
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 16 December 2021
Sec. Genetics of Common and Rare Diseases
Volume 12 - 2021 | https://doi.org/10.3389/fgene.2021.781189
 Valeria Lo Faro1,2
Valeria Lo Faro1,2 Ilja M. Nolte3
Ilja M. Nolte3 Jacoline B. Ten Brink2
Jacoline B. Ten Brink2 Harold Snieder3
Harold Snieder3 Nomdo M. Jansonius1
Nomdo M. Jansonius1 Arthur A. Bergen2,4*
Arthur A. Bergen2,4* Lifelines Cohort Study
Lifelines Cohort StudyBackground and purpose: Primary open-angle glaucoma (POAG) is an optic neuropathy characterized by death of retinal ganglion cells and atrophy of the optic nerve head. The susceptibility of the optic nerve to damage has been shown to be mediated by mitochondrial dysfunction. In this study, we aimed to determine a possible association between mitochondrial SNPs or haplogroups and POAG.
Methods: Mitochondrial DNA single nucleotide polymorphisms (mtSNPs) were genotyped using the Illumina Infinium Global Screening Array-24 (GSA) 700K array set. Genetic analyses were performed in a POAG case-control study involving the cohorts, Groningen Longitudinal Glaucoma Study-Lifelines Cohort Study and Amsterdam Glaucoma Study, including 721 patients and 1951 controls in total. We excluded samples not passing quality control for nuclear genotypes and samples with low call rate for mitochondrial variation. The mitochondrial variants were analyzed both as SNPs and haplogroups. These were determined with the bioinformatics software HaploGrep, and logistic regression analysis was used for the association, as well as for SNPs.
Results: Meta-analysis of the results from both cohorts revealed a significant association between POAG and the allele A of rs2853496 [odds ratio (OR) = 0.64; p = 0.006] within the MT-ND4 gene, and for the T allele of rs35788393 (OR = 0.75; p = 0.041) located in the MT-CYB gene. In the mitochondrial haplogroup analysis, the most significant p-value was reached by haplogroup K (p = 1.2 × 10−05), which increases the risk of POAG with an OR of 5.8 (95% CI 2.7–13.1).
Conclusion: We identified an association between POAG and polymorphisms in the mitochondrial genes MT-ND4 (rs2853496) and MT-CYB (rs35788393), and with haplogroup K. The present study provides further evidence that mitochondrial genome variations are implicated in POAG. Further genetic and functional studies are required to substantiate the association between mitochondrial gene polymorphisms and POAG and to define the pathophysiological mechanisms of mitochondrial dysfunction in glaucoma.
Primary open-angle glaucoma (POAG) is a complex and chronic eye disease characterized by progressive death of retinal ganglion cells (RGCs), which manifests itself initially as visual field loss. Untreated POAG ultimately leads to irreversible blindness (Tham et al., 2014). High intraocular pressure (IOP) is the first and major risk factor identified in patients with POAG (Shaffer, 1996), in addition other known risk factors are advanced age, myopia, ethnicity, and positive family history for POAG (Gramer and Grehn, 2012). Nowadays, the only efficacious therapy for protecting the RCGs (from which the axons form the optic nerve) in POAG is directed towards decreasing IOP. POAG is also defined as a genetically complex disease because many genes have been associated with this condition (Thorleifsson et al., 2010; Burdon et al., 2011; Ramdas et al., 2011; Wiggs et al., 2011; Cao et al., 2012; Osman et al., 2012; Liu et al., 2013; Chen et al., 2014; Gharahkhani et al., 2014; Springelkamp et al., 2015; Trikha et al., 2015; Choquet et al., 2018; Pasquale, 2019). Rearrangements in the DNA and multiple disease genes have been implicated in the pathogenesis of POAG (Davis et al., 2011; Janssen et al., 2013; Liu et al., 2014; Lo Faro et al., 2021). Known disease genes include myocilin (MYOC), optineurin (OPTN), WD repeat domain 36 (WDR36), cytochrome P450 family 1 subfamily B polypeptide 1 (CYP1B1), and TANK-binding kinase 1 (TBK1) (Janssen et al., 2013). So far, more than 120 chromosomal loci have been discovered through genome-wide association studies (Burdon et al., 2011; Ramdas et al., 2011; Choquet et al., 2018; Gharahkhani et al., 2021). Even so, the disease genes and genetic risk factors identified only explain a small proportion of POAG heritability, and contribute relatively little to understanding the pathogenetic mechanisms. To date, the majority of the genetic studies aim to identify the causes underlying POAG by focusing on the nuclear genome. We and other researchers have postulated that at least part of the remaining POAG heritability might be found in variants in the mitochondrial DNA (mtDNA) (Lascaratos et al., 2012; Osiewacz, 2014; Williams et al., 2017).
The mitochondrion is an organelle mainly involved in the production of cellular energy with distinct extrachromosomal circular and double-stranded molecules of DNA. The mtDNA is transmitted through the maternal germline and it has a unique subcellular transcription and replication machinery (Vincent and Picard et al., 2018). Mutations in the mtDNA have previously been implicated in cellular energy deficits that lead to ocular degenerative disease (Yu-Wai-Man et al., 2011). Indeed, the mitochondria can be considered as a “power plant” for the cell, then it is expected that mitochondrial disorders tend to affect more frequently tissues with high energy demand, such as the retina, brain, muscles, heart, and endocrine systems (Wallace, 2010). A characteristic of the mitochondrial genome (mtGenome) is that it accumulates mutations at a notably faster rate than the nuclear genome. As a result, mtDNA is highly polymorphic. Most likely, this characteristic can be explained by two processes: the lack of protective histones and repair mechanisms, which increase the replication errors, and, given the proximity of mtDNA with the respiratory chain complexes, the exposure to reactive oxygen species (ROS) (Wallace, 2010; Lascaratos et al., 2012).
Many mitochondrial single-nucleotide polymorphisms (mtSNPs) have become fixed in a variety of populations during human evolution (Yu-Wai-Man et al., 2011; Collins et al., 2016). Due to the exclusive maternal inheritance of mtDNA and the fact that the mtGenome does not recombine, mtSNPs are accumulating and co-transmitted through the maternal lineages (Andersen and Balding et al., 2018). This characteristic allows tracing specific, ancestral patterns of human migration that occurred millennia ago, from Africa to other continents. These specific polymorphic SNP-sets accumulating on the mtDNA, allowed researchers to classify human populations into various mtDNA ‘‘haplogroups”. There are a total of nine known haplogroups that identify individuals with European ancestry. These are named H, I, J, K, T, U, V, W, and X, where haplogroup H represents about 40–45% of the total. According to recent studies, specific haplogroups can influence the development of diseases such as POAG, primary angle-closure glaucoma, exfoliation glaucoma, as well as cancer, diabetes, and late-onset neurodegenerative conditions (Herrnstadt et al., 2002; Abu-Amero et al., 2008; van Oven and Kayser, 2009; Abu-Amero et al., 2011a; Abu-Amero et al., 2011b; Urzua-Traslavina and Carlos, 2014). The association between mitochondrial haplogroups and POAG has been investigated in few studies, with conflicting results: Andrews et al. (2006), in a case-control comparison of 140 POAG patients and 75 healthy individuals from England, did not find a difference in the haplogroup distribution between cases and controls (Andrews et al., 2006). In contrast, Collins et al. (2016) found that in African populations the haplogroups L1c2, L1c2b, and L2 were risk factors for POAG (Collins et al., 2016).
An important aspect in the pathophysiology of POAG and similar optic neuropathies is represented by the increased apoptosis of RGCs, and by the functional decay of the trabecular meshwork (TM) (Izzotti et al., 2011; Almasieh et al., 2012). RGCs contain a high number of mitochondria related to their high energy demand. They are especially vulnerable to oxidative damage caused by mitochondrial dysfunction (Sanchez et al., 2016). Apart from POAG, several other optic neuropathies show axonal RGC loss correlated with mitochondrial dysfunction (Howell, 1997; Howell, 2003). Two examples are Leber’s Hereditary Optic Neuropathy (LHON; OMIM 535000) and autosomal dominant optic atrophy (DOA; OMIM 165500). While LHON is caused by three well-known pathogenic mtDNA mutations in the MT-ND1, MT-ND4, and MT-ND6 genes, DOA is caused by pathogenic mutations within the nuclear OPA1 gene, which codes for a mitochondrial wall membrane protein (Yu-Wai-Man et al., 2009; Wallace, 2010; Yu-Wai-Man et al., 2011).
