- 1Department of Nephrology, Xijing Hospital, Fourth Military Medical University, Xi’an, China
- 2State Key Laboratory of Cancer Biology, Fourth Military Medical University, Xi’an, China
Background: Circular RNAs (circRNAs), which have broad posttranscriptional regulatory potencies, are involved in the pathogenesis of fibrotic diseases and are promising diagnostic biomarkers and therapeutic targets. However, their specific roles in renal fibrosis remain elusive.
Methods: A robust unilateral renal ischemia reperfusion injury (UIRI) mouse model was established to recapitulate the pathophysiology of renal fibrosis. The expression of circRNAs, miRNAs, and mRNAs was profiled by high-throughput RNA sequencing technology.
Results: In total, 4983 circRNAs, 216 miRNAs, and 6371 mRNAs were differentially expressed in UIRI-induced fibrotic kidneys. Candidate circRNAs and miRNAs were validated by RT–qPCR in both UIRI and unilateral ureteral obstruction mouse models. Bioinformatic analysis indicated that the parental genes of the differentially expressed circRNAs were predominantly implicated in focal adhesion, adhesion junctions, and regulation of actin cytoskeleton pathways. Through circRNA-miRNA-mRNA construction, we identified two hub genes, circSlc8a1 and circApoe, that targeted a large number of differentially expressed miRNAs and mRNAs related to metabolism and cytokine–cytokine receptor pathways, respectively.
Conclusion: CircRNAs were dysregulated in the UIRI model and might be potentially involved in the pathogenesis of renal fibrosis. Research efforts should focus on unravelling the functions of aberrantly expressed circRNAs in renal fibrosis to uncover biomarkers that would enable early diagnosis and the design of prompt therapeutic interventions to prevent disease progression.
Introduction
Chronic kidney disease (CKD) affects 8–16% of the world population and contributes significantly to global mortality and morbidity (Nelson et al., 2019). Currently, strategies for the early recognition and prevention of the progression of CKD are lacking. Renal ischemia reperfusion injury (IRI) triggered by cardiac surgery, sepsis, and transplantation, among other influencing factors, is not only the most common risk factor for acute kidney injury (AKI) (Ronco et al., 2019) but also the main cause of the AKI to CKD transition (Ó hAinmhire and Humphreys, 2017). Severe IRI primes for the persistent injury and maladaptive repair, manifesting as renal dysfunction, loss of nephrons, microvascular rarefaction, and extracellular matrix deposition, ultimately lead to glomerular and tubulointerstitial fibrosis or renal fibrosis (Shiva et al., 2020). Consequently, further in-depth exploration of the molecular mechanism of IRI-induced renal fibrosis is needed to facilitate the discovery of potential biomarkers and new therapeutic targets for CKD.
Noncoding RNAs (ncRNAs), vital components of epigenetics that make up of more than 90% of the transcriptome, are delicately regulated and play pivotal roles in a range of kidney disorders (Fan et al., 2020; Lin et al., 2020; Chen et al., 2021; Ren and Wang, 2021). Noncoding RNAs are classified into three major categories: microRNAs, long ncRNAs (lncRNAs), and circular RNAs (circRNAs), based on their length and structure. Unlike other ncRNAs, knowledge about circRNAs in kidney diseases is still in the nascent stage. Studies have reported dysregulation of circRNAs in AKI (Xu et al., 2020a), diabetic nephropathy (Bai et al., 2020), immunoglobulin A nephropathy (Luan et al., 2021), autoimmune kidney disease (Zhang et al., 2021), and renal malignancies (Cen et al., 2021). While circRNAs are believed to participate in the fibrotic process (Liu et al., 2019), their specific functions in renal fibrosis remain largely uncharacterized.
CircRNAs are covalently closed loops formed by head-to-tail back-splicing mechanism. They exert their functions through multiple mechanisms, including transcriptional regulation, miRNA sponging, translation into peptides, or interacting with proteins (Zhou et al., 2020). The most explored function of circRNAs is their role as competitive endogenous RNAs (ceRNAs) by sponging miRNAs. A single miRNA can potentially target hundreds or thousands of mRNAs. Thus, the circRNA-miRNA-mRNA network could regulate crucial functions in numerous biological processes, including development, differentiation, proliferation, stress responses, and apoptosis. Moreover, circRNAs are also relatively abundant in tissues and body fluids and highly conserved between species, thereby promoting the translation of findings in rodent models to humans. CircRNAs have a long half-life time and are resistant to exonuclease degradation, therefore holding great promise as ideal biomarkers for disease diagnosis, treatment, and prognosis (Verduci et al., 2021).
In this study, we used the unilateral renal ischemia reperfusion injury (UIRI) and unilateral ureteric obstruction (UUO) mouse models to reproduce the pathological changes of CKD. High-throughput RNA sequencing was performed on kidney tissues obtained from the UIRI mouse model to identify the expression profiles of circular RNAs, miRNAs, and mRNAs. To validate our sequencing results, we performed real-time quantitative PCR of the differentially expressed circRNAs and miRNAs in both the UIRI and UUO mouse models. We then conducted Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analyses to investigate the potential mechanisms associated with renal fibrosis. Our research based on high-throughput sequencing technology and fundamental experiments broadens the understanding of the role of circRNAs in IRI-induced renal fibrosis and provides a gateway for the development of potential therapeutic interventions for CKD.
Materials and Methods
Animal Models
All animal experiments were approved by the Committee on Ethical Animal Care and Use of the Fourth Military Medical University and performed in accordance with the committee guidelines. C57BL/6J male mice were used in this experiment and maintained on a 12-h light and 12-h dark cycle with free access to standard food and water before and after surgery.
Unilateral Renal Ischemia Reperfusion Model
Three mice as biological replicates were set as either the sham or UIRI group for RNA sequencing. Another five to seven mice who underwent the same surgical procedure comprised the validation group. UIRI surgery was performed as previously described (Jao et al., 2019). Briefly, mice (body weight 20–25 g) were anaesthetized with pentobarbital sodium. Body temperature was maintained at 37°C by a temperature-controlled heating system. A flank incision was made and ischemic injury was induced in the right renal pedicle by nontraumatic vascular clamp for 45 min. The clamp was then removed, and reperfusion of the kidney was visually confirmed by a change in renal color from black purple to pink. One milliliter of saline solution was supplemented, and the flank incisions were sutured in 2 layers. Control animals underwent the same procedure without clamping the right renal pedicle. Mice were euthanized on day 14 after UIRI.
Unilateral Ureteric Obstruction
UUO, as a canonical renal fibrosis model, was built as the validation group to verify the differentially expressed (DE) circRNAs and miRNAs. Six- to eight-week-old mice (18–20 g) (n = 7 in the sham group and 8 in the UUO group) underwent a midline incision. The left ureter was identified and ligated using two 4.0 silk sutures. The midline abdominal incision was sutured in 2 layers. Control mice underwent the same procedure without ligation of the ureter. Mice were euthanized after 14 days post UUO surgery.
Circular RNA Library Construction and Sequencing
RNA was extracted from kidney tissues using TRIzol reagents (Thermo Fisher, United States). The concentration, quality, and integrity of RNA were determined using the Agilent RNA 6000 nano kit and reagents and the Agilent 2100 Bioanalyzer (Applied Biosystems, Carlsbad, CA, United States), respectively. Standard cDNA libraries were sequenced using the DNBSEQ® platform (BGI-Genomics, BGI-Shenzhen, Shenzhen, China) and 100 bp paired-end (PE100) sequencing was performed on all samples.
Identification of CircRNAs
Raw reads with adaptors, excessive N content of unknown bases, and low-quality tags were removed using SOAPnuke software (v1.5.6), and the remaining clean reads were used in the subsequent analyses. First, we used Hierarchical Indexing for Spliced Alignment of Transcripts (HISAT2, versionv2.0.4) and Bowtie 2 to map paired-end read to the latest UCSC transcript set (Mus_musculus, GCF_000001635.26_GRCm38.p6). CircRNAs were annotated by computational detection using two separate circRNA algorithms: CIRI and find_circ. Two or more backsplice spanning reads were required to confirm circRNAs in individual samples. The final results are the union of data from the two pipelines. CircRNA expression was normalized using RPB (junction reads per billion mapped reds). DEGseq was used to identify differentially expressed circRNAs. CircRNAs meeting the condition of an adjusted p value (for DEG seq < 0.001) and fold change ≥2 were considered to be differentially expressed. When the clustering of junction reads and recording of each circRNA candidate are finished, CIRI scans the SAM alignment one more time to detect additional junction reads. CIRI simultaneously performs further filtering to remove the false-positive candidates generated from the reads incorrectly mapped to the homologous genes or repetitive sequences. The circRNA identification was done by using find-circ software, meeting the following conditions: GU/AG, on the sides of splice site; clear breakpoint; two mismatches; appearance of breakpoint in the position within 2 nucleotides; more than two reads supporting the junction; and a score of blasting to the right position of short sequence that is at least 35 higher than blasting to the other positions. Finally, identified circRNAs were outputted with annotation information.
