
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 17 January 2022
Sec. Genomics of Plants and the Phytoecosystem
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.783482
 Xian-bo Zheng†
Xian-bo Zheng† Yao Wu†
Yao Wu† Hao Wang
Hao Wang Shang-wei Song
Shang-wei Song Tuan-hui Bai
Tuan-hui Bai Jian Jiao
Jian Jiao Chun-hui Song
Chun-hui Song Hong-guang Pang
Hong-guang Pang Miao-miao Wang*
Miao-miao Wang*Zinc finger-homeodomain (ZF-HD) transcription factors play an important role in the regulation of plant growth and development, as well as the regulation of stress responses. Studies on the ZF-HD family genes have been conducted in many plants, however, the characteristics of this family in apple (Malus domestica) fruit remains to be poorly understood. In this study, we identified nineteen ZF-HD family genes in apple at the whole-genome scale, which were unevenly located on ten chromosomes. These MdZF-HD genes were phylogenetically divided into two subfamilies: zinc finger-homeodomain (ZHD) and MINI ZINC FINGER (MIF), and the ZHD subfamily was further classified into five groups (ZHDI–ZHDV). Analysis of the gene structures showed that most MdZF-HD genes lack introns. Gene expression analysis indicated that nine selected MdZF-HD genes were differentially responsive to 1-MCP (1-methylcyclopropene) treatment during the postharvest storage of “Qinguan” apple fruit. Moreover, the transcripts of six genes were further validated in “Golden Delicious” apple fruit, and five genes (MdZHD1/2/6/10/11) were significantly repressed and one gene (MdZHD7) was slightly induced by ethylene treatment. These results indicated that these six MdZF-HD genes may involve in the regulation of ethylene induced ripening process of postharvest apple fruit. These findings provide new clues for further functional investigation of ZF-HD genes, such as their roles in the regulation of fruit ripening.
Varieties of regulatory proteins manipulate a series of developmental processes in plants. Among them, transcription factors (TFs) have been shown to play important roles in regulating the different biological processes of plant growth, flowering, fruiting, and fruit ripening (Mitsuda and Ohme-Takagi, 2009). Recently, zinc finger-homeodomain (ZF-HD), a plant specific transcription factor family, has attracted increasing attention due to its role in regulating plant growth and development as well as in response to a variety of biotic and abiotic stresses (Agarwal and Jha, 2010; Zhang et al., 2015; Khatun et al., 2017). ZF-HD transcription factors are mainly composed of a N-terminal C2H2-type zinc finger domain (ZF) and a C-terminal homeodomain (HD) domain (Tran et al., 2006). According to phylogeny, the ZF-HD gene family can be divided into zinc finger-homeodomain (ZHD) and MINI ZINC FINGER (MIF) subfamilies (Vision et al., 2000; Schoof et al., 2004). The MIF proteins contain the ZF domain but lack the HD domain (Hu and Ma, 2006), and the evolutionary relationship of ZHDs and MIFs remain unclear. Zinc finger protein has a local polypeptide structure and is formed by cysteine/histidine binding zinc ions (Tan and Irish, 2006). The ZF domain is rarely involved in DNA binding, but can enhance the protein-DNA interactions mediated by the HD domain (Windhövel et al., 2001). HD is a DNA binding domain, which is composed of a highly conserved basic sequence with a length of about 180 bp and can encode 60 amino acid sequences (Mukherjee et al., 2009). HD proteins are mostly correlated to other domains or motifs as well as protein-protein interactions (Ariel et al., 2007).
Currently, ZF-HD has been reported in many plant species, after the first discovery in the C4 plant Flaveria trinervia (Windhövel et al., 2001). For example, seventeen ZF-HD genes were identified in the model plant Arabidopsis thaliana (Hu et al., 2008), 31 members in Chinese cabbage (Wang et al., 2016), 22 members in tomato (Khatun et al., 2017), 37 members from cotton (Abdullah et al., 2018), 20 members from bitter buckwheat (Liu et al., 2019), 18 members in tea tree (Zhou et al., 2021), and 10 members from cucumber (Lai et al., 2021). Several studies show that ZF-HD genes can act as regulators in response to various stresses as well as during flower or fruit development (Tan and Irish, 2006; Zhou et al., 2021). In Arabidopsis, ZFHD1 can be induced by drought, high salinity, low temperature and abscisic acid (ABA), and can also bind to the promoter of ERD1 (EARLY RESPONSE TO DEHYDRATION STREES 1) gene (Tran et al., 2006). In soybean, GmZF-HD1 and GmZF-HD2 were up-regulated in response to pathogen infection and can bind to the promoter of GmCaM4 gene (Park et al., 2007). In rice, the overexpression of OsZF-HD1 gene leads to curl and drooping of rice leaves (Xu et al., 2014). In Camellia sinensis, the transcription level of CsZF-HD5 is very high in flower tissues, suggesting that CsZF-HD5 is closely related to flower development (Zhou et al., 2021). Furthermore, four SlZHD genes (SlZHD1/19/20/22) were highly expressed in mature tomato fruit (Khatun et al., 2017). Although ZF-HD family genes have been widely investigated in several model plants and some other species, the comprehensive analysis of ZF-HD family in apple has rarely been reported.
Apple (Malus domestica) is an important economic crop cultivated worldwide. As a climacteric fruit, the ripening process of apple fruit is highly dependent on ethylene (Yue et al., 2020). The rates of postharvest ripening and softening process critically impact the shelf life of apple fruit. Therefore, it is of significance to study the molecular mechanisms of the ripening process for apple fruit. In this paper, a genome-wide analysis of the MdZF-HD genes based on the apple genome data was conducted to explore their potential roles in regulating the postharvest ripening process. The phylogenetic relationships, gene structures, chromosome locations and replication events of the ZF-HD genes in apple were introduced in detail. Furthermore, the expression profiles of MdZF-HD genes in response to ethylene or 1-MCP (1-methylcyclopropene, the ethylene receptor inhibitor) treatment in apple fruit were analyzed by quantitative real time PCR (qRT-PCR) technique. This study is expected to provide valuable clues for the functional investigation of ZF-HD family genes in the regulation of apple fruit ripening.
Two cultivars of apple (Malus domectica) fruit were selected, including one late-ripening cultivar “Qinguan” and one mid-ripening cultivar “Golden Delicious.” Mature “Qinguan” apple fruit were harvested from a commercial orchard at Lingbao (Henan, China) in 2018. Each picked fruit was inspected to be free from mechanical damages, diseases and insect pests. The fruits were divided into two batches for two different treatments. Each batch contained three replicates of approximately 270 fruits. The fruits were treated with 1-MCP (1 μL L−1, 20°C, 24 h), or air as the control group (20°C, 24 h) in 25-L airtight containers. The weight of apple fruit in each containers was about 6 kg. For the 1-MCP fumigation treatment, 1.22 mg 1-MCP powder (effective mass fraction is 3.30%) was dissolved in 1 mL distilled water about 40°C in 1.5 mL centrifuge tube. The tube containing the 1-MCP reagent was put to the bottom of the containers, and the lid was opened exactly before the containers was sealed.
To verify the effect of 1-MCP treatment, and to confirm the roles of ethylene in fruit ripening, mature “Golden Delicious” apple fruit were collected from a commercial orchard at Luoning (Henan, China) in 2019. The 1-MCP treatment were the same as in 2018, and the fruits were treated with ethylene (100 μL L−1, 20°C, 24 h), with those treated in air as control group (20°C, 24 h) in 25-L airtight containers. The fruits after treatment were transferred to storage in air with relative humidity of 85–90% at 20°C. The sampling points were 0, 7, 14, 21 and 28 days, respectively.