Next to mitochondrial damage in the RGCs, Izzotti and coworkers investigated potential mitochondrial damage in the TM, a tissue involved in POAG via its influence on IOP. By comparing mtDNA deletions in TM from glaucoma patients and controls by qRT-PCR, the authors observed that oxidative damage arising from mitochondrial failure plays a role in the functional decay of the TM (Izzotti et al., 2011). Another, independent line of evidence for mitochondrial involvement in POAG has come from investigations of glaucoma-prone mice: retinal levels of nicotinamide adenine dinucleotide (NAD) decreased with age, rendering neurons vulnerable to disease-related insults. Interestingly, the administration of a redox metabolite NAD+ and gene therapy of the expression of Nmnat1, a key NAD + producing enzyme, had a protective effect. At the highest dose tested, 93% of eyes did not develop glaucoma symptomatology, compared to 50% for the control group (Williams et al., 2017).
Given the hypothesis that RGCs degeneration and functional decay of TM in POAG are influenced by mitochondrial dysfunction, we aimed to explore whether POAG is associated with variations in the mtGenome. To this purpose, we conducted two different association analyses further described below. First, we analyzed mtDNA SNPs in order to detect genetic variations potentially associated with the disease. Second, we analyzed the role of major haplogroups.
We performed our association analysis using two case-control studies. The first case-control study consisted of glaucoma cases from the Groningen Longitudinal Glaucoma Study (GLGS), of which a subset of the POAG patients (see below) was genotyped (n = 592) (Heeg et al., 2005). The controls (n = 1841) were selected from the Lifelines Cohort Study and Biobank (LL) and came from the same geographical region as the GLGS cases. This cohort is addressed in this study as the GLGS-LL cohort. They were age-matched with a 1:3 ratio, using the R package MatchIt with nearest-neighbor matching (Ho et al., 2011). The second case-control study consisted of glaucoma cases (n = 129) and controls (n = 110) from the Amsterdam Glaucoma Study (AGS) (Ramdas et al., 2011). All the participants were Dutch and of European-ancestry.
The original GLGS cohort has been described in detail by Heeg and colleagues (Heeg et al., 2005). After the inclusion of the initial cohort in 2000–2001, the GLGS continued as a dynamic population, that is, new participants were added during follow-up. We included glaucoma patients who visited the outpatient department of the UMCG in 2015 and who gave written informed consent for a blood sample being taken for genetic analyses. In the GLGS, glaucoma patients showed glaucomatous visual field (VF) loss in at least one eye. For glaucomatous baseline VF loss, two consecutive tests had to be abnormal in at least one eye. Defects had to be compatible with glaucoma and without any other explanation. A VF test before the two baseline tests was discarded to reduce the influence of learning. Those with pseudoexfoliative or pigment dispersion glaucoma or a history of angle-closure or secondary glaucoma were excluded for the current analysis, leaving only POAG patients.
The LL is a multi-disciplinary prospective population-based cohort study examining in a unique three-generation design the health and health-related behaviours of 167,729 persons living in the North of Netherlands. It employs a broad range of investigative procedures in assessing the biomedical, socio-demographic, behavioural, physical and psychological factors which contribute to the health and disease of the general population, with a special focus on multi-morbidity and complex genetics. Participants completed questionnaires, underwent physical examinations, and biological samples including DNA were collected (Scholtens et al., 2015). For the current study, we only included participants without glaucoma and aged 60 years or older. Glaucoma phenotype was defined using a questionnaire-based glaucoma proxy, a classification algorithm built on questions regarding self-reported eye diseases and glaucoma-specific visual complaints (Neustaeter et al., 2020). Participants were classified as having definite, probable, or possible glaucoma, or as healthy. Lifelines controls used in this study were those individuals classified by the proxy as healthy and from whom the genotyping data was available.
The AGS study includes glaucoma cases and healthy controls collected from eye clinics, meetings of the glaucoma patients’ association, nursing homes, and fairs for the elderly from all over the Netherlands. The AGS patients underwent ophthalmoscopy and biomicroscopy with a 90-diopter lens, and digital stereo images of the optic nerve head were taken after mydriatic drops. POAG cases had to have glaucomatous optic neuropathy vertical cup-disc ratio (VCDR) > 0.7 with corresponding glaucomatous visual field loss in at least one eye or a VCDR ≥ 0.8 when no visual field was available (Ramdas et al., 2011). Control subjects from the AGS cohort were selected from unrelated individuals, aged 60 years or older with a VCDR ≤ 0.6 on ophthalmoscopy and fundus photography, and without eye abnormalities.
Genomic DNA was extracted from the peripheral blood and all individuals were genotyped using the Illumina Infinium Global Screening Array® (GSA) MultiEthnic Disease beadchip version. This array contains approximately 700,000 SNPs and combines multi-ethnic genome-wide content, curated clinical research variants, and quality control (QC) markers. Specifically, for the mtDNA, this array covers 140 mtDNA SNPs. For these latter SNPs, the raw probe intensities were combined in one dataset and called together with Opticall using the -MT option (Shah et al., 2012). To obtain position, strand orientation, and reference allele of mtDNA for our data, we aligned the genotypes with the Cambridge Reference Sequence (rCRS) (ENCODE Project Consortium 2012). For further analysis, we considered only variants that could be mapped perfectly with the reference panel. Next, the following quality control (QC) criteria were applied to SNPs level: 95% call rate per mtDNA SNP in the combined set of case and control individuals and heterozygote mitochondrial genotypes were set to missing, allowing only homozygous calls. At DNA sample level the quality control was first conducted in the autosomal chromosomes and exclusion criteria were 1) duplicated sample, 2) excessive heterozygosity rate, 3) sex discordance, 4) the presence of first and/or second-degree relatives (pi-hat >0.20), and 5) non-European ancestry.
The genotyping datasets of AGS and GLGS-LL were then imputed in IMPUTE2 (Howie et al., 2009). Before the imputation, monomorphic variants were removed and all samples were assigned to male sex to allow haploid imputation. Then we followed instructions for the imputation of chromosome X. The reference panel used contained 36,960 sequences aligned to mtDNA sequences (McInerney et al., 2021). Variants with imputation quality score less than 0.3 and monomorphic variants were excluded.
To test for association of the mtDNA SNP markers with glaucoma, logistic regressions were conducted separately for GLGS-LL and AGS, with POAG as outcome and SNP as independent variable, adjusting for age and sex. We choose to analyze the two cohorts separately to avoid risk of batch effects and false positive results caused by population stratification. Only SNPs with a minor allele frequency (MAF) > 1% were included in this analysis. To estimate the risks of POAG, odds ratios (ORs) and 95% confidence intervals (CIs) were calculated. Analyses were also performed stratified by sex. The genetic analyses were conducted using PLINK v1.90 (Purcell et al., 2007). Subsequently, we combined the results of the two cohorts by a fixed effects inverse variance weighted meta-analysis using METAL software in which double genomic control was applied (Willer et al., 2010).
A second association analysis was done in the two cohorts separately on reconstructed haplogroups. Since the mtDNA does not recombine, it behaves like a single locus with many alleles making all variants correlated with each other. Mitochondrial haplogroups were estimated from the directly genotyped variants and the haplogroup of each individual was determined with HaploGrep, available at https://haplogrep.i-med.ac.at/ (van Oven and Kayser, 2009; Kloss-Brandstätter et al., 2011). All 140 mtDNA SNPs were entered for the haplogroup determination, and each individual’s haplogroup was determined based on PhyloTree build 17 (implemented in HaploGrep 2.1.21.jar). We only included haplogroup assignments with a quality score above 80% (determined by HaploGrep), and with a frequency above 1% (van Oven and Kayser, 2009; Kloss-Brandstätter et al., 2011). For the association test, sub-haplogroups were first assigned to their respective major haplogroups. We tested each haplogroup against haplogroup H, which is the most common European haplogroup (22.9% in our dataset; see Discussion section), as the reference using logistic regression, adjusting for age and sex (Torroni and Wallace et al., 1994). Chi-square analysis was conducted to determine the effect of specified sub-haplogroup K (K1, K1a1, K1a11, K1a1b2a1, K1a24a, K1a4a, K1a4a1a2a, K1b2a, and K1c1).
Given the hypothesis-free approach of our exploratory study and the risk to test not independent SNPs due to high linkage disequilibrium in mtDNA, we reported nominal significant p-values (≤0.05) (Andersen and Balding et al., 2018). The analyses were performed using RStudio.
In order to evaluate gene expression of significant SNPs, we queried the EyeIntegration database v1.05 (https://eyeintegration.nei.nih.gov/) in cornea, retina, and retinal pigment epithelium (RPE) (Bryan et al., 2018). This database is created by investigators at the National Eye Institute (National Institutes of Health) and contains publicly deposited RNA-seq datasets from human ocular tissues (Bryan et al., 2018). Gene correlation networks were constructed using the kWithin metric to measure the connectivity. Genes with higher connectivity are, theoretically, more likely to be important in gene regulation as perturbations in them will affect the system more than less connected genes. Identified genes, either those closest to significant SNPs or resulting from the gene correlation network, were queried in the Online Mendelian Inheritance in Man (OMIM) database, to identify associated phenotypes (Hamosh, 2004).
