
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Genet. , 05 January 2022
Sec. Statistical Genetics and Methodology
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.783078
This article is part of the Research Topic Systems Genetics of Human Complex Diseases, Volume II View all 24 articles
 Qiaoli Zeng1,2,3†
Qiaoli Zeng1,2,3† Dehua Zou2,3,4†
Dehua Zou2,3,4† Shanshan Gu3,5†
Shanshan Gu3,5† Fengqiong Han6
Fengqiong Han6 Shilin Cao7*
Shilin Cao7* Yue Wei8*
Yue Wei8* Runmin Guo1,2,3,9*
Runmin Guo1,2,3,9*Background: CDK5 regulatory subunit associated protein 1 like 1 (CDKAL1) is a major pathogenesis-related protein for type 2 diabetes mellitus (T2DM). Recently, some studies have investigated the association of CDKAL1 susceptibility variants, including rs4712523, rs4712524, and rs9460546 with T2DM. However, the results were inconsistent. This study aimed to evaluate the association of CDKAL1 variants and T2DM patients.
Methods: A comprehensive meta-analysis was performed to assess the association between CDKAL1 SNPs and T2DM among dominant, recessive, additive, and allele models.
Results: We investigated these three CDKAL1 variants to identify T2DM risk. Our findings were as follows: rs4712523 was associated with an increased risk of T2DM for the allele model (G vs A: OR = 1.172; 95% CI: 1.103–1.244; p < 0.001) and dominant model (GG + AG vs AA: OR = 1.464; 95% CI: 1.073–1.996; p = 0.016); rs4712524 was significantly associated with an increased risk of T2DM for the allele model (G vs A: OR = 1.146; 95% CI: 1.056–1.245; p = 0.001), additive model (GG vs AA: OR = 1.455; 95% CI: 1.265–1.673; p < 0.001) recessive model (GG vs AA + AG: OR = 1.343; 95% CI: 1.187–1.518; p < 0.001) and dominant model (GG + AG vs AA: OR = 1.221; 95% CI: 1.155–1.292; p < 0.001); and rs9460546 was associated with an increased risk of T2DM for the allele model (G vs T: OR = 1.215; 95% CI: 1.167–1.264; p = 0.023). The same results were found in the East Asian subgroup for the allele model.
Conclusions: Our findings suggest that CDKAL1 polymorphisms (rs4712523, rs4712524, and rs9460546) are significantly associated with T2DM.
Type 2 diabetes mellitus (T2DM) is a complex disease characterized by insulin resistance in peripheral tissues and dysregulated insulin secretion by pancreatic β-cells (Li et al., 2020). The incidence of T2DM in adults has been increasing over recent decades (Yang et al., 2010; Tian et al., 2019) and is estimated to increase to over 700 million by 2045 (Saeedi et al., 2019; Li et al., 2020). T2DM is caused by genetic and environmental factors (Tian et al., 2019; Wu et al., 2014). Genetic variants are thought to be involved in the development of T2DM. Genome-wide association studies have indicated that some single nucleotide polymorphisms (SNPs) are critical risk factors for T2DM (Tian et al., 2019).
CDK5 regulatory subunit associated protein 1 like 1 (CDKAL1) is a crucial pathogenesis-related protein for T2DM. The CDKAL1 gene encodes cyclin-dependent kinase 5 regulatory subunit-associated protein 1 (CDK5RAP1)-like 1. Cyclin-dependent kinase 5 (CDK5) is a serine/threonine protein kinase that contributes to the glucose-dependent regulation of insulin secretion (Li et al., 2020); therefore, it plays a critical role in the pathophysiology of β-cell dysfunction and predisposition to T2DM (Li et al., 2020; Wei et al., 2005; Ubeda et al., 2006). The associations of many SNPs in CDKAL1 with T2DM have been examined in some meta-analyses, but no published meta-analysis has evaluated the role of CDKAL1 rs4712523, rs4712524 and rs9460546 variants in the susceptibility to T2DM. Several studies have examined the association between CDKAL1 polymorphisms (rs4712523, rs4712524 and rs9460546) and T2DM risk, but some findings were failed to replicate. Therefore, performing a meta-analysis is needed to evaluate the association between CDKAL1 polymorphisms (rs4712523, rs4712524, and rs9460546) and T2DM.
This meta-analysis was conducted according to Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines.
The Google Scholar, PubMed and Chinese National Knowledge Infrastructure databases were systematically searched for relevant studies using the following terms:
1 “CDKAL1” or “rs4712523” or “polymorphism” and “T2DM”;
2 “CDKAL1” or “rs4712524” or “polymorphism” and “T2DM”;
3 “CDKAL1”, or “rs9460546” or “polymorphism” and “T2DM”, respectively.
The search was performed with no date or language restrictions. All the studies were evaluated by reading the title and abstract and excluding irrelevant studies. The full texts of eligible studies were then assessed by reading the full text to confirm inclusion in the study.
The inclusion criteria of the studies were as follows: 1) case-control/cohort studies; 2) studies that evaluated the association between CDKAL1 SNPs (rs4712523, rs4712524, and rs9460546) and T2DM; 3) adequate raw data or sufficient data to calculate odds ratios (ORs) with corresponding 95% confidence intervals (CIs); 4) a T2DM diagnosis based on the clinical criteria of the World Health Organization.
The exclusion criteria were as follows: 1) not a case-control/cohort study; 2) not related to CDKAL1 SNPs (rs4712523, rs4712524, and rs9460546) and T2DM; 3) insufficient data; 4) NDM data not in Hardy-Weinberg equilibrium (HWE).
Two authors independently extracted the following data from the included studies: first author, ethnicity, year of publication, numbers of T2DM patients and NDM controls, distribution of alleles and genotypes, and ORs with 95% CIs of the allele distribution.
