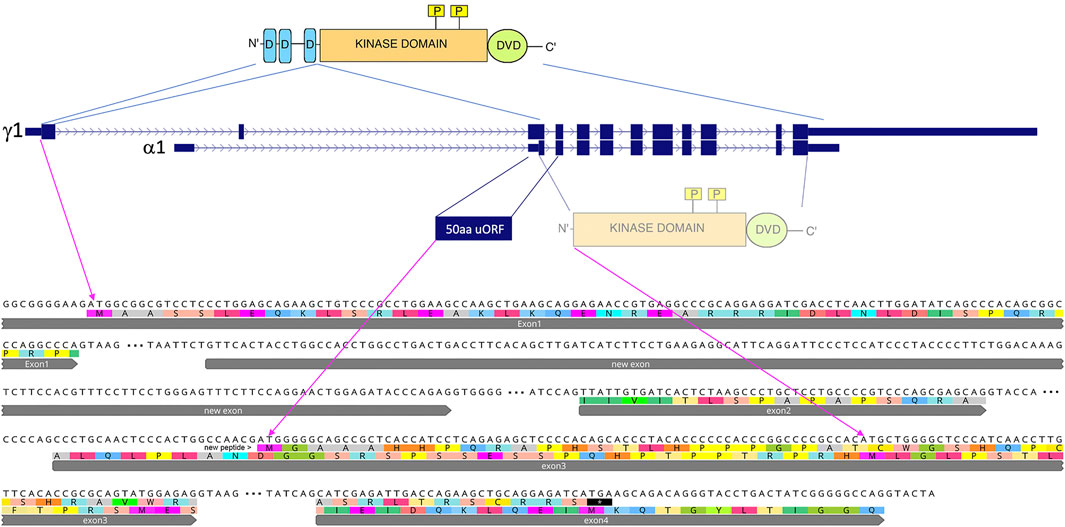
FIGURE 3. Comparison of the main Map2k7 transcripts and their coding potentials. Transcript depictions are taken from the UCSC browser annotations (see also Figure 2), whereby only the two main transcripts are shown, the conserved one (γ1) and the new one starting from the new promoter (α1). The protein domains of the Map2k7 functional kinase are depicted on the top, including the JNK-binding sites (D-domains in blue), the kinase domain (in orange) with its functional phosphorylation sites (yellow), as well as the C-terminal DVD domain (green). The first AUG in the new transcript is in the third exon, and it has a 50aa ORF, which would represent a de novo generated protein. If this AUG would not be used, the next AUG in a different frame could potentially lead to the expression of a truncated version of the Map2k7 protein, containing only the kinase and the DVD domains. The corresponding coding exon sequences (up to exon 4 for the γ1 transcript) are depicted below (flanked by their splice sites, introns are depicted as three dots) with the translations in the two alternative reading frames.
Based on the phylogenetic tree of wild mice (Guenet and Bonhomme, 2003; Chevret et al., 2005), we can infer that the new 1.6 kb transcript has arisen before the branching of M. spicilegus and M. spretus at least 2 million years ago, but not more than 6 million years ago, since it is not present in the outgroups M. matheyi and Apodemus (Figure 2). Note that while these outgroups show some RNAseq reads mapping to this region, thus indicating general low-level transcription, none of these reads are spliced at the exon-intron boundary seen in the other species. The new promoter activity has secondarily disappeared in the lineage of M. m. musculus and M. m. castaneus, apparently because of a crucial T/A substitution (see below).
Promoter Analysis
To further understand the cis-regulation of the spermatid specific 1.6 kb Map2k7α1 transcript, we analyzed genomic sequences in a 500 bp window upstream of the presumptive transcription start site in different wild populations, subspecies, and species of M. m. domesticus, M. m. musculus, M. m. castaneus, M. spretus and M. spicilegus (Figure 4). Only one SNP at -84 bp (with respect to the start site of transcription at chr8:4,240,136—see legend Figure 4 and Supplementary Figure S2 for alternative start sites) is correlated with the expression of the spermatid specific isoform in the different populations. An adenine is found in populations that show an expression of the new transcript, whereas thymine is present in those without expression (Figure 4).
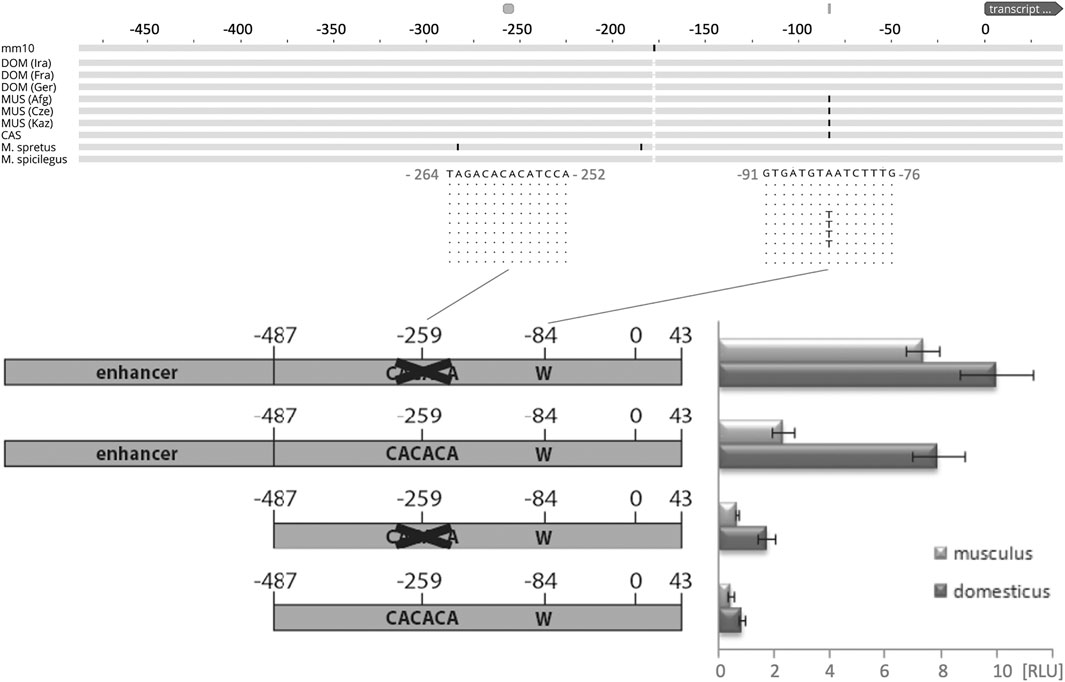
FIGURE 4. Functional test of the promoter region driving the new transcript. The top shows the alignment scheme of genomic sequences from populations, sub-species, and species based on the genome data described in (Harr et al., 2016), aligned to the mouse GRCm38/mm10 reference sequence. The fragment shown represents the one used for the promoter studies - only replacements with respect to the reference are marked, the two relevant regions discussed in the text are enlarged with their respective sequences. DOM represents M. m. domesticus populations, MUS represents M. m. musculus populations, CAS represents a M. m. castaneus population. T/A is the only mutation that correlates with the expression of the new promoter. Note that the transcriptional start site, marked as “0”, is located between the annotated site for this transcript, which would be 31 bp further upstream, and the site from which the bulk of the transcripts generated in the RNASeq experiment starts, which is 8 bp downstream (see Supplementary Figure S2 for a corresponding sequence depiction of this region). The bottom shows the scheme of the four constructs tested in cell culture and their expression levels measured as relative luminescence units [RLU] (see Methods). Error bars indicate standard deviations from eight replicates; all differences are significant (p < 0.05, t-test).