The study followed the tenets of the Declaration of Helsinki and the ethics board of the University Medical Center of Amsterdam (UMC) approved this research (METc submission # 2013_327). All participants provided written informed consent.
A total of 2,672 individuals were included. Table 1 shows the demographics of both cohorts.
In total, 140 mtSNPs were initially screened on the DNA of our case control populations. Forty SNPs in the GLGS-LL dataset and 39 SNPs in the AGS cohort passed the quality control criteria previously mentioned (see Subjects and Methods section) and were used for further analysis. In the GLGS-LL cohort we excluded 51 SNPs that were monomorphic or had a low MAF. We excluded 49 additional ones for a relatively high missing genotype rate. In the AGS cohort, 101 mtDNA SNPs were monomorphic or had a low MAF but we did not exclude any SNP for high missing genotype rate. After imputation, 69 and 63 variants were retained in the GLGS-LL and AGS cohorts, respectively.
Logistic regression analyses were conducted separately in the GLGS-LL and AGS datasets, where POAG was modelled as the dependent variable and the genotyped variants as the independent variable (Table 2).
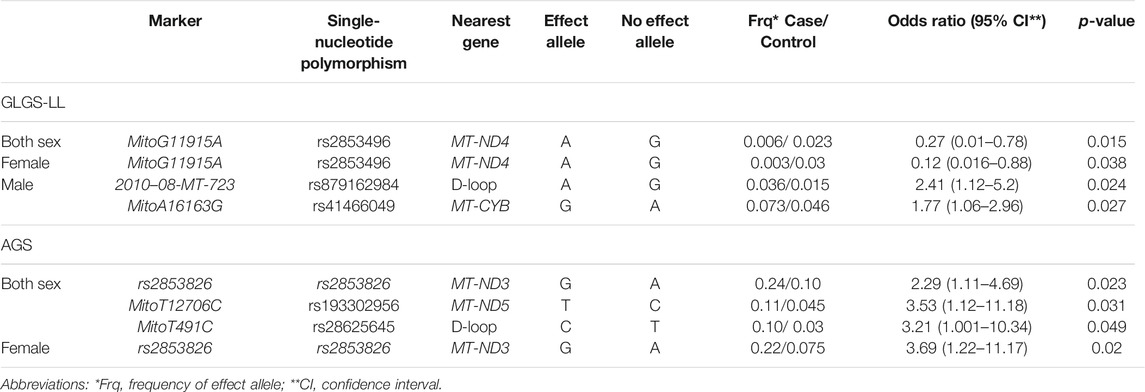
TABLE 2. Significant mtDNA single-nucleotide polymorphisms associated with POAG in the GLGS-LL and AGS cohorts, separated for sex.
These results were then meta-analyzed for 32 SNPs in common between the two datasets, revealing nominal associations in the mtDNA SNPs rs2853496 (p = 0.006) and rs35788393 (p = 0.041) with POAG, in the sex combined cohorts (Table 3).
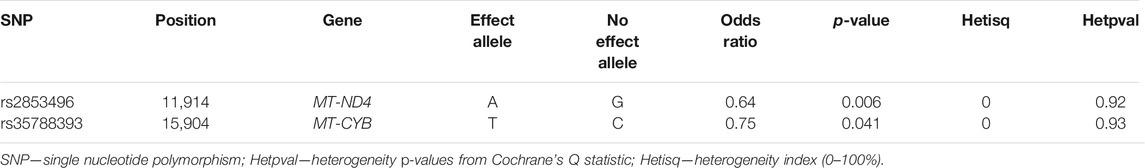
TABLE 3. Nominal significant mtDNA SNPs associated with POAG from the meta-analysis of the GLGS-LL and AGS cohorts, conducted in both sexes.
In addition to single SNP association analyses, we also conducted an association test between mitochondrial haplogroups and POAG. The inclusion threshold for this analysis (see methods section) was reached by a total of 235 individuals, i.e., 143 cases and 92 controls from the two cohorts that fulfilled the quality criteria. The identified haplogroups were all assigned to one of the European major haplogroups (H, K, and U). Table 4 summarizes haplogroup frequency distributions for the cases and controls.
The haplogroup K showed an increased risk effect for POAG, with an OR = 5.8 (95% CI = 2.7–13.1; p = 1.2 × 10−05) and the haplogroup U an OR = 2.6 (95% CI = 1.3–5.2; p = 0.005). We further observed that the haplogroup U was the most common haplogroup (45.5%), followed by haplogroups K (31.4%) and H (22.9%). None of these K sub-haplogroups showed any statistical significance (data not shown). The haplogroup U is phylogenetically connected with the haplogroup K, and was also significantly associated with increased risk of POAG. Therefore, we repeated our analysis combining the haplogroups U and K in the UK cluster and found that UK frequency also differed significantly from the control group (odds ratio 3.5, 95% CI = 1.8–6.7; p = 0.00012).
We queried the two genes located closest to the two identified SNPs (MT-CYB and MT-ND4) for expression in ocular and nonocular tissues in the EyeIntegration database. We found the highest expression of both MT-CYB and MT-ND4 in the following tissues, respectively: adult retina of 19.56 log2 (TPM + 1) and 20.60 log2 (TPM + 1), and RPE of 19.59 log2 (TPM + 1) and 20.03 log2 (TPM + 1). The lowest gene expression was reported in the cornea [11.50 log2 (TPM + 1) and 11.97 log2 (TPM + 1)] (Figure 1).
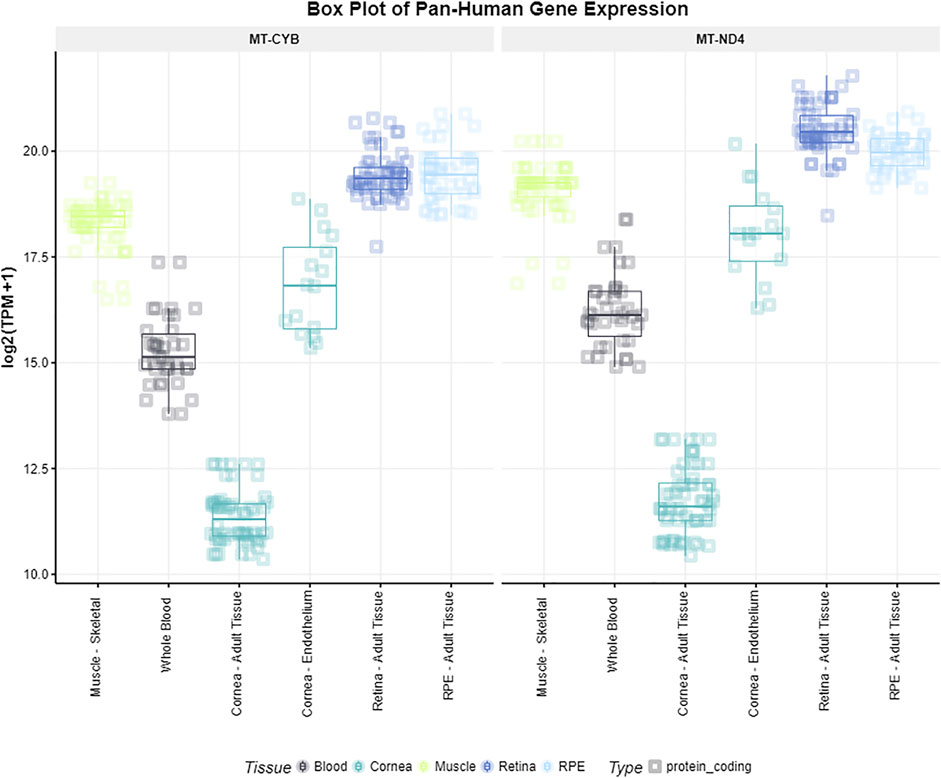
FIGURE 1. Gene expression of MT-CYB and MT-ND4 in different tissues. Gene expression levels of the genes MT-CYB and MT-ND4 according to the EyeIntegration database v1.05 for muscle skeletal, whole blood, cornea, retina and RPE tissues.
Gene expression levels of the genes MT-CYB and MT-ND4 according to the EyeIntegration database v1.05 for muscle skeletal, whole blood, cornea, retina and RPE tissues.
We also queried in the EyeIntegration database v1.05 the retina network to examine which genes were the most connected in this network. In the retina network, MT-CYB and MT-ND4 genes have high connectivity with the POMGNT1 gene (kWithin = 18.447). Genes with higher connectivity are, theoretically, more likely to be important in gene regulation because perturbations in them will affect more the system compared to the effect in less connected genes. When we queried the POMGNT1 gene in the OMIM database, we identified eye phenotypes linked to muscular dystrophy-dystroglycanopathy, in which patients have congenital glaucoma and retinitis pigmentosa (Parton, 2003).