At each sampling time, twelve fruits from three replicate samples (four fruits in each) were collected from each batch. The outer pericarp (without skin) were cut into pieces and immediately frozen in liquid nitrogen and then stored at −80°C until future use.
Apple genome annotation information and genome sequence were sourced from the Rosaceae genome website GDR (https://www.rosaceae.org/). The HMM (Hidden Markov Model) configuration profiles of ZF-HD (PF04770) was downloaded from the Pfam 34.0 database (https://pfam.xfam.org/) and perform sexual screening through e value < 0.01. The molecular mass, isoelectric point and other physical and chemical properties of the identified nineteen apple ZF-HD proteins were obtained by using the tools of the ExPASy 3.0 (https://web.expasy.org/compute_pi/) website (Duvaud et al., 2021). The prediction of subcellular location on the Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/) was also conducted (Chou and Shen, 2008).
The protein sequences of the Arabidopsis ZF-HD family were downloaded from the PlantTFDB database v5.0 (http://planttfdb.gao-lab.org/) and the MEGAX (v. 10.2.4) software was used to construct a phylogenetic tree by the Neighbor-joining (NJ) method {Formatting Citation} (Jin et al., 2017; Kumar et al., 2018). The evolution standard Bootstrap value is 1000. The evolutionary tree is optimized by EvolView v2 (https://www.evolgenius.info/evolview/) (He et al., 2016).
The conserved motifs of the apple ZF-HD proteins were analyzed by Multiple Em for Motif Elicitation (MEME) online server (version 5.3.3, https://meme-suite.org/meme/tools/meme) (Bailey et al., 2009). The structures of the apple ZF-HD genes were visualized using TBtools software (Chen et al., 2020). Besides, the arrangement of the introns and exons of the nineteen MdZF-HD genes were obtained visually.
The apple chromosome file information and the GFF file configuration information were used to obtain the chromosome interval information of the apple ZF-HD gene family. The visualization was achieved by TBtools. The Genome Collinearity Analysis Toolkit (MCScanX) was used to analyze the collinearity between each pair of chromosomes (Wang et al., 2013).
The 2000 bp upstream sequence of the start codon (ATG) of each MdZF-HD gene was extracted from the Malus domestica genome database (GDDH13 1.1, https://iris.angers.inra.fr/gddh13/the-apple-genome-downloads.html) (Daccord et al., 2017) and submitted to the PlantCARE server (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to analyze the distribution of cis-elements of the ZF-HD family genes (Lescot et al., 2002).
Total RNA was extracted from frozen fruit flesh (1.0 g) by the method described by Wang et al. (2021). The PrimeScriptTM RT reagent Kit with gDNA Eraser (TaKaRa) was used to remove the contaminated gDNA. cDNA was synthesized from 1.0 µg DNA-free RNA, using the Reverse Transcription System (TaKaRa). At each sampling point, three biological replicates were performed for RNA extraction.
Oligonucleotide primers used for real-time quantitative PCR analysis were designed with Primer3 (version 0.4.0, https://bioinfo.ut.ee/primer3-0.4.0/). Gene specificity of the primers was determined by melting curves and PCR products resequencing. The sequences of primers used for PCR analysis are listed in Supplementary Table S1. The apple Actin gene, a housekeeping gene, was chosen to monitor the abundance of mRNA (Wang et al., 2021).
Real-time PCR was performed using an ABI PRISM 7500 instrument (Applied Biosystems). The PCR protocols were the same as our previous reports (Wang et al., 2021), with SYBRTM Select PCR Master Mix (Applied Biosystems). The relative expression levels of genes were calculated by the 2- ∆∆Ct method (Livak and Schmittgen, 2001).
The full-length coding sequences of three selected ZF-HD genes (MdZHD2/6/7) without the stop codons were amplified with primers (described in Supplementary Table S2) and constructed into the GFP vector (Li S. J. et al., 2017). 35S-MdZHD2-GFP, 35S-MdZHD6-GFP and 35S-MdZHD7-GFP were transiently expressed in tobacco (Nicotiana benthamiana) leaves by Agrobacterium-mediated infiltration (GV3101) (Zeng et al., 2019). The tobacco plants were grown in an artificial climate room at 22°C with daylight extension to 16 h. The green fluorescent protein (GFP) fluorescence of tobacco leaves was imaged 3 days after infiltration using the Laser Scanning Confocal Microscope (Nikon, AIR HD25, Japan).
Statistical significance of differences were calculated using the Student´s t-test by DPS 7.05 (Zhejiang University, Hangzhou, China). Figures were drawn with PRISM 8 (Graphpad, San Diego, CA, United States).
Nineteen ZF-HD family candidate genes were finally obtained by using the NCBI conserved domain database CDD and smart website for double verification of the conserved structure of the protein, and the genes were named MdZHD1-MdZHD15 and MdMIF1-MdMIF4 based on the chromosome location information (Table 1). The CDS lengths of the family members ranged from 273 bp (MdMIF2/4) to 1131 bp (MdZHD13). The lengths of MdZF-HD proteins were 90–376 amino acids (AA), and the molecular weight (MW) varied from 9.62 to 41.65 KDa. Besides, the predicted isoelectric points (pIs) of MdZF-HD proteins ranged from 6.51 (MdZHD12) to 9.44 (MdZHD2). The results indicated that except for the acidic proteins (MdZHD5/11/12/15), the other MdZF-HD proteins were basic proteins. The subcellular localization prediction showed that all the MdZHD proteins (MdZHD1-15) were located in the nucleus, while the MdMIF proteins (MdMIF1-4) were located in the mitochondria. This is consistent with the previous research that most ZF-HD proteins located in the nucleus (Wang et al., 2016).
To gain insights into the evolutionary relationship of the ZF-HD family proteins in apple, a NJ phylogenetic tree consisting of Arabidopsis (17 genes) and apple (19 genes) was constructed (Figure 1). The sequences of protein used for phylogenetic tree analysis were listed in Supplementary Table S3. According to the ZF-HD family classification of Arabidopsis, the apple ZF-HD gene family was phylogenetically divided into two subfamilies: ZHD and MIF. ZHD was further divided into five parts, including ZHDI (MdZHD1/5/11/12), ZHDII (MdZHD13), ZHDIII (MdZHD2/4/9/10), ZHDIV (MdZHD6/7/14/15) and ZHDV (MdZHD3/8) (Figure 1). Among these, the ZHDII subfamily of apple has the least gene with only one.
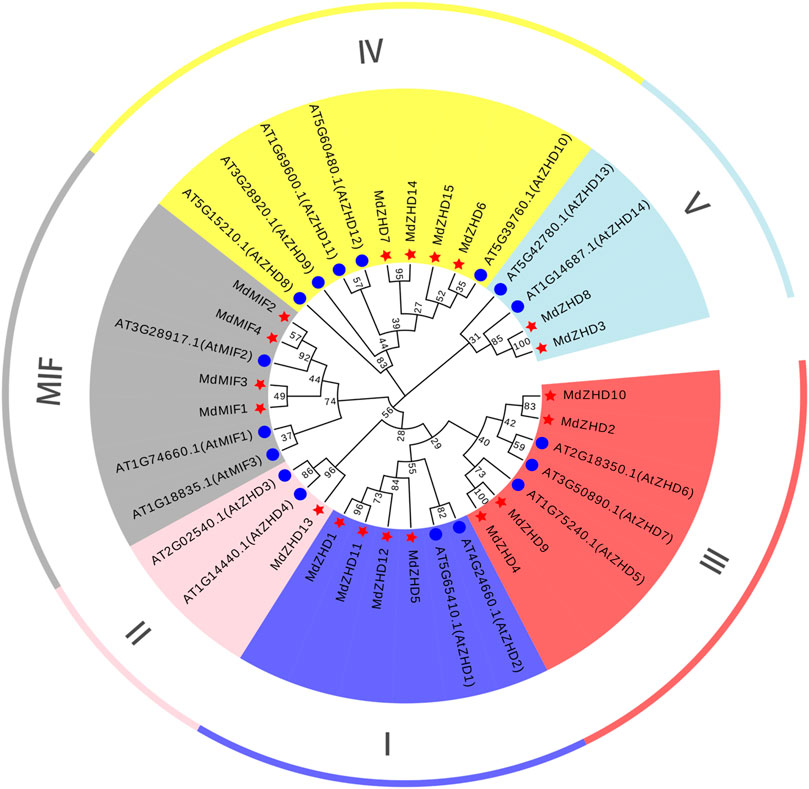
FIGURE 1. The phylogenetic tree of ZF-HD proteins from apple (Malus domestica, MdZHD) and Arabidopsis (Arabidopsis thaliana, AtZHD). The tree was constructed by the Neighbor-joining (NJ) method using a MEGAX software with 1000 bootstrap replications. The MdZHD member was accompanyed with a red pentagram. Six subfamilies were identified in apple and were distinguished in different color: ZHD I, ZHD II, ZHD III, ZHD IV, ZHD V, and mini zinc finger (MIF).