Empirical data provide evidence that specific transcription activity in reproductive tissues and particularly in spermatogenesis is regulated by very short proximal promoters (Zambrowicz et al., 1993; Li et al., 1998; Reddi et al., 1999; Blaise et al., 2001; Topaloglu et al., 2001; Han et al., 2004; Scieglinska et al., 2004; Somboonthum et al., 2005; Acharya et al., 2006). Those studies demonstrated that proximal promoters shorter than 300 bp, or even less than 100 bp, are sufficient to drive spermatid-specific expression in mice. Additionally, it was shown that a 5′-CACACA motif ∼170 bp upstream of the transcription start serves as an insulator in the spermatid specific expression of the SP-10 gene (acrosomal vesicle protein 1—Acrv1), and it was suggested that insulators might generally play an important role in maintaining spermatid specific transcription (Reddi et al., 2003; Acharya et al., 2006; Abhyankar et al., 2007; Reddi et al., 2007).
Such a 5′- CACACA motif can be found at around -259 base pairs upstream the transcription start site of the testis-specific Map2k7-α1 RNA (Figure 4). These considerations raise the question, whether a short sequence carrying the −84 A mutation in combination with the -259 5′- CACACA motif would meet the requirements to serve as a testis specific promoter. Therefore, we tested a fragment representing the genomic sequence between −487/+43 (and thus larger than the expected size of the promoter region) of the new Map2k7 testis promoter in cell culture-based luciferase expression assays (Figure 4).
The expression of most interest in this context is restricted to late spermatids. Culturing these types of cells is very difficult due to its haploid post-meiotic stage with condensed chromatin. A well-established spermatid cell culture model is not available, and alternative cell lines have the disadvantage that they will most likely not recognize the spermatid-specific Map2k7 promoter. Therefore, we have chosen the widely used NIH/3T3 fibroblast cell line in the absence of better options for this experiment.
It cannot be expected that the tested fragment is sufficient to drive luciferase expression in non-spermatid cells, but the expression level can likely be raised by deleting the 5′-CACACA motif at −259, if the assumption is correct, that this sequence maintains spermatid specific transcription by acting as an insulator in other cells. Thus, −487/+43 fragments lacking the 5′-CACACA motif at −259 were generated as well. It can be assumed that the −487/+43 fragments do not contain enhancer elements that promote expression in fibroblasts. Therefore, a CMV enhancer was ligated upstream to both versions. All four constructs (wild type, wild type with deleted insulator, CMV enhancer + wild type, CMV enhancer + wild type with deleted insulator) were created as M. m. domesticus variants with an adenine at position −84 and as M. m. musculus variants with a thymine at position −84. We found that the M. m. domesticus variant generates significantly higher signals compared to M. m. musculus in every combination. The different replicates are consistent, indicated by relatively small standard deviations (Figure 4). Deletion of the 5′-CACACA motif at −259 indeed increases the expression strength in both variants. The presence of an enhancer potentiates the effects as expected. These data provide strong evidence that the adenine at position -84 enhances the activity of the basal promoter. For this reason, it can be supposed that a major contribution of this mutation to the expression difference between M. m. domesticus and M. m. musculus in late spermatids is likely. The sequence 5′-CACACA represses the action of an adjacent enhancer to a certain extent. This finding supports the hypothesis that it acts as an insulator in the spermatid-specific Map2k7 promoter.
Functional Analysis
To assess a possible functional role of the new spermatid specific Map2k7 promoter, a knockout mouse was generated in which a fragment of 593 bp including the promoter was deleted in the M. m. domesticus background (see Methods and targeting strategy in Supplementary Figure S2). The knockout was designed in a way that it should not interfere with the conserved Map2k7 transcript.
The knockout animals were fully viable and showed no overt phenotype. KO animals are, on average, a bit heavier but have lower normalized testis weights (Table 1). Given the specific expression of the new transcripts during a crucial phase of sperm maturation, we also assessed sperm motility phenotypes. KO animals have fewer motile sperm and fewer progressive sperm (Table 1). All differences are significant at p < 0.05 (t-test, 2 sided).
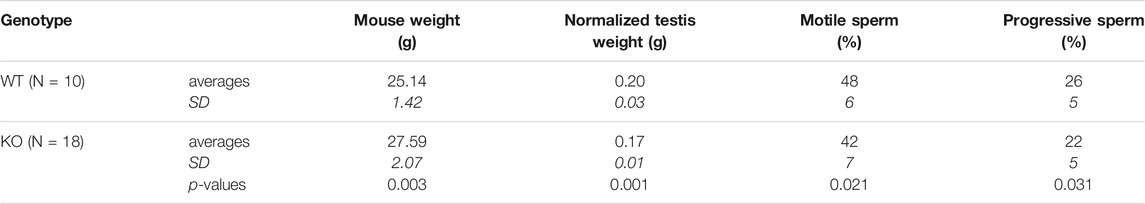
TABLE 1. Testis and sperm analysis for the Map2k7-α1 promotor KO animals versus wild type (WT) animals. Testis weights were normalized with mouse body weights. Sperm analysis was done with a CEROS Sperm Analyzer (Hamilton Thorne, Beverly MA) setup. Two-sided t-tests were done to calculate the p-values. SD is standard deviation (in italics).
RNASeq Analysis
A comparative RNASeq analysis with RNA from knockout mice versus wildtype mice was used to determine whether the 1.6 kb Map2k7-α1 testis-specific transcript influences the expression of other genes. The RNA was collected from three different tissues of the male reproductive organs, the testis, the caput epididymis, and the cauda, with eight biological replicates each. The testis is the place of the primary sperm production. The sperm from the testis move through the caput epididymis, where they mature and are eventually stored in the cauda. While the chromatin of post-meiotic sperm is condensed, there is still some transcriptome turnover (Ren et al., 2017), and the epididymal cells contribute to this transcriptome turnover as well (Shi et al., 2021).
The overall analysis of the transcriptome data in the PCA showed that the samples from each of the three tissues were very different, implying that there is indeed a major turnover of RNA between these stages, either due to differential stability, or to new transcription. On the other hand, differences between wild type and knockout are much smaller (Figure 5A). Still, since we used eight replicates for each tissue, we have a very high sensitivity to detect even small transcriptome changes (Xie et al., 2020). Accordingly, we find thousands of genes with significant expression differences (i.e., padj values < 0.05 in the DeSeq2 analysis), but mostly with relatively low log2fold-changes (Figures 5B–D). Interestingly, however, the cauda samples show a set of genes with very high positive log2fold changes (see set of dots on the top of the panel for cauda in Figure 5D).