In this study, we investigated the possible involvement of genetic variation in mitochondria in POAG, by performing an association analysis for mitochondrial SNPs and haplogroups in 721 patients with POAG and 1951 healthy individuals. Based on evidence derived from combined analysis of our datasets, we concluded that two mtSNPs (rs2853496 and rs35788393) are nominal associated with POAG. Our data suggest that the A allele of rs2853496, within the MT-ND4 gene, and the T allele of rs35788393, located in the MT-CYB gene, have a protective effect. With respect to mitochondrial haplogroups, our analyses identified haplogroup K as highly associated with an increased risk of POAG (OR = 5.8; 95% CI = 2.7–13.1; p = 1.2 × 10−5).
Our findings are consistent with evidence from the literature that suggest a potential role of the mtGenome, and more specifically of the genes MT-ND4 and MTCYB in optic neuropathies or glaucoma (Cortopassi and Arnheim et al., 1990; Votruba et al., 2004; Abu-Amero et al., 2006). The MT-ND4 gene is a protein-coding gene located in the mtDNA, encoding for subunit 4 of complex I (NADH ubiquinone oxidoreductase) (MT-ND4, 2021). The complex I is the first enzyme of the respiratory chain, a vulnerable site to oxidative stress, also involved in cellular functions like apoptosis (Ferguson et al., 1976). SNPs in subunit 4 of MT-ND4 can affect the first step of the electron transport chain. Therefore, these mutations may have an impact on mitochondrial respiratory chain function and could result in an alteration of the cellular energy metabolism.
Genetic variations in the MT-ND4 are implicated in other optic neuropathies. This is the case of LHON, where one of the most prevalent variants that accounts for more than 70% of all cases is the m.11778G > A, located in the MT-ND4 gene (Yu-Wai-Man et al., 2014; Mancuso and Klopstock et al., 2019). LHON is one of the most common inherited optic neuropathies and it is characterized by bilateral optic atrophy and loss of central vision due to loss of RGCs (Sadun, 2002; Yu-Wai-Man et al., 2011). MtDNA mutations associated with LHON have also been described in animal models: mice with a mutation in the mt-Nd4 gene show nerve atrophy and RGCs degeneration. Both conditions are also characteristics of LHON in humans (Divi et al., 2007; Koilkonda and Guy, 2011). In contrast to the mitochondrial mutations identified in LHON cases, in the mitochondrial genome of POAG patients the majority of the mutations were somatic transversions (a replacement of a purine with a pyrimidine, or vice versa), caused by the accumulation of oxidative stress over time (Abu-Amero et al., 2006).
In this study, we also report an association between POAG and the mtDNA variation rs35788393 in the MT-CYB gene. The protein product of this gene is involved in the oxidative phosphorylation system. This system is composed of five complexes where the MT-CYB gene encodes for the cytochrome b of complex III (that catalyzes the transfer of electrons from ubiquinol to cytochrome c), the only one solely encoded by a mitochondrial gene (Chaussenot et al., 2018). Pathogenic mutations in the MT-CYB gene can disrupt the normal activity of the electron transport chain and affect the production of ATP by increasing the production of ROS. This leads to damage of cellular proteins, lipids and nucleic acids via oxidation reactions (West et al., 2011). So far, mutations in MT-CYB have been associated with LHON, retinitis pigmentosa and cataract (Brown et al., 1992; Wibrand et al., 2001; Schuelke et al., 2002; Ronchi et al., 2011). Since the MT-CYB gene is involved in the production of ATP in the electron transport chain, it is pivotal to explore the possible role of this gene in POAG in future studies.
Genetic variations in both MT-CYB and MT-ND4 genes are able to destabilize the so-called mitochondrial super complex: the physical interaction between mitochondrial complex-I and complex-III. The destabilization of this complex leads to the loss of complex I activity, the major entry point for electrons to the respiratory chain (Hudson et al., 2007). In transgenic mice, loss of complex-I activity showed an increase of ROS levels in the RCGs and optic nerve degeneration (Qi et al., 2003). In human, glaucomatous TM cells have been reported to be more sensitive to the inhibition of complex-I (Banerjee et al., 2013). Indeed, damages in complex-I were observed to contribute to the progressive loss of TM cells in POAG patients due to the excessive mitochondrial ROS production and to the attenuation of the mitochondrial membrane. This decrease reduces the ATP synthesis, driving the cells towards apoptosis (He et al., 2008). Another study comparing both POAG and LHON lymphoblasts found an impairment of the complex-I, where functional defects of this complex were milder in POAG than LHON. This is in accordance with the less severe development of the disease in POAG (Van Bergen et al., 2015). However, more comprehensive investigations are still necessary to define the regulatory function of complex-I that in turn might lead to the increase of the oxidative stress and favor the glaucomatous condition.
To add biological context to our study, we also evaluated bioinformatically which genes were the most highly connected with MT-CYB and MT-ND4 in the retina network (Bryan et al., 2018). Genes highly connected indicate that they are more likely to have an effect in gene regulation. By querying the retina network, we implicated the POMGNT1 gene (protein O-mannose beta-1,2-N-acetylglucosaminyltransferase-1). POMGNT1 synthesizes a unique O-mannose sugar chain on α-dystroglycan, the extracellular protein that binds laminin α2 in the extracellular matrix. Reported mutations in the POMGNT1 gene have enlightened its role in four genetic muscular dystrophy disease entities: 1) Walker–Warburg syndrome (OMIM #253280), 2) the muscle-eye-brain disease (OMIM #253280)—for which patients show ocular symptoms as retinal degeneration, optic atrophy and congenital glaucoma—3) congenital muscular dystrophy with mental retardation (OMIM #613151), and 4) retinitis pigmentosa (OMIM #606822) (Pihko et al., 1995; Yoshida et al., 2001; Godfrey et al., 2007; Clement et al., 2008; Wang et al., 2016; Xu et al., 2016). The dystroglycan gene (DAG1) encodes for α-dystroglycan and β-dystroglycan (Holt et al., 2000). In the retina, dystroglycan is highly expressed in Müller glial, rods and cones at the outer plexiform layer, and plays an important role in retinal function and survival (Schmitz et al., 1993; Montanaro et al., 1995; Blank et al., 1999; Jastrow et al., 2006). The DAG1 gene is also linked to Muscular dystrophy-dystroglycanopathy (congenital with brain and eye anomalies, OMIM #616538). Using zebrafish animal models, Gupta et al. demonstrated that dystroglycan deficiency caused abnormal development of ganglion cells, lens, and cornea (Gupta et al., 2011). In addition, in a consanguineous Israeli-Arab family a homozygous loss-of-function mutation in the DAG1 gene was detected in infants with a congenital phenotype consistent with Walker–Warburg syndrome. Ocular features in those infants included bilateral corneal opacity and glaucoma (Riemersma et al., 2015). Taken together, these findings suggest that dystroglycan deficiency is strongly correlated with eye abnormalities, including glaucoma. In our view, further studies on the role of DAG1 and POMGNT1 genes in the pathomechanisms underlying POAG are warranted.
Another part of our current investigation in POAG was focused on the analysis of potentially associated mtDNA haplogroups. In order to interpret our results, it is important to consider the variations reported in the population distributions of mitochondrial haplotypes in comparison with the frequencies identified in our study (Torroni and Wallace, 1994). A study conducted in the Netherlands in 680 individuals randomly selected identified that the most common haplogroup was H (45.3%), followed by haplogroups U (25.6%), T (11.6%), J (10.7%) and K (6.3%) (Chaitanya et al., 2016). These frequencies, when present in our POAG dataset, differed from those reported in the general population. Regarding the haplogroup H, it is important to point out that it shows a complex variation with many sub-lineages. In our study, we used data generated by a SNP-chip array, which is able to detect sites that are polymorphic in populations. Therefore, the differences in frequency reported here for the haplogroup H can be attributed to the absence of sites that allows an accurate classification (Loogväli et al., 2004; Pereira et al., 2005). In our study, haplogroup K was the most significant association with POAG. Considering that haplogroup K occurs approximately in 8% of European and 6% in Dutch individuals, we reported a higher frequency of this haplogroup in our POAG cases (24.6%) compared to controls (6.8%). In line with our findings, a meta-analysis conducted in 3,613 individuals affected by LHON from 159 European pedigrees indicated that the risk of visual loss was higher in carriers of the mitochondrial haplogroup K: individual carriers of haplogroup K were more exposed to experience visual loss, whereas individuals carriers of haplogroup H had a lower risk of visual loss (Hudson et al., 2007). The haplogroup association in our study showed a similar outcome: individuals with haplogroup K had a higher risk to develop POAG compared to individuals belonging to haplogroup H. We also observed that the UK cluster shows the same direction of risk. Interestingly, studies conducted in cybrids showed that haplogroup UK are associated with less levels of mitochondrial protein synthesis and respiratory complex IV activities than cybrids from haplogroup H (Gómez-Durán et al., 2010). Therefore, also supported by previous studies, we speculate on the possible link of the mtGenome in the pathogenesis of POAG (Abu-Amero et al., 2011b; Bosley et al., 2011; Collins et al., 2016; Singh et al., 2018). In contrast, a few studies reported that mitochondrial haplogroups did not contribute to the pathogenesis of POAG. Negative associations were found in POAG cohorts from the north east of England, Saudi Arabia and Ghana (Andrews et al., 2006; Abu-Amero et al., 2008, 2012). Combining our data with those of the literature, there is evidence that mitochondrial haplotypes K plays a role in the pathogenesis of POAG, at least in some populations.
