- 1Department of TCM, Tianjin University of TCM, Tianjin, China
- 2Acupuncture Department, The First Affiliated Hospital of Tianjin University of TCM, Tianjin, China
- 3Department of Cardiology, The Second Affiliated Hospital of Tianjin University of TCM, Tianjin, China
Myocardial infarction (MI) is a complicated pathology triggered by numerous environmental and genetic factors. Understanding the effect of epigenetic regulation mechanisms on the cardiovascular disease would advance the field and promote prophylactic methods targeting epigenetic mechanisms. Genetic screening guides individualised MI therapies and surveillance. The present review reported the latest development on the epigenetic regulation of MI in terms of DNA methylation, histone modifications, and microRNA-dependent MI mechanisms and the novel therapies based on epigenetics.
1 Introduction
Myocardial infarction (MI) is a common cause of global morbidity and mortality and has caused nearly half of all deaths across Europe, more than 24 million deaths in the United States, and approximately 7–18% of the global 1-year mortality (Nichols et al., 2014; Reed et al., 2017). Atherosclerosis develops in the younger age group and persists for several decades, resulting in MI or other lethal cardiovascular diseases such as heart failure, stroke, and sudden death (Weintraub et al., 2011). Lifestyles changes and effective therapeutic strategies such as diet, abstinence from cigarettes and alcohol, percutaneous coronary intervention, and coronary artery bypass graft resulted in a considerable reduction in MI-induced mortality (Cokkinos and Pantos, 2007; Nichols et al., 2014; Xue et al., 2021). Although patients with MI increased, the life expectancy of patients is almost unaffected (Shibata et al., 2015; Thygesen et al., 2018). A New England Journal of Medicine study reported no evidence comparing an initial invasive strategy to an initial conservative approach to decrease cardiovascular risk or deaths in a median of 3.2 years (Maron et al., 2020). Thus, finding novel biomarkers is necessary for early MI detection. Patients with coronary artery disease (CAD) or MI repeatedly exhibited positive family history (Mayer et al., 2007). Familial genetic defects with an autosomal dominant form resulted in MI in humans (Wang et al., 2003). MI is a complex disease involving both environmental and genetic factors and their interactions. Genetic polymorphisms for numerous genes through atherosclerosis, inflammation and thrombogenesis pathways may account for the susceptibility to MI and severe CAD consequences (Chen et al., 2007). With improved resequencing technology, the gene identification and confirmation methodology can be used for reference in CAD. This enables researchers to better quantify CAD risk in early life, formulate more efficient therapeutic approaches, and reduce the individual probability of developing MI (Damani and Topol, 2007). Genetic linkage studies were performed in both human and animal models to identify these gene polymorphisms. Several genetic association studies have revealed numerous genes and biological pathways (Newton-Cheh and O’Donnell, 2004).
Research work on epigenetics and epigenomics in this field has made remarkable progress and attracted numerous geneticists, molecular biologists, oncologists, and cardiologists. Advancements in epigenetic areas have offered a fresh perspective on human diseases and ushered a new era in genomics by exploring the role of environmental interaction and genetic heritability in disease pathology (Portela and Esteller, 2010; Cao et al., 2014). The heritability of cardiovascular diseases such as myocardial infarction can vary depending on sex, age, and environmental and lifestyle conditions (Saban et al., 2014; Dorn and Matkovich, 2015; Sen et al., 2016; Cavalli and Heard, 2019; Asllanaj et al., 2020; Deegan et al., 2021). Epigenetics is currently a popular biological research area. The term “epigenetics” generally suggests all genetic variations of gene expression regulation except nucleotide sequence and chromatin organisation depending on DNA sequences (Egger et al., 2004; Abi Khalil, 2014). Thus, epigenetic mechanisms related to gene expression regulation are chromatin-based and not involving any DNA sequence changes (Cao et al., 2014). Epigenetic inheritance is a critical mechanism that maintains the dynamic and stable propagation of gene activity states from cells of the last generation to those of the following generation (Kim, 2013; Abi Khalil, 2014).
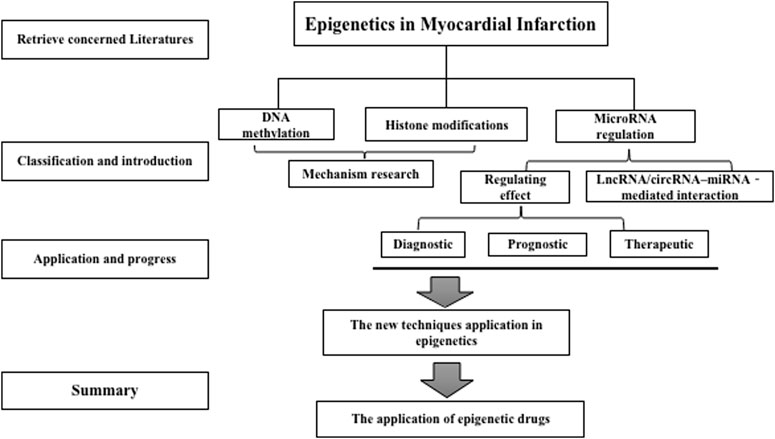
Epigenetic regulatory processes encompass diverse molecular mechanisms such as DNA methylation (DNAm), histone post-translational modifications, and RNA-based mechanisms such as long non-coding RNAs, lncRNAs, and microRNAs (Kim et al., 2009). In several cases, the epigenetic changes reflect responses to environmental and lifestyle factors, resulting in persistent dynamic changes in gene expression that affect the course of cardiovascular disease. Epigenetic regulators have been increasingly targeted in cancer therapeutics. Thus, epigenetic regulatory mechanisms for cancer and CAD must be explored and are significant in the oncology and cardiology fields (Ding et al., 2018). The epigenome expression can fundamentally differ from different cell types, possibly modulating single cell gene expression by organising nuclear architecture in chromosomes, suppressing or promoting transcription factor access to DNA, and regulating gene expression (Wang and Chang, 2018). Epigenetics dysregulation is considered the cause of many human disorders, such as severe cardiovascular diseases, due to the significance of differential gene regulation in cellular differentiation and application function (Abi Khalil, 2014; Yang et al., 2015; Prasher et al., 2020). Through the search of PubMed, we have summarized a large amount of literature related to epigenetics, aiming to gain insight into their potential application in MI (In Figure 1 -Flow Chart). The present review focused on the crucial role of epigenetic regulatory mechanisms in MI.
2 DNA Methylation and MI
Normal DNA methylation, among the central mechanisms regulating gene expression, can decide a severe cardiovascular event (Nurnberg et al., 2020). However, aberrant DNAm with genome-wide hypomethylation and CpG island hypermethylation is also observed in CAD (Breton et al., 2014; Sharma et al., 2014). The development of epigenetic epidemiology increases the probability to investigate the correlations of genomic coding, modifiable exposures, and disease phenotype manifestations. As a vital epigenetic modification type, DNAm plays a significant role as a potential mechanism of such correlations (Zhong et al., 2016). DNA methylation represents a pre-transcriptional modification that can alter the transcriptional process by adding methyl groups onto specific DNA nucleotides (Ma et al., 2014). The process leads to inactive gene expression because the methyl binding protein binds transcriptional factors and DNA. Hypomethylation is more common in most diseases than hypermethylation (Movassagh et al., 2011). DNAm, together with genetic mechanisms, is crucial for natural evolution and maintenance under specific gene expression patterns among mammals. Simultaneously, it is also dynamic and reversible for the regulating effect of genetic mechanisms (Fardi et al., 2018). DNAm pattern changes and the resulting differentially methylated regions have focused on numerous studies on normal development and disease (Ziller et al., 2013). Several studies report the value of epigenetic processes as disease biomarkers, with multiple studies associating DNAm with clinical events (Locke et al., 2019).
DNAm is the most promising target for accurate diagnosis, prognosis, and treatment (Koch et al., 2018). A lack of DNAm markers has been successfully translated into clinical applications. However, the recent improvements in DNA sequencing and other molecular biomedical science technologies result in DNA methylation-based biomarkers exhibiting colossal potential for transforming the treatment and observation of diseases like MI and cancer (Gallardo-Gómez et al., 2018). Several reports reported the role of DNAm in regulating cardiovascular risk factors and myocardial protection in MI, particularly those related to lipid metabolism and myocardial protection against ischaemia (for the detailed description, Table 1). Talens et al. (2012) reported that the risk and developmental components of MI in women are linked to DNA methylation marks at specific loci that were earlier sensitive to prenatal conditions. Aldehyde dehydrogenase 2 (ALDH2) is crucial for protection against myocardial ischaemia. Regulatory T (Treg) cells have been shown to play a protective role in experimental atherosclerosis. Demethylation of the DNA encoding the transcription factor forkhead box P3 (FOXP3) was found to be essential for the stable maintenance of the suppressive properties of Tregs. Lei et al. (Jia et al., 2013)demonstrated that reduction in Treg cells is associated with ACS in atherosclerotic patients. Epigenetic suppression of FOXP3 might lead to down-regulation of Treg cells, and in turn increase the risk of ACS. The Notch pathway plays a key role in stimulating mammalian cardiomyocyte proliferation during development and in the early postnatal life; Analysis of Notch-responsive promoters in adult cardiomyocytes showed marks of repressed chromatin and irreversible CpG DNA methylation (Felician et al., 2014). Wang et al. (2015) proved the correlation between aberrant hypermethylation at CpG sites in animal models in ALDH2 promoter upstream sequence and myocardial ischaemia injury that possibly lead to ALDH2 downregulation after MI. The modulative effects of DNAm on cardiac function, carcinogenesis, and recovery after ischaemic injury, thrombosis, and altered endothelial function in patients with MI have also been investigated. DNA methylation significantly changes following MI. The gene expression proves the correlation of cardiac injury-related epigenetic changes with branched-chain amino acid metabolism (Ward-Caviness et al., 2018). Mathias et al. (Rask-Andersen et al., 2016) observed more than a hundred significant genes for MI pathogenesis or recovery. Similarly, Farzana et al. (Yousuf et al., 2020) reported that hypermethylation of the ABO gene promoter seemingly increases the AMI risk in the hospitalised MI population. Otherwise, gestational diabetes mellitus induced offspring cardiac oxidative stress and DNA hypermethylation, resulting in an epigenetic down-regulation of Sirt1 gene and aberrant development of heart ischemia-sensitive phenotype, which suggests that Sirt 1-mediated signaling is the potential therapeutic target for the heart ischemic disease in offspring (Chen et al., 2019). Not only that, Diabetes increases the vulnerability of STEMI patients to post-MI HF by down-regulating SLN promoter methylation, which further regulates SERCA2a activity via increasing cardiac SLN expression (Liu Z. et al., 2020). Interleukin-6 (IL-6) is implicated in the pathogenesis of coronary heart disease, and IL-6 expression has associated with the level of DNA methylation of its gene promoter. There are two findings suggest that an increase in IL-6 gene expression and its DNA hypomethylation promoter are associated with acute myocardial infarction and CABG surgery patients (Zuo et al., 2016; Mohammadpanah et al., 2020).
Studies have revealed the complementary expression patterns of lipid metabolism, calcium regulation, and methylation of related genes in peripheral blood leucocyte samples of patients with MI (Agha et al., 2019). These CpGs sites and genes stress the correlation of ion regulation, lipid metabolism, and inflammation in the MI biological mechanisms (Fernández-Sanlés et al., 2021). Thus, the new DNA methylation sequencing technology can identify potential target sites related to the aberrant epigenetic regulation of MI (Koseler et al., 2020). Additionally, the sites stated by two studies (Guarrera et al., 2015; Nakatochi et al., 2017) are candidates for further assessment as underlying MI biomarkers. These results exhibited that DNA methylation could be used as a major molecular process linking genetic variations to MI susceptibility.
3 Histone Modifications and HDACs in MI
Histone modification is the primary mechanism in epigenetic regulation, including post-transcriptional modifications, and the most common modifications are phosphorylation, acetylation, methylation, and ubiquitination (Tingare et al., 2013). Such post-transcriptional modifications exert vital biological functions on multiple cellular processes such as cell cycle and metabolism control, DNA repair, and gene transcription (Tang and Zhuang, 2019). Histone deacetylases (HDACs) belonging to transcriptional regulators can serve as a post-translational modifier with different cardiac pathophysiology roles. The basic experiment exhibited that HDAC inhibitors benefit against arrhythmia, MI, cardiac remodelling, hypertension, and fibrosis (Eom and Kook, 2014). Additionally, HDACs are strongly associated with other vascular disorders such as neointima formation, atherosclerosis, and vascular calcification (McKinsey, 2011). Zhang L. et al. (2018) reported acute HDAC effects as positive and negative regulators for pathological cardiac remodelling. Wang J. et al. (2020) uncovered the histone modification profile in the early stage of MI of mice and proved that the modulation of histone modifications could involve inflammation and angiogenesis through adjusting promoters and super enhancers and joining cardiac remodelling pathological processes.
Additionally, the protective effect and therapeutic potential of HDACs were verified by cardiac disease pathogenesis, including suppressing cardiac fibrosis; enhancing angiogenesis; preventing electrical remodelling; and regulating apoptosis, autophagy, and cell cycle arrest (Chun, 2020). Studies exhibited that HDAC enzyme suppression has become a potential candidate for decreasing reperfusion impairment (Xie et al., 2019). Ting et al. (Zhao et al., 2007) reported the use of trichostatin A (TSA) as an efficient HDAC inhibitor to imitate early pharmacologic preconditioning. TSA significantly improved post-ischaemic ventricular function recovery and reduced infarct size during early and delayed preconditioning.
The vital role of HDACs in CVD was greatly emphasized in the past. However, few studies have focused on the association between MI and HDACs (Chen X. et al., 2020). Several studies described the effect of HDACs on vascular dysfunction and MI. Although HDAC could prevent the pathological process of MI in most cases, some HDACs might exacerbate it. Thus, the present study summarised the mechanism and treatment with HDACs, discussed the use of available medicine, and suggested a direction for future clinical studies. The fundamental mechanisms of HDAC action include induction of cardiomyocyte autophagy, augmentation of cardiac remodelling, enhancement of myocardial repairs, and improvement of myocardial ischaemic injury. HDAC inhibitor prevented post-MI cardiac remodelling and depended upon the recovery of autophagosome processing for cardiac fibroblasts. Both clinical trials and animal studies indicated that the HDAC inhibitor TSA could reverse hypoxia-induced impaired autophagic flux and resulted in a 40% reduction in cell death (Wang Y. et al., 2018). Another anticancer, HDAC inhibitor SAHA decreased the myocardial infarct size in an animal model by autophagic flux induction (Xie et al., 2014). HDAC suppression facilitated cardiac repairs and neovascularisation of the infarcted myocardium. Zhang et al. (2012a) proved that c-kit + cardiac stem cell (CSC) preconditioning through HDAC inhibition with trichostatin could substantially increase c-kit + CSC-derived myocytes and microvessels and reinforce in vivo functional recovery of MI. However, it is still unclear if specific HDAC4 suppression can modulate CSCs to promote myocardial repair and maintain cardiac performance. HDAC inhibition facilitated c-kit + CSCs to be differentiated into cardiac lineage commitments in vitro, whereas HDAC4 overexpression weakened c-kit + CSC-derived cardiogenesis (Zhang et al., 2014).
Additionally, some studies reported that gut microbiota possibly affected the post-MI acetylation levels and tissue repair by influencing butyric acid production (Song et al., 2021). These results prove the role of HDAC4 inhibition in promoting CSC-derived cardiac regeneration and improving cardiac function recovery. Zhang et al. (2018c) demonstrated for the first time that transgenic HDAC overexpression is crucial for the regulation of cardiac function and remodelling.
