- 1College of Agriculture, Yangtze University, Jingzhou, China
- 2Key Laboratory for Biological Sciences and Genetic Improvement of Oil Crops, Oil Crops Research Institute of Chinese Academy of Agricultural Sciences, Ministry of Agriculture and Rural Affairs, Wuhan, China
- 3Fujian Provincial Key Laboratory of Crop Molecular and Cell Biology, Center of Legume Crop Genetics and Systems Biology/College of Agriculture, Oil Crops Research Institute, Fujian Agriculture and Forestry University (FAFU), Fuzhou, China
TCP proteins are plant-specific transcription factors that have multipurpose roles in plant developmental procedures and stress responses. Therefore, a genome-wide analysis was performed to categorize the TCP genes in the rapeseed genome. In this study, a total of 80 BnTCP genes were identified in the rapeseed genome and grouped into two main classes (PCF and CYC/TB1) according to phylogenetic analysis. The universal evolutionary analysis uncovered that BnTCP genes had experienced segmental duplications and positive selection pressure. Gene structure and conserved motif examination presented that Class I and Class II have diverse intron-exon patterns and motifs numbers. Overall, nine conserved motifs were identified and varied from 2 to 7 in all TCP genes; and some of them were gene-specific. Mainly, Class II (PCF and CYC/TB1) possessed diverse structures compared to Class I. We identified four hormone- and four stress-related responsive cis-elements in the promoter regions. Moreover, 32 bna-miRNAs from 14 families were found to be targeting 21 BnTCPs genes. Gene ontology enrichment analysis presented that the BnTCP genes were primarily related to RNA/DNA binding, metabolic processes, transcriptional regulatory activities, etc. Transcriptome-based tissue-specific expression analysis showed that only a few genes (mainly BnTCP9, BnTCP22, BnTCP25, BnTCP48, BnTCP52, BnTCP60, BnTCP66, and BnTCP74) presented higher expression in root, stem, leaf, flower, seeds, and silique among all tested tissues. Likewise, qRT-PCR-based expression analysis exhibited that BnTCP36, BnTCP39, BnTCP53, BnTCP59, and BnTCP60 showed higher expression at certain time points under various hormones and abiotic stress conditions but not by drought and MeJA. Our results opened the new groundwork for future understanding of the intricate mechanisms of BnTCP in various developmental processes and abiotic stress signaling pathways in rapeseed.
Introduction
Transcription factors (TFs) contribute to plant growth and development, regulate gene expression, and play a significant role in several cellular/biological processes (Hoang et al., 2017; Karaaslan et al., 2020). The Teosinte Branched 1, Cycloidea, and Proliferating Cell Factors (TCP) gene family is a group of plant-specific TFs limited to higher plants (Cubas et al., 1999). These TFs have numerous roles in controlling plant growth and developmental procedures by directing cell proliferation (Cubas et al., 1999; Martín-Trillo and Cubas, 2010). Notably, these TFs are categorized by an extremely conserved 59-amino-acid basic helix–loop–helix (bHLH) motif at the N-terminus nominated as the TCP domain (Aggarwal et al., 2010). This domain is accountable for nuclear targeting, DNA binding and also contributes to protein-protein interactions (Kosugi and Ohashi, 2002). Due to the dissimilarity in the TCP domain, TCP gene family members are grouped into two main classes, i.e., Class I (PCF) and Class II (CYC/TB1 and CIN sub-classes) (Navaud et al., 2007; Viola et al., 2012). Along with the TCP domain, several members of Class II retain 18–20-residue arginine-rich motifs (Viola et al., 2012; Sarvepalli and Nath, 2018). This supposed R domain was anticipated to develop a hydrophilic-helix or α-coiled-coil structure that facilitates the protein-protein interactions in plants (Cubas et al., 1999).
Over the past few years, TCP gene families have been reported in several plant species. For example, 16 TCP members in Moso bamboo (Phyllostachys edulis) (Liu et al., 2018); 18 TCP members in sweet potato (Ipomoea batatas L.) (Ren et al., 2021); 23 TCP members in orchids (Phalaenopsis equestris) (Lin et al., 2016); 24 TCP members in Arabidopsis thaliana (Yao et al., 2007); 27 TCP members in cucumber (Cucumis sativus L.) (Wen et al., 2020); 28 TCP members in rice (Oryza sativa L.) (Yao et al., 2007); 38 TCP members in cotton (Gossypium raimondii L.) (Ma et al., 2014); 39 TCP members in turnips (Brassica rapa ssp. rapa) (Du et al., 2017); 63 TCP members in Brassica juncea var. tumida (He et al., 2020), 73 TCP members in allotetraploid cotton (Gossypium hirsutum L.) (Yin et al., 2018), etc. Several studies have shown that TCP genes play significant roles in plant growth and developmental processes, including trichome formation (Wang et al., 2013), seed germination (Zhang et al., 2019), petal development (van Es et al., 2018), floral development (Li et al., 2019), shoot branching (Braun et al., 2012), and leaf anatomical morphology (Ren et al., 2021). These findings indicate the diverse role of TCP genes in regulating plant developmental processes.
Likewise, several TCP genes expression is also regulated by various hormones and abiotic stress treatments. For instance, in cotton, the GhTCP14 gene is considered a vital switch in an auxin-facilitated extension of fiber cells (Wang et al., 2013). In A. thaliana, the expression level of AtTCP15 was induced by gibberellic acid (Gastaldi et al., 2020), and AtTCP1 was positively regulated by brassinosteroids treatment (Gao et al., 2015). Furthermore, the different TCP genes also participate in ethylene, strigolactone, and cytokinins signaling pathways (Braun et al., 2012; van Es et al., 2018). Under abiotic stress conditions, the expression level of AtTCP15 was induced by heat stress (Wu et al., 2016), and TCP20/22 was positively regulated by the circadian clock in A. thaliana (Wu et al., 2016). In cotton, several GhTCP genes were significantly up-regulated under heat, salt, and drought stresses (Yin et al., 2018). In cucumber, many CsTCP genes significantly responded to temperature and photoperiod and were also induced by gibberellic acid and ethylene treatments (Wen et al., 2020). TCP genes are also expressed in different tissues, such as many TCP genes mainly expressed in flower, leaf, and stem, flower, bud differentiation, and gametophyte development in Prunus mume (Zhou et al., 2016).
Rapeseed (Brassica napus L.) is the second most crucial oilseed crop and possesses a complex genome. However, its growth and productivity are significantly affected at physiological, biochemical, and molecular levels in response to various plant hormones and abiotic stress conditions (Raza et al., 2021a; He et al., 2021; Raza, 2021; Su et al., 2021). To date, the TCP gene family and key TCP genes responding to abiotic stress have not been fully characterized in rapeseed. Consequently, this is the first genome-wide study to identify the TCP genes in the rapeseed genome. To boost our understanding into TCP genes evolution in rapeseed, their phylogenetic relationships, synteny study, gene structures, conserved motifs, cis-elements, miRNA predictions, and functional annotation were evaluated. Additionally, their expression profile in numerous tissues/organs and under various phytohormones and abiotic stress conditions were largely appraised, which deeply improved our understanding of the TCP genes in rapeseed.
Material and Methods
Identification and Characterization of TCP Genes in Rapeseed
As described in our recent studies (Raza et al., 2021a; Su et al., 2021), two methods were used to recognize TCP genes in the B. napus genome, i.e., BLASTP and the Hidden Markov Model (HMM). The genome of B. napus (ZhongShuang 11; ZS11 variety) was downloaded from the BnPIR database (http://cbi.hzau.edu.cn/bnapus/index.php). For BLASTP, the amino acid sequences of 24 AtTCPs were used as a query with an e-value set to 1e−5. The amino acid sequences of 24 AtTCPs were attained from the TAIR Arabidopsis genome database (http://www.arabidopsis.org/). Secondly, the local software HMMER 3.1 (http://www.hmmer.org/) was used to search the TCP genes with default constraints. Then, the HMM file of the TCP domain (PF03634) was downloaded from the Pfam protein domain database (http://pfam.xfam.org/). Finally, 80 BnTCP genes were found by merging the two processes in the rapeseed genome. Likewise, TCP genes were also identified in Brassica rapa and Brassica oleracea genomes using the same method. Their genome sequences were downloaded from JGI Phytozome 12.0 database (https://phytozome.jgi.doe.gov/pz/portal.html).
The physico-chemical properties of BnTCPs were evaluated with the help of ProtParam (http://web.expasy.org/protparam/). The subcellular localization of BnTCP proteins was prophesied from the WoLF PSORT server (https://wolfpsort.hgc.jp/). Genes structures were created via TBtools software (V 1.068; https://github.com/CJ-Chen/TBtools). The conserved motifs of BnTCP protein sequences were recognized employing the MEME website (https://meme-suite.org/meme/db/motifs).
Phylogenetic Tree and Synteny Analysis of BnTCP Proteins
To explore the evolutionary relationship of the BnTCPs, a phylogenetic tree between B. napus, B. oleracea, B. rapa, and A. thaliana protein sequences were constructed. The sequence alignment was executed using MEGA 7 software (https://megasoftware.net/home). The neighbor-joining process was accomplished to create a phylogenetic tree with 1,000 bootstrap replicates using the Evolview v3 website (https://www.evolgenius.info/evolview) to display the phylogenetic tree. Synteny relationships of TCP genes between B. napus, B. oleracea, B. rapa, and A. thaliana were performed using the python-package, JCVI (https://github.com/tanghaibao/jcvi) and Multiple Collinearity Scan Toolkit (MCScanX; https://github.com/wyp1125/MCScanX). The Ka/Ks ratios of all TCPs were calculated using KaKs_Calculator 2.0 (https://sourceforge.net/projects/kakscalculator2/).
Analysis of Cis-Regulatory Elements in the BnTCPs Promoters
To investigate the putative cis-elements in the BnTCPs promoters, we extracted the 2Kb sequence upstream of start codons in the B. napus genome. Moreover, the promoter sequence of each gene was examined applying the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), and the figure was drawn with the help of TBtools software.
Prediction of Putative miRNA Targeting BnTCP Genes and GO Annotation Analysis
The CDS of BnTCPs was employed to distinguish possible target miRNAs in the psRNATarget database (https://www.zhaolab.org/psRNATarget/home) with default parameters. The interaction network figure between the miRNAs and BnTCP genes was constructed with Cytoscape software (V3.8.2; https://cytoscape.org/download.html). Gene ontology (GO) annotation analysis was accomplished by submitting all BnTCPs protein sequences to the eggNOG website (http://eggnog-mapper.embl.de/). At the same time, GO enrichment analysis was performed with TBtools software.
Expression Profiling of BnTCP Genes in Diverse Tissues
For tissue-specific expression profiling, the rapeseed RNA-seq data was downloaded from the National Genomics Data Center (https://ngdc.cncb.ac.cn/gsa/; BioProject ID: PRJCA001495). The data were analyzed as described in a recent study (Raza et al., 2021a). In short, Cuffquant and Cuffnorm were used to produce normalized counts in transcripts per million (TPM) values. Due to the large difference in the expression profiles, we used the log10 method to calculate the expression results to better visualize differently expressed genes in diverse tissues. Based on log10 values, the expression heat map was created using GraphPad Prism 9.0.0 software (https://www.graphpad.com/).
Plant Material and Stress Conditions
A widely used variety, ZhongShuang 11 (ZS11), was used for stress treatments. Seeds of ZS11 were provided by the OCRI-CAAS, Wuhan, China. The stress treatments were carried out as described in our recent work (Raza et al., 2021a). All the treatments were executed with three biological replicates. All the samples were immediately frozen in liquid nitrogen and were stored at −80°C for further experimentation.
RNA Extraction and qRT-PCR Analysis
Total RNA was extracted using TransZol Up Plus RNA Kit (TransGen Biotech, Beijing, China), and cDNA was synthesized using cDNA SuperMix kit (TransGen Biotech, Beijing, China) according to developer instructions. The complete data of qRT-PCR reaction has been explained in our recent work (Raza et al., 2021a). The expression data were investigated using the 2−ΔΔCT method as described by Wang et al. (2014). All the primers used for qRT-PCR are listed in Supplementary Table S1. The graphs were developed using GraphPad Prism 9.0.0 software.
Results
Identification and Characterization of TCP Genes in Rapeseed
To identify TCP family genes in rapeseed, 24 AtTCP protein sequences were used as queries for a BlastP search against the rapeseed genome. As a result, a sum of 80 BnTCP genes was identified with the TCP domain (Table 1). Hereafter, these genes are named as “BnTCP1–BnTCP80.” Among which 38 genes were positioned in the A subgenome, and 42 genes were positioned in the C subgenome (Table 1). Comprehensive data of the 80 BnTCP genes are shown in Table 1. Briefly, gene length varied from 531 bp (BnTCP10) to 9,673 bp (BnTCP40), the CDS length varied from 531 bp (BnTCP10) to 5,193 bp (BnTCP40), and the amino acid length varied from 176 (BnTCP10) to 1,730 (BnTCP40) amino acids. The number of exons varied from one (BnTCP3–BnTCP6, BnTCP8–BnTCP10, BnTCP12–BnTCP15) to 11 (BnTCP40) (Table 1). Notably, only two genes (BnTCP40 and BnTCP66) had the highest number of introns (11 and 8, respectively), and several genes lack introns, such as BnTCP3-BnTCP6, BnTCP8–BnTCP10, BnTCP12–BnTCP15 (Table 1). The forecasted molecular weights of the 80 BnTCP proteins extended from 18.60 kDa (BnTCP10) to 194.78 kDa (BnTCP40), and the isoelectric points prolonged from 5.31 (BnTCP28/45) to 11.12 (BnTCP49). The differences in molecular weights and isoelectric points are mainly due to the unusual high content of basic amino acids and post-translational modifications. The results of in silico subcellular localization prophesied that two BnTCP proteins were located on the peroxisome, four BnTCP proteins on the chloroplast, seven BnTCP proteins on the mitochondria, and remaining 67 BnTCP proteins were located on the nucleus (Table 1). Meanwhile, 24 genes (AtTCP1–AtTCP24) from A. thaliana, 39 genes (BraTCP1–BraTCP39) from B. rapa, and 40 genes (BoTCP1–BoTCP41) from B. oleracea genomes were also identified (Supplementary Table S2).
Phylogenetic Relationship of TCP Genes
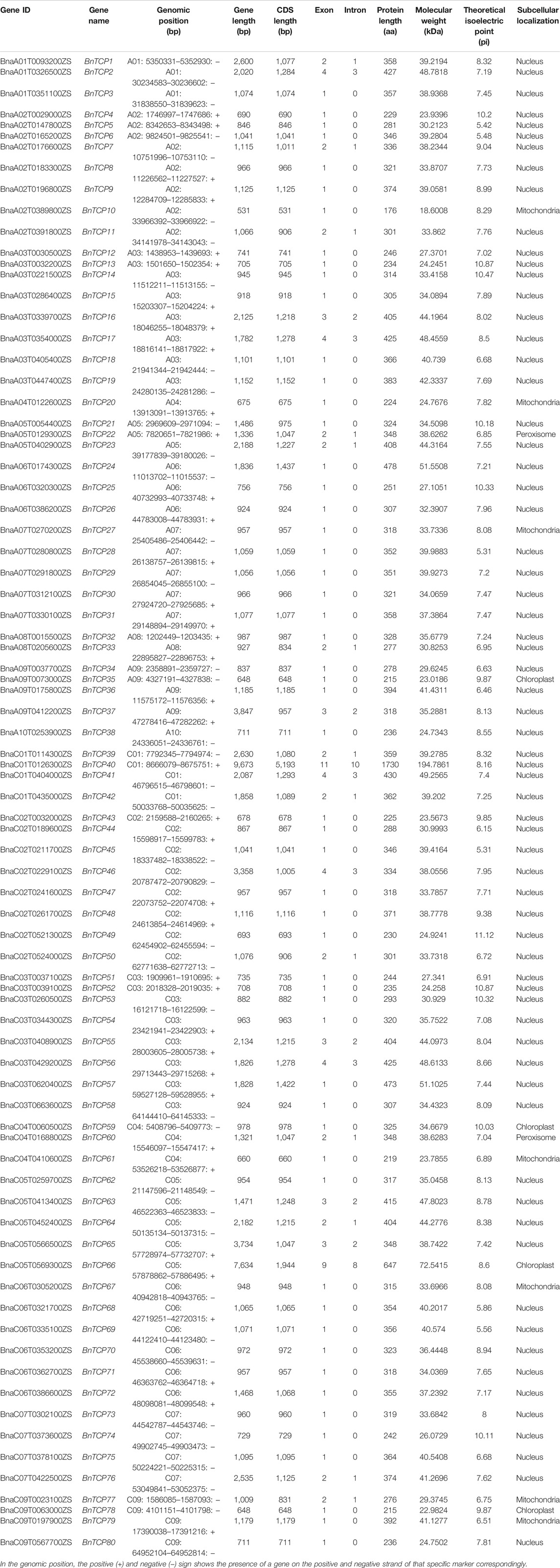
To unreveal the evolutionary relationship between the BnTCPs, BoTCPs, BraTCPs, and AtTCPs genes, an unrooted phylogenetic tree was built by a multiple sequence alignment of the prophesied TCP protein sequences from B. napus, B. oleracea, B. rapa, and A. thaliana (Figure 1). Based on the standard classification of Arabidopsis TCP genes (Yao et al., 2007), the 184 TCPs genes from four plant species were distributed into two main classes, i.e., Class I (PCF) and Class II. Further, Class II was further divided into two sub-classes (CYC/TB1 and CIN) (Figure 1). The TCPs from the four plant species were scattered in nearly all clades, suggesting that the TCP family genes differentiated before the separation of these ancestral plant species. The findings discovered that Class I (PCF) was encompassed 91 TCP members (39 BnTCPs, 19 BoTCPs, 20 BraTCPs, and 13 AtTCPs). Class II contained a sum of 92 TCP members. Particularly, sub-class CYC/TB1 was comprised of 31 TCP members (14 BnTCPs, eight BoTCPs, six BraTCPs, and three AtTCPs), and CIN was comprised of 61 TCP members (27 BnTCPs, 13 BoTCPs, 13 BraTCPs, and eight AtTCPs) (Figure 1; Supplementary Table S3). In a nutshell, TCPs grouping into the same sub-class may retain corresponding functions. It is worth highlighting that BnTCP genes were almost equally scattered in both classes with homologs from other plant species (Figure 1; Supplementary Table S3). Mainly, Class I (PCF) was observed to have a greater number of BnTCPs (39), followed by CIN (27) and CYC/TB1 (14). Moreover, it was observed that the BnTCPs have a more closely phylogenetic relationship with the BoTCPs and BraTCPs in each class.
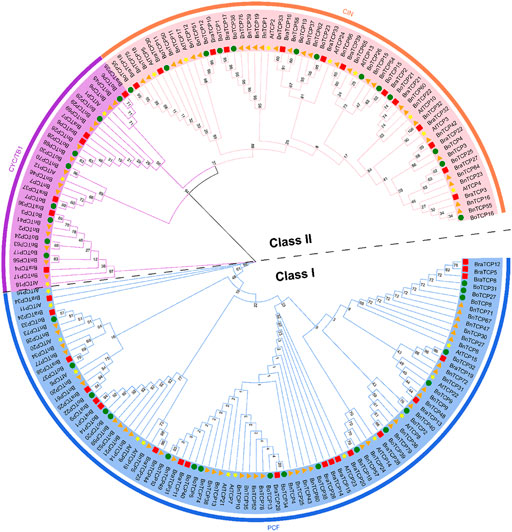
FIGURE 1. A neighbor-joining phylogenetic tree assessment of TCP genes from B. napus, B. oleracea, B. rapa, and A. thaliana. Overall, 80 BnTCPs from B. napus (orange triangle), 40 BoTCPs from B. oleracea (green circles), 39 BraTCPs from B. rapa (red boxes), and 24 AtTCPs from A. thaliana (yellow stars) were clustered into two major classes, denoted by exclusive colors.
Chromosomal Locations and Synteny Evaluation of TCP Genes
The chromosol location of 38 BnTCPs gene pairs was observed (Figure 2; Supplementary Table S4), and 18 out of 19 chromosomes held BnTCPs genes (Figure 2). Mainly, chromosomes A04 and A10 possessed only one gene, chromosome A08 possessed two genes, chromosomes A01, A05, A06, C01, C04, and C09 possessed three genes, chromosomes A07, A09, and C07 possessed four genes, chromosomes A02 and C05 possessed five genes, chromosome C06 possessed six genes, chromosomes C02 and C03 possessed eight genes. Chromosome A03 had the highest number of genes (10). Astonishingly, the C08 chromosome did not contain any BnTCP genes (Figure 2). The dataset displays that segmental duplications have contributed to the development of BnTCP genes (Supplementary Table S4). Notably, one putative paralog pair, i.e., BnTCP16/BnTCP55, was produced by tandem duplication (Supplementary Table S4). These results showed the large-scale segmental duplication events involved in the evolution of the BnTCP gene family in rapeseed.
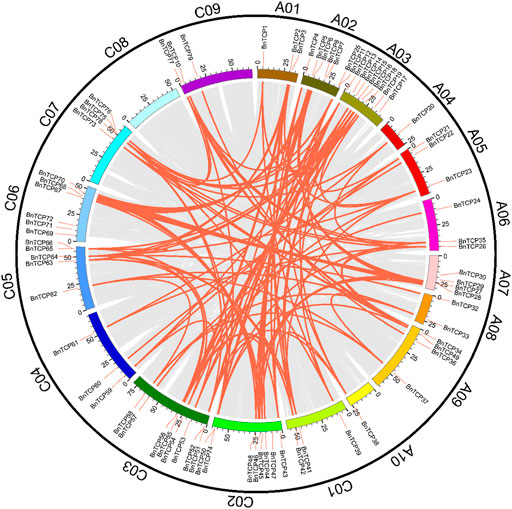
FIGURE 2. Chromosomal locations and inter-chromosomal relations of BnTCP genes. Grey lines in the background present all syntenic blocks in the B. napus genome, and the red lines present syntenic BnTCPs gene pairs.
Collinearity analysis was used to appraise the evolutionary relationship of the TCP genes between B. napus and the other heritable plant species (Figure 3; Supplementary Table S4). In general, in the A subgenome, several B. napus genes showed syntenic networks with various BraTCPs, and AtTCP. Notably, one BnTCP gene at chromosome A10 also displayed a syntenic relationship with a BoTCP gene at chromosome C09. Likewise, in the C subgenome, many B. napus genes revealed syntenic connections with different BraTCPs, and AtTCPs (Figure 3). Mainly, three BnTCP genes at chromosome C03 and C05 also showed a syntenic relationship with three BrTCP genes at chromosome A03, A05 and A08 (Figure 3). On the other hand, several homologues of A. thaliana, B. rapa, and B. oleracea showed a syntenic alliance with BnTCPs genes in the rapeseed genome, suggesting that whole-genome or segmental duplication events represented a crucial role in the evolution of BnTCPs gene family in the rapeseed genome (Figure 3; Supplementary Table S4).
FIGURE 3. Collinearity analysis of TCP genes in B. napus, B. rapa, B. oleracea, and A. thaliana chromosomes. Grey lines in the background display the collinear blocks within B. napus and other plant genomes. However, the red and green lines signify the syntenic TCP gene pairs. Genes located on the B. napus A subgenome are syntenic with B. rapa, B. oleracea, and A. thaliana, while genes located on B. napus C subgenome are mainly syntenic with B. rapa, B. oleracea, and A. thaliana.
The Ka/Ks ratio is regarded as a considerable indicator in assessing the sequence evolution in terms of selection pressures and duplication types (Hurst, 2002). Hence, to comprehend the evolutionary history of the BnTCPs, the Ka, Ks, and Ka/Ks ratio was determined for B. napus and the other three plant species (Supplementary Table S4). The calculations disclosed that all of the duplicated BnTCP gene pairs had a Ka/Ks ratio of <1 (Supplementary Table S4), indicating that the BnTCP genes may have undergone intense purifying selective pressure during the evolution process (Supplementary Table S4). Similar outcomes were also examined in B. rapa, B. oleracea, and A. thaliana (Supplementary Table S4).
Assessment of Gene Structures and Conserved Motifs of BnTCP Genes
The exon-intron configurations of the BnTCP genes were examined to boost our knowledge of the evolution of the rapeseed TCP family genes. The results showed that the number of introns and exons ranged from 0 to 10 and 1–11, respectively (Figure 4B; Table 1). Overall, 55 genes have one exon and zero intron; 13 genes have two exons and one intron; five genes have three exons and two introns; five genes have four exons and three introns; one gene has nine exons and eight introns; and one gene has 11 exons and 10 introns (Figure 4; Table 1). Notably, Class I (PCF) and Class II (CIN and CYC/TB1) possessed almost similar structures except for a few genes (Figure 4B). Losses or gains of exons were discovered throughout the evolution of the TCP family genes. Our outcomes advised that TCP genes retained a comparatively constant exon-intron arrangement during the evolution of the rapeseed genome. Moreover, BnTCP gene members inside a Class had extremely matching gene structures, constant with their phylogenetic groups.
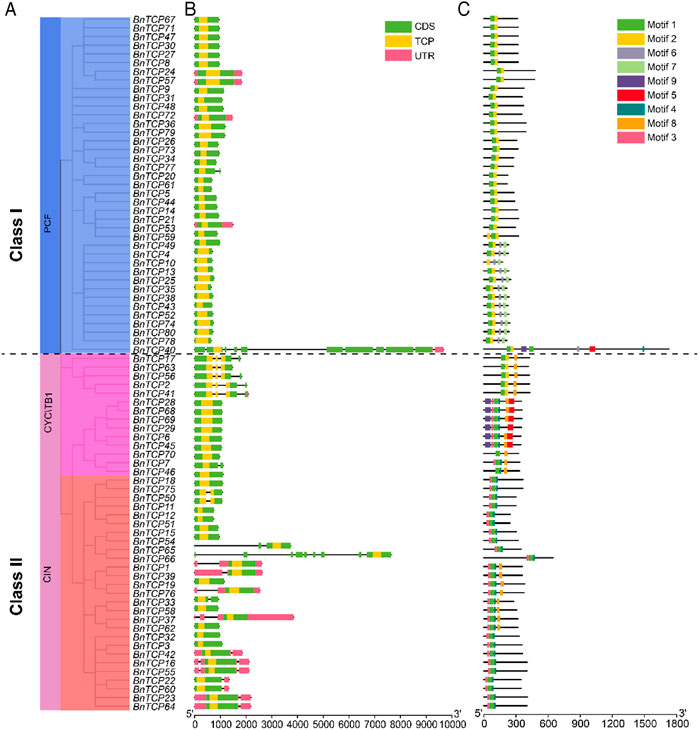
FIGURE 4. The gene structure and motif analysis of BnTCPs genes. (A) Based on phylogenetic relationships, the BnTCPs were clustered into two major classes. (B) Gene structure of BnTCPs genes. The pink color signifies UTR regions, the light green color denotes exon or CDS, the yellow color represents TCP domain, and the black horizontal line symbolizes introns. (C) Conserved motif compositions detected in BnTCPs. Different color boxes represent different motifs.
Furthermore, the full-length protein sequences were examined to identify their conserved motifs (Figure 4C). The conserved motif of the TCP genes varied from 2 to 7. Overall, nine conserved motifs were identified, and the complete data (such as name, sequence, width, and E-value) of discovered motifs are presented in Supplementary Table S5. The motif dispersals were also comparable within the Classes (Figure 4C). For example, motifs 1, 2, 6, and 7 were specific to Class I; however, motifs 6 were also present in some genes belonging to Class II (mainly CYC/TB1). In Class I, only one gene (BnTCP40) was found to have a greater number of motifs (seven). In Class II (mainly CYC/TB1), motifs 5 and 9 were specific and absent in other genes. Likewise, motifs 4, 8, and 3 were specific to Class II (CIN and CYC/TB1) (Figure 4C). In short, the class organization’s consistency was convincingly sustained by evaluating the conserved motifs composition, gene structures, and phylogenetic interactions, indicating that the TCP proteins have tremendously well-maintained amino acid remains and members within a class may possess parallel roles.
Examination of Cis-Elements in Promoters of BnTCP Genes
The cis-acting elements in the gene promoter generally regulate gene expression levels and functions. Therefore, cis-acting elements in BnTCPs promoter localities were analyzed to characterize the gene functions and regulatory roles. The comprehensive data of cis-elements are shown in Supplementary Table S6. Overall, we focused on and identified eight types of elements. Particularly, four phytohormone-correlated [(abscisic acid (ABA), auxin, methyl jasmonate (MeJA), and gibberellin (GA)] responsive elements consist of ABRE, TGA-element, TGACG-motif, CGTCA-motif, P-box, GARE-motif, TATC-box, were distinguished (Figure 5; Supplementary Table S6). Specifically, several phytohormone-correlated elements were anticipated to be limited to some genes and extensively dispersed (Figure 5; Supplementary Table S6), indicating the essential role of these genes in phytohormone-arbitration.
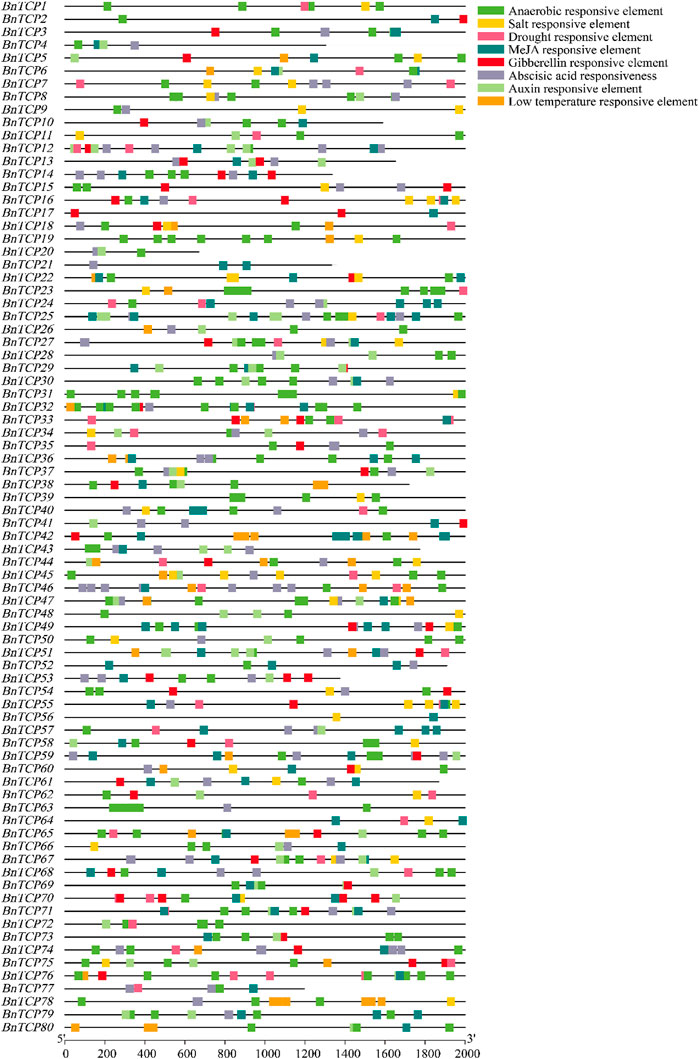
FIGURE 5. Cis-elements in the promoter regions of the BnTCP genes are linked with different hormone- and stress-responsive elements. Different color boxes show different identified elements. See Supplementary Table S6 for more information.
Additionally, we identified four stress-associated (salt, drought, low-temperature, and anaerobic) responsive elements, including TCA-element, MBS, LTR, ARE, suggesting their contribution to stress stimulation (Figure 5; Supplementary Table S6). TCA-element associated with salt stress responsiveness was mainly present in 38 BnTCP genes and widely distributed in four genes (BnTCP16, BnTCP22, BnTCP45, and BnTCP55) (Figure 5; Supplementary Table S6). Drought stress responsive element (MBS) was mainly present in 36 genes and widely circulated in 12 genes (Figure 5; Supplementary Table S6). Low-temperature responsive element (LTR) was particularly present in 26 genes and widely scattered in six genes (BnTCP42, BnTCP44, BnTCP46, BnTCP65, BnTCP78, and BnTCP80) (Figure 5; Supplementary Table S6). ARE element associated with anaerobic responsiveness was mainly present in 74 genes and largely dispersed in 35 genes (Figure 5; Supplementary Table S6). Generally, these findings recommended that BnTCPs expression profiles may differ under phytohormone and abiotic stress conditions.
Genome-Wide Prediction of miRNA Targeting BnTCP Genes
During the past few years, various investigations have uncovered that miRNA-induced regulation participates in the stress responses in plants. Consequently, for a profound knowledge of miRNA-mediated post-transcriptional adjustment of BnTCP genes, we forecasted 32 miRNAs targeting 21 BnTCP genes (Figure 6A; Supplementary Table S7). Some of the miRNA-targeted sites are shown in Figure 6B, while the comprehensive data of all miRNAs targeted genes/sites is given in Supplementary Table S7. Overall, the results demonstrated that one member of the bna-miR1140 family targeted one gene (BnTCP57); one member of the bna-miR161 family targeted one gene (BnTCP32); one member of the bna-miR162 family targeted one gene (BnTCP77); one member of the bna-miR2111a-3p targeted one gene (BnTCP39); one member of the bna-miR6031 targeted one gene (BnTCP61); one member of the bna-miR6033 targeted one gene (BnTCP57); one member of bna-miR396 family targeted three genes (BnTCP28, BnTCP34, and BnTCP68); and one member of bna-miR159 family targeted 17 genes. Two members of the bna-miR171 family targeted one gene (BnTCP39); two members of the bna-miR6029 family targeted two genes (BnTCP6 and BnTCP45); two members of the bna-miR6030 family targeted two genes (BnTCP24 and BnTCP57); and two members of the bna-miR394 family targeted two genes (BnTCP40 and BnTCP56). Three members of the bna-miR395 family targeted four genes (BnTCP5, BnTCP61, BnTCP20, and BnTCP44). Four members of the bna-miR172 family targeted five genes (BnTCP11, BnTCP19, BnTCP58, BnTCP62, and BnTCP76) (Figure 6A; Supplementary Table S7). In general, BnTCP5, BnTCP19, BnTCP20, BnTCP39, BnTCP44, BnTCP57, BnTCP58, BnTCP61, and BnTCP76 were predicted to be targeted by a greater number (three and four) of miRNAs (Figure 6A; Supplementary Table S7). In further examination, the expression levels of these miRNAs and their targeted genes require validation to preside their biological functions in the rapeseed genome.
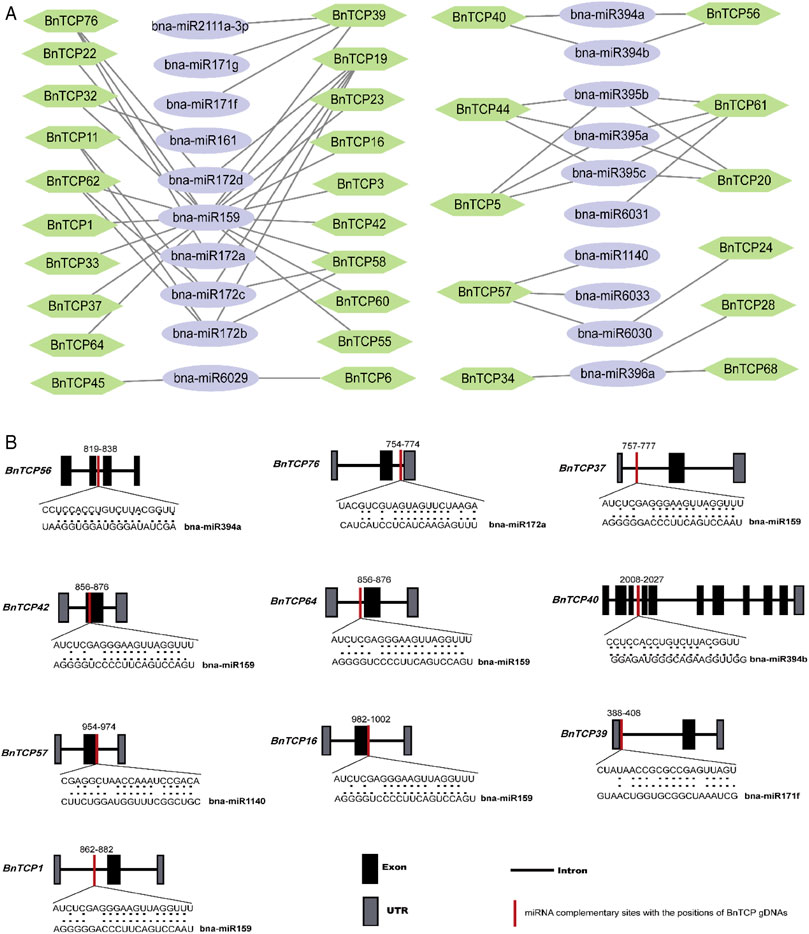
FIGURE 6. miRNA targeting BnTCP genes. (A) Network figure of anticipated miRNA targeting BnTCPs genes. Green hexagon colors correspond to BnTCPs genes, and bluish ellipse shapes represent miRNAs. (B) The schematic illustration signifies the BnTCPs targeted by miRNAs. The RNA sequence of each complementary site from 5′ to 3′ and the predicted miRNA sequence from 3′ to 5′ are exposed in the long-drawn-out areas. See Supplementary Table S7 for the detailed data of all predicted miRNAs.
Functional Annotation Study of BnTCP Genes
To further recognize the contribution of BnTCP genes, we performed GO annotation and enrichment analysis based on biological process (BP), molecular function (MF), and cellular component (CC) classes. These terms can boost our understanding of the diverse function of genes. The results of BP, MF, and CC exhibited several significantly enriched terms (Supplementary Table S8). For instance, in GO-BP class, 49 highly enriched terms were identified, including cellular process (GO: 0009987), RNA biosynthetic process (GO: 0032774), regulation of nucleic acid-templated transcription (GO: 1903506), heterocycle metabolic process (GO: 0046483), organic substance biosynthetic process (GO: 1901576), regulation of metabolic process (GO:0019222), nitrogen compound metabolic process (GO: 0006807) (Supplementary Table S8). The GO-CC enrichment outcomes distinguished ten highly enriched terms, including intracellular part (GO: 0044424), cell part (GO: 0044464), cellular_component (GO: 0005575), organelle (GO: 0043226), membrane-bounded organelle (GO: 0043227), intracellular (GO: 0005622), nucleus (GO: 0005634) (Supplementary Table S8). Similarly, the GO-MF results projected eight significantly enriched terms such as transcription regulator activity (GO: 0140110), molecular_function (GO: 0003674), DNA binding transcription factor activity (GO: 0003700), organic cyclic compound binding (GO: 0097159), nucleic acid binding (GO: 0003676) (Supplementary Table S8). Overall, GO enrichment analysis validates the functional role of BnTCP genes in several cellulars, molecular, and biological processes related to RNA/DNA binding, metabolic processes, transcriptional regulatory activities associated with rapeseed growth and developmental processes.
Expression Profiles of BnTCP Genes Under Various Tissues
Tissue-specific expression profiles of BnTCPs genes were examined in six diverse tissues and organs, i.e., roots, stems, leaves, flowers, seeds, and silique using RNA-seq data from B. napus (ZS11 variety) (BioProject ID: PRJCA001495). Overall, the results showed that only a few genes showed higher expression in some specific tissues (Figure 7). For instance, some genes showed higher expression levels in the root, including BnTCP9, BnTCP13, BnTCP25, BnTCP31, BnTCP35, BnTCP36, BnTCP48, BnTCP52, BnTCP72, and BnTCP74). In stem, a few genes showed higher expression levels, including BnTCP9, BnTCP26, BnTCP31, BnTCP36, BnTCP48, BnTCP52, BnTCP66, BnTCP67, BnTCP72, BnTCP73, and BnTCP74). In leaf, BnTCP9, BnTCP13, BnTCP16, BnTCP19, BnTCP22, BnTCP23, BnTCP35, BnTCP39, BnTCP52, BnTCP55, BnTCP60, BnTCP64, BnTCP65, BnTCP66, BnTCP67, BnTCP74, and BnTCP76 genes display higher expression patterns. In flower, BnTCP16, BnTCP19, BnTCP23, BnTCP35, BnTCP42, BnTCP55, BnTCP60, BnTCP64, BnTCP52, BnTCP55, BnTCP60, BnTCP64, BnTCP66, BnTCP74, BnTCP76, and BnTCP79 genes exhibited higher expression patterns. In seeds, several genes showed higher expression in seeds at different time points, such as BnTCP3, BnTCP9, BnTCP22, BnTCP25, BnTCP35, BnTCP36 BnTCP60, BnTCP74, BnTCP78, and BnTCP79). Likewise, in silique, many genes including BnTCP9, BnTCP22, BnTCP24, BnTCP25, BnTCP27, BnTCP30, BnTCP31, BnTCP35, BnTCP39, BnTCP48, BnTCP52, BnTCP60, BnTCP64, BnTCP66, BnTCP67, BnTCP71, BnTCP74, BnTCP78, and BnTCP80, showed higher expression patterns at different time points (Figure 7). Notably, some genes also showed moderate expression levels in various tissues. Overall, expression data show that only a few genes may significantly contribute to rapeseed growth and development. Therefore, the precise function of these genes could be discovered in future investigations.
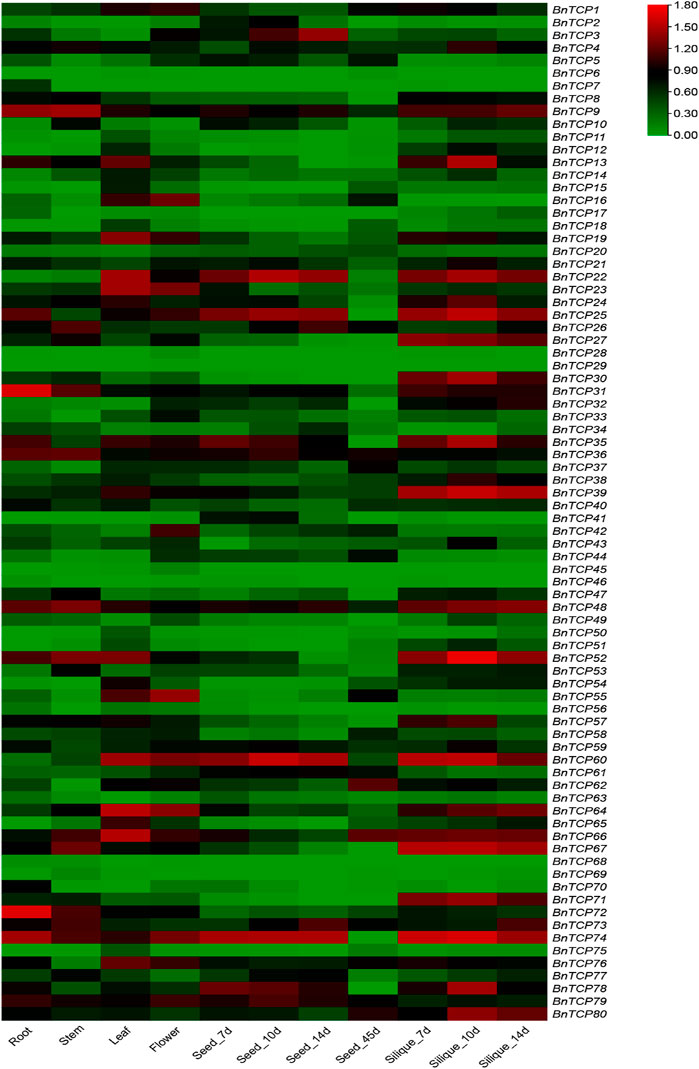
FIGURE 7. Expression profiling of BnTCP genes in various developmental organs/tissues. The 7, 10, 14, and 45 d tags specified the time-points when the samples were harvested. The red, black, and green colors display high to low expression levels. The expression heat map was created by taking log10 of transcripts per million (TPM) values.
Expression Profiles of BnTCP Genes Under Phytohormones and Abiotic Stress Conditions
In this study, qRT-PCR-based expression profiles of ten randomly selected BnTCPs genes were evaluated in response to abiotic (cold, waterlogging, salinity, and drought), and phytohormones (ABA, GA, IAA, and MeJA) stresses at different time points (Figures 8, 9). Under abiotic stress conditions, most of the genes exhibited comparatively low expression levels, apart from some genes. For example, in response to cold stress, BnTCP53 showed significantly higher expression at 4 h (1.521 fold), 6 h (2.081 fold), and 8 h (1.616 fold); BnTCP59 showed higher expression at 2 h (2.312 fold), 4 h (2.409 fold), 6 h (2.873 fold), and 8 h (2.187 fold); and BnTCP60 showed higher expression mainly at 6 h (1.254 fold), and 8 h (1.598 fold), compared to CK. Under waterlogging stress, BnTCP39 showed higher expression at 2 h (11.684 fold), 8 h (13.369 fold); and BnTCP53 showed higher expression at 2 h (10.865 fold), compared to CK. Under salt stress, almost all genes showed relatively higher expression at CK; however, BnTCP53 display substantially higher expression at 2 h (1.402 fold), 4 h (1.395 fold), 6 h (1.109 fold), and 8 h (1.197 fold), compared to CK and other genes. In response to drought stress, only BnTCP7 was up-regulated at 4 h (29.946 fold), 6 h (31.192 fold), and 8 h (28.428 fold) (Figure 8).
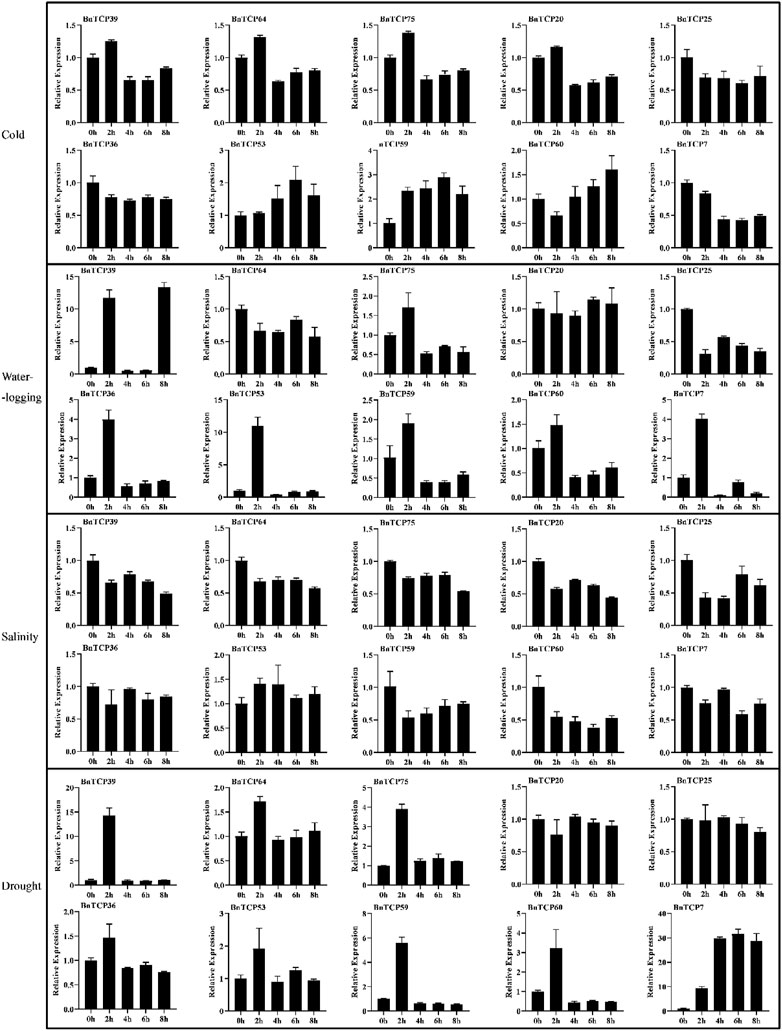
FIGURE 8. Expression patterns of the BnTCP genes under different abiotic stress conditions. The 0 h (CK), 2, 4, 6, and 8 h labels presented the time points (hours) when the samples were harvested for expression study after the stress treatment. Error bars represent the SD (n = 3).
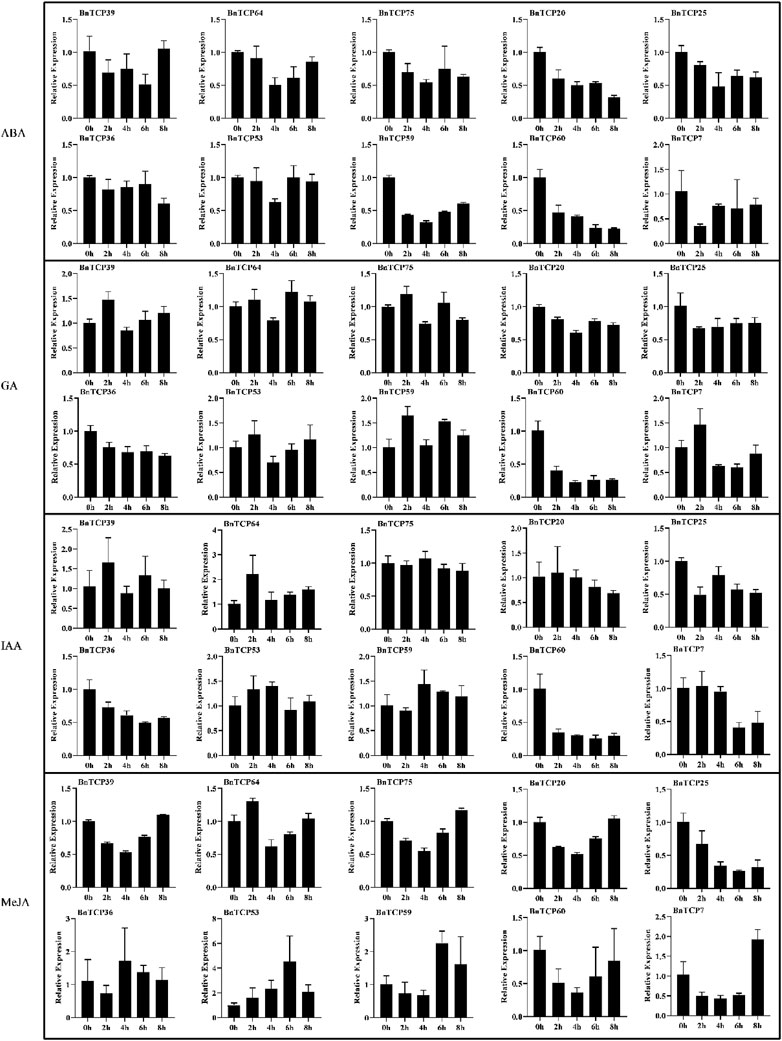
FIGURE 9. Expression patterns of the BnTCP genes under different hormones treatments. The 0 h (CK), 2, 4, 6, and 8 h labels presented the time points (hours) when the samples were harvested for expression study after the stress treatment. Error bars represent the SD (n = 3).
Under phytohormones treatment, almost all genes showed significantly higher expression except MeJA. Particularly, BnTCP39, BnTCP64, BnTCP75, BnTCP25, BnTCP36, BnTCP53, and BnTCP7 display higher expression levels in response to ABA and GA treatments. These genes also possess ABA and GA-associated cis-elements in their promoter regions (Supplementary Table S6). Furthermore, BnTCP39 under IAA treatment showed higher expression at 2 h (1.568 fold); BnTCP64 significantly showed higher expression at 2 h (2.209 fold), 6 h (1.384 fold), and 8 h (1.590 fold) compared to CK. Notably, BnTCP53 displays higher expression at 6 h (4.487 fold) in response to MeJA treatment than CK (Figure 9). However, BnTCP60 did not reveal any significant expression to any stress conditions except cold stress, and BnTCP53 was found to be more active in response to both hormones and abiotic conditions. These results are also consistent with the identified cis-elements in the promoter regions of genes.
Discussion
Characterization and Evolution of TCP Gene Family in Rapeseed
Rapeseed is an allotetraploid crop that practices broad genome replication and combination activities (Song et al., 2020). Nevertheless, rapeseed yield is pretentious by numerous environmental factors such as cold, heat, drought, salinity, and heavy metals toxicity (Raza et al., 2021a; Raza et al., 2021b; He et al., 2021; Raza, 2021; Su et al., 2021). The TCP TFs are antique plant-specific proteins (Cubas et al., 1999). So far, these TFs are yet to be reported in unicellular algae; and moss, pluricellular green algae, lycophyte, and ferns possess five-six TCP genes in their genomes (Martín-Trillo and Cubas, 2010). Owing to the development, duplications, and divergence of TCP genes, the gymnosperms and angiosperms plants possess bigger TCP gene families containing more than ten TCP members (Martín-Trillo and Cubas, 2010). Over the past few years, TCP gene families have been reported in various crop plants such as Arabidopsis thaliana (Yao et al., 2007), Moso bamboo (Liu et al., 2018), sweet potato (Ren et al., 2021), cucumber (Wen et al., 2020), rice (Yao et al., 2007), Brassica rapa (Du et al., 2017); Brassica juncea (He et al., 2020); and cotton (Ma et al., 2014; Yin et al., 2018), etc. To our best knowledge, the TCP gene family has not been fully characterized in the rapeseed genome. The accessibility of the rapeseed genome sequences offers resources for genome-wide identification of rapeseed genes (Song et al., 2020). Here, we recognized 80 BnTCP genes in the rapeseed genome. Notably, this is the largest TCP gene family found in the rapeseed genome due to whole-genome duplication (WGD; polyploidy). Previously, the highest number of TCP genes (73 members) was reported in allotetraploid cotton (Yin et al., 2018).
In the rapeseed genome, the BnTCP gene family may be evolved by WGD or segmental events, together with numerous tandem duplications. Notably, WGD differs from tandem duplication in that WGD enhances the quantity of all genes altogether and produces duplicate, theoretically redundant copies of all the genes within a genome. In contrast, tandem duplication is another vital way for gene expansion. Genes expanded by tandem duplication are always scattered all together as a cluster in chromosomes (Sémon and Wolfe, 2007; Fang et al., 2012). Gene duplication events of TCP genes were also spotted in several plant species (Ma et al., 2014; Lin et al., 2016; Yin et al., 2018; Wen et al., 2020). Apart from duplication events, exon-intron associations and numbers can also explain the evolutionary story of the gene family (Xu et al., 2012; Raza et al., 2021a; Su et al., 2021). The BnTCPs gene structure compared with the same group presented that TCP genes contribute to analogous exon/intron allocation in terms of exon and intron numbers; in the meantime, BnTCPs with the same class exhibited comparable motif patterns. These results imply that these TCP members may play identical roles in response to a variety of abiotic stresses. Similar observations have been reported in previous TCP gene families where both classes showed comparable diverse intron/exon distribution and motifs patterns (Ma et al., 2014; Lin et al., 2016; Yin et al., 2018; Wen et al., 2020).
The Contribution of TCP Genes to Stress Responses and Tolerance Mechanisms
To get further insights into the contribution of BnTCP genes against various environmental cues, cis-elements in the promoter regions were envisaged. The findings demonstrated that two types of cis-elements were documented, i.e., stress (salt, drought, low-temperature, and anaerobic) and hormones (auxin, ABA, MeJA, and GA) associated. Consistent with earlier studies, cis-elements participate in the plant stress responses (Yamaguchi-Shinozaki and Shinozaki, 1994; Liu et al., 2014).
For instance, in rice, TCA-element between −563 and −249 bp upstream of OsPIANK1 was found to be associated with resistance to Magnaporthe oryzae infection and exogenous SA treatment (Mou et al., 2013). ABREs are a class of cis-elements capable of binding to sturdily conserved ABA-dependent transcription factors, which exist in the promoter region of numerous stress-resistant genes and normalize the expression level of associated genes under abiotic stress conditions (Choi et al., 2000; Barbosa et al., 2013; Fujita et al., 2013). In wheat, the expression pattern of TaGAPC1 was regulated under osmotic and salinity stress conditions. The promoter region of TaGAPC1 contains various stress-related cis-regulatory elements (including MBS, DRE, GT1, and LTR), and methylation changes were observed during the stress treatments, leading to stress tolerance and regulation in gene expression (Fei et al., 2017). In the current study, these results were further authenticated by the GO annotation evaluation. For instance, GO enrichment analysis validates the functional role of BnTCP genes in several cellulars, molecular, and biological processes associated with RNA/DNA binding, metabolic processes, transcriptional regulatory activities related to rapeseed growth and developmental processes under stress conditions. Likewise, numerous scholars stated comparable outcomes in diverse crop plant species where TCP genes were found to play a substantial part in plant growth and developmental processes and responses to different abiotic stress episodes (Yao et al., 2007; Lin et al., 2016; Zhou et al., 2016; Wen et al., 2020). These results can enhance our knowledge of BnTCP genes under different stress conditions and rapeseed developmental processes at a molecular level.
Moreover, 10 randomly selected BnTCP genes’ expression profiling was appraised against various hormones and abiotic stress treatments. Particularly, BnTCP7, BnTCP53, and BnTCP59 exhibited higher expression in response to cold, waterlogging, drought, and salinity stresses. Under hormones treatment, many genes showed higher expression, mainly by ABA, IAA, and GA treatments. These results are in agreement with recent reports where several TCP genes displayed higher expression levels under stress conditions. For example, in cotton, 41 GhTCP genes responded significantly to heat, salinity, and drought stresses (Yin et al., 2018). In cucumber, several TCP genes were up-regulated in response to photoperiod and temperature stresses; likewise, some genes were also induced by phytohormones like ethylene and gibberellin (Wen et al., 2020). In Moso bamboo, several TCP genes were substantially up-regulated in response to various hormones such as ABA, MeJA, and SA (Liu et al., 2018). The overexpression of maize ZmTCP42 gene in A. thaliana led to sensitivity to ABA in seed germination and improved drought stress tolerance in transgenic A. thaliana (Ding et al., 2019). In another study, the overexpression of the MeTCP4 gene regulated the cold tolerance by mediating ROS production and scavenging in transgenic A. thaliana. It also enhances the expression levels of stress- and ROS-scavenging-associated genes in the cold stressed A. thaliana plants (Cheng et al., 2019). Overexpressed PeTCP10 gene improved salt and ABA tolerance of transgenic A. thaliana at the vegetative growth phase (Xu et al., 2021). These results indicate that TCP genes pointedly contribute to hormone signaling pathways and abiotic stress tolerance in various plant species.
Expression Pattern of TCP Genes in Different Developmental Tissues
In the present study, tissue-specific expression profiles of 80 BnTCPs genes were assessed in six unique tissues using a publicly available RNA-seq dataset (https://ngdc.cncb.ac.cn/gsa/; BioProject ID: PRJCA001495). The aforementioned studies have illustrated that TCP genes may exhibit diverse expression profiles in various developmental tissues. For instance, the RNA-seq data was used to analyze the tissue-specific expression levels in cotton. The results showed that many GhTCP genes exhibited higher expression in various tissues, including mature leaves, stem, root, torus, petal, stamen, pistil, calycle (bracts), ovules, and fibers (Yin et al., 2018). In cucumber, qRT-PCR-based expression analysis showed that almost all 27 CsTCP genes exhibited higher expression in diverse tissues comprising root, stem, leaf, cotyledon, tendril, and flower (Wen et al., 2020). In B. rapa, some BrTCP genes showed higher expression in roots, leaves, and flower buds (Du et al., 2017). These results agree with our results where some TCP genes showed higher expression in all studied tissues (roots, stems, leaves, flowers, seeds, and silique), demonstrating that these genes may participate in rapeseed growth and development.
miRNA: Key Performers in the Adaptation of Stress Responses
MicroRNAs (miRNAs) are a class of small-non-coding RNAs produced from single-strand hairpin RNA precursors. These miRNAs control gene expression through binding to complementary sequences within target mRNAs (Shen et al., 2015; Wani et al., 2020). Substantial development has been made in discovering the targets of rapeseed miRNAs that act to diverse stresses and also participate in developmental processes (Buhtz et al., 2008; Huang et al., 2010; Körbes et al., 2012; Zhou et al., 2012; Shen et al., 2015; Chen et al., 2018; Fu et al., 2019). In the present study, we anticipated 32 miRNAs targeting 21 BnTCPs genes. Recent reports support our results, e.g., five TCP genes, including TCP2, TCP3, TCP4, TCP10, and TCP24, were targeted by miR319 and have been involved in normalizing leaf morphogenesis in A. thaliana (Schommer et al., 2008). In cotton, three TCP (GhTCP21, GhTCP31, and GhTCP54) genes were targeted by miR319 (Wen et al., 2020). The overexpression of the miR319-associated TCP gene improves the growth, and it also increases the tolerance to salinity and drought stresses in transgenic bentgrass (Agrostis stolonifera L.) (Zhou et al., 2013).
The overexpression of soybean miR172c improves tolerance to drought and salinity stress, and it also enhances sensitivity to ABA in transgenic A. thaliana plants (Li et al., 2016). Recently, Cheng et al. (2021) found that miR172s (mainly miR172a and miR172b) act as a positive regulator of salt tolerance in rice and wheat. Further, it also maintains the ROS homeostasis during salt stress, generally by balancing the expression of several ROS-scavenging-related genes in both wheat and rice (Cheng et al., 2021). The transgenic B. napus plants overexpressing miR395 indicated a lower degree of cadmium-induced oxidative stress compared to wild-type plants (Zhang et al., 2013). Another miRNA (miR394) was reported to enhance the multiple abiotic stress tolerance, including salinity and drought (Song et al., 2013) and cold stress (Song et al., 2016), in A. thaliana plants by ABA-dependent manner. These discoveries stipulated that miRNA might play critical roles in plant growth, development, and abiotic stress tolerance by regulating the expression levels of their target and stress-responsive genes.
Conclusion
In summary, for the first time, we identified 80 putative BnTCP genes in the rapeseed genome, which are distributed on 18 chromosomes. Genome-wide in silico analysis such as characterization, evolution, gene structures, conserved motifs, cis-elements, miRNA prediction, and GO annotation was performed to gain insights into TCP genes in rapeseed. Furthermore, their expression profiling was also appraised in different tissues and against diverse hormones and abiotic stresses. In a nutshell, this study laid the basis for additional functional studies (such as overexpression, knockout via CRISPR/Cas system, etc.) of the BnTCP genes in rapeseed breeding programs under adverse environmental conditions.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
YW, AR, and WC conceived the idea, analyzed the data, and wrote the manuscript. QH, WW, XZ, HC, and JL supervised the research, reviewed and improved the manuscript. All authors have read and approved the final version of the manuscript.
Funding
This work was supported by the Central Public-interest Scientific Institution Basal Research Fund (Y2020YJ03), the Science and Technology Innovation Project of the Chinese Academy of Agricultural Sciences (CAAS-ZDRW202105), and China Agriculture Research System of MOF and MARA (CARS-12) supported this work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.794297/full#supplementary-material
References
Aggarwal, P., Das Gupta, M., Joseph, A. P., Chatterjee, N., Srinivasan, N., and Nath, U. (2010). Identification of Specific DNA Binding Residues in the TCP Family of Transcription Factors in Arabidopsis. Plant Cell 22 (4), 1174–1189. doi:10.1105/tpc.109.066647
Barbosa, E. G. G., Leite, J. P., Marin, S. R. R., Marinho, J. P., de Fátima Corrêa Carvalho, J., Fuganti-Pagliarini, R., et al. (2013). Overexpression of the ABA-Dependent AREB1 Transcription Factor from Arabidopsis thaliana Improves Soybean Tolerance to Water Deficit. Plant Mol. Biol. Rep. 31 (3), 719–730. doi:10.1007/s11105-012-0541-4
Braun, N., de Saint Germain, A., Pillot, J.-P., Boutet-Mercey, S., Dalmais, M., Antoniadi, I., et al. (2012). The Pea TCP Transcription Factor PsBRC1 Acts Downstream of Strigolactones to Control Shoot Branching. Plant Physiol. 158 (1), 225–238. doi:10.1104/pp.111.182725
Buhtz, A., Springer, F., Chappell, L., Baulcombe, D. C., and Kehr, J. (2008). Identification and Characterization of Small RNAs from the Phloem of Brassica Napus. Plant J. 53 (5), 739–749. doi:10.1111/j.1365-313x.2007.03368.x
Chen, L., Chen, L., Zhang, X., Liu, T., Niu, S., Wen, J., et al. (2018). Identification of miRNAs that Regulate Silique Development in Brassica Napus. Plant Sci. 269, 106–117. doi:10.1016/j.plantsci.2018.01.010
Cheng, Z., Lei, N., Li, S., Liao, W., Shen, J., and Peng, M. (2019). The Regulatory Effects of MeTCP4 on Cold Stress Tolerance in Arabidopsis thaliana: a Transcriptome Analysis. Plant Physiol. Biochem. 138, 9–16. doi:10.1016/j.plaphy.2019.02.015
Cheng, X., He, Q., Tang, S., Wang, H., Zhang, X., and Lv, M. (2021). The miR172/IDS1 Signaling Module Confers Salt Tolerance through Maintaining ROS Homeostasis in Cereal Crops. New Phytol. 230 (3), 1017–1033. doi:10.1111/nph.17211
Choi, H.-i., Hong, J.-h., Ha, J.-o., Kang, J.-y., and Kim, S. Y. (2000). ABFs, a Family of ABA-Responsive Element Binding Factors. J. Biol. Chem. 275 (3), 1723–1730. doi:10.1074/jbc.275.3.1723
Cubas, P., Lauter, N., Doebley, J., and Coen, E. (1999). The TCP Domain: a Motif Found in Proteins Regulating Plant Growth and Development. Plant J. 18 (2), 215–222. doi:10.1046/j.1365-313x.1999.00444.x
Ding, S., Cai, Z., Du, H., and Wang, H. (2019). Genome-wide Analysis of TCP Family Genes in Zea mays L. Identified a Role for ZmTCP42 in Drought Tolerance. Ijms 20 (11), 2762. doi:10.3390/ijms20112762
Du, J., Hu, S., Yu, Q., Wang, C., Yang, Y., Sun, H., et al. (2017). Genome-wide Identification and Characterization of BrrTCP Transcription Factors in Brassica Rapa Ssp. Rapa. Front. Plant Sci. 8, 1588. doi:10.3389/fpls.2017.01588
Fang, L., Cheng, F., Wu, J., and Wang, X. (2012). The Impact of Genome Triplication on Tandem Gene Evolution in Brassica Rapa. Front. Plant Sci. 3, 261. doi:10.3389/fpls.2012.00261
Fei, Y., Xue, Y., Du, P., Yang, S., and Deng, X. (2017). Expression Analysis and Promoter Methylation under Osmotic and Salinity Stress of TaGAPC1 in Wheat (Triticum aestivum L). Protoplasma 254 (2), 987–996. doi:10.1007/s00709-016-1008-5
Fu, Y., Mason, A. S., Zhang, Y., Lin, B., Xiao, M., Fu, D., et al. (2019). MicroRNA-mRNA Expression Profiles and Their Potential Role in Cadmium Stress Response in Brassica Napus. BMC Plant Biol. 19 (1), 570. doi:10.1186/s12870-019-2189-9
Fujita, Y., Yoshida, T., and Yamaguchi-Shinozaki, K. (2013). Pivotal Role of the AREB/ABF-SnRK2 Pathway in ABRE-Mediated Transcription in Response to Osmotic Stress in Plants. Physiol. Plantarum 147 (1), 15–27. doi:10.1111/j.1399-3054.2012.01635.x
Gao, Y., Zhang, D., and Li, J. (2015). TCP1 Modulates DWF4 Expression via Directly Interacting with the GGNCCC Motifs in the Promoter Region of DWF4 in Arabidopsis thaliana. J. Genet. Genomics 42 (7), 383–392. doi:10.1016/j.jgg.2015.04.009
Gastaldi, V., Lucero, L. E., Ferrero, L. V., Ariel, F. D., and Gonzalez, D. H. (2020). Class-I TCP Transcription Factors Activate the SAUR63 Gene Subfamily in Gibberellin-dependent Stamen Filament Elongation. Plant Physiol. 182 (4), 2096–2110. doi:10.1104/pp.19.01501
He, J., He, X., Chang, P., Jiang, H., Gong, D., and Sun, Q. (2020). Genome-wide Identification and Characterization of TCP Family Genes in Brassica Juncea Var. Tumida. PeerJ 8, e9130. doi:10.7717/peerj.9130
He, H., Lei, Y., Yi, Z., Raza, A., Zeng, L., Yan, L., et al. (2021). Study on the Mechanism of Exogenous Serotonin Improving Cold Tolerance of Rapeseed (Brassica Napus L.) Seedlings. Plant Growth Regul. 94, 161–170. doi:10.1007/s10725-021-00700-0
Hoang, X. L. T., Nhi, D. N. H., Thu, N. B. A., Thao, N. P., and Tran, L. P. (2017). Transcription Factors and Their Roles in Signal Transduction in Plants under Abiotic Stresses. Curr. Genomics 18 (6), 483–497. doi:10.2174/1389202918666170227150057
Huang, S. Q., Xiang, A. L., Che, L. L., Chen, S., Li, H., Song, J. B., et al. (2010). A Set of miRNAs from Brassica Napus in Response to Sulphate Deficiency and Cadmium Stress. Plant Biotechnol. J. 8 (8), 887–899. doi:10.1111/j.1467-7652.2010.00517.x
Hurst, L. D. (2002). The Ka/Ks Ratio: Diagnosing the Form of Sequence Evolution. Trends Genet. 18 (9), 486–487. doi:10.1016/s0168-9525(02)02722-1
Karaaslan, E. S., Wang, N., Faiß, N., Liang, Y., Montgomery, S. A., Laubinger, S., et al. (2020). Marchantia TCP Transcription Factor Activity Correlates with Three-Dimensional Chromatin Structure. Nat. Plants 6 (10), 1250–1261. doi:10.1038/s41477-020-00766-0
Körbes, A. P., Machado, R. D., Guzman, F., Almerão, M. P., de Oliveira, L. F. V., Loss-Morais, G., et al. (2012). Identifying Conserved and Novel microRNAs in Developing Seeds of Brassica Napus Using Deep Sequencing. PLoS One 7 (11), e50663. doi:10.1371/journal.pone.0050663
Kosugi, S., and Ohashi, Y. (2002). DNA Binding and Dimerization Specificity and Potential Targets for the TCP Protein Family. Plant J. 30 (3), 337–348. doi:10.1046/j.1365-313x.2002.01294.x
Li, W., Wang, T., Zhang, Y., and Li, Y. (2016). Overexpression of Soybean miR172c Confers Tolerance to Water Deficit and Salt Stress, but Increases ABA Sensitivity in Transgenic Arabidopsis thaliana. Exbotj 67 (1), 175–194. doi:10.1093/jxb/erv450
Li, D., Zhang, H., Mou, M., Chen, Y., Xiang, S., Chen, L., et al. (2019). Arabidopsis Class II TCP Transcription Factors Integrate with the FT-FD Module to Control Flowering. Plant Physiol. 181 (1), 97–111. doi:10.1104/pp.19.00252
Lin, Y.-F., Chen, Y.-Y., Hsiao, Y.-Y., Shen, C.-Y., Hsu, J.-L., Yeh, C.-M., et al. (2016). Genome-wide Identification and Characterization ofTCPgenes Involved in Ovule Development ofPhalaenopsis Equestris. Exbotj 67 (17), 5051–5066. doi:10.1093/jxb/erw273
Liu, J.-H., Peng, T., and Dai, W. (2014). Critical Cis-Acting Elements and Interacting Transcription Factors: Key Players Associated with Abiotic Stress Responses in Plants. Plant Mol. Biol. Rep. 32 (2), 303–317. doi:10.1007/s11105-013-0667-z
Liu, H.-L., Wu, M., Li, F., Gao, Y.-M., Chen, F., and Xiang, Y. (2018). TCP Transcription Factors in Moso Bamboo (Phyllostachys Edulis): Genome-wide Identification and Expression Analysis. Front. Plant Sci. 9, 1263. doi:10.3389/fpls.2018.01263
Ma, J., Wang, Q., Sun, R., Xie, F., Jones, D. C., and Zhang, B. (2014). Genome-wide Identification and Expression Analysis of TCP Transcription Factors in Gossypium Raimondii. Sci. Rep. 4 (1), 6645. doi:10.1038/srep06645
Martín-Trillo, M., and Cubas, P. (2010). TCP Genes: a Family Snapshot Ten Years Later. Trends Plant Sci. 15 (1), 31–39. doi:10.1016/j.tplants.2009.11.003
Mou, S., Liu, Z., Guan, D., Qiu, A., Lai, Y., and He, S. (2013). Functional Analysis and Expressional Characterization of rice Ankyrin Repeat-Containing Protein, OsPIANK1, in Basal Defense against Magnaporthe Oryzae Attack. PLoS One 8 (3), e59699. doi:10.1371/journal.pone.0059699
Navaud, O., Dabos, P., Carnus, E., Tremousaygue, D., and Hervé, C. (2007). TCP Transcription Factors Predate the Emergence of Land Plants. J. Mol. Evol. 65 (1), 23–33. doi:10.1007/s00239-006-0174-z
Raza, A., Su, W., Gao, A., Mehmood, S. S., Hussain, M. A., Nie, W., et al. (2021a). Catalase (CAT) Gene Family in Rapeseed (Brassica Napus L.): Genome-wide Analysis, Identification, and Expression Pattern in Response to Multiple Hormones and Abiotic Stress Conditions. Ijms 22 (8), 4281. doi:10.3390/ijms22084281
Raza, A., Su, W., Hussain, M. A., Mehmood, S. S., Zhang, X., Cheng, Y., et al. (2021b). Integrated Analysis of Metabolome and Transcriptome Reveals Insights for Cold Tolerance in Rapeseed (Brassica Napus L.). Front. Plant Sci. 12, 721681. doi:10.3389/fpls.2021.721681
Raza, A. (2021). Eco-physiological and Biochemical Responses of Rapeseed (Brassica Napus L.) to Abiotic Stresses: Consequences and Mitigation Strategies. J. Plant Growth Regul. 40, 1368–1388. doi:10.1007/s00344-020-10231-z
Ren, L., Wu, H., Zhang, T., Ge, X., Wang, T., Zhou, W., et al. (2021). Genome-Wide Identification of TCP Transcription Factors Family in Sweet Potato Reveals Significant Roles of miR319-Targeted TCPs in Leaf Anatomical Morphology. Front. Plant Sci. 12, 686698. doi:10.3389/fpls.2021.686698
Sarvepalli, K., and Nath, U. (2018). CIN-TCP Transcription Factors: Transiting Cell Proliferation in Plants. Iubmb Life 70 (8), 718–731. doi:10.1002/iub.1874
Schommer, C., Palatnik, J. F., Aggarwal, P., Chételat, A., Cubas, P., Farmer, E. E., et al. (2008). Control of Jasmonate Biosynthesis and Senescence by miR319 Targets. Plos Biol. 6 (9), e230. doi:10.1371/journal.pbio.0060230
Sémon, M., and Wolfe, K. H. (2007). Consequences of Genome Duplication. Curr. Opin. Genet. Dev. 17 (6), 505–512. doi:10.1016/j.gde.2007.09.007
Shen, E., Zou, J., Hubertus Behrens, F., Chen, L., Ye, C., Dai, S., et al. (2015). Identification, Evolution, and Expression Partitioning of miRNAs in allopolyploidBrassica Napus. Exbotj 66 (22), 7241–7253. doi:10.1093/jxb/erv420
Song, J. B., Gao, S., Sun, D., Li, H., Shu, X. X., and Yang, Z. M. (2013). miR394 and LCR Are Involved in Arabidopsis Salt and Drought Stress Responses in an Abscisic Acid-dependent Manner. BMC Plant Biol. 13 (1), 210–216. doi:10.1186/1471-2229-13-210
Song, J. B., Gao, S., Wang, Y., Li, B. W., Zhang, Y. L., and Yang, Z. M. (2016). miR394 and its Target Gene LCR Are Involved in Cold Stress Response in Arabidopsis. Plant Gene 5, 56–64. doi:10.1016/j.plgene.2015.12.001
Song, J.-M., Guan, Z., Hu, J., Guo, C., Yang, Z., Wang, S., et al. (2020). Eight High-Quality Genomes Reveal Pan-Genome Architecture and Ecotype Differentiation of Brassica Napus. Nat. Plants 6 (1), 34–45. doi:10.1038/s41477-019-0577-7
Su, W., Raza, A., Gao, A., Jia, Z., Zhang, Y., Hussain, M. A., et al. (2021). Genome-Wide Analysis and Expression Profile of Superoxide Dismutase (SOD) Gene Family in Rapeseed (Brassica Napus L.) under Different Hormones and Abiotic Stress Conditions. Antioxidants 10 (8), 1182. doi:10.3390/antiox10081182
van Es, S. W., Silveira, S. R., Rocha, D. I., Bimbo, A., Martinelli, A. P., Dornelas, M. C., et al. (2018). Novel Functions of the Arabidopsis Transcription factorTCP5in Petal Development and Ethylene Biosynthesis. Plant J. 94 (5), 867–879. doi:10.1111/tpj.13904
Viola, I. L., Reinheimer, R., Ripoll, R., Manassero, N. G. U., and Gonzalez, D. H. (2012). Determinants of the DNA Binding Specificity of Class I and Class II TCP Transcription Factors. J. Biol. Chem. 287 (1), 347–356. doi:10.1074/jbc.m111.256271
Wang, M.-Y., Zhao, P.-M., Cheng, H.-Q., Han, L.-B., Wu, X.-M., Gao, P., et al. (2013). The Cotton Transcription Factor TCP14 Functions in Auxin-Mediated Epidermal Cell Differentiation and Elongation. Plant Physiol. 162 (3), 1669–1680. doi:10.1104/pp.113.215673
Wang, Z., Chen, Y., Fang, H., Shi, H., Chen, K., Zhang, Z., et al. (2014). Selection of Reference Genes for Quantitative Reverse-Transcription Polymerase Chain Reaction Normalization in Brassica Napus under Various Stress Conditions. Mol. Genet. Genomics 289 (5), 1023–1035. doi:10.1007/s00438-014-0853-1
Wani, S. H., Kumar, V., Khare, T., Tripathi, P., Shah, T., Ramakrishna, C., et al. (2020). miRNA Applications for Engineering Abiotic Stress Tolerance in Plants. Biologia 75 (7), 1063–1081. doi:10.2478/s11756-019-00397-7
Wen, H., Chen, Y., Du, H., Zhang, L., Zhang, K., He, H., et al. (2020). Genome-Wide Identification and Characterization of the TCP Gene Family in Cucumber (Cucumis Sativus L.) and Their Transcriptional Responses to Different Treatments. Genes 11 (11), 1379. doi:10.3390/genes11111379
Wu, J. F., Tsai, H. L., Joanito, I., Wu, Y. C., Chang, C. W., Li, Y. H., et al. (2016). LWD-TCP Complex Activates the Morning Gene CCA1 in Arabidopsis. Nat. Commun. 7 (1), 13181. doi:10.1038/ncomms13181
Xu, G., Guo, C., Shan, H., and Kong, H. (2012). Divergence of Duplicate Genes in Exon-Intron Structure. Proc. Natl. Acad. Sci. 109 (4), 1187–1192. doi:10.1073/pnas.1109047109
Xu, Y., Liu, H., Gao, Y., Xiong, R., Wu, M., Zhang, K., et al. (2021). The TCP Transcription Factor PeTCP10 Modulates Salt Tolerance in Transgenic Arabidopsis. Plant Cel Rep. 40 (10), 1971–1987. doi:10.1007/s00299-021-02765-7
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A Novel Cis-Acting Element in an Arabidopsis Gene Is Involved in Responsiveness to Drought, Low-Temperature, or High-Salt Stress. Plant Cell 6 (2), 251–264. doi:10.1105/tpc.6.2.251
Yao, X., Ma, H., Wang, J., and Zhang, D. (2007). Genome-Wide Comparative Analysis and Expression Pattern of TCP Gene Families in Arabidopsis thaliana and Oryza Sativa. J. Integr. Plant Biol. 49 (6), 885–897. doi:10.1111/j.1744-7909.2007.00509.x
Yin, Z., Li, Y., Zhu, W., Fu, X., Han, X., Wang, J., et al. (2018). Identification, Characterization, and Expression Patterns of Tcp Genes and Microrna319 in Cotton. Ijms 19 (11), 3655. doi:10.3390/ijms19113655
Zhang, L. W., Song, J. B., Shu, X. X., Zhang, Y., and Yang, Z. M. (2013). miR395 Is Involved in Detoxification of Cadmium in Brassica Napus. J. Hazard. Mater. 250-251, 204–211. doi:10.1016/j.jhazmat.2013.01.053
Zhang, W., Cochet, F., Ponnaiah, M., Lebreton, S., Mathéron, L., Pionneau, C., et al. (2019). The MPK 8‐ TCP 14 Pathway Promotes Seed Germination in Arabidopsis. Plant J. 100 (4), 677–692. doi:10.1111/tpj.14461
Zhou, Z. S., Song, J. B., and Yang, Z. M. (2012). Genome-wide Identification of Brassica Napus microRNAs and Their Targets in Response to Cadmium. J. Exp. Bot. 63 (12), 4597–4613. doi:10.1093/jxb/ers136
Zhou, M., Li, D., Li, Z., Hu, Q., Yang, C., Zhu, L., et al. (2013). Constitutive Expression of a miR319 Gene Alters Plant Development and Enhances Salt and Drought Tolerance in Transgenic Creeping Bentgrass. Plant Physiol. 161 (3), 1375–1391. doi:10.1104/pp.112.208702
Keywords: abiotic stress, cis-elements, evolution, genomics, miRNA, phytohomones, segmental duplication
Citation: Wen Y, Raza A, Chu W, Zou X, Cheng H, Hu Q, Liu J and Wei W (2021) Comprehensive In Silico Characterization and Expression Profiling of TCP Gene Family in Rapeseed. Front. Genet. 12:794297. doi: 10.3389/fgene.2021.794297
Received: 13 October 2021; Accepted: 01 November 2021;
Published: 17 November 2021.
Edited by:
Reyazul Rouf Mir, Sher-e-Kashmir University of Agricultural Sciences and Technology, IndiaReviewed by:
Muhammad Azhar Nadeem, Sivas University of Science and Technology, TurkeyManu Kumar, Dongguk University Seoul, South Korea
Muhammad Waseem, South China Agricultural University, China
Muhammad Azhar Hussain, Hainan University, China
Copyright © 2021 Wen, Raza, Chu, Zou, Cheng, Hu, Liu and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia Liu, bGl1amlhMDJAY2Fhcy5jbg==; Wenliang Wei, d2h3ZW5saWFuZ0AxNjMuY29t
†These authors have contributed equally to this work
 Yunfei Wen
Yunfei Wen Ali Raza
Ali Raza Wen Chu
Wen Chu Xiling Zou
Xiling Zou Hongtao Cheng
Hongtao Cheng Qiong Hu
Qiong Hu Jia Liu
Jia Liu Wenliang Wei
Wenliang Wei