Four genetic models were evaluated in rs4712523 and rs4712524: the dominant model (GG + AG vs AA), recessive model (GG vs AA + AG), additive model (GG vs AA) and allele model (G vs A). Additionally, the allele model (G vs T) was evaluated in rs9460546. Genetic heterogeneity was estimated using Q-test and I2 test. Lower heterogeneity was defined as I2 <50% and p > 0.01, using the fixed effects model (Mantel–Haenszel) to calculate ORs with corresponding 95% CIs. Otherwise, the random effects model (Mantel–Haenszel) was used. The significance of the ORs was evaluated using the Z test. Begg’s and Egger’s tests were used to determine publication bias. STATA v.14.0 software (Stata Corporation, Texas, United States) was used to perform all statistical analyses.
A total of 179 potential studies were searched using the inclusion and exclusion criteria. Figure 1 shows a flow chart of the study selection process. Twelve articles, including 7 in English and 5 in Chinese, had rs4712523 data. Eight articles, including 5 in English, 2 in Chinese and 1 in Russian, had rs4712524 data. Five articles, including 5 in English, had rs9460546 data. The characteristics of each included study are shown in Tables 1−3.
High heterogeneity among studies (Scott et al., 2007; Rung et al., 2009; Takeuchi et al., 2009; Long et al., 2012; Lu et al., 2012; Gong, 2016; Li et al., 2013; Ren et al., 2013; Rao et al., 2016; Qian, 2019; Tian et al., 2019; Liju et al., 2020) was detected in the allele model (G vs A: I2 = 84.4%; p < 0.001), additive model (GG vs AA: I2 = 84.6%; p < 0.001), recessive model (GG vs AA + AG: I2 = 73.8%; p = 0.002), and dominant model (GG + AG vs AA: I2 = 86.1%; p < 0.001) (Figure 2).
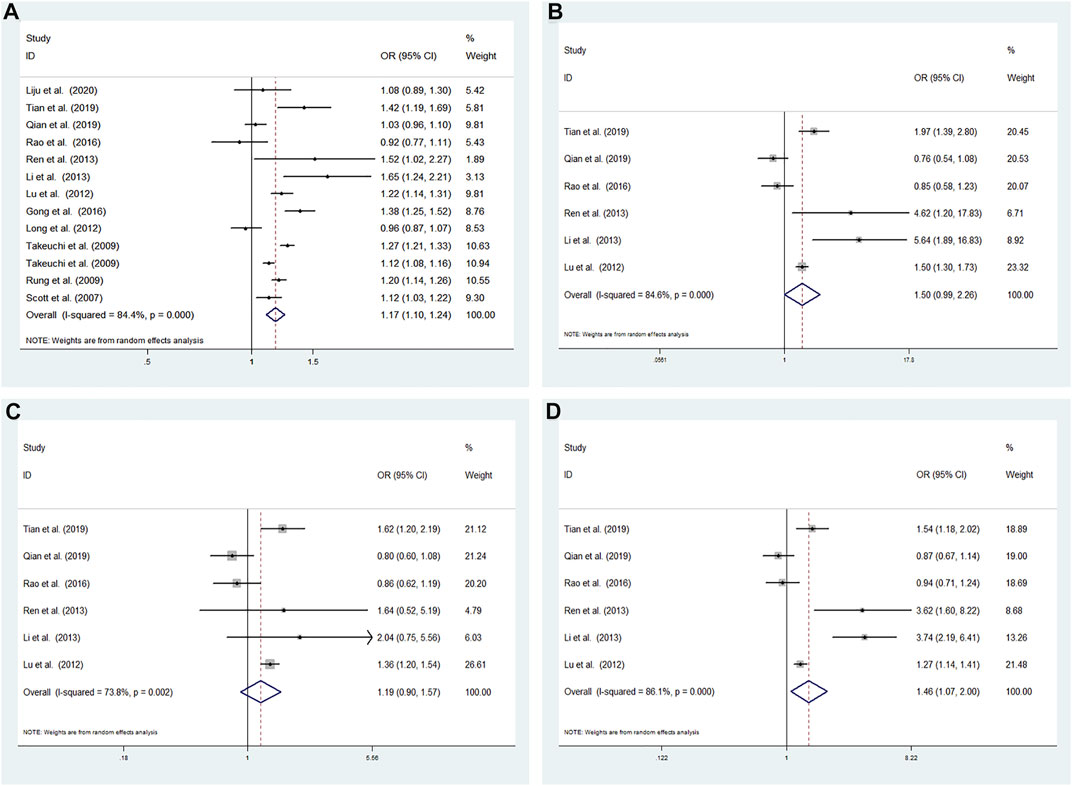
FIGURE 2. Meta-analysis using a random effects model for the association between the CDKALl rs4712523 polymorphism and T2DM susceptibility (A) Allele model, G vs A (B) Additive model, GG vs AA (C) Recessive model, GG vs AA + AG (D) Dominant model, GG + AG vs AA. OR: odds ratio, CI: confidence interval, I-squared: measure to quantify the degree of heterogeneity in meta-analyses.
High heterogeneity among studies (Unoki et al., 2008; Lu et al., 2012; Rao et al., 2016; Li, 2018; Tian et al., 2019; Azarova, 2020; Li et al., 2020; Liju et al., 2020) was detected in the allele model (G vs A: I2 = 75.1%; p < 0.001). A moderate degree of heterogeneity among studies was detected under the additive model (GG vs AA: I2 = 58.7%; p = 0.024) and recessive model (GG vs AA + AG: I2 = 57.8%; p = 0.027). Low heterogeneity among studies was detected under the dominant model (GG + AG vs AA: I2 = 31.8%; p = 0.185) (Figure 3).