Although HDAC activation could serve as a regulator of cardiac function in MI, activated HDAC overexpression augmented remodelling. Santhosh et al. (Mani et al., 2015) proved that HDAC inhibition could stimulate myogenesis and angiogenesis under an incubated embryonic stem cell model. HDAC inhibition prevents cardiac remodelling through the stimulation of endogenous regeneration. Additionally, HDAC inhibition improved post-MI myocardial functional recovery through the prevention of myocardial remodelling and a decrease in myocardial and serum tumour necrosis factor α (Zhang et al., 2012b). Thus, HDAC inhibition maintains cardiac performance and relieves myocardial remodelling through the simulation of endogenous cardiac regeneration. Lin et al. (2020) proved that HDAC inhibition could stimulate proteasome-dependent degradation of HDAC4, which may be related to HDAC4 sumoylation to provoke such protective effects. Du et al. (2015) discovered that HDAC inhibition avoids cell death, promotes cell-viability, and decreases ROS production and apoptosis of cardiomyocytes under exposure to H/R. These studies offer a novel understanding of the molecular mechanism of HDAC inhibition and the potential development of specific HDAC inhibitors as new MI therapies.
4 Non-coding RNAs and MI
Although more than 90% of human genomes cannot encode proteins, they exhibit high transcriptional activity and generate a broad spectrum for non-coding RNAs having regulatory and structural functions (Mattick et al., 2010). MicroRNAs (miRNAs), small interference RNAs (siRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) exert regulatory functions or diagnostic potential against CVDs (Poller et al., 2018). All non-coding RNAs are MI biomarkers (Wang and Jing, 2018). The present study also analysed these ncRNAs and associated interactions in regulating cardiomyocyte apoptosis, inflammation, angiogenesis, and fibrosis following the acute setting to understand their potential in acute MI treatment (Guo Y. et al., 2017; Poller et al., 2018). The present review summarises the latest advances and future applications for non-coding RNAs as MI biomarkers and focuses on the diagnostic value, prognostic potential and therapeutic effect in such RNAs. Several animals and clinical studies demonstrated the diagnostic value, prognostic potential and therapeutic effect for MI associated miRNAs.
4.1 miRNAs and MI
4.1.1 miRNAs as Diagnostic Biomarkers of MI
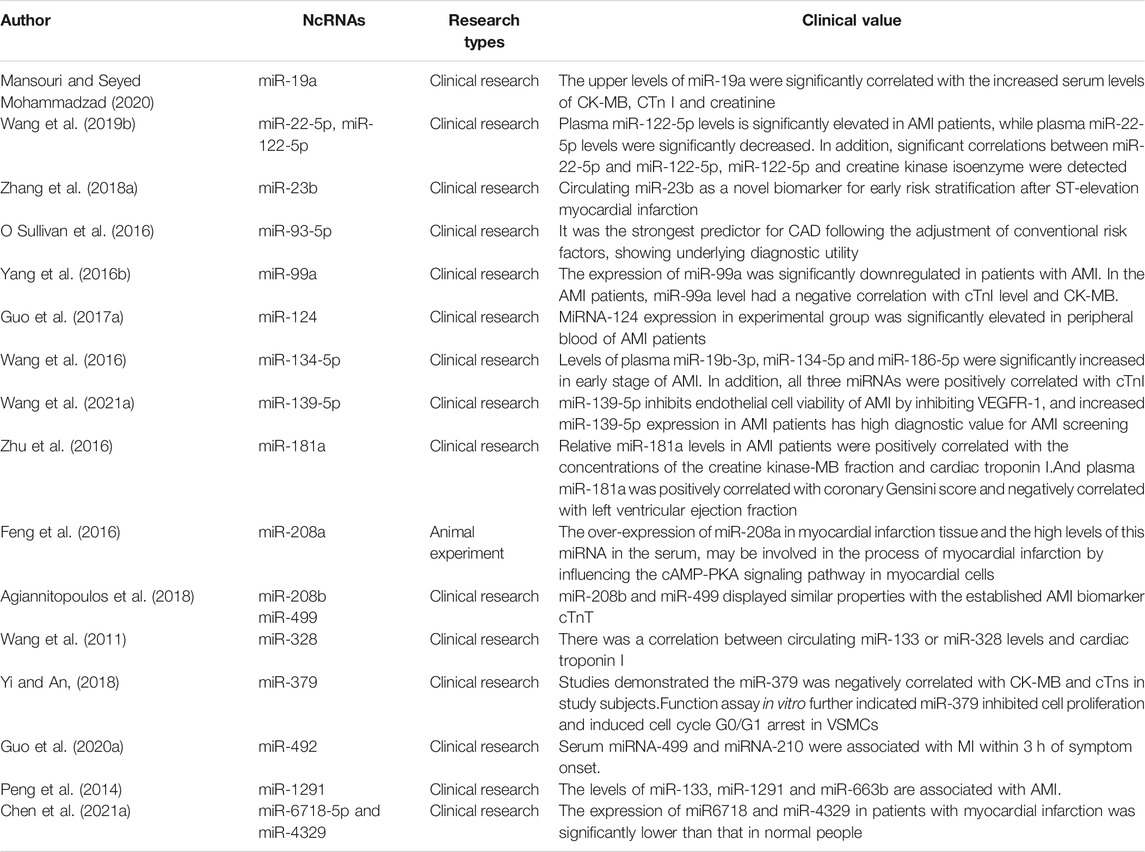
Despite the difference in sensitivity and accuracy among circulating miRNAs, some new circulating miRNAs containing unique release kinetics can be used as promising candidates for acute MI diagnostic biomarkers. Most of the circulating miRNAs could function as diagnostic biomarkers of acute MI (In Table2). In AMI patients, the upper levels of miR-19a (Mansouri and Seyed Mohammadzad, 2020), miR-22-5p,miR-122-5p (Wang Y. et al., 2019), miR-23b (Zhang J. et al., 2018), miR93-5p (O Sullivan et al., 2016), miRNA-124 (Guo ML. et al., 2017), miR-134-5p, miR-186-5p (Wang et al., 2016), miR-139-5p (Wang C. et al., 2021), miR-181a (Zhu et al., 2016),miR-208b, miR-499 (Agiannitopoulos et al., 2018),miR-328, miR-492 (Guo LL. et al., 2020), miR-1291and miR-663b (Peng et al., 2014) were significantly correlated with the increased serum levels of CK-MB and cTnI. On the contrary, the level of miR-99a (Yang SY. et al., 2016), miR-379 (Yi and An, 2018), miR6718 and miR-4329 (Chen S. et al., 2021) had a negative correlation with cTnI level and CK-MB in the AMI patients. Besides, miR-139-5p inhibited endothelial cell viability of AMI by inhibiting VEGFR-1, and increased miR-139-5p expression in AMI patients has high diagnostic value for AMI screening (Wang C. et al., 2021). Correlation analysis showed that plasma miR-181a was positively correlated with coronary Gensini score and negatively correlated with left ventricular ejection fraction. Relative miR-181a levels in AMI patients were positively correlated with the concentrations of the creatine kinase-MB fraction and cardiac troponin I (Zhu et al., 2016). The over-expression of miR-208a in myocardial infarction tissue and the high levels of this miRNA in the serum, may be involved in the process of myocardial infarction by influencing the cAMP-PKA signaling pathway in myocardial cells (Feng et al., 2016). At the same time, some circulating miRNAs were used for ischaemic risk stratification (He et al., 2017; Hromadka et al., 2021) prediction of the major adverse cardiovascular events after AMI future occurrence rate of MACE (Liu et al., 2017; Guo X. et al., 2020) or prognostic value of left ventricular (LV) dysfunction and symptoms of heart failure following acute MI (Maciejak et al., 2018). These results verified that constructing a complete network for circulating miRNAs after MI allows rapid MI diagnosis and opens novel therapeutic opportunities of MI, thus providing personalised therapies for patients at MI risk (Wang and Jing, 2018).
4.1.2 The Prognostic Value of MI Associated miRNAs
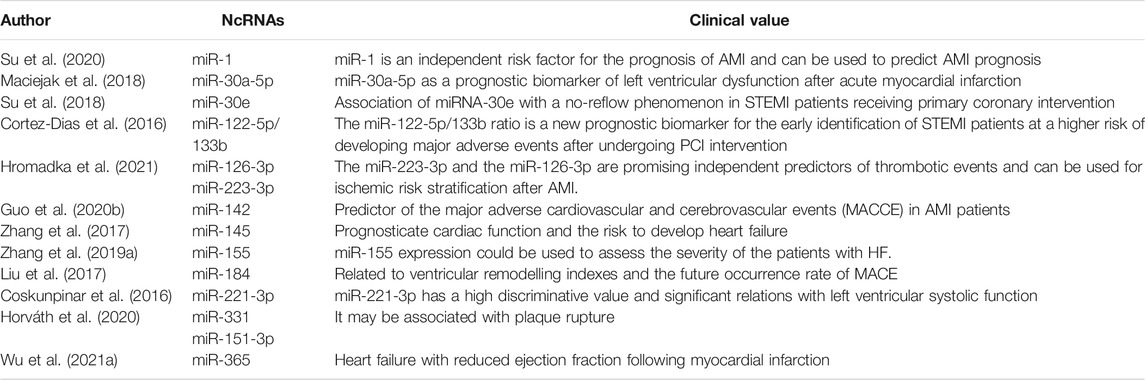
The prognosis prediction of myocardial infarction is beneficial to delay the progression of heart failure, reduce the mortality of cardiovascular events, and prolong the survival time of patients. We found that mRNAs could be used not only as an independent factor of cardiovascular risk events, but also as a predictor of the development of myocardial infarction (In Table 3). According to the literature, it has been confirmed that miR-1 (Su et al., 2020), miR-30a-5p (Maciejak et al., 2018), miR-223-3p and miR-126-3p (Hromadka et al., 2021) can be used to predict AMI prognosis after MI. Numerous clinical researches in MI patients identified miR-30e (Su et al., 2018), miR-142 (Guo X. et al., 2020), miR-184 (Liu et al., 2017) and miR-221-3p (Coskunpinar et al., 2016), in particular, as the most significantly changing miRNAs in MI, miR-142 and miR-184 over-expression analysis showed that aberrant their levels effect the future occurrence rate of MACE and the function of cardiovascular. Furthermore, miR-145 (Zhang et al., 2017), miR-155 (Zhang B. et al., 2019) and miR-365 (Wu H.-B. et al., 2021) expression also could be used to assess the severity of the patients with HF and prognosticate cardiac function and the risk to develop heart failure.
4.1.3 The Therapeutic Application of MI Associated miRNAs
Cardiac injury was accompanied by dynamic changes in the expression of miRNAs (In Table 4). Related studies have successively reported on the therapeutic effect of MI in patients with myocardial infarction. Li S. et al. (2018) found that downregulation of phosphatase and tensin homolog (PTEN), by the PTEN inhibitor bpV, increased miRNA-23a expression and suppressed the Bax/Bcl-2 protein expression ratio, caspase-3 activity level and p53 protein expression. It indicated that the expression of miRNA-23a may regulate AMI through targeting PTEN in patients and in vitro. Studies (Bonauer et al., 2009) have shown that the miR-17approximately92 cluster is highly expressed in human endothelial cells and that miR-92a, a component of this cluster, controls the growth of new blood vessels. Besides, miR-92a appears to target mRNAs corresponding to several proangiogenic proteins, including the integrin subunit alpha5. It may serve as a valuable therapeutic target in the setting of ischemic disease.
In animal and cell experiments, there are three miRNAs have been shown to potentially treat MI, including miR-144, miRNA-532 and miR-539 (Bayoumi et al., 2017; Hui et al., 2017; Li J. et al., 2018). Interestingly, miR-144 provides potent acute cardioprotection in an ischemia/reperfusion injury model and Intravenous miR-144 has potent effects on post-MI remodeling. MiRNA-532 protects the heart in acute myocardial infarction, and represses prss23, a positive regulator of endothelial-to-mesenchymal transition. Overexpression of miR-539 plays a role in the degree of myocardial infarction. The results of experiments demonstrated an increase in the expression of miR-539 and a decrease in the expression of MEK, which led not only to suppressed proliferation but also to apoptosis and autophagy of H9C2 cells. Although other miRNAs also have been proposed to have anti-myocardial infarction effects, it still needs further experimental verification (Xiang and Yang, 2020; Zhu et al., 2021b; Hu et al., 2021).
4.2 Regulation of Fibrosis in Infarct Regions by MiRNAs
Major processes leading to post-infarction injury and following remodelling responses are controlled by miRNAs. For example, miRNAs may assist or prohibit cardiomyocyte cell necrosis, modulate post-ischaemic neovascularisation, and control cardiac fibrosis (In Figure 2A).
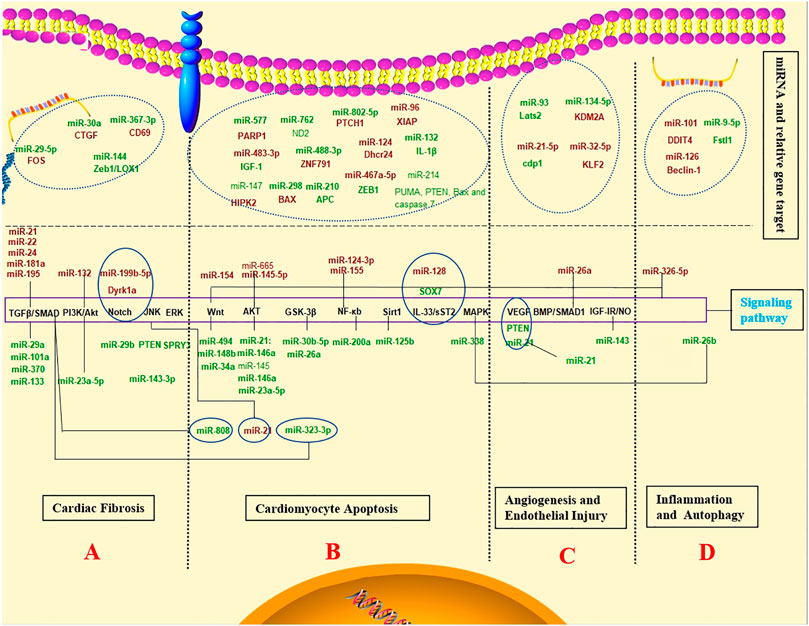
FIGURE 2. The diagnostic value, prognostic potential and therapeutic effect for MI associated miRNAs. Schematic representation of the activated pathway program in MI. The regulation mechanism of miRNAs networks in MI. (Red and green labels correspond with induced or repressed molecules in MI, respectively).
Some non-beneficial miRNAs regulate cardiac fibrosis to promote remodelling. miR-21, miR-22, miR-24, miR-133, miR-181a, and miR-195 can be upregulated in response to MI and are involved in cardiac fibrosis by tumour growth factor-β (TGF-β) signalling pathway (Hong et al., 2016; Chen et al., 2017; Yuan et al., 2017, 7; Chen P. et al., 2018; Yu et al., 2019; Wang DM. et al., 2020). On the other hand, miR-29a, miR-101a, and miR-370 inhibition protect against cardiac injury following MI (Xiao et al., 2017; Yuan and Gao, 2017). Similarly, miR-29b-3p degraded the pro-fibrosis effect from TGF-β1 through FOS targeting, which provided a promising therapy for post-MI cardiac fibrosis (Xue et al., 2020). CTGF expression was possibly inhibited by MiR-30a through direct combination with the 3′-UTR site of CTGF following MI, reducing collagen generation in myocardia, inhibiting myocardial fibrosis, and improving cardiac function (Chen L. et al., 2018). Yuan et al. (Yuan et al., 2019) discovered the involvement of miR-144 in extracellular matrix remodelling after MI, in which its loss results in enhanced myocardial fibrosis and damaged functional recovery. In animal experiments, miR-29b and miR-199b-5p have been inhibited myocardial fibrosis and cardiac hypertrophy by activating the Notch signaling pathway and protected myocardium against myocardial infarction (Duygu et al., 2017; Liu et al., 2019). Additionally, miR-143-3p and miR-494 promoted fibrosis through different signalling pathways such as ERK, JNK, and Wnt pathways (Li C. et al., 2019; Su et al., 2019). The regulation of these miRNAs can provide novel therapies for MI.