Small RNA Library Construction and Sequencing
Small RNAs were isolated from total RNA using PAGE gels by selecting 18–30 nt (14–30 ssRNA Ladder Marker, TAKARA) stripes and recycling. They were purified and ligated to 3 and 5′ adapters. Subsequently, cDNAs were generated by reverse transcription, and PCR amplification was performed to obtain sufficient fragments. The PCR products were purified and tested to ensure the quality of the library prior to sequencing. The expression level of small RNAs was calculated by counting the absolute numbers of molecules using unique molecular identifiers. Differential expression analysis was performed using the DEGseq, and a Q value < 0.001 and an absolute value of |log2 (fold change)| > 1 were set as the default thresholds by which to judge the significance of expression differences.
Immunoblotting
Frozen kidney tissues were homogenized in ice-cold radioimmunoprecipitation assay buffer (Sigma–Aldrich) containing protease inhibitor cocktails (Millipore). Equal amounts of protein were electrophoresed in 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gels. The resolved proteins were then transferred to polyvinylidene difluoride membranes (Roche, IN, United States). The membranes were blocked in fat-free milk at room temperature and incubated overnight at 4°C with a polyclonal rabbit anti-E-cadherin antibody (1:500 dilution, Bioworld) or a polyclonal rabbit anti-vimentin antibody (1:1,000, Abcam). Blots were washed with Tris-buffered saline and Tween 20 and incubated with horseradish peroxidase-labeled goat anti-rabbit antibody (1:5,000, Sangon Biotech, Shanghai, China) at room temperature for 1 h with gentle shaking. Blots were developed by enhanced chemiluminescence (ECL, Zeta, MA, United States) and visualized with BIO-RAD (BIO-RAD Gel Doc XR+, United States). The protein bands were quantified based on optical densities using ImageJ (version 1.34) and normalized to β-actin.
Renal Histology
Kidney samples were fixed in 4% paraformaldehyde, embedded in paraffin, and cut into 4-µm-thick sections. The tissue sections were deparaffinized, rehydrated, and stained with hematoxylin and eosin (H&E) and periodic acid-Schiff (PAS) reagents to determine histopathological abnormalities. Collagen was stained with Sirius red, and fibrosis was evaluated by Masson staining. Slides were subsequently scanned by PANNORAMIC 250 (3DHISTECH company, Budapest, Hungary) to examine glomerular, tubular morphology, and collagen deposition.
Real-Time Quantitative PCR Validation
Total RNA was isolated from kidney tissues using TRIzol reagent. To quantify the amount of circRNA, cDNA was synthesized with the Maxima Reverse Transcriptase (Thermo Scientific, EP0743). RT–qPCR was performed using TB Green® Premix Ex Taq™ II (Takara, Japan) reagent in a LightCycler 480II (Roche Applied Science) according to the manufacturer’s instructions. In particular, divergent primers annealing across the junction sites of circRNAs were used to determine their abundance. The specific primers for circRNAs and miRNAs were designed by Sangon Company (Shanghai), and the sequences were shown in Table 1. The housekeeping gene β-actin was used as an internal control for circRNAs, and U6 was used for miRNAs. The results were calculated using the 2−ΔΔCq method. All experiments were performed in triplicate.
Sanger Sequencing
To obtain more reliable PCR results, PCR amplification products were separated by electrophoresis in agarose gels. Briefly, the samples were loaded into precast wells in the gels and then subjected to electrophoresis. Afterward, the PCR products were stained with ethidium bromide, visualized under ultraviolet light, and dissected for subsequent sequencing experiments. For Sanger sequencing, the specific PCR products were extracted and purified directly with the Mag-MK Gel DNA Purification Kit (Sangon Biotech, Shanghai, China) according to the manufacturer’s instructions. The sequences of the products were determined using ABI 3730 DNA sequencers (Applied Biosystems, CA, United States).
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Pathway Analysis
GO enrichment analysis was performed on the DE target candidates of DE circRNAs and miRNAs with R software (ClusterProfiler package). KEGG is a database resource for understanding the high-level functions and utilities of biological systems (http://www.genome.jp/kegg/). We used R software for the KEGG enrichment analysis.
ceRNA Network Construction
MiRanda (http://www.microrna.org/microrna/), RNAhybrid (http://bibiserv. cebitec.uni-bielefeld.de/rnahybrid), and TargetScan (http://www.targetscan.org/) were used to predict the target miRNAs of circRNAs and the target mRNAs of miRNAs. For ceRNA construction, we took the intersection of the target miRNAs identified by the 3 databases above and the differentially expressed miRNAs in our samples. These miRNAs were identified as hub miRNAs. Then we screened miRanda, RNAhybrid, and TargetScan for the targeted mRNAs of the hub miRNAs, and we used the overlap with our sequenced DE mRNAs as the final results. The network was built and visually displayed using Cytoscape software (v3.9.0) on the basis of circRNA--miRNA-mRNA pairs. Different colors represent different expression levels for individual genes.
Statistical Analysis
All data were obtained from at least three independent experiments and analyzed using SPSS 25.0 (SPSS, Inc., IL) and GraphPad Prism v 8.0 (GraphPad Software, CA). Continuous data are presented as the mean ± standard deviation and were compared using Student’s t tests. A p value <0.05 was the significance threshold.
Results
Validation of IRI-Induced Renal Fibrosis in Mice
IRI is the injury-specific process triggered by a transient cessation of blood flow, followed by the re-establishment of renal perfusion (Pefanis et al., 2019). Compared to the bilateral IRI (BIRI), UIRI markedly decreases the postoperative death rate, and thus is an ideal animal model to investigate the dynamic pathophysiological mechanism of the AKI to CKD transition and subsequent progression to the end-stage renal disease (Tod et al., 2020). Warm long-term (45 min) ischemia induces a severe injury phenotype of progressive renal parenchymal atrophy and extensive tubulointerstitial fibrosis following incomplete recovery (Fernández et al., 2020; Holderied et al., 2020; Polichnowski et al., 2020). Renal histopathology showed intact structures from control mice who had undergone the sham operation. By contrast, kidney tissues from IRI mice exhibited the extensive renal tubular atrophy, glomerular sclerosis, cast formation, tubule dilation, and inflammatory cell infiltration through HE and PAS staining. Masson and Sirus Red staining revealed excessive extracellular matrix deposition and collagen production (Figure 1A). Immunohistochemistry showed increased expression of the fibrotic markers Collagen I and fibronectin, as well as the fibroblast/myofibroblast markers actin alpha 2, smooth muscle, and platelet-derived growth factor receptor-beta (PDGFR-beta) (Figure 1B). The epithelial marker E-cadherin was decreased, and the mesenchymal marker vimentin was increased in the UIRI group compared to the sham group (Figure 1C). The mRNA levels of fibrotic markers, including transforming growth factor beta 1 (Tgfb1), tissue inhibitor of metalloproteinase (Timp1), fibronectin (Fn), and collagen1a (Col1a), were also elevated significantly after UIRI (Figure 1D). These results indicated that at 2 weeks after UIRI, renal histopathologic examination exhibited signs of extensive tubulointerstitial fibrosis, suggesting that the transition to CKD was already occurring.