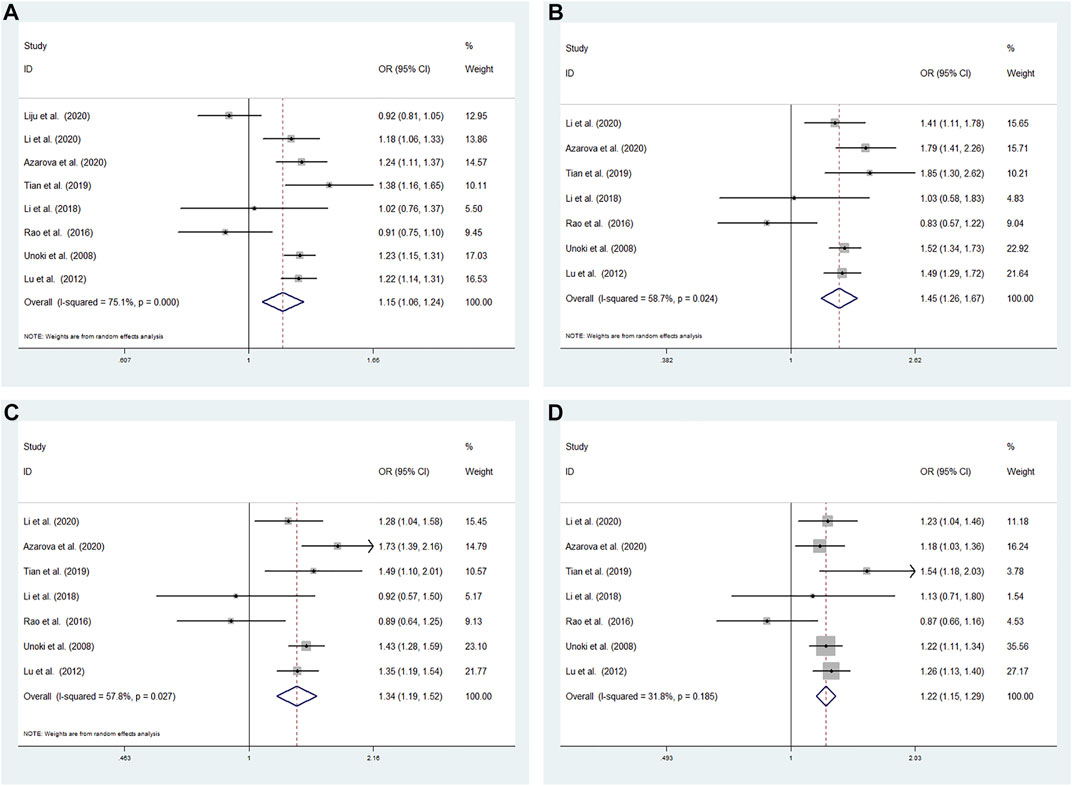
FIGURE 3. Meta-analysis for the association between the CDKALl rs4712524 polymorphism and T2DM susceptibility (A) Allele model, G vs A (random effects model) (B) Additive model, GG vs AA (random effects model) (C) Recessive model, GG vs AA + AG (random effects model) (D) Dominant model, GG + AG vs AA (fixed effects model). OR: odds ratio, CI: confidence interval, I-squared: measure to quantify the degree of heterogeneity in meta-analyses.
Low heterogeneity among studies (Herder et al., 2008; Unoki et al., 2008; Hu et al., 2009; Maller et al., 2012; Li et al., 2020) was detected in the allele model (G vs T: I2 = 37.0%; p = 0.174) (Figure 4).
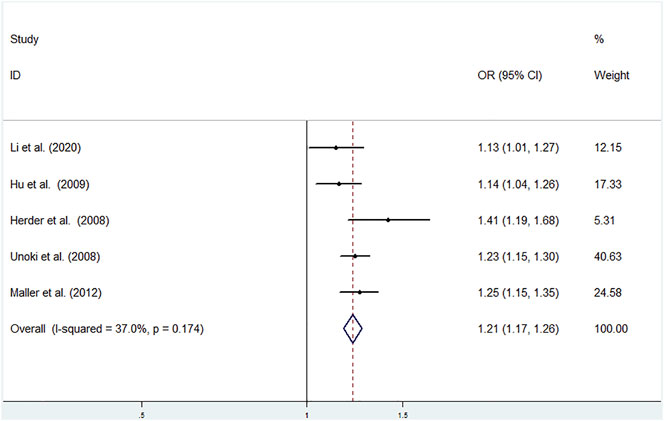
FIGURE 4. Meta-analysis using a fixed effects model for the association between the CDKAL1 rs9460546 polymorphism and T2DM susceptibility (Allele model, G vs T). OR: odds ratio, CI: confidence interval, I-squared: measure to quantify the degree of heterogeneity in meta-analyses.
A significant difference was found between T2DM patients and NDM controls for the allele model (G vs A: OR = 1.172; 95% CI: 1.103–1.245; p < 0.001) and dominant model (GG + AG vs AA: OR = 1.464; 95% CI: 1.073–1.996; p = 0.016). No significant associations were found under the additive model (GG vs AA: OR = 1.495; 95% CI: 0.990–2.257; p = 0.056) and recessive model (GG vs AA + AG: OR = 1.188; 95% CI: 0.900–1.568; p = 0.223) using a random effects model (Figure 2).
A random effects model was used to analyze the allele, additive and recessive models, and the dominant model was analyzed using a fixed effects model. A significant difference was found between T2DM patients and NDM controls for the allele model (G vs A: OR = 1.146; 95% CI: 1.056–1.245; p = 0.001), additive model (GG vs AA: OR = 1.455; 95% CI: 1.265–1.673; p < 0.001) recessive model (GG vs AA + AG: OR = 1.343; 95% CI: 1.187–1.518; p < 0.001) and dominant model (GG + AG vs AA: OR = 1.221; 95% CI: 1.155–1.292; p < 0.001) (Figure 3).