4.3 miRNAs in Cardiomyocyte Apoptosis
The relevant miRNA mechanisms in cardiomyocyte apoptosis are summarised in In Figures 2B. The Wnt/β-catenin and PI3K/AKT pathways constitute two major signalling pathways to inhibit apoptosis, which can be constantly activated by activating the pro-apoptotic pathway following acute MI. Some miRNAs protect cardiomyocytes from apoptosis following acute MI through activation of Wnt/β-catenin and PI3K/AKT pathways and their downstream regulators. MiR-30b-5p participates in myocardial cell proliferation and apoptosis by modulation of the Wnt/β-catenin signalling pathway, possibly providing the new underlying target to diagnose MI in the clinic (Chi et al., 2020). MiR-34a affects myocardial cell apoptosis by regulating the activation and inactivation of the Wnt/β-catenin signalling pathway (Li JH. et al., 2019). The miR-148b inhibition reinforced the antioxidative capacity and myocardial cell survival to inhibit apoptosis by activating the Wnt/β-catenin signalling pathway, improving myocardial I/R injury (Yang et al., 2019). Moreover, miR-154 can activate the Wnt/β-catenin signalling pathway, eventually promoting myocardial apoptosis (Sun HY. et al., 2019). Additionally, miR-23a-5p-PI3K/Akt axis regulated apoptosis in MI. Thus, the new axis was incorporated as an underlying indicator for detecting ischaemic heart disease and therapeutic intervention (Huang J. et al., 2020). Furthermore, the apoptosis-associated protein expression levels rose significantly in H9c2 cells transfected with miR-145-5p mimic. MiR-145-5p may inactivate the PI3K/Akt pathway to assist MI cell apoptosis (Huangfu et al., 2020). Other miRNAs can also promote or inhibit myocardial apoptosis after acute MI via different pathways and relevant targets. The miRNA-21 expression experienced upregulation in the serum of elderly patients with acute MI, which suppressed TNF-a caused apoptosis in HCM through activation of the JNK/p38/caspase-3 signalling pathway (Wang Z.-H. et al., 2017). MiR-26a activates the GSK-3β signalling pathway to inhibit myocardial cell apoptosis after acute MI (Lu and Lu, 2020). MiRNA-145 suppresses myocardial infarction-induced apoptosis through autophagy-related to the Akt3/mTOR signalling pathway in vivo and in vitro (Yan et al., 2018). MiR-214 and miR-203 have abilities to alleviate MI-caused injury on myocardium tissues and reduce mitochondria-mediated apoptosis, which might be a possible mechanism in protecting against AMI injury (Yang X. et al., 2016; Zhang J. et al., 2019). Exosomal miR-338 can inhibit cardiomyocyte apoptosis and improve cardiac function in rats suffering MI by regulating the MAP3K2/JNK signalling pathway (Fu et al., 2020).
Conversely, parts of miRNAs could promote myocardial apoptosis. MiR-96 facilitated acute MI progression by directly targeting XIAP and suppressing XIAP anti-apoptotic function, providing a new therapeutic target to treat acute MI (Wang J. et al., 2021). A mice model exhibited an increase in MI by miR-467a-5p through ZEB1 expression regulation (Huang et al., 2021). miR-665 downregulation protected from cardiomyocyte ischaemia/reperfusion injury-induced ROS accumulation and apoptosis by activating Pak1/Akt signalling of MI (Liu C. et al., 2020).
4.3.1 MiRNAs in Angiogenesis and Endothelial Injury
Numerous clinical studies have attempted to stimulate angiogenesis to combat ischaemic pathologies and tissue injury. These studies have primarily focused on the intra-arterial introduction of a range of angiogenic growth factors such as VEGF (Yang F. et al., 2016), insulin-like growth factor 1 receptor (IGF) (Geng et al., 2020), and HGF (Fan et al., 2018) to promote neovascularisation and tissue perfusion in subjects with MI. Additionally, Liao et al. (Liao et al., 2021) observed that cardiac telocyte suppressed cardiac microvascular endothelial cell apoptosis by exosomal miRNA-21-5p-targeted Cdip1 silencing to ameliorate angiogenesis of MI. Another study from China demonstrated that miR-134-5p silencing facilitated myocardial angiogenesis and suppressed myocardial apoptosis through KDM2A upregulation in MI mice (Li X. et al., 2020). MiR-93 may promote angiogenesis and weaken remodelling by inactivating the Hippo/Yap pathway through Lats2 targeting (Ma et al., 2020). Endothelial injury is crucial for numerous physiological processes and is closely related to tissue repair and recovery after an injury caused by pathological conditions (Icli et al., 2020). Cellular and molecular mechanisms can assist the formulation of novel cardiac cell therapies for the functional and structural regeneration of impaired myocardium. (In Figure 2C).
4.3.2 MicroRNAs Regulate Inflammation and Autophagy
Autophagy is a well-organised homeostatic cellular process responsible for removing damaged organelles and intracellular pathogens. Furthermore, it can modulate the innate and adaptive immune systems and suppress gene expression by targeting messenger RNAs for translational repression. The present study summarised the regulation of different non-coding RNAs in autophagy and other mechanisms (In Figure 2D). Several studies indicated that miRNAs regulate autophagy through different pathways and exhibit a significant influence on MI treatment. Li Q. et al. (2020) referred to MI attenuated by miR-101 induced injury through targeting DDIT4 to modulate autophagy, which implicated miR-101 or DDIT4 as targets for MI. Likewise, miR-126 downregulation will lead to the overactivation of myocardial autophagy induced by Beclin-1, an autophagy-related protein (Shi et al., 2020). MiR-21 suppresses the inflammatory responses in the early phase of MI through targeting KBTBD7 and attenuating MKK3/6 activation of immune cells, thus avoiding excessive scar formation and improving cardiac function (Yang et al., 2018, 7). One study found that miR-26b alleviates inflammatory response and myocardial remodelling in mice with MI by suppressing the MAPK pathway by binding to PTGS2 (Ge et al., 2019). These inflammatory and autophagy miRNAs might be potent therapeutic targets in the setting of MI.
4.4 LncRNA/circRNA–miRNA-Mediated Interaction
The development of human genome sequencing and annotation technologies has indicated that the human genome comprises numerous non-coding lncRNA regions (Castellanos-Rubio and Ghosh, 2019). lncRNAs refer to RNA molecules over 200 bp in length without protein-coding potential (Beermann et al., 2016). Additionally, the new regulatory mechanism for lncRNA/circRNA, miRNA, and mRNA has aroused concerns (Hansen et al., 2013; Schmitz et al., 2016). The interaction of lncRNAs and circRNAs with miRNAs influences related mRNA expression. As we know, lncRNAs and circRNAs both contain complementary binding sites to miRNAs and act as endogenous miRNA sponges; miRNAs in turn interact with mRNAs, serving as negative regulators of protein expression. Therefore, LncRNAs and circRNAs function as molecular regulators by determining gene expression.
lncRNAs take up a large proportion of genes that have differential expression in response to different stress stimuli. After being induced, lncRNAs will regulate downstream cellular processes such as feedback regulation for essential stress response proteins (Valadkhan and Valencia-Hipólito, 2016). Although the significance of lncRNA molecules during various biological processes has been recognised, several details remain unclear. Presently, experiments concerning the functional role of lncRNAs were performed under experimental animal models or by in vitro assays. Some studies revealed the active role of lncRNAs in cell autophagy (Liang et al., 2020; Zhang and He, 2020; Li J. et al., 2021), apoptosis (Zhu et al., 2018; Gong et al., 2019; Zhang D. et al., 2019, Zhang M. et al., 2019, Zhang Y. et al., 2019; Zhou et al., 2019; Huang L. et al., 2020; Liao et al., 2020; Lv et al., 2020; Yan et al., 2020; Zhang Y. et al., 2020; Zhou et al., 2020; Chen Y. et al., 2021; Liu et al., 2021; Zhu et al., 2021a, 1), cardiac fibrosis (Wang X. et al., 2018, 30; Huang et al., 2019; Sun F. et al., 2019; Zhang JC. et al., 2019, 21; Lang et al., 2021; Zhang H. et al., 2021, 155–5), cardiac remodelling (Liu B. et al., 2020; Zhang B.f. et al., 2020), inflammation, and angiogenesis (Chen ZL. et al., 2020; Zhao et al., 2020) (In Figure 3).
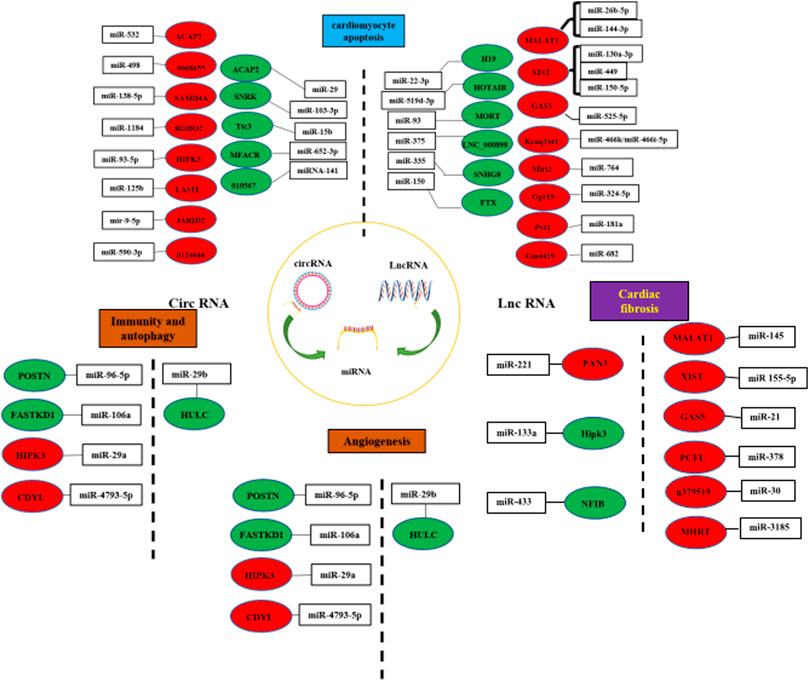
FIGURE 3. LncRNA/circRNA–miRNA-mediated interaction. (LncRNAs/circRNAs interact with miRNAs to modulate cardiomyocyte apoptosis, cardiac fibrosis, anglognesis, immunity and autophagy. Pro-factor lncRNAs/circRNAs are marked in blue. Anti-factor lncRNAs/circRNAs are marked in red.) (The left side of the dashed line represents circRNA, and the right side represents LncRNA).
Further research on the role of lncRNA in MI and cardiovascular events can deepen the understanding of the lncRNA network, contributing to the regulation of gene expression changes underlying MI, and assist the development of specific therapies based on the interference of miRNAs and lncRNA vital to MI.
4.5 CircRNAs and MI
CirRNAs are crucial for the physiology and pathology of biological systems and are involved in disease development. CircRNAs refer to a type of non-coding RNAs with higher stability than linear RNAs because they form a covalently closed continual loop with resistance against RNase R activity (Gao et al., 2015). Non-coding RNAs were optimal regulators of the cardiovascular system, and circRNAs were associated with CVDs (Wang W. et al., 2019).
Under component derivation, circRNAs can be usually classified into three categories, namely exon-derived circRNA (ecircRNA), lariat intron-derived circRNA (ciRNA), and exon-derived circRNA with retained introns (EIciRNA) (Li et al., 2015). Most known circRNAs originate from exons (Bei et al., 2018). CircRNAs maintain high stability and are abundantly expressed, making them better biomarkers relative to linear RNAs (Zhou et al., 2018). Simultaneously, with progress in bioinformatics and high-throughput sequencing technology, circRNAs have become a research direction for multiple biological functions and provide novel diagnostic methods and therapies for CVDs (Sun J.-Y. et al., 2020).
The present study addresses the regulatory role and functions of circRNAs, discusses the latest studies, and investigates the role and the regulatory mechanism of circRNAs in MI. Moreover, the roles of circRNAs in multiple MI such as myocardial apoptosis (Wang K. et al., 2017; Cai et al., 2019; Chai et al., 2020; Liu X. et al., 2020, 29; Wang Y. et al., 2020; Zhai et al., 2020; Chen T.-P. et al., 2021; Wu Y. et al., 2021; Zhang J. et al., 2021; Zhao B. et al., 2021; Zhu Y. et al., 2021), autophagy regulation, inflammatory response (Hu et al., 2020; Zhu Y. et al., 2020; Bian et al., 2021; Cai et al., 2021, 3), improvement of fibrosis (Zhu et al., 2019; Li F. et al., 2020; Sun L.y. et al., 2020; Si et al., 2020), and effects of ventricular remodelling (Garikipati et al., 2019; Cheng et al., 2020) (Gao et al., 2020; Zhang M. et al., 2020; Tan et al., 2021, 4; Zhao Q. et al., 2021) have been summarised (Figure 3). circRNAs mediate the fundamental physiological and pathological MI processes. Furthermore, since their dynamic changes can exhibit various disease stages, they are defined as ideal biomarkers. Thus, the present study also summed up the latest development of the role of circRNAs in MI and for convenience of reference.
5 The New Techniques and Target Drugs of MI in Epigenetics
The advent of high throughput epigenome mapping technologies has ushered in a new era of multi-omics where powerful tools can now delineate and record different layers of genomic output. Epigenetics play a central role in the regulation of many important biological processes. Despite significant technological advances for epigenetic profiling, there is still a need for a systematic understanding of how epigenetics shapes biological pathway, and disease pathogenesis (Angarica and Del Sol, 2017). DNA methylation and histone modifications and numerous techniques had been invented to analyze epigenetic processes not only at the level of specific genes, but also to analyze epigenetic changes that occur in defined regions of the genome as well as genome-wide. These technologies that are currently driving the field of epigenetics will greatly facilitate continued expansion of this exponentially growing discipline of genetics. A major breakthrough in the analysis of DNA methylation occurred with the development of bisulfite methylation sequencing (Wang and Chang, 2018). It used to be a gold-standard for detection of DNA methylation largely because it allows identification of 5-methylcytosine. This leading method of DNA methylation analysis has led to numerous subsequent methods such as Methylation Specific PCR and so on. Many proteins interact with RNA to modulate RNA-based epigenetic processes. Reaserches usually used the tools available to detect direct and indirect interactions between specific proteins and RNA in vivo. This is best achieved through the RNA immunoprecipitation technique (RIP). The uses of the RIP technique are vast and may be applied to epigenetics to help unravel the increasingly appreciated role of RNA in epigenetic processes (Tollefsbol, 2011). Beside, with the rapid development of technology, a number of epigenetic tests have emerged, such as Infinium Methylation450/850 BeadChips (450/850K), Methylated DNA immunoprecipitation-sequencing (MeDIP-Seq), Methylation-specificPCR (MSP),Pyrosequencing; Reduced representation bisulfite sequencing (RRBS), EWAS and so on (Feng and Lou, 2019). Researches should strictly choose appropriate detection methods according to the research direction. The availability of ultra-deep sequencing of genomic will transform the medical in analysis of the causes of disease, development of new drugs and diagnostics fields in the near future (Pareek et al., 2011).