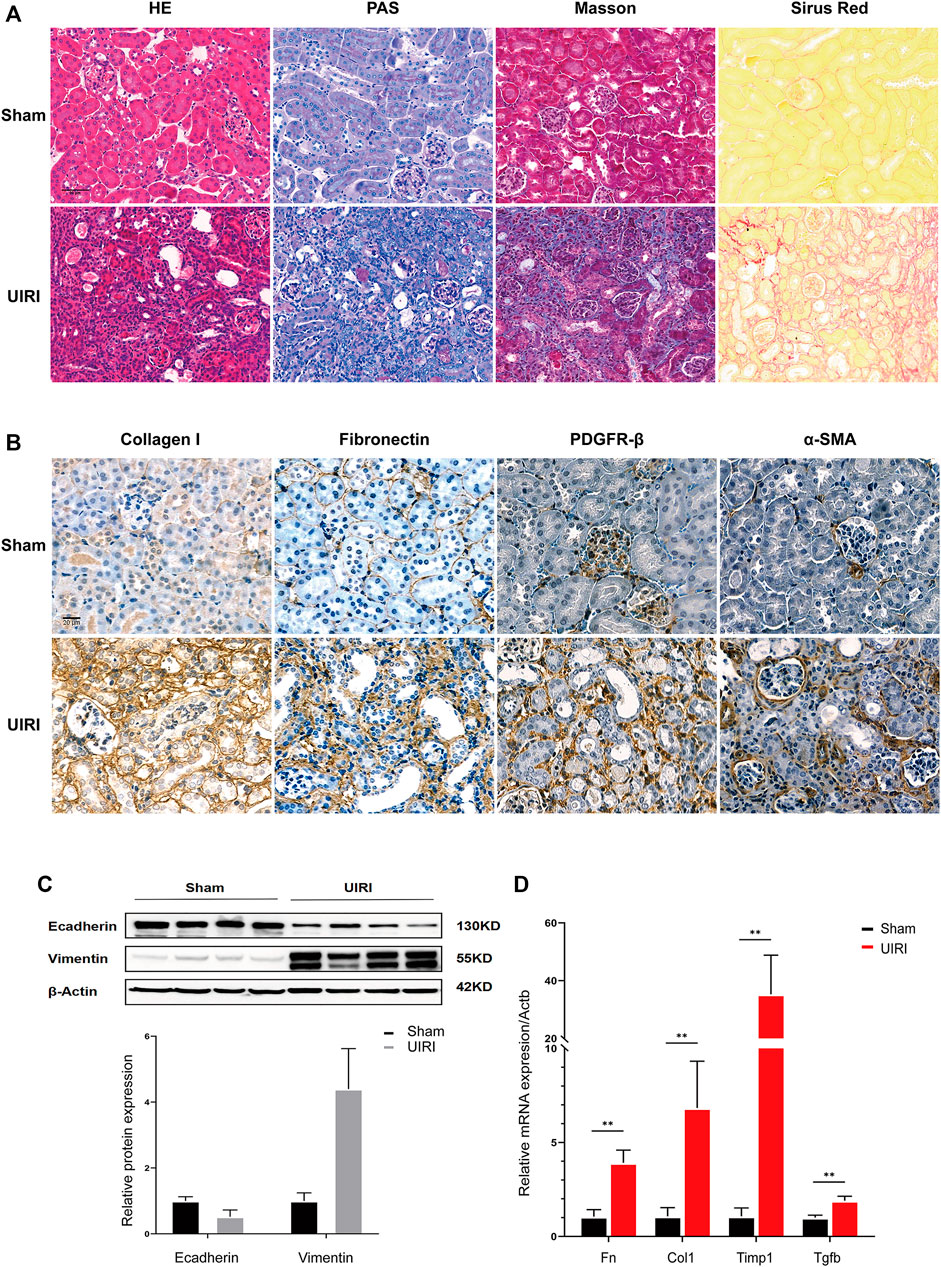
FIGURE 1. Validation of the transition of UIRI injury to the chronic phase characterized as extensive renal fibrosis. (A) Kidney tissues 14 days after IRI displayed tubular necrosis, atrophy, dilation, cast formation, and inflammatory cell infiltration in HE and PAS staining. UIRI kidney tissues showed extensive, severe histological deposition of collagen in kidney tissues in Sirius Red and Masson’s trichrome staining. (B) Immunohistological staining showed increased collagen I and fibronectin expression and PDGFR-β+, α-SMA+ fibroblasts/myofibroblasts in kidney tissues after UIRI compared to the sham group. Scale bar = 50 µm. (C) Representative Western blots showing the change in epithelial marker E-cadherin and mesenchymal marker vimentin protein expression in kidney tissues after UIRI. (D) The relative expression levels of fibrotic factors Fn, Col1, Timp1, and Tgfb by the RT–qPCR. **p < 0.01 vs. sham group. Fn, fibronectin; Col1, collagen I; Timp1, tissue inhibitor of metallopoteinase 1; Tgfb, transforming growth factor-beta.
CircRNA Expression Profiles in the UIRI-Induced Renal Fibrosis
The RNA-seq data analyzed by CIRI and Find_circ software showed that 3409 and 3200 circRNAs were identified in mouse kidney tissues from the sham and UIRI groups (n = 3 in each group), respectively. According to the chromosomal location of circRNAs, we concluded that 70.33% of circRNAs were derived from exonic regions and 18.73% were derived from intronic regions, whereas only 6.5% of circRNAs were derived from intergenic regions. Exon-intron (EIcircRNA) and intergenic circRNA accounted for 4.65 and 6.29%, respectively. The Circos plot illustrated their distribution in the chromosomes (Figure 2D). According to a predefined cutoff value, we identified 2425 upregulated and 2558 downregulated circRNAs (log2|fold change|>1, p value <0.05) in the UIRI group compared to the sham group (Supplementary Material S1). The heatmap (Figure 2A) and volcano plot (Figure 2B) revealed the DE circRNAs between the sham group and UIRI group. The ten most upregulated and downregulated circRNAs are listed in Table 2. As there is no existing public database of circRNA expression profiles in human CKD or animal models, we compared our sequencing data with the circAtlas database (http://circatlas.biols.ac.cn/), which contains the circRNA expression data of mouse normal kidney tissues. Among the top 30 circRNAs in mouse kidney from circAtlas database, 28 were identified among the 100 most highly expressed circRNAs in our sequencing data (Figures 2E,F). Our data and that of circAtlas suggested that the most enriched circRNA in mouse kidney tissue was circSlc8a1 (mmu_circ_0000823), derived from the exon 2 within the Slc8a1 locus (reference genome NCBI37/mm9).
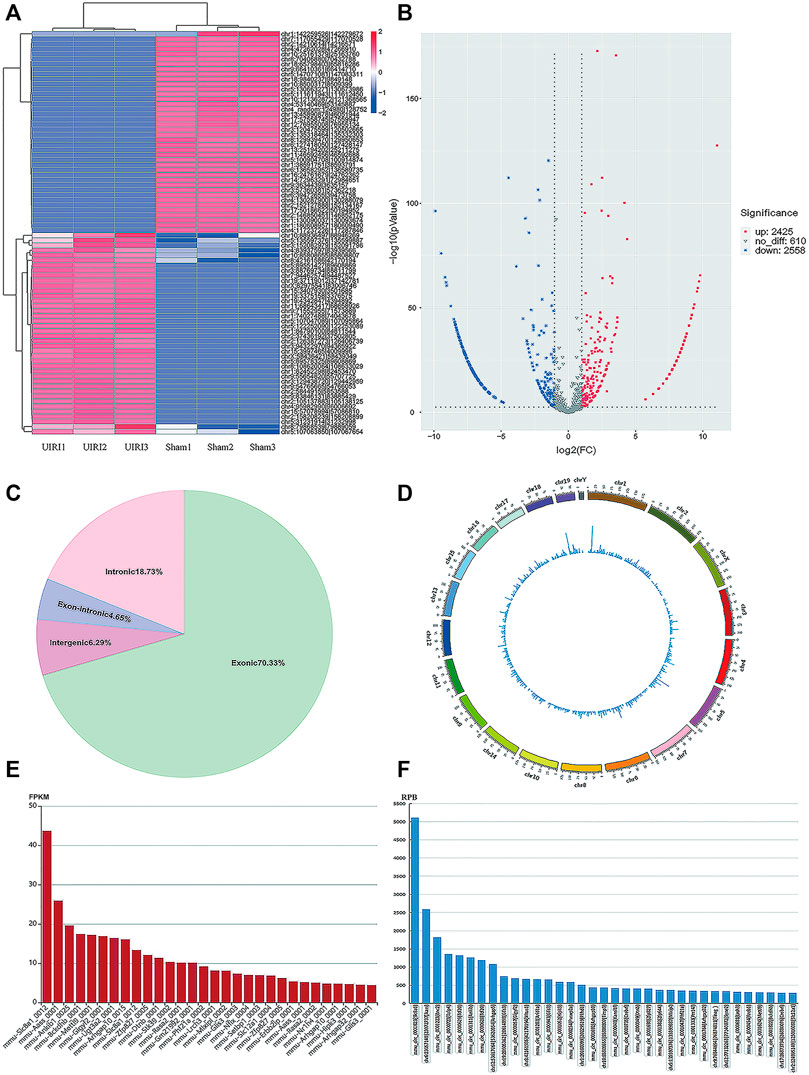
FIGURE 2. CircRNA expression profiles of the UIRI group. (A) The heatmap of representative differentially expressed (DE) circRNAs. (B) Volcano plot showing significantly DE circRNAs. Red and blue pixels represent upregulated and downregulated genes, respectively, while gray pixels indicate genes with no significant difference. An absolute value of fold-change > 2.0 and p < 0.05 were considered as significantly different. (C) The subtypes of the DE circRNAs. (D) Circos plot of DE circRNAs illustrating their distribution on the chromosome. (E) The top 30 most abundant circRNAs in mouse kidneys from the circAtlas database. (F) The 35 most enriched circRNAs in our sequencing data for normal mouse kidneys. **p < 0.01 vs. sham group.