A significant difference was found between T2DM patients and NDM controls for the allele model (G vs T: OR = 1.215; 95% CI: 1.167–1.264; p = 0.023) using a fixed effects model (Figure 4).
We performed subgroup analysis according to ethnicity to evaluate the association between rs4712523 and T2DM susceptibility in the allele model. Rs35767 was significantly related to the risk of T2DM in the East Asian (G vs A: OR = 1.241; 95% CI: 1.123–1.371; p < 0.001) and others subgroup (G vs A: OR = 1.108; 95% CI: 1.039–1.180; p = 0.002) using a random effects model (Figure 5A).
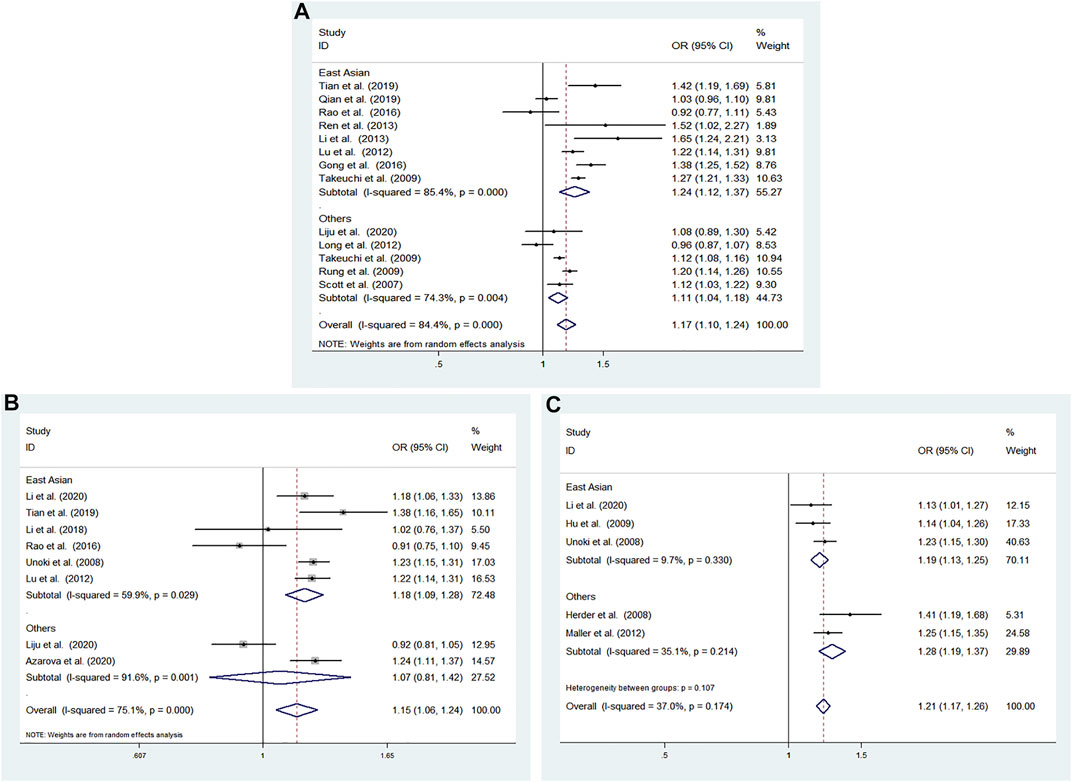
FIGURE 5. Association between the CDKALl variants and T2DM susceptibility in the subgroup for the allele model (A) rs4712523: G vs A (random effects model) (B) rs4712524: G vs A (random effects model) (C) rs9460546: G vs T (fixed effects model). OR: odds ratio, CI: confidence interval, I-squared: measure to quantify the degree of heterogeneity in meta-analyses.
We performed subgroup analysis according to ethnicity to evaluate the association between rs4712524 and T2DM susceptibility in the allele model. Rs4712524 was significantly related to the risk of T2DM in the East Asian (G vs A: OR = 1.182; 95% CI: 1.095–1.277; p < 0.001), but no significant associations were found in others subgroup (G vs A: OR = 1.071; 95% CI: 0.807–1.423; p = 0.634) using a random effects model (Figure 5B).
We performed subgroup analysis according to ethnicity to evaluate the association between rs9460546 and T2DM susceptibility in the allele model. Rs9460546 was significantly related to the risk of T2DM in the East Asian (G vs T: OR = 1.189; 95% CI: 1.134–1.247; p < 0.001) and others subgroup (G vs T: OR = 1.277; 95% CI: 1.188–1.373; p < 0.001) using a fixed effects model (Figure 5C).
According to Begg’s and Egger’s tests, no significant publication bias was found in each of the genetic models (all p > 0.05, data not shown), and the funnel plots are shown in Figures 6–9.