In order to further investigate the diversity of the apple ZF-HD family genes, MEME web server was used to analyze the conserved motifs of the MdZF-HD proteins. From the results of the MEME analysis, ten conserved motifs were identified (Figure 2) (Supplementary Figure S1). All MdZF-HD proteins contain motif1 and motif3, indicating that motif1and motif3 are the specific motifs of the ZF-HD gene family (Figure 2B). It is worth noting that all members of the MdZHD subfamily contain motif1, motif2, motif3 and motif4, while the MdMIF proteins only contain motif1 and motif3. This result is consistent with the previous report (Hu and Ma, 2006; Hu et al., 2018) that the MIF subfamily harbors only a ZF domain but lacks HD (Supplementary Figure S2). Thus, motif2 and motif4 are the specific motifs for MdZHD subfamily and these motifs all appear in pairs. Besides, the types and numbers of motifs were identical between MdZHD4 and MdZHD9, MdZHD1 and MdZHD11, MdZHD2 and MdZHD10, MdZHD6 and MdZHD15, respectively. These results indicated that the ZF-HD members in the same subgroup contained the similar motifs. Furthermore, compared with the MdMIF subfamily, the different motifs existed among different members of the MdZHD subfamily supply evidence of their functional diversity. For example, MdZHD13 only had the motif1, motif2, motif3 and motif4, indicating that the MdZF-HD family genes differed in the evolutionary degree.
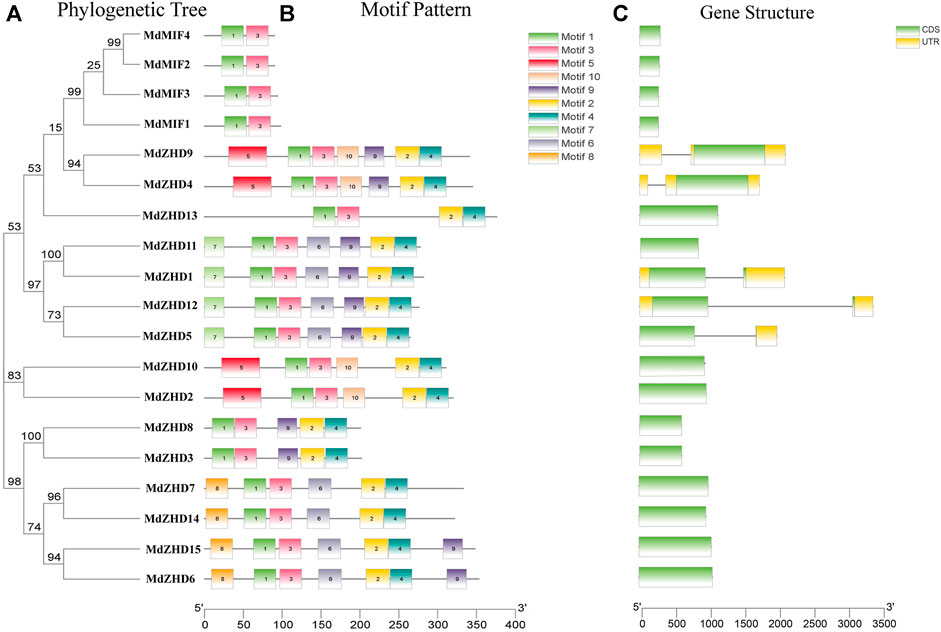
FIGURE 2. Phylogenetic relationships (A), conserved motifs (B) and gene structures (C) of MdZF-HD genes. Different colored boxes represent different kinds of motifs. CDS sequences are represented by green rounded rectangles, introns are represented by gray lines, and UTRs are represented by yellow boxes.
To further understand the composition of the MdZF-HD gene structure, we obtained the exon and intron structure of the gene through an annotation file. Among the nineteen MdZF-HD family genes, only five MdZF-HD genes contain introns, and each of them contains one intron (Figure 2C). It means that the function of the genes that lack introns is relatively conserved, which is consistent with the previous reports (Windhövel et al., 2001; Wang et al., 2016; Liu et al., 2019) that most MdZF-HD family genes lack introns.
As shown in Figure 3, nineteen MdZF-HD genes were distributed on ten chromosomes in the apple genome. Chromosome 1 (Chr01), Chr02, Chr13 and Chr16 each had one MdZF-HD gene; Chr06, Chr08, Chr09, Chr14 and Chr17 contained two MdZF-HD genes, respectively; while Chr15 harbored the largest number of MdZF-HD genes (five genes) (Figure 3). The distribution of the genes was uneven. This suggests that the genes play an important role in different transcription initiations.
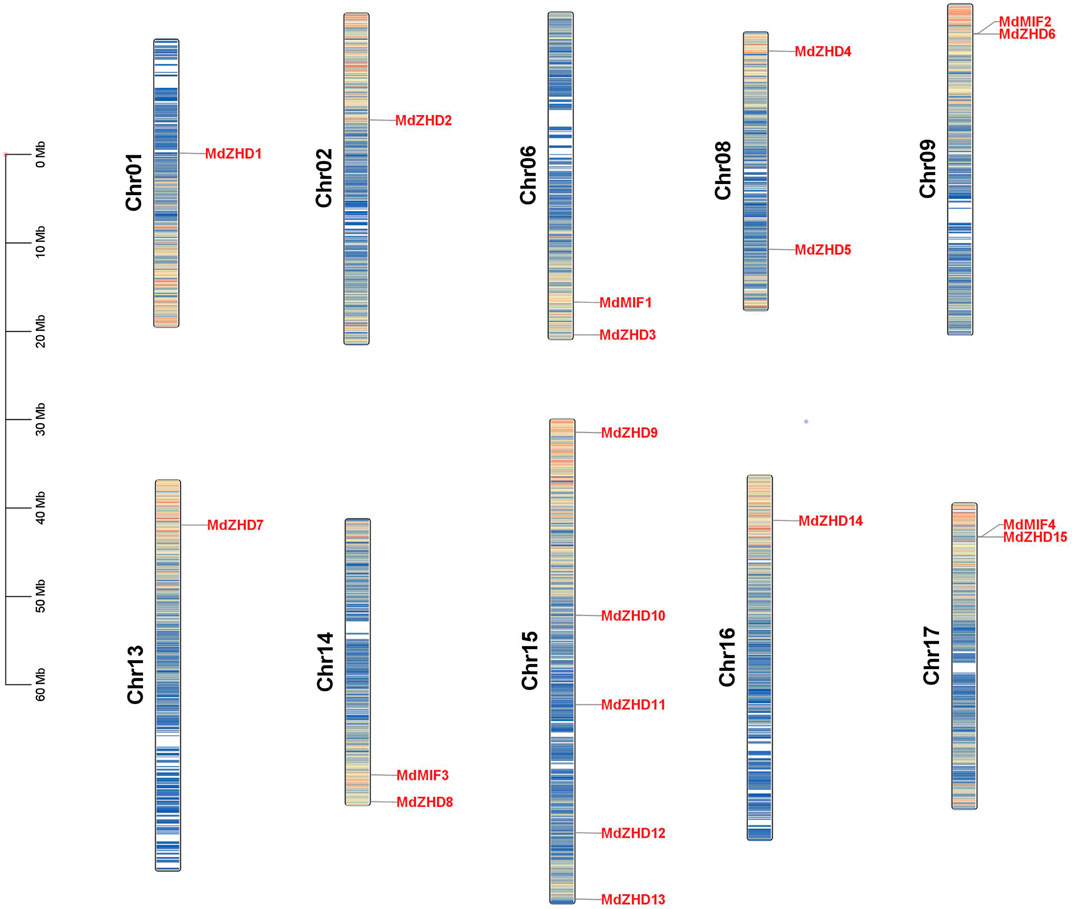
FIGURE 3. Distribution of MdZF-HD genes along apple chromosomes. The scale on the left stands for the length of each chromosome shown in mega base (Mb).