Gene Ontology (GO) was used to annotate and classify the transcripts of circRNAs based on the GO hierarchical categories of biological process, cellular component, and molecular function. The number of parental genes corresponding to GO entries was determined, and the -log10 (p value) was used as the enrichment score. For biological process, the term that contained the most genes was the positive regulation of transcription by RNA polymerase II (GO:0045944, count = 270), and the most significantly enriched term was phosphorylation (GO:0016310, p = 2.03E-24). For cellular component and molecular function, the terms that contained the most genes and most enriched were cytoplasm (GO:0005737, count = 1,259, p = 5.36E-69) and protein binding (GO:0005515, count = 988, p = 2.21E-78) (Figure 3A). The KEGG results indicated that the parental genes of the DE circRNAs were involved in focal adhesion (04510, Gene number = 62, p = 4.06E-9), adhesion junctions (04520, Gene number = 22, p = 4.22E-8), regulation of actin cytoskeleton (04810, Gene number = 60, p = 9.50E-8), thyroid hormone signaling pathway (04919, gene number = 38, p = 9.46E-7), and phosphatidylinositol signaling system pathway (04070, gene number = 35, p = 1.03E-6).
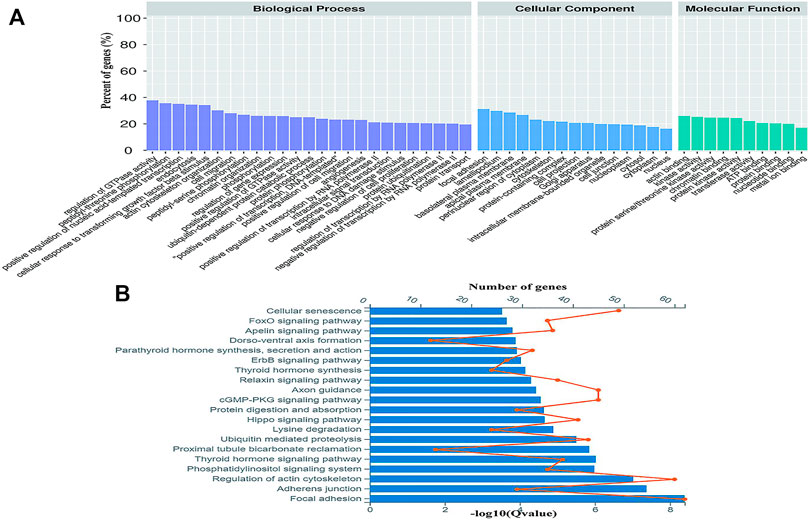
FIGURE 3. Gene ontology (GO) and Encyclopedia of Genes and Genomes (KEGG) analyses of the parental genes of DE circRNAs. (A) Gene ontology analysis on parental genes of the DE circRNAs. The vertical axis shows the annotated functions of the target genes. The horizontal axis shows the number of genes in each cluster. (B) Kyoto Encyclopedia of Genes and Genomes analysis of parental genes of the DE circRNAs. The orange node represents the number of the DE circRNAs in each pathway and the value of the blue bars shows the -log10 (Q value).
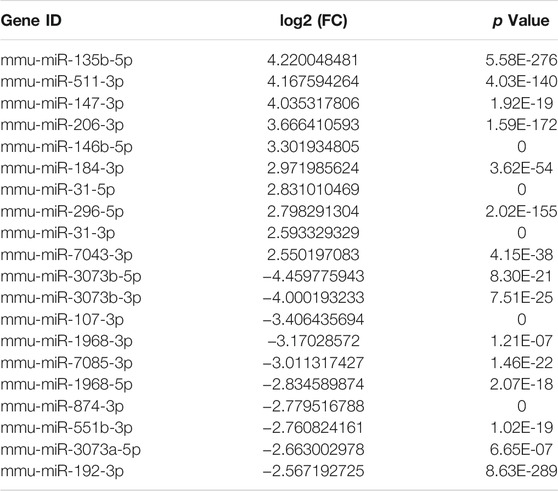
miRNA Profiling of UIRI- Induced Renal Fibrosis
The miRNA expression profiles of the UIRI were also detected. Differentially expressed miRNAs (DEMs) are shown in the hierarchical clustering heatmap (Figure 4A) and volcano plot (Figure 4B). A total of 216 DEMs were identified, among which 125 were upregulated and 91 were downregulated (log2|fold change|>1 and q value < 0.001) (Supplementary Material S2). The 10 most upregulated and downregulated miRNAs are listed in Table 3. To gain insight into the underlying mechanism of IRI-induced renal fibrosis, GO and KEGG analyses were conducted to functionally analyze the target mRNAs of DEMs, which were simultaneously predicted by Miranda, TargetScan, and RNAhybrid and screened by our mRNA sequencing data from the same tissue samples (Supplementary Material S3). The most enriched GO term for the cellular component was plasma membrane (GO:0005886, p = 5.46E-40). The most enriched terms for the molecular function and biological process were protein binding (GO:0005515, p = 2.87E-56) and cell adhesion (GO:0007155, p = 1.74E-54) (Figure 4C). Moreover, the axon guidance pathway (4360) was the most enriched pathway in the KEGG database (Figure 4D).
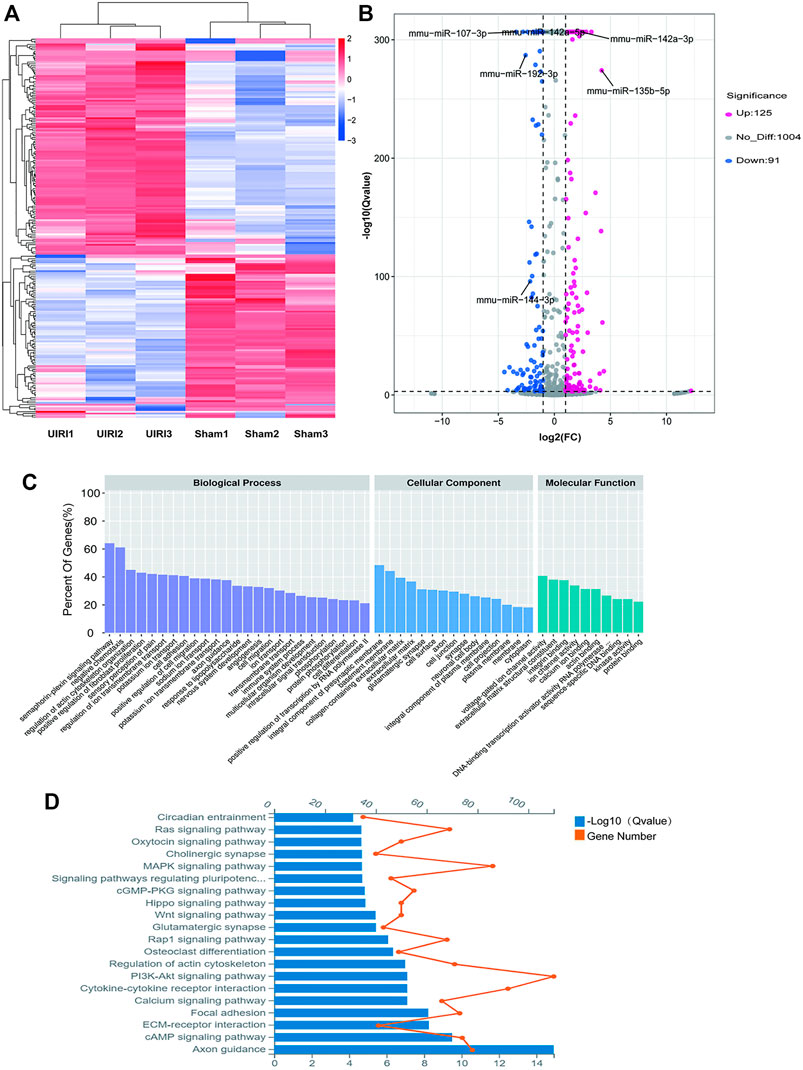
FIGURE 4. Expression profiles and the enrichment analyses of the DE miRNAs. (A) The heatmap of the representative DE miRNAs. (B) Volcano plot showing significantly DE miRNAs. Red and blue pixels represent upregulated and downregulated genes, respectively; gray pixels indicate genes with no significant difference. An absolute value of fold-change > 2.0 and Q value < 0.001 were considered as significantly different. (C) GO analysis of the targets of the DE miRNAs. (D) KEGG analysis of the targets of DE miRNAs.