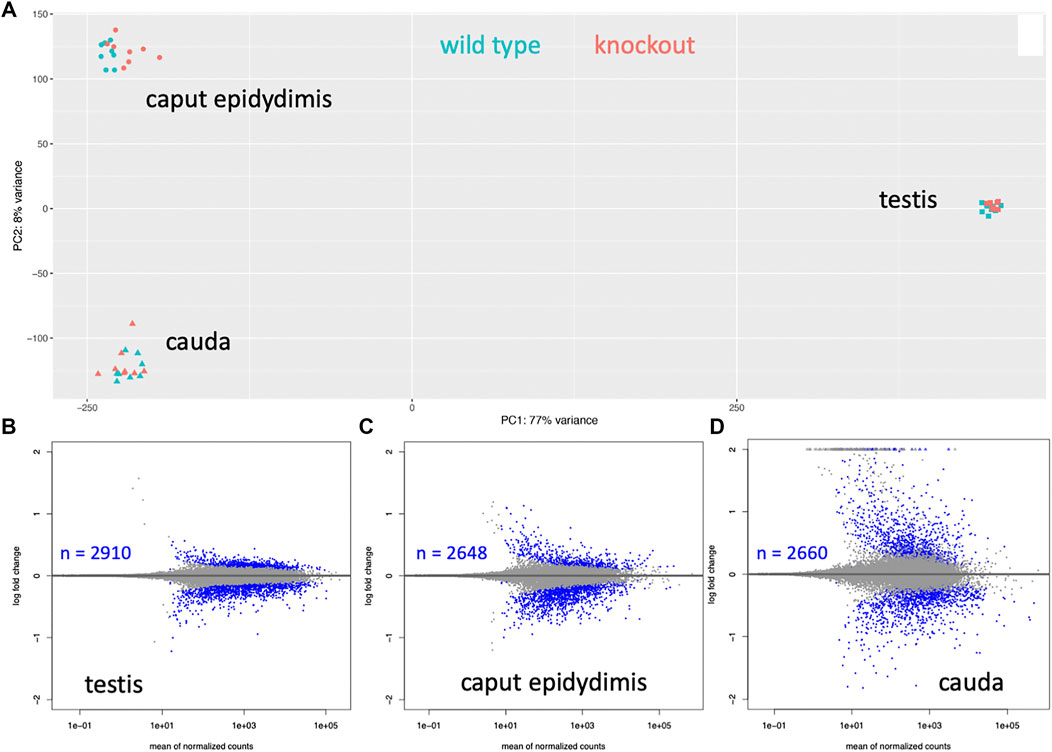
FIGURE 5. Whole transcriptomes analysis of three tissues from wild type versus knockout mice. (A) Overall PCA comparison. Strong differentiation is seen between the tissue samples, implying that those transcript sets are very different. The differences between wild type and knockouts are much smaller for each tissue. (B–D) Significantly differentially expressed genes. A dot represents each gene, genes with padj < 0.05 values are plotted as blue dots. The number of significantly differentially expressed genes is provided as an inset for each tissue.
First, we asked whether kinase signaling processes are specifically affected, which would suggest that the shortened protein of the Map2k7-α1 transcript could be involved in signaling, although lacking the D-domains that mediate the interaction with JNK (Tournier et al., 1999). had shown a residual kinase activity for this protein, but only under over-expression conditions in cell culture. However, the top biological process GO terms among the significant genes do not include “kinase signaling”, “signal transduction” or “Jnk cascade” in either of the data sets [based on a GO analysis with Panther (Mi et al., 2013)—see Supplementary Table S1]. Instead, the top GO terms indicate involvement in the meiotic division and chromosome segregation for testis, an involvement in extracellular matrix organization for the caput epididymis, and an involvement in peptide biosynthetic processes for the cauda (Supplementary Tables S1B,D,F). Hence, it is unlikely that the primary function of the Map2k7-α1 transcript is related to residual kinase signaling activity.
To get a further insight into the functional changes in the knockout animals, we focused on the genes that are most highly expressed in the respective tissues (based on the length-normalized baseMean counts of the RNASeq data), since even small concentration changes in such genes could have a marked influence on the phenotype.
For most of the highly expressed genes that we identified in the significant gene lists for testis and cauda epididymis, one can retrieve studies with functional information from knockout experiments in mice, and almost all of these find an effect on sperm maturation and/or sperm mobility (Table 2). Hence, while the relative expression changes are not large, the effect of the Map2k7-α1 transcript knockout may well be mediated via these genes. Interestingly, for the cauda, we found a rather different pattern. Most of the top expressed genes in the list are not specific for the cauda, but are more broadly expressed (e.g., an enzyme, actin, and a ribosomal protein). Interestingly, several code for immunity proteins, including sperm-specific antimicrobial peptides. Another major difference in the cauda transcriptome is a set of 37 genes with very high log2fold changes, i.e., expression at a much higher level in the knockout than in the wild type. Intriguingly, most of these are generally expressed motor proteins, and the role of such proteins in spermiogenesis has only recently been fully recognized (Wu et al., 2021).
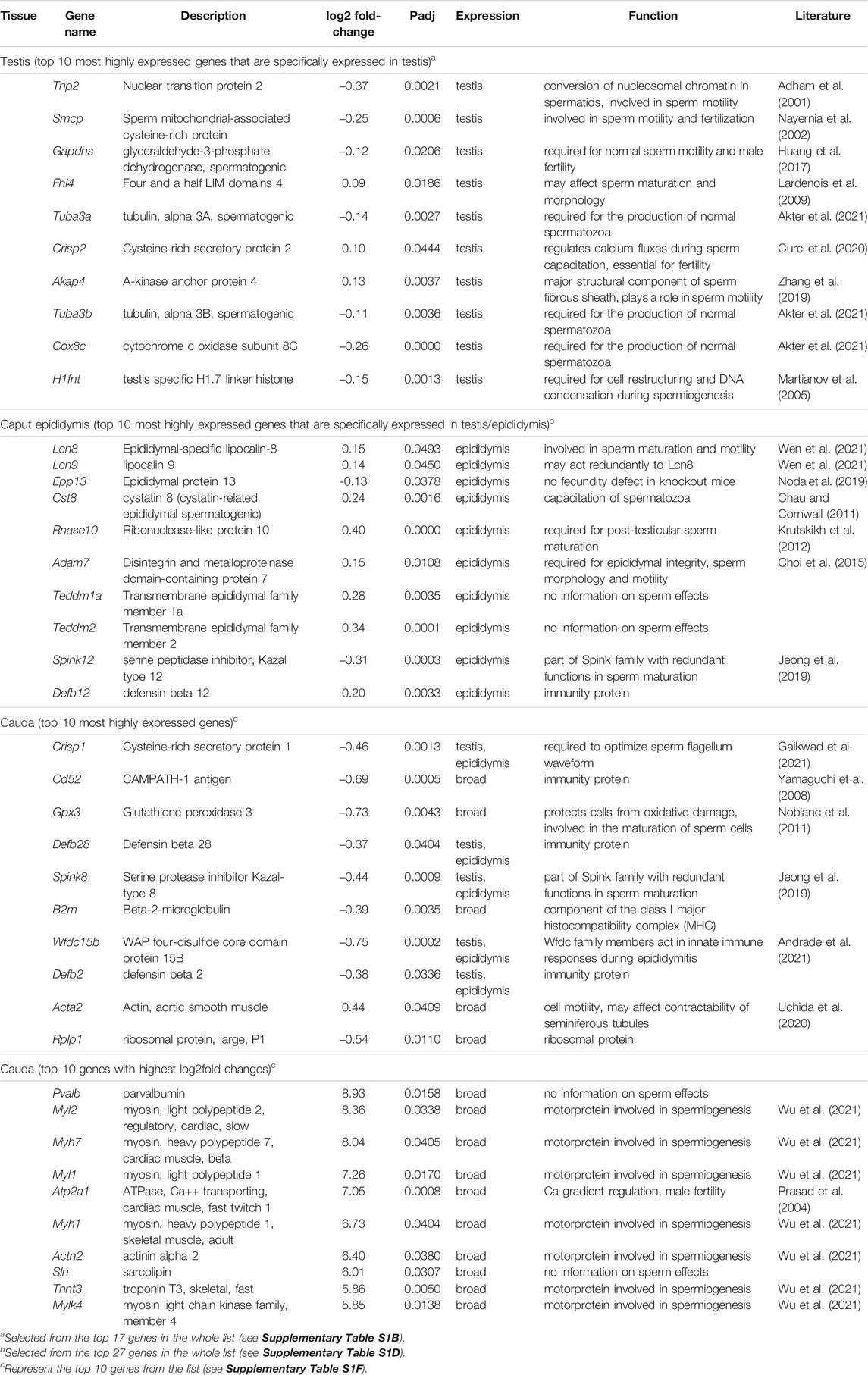
TABLE 2. List of top significant genes in the RNASeq analysis for the three tissue samples. Named genes (i.e., predicted genes were not included) were retrieved from the DeSeq2 output ranked according to baseMean/mRNA length and expression characteristics (see Supplementary Table S1 for full results).