DNA methylation, histone modification, nucleosome remodeling, and RNA-mediated targeting regulate many biological processes that are fundamental to the genesis of cancer. Along with the promising clinical and preclinical results seen with epigenetic drugs against chromatin regulators, signifies that it is the central role of epigenetics in cancer (Dawson and Kouzarides, 2012). Most of the drug research and development carried out from the perspective of epigenetics are related to tumors (Asano, 2020). At the same time, dietary intake has also presented significant influence on human health and disease development and nutritional modifications have proven important in prevention, but also the treatment of disease (Lundstrom, 2019). There are many epigenetic drugs have been identified in the past decade that effectively prevented or treated atherosclerosis and myocardial ischemia in several translational animal models, raising the possibility to combat coronary heart disease by targeting epigenetic processes also in humans. We have summarized several epigenetic therapy agents and strategies that may be associated with myocardial infarction by searching published reviews, including DNMT inhibitors, TET2 activators, Histone deacetylase inhibitors (Pickell et al., 2020), Sirtuin activating compounds, EZH2 inhibitors, BET inhibitor and other target epigenetic processes in atherosclerosis and associated vascular diseases (Schiano et al., 2015; Voelter-Mahlknecht, 2016; Xu et al., 2019). Interestingly, some dietary compounds, including polyphenols, cocoa, and folic acid, can modulate DNA methylation status, whereas statins may promote epigenetic-based control in CVD prevention through histone modifications (Voelter-Mahlknecht, 2016). Unfortunately, according to our knowledge, no epigenetically active agents or drugs targeting histone acetylation and/or methylation have thus far entered clinical trials for MI, nor have any of the latter been approved by the US Food and Drug Administration. The complex relationship between epigenetic regulation and MI development clearly demands further studies (Voelter-Mahlknecht, 2016).
6 Conclusion and Perspectives
MI exhibits the maximum morbidity, mortality, and effect on life quality among CVDs worldwide. Considerable progress has been attained in the discovery of MI genetic bases. Though the prospects entailed by understanding and controlling transcription through studies on histone and DNA modifications has received extensive attention, the reading of histone marks once placed shouldn’t been ignored in. executing gene expression, including bromodomain extra-terminal ((BET) (Borck et al., 2020). Some studies suggest that BET-containing family of epigenetic reader proteins, including BRD2, BRD3, BRD4 and the testis-restricted BRDT, provides a robust example of how epigenetic reader proteins can orchestrate transcriptional programs, provide new insight into mechanism of action and regulating effect and offer potentially novel therapeutic strategies in cardiovascular (Lin and Du, 2020; Li L. et al., 2021). Inhibition of BET epigenetic reader proteins might thus represent a promising therapeutic strategy to prevent adverse vascular remodelling (Dutzmann et al., 2021). BRD4, as a BET family member, plays an important role in critical biological processes. WU et al. found that BRD4 expression was up-regulated in human and mouse hypertrophied hearts, and importantly these effects were modulated by reactive oxygen species generation (Zhu W. et al., 2020). In one study, it has been reported that BETs are critical effectors of pathologic cardiac remodeling via their ability to co-activate defined stress-induced transcriptional programs in the heart (Auguste et al., 2020). Taken together, with the in-depth study of epigenetics, the secrets of related mechanisms will gradually be revealed.
The research on MI genetics contributes to early detection and the ability to provide personalised medical care. The MI pathophysiology would be progressively deciphered, demonstrating that genetics and epigenetics expedited MI onset and progression, enriching candidate methods (Nakatochi et al., 2017). Interactions between the genetic, epigenetic, and environmental factors constitute the critical factors of MI onset. The emergence of new genetic methods such as genome wide association analyses (GWAS) avoided some of these restrictions. GWAS analyses have exhibited that although different nationalities have different susceptibility genes and degrees of MI, multiple crucial loci have been identified for MI by GWAS (Takeuchi et al., 2012; Wakil et al., 2016). Epigenetic studies in cardiovascular medicine will improve our understanding of the molecular pathogenesis of MI and most importantly, facilitate novel biomarker identification, improved disease prevention, and new therapeutic strategies in managing MI. Future research should clarify how epigenetic mechanisms affect the MI process and prognosis to identify new drug targets and therapeutic strategies for MI. Although there is no specific drug for the epigenetic action of MI in clinic, currently available therapies, such as those using statins to promote epigenetic-based control in cardiovascular disease prevention through histone modifications, are already moving towards an exploitation of these mechanisms.
Author Contributions
JC and ZL wrote the manuscript with support from XG and BW. LM and SG handled the reference resource. HF, AL, and CW realized tables and images. All authors read, provided feedback, and approved the final protocol manuscript.
Funding
The research was funded by the National Natural Sciences Foundation of China (Project No. 81873149).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abi Khalil, C. (2014). The Emerging Role of Epigenetics in Cardiovascular Disease. Ther. Adv. Chronic Dis. 5, 178–187. doi:10.1177/2040622314529325
Agha, G., Mendelson, M. M., Ward-Caviness, C. K., Joehanes, R., Huan, T., Gondalia, R., et al. (2019). Blood Leukocyte DNA Methylation Predicts Risk of Future Myocardial Infarction and Coronary Heart Disease. Circulation 140, 645–657. doi:10.1161/CIRCULATIONAHA.118.039357
Agiannitopoulos, K., Pavlopoulou, P., Tsamis, K., Bampali, K., Samara, P., Nasioulas, G., et al. (2018). Expression of miR-208b and miR-499 in Greek Patients with Acute Myocardial Infarction. In Iv 32, 313–318. doi:10.21873/invivo.11239
Angarica, V. E., and Del Sol, A. (2017). Bioinformatics Tools for Genome-wide Epigenetic Research. Adv. Exp. Med. Biol. 978, 489–512. doi:10.1007/978-3-319-53889-1_25
Asano, T. (2020). Drug Resistance in Cancer Therapy and the Role of Epigenetics. J. Nippon Med. Sch. 87, 244–251. doi:10.1272/jnms.JNMS.2020_87-508
Asllanaj, E., Zhang, X., Ochoa Rosales, C., Nano, J., Bramer, W. M., Portilla-Fernandez, E., et al. (2020). Sexually Dimorphic DNA-Methylation in Cardiometabolic Health: A Systematic Review. Maturitas 135, 6–26. doi:10.1016/j.maturitas.2020.02.005
Auguste, G., Rouhi, L., Matkovich, S. J., Coarfa, C., Robertson, M. J., Czernuszewicz, G., et al. (2020). BET Bromodomain Inhibition Attenuates Cardiac Phenotype in Myocyte-specific Lamin A/C-deficient Mice. J. Clin. Invest. 130, 4740–4758. doi:10.1172/JCI135922
Bayoumi, A. S., Teoh, J.-P., Aonuma, T., Yuan, Z., Ruan, X., Tang, Y., et al. (2017). MicroRNA-532 Protects the Heart in Acute Myocardial Infarction, and Represses Prss23, a Positive Regulator of Endothelial-To-Mesenchymal Transition. Cardiovasc. Res. 113, 1603–1614. doi:10.1093/cvr/cvx132
Beermann, J., Piccoli, M.-T., Viereck, J., and Thum, T. (2016). Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 96, 1297–1325. doi:10.1152/physrev.00041.2015
Bei, Y., Yang, T., Wang, L., Holvoet, P., Das, S., Sluijter, J. P. G., et al. (2018). Circular RNAs as Potential Theranostics in the Cardiovascular System. Mol. Ther. - Nucleic Acids 13, 407–418. doi:10.1016/j.omtn.2018.09.022
Bian, Y., Pang, P., Li, X., Yu, S., Wang, X., Liu, K., et al. (2021). CircHelz Activates NLRP3 Inflammasome to Promote Myocardial Injury by Sponging miR-133a-3p in Mouse Ischemic Heart. J. Mol. Cell Cardiol. 158, 128–139. doi:10.1016/j.yjmcc.2021.05.010
Bonauer, A., Carmona, G., Iwasaki, M., Mione, M., Koyanagi, M., Fischer, A., et al. (2009). MicroRNA-92a Controls Angiogenesis and Functional Recovery of Ischemic Tissues in Mice. Science 324, 1710–1713. doi:10.1126/science.1174381
Borck, P. C., Guo, L.-W., and Plutzky, J. (2020). BET Epigenetic Reader Proteins in Cardiovascular Transcriptional Programs. Circ. Res. 126, 1190–1208. doi:10.1161/CIRCRESAHA.120.315929
Breton, C. V., Park, C., Siegmund, K., Gauderman, W. J., Whitfield-Maxwell, L., Hodis, H. N., et al. (2014). NOS1 Methylation and Carotid Artery Intima-media Thickness in Children. Circ. Cardiovasc. Genet. 7, 116–122. doi:10.1161/CIRCGENETICS.113.000320
Cai, L., Qi, B., Wu, X., Peng, S., Zhou, G., Wei, Y., et al. (2019). Circular RNA Ttc3 Regulates Cardiac Function after Myocardial Infarction by Sponging miR-15b. J. Mol. Cell Cardiol. 130, 10–22. doi:10.1016/j.yjmcc.2019.03.007
Cai, X., Li, B., Wang, Y., Zhu, H., Zhang, P., Jiang, P., et al. (2021). CircJARID2 Regulates Hypoxia-Induced Injury in H9c2 Cells by Affecting miR-9-5p-Mediated BNIP3. J. Cardiovasc. Pharmacol. 78, e77–e85. doi:10.1097/FJC.0000000000001033
Cao, Y., Lu, L., Liu, M., Li, X. C., Sun, R. R., Zheng, Y., et al. (2014). Impact of Epigenetics in the Management of Cardiovascular Disease: a Review. Eur. Rev. Med. Pharmacol. Sci. 18, 3097–3104.
Castellanos-Rubio, A., and Ghosh, S. (2019). Disease-Associated SNPs in Inflammation-Related lncRNAs. Front. Immunol. 10, 420. doi:10.3389/fimmu.2019.00420
Cavalli, G., and Heard, E. (2019). Advances in Epigenetics Link Genetics to the Environment and Disease. Nature 571, 489–499. doi:10.1038/s41586-019-1411-0
Chai, Q., Zheng, M., Wang, L., Wei, M., Yin, Y., Ma, F., et al. (2020). Circ_0068655 Promotes Cardiomyocyte Apoptosis via miR-498/PAWR Axis. Tissue Eng. Regen. Med. 17, 659–670. doi:10.1007/s13770-020-00270-8
Chen, L., Ji, Q., Zhu, H., Ren, Y., Fan, Z., and Tian, N. (2018a). miR-30a Attenuates Cardiac Fibrosis in Rats with Myocardial Infarction by Inhibiting CTGF. Exp. Ther. Med. 15, 4318–4324. doi:10.3892/etm.2018.5952
Chen, P., Pan, J., Zhang, X., Shi, Z., and Yang, X. (2018b). The Role of MicroRNA-181a in Myocardial Fibrosis Following Myocardial Infarction in a Rat Model. Med. Sci. Monit. 24, 4121–4127. doi:10.12659/MSM.908056
Chen, S., Fang, H., Liu, R., Fang, Y., Wu, Z., and Xie, P. (2021a). miR‐6718‐5p and miR‐4329 Can Be Used as Potential Biomarkers for Acute Myocardial Infarction. J. Card. Surg. 36, 3721–3728. doi:10.1111/jocs.15868
Chen, T.-P., Zhang, N.-J., Wang, H.-J., Hu, S.-G., and Geng, X. (2021b). Knockdown of circROBO2 Attenuates Acute Myocardial Infarction through Regulating the miR-1184/TRADD axis. Mol. Med. 27, 21. doi:10.1186/s10020-021-00275-6
Chen, X., He, Y., Fu, W., Sahebkar, A., Tan, Y., Xu, S., et al. (2020a). Histone Deacetylases (HDACs) and Atherosclerosis: A Mechanistic and Pharmacological Review. Front. Cel Dev. Biol. 8, 581015. doi:10.3389/fcell.2020.581015
Chen, Y., Li, S., Zhang, Y., Wang, M., Li, X., Liu, S., et al. (2021c). The lncRNA Malat1 Regulates Microvascular Function after Myocardial Infarction in Mice via miR-26b-5p/Mfn1 axis-mediated Mitochondrial Dynamics. Redox Biol. 41, 101910. doi:10.1016/j.redox.2021.101910
Chen, Y., Rollins, J., Paigen, B., and Wang, X. (2007). Genetic and Genomic Insights into the Molecular Basis of Atherosclerosis. Cel Metab. 6, 164–179. doi:10.1016/j.cmet.2007.07.001
Chen, Z., Gong, L., Zhang, P., Li, Y., Liu, B., Zhang, L., et al. (2019). Epigenetic Down-Regulation of Sirt 1 via DNA Methylation and Oxidative Stress Signaling Contributes to the Gestational Diabetes Mellitus-Induced Fetal Programming of Heart Ischemia-Sensitive Phenotype in Late Life. Int. J. Biol. Sci. 15, 1240–1251. doi:10.7150/ijbs.33044
Chen, Z. L., Chen, Y. X., Zhou, J., Li, Y., Gong, C. Y., and Wang, X. B. (2020b). LncRNA HULC Alleviates HUVEC Inflammation and Improves Angiogenesis after Myocardial Infarction through Down-Regulating miR-29b. Eur. Rev. Med. Pharmacol. Sci. 24, 6288–6298. doi:10.26355/eurrev_202006_21527
Chen, Z., Lu, S., Xu, M., Liu, P., Ren, R., and Ma, W. (2017). Role of miR-24, Furin, and Transforming Growth Factor-Β1 Signal Pathway in Fibrosis after Cardiac Infarction. Med. Sci. Monit. 23, 65–70. doi:10.12659/msm.898641
Cheng, N., Wang, M.-Y., Wu, Y.-B., Cui, H.-M., Wei, S.-X., Liu, B., et al. (2020). Circular RNA POSTN Promotes Myocardial Infarction-Induced Myocardial Injury and Cardiac Remodeling by Regulating miR-96-5p/BNIP3 Axis. Front. Cel Dev. Biol. 8, 618574. doi:10.3389/fcell.2020.618574
Chi, F., Feng, L., Li, Y., Zhao, S., Yuan, W., Jiang, Y., et al. (2020). MiR-30b-5p Promotes Myocardial Cell Apoptosis in Rats with Myocardial Infarction through Regulating Wnt/β-Catenin Signaling Pathway. Minerva Med. doi:10.23736/S0026-4806.20.06565-9
Chun, P. (2020). Therapeutic Effects of Histone Deacetylase Inhibitors on Heart Disease. Arch. Pharm. Res. 43, 1276–1296. doi:10.1007/s12272-020-01297-0
Cokkinos, D. V., and Pantos, C. (2007). Myocardial protection in Man-From Research Concept to Clinical Practice. Heart Fail. Rev. 12, 345–362. doi:10.1007/s10741-007-9030-5
Cortez-Dias, N., Costa, M. C., Carrilho-Ferreira, P., Silva, D., Jorge, C., Calisto, C., et al. (2016). Circulating miR-122-5p/miR-133b Ratio Is a Specific Early Prognostic Biomarker in Acute Myocardial Infarction. Circ. J. 80, 2183–2191. doi:10.1253/circj.CJ-16-0568
Coskunpinar, E., Cakmak, H. A., Kalkan, A. K., Tiryakioglu, N. O., Erturk, M., and Ongen, Z. (2016). Circulating miR-221-3p as a Novel Marker for Early Prediction of Acute Myocardial Infarction. Gene 591, 90–96. doi:10.1016/j.gene.2016.06.059
Damani, S. B., and Topol, E. J. (2007). Future Use of Genomics in Coronary Artery Disease. J. Am. Coll. Cardiol. 50, 1933–1940. doi:10.1016/j.jacc.2007.07.062
Dawson, M. A., and Kouzarides, T. (2012). Cancer Epigenetics: from Mechanism to Therapy. Cell 150, 12–27. doi:10.1016/j.cell.2012.06.013
Deegan, D. F., Nigam, P., and Engel, N. (2021). Sexual Dimorphism of the Heart: Genetics, Epigenetics, and Development. Front. Cardiovasc. Med. 8, 668252. doi:10.3389/fcvm.2021.668252
Ding, Y.-N., Tang, X., Chen, H.-Z., and Liu, D.-P. (2018). Epigenetic Regulation of Vascular Aging and Age-Related Vascular Diseases. Adv. Exp. Med. Biol. 1086, 55–75. doi:10.1007/978-981-13-1117-8_4
Dorn, G. W., and Matkovich, S. J. (2015). Epitranscriptional Regulation of Cardiovascular Development and Disease. J. Physiol. 593, 1799–1808. doi:10.1113/jphysiol.2014.283234
Du, J., Zhang, L., Zhuang, S., Qin, G. J., and Zhao, T. C. (2015). HDAC4 Degradation Mediates HDAC Inhibition-Induced Protective Effects against Hypoxia/Reoxygenation Injury. J. Cel. Physiol. 230, 1321–1331. doi:10.1002/jcp.24871