Validation of the Candidate CircRNAs and miRNAs by Real-Time Quantitative PCR and Sanger Sequencing
According to the fold change and expression abundance of the deep sequencing results, upregulated miRNAs (miR-142a-5p, miR-142a-3p, miR-135b-5p) and three downregulated miRNAs (miR-144-3p, miR-107-3p, miR-192-3p) were selected for validation using RT–qPCR in both the UIRI model with an expanded sample size and the UUO model, which is another classic fibrosis model. The RT–qPCR results of the five selected upregulated (circCol1a2, circKcnt2, circDcdc2, circApoe, and circRftn1) and downregulated circRNAs (circSlc8a1, circTinag, circMettl9, and circEgf), were in accordance with those in the RNA sequencing (Figures 5B,C). Sanger sequencing of the PCR products of circRNAs, including circSlc8a1, circOxct1, circMettl9, circDcdc2, and circKcnt2, verified the back splicing junction sites (Figure 5D). Taken together, these results confirmed the accuracy of our sequencing data.
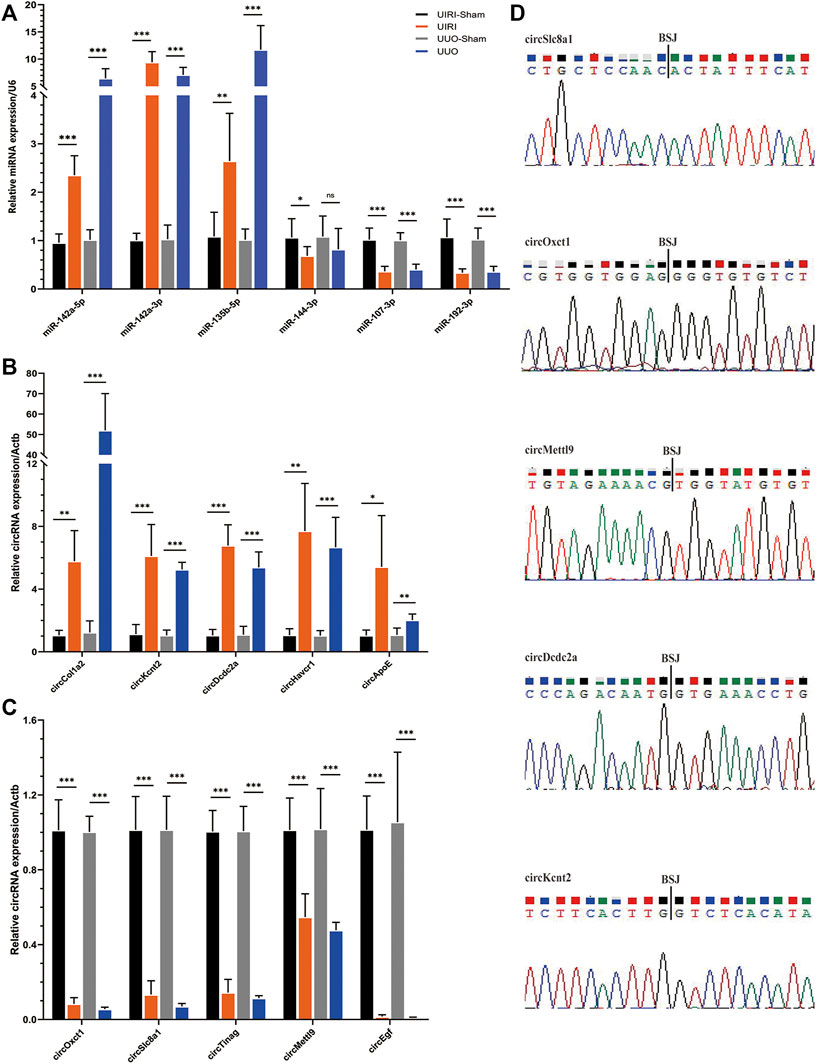
FIGURE 5. Validation of the circRNA and miRNA expression. (A) The relative expression levels of the selected miRNAs detected by the RT–qPCR method. (B, C) The results of the five upregulated and downregulated candidate circRNA expression levels by RT–qPCR analysis. *p < 0.05, **p < 0.01, and ***p < 0.001 vs. sham group. (D) The back-splicing positions tested by Sanger sequencing. BSJ, back splicing junction.
The CircRNA-miRNA-mRNA Network Construction
TargetScan, miRanda, and RNAhybrid were utilized to predict the DEMs targeted by the DE circRNAs and mRNAs targeted by DEMs. In this network, we identified two hub genes, circSlc8a1 (mmu_circ_0000823) and circApoe (mmu_circ_0014064). CircSlc8a1 is the most enriched circRNA in normal mouse renal tissue and was significantly downregulated in the IRI- and UUO-induced renal fibrosis, targeting 13 significantly upregulated miRNAs and the downstream corresponding downregulated mRNAs. The differentially downregulated targeted mRNAs of those 13 miRNAs were mainly enriched in the metabolic pathway, as indicated in Figure 6. CircApoe was significantly upregulated and could sponge 5 significantly downregulated miRNAs, with their target mRNAs enriched in the cytokine–cytokine receptor pathway (Figure 7).
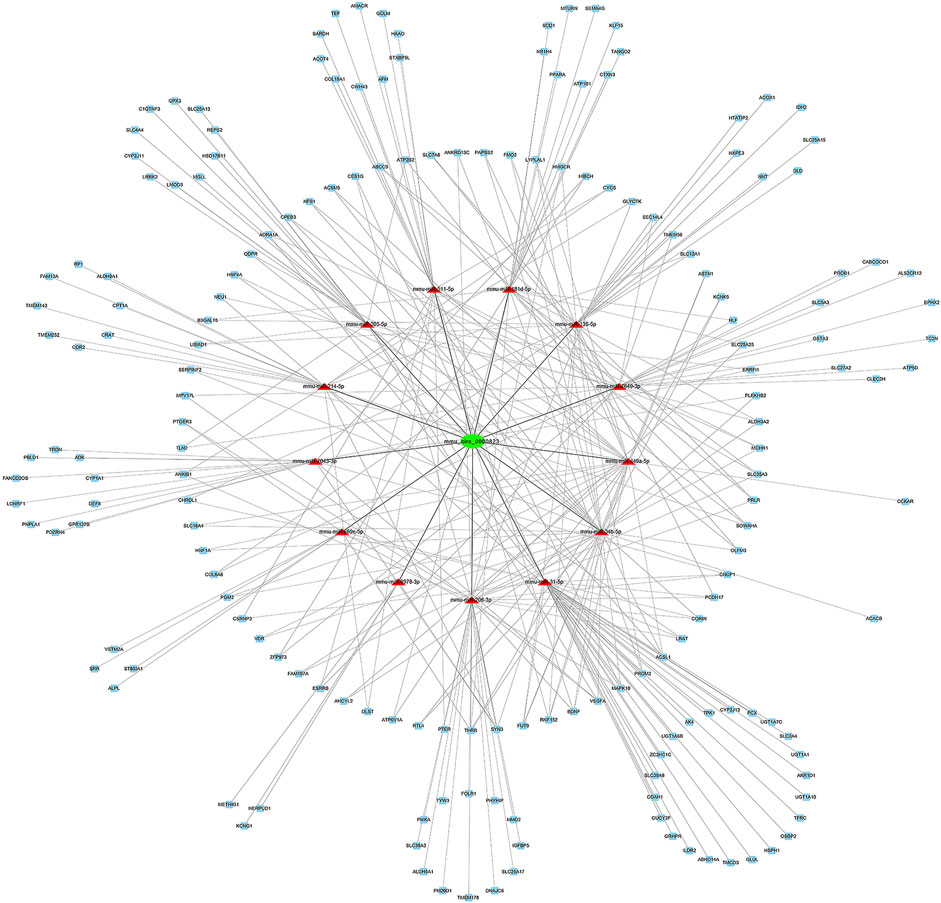
FIGURE 6. The predicted mmu_circ_0000823 targeted ceRNA network. The colors of green and blue nodes represent the expression levels of the DE genes that were downregulated, while the red nodes represent the upregulated genes. mRNAs in the outer circle can be regulated by a single miRNA, while mRNAs in the inner circle can be targeted by multiple miRNAs.