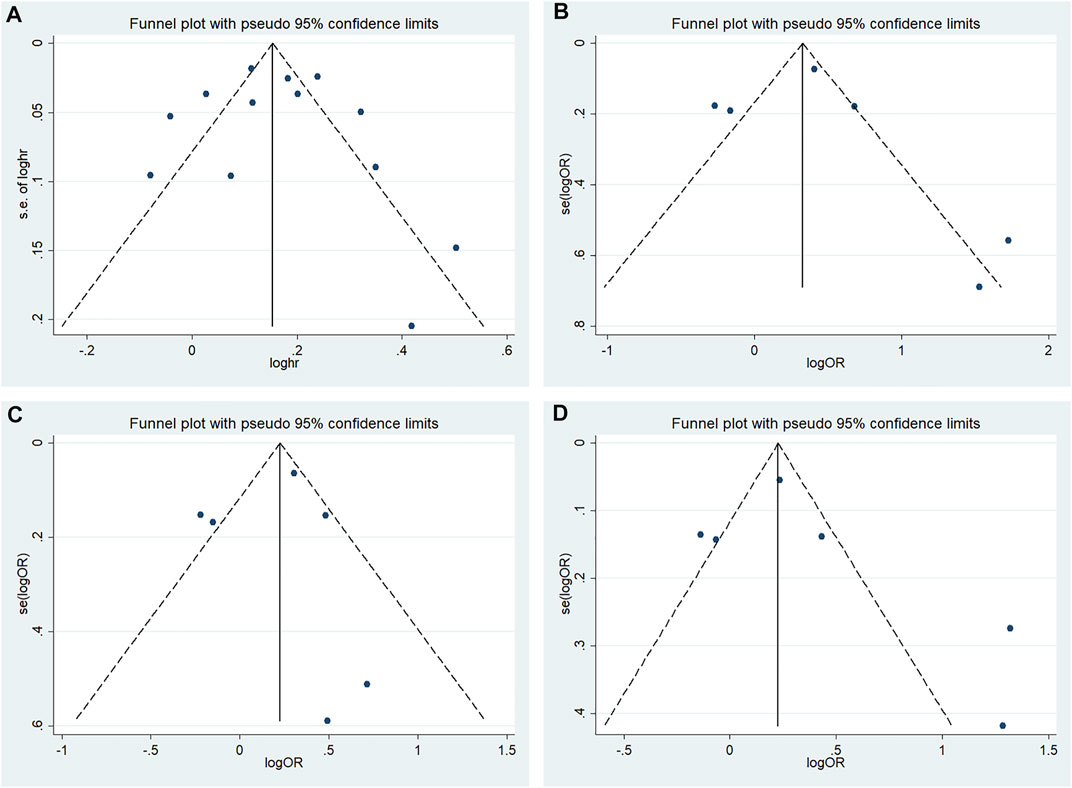
FIGURE 6. Funnel plot of the odds ratios in the CDKALl rs4712523 meta-analysis (A) Allele model, G vs A (B) Additive model, GG vs AA (C) Recessive model, GG vs AA + AG (D) Dominant model, GG + AG vs AA.
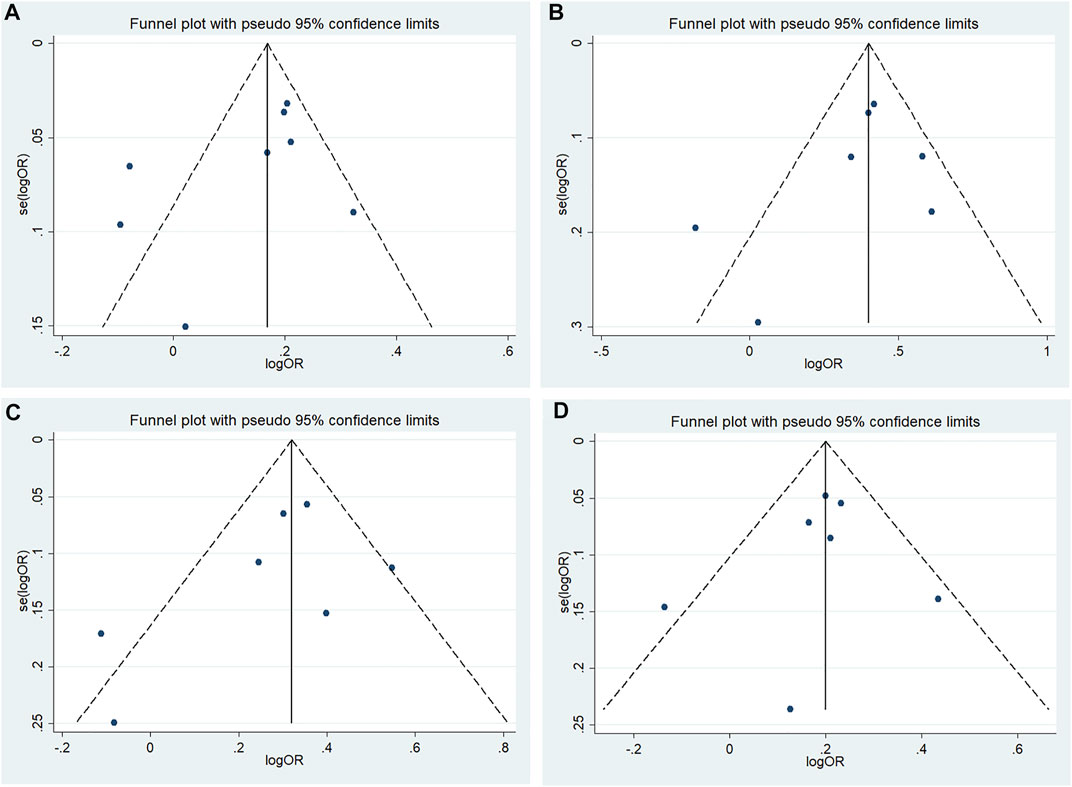
FIGURE 7. Funnel plot of the odds ratios in the CDKALl rs4712524 meta-analysis (A) Allele model, G vs A (B) Additive model, GG vs AA (C) Recessive model, GG vs AA + AG (D) Dominant model, GG + AG vs A.
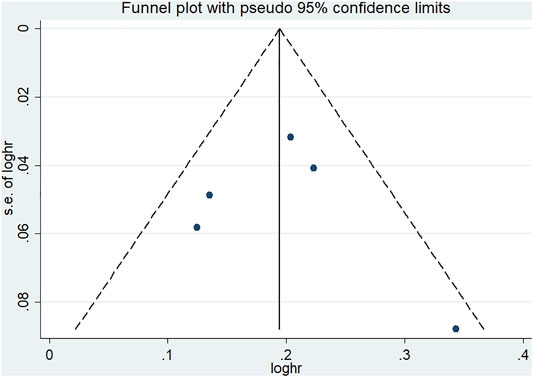
FIGURE 8. Funnel plot of the odds ratios in the CDKALl rs9460546 meta-analysis for the allele model (G vs T).
