
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Genet., 04 November 2021
Sec. Evolutionary and Population Genetics
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.785218
It has been argued that DNA repair by homologous recombination in the context of endonuclease-mediated cleavage can cause mutations. To better understand this phenomenon, we examined homologous recombination following endonuclease cleavage in a native genomic context: the movement of self-splicing introns in the mitochondrial genomes of Metschnikowia yeasts. Self-splicing mitochondrial introns are mobile elements, which can copy and paste themselves at specific insertion sites in mitochondrial DNA using a homing endonuclease in conjunction with homologous recombination. Here, we explore the mutational effects of self-splicing introns by comparing sequence variation within the intron-rich cox1 and cob genes from 71 strains (belonging to 40 species) from the yeast genus Metschnikowia. We observed a higher density of single nucleotide polymorphisms around self-splicing-intron insertion sites. Given what is currently known about the movement of organelle introns, it is likely that their mutational effects result from the high binding affinity of endonucleases and their interference with repair machinery during homologous recombination (or, alternatively, via gene conversion occurring during the intron insertion process). These findings suggest that there are fitness costs to harbouring self-splicing, mobile introns and will help us better understand the risks associated with modern biotechnologies that use endonuclease-mediated homologous recombination, such as CRISPR-Cas9 gene editing.
The ability to safely modify the DNA of living organisms may soon be within our grasp thanks to CRISPR-Cas9. Despite its immense promise, CRISPR-Cas9 gene editing has not yet been approved for widespread therapeutic use because of its potential for introducing unwanted mutations (Kosicki et al., 2018). Recent reports have proposed that homologous recombination following endonuclease cleavage is responsible for CRISPR-Cas9’s on-target mutagenicity. This is because DNA-binding proteins, such as endonucleases, interfere with double-strand break repair by competing with the repair machinery for the target site (Reijns et al., 2015; Kaiser et al., 2016; Sabarinathan et al., 2016). Moreover, it has been shown that mutation rates are higher in regions of high recombination and that indels are often associated with a high density of single nucleotide polymorphisms (SNPs) (Lercher and Hurst, 2002; Tian et al., 2008).
An excellent model for studying the relationship between endonuclease activity and on-target mutagenicity in a native genomic context is the movement of self-splicing introns in mitochondrial genomes. Self-splicing introns are a class of mobile element commonly found in the mitochondrial DNAs (mtDNAs) of plants, fungi, and protists (Lambowitz and Belfort, 1993; Burger et al., 2003; Lang et al., 2007; Brown et al., 2014; Smith and Keeling, 2015; Megarioti and Kouvelis, 2020). These elements can copy and paste themselves at specific insertion sites in the host mitochondrial genome via their ability to form a complex secondary structure, which functions as a homing endonuclease (Cech et al., 1994; Lambowitz and Zimmerly, 2004). The homing endonuclease recognizes and binds to its target site with high affinity, inducing a double-stranded break (DSB). The break is then repaired by homologous recombination using the intron-containing strand as a template, resulting in intron movement (Cech et al., 1994; Lambowitz and Zimmerly, 2004).
Although self-splicing mitochondrial introns are found in a diversity of taxa (Del Vasto et al., 2015; Mower, 2020; Wideman et al., 2020), they are particularly prevalent within the yeast genus Metschnikowia (Lee et al., 2020), many members of which are commonly found in the guts of beetles inhabiting morning glories and other flowers around the world. Metschnikowia mtDNAs harbour among the largest numbers of organelle self-splicing introns of all eukaryotes. The cox1 and cob genes from Metschnikowia mtDNAs are especially intron rich, with the number of introns varying from 0–20 within cox1 and 0–13 within cob. These introns can be found at 25 and 13 unique intron insertion sites across cox1 and cob, respectively (Supplementary Tables S1A, B), making this genus the largest known reservoir of self-splicing, mobile organelle introns (Férandon et al., 2010; Lee et al., 2020). For detailed summary statistics on the mitochondrial introns of Metschnikowia yeasts, including the variation in intron number across species/strains, the types of introns (e.g., group I or group II), and the presence/absence of intronic homing endonucleases, please refer to Supplemental Information of Lee et al. (2020).