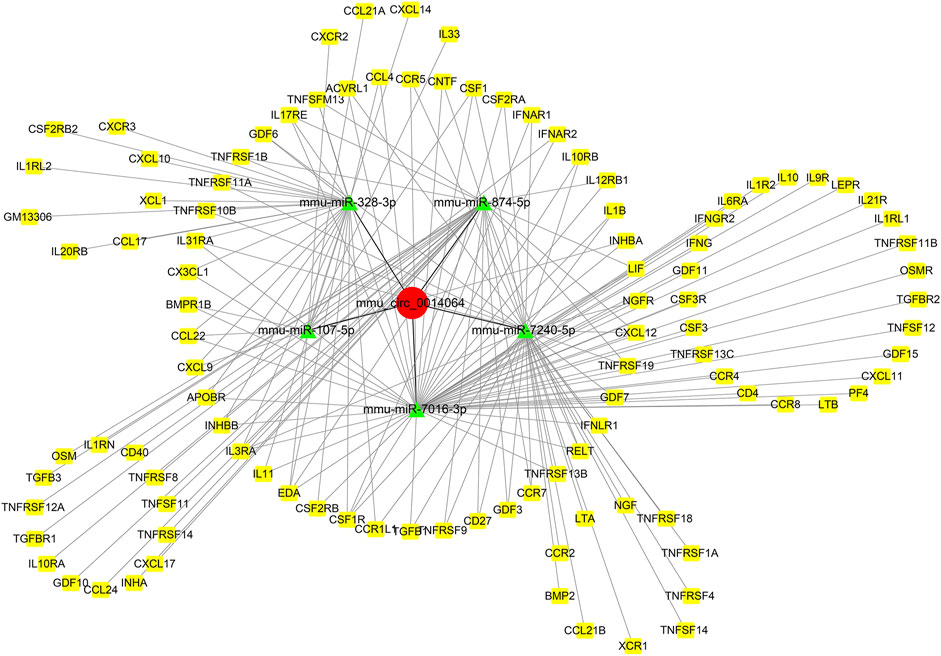
FIGURE 7. The predicted mmu_circ_0014064-based regulatory network. Green nodes represent downregulated DE miRNAs, while the red and yellow nodes represent upregulated genes.
Discussion
Renal fibrosis is one of the most characterized pathological features of CKD of various etiologies. Progressive fibrosis eventually leads to renal dysfunction, leaving only intensive treatment options of dialysis or transplantation for patients with CKD. Despite the identification of multiple regulatory signaling pathways in past decades, no effective therapeutic strategies have been developed to halt the progression of renal fibrosis. CircRNA is fine-tuned and controlled in many physiological and pathological conditions, and could trigger a significant breakthrough in CKD prevention. Mounting evidence has shown that circRNA expression is altered in kidney disorders (van Zonneveld et al., 2021; Yu et al., 2021). However, the identification and function of circRNAs are largely unknown in the context of renal fibrosis. To address this knowledge gap, we performed systemic transcriptome sequencing of circRNAs, miRNAs, and mRNAs in mouse kidney tissues with UIRI-induced renal fibrosis. We identified a total of 4983 circRNAs, 216 miRNAs, and 6371 mRNAs as differentially expressed. Through comprehensive data mining and bioinformatics analysis, we found two hub circRNAs and their related action network based on our own sequencing data. Our research thus contributes to the body of knowledge that helps us to understand the mechanism of renal fibrosis induced by ischemia reperfusion injury.
Ten candidate circRNAs and six miRNAs were validated by RT–qPCR. The RT–qPCR results showed a consistent expression trend with the sequencing data. The upregulation of miR-142a-5p, miR-142a-3p, and miR-135b-5p, and downregulation of miR-107-3p and miR-192-3p, have been implicated in previous reports as key genes and potential biomarkers in CKD (Jaswani et al., 2017; Gholaminejad et al., 2018; Min et al., 2018; Zhu et al., 2018; Wang et al., 2020). Among the candidate circRNAs, circKcnt2 is highly conserved among species and was reported to regulate the Group 3 innate lymphoid cell functions by inhibiting their activation and resolution of inflammation in colitis (Liu et al., 2020a). However, the primary expressing cell of circKcnt2 in fibrotic kidneys still needs to be clarified. Liu et al. found that the CircOxct1/miR-136/SMAD4 axis inhibited epithelial to mesenchymal transition (EMT) in gastric cancer by antagonizing the TGF-beta pathway (Liu et al., 2020b). Notably, circSlc8a1 (mmu_circ_0000823), highly conserved among different species (Mus musculus versus Homo sapiens: 88.7% homology), was the most abundant among all of the DE circRNAs and was significantly downregulated in UIRI-induced fibrotic kidneys. CircSLC8A1 was reported to be significantly downregulated in cancers, exerting a tumor suppression function via the ceRNA mechanism. In prostate cancer, circSLC8A1 directly interacted with miR-21 to inhibit the proliferation, angiogenesis, cell migration, and EMT behaviors of tumor cells (Wang et al., 2021). In another study, circSlc8a1 was also significantly reduced in bladder cancer tissues and cell lines. Overexpression of circSLC8A1 inhibited tumor migration, invasion, and proliferation both in vitro and in vivo. Mechanistically, circSLC8A1 upregulated PTEN by sponging miR-130b/miR-494 (Lu et al., 2019). Interestingly, the downstream targets of circSlc8a1, miR-21, and miR-130b were both dysregulated in our sequencing results and have been reported as biomarkers in CKD (Zhong et al., 2011; Ma et al., 2019). Further bioinformatic analysis of the circSlc8a1-related ceRNA network among the DE miRNAs and mRNAs suggested that it might affect the metabolism of the kidney and be involved in the pathogenesis of renal fibrosis. CircApoe was another hub gene targeting five DE miRNAs and was involved in the cytokine–cytokine receptor pathway. The expression and functions of the rest of the validated circRNAs, including circTinag, circMettl9, circDcdc2a, etc., have been incompletely investigated. Our findings based on bioinformatic predictions and sequencing data provide a more precise scope for subsequent research.
The relationship between aberrantly expressed circRNAs and kidney fibrosis has been debated in several studies. Wen et al. found that circACTR2 was upregulated in glucose-stressed HK-2 cells and promoted renal fibrosis by affecting pyroptosis (Wen et al., 2020). In a UUO mouse model and in TGF-beta-stimulated mouse renal tubular epithelial cells, circRNA_30032 was significantly upregulated and exacerbated renal fibrosis via the miR-96-5p/HBEGF/KRAS signaling pathway (Yi et al., 2021). In a db/db mouse model of type 2 diabetes and renal tubular cells stimulated by high glucose, circEIF4G2 was significantly upregulated (Xu et al., 2020b), while circRNA_010383 was markedly downregulated (Peng et al., 2021). CircEIF4G2 promotes renal fibrosis via the miR-218/SERBP1 pathway. Overexpression of circRNA_010383 inhibited the proteinuria and the accumulation of the extracellular matrix. These findings suggested that circRNAs functioning as ceRNAs by sponging miRNAs are crucial regulators in renal fibrosis. Based on the circRNA-related regulatory network, we identified two hub genes, circSlc8a1 and circApoe, which might be involved in the pathogenesis of renal fibrosis by sponging multiple DE miRNAs and their downstream targeted DE mRNAs. Metabolic disturbance has been increasingly recognized as an important pathogenic process that underlies renal fibrosis, and development of drugs for metabolic reprogramming will be a promising anti-fibrotic therapy (Zhao et al., 2020; Zhu et al., 2021). This study might provide a new perspective on renal fibrosis and contribute to the development of novel therapeutics for CKD.