Discussion
Based on comparative genomic and functional analysis, we show here that a new intra-intronic promoter has arisen in the mouse lineage 2–6 million years ago and has led to the evolution of a functionally new transcript within an otherwise highly conserved gene. The transcript is specific to the testis, and knockout combined with transcriptome analysis shows that it is functionally involved in sperm maturation. Interestingly, the new promoter activity also got secondarily lost in some mouse lineages, apparently due to acquiring a disabling mutation in the promoter region.
The emergence of evolutionary novelties out of regulatory changes is by now well documented in many species [see (Tautz, 2000; Wray, 2007; Carroll, 2008; Romero et al., 2012; Osada et al., 2017; Signor and Nuzhdin, 2018; Mattioli et al., 2020; He et al., 2021) for a subset of relevant papers and reviews]. In fact, it is so abundant that it often constitutes the first measurable differences in population and species divergence, including diverging mouse populations (Bryk et al., 2013), which raises the question of whether much of is initially neutrally evolving or could be functional (Fay and Wittkopp, 2008; Staubach et al., 2010; Hodgins-Davis et al., 2019; Hill et al., 2021). Given the evolutionary volatility of the Map2k7-α1 transcript, with its fast secondary loss after its initial emergence, one would typically have considered it to be mostly neutral and therefore subject to random fixation or loss. However, our data show that it has a clear functional role in spermatogenesis. Therefore, the high evolutionary dynamics of this transcript is more likely explained by the general effects of sexual selection that would be particularly effective in the germline and the gonads (Kleene, 2005).
There are several possibilities of how the Map2k7-α1 transcript could function. The first is that it leads to translating a truncated protein that codes only for Map2k7 kinase but does not bind specifically to JNK. It could therefore phosphorylate other signaling proteins but in an unspecific manner. This would likely be detrimental rather than advantageous for the cells. Also, since we do not find GO terms that relate to signaling processes in the transcriptome analysis of knockout mice, we assume that the truncated protein is not expressed, or at least, not functional.
The second possibility is that Map2k7-α1 acts as a non-coding RNA. There are multiple ways non-coding RNAs can regulate other genes or gene complexes (Gil and Ulitsky, 2020; Statello et al., 2021), and some have been implicated in male infertility (Joshi and Rajender, 2020). In a previous study, we identified a testis-specific new transcript that has also emerged via a new promoter acquisition and for which we could infer that it acts as lncRNA in spermiogenesis (Heinen et al., 2009). However, given that most of the Map2k7-α1 RNA overlaps with the functional Map2k7 transcripts, it would seem unlikely that it could have assumed such a function as non-coding RNA as a whole since most of its RNA is actually potentially coding. Only the new exon that emerged out of intronic sequences might have such a function.
On the other hand, the first AUG in this new transcript is embedded in an optimal Kozak-consensus-sequence (Kozak, 1987), and one would therefore expect that it leads to the translation of a 50aa ORF. The resulting peptide does not match with any other protein or domain in the data bases, since it is actually produced out of an alternative reading frame of the Map2k7 gene. A structure prediction analysis is therefore currently not possible, because even the most refined algorithms require appropriate comparative data from other species (Diwan et al., 2021). This is generally a problem for all de novo emerged proteins, although comparative analysis of large numbers of them has suggested that they more likely have a higher intrinsic disorder, which makes them better tolerated in cells (Tretyachenko et al., 2017; Wilson et al., 2017; Fajardo and Tautz, 2021; James et al., 2021). Still, for at least one de novo emerged protein foldability has been experimentally confirmed (Bungard et al., 2017).
While it has long been thought that proteins that emerge out of such more or less random sequences would not be functional, it has by now become clear that the de novo evolution of proteins is well possible (Tautz and Domazet-Loso, 2011; Van Oss and Carvunis, 2019; Zhang et al., 2019). In fact, we have recently described such a case of a very recent de novo emergence of a protein that regulates pregnancy cycles in mice (Xie et al., 2019). In that case, we could prove the protein’s direct function in a knockout mouse that carried only a frameshift mutation in the protein. In the current study, we deleted the whole transcript, implying that it is not fully proven that it is the translated peptide that conveys the function. But studies with random peptide sequences in E. coli and plants have shown that a substantial fraction of them can directly affect their hosts (Bao et al., 2017; Neme et al., 2017; Fajardo and Tautz, 2021). Therefore, it seems possible that the new peptide encoded by the Map2k7-α1 transcript is indeed a de novo protein with a function. Hence, this would be a case where a pre-existing potentially functional sequence was “waiting” for a promoter emergence to allow it to become functional via overprinting of an existing reading frame (Rancurel et al., 2009; Neme and Tautz, 2013; Carter, 2021).
The ORF is actually already present in the outgroup species but would not be expected to be translated in these species. The fact that it got secondarily lost in the M. m. musculus subspecies is in line with the observation of fast gain and loss cycles of de novo evolved transcripts and proteins (Carvunis et al., 2012; Neme and Tautz, 2014; Palmieri et al., 2014). Nevertheless, our functional data show that the new transcript (and/or peptide) can still be functional despite this evolutionary instability.
Author’s Note
Part of this work was done in the framework of the PhD thesis of the first author (Heinen, 2008).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: European Nucleotide Archive (accession: PRJEB39625).
Ethics Statement
Ethical review and approval was not required for the animal study because the work did not involve in vivo experiments with animals. Mouse samples were taken from mice derived from the maintenance of the mouse strain collections at our institute (Harr et al., 2016). Maintenance and handling of mice in the facility were conducted in accordance with German animal welfare law (Tierschutzgesetz) and FELASA guidelines. Permits for keeping mice were obtained from the local veterinary office “Veterinäramt Kreis Plön” (permit number: 1401-144/PLÖ-004697). The respective animal welfare officer at the University of Kiel was informed about the sacrifice of the animals for this study.
Author Contributions
TH: conceptualization, experimental work, data analysis, paper writing, CX: data analysis, MK: data analysis, DS: experimental work, data analysis, SK: experimental work, data analysis, DT: conceptualization, data analysis, paper writing.