Large variations in intron number can lead to remarkable differences in gene and genome size (Del Vasto et al., 2015; Mower, 2020; Wideman et al., 2020). Among Metschnikowia mtDNAs, there is a 30-fold size range for cox1 and a 20-fold size range for cob, which encompasses the entire known size variation for both genes across the eukaryotic domain (Lee et al., 2020). More astonishing is that even within members of the same species, the size of cox1 has been found to vary by up to 5 kb (Lee et al., 2020) — and keep in mind that the mature cox1 mRNA from Metschnikowia species is less than 1.6 kb (Figure 1). Metschnikowia mitogenomes have other notable features, including extreme differences in intergenic content and genomic structure (e.g., linear vs circular mitochondrial chromosomes), both between and within species.
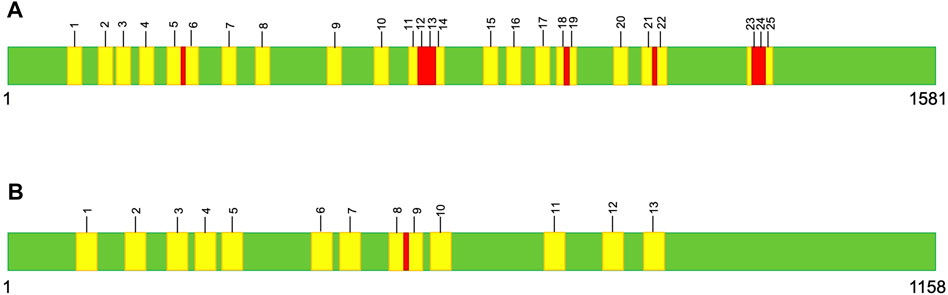
FIGURE 1. Graphical representation of the cox1 (A) and cob (B) consensus sequences, which are 1,581 nt and 1,158 nt, respectively. Self-splicing-intron insertion sites are numbered and the 10 nt windows are highlighted in yellow. Regions where a single SNP occurred in two overlapping windows are highlighted in red.
The co-occurrence of mtDNA diversity and large numbers of self-splicing introns in Metschnikowia may not be coincidental (Lee et al., 2020). If the movement of self-splicing introns is mutagenic, then this could help explain why Metschnikowia mitogenomes are so diverse. To explore this idea further, we studied the relationship between SNPs and their proximity to self-splicing-intron insertion sites within cox1 and cob from 71 different species/strains of Metschnikowia. These two genes are ideal candidates for investigating this topic as they are both intron-dense and highly conserved, making sequence alignments and comparisons relatively straightforward (Chandrasekharan and Simmons, 2004; Linacre and Lee, 2005).
We predict that SNP abundance will increase close to self-splicing intron insertion sites in cox1 and cob. If true, this would support the notion that the movement of mitochondrial introns is mutagenic and that this mutagenicity may be connected to endonuclease interference of homologous recombination or gene conversion. We should stress that this study is partly inspired by the work of Repar and Warnecke (2017) who found elevated levels of SNPs close to the insertion sites of self-splicing introns in the mitochondrial genomes of Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Lachancea kluyveri. These findings ultimately led the authors to conclude that intron mobility is a direct driver of host genetic diversity.