Dutzmann, J., Haertlé, M., Daniel, J.-M., Kloss, F., Musmann, R.-J., Kalies, K., et al. (2021). BET Bromodomain-Containing Epigenetic Reader Proteins Regulate Vascular Smooth Muscle Cell Proliferation and Neointima Formation. Cardiovasc. Res. 117, 850–862. doi:10.1093/cvr/cvaa121
Duygu, B., Poels, E. M., Juni, R., Bitsch, N., Ottaviani, L., Olieslagers, S., et al. (2017). miR-199b-5p Is a Regulator of Left Ventricular Remodeling Following Myocardial Infarction. Non-coding RNA Res. 2, 18–26. doi:10.1016/j.ncrna.2016.12.002
Egger, G., Liang, G., Aparicio, A., and Jones, P. A. (2004). Epigenetics in Human Disease and Prospects for Epigenetic Therapy. Nature 429, 457–463. doi:10.1038/nature02625
Eom, G. H., and Kook, H. (2014). Posttranslational Modifications of Histone Deacetylases: Implications for Cardiovascular Diseases. Pharmacol. Ther. 143, 168–180. doi:10.1016/j.pharmthera.2014.02.012
Fan, Z. G., Qu, X. L., Chu, P., Gao, Y. L., Gao, X. F., Chen, S. L., et al. (2018). MicroRNA-210 Promotes Angiogenesis in Acute Myocardial Infarction. Mol. Med. Rep. 17, 5658–5665. doi:10.3892/mmr.2018.8620
Fardi, M., Solali, S., and Farshdousti Hagh, M. (2018). Epigenetic Mechanisms as a New Approach in Cancer Treatment: An Updated Review. Genes Dis. 5, 304–311. doi:10.1016/j.gendis.2018.06.003
Felician, G., Collesi, C., Lusic, M., Martinelli, V., Ferro, M. D., Zentilin, L., et al. (2014). Epigenetic Modification at Notch Responsive Promoters Blunts Efficacy of Inducing Notch Pathway Reactivation after Myocardial Infarction. Circ. Res. 115, 636–649. doi:10.1161/CIRCRESAHA.115.304517
Feng, G., Yan, Z., Li, C., and Hou, Y. (2016). microRNA-208a in an Early Stage Myocardial Infarction Rat Model and the Effect on cAMP-PKA Signaling Pathway. Mol. Med. Rep. 14, 1631–1635. doi:10.3892/mmr.2016.5402
Feng, L., and Lou, J. (2019). DNA Methylation Analysis. Methods Mol. Biol. 1894, 181–227. doi:10.1007/978-1-4939-8916-4_12
Fernández-Sanlés, A., Sayols-Baixeras, S., Subirana, I., Sentí, M., Pérez-Fernández, S., de Castro Moura, M., et al. (2021). DNA Methylation Biomarkers of Myocardial Infarction and Cardiovascular Disease. Clin. Epigenet 13, 86. doi:10.1186/s13148-021-01078-6
Fu, D. L., Jiang, H., Li, C. Y., Gao, T., Liu, M. R., and Li, H. W. (2020). MicroRNA-338 in MSCs-Derived Exosomes Inhibits Cardiomyocyte Apoptosis in Myocardial Infarction. Eur. Rev. Med. Pharmacol. Sci. 24, 10107–10117. doi:10.26355/eurrev_202010_23230
Gallardo-Gómez, M., Moran, S., Páez de la Cadena, M., Martínez-Zorzano, V. S., Rodríguez-Berrocal, F. J., Rodríguez-Girondo, M., et al. (2018). A New Approach to Epigenome-wide Discovery of Non-invasive Methylation Biomarkers for Colorectal Cancer Screening in Circulating Cell-free DNA Using Pooled Samples. Clin. Epigenet 10, 53. doi:10.1186/s13148-018-0487-y
Gao, W.-Q., Hu, X.-M., Zhang, Q., Yang, L., Lv, X.-Z., Chen, S., et al. (2020). Downregulation of circFASTKD1 Ameliorates Myocardial Infarction by Promoting Angiogenesis. Aging 13, 3588–3604. doi:10.18632/aging.202305
Gao, Y., Wang, J., and Zhao, F. (2015). CIRI: an Efficient and Unbiased Algorithm for De Novo Circular RNA Identification. Genome Biol. 16, 4. doi:10.1186/s13059-014-0571-3
Garikipati, V. N. S., Verma, S. K., Cheng, Z., Liang, D., Truongcao, M. M., Cimini, M., et al. (2019). Circular RNA CircFndc3b Modulates Cardiac Repair after Myocardial Infarction via FUS/VEGF-A axis. Nat. Commun. 10, 4317. doi:10.1038/s41467-019-11777-7
Ge, Z.-W., Zhu, X.-L., Wang, B.-C., Hu, J.-L., Sun, J.-J., Wang, S., et al. (2019). MicroRNA-26b Relieves Inflammatory Response and Myocardial Remodeling of Mice with Myocardial Infarction by Suppression of MAPK Pathway through Binding to PTGS2. Int. J. Cardiol. 280, 152–159. doi:10.1016/j.ijcard.2018.12.077
Geng, T., Song, Z.-Y., Xing, J.-X., Wang, B.-X., Dai, S.-P., and Xu, Z.-S. (2020). Exosome Derived from Coronary Serum of Patients with Myocardial Infarction Promotes Angiogenesis through the miRNA-143/IGF-IR Pathway. Int. J. Nanomedicine 15, 2647–2658. doi:10.2147/IJN.S242908
Gong, X., Zhu, Y., Chang, H., Li, Y., and Ma, F. (2019). Long Noncoding RNA MALAT1 Promotes Cardiomyocyte Apoptosis after Myocardial Infarction via Targeting miR-144-3p. Biosci. Rep. 39, BSR20191103. doi:10.1042/BSR20191103
Guarrera, S., Fiorito, G., Onland-Moret, N. C., Russo, A., Agnoli, C., Allione, A., et al. (2015). Gene-specific DNA Methylation Profiles and LINE-1 Hypomethylation Are Associated with Myocardial Infarction Risk. Clin. Epigenet 7, 133. doi:10.1186/s13148-015-0164-3
Guo, L. L., Chen, H. H., Qu, F. C., and Lu, Q. H. (2020a). Clinical Significance of miR-492 in Peripheral Blood of Acute Myocardial Infarction. Eur. Rev. Med. Pharmacol. Sci. 24, 9041–9045. doi:10.26355/eurrev_202009_22849
Guo, M. L., Guo, L. L., and Weng, Y. Q. (2017a). Implication of Peripheral Blood miRNA-124 in Predicting Acute Myocardial Infarction. Eur. Rev. Med. Pharmacol. Sci. 21, 1054–1059. doi:10.1111/jcmm.13032
Guo, X., Chen, Y., Lu, Y., Li, P., Yu, H., Diao, F.-R., et al. (2020b). High Level of Circulating microRNA-142 Is Associated with Acute Myocardial Infarction and Reduced Survival. Ir J. Med. Sci. 189, 933–937. doi:10.1007/s11845-020-02196-5
Guo, Y., Luo, F., Liu, Q., and Xu, D. (2017b). Regulatory Non-coding RNAs in Acute Myocardial Infarction. J. Cel. Mol. Med. 21, 1013–1023. doi:10.1111/jcmm.13032
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA Circles Function as Efficient microRNA Sponges. Nature 495, 384–388. doi:10.1038/nature11993
He, Y., Zhong, J., Huang, S., Shui, X., Kong, D., Chen, C., et al. (2017). Elevated Circulating miR-126-3p Expression in Patients with Acute Myocardial Infarction: its Diagnostic Value. Int. J. Clin. Exp. Pathol. 10, 11051–11056.
Hong, Y., Cao, H., Wang, Q., Ye, J., Sui, L., Feng, J., et al. (2016). MiR-22 May Suppress Fibrogenesis by Targeting TGFβR I in Cardiac Fibroblasts. Cell Physiol Biochem 40, 1345–1353. doi:10.1159/000453187
Horváth, M., Horváthová, V., Hájek, P., Štěchovský, C., Honěk, J., Šenolt, L., et al. (2020). MicroRNA-331 and microRNA-151-3p as Biomarkers in Patients with ST-Segment Elevation Myocardial Infarction. Sci. Rep. 10, 5845. doi:10.1038/s41598-020-62835-w
Hromadka, M., Motovska, Z., Hlinomaz, O., Kala, P., Tousek, F., Jarkovsky, J., et al. (2021). MiR-126-3p and MiR-223-3p as Biomarkers for Prediction of Thrombotic Risk in Patients with Acute Myocardial Infarction and Primary Angioplasty. J. Pers. Med. 11, 508. doi:10.3390/jpm11060508
Hu, H.-L., Gao, S.-H., and Pan, J. (2021). MiR-135b Might Be a Potential Therapeutic Target in the Treatment of Myocardial Infarction. Int. J. Cardiol. 322, 250. doi:10.1016/j.ijcard.2020.08.084
Hu, X., Ma, R., Cao, J., Du, X., Cai, X., and Fan, Y. (2020). CircSAMD4A Aggravates H/R‐induced Cardiomyocyte Apoptosis and Inflammatory Response by Sponging miR‐138‐5p. J. Cel Mol Med. doi:10.1111/jcmm.16093
Huang, J., Jiang, R., Chu, X., Wang, F., Sun, X., Wang, Y., et al. (2020a). Overexpression of microRNA ‐23a‐5p Induces Myocardial Infarction by Promoting Cardiomyocyte Apoptosis through Inhibited of PI3K/AKT Signalling Pathway. Cell Biochem Funct 38, 1047–1055. doi:10.1002/cbf.3536
Huang, L., Guo, B., Liu, S., Miao, C., and Li, Y. (2020b). Inhibition of the LncRNA Gpr19 Attenuates Ischemia‐reperfusion Injury after Acute Myocardial Infarction by Inhibiting Apoptosis and Oxidative Stress via the miR‐324‐5p/Mtfr1 axis. IUBMB Life 72, 373–383. doi:10.1002/iub.2187
Huang, S., Zhang, L., Song, J., Wang, Z., Huang, X., Guo, Z., et al. (2019). Long Noncoding RNA MALAT1 Mediates Cardiac Fibrosis in Experimental Postinfarct Myocardium Mice Model. J. Cel Physiol 234, 2997–3006. doi:10.1002/jcp.27117
Huang, W., Cao, Y., Chen, C., Wu, X., Sheng, Z., and Li, A. (2021). MiR-467a-5p Aggravates Myocardial Infarction by Modulating ZEB1 Expression in Mice. J. Mol. Histol. 52, 767–780. doi:10.1007/s10735-021-09978-w
Huangfu, F. T., Tang, L. Q., Wang, H. Q., Zhao, X., and Yang, M. (2020). MiR-145-5p Promotes Myocardial Cell Apoptosis in Rats with Myocardial Infarction through PI3K/Akt Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 24, 12904–12911. doi:10.26355/eurrev_202012_24194
Hui, J., Huishan, W., Tao, L., Zhonglu, Y., Renteng, Z., and Hongguang, H. (2017). miR-539 as a Key Negative Regulator of the MEK Pathway in Myocardial Infarction. Herz 42, 781–789. doi:10.1007/s00059-016-4517-2
Icli, B., Li, H., Pérez-Cremades, D., Wu, W., Ozdemir, D., Haemmig, S., et al. (2020). MiR-4674 Regulates Angiogenesis in Tissue Injury by Targeting p38K Signaling in Endothelial Cells. Am. J. Physiology-Cell Physiol. 318, C524–C535. doi:10.1152/ajpcell.00542.2019
Jia, L., Zhu, L., Wang, J. Z., Wang, X. J., Chen, J. Z., Song, L., et al. (2013). Methylation of FOXP3 in Regulatory T Cells Is Related to the Severity of Coronary Artery Disease. Atherosclerosis 228, 346–352. doi:10.1016/j.atherosclerosis.2013.01.027
Kim, G. H. (2013). MicroRNA Regulation of Cardiac Conduction and Arrhythmias. Translational Res. 161, 381–392. doi:10.1016/j.trsl.2012.12.004
Kim, J. K., Samaranayake, M., and Pradhan, S. (2009). Epigenetic Mechanisms in Mammals. Cell. Mol. Life Sci. 66, 596–612. doi:10.1007/s00018-008-8432-4
Koch, A., Joosten, S. C., Feng, Z., de Ruijter, T. C., Draht, M. X., Melotte, V., et al. (2018). Analysis of DNA Methylation in Cancer: Location Revisited. Nat. Rev. Clin. Oncol. 15, 459–466. doi:10.1038/s41571-018-0004-4
Koseler, A., Ma, F., Kilic, I. D., Morselli, M., Kilic, O., and Pellegrini, M. (2020). Genome-wide DNA Methylation Profiling of Blood from Monozygotic Twins Discordant for Myocardial Infarction. In Vivo 34, 361–367. doi:10.21873/invivo.11782
Lang, M., Ou, D., Liu, Z., Li, Y., Zhang, X., and Zhang, F. (2021). LncRNA MHRT Promotes Cardiac Fibrosis via miR-3185 Pathway Following Myocardial Infarction. Int. Heart J. 62, 891–899. doi:10.1536/ihj.20-298
Li, C., Li, J., Xue, K., Zhang, J., Wang, C., Zhang, Q., et al. (2019a). MicroRNA-143-3p Promotes Human Cardiac Fibrosis via Targeting Sprouty3 after Myocardial Infarction. J. Mol. Cell Cardiol. 129, 281–292. doi:10.1016/j.yjmcc.2019.03.005
Li, F., Long, T.-Y., Bi, S.-S., Sheikh, S. A., and Zhang, C.-L. (2020a). circPAN3 Exerts a Profibrotic Role via Sponging miR-221 through FoxO3/ATG7-Activated Autophagy in a Rat Model of Myocardial Infarction. Life Sci. 257, 118015. doi:10.1016/j.lfs.2020.118015
Li, J., Cai, S. X., He, Q., Zhang, H., Friedberg, D., Wang, F., et al. (2018a). Intravenous miR-144 Reduces Left Ventricular Remodeling after Myocardial Infarction. Basic Res. Cardiol. 113, 36. doi:10.1007/s00395-018-0694-x
Li, J. H., Dai, J., Han, B., Wu, G. H., and Wang, C. H. (2019b). MiR-34a Regulates Cell Apoptosis after Myocardial Infarction in Rats through the Wnt/β-Catenin Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 2555–2562. doi:10.26355/eurrev_201903_17404
Li, J., Tong, Y., Zhou, Y., Han, Z., Wang, X., Ding, T., et al. (2021a). LncRNA KCNQ1OT1 as a miR-26a-5p Sponge Regulates ATG12-Mediated Cardiomyocyte Autophagy and Aggravates Myocardial Infarction. Int. J. Cardiol. 338, 14–23. doi:10.1016/j.ijcard.2021.05.053
Li, L., Xie, W., Gui, Y., and Zheng, X. L. (2021b). Bromodomain‐containing Protein 4 and its Role in Cardiovascular Diseases. J. Cel Physiol 236, 4829–4840. doi:10.1002/jcp.30225
Li, Q., Gao, Y., Zhu, J., and Jia, Q. (2020b). MiR-101 Attenuates Myocardial Infarction-Induced Injury by Targeting DDIT4 to Regulate Autophagy. Curr. Neurovasc Res. 17, 123–130. doi:10.2174/1567202617666200211113016
Li, S., Ren, J., and Sun, Q. (2018b). The Expression of microRNA-23a Regulates Acute Myocardial Infarction in Patients and In�vitro through Targeting PTEN. Mol. Med. Rep. 17, 6866–6872. doi:10.3892/mmr.2018.8640
Li, X., Wei, C., Zhang, Z., Jin, Q., and Xiao, X. (2020c). MiR-134-5p Regulates Myocardial Apoptosis and Angiogenesis by Directly Targeting KDM2A after Myocardial Infarction. Int. Heart J. 61, 815–821. doi:10.1536/ihj.19-468
Li, Z., Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., et al. (2015). Exon-intron Circular RNAs Regulate Transcription in the Nucleus. Nat. Struct. Mol. Biol. 22, 256–264. doi:10.1038/nsmb.2959
Liang, H., Su, X., Wu, Q., Shan, H., Lv, L., Yu, T., et al. (2020). LncRNA 2810403D21Rik/Mirf Promotes Ischemic Myocardial Injury by Regulating Autophagy through Targeting Mir26a. Autophagy 16, 1077–1091. doi:10.1080/15548627.2019.1659610
Liao, B., Dong, S., Xu, Z., Gao, F., Zhang, S., and Liang, R. (2020). LncRNA Kcnq1ot1 Renders Cardiomyocytes Apoptosis in Acute Myocardial Infarction Model by Up-Regulating Tead1. Life Sci. 256, 117811. doi:10.1016/j.lfs.2020.117811
Liao, Z., Chen, Y., Duan, C., Zhu, K., Huang, R., Zhao, H., et al. (2021). Cardiac Telocytes Inhibit Cardiac Microvascular Endothelial Cell Apoptosis through Exosomal miRNA-21-5p-Targeted Cdip1 Silencing to Improve Angiogenesis Following Myocardial Infarction. Theranostics 11, 268–291. doi:10.7150/thno.47021
Lin, C.-F., Hsu, K.-C., HuangFu, W.-C., Lin, T. E., Huang, H.-L., and Pan, S.-L. (2020). Investigating the Potential Effects of Selective Histone Deacetylase 6 Inhibitor ACY1215 on Infarct Size in Rats with Cardiac Ischemia-Reperfusion Injury. BMC Pharmacol. Toxicol. 21, 21. doi:10.1186/s40360-020-0400-0
Lin, S., and Du, L. (2020). The Therapeutic Potential of BRD4 in Cardiovascular Disease. Hypertens. Res. 43, 1006–1014. doi:10.1038/s41440-020-0459-4
Liu, B., Cheng, Y., Tian, J., Zhang, L., and Cui, X. (2020a). Upregulated lncRNA Pvt1 May Be Important for Cardiac Remodeling at the Infarct Border Zone. Mol. Med. Rep. 22, 2605–2616. doi:10.3892/mmr.2020.11371
Liu, C., Tang, M., Zhang, X., Li, J., and Cao, G. (2020b). Knockdown of miR-665 Protects against Cardiomyocyte Ischemia/Reperfusion Injury-Induced ROS Accumulation and Apoptosis through the Activation of Pak1/Akt Signaling in Myocardial Infarction. Int. Heart J. 61, 347–354. doi:10.1536/ihj.19-416
Liu, X., Wang, M., Li, Q., Liu, W., Song, Q., and Jiang, H. (2020c). CircRNA ACAP2 Induces Myocardial Apoptosis after Myocardial Infarction by Sponging miR-29. Minerva Med. doi:10.23736/S0026-4806.20.06600-8
Liu, Y., Wang, H., Wang, X., and Xie, G. (2019). MiR-29b Inhibits Ventricular Remodeling by Activating Notch Signaling Pathway in the Rat Myocardial Infarction Model. Heart Surg. Forum 22, E019–E023. doi:10.1532/hsf.2079
Liu, Y., Zhou, P., Wang, F., Zhang, X., Yang, D., Hong, L., et al. (2021). Inhibition of lncRNA SNHG8 Plays a Protective Role in Hypoxia-Ischemia-Reoxygenation-Induced Myocardial Injury by Regulating miR-335 and RASA1 Expression. Mol. Med. Rep. 24, 597. doi:10.3892/mmr.2021.12236
Liu, Z. H., Sun, X. P., Zhou, S. L., and Wang, H. X. (2017). Research on the Relations between the Variation of miRNA-184 before and after Treatment of Acute Myocardial Infarction and Prognosis. Eur. Rev. Med. Pharmacol. Sci. 21, 843–847.