To explore the possible relationships and potential repeated events in the MdZF-HD family, we then analyzed the collinearity of the MdZF-HD family. A total of 25 repeated events were identified, including two tandem repeated events and 23 fragments repeated events (Figure 4). The result shows that the MdZF-HD genes have relatively conservative and similar functions during the evolutionary process. This is also similar to the previous studies, and the genes in the ZF-HD family are highly overlapping (Wang et al., 2016). In addition, there are repeated events between MdMIF and MdMIF genes in the MIF subgroup, as well as MdMIF and MdZHD genes. The repeated events indicated that the MIF gene family may originate from a ZF-HD gene by losing the homologous domain. Accordingly, the high functional redundancy and the gene replication in this family may be due to genome replication; the functional redundancy is thus deduced to be normal in the ZHD subfamily.
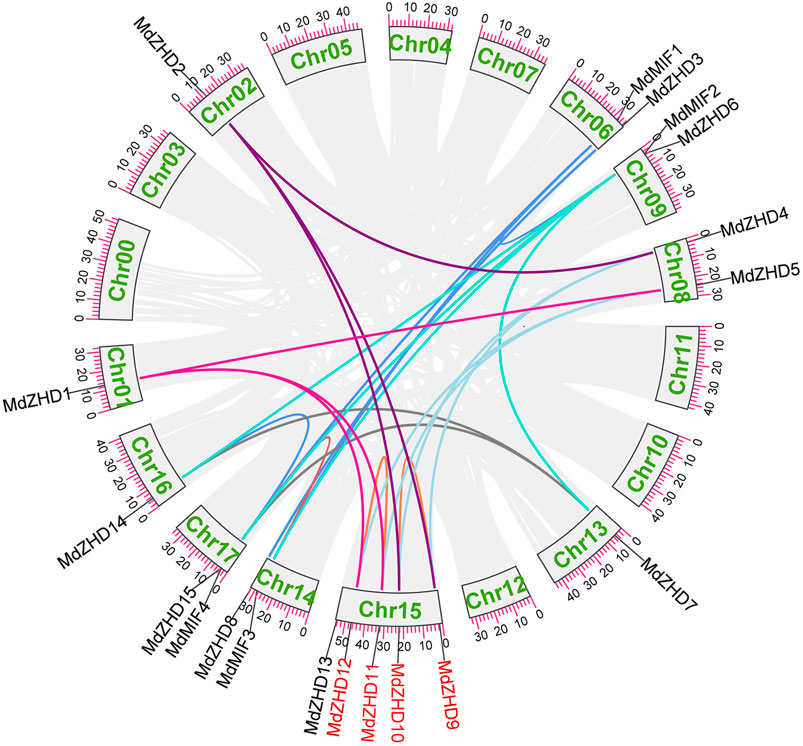
FIGURE 4. Homologous relationships and gene replications of the ZF-HD genes in apple. The different colored lines indicate the segmental duplicated genes, while the red gene names highlight the tandem duplicated genes.
The cis-elements are important regulators during plant growth and development, hormone responses as well as in response to biotic and abiotic stresses (Latchman, 1997; Yamaguchi-Shinozaki and Shinozaki, 2005). In order to explore the potential functions and the regulatory patterns of MdZF-HD family genes, we extracted a 2000 bp fragments upstream of the start codon (ATG) of each MdZF-HD gene for cis-elements analysis (Supplementary Table S4). The results indicated that the identified cis-elements can be roughly divided into four categories: stress responses (anoxic specific inducibility, anaerobic induction, defense and stress, low temperature), hormone responses (auxin, MeJA, gibberellin, abscisic acid, salicylic acid and ethylene)), the binding sites (protein binding sites, MYB, MYBHv), and development related responses (cell cycle and meristem expression) (Figure 5A).
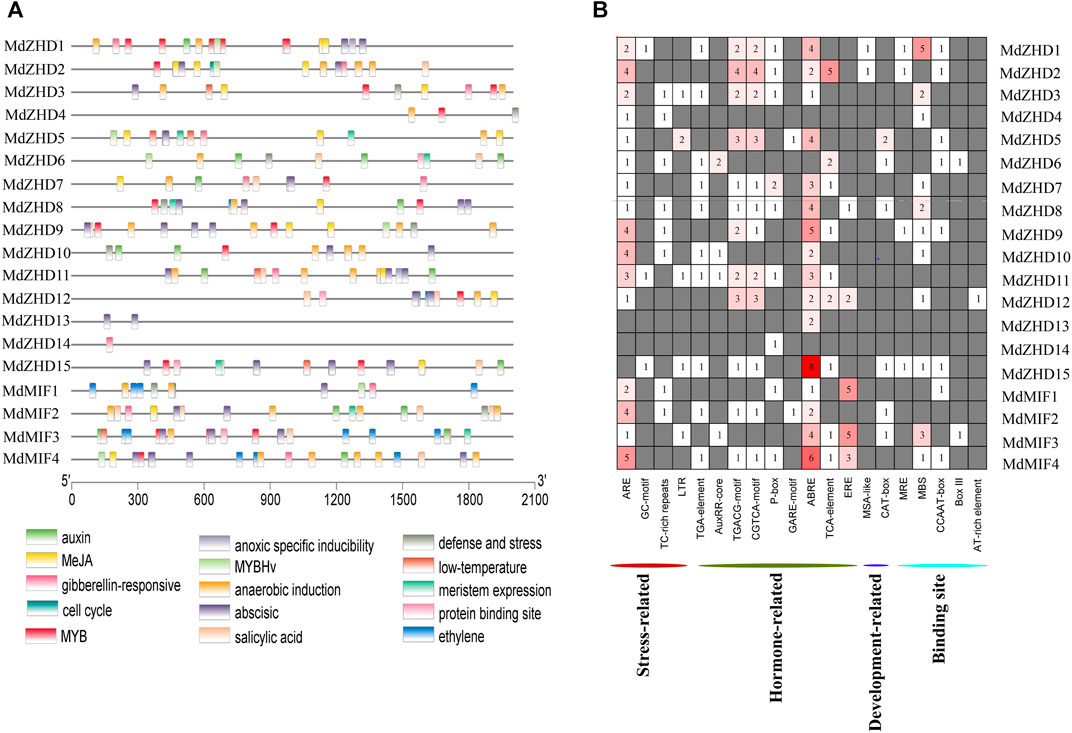
FIGURE 5. Cis-element analysis of the promoters of MdZF-HD genes. (A) The different colored rectangles show the different types of cis-elements and their positions in each MdZF-HD gene. (B) Heatmap of the numbers of cis-elements in the promoters of MdZF-HD genes.