Annotated Metschnikowia mitochondrial genomes (Lee et al., 2020) were downloaded from GenBank (accessions MT421951-MT449704) and uploaded to Geneious Prime v. 2020.1.2 (Biomatter Ltd., Auckland, New Zealand). These genomes were generated as part of a large-scale collaborative effort to explore Metschnikowia diversity, natural history and phylogenetics (Lachance et al., 2016), and include Metschnikowia strains that have been collected intensively over 3 decades across a broad array of biogeographic zones (Lachance, 2011; de Oliveira Santos et al., 2015). The 71 cox1 and cob genes were extracted, their introns were removed, and their coding sequences were aligned with MUSCLE (Edgar, 2004), implemented in Geneious using default settings. An initial pre-assessment of the alignments was performed to confirm that the coding regions and intron insertion sites were properly annotated. A few minor corrections were made, and the final alignments were consistent with previous analyses: 25 and 13 intron insertion sites across the cox1 and cob alignments, respectively (Lee et al., 2020).
SNPs were evaluated in Geneious, but each polymorphic site within the alignments was also checked and counted manually. We used the chi-squared test of independence to determine if the number of SNPs within a given window was significantly greater than the number outside that window. To conduct this test, a series of 2x2 contingency tables for each window size were constructed in Excel for the cox1 and cob alignments (Table 1). From the tables, the expected values were calculated, which were then used in the CHISQ.TEST function in Excel to obtain the p-values (Table 1).
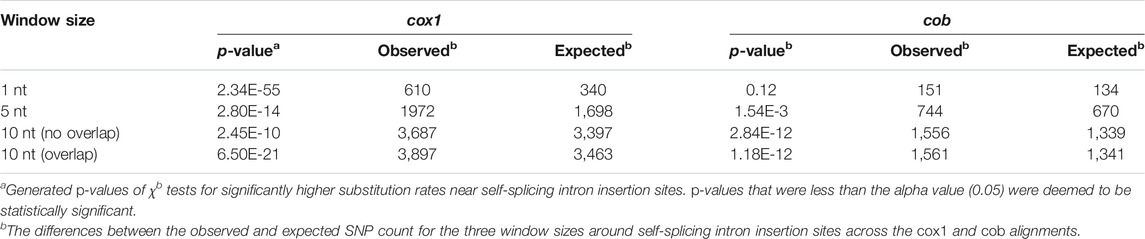
TABLE 1. Statistical significance of the occurrence of SNPs near self-splicing intron insertion sites in the cox1 and cob genes from 71 strains of Metschnikowia.
To investigate the potential mutational effects of self-splicing intron movement in the mitogenomes of Metschnikowia, we recorded the number of SNPs in the cox1 and cob genes across 71 species/strains. We binned the SNPs into the following window sizes: one nucleotide (nt), 5 nt, and 10 nt to the left and right of each intron insertion site. The number of SNPs that fell outside of these windows were also counted. Note: for the 10 nt windows, there were instances where a single SNP was present in two different windows (Figure 1). To account for this, two separate analyses were performed: one where every overlapping variant was counted twice and one where they were only counted once (Table 1).
For all parameters, except for the 1 nt window of the cob alignment, the observed p-values were less than 0.05 (Table 1), indicating that the null hypothesis (that there is no relationship between SNP density and self-splicing intron movement) could be rejected with confidence. To confirm that our findings were indicative of a higher density of SNPs within the chosen parameters (rather than lower), we calculated the observed versus the expected values (referring to the number of SNPs that we would expect to see within the windows under random conditions) (Table 1). We found that the observed values were higher than the expected values, confirming that there were more SNPs in these regions than predicted under a random model. Together, these data suggest that there is a relationship between mutagenicity and self-splicing intron movement, which may be connected to how homing endonucleases bind to their template strands while competing with mtDNA repair machinery (Reijns et al., 2015; Kaiser et al., 2016; Sabarinathan et al., 2016), or might be a consequence of gene conversion during the intron insertion process, which could affect the exonic nucleotides around insertion sites (Repar and Warnecke, 2017).