Liu, Z., Zhang, Y., Qiu, C., Zhu, H., Pan, S., Jia, H., et al. (2020d). Diabetes Mellitus Exacerbates post‐myocardial Infarction Heart Failure by Reducing Sarcolipin Promoter Methylation. ESC Heart Fail. 7, 1935–1948. doi:10.1002/ehf2.12789
Locke, W. J., Guanzon, D., Ma, C., Liew, Y. J., Duesing, K. R., Fung, K. Y. C., et al. (2019). DNA Methylation Cancer Biomarkers: Translation to the Clinic. Front. Genet. 10, 1150. doi:10.3389/fgene.2019.01150
Lu, S., and Lu, Y. (2020). MiR-26a Inhibits Myocardial Cell Apoptosis in Rats with Acute Myocardial Infarction through GSK-3β Pathway. Eur. Rev. Med. Pharmacol. Sci. 24, 2659–2666. doi:10.26355/eurrev_202003_20535
Lundstrom, K. (2019). Epigenetics, Nutrition, Disease and Drug Development. Curr. Drug Discov. Technol. 16, 386–391. doi:10.2174/1570163815666180419154954
Lv, J., Zhu, Y., and Yao, S. (2020). LncRNAMORT Is Upregulated in Myocardial Infarction and Promotes the Apoptosis of Cardiomyocyte by Downregulating miR-93. BMC Cardiovasc. Disord. 20, 247. doi:10.1186/s12872-020-01522-0
Ma, B., Wilker, E. H., Willis-Owen, S. A. G., Byun, H.-M., Wong, K. C. C., Motta, V., et al. (2014). Predicting DNA Methylation Level across Human Tissues. Nucleic Acids Res. 42, 3515–3528. doi:10.1093/nar/gkt1380
Ma, C., Peng, P., Zhou, Y., Liu, T., Wang, L., and Lu, C. (2020). MicroRNA-93 P-romotes A-ngiogenesis and A-ttenuates R-emodeling via I-nactivation of the Hippo/Yap P-athway by T-argeting Lats2 after M-yocardial I-nfarctionω. Mol. Med. Rep. 22, 483–493. doi:10.3892/mmr.2020.11085
Maciejak, A., Kostarska-Srokosz, E., Gierlak, W., Dluzniewski, M., Kuch, M., Marchel, M., et al. (2018). Circulating miR-30a-5p as a Prognostic Biomarker of Left Ventricular Dysfunction after Acute Myocardial Infarction. Sci. Rep. 8, 9883. doi:10.1038/s41598-018-28118-1
Mani, S. K., Kern, C. B., Kimbrough, D., Addy, B., Kasiganesan, H., Rivers, W. T., et al. (2015). Inhibition of Class I Histone Deacetylase Activity Represses Matrix Metalloproteinase-2 and -9 Expression and Preserves LV Function Postmyocardial Infarction. Am. J. Physiology-Heart Circulatory Physiol. 308, H1391–H1401. doi:10.1152/ajpheart.00390.2014
Mansouri, F., and Seyed Mohammadzad, M. H. (2020). Molecular miR-19a in Acute Myocardial Infarction: Novel Potential Indicators of Prognosis and Early Diagnosis. Asian Pac. J. Cancer Prev. 21, 975–982. doi:10.31557/APJCP.2020.21.4.975
Maron, D. J., Hochman, J. S., Reynolds, H. R., Bangalore, S., O'Brien, S. M., Boden, W. E., et al. (2020). Initial Invasive or Conservative Strategy for Stable Coronary Disease. N. Engl. J. Med. 382, 1395–1407. doi:10.1056/NEJMoa1915922
Mattick, J. S., Taft, R. J., and Faulkner, G. J. (2010). A Global View of Genomic Information - Moving beyond the Gene and the Master Regulator. Trends Genet. 26, 21–28. doi:10.1016/j.tig.2009.11.002
Mayer, B., Erdmann, J., and Schunkert, H. (2007). Genetics and Heritability of Coronary Artery Disease and Myocardial Infarction. Clin. Res. Cardiol. 96, 1–7. doi:10.1007/s00392-006-0447-y
McKinsey, T. A. (2011). The Biology and Therapeutic Implications of HDACs in the Heart. Handb Exp. Pharmacol. 206, 57–78. doi:10.1007/978-3-642-21631-2_4
Mohammadpanah, M., Heidari, M. M., Khatami, M., and Hadadzadeh, M. (2020). Relationship of Hypomethylation CpG Islands in Interleukin-6 Gene Promoter with IL-6 mRNA Levels in Patients with Coronary Atherosclerosis. J. Cardiovasc. Thorac. Res. 12, 221–228. doi:10.34172/jcvtr.2020.37
Movassagh, M., Choy, M.-K., Knowles, D. A., Cordeddu, L., Haider, S., Down, T., et al. (2011). Distinct Epigenomic Features in End-Stage Failing Human Hearts. Circulation 124, 2411–2422. doi:10.1161/CIRCULATIONAHA.111.040071
Nakatochi, M., Ichihara, S., Yamamoto, K., Naruse, K., Yokota, S., Asano, H., et al. (2017). Epigenome-wide Association of Myocardial Infarction with DNA Methylation Sites at Loci Related to Cardiovascular Disease. Clin. Epigenet 9, 54. doi:10.1186/s13148-017-0353-3
Newton-Cheh, C., and O’Donnell, C. J. (2004). Sex Differences and Genetic Associations with Myocardial Infarction. JAMA 291, 3008–3010. doi:10.1001/jama.291.24.3008
Nichols, M., Townsend, N., Scarborough, P., and Rayner, M. (2014). Cardiovascular Disease in Europe 2014: Epidemiological Update. Eur. Heart J. 35, 2929. doi:10.1093/eurheartj/ehu378
Nurnberg, S. T., Guerraty, M. A., Wirka, R. C., Rao, H. S., Pjanic, M., Norton, S., et al. (2020). Genomic Profiling of Human Vascular Cells Identifies TWIST1 as a Causal Gene for Common Vascular Diseases. Plos Genet. 16, e1008538. doi:10.1371/journal.pgen.1008538
O′Sullivan, J. F., Neylon, A., McGorrian, C., and Blake, G. J. (2016). miRNA-93-5p and Other miRNAs as Predictors of Coronary Artery Disease and STEMI. Int. J. Cardiol. 224, 310–316. doi:10.1016/j.ijcard.2016.09.016
Pareek, C. S., Smoczynski, R., and Tretyn, A. (2011). Sequencing Technologies and Genome Sequencing. J. Appl. Genet. 52, 413–435. doi:10.1007/s13353-011-0057-x
Peng, L., Chun-guang, Q., Bei-fang, L., Xue-zhi, D., Zi-hao, W., Yun-fu, L., et al. (2014). Clinical Impact of Circulating miR-133, miR-1291 and miR-663b in Plasma of Patients with Acute Myocardial Infarction. Diagn. Pathol. 9, 89. doi:10.1186/1746-1596-9-89
Pickell, Z., Williams, A. M., Alam, H. B., and Hsu, C. H. (2020). Histone Deacetylase Inhibitors: A Novel Strategy for Neuroprotection and Cardioprotection Following Ischemia/Reperfusion Injury. J. Am. Heart Assoc. 9, e016349. doi:10.1161/JAHA.120.016349
Poller, W., Dimmeler, S., Heymans, S., Zeller, T., Haas, J., Karakas, M., et al. (2018). Non-coding RNAs in Cardiovascular Diseases: Diagnostic and Therapeutic Perspectives. Eur. Heart J. 39, 2704–2716. doi:10.1093/eurheartj/ehx165
Portela, A., and Esteller, M. (2010). Epigenetic Modifications and Human Disease. Nat. Biotechnol. 28, 1057–1068. doi:10.1038/nbt.1685
Prasher, D., Greenway, S. C., and Singh, R. B. (2020). The Impact of Epigenetics on Cardiovascular Disease. Biochem. Cel Biol. 98, 12–22. doi:10.1139/bcb-2019-0045
Rask-Andersen, M., Martinsson, D., Ahsan, M., Enroth, S., Ek, W. E., Gyllensten, U., et al. (2016). Epigenome-wide Association Study Reveals Differential DNA Methylation in Individuals with a History of Myocardial Infarction. Hum. Mol. Genet. 25, ddw302–4748. doi:10.1093/hmg/ddw302
Reed, G. W., Rossi, J. E., and Cannon, C. P. (2017). Acute Myocardial Infarction. The Lancet 389, 197–210. doi:10.1016/S0140-6736(16)30677-8
Saban, K., Mathews, H. L., DeVon, H. A., and Janusek, L. W. (2014). Epigenetics and Social Context: Implications for Disparity in Cardiovascular Disease. Aging Dis. 5, 346–355. doi:10.14336/AD.2014.0500346
Schiano, C., Vietri, M. T., Grimaldi, V., Picascia, A., De Pascale, M. R., and Napoli, C. (2015). Epigenetic-related Therapeutic Challenges in Cardiovascular Disease. Trends Pharmacol. Sci. 36, 226–235. doi:10.1016/j.tips.2015.02.005
Schmitz, S. U., Grote, P., and Herrmann, B. G. (2016). Mechanisms of Long Noncoding RNA Function in Development and Disease. Cel. Mol. Life Sci. 73, 2491–2509. doi:10.1007/s00018-016-2174-5
Sen, P., Shah, P. P., Nativio, R., and Berger, S. L. (2016). Epigenetic Mechanisms of Longevity and Aging. Cell 166, 822–839. doi:10.1016/j.cell.2016.07.050
Sharma, P., Garg, G., Kumar, A., Mohammad, F., Kumar, S. R., Tanwar, V. S., et al. (2014). Genome Wide DNA Methylation Profiling for Epigenetic Alteration in Coronary Artery Disease Patients. Gene 541, 31–40. doi:10.1016/j.gene.2014.02.034
Shi, C. C., Pan, L. Y., Peng, Z. Y., and Li, J. G. (2020). MiR-126 Regulated Myocardial Autophagy on Myocardial Infarction. Eur. Rev. Med. Pharmacol. Sci. 24, 6971–6979. doi:10.26355/eurrev_202006_21689
Shibata, T., Kawakami, S., Noguchi, T., Tanaka, T., Asaumi, Y., Kanaya, T., et al. (2015). Prevalence, Clinical Features, and Prognosis of Acute Myocardial Infarction Attributable to Coronary Artery Embolism. Circulation 132, 241–250. doi:10.1161/CIRCULATIONAHA.114.015134
Si, X., Zheng, H., Wei, G., Li, M., Li, W., Wang, H., et al. (2020). circRNA Hipk3 Induces Cardiac Regeneration after Myocardial Infarction in Mice by Binding to Notch1 and miR-133a. Mol. Ther. - Nucleic Acids 21, 636–655. doi:10.1016/j.omtn.2020.06.024
Song, T., Guan, X., Wang, X., Qu, S., Zhang, S., Hui, W., et al. (2021). Dynamic Modulation of Gut Microbiota Improves post‐myocardial Infarct Tissue Repair in Rats via Butyric Acid‐mediated Histone Deacetylase Inhibition. FASEB j. 35, e21385. doi:10.1096/fj.201903129RRR
Su, Q., Lv, X.-W., Sun, Y.-H., Ye, Z.-L., Kong, B.-H., and Qin, Z.-B. (2019). MicroRNA-494 Inhibits the LRG1 Expression to Induce Proliferation and Migration of VECs in Rats Following Myocardial Infarction. Mol. Ther. - Nucleic Acids 18, 110–122. doi:10.1016/j.omtn.2019.08.007
Su, Q., Ye, Z., Sun, Y., Yang, H., and Li, L. (2018). Relationship between circulating miRNA-30e and no-reflow phenomenon in STEMI patients undergoing primary coronary intervention. Scand. J. Clin. Lab. Invest. 78, 318–324. doi:10.1080/00365513.2018.1467571
Su, T., Shao, X., Zhang, X., Yang, C., and Shao, X. (2020). Value of Circulating miRNA-1 Detected within 3 H after the Onset of Acute Chest Pain in the Diagnosis and Prognosis of Acute Myocardial Infarction. Int. J. Cardiol. 307, 146–151. doi:10.1016/j.ijcard.2019.09.050
Sun, F., Zhuang, Y., Zhu, H., Wu, H., Li, D., Zhan, L., et al. (2019a). LncRNA PCFL Promotes Cardiac Fibrosis via miR-378/GRB2 Pathway Following Myocardial Infarction. J. Mol. Cell Cardiol. 133, 188–198. doi:10.1016/j.yjmcc.2019.06.011
Sun, H. Y., Wang, X. L., Ma, L. C., Yang, M., Yang, H. J., Huang, H. W., et al. (2019b). Influence of MiR-154 on Myocardial Apoptosis in Rats with Acute Myocardial Infarction through Wnt/β-Catenin Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 818–825. doi:10.26355/eurrev_201901_16896
Sun, J.-Y., Shi, Y., Cai, X.-Y., and Liu, J. (2020a). Potential Diagnostic and Therapeutic Value of Circular RNAs in Cardiovascular Diseases. Cell Signal. 71, 109604. doi:10.1016/j.cellsig.2020.109604
Sun, L. y., Zhao, J. c., Ge, X. m., Zhang, H., Wang, C. m., and Bie, Z. d. (2020b). Circ_LAS1L Regulates Cardiac Fibroblast Activation, Growth, and Migration through miR‐125b/SFRP5 Pathway. Cel Biochem Funct 38, 443–450. doi:10.1002/cbf.3486