In total, eight types of cis-elements responsive to different hormones including auxin response (AuxRR-core, TGA-element), MeJA response (CGTCA/TGACG-motif), gibberellin response (P-box), abscisic acid response (ABRE), salicylic acid response (TCA-element) and ethylene response elements (ERE) were found in the promoters of all MdZF-HD genes except MdZHD4 (Figure 5B; Supplementary Table S5). Notably, ABRE was the most abundant cis-elements of these hormone responsiveness, and sixteen out of the nineteen MdZF-HD promoter regions contained at least one ABRE element. In addition, ERE was most distributed in the promoters of MdMIF genes (Figure 5B), indicating that MdMIF genes may be more responsive to ethylene. Furthermore, ARE was found to be distributed in almost all promoter regions of MdZF-HD genes, except for MdZHD13/14/15 (Figure 5B), suggesting that the MdZF-HD genes may be responsive to anaerobic stress. These findings indicated that MdZF-HD genes may play a certain role in the regulation of gene expressions in response to hormones and abiotic stresses.
To explore the relationship between the ZF-HD family genes and apple fruit ripening, the expression levels of the MdZF-HD genes in response to 1-MCP treatment in “Qinguan” fruit were analyzed by qRT-PCR. Based on the results, nine MdZF-HD genes were differentially expressed, as shown in Figure 6. Except for MdZHD7 and MdMIF2, all the other genes (including MdZHD1/2/5/6/10/11/15) were significantly up-regulated by 1-MCP treatment and reached a peak after storage for 21 days, which showed negative association with apple fruit postharvest ripening and softening. Nevertheless, the expressions of MdZHD7 and MdMIF2 genes were obviously repressed by the 1-MCP treatment, which showed positive association with apple fruit postharvest ripening and softening (Figure 6).
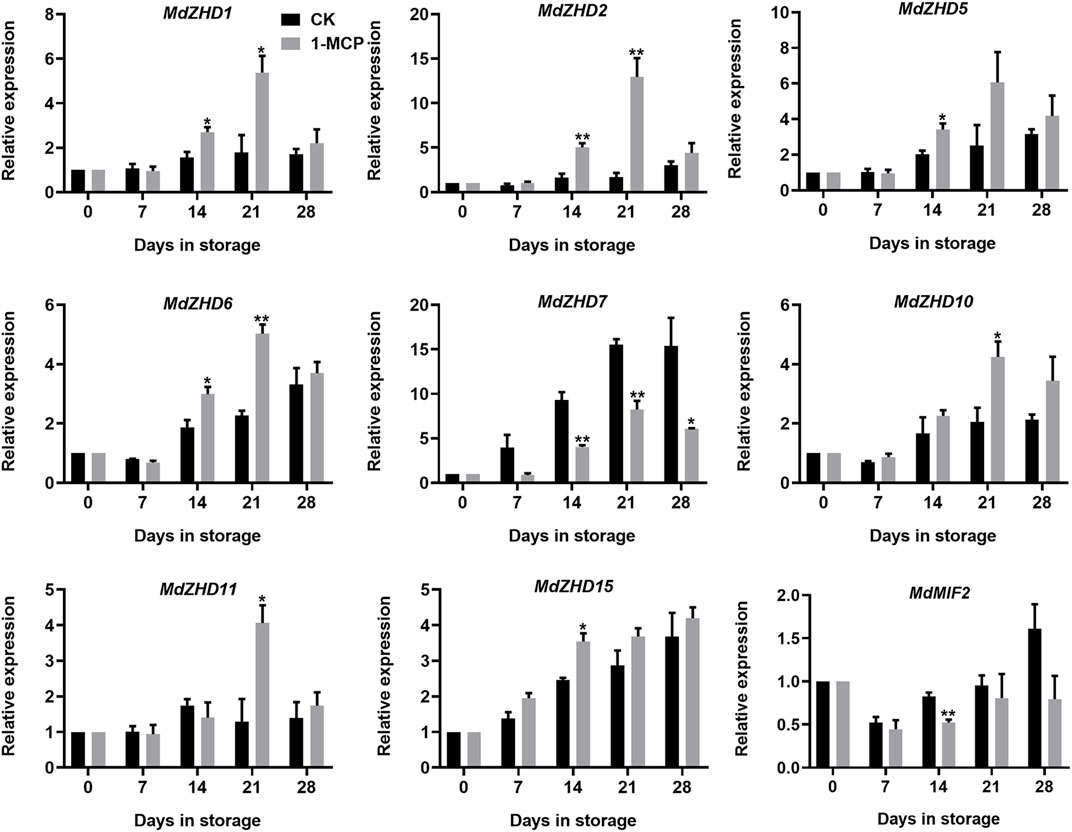
FIGURE 6. Expression patterns of ZF-HD genes in response to 1-MCP treatment during “Qinguan” apple fruit ripening. “Qinguan” fruit were treated with 1-MCP (1 μL L−1) or air (control/CK) for 24 h at 20°C. The values of day 0 fruit were set as 1. Black columns and gray columns represent the expression levels of the genes transcripts in control (CK) and 1-MCP treated fruit, respectively. Error bars represent SEs from three biological replicates (*p < 0.05; **p < 0.01).
In order to verify the expression patterns of these candidate MdZF-HD genes in response to ethylene, six MdZF-HDs with higher significance levels were further analyzed in “Golden Delicious” apple fruit. Similar to the expression levels of these MdZF-HD genes in response to 1-MCP treatment in “Qinguan” fruit, MdZHD1/2/6/10/11 were also significantly induced in the 1-MCP treated “Golden Delicious” apple fruit and peaked at 7 or 14 days in storage, and were mostly inhibited by ethylene treatment (Figure 7). In comparison, the expression patterns of MdZHD7 was significantly down-regulated in the 1-MCP treated “Golden Delicious” apple fruit, and were slightly induced by ethylene treatment (Figure 7). To sum up, these six MdZF-HD genes could be potential candidates regulating the ethylene induced ripening and softening of postharvest apple fruit.
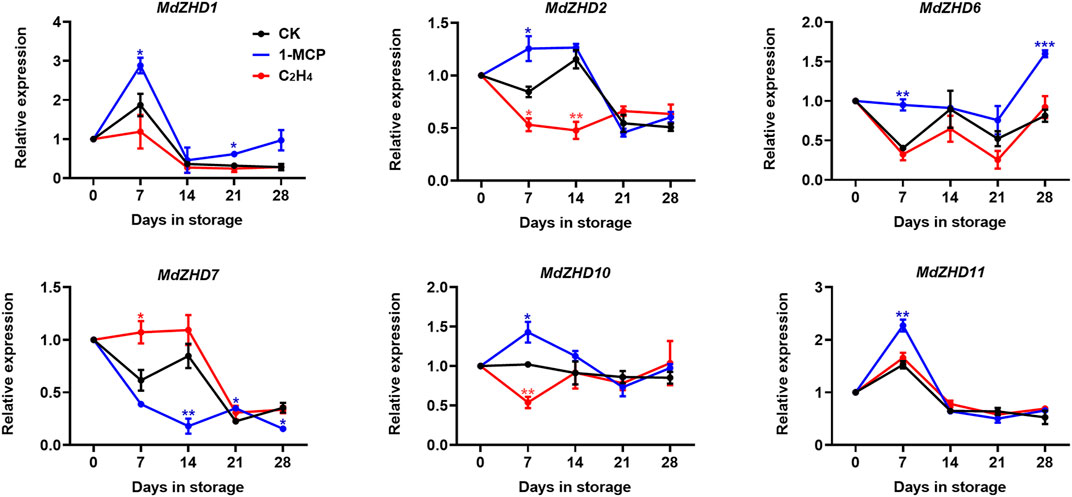
FIGURE 7. Expression patterns of ZF-HD genes in response to ethylene or 1-MCP treatment during “Golden Delicious” apple fruit ripening. “Golden Delicious” fruit were treated with 1-MCP (1 μL L−1), ethylene (C2H4, 100 μL L−1), and air (control/CK) for 24 h at 20°C. The values of day 0 fruit were set as 1. Black lines, blue lines and red lines represent the expression levels of the genes transcripts in control (CK), 1-MCP and ethylene treated fruit, respectively. Error bars represent SEs from three biological replicates (*p < 0.05; **p < 0.01; ***p < 0.001).