In simpler terms, we found that there was an average of 1.2x more SNPs in the cox1 alignment and 1.5x more in the cob alignment within 10 nt of mobile intron insertion sites than throughout the rest of the sequences. While our values did achieve statistical significance (indicating that there are more SNPs in these areas than expected under random circumstances), they also indicate that there is significant variation outside of our selected windows as well, and that forces other than intron movement are likely contributing to the observed SNPs. Keeping in mind the huge reservoir of mitochondrial introns within this genus, there is a strong possibility that introns recently existed in other regions of the genes but have recently been lost. If true, this could have potentially led to some of the SNPs observed outside of the defined windows. We reiterate our call for more research on both the potential mutagenicity of self-splicing intron movement and the high amount of mtDNA variability exhibited by species of the genus Metschnikowia.
To explore whether a particular site within the 10 nt window experienced a higher incidence of mutation, we plotted the average number of SNPs for each of the 20 nt positions (10 to the left and 10 to the right) across the 25 and 13 intron insertion sites of cox1 and cob, respectively. Position 7 to the left of the insertion site showed the highest average number of SNPs for both gene alignments (Figures 2A,B), suggesting that this site may be particularly prone to mutations during intron movement. However, one should not place too much weight on this observation as there is obvious variability associated with the genetic code (i.e., the 3 nt periodicity), with introns inserting between codons (phase 0) occurring 68 and 69% of the time in cox1 and cob, respectively.
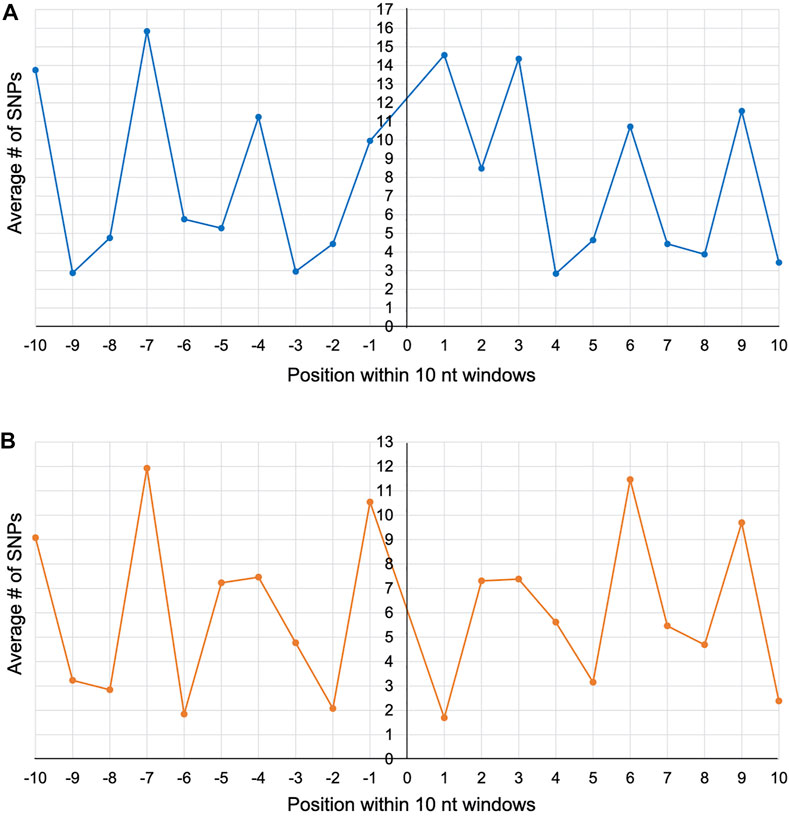
FIGURE 2. Line graphs depicting the average number of SNPs per nucleotide position within 10 nt windows around the self-splicing intron insertion sites across the cox1 (A) and cob (B) alignments. The horizontal axis represents the nucleotide position (from -10 to +10) and the vertical axis represents the average number of SNPs.