Our study still has some limitations. Future research should evaluate the quality of our sequencing data by validating more DE circRNAs. The highly conserved DE circRNAs we identified should be examined in kidney tissues from CKD patients to profile their expression levels in the context of renal diseases. The functions of the DE circRNAs need further elucidation, and a direct relationship between circRNAs and their targeted miRNAs should be confirmed. Although the function of circRNAs as miRNA sponges has been the most studied, unearthing other roles, such as encoding peptides or interacting with proteins (Huang et al., 2020; Jarlstad Olesen and S. Kristensen, 2021), will also be critical to deepen our understanding of this new star in the transcriptome.
In summary, the present study offers a systematic perspective on the expression of circRNAs, miRNAs, and mRNAs in IRI-induced renal fibrosis. The results provided convincing evidence that the identified circRNAs and miRNAs are potential biomarkers for renal fibrosis. However, further studies on the target verification and functional analysis of these circRNAs are needed to furnish conclusive evidence to explain the regulatory mechanism of circRNAs in CKD. To better characterize the pathophysiological role of circRNAs, studies should be tailored to (1) further verify circRNA expressions in CKD patients, not only in animal models; (2) delve deeply into the their functions in renal fibrosis by in vitro and in vivo experiments; (3) explore the cellular molecular mechanism through which the circRNAs exert their functions and try to assess their value as treatment targets; and (4) aim to employ them as diagnostic and prognostic biomarkers in CKD.
Abbreviations; AKI, acute kidney injury; ceRNA, competitive endogenous RNA; circRNA, circular RNA; CKD, chronic kidney disease; DE, differentially expressed; DEMs, differentially-expressed miRNAs; GO, Gene Ontology; IRI, ischemia reperfusion injury; KEGG, Kyoto Encyclopedia of Genes and Genomes; lncRNA, long ncRNA; miRNA, microRNA; ncRNA, noncoding RNA; UIRI, unilateral renal ischemia reperfusion; UUO, unilateral ureteric obstruction.; Ethical Approval and Consent to participate; The animal study was reviewed and approved by the Experimental Animal Ethics Committee of Teaching and Research Support Center at the Fourth Military Medical University, approval number IACUC-2019014.
Data Availability Statement
The original contributions presented in the study are publicly available in NCBI under accession number PRJNA747937.
Ethics Statement
The animal study was reviewed and approved by the Experimental Animal Ethics Committee of teaching and Research Support Center in the Fourth Military Medical University, approval number (IACUC-2019014).
Author Contributions
LeW wrote the manuscript, analyzed the data, and conducted most experiments. LL designed the study. ZY and YZ contributed to the writing. XB and LiW performed the animal experiments. SS and MB critically revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China grants No.82170722, No.81870470, No.81670655, and No. 82070699.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.793182/full#supplementary-material
References
Bai, S., Xiong, X., Tang, B., Ji, T., Li, X., Qu, X., et al. (2020). Exosomal circ_DLGAP4 Promotes Diabetic Kidney Disease Progression by Sponging miR-143 and Targeting ERBB3/NF-Κb/mmp-2 axis. Cell Death Dis 11 (11), 1008. doi:10.1038/s41419-020-03169-3
Cen, J., Liang, Y., Huang, Y., Pan, Y., Shu, G., Zheng, Z., et al. (2021). Circular RNA circSDHC Serves as a Sponge for miR-127-3p to Promote the Proliferation and Metastasis of Renal Cell Carcinoma via the CDKN3/E2F1 axis. Mol. Cancer 20 (1), 19. doi:10.1186/s12943-021-01314-w
Chen, X.-T., Li, Z.-W., Zhao, X., Li, M.-L., Hou, P.-F., Chu, S.-F., et al. (2021). Role of Circular RNA in Kidney-Related Diseases. Front. Pharmacol. 12, 615882. doi:10.3389/fphar.2021.615882
Fan, Y., Chen, H., Huang, Z., Zheng, H., and Zhou, J. (2020). Emerging Role of miRNAs in Renal Fibrosis. RNA Biol. 17 (1), 1–12. doi:10.1080/1547628610.1080/15476286.2019.1667215
Fernández, A. R., Sánchez-Tarjuelo, R., Cravedi, P., Ochando, J., and López-Hoyos, M. (2020). Review: Ischemia Reperfusion Injury-A Translational Perspective in Organ Transplantation. Ijms 21 (22), 8549. doi:10.3390/ijms21228549
Gholaminejad, A., Abdul Tehrani, H., and Gholami Fesharaki, M. (2018). Identification of Candidate microRNA Biomarkers in Renal Fibrosis: a Meta-Analysis of Profiling Studies. Biomarkers 23 (8), 713–724. doi:10.1080/1354750X.2018.1488275
Holderied, A., Kraft, F., Marschner, J. A., Weidenbusch, M., and Anders, H.-J. (2020). "Point of No Return" in Unilateral Renal Ischemia Reperfusion Injury in Mice. J. Biomed. Sci. 27 (1), 34. doi:10.1186/s12929-020-0623-9
Huang, A., Zheng, H., Wu, Z., Chen, M., and Huang, Y. (2020). Circular RNA-Protein Interactions: Functions, Mechanisms, and Identification. Theranostics 10 (8), 3503–3517. doi:10.7150/thno.42174
Jao, T.-M., Nangaku, M., Wu, C.-H., Sugahara, M., Saito, H., Maekawa, H., et al. (2019). ATF6α Downregulation of PPARα Promotes Lipotoxicity-Induced Tubulointerstitial Fibrosis. Kidney Int. 95 (3), 577–589. doi:10.1016/j.kint.2018.09.023
Jarlstad Olesen, M. T., and S. Kristensen, L. (2021). Circular RNAs as microRNA Sponges: Evidence and Controversies. Essays Biochem. 65 (4), 685–696. doi:10.1042/EBC20200060
Jaswani, P., Prakash, S., Agrawal, S., Prasad, N., and Sharma, R. K. (2017). Predicting miRNA Association with Corresponding Target Genes and Single Nucleotide Polymorphisms in Altered Renal Pathophysiology. Mirna 6 (3), 213–221. doi:10.2174/2211536606666171016151846
Lin, J., Jiang, Z., Liu, C., Zhou, D., Song, J., Liao, Y., et al. (2020). Emerging Roles of Long Non-coding RNAs in Renal Fibrosis. Life 10 (8), 131. doi:10.3390/life10080131
Liu, B., Ye, B., Zhu, X., Yang, L., Li, H., Liu, N., et al. (2020). An Inducible Circular RNA circKcnt2 Inhibits ILC3 Activation to Facilitate Colitis Resolution. Nat. Commun. 11 (1), 4076. doi:10.1038/s41467-020-17944-5
Liu, J., Dai, X., Guo, X., Cheng, A., Mac, S. M., and Wang, Z. (2020). Circ-OXCT1 Suppresses Gastric Cancer EMT and Metastasis by Attenuating TGF-β Pathway through the Circ-OXCT1/miR-136/smad4 Axis. Ott Vol. 13, 3987–3998. doi:10.2147/OTT.S239789
Liu, Z., Wang, Y., Shu, S., Cai, J., Tang, C., and Dong, Z. (2019). Non-coding RNAs in Kidney Injury and Repair. Am. J. Physiology-Cell Physiol. 317 (2), C177–C188. doi:10.1152/ajpcell.00048.2019