Takeuchi, F., Yokota, M., Yamamoto, K., Nakashima, E., Katsuya, T., Asano, H., et al. (2012). Genome-wide Association Study of Coronary Artery Disease in the Japanese. Eur. J. Hum. Genet. 20, 333–340. doi:10.1038/ejhg.2011.184
Talens, R. P., Jukema, J. W., Trompet, S., Kremer, D., Westendorp, R. G. J., Lumey, L. H., et al. (2012). Hypermethylation at Loci Sensitive to the Prenatal Environment Is Associated with Increased Incidence of Myocardial Infarction. Int. J. Epidemiol. 41, 106–115. doi:10.1093/ije/dyr153
Tan, J., Pan, W., Chen, H., Du, Y., Jiang, P., Zeng, D., et al. (2021). Circ_0124644 Serves as a ceRNA for miR-590-3p to Promote Hypoxia-Induced Cardiomyocytes Injury via Regulating SOX4. Front. Genet. 12, 667724. doi:10.3389/fgene.2021.667724
Tang, J., and Zhuang, S. (2019). Histone Acetylation and DNA Methylation in Ischemia/reperfusion Injury. Clin. Sci. (Lond) 133, 597–609. doi:10.1042/CS20180465
Thygesen, K., Alpert, J. S., Jaffe, A. S., Chaitman, B. R., Bax, J. J., Morrow, D. A., et al. (2018). Fourth Universal Definition of Myocardial Infarction (2018). Circulation 138, e618–e651. doi:10.1161/CIR.0000000000000617
Tingare, A., Thienpont, B., and Roderick, H. L. (2013). Epigenetics in the Heart: the Role of Histone Modifications in Cardiac Remodelling. Biochem. Soc. Trans. 41, 789–796. doi:10.1042/BST20130012
Tollefsbol, T. O. (2011). Advances in Epigenetic Technology. Methods Mol. Biol. 791, 1–10. doi:10.1007/978-1-61779-316-5_1
Valadkhan, S., and Valencia-Hipólito, A. (2015). lncRNAs in Stress Response. Curr. Top. Microbiol. Immunol. 394, 203–236. doi:10.1007/82_2015_489
Voelter-Mahlknecht, S. (2016). Epigenetic Associations in Relation to Cardiovascular Prevention and Therapeutics. Clin. Epigenet 8, 4. doi:10.1186/s13148-016-0170-0
Wakil, S. M., Ram, R., Muiya, N. P., Mehta, M., Andres, E., Mazhar, N., et al. (2016). A Genome-wide Association Study Reveals Susceptibility Loci for Myocardial Infarction/coronary Artery Disease in Saudi Arabs. Atherosclerosis 245, 62–70. doi:10.1016/j.atherosclerosis.2015.11.019
Wang, C., and Jing, Q. (2018). Non-coding RNAs as Biomarkers for Acute Myocardial Infarction. Acta Pharmacol. Sin 39, 1110–1119. doi:10.1038/aps.2017.205
Wang, C., Yang, D., Xu, C., and Duan, H. (2021a). MicroRNA-139-5p Inhibits Vascular Endothelial Cell Viability and Serves as a Diagnostic Biomarker in Acute Myocardial Infarction Patients. Exp. Gerontol. 152, 111453. doi:10.1016/j.exger.2021.111453
Wang, D. M., Jin, J. J., Tian, L. M., and Zhang, Z. (2020a). MiR-195 Promotes Myocardial Fibrosis in MI Rats via Targeting TGF-β1/Smad. J. Biol. Regul. Homeost Agents 34, 1325–1332. doi:10.23812/20-201-A
Wang, J., Dong, G., Chi, W., and Nie, Y. (2021b). MiR-96 Promotes Myocardial Infarction-Induced Apoptosis by Targeting XIAP. Biomed. Pharmacother. 138, 111208. doi:10.1016/j.biopha.2020.111208
Wang, J., Lin, B., Zhang, Y., Ni, L., Hu, L., Yang, J., et al. (2020b). The Regulatory Role of Histone Modification on Gene Expression in the Early Stage of Myocardial Infarction. Front. Cardiovasc. Med. 7, 594325. doi:10.3389/fcvm.2020.594325
Wang, K.-J., Zhao, X., Liu, Y.-Z., Zeng, Q.-T., Mao, X.-B., Li, S.-N., et al. (2016). Circulating MiR-19b-3p, MiR-134-5p and MiR-186-5p Are Promising Novel Biomarkers for Early Diagnosis of Acute Myocardial Infarction. Cel Physiol Biochem 38, 1015–1029. doi:10.1159/000443053
Wang, K. C., and Chang, H. Y. (2018). Epigenomics. Circ. Res. 122, 1191–1199. doi:10.1161/CIRCRESAHA.118.310998
Wang, K., Gan, T.-Y., Li, N., Liu, C.-Y., Zhou, L.-Y., Gao, J.-N., et al. (2017a). Circular RNA Mediates Cardiomyocyte Death via miRNA-dependent Upregulation of MTP18 Expression. Cell Death Differ 24, 1111–1120. doi:10.1038/cdd.2017.61
Wang, L., Fan, C., Topol, S. E., Topol, E. J., and Wang, Q. (2003). Mutation of MEF2A in an Inherited Disorder with Features of Coronary Artery Disease. Science 302, 1578–1581. doi:10.1126/science.1088477
Wang, P., Shen, C., Diao, L., Yang, Z., Fan, F., Wang, C., et al. (2015). Aberrant Hypermethylation of Aldehyde Dehydrogenase 2 Promoter Upstream Sequence in Rats with Experimental Myocardial Infarction. Biomed. Res. Int. 2015, 1–13. doi:10.1155/2015/503692
Wang, R., Li, N., Zhang, Y., Ran, Y., and Pu, J. (2011). Circulating microRNAs Are Promising Novel Biomarkers of Acute Myocardial Infarction. Intern. Med. 50, 1789–1795. doi:10.2169/internalmedicine.50.5129
Wang, W., Wang, Y., Piao, H., Li, B., Huang, M., Zhu, Z., et al. (2019a). Circular RNAs as Potential Biomarkers and Therapeutics for Cardiovascular Disease. PeerJ 7, e6831. doi:10.7717/peerj.6831
Wang, X., Yong, C., Yu, K., Yu, R., Zhang, R., Yu, L., et al. (2018a). Long Noncoding RNA (lncRNA) N379519 Promotes Cardiac Fibrosis in Post-Infarct Myocardium by Targeting miR-30. Med. Sci. Monit. 24, 3958–3965. doi:10.12659/MSM.910000
Wang, Y., Chang, W., Zhang, Y., Zhang, L., Ding, H., Qi, H., et al. (2019b). Circulating miR‐22‐5p and miR‐122‐5p Are Promising Novel Biomarkers for Diagnosis of Acute Myocardial Infarction. J. Cel Physiol 234, 4778–4786. doi:10.1002/jcp.27274
Wang, Y., Chen, P., Wang, L., Zhao, J., Zhong, Z., Wang, Y., et al. (2018b). Inhibition of Histone Deacetylases Prevents Cardiac Remodeling after Myocardial Infarction by Restoring Autophagosome Processing in Cardiac Fibroblasts. Cel Physiol Biochem 49, 1999–2011. doi:10.1159/000493672
Wang, Y., Zhao, R., Shen, C., Liu, W., Yuan, J., Li, C., et al. (2020c). Exosomal CircHIPK3 Released from Hypoxia-Induced Cardiomyocytes Regulates Cardiac Angiogenesis after Myocardial Infarction. Oxidative Med. Cell Longevity 2020, 1–19. doi:10.1155/2020/8418407
Wang, Z.-H., Sun, X.-Y., Li, C.-L., Sun, Y.-M., Li, J., Wang, L.-F., et al. (2017b). miRNA-21 Expression in the Serum of Elderly Patients with Acute Myocardial Infarction. Med. Sci. Monit. 23, 5728–5734. doi:10.12659/msm.904933
Ward-Caviness, C. K., Agha, G., Chen, B. H., Pfeiffer, L., Wilson, R., Wolf, P., et al. (2018). Analysis of Repeated Leukocyte DNA Methylation Assessments Reveals Persistent Epigenetic Alterations after an Incident Myocardial Infarction. Clin. Epigenet 10, 161. doi:10.1186/s13148-018-0588-7
Weintraub, W. S., Daniels, S. R., Burke, L. E., Franklin, B. A., Goff, D. C., Hayman, L. L., et al. (2011). Value of Primordial and Primary Prevention for Cardiovascular Disease. Circulation 124, 967–990. doi:10.1161/CIR.0b013e3182285a81
Wu, H.-B., Wang, Y.-C., Yao, W.-J., Xie, Y.-T., Wang, X.-C., and Du, R.-P. (2021a). MicroRNA-365 Expression in the Serum of Patients with Heart Failure with Reduced Ejection Fraction after Myocardial Infarction. Am. J. Emerg. Med. 45, 645–646. doi:10.1016/j.ajem.2020.11.017
Wu, Y., Wu, M., Yang, J., Li, Y., Peng, W., Wu, M., et al. (2021b). Silencing CircHIPK3 Sponges miR-93-5p to Inhibit the Activation of Rac1/PI3K/AKT Pathway and Improves Myocardial Infarction-Induced Cardiac Dysfunction. Front. Cardiovasc. Med. 8, 645378. doi:10.3389/fcvm.2021.645378
Xiang, Z., and Yang, J. (2020). The Therapeutic Potential of miR-135b in Myocardial Infarction: Anti-inflammatory Trials May Be Enlightening. Int. J. Cardiol. 312, 99. doi:10.1016/j.ijcard.2020.03.030
Xiao, L., He, H., Ma, L., Da, M., Cheng, S., Duan, Y., et al. (2017). Effects of miR-29a and miR-101a Expression on Myocardial Interstitial Collagen Generation after Aerobic Exercise in Myocardial-Infarcted Rats. Arch. Med. Res. 48, 27–34. doi:10.1016/j.arcmed.2017.01.006
Xie, M., Kong, Y., Tan, W., May, H., Battiprolu, P. K., Pedrozo, Z., et al. (2014). Histone Deacetylase Inhibition Blunts Ischemia/Reperfusion Injury by Inducing Cardiomyocyte Autophagy. Circulation 129, 1139–1151. doi:10.1161/CIRCULATIONAHA.113.002416
Xie, M., Tang, Y., and Hill, J. A. (2019). HDAC Inhibition as a Therapeutic Strategy in Myocardial Ischemia/reperfusion Injury. J. Mol. Cell Cardiol. 129, 188–192. doi:10.1016/j.yjmcc.2019.02.013
Xu, S., Kamato, D., Little, P. J., Nakagawa, S., Pelisek, J., and Jin, Z. G. (2019). Targeting Epigenetics and Non-coding RNAs in Atherosclerosis: from Mechanisms to Therapeutics. Pharmacol. Ther. 196, 15–43. doi:10.1016/j.pharmthera.2018.11.003
Xue, Y., Fan, X., Yang, R., Jiao, Y., and Li, Y. (2020). miR-29b-3p Inhibits post-infarct Cardiac Fibrosis by Targeting FOS. Biosci. Rep. 40, BSR20201227. doi:10.1042/BSR20201227
Xue, Y., Shen, J., Hong, W., Zhou, W., Xiang, Z., Zhu, Y., et al. (2021). Risk Stratification of ST-Segment Elevation Myocardial Infarction (STEMI) Patients Using Machine Learning Based on Lipid Profiles. Lipids Health Dis. 20, 48. doi:10.1186/s12944-021-01475-z
Yan, L., Guo, N., Cao, Y., Zeng, S., Wang, J., Lv, F., et al. (2018). miRNA-145 Inhibits Myocardial Infarction-induced A-poptosis through A-utophagy via Akt3/mTOR S-ignaling P-athway In V-itro and In V-ivo. Int. J. Mol. Med. 42, 1537–1547. doi:10.3892/ijmm.2018.3748
Yan, M., Liu, Q., Jiang, Y., Wang, B., Ji, Y., Liu, H., et al. (2020). Long Non-coding RNA LNC_000898 Alleviates Cardiomyocyte Apoptosis and Promotes Cardiac Repair after Myocardial Infarction via Modulating miR-375/PDK1 axis. J. Cardiovasc. Pharmacol. 76, 77–85. doi:10.1097/FJC.0000000000000845
Yang, F., Liu, W., Yan, X., Zhou, H., Zhang, H., Liu, J., et al. (2016a). Effects of Mir-21 on Cardiac Microvascular Endothelial Cells after Acute Myocardial Infarction in Rats: Role of Phosphatase and Tensin Homolog (PTEN)/Vascular Endothelial Growth Factor (VEGF) Signal Pathway. Med. Sci. Monit. 22, 3562–3575. doi:10.12659/msm.897773
Yang, J., Xu, W.-W., and Hu, S.-J. (2015). Heart Failure: Advanced Development in Genetics and Epigenetics. Biomed. Res. Int. 2015, 1–11. doi:10.1155/2015/352734
Yang, L., Wang, B., Zhou, Q., Wang, Y., Liu, X., Liu, Z., et al. (2018). MicroRNA-21 Prevents Excessive Inflammation and Cardiac Dysfunction after Myocardial Infarction through Targeting KBTBD7. Cell Death Dis 9, 769. doi:10.1038/s41419-018-0805-5
Yang, M., Kong, D. Y., and Chen, J. C. (2019). Inhibition of miR‐148b Ameliorates Myocardial Ischemia/reperfusion Injury via Regulation of Wnt/β‐catenin Signaling Pathway. J. Cel Physiol 234, 17757–17766. doi:10.1002/jcp.28401
Yang, S. Y., Wang, Y. Q., Gao, H. M., Wang, B., and He, Q. (2016b). The Clinical Value of Circulating miR-99a in Plasma of Patients with Acute Myocardial Infarction. Eur. Rev. Med. Pharmacol. Sci. 20, 5193–5197.