Funding
This work was funded by institutional funds of the MPG to DT.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Heike Harre for help with the sperm counting and phenotyping, Cornelia Burghardt for RNA extraction and the mouse team lead by Christine Pfeifle for expert support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.812139/full#supplementary-material
References
Abhyankar, M. M., Urekar, C., and Reddi, P. P. (2007). A Novel CpG-free Vertebrate Insulator Silences the Testis-specific SP-10 Gene in Somatic Tissues. J. Biol. Chem. 282, 36143–36154. doi:10.1074/jbc.m705811200
Acharya, K. K., Govind, C. K., Shore, A. N., Stoler, M. H., and Reddi, P. P. (2006). cis-requirement for the Maintenance of Round Spermatid-specific Transcription. Develop. Biol. 295, 781–790. doi:10.1016/j.ydbio.2006.04.443
Adham, I. M., Nayernia, K., Burkhardt-Gottges, E., Topaloglu, O., Dixkens, C., Holstein, A. F., et al. (2001). Teratozoospermia in Mice Lacking the Transition Protein 2 (Tnp2). Mol. Hum. Reprod. 7, 513–520. doi:10.1093/molehr/7.6.513
Akter, M. S., Hada, M., Shikata, D., Watanabe, G., Ogura, A., and Matoba, S. (2021). CRISPR/Cas9-based Genetic Screen of SCNT-Reprogramming Resistant Genes Identifies Critical Genes for Male Germ Cell Development in Mice. Sci. Rep. 11, 15438. doi:10.1038/s41598-021-94851-9
Alwine, J. C., Kemp, D. J., and Stark, G. R. (1977). Method For Detection Of Specific Rnas In Agarose Gels By Transfer To Diazobenzyloxymethyl-Paper And Hybridization With Dna Probes. Proc. Natl. Acad. Sci. 74, 5350–5354. doi:10.1073/pnas.74.12.5350
Andrade, A. D., Almeida, P. G. C., Mariani, N. A. P., Freitas, G. A., Kushima, H., Filadelpho, A. L., et al. (2021). Lipopolysaccharide-induced Epididymitis Modifies the Transcriptional Profile of Wfdc Genes in Mice†. Biol. Reprod. 104, 144–158. doi:10.1093/biolre/ioaa189
Bao, Z., Clancy, M. A., Carvalho, R. F., Elliott, K., and Folta, K. M. (2017). Identification of Novel Growth Regulators in Plant Populations Expressing Random Peptides. Plant Physiol. 175, 619–627. doi:10.1104/pp.17.00577
Blaise, R., Guillaudeux, T., Tavernier, G., Daegelen, D., Evrard, B., Mairal, A., et al. (2001). Testis Hormone-Sensitive Lipase Expression in Spermatids Is Governed by a Short Promoter in Transgenic Mice. J. Biol. Chem. 276, 5109–5115. doi:10.1074/jbc.m009103200
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30, 2114–2120. doi:10.1093/bioinformatics/btu170
Bryk, J., Somel, M., Lorenc, A., and Teschke, M. (2013). Early Gene Expression Divergence between Allopatric Populations of the House Mouse ( Mus musculus Domesticus). Ecol. Evol. 3, 558–568. doi:10.1002/ece3.447
Bungard, D., Copple, J. S., Yan, J., Chhun, J. J., Kumirov, V. K., Foy, S. G., et al. (2017). Foldability of a Natural De Novo Evolved Protein. Structure 25, 1687–1696. doi:10.1016/j.str.2017.09.006
Carroll, S. B. (2008). Evo-devo and an Expanding Evolutionary Synthesis: A Genetic Theory of Morphological Evolution. Cell 134, 25–36. doi:10.1016/j.cell.2008.06.030
Carter, C. W. (2021). Simultaneous Codon Usage, the Origin of the Proteome, and the Emergence of De-novo Proteins. Curr. Opin. Struct. Biol. 68, 142–148. doi:10.1016/j.sbi.2021.01.004
Carvunis, A.-R., Rolland, T., Wapinski, I., Calderwood, M. A., Yildirim, M. A., Simonis, N., et al. (2012). Proto-genes and De Novo Gene Birth. Nature 487, 370–374. doi:10.1038/nature11184
Castro, J. F., and Tautz, D. (2021). The Effects of Sequence Length and Composition of Random Sequence Peptides on the Growth of E. coli Cells. Genes 12, 1913. doi:10.3390/genes12121913
Chang, L., Jones, Y., Ellisman, M. H., Goldstein, L. S. B., and Karin, M. (2003). JNK1 Is Required for Maintenance of Neuronal Microtubules and Controls Phosphorylation of Microtubule-Associated Proteins. Develop. Cel 4, 521–533. doi:10.1016/s1534-5807(03)00094-7
Chang, L., and Karin, M. (2001). Mammalian MAP Kinase Signalling Cascades. Nature 410, 37–40. doi:10.1038/35065000
Chau, K. M., and Cornwall, G. A. (2011). Reduced Fertility In Vitro in Mice Lacking the Cystatin CRES (Cystatin-Related Epididymal Spermatogenic): Rescue by Exposure of Spermatozoa to Dibutyryl cAMP and Isobutylmethylxanthine1. Biol. Reprod. 84, 140–152. doi:10.1095/biolreprod.110.084855
Chevret, P., Veyrunes, F., and Britton-Davidian, J. (2005). Molecular Phylogeny of the Genus Mus (Rodentia: Murinae) Based on Mitochondrial and Nuclear Data. Biol. J. Linn. Soc. 84, 417–427. doi:10.1111/j.1095-8312.2005.00444.x
Choi, H., Han, C., Jin, S., Kwon, J. T., Kim, J., Jeong, J., et al. (2015). Reduced Fertility and Altered Epididymal and Sperm Integrity in Mice Lacking ADAM7. Biol. Reprod. 93, 70. doi:10.1095/biolreprod.115.130252
Curci, L., Brukman, N. G., Weigel Muñoz, M., Rojo, D., Carvajal, G., Sulzyk, V., et al. (2020). Functional Redundancy and Compensation: Deletion of Multiple Murine Crisp Genes Reveals Their Essential Role for Male Fertility. FASEB j. 34, 15718–15733. doi:10.1096/fj.202001406r
Deng, Y., Ren, X., Yang, L., Lin, Y., and Wu, X. (2003). A JNK-dependent Pathway Is Required for TNFα-Induced Apoptosis. Cell 115, 61–70. doi:10.1016/s0092-8674(03)00757-8
Diwan, G. D., Gonzalez-Sanchez, J. C., Apic, G., and Russell, R. B. (2021). Next Generation Protein Structure Predictions and Genetic Variant Interpretation. J. Mol. Biol. 433, 167180. doi:10.1016/j.jmb.2021.167180
Dong, C., Yang, D. D., Tournier, C., Whitmarsh, A. J., Xu, J., Davis, R. J., et al. (2000). JNK Is Required for Effector T-Cell Function but Not for T-Cell Activation. Nature 405, 91–94. doi:10.1038/35011091
English, J., Pearson, G., Wilsbacher, J., Swantek, J., Karandikar, M., Xu, S., et al. (1999). New Insights into the Control of MAP Kinase Pathways. Exp. Cel Res. 253, 255–270. doi:10.1006/excr.1999.4687
Fay, J. C., and Wittkopp, P. J. (2008). Evaluating the Role of Natural Selection in the Evolution of Gene Regulation. Heredity 100, 191–199. doi:10.1038/sj.hdy.6801000
Fleming, Y., Armstrong, C. G., Morrice, N., Paterson, A., Goedert, M., and Cohen, P. (2000). Synergistic Activation of Stress-Activated Protein Kinase 1/c-Jun N-Terminal Kinase (SAPK1/JNK) Isoforms by Mitogen-Activated Protein Kinase Kinase 4 (MKK4) and MKK7. Biochem. J. 352, 145–154. doi:10.1042/bj3520145
Foltz, I. N., Gerl, R. E., Wieler, J. S., Luckach, M., Salmon, R. A., and Schrader, J. W. (1998). Human Mitogen-Activated Protein Kinase Kinase 7 (MKK7) Is a Highly Conserved C-Jun N-Terminal Kinase/stress-Activated Protein Kinase (JNK/SAPK) Activated by Environmental Stresses and Physiological Stimuli. J. Biol. Chem. 273, 9344–9351. doi:10.1074/jbc.273.15.9344
Frazer, K. A., Eskin, E., Kang, H. M., Bogue, M. A., Hinds, D. A., Beilharz, E. J., et al. (2007). A Sequence-Based Variation Map of 8.27 Million SNPs in Inbred Mouse Strains. Nature 448, 1050–1053. doi:10.1038/nature06067
Gaikwad, A. S., Nandagiri, A., Potter, D. L., Nosrati, R., O'Connor, A. E., Jadhav, S., et al. (2021). CRISPs Function to Boost Sperm Power Output and Motility. Front Cel Dev Biol 9, 693258. doi:10.3389/fcell.2021.693258
Gil, N., and Ulitsky, I. (2020). Regulation of Gene Expression by Cis-Acting Long Non-coding RNAs. Nat. Rev. Genet. 21, 102–117. doi:10.1038/s41576-019-0184-5
Goossens, E., De Block, G., and Tournaye, H. (2008). Computer-assisted Motility Analysis of Spermatozoa Obtained after Spermatogonial Stem Cell Transplantation in the Mouse. Fertil. Sterility 90, 1411–1416. doi:10.1016/j.fertnstert.2007.08.035
Guénet, J.-L., and Bonhomme, F. (2003). Wild Mice: an Ever-Increasing Contribution to a Popular Mammalian Model. Trends Genet. 19, 24–31. doi:10.1016/s0168-9525(02)00007-0
Han, S., Xie, W., Kim, S. H., Yue, L., and DeJong, J. (2004). A Short Core Promoter Drives Expression of the ALF Transcription Factor in Reproductive Tissues of Male and Female Mice1. Biol. Reprod. 71, 933–941. doi:10.1095/biolreprod.104.030247
Harr, B., Karakoc, E., Neme, R., Teschke, M., Pfeifle, C., Pezer, Ž., et al. (2016). Genomic Resources for Wild Populations of the House Mouse, Mus musculus and its Close Relative Mus spretus. Sci. Data 3, 160075. doi:10.1038/sdata.2016.75
Harr, B., Voolstra, C., Heinen, T. J. A. J., Baines, J. F., Rottscheidt, R., Ihle, S., et al. (2006). A Change of Expression in the Conserved Signaling Gene MKK7 Is Associated with a Selective Sweep in the Western House Mouse Mus musculus Domesticus. J. Evol. Biol 19, 1486–1496. doi:10.1111/j.1420-9101.2006.01130.x
He, F., Steige, K. A., Kovacova, V., Göbel, U., Bouzid, M., Keightley, P. D., et al. (2021). Cis-regulatory Evolution Spotlights Species Differences in the Adaptive Potential of Gene Expression Plasticity. Nat. Commun. 12, 3376. doi:10.1038/s41467-021-23558-2
Heinen, T. J. A. J. (2008). Characterization of Genes Involved in Recent Adaptation. Cologne, Germany: Universität zu Köln.