Our study had several strengths and limitations. Strong points are: the AGS and GLGS datasets are well defined in terms of diagnosis, design, and method of investigation. In fact, these datasets are clinical cohorts, in which POAG patients received diagnoses by experienced physicians, following strict criteria. In addition, the cohorts used in this study were genotyped on the same array and have been processed applying the same quality control procedures. There are also a number of limitations: first, by nature of the study, we focused only on homoplasmic mtDNA variations, detected in blood but not in potentially relevant ocular tissues. Furthermore, we cannot exclude potential additional influence of nuclear DNA of mitochondrial origin, other genetic variations elsewhere or non-genetic factors. Second, we performed our analyses in samples of European ancestry and for this reason our findings are not, without more research, transferable to other populations. Third, replications in other populations of our results are necessary to corroborate their association with POAG.
In our study, we identified associations between mitochondrial polymorphisms in the MT-ND4 and MT-CYB genes and POAG, and reported that individuals carrying mtDNA haplogroup K were at the highest risk of developing this eye disease. Our findings support the hypothesis that mitochondria have a role in the pathogenesis of POAG. Nonetheless, further genetic and functional studies are still required to highlight the role of mitochondrial genes, in relation to POAG pathophysiology.
Raul Aguirre-Gamboa, Department of Genetics, University of Groningen, University Medical Center Groningen, Netherlands; Patrick Deelen, Department of Genetics, University of Groningen, University Medical Center Groningen, Netherlands; Lude Franke, Department of Genetics, University of Groningen, University Medical Center Groningen, Netherlands; Jan A. Kuivenhoven, Department of Pediatrics, University of Groningen, University Medical Center Groningen, Netherlands; Esteban A. Lopera Maya, Department of Genetics, University of Groningen, University Medical Center Groningen, Netherlands; Ilja M. Nolte, Department of Epidemiology, University of Groningen, University Medical Center Groningen, Netherlands; Serena Sanna, Department of Pediatrics, University of Groningen, University Medical Center Groningen, Netherlands; Harold Snieder, Department of Epidemiology, University of Groningen, University Medical Center Groningen, Netherlands; Morris A. Swertz, Department of Genetics, University of Groningen, University Medical Center Groningen, Netherlands; Peter M. Visscher, Department of Epidemiology, University of Groningen, University Medical Center Groningen, Netherlands, and Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland, Australia; Judith M Vonk, Department of Epidemiology, University of Groningen, University Medical Center Groningen, Netherlands; Cisca Wijmenga, Department of Genetics, University of Groningen, University Medical Center Groningen, Netherlands.
The data that support the findings of this study are available from Lifelines Biobank but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of Lifelines.
The studies involving human participants were reviewed and approved by the ethics board of the University Medical Center of Amsterdam (METc submission # 2013_327). The data analyzed in this study was also obtained from the Lifelines biobank, under project application number OV18_0463. Requests to access this dataset should be directed to Lifelines Research Office (cmVzZWFyY2hAbGlmZWxpbmVzLm5s). The patients/participants provided their written informed consent to participate in this study.
VL, IN, and AB conceptualized, designed the experimental setup, performed genetic analyses, and wrote the main manuscript text. JT collected the data and wrote the main manuscript. VL, AB, IN, HS, and NJ supervised the study, interpreted the results and revised the final draft of the manuscript. All authors have read and agreed to the published version of the manuscript.
This project was funded by European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 675033 (EGRET plus). Additional funding was provided by the Rotterdamse Stichting Blindenbelangen (Grant ID B20150036). The funding organization had no role in the design, conduct, analysis, or publication of this research. The Lifelines Biobank initiative has been made possible by subsidy from the Dutch Ministry of Health, Welfare and Sport, the Dutch Ministry of Economic Affairs, the University Medical Center Groningen (UMCG the Netherlands), University of Groningen and the Northern Provinces of Netherlands. The Lifelines Biobank initiative has been made possible by funding from the Dutch Ministry of Health,Welfare and Sport, the Dutch Ministry of Economic Affairs, the University Medical Center Groningen(UMCG Netherlands), University of Groningen and the Northern Provinces of the Netherlands. The generation and management of GWAS genotype data for the Lifelines Cohort Study is supported by the UMCG Genetics Lifelines Initiative (UGLI). UGLI is partly supported by a Spinoza Grant from NWO, awarded to Cisca Wijmenga.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors wish to acknowledge the services of the Lifelines Cohort Study, the contributing research centers delivering data to Lifelines, and all the study participants.
Abu-Amero, K. K., Cabrera, V. M., Larruga, J. M., Osman, E. A., González, A. M., and Al-Obeidan, S. A. (2011a). Eurasian and Sub-saharan African Mitochondrial DNA Haplogroup Influences Pseudoexfoliation Glaucoma Development in Saudi Patients. Mol. Vis. 17, 543–547.
Abu-Amero, K. K., González, A. M., Osman, E. A., Larruga, J. M., Cabrera, V. M., and Al-Obeidan, S. A. (2011b). Mitochondrial DNA Lineages of African Origin Confer Susceptibility to Primary Open-Angle Glaucoma in Saudi Patients. Mol. Vis. 17, 1468–1472.
Abu-Amero, K. K., Hauser, M. A., Mohamed, G., Liu, Y., Gibson, J., Gonzalez, A. M., et al. (2012). Mitochondrial Genetic Background in Ghanaian Patients with Primary Open-Angle Glaucoma. Mol. Vis. 18, 1955–1959.
Abu-Amero, K. K., Morales, J., Bosley, T. M., Mohamed, G. H., and Cabrera, V. M. (2008). The Role of Mitochondrial Haplogroups in Glaucoma: A Study in an Arab Population. Mol. Vis. 14, 518–522.
Abu-Amero, K. K., Morales, J., and Bosley, T. M. (2006). Mitochondrial Abnormalities in Patients with Primary Open-Angle Glaucoma. Invest. Ophthalmol. Vis. Sci. 47 (6), 2533–2541. doi:10.1167/iovs.05-1639
Almasieh, M., Wilson, A. M., Morquette, B., Cueva Vargas, J. L., and Di Polo, A. (2012). The Molecular Basis of Retinal Ganglion Cell Death in Glaucoma. Prog. Retin. Eye Res. 31, 152–181. doi:10.1016/j.preteyeres.2011.11.002
Andersen, M. M., and Balding, D. J. (2018). How Many Individuals Share a Mitochondrial Genome. Plos Genet. 14, e1007774. doi:10.1371/journal.pgen.1007774
Andrews, R., Ressiniotis, T., Turnbull, D. M., Birch, M., Keers, S., Chinnery, P. F., et al. (2006). The Role of Mitochondrial Haplogroups in Primary Open Angle Glaucoma. Br. J. Ophthalmol. 90 (4), 488–490. doi:10.1136/bjo.2005.084335
Banerjee, D., Banerjee, A., Mookherjee, S., Vishal, M., Mukhopadhyay, A., Sen, A., et al. (2013). Mitochondrial Genome Analysis of Primary Open Angle Glaucoma Patients. PLoS ONE 8 (8), e70760. doi:10.1371/journal.pone.0070760
Blank, M., Koulen, P., Blake, D. J., and Kröger, S. (1999). Dystrophin and Beta-Dystroglycan in Photoreceptor Terminals from Normal and mdx3Cvmouse Retinae. Eur. J. Neurosci. 11, 2121–2133. doi:10.1046/j.1460-9568.1999.00636.x
Bosley, T. M., Hellani, A., Spaeth, G. L., Myers, J., Katz, L. J., Moster, M. R., et al. (2011). Down-Regulation of OPA1 in Patients with Primary Open Angle Glaucoma. Mol. Vis. 17, 1074–1079.