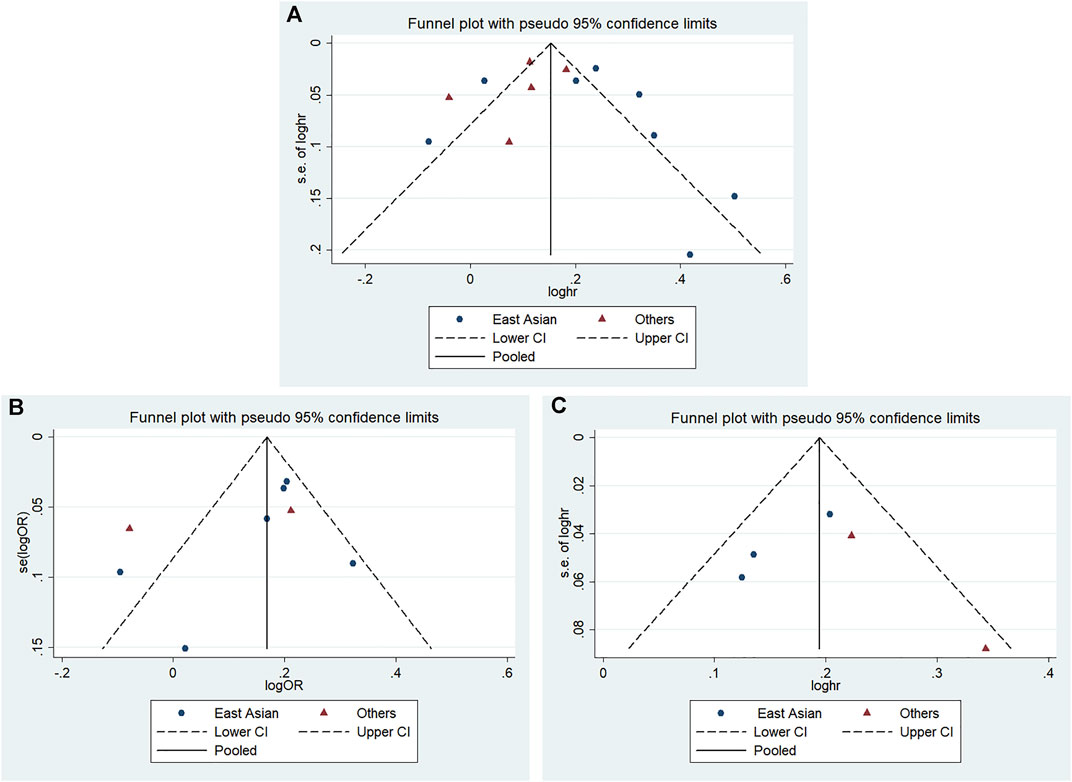
FIGURE 9. Funnel plot of the odds ratios in the CDKALl variants in the subgroup meta-analysis for the allele model (A) rs4712523: G vs A (B) rs4712524: G vs A (C) rs9460546: G vs T.
CDKAL1 is a key pathogenesis-related protein for T2DM (Tian et al., 2019). Genetic variants may play an essential role in T2DM susceptibility. In this meta-analysis, three SNPs (rs4712523, rs4712524, and rs9460546) from previous studies were evaluated to determine the association of CDKAL1 polymorphisms with T2DM. CDKAL1 polymorphisms (rs4712523, rs4712524, and rs9460546) showed a significant association with T2DM. Our results were consistent with some previous study findings.
The results revealed that the G allele and GG + AG genotypes of rs4712523 were associated with an increased risk of T2DM. Nine of the thirteen previous studies investigated rs4712523 showed an association between the G allele and T2DM (Scott et al., 2007; Rung et al., 2009; Takeuchi et al., 2009; Long et al., 2012; Lu et al., 2012; Gong, 2016; Li et al., 2013; Ren et al., 2013; Tian et al., 2019), and four studies found an association between the GG + AG genotypes and T2DM (Lu et al., 2012; Li et al., 2013; Ren et al., 2013; Tian et al., 2019). In addition, the rs4712524 G allele, GG and GG + AG genotypes were associated with an increased risk of T2DM susceptibility. That have been confirmed previous observations (Unoki et al., 2008; Lu et al., 2012; Tian et al., 2019; Azarova, 2020; Li et al., 2020). Additionally, the results showed that rs9460546 G allele was associated with T2DM susceptibility. Markedly, all five studies found that the rs9460546 G allele was associated with T2DM in various populations (Herder et al., 2008; Unoki et al., 2008; Hu et al., 2009; Maller et al., 2012; Li et al., 2020). Moreovr, rs4712523, rs4712524, and rs9460546 showed a significant association with T2DM in the East Asian subgroup for the allele model. In general, Our results have confirmed previous observations suggesting that CDKAL1 may play a role in T2DM. But it is worth noting that high heterogeneity among studies was detected in rs4712523 and rs4712524 likely because of the difference in country, ethnicity, genetic background and environmental factors. Subgroup analyses were performed by ethnicity in the allele model, and the subgroup still had high heterogeneity. Importantly, the high heterogeneity among studies might have affected our data.
CDKAL1 expression in human pancreatic β-cells increases insulin secretion by inhibiting CDK5 (Li et al., 2020; Wei et al., 2005; Ubeda et al., 2006; Ching et al., 2002). Subsequently, several studies have shown the association of genetic variants in CDKAL1 with defects in proinsulin conversion and the insulin response following glucose stimulation (Pascoe et al., 2007; Steinthorsdottir et al., 2007; Tian et al., 2019). Thus, CDKAL1 is involved in the development of T2DM. Genome-wide association studies have identified several SNPs in the CDKAL1 gene associated with T2D (Saxena et al., 2007; Scott et al., 2007; Tian et al., 2019). Our results confirmed the significant association between CDKAL1 SNPs and T2DM susceptibility. However, the mechanisms must be verified in functional studies. Our association results provide reference data to identify new biomarkers of T2DM that could contribute to the diagnosis of T2DM.