The subcellular locations of three candidate ripening related MdZF-HD proteins (MdZHD2/6/7) were examined in tobacco (Nicotiana benthamiana) leaves by using GFP tagging. The signals of the control GFP was detected in both the nucleus and cell membrane, while MdZHD2/6/7 showed strong fluorescence signals in the nucleus, with the except that MdZHD7 also gave signals in the cell membrane (Figure 8).
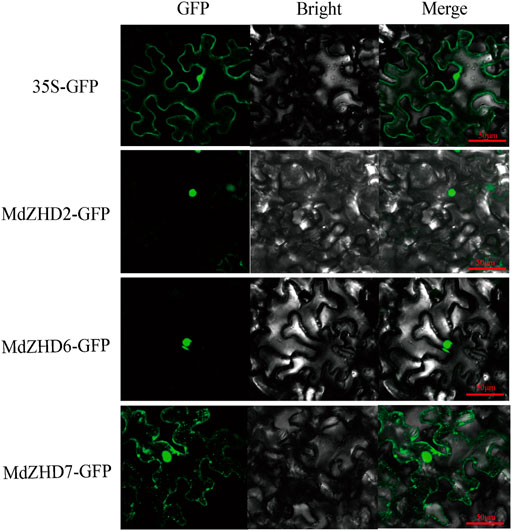
FIGURE 8. Subcellular localization of MdZHD2/6/7-GFP in tobacco leaves. Agrobacterium-mediated transformation was expressed in tobacco epidermal cells and the signal of green fluorescent protein (GFP) was observed by laser confocal microscope. Bars = 50 µm.
Previous studies have shown that the ZF-HD family genes play important roles in regulating the plant growth and development, and can enhance the resistance to stress conditions (Abdullah et al., 2018; Hu et al., 2018). To date, ZF-HD genes have been characterized in many plant species, such as Arabidopsis, tomato, tea tree and cucumber (Hu and Ma, 2006; Khatun et al., 2017; Lai et al., 2021; Zhou et al., 2021). Nevertheless, the evolutionary characteristics and function of the apple ZF-HD genes have been poorly understood, notably in regulating fruit ripening.
In present study, a comprehensive investigation of apple ZF-HD gene family was conducted, and nineteen ZF-HD genes including fifteen ZHDs and four MIFs were identified in apple (Table 1). The number of ZF-HD genes identified in apple is inconsistent with a previous report (Shalmani et al., 2019), on account of the different genome database we used. The number of the ZF-HD genes in apple was slightly higher than that of Arabidopsis thaliana (Hu et al., 2008), tea tree (Zhou et al., 2021) and cucumber (Lai et al., 2021). Besides, these nineteen MdZF-HD genes and seventeen AtZF-HD genes were constructed as evolutionary trees. According to the classification of Arabidopsis, the apple ZF-HD proteins can be divided into two subfamilies (ZHD and MIF), while MdZHD can be further classified into five subgroups (ZHDⅠ–ZHDV) (Figure 1), which is also consistent with the classification of ZF-HD proteins in other plants (Khatun et al., 2017; Zhou et al., 2021). After that, we analyzed the conserved motifs of MdZF-HD family proteins, and the conserved motifs between the ZHD and MIF subfamilies were significantly different, but similar conserved motifs were found among the same subfamilies (Figures 2A,B). All MdZF-HD proteins have motif1 and motif3, suggesting that ZF-HD proteins are likely to be highly conserved during evolution. ZF-HD genes have high plant specificity and most do not have introns (Figure 2C), indicating that ZF-HD is a relatively new family. Notably, genes containing introns (MdZHD1/4/5/9/12) are longer than those without introns in the MdZF-HD family, which is consistent with the previous reports that the number of introns is positively correlated with gene length (Zhu et al., 2020; Zhou et al., 2021). Duplication of genes can increase the number of genes (Vision et al., 2000; Blanc et al., 2003; Cannon et al., 2004; Abdullah et al., 2018). Gene replication, including fragment replication and tandem replication, is a crucial factor in the biological evolution of many plants (Kong et al., 2007). In this study, 23 fragment replications and two tandem replications were found in the chromosome distribution of ZF-HD genes in apple (Figure 4), indicating that the gene fragment duplications contributed to the amplification of the ZF-HD gene family in apple.
Promoter region analysis of the MdZF-HD genes identified several cis-elements related to phytohormones and abiotic stresses (Figure 5). Based on the previous studies, ZF-HD family genes were mostly found to participate in the responses to abiotic stresses (Tan and Irish, 2006; Figueiredo et al., 2012; Wang et al., 2016; Liu et al., 2021). For instance, ZF-HD family genes from Arabidopsis, tomato, cucumber, and also apple were shown to be up-regulated by various kinds of stress conditions, such as drought, salt, cold, heat, and phytohormones including GA and ABA (Shan et al., 2012; Zhang et al., 2015; Khatun et al., 2017; Shalmani et al., 2019; Lai et al., 2021). However, the roles of ZF-HD genes in regulating fruit ripening and softening have rarely been reported. In this study, nine selected MdZF-HD genes were differentially expressed during the postharvest ripening process of “Qinguan” apple fruit (Figure 6). Among them, MdZHD7 and MdMIF2, were significantly downregulated by the 1-MCP treatment, which showed positive correlation with the postharvest ripening of apple fruit. In comparison, mRNAs from other seven genes (MdZHD1/2/5/6/10/11/15), especially MdZHD1/2/6, displayed increase in abundance in response to 1-MCP treatment during the postharvest storage of “Qinguan” apple fruit, which showed negative association with apple fruit ripening. In addition, six candidate MdZF-HD genes with higher significance levels were further analyzed in another cultivar “Golden Delicious,” and showed similar expression patterns in response to the1-MCP treatment. Of the six MdZF-HD genes, five genes (MdZHD1/2/6/10/11) were repressed and one gene (MdZHD7) was slightly induced in response to ethylene treatment (Figure 7), implying that they may be involved in the regulation of the ethylene induced ripening of postharvest apple fruit. The detailed regulatory mechanisms remain to be further investigated. Furthermore, the subcellular localizations of three selected ZHDs (MdZHD2/6/7) in nucleus were consistent with the prediction (Table 1; Figure 8), and the MdZHD7 protein was also located in the cell membrane. The results showed that these three genes can be located in the nucleus, indicating that they may have functions as transcription factors. Similar to this phenomenon, some transcription factors are not only located in the nucleus. For instance, the TaMIF4-5D in Triticum aestivum was located in the nucleus and cell membrane (Niu et al., 2021); the CitNAC62 in citrus was not located in the nucleus and its subcellular location was within plastids (Li T. et al., 2017).