Brown, M. D., Voljavec, A. S., Lott, M. T., Torroni, A., Yang, C. C., and Wallace, D. C. (1992). Mitochondrial DNA Complex I and III Mutations Associated with Leber's Hereditary Optic Neuropathy. Genetics 130 (1), 163–173. doi:10.1093/genetics/130.1.163
Bryan, J. M., Fufa, T. D., Bharti, K., Brooks, B. P., Hufnagel, R. B., and McGaughey, D. M. (2018). Identifying Core Biological Processes Distinguishing Human Eye Tissues with Precise Systems-Level Gene Expression Analyses and Weighted Correlation Networks. Hum. Mol. Genet. 27 (19), 3325–3339. doi:10.1093/hmg/ddy239
Burdon, K. P., Macgregor, S., Hewitt, A. W., Sharma, S., Chidlow, G., Mills, R. A., et al. (2011). Genome-Wide Association Study Identifies Susceptibility Loci for Open Angle Glaucoma at TMCO1 and CDKN2B-AS1. Nat. Genet. 43, 574–578. doi:10.1038/ng.824
Cao, D., Jiao, X., Liu, X., Hennis, A., Leske, M. C., Nemesure, B., et al. (2012). CDKN2B Polymorphism Is Associated with Primary Open-Angle Glaucoma (POAG) in the Afro-Caribbean Population of Barbados, West Indies. PLoS ONE 7 (6), e39278. doi:10.1371/journal.pone.0039278
Chaitanya, L., van Oven, M., Brauer, S., Zimmermann, B., Huber, G., Xavier, C., et al. (2016). High-Quality mtDNA Control Region Sequences from 680 Individuals Sampled across the Netherlands to Establish a National Forensic mtDNA Reference Database. Forensic Sci. Int. Genet. 21, 158–167. doi:10.1016/j.fsigen.2015.12.002
Chaussenot, A., Rouzier, C., Fragaki, K., Sacconi, S., Ait-El-Mkadem, S., Paquis-Flucklinger, V., et al. (2018). MT-CYB Deletion in an Encephalomyopathy with Hyperintensity of Middle Cerebellar Peduncles. Neurol. Genet. 4 (5), e268. doi:10.1212/nxg.0000000000000268
Chen, Y., Lin, Y., Vithana, E. N., Jia, L., Zuo, X., Wong, T. Y., et al. (2014). Common Variants Near ABCA1 and in PMM2 Are Associated with Primary Open-Angle Glaucoma. Nat. Genet. 46, 1115–1119. doi:10.1038/ng.3078
Choquet, H., Paylakhi, S., Kneeland, S. C., Thai, K. K., Hoffmann, T. J., Yin, J., et al. (2018). A Multiethnic Genome-wide Association Study of Primary Open-Angle Glaucoma Identifies Novel Risk Loci. Nat. Commun. 9 (1), 2278. doi:10.1038/s41467-018-04555-4
Clement, E., Mercuri, E., Godfrey, C., Smith, J., Robb, S., Kinali, M., et al. (2008). Brain Involvement in Muscular Dystrophies with Defective Dystroglycan Glycosylation. Ann. Neurol. 64 (5), 573–582. doi:10.1002/ana.21482
Collins, D. W., Gudiseva, H. V., Trachtman, B., Bowman, A. S., Sagaser, A., Sankar, P., et al. (2016). Association of Primary Open-Angle Glaucoma with Mitochondrial Variants and Haplogroups Common in African Americans. Mol. Vis. 22, 454–471.
Cortopassi, G. A., and Arnheim, N. (1990). Detection of a Specific Mitochondrial DNA Deletion in Tissues of Older Humans. Nucl. Acids Res. 18 (23), 6927–6933. doi:10.1093/nar/18.23.6927
Davis, L. K., Meyer, K. J., Schindler, E. I., Beck, J. S., Rudd, D. S., Grundstad, A. J., et al. (2011). Copy Number Variations and Primary Open-Angle Glaucoma. Invest. Ophthalmol. Vis. Sci. 52 (10), 7122–7133. doi:10.1167/iovs.10-5606
Divi, R. L., Leonard, S. L., Kuo, M. M., Nagashima, K., Thamire, C., St Claire, M. C., et al. (2007). Transplacentally Exposed Human and Monkey Newborn Infants Show Similar Evidence of Nucleoside Reverse Transcriptase Inhibitor-Induced Mitochondrial Toxicity. Environ. Mol. Mutagen 48 (3-4), 201–209. doi:10.1002/em.20201
ENCODE Project Consortium (2012). An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 489 (7414), 57–74. doi:10.1038/nature11247
Ferguson, S. J., John, P., Lloyd, W. J., Radda, G. K., and Whatley, F. R. (1976). The ATPase as an Irreversible Component in Electron Transport Linked ATP Synthesis. FEBS Lett. 62, 272. doi:10.1016/0014-5793(76)80073-7
Gharahkhani, P., Burdon, K. P., Burdon, K. P., Fogarty, R., Sharma, S., Hewitt, A. W., et al. Wellcome Trust Case Control Consortium (2014). Common Variants Near ABCA1, AFAP1 and GMDS Confer Risk of Primary Open-Angle Glaucoma. Nat. Genet. 46, 1120–1125. doi:10.1038/ng.3079
Gharahkhani, P., Jorgenson, E., Jorgenson, E., Hysi, P., Khawaja, A. P., Pendergrass, S., et al. (2021). Genome-Wide Meta-Analysis Identifies 127 Open-Angle Glaucoma Loci with Consistent Effect across Ancestries. Nat. Commun. 12 (1), 1258. doi:10.1038/s41467-020-20851-4
Godfrey, C., Clement, E., Mein, R., Brockington, M., Smith, J., Talim, B., et al. (2007). Refining Genotype Phenotype Correlations in Muscular Dystrophies with Defective Glycosylation of Dystroglycan. Brain 130, 2725–2735. doi:10.1093/brain/awm212
Gómez-Durán, A., Pacheu-Grau, D., López-Gallardo, E., Díez-Sánchez, C., Montoya, J., López-Pérez, M. J., et al. (2010). Unmasking the Causes of Multifactorial Disorders: OXPHOS Differences between Mitochondrial Haplogroups. Hum. Mol. Genet. 19 (17), 3343–3353. doi:10.1093/hmg/ddq246
Gramer, E., and Grehn, F. (2012). Pathogenesis and Risk Factors of Glaucoma. Springer Science & Business Media.
Gupta, V., Kawahara, G., Gundry, S. R., Chen, A. T., Lencer, W. I., Zhou, Y., et al. (2011). The Zebrafish Dag1 Mutant: A Novel Genetic Model for Dystroglycanopathies. Hum. Mol. Genet. 20, 1712–1725. doi:10.1093/hmg/ddr047
Hamosh, A. (2004). Online Mendelian Inheritance in Man (OMIM), a Knowledgebase of Human Genes and Genetic Disorders. Nucleic Acids Res. 33, D514–D517. doi:10.1093/nar/gki033
He, Y., Leung, K. W., Zhang, Y.-H., Duan, S., Zhong, X.-F., Jiang, R.-Z., et al. (2008). Ru-Zhang Jiang, Zhan Peng, Joyce Tombran-Tink, and Jian Ge.Mitochondrial Complex I Defect Induces ROS Release and Degeneration in Trabecular Meshwork Cells of POAG Patients: Protection by Antioxidants. Invest. Ophthalmol. Vis. Sci. 49 (4), 1447–1458. doi:10.1167/iovs.07-1361
Heeg, G. P., Blanksma, L. J., Peter, L. L. J., Hardus, P. L. L. J., and Jansonius, N. M. (2005). The Groningen Longitudinal Glaucoma Study. I. Baseline Sensitivity and Specificity of the Frequency Doubling Perimeter and the GDx Nerve Fibre Analyser. Acta Ophthalmologica Scand. 83 (1), 46–52. doi:10.1111/j.1600-0420.2005.00423.x
Herrnstadt, C., Elson, J. L., Fahy, E., Preston, G., Turnbull, D. M., Anderson, C., et al. (2002). Reduced-Median-Network Analysis of Complete Mitochondrial DNA Coding-Region Sequences for the Major African, Asian, and European Haplogroups. Am. J. Hum. Genet. 70 (5), 1152–1171. doi:10.1086/339933
Ho, D. E., Imai, K., King, G., and Stuart, E. A. (2011). MatchIt: Nonparametric Preprocessing for Parametric Causal Inference. J. Stat. Soft. 42, 1–28. doi:10.18637/jss.v042.i08
Holt, K. H., Crosbie, R. H., Venzke, D. P., and Campbell, K. P. (2000). Biosynthesis of Dystroglycan: Processing of a Precursor Propeptide. FEBS Lett. 468 (1), 79–83. doi:10.1016/s0014-5793(00)01195-9