Heinen, T. J. A. J., Staubach, F., Häming, D., and Tautz, D. (2009). Emergence of a New Gene from an Intergenic Region. Curr. Biol. 19, 1527–1531. doi:10.1016/j.cub.2009.07.049
Hill, M. S., Vande Zande, P., and Wittkopp, P. J. (2021). Molecular and Evolutionary Processes Generating Variation in Gene Expression. Nat. Rev. Genet. 22, 203–215. doi:10.1038/s41576-020-00304-w
Hodgins-Davis, A., Duveau, F., Walker, E. A., and Wittkopp, P. J. (2019). Empirical Measures of Mutational Effects Define Neutral Models of Regulatory Evolution inSaccharomyces Cerevisiae. Proc. Natl. Acad. Sci. USA 116, 21085–21093. doi:10.1073/pnas.1902823116
Holland, P. M., Suzanne, M., Campbell, J. S., Noselli, S., and Cooper, J. A. (1997). MKK7 Is a Stress-Activated Mitogen-Activated Protein Kinase Kinase Functionally Related to Hemipterous. J. Biol. Chem. 272, 24994–24998. doi:10.1074/jbc.272.40.24994
Howe, K. L., Achuthan, P., Allen, J., Allen, J., Alvarez-Jarreta, J., Amode, M. R., et al. (2021). Ensembl 2021. Nucleic Acids Res. 49, D884–D891. doi:10.1093/nar/gkaa942
Huang, Z., Danshina, P. V., Mohr, K., Qu, W., Goodson, S. G., O’Connell, T. M., et al. (2017). Sperm Function, Protein Phosphorylation, and Metabolism Differ in Mice Lacking Successive Sperm-specific Glycolytic Enzymes†. Biol. Reprod. 97, 586–597. doi:10.1093/biolre/iox103
James, J. E., Willis, S. M., Nelson, P. G., Weibel, C., Kosinski, L. J., and Masel, J. (2021). Universal and Taxon-specific Trends in Protein Sequences as a Function of Age. Elife 10, e57347. doi:10.7554/eLife.57347
Jeong, J., Lee, B., Kim, J., Kim, J., Hong, S. H., Kim, D., et al. (2019). Expressional and Functional Analyses of Epididymal SPINKs in Mice. Gene Expr. Patterns 31, 18–25. doi:10.1016/j.gep.2018.12.001
Joshi, M., and Rajender, S. (2020). Long Non-coding RNAs (lncRNAs) in Spermatogenesis and Male Infertility. Reprod. Biol. Endocrinol. 18, 103. doi:10.1186/s12958-020-00660-6
Kent, W. J., Sugnet, C. W., Furey, T. S., Roskin, K. M., Pringle, T. H., Zahler, A. M., et al. (2002). The Human Genome Browser at UCSC. Genome Res. 12, 996–1006. doi:10.1101/gr.229102
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 12, 357–360. doi:10.1038/nmeth.3317
Kishimoto, H., Nakagawa, K., Watanabe, T., Kitagawa, D., Momose, H., Seo, J., et al. (2003). Different Properties of SEK1 and MKK7 in Dual Phosphorylation of Stress-Induced Activated Protein Kinase SAPK/JNK in Embryonic Stem Cells. J. Biol. Chem. 278, 16595–16601. doi:10.1074/jbc.m213182200
Kleene, K. C. (2005). Sexual Selection, Genetic Conflict, Selfish Genes, and the Atypical Patterns of Gene Expression in Spermatogenic Cells. Develop. Biol. 277, 16–26. doi:10.1016/j.ydbio.2004.09.031
Kozak, M. (1987). An Analysis of 5'-noncoding Sequences from 699 Vertebrate Messenger RNAs. Nucl. Acids Res. 15, 8125–8148. doi:10.1093/nar/15.20.8125
Krutskikh, A., Poliandri, A., Cabrera‐Sharp, V., Dacheux, J. L., Poutanen, M., and Huhtaniemi, I. (2012). Epididymal Protein Rnase10 Is Required for post‐testicular Sperm Maturation and Male Fertility. FASEB j. 26, 4198–4209. doi:10.1096/fj.12-205211
Lardenois, A., Chalmel, F., Demougin, P., Kotaja, N., Sassone-Corsi, P., and Primig, M. (2009). Fhl5/Act, a CREM-Binding Transcriptional Activator Required for normal Sperm Maturation and Morphology, Is Not Essential for Testicular Gene Expression. Reprod. Biol. Endocrinol. 7, 133. doi:10.1186/1477-7827-7-133
Lei, K., Nimnual, A., Zong, W.-X., Kennedy, N. J., Flavell, R. A., Thompson, C. B., et al. (2002). The Bax Subfamily of Bcl2-Related Proteins Is Essential for Apoptotic Signal Transduction by C-Jun NH 2 -Terminal Kinase. Mol. Cel Biol 22, 4929–4942. doi:10.1128/mcb.22.13.4929-4942.2002
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map Format and SAMtools. Bioinformatics 25, 2078–2079. doi:10.1093/bioinformatics/btp352
Li, S., Zhou, W., Doglio, L., and Goldberg, E. (1998). Transgenic Mice Demonstrate a Testis-specific Promoter for Lactate Dehydrogenase, LDHC. J. Biol. Chem. 273, 31191–31194. doi:10.1074/jbc.273.47.31191
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 30, 923–930. doi:10.1093/bioinformatics/btt656
Love, M. I., Huber, W., and Anders, S. (2014). Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 15, 550. doi:10.1186/s13059-014-0550-8
Martianov, I., Brancorsini, S., Catena, R., Gansmuller, A., Kotaja, N., Parvinen, M., et al. (2005). Polar Nuclear Localization of H1T2, a Histone H1 Variant, Required for Spermatid Elongation and DNA Condensation during Spermiogenesis. Proc. Natl. Acad. Sci. 102, 2808–2813. doi:10.1073/pnas.0406060102
Mattioli, K., Oliveros, W., Gerhardinger, C., Andergassen, D., Maass, P. G., Rinn, J. L., et al. (2020). Cis and Trans Effects Differentially Contribute to the Evolution of Promoters and Enhancers. Genome Biol. 21, 210. doi:10.1186/s13059-020-02110-3
Mi, H., Muruganujan, A., and Thomas, P. D. (2013). PANTHER in 2013: Modeling the Evolution of Gene Function, and Other Gene Attributes, in the Context of Phylogenetic Trees. Nucleic Acids Res. 41, D377–D386. doi:10.1093/nar/gks1118
Moriguchi, T., Toyoshima, F., Masuyama, N., Hanafusa, H., Gotoh, Y., and Nishida, E. (1997). A Novel SAPK/JNK Kinase, MKK7, Stimulated by TNFalpha and Cellular Stresses. Embo J. 16, 7045–7053. doi:10.1093/emboj/16.23.7045
Nayernia, K., Adham, I. M., Burkhardt-Göttges, E., Neesen, J., Rieche, M., Wolf, S., et al. (2002). Asthenozoospermia in Mice with Targeted Deletion of the Sperm Mitochondrion-Associated Cysteine-Rich Protein ( Smcp ) Gene. Mol. Cel Biol 22, 3046–3052. doi:10.1128/mcb.22.9.3046-3052.2002