Nineteen ZF-HD family genes were newly identified in apple, and their phylogenetic relationships, gene structures, conserved motifs, subcellular localizations, as well as their expression patterns in response to ethylene or 1-MCP treatment during the postharvest storage of apple fruit were analyzed. Besides, the expressions of several MdZF-HD genes in apple fruit of two cultivars were obviously altered in response to ethylene or 1-MCP treatment. Our findings may supply valuable clues for identifying the potential roles of MdZF-HD genes in regulating the fruit ripening.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
M-MW designed and supervised the experiment. YW and HW conducted the bioinformatics analysis, carried out the experiments, and prepared the original manuscript. S-WS, T-HB, JJ, C-HS and H-gp helped with revisions to the manuscript. M-MW and X-BZ provided support for the funding, and revised the manuscript. All authors contributed to this article and agreed to the submission.
This work was financially supported by the National Natural Science Foundation of China (32102448), the Key R & D and Promotion Projects in Henan Province (212102110401), the Young Talents Project of Henan Agricultural University (30500423), the Innovation Team Project of Henan University (19IRTSTHN009), the Special Fund for Henan Agriculture Research System (Z2014-11-03).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.783482/full#supplementary-material
Abdullah, M., Cheng, X., Cao, Y., Su, X., Manzoor, M. A., Gao, J., et al. (2018). Zinc Finger-Homeodomain Transcriptional Factors (ZHDs) in upland Cotton (Gossypium Hirsutum): Genome-Wide Identification and Expression Analysis in Fiber Development. Front. Genet. 9, 1–13. doi:10.3389/fgene.2018.00357
Agarwal, P. K., and Jha, B. (2010). Transcription Factors in Plants and ABA Dependent and Independent Abiotic Stress Signalling. Biol. Plant. 54, 201–212. doi:10.1007/s10535-010-0038-7
Ariel, F. D., Manavella, P. A., Dezar, C. A., and Chan, R. L. (2007). The True Story of the HD-Zip Family. Trends Plant Sci. 12, 419–426. doi:10.1016/j.tplants.2007.08.003
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME Suite: Tools for Motif Discovery and Searching. Nucleic Acids Res. 37, W202–W208. doi:10.1093/nar/gkp335
Blanc, G., Hokamp, K., and Wolfe, K. H. (2003). A Recent Polyploidy Superimposed on Older Large-Scale Duplications in the Arabidopsis Genome. Genome Res. 13, 137–144. doi:10.1101/gr.751803
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis thaliana. BMC Plant Biol. 4, 10–21. doi:10.1186/1471-2229-4-10
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Chou, K.-C., and Shen, H.-B. (2008). Cell-PLoc: a Package of Web Servers for Predicting Subcellular Localization of Proteins in Various Organisms. Nat. Protoc. 3, 153–162. doi:10.1038/nprot.2007.494
Daccord, N., Celton, J.-M., Linsmith, G., Becker, C., Choisne, N., Schijlen, E., et al. (2017). High-Quality De Novo Assembly of the Apple Genome and Methylome Dynamics of Early Fruit Development. Nat. Genet. 49, 1099–1106. doi:10.1038/ng.3886
Duvaud, S., Gabella, C., Lisacek, F., Stockinger, H., Ioannidis, V., and Durinx, C. (2021). Expasy, the Swiss Bioinformatics Resource portal, as Designed by its Users. Nucleic Acids Res. 49, W216–W227. doi:10.1093/nar/gkab225
Figueiredo, D. D., Barros, P. M., Cordeiro, A. M., Serra, T. S., Lourenço, T., Chander, S., et al. (2012). Seven Zinc-Finger Transcription Factors Are Novel Regulators of the Stress Responsive Gene OsDREB1B. J. Exp. Bot. 63, 3643–3656. doi:10.1093/jxb/err313
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W.-H., and Hu, S. (2016). Evolview V2: an Online Visualization and Management Tool for Customized and Annotated Phylogenetic Trees. Nucleic Acids Res. 44, W236–W241. doi:10.1093/nar/gkw370
Hu, J., Gao, Y., Zhao, T., Li, J., Yao, M., and Xu, X. (2018). Genome-Wide Identification and Expression Pattern Analysis of Zinc-Finger Homeodomain Transcription Factors in Tomato Under Abiotic Stress. J. Amer. Soc. Hort. Sci. 143, 14–22. doi:10.21273/JASHS04245-17
Hu, W., DePamphilis, C. W., and Ma, H. (2008). Phylogenetic Analysis of the Plant-Specific Zinc Finger-Homeobox and Mini Zinc Finger Gene Families. J. Integr. Plant Biol. 50, 1031–1045. doi:10.1111/j.1744-7909.2008.00681.x
Hu, W., and Ma, H. (2006). Characterization of a Novel Putative Zinc Finger GeneMIF1: Involvement in Multiple Hormonal Regulation of Arabidopsis Development. Plant J. 45, 399–422. doi:10.1111/j.1365-313X.2005.02626.x
Jin, J., Tian, F., Yang, D.-C., Meng, Y.-Q., Kong, L., Luo, J., et al. (2017). PlantTFDB 4.0: Toward a Central Hub for Transcription Factors and Regulatory Interactions in Plants. Nucleic Acids Res. 45, D1040–D1045. doi:10.1093/nar/gkw982
Khatun, K., Nath, U. K., Robin, A. H. K., Park, J.-I., Lee, D.-J., Kim, M.-B., et al. (2017). Genome-Wide Analysis and Expression Profiling of Zinc Finger Homeodomain (ZHD) Family Genes Reveal Likely Roles in Organ Development and Stress Responses in Tomato. BMC Genomics. 18, 695. doi:10.1186/s12864-017-4082-y
Kong, H., Landherr, L. L., Frohlich, M. W., Leebens-Mack, J., Ma, H., and dePamphilis, C. W. (2007). Patterns of Gene Duplication in the Plant SKP1 Gene Family in Angiosperms: Evidence for Multiple Mechanisms of Rapid Gene Birth. Plant J. 50, 873–885. doi:10.1111/j.1365-313X.2007.03097.x
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Lai, W., Zhu, C., Hu, Z., Liu, S., Wu, H., and Zhou, Y. (2021). Identification and Transcriptional Analysis of Zinc Finger-Homeodomain (ZF-HD) Family Genes in Cucumber. Biochem. Genet. 59, 884–901. doi:10.1007/s10528-021-10036-z
Latchman, D. S. (1997). Transcription Factors: An Overview. Int. J. Biochem. Cell Biol. 29, 1305–1312. doi:10.1016/S1357-2725(97)00085-X
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 30, 325–327. doi:10.1093/nar/30.1.325
Li, S.-J., Yin, X.-R., Wang, W.-L., Liu, X.-F., Zhang, B., and Chen, K.-S. (2017). Citrus CitNAC62 Cooperates With CitWRKY1 to Participate in Citric Acid Degradation via Up-Regulation of CitAco3. J. Exp. Bot. 68, 3419–3426. doi:10.1093/jxb/erx187
Li, T., Xu, Y., Zhang, L., Ji, Y., Tan, D., Yuan, H., et al. (2017). The Jasmonate-Activated Transcription Factor MdMYC2 Regulates ETHYLENE RESPONSE FACTOR and Ethylene Biosynthetic Genes to Promote Ethylene Biosynthesis during Apple Fruit Ripening. Plant Cell. 29, 1316–1334. doi:10.1105/tpc.17.00349