Howell, N. (1997). Leber Hereditary Optic Neuropathy: Mitochondrial Mutations and Degeneration of the Optic Nerve. Vis. Res. 37, 3495–3507. doi:10.1016/s0042-6989(96)00167-8
Howell, N. (2003). LHON and Other Optic Nerve Atrophies: The Mitochondrial Connection. Dev. Ophthalmol. 37, 94–108. doi:10.1159/000072041
Howie, B. N., Donnelly, P., and Marchini., J. (2009). A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-wide Association Studies. Plos Genet. 5 (6), e1000529. doi:10.1371/journal.pgen.1000529
Hudson, G., Carelli, V., Spruijt, L., Gerards, Mike., Mowbray, C., Achilli, A., et al. (2007). Clinical Expression of Leber Hereditary Optic Neuropathy Is Affected by the Mitochondrial DNA-Haplogroup Background. Am. J. Hum. Genet. 81 (2), 228–233. doi:10.1086/519394
Izzotti, A., Longobardi, M., Cartiglia, C., and Saccà, C. S. (2011). Title: Mitochondrial Damage in the Trabecular Meshwork Occurs Only in Primary Open-Angle Glaucoma and in Pseudoexfoliative Glaucoma. PLoS One 6 (1), e14567 doi:10.1371/journal.pone.0014567
Janssen, S. F., Gorgels, T. G., Ramdas, W. D., Klaver, C. C., van Duijn, C. M., Jansonius, N. M., et al. (2013). The Vast Complexity of Primary Open Angle Glaucoma: Disease Genes, Risks, Molecular Mechanisms and Pathobiology. Prog. Retin. Eye Res. 37, 31–67. doi:10.1016/j.preteyeres.2013.09.001
Jastrow, H., Koulen, P., Altrock, W. D., and Kröger, S. (2006). Identification of a β-Dystroglycan Immunoreactive Subcompartment in Photoreceptor Terminals. Invest. Ophthalmol. Vis. Sci. 47 (1), 17–24. doi:10.1167/iovs.05-0597
Kloss-Brandstätter, A., Pacher, D., Schönherr, S., Weissensteiner, H., Binna, R., Specht, G., et al. (2011). HaploGrep: a Fast and Reliable Algorithm for Automatic Classification of Mitochondrial DNA Haplogroups. Hum. Mutat. 32, 25–32. doi:10.1002/humu.21382
Koilkonda, R. D., and Guy, J. (2011). Leber's Hereditary Optic Neuropathy-Gene Therapy: From Benchtop to Bedside. J. Ophthalmol. 2011, 1–16. doi:10.1155/2011/179412
Lascaratos, G., Garway-Heath, D. F., Willoughby, C. E., Chau, K.-Y., Schapira, A. H. V., and Schapira, V. (2012). Mitochondrial Dysfunction in Glaucoma: Understanding Genetic Influences. Mitochondrion 12, 202–212. doi:10.1016/j.mito.2011.11.004
Liu, Y., Garrett, M. E., Yaspan, B. L., Bailey, J. C., Loomis, S. J., Brilliant, M., et al. (2014). DNA Copy Number Variants of Known Glaucoma Genes in Relation to Primary Open-Angle Glaucoma. Invest. Ophthalmol. Vis. Sci. 55 (12), 8251–8258. doi:10.1167/iovs.14-15712
Liu, Y., Hauser, M. A., Akafo, S. K., Qin, X., Miura, S., Gibson, J. R., et al. (2013). Investigation of Known Genetic Risk Factors for Primary Open Angle Glaucoma in Two Populations of African Ancestry. Invest. Ophthalmol. Vis. Sci. 54 (9), 6248–6254. doi:10.1167/iovs.13-12779
Lo Faro, V., ten Brink, J. B., Snieder, H., Jansonius, N. M., and Bergen, A. A. (2021). Genome-Wide CNV Investigation Suggests a Role for Cadherin, Wnt, and P53 Pathways in Primary Open-Angle Glaucoma. BMC Genomics 22 (1), 590. doi:10.1186/s12864-021-07846-1
LoogväliEva-Liis, E.-L., Roostalu, U., Malyarchuk, B. A., Derenko, M. V., Kivisild, T., Metspalu, E., et al. (2004). Disuniting Uniformity: A Pied Cladistic Canvas of mtDNA Haplogroup H in Eurasia. Mol. Biol. Evol. 21, 2012–2021. doi:10.1093/molbev/msh209
Lopez Sanchez, M. I. G., Crowston, J. G., Mackey, D. A., and Trounce, I. A. (2016). Emerging Mitochondrial Therapeutic Targets in Optic Neuropathies. Pharmacol. Ther. 165, 132–152. doi:10.1016/j.pharmthera.2016.06.004
Mancuso, M., and Klopstock, T. (2019). Diagnosis and Management of Mitochondrial Disorders. Springer.
McInerney, T. W., Fulton-Howard, B., Patterson, C., Paliwal, D., Jermiin, L. S., Patel, H. R., et al. (2021). A Globally Diverse Reference Alignment and Panel for Imputation of Mitochondrial DNA Variants. BMC Bioinformatics 22 (1), 417. doi:10.1186/s12859-021-04337-8
Montanaro, F., Carbonetto, S., Campbell, K. P., and Lindenbaum, M. (1995). Dystroglycan Expression in the Wild Type Andmdx Mouse Neural Retina: Synaptic Colocalization with Dystrophin, Dystrophin-Related Protein but Not Laminin. J. Neurosci. Res. 42, 528–538. doi:10.1002/jnr.490420411
MT-ND4 (2021). MT-ND4 Mitochondrially Encoded NADH Dehydrogenase 4 [Homo Sapiens (Human)] - Gene - NCBI. Availableat: https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=ShowDetailView&TermToSearch=4538 (Accessed July 5, 2021).
Neustaeter, A., Vehof, J., Snieder, H., and Jansonius, N. M. (2020). Glaucoma in Large-Scale Population-Based Epidemiology: A Questionnaire-Based Proxy. Eye 35, 508–516. doi:10.1038/s41433-020-0882-4
Osman, W., Low, S.-K., Takahashi, A., Kubo, M., and Nakamura, Y. (2012). A Genome-wide Association Study in the Japanese Population Confirms 9p21 and 14q23 as Susceptibility Loci for Primary Open Angle Glaucoma. Hum. Mol. Genet. 21, 2836–2842. doi:10.1093/hmg/dds103
Parton, M. J. (2003). Online Mendelian Inheritance in Man OMIM: www.ncbi.nlm.nih.Gov/entrez. J. Neurol. Neurosurg. Psychiatry 74, 703. doi:10.1136/jnnp.74.6.703
Pasquale, L. R. (2019). Genetic Insights into Primary Open-Angle Glaucoma, 301–305. doi:10.1007/978-981-13-2137-5_44
Pereira, L., Richards, M., Goios, A., Alonso, A., Albarrán, A., Garcia, O., et al. (2005). High-Resolution mtDNA Evidence for the Lateglacial Resettlement of Europe from an Iberian Refugium. Genome Res. 15 (1), 19–24. doi:10.1101/gr.3182305
Pihko, H., Lappi, M., Raitta, C., Sainio, K., Valanne, L., Somer, H., et al. (1995). Ocular Findings in Muscle-Eye-Brain (MEB) Disease: A Follow-Up Study . Brain Dev. 17 (1), P57–P61. doi:10.1016/0387-7604(94)00101-3
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 81, 559–575. doi:10.1086/519795
Qi, X., Lewin, A. S., Hauswirth, W. W., and Guy, J. (2003). Suppression of Complex I Gene Expression Induces Optic Neuropathy. Ann. Neurol. 53 (2), 198–205. doi:10.1002/ana.10426
Ramdas, W. D., van Koolwijkvan Koolwijk, L. M. E., Lemij, H. G., Pasutto, F., Cree, A. J., Thorleifsson, G., et al. (2011). Common Genetic Variants Associated with Open-Angle Glaucoma. Hum. Mol. Genet. 20 (12), 2464–2471. doi:10.1093/hmg/ddr120
Riemersma, M., Mandel, H., van Beusekom, E., Gazzoli, I., Roscioli, T., Eran, A., et al. (2015). Absence of - and -Dystroglycan Is Associated with Walker-Warburg Syndrome. Neurology 84, 2177–2182. doi:10.1212/wnl.0000000000001615
Ronchi, D., Cosi, A., Tonduti, D., Orcesi, S., Bordoni, A., Fortunato, F., et al. (2011). Clinical and Molecular Features of an Infant Patient Affected by Leigh Disease Associated to m.14459G>A Mitochondrial DNA Mutation: A Case Report. BMC Neurol. 11, 85. doi:10.1186/1471-2377-11-85
Sadun, A. A. (2002). Mitochondrial Optic Neuropathies. J. Neurol. Neurosurg. Psychiatry 72 (4), 423–425.