Neme, R., Amador, C., Yildirim, B., McConnell, E., and Tautz, D. (2017). Random Sequences Are an Abundant Source of Bioactive RNAs or Peptides. Nat. Ecol. Evol. 1, 0217. doi:10.1038/s41559-017-0127
Neme, R., and Tautz, D. (2014). Evolution: Dynamics of De Novo Gene Emergence. Curr. Biol. 24, R238–R240. doi:10.1016/j.cub.2014.02.016
Neme, R., and Tautz, D. (2016). Fast Turnover of Genome Transcription across Evolutionary Time Exposes Entire Non-coding DNA to De Novo Gene Emergence. Elife 5, e09977. doi:10.7554/eLife.09977
Neme, R., and Tautz, D. (2013). Phylogenetic Patterns of Emergence of New Genes Support a Model of Frequent De Novo Evolution. Bmc Genomics 14, 117. doi:10.1186/1471-2164-14-117
Nishina, H., Wada, T., and Katada, T. (2004). Physiological Roles of SAPK/JNK Signaling Pathway. J. Biochem. 136, 123–126. doi:10.1093/jb/mvh117
Noblanc, A., Kocer, A., Chabory, E., Vernet, P., Saez, F., Cadet, R., et al. (2011). Glutathione Peroxidases at Work on Epididymal Spermatozoa: An Example of the Dual Effect of Reactive Oxygen Species on Mammalian Male Fertilizing Ability. J. Androl. 32, 641–650. doi:10.2164/jandrol.110.012823
Noda, T., Sakurai, N., Nozawa, K., Kobayashi, S., Devlin, D. J., Matzuk, M. M., et al. (2019). Nine Genes Abundantly Expressed in the Epididymis Are Not Essential for Male Fecundity in Mice. Andrology 7, 644–653. doi:10.1111/andr.12621
Osada, N., Miyagi, R., and Takahashi, A. (2017). Cis- and Trans-regulatory Effects on Gene Expression in a Natural Population of Drosophila melanogaster. Genetics 206, 2139–2148. doi:10.1534/genetics.117.201459
Palmieri, N., Kosiol, C., and Schlötterer, C. (2014). The Life Cycle of Drosophila Orphan Genes. Elife 3, e01311. doi:10.7554/eLife.01311
Prasad, V., Okunade, G. W., Miller, M. L., and Shull, G. E. (2004). Phenotypes of SERCA and PMCA Knockout Mice. Biochem. Biophysical Res. Commun. 322, 1192–1203. doi:10.1016/j.bbrc.2004.07.156
Rancurel, C., Khosravi, M., Dunker, A. K., Romero, P. R., and Karlin, D. (2009). Overlapping Genes Produce Proteins with Unusual Sequence Properties and Offer Insight into De Novo Protein Creation. J. Virol. 83, 10719–10736. doi:10.1128/jvi.00595-09
Reddi, P. P., Flickinger, C. J., and Herr, J. C. (1999). Round Spermatid-specific Transcription of the Mouse SP-10 Gene Is Mediated by a 294-Base Pair Proximal Promoter1. Biol. Reprod. 61, 1256–1266. doi:10.1095/biolreprod61.5.1256
Reddi, P. P., Shore, A. N., Shapiro, J. A., Anderson, A., Stoler, M. H., and Acharya, K. K. (2003). Spermatid-specific Promoter of the SP-10 Gene Functions as an Insulator in Somatic Cells. Develop. Biol. 262, 173–182. doi:10.1016/s0012-1606(03)00349-x
Reddi, P. P., Urekar, C. J., Abhyankar, M. M., and Ranpura, S. A. (2007). Role of an Insulator in Testis-specific Gene Transcription. Ann. N.Y Acad. Sci. 1120, 95–103. doi:10.1196/annals.1411.012
Ren, X., Chen, X., Wang, Z., and Wang, D. (2017). Is Transcription in Sperm Stationary or Dynamic? J. Reprod. Develop. 63, 439–443. doi:10.1262/jrd.2016-093
Romero, I. G., Ruvinsky, I., and Gilad, Y. (2012). Comparative Studies of Gene Expression and the Evolution of Gene Regulation. Nat. Rev. Genet. 13, 505–516. doi:10.1038/nrg3229
Russell, L., Ettlin, R., Sinha Hikim, A., and Clegg, E. (1990). Histological and Histopathological Evaluation of the Testis. St. Louis: Cache River Press.
Sabapathy, K., Jochum, W., Hochedlinger, K., Chang, L., Karin, M., and Wagner, E. F. (1999). Defective Neural Tube Morphogenesis and Altered Apoptosis in the Absence of Both JNK1 and JNK2. Mech. Develop. 89, 115–124. doi:10.1016/s0925-4773(99)00213-0
Scieglinska, D., Vydra, N., Krawczyk, Z., and Widlak, W. (2004). Location of Promoter Elements Necessary and Sufficient to Direct Testis-specific Expression of the Hst70/Hsp70.2 Gene. Biochem. J. 379, 739–747. doi:10.1042/BJ20031842
Shi, J. W., Fok, K. L., Dai, P. Y., Qiao, F., Zhang, M. Y., Liu, H. G., et al. (2021). Spatio-temporal Landscape of Mouse Epididymal Cells and Specific Mitochondria-Rich Segments Defined by Large-Scale Single-Cell RNA-Seq. Cel Discov. 7, 34. doi:10.1038/s41421-021-00260-7
Signor, S. A., and Nuzhdin, S. V. (2018). The Evolution of Gene Expression in Cis and Trans. Trends Genet. 34, 532–544. doi:10.1016/j.tig.2018.03.007
Somboonthum, P., Ohta, H., Yamada, S., Onishi, M., Ike, A., Nishimune, Y., et al. (2005). cAMP-Responsive Element in TATA-Less Core Promoter Is Essential for Haploid-specific Gene Expression in Mouse Testis. Nucleic Acids Res. 33, 3401–3411. doi:10.1093/nar/gki652
Statello, L., Guo, C.-J., Chen, L.-L., and Huarte, M. (2021). Gene Regulation by Long Non-coding RNAs and its Biological Functions. Nat. Rev. Mol. Cel Biol 22, 96–118. doi:10.1038/s41580-020-00315-9
Staubach, F., Teschke, M., Voolstra, C. R., Wolf, J. B. W., and Tautz, D. (2010). A Test Of The Neutral Model Of Expression Change In Natural Populations Of House Mouse Subspecies. Evolution 64, 549–560. doi:10.1111/j.1558-5646.2009.00818.x
Takekawa, M., Tatebayashi, K., and Saito, H. (2005). Conserved Docking Site Is Essential for Activation of Mammalian MAP Kinase Kinases by Specific MAP Kinase Kinase Kinases. Mol. Cel 18, 295–306. doi:10.1016/j.molcel.2005.04.001
Tautz, D., and Domazet-Lošo, T. (2011). The Evolutionary Origin of Orphan Genes. Nat. Rev. Genet. 12, 692–702. doi:10.1038/nrg3053
Tautz, D. (2000). Evolution of Transcriptional Regulation. Curr. Opin. Genet. Develop. 10, 575–579. doi:10.1016/s0959-437x(00)00130-1
Tautz, D., and Pfeifle, C. (1989). A Non-radioactive In Situ Hybridization Method for the Localization of Specific RNAs in Drosophila Embryos Reveals Translational Control of the Segmentation Gene Hunchback. Chromosoma 98, 81–85. doi:10.1007/bf00291041