These findings raise an intriguing question: If the movement of self-splicing introns is mutagenic, why have Metschnikowia species accumulated so many mitochondrial introns? In other words, why would this genus retain something that arguably imposes a high mutational burden? The simplest answer is that Metschnikowia may not be able rid itself of these selfish elements. This could be a consequence of where the insertion sites are located as well as recurring horizontal gene transfer events involving the lateral exchange of mitochondrial introns between species and strains (Bhattacharya et al., 2001; Haugen et al., 2005).
Mitochondrial intron insertion sites tend to occur in highly conserved, constitutively expressed genes, like cox1 and cob (Repar and Warnecke, 2017). Changes to the intron insertion sites of such genes can have deleterious consequences, meaning their sequences are typically maintained among close relatives. Self-splicing introns can, however, be lost from a genome through random deletion or gene conversion events (Goddard and Burt, 1999). But there is growing evidence of extensive horizontal gene transfer between yeast populations, meaning that even if an individual rids itself of a mitochondrial intron there is the potential that it will get it back from a neighbour (Bhattacharya et al., 2001; Haugen et al., 2005; Repar and Warnecke 2017).
There is also the possibility that Metschnikowia yeasts have become physiologically reliant on mitochondrial introns. Rudan et al. (2018) investigated two strains of S. cerevisiae: a wild-type strain with mobile introns and an artificial strain with the introns removed. They found that mitochondrial morphology was altered in the intron-less strain, resulting in a highly stressed cellular phenotype with lower fitness. It was proposed that these physiological changes were the consequence of overly efficient transcription of cox1 and cob (their transcripts were in excess in the intron-less strain). One explanation for these observations is that mitochondrial introns can lead to inefficient splicing; thus, in genes where introns abound transcription needs to be upregulated, but removal of the introns can result in an overabundance of transcripts. This evolutionary lock-in event may partly explain the longstanding persistence of self-splicing introns in mitochondrial genomes, despite their potential deleterious effects.
Finally, it is noteworthy that the Metschnikowia species explored here are all insect symbionts (mostly of beetles). Although speculative, this symbiotic lifestyle could potentially reduce their reliance on proper mitochondrial function, as their insect vectors constantly make sure that they occupy the proper habitat. It may also greatly impact the population genetics of these yeasts, potentially increasing random genetic drift.
We observed a higher density of SNPs near to self-splicing intron insertion sites, which we argue is the result of mitochondrial intron movement. The mutagenicity of self-splicing introns may result from incorrect mtDNA repair following homologous recombination due to the persistent presence and high binding affinity of endonucleases. These findings provide further support for the notion that, while once considered to have no ill effects (Werren, 2011), self-splicing introns impose a mutational burden on their host genomes (Mueller et al., 1996; Repar and Warnecke, 2017). We hope that this work will encourage more research on endonuclease binding and its potential interactions with host-cell repair machinery. A better understanding of this phenomenon could be crucial to treating diseases via CRISPR-Cas9 gene editing.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