This meta-analysis has a few limitations. First, because of the limited examination of CDKAL1 variants in T2DM, the included studies had comparatively small sample sizes, which might affect the results of the meta-analysis because of insufficient statistical power. Thus, studies must be performed across different geographical and ethnic groups. Additionally, the factors of T2DM might be complex, with the contribution of genetic, environmental and dietary habits. Therefore, further study is required to evaluate whether other risk factors together with the CDKAL1 gene influence T2DM susceptibility.
To our knowledge, this study is the first to assess the role of CDKAL1 polymorphisms (rs4712523, rs4712524, and rs9460546) in T2DM. Significant associations were found between the CDKAL1 rs4712523, rs4712524, and rs9460546 polymorphisms and susceptibility to T2DM.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
QZ, DZ and SG were responsible for the study design, statistical analysis, and manuscript preparation. QZ and FH managed the literature searches and analyses. The study was supervised by SC, YW and RG.
Support for this work includes funding from the National Natural Science Foundation of China (81873649); Doctoral scientific research Initiate funding project of Shunde Women and Children’s Hospital of Guangdong Medical University (Maternity and Child Healthcare Hospital of Shunde Foshan) (2020BSQD007); Guangdong Medical University Research Foundation (GDMUM2020008 and GDMUM2020012); Medical Research Project of Foshan Health Bureau (20210188 and 20210289).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Azarova, L. (2020). POLYMORPHIC VARIANTS rs4712524 AND rs6931514 IN THE CDKAL1 GENE: ASSOCIATION WITH THE RISK OF DEVELOPMENT OF TYPE 2 DIABETES MELLITUS IN RUSSIAN POPULATIONE. Естественные и технические науки (in Russian) 9. doi:10.25633/ETN.2020.09.02
Ching, Y.-P., Pang, A. S. H., Lam, W.-H., Qi, R. Z., and Wang, J. H. (2002). Identification of a Neuronal Cdk5 Activator-Binding Protein as Cdk5 Inhibitor. J. Biol. Chem. 277 (18), 15237–15240. doi:10.1074/jbc.C200032200
Gong, X. (2016). “The Genetic Diversity Analysis of Candidate Genes Associated with Type 2 Diabetes in Xinjiang Ethnic Minority Populations,” in A Dissertation Submitted to Xinjiang Medical University (Xinjiang: Xinjiang Medical University) (in Chinese).
Herder, C., Rathmann, W., Strassburger, K., Finner, H., Grallert, H., Huth, C., et al. (2008). Variants of thePPARG,IGF2BP2,CDKAL1,HHEX, andTCF7L2Genes Confer Risk of Type 2 Diabetes Independently of BMI in the German KORA Studies. Horm. Metab. Res. 40 (10), 722–726. doi:10.1055/s-2008-1078730
Hu, C., Zhang, R., Wang, C., Wang, J., Ma, X., Lu, J., et al. (2009). PPARG, KCNJ11, CDKAL1, CDKN2A-Cdkn2b, IDE-KIF11-HHEX, IGF2BP2 and SLC30A8 Are Associated with Type 2 Diabetes in a Chinese Population. PLoS One 4 (10), e7643. doi:10.1371/journal.pone.0007643
Li, C., Shen, K., Yang, M., Yang, Y., Tao, W., He, S., et al. (2020). Association between Single Nucleotide Polymorphisms in CDKAL1 and HHEX and Type 2 Diabetes in Chinese Population. Dmso 13, 5113–5123. doi:10.2147/DMSO.S288587
Li, X., Su, Y., Yan, Z., Gu, L., Li, C., and Li, A. (2013). CDKALl Rs4712523 Polymorphism Is Associatedwith Type 2 Diabetes in Han Population in Inner Mongolia. Basic Clin. Med. 33, 3 (in Chinese). doi:10.16352/j.issn.1001-6325.2013.03.017
Li, Y. (2018). “Association between Genetic Polymorphisms and Comorbidity of Coronary Heart Disease and Type 2 Diabetes in the Elderly,” in A Dissertation Submitted to Peking Union Medical College (Beijing: Peking Union Medical College) (in Chinese).
Liju, S., Chidambaram, M., Mohan, V., and Radha, V. (2020). Impact of Type 2 Diabetes Variants Identified through Genome-wide Association Studies in Early-Onset Type 2 Diabetes from South Indian Population. Genomics Inform. 18 (3), e27. doi:10.5808/GI.2020.18.3.e27
Long, J., Edwards, T., Signorello, L. B., Cai, Q., Zheng, W., Shu, X.-O., et al. (2012). Evaluation of Genome-wide Association Study-Identified Type 2 Diabetes Loci in African Americans. Am. J. Epidemiol. 176 (11), 995–1001. doi:10.1093/aje/kws176
Lu, F., Qian, Y., Li, H., Dong, M., Lin, Y., Du, J., et al. (2012). Genetic Variants on Chromosome 6p21.1 and 6p22.3 Are Associated with Type 2 Diabetes Risk: a Case-Control Study in Han Chinese. J. Hum. Genet. 57 (5), 320–325. doi:10.1038/jhg.2012.25
Maller, J. B., McVean, G., McVean, G., Byrnes, J., Vukcevic, D., Palin, K., et al. (2012). Bayesian Refinement of Association Signals for 14 Loci in 3 Common Diseases. Nat. Genet. 44 (12), 1294–1301. doi:10.1038/ng.2435
Pascoe, L., Tura, A., Patel, S. K., Ibrahim, I. M., Ferrannini, E., Zeggini, E., et al. (2007). Common Variants of the Novel Type 2 Diabetes Genes CDKAL1 and HHEX/IDE Are Associated with Decreased Pancreatic -Cell Function. Diabetes 56 (12), 3101–3104. doi:10.2337/db07-0634
Qian, X. (2019). “Associations of Polymorphisms of CDKALI Gene with Type 2 Diabetes Mellitus and Diabetes-Related Traits,” in A Thesis Submitted to Zhengzhou University for the Degree of Master (Zhengzhou: Zhengzhou University) (in Chinese).