Yang, X., Qin, Y., Shao, S., Yu, Y., Zhang, C., Dong, H., et al. (2016c). MicroRNA-214 Inhibits Left Ventricular Remodeling in an Acute Myocardial Infarction Rat Model by Suppressing Cellular Apoptosis via the Phosphatase and Tensin Homolog (PTEN). Int. Heart J. 57, 247–250. doi:10.1536/ihj.15-293
Yi, J., and An, Y. (2018). Circulating miR-379 as a Potential Novel Biomarker for Diagnosis of Acute Myocardial Infarction. Eur. Rev. Med. Pharmacol. Sci. 22, 540–546. doi:10.26355/eurrev_201801_14207
Yousuf, F. A., Kazmi, K., Iqbal, J., Ahmed, N., and Iqbal, M. P. (2020). Higher DNA Methylation of ABO Gene Promoter Is Associated with Acute Myocardial Infarction in a Hospital-Based Population in Karachi. Pak J. Med. Sci. 36, 505–510. doi:10.12669/pjms.36.3.1406
Yu, B. T., Yu, N., Wang, Y., Zhang, H., Wan, K., Sun, X., et al. (2019). Role of miR-133a in Regulating TGF-Β1 Signaling Pathway in Myocardial Fibrosis after Acute Myocardial Infarction in Rats. Eur. Rev. Med. Pharmacol. Sci. 23, 8588–8597. doi:10.26355/eurrev_201910_19175
Yuan, H., and Gao, J. (2017). The Role of miR-370 in Fibrosis after Myocardial Infarction. Mol. Med. Rep. 15, 3041–3047. doi:10.3892/mmr.2017.6397
Yuan, J., Chen, H., Ge, D., Xu, Y., Xu, H., Yang, Y., et al. (2017). Mir-21 Promotes Cardiac Fibrosis after Myocardial Infarction via Targeting Smad7. Cel Physiol Biochem 42, 2207–2219. doi:10.1159/000479995
Yuan, X., Pan, J., Wen, L., Gong, B., Li, J., Gao, H., et al. (2019). MiR-144-3p Enhances Cardiac Fibrosis after Myocardial Infarction by Targeting PTEN. Front. Cel Dev. Biol. 7, 249. doi:10.3389/fcell.2019.00249
Zhai, C., Qian, G., Wu, H., Pan, H., Xie, S., Sun, Z., et al. (2020). Knockdown of Circ_0060745 Alleviates Acute Myocardial Infarction by Suppressing NF‐κB Activation. J. Cel. Mol. Med. 24, 12401–12410. doi:10.1111/jcmm.15748
Zhang, B. f., Jiang, H., Chen, J., Hu, Q., Yang, S., Liu, X. p., et al. (2020a). LncRNA H19 Ameliorates Myocardial Infarction‐induced Myocardial Injury and Maladaptive Cardiac Remodelling by Regulating KDM3A. J. Cel Mol Med 24, 1099–1115. doi:10.1111/jcmm.14846
Zhang, B., Li, B., Qin, F., Bai, F., Sun, C., and Liu, Q. (2019a). Expression of Serum microRNA-155 and its Clinical Importance in Patients with Heart Failure after Myocardial Infarction. J. Int. Med. Res. 47, 6294–6302. doi:10.1177/0300060519882583
Zhang, D., Wang, B., Ma, M., Yu, K., Zhang, Q., and Zhang, X. (2019b). lncRNA HOTAIR Protects Myocardial Infarction Rat by Sponging miR-519d-3p. J. Cardiovasc. Trans. Res. 12, 171–183. doi:10.1007/s12265-018-9839-4
Zhang, H., Ma, J., Liu, F., and Zhang, J. (2021a). Long Non-coding RNA XIST P-romotes the P-roliferation of C-ardiac F-ibroblasts and the A-ccumulation of E-xtracellular M-atrix by S-ponging microRNA-155-5p. Exp. Ther. Med. 21, 477. doi:10.3892/etm.2021.9908
Zhang, J., and He, J. F. (2020). LncRNA-MALAT1 Influences Myocardial Infarction by Regulating miR-30a/beclin-1 Pathway. Eur. Rev. Med. Pharmacol. Sci. 24, 885–892. doi:10.26355/eurrev_202001_20073
Zhang, J. C., Xia, L., Jiang, Y., Wu, D. Q., Liu, S. C., Zhou, X. N., et al. (2019d). Effect of lncRNA GAS5 on Rats with Acute Myocardial Infarction through Regulating miR-21. Eur. Rev. Med. Pharmacol. Sci. 23, 8573–8579. doi:10.26355/eurrev_201910_19173
Zhang, J., Li, Y., and Zhao, Q. (2018a). Circulating miR-23b as a Novel Biomarker for Early Risk Stratification after ST-Elevation Myocardial Infarction. Med. Sci. Monit. 24, 1517–1523. doi:10.12659/msm.908060
Zhang, J., Pan, J., Yang, M., Jin, X., Feng, J., Wang, A., et al. (2019c). Upregulating MicroRNA-203 Alleviates Myocardial Remodeling and Cell Apoptosis through Downregulating Protein Tyrosine Phosphatase 1B in Rats with Myocardial Infarction. J. Cardiovasc. Pharmacol. 74, 474–481. doi:10.1097/FJC.0000000000000733
Zhang, J., Tang, Y., Zhang, J., Wang, J., He, J., Zhang, Z., et al. (2021b). CircRNA ACAP2 Is Overexpressed in Myocardial Infarction and Promotes the Maturation of miR-532 to Induce the Apoptosis of Cardiomyocyte. J. Cardiovasc. Pharmacol. 78, 247–252. doi:10.1097/FJC.0000000000001065
Zhang, L., Chen, B., Zhao, Y., Dubielecka, P. M., Wei, L., Qin, G. J., et al. (2012a). Inhibition of Histone Deacetylase-Induced Myocardial Repair Is Mediated by C-Kit in Infarcted Hearts. J. Biol. Chem. 287, 39338–39348. doi:10.1074/jbc.M112.379115
Zhang, L., Qin, X., Zhao, Y., Fast, L., Zhuang, S., Liu, P., et al. (2012b). Inhibition of Histone Deacetylases Preserves Myocardial Performance and Prevents Cardiac Remodeling through Stimulation of Endogenous Angiomyogenesis. J. Pharmacol. Exp. Ther. 341, 285–293. doi:10.1124/jpet.111.189910
Zhang, L., Wang, H., Zhao, Y., Wang, J., Dubielecka, P. M., Zhuang, S., et al. (2018b). Myocyte-specific Overexpressing HDAC4 Promotes Myocardial Ischemia/reperfusion Injury. Mol. Med. 24, 37. doi:10.1186/s10020-018-0037-2
Zhang, L. X., DeNicola, M., Qin, X., Du, J., Ma, J., Tina Zhao, Y., et al. (2014). Specific Inhibition of HDAC4 in Cardiac Progenitor Cells Enhances Myocardial Repairs. Am. J. Physiology-Cell Physiol. 307, C358–C372. doi:10.1152/ajpcell.00187.2013
Zhang, L. X., Du, J., Zhao, Y. T., Wang, J., Zhang, S., Dubielecka, P. M., et al. (2018c1985). Transgenic Overexpression of Active HDAC4 in the Heart Attenuates Cardiac Function and Exacerbates Remodeling in Infarcted Myocardium. J. Appl. Physiol. 125, 1968–1978. doi:10.1152/japplphysiol.00006.2018
Zhang, M., Liu, H. Y., Han, Y. L., Wang, L., Zhai, D. D., Ma, T., et al. (2019e). Silence of lncRNA XIST Represses Myocardial Cell Apoptosis in Rats with Acute Myocardial Infarction through Regulating miR-449. Eur. Rev. Med. Pharmacol. Sci. 23, 8566–8572. doi:10.26355/eurrev_201910_19172
Zhang, M., Cheng, Y.-J., Sara, J. D., Liu, L.-J., Liu, L.-P., Zhao, X., et al. (2017). Circulating MicroRNA-145 Is Associated with Acute Myocardial Infarction and Heart Failure. Chin. Med. J. (Engl) 130, 51–56. doi:10.4103/0366-6999.196573
Zhang, M., Wang, Z., Cheng, Q., Wang, Z., Lv, X., Wang, Z., et al. (2020b). Circular RNA (circRNA) CDYL Induces Myocardial Regeneration by ceRNA after Myocardial Infarction. Med. Sci. Monit. 26, e923188. doi:10.12659/MSM.923188
Zhang, Y., Fan, X., and Yang, H. (2020c). Long Noncoding RNA FTX Ameliorates Hydrogen Peroxide-Induced Cardiomyocyte Injury by Regulating the miR-150/KLF13 axis. Open Life Sci. 15, 1000–1012. doi:10.1515/biol-2020-0100
Zhang, Y., Hou, Y. m., Gao, F., Xiao, J. w., Li, C. c., and Tang, Y. (2019f). lncRNA GAS5 Regulates Myocardial Infarction by Targeting the miR‐525‐5p/CALM2 axis. J. Cel Biochem 120, 18678–18688. doi:10.1002/jcb.29156
Zhao, B., Li, G., Peng, J., Ren, L., Lei, L., Ye, H., et al. (2021a). CircMACF1 Attenuates Acute Myocardial Infarction through miR-500b-5p-EMP1 Axis. J. Cardiovasc. Trans. Res. 14, 161–172. doi:10.1007/s12265-020-09976-5
Zhao, G., Hailati, J., Ma, X., Bao, Z., Bakeyi, M., and Liu, Z. (2020). LncRNA Gm4419 Regulates Myocardial Ischemia/Reperfusion Injury through Targeting the miR-682/TRAF3 Axis. J. Cardiovasc. Pharmacol. 76, 305–312. doi:10.1097/FJC.0000000000000867
Zhao, Q., Li, W., Pan, W., and Wang, Z. (2021b). CircRNA 010567 Plays a Significant Role in Myocardial Infarction via the Regulation of the miRNA-141/DAPK1 axis. J. Thorac. Dis. 13, 2447–2459. doi:10.21037/jtd-21-212
Zhao, T., Cheng, G., Zhang, L., Tseng, Y., and Padbury, J. (2007). Inhibition of Histone Deacetylases Triggers Pharmacologic Preconditioning Effects against Myocardial Ischemic Injury. Cardiovasc. Res. 76, 473–481. doi:10.1016/j.cardiores.2007.08.010
Zhong, J., Agha, G., and Baccarelli, A. A. (2016). The Role of DNA Methylation in Cardiovascular Risk and Disease. Circ. Res. 118, 119–131. doi:10.1161/CIRCRESAHA.115.305206
Zhou, J., Li, D., Yang, B. P., and Cui, W. J. (2020). LncRNA XIST Inhibits Hypoxia-Induced Cardiomyocyte Apoptosis via Mediating miR-150-5p/Bax in Acute Myocardial Infarction. Eur. Rev. Med. Pharmacol. Sci. 24, 1357–1366. doi:10.26355/eurrev_202002_20193
Zhou, Q., Zhang, Z., Bei, Y., Li, G., and Wang, T. (2018). Circular RNAs as Novel Biomarkers for Cardiovascular Diseases. Adv. Exp. Med. Biol. 1087, 159–170. doi:10.1007/978-981-13-1426-1_13
Zhou, T., Qin, G., Yang, L., Xiang, D., and Li, S. (2019). LncRNA XIST Regulates Myocardial Infarction by Targeting miR‐130a‐3p. J. Cel Physiol 234, 8659–8667. doi:10.1002/jcp.26327
Zhu, F., Li, Q., Li, J., Li, B., and Li, D. (2021a). Long Noncoding Mirt2 Reduces Apoptosis to Alleviate Myocardial Infarction through Regulation of the miR-764/PDK1 axis. Lab. Invest. 101, 165–176. doi:10.1038/s41374-020-00504-2
Zhu, F., Liu, Y., and Tang, K. (2021b). MicroRNA-26b Might Be a Novel Therapeutic Target in the Treatment of Myocardial Infarction. Int. J. Cardiol. 323, 261. doi:10.1016/j.ijcard.2020.09.007
Zhu, J., Gu, H., Lv, X., Yuan, C., Ni, P., and Liu, F. (2018). LINC-PINT Activates the Mitogen-Activated Protein Kinase Pathway to Promote Acute Myocardial Infarction by Regulating miR-208a-3p. Circ. J. 82, 2783–2792. doi:10.1253/circj.CJ-18-0396
Zhu, J., Yao, K., Wang, Q., Guo, J., Shi, H., Ma, L., et al. (2016). Circulating miR-181a as a Potential Novel Biomarker for Diagnosis of Acute Myocardial Infarction. Cel Physiol Biochem 40, 1591–1602. doi:10.1159/000453209
Zhu, W., Wu, R.-D., Lv, Y.-G., Liu, Y.-M., Huang, H., and Xu, J.-Q. (2020a). BRD4 Blockage Alleviates Pathological Cardiac Hypertrophy through the Suppression of Fibrosis and Inflammation via Reducing ROS Generation. Biomed. Pharmacother. 121, 109368. doi:10.1016/j.biopha.2019.109368
Zhu, Y., Pan, W., Yang, T., Meng, X., Jiang, Z., Tao, L., et al. (2019). Upregulation of Circular RNA CircNFIB Attenuates Cardiac Fibrosis by Sponging miR-433. Front. Genet. 10, 564. doi:10.3389/fgene.2019.00564
Zhu, Y., Zhao, P., Sun, L., Lu, Y., Zhu, W., Zhang, J., et al. (2021c). Overexpression of circRNA SNRK Targets miR-103-3p to Reduce Apoptosis and Promote Cardiac Repair through GSK3β/β-Catenin Pathway in Rats with Myocardial Infarction. Cell Death Discov. 7, 84. doi:10.1038/s41420-021-00467-3
Zhu, Y., Zou, C., Jia, Y., Zhang, H., Ma, X., and Zhang, J. (2020b). Knockdown of Circular RNA circMAT2B Reduces Oxygen-Glucose Deprivation-Induced Inflammatory Injury in H9c2 Cells through Up-Regulating miR-133. Cell Cycle 19, 2622–2630. doi:10.1080/15384101.2020.1814025
Ziller, M. J., Gu, H., Müller, F., Donaghey, J., Tsai, L. T.-Y., Kohlbacher, O., et al. (2013). Charting a Dynamic DNA Methylation Landscape of the Human Genome. Nature 500, 477–481. doi:10.1038/nature12433
Keywords: epigenetics, DNA methylation DNA, histone modifications, non-coding RNAs rna, micro-RNA, cardiovascular, myocardial infarction
Citation: Chen J, Liu Z, Ma L, Gao S, Fu H, Wang C, Lu A, Wang B and Gu X (2021) Targeting Epigenetics and Non-coding RNAs in Myocardial Infarction: From Mechanisms to Therapeutics. Front. Genet. 12:780649. doi: 10.3389/fgene.2021.780649
Received: 21 September 2021; Accepted: 30 November 2021;
Published: 20 December 2021.
Edited by:
Ping Wang, Tianjin Medical University, ChinaReviewed by:
Andrea Caporali, University of Edinburgh, United KingdomLiang Chen, Chinese Academy of Medical Sciences and Peking Union Medical College, China
Copyright © 2021 Chen, Liu, Ma, Gao, Fu, Wang, Lu, Wang and Gu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xufang Gu, bDIwMTkzMDEzMTEwNEB5ZWFoLm5ldA==
†These authors have contributed equally to this work and share the first authorship
 Jinhong Chen
Jinhong Chen Zhichao Liu
Zhichao Liu Li Ma1
Li Ma1