This study was conceptualized by DS. ST performed the analyses under the supervision of DS and M-AL. ST wrote the initial draft of the manuscript. All authors edited and contributed to the final draft.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
MAL and DRS acknowledge Discovery Grants from the Natural Sciences and Engineering Research Counsel of Canada (NSERC).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.785218/full#supplementary-material
Bhattacharya, D., Cannone, J. J., and Gutell, R. R. (2001). Group I Intron Lateral Transfer between Red and Brown Algal Ribosomal RNA. Curr. Genet. 40, 82–90. doi:10.1007/s002940100227
Brown, G. G., Colas des Francs-Small, C., and Ostersetzer-Biran, O. (2014). Group II Intron Splicing Factors in Plant Mitochondria. Front. Plant Sci. 5, 35. doi:10.3389/fpls.2014.00035
Burger, G., Forget, L., Zhu, Y., Gray, M. W., and Lang, B. F. (2003). Unique Mitochondrial Genome Architecture in Unicellular Relatives of Animals. Proc. Natl. Acad. Sci. 100, 892–897. doi:10.1073/pnas.0336115100
Cech, T. R., Damberger, S. H., and Gutell, R. R. (1994). Representation of the Secondary and Tertiary Structure of Group I Introns. Nat. Struct. Mol. Biol. 1, 273–280. doi:10.1038/nsb0594-273
Chandrasekharan, N., and Simmons, D. L. (2004). The Cyclooxygenases. Genome Biol. 5, 241. doi:10.1186/gb-2004-5-9-241
de Oliveira Santos, A. R., Perri, A. M., Andrietta, M. D. G. S., Rosa, C. A., and Lachance, M.-A. (2015). The Expanding Large-Spored Metschnikowia Clade: Metschnikowia Matae Sp. nov., a Yeast Species with Two Varieties from the Brazilian Atlantic Forest. Antonie van Leeuwenhoek 108, 753–763. doi:10.1007/s10482-015-0531-2
Del Vasto, M., Figueroa-Martinez, F., Featherston, J., González, M. A., Reyes-Prieto, A., Durand, P. M., et al. (2015). Massive and Widespread Organelle Genomic Expansion in the Green Algal Genus Dunaliella. Genome Biol. Evol. 7, 656–663. doi:10.1093/gbe/evv027
Edgar, R. C. (2004). MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinf. 5, 113. doi:10.1186/1471-2105-5-113
Férandon, C., Moukha, S., Callac, P., Benedetto, J. P., Castroviejo, M., and Barroso, G. (2010). The Agaricus Bisporus Cox1 Gene: the Longest Mitochondrial Gene and the Largest Reservoir of Mitochondrial Group I Introns. PLoS One 5, e14048. doi:10.1371/journal.pone.0014048
Goddard, M. R., and Burt, A. (1999). Recurrent Invasion and Extinction of a Selfish Gene. Proc. Natl. Acad. Sci. 96, 13880–13885. doi:10.1073/pnas.96.24.13880
Haugen, P., Simon, D. M., and Bhattacharya, D. (2005). The Natural History of Group I Introns. Trends Genet. 21, 111–119. doi:10.1016/j.tig.2004.12.007
Kaiser, V. B., Taylor, M. S., and Semple, C. A. (2016). Mutational Biases Drive Elevated Rates of Substitution at Regulatory Sites across Cancer Types. Plos Genet. 12, e1006207. doi:10.1371/journal.pgen.1006207
Kosicki, M., Tomberg, K., and Bradley, A. (2018). Repair of Double-Strand Breaks Induced by CRISPR-Cas9 Leads to Large Deletions and Complex Rearrangements. Nat. Biotechnol. 36, 765–771. doi:10.1038/nbt.4192
Lachance, M.-A., Hurtado, E., and Hsiang, T. (2016). A Stable Phylogeny of the Large-sporedMetschnikowiaclade. Yeast 33, 261–275. doi:10.1002/yea.3163
Lachance, M.-A. (2011). “Metschnikowia,” in The Yeasts, A Taxonomic Study. Editors C. P. Kurtzman, J. W. Fell, and T. Boekhout T. (Amsterdam: Elsevier), 575–620. doi:10.1016/b978-0-444-52149-1.00046-x
Lambowitz, A. M., and Belfort, M. (1993). Introns as mobile Genetic Elements. Annu. Rev. Biochem. 62, 587–622. doi:10.1146/annurev.bi.62.070193.003103