Rao, P., Yu, X., Gai, S., Wang, H., Fang, H., Wang, Y., et al. (2016). Association of IGF2BP2, CDKAL1 Gene Polymorphism and Gene Environment Interaction with Type 2 Diabetes Mellitus. Mod. instrument Med. Treat. 22, 2 (in Chinese). doi:10.11876/mimt201602008
Ren, X., Yan, Z., Li, X., Su, Y., and Zhang, S. (2013). Association of CDKAL1 Rs4712523 Polymorphism with Susceptibility to the Blood Lipid of Type 2 Diabetes in Han Population of Inner Mongolia. Chin. J. Clinicians 7, 17 (in Chinese). doi:10.3877/cma.j.issn.1674-0785.2013.17.013
Rung, J., Cauchi, S., Albrechtsen, A., Shen, L., Rocheleau, G., Cavalcanti-Proença, C., et al. (2009). Genetic Variant Near IRS1 Is Associated with Type 2 Diabetes, Insulin Resistance and Hyperinsulinemia. Nat. Genet. 41 (10), 1110–1115. doi:10.1038/ng.443
Saeedi, P., Petersohn, I., Salpea, P., Malanda, B., Karuranga, S., Unwin, N., et al. (2019). Global and Regional Diabetes Prevalence Estimates for 2019 and Projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th Edition. Diabetes Res. Clin. Pract. 157, 107843. doi:10.1016/j.diabres.2019.107843
Saxena, R., Voight, B. F., Lyssenko, V., Burtt, N. P., de Bakker, P. I. W., Chen, H., et al. (2007). Genome-Wide Association Analysis Identifies Loci for Type 2 Diabetes and Triglyceride Levels. Science 316 (5829), 1331–1336. doi:10.1126/science.1142358
Scott, L. J., Mohlke, K. L., Bonnycastle, L. L., Willer, C. J., Li, Y., Duren, W. L., et al. (2007). A Genome-wide Association Study of Type 2 Diabetes in Finns Detects Multiple Susceptibility Variants. Science 316 (5829), 1341–1345. doi:10.1126/science.1142382
Steinthorsdottir, V., Thorleifsson, G., Reynisdottir, I., Benediktsson, R., Jonsdottir, T., Walters, G. B., et al. (2007). A Variant in CDKAL1 Influences Insulin Response and Risk of Type 2 Diabetes. Nat. Genet. 39 (6), 770–775. doi:10.1038/ng2043
Takeuchi, F., Serizawa, M., Yamamoto, K., Fujisawa, T., Nakashima, E., Ohnaka, K., et al. (2009). Confirmation of Multiple Risk Loci and Genetic Impacts by a Genome-wide Association Study of Type 2 Diabetes in the Japanese Population. Diabetes 58 (7), 1690–1699. doi:10.2337/db08-1494
Tian, Y., Xu, J., Huang, T., Cui, J., Zhang, W., Song, W., et al. (2019). A Novel Polymorphism (Rs35612982) in CDKAL1 Is a Risk Factor of Type 2 Diabetes: A Case-Control Study. Kidney Blood Press. Res. 44 (6), 1313–1326. doi:10.1159/000503175
Ubeda, M., Rukstalis, J. M., and Habener, J. F. (2006). Inhibition of Cyclin-dependent Kinase 5 Activity Protects Pancreatic Beta Cells from Glucotoxicity. J. Biol. Chem. 281 (39), 28858–28864. doi:10.1074/jbc.M604690200
Unoki, H., Takahashi, A., Kawaguchi, T., Hara, K., Horikoshi, M., Andersen, G., et al. (2008). SNPs in KCNQ1 Are Associated with Susceptibility to Type 2 Diabetes in East Asian and European Populations. Nat. Genet. 40 (9), 1098–1102. doi:10.1038/ng.208
Wei, F.-Y., Nagashima, K., Ohshima, T., Saheki, Y., Lu, Y.-F., Matsushita, M., et al. (2005). Cdk5-dependent Regulation of Glucose-Stimulated Insulin Secretion. Nat. Med. 11 (10), 1104–1108. doi:10.1038/nm1299
Wu, Y., Ding, Y., Tanaka, Y., and Zhang, W. (2014). Risk Factors Contributing to Type 2 Diabetes and Recent Advances in the Treatment and Prevention. Int. J. Med. Sci. 11 (11), 1185–1200. doi:10.7150/ijms.10001
Keywords: type 2 diabetes mellitus, CDKAL1, polymorphisms, susceptibility, meta-analysis
Citation: Zeng Q, Zou D, Gu S, Han F, Cao S, Wei Y and Guo R (2022) Different Associations Between CDKAL1 Variants and Type 2 Diabetes Mellitus Susceptibility: A Meta-analysis. Front. Genet. 12:783078. doi: 10.3389/fgene.2021.783078
Received: 25 September 2021; Accepted: 13 December 2021;
Published: 05 January 2022.
Edited by:
Liangcai Zhang, Janssen Research and Development, United StatesReviewed by:
Dalin Li, Cedars Sinai Medical Center, United StatesCopyright © 2022 Zeng, Zou, Gu, Han, Cao, Wei and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shilin Cao, MzMwMzM5NTBAcXEuY29t; Yue Wei, d2VpeXVlMTM4QDE2My5jb20=; Runmin Guo, cnVubWluLmd1b0BnZG11LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.