Schmitz, F., Holbach, M., and Drenckhahn, D. (1993). Colocalization of Retinal Dystrophin and Actin in Postsynaptic Dendrites of Rod and Cone Photoreceptor Synapses. Histochemistry 100 (6), 473–479. doi:10.1007/bf00267828
Scholtens, S., Smidt, N., Swertz, M. A., Bakker, S. J., Dotinga, A., Vonk, J. M., et al. (2015). Cohort Profile: LifeLines, a Three-Generation Cohort Study and Biobank. Int. J. Epidemiol. 44, 1172–1180. doi:10.1093/ije/dyu229
Schuelke, M., Krude, H., Finckh, B., Mayatepek, E., Janssen, A., Schmelz, M., et al. (2002). Septo-optic Dysplasia Associated with a New Mitochondrialcytochrome B Mutation. Ann. Neurol. 51 (3), 388–392. doi:10.1002/ana.10151
Shaffer, R. N. (1996). The Centennial History of Glaucoma (1896–1996). Ophthalmology 103, S40–S50. doi:10.1016/s0161-6420(96)30763-x
Shah, T. S., Liu, J. Z., Floyd, J. A. B., Morris, J. A., Wirth, N., Barrett, J. C., et al. (2012). OptiCall: A Robust Genotype-Calling Algorithm for Rare, Low-Frequency and Common Variants. Bioinformatics 28 (12), 1598–1603. doi:10.1093/bioinformatics/bts180
Singh, L. N., Crowston, J. G., Lopez Sanchez, M. I. G., Van Bergen, N. J., Kearns, L. S., Hewitt, A. W., et al. (2018). Mitochondrial DNA Variation and Disease Susceptibility in Primary Open-Angle Glaucoma. Invest. Ophthalmol. Vis. Sci. 59 (11), 4598–4602. doi:10.1167/iovs.18-25085
Springelkamp, H., Iglesias, A. I., Cuellar-Partida, G., Amin, N., Burdon, K. P., van Leeuwen, E. M., et al. (2015). ARHGEF12 Influences the Risk of Glaucoma by Increasing Intraocular Pressure. Hum. Mol. Genet. 24, 2689–2699. doi:10.1093/hmg/ddv027
Tham, Y.-C., Li, X., Wong, T. Y., Quigley, H. A., Aung, T., and Cheng, C.-Y. (2014). Global Prevalence of Glaucoma and Projections of Glaucoma Burden through 2040. Ophthalmology 121 (11), 2081–2090. doi:10.1016/j.ophtha.2014.05.013
Thorleifsson, G., Walters, G. B., Hewitt, A. W., Masson, G., Helgason, A., DeWan, A., et al. (2010). Common Variants Near CAV1 and CAV2 Are Associated with Primary Open-Angle Glaucoma. Nat. Genet. 42, 906–909. doi:10.1038/ng.661
Torroni, A., and Wallace, D. C. (1994). Mitochondrial DNA Variation in Human Populations and Implications for Detection of Mitochondrial DNA Mutations of Pathological Significance. J. Bioenerg. Biomembr 26 (3), 261–271. doi:10.1007/bf00763098
Trikha, S., Saffari, E., Nongpiur, M., Baskaran, M., Ho, H., Li, Z., et al. (2015). A Genetic Variant in TGFBR3-CDC7 Is Associated with Visual Field Progression in Primary Open-Angle Glaucoma Patients from Singapore. Ophthalmology 122, 2416–2422. doi:10.1016/j.ophtha.2015.08.016
Urzua-Traslavina, C. G., and Carlos, G. (2014). Relationship of Mitochondrial DNA Haplogroups with Complex Diseases. J. Genet. Genome Res. 1, 011. doi:10.23937/2378-3648/1410011
Van Bergen, N. J., Crowston, J. G., Craig, J. E., Burdon, K. P., Kearns, L. S., Sharma, S., et al. (2015). Measurement of Systemic Mitochondrial Function in Advanced Primary Open-Angle Glaucoma and Leber Hereditary Optic Neuropathy. PLoS ONE 10 (10), e0140919. doi:10.1371/journal.pone.0140919
van Oven, M., and Kayser, M. (2009). Updated Comprehensive Phylogenetic Tree of Global Human Mitochondrial DNA Variation. Hum. Mutat. 30 (2), E386–E394. doi:10.1002/humu.20921
Vincent, A. E., and Picard, M. (2018). Multilevel Heterogeneity of Mitochondrial Respiratory Chain Deficiency. J. Pathol. 246, 261–265. doi:10.1002/path.5146
Votruba, M. (2004). Molecular Genetic Basis of Primary Inherited Optic Neuropathies. Eye 18 (11), 1126–1132. doi:10.1038/sj.eye.6701570
Wallace, D. C. (2010). Mitochondrial DNA Mutations in Disease and Aging. Environ. Mol. Mutagen 51 (5), 440–450. doi:10.1002/em.20586
Wang, N. H.-H., Chen, S.-J., Yang, C.-F., Chen, H.-W., Chuang, H.-P., Lu, Y.-H., et al. (2016). Homozygosity Mapping and Whole-Genome Sequencing Links a Missense Mutation inPOMGNT1to Autosomal Recessive Retinitis Pigmentosa. Invest. Ophthalmol. Vis. Sci. 57, 3601. doi:10.1167/iovs.16-19463
West, A. P., Shadel, G. S., and Ghosh, S. (2011). Mitochondria in Innate Immune Responses. Nat. Rev. Immunolimmunology 11 (6), 389–402. doi:10.1038/nri2975
Wibrand, F., Ravn, K., Schwartz, M., Rosenberg, T., Horn, N., and Vissing, J. (2001). Multisystem Disorder Associated with a Missense Mutation in the Mitochondrial Cytochromeb Gene. Ann. Neurol. 50 (4), 540–543. doi:10.1002/ana.1224
Wiggs, J. L., Hee KangMirel, J., Yaspan, B. L., Mirel, D. B., Laurie, C., Crenshaw, A., et al. (2011). Common Variants Near CAV1 and CAV2 Are Associated with Primary Open-Angle Glaucoma in Caucasians from the USA. Hum. Mol. Genet. 20 (23), 4707–4713. doi:10.1093/hmg/ddr382
Willer, C. J., Li, Y., and Abecasis, G. R. (2010). METAL: Fast and Efficient Meta-Analysis of Genomewide Association Scans. Bioinformatics 26, 2190–2191. doi:10.1093/bioinformatics/btq340
Williams, P. A., Harder, J. M., Foxworth, N. E., Cochran, K. E., Philip, V. M., Porciatti, V., et al. (2017). Vitamin B 3 Modulates Mitochondrial Vulnerability and Prevents Glaucoma in Aged Mice. Science 355 (6326), 756–760. doi:10.1126/science.aal0092
Xu, M., Yamada, T., Sun, Z., Eblimit, A., Lopez, I., Wang, F., et al. (2016). Mutations inPOMGNT1cause Non-syndromic Retinitis Pigmentosa. Hum. Mol. Genet. 25 (8), 1479–1488. doi:10.1093/hmg/ddw022
Yoshida, A., Kobayashi, K., Manya, H., Taniguchi, K., Kano, H., Mizuno, M., et al. (2001). Muscular Dystrophy and Neuronal Migration Disorder Caused by Mutations in a Glycosyltransferase, POMGnT1. Develop. Cel 1, 717–724. doi:10.1016/s1534-5807(01)00070-3
Yu-Wai-Man, P., Griffiths, P. G., Hudson, G., and Chinnery, P. F. (2009). Inherited Mitochondrial Optic Neuropathies. J. Med. Genet. 46 (3), 145–158. doi:10.1136/jmg.2007.054270
Yu-Wai-Man, P., Griffiths, P. G., and Chinnery, P. F. (2011). Mitochondrial Optic Neuropathies - Disease Mechanisms and Therapeutic Strategies. Prog. Retin. Eye Res. 30 (2), 81–114. doi:10.1016/j.preteyeres.2010.11.002
Keywords: mitochondrial polymorphism, mitochondrial haplogroup, primary open-angle glaucoma (POAG), MT-CYB, MT-ND4, haplogroup K, genetic association study
Citation: Lo Faro V, Nolte IM, Ten Brink JB, Snieder H, Jansonius NM, Bergen AA and Lifelines Cohort Study (2021) Mitochondrial Genome Study Identifies Association Between Primary Open-Angle Glaucoma and Variants in MT-CYB, MT-ND4 Genes and Haplogroups. Front. Genet. 12:781189. doi: 10.3389/fgene.2021.781189
Received: 22 September 2021; Accepted: 29 November 2021;
Published: 16 December 2021.
Edited by:
Enrico Baruffini, University of Parma, ItalyReviewed by:
Mercedes Fernandez-Moreno, Institute of Biomedical Research of A Coruña (INIBIC), SpainCopyright © 2021 Lo Faro, Nolte, Ten Brink, Snieder, Jansonius, Bergen and Lifelines Cohort Study. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arthur A. Bergen, YWFiZXJnZW5AYW1zdGVyZGFtdW1jLm5s
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.