Liu, H., Yang, Y., and Zhang, L. (2021). Zinc Finger-Homeodomain Transcriptional Factors (ZF-HDs) in Wheat (Triticum aestivum L.): Identification, Evolution, Expression Analysis and Response to Abiotic Stresses. Plants. 10, 593. doi:10.3390/plants10030593
Liu, M., Wang, X., Sun, W., Ma, Z., Zheng, T., Huang, L., et al. (2019). Genome-Wide Investigation of the ZF-HD Gene Family in Tartary Buckwheat (Fagopyrum Tataricum). BMC Plant Biol. 19, 248. doi:10.1186/s12870-019-1834-7
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 25, 402–408. doi:10.1006/meth.2001.1262
Mitsuda, N., and Ohme-Takagi, M. (2009). Functional Analysis of Transcription Factors in Arabidopsis. Plant Cell Physiol. 50, 1232–1248. doi:10.1093/pcp/pcp075
Mukherjee, K., Brocchieri, L., and Bürglin, T. R. (2009). A Comprehensive Classification and Evolutionary Analysis of Plant Homeobox Genes. Mol. Biol. Evol. 26, 2775–2794. doi:10.1093/molbev/msp201
Niu, H., Xia, P., Hu, Y., Zhan, C., Li, Y., Gong, S., et al. (2021). Genome-wide Identification of ZF-HD Gene Family in Triticum aestivum: Molecular Evolution Mechanism and Function Analysis. PLoS One. 16, e0256579. doi:10.1371/journal.pone.0256579
Park, H. C., Kim, M. L., Lee, S. M., Bahk, J. D., Yun, D.-J., Lim, C. O., et al. (2007). Pathogen-Induced Binding of the Soybean Zinc Finger Homeodomain Proteins GmZF-HD1 and GmZF-HD2 to Two Repeats of ATTA Homeodomain Binding Site in the Calmodulin Isoform 4 (GmCaM4) Promoter. Nucleic Acids Res. 35, 3612–3623. doi:10.1093/nar/gkm273
Schoof, H., Ernst, R., Nazarov, V., Pfeifer, L., Mewes, H. W., and Mayer, K. F. X. (2004). MIPS Arabidopsis Thaliana Database (MAtDB): An Integrated Biological Knowledge Resource for Plant Genomics. Nucleic Acids Res. 32, 373D–376D. doi:10.1093/nar/gkh068
Shalmani, A., Muhammad, I., Sharif, R., Zhao, C., Ullah, U., Zhang, D., et al. (2019). Zinc Finger-Homeodomain Genes: Evolution, Functional Differentiation, and Expression Profiling Under Flowering-Related Treatments and Abiotic Stresses in Plants. Evol. Bioinform Online. 15, 117693431986793–16. doi:10.1177/1176934319867930
Shan, X., Yan, J., and Xie, D. (2012). Comparison of Phytohormone Signaling Mechanisms. Curr. Opin. Plant Biol. 15, 84–91. doi:10.1016/j.pbi.2011.09.006
Tan, Q. K.-G., and Irish, V. F. (2006). The Arabidopsis Zinc Finger-Homeodomain Genes Encode Proteins With Unique Biochemical Properties That Are Coordinately Expressed During Floral Development. Plant Physiol. 140, 1095–1108. doi:10.1104/pp.105.070565
Tran, L.-S. P., Nakashima, K., Sakuma, Y., Osakabe, Y., Qin, F., Simpson, S. D., et al. (2006). Co-expression of the Stress-Inducible Zinc finger Homeodomain ZFHD1 and NAC Transcription Factors Enhances Expression of the ERD1 Gene in Arabidopsis. Plant J. 49, 46–63. doi:10.1111/j.1365-313X.2006.02932.x
Vision, T. J., Brown, D. G., and Tanksley, S. D. (2000). The Origins of Genomic Duplications in Arabidopsis. Science. 290, 2114–2117. doi:10.1126/science.290.5499.2114
Wang, M.-M., Li, T.-X., Wu, Y., Song, S.-W., Bai, T.-H., Jiao, J., et al. (2021). Genome-Wide Identification of microRNAs Involved in the Regulation of Fruit Ripening in Apple (Malus Domestica). Scientia Horticulturae. 289, 110416. doi:10.1016/j.scienta.2021.110416
Wang, W., Wu, P., Li, Y., and Hou, X. (2016). Genome-Wide Analysis and Expression Patterns of ZF-HD Transcription Factors Under Different Developmental Tissues and Abiotic Stresses in Chinese Cabbage. Mol. Genet. Genomics. 291, 1451–1464. doi:10.1007/s00438-015-1136-1
Wang, Y., Li, J., and Paterson, A. H. (2013). MCScanX-Transposed: Detecting Transposed Gene Duplications Based on Multiple Colinearity Scans. Bioinformatics. 29, 1458–1460. doi:10.1093/bioinformatics/btt150
Windhövel, A., Hein, I., Dabrowa, R., and Stockhaus, J. (2001). Characterization of a Novel Class of Plant Homeodomain Proteins that Bind to the C4 Phosphoenolpyruvate Carboxylase Gene of Flaveria Trinervia. Plant Mol. Biol. 45, 201–214. doi:10.1023/A:1006450005648
Xu, Y., Wang, Y., Long, Q., Huang, J., Wang, Y., Zhou, K., et al. (2014). Overexpression of OsZHD1, a Zinc Finger Homeodomain Class Homeobox Transcription Factor, Induces Abaxially Curled and Drooping Leaf in rice. Planta. 239, 803–816. doi:10.1007/s00425-013-2009-7
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2005). Organization of Cis-Acting Regulatory Elements in Osmotic- and Cold-Stress-Responsive Promoters. Trends Plant Sci. 10, 88–94. doi:10.1016/j.tplants.2004.12.012
Yue, P., Lu, Q., Liu, Z., Lv, T., Li, X., Bu, H., et al. (2020). Auxin‐Activated MdARF5 Induces the Expression of Ethylene Biosynthetic Genes to Initiate Apple Fruit Ripening. New Phytol. 226, 1781–1795. doi:10.1111/nph.16500
Zeng, H., Xie, Y., Liu, G., Wei, Y., Hu, W., and Shi, H. (2019). Agrobacterium-Mediated Gene Transient Overexpression and Tobacco Rattle Virus (TRV)-based Gene Silencing in Cassava. Int. J. Mol. Sci. 20, 3976. doi:10.3390/ijms20163976
Zhang, Y. T., Zhang, Y. L., Chen, S. X., Yin, G. H., Yang, Z. Z., Lee, S., et al. (2015). Proteomics of Methyl Jasmonate Induced Defense Response in Maize Leaves against Asian Corn Borer. BMC Genomics. 16, 224. doi:10.1186/s12864-015-1363-1
Zhou, C., Zhu, C., Xie, S., Weng, J., Lin, Y., Lai, Z., et al. (2021). Genome-Wide Analysis of Zinc Finger Motif-Associated Homeodomain (ZF-HD) Family Genes and Their Expression Profiles Under Abiotic Stresses and Phytohormones Stimuli in tea Plants (Camellia Sinensis). Scientia Horticulturae. 281, 109976. doi:10.1016/j.scienta.2021.109976
Keywords: apple, ZF-HD, genome-wide, gene expression, fruit ripening
Citation: Zheng X-b, Wu Y, Wang H, Song S-w, Bai T-h, Jiao J, Song C-h, Pang H-g and Wang M-m (2022) Genome-Wide Investigation of the Zinc Finger-Homeodomain Family Genes Reveals Potential Roles in Apple Fruit Ripening. Front. Genet. 12:783482. doi: 10.3389/fgene.2021.783482
Received: 26 September 2021; Accepted: 22 December 2021;
Published: 17 January 2022.
Edited by:
Ahmed Sallam, Assiut University, EgyptReviewed by:
Hamed Bostan, National Institute of Environmental Health Sciences (NIEHS), United StatesCopyright © 2022 Zheng, Wu, Wang, Song, Bai, Jiao, Song, Pang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Miao-miao Wang, d21tMjAxOEBoZW5hdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.