Lambowitz, A. M., and Zimmerly, S. (2004). Mobile Group II Introns. Annu. Rev. Genet. 38, 1–35. doi:10.1146/annurev.genet.38.072902.091600
Lang, B. F., Laforest, M.-J., and Burger, G. (2007). Mitochondrial Introns: a Critical View. Trends Genet. 23, 119–125. doi:10.1016/j.tig.2007.01.006
Lee, D. K., Hsiang, T., Lachance, M.-A., and Smith, D. R. (2020). The Strange Mitochondrial Genomes of Metschnikowia Yeasts. Curr. Biol. 30, R800–R801. doi:10.1016/j.cub.2020.05.075
Lercher, M. J., and Hurst, L. D. (2002). Human SNP Variability and Mutation Rate Are Higher in Regions of High Recombination. Trends Genet. 18, 337–340. doi:10.1016/s0168-9525(02)02669-0
Linacre, A., and Lee, J. C. (2016). Species Determination: The Role and Use of the Cytochrome B Gene. Methods Mol. Biol. 1420, 287–296. doi:10.1007/978-1-4939-3597-0_20
Megarioti, A. H., and Kouvelis, V. N. (2020). The Coevolution of Fungal Mitochondrial Introns and Their Homing Endonucleases (GIY-YIG and LAGLIDADG). Genome Biol. Evol. 12, 1337–1354. doi:10.1093/gbe/evaa126
Mower, J. P. (2020). Variation in Protein Gene and Intron Content Among Land Plant Mitogenomes. Mitochondrion 53, 203–213. doi:10.1016/j.mito.2020.06.002
Mueller, J. E., Smith, D., and Belfort, M. (1996). Exon Coconversion Biases Accompanying Intron Homing: Battle of the Nucleases. Genes Dev. 10, 2158–2166. doi:10.1101/gad.10.17.2158
Reijns, M. A. M., Kemp, H., Ding, J., Marion de Procé, S., Jackson, A. P., and Taylor, M. S. (2015). Lagging-strand Replication Shapes the Mutational Landscape of the Genome. Nature 518, 502–506. doi:10.1038/nature14183
Repar, J., and Warnecke, T. (2017). Mobile Introns Shape the Genetic Diversity of Their Host Genes. Genetics 205, 1641–1648. doi:10.1534/genetics.116.199059
Rudan, M., Bou Dib, P., Musa, M., Kanunnikau, M., Sobočanec, S., Rueda, D., et al. (2018). Normal Mitochondrial Function in Saccharomyces cerevisiae Has Become Dependent on Inefficient Splicing. eLife 7, e35330. doi:10.7554/eLife.35330
Sabarinathan, R., Mularoni, L., Deu-Pons, J., Gonzalez-Perez, A., and López-Bigas, N. (2016). Nucleotide Excision Repair Is Impaired by Binding of Transcription Factors to DNA. Nature 532, 264–267. doi:10.1038/nature17661
Smith, D. R., and Keeling, P. J. (2015). Mitochondrial and Plastid Genome Architecture: Reoccurring Themes, but Significant Differences at the Extremes. Proc. Natl. Acad. Sci. USA 112, 10177–10184. doi:10.1073/pnas.1422049112
Tian, D., Wang, Q., Zhang, P., Araki, H., Yang, S., Kreitman, M., et al. (2008). Single-nucleotide Mutation Rate Increases Close to Insertions/deletions in Eukaryotes. Nature 455, 105–108. doi:10.1038/nature07175
Werren, J. H. (2011). Selfish Genetic Elements, Genetic Conflict, and Evolutionary Innovation. Proc. Natl. Acad. Sci. 108, 10863–10870. doi:10.1073/pnas.1102343108
Keywords: endonuclease, homologous recombination, selfish elements, yeast, mobile intron
Citation: Thompson SRL, Lee DK, Lachance M-A and Smith DR (2021) Mutational Effects of Mobile Introns on the Mitochondrial Genomes of Metschnikowia Yeasts. Front. Genet. 12:785218. doi: 10.3389/fgene.2021.785218
Received: 28 September 2021; Accepted: 19 October 2021;
Published: 04 November 2021.
Edited by:
Ben-Yang Liao, National Health Research Institutes, TaiwanReviewed by:
Wenfeng Qian, Institute of Genetics and Developmental Biology (CAS), ChinaCopyright © 2021 Thompson, Lee, Lachance and Smith. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Roy Smith, ZHNtaXQyNDJAdXdvLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.