Topaloglu, Ö., Schlüter, G., Nayernia, K., and Engel, W. (2001). A 74-bp Promoter of the Tnp2 Gene Confers Testis- and Spermatid-specific Expression in Transgenic Mice. Biochem. Biophysical Res. Commun. 289, 597–601. doi:10.1006/bbrc.2001.5999
Tournier, C., Dong, C., Turner, T. K., Jones, S. N., Flavell, R. A., and Davis, R. J. (2001). MKK7 Is an Essential Component of the JNK Signal Transduction Pathway Activated by Proinflammatory Cytokines. Genes Dev. 15, 1419–1426. doi:10.1101/gad.888501
Tournier, C., Whitmarsh, A. J., Cavanagh, J., Barrett, T., and Davis, R. J. (1997). Mitogen-activated Protein Kinase Kinase 7 Is an Activator of the C-Jun NH2-terminal Kinase. Proc. Natl. Acad. Sci. 94, 7337–7342. doi:10.1073/pnas.94.14.7337
Tournier, C., Whitmarsh, A. J., Cavanagh, J., Barrett, T., and Davis, R. J. (1999). The MKK7 Gene Encodes a Group of C-Jun NH 2 -Terminal Kinase Kinases. Mol. Cel Biol 19, 1569–1581. doi:10.1128/mcb.19.2.1569
Tretyachenko, V., Vymětal, L., Bednárová, V., Kopecký, K., Hofbauerová, H., Jindrová, M., et al. (2017). Random Protein Sequences Can Form Defined Secondary Structures and Are Well-Tolerated In Vivo. Sci. Rep. 7, 15449. doi:10.1038/s41598-017-15635-8
Turner, L. M., Schwahn, D. J., and Harr, B. (2012). Reduced Male Fertility Is Common But Highly Variable In Form And Severity In A Natural House Mouse Hybrid Zone. Evolution 66, 443–458. doi:10.1111/j.1558-5646.2011.01445.x
Uchida, A., Sakib, S., Labit, E., Abbasi, S., Scott, R. W., Underhill, T. M., et al. (2020). Development and Function of Smooth Muscle Cells Is Modulated by Hic1 in Mouse Testis. Development 147, dev185884. doi:10.1242/dev.185884
Van Oss, S. B., and Carvunis, A. R. (2019). De Novo gene Birth. Plos Genet. 15, e1008160. doi:10.1371/journal.pgen.1008160
Wada, T., Joza, N., Cheng, H.-y. M., Sasaki, T., Kozieradzki, I., Bachmaier, K., et al. (2004). MKK7 Couples Stress Signalling to G2/M Cell-Cycle Progression and Cellular Senescence. Nat. Cel Biol 6, 215–226. doi:10.1038/ncb1098
Wang, X., Destrument, A., and Tournier, C. (2007). Physiological Roles of MKK4 and MKK7: Insights from Animal Models. Biochim. Biophys. Acta (Bba) - Mol. Cel Res. 1773, 1349–1357. doi:10.1016/j.bbamcr.2006.10.016
Waterston, R. H., Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., et al. (2002). Initial Sequencing and Comparative Analysis of the Mouse Genome. Nature 420, 520–562. doi:10.1038/nature01262
Wen, Z., Liu, D., Zhu, H., Sun, X., Xiao, Y., Lin, Z., et al. (2021). Deficiency for Lcn8 Causes Epididymal Sperm Maturation Defects in Mice. Biochem. Biophysical Res. Commun. 548, 7–13. doi:10.1016/j.bbrc.2021.02.052
Wilson, B. A., Foy, S. G., Neme, R., and Masel, J. (2017). Young Genes Are Highly Disordered as Predicted by the Preadaptation Hypothesis of De Novo Gene Birth. Nat. Ecol. Evol. 1, 0146. doi:10.1038/s41559-017-0146
Wray, G. A. (2007). The Evolutionary Significance of Cis-Regulatory Mutations. Nat. Rev. Genet. 8, 206–216. doi:10.1038/nrg2063
Wu, S., Li, H., Wang, L., Mak, N., Wu, X., Ge, R., et al. (2021). “Motor Proteins and Spermatogenesis,” in Molecular Mechanisms in Spermatogenesis. Editors C Cheng, and F Sun (Berlin: Springer). doi:10.1007/978-3-030-77779-1_7
Xie, C., Bekpen, C., Künzel, S., Keshavarz, M., Krebs-Wheaton, R., Skrabar, N., et al. (2019). A De Novo Evolved Gene in the House Mouse Regulates Female Pregnancy Cycles. Elife 8, e44392. doi:10.7554/eLife.44392
Xie, C., Bekpen, C., Künzel, S., Keshavarz, M., Krebs-Wheaton, R., Skrabar, N., et al. (2020). Dedicated Transcriptomics Combined with Power Analysis lead to Functional Understanding of Genes with Weak Phenotypic Changes in Knockout Lines. Plos Comput. Biol. 16, e1008354. doi:10.1371/journal.pcbi.1008354
Yamaguchi, R., Yamagata, K., Hasuwa, H., Inano, E., Ikawa, M., and Okabe, M. (2008). Cd52, Known as a Major Maturation-Associated Sperm Membrane Antigen Secreted from the Epididymis, Is Not Required for Fertilization in the Mouse. Genes to Cells 13, 851–861. doi:10.1111/j.1365-2443.2008.01210.x
Yang, S.-H., Sharrocks, A. D., and Whitmarsh, A. J. (2003). Transcriptional Regulation by the MAP Kinase Signaling Cascades. Gene 320, 3–21. doi:10.1016/s0378-1119(03)00816-3
Zambrowicz, B. P., Harendza, C. J., Zimmermann, J. W., Brinster, R. L., and Palmiter, R. D. (1993). Analysis of the Mouse Protamine 1 Promoter in Transgenic Mice. Proc. Natl. Acad. Sci. 90, 5071–5075. doi:10.1073/pnas.90.11.5071
Zhang, L., Ren, Y., Yang, T., Li, G., Chen, J., Gschwend, A. R., et al. (2019). Rapid Evolution of Protein Diversity by De Novo Origination in Oryza. Nat. Ecol. Evol. 3, 679–690. doi:10.1038/s41559-019-0822-5
Keywords: regulatory evolution, new promotor, kinase, testis, sperm maturation
Citation: Heinen T, Xie C, Keshavarz M, Stappert D, Künzel S and Tautz D (2022) Evolution of a New Testis-Specific Functional Promoter Within the Highly Conserved Map2k7 Gene of the Mouse. Front. Genet. 12:812139. doi: 10.3389/fgene.2021.812139
Received: 09 November 2021; Accepted: 08 December 2021;
Published: 05 January 2022.
Edited by:
Li Zhang, Chinese Institute for Brain Research, Beijing (CIBR), ChinaReviewed by:
Shengqian Xia, University of Chicago, United StatesBharat Ravi Iyengar, University of Münster, Germany
Copyright © 2022 Heinen, Xie, Keshavarz, Stappert, Künzel and Tautz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diethard Tautz, dGF1dHpAZXZvbGJpby5tcGcuZGU=
 Tobias Heinen
Tobias Heinen Chen Xie
Chen Xie Maryam Keshavarz
Maryam Keshavarz Dominik Stappert
Dominik Stappert Sven Künzel
Sven Künzel Diethard Tautz
Diethard Tautz