Lu, Q., Liu, T., Feng, H., Yang, R., Zhao, X., Chen, W., et al. (2019). Circular RNA circSLC8A1 Acts as a Sponge of miR-130b/miR-494 in Suppressing Bladder Cancer Progression via Regulating PTEN. Mol. Cancer 18 (1), 111. doi:10.1186/s12943-019-1040-0
Luan, R., Tian, G., Ci, X., Zheng, Q., Wu, L., and Lu, X. (2021). Differential Expression Analysis of Urinary Exosomal Circular RNAs in Patients with IgA Nephropathy. Nephrology 26 (5), 432–441. doi:10.1111/nep.13855
Ma, Y., Shi, J., Wang, F., Li, S., Wang, J., Zhu, C., et al. (2019). MiR‐130b Increases Fibrosis of HMC Cells by Regulating the TGF‐β1 Pathway in Diabetic Nephropathy. J. Cel Biochem 120 (3), 4044–4056. doi:10.1002/jcb.27688
Min, Q.-H., Chen, X.-M., Zou, Y.-Q., Zhang, J., Li, J., Wang, Y., et al. (2018). Differential Expression of Urinary Exosomal microRNAs in IgA Nephropathy. J. Clin. Lab. Anal. 32 (2), e22226. doi:10.1002/jcla.22226
Nelson, R. G., Grams, M. E., Ballew, S. H., Sang, Y., Azizi, F., Chadban, S. J., et al. (2019). Development of Risk Prediction Equations for Incident Chronic Kidney Disease. JAMA 322 (21), 2104–2114. doi:10.1001/jama.2019.17379
Ó hAinmhire, E., and Humphreys, B. D. (2017). Fibrotic Changes Mediating Acute Kidney Injury to Chronic Kidney Disease Transition. Nephron 137 (4), 264–267. doi:10.1159/000474960
Pefanis, A., Ierino, F. L., Murphy, J. M., and Cowan, P. J. (2019). Regulated Necrosis in Kidney Ischemia-Reperfusion Injury. Kidney Int. 96 (2), 291–301. doi:10.1016/j.kint.2019.02.009
Peng, F., Gong, W., Li, S., Yin, B., Zhao, C., Liu, W., et al. (2021). circRNA_010383 Acts as a Sponge for miR-135a and its Downregulated Expression Contributes to Renal Fibrosis in Diabetic Nephropathy. Diabetes 70 (2), db200203–615. doi:10.2337/db200203
Polichnowski, A. J., Griffin, K. A., Licea-Vargas, H., Lan, R., Picken, M. M., Long, J., et al. (2020). Pathophysiology of Unilateral Ischemia-Reperfusion Injury: Importance of Renal Counterbalance and Implications for the AKI-CKD Transition. Am. J. Physiology-Renal Physiol. 318 (5), F1086–F1099. doi:10.1152/ajprenal.00590.2019
Ren, H., and Wang, Q. (2021). Non-Coding RNA and Diabetic Kidney Disease. DNA Cel Biol. 40 (4), 553–567. doi:10.1089/dna.2020.5973
Ronco, C., Bellomo, R., and Kellum, J. A. (2019). Acute Kidney Injury. The Lancet 394 (10212), 1949–1964. doi:10.1016/S0140-6736(19)32563-2
Shiva, N., Sharma, N., Kulkarni, Y. A., Mulay, S. R., and Gaikwad, A. B. (2020). Renal Ischemia/reperfusion Injury: An Insight on In Vitro and In Vivo Models. Life Sci. 256, 117860. doi:10.1016/j.lfs.2020.117860
Tod, P., Bukosza, E. N., Róka, B., Kaucsár, T., Fintha, A., Krenács, T., et al. (2020). Post-ischemic Renal Fibrosis Progression Is Halted by Delayed Contralateral Nephrectomy: The Involvement of Macrophage Activation. Ijms 21 (11), 3825. doi:10.3390/ijms21113825
van Zonneveld, A. J., Kölling, M., Bijkerk, R., and Lorenzen, J. M. (2021). Circular RNAs in Kidney Disease and Cancer. Nat. Rev. Nephrol. 17 (12), 814–826. doi:10.1038/s41581-021-00465-9
Verduci, L., Tarcitano, E., Strano, S., Yarden, Y., and Blandino, G. (2021). CircRNAs: Role in Human Diseases and Potential Use as Biomarkers. Cel Death Dis 12 (5), 468. doi:10.1038/s41419-021-03743-3
Wang, D., Yan, S., Wang, L., Li, Y., and Qiao, B. (2021). circSLC8A1 Acts as a Tumor Suppressor in Prostate Cancer via Sponging miR-21. Biomed. Res. Int. 2021, 1–9. doi:10.1155/2021/6614591
Wang, Z., Liu, Z., Yang, Y., and Kang, L. (2020). Identification of Biomarkers and Pathways in Hypertensive Nephropathy Based on the ceRNA Regulatory Network. BMC Nephrol. 21 (1), 476. doi:10.1186/s12882-020-02142-8
Wen, S., Li, S., Li, L., and Fan, Q. (2020). circACTR2: A Novel Mechanism Regulating High Glucose-Induced Fibrosis in Renal Tubular Cells via Pyroptosis. Biol. Pharm. Bull. 43 (3), 558–564. doi:10.1248/bpb.b19-00901
Xu, B., Wang, Q., Li, W., Xia, L., Ge, X., Shen, L., et al. (2020). Circular RNA circEIF4G2 Aggravates Renal Fibrosis in Diabetic Nephropathy by Sponging miR‐218. J. Cel Mol Med. doi:10.1111/jcmm.16129
Xu, Y., Jiang, W., Zhong, L., Li, H., Bai, L., Chen, X., et al. (2020). circ‐AKT3 Aggravates Renal Ischaemia‐reperfusion Injury via Regulating miR‐144‐5p/Wnt/β‐catenin Pathway and Oxidative Stress. J. Cel Mol Med. doi:10.1111/jcmm.16072
Yi, L., Ai, K., Li, H., Qiu, S., Li, Y., Wang, Y., et al. (2021). CircRNA_30032 Promotes Renal Fibrosis in UUO Model Mice via miRNA-96-5p/HBEGF/KRAS axis. Aging 13 (9), 12780–12799. doi:10.18632/aging.202947
Yu, J., Xie, D., Huang, N., and Zhou, Q. (2021). Circular RNAs as Novel Diagnostic Biomarkers and Therapeutic Targets in Kidney Disease. Front. Med. 8, 714958. doi:10.3389/fmed.2021.714958
Zhang, C., Gao, C., Di, X., Cui, S., Liang, W., Sun, W., et al. (2021). Hsa_circ_0123190 Acts as a Competitive Endogenous RNA to Regulate APLNR Expression by Sponging Hsa-miR-483-3p in Lupus Nephritis. Arthritis Res. Ther. 23 (1), 24. doi:10.1186/s13075-020-02404-8
Zhao, X., Kwan, J. Y. Y., Yip, K., Liu, P. P., and Liu, F.-F. (2020). Targeting Metabolic Dysregulation for Fibrosis Therapy. Nat. Rev. Drug Discov. 19 (1), 57–75. doi:10.1038/s41573-019-0040-5
Zhong, X., Chung, A. C. K., Chen, H.-Y., Meng, X.-M., and Lan, H. Y. (2011). Smad3-Mediated Upregulation of miR-21 Promotes Renal Fibrosis. Jasn 22 (9), 1668–1681. doi:10.1681/ASN.2010111168
Zhou, W.-Y., Cai, Z.-R., Liu, J., Wang, D.-S., Ju, H.-Q., and Xu, R.-H. (2020). Circular RNA: Metabolism, Functions and Interactions with Proteins. Mol. Cancer 19 (1), 172. doi:10.1186/s12943-020-01286-3
Zhu, K., Zheng, T., Chen, X., and Wang, H. (2018). Bioinformatic Analyses of Renal Ischaemia-Reperfusion Injury Models: Identification of Key Genes Involved in the Development of Kidney Disease. Kidney Blood Press. Res. 43 (6), 1898–1907. doi:10.1159/000496001
Keywords: ceRNA network, chronic kidney disease, unilateral ischemia reperfusion injury, RNA-seq, circRNA
Citation: Wei L, Yu Z, Liu L, Zhou Y, Bai X, Wang L, Bai M and Sun S (2022) Integrated Analysis of the CircRNA-Based ceRNA Network in Renal Fibrosis Induced by Ischemia Reperfusion Injury. Front. Genet. 12:793182. doi: 10.3389/fgene.2021.793182
Received: 11 October 2021; Accepted: 29 December 2021;
Published: 10 February 2022.
Edited by:
Ratnakar Tiwari, Northwestern University, United StatesReviewed by:
Nabin Poudel, University of Virginia, United StatesTomohito Doke, University of Pennsylvania, United States
Copyright © 2022 Wei, Yu, Liu, Zhou, Bai, Wang, Bai and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiren Sun, c3Vuc2hpcmVuQG1lZG1haWwuY29tLmNu
†These authors have contributed equally to this work
 Lei Wei
Lei Wei Zhixiang Yu
Zhixiang Yu Limin Liu
Limin Liu Ying Zhou
Ying Zhou Xiao Bai
Xiao Bai Liya Wang
Liya Wang Ming Bai
Ming Bai Shiren Sun
